Abstract
The glucocorticoid and progesterone receptors (GR and PR) are closely related members of the steroid receptor family. Despite sharing similar structural and functional characteristics; the cognate hormones display very distinct physiological responses. In mammary epithelial cells, PR activation is associated with the incidence and progression of breast cancer, whereas the GR is related to growth suppression and differentiation. Despite their pharmacological relevance, only a few studies have compared GR and PR activities in the same system. Using a PR+/GR+ breast cancer cell line, here we report that either glucocorticoid-free or dexamethasone (DEX)-activated GR inhibits progestin-dependent gene expression associated to epithelial-mesenchymal-transition and cell proliferation. When both receptors are activated with their cognate hormones, PR and GR can form part of the same complex according to co-immunoprecipitation, quantitative microscopy and sequential ChIP experiments. Moreover, genome-wide studies in cells treated with either DEX or R5020, revealed the presence of several regions co-bound by both receptors. Surprisingly, GR also binds novel genomic sites in cells treated with R5020 alone. This progestin-induced GR binding was enriched in REL DNA motifs and located close to genes coding for chromatin remodelers. Understanding GR behavior in the context of progestin-dependent breast cancer could provide new targets for tumor therapy.
INTRODUCTION
Steroid hormones regulate a wide range of physiological processes through their binding to ligand-regulated transcription factors, including the estrogen receptor (ER), progesterone receptor (PR) and the glucocorticoid receptor (GR). In particular, their combined action modulates the development and differentiation of the mammary gland (1). Consistently with this pivotal role, their activity is also linked to breast cancer (2–4).
In ER+/PR+ breast cancer cells, increased circulating levels of progestins and estrogens and/or over-expression of their receptors lead to an uncontrolled cellular division (5,6). While the proliferating role of estrogens is well understood, widespread controversy exists regarding progestin actions. Although progestins are involved in driving cell proliferation, thus favoring breast cancer development, they may be safely and effectively used in treating ER-dependent breast cancer (6,7). In contrast, glucocorticoids are known to be involved in cellular differentiation in the post-natal mammary gland (8,9), while in proliferating cells—along pregnancy or in tumor cells—these hormones induce the expression of cell-cycle inhibitors (8) and mesenchymal-to-epithelial transition (10).
The functional crosstalk between GR and ER has been widely studied (7,11–14). Glucocorticoids exert an antagonistic effect on estrogen-dependent cell growth in ER+/GR+ breast and uterine carcinoma cells (15,16) and reduce MCF-7 cell proliferation by more than 30% compared to untreated cells (17). In contrast to ER and GR studies, little is known about the influence of GR on PR transcriptional activity. These receptors share many similar structural characteristics, although the regulation of their quaternary structure may differ (18). With a 90% sequence identity between their DNA binding domains (DBD), they have similar capacity to bind their responsive elements in chromatin. PR and GR are also able to interact with the same members of the p160 cofactor family [with histone acetyltransferase activity (19)] and with similar chromatin remodelers [e.g. SWI/SNF, P/CAF and/or SAGA (20,21)]. Even with a 55% sequence identity between their ligand binding domains, some steroids are able to bind both PR and GR (22), suggesting a potential crosstalk between the two pathways. However, in cells expressing both GR and PR, glucocorticoids and progestins exert very distinct and, in some situations opposite physiological responses. For example, the association of progestins with the incidence and progression of breast cancer contrasts with the growth suppressive action of glucocorticoids in ER+/PR+ mammary cancer cells (23–25). Moreover, while GR and PR can both activate and repress target genes (26), the relevant features that make these receptors and their actions different are still unknown.
To date, only a few studies have been performed comparing the GR and PR responses in the same system (25,27–29), which is limited by the tissue-specific expression pattern of both receptors. Particularly, microarray analysis in the T47D/A1–2 cell line, which expresses similar amounts of both receptors, revealed that the two hormones differentially regulate overlapping but also distinct sets of genes (25).
A potential molecular interaction between GR and PR has also remained largely unexplored. In the GR+ MDA-MB-231 breast cancer cell line, transfection with PR has shown that corticosterone, the endogenous glucocorticoid, induces progesterone-like morphological changes (30). This suggests that glucocorticoids can regulate cell morphology through the PR regulated pathway. On the other hand, little information is available on the effect of progesterone treatment on GR activity in breast cancer cell models (12,31).
Understanding how these receptors behave in breast cancer is relevant not only from a physiological but also from a pharmacological perspective. Due to the extensive use of glucocorticoids as a palliative option for the treatment of breast cancer and the activation of GR by synthetic progestins used in hormone replacement therapies, we decided to focus our study on the influence of GR on the PR-dependent breast cancer cell proliferation and dedifferentiation (31). We used the T47D/A1-2 human breast cancer cell line that expresses comparable levels of GR and PR (27).
We found that GR expression, even in the absence of glucocorticoids, inhibits PR-dependent cell proliferation through the modulation of key PR-target genes. When GR is activated by the synthetic glucocorticoid dexamethasone (DEX), the antagonistic effect increases and most likely involves the formation of GR-PR protein complexes. PR and GR ChIP-seq analyses reveal overlapping binding sites genome-wide and sequential-ChIP on key enhancer sites confirms co-recruitment to shared sites. Unexpectedly, we found that glucocorticoid-free GR can bind to a pattern of unique binding sites upon stimulation with the PR-agonist R5020, which could explain, at least in part, the interference with the PR-dependent transcriptional response.
MATERIALS AND METHODS
Cell culture and treatments
T47D-WT breast cancer cells carrying one stably integrated copy of the luciferase reporter gene driven by the MMTV promoter (32) and its derivative clone T47D/A1-2 that also expresses a functional full length rGR (27) were routinely grown in Dulbecco's Modified Eagle's medium (DMEM, Thermo Fisher Scientific). MCF-7L cells were kindly provided by Carol Lange although originally obtained from C. Kent Osborne (Baylor College of Medicine, Houston, TX, USA) (33). These cells were maintained in Dulbecco's Modification of Eagle's Medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 mg/ml streptomycin and 1.4 U/ml Insulin (Novo Nordisk® Pharma). For experiments, all cells were plated in DMEM medium without phenol red supplemented with 10% dextran-coated charcoal-treated FBS (DCC/FBS) and 48-h later medium was replaced by fresh medium without serum. After 24 h in serum-free conditions, cells were incubated with R5020 (Sigma) 10 nM and/or DEX (Sigma) 10 nM for different lengths of times at 37°C according to each experiment.
RNA interference experiments
siRNA of GR from Dharmacon (ON-TARGETplus SMARTpool Rat Nr3c1 siRNA L-089504-02-0005 or SMARTpool siGENOME Human NR3C1 siRNA M-003424-03-0005) was transfected into the T47D/A1-2 or MCF-7L cells, respectively using Lipofectamine 2000 (Invitrogen). After 48 h, the medium was replaced by fresh medium without serum. After one day in serum-free conditions, cells were incubated with R5020 10 nM, and/or DEX 10 nM or vehicle (ethanol) for different times at 37°C. The downregulation of GR expression was determined by western blot using GR antibody (MA1-510, Thermo Fisher Scientific).
Cell proliferation assay
T47D-WT and T47D/A1-2 cells transfected with control or GR siRNAs were cultured as described above. Cells (1 × 104) were plated in a 96-well plate in the presence or absence of 10 nM R5020 or DEX. The cell proliferation ELISA BrdU Colorimetric assay (Roche) was performed according to the manufacturer's instructions. Figures show the percentage increase of proliferation in the presence versus absence of R5020 or DEX. The experiments were performed in quintuplicate.
Cell-cycle analysis
The effects of R5020 or DEX on cell cycle distribution were examined using PI flow cytometry assay. T47D-WT; T47D/A1-2 and MCF-7L cells were seeded in DMEM medium without phenol red supplemented with 10% DCC/FBS. After an overnight incubation, cells were treated with R5020 10 nM and/or DEX 10 nM for 18 h. Detached and adherent cells were collected and fixed in 70% ethanol in phosphate-buffered saline (PBS) at −20°C overnight. Cells were then resuspended in PBS containing 20 μg/ml PI and 100 μg/ml RNAse and incubated for 30 min at 37°C in the dark. Samples were then analyzed on a BD FACSAria II flow cytometer (BD Biosciences) using BD FACSDiva6.0 software.
Flow cytometry
For cell surface marker analysis, cells were washed once with PBS and then harvested with 0.05% trypsin/0.025% EDTA. Detached cells were washed with PBS containing 0.5% BSA (wash buffer) and resuspended in the wash buffer (106 cells/100 μl). Combinations of fluorochrome-conjugated monoclonal antibodies obtained from BD Biosciences against human CD44 (APC; cat. # 560890) and CD24 (PE; cat. #560991), were added to the cell suspension at concentrations recommended by the manufacturer and incubated at 4°C in the dark for 30 min. The labeled cells were washed in wash buffer and immediately were subjected to fluorescence-activated cell sorting analysis using BD FACS Calibur (34).
Subcellular fractions separation and Co-IP assay
T47D/A1-2 cells were incubated for 1 h with R5020 and DEX. For subcellular fraction separation, cells were centrifuged for 5 min at 2000 rpm and washed with ice-cold PBS buffer. Cells were then incubated in 300 μl of hypotonic buffer (10 mM Tris–HCl pH 6.7; 0.2 mM MgCl2; 1 mM EGTA; protease inhibitors) for 5 min on an ice-water bath and lysed by douncing homogenization. Samples were centrifuged at 1500 rpm for 10 min at 4°C and supernatants were referred as cytoplasmic fraction. Pellets were washed twice in ice-cold PBS buffer, resuspended in 250 μl of lysis buffer (20 mM Tris–HCl pH 6.7; 70 mM NaCl; 10% glycerol; 1% Triton X-100; 0.5% Nonidet P-40; protease inhibitor), and incubated on ice-water bath for 30 min. Samples were sonicated three times for 10 s at 20% output with a Branson 450 Sonifier. After the addition of 0.5 M NaCl, the incubation was continued for additional 30 min. Samples were centrifuged at 12 000 rpm for 5 min at 4°C, and supernatants were referred to as nuclear fraction. For co-immunoprecipitation assay (Co-IP), nuclear fraction (1 mg protein) was precleared at 4°C with Dynabeads® Protein G (Thermo Fisher Scientific) for 1 h before incubating overnight at 4°C with anti-PR antibody (H-190, sc-7208, Santa Cruz Biotechnology) or an unspecific control antibody (normal rabbit IgG, sc-2017, Santa Cruz Biotechnology). Complexes were incubated with Dynabeads® Protein G (Thermo Fisher Scientific) for 3 h at 4°C. The beads were washed by rocking for 5 min in nuclear lysis buffer and the immunoprecipitated (IP) proteins were eluted by boiling in sodium dodecyl sulfate-sample buffer. Inputs and IPs were analyzed by western blot using PR antibody (H-190, sc-7208, Santa Cruz Biotechnology) and GR antibody (MA1-510, Thermo Fisher Scientific).
