Abstract
The initiation of eukaryotic DNA replication occurs in two discrete stages1: the minichromosome maintenance (MCM) complex is first loaded as a head-to-head double hexamer (DH) encircling duplex origin DNA during G1 phase; ‘firing factors’ then convert each DH into two active CMG (Cdc45-MCM-GINS) helicases during S phase. This second stage requires separation of the two origin DNA strands and remodelling of DH so that each MCM hexamer encircles a single DNA strand. We show here that MCM, which hydrolyses ATP during DH formation2,3, remains stably bound to ADP in the DH. Firing factors trigger ADP release; subsequent ATP binding promotes stable CMG assembly. CMG assembly is accompanied by initial DNA untwisting and separation of DH into two discrete but inactive CMG helicases. Mcm10, together with ATP hydrolysis, then triggers further DNA untwisting and helicase activation. After activation, the two CMG helicases translocate in an ‘N-terminus first’ direction, and in doing so pass each other within the origin, requiring that each is bound entirely to single-stranded DNA. Our experiments elucidate the mechanism of eukaryotic replicative helicase activation, which we propose provides a fail-safe mechanism for bidirectional replisome establishment.
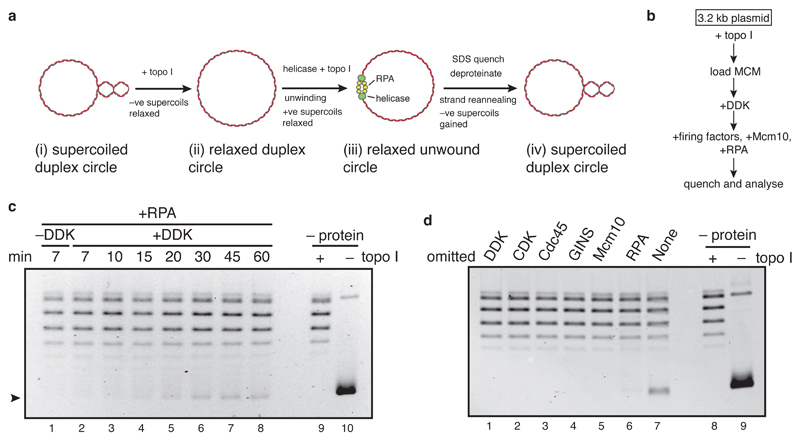
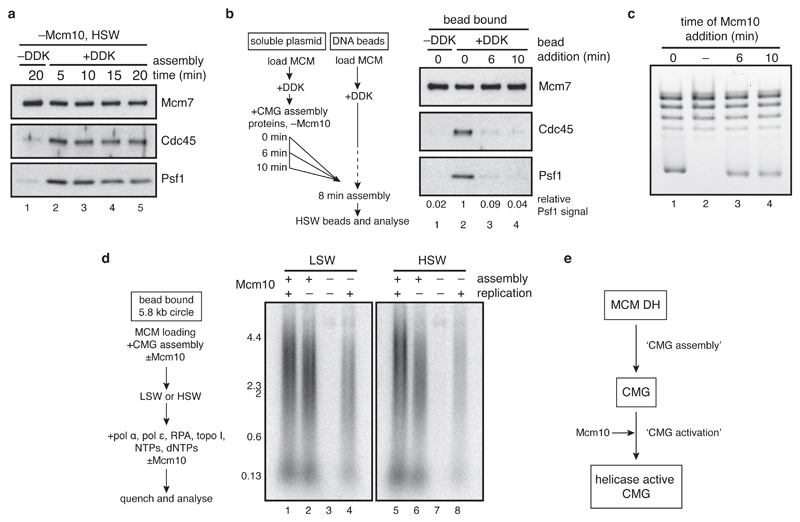
Previous studies have focussed on the activities of CMG preformed by co-overexpression of individual subunits4,5. We sought to understand how CMG is assembled and activated during the initiation of DNA replication, using purified budding yeast proteins6. We first used a DNA topology-based assay7 (Figure 1a) to study DNA unwinding by CMG. In the presence of topoisomerase I (topo I), the DNA linking number of a covalently closed circular DNA molecule decreases by one for each helical turn untwisted, allowing changes in DNA unwinding to be inferred quantitatively from changes in DNA supercoiling. MCM was loaded onto a relaxed, circular plasmid in solution, phosphorylated with Dbf4-dependent kinase (DDK), and incubated with firing factors (defined as: CDK, Cdc45, GINS, Sld2,3,7, Dpb11, DNA polymerase ε, Mcm10), RPA, and topo I in the presence of ATP (Figure 1b). After incubation, DNA was isolated and plasmid topology examined by native agarose gel electrophoresis. A fraction of the relaxed plasmid DNA became supercoiled in a time-dependent manner (Figure 1c), but not when either DDK, CDK, Cdc45, GINS, Mcm10 or RPA were omitted (Figure 1d). These results show that CMG assembled from the DH can unwind DNA even when uncoupled from DNA synthesis, consistent with previous experiments6,8. Mcm10 is required for unwinding (Figure 1d) but not CMG formation1,6. Mcm10 supported extensive unwinding and DNA synthesis even when added after CMG assembly was finished (Extended Data Figures 1a-d). Thus, Mcm10 activates CMG helicase in a distinct step after CMG assembly (Extended Data Figure 1e).
Figure 1. Analysis of replicative helicase activation with a DNA unwinding assay.
a, b, outline of the assay. c, time course of unwinding. Purified DNA products were separated on a native agarose gel and stained with ethidium bromide. No loading or firing factors were added to ‘–protein’ reactions. d, as in c, with proteins omitted as indicated. Reactions were quenched after 40 min.
