Abstract
Thiopurines are a standard treatment for childhood leukemia, but like all chemotherapeutics their use is limited by inherent or acquired resistance in patients. Recently, the nucleoside diphosphate hydrolase NUDT15 has received attention based on its ability to hydrolyze the thiopurine effector metabolites 6-thio-deoxy-GTP (dGTP) and 6-thio-GTP, thereby limiting the efficacy of thiopurines. In particular, increasing evidence suggests an association between the NUDT15 missense variant, R139C and thiopurine sensitivity. In this study, we elucidated the role of NUDT15 and NUDT15 R139C in thiopurine metabolism. In vitro and cellular results argued that 6-thio-dGTP and 6-thio-GTP are favored substrates for NUDT15, a finding supported by a crystallographic determination of NUDT15 in complex with 6-thio-GMP. We found that NUDT15 R139C mutation did not affect enzymatic activity but instead negatively influenced protein stability, likely due to a loss of supportive intramolecular bonds that caused rapid proteasomal degradation in cells. Mechanistic investigations in cells indicated that NUDT15 ablation potentiated induction of the DNA damage checkpoint and cancer cell death by 6-thioguanine. Taken together, our results defined how NUDT15 limits thiopurine efficacy and how genetic ablation via the R139C missense mutation confers sensitivity to thiopurine treatment in patients.
Keywords: NUDT15, thiopurine, 6-thio-dGTP, 6-thio-GTP, NUDT15 R139C
Introduction
Interfering with nucleotide metabolism is one of the most successful treatment strategies against cancer. Nucleoside analogues target rapidly proliferating cancer cells by disrupting processes related to RNA and DNA synthesis. Even after half a century of clinical research (1), thiopurines remain one of the most effective maintenance therapies against childhood leukemia (acute lymphoblastic leukemia (ALL)) (2) and are also used as anti-inflammatory and immunosuppressant drugs (3).
Three thiopurines are used in the clinic; 6-thioguanine (6-TG), azathioprine (AZA-T) and 6-mercaptopurine (6-MP). The metabolism of thiopurines is complex and involves a number of enzymes and intermediates that may improve or impair effective treatment (4). The nucleoside triphosphates, 6-thio-GTP and 6-thio-dGTP, are the primary active metabolites. 6-thio-dGTP is a substrate for DNA polymerases leading to substitution of 0.01 to 0.1% of canonical guanine bases for thiobases (5–8). Incorporation of 6-thio-dGTP is neither particularly toxic nor mutagenic (6, 9–11). However, methylation, most likely by S-adenosylmethionine (SAM) (12), and a second round of replication generates the Me-6-thio-dG:T mispair, which is detected by the mismatch repair (MMR) machinery (12, 13). Processing of this lesion presumably leads to futile attempts of repair, irreparable DNA damage and cell death (14–16). Since this cascade requires two rounds of DNA replication, the anti-proliferative effects of thiopurines are noticeably delayed (14). MMR-deficient cells are significantly less sensitive to thiopurines (14), yet residual sensitivity in these cells indicates that thiopurines may act via additional mechanisms. The complexity of thiopurine metabolism complicates treatment regimens and imposes a restrictive therapeutic window that requires close monitoring (2, 3, 10).
In searching for novel thiopurine sensitivity factors, several research groups have identified a missense variant in NUDT15 (rs116855232) that results in an arginine to cysteine substitution at position 139 (R139C) and thiopurine intolerance. Inflammatory bowel disease (IBD) patients with this mutation showed increased side effects after thiopurine treatment (17, 18). Accordingly, this NUDT15 variant was found to influence the sensitivity to 6-MP in children with ALL (19–21). NUDT15 (also known as MutT homolog 2 (MTH2)) belongs to the NUDIX hydrolase family and was initially described as an oxidized nucleotide sanitation enzyme, similar to MTH1 (NUDT1) (22). A comparison of MTH1 and NUDT15 revealed that NUDT15 had minimal activity towards oxidized nucleotides in vitro and in cells. However, a substrate screen identified that NUDT15 could hydrolyze 6-thio-dGTP and 6-thio-GTP, which, like the pharmacogenetic studies, suggested a possible role for NUDT15 in thiopurine metabolism (23).
Herein, we present that NUDT15 is a key enzyme in thiopurine metabolism and tempers the activity of thiopurine drugs by catalyzing the hydrolysis of 6-thio-dGTP and 6-thio-GTP. We resolve the first crystal structure of NUDT15 in complex with a reaction product, 6-thio-GMP, thereby elucidating key determinants for the observed substrate selectivity. With in vitro and cellular experiments, we explore the activity of NUDT15 R139C and find that the protein maintains catalytic activity but is unstable under physiological conditions. In cells, NUDT15 R139C is rapidly degraded, likely causing thiopurine sensitivity in patients with this mutation. In line with this, depletion with NUDT15-specific RNAi in cancer cells increases the sensitivity to 6-TG. Altogether, our data suggest that NUDT15 is integral to thiopurine metabolism and acts as a barrier to therapeutic efficacy.