Subcellular localization and cross-correlation analysis
T47D-WT cells were transfected with 2 μg of peGFP-PR (kindly provided by Dr Carol Lange, Departments of Medicine and Pharmacology, University of Minnesota Masonic Cancer Center, Minneapolis, MN, USA) (35) and pmCherry-GR (36) and incubated with vehicle, R5020 10 nM and/or DEX 10 nM for at least 1 h. Images were obtained in a FV1000 confocal laser scanning microscope (Olympus), with an Olympus UPlanSApo 60× oil immersion objective (NA = 1.35). eGFP and mCherry were excited with a multi-line Ar laser at 488 nm and a He-Ne green laser at 543 nm (average power at the sample, 700 nW), respectively. Fluorescence was detected with a photomultiplier set in the pseudo photon-counting detection mode. The pixel size was set at 83 nm. Analysis of subcellular distribution was performed using ImageJ (NIH, USA) software. The nucleus and the cytoplasm regions were manually selected for each individual cell and the nucleus/cytoplasm intensity ratio was then calculated (n = 19–26). Cross-brightness (Bcc) measurements were done as previously described (36). Briefly, for each studied cell a time stack of 200 images (256 × 256 pixels) was taken in the microscope setup mentioned above, setting the pixel dwell time to 10 μs. The frame time was 0.9 s. Each stack was further analyzed using the ‘N&B’ routine of the ‘GLOBALS for Images’ software developed at the Laboratory for Fluorescence Dynamics (University of California, Irvine, CA, USA). Bcc was determined following the procedures described by Digman et al. (37) for every pixel of the nucleus. This parameter was calculated from the cross-variance between eGFP and mCherry channels normalized by the product of their mean intensities. Therefore, Bcc describes cross-correlated fluctuations in both channels and acquires positive values when both fluorescent molecules are present in the same complex. An average Bcc was determined from the pixels of the nucleus and this analysis was performed for 50 cells in each condition.
ChIP-seq
All ChIP-seq experiments were performed with the following conditions of culture and treatments: T47D/A1-2 cells were grown 48 h in DMEM without phenol red supplemented with 10% DCC/FBS and synchronized in G0/G1 by 24 h of serum starvation. R5020 or DEX was added to the medium at 10 nM, and cells were harvested after 1 h of hormone treatment. ChIP-DNA was purified and subjected to deep sequencing using the Solexa Genome Analyzer (Illumina, San Diego, CA, USA). Replicates for ChIP-seq experiments of the GR treated with or without DEX for 1 h were obtained from a previous publication (38) (Supplementary Table S1). The raw sequence reads were aligned and processed as previously described (39,40). In brief, sequence reads were aligned to human (UCSC hg19) genome using Bowtie (41). Only sequences uniquely aligned with ≤1 mismatch were retained. Post-alignment processing of sequence reads included in silico extension and signal normalization based on the number of million mapped reads. Reads were extended to a final length equal to MACS fragment size estimation (42), and only unique reads were retained. For signal normalization, the number of reads mapping to each base in the genome was counted using the genomeCoverageBed command from BedTools (43). Processed files were visualized in the UCSC genome browser (44). Transcription factor enrichment sites were detected with MACS (42) using default parameters and a P-value of 1E-10. A control dataset derived by sequencing input DNA samples was used to define a background model. Heatmap and aggregation plots: To compute the heatmap and aggregation (averageogram) plots we divided the bed files containing the regions of interest (spanning ±3 Kb from the center of the TF peaks for the averageograms, or ±6 Kb in the case of the heatmaps) in 100 bp bins. The coverage signal from the GR and PR aligned read bed files for the different cell lines (PR: T47D-A1/2 and wild-type; GR: T47D-A1/2) and conditions (i.e. hormone treated or not) was obtained for each bin using the coverageBed command from BedTools (43). For quantile normalization of the results obtained from each condition (defined by the combination of TF, hormone treatment and cell line), we produced 100 bp bins across all the peaks called in each condition and calculated the signal coverage of the ChIP-seq dataset in the same condition in which the peaks were called. The 95th quantile obtained from the binned peaks in each TF and cell line was used to adjust the bin values obtained for the aggregation plots. The heatmaps were produced with Cluster3 software (45) and plotted using Treeview (46). Motif Discovery: De novo motif discovery was performed with HOMER (47) using a window of 200 bp around peaks and setting motifs length to 8, 10, 12 and 14 bp. The best scoring motif was chosen from each analysis to represent the DNA binding domain for each of the factors profiled. The co-enriched motifs obtained for the different lengths were independently analyzed for consistency of the results and manually curated to exclude redundant motifs. The top scoring motifs, ranked by P-value, are shown in each case. Matching DNA binding motifs were associated to the de novo recovered matrix when the HOMER score was 0.65 or higher. In Supplementary Figures S3A and 7B, the percentage of occurrence of the motifs are the defaults calculated by the HOMER software, and represent the number (expressed as percentage) of GR- or PR-bound regions (under the corresponding stimuli) in which the motif is found. As a reference, the number of background regions (defined by default in the HOMER software) in which the same motif is found, are shown separated by a bar. For the bar-plots showing the occurrence of de novo recovered motifs at selected regions (Figures 2B and 6D), the de novo motif matrices recovered by HOMER were added to the list of known motifs used by HOMER, and the motif analysis was re-run using this file. The results then show the number of selected regions (expressed as percentage) in which the corresponding motif was found in the regions analyzed, and the corresponding background regions used. Functional annotation: Functional annotation of genes associated with TF binding regions was performed using GREAT with default settings (48).
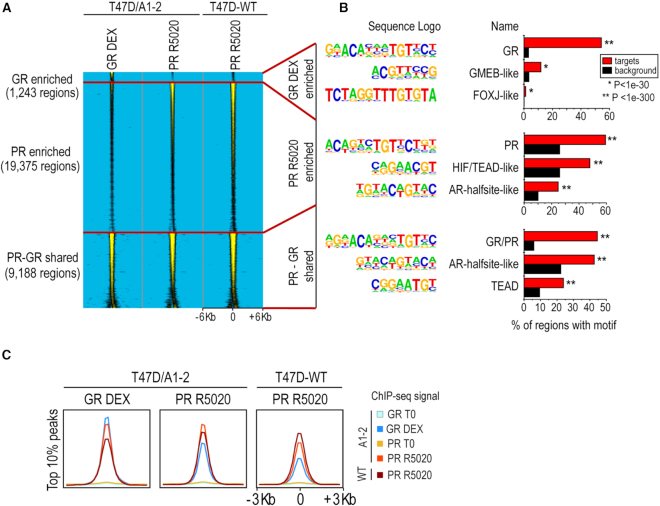
Figure 2.
GR and PR share many binding sites. (A) Normalized GR and PR ChIP-seq signals from T47D/A1-2 cells treated for 1 h with DEX 10 nM (GR DEX) or R5020 10 nM (PR R5020). Data are represented as a heatmap clustered in GR enriched, PR enriched and GR-PR shared binding sites, centered (±6 Kb) at the GR DEX (for GR enriched regions) or PR R5020 (for PR shared and enriched regions) peaks. The normalized PR ChIP-seq signal from T47D-WT treated for 1 h with R5020 10 nM is also shown. (B) Selected top de novo-motifs recovered from GR-enriched; PR-enriched and PR-GR shared binding sites. See Methods for details. (C) Aggregation plots showing GR and PR (with or without hormone treatment, as indicated on the right) ChIP-seq signal enrichments centered (+/- 3Kb) at the peaks with the highest score (top 10%), as analyzed by the peak finding algorithm (see Methods). The set of peaks used for each plot corresponds to the ChIP-seq samples indicated in the upper label of each box. As a control, the signal enrichment for PR-R5020, profiled by ChIP-seq in T47D-WT cells is presented in all plots. The last box shows the aggregation plots obtained for the top 10% PR-R5020 regions profiled in T47D-WT cells.
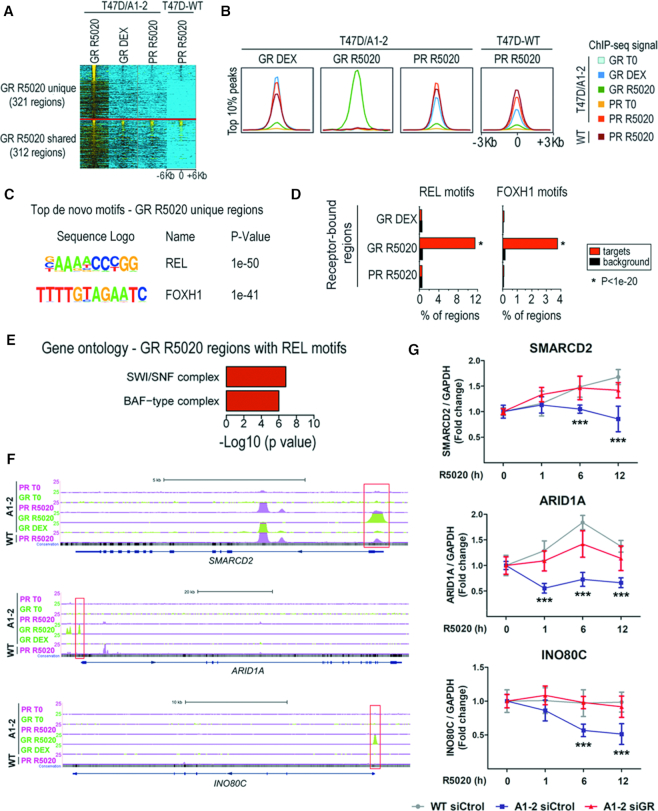
Figure 6.