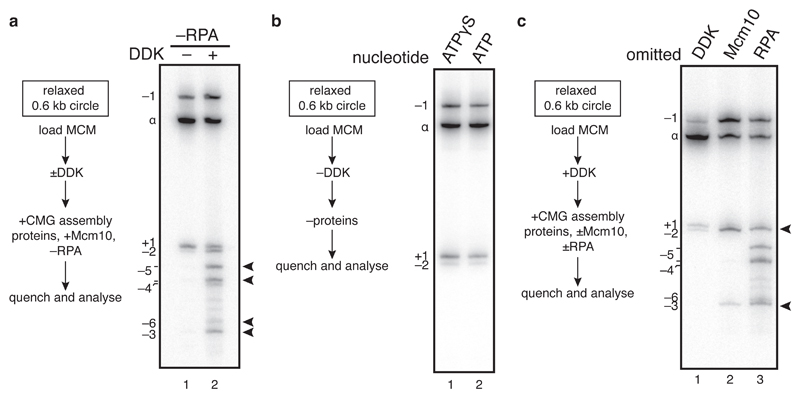
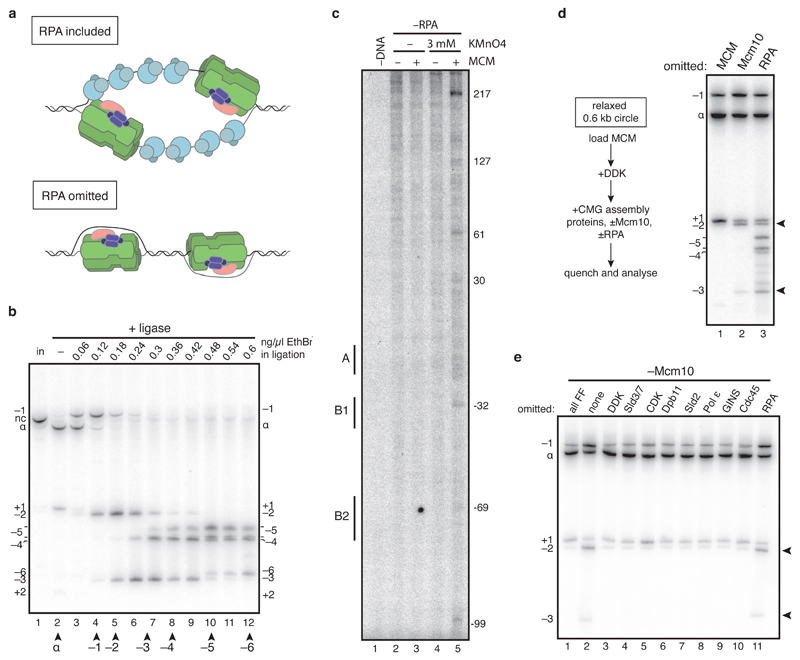
The amount of supercoiling without RPA should reflect DNA untwisting which is constrained by the CMG itself (see Extended Data Figure 2a). To assess this, we constructed a covalently closed, circular 616 base pairs (bp) radiolabeled DNA molecule, which allows us to quantify even small changes in topoisomer distribution relative to the relaxed ground state (α) (Extended Data Figure 2b). CMG assembly and activation in the absence of RPA shifted a proportion of the 4 starting topoisomers (α+1, α, α-1, α-2) to a new set of supercoils, α-3, α-4, α-5 and α-6 indicating that each circle had been unwound by 3-4 helical turns (Figure 2a, lane 2). This is likely due to genuine unwinding, because thymine residues in DNA became reactive to potassium permanganate (KMnO4) across a wide region (Extended Data Figure 2c). Thus, each of the two activated CMG helicases constrains approximately 1.5-2 helical turns of unwound DNA.
Figure 2. Origin unwinding takes place in two steps.
a, active CMG was assembled on a radiolabelled 616 bp ARS1 circle without RPA for 40 min and products separated on a native bis-polyacrylamide gel. b, as in a, except all firing factors were omitted. MCM loading does not occur in ATPγS. c, as in a, with proteins omitted as indicated.
Based on the kinked central channel at the interface between hexamers9,10, it was proposed that duplex DNA may be distorted by the DH10. However, the topoisomer distribution was identical when circles were incubated with loading factors and topo I in ATPγS (no DH assembly) or ATP (DH assembly) (Figure 2b), indicating that DNA in the DH is not significantly untwisted, consistent with recent cryo-electron microscopy (cryo-EM) structures of DH with DNA11,12. By contrast, incubation of DH with a full complement of firing factors lacking Mcm10 shifted a proportion of circles to more negatively supercoiled topoisomers (Figure 2c, lane 2, α-2 and α-3). Supercoiling required MCM loading (Extended Data Figure 2d) and each of the firing factors (Extended Data Figure 2e), indicating that it takes place during CMG assembly. This shift corresponds to a decrease in linking number of 1.3 (see methods), indicating that each MCM hexamer untwists ~0.6 - 0.7 turns of DNA. Origin melting therefore proceeds via two separable steps: untwisting of 0.6 - 0.7 turns per MCM hexamer during CMG assembly, and approximately one further turn when CMG is activated by Mcm10.
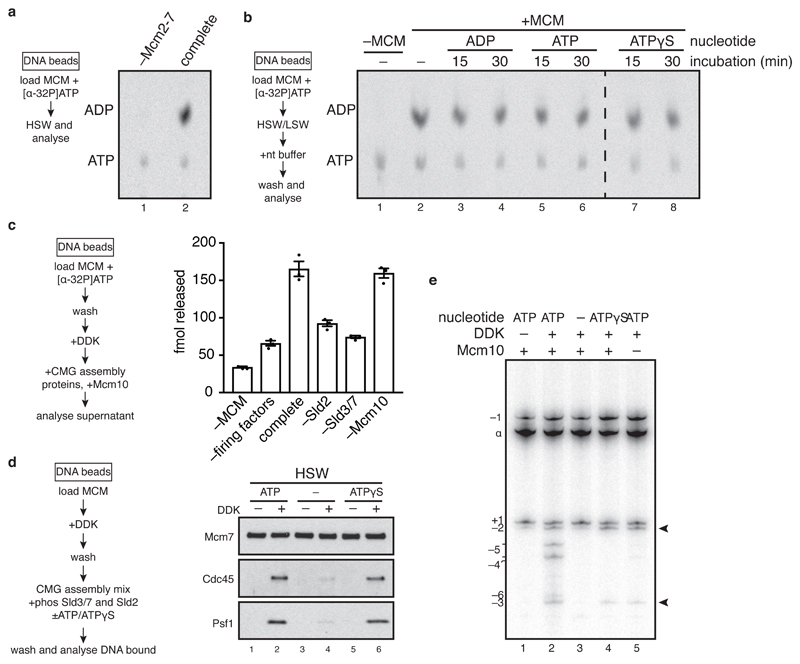
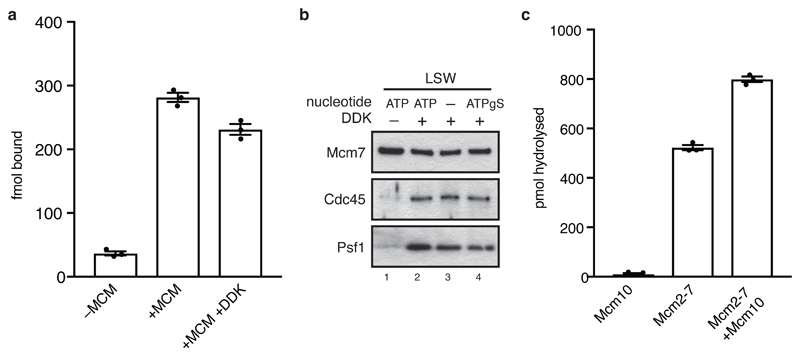
ATP hydrolysis by MCM is required for DH formation2,3, but nothing is known about downstream roles for nucleotide in helicase activation. To examine this, we first analyzed nucleotide binding and release by the DH. MCM loaded in the presence of [α-32P]ATP and washed with high salt was bound to ADP with only background levels of ATP (Figure 3a). As shown in Figure 3b, without further activation, ADP remained bound and did not exchange with unlabeled ADP, ATP or ATPγS over 30 minutes. To determine whether bound ADP is exchanged during helicase activation, MCM primed with radiolabelled ADP as above was used as a substrate for CMG assembly. Phosphorylation of MCM by DDK had little effect on the amount of ADP bound (Extended Data Figure 3a), but ADP was released from the DH into the supernatant of a full helicase activation reaction (Figure 3c, ‘complete’). This was reduced without Sld2 or Sld3/7, but not without Mcm10 (Figure 3c), indicating that ADP release takes place during CMG assembly.