Materials and Methods
Protein production
The construct pNIC28hNUDT15 for bacterial expression of NUDT15 was a gift from the Structural Genome Consortium (Stockholm, Sweden). NUDT15 WT was expressed and purified as earlier described (23). Site-directed mutagenesis for the R139C, R139S, R139A and R139K mutants was performed as described by Li et al (24). The mutants were expressed from pNIC28 in BL21DE3 at 37 °C and grown for 4 hours after induction by 1 mM IPTG before harvesting by centrifugation. Bacteria were lysed using Bugbuster (Merck-Millipore), fortified by benzonase (2.5U/mL, Merck-Millipore) and cOmplete™ Mini, EDTA-free protease inhibitor (Roche) was added to the cleared lysate. NUDT15 R139C was purified using HisTrap HP (GE Healthcare) and 100 mM HEPES (pH 7.5), 500 mM NaCl, 10 mM imidazole and 10% glycerol as starting buffer. Proteins were eluted using an imidazole gradient (0 to 500 mM), and the His-tag was removed by using TEV protease and passing the protein over a HisTrap HP column. Following dialysis with ion exchange chromatography starting buffer (20 mM HEPES (pH 7.5), 20 mM NaCl, 10% glycerol), NUDT15 R139C was further purified by MonoQ HP (GE Healthcare) and eluted with a NaCl gradient (10-500 mM). All purification was performed in the absence of reducing agent. SDS-PAGE and Coomassie staining confirmed purity of the proteins, and protein concentration was determined by NanoDrop (Thermo Fisher Scientific) A280 measurement.
Preparation of enzymatic reactions for HPLC
HPLC analysis was performed similar to previously reported conditions (25). Briefly, NUDT15 WT (10 nM) was incubated with 6-thio-GTP or 6-thio-dGTP (50 μM) in buffer containing 100 mM Tris-HCl (pH 7.5), 40 mM NaCl, 10 mM MgCl2, 1 mM DTT and 0.005 % Tween-20. Mixtures were incubated at 37 °C and stirred using magnetic stir bars. At each time point, 40 μL reaction mixture was added to 60 μL of -20 °C methanol. Samples were stored at -20 °C for at least 20 minutes before centrifugation. The supernatant (90 μL) was moved to fresh 1.5 mL tubes and evaporated by vacuum centrifugation at 60 °C. The samples were resuspended in 90 μL ddH2O, moved into a sealed 96-well plate, and loaded for HPLC analysis (samples were kept at 4 °C prior to injection).
Determination of kinetic parameters of WT and R139C NUDT15
NUDT15-mediated hydrolysis of 6-thio-dGTP, 6-thio-GTP, dGTP and GTP was monitored after 10, 20 and 30 minutes incubation at 22 °C in assay buffer (100 mM TrisAcetate pH 7.5, 40 mM NaCl, 10 mM MgAcetate, 1 mM DTT). For determination of kinetic parameters, initial rates were determined using reaction buffer, 4 nM NUDT15 WT or R139C and concentrations of the substrates ranging from 0-40 μM for 6-thio-dGTP and 6-thio-GTP and 0-50 μM for dGTP and GTP (or 0-400 μM for R139C). NUDT15 R139C reactions were performed in the absence or presence of 1 mM DTT. Formed pyrophosphate (PPi) was detected using PPiLight™ Inorganic Pyrophosphate Assay (Lonza) as described previously (23).
Crystallization and structure determination
Full length NUDT15 (15 mg/mL) was crystallized with 2 mM 6-thio-GTP in 15 mM HEPES, 225 mM NaCl, 7.5% Glycerol, and 1.5 mM TCEP pH 7.5. Hanging drop vapour diffusion experiments at 4 °C were performed, and NUDT15 was mixed with reservoir solution (30% PEG3350, 0.1 M Tris pH 8.5, and 0.24 M MgCl) in a 1:1 ratio. Diffraction quality crystals appeared overnight and grew to full size within 3 days. The crystals were extracted quickly without additional cryoprotectant, and flash frozen in liquid nitrogen. Data collection was performed at beam line ID29 at ESRF, Grenoble, at 100 K and wavelength 1.072 Å equipped with a Pilatus 3 6M detector (Dectris, Switzerland). Data reduction and processing were carried out using Mosflm (25) and Aimless (26) program from the CCP4 suite (27). The structure was solved by molecular replacement of the template structure file with PDB ID 5BON using Phaser (28). The resultant models were refined using Refmac5 (29) and Arp/wARP (30) was used for initial addition of the water molecules. Manual adjustments of the model were carried out using Coot (31). Validation was conducted with Molprobity (32). Relevant statistics can be found in Supplementary Table S1. All figures were drawn with PyMOL (Schrödinger, LLC, New York).
Cell culture and treatments
Cells were cultured in a humidified incubator at 37°C with 5% CO2. HCT116 and HCT116 3-6 human colon carcinoma cells were obtained from Dr. Bert Vogelstein (2001, Johns Hopkins) and HEK293T transformed human embryonic kidney cells from ATCC (2010). For HCT116 and HCT116 3-6, McCoy’s 5A GlutaMAX™ (Life Technologies) was used and for HEK293T DMEM GlutaMAX™ (Life Technologies). All media was supplemented with 10% fetal bovine serum, penicillin (50 U/mL) and streptomycin (50 μg/mL). Mycoplasma contamination was screened using the MycoAlert™ Mycoplasma Detection Kit (Lonza).
Doxycycline hydrochloride (Sigma Aldrich) was dissolved in MilliQ H2O and used at 1 μg/mL. 6-thioguanine (6-TG, Sigma Aldrich) was dissolved in DMSO to a stock concentration of 10 mM immediately before use and was protected from light. MG-132 (Z-Leu-Leu-Leu-al, Sigma Aldrich) was dissolved in DMSO.
Lentiviral transfection
HEK293T cells were transfected with lentiviral particles as described before (33). Selection for Tet-pLKO-puro-containing cells was achieved by using 1 μg/mL puromycin (Sigma Aldrich) and pInducer20-containing cells with 400 μg/mL neomycin (G418, Sigma Aldrich).