GR can bind an unexpected and unique pattern of binding sites after R5020 stimulation. (A) Normalized GR and PR binding ChIP-seq signals from T47D/A1-2 and T47D-WT cells treated for 1 h with R5020 (GR R5020, PR R5020) or DEX (GR DEX), as indicated by the top labels, is represented as a heatmap clustered in GR R5020 unique and GR R5020 shared binding sites, centered (±6 Kb) at the GR R5020 peaks. (B) Aggregation plots showing GR and PR (with or without cognate hormone treatment, as well as GR-R5020) ChIP-seq signal enrichments centered (±3 Kb) at the peaks with the highest score (top 10%), as analyzed by the peak finding algorithm (see ‘Materials and Methods’ section). The set of peaks used for each plot corresponds to the ChIP-seq samples indicated in the upper label of each box. As a control, the signal enrichment for PR-R5020, profiled by ChIP-seq in T47D-WT cells, is presented in all plots. The last box shows the aggregation plots obtained for the top 10% PR-R5020 regions profiled in T47D-WT cells. (C) Top de novo–motifs recovered from GR R5020 unique binding sites revealed a strong enrichment for REL and FOXH1 recognition motifs. (D) Direct interrogation for the presence of these motifs performed on all the datasets presented in this study, revealed that REL and FOXH1 motifs were exclusively enriched in GR-R5020 regions. See ‘Materials and Methods’ section for details. (E) Gene ontology analysis of the GR R5020 unique regions with REL motifs revealed enrichments for genes associated with chromatin remodeling complexes. (F) PR and GR binding profiles for the indicated samples in the vicinity of the SMARCD2, ARID1A and INO80C genes. Red boxes show the nuclear receptor binding site validated by ChIP. (G) Gene expression changes (measured by RT-qPCR for selected genes) induced by R5020 10 nM for 1, 6 and 12 h in T47D-WT and T47D/A1-2 cells transfected with control (Ctrol) or GR siRNAs. Values were normalized with GAPDH expression. Means ± S.D. from three independent experiments performed in duplicate are shown. ***, P ≤ 0.001.
ChIP and sequential ChIP assays in cultured cells
ChIP assays were performed as described (49) using anti-PR antibody (H-190, sc-7208, Santa Cruz Biotechnology), a combination of anti-GR antibodies (P-20, sc-1002 and M-20, sc-1004, Santa Cruz Biotechnology) or a non-specific control antibody. Quantification of ChIP was performed by real-time polymerase chain reaction (PCR) using Applied Biosystems StepOnePlus™ (Thermo Fisher Scientific). The fold enrichment of the target sequence in the IP DNA compared with the input (Ref) fractions was calculated using the comparative Ct (the number of cycles required to reach a threshold concentration) method with the equation 2Ct(IP) - Ct(Ref) and referred as % of input. The sequences of the PCR primers are available upon request. For sequential ChIP or re-ChIP assays, immunoprecipitation was washed sequentially as described previously (49). Complexes were eluted with 10 mM Dithiothreitol (DTT) for 30 min at 37°C, diluted 50 times with dilution buffer and then subjected to a second round of immunoprecipitation with the indicated antibodies or with IgG.
RNA extraction and RT–PCR
Total RNA was prepared, and cDNA was generated as previously described (50). Quantification of gene products was performed by real-time PCR. Each value calculated using the standard curve method was corrected by the human Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and expressed as relative RNA abundance over time zero. Primer sequences are available on request.
Statistical analysis
Results were expressed as means ± S.D., as it is indicated in the figure legends. Student's t-test and one-way or two-way ANOVA tests followed by Tukey's post-hoc test or Bonferroni post-test were used to detect significant differences among treatments. Statistical analyses were performed using GraphPad Prism-5 (GraphPad Software). Differences were considered statistically significant if P ≤ 0.05.
RESULTS
GR expression affects progestin-mediated cell proliferation and dedifferentiation
To test GR influence on progestin action, we compared two PR+ cell line models: the T47D-MTVL (WT) cells and its derivative clone T47D/A1-2, which also expresses a functional full length GR (27,51). Even though T47D-MTVL (WT) cells have been characterized as PR+ and GR low (32,52), in our hands GR levels in these cells are undetectable by Western blot (Supplementary Figure S1A) and, more importantly, glucocorticoids fail to activate the MMTV promoter [Supplementary Figure S1B and (27)]. Thus, we will consider T47D-MTVL (WT) cells as ‘GR functionally negative’ hereafter.
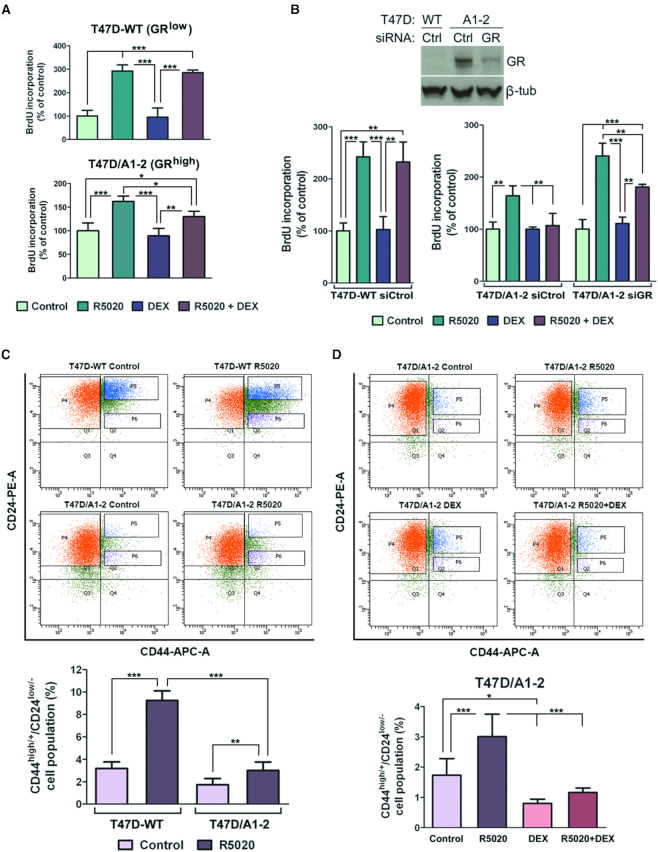
Given the known association of progestins with the incidence and progression of breast cancer and the inhibition of cell growth exerted by glucocorticoids (8,12,53), we first evaluated whether GR expression and/or activation affected progestin-dependent cell proliferation in breast cancer cells. Hormone-dependent cell proliferation was assessed by quantifying BrdU incorporation in either T47D-WT or T47D/A1-2 cells. We observed a clear progestin-dependent increase in proliferation of T47D-WT cells (3-fold versus untreated cells) upon 14 h addition of the synthetic PR agonist, R5020 (Figure 1A, upper panel and Supplementary Figure S1C, left panel). Intriguingly, no progestin-dependent BrdU incorporation was detected in T47D/A1-2 cells treated with R5020 for 14 h (Supplementary Figure S1C, right panel). However, by extending hormone treatment to 18 h we observed an increase in proliferation (∼2-fold), indicating a slower growing-rate of this GR+ cell line compared to WT (Figure 1A, lower panel and Supplementary Figure S1C, right panel). Consistently, the progestin effect was significantly impaired when the synthetic glucocorticoid DEX was added (Figure 1A lower panel). The inhibitory effect is GR-dependent, as only T47D/A1-2 cells respond to DEX. The glucocorticoid alone had no effect compared with control (Figure 1A). These results were confirmed by FACS analysis performed in both cell lines stained with propidium iodide. The percentage of cells accumulated in S phase was significantly lower in T47D/A1-2 cells treated with R5020 compared to T47D-WT (14.7 versus 21.8%, respectively) (Supplementary Figure S2A) reinforcing the idea that GR expression affects progestin-dependent cell proliferation. Moreover, DEX alone did not affect S phase accumulation in T47D/A1-2 cells (6.9 versus 7% in untreated cells) but inhibited R5020 mediated action (12.7 versus 15.2% in R5020 treated cells, Supplementary Figure S2B).
Figure 1.
GR interferes with progestin-induced cell proliferation and dedifferentiation. (A and B) Cell proliferation was evaluated by using ELISA BrdU colorimetric assay (Roche) in T47D-WT (GRlow) and T47D/A1-2 (GRhigh) growing cells incubated with R5020 10 nM, DEX 10 nM or both hormones. (A) Cells were harvested at 14 h (upper panel) or 18 h (lower panel) after hormone treatment. (B) Previous to hormone treatment, cells were transfected with control (Ctrol) or GR siRNAs. GR expression was analyzed by western blotting using a specific GR antibody (upper panel). Values are expressed as percentage of increased proliferation relative to untreated cells (Control). Means ± S.D. from five independent experiments are shown. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. (C andD) T47D-WT and T47D/A1-2 cells were treated with R5020 10 nM (C and D) or/and DEX 10 nM (D). After 24 h, cells were harvested and incubated with antibodies against human CD44-APC and CD24-PE and analyzed by flow cytometry. The upper figures show a representative expression pattern of CD44 and CD24 obtained by FACS. Quadrant 2, Panel 6 (Q2, P6) indicates CD44high/CD24low labeled cells. In the lower figures values are expressed as percentage of CD44high/CD24low subpopulation. Means ± S.D. from three independent experiments are shown. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05.
To test whether the presence of GR is solely responsible for the observed inhibitory effect on progestin-dependent cell proliferation, we monitored cell proliferation upon transfection of T47D/A1-2 cells with small interfering RNA (siRNA) for GR (Figure 1B). GR depletion lead to WT levels of BrdU incorporation in T47D/A1-2 cells treated with the progestin alone. Also, GR knock-down partially reverted DEX mediated inhibition in cells treated with both hormones (Figure 1B, right panel). In conclusion, while GR expression seems to impair progestin-dependent cell proliferation in mammary tumor epithelial cells, GR activation with DEX completely abrogates it.
Next, we asked whether GR can also affect the progestin-dependent dedifferentiation process occurring in T47D-WT cells (34). For this purpose, we evaluated the hormone-dependent expression patterns of the dedifferentiation markers CD44 and CD24 by flow cytometry and quantified the proportion of CD44high/CD24low population, a hallmark of dedifferentiation. While the CD44high/CD24low phenotype was lower in T47D/A1-2 cells compared to T47D-WT in the absence of any stimuli (1.7 versus 3.2%, respectively), R5020 treatment significantly increased the percentage to 9.2% in T47D-WT cells (Figure 1C) but only to 3.0% in the T47D/A1-2 cells. These results indicate that GR presence impairs cell dedifferentiation. Moreover, as shown in Figure 1D, GR activation with DEX significantly diminished both the percentage of CD44high/CD24low in untreated cells or treated with R5020 to values comparable to T47D/A1-2 control cells (0.8 and 1.2%, respectively), supporting the idea that GR interferes with PR-mediated cell dedifferentiation.
GR and PR share many genomic binding regions
To explore potential mechanisms involved in the GR inhibitory effect on PR activity, we characterized PR and GR binding by chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) upon treatment with their respective hormones (PR-R5020 and GR-DEX) in T47D/A1-2 cells. We performed deep sequence coverage for all experiments (Supplementary Table S1) and called peaks using a stringent P-value cutoff (1E-10) to ensure high confidence in the identification of GR and PR binding sites.