Figure 3. CMG assembly and activation are coupled to ATP binding and hydrolysis.
a, MCM loading reactions containing [α32P]ATP were washed with high salt (buffer A + NaCl) and analysed by thin layer chromatography. b, as in a. except washed reactions were incubated with nucleotide as indicated and washed again before analysis. c, DHs assembled on bead-immobilised DNA with [α32P]ATP were used in CMG assembly reactions, the supernatants from which were analysed by scintillation counting. Error bars show standard error of the mean. d, immunoblots of CMG assembly reactions carried out as in Extended Data Figure 1a, except CDK was omitted, Sic1 was added, Sld2 and Sld3/7 prephosphorylated with CDK were used, and the nucleotide indicated was added. Reactions were quenched 15 min after firing factor addition. e, as in d. except reactions were carried out on soluble ARS1 circles and analysed as in Figure 2a. ATP was removed after DDK phosphorylation using a spin column.
To examine the role of ATP binding and hydrolysis in CMG assembly, DH was loaded on bead-immobilised DNA, phosphorylated with DDK and washed to remove ATP and DDK. This DDK-phosphorylated DH was then incubated with firing factors, including Sld2 and Sld3/7 which had been phosphorylated by CDK and repurified to remove ATP and CDK. Cdc45 and GINS (Psf1) were recruited (low salt wash) equally efficiently in a DDK-dependent manner in the presence or absence of nucleotide (Extended Data Figure 3b, lane 3); however, formation of CMG (high salt wash) did not occur without added nucleotide (Figure 3d, lanes 2 and 4). ATPγS supported stable CMG formation (Figure 3d, lane 6) indicating that CMG assembly does not require ATP hydrolysis. To analyze the effect of nucleotide on DNA untwisting (Figure 2), reactions were performed as in Figure 3d except soluble DNA circles were used as template and ATP was removed from reactions by spin column. Whereas no supercoiling was detected without ATP (Figure 3e, lane 3), assembly of CMG in reactions containing ATPγS and Mcm10 generated the same amount of supercoiling as reactions containing ATP but lacking Mcm10 (Figure 3e, lane 5 and Figure 2c). Together, these data show that CMG assembly and the initial untwisting of DNA are coupled to ADP release and ATP binding, whilst CMG activation by Mcm10 requires ATP hydrolysis. Mcm10 can stimulate ATP turnover by soluble MCM (Extended Data Figure 3c), so may trigger CMG activation by promoting ATP hydrolysis.
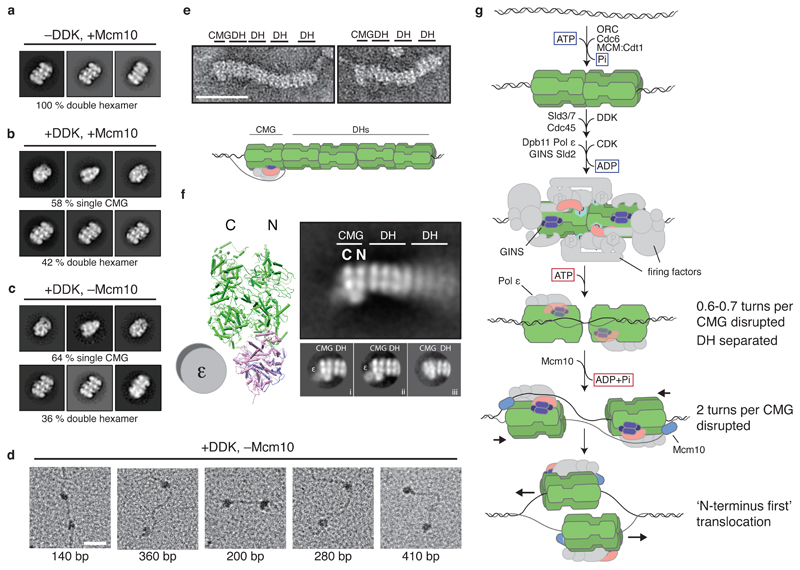
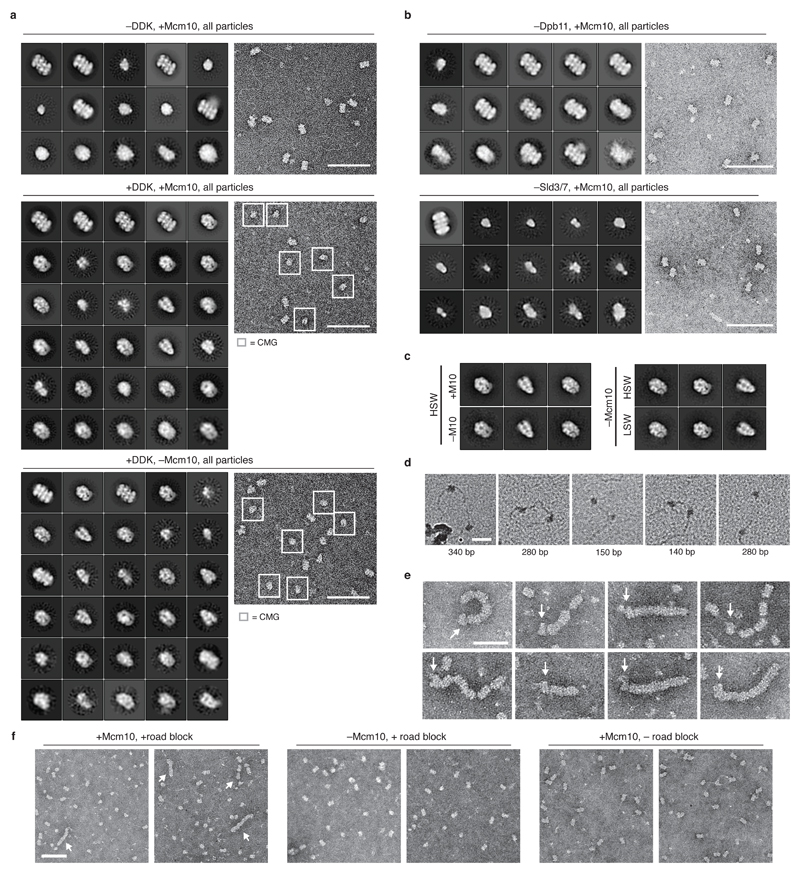
To characterize structural changes during these processes, products assembled on bead-immobilised DNA were washed with high salt, released from beads by restriction enzyme digestion and analysed by EM after negative staining. MCM-containing particles were only seen as DHs in reactions lacking DDK, Sld3/7 or Dpb11 (Figure 4a, Extended Data Figures 4a and 4b). By contrast, in a complete reaction, nearly 60% of MCM-containing particles resembled discrete, single CMGs, indicating that approximately 40% of the input DHs had been activated (Figure 4b and Extended Data Figure 4a). In a reaction containing all firing factors except Mcm10, we observed the same proportion of discrete, single CMGs, after either high or low salt wash (Figure 4c, Extended Data Figures 4a and 4c). Double hexamer separation therefore takes place during CMG assembly, before CMG activation. We did not observe double CMGs in these reactions, and pairs of CMG-sized particles that co-localised on the same DNA molecule were separated by up to ~400 base pairs (Figure 4d and Extended Data Figure 4d) indicating that inactive CMGs move apart before activation.