Western blotting
Cells were lysed and prepared for western blotting as described previously (23). Primary and secondary antibodies are listed in the Supplementary Methods. IRDye secondary antibodies (LI-COR) were used and blots were visualized and analyzed using an Odyssey® Fc Imager and Image Studio™ Software (LI-COR).
Thermal stability assay
Protein unfolding was detected by differential scanning fluorimetry (DSF) (34). The conditions used were 4 μM enzyme in assay buffer and SYPRO® Orange (Thermo Fisher Scientific) with or without 1 mM TCEP. A CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) was used to increase the temperature from 25-95 °C in 1 °C/min increments and fluorescence intensity was measured at each step. The melting temperature (Tm) was calculated by CFX Manager Software (Bio-Rad).
Clonogenic survival assay
Clonogenic survival experiments were based on those from Meyers et al. (35). Cells were treated with doxycycline for 48 hours and then re-plated as single cells. The following day, 6-TG or DMSO was added (containing doxycycline). Medium was replaced every 3 days containing fresh 6-TG/DMSO and doxycycline. After 9 days, the colonies (>50 cells) were fixed/stained with methylene blue (4 g/L) in methanol and scored by eye. The plating efficiencies (PE) were determined from control cells (DMSO treated) and used to calculate the surviving fraction (SF) after 6-TG treatments.
Site-directed mutagenesis primers, HPLC instrument settings, circular dichroism, size-exclusion chromatography, cloning of lentiviral constructs, RT-qPCR, primary antibodies, siRNA transfection and resazurin assay methods can be found in the Supplementary Methods.
Results
Wild-type and R139C mutant NUDT15 efficiently hydrolyze 6-thio-(d)GTP
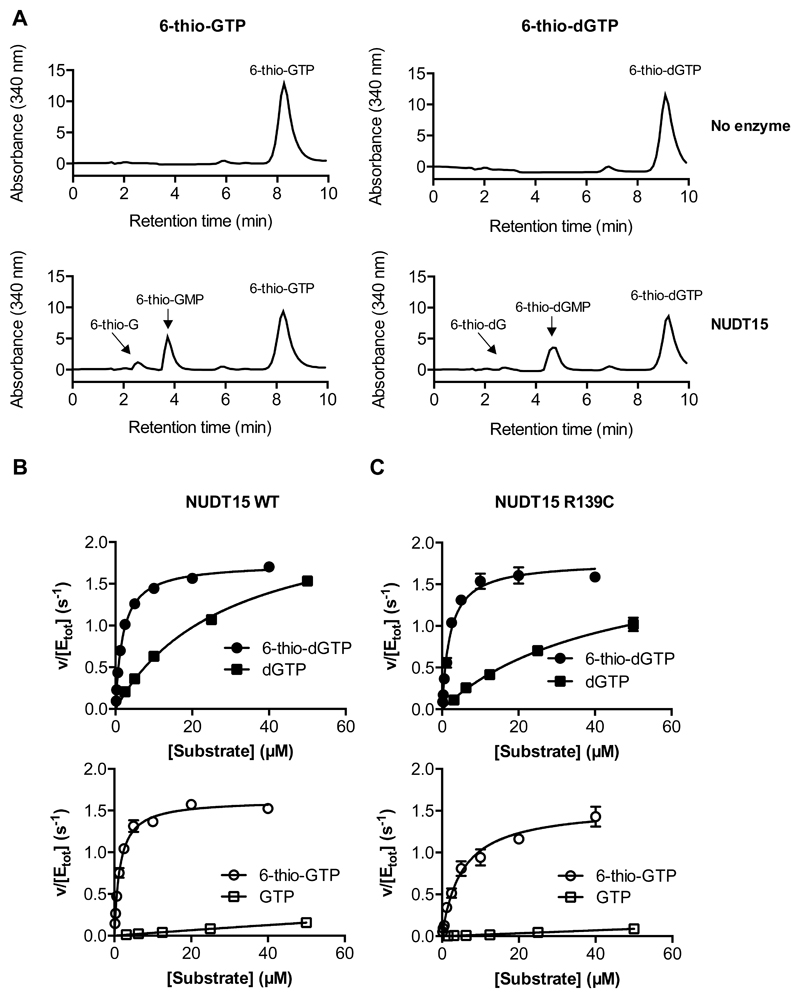
With substantial clinical evidence and preliminary biochemical data supporting a role for NUDT15 in thiopurine metabolism (23), we conducted thorough biochemical analyses with NUDT15 wild-type (WT) and the protein derivative R139C expressed and purified from bacterial lysates under non-reducing conditions (Supplementary Fig. S1). To analyze the formed products, NUDT15 activity towards 6-thio-dGTP and 6-thio-GTP was assessed by high performance liquid chromatography (HPLC). NUDT15 was able to hydrolyze 6-thio-dGTP and 6-thio-GTP to their corresponding monophosphate species (Fig. 1A). Interestingly, 6-thioguanosine was also detected as a product, albeit in minor amounts. Thus, it appears that NUDT15 may have some ambiguity with respect to which phosphate bond is hydrolyzed in thioguanosine triphosphates.
Figure 1. NUDT15 hydrolyzes 6-thio-(d)GTP to 6-thio-(d)GMP, prefers thionylated over canonical (d)GTP and maintains enzymatic activity with the R139C mutation.
(A) HPLC analysis demonstrates that NUDT15 hydrolyzes 6-thio-GTP to 6-thio-GMP and 6-thio-G (left) and 6-thio-dGTP to 6-thio-dGMP and 6-thio-dG (right). The spectra indicate 50 μM substrate, alone (No enzyme; upper panels) or after 30 minutes incubation with 10 nM WT NUDT15 (NUDT15; lower panels). Representative spectra are shown.