Consistent with previous reports (13,54–57), our data showed that most of the GR and PR binding events (>95%) depend on hormone activation, (Supplementary Table S1). In fact, we found many more binding events for PR than for GR (28 563 and 10 640 respectively) upon 1 h treatment with their specific hormones. Of note, most PR binding events (23 602, i.e. >80%) in T47D/A1-2 cells correspond to PR R5020 binding sites identified in T47D-WT cells (57). Moreover, an important fraction of the GR binding regions upon DEX treatment (>88%) mapped to regions that were also bound by PR upon R5020 stimulation in T47D/A1-2. Consistently, almost half of the activated PR binding regions (>32%) were also bound by the GR upon DEX treatment. Taken together, these results prompted us to identify and define ‘GR enriched’, ‘PR enriched’ and ‘PR-GR shared’ genomic regions in T47D/A1-2 cells, as those sites that present only GR or PR ChIP-seq signal, or both. As depicted in the heatmap of Figure 2A, the GR clearly bound to GR enriched regions while the PR was largely absent at these sites in both T47D/A1-2 and WT cells. Conversely, PR bound to PR enriched regions in both epithelial mammary cell types, while there was no signal from the GR ChIP-seq in the vicinity of these regions. At last, both GR and PR bound to the PR-GR shared regions. Noteworthy, the PR ChIP-seq signal profiled in T47D/A1-2 cells was faithfully recapitulated by the PR ChIP-seq performed in T47D-WT cells, further reinforcing the suitability of the T47D/A1-2 as model to study the GR effects in the T47D breast cancer cells.
De novo motif analyses showed that both PR and GR bound to their known DNA-binding motif upon stimulation with their respective hormones (Supplementary Figure S3A). These binding motifs are almost the same for both receptors, thus accounting for the large amount of shared binding sites in T47D/A1-2 cells. Also, several of the significantly co-enriched motifs were shared between GR and PR binding sites, although present at different proportions (e.g. AR-halfsite, FOX and AP-like motifs, Supplementary Figure S3A).
To gain further insights into the DNA-binding motifs specifically enriched in the non-shared receptor binding regions, we performed de novo motif analyses on the GR and PR enriched binding sites (i.e. the 1243 GR-DEX regions that are not bound by the PR upon R5020 stimulation, and the 19,375 PR-R5020 regions not shared with GR-DEX, respectively, Figure 2A), and compared them with the 9188 PR/GR shared regions. The global de novo analysis (using by default random genomic regions to calculate the motif enrichment) showed that the top motif enriched in the GR enriched, the PR enriched and the PR-GR shared regions was highly similar (Figure 2B). There was only one base different with the top motif enriched in the PR enriched regions, which did not present a strong G in the sixth position upstream the center of the palindrome (Supplementary Figure S3B). In contrast, co-enriched motifs identified in this analysis with default settings were largely different for the GR and PR enriched regions (Figure 2B; Supplementary Tables S2 and 3).
To gain further insights into the genomic occupancy of the steroid receptors profiled, we generated aggregation plots centered at the peaks obtained for the cognate hormone-stimulated receptor datasets. Results presented in Supplementary Figure S3C show the average GR (in T47D/A1-2 cells) and PR ChIP-seq signals (in both T47D-WT and T47D/A1-2 cells) centered at either the 10 640 GR-DEX (in T47D/A1-2 cells), the 28 563 PR-R5020 (in T47D/A1-2 cells) or the 43 398 PR-R5020 (in T47D-WT) peaks. This analysis revealed strong and shared ChIP-seq signals for the GR-DEX and PR-R5020 at the opposite receptor peaks (Supplementary Figure S3C). To directly interrogate whether protein occupancy was similarly high at the strongest regions of the opposite receptor, we restricted the regions used for the aggregation plots to the top 10% regions with the strongest PR-R5020 (either in T47D/A1-2 or T47D-WT) and GR-DEX signals. These analyses also revealed that the strongest PR binding sites upon R5020 treatment are indeed strong GR binding sites after DEX stimulation (Figure 2C).
Most importantly, these results are still valid when using PR binding sites in T47D-WT cells. Thus, PR and GR binding sites are largely shared throughout the genome of T47D/A1-2 cells and present similar occupancy enrichments at the strongest binding regions, as depicted in the genome browser screenshot examples of Supplementary Figure S4A, showing GR and PR binding at several inter- and intra-genic regions in the vicinity of selected genes. In particular, we observed such shared binding sites on some genes known to be regulated by R5020 in T47D-MTVL (WT) (56,58–59) and also involved in either epithelial-to-mesenchymal transition and cell differentiation (i.e. STAT5A, ELF5 and SNAI1) (60,61), and cell cycling/proliferation (i.e.GREB1) (62,63) (Supplementary Figure S4A).
GR alters PR-mediated transcriptional response in a glucocorticoid dependent and independent manner
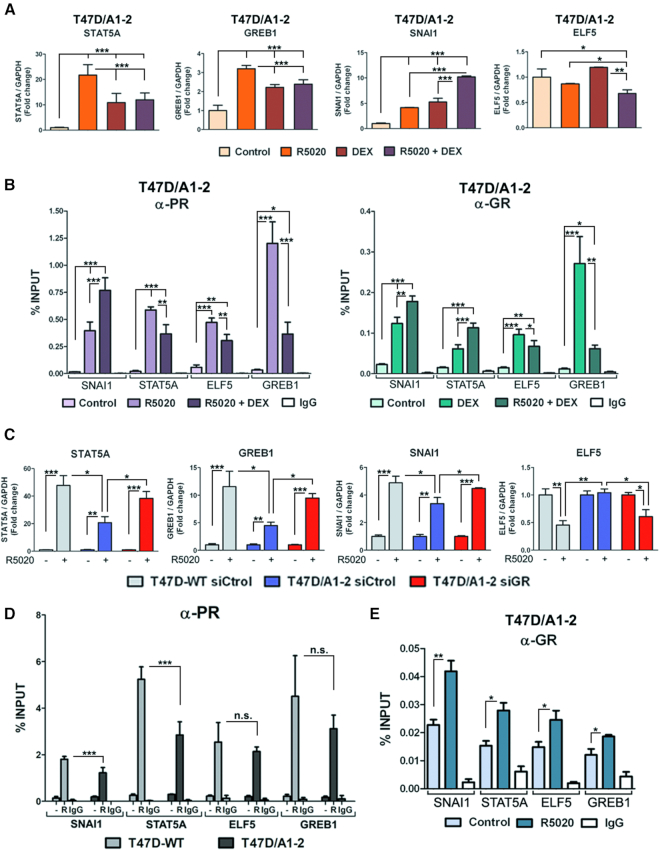
Given the observation that both PR and GR are bound to regions located nearby some genes relevant to PR-downstream activity (Supplementary Figure S4A, red boxes), we hypothesized that these genes may be transcriptionally modulated by GR. When T47D/A1-2 cells were incubated with each hormone alone, the expression of STAT5A, GREB1 and SNAI1 was induced; however, neither R5020 nor DEX affected ELF5 expression (Figure 3A). Interestingly, when T47D/A1-2 cells were co-incubated with both hormones, GR activation by DEX inhibited the R5020-dependent induction of STAT5A and GREB1 (Figure 3A). This negative modulation was not observed on SNAI1 and ELF5 expression, wherein the presence of DEX potentiated R5020-mediated SNAI1 increase while downregulating ELF5 transcript levels. At the genomic level, R5020-dependent PR recruitment to the regulatory regions located in the vicinity of STAT5A, ELF5 and GREB1 significantly diminished when cells were co-treated with both hormones, while PR binding increased at the regulatory regions associated with SNAI1 (Figure 3B, left panel). Oppositely, DEX-dependent GR recruitment to the same regions was also affected by the presence of the progestin (Figure 3B, right panel). In fact, R5020 potentiates binding of the activated GR to regulatory regions associated with SNAI1 and STAT5A while it hinders GR recruitment to ELF5 and GREB1. Taken together, these results indicate that GR and PR are able to modulate each other′s activities upon activation with their cognate hormones. The functional crosstalk between both receptors would be mediated, at least in part, through their recruitment to shared regulatory regions.
Figure 3.
GR affects PR binding and transcriptional activity of some progestin-response genes. (A) Gene expression changes measured by RT-qPCR for selected genes in T47D/A1-2 cells untreated (control) or treated with R5020 10 nM, DEX 10 nM or both hormones for 6 h. Values were normalized with GAPDH expression. Means ± S.D. from three independent experiments performed in duplicate are shown. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. (B) Chromatin occupancy analysis measured by ChIP-qPCR for selected regions of PR (left panel) and GR (right panel) in T47D/A1-2 cells treated for 1 h with R5020 10 nM, DEX 10 nM or both hormones. IgG was used as a negative control. Data are presented as percent input and histograms show the mean ± S.D. of three experiments performed in duplicate. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. (C) Gene expression changes measured by RT-qPCR for selected genes in T47D-WT and T47D/A1-2 cells transfected with control (Ctrol) or GR siRNAs and then treated with R5020 10 nM for 6 h. Means ± S.D. from three independent experiments performed in duplicate are shown. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. (D) Chromatin occupancy analysis measured by ChIP-qPCR for selected regions of PR in T47D/A1-2 and T47D-WT cells treated for 1 h with R5020 10 nM. IgG was used as a negative control. Data are presented as percent input and the histograms show the mean ± S.D. of three experiments performed in duplicate. ***, P ≤ 0.001; n.s.: not significant. R: R5020. (E) Chromatin occupancy analysis measured by ChIP-qPCR for selected regions of GR in T47D-WT and T47D/A1-2 cells treated for 1 h with R5020 10 nM. IgG was used as a negative control. Data are presented as percent input and the histograms show the mean ± S.D. of three experiments performed in duplicate. **, P ≤ 0.01; *, P ≤ 0.05.
Since GR appears to be sufficient to modulate PR activity in the absence of a glucocorticoid stimulus (Figure 1), we wondered whether GR expression could affect the transcriptional levels of the genes originally selected based on shared GR-PR binding. Strikingly, STAT5A, GREB1 and SNAI1 genes exhibit a diminished R5020-dependent transcriptional activity in T47D/A1-2 compared to WT cells. Conversely, ELF5, which is repressed by R5020, did not show a significant level of transcriptional repression in the T47D/A1-2 compared to T47D-WT cells (Figure 3C). Indeed, depletion of GR by siRNA knockdown restores the progestin-dependent expression of STAT5A, GREB1, SNAI1 and ELF5 which reached comparable levels to those observed in WT cells (Figure 3C). Other relevant genes such as EGFR and BCL-X, also known to be regulated by progestins (56,64) exhibited a similar transcriptional activity in response to R5020 in both cell lines (Supplementary Figure S4B), pointing to a gene-specific GR-PR modulation.