Figure 4. Structural characterisation of replicative helicase activation.
a-c, representative reference free class averages of helicase activation reactions washed with high salt (buffer A + KCl). DH and CMG classes are shown. All particle classes are presented in Extended Data Figure 4a. d, helicase activation reactions lacking Mcm10 were washed with high salt and positively stained to visualise DNA. Examples of two CMG-sized particles co-localised with a single DNA fragment are shown. The approximate base pair distance between particles is indicated. Scale bar represents 50 nm. e, examples of CMGs neighbouring DH ‘trains’ in high salt washed reactions on roadblocked DNA. Scale bar is 50 nm. f, annotated reference free class average from 469 train ends. CMG structure (from PDB file 3jc5) is included for reference. 2D classification of train tip particles into several classes (i-iii) reveals doughnut-shaped polymerase ε density on a subset of CMGs (i and ii). g, model of eukaryotic replicative helicase assembly and activation.
CMG translocates 3’ to 5’ along the leading strand template1, but its orientation at the fork is uncertain. The MCM hexamer comprises two rings: one formed from the six C-terminal AAA+ domains and one formed from the N-terminal domains. The hexamers in the DH are attached by their N-terminal domains, with the C-terminal domains on the outside. Therefore, if the C-terminal MCM ring is at the front of the helicase13–15, then the two activated CMGs immediately move away from each other after initiation. If, however, the N-terminal ring is at the front16, then the two hexamers must first pass each other during initiation. To address this, we used a 3 kb substrate containing covalent protein roadblocks at the end of each leading strand. We loaded an excess of DHs relative to DNA, activated a subset of these with the full set of firing factors and analysed products by EM after negative staining. Figure 4e and Extended Data Figure 4e show images of multiple, adjacent DHs in ‘trains’ with a single CMG at one end. These were not seen if either Mcm10 or the roadblock was omitted (Extended Data Figure 4f) indicating that active CMG pushes DHs, which are free to slide9, and the roadblock prevents them from sliding off the DNA end. Comparing 2D averages of trains’ ends with the structure of the CMG and its 2D projections (Figure 4f and Supplementary Video 1) indicates that the CMG translocates with the N-terminus of MCM in front of the helicase. In agreement with this orientation, a ‘doughnut’-shaped density characteristic of polymerase ε, which binds the CMG C-terminus17,18, was observed in a subset of CMGs opposite from the DH (Figure 4f, i and ii, Supplementary Video 1).
Our results lead us to propose the model summarized in Figure 4g. ADP formed during DH assembly remains stably bound to MCM; based on recent structural studies, probably at only a subset of MCM active sites11,12. ADP is released in response to firing factors and subsequent ATP binding by MCM triggers CMG assembly, during which DH separation takes place. The position of GINS in the CMG is sterically incompatible with the DH19, suggesting these processes occur concomitantly. This step is also accompanied by the first stage of origin melting, when 0.6-0.7 helical turns are unwound per CMG. The earliest steps of origin melting by SV40 large T antigen7 and E.coli DnaA20 are also triggered by ATP binding, suggesting this is a conserved feature of replication initiation. The CMG is more than 10nm in length but at this stage contains less than 5nm of ssDNA (6-7 bp fully stretched). Consequently, the lagging strand template cannot yet be entirely excluded from the MCM central channel. Full strand exclusion is required for CMGs to pass one another, indicating that the initial separation of hexamers must take place in the C-terminal direction. CMGs can separate by hundreds of base pairs in vitro without Mcm10 (Figure 4d), but this movement may be restricted in vivo by nucleosomes. Moreover, Mcm10 can bind to the DH before firing factor recruitment21, which may facilitate immediate activation of CMG. Each active CMG constrains approximately two turns of untwisted DNA, which is long enough (~15nm) to be completely excluded from the MCM central channel. Mcm10 binds ssDNA avidly, so may play a direct role in this exclusion process22. The order and timing of firing factor release is unknown, but Mcm10 has a subsequent role in elongation8, suggesting it may remain bound to the active helicase. Subsequent crossing of CMGs ‘N-terminus first’ ensures that all origin DNA will be unwound, and may help to coordinate assembly of the two leading strand replisomes to ensure this occurs only at origins. Furthermore, the requirement that two helicases can only pass one another when both are bound around ssDNA provides a fail-safe mechanism for bidirectional DNA replication, preventing CMGs from escaping the origin until both helicases are active. The ability of active CMG to push inactive DHs ahead of the fork (Figure 4e,f) may be important to remove unfired DHs from replicated DNA and to rescue stalled forks, but may also necessitate a pathway for DH removal prior to termination.
Methods
Protein purification
All proteins were purified as described6.
DNA templates
Bead-immobilised linear and circular DNA templates were as described6. All plasmid-based assays used pBS/ARS1WTA, a 3.2 kb plasmid containing ARS123. To assemble 616 bp ARS1 circles, a 610 bp fragment around ARS1 was PCR amplified from this vector using oligonucleotides oMD171 and oMD172, which introduce recognition sites for EcoRI at both fragment ends. 1.8 μg DNA was digested with 200 U EcoRI in 100 μl for 3 h at 37°C, spin column purified (Roche), and dephosphorylated with 5 U Antarctic phosphatase for 15 min at 37°C. Phosphatase was inactivated at 70°C for 5 min, and DNA ends phosphorylated with PNK and [γ-32P]ATP. PNK was heat inactivated for 20 min at 65°C, the sample desalted over a G50 spin column (GE) and ligated at a concentration of 180 ng/ml overnight at 10°C with 20 U/ml T4 DNA ligase. The ligation reaction was concentrated 5-10 fold through a 10 kDa cutoff spin filter (Millipore), ethanol precipitated and run on a 1x tris-borate-EDTA (TBE) 1.5 % agarose gel. DNA corresponding to supercoiled 616 bp circles was excised and electroeluted for 1 h in 0.1x TBE. The sample was ethanol precipitated and resuspended in 1x Tris-EDTA (TE) before use.
For the roadblocked DNA template used in Figure 4e and 4f, a 2.8 kb fragment containing ARS1 was amplified using oligonucleotides oMD203 and oMD204, which introduce a single recognition sequence for the methyltransferase HpaII on each end of the ARS1 fragment. PCR product was digested with XhoI and cloned into bluescript ks+ digested with XhoI and SmaI to make pMD142. pMD142 was used as a template for PCR with oMD215, which was biotinylated at the 5’ end, and oMD208. Methyltransferase HpaII was purified and coupled to this PCR product as described24 and the coupled DNA product was immobilised on M280 streptavidin resin essentially as described6, except resin was washed twice with 10 mM Tris pH 7.2, 1 mM EDTA and 1 M NaCl, twice with 10 mM Hepes pH 7.6, 1 mM EDTA, 1 M KOAc and twice with 10 mM Hepes 7.6, 1 mM EDTA and resuspended in half the starting resin volume with 10 mM Hepes 7.6, 1 mM EDTA.