(B and C) Saturation curves for 6-thio-dGTP and dGTP (filled circles, filled squares; top) and 6-thio-GTP and GTP (empty circles, empty squares; bottom) with NUDT15 WT (B) or NUDT15 R139C (C). The data is presented as v (hydrolyzed substrate (μM) per second) per total enzyme concentration (μM). The graphs show representative data from at least two independent experiments and the standard deviation.
We measured time-dependent hydrolysis of dGTP at 5 μM, close to reported cellular concentrations (36, 37), as well as with GTP, 6-thio-dGTP, and 6-thio-GTP by analyzing PPi formation. Pronounced hydrolysis of 6-thio-dGTP and 6-thio-GTP, but undetectable activity with dGTP and GTP, was observed (Supplementary Fig. S2), indicating a preference for the thionylated substrates.
To better understand substrate preferences we performed in-depth kinetic analyses with increasing concentrations of 6-thio-dGTP, dGTP, 6-thio-GTP and GTP to better understand the substrate preferences of NUDT15 WT and R139C, using the PPiLight™ assay. NUDT15 showed higher affinity for thioguanosine triphosphates over canonical guanine substrates with KM = 1.9 and 1.8 μM for 6-thio-dGTP and 6-thio-GTP, respectively, compared to 43 and 254 μM for dGTP and GTP, respectively (Fig. 1B, Table 1). Catalytic turnover (kcat) was similar for all substrates (Table 1). A comparison of the catalytic efficiencies (kcat/KM) shows that 6-thio-dGTP and 6-thio-GTP are preferred substrates for NUDT15 over canonical guanine substrates, with 243- to 290-fold higher catalytic efficiency than for GTP and 13- to 15-fold higher efficiency than for dGTP, respectively (Table 1).
Table 1. Comparison of kinetic parameters for NUDT15 WT and R139C under reducing conditions.
The kinetic parameters for NUDT15 WT and R139C for dGTP, 6-thio-dGTP, GTP and 6-thio-GTP under reducing conditions.
| kcat (s-1) | KM (μM) | kcat/KM (M-1s-1) | |||||
|---|---|---|---|---|---|---|---|
| Substrate | WT | R139C | WT | R139C | WT | R139C | |
| dGTP | 2.58 ± 0.55 | 2.67 ± 0.06 | 43.3 ± 4.4 | 75.1± 4.3 | 59220 | 35552 | 1.7 |
| 6-thio-dGTP | 1.44 ± 0.44 | 1.78 ± 0.04 | 1.9 ± 0.05 | 2.11± 0.2 | 761140 | 843601 | 0.9 |
| GTP | 0.8 ± 0.17 | 1.27 ± 0.12 | 254 ± 25 | 683 ± 90 | 3140 | 1859 | 1.7 |
| 6-thio-GTP | 1.6 ± 0.11 | 1.51 ± 0.04 | 1.8 ± 0.46 | 4.9 ± 0.5 | 909000 | 308163 | 2.9 |
Because patients with the R139C mutation are particularly sensitive to thioguanine treatments, we hypothesized that the enzyme is unable to hydrolyze thioguanosine triphosphates efficiently. Surprisingly, NUDT15 R139C could also hydrolyze all species with similar activity as the WT protein when activity was measured under reducing conditions at 22°C (Fig. 1C). Kinetic analyses indicated similar KM values and catalytic turnover rates for WT and mutant NUDT15 (Table 1). The resultant catalytic efficiency was only marginally affected by the R139C mutation with respect to the thioguanine nucleotides. In fact, NUDT15 R139C had slightly higher activity than WT with 6-thio-dGTP as the substrate, and only 3-fold lower catalytic efficiency with 6-thio-GTP.
6-thio-GMP binding to NUDT15
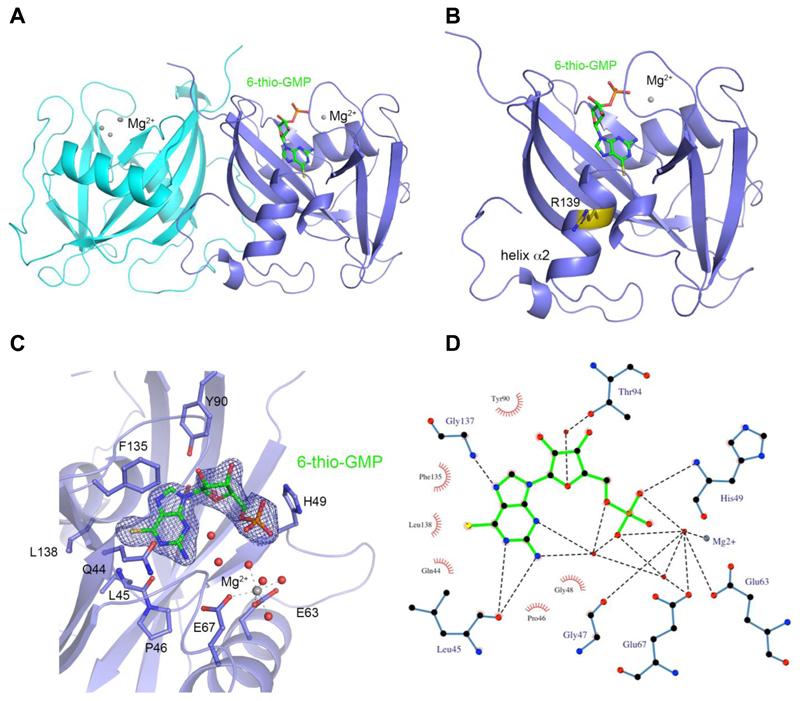