The crosstalk between GR and PR was not only constraint to T47D cells, as similar results were obtained in MCF-7L, another breast-cancer cell line (Supplementary Figure S5). These cells, contrary to the original MCF-7, express endogenous PR and GR independently of ER activity (65). RT-qPCRs of selected genes show that GR activation by DEX inhibited the R5020-dependent induction of STAT5A, SNAI1A and EGFR, wherein the presence of DEX potentiated R5020-mediated GREB1 and ELF5 increase (Supplementary Figure S5A). On the other hand, R5020 affects DEX-dependent induction of Bcl-X. These results were confirmed by depletion of GR by siRNA knockdown, where the progestin-dependent expression of those genes was restored in the absence of GR (Supplementary Figure S5B). Moreover, cell cycle analyses performed in MCF-7L cells treated for 18 h with R5020 show that the percentage of cells accumulated in S phase was significantly higher compared to untreated cells (13.7 ± 0.7% versus 10.2 ± 0.3%) (Supplementary Figure S5C). DEX alone did not affect S phase accumulation (10.3 ± 0.6%) but inhibited R5020 mediated action (10.9 ± 0.9%). Together, these findings support the idea that both receptors are able to modulate each other’s activities upon activation with their cognate hormones in breast cancer cells.
The recruitment of PR to the regulatory regions associated with the selected genes was assessed by ChIP-qPCR assays. Results show significantly reduced PR recruitment in T47D/A1-2 cells compared to T47D-WT upon R5020 stimulation in SNAI1 and STAT5A regulatory regions. On the other hand, we detected no changes in the regions associated with the ELF5 and GREB1 genes (Figure 3D). Intriguingly, despite DEX is not affecting PR recruitment (Supplementary Figure S4C), GR binding to these regulatory regions significantly increased in cells treated with the progestin-alone compared to untreated cells (Figure 3E), hinting at a possible R5020-dependent GR activation. Collectively, these results support the idea that both glucocorticoid-bound and glucocorticoid-free GR hinders PR-mediated transcriptional regulation of a certain subset of genes, either by impairing the recruitment of PR to those shared regulatory regions, or by affecting unidentified downstream processes.
GR and PR co-bind at a subset of genomic sites
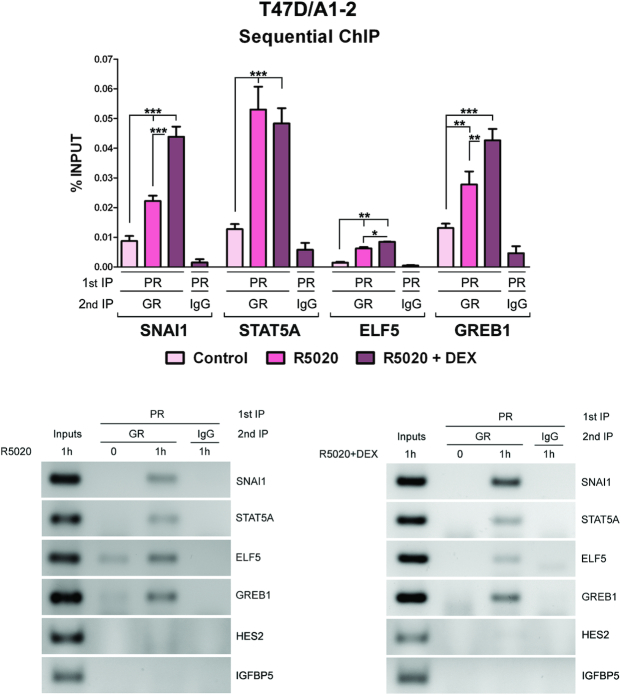
The above findings led us to hypothesize that GR and PR may simultaneously co-bind at these regions. To test this hypothesis, we performed sequential ChIP for the selected PR/GR binding regions associated with STAT5A, GREB1, SNAI1 and ELF5 genes in T47D/A1-2 cells. Interestingly, R5020 is sufficient to trigger GR and PR co-binding at these regions (Figure 4). Co-stimulation with both hormones further increased GR/PR co-recruitment at all loci, except at STAT5A. Sequential ChIP analysis performed in regions containing PR enriched binding sites (i.e. regions associated with HES2 and IGFBP5 genes, Supplementary Figure S6) did not evidence PR/GR co-recruitment after the exposure of R5020 alone or R5020 plus DEX (Figure 4). Taken together, these results indicate that GR binding is mainly directed to regions that are also targets of the PR upon R5020 stimulation. Moreover, for a subset of these regions, increased co-occupation with both receptors is observed upon simultaneous hormone treatment, suggesting that the activated GR and PR integrate the same protein complex. Although only explored at selected regions, in this work we predict co-binding could occur at many genomic loci given the large fraction of PR and GR shared binding regions (Figure 2).
Figure 4.
PR and GR can simultaneously bind the same chromatin binding sites. Sequential ChIP-qPCR analysis (measured by PR followed by GR ChIP for selected regions) in T47D/A1-2 cells treated 1 h with R5020 10 nM or R5020 plus DEX 10 nM. IgG was used as a negative control. Data are presented as percent input (upper panel). Histograms show the mean ± S.D. of three experiments performed in duplicate. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. Bottom panels show the results in agarose gel of one representative experiment.
GR and PR can be part of the same complex
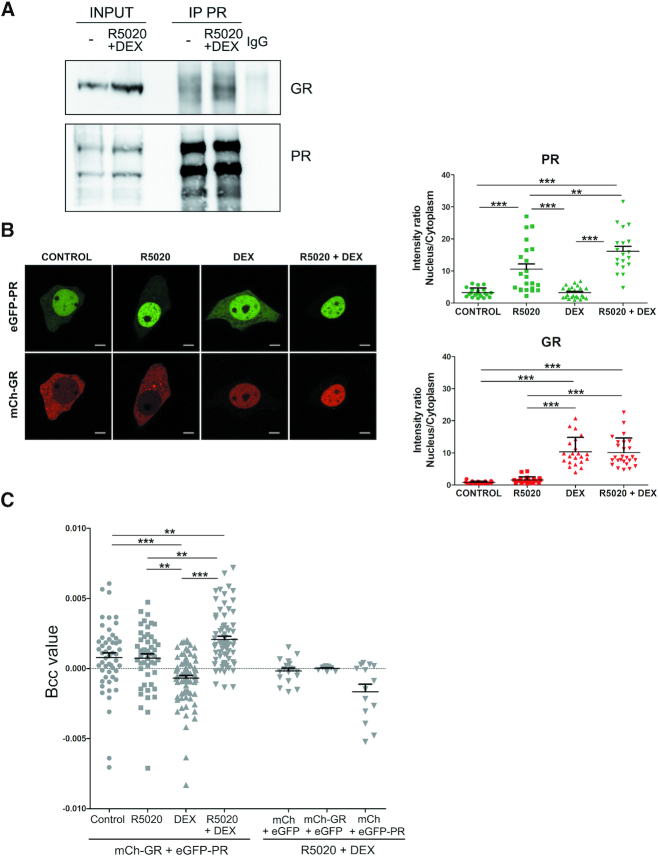
Bearing in mind previous reports showing steroid receptors ability to interact with each other (66–68), we sought to evaluate whether GR could physically interact with PR. To identify a potential association between GR and PR in the presence or in the absence of hormones, we performed Co-IP experiments using PR or GR specific antibodies from nuclear extracts of T47D/A1-2 cells treated with both agonists. Our results indicated that both PR and GR co-precipitate after 1 h treatment with R5020 and DEX (Figure 5A, compare IP PR: control (-) versus R5020+DEX).
Figure 5.
PR and GR can be part of the same complex. (A) Nuclear extracts from T47D/A1-2 cells treated or not with R5020 10 nM and DEX 10 nM for 1 h were immunoprecipitated with α-PR specific antibody. Rabbit IgG was used as a negative control (IgG). The immunoprecipitates (IP) and inputs lysates were analyzed by western blotting with α-GR or α-PR as indicated. Blots correspond to one representative from three independent experiments. (B and C) T47D-WT cells transiently expressing mCherry-GR and GFP-PR were treated with R5020 10 nM and/or DEX 10 nM for 1 h and imaged by confocal microscopy. (B) Subcellular distribution of mCherry-GR and GFP-PR in each condition. Representative cells are shown (Scale bar: 7 μm) (left panel). Fluorescence intensity ratios between nucleus and cytoplasm for each receptor in the indicated conditions are shown (right panels). ***, P ≤ 0.001; **, P ≤ 0.01. (C) Cross-correlation analysis of the fluorescence fluctuations was performed. A scatter plot with the mean ± S.E.M. of the cross-brightness (Bcc) values in the nuclei (n = 50) is shown. eGFP and mCherry expression vectors were used as a negative control. ***, P ≤ 0.001; **, P ≤ 0.01.
To directly evaluate GR and PR interactions in living cells, we run imaging and fluorescence fluctuation cross-correlation experiments (37) by co-transfecting T47D-WT cells with peEGFP-PR (35) and pmCherry-GR (36) expression vectors. Figure 5B displays representative images of these cells (left panel) and shows the intensity ratios between the nucleus and cytoplasm for each receptor (right panel). Non-stimulated PR mainly localizes to the nucleus and GR mostly distributes in the cytoplasm, while nuclear localization of both receptors significantly increases upon 30 min stimulation with their respective hormones. We did not observe GR nuclear translocation after R5020 10 nM stimulation.
Analysis of intensity fluctuations showed no cross-correlation (cross-brightness [Bcc] = 0) in cells expressing eGFP and mCherry (Figure 5C), as expected from the absence of interaction between these two molecules. We obtained similar results with cells expressing eGFP-PR and mCherry or mCherry-GR and eGFP (Figure 5C), where interaction was also not expected. Cells transfected with mCherry-GR and eGFP-PR also showed a mean Bcc value that did not differ from that obtained in cells expressing eGFP and mCherry, when untreated or treated with either hormone (Figure 5C). Nevertheless, and crucially, cells treated with both hormones simultaneously showed a positive and significantly greater mean Bcc value (Figure 5C), suggesting the presence of protein-complexes containing both activated receptors. Taken together these results indicate that both receptors can be part of the same complex in living breast cancer cells.
GR mapping upon R5020 stimulation reveals an unexpected pattern of binding sites
The results presented in the previous sections showed that, in the absence of glucocorticoids and upon R5020 stimulation, GR expression interferes with the recruitment of PR to certain GR/PR shared regions, affecting PR-mediated gene expression regulation. To gain insights into the potential mechanisms involved, we profiled by ChIP-seq the genome-wide binding pattern of GR in T47D/A1-2 cells treated with R5020 alone (Figure 6). We found a total of 633 consistent binding regions between replicates, wherein almost half are also bound by GR and/or PR upon stimulation with their corresponding hormones (Figure 6A). The other half (321 peaks) are not bound by GR or PR in either non-stimulated cells or in cells stimulated with the cognate hormone for each receptor (Figure 6A). Thus, these regions are GR unique binding sites that appear selectively upon R5020 stimulation. We refer hereon to these regions as ‘GR R5020 unique’.