Oligonucleotides used
The sequences (5’ to 3’) of oligonucleotides used in this study are: oMD167 – CGGAGGTGTGGAGAC, oMD171 – TAGTAGGAATTCAAGCAGGTGGGACAGG, oMD172 – TAGTAGGAATTCGCGAAAAGACGATAAATACAAG, oMD203 – GGTGTATGCATGCTACTGTTTCTCGAGGTGTGAAAGTGGGGTCTCAT CCTCAGCATCCGGTACCTCAGCGGTAGTTATAAGAAAGAGACCGAGTTAG, oMD204 – GAGCCTGAATCCTCAGCATCCGGTACCTCAGCAAGAGTATTGGCGAT GACGAAAC, oMD208 – CAGGAAACAGCTATGACCATG and oMD215 – Biotin-CGAAAAACCGTCTATCAGGGCGATG.
Assigning relative supercoiling states
To assign the relative supercoiling state of different 616 bp circle topoisomers, 2 fmol/μl 616 bp DNA circles were incubated at 65°C for 30 min with 0.25 U/μl Nb.BsrDI enzyme, which specifically recognises and nicks a single site on the circle. Nb.BsrDI was heat inactivated at 80°C for 20 min, DNA extracted once with phenol:chloroform:isoamylalcohol (25:24:1) and ethanol precipitated. The DNA pellet was resuspended in 1x TE, and 3 fmol ligated in the presence of the ethidium bromide concentrations indicated at 10-12°C overnight with 10 U/μl T4 DNA ligase (NEB). Ligated DNA was phenol:chloroform extracted, ethanol precipitated and the DNA pellet resuspended in 1x TE prior to analysis by electrophoresis. Final DNA circles are increasingly negatively supercoiled the more ethidium bromide that is present during the ligation step. Topoisomers were therefore assigned relative to the ground state (α, the most prevalent topoisomer when ethidium bromide was omitted) by tracking the order in which bands peaked as the ethidium bromide concentration increased25. The non-linear relationship between increased negative supercoiling and electrophoretic mobility (for example, compare mobility of topoisomers α-3 and α-4) has been seen previously26 and may reflect extrusion of cruciform DNA, which is favoured as linking number decreases27.
Unwinding assays
25 fmol plasmid DNA or 5 fmol 616 bp DNA was relaxed in 25 mM HEPES-KOH pH 7.6, 100 mM K-glutamate, 10 mM magnesium acetate, 0.02% NP-40-S, 5% glycerol, 2 mM DTT, 5 mM ATP (loading buffer), with 20 nM Topo I for 30 min at 30°C. 5 nM ORC, 50 nM Cdc6 and 100 nM Mcm2-7:Cdt1 were added for 20 minutes at 30°C, the reaction supplemented with 50-100 nM DDK, and incubation continued for a further 30 min at 30°C. Buffer was added to give a final concentration 250 mM K-glutamate, 25 mM Hepes, 10 mM Mg-acetate, 0.02 % NP-40-S, 8 % glycerol, 400 μg/ml BSA, 5 mM ATP, 1 mM DTT and 25 nM Topo I (buffer CMG). A mix of firing factors was assembled immediately before use and added at time 0, to a final concentration 50 nM Dpb11, 200 nM GINS, 50 nM Cdc45, 30 nM Pol ε, 50 nM A-Cdk2 (cyclin A/Cdk2), 2.5 nM Mcm10, 30 nM Sld3/7, 55 nM Sld2 (firing factor mix). After 40 min at 25°C (for plasmid DNA) or 30°C (for small circles), the reaction was quenched with 13 mM EDTA, 0.3 % SDS, 0.1 mg/ml Proteinase K (Merck) (stop mix), and incubated at 42°C for 20 min. Sample was extracted once with phenol:chloroform:isoamylalcohol (25:24:1), ethanol precipitated, and the DNA pellet resuspended in 1x TE for analysis.
Modified unwinding assays
The experiment in Figure 2b was carried out as per ‘unwinding assays’, except no DDK was used, and no firing factor mix was added after dilution into buffer CMG. The experiment in Figure 3e was carried out as per ‘unwinding assays’, with the following modifications: ATP concentration was reduced to 1 mM for the loading and DDK phosphorylation steps. After phosphorylation, reactions were passed over a G50 spin column (GE healthcare) washed 4x with 25 mM Hepes 7.6, 5 mM Mg-acetate, 10 % (v/v) glycerol and 0.02 % NP-40-S (buffer A) supplemented with 0.1 M K-glutamate. CDK was excluded from the firing factor mix; prephosphorylated Sld2 was used at a final concentration of 10-15 nM, and prephosphorylated Sld3/7 at 10-20 nM. Prephosphorylation procedure is described below. Sic1 was added to a final concentration of 145 nM.
Gel electrophoresis
For plasmid-based unwinding assays, DNA was run on native 1.5 % agarose tris-acetate-EDTA (TAE) gels, at 1.5 V/cm for 16 h. Gels were stained with 0.5 μg/ml ethidium bromide for 1 h at room temperature, and destained with 1 mM Mg-sulphate for 1 h before imaging. For 616 bp circle unwinding assays, DNA was run on native 3.5 % bis-polyacrylamide 1x TBE gels at 4.5 V/cm for 20 h.
Nucleotide binding analysis
MCM loading reactions were carried out on 60 ng of immobilised 2.8 kb fragment of ARS16 in loading buffer with 500 μM ATP, 0.5 μCi/μl [α-32P]ATP, 37.5 nM ORC, 50 nM Cdc6 and 100 nM Mcm2-7:Cdt1. After 30 min at 30°C, beads were washed twice with buffer A supplemented with 0.5 M NaCl (buffer A + NaCl) and once with buffer A supplemented with 0.25 M K-glutamate and 2 mM CaCl2 (buffer A + CaCl2). DNA bound complexes were released by cleavage with buffer A + CaCl2 supplemented with 60 U/μl micrococcal nuclease (NEB) (buffer MNase) for 5 min at 30°C. Cleaved samples were spotted onto PEI-cellulose TLC plates (Camlab), which were developed in 0.6 M Na2HPO4/NaH2PO4 pH 3.5. For the nucleotide competition experiments in Figure 3b, after the NaCl washes above, DNA-bound complexes were washed once with buffer A + 0.1 M K-glutamate, incubated at 30°C for 15 or 30 min in buffer A + 0.1 M K-glutamate and 5 mM of the appropriate nucleotide. Samples were then washed once with buffer A + CaCl2 and MNase cleaved and analysed as above.