To understand the observed substrate specificity of NUDT15 for thioguanine substrates, we crystallized NUDT15 in the presence of Mg2+ and 6-thio-GTP. We determined the 1.7 Å resolution structure of NUDT15 in complex with the product, 6-thio-GMP, and Mg2+ (Fig. 2A, B). The overall structure is highly similar to the NUDT15 apo structure with RMSDs between 0.34 – 0.6 Å. There are no major structural differences, except in the loop formed by residues Arg34-Gly41 (Supplementary Fig. S3). Molecule A of the NUDT15 dimer has bound the hydrolysis product 6-thio-GMP and one Mg2+ ion coordinated by NUDIX box residues (Fig. 2C). Molecule B of the dimer did not bind 6-thio-GMP and instead displayed coordination of four Mg2+ ions as previously observed in the apo structure (23). Hydrogen bonding between the thioguanine base and the backbones of Leu45 and Gly137 coordinate the substrate in the active site (Fig. 2D). The α-phosphate of 6-thio-GMP forms one direct hydrogen bond to His49. In addition, multiple water-mediated hydrogen bonds between 6-thio-GMP and surrounding residues contribute to the binding (Fig. 2C, D). The 6-thio moiety is well accommodated in a hydrophobic pocket formed by Phe135, Leu138, Gln44, and Leu45 of NUDT15, explaining the lower KM values for 6-thio-GTP and 6-thio-dGTP compared to the canonical guanosine triphosphates (Table 1). The steric fit and the hydrophobic interactions of the sulfur further stabilize the binding (Fig. 2D).
Figure 2. Structure of NUDT15 with bound 6-thio-GMP and Mg2+ coordination.
(A) NUDT15 dimer in cartoon representation with hydrolysis product 6-thio-GMP and one Mg2+ ion bound in the active site of molecule A (slate blue) and molecule B (cyan) in the unbound state with four coordinated Mg2+ ions.
(B) Molecule A of NUDT15 dimer with 6-thio-GMP bound highlighting the position of R139 mutation site in yellow.
(C) Binding pocket of 6-thio-GMP and Mg2+ coordination with relevant residues labeled in stick format and the 2Fo-Fc electron density map of 6-thio-GMP in blue mesh.
(D) Ligplot+ representation of 6-thio-GMP interactions with hydrogen bonds shown as dashed lines and hydrophobic interactions represented as an arc with spokes radiating towards the ligand atoms they contact. The bonding lengths are indicated in angstroms (Å).
NUDT15 R139C is expressed but rapidly degraded in cells
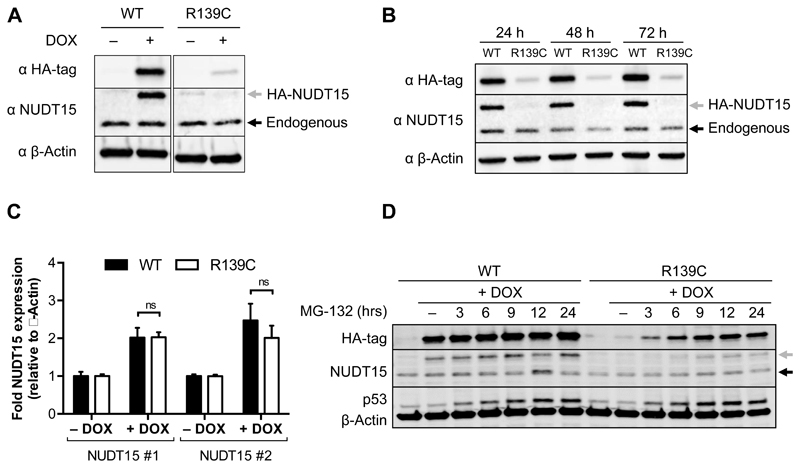
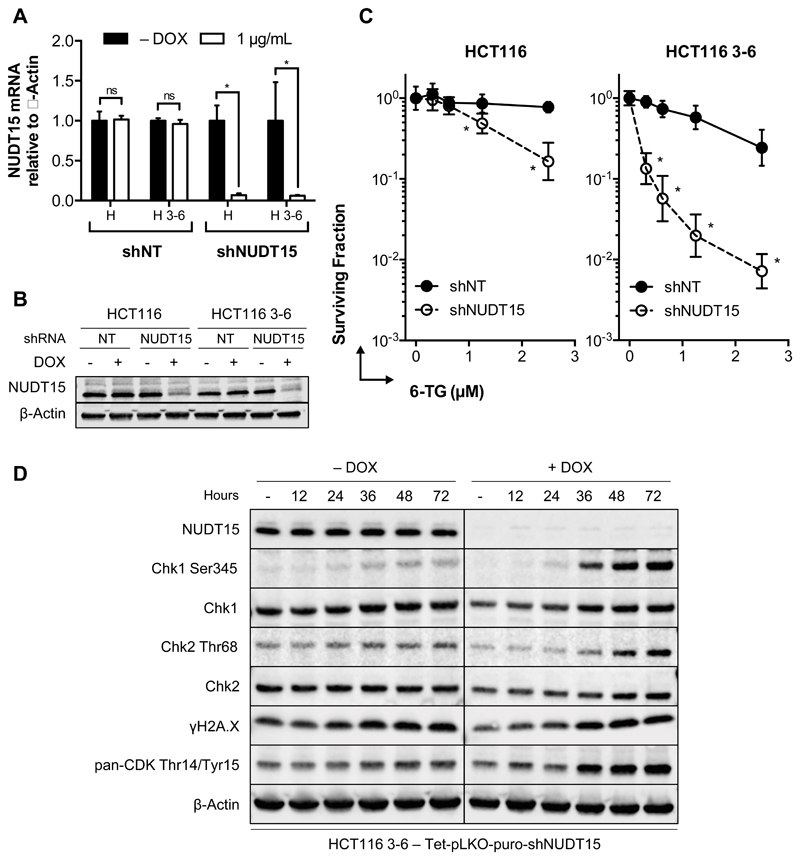
We next sought to understand how R139C protein can be active in vitro but induce thiopurine sensitivity in its carriers. Therefore, we overexpressed HA-tagged WT or R139C NUDT15 using doxycycline-inducible expression constructs in HCT116 cells, which are carrying endogenous WT NUDT15. Overexpression of the HA-tagged proteins was assessed with an anti-HA or anti-NUDT15 antibody. When analyzing protein levels, overexpressed NUDT15 WT was robustly induced upon doxycycline addition, but the overexpressed R139C mutant was barely detectable (Fig. 3A). No alterations of endogenous NUDT15 were observed. This was not a time-dependent event, as overexpressed WT and R139C protein did not fluctuate over the course of 72 hours (Fig. 3B). Interestingly, measurements of total NUDT15 mRNA showed roughly equal expression after induction by doxycycline over 72 hours (Fig. 3C), indicating a stark deviation from the protein levels. This suggested that something inherent to the R139C protein was limiting its accumulation.