In agreement with these observations, the regions bound by GR upon R5020 treatment are moderately enriched for GR and PR occupancy upon stimulation with DEX or R5020, respectively, as depicted by the corresponding normalized ChIP-seq signals (Supplementary Figure S7A). Focusing on the receptor enrichments at the top 10% regions with higher occupancy for each of the GR and PR datasets analyzed in this study, we found that while there was extensive co-enrichment of PR and GR occupancy upon stimulation with their respective hormones, this was not the case at the GR R5020-bound regions (Figure 6B). Thus, highly occupied regions by GR upon R5020 treatment are neither enriched in signal from the GR-DEX or PR-R5020 (Figure 6B). These results indicate that, unexpectedly, novel and highly occupied GR binding regions appear after R5020 treatment. These are genomic sites that are not occupied by either activated GR or PR upon cell treatment with their corresponding hormones.
R5020 treatment drives novel GR recruitment to unique genomic regions
To further explore the reasons for differential binding of GR upon R5020 treatment, we performed de novo motif analysis on these set of regions. Unexpectedly, the top de novo motif recovered when analyzing all the GR R5020 peaks was REL and not the GR motif (Supplementary Figure S7B and Table S4). The GR motif itself was not significant enrichment in this analysis. In fact, constraining the de novo motif analysis using the GR R5020 unique regions (i.e. those that were not bound by GR or PR in the other datasets profiled in this study, Figure 6A) did not change the results (Figure 6C; Supplementary Figure S7C and Table S5).The GR motif was, again, not enriched in this dataset. Conversely, the GR motif was significant enriched and was recovered as the top result when the de novo motif analysis was performed in the complementary GR R5020 regions (i.e. the 312 regions that are not GR R5020 unique, Figure 6A and Supplementary Table S6). Taken together, these results suggest that GR-binding at the GR-R5020 unique regions occurs tethered to other proteins that would recognize REL motifs such as NFκB-like transcription factors (69). Alternatively, it could also suggest direct binding to these motifs, as recently reported (70). To gain further insights into these findings, we directly interrogated all the GR and PR datasets used in this study for enrichments of the two top de novo motifs recovered from GR R5020 unique regions. Interestingly, the REL and FOXH1 motifs were only enriched in the GR R5020 regions (Figure 6D). Taken together, these results suggest a novel mechanism for GR interference with the PR pathway in which R5020 treatment drives novel GR tethering/direct binding to unique genomic regions, otherwise not bound by the GR, which could potentially be co-bound by REL and FOXH1 proteins.
We, at last focused in the GR R5020 unique binding sites that contain REL motifs (82 regions). A gene ontology analysis of the nearby genes revealed a significant enrichment for proteins involved in the SWI/SNF and other chromatin remodeler complexes (Figure 6E). Such genes, encoding for well-known chromatin remodelers as SMARCD2, ARID1A and INO80C, did present clear GR R5020 peaks at their promoter and/or enhancer associated regions that were not otherwise bound by the GR or the PR in any of the tested conditions (Figure 6F). This receptor-selective binding profile was further validated by ChIP-qPCR, in which we detected binding of GR without co-binding of PR at SMARCD2, ARID1A and INO80C regions (Supplementary Figure S7C-D). The expression of SMARCD2 and ARID1A genes was increased in T47D-WT but not in T47D/A1-2 cells when treated with R5020 (Figure 6G). In contrast, the expression of INO80C was not modified after 12 h of treatment with R5020 in T47D-WT cells. However, it was inhibited in T47D/A1-2 cells. Depletion of GR by siRNA knockdown restored the progestin-dependent transcriptional activity of these genes (Figure 6G). Taken together, this provocative finding suggests that upon R5020 addition the GR is able to bind novel, unique genomic regions not bound otherwise by GR upon stimulation with its cognate hormone. These exclusive binding events could involve a direct interaction of the receptor to the DNA or be facilitated by GR tethering to other transcription factors.
DISCUSSION
In a physiological context, cells are contained in a complex environment and are bathed by a composite milieu where different hormones and nutrients coexist. Hence, the cellular response that evolves after the hormonal intake derives from multiple and simultaneous activation of hormone receptors which can mutually influence each other. In fact, functional crosstalk among different steroid receptors that participate in the development and the differentiation of the mammary gland has been widely proposed (1,14,16) but the molecular mechanisms associated with these receptor cross-actions are still poorly understood.
In this study, we assessed the crosstalk between PR and GR pathways in a human breast cancer cell line (T47D/A1-2) that co-expresses similar levels of both functional receptors. When GR is activated with DEX, progestin-dependent cell cycling/proliferation is completely abrogated (Figure 1). These results complement those from Wan & Nordeen carried out in the same cell line (25) and are similar to those observed in MCF-7 cells (12).
Invaluable information provided by global genomic studies achieved on the functionality of these receptors has been accumulated during the last decade (38,56,71). For example, Wan and Nordeen compared glucocorticoid- and progestin-regulated gene expression in T47D/A1-2 cells (25). Their global analysis revealed that the two hormones separately regulate many common genes, including some which are differentially regulated. In fact, the differentially regulated group was almost as large as the set of genes regulated by both hormones. These observations are consistent with the results from our ChIP-seq experiments performed in cells treated with either R5020 or DEX alone (Figure 2), in which an important fraction of GR binding regions is shared with the PR, and they both show enrichments for known and common DNA-binding motifs (Supplementary Figure S3). This suggests that in cells treated with both hormones, PR and GR would have the ability to either displace each other or to be recruited directly and simultaneously to the same receptor binding site.
The genes studied in our experimental model were previously identified as PR-target genes in other systems (56,58–63). Interestingly, we found shared PR/GR binding sites nearby these genes (Figure 3). We therefore used this set of potential enhancers as a proxy to identify the mechanisms involved in GR-mediated inhibition of PR action. ChIP-qPCR results revealed a complex gene-specific effect. For SNAI1, both GR and PR independently bind to the promoter region and consistently, both receptors increase SNAI1 transcription. Interestingly, co-activation, and at least a fraction of co-binding promotes a synergistic effect on both binding and mRNA levels (Figures 3 and 4). On the other hand, GR inhibits PR-dependent STAT5A expression at least partially by decreasing PR binding to the putative enhancer (Figure 3). A similar transcriptional effect is observed in GREB1, however, both GR and PR decrease their binding at the shared sites. Together, these examples illustrate the complexity of the crosstalk at the genomic level.
The role of steroid receptors crosstalk in cancer is becoming increasingly relevant (72), although the mechanisms involved are still a matter of debate. For example, heterodimerization has been proposed for GR–ER (11,13,73); GR-MR (74) and GR–AR (75). Other mechanisms beyond protein–protein complex formation involves assisted loading (76), where dynamic binding of one receptor can lead to more accessible chromatin, allowing access of the second receptor, resulting in additional chromatin remodeling and recruitment of additional transcription factors that can regulate cooperative gene expression, as in the case of GR assisting ER binding in mammary cells (13,14). Our data from co-immunoprecipitation assays and cross-correlation analysis in living cells (Figure 5) suggest that GR and PR can form part of the same complex, although a direct physical interaction has yet to be confirmed. Since higher oligomerization states have been described for both PR and GR (18), we propose that these receptors can form transcriptional active heterocomplexes. Several questions arise from this hypothesis: Are GR–PR complexes able to bind PR- or GR-specific response elements? Do these heterocomplexes affect the dynamics of recruitment of each receptor to their sequence-specific DNA binding sites and consequently modulate each other′s transcriptional activity? How would the transcription outcome be affected when PR–GR heterocomplexes are recruited to those sites instead of GR–GR or PR–PR homo-oligomers? Particularly, at specific regions associated with the above-mentioned progestin–target genes, sequential ChIPs performed upon co-stimulation showed that both activated receptors bind simultaneously to these genomic sites (Figure 4), supporting the existence of PR–GR heterocomplexes. The capability of these PR–GR complexes to regulate gene expression would depend on their target gene context. In fact, GR activation by DEX inhibited STAT5A and GREB1 but increased SNAI1 and ELF5 R5020-dependent gene regulation in T47D/A1-2 cells while in MCF-7L, DEX treatment inhibited R5020-dependent expression of STAT5A; SNAI1 and EGFR but increased GREB1 and ELF5. Thus our findings suggest a different transcriptional outcome for PR-GR compared with PR–PR complexes in breast cancer cells. Differences in the gene expression outcome could be due to distinct epigenetic states at the receptor's binding sites.
Surprisingly, we also observed that the presence of GR, even in the absence of glucocorticoids, decreased progestin-dependent cell proliferation (Figure 1). This effect seems to occur at the genomic level, as PR binding is reduced at some enhancers (Figure 3). Since GR in the absence of ligand has almost no biological activity, we hypothesized that R5020 could modulate the GR, most likely activating the receptor (77). In fact, GR ChIP-seq of cells treated with R5020-only revealed an intriguing binding profile, with slightly more than six hundred peaks identified with high confidence (Supplementary Figure S7). These binding regions are enriched in REL and FOXH1 DNA binding motifs, while these motifs are absent in GR or PR activated with their respective hormones. Such novel GR-binding profile could be mediated by other factors that might include NFĸB-like transcription factors, since PR was not recruited to those regions. In this sense, among the different mechanisms that explain the functional crosstalk between the GR and NFĸ-B signaling pathways (69), a physical interaction between GR and NFĸ-B p65 subunit could mediate GR recruitment to REL motifs. Alternatively, it was recently suggested that GR can bind directly to ĸ-B response elements to repress gene expression (70).
Gene ontology analysis of the genes associated to GR R5020 unique binding sites containing REL motifs revealed a significant enrichment for proteins involved in the SWI/SNF and other chromatin remodeler complexes. Indeed, the expression genes associated with these remodeler complexes, such as SMARCD2, ARID1A and INO80C, was inhibited in response to R5020 in cells expressing GR (Figure 6). We therefore suggest a potential second-target effect of GR on PR activity, where progestins—via GR—modulate the amount of chromatin remodeler's availability.