Recruitment assays
MCM loading, DDK phosphorylation and CMG assembly steps were carried out as described in the ‘unwinding assays’ section, with the following modifications: Each 20 μl CMG assembly reaction used approx. 60 ng linear DNA or 40 ng plasmid DNA immobilised on M-280 streptavidin magnetic beads (Invitrogen)6. The concentration of ORC was increased to 37.5 nM. Supernatant was removed after DDK phosphorylation and DNA beads were resuspended in buffer CMG without Topo I. After CMG assembly and activation for 8 min, beads were washed twice with 200 μl buffer A with 0.3 M KCl (buffer A + KCl) or twice with buffer A with 0.25 M K-glutamate (Buffer A + 0.25 M K-glutamate). After one further wash with 200 μl buffer A + CaCl2, beads were resuspended in buffer MNase, and cleaved for 5 min at 30°C. Supernatant was supplemented with 1/3 volume of 4x SDS-loading buffer, heated at 95°C for 3 min. Proteins were separated through 4–12% bis-tris polyacrylamide gels (Bio-Rad) and analysed by immunoblotting.
Modified recruitment assays
For the experiment in Figure 2e, MCM loading and DDK phosphorylation was carried out in parallel on 16 fmol ARS1 plasmid in solution or randomly biotinylated, and immobilised on Steptavidin M-280 resin (Invitrogen). Buffer CMG was added to four soluble reactions, two of which were immediately used to resuspend resin-immobilised reactions for samples 1 and 2. Firing factor mix without Mcm10 was added to all four samples at time 0. After 6 and 10 minutes, the two remaining soluble reactions were used to resuspend resin-immobilised reactions for samples 3 and 4 respectively. 8 minutes after addition of the soluble reaction to beads, resin was washed twice with 200 μl buffer A + KCl, once with 200 μl buffer A + CaCl2, and DNA cleaved with buffer MNase for 5 min at 30°C.
Recruitment assays with prephosphorylated Sld2 and Sld3/7 involved the following modifications: the concentration of ATP was reduced to 1 mM for loading and DDK phosphorylation steps, after which beads were washed 3x with buffer A + 0.25 M K-glutamate. CDK was excluded from the firing factor mix and prephosphorylated Sld2 and Sld3/7 were used at 10-15 nM, and 10-20 nM respectively. Sic1 was added to a final concentration of 145 nM.
For the EM assays in Figure 4e and 4f, 120 ng linear, HpaII coupled DNA immobilized on M-280 streptavidin magnetic beads was used per 20 μl CMG assembly reaction, and Mcm2-7:Cdt1 was used at 200 nM during MCM loading.
Nucleotide release analysis
Loading of MCM was carried out with [α-32P]ATP as described in ‘nucleotide binding analysis’. After 20 min at 30°C, DDK was added to 100 nM. After a further 30 min at 30°C, resin was washed twice with buffer A + NaCl, once with buffer A + 0.1 M K-glutamate and resuspended in buffer CMG without Topo I. At time 0, firing factor mix (described in ‘unwinding assays’) was added. After 15 min at 30°C, supernatant was collected, supplemented with 12.5 mM EDTA, mixed with 5 ml scintillation fluid and measured in a scintillation counter.
Protein prephosphorylation
Immediately prior to phosphorylation, FLAG-tagged Sld3/7 was diluted to 30 nM in 40 mM HEPES-KOH pH 7.6, 8 % glycerol, 400 μg/ml BSA, 0.02 % NP-40-S, 10 mM Mg-acetate, 2 mM DTT, 5 mM ATP with 310 mM K-glutamate (buffer PP + 310 mM K-glutamate). Sld2 was diluted to 120 nM in buffer PP + 235 mM K-glutamate. Budding yeast S-phase CDK (S-CDK) was added to 10 nM, and reactions incubated for 8 min at 25°C before addition of Sic1 to 220 nM. After 2 min incubation at 25°C, Sld2 mix was diluted 4x in buffer A + 0.5 M KCl. 5 μl magnetic anti-FLAG M2 resin (Sigma) washed with buffer A + 0.5 M KCl was added, and each sample incubated at 4°C for 30 min with rotation. Resin was washed 5x with 300 μl buffer A + 0.5 M KCl (Sld3/7) or +0.35 M KCl (Sld2), and resuspended in 10 μl of the same buffer supplemented with 0.25 mg/ml FLAG peptide. After shaking at 4°C for 30 min, supernatant was collected, aliquoted and frozen in liquid nitrogen for storage.
Electron microscopy sample preparation
For positive and negative stain, CMG assembly was carried out as described in ‘Recruitment assays’. Reactions were washed twice with 200 μl buffer A + KCl, once with 100 μl 25 mM Hepes, 5 mM Mg-acetate, 250 mM K-glutamate (buffer EM) and DNA-bound complexes released from beads by restriction enzyme cleavage in 5-10 μl buffer EM supplemented with 0.1 U/μl MseI (NEB) for 10 min at 30°C, giving rise to an average DNA fragment size of 1.5-2 kb. Negative stain sample preparation was performed on 400 mesh copper grids (Agar Scientific) with floated carbon that had been freshly evaporated onto cleaved mica using a Q150TE coater (Quorom Technologies). Grids were glow-discharged for 30 s at 45 mA (Electron Microscopy Sciences). 3 μl drops of sample were applied to the grids and left to incubate for 1 min. Excess sample was blotted away and staining was performed on four separate 70 μl 2% uranyl formate drops by stirring for 5, 10, 15 and 20 s, respectively. Excess stain was blotted away and grids were stored before imaging. For positive stain, two-week old carbon coated 400 mesh copper grids (Agar Scientific) were glow discharged for 10 s at 45 mA. 3 μl sample was applied and incubated for 30 s. Half the sample solution was blotted away before staining on a single 75 μl 2% uranyl acetate drop for 30 s. Stain was washed away by stirring the grid on four 75 μl ddH2O drops for 5 s each before blotting the grid to dryness.
Electron microscopy data acquisition
Data collection of negative stain grids was performed on a Tecnai LaB6 G2 Spirit transmission electron microscope (FEI) operating at 120 keV (EM STP, The Francis Crick institute). Micrographs were collected using a 2Kx2K GATAN Ultrascan 100 camera at a nominal magnification of 30,000 (3.45 Å pixel size) or 21,000 (4.92 Å pixel size, train classes) within a -0.5 to 2.5 μm defocus range. Analysis of positive stain grids was performed on a Tecnai G2 F20 TWIN electron microscope operating at 200 keV (FEI; Electron Microscopy Centre, Imperial College London) equipped with a Falcon II direct electron detector (FEI). Micrographs were collected at a nominal magnification of 50,000 (2.05 Å pixel size) in a defocus range from -3 to -6 μm.
Electron microscopy single particle analysis
Negative stain particles were semi-automatically picked using EMAN228, version 2.07 and the rest of the image processing was performed using RELION29, version 1.4. Particles were extracted with a box size of 128x128 pixels (except the large train class in Figure 4e, which was extracted with a box size of 250x250 pixels) from CTF corrected (CTFFIND330) micrographs and subjected to reference free two-dimensional classification with the --only_flip_phases additional argument. Comparisons of 2D classes from different samples was performed using the multi reference alignment function in IMAGIC31.