Figure 3. NUDT15 R139C is expressed but undergoes proteasomal degradation in cells.
(A) Western blot after 72 hours doxycycline-induced overexpression of HA-tagged wild-type (WT) or R139C mutant.
(B) Protein levels for overexpressed WT and R139C NUDT15 do not change appreciably over the course of 72 hours. HA-tagged expression was induced in HCT116 cells for 24, 48 or 72 hours and harvested for western blotting.
(C) RT-qPCR analyzing the total mRNA levels of NUDT15 relative to β-Actin for HCT116 cells expressing WT NUDT15 (black) or the R139C mutant (white). Cells were treated for 72 hours with 1 μg/mL doxycycline. Asterisks (*) denote statistical significance (p < 0.05) by multiple t-test analysis (GraphPad Prism). Each condition was performed in triplicate and two independent experiments. The standard deviations are shown.
(D) HCT116 cells overexpressing HA-WT or HA-R139C NUDT15 after 72 hours of doxycycline induction were treated with 5 μM MG-132 for 3, 6, 9, 12 or 24 hours. In (A), (B), and (D) the grey arrows indicate HA-tagged expression constructs and the black arrows endogenous NUDT15 (WT). Representative experiments are shown.
In the cell, chaperone proteins first mediate the fate of structurally unstable or misfolded proteins, but, ultimately, chronically misfolded proteins are either degraded by autophagy or the proteasome system to prevent detrimental aggregation (38). To determine if NUDT15 R139C is degraded by the proteasome, we performed a time-course experiment with the proteasome inhibitor, MG-132. After 72 hours of doxycycline-induced overexpression of NUDT15 WT or R139C doxycycline was removed. 5 μM MG-132 was added for the indicated time points starting with the longest treatment and all samples were harvested at the same time-point (Fig. 3D). Accrual of p53, which rapidly accumulates following proteasome inhibition, was monitored as a control (39). While inhibition of the proteasome did not lead to increased accumulation of overexpressed NUDT15 WT protein, treatment with MG-132 led to a clear and distinct accumulation of R139C in a time-dependent manner (Fig. 3D). Thus, while the NUDT15 R139C mutant is expressed normally in cells, the protein is rapidly degraded.
The R139C mutation decreases the thermal stability of NUDT15
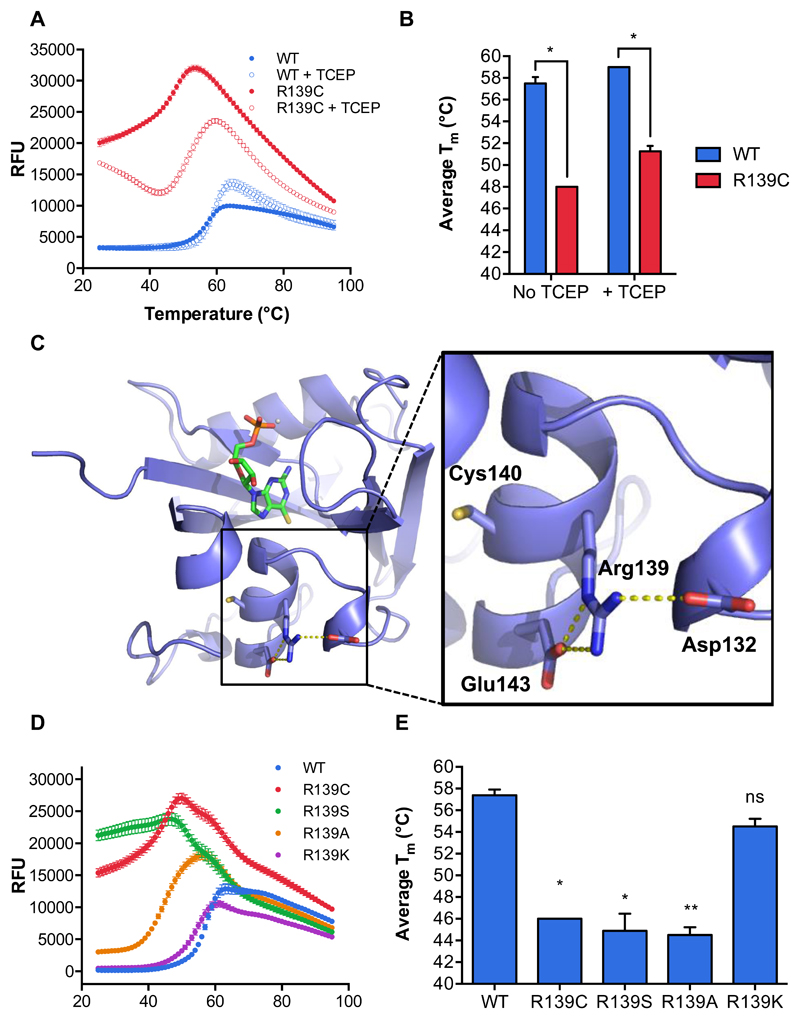
Since NUDT15 R139C showed activity under reducing conditions at 22°C in vitro but little protein was detected in a cellular setting, we investigated protein stability in vitro using a Differential Scanning Fluorimetry (DSF) assay and compared non-reducing and reducing conditions (Fig. 4A, B). NUDT15 WT had a melting point (Tm) of 59 and 58 °C with or without TCEP, respectively. This was much higher than the NUDT15 R139C mutant, which had a melting point of 48°C without TCEP and 51 °C with TCEP. Interestingly, NUDT15 R139C showed higher background fluorescence levels, which indicates that the SYPRO® Orange has more hydrophobic surface to interact with (Fig. 4A). This may suggest that the protein exists in a more open form, where more of the NUDT15 R139C interior is accessible, and could be due to alterations in secondary structure or disruption of protein dimerization. Circular dichroism experiments, however, indicated that the R139C secondary structures did not deviate significantly from WT protein (Supplementary Fig. S4A). In addition, R139C was still able to form a homodimer (23), as seen by size-exclusion chromatography (Supplementary Fig. S4B). Thus, NUDT15 R139C is less stable than the WT protein.