In conclusion, GR negatively modulates PR activity through at least two independent mechanisms, depending on which hormones are present in the system. First, in the absence of glucocorticoids, R5020 activates both PR and GR. This, nevertheless, would trigger PR binding to ∼28 500 sites and only ∼630 peaks for GR, half of which are shared with PR. The small number of GR binding sites may be explained by the low amount of GR molecules in the nucleus, as R5020 10 nM failed to induce translocation of the receptor (Figure 2B). We therefore propose that a redistribution of the nuclear pool of GR molecules are recruited to these new binding sites. Some of these shared sites may contain GR/PR complexes (as in SNAI1 and STAT5A enhancers) and ultimately lead to a decrease in PR binding. However, this is not a general mechanism, as some co-binding events do not affect PR binding (as in ELF5 and GREB1). The GR-R5020 unique sites are not enriched in canonical GREs, suggesting alternative modes of action. Our data points to REL as a candidate transcription factor, however, further experiments are needed to confirm this hypothesis. GR–R5020-dependent binding produces transcriptional regulation, as observed in the expression of several chromatin remodelers. These findings open the possibility of another type of regulation of GR function and reveal a potential pharmacological interest of the R5020–GR complex. Second, in the presence of DEX and R5020, GR–DEX complexes most likely take over the GR–R5020 complexes and hence the modulation of PR activity by GR would be different. GR–DEX complexes bind many more sites than GR–R5020 and many regions are shared between GR and PR, which are in general enriched in hormone responsive elements. We detected an increase in PR-GR interaction both in the nucleoplasm and at individual loci. Taken together, the heterogeneity in the behavior at specific regulatory regions could contribute to an overall reduction in PR activity, evidenced by cell proliferation and differentiation assays.
Steroid receptors are among the most promising targets for translational research. An overwhelming majority of past work investigating hormone-driven cancers have been focused on individual receptors and single hormone treatment conditions. Although this has allowed us to understand steroid receptor function, physiologically, we need to take into account multiple stimuli (78). In this sense, the study of the steroid receptor interactions in hormone-dependent cancers is particularly relevant. Here, we explored different potential mechanisms wherein GR affect PR activity after progestin′s treatment both in the absence and in the presence of DEX, a glucocorticoid widely used as palliative in different breast cancer therapies. Although the exact role of these mechanisms requires further investigation, understanding this crosstalk could explain the better prognosis of PR+/ER+/GR+ breast tumors and may contribute to the development of new endocrine combined therapies.
DATA AVAILABILITY
ChIP-seq data are submitted to the NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database, accession code: GSE126859.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Steven K Nordeen (University of Colorado, Department of Pathology) for the T47D/A1-2 cells used throughout this study; Dr C. Kent Osborne (Baylor College of Medicine, Houston, TX, USA) for the MCF-7L cell line; Dr Carol Lange, Departments of Medicine and Pharmacology, University of Minnesota Masonic Cancer Center, Minneapolis, USA, for the peGFP-PR expression vectors and Dr Miguel Beato, Centre de Regulació Genomica, Barcelona, Spain for kindly providing the necessary materials and facilities to perform ChIP-seq assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness, Spain [SAF2016-75006P]; CONICET [PIP112-200801-00859]; Agencia Nacional de Programación Científica y Tecnológica [Préstamo BID-PICT2014-0630]; University of Buenos Aires, Argentina [20820180100745BA]. CONICET-Argentina (M.F.O., S.A.R.S., V.L, D.M.P, A.P.); Spanish National Research Council (CSIC), Spain (to G.P.V.); IUBMB Short-term Scholarship (to M.F.O, M.S.M.); EMBO Short-term Scholarship (to M.F.O.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Brisken C., O’Malley B.. Hormone action in the mammary gland. Cold Spring Harb. Perspect. Biol. 2010; 2:a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aupperlee M., Kariagina A., Osuch J., Haslam S.Z.. Progestins and breast cancer. Breast Dis. 2005; 24:37–57. [DOI] [PubMed] [Google Scholar]
- 3. Vilasco M., Communal L., Mourra N., Courtin A., Forgez P., Gompel A.. Glucocorticoid receptor and breast cancer. Breast Cancer Res. Treat. 2011; 130:1–10. [DOI] [PubMed] [Google Scholar]
- 4. Santen R., Cavalieri E., Rogan E., Russo J., Guttenplan J., Ingle J., Yue W.. Estrogen mediation of breast tumor formation involves estrogen receptor-dependent, as well as independent, genotoxic effects. Ann. N.Y. Acad. Sci. 2009; 1155:132–140. [DOI] [PubMed] [Google Scholar]
- 5. Lange C.A., Sartorius C.A., Abdel-Hafiz H., Spillman M.A., Horwitz K.B., Jacobsen B.M.. Progesterone receptor action: translating studies in breast cancer models to clinical insights. Adv. Exp. Med. Biol. 2008; 630:94–111. [PubMed] [Google Scholar]
- 6. Carroll J.S., Hickey T.E., Tarulli G.A., Williams M., Tilley W.D.. Deciphering the divergent roles of progestogens in breast cancer. Nat. Rev. Cancer. 2017; 17:54–64. [DOI] [PubMed] [Google Scholar]
- 7. Mohammed H., Russell I.A., Stark R., Rueda O.M., Hickey T.E., Tarulli G.A., Serandour A.A., Birrell S.N., Bruna A., Saadi A. et al.. Progesterone receptor modulates ERalpha action in breast cancer. Nature. 2015; 523:313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoijman E., Rocha-Viegas L., Kalko S.G., Rubinstein N., Morales-Ruiz M., Joffe E.B., Kordon E.C., Pecci A.. Glucocorticoid alternative effects on proliferating and differentiated mammary epithelium are associated to opposite regulation of cell-cycle inhibitor expression. J. Cell Physiol. 2012; 227:1721–1730. [DOI] [PubMed] [Google Scholar]
- 9. Bertucci P.Y., Quaglino A., Pozzi A.G., Kordon E.C., Pecci A.. Glucocorticoid-induced impairment of mammary gland involution is associated with STAT5 and STAT3 signaling modulation. Endocrinology. 2010; 151:5730–5740. [DOI] [PubMed] [Google Scholar]
- 10. Voutsadakis I.A. Epithelial-Mesenchymal Transition (EMT) and regulation of EMT factors by steroid nuclear receptors in breast cancer: a review and in silico investigation. J. Clin. Med. 2016; 5:11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karmakar S., Jin Y., Nagaich A.K.. Interaction of glucocorticoid receptor (GR) with estrogen receptor (ER) alpha and activator protein 1 (AP1) in dexamethasone-mediated interference of ERalpha activity. J. Biol. Chem. 2013; 288:24020–24034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hegde S.M., Kumar M.N., Kavya K., Kumar K.M., Nagesh R., Patil R.H., Babu R.L., Ramesh G.T., Sharma S.C.. Interplay of nuclear receptors (ER, PR, and GR) and their steroid hormones in MCF-7 cells. Mol. Cell Biochem. 2016; 422:109–120. [DOI] [PubMed] [Google Scholar]
- 13. Miranda T.B., Voss T.C., Sung M.H., Baek S., John S., Hawkins M., Grontved L., Schiltz R.L., Hager G.L.. Reprogramming the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res. 2013; 73:5130–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. West D.C., Pan D., Tonsing-Carter E.Y., Hernandez K.M., Pierce C.F., Styke S.C., Bowie K.R., Garcia T.I., Kocherginsky M., Conzen S.D.. GR and ER coactivation alters the expression of differentiation genes and associates with improved ER+ breast cancer outcome. Mol. Cancer Res. 2016; 14:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bolt M.J., Stossi F., Newberg J.Y., Orjalo A., Johansson H.E., Mancini M.A.. Coactivators enable glucocorticoid receptor recruitment to fine-tune estrogen receptor transcriptional responses. Nucleic Acids Res. 2013; 41:4036–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou F., Bouillard B., Pharaboz-Joly M.O., Andre J.. Non-classical antiestrogenic actions of dexamethasone in variant MCF-7 human breast cancer cells in culture. Mol. Cell Endocrinol. 1989; 66:189–197. [DOI] [PubMed] [Google Scholar]
- 17. Buxant F., Kindt N., Laurent G., Noel J.C., Saussez S.. Antiproliferative effect of dexamethasone in the MCF-7 breast cancer cell line. Mol. Med. Rep. 2015; 12:4051–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Presman D.M., Ganguly S., Schiltz R.L., Johnson T.A., Karpova T.S., Hager G.L.. DNA binding triggers tetramerization of the glucocorticoid receptor in live cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8236–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J., Li Q.. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol. Endocrinol. 2003; 17:1681–1692. [DOI] [PubMed] [Google Scholar]
- 20. McKenna N.J., O’Malley B.W.. SnapShot: NR coregulators. Cell. 2010; 143:172–172. [DOI] [PubMed] [Google Scholar]
- 21. Vicent G.P., Zaurin R., Nacht A.S., Li A., Font-Mateu J., Le Dily F., Vermeulen M., Mann M., Beato M.. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLos Genet. 2009; 5:e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Africander D., Verhoog N., Hapgood J.P.. Molecular mechanisms of steroid receptor-mediated actions by synthetic progestins used in HRT and contraception. Steroids. 2011; 76:636–652. [DOI] [PubMed] [Google Scholar]
- 23. Goya L., Maiyar A.C., Ge Y., Firestone G.L.. Glucocorticoids induce a G1/G0 cell cycle arrest of Con8 rat mammary tumor cells that is synchronously reversed by steroid withdrawal or addition of transforming growth factor-alpha. Mol. Endocrinol. 1993; 7:1121–1132. [DOI] [PubMed] [Google Scholar]
- 24. Horwitz K.B. The molecular biology of RU486. Is there a role for antiprogestins in the treatment of breast cancer. Endocr. Rev. 1992; 13:146–163. [DOI] [PubMed] [Google Scholar]
- 25. Wan Y., Nordeen S.K.. Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol. Endocrinol. 2002; 16:1204–1214. [DOI] [PubMed] [Google Scholar]
- 26. Buser A.C., Obr A.E., Kabotyanski E.B., Grimm S.L., Rosen J.M., Edwards D.P.. Progesterone receptor directly inhibits beta-casein gene transcription in mammary epithelial cells through promoting promoter and enhancer repressive chromatin modifications. Mol. Endocrinol. 2011; 25:955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nordeen S.K., Kuhnel B., Lawler-Heavner J., Barber D.A., Edwards D.P.. A quantitative comparison of dual control of a hormone response element by progestins and glucocorticoids in the same cell line. Mol. Endocrinol. 1989; 3:1270–1278. [DOI] [PubMed] [Google Scholar]
- 28. Shao R., Egecioglu E., Weijdegard B., Ljungstrom K., Ling C., Fernandez-Rodriguez J., Billig H.. Developmental and hormonal regulation of progesterone receptor A-form expression in female mouse lung in vivo: interaction with glucocorticoid receptors. J. Endocrinol. 2006; 190:857–870. [DOI] [PubMed] [Google Scholar]