Electron Microscopy positive stain image analysis
CMGs were distinguished from MCM DHs on positive stain micrographs by manually measuring particle length parallel to the DNA axis using ImageJ. Examples where two particles were associated with the same fragment of DNA were analysed, and 84 % of particles found to correspond to CMG (57/68). DNA fragments containing two CMG-sized particles are shown.
Potassium permanganate footprinting
CMG assembly and activation on small DNA circles were performed as in ‘unwinding assays’ with the following modifications. The buffer for DNA relaxation and MCM loading was 25 mM Tris-Cl pH 7.2, 100 mM K-glutamate, 10 mM magnesium acetate, 0.02% NP-40-S, 5 mM ATP. After phosphorylation with DDK, buffer was added to give a final concentration 250 mM K-glutamate, 25 mM Tris-Cl, 10 mM Mg-acetate, 0.02 % NP-40-S, 400 μg/ml BSA, 5 mM ATP and 25 nM Topo I. After 10 min CMG assembly at 30°C, KMnO4 was added to 3 mM for 4 min before the reaction was quenched with 1 M beta-mercaptoethanol (Sigma) and stop mix, and DNA processed as described in ‘unwinding assays’. The DNA pellet was resuspended in 48 μl 1x cutsmart buffer (NEB), digested with 40 U EcoRI-HF (NEB) for 20 min at 37°C, extracted once with phenol:chloroform:isoamylalcohol (25:24:1), ethanol precipitated, and analysed by primer extension. Primer extension reactions contained 32P end-labelled primer oMD167 and 70 U/ml Vent (exo-) DNA polymerase (NEB), and were carried out for 26 cycles. Reactions were quenched with stop mix, ethanol precipitated and separated on a denaturing 5 % urea-bis-polyacrylamide gel.
DNA replication assay
For the experiment in Extended Data Figure 1d, CMG assembly was carried out for 10 min as described in ‘recruitment assays’, using a randomly biotinylated 5.6 kb ARS1 plasmid immobilized on M280 steptavidin resin6. After CMG assembly in the presence or absence of Mcm10, resin was washed twice with Buffer A + KCl (‘HSW’) or Buffer A + 0.25 M K-glutamate (‘LSW’), and once with Buffer A + 0.25 M K-glutamate. Beads were resuspended in 25 mM Hepes 7.6, 5 mM MgOAc, 0.02 % NP-40S, 125 mM K-glutamate, 2 mM ATP, 1 mM DTT, 200 μM CTP, UTP, GTP, 40 μM each dNTP, 40 nM [α-32P]dCTP (Perkin Elmer), 50 nM RPA, 30 nM Polymerase ε, 40 mM Polymerase α, 25 nM Topo I and 5 nM Mcm10 or Mcm10 buffer, and incubated for 45 min at 30°C. Reactions were stopped and processed as described6.
MCM ATPase assay
Mcm2-7 was diluted to 0.5 μM in buffer A + 0.5 M K-glutamate, containing 1 μM Mcm10. ATP was added to 100 μM, including 0.125 μCi/μl [α-32P]ATP. After 30 min at 30°C, EDTA was added to 15 mM and sample spotted onto PEI-cellulose TLC plates (Camlab), which were developed in 0.6 M Na2HPO4/NaH2PO4 pH 3.5.
Estimating linking number shift
When Mcm10 is omitted from an otherwise complete reaction, CMG is assembled on a fraction of circles, causing a shift in linking number by an unknown amount. If we denote Yk as the abundance of a topoisomer k in the starting distribution of circles without CMG assembly (such as –DDK, lane 1 of Figure 2c), when CMG is assembled, a proportion (a) of circles shift linking number by λ, such that for any k, a of k gets shifted to another state (k-λ), while the remaining (1-a) remains in state k. We can then model Xk, the abundance of k in the –Mcm10 reaction, as Xk = Yk+λ a + Yk(1-a), where Yk+λ a is the fraction of topoisomer k+λ that moves into state k in the –Mcm10 reaction, and Yk(1-a) is the amount of topoisomer k that remains in the –Mcm10 reaction after a fraction has moved to state k-λ. This can be re-arranged to (Xk-Yk) = a(Yk+λ – Yk) which, given λ, can be solved by linear regression through the origin. Iterating through all possible values of λ on a grid, we choose the one for which residual mean square error was least. To enable fractional offsets to be calculated, we interpolated the original data using a cubic smoothing spline to give us estimated abundances at the resolution of tenths of an integer. A λ value of 1.3 at an efficiency (a) of 43 % gave the best fit to the measured abundance of topoisomers in the –Mcm10 sample, with an R2 value of 0.996. This is compared with shifts of 1.1, 1.2, 1.4 and 1.5, which gave R2 values of 0.9716998, 0.9889401, 0.9888459 and 0.9653362 respectively.
Statistics and Reproducibility
The experiments in Figures 1d, 2a, 2c, 3a, 3b, 3e and 4a-c were performed at least three times, whilst the experiments in Figures 1c, 2b, 3d and 4d-e were performed twice. The experiments in Extended Data Figures 1a, 2c-e and 4c were performed at least three times, whilst the experiments in Extended Data Figures 1b-d, 2b, 3b and 4d-f were performed twice. For Figure 3c, and Extended Data Figures 3a and 3c, n=3 independent experiments.
Extended Data
Extended Data Figure 1.
CMG assembly and activation are separable steps. a, to determine when CMG assembly saturates in our reactions, CMG assembly was carried out on bead-immobilised ARS1 DNA and washed with high salt wash buffer (HSW, buffer A +KCl) at the times indicates. The data show that no new CMG assembly takes place after 5 minutes. b, to confirm this, MCMs were loaded in parallel onto a bead-immobilised ARS1 DNA fragment and a soluble ARS1 plasmid, and phosphorylated with DDK. A firing factor mix that was complete except for Mcm10 was added to the soluble reaction only, which was then added to the bead-immobilised MCMs at the times indicated after firing factor addition to the soluble reaction. After 8 min, beads were washed with high salt (buffer A + KCl) and bound proteins analysed by immunoblotting. Psf1 signal relative to lane 2 is indicated. The experiment confirms that no CMG assembly can take place 5 minutes after firing factors have been added. c, to test whether Mcm10 can trigger DNA unwinding even after CMG assembly was finished, reactions were set up as in Figure 1d, except Mcm10 was omitted until the times indicated after firing factor addition. Mcm10 triggered robust unwinding, even when added more than 5 minutes after firing factors. Mcm10 can therefore activate preassembled CMG for DNA unwinding. d, to test whether Mcm10 can activate preassembled CMG for replication, CMG was assembled on an immobilised ARS1 plasmid ±Mcm10. Beads were washed with low (Buffer A + 0.25 M K-glutamate) or high (Buffer A + KCl) salt buffer, and replication proteins ±Mcm10 and cofactors including radiolabelled dCTP were added. Mcm10 enabled DNA replication even when CMG had been washed to remove excess firing factors. e, schematic outlining the CMG assembly and CMG activation steps described here.