Figure 4. Mutations of Arg139 decrease the thermal stability of NUDT15.
(A) A representative DSF melting curve demonstrating the stability of NUDT15 WT (blue) and the R139C mutant (red) in the absence or presence of the reducing agent, TCEP (filled or empty circles, respectively). Relative Fluorescence Units (RFU) represents fluorescence of SYPRO® Orange at 570 nm.
(B) Melting points of NUDT15 WT (blue) and NUDT15 R139C (red) in the absence or presence of TCEP from experiments shown in (A). Melting points were calculated by determining the minima of the negative derivative of the melting curve (∂RFU/∂T). The asterisk (*) denotes a statistically significant difference (p < 0.05) by multiple t-test analysis (GraphPad Prism). Each condition was performed in quadruplicate and the standard deviations are shown.
(C) A detailed view of intramolecular bonds formed by Arg139 with 6-thio-GMP and how R139C may influence NUDT15 stability. Asp132, Arg139, Cys140 and Glu143 are labeled and hydrogen bonding between Arg139 and Glu143 and ionic interactions between Arg139 and Asp132 are illustrated by yellow dotted lines (PyMOL).
(D) A representative DSF melting curve showing the stability of NUDT15 WT (blue), R139C (red), R139S (green), R139A (orange) and R139K (purple) under non-reducing conditions. Relative Fluorescence Units (RFU) represents fluorescence of SYPRO® Orange at 570 nm.
(E) Melting points of NUDT15 WT, R139C, R139S, R139A and R139K from experiments in (D). Melting points were calculated as in (B). Statistical significance (ns, not significant; *, p < 0.05; **, p < 0.01) was calculated by t-test analysis and comparing each mutant to WT NUDT15 (GraphPad Prism). n=2 independent experiments performed in quadruplicate. The standard deviations are shown.
Catalytic activity of NUDT15 R139C is modestly influenced by redox state
Since NUDT15 R139C melting temperature is slightly influenced by non-reducing conditions, we explored potential sources of instability. Interestingly, a second cysteine residue is located adjacent to residue 139 at position 140, which could poise these residues to form a disulfide bridge.
If a disulfide bond occurs, we might expect a decrease in NUDT15 R139C enzymatic activity under non-reducing conditions. To test this, NUDT15 R139C was subjected to kinetic analysis in the absence or presence of reducing agent (Supplementary Fig. S5A, B). Surprisingly, the R139C mutant maintained activity under non-reducing conditions, and there was only a 2.3- to 2.5-fold increase in catalytic efficiency when dithiothreitol (DTT) was added (Supplementary Fig. S5C). Thus, with little difference between the activity of WT and R139C NUDT15 under reducing conditions, it was not apparently clear, from a biochemical standpoint, what contribution a potential disulfide bond has to R139C stability.