- 29. Vegeto E., Shahbaz M.M., Wen D.X., Goldman M.E., O’Malley B.W., McDonnell D.P.. Human progesterone receptor A form is a cell- and promoter-specific repressor of human progesterone receptor B function. Mol. Endocrinol. 1993; 7:1244–1255. [DOI] [PubMed] [Google Scholar]
- 30. Leo J.C., Guo C., Woon C.T., Aw S.E., Lin V.C.. Glucocorticoid and mineralocorticoid cross-talk with progesterone receptor to induce focal adhesion and growth inhibition in breast cancer cells. Endocrinology. 2004; 145:1314–1321. [DOI] [PubMed] [Google Scholar]
- 31. McNamara K.M., Kannai A., Sasano H.. Possible roles for glucocorticoid signalling in breast cancer. Mol. Cell Endocrinol. 2018; 466:38–50. [DOI] [PubMed] [Google Scholar]
- 32. Truss M., Bartsch J., Schelbert A., Hache R.J., Beato M.. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995; 14:1737–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arteaga C.L., Coronado E., Osborne C.K.. Blockade of the epidermal growth factor receptor inhibits transforming growth factor alpha-induced but not estrogen-induced growth of hormone-dependent human breast cancer. Mol. Endocrinol. 1988; 2:1064–1069. [DOI] [PubMed] [Google Scholar]
- 34. Cittelly D.M., Finlay-Schultz J., Howe E.N., Spoelstra N.S., Axlund S.D., Hendricks P., Jacobsen B.M., Sartorius C.A., Richer J.K.. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene. 2013; 32:2555–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diep C.H., Charles N.J., Gilks C.B., Kalloger S.E., Argenta P.A., Lange C.A.. Progesterone receptors induce FOXO1-dependent senescence in ovarian cancer cells. Cell Cycle. 2013; 12:1433–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Presman D.M., Ogara M.F., Stortz M., Alvarez L.D., Pooley J.R., Schiltz R.L., Grontved L., Johnson T.A., Mittelstadt P.R., Ashwell J.D. et al.. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 2014; 12:e1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Digman M.A., Wiseman P.W., Choi C., Horwitz A.R., Gratton E.. Stoichiometry of molecular complexes at adhesions in living cells. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2170–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoffman J.A., Trotter K.W., Ward J.M., Archer T.K.. BRG1 governs glucocorticoid receptor interactions with chromatin and pioneer factors across the genome. Elife. 2018; 7:e35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cebola I., Rodriguez-Segui S.A., Cho C.H., Bessa J., Rovira M., Luengo M., Chhatriwala M., Berry A., Ponsa-Cobas J., Maestro M.A. et al.. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 2015; 17:615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pasquali L., Gaulton K.J., Rodriguez-Segui S.A., Mularoni L., Miguel-Escalada I., Akerman I., Tena J.J., Moran I., Gomez-Marin C., van de Bunt M. et al.. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014; 46:136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Hoon M.J., Imoto S., Nolan J., Miyano S.. Open source clustering software. Bioinformatics. 2004; 20:1453–1454. [DOI] [PubMed] [Google Scholar]
- 46. Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 47. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vicent G.P., Nacht A.S., Font-Mateu J., Castellano G., Gaveglia L., Ballare C., Beato M.. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 2011; 25:845–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vicent G.P., Ballare C., Nacht A.S., Clausell J., Subtil-Rodriguez A., Quiles I., Jordan A., Beato M.. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell. 2006; 24:367–381. [DOI] [PubMed] [Google Scholar]
- 51. Lambert J.R., Nordeen S.K.. CBP recruitment and histone acetylation in differential gene induction by glucocorticoids and progestins. Mol. Endocrinol. 2003; 17:1085–1094. [DOI] [PubMed] [Google Scholar]
- 52. Mikosz C.A., Brickley D.R., Sharkey M.S., Moran T.W., Conzen S.D.. Glucocorticoid receptor-mediated protection from apoptosis is associated with induction of the serine/threonine survival kinase gene, sgk-1. J. Biol. Chem. 2001; 276:16649–16654. [DOI] [PubMed] [Google Scholar]
- 53. Beleut M., Rajaram R.D., Caikovski M., Ayyanan A., Germano D., Choi Y., Schneider P., Brisken C.. Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A.. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011; 43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grontved L., John S., Baek S., Liu Y., Buckley J.R., Vinson C., Aguilera G., Hager G.L.. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013; 32:1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ballare C., Castellano G., Gaveglia L., Althammer S., Gonzalez-Vallinas J., Eyras E., Le Dily F., Zaurin R., Soronellas D., Vicent G.P. et al.. Nucleosome-driven transcription factor binding and gene regulation. Mol. Cell. 2013; 49:67–79. [DOI] [PubMed] [Google Scholar]
- 57. Nacht A.S., Pohl A., Zaurin R., Soronellas D., Quilez J., Sharma P., Wright R.H., Beato M., Vicent G.P.. Hormone-induced repression of genes requires BRG1-mediated H1.2 deposition at target promoters. EMBO J. 2016; 35:1822–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wierer M., Verde G., Pisano P., Molina H., Font-Mateu J., Di Croce L., Beato M.. PLK1 signaling in breast cancer cells cooperates with estrogen receptor-dependent gene transcription. Cell Rep. 2013; 3:2021–2032. [DOI] [PubMed] [Google Scholar]
- 59. Kougioumtzi A., Tsaparas P., Magklara A.. Deep sequencing reveals new aspects of progesterone receptor signaling in breast cancer cells. PLoS One. 2014; 9:e98404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hilton H.N., Kalyuga M., Cowley M.J., Alles M.C., Lee H.J., Caldon C.E., Blazek K., Kaplan W., Musgrove E.A., Daly R.J. et al.. The antiproliferative effects of progestins in T47D breast cancer cells are tempered by progestin induction of the ETS transcription factor Elf5. Mol. Endocrinol. 2010; 24:1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee H.J., Gallego-Ortega D., Ledger A., Schramek D., Joshi P., Szwarc M.M., Cho C., Lydon J.P., Khokha R., Penninger J.M. et al.. Progesterone drives mammary secretory differentiation via RankL-mediated induction of Elf5 in luminal progenitor cells. Development. 2013; 140:1397–1401. [DOI] [PubMed] [Google Scholar]
- 62. Haines C.N., Braunreiter K.M., Mo X.M., Burd C.J.. GREB1 isoforms regulate proliferation independent of ERalpha co-regulator activities in breast cancer. Endocr. Relat. Cancer. 2018; 25:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hodgkinson K.M., Vanderhyden B.C.. Consideration of GREB1 as a potential therapeutic target for hormone-responsive or endocrine-resistant cancers. Expert Opin. Ther. Targets. 2014; 18:1065–1076. [DOI] [PubMed] [Google Scholar]
- 64. Bertucci P.Y., Nacht A.S., Allo M., Rocha-Viegas L., Ballare C., Soronellas D., Castellano G., Zaurin R., Kornblihtt A.R., Beato M. et al.. Progesterone receptor induces bcl-x expression through intragenic binding sites favoring RNA polymerase II elongation. Nucleic Acids Res. 2013; 41:6072–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Daniel A.R., Gaviglio A.L., Knutson T.P., Ostrander J.H., D’Assoro A.B., Ravindranathan P., Peng Y., Raj G.V., Yee D., Lange C.A.. Progesterone receptor-B enhances estrogen responsiveness of breast cancer cells via scaffolding PELP1- and estrogen receptor-containing transcription complexes. Oncogene. 2015; 34:506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cowley S.M., Hoare S., Mosselman S., Parker M.G.. Estrogen receptors alpha and beta form heterodimers on DNA. J. Biol. Chem. 1997; 272:19858–19862. [DOI] [PubMed] [Google Scholar]
- 67. Trapp T., Holsboer F.. Heterodimerization between mineralocorticoid and glucocorticoid receptors increases the functional diversity of corticosteroid action. Trends Pharmacol. Sci. 1996; 17:145–149. [DOI] [PubMed] [Google Scholar]
- 68. Liu W., Wang J., Sauter N.K., Pearce D.. Steroid receptor heterodimerization demonstrated in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:12480–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Petta I., Dejager L., Ballegeer M., Lievens S., Tavernier J., De Bosscher K., Libert C.. The interactome of the glucocorticoid receptor and its influence on the actions of glucocorticoids in combatting inflammatory and infectious diseases. Microbiol. Mol. Biol. Rev. 2016; 80:495–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hudson W.H., Vera I.M.S., Nwachukwu J.C., Weikum E.R., Herbst A.G., Yang Q., Bain D.L., Nettles K.W., Kojetin D.J., Ortlund E.A.. Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappaB response elements. Nat. Commun. 2018; 9:1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reddy T.E., Pauli F., Sprouse R.O., Neff N.F., Newberry K.M., Garabedian M.J., Myers R.M.. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009; 19:2163–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dhiman V.K., Bolt M.J., White K.P.. Nuclear receptors in cancer - uncovering new and evolving roles through genomic analysis. Nat. Rev. Genet. 2018; 19:160–174. [DOI] [PubMed] [Google Scholar]
- 73. Vahrenkamp J.M., Yang C.H., Rodriguez A.C., Almomen A., Berrett K.C., Trujillo A.N., Guillen K.P., Welm B.E., Jarboe E.A., Janat-Amsbury M.M. et al.. Clinical and genomic crosstalk between glucocorticoid receptor and estrogen receptor alpha in endometrial cancer. Cell Rep. 2018; 22:2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Savory J.G., Prefontaine G.G., Lamprecht C., Liao M., Walther R.F., Lefebvre Y.A., Hache R.J.. Glucocorticoid receptor homodimers and glucocorticoid-mineralocorticoid receptor heterodimers form in the cytoplasm through alternative dimerization interfaces. Mol. Cell Biol. 2001; 21:781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen S., Wang J., Yu G., Liu W., Pearce D.. Androgen and glucocorticoid receptor heterodimer formation. A possible mechanism for mutual inhibition of transcriptional activity. J. Biol. Chem. 1997; 272:14087–14092. [DOI] [PubMed] [Google Scholar]
- 76. Swinstead E.E., Paakinaho V., Presman D.M., Hager G.L.. Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: A new perspective: Multiple transcription factors can effect chromatin pioneer functions through dynamic interactions with ATP-dependent chromatin remodeling factors. Bioessays. 2016; 38:1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Svec F., Yeakley J., Harrison R.W. 3rd.. Progesterone enhances glucocorticoid dissociation from the AtT-20 cell glucocorticoid receptor. Endocrinology. 1980; 107:566–572. [DOI] [PubMed] [Google Scholar]
- 78. Truong T.H., Lange C.A.. Deciphering steroid receptor crosstalk in hormone-driven cancers. Endocrinology. 2018; 159:3897–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data are submitted to the NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database, accession code: GSE126859.