Extended Data Figure 2.
a, models of DNA unwinding ±RPA. b, to define the relative position of different topoisomers of radiolabelled 616 bp DNA circles containing ARS1 (used to analyse small changes in DNA supercoiling in the unwinding assay), nicked circles (nc, lane 1) were ligated closed under the ethidium bromide concentrations indicated. The supercoiling states of different bands of covalently closed DNA were determined relative to the ground state (α) by tracking the order in which bands peaked as ethidium bromide concentration increased and DNA was increasingly negatively supercoiled (see methods for further details). Two bands peak at the same position for α-5 and likely represent alternative configurations of the α-5 topoisomer. c, primer extension reactions reading the T-rich strand of the ARS-consensus sequence (ACS) of ARS1 were carried out on 616 bp ARS1 DNA treated with potassium permanganate as indicated after CMG assembly in the absence of RPA. Reactions were separated on 5 % sequencing gels, dried and analysed by autoradiography. Base pair numbering is relative to the 5’ end of the T-rich strand of the ACS. d, as in Figure 2c. Lane 1 shows that MCM loading is required for all shifts in topoisomer distribution. Compared with other control samples, such as –DDK, topoisomer distribution is subtly different without MCM; this is not due to loading, which, as shown in Figure 2b, does not affect topoisomer distribution. e, as in Figure 2a, except Mcm10 was omitted from all reactions. No proteins except topoisomerase were added to the reaction in lane 1 after MCM loading. No detectable change in supercoiling relative to when no firing factors were added (lane 1) was observed when each individual firing factor was omitted, suggesting DNA untwisting without Mcm10 takes place during CMG assembly.
Extended Data Figure 3.
a, DHs assembled on bead-immobilised DNA using [α32P]ATP were treated with DDK as indicated and analysed by scintillation counting. Error bars represent standard error of the mean. b, immunoblots of CMG assembly reactions carried out as in Figure 3d and washed with low salt wash buffer (Buffer A + 0.25 M K-glutamate). c, ATPase assays using [α32P]ATP, single MCM hexamers and Mcm10 as indicated were quantified after thin layer chromatography. Error bars represent standard error of the mean.
Extended Data Figure 4.
a, example micrographs and complete sets of reference free class averages of the helicase activation reactions indicated, washed with high salt wash buffer (buffer A + KCl). –DDK, +Mcm10, 7410 of 23092 total particles were DH. +DDK, +Mcm10, of 43320 total particles, 14668 and 10492 were CMG and DH respectively. +DDK, –Mcm10, of 12920 total particles, 3984 and 2226 were CMG and DH respectively. Classes are positioned with respect to the abundance of source particles, with the most abundant class in the top left-hand corner, and abundance decreasing from left to right and top to bottom. b, as in a. with representative source micrographs. 5032/6815 and 2049/20904 particles were DH when Dpb11 or Sld3/7 were omitted respectively. Scale bar represents 100 nm. c, comparison of CMG formed under the conditions indicated. HSW is buffer A + KCl, LSW is buffer A + 0.25 M K-glutamate. d, as in Figure 4d. e, as in Figure 4e. Arrows mark position of CMG. f, representative crops from micrographs imaging the samples indicated. MCM trains are marked with arrows. Trains were not observed when Mcm10 or the protein road block was omitted. Scale bar is 100 nm.
Supplementary Material
Acknowledgements
We are grateful to Karim Labib for anti-Psf1 antibody, Gavin Kelly (the Francis Crick Institute, Bioinformatics) for help with mathematical modelling, the Francis Crick Institute Fermentation Facility for cell production and Lucy Collinson, Raffaella Carzaniga (the Francis Crick Institute, Electron Microscopy) and Tillmann Pape (Electron Microscopy Centre, Imperial College) for EM support. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001065 and FC001066), the UK Medical Research Council (FC001065 and FC001066), and the Wellcome Trust (FC001065 and FC001066). This work was also funded by a Wellcome Trust Senior Investigator Award (106252/Z/14/Z) and a European Research Council Advanced Grant (669424-CHROMOREP) to J.F.X.D.
Footnotes
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
Author Contributions
All authors conceived the electron microscopy experiments; M.E.D prepared the samples and F.A.A performed the imaging. M.E.D and J.F.X.D conceived all other experiments and M.E.D performed them. M.E.D and J.F.X.D wrote the paper with input from F.A.A and A.C.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Bell SP, Labib K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coster G, Frigola J, Beuron F, Morris EP, Diffley JFX. Origin Licensing Requires ATP Binding and Hydrolysis by the MCM Replicative Helicase. Mol Cell. 2014;55:666–677. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang S, Warner MD, Bell SP. Multiple functions for Mcm2-7 ATPase motifs during replication initiation. Mol Cell. 2014;55:655–665. doi: 10.1016/j.molcel.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JFX. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean FB, Hurwitz J. Simian virus 40 large T antigen untwists DNA at the origin of DNA replication. J Biol Chem. 1991;266:5062–5071. [PubMed] [Google Scholar]
- 8.Looke M, Maloney MF, Bell SP. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31:291–305. doi: 10.1101/gad.291336.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 A. Nature. 2015;524:186–91. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 11.Noguchi Y, et al. Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc Natl Acad Sci U S A. 2017;114:E9529–E9538. doi: 10.1073/pnas.1712537114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abid Ali F, et al. Cryo-EM Structure of a Licensed DNA Replication Origin. Nature Commun. 2017 doi: 10.1038/s41467-017-02389-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 14.Costa A, et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. eLife. 2014;3:e03273. doi: 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Froelich CA, Kang S, Epling LB, Bell SP, Enemark EJ. A conserved MCM single-stranded DNA binding element is essential for replication initiation. eLife. 2014;3:e01993. doi: 10.7554/eLife.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgescu R, et al. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci U S A. 2017;114:E697–E706. doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou JC, et al. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc Natl Acad Sci U S A. 2017;114:4141–4146. doi: 10.1073/pnas.1700530114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nature communications. 2016;7 doi: 10.1038/ncomms10708. 10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas ME, Diffley JFX. Recruitment of Mcm10 to Sites of Replication Initiation Requires Direct Binding to the Minichromosome Maintenance (MCM) Complex. J Biol Chem. 2016;291:5879–5888. doi: 10.1074/jbc.M115.707802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson PD, et al. Domain architecture and biochemical characterization of vertebrate Mcm10. J Biol Chem. 2008;283:3338–3348. doi: 10.1074/jbc.M706267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 24.Coster G, Diffley JFX. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science. 2017;357:314–318. doi: 10.1126/science.aan0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shore D, Baldwin RL. Energetics of DNA twisting. II. Topoisomer analysis. J Mol Biol. 1983;170:983–1007. doi: 10.1016/s0022-2836(83)80199-5. [DOI] [PubMed] [Google Scholar]
- 26.Zivanovic Y, Goulet I, Prunell A. Properties of supercoiled DNA in gel electrophoresis. The V-like dependence of mobility on topological constraint. DNA-matrix interactions. J Mol Biol. 1986;192:645–660. doi: 10.1016/0022-2836(86)90282-2. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh TS, Wang JC. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975;14:527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- 28.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 31.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. Journal of structural biology. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.