NUDT15 Arg139 mutants display varying degrees of thermal stability likely due to loss of key intramolecular bonding
Residue 139 is contained within helix α2, whose N terminus makes up the base of the active site (Fig. 2B, C). We hypothesized that perturbations in this helix could influence the stability of NUDT15. Arg139 is able to make key hydrogen bonds within helix α2 (Glu143) and a potentially crucial ionic interaction with Asp132, which is adjacent to helix α2 (Fig. 4C). Deviations from arginine at position 139 could, therefore, have significant structural implications. In addition, Arg139 is proximal to the negatively charged absolute C-terminus (Leu164), further requiring a positive charge to stabilize this network (Fig. 2B). To assess whether the loss of Arg139 or the introduction of a cysteine disturbs NUDT15 protein stability, the mutants R139A, R139S, R139C and R139K were produced under non-reducing conditions and their stability was assayed using DSF (Supplementary Fig. S1, Fig. 4D, E). Compared to WT, only the R139K mutant had a similar melting point (Tm = 54.5 °C). This can be explained by the fact that lysine is able to make the same intramolecular ionic bond as arginine (Fig. 4C, Supplementary Fig. S6). The R139C, R139S and R139A mutants had significantly lower melting points (Tm = 46, 44.9, and 44.5 °C, respectively). While their contributions to hydrogen bonding within helix α2 vary, each of these residues would be unable to make the ionic connection to Asp132, implying that this interaction is important for NUDT15 stability. Taken together, the data suggest that R139 has a stabilizing effect on helix α2 in NUDT15, and the alteration of this residue is likely to contribute to inherent instability.
NUDT15 depletion sensitizes cancer cells to thiopurine treatment
Since our in vitro experiments showed activity of NUDT15 with 6-thio-dGTP and 6-thio-GTP, we wanted to investigate whether depletion of the protein increases efficacy of 6-thioguanine in a cellular setting, similar to the conditions expected in patients carrying the R139C mutant. Therefore, we depleted NUDT15 in HCT116 and HCT116 3-6 cells using doxycycline-inducible NUDT15-specific shRNA (shNUDT15) and compared it with a non-targeting shRNA control. HCT116 cells and their isogenic, MMR-proficient counterpart, HCT116 3-6 (40), have been used extensively to study MMR-dependent mechanisms, such as those involving thiopurines (41). shNUDT15 efficiently silenced endogenous NUDT15 mRNA and protein in both cell lines after a 72-hour incubation with doxycycline (Fig. 5A and 5B). Subsequently, clonogenic survival was assessed with increasing doses of 6-TG. As expected, MMR-deficient HCT116 cells were more resistant to 6-TG while MMR-proficient HCT116 3-6 cells were more sensitive. Depletion of NUDT15 dramatically increased the sensitivity of HCT116 3-6 cells to 6-TG, and, to a lesser extent, parental HCT116 cells (Fig. 5C). This sensitization was also seen with three different siRNA oligos against NUDT15 by resazurin assay, which confirmed target specificity (Supplementary Fig. S7).
Figure 5. NUDT15 depletion sensitizes cells to 6-TG.
(A) NUDT15 mRNA levels after addition of doxycycline to shRNA-containing HCT116 (H) or HCT116 3-6 (H 3-6) cells for 72 hours. Doxycycline (DOX) was added (white bars) or not (black bars) to HCT116 and HCT116 3-6 cells containing a non-targeting control vector (shNT) or NUDT15 specific shRNA (shNUDT15). NUDT15 mRNA levels were normalized to β-Actin for each sample. n=2 independent experiments performed in triplicate. Asterisks (*) denote statistical significance (p < 0.05) by multiple t-test analysis (GraphPad Prism). The standard deviations are shown.
(B) NUDT15 protein levels of experiments performed as in (A). β-Actin is used as a loading control.
(C) Clonogenic survival assay with 6-TG in HCT116 (left) or HCT116 3-6 cells (right) comparing the effect of NUDT15 depletion. NUDT15-specific shRNA (empty circles) and a non-targeting control (filled circles) were compared. Experiments were performed in triplicate and n=2 replicates. Asterisks (*) denote statistical significance (p < 0.05) by multiple t-test analysis (GraphPad Prism). The standard deviations are shown.
(D) Western blot depicting the effect of NUDT15 depletion on ATR-Chk1 checkpoint activation in HCT116 3-6 cells. Cells were depleted of NUDT15 over 72 hours by doxycycline induction (or not) and exposed to 150 nM 6-TG over the described time (in hours). A representative experiment is shown.
Futile repair cycling and prolonged G2 arrest are signatures of 6-TG DNA incorporation in MMR-proficient cells (11, 12), which activates the ATR-Chk1 checkpoint and accounts for most of the toxic effects of thiopurines (14, 15). To determine if NUDT15 knockdown cells are more sensitive to 6-TG-induced checkpoint activation, HCT116 3-6 cells were depleted of NUDT15 and submitted to a time course treatment with low-dose 6-TG (Fig. 5D). At 150 nM 6-TG, cells without doxycycline-induced depletion of NUDT15 (– DOX) had minor increases in Chk1 and Chk2 phosphorylation, γH2A.X, and CDK inhibitory phosphorylation, an indicator of cells in G2-phase (42). Depletion of NUDT15 had a striking increase on these markers, indicating a much stronger response to 6-TG treatment. Importantly, Chk1 phosphorylation was followed by Chk2 phosphorylation and occurred approximately two cell cycles after addition of 6-TG, as described previously (42). Altogether, these experiments show that NUDT15 reduces the efficacy of 6-TG treatment.
Discussion
After a half-century in clinical applications, thiopurines are still a cornerstone therapy for childhood ALL and autoimmune disorders, such as IBD (2, 3, 10). Several pharmacogenetic studies have linked NUDT15 Arg139 mutations to thiopurine sensitivity in leukemia and IBD patients (17–21). Further supporting a role for NUDT15 in thiopurine metabolism was the identification of 6-thio-dGTP and 6-thio-GTP as substrates(23).
Our previously reported substrate screen performed at 50 μM concentration identified that NUDT15 hydrolyses 6-thio-dGTP, 6-thio-GTP and dGTP (23), which we now know to be saturating. The in-depth kinetic analysis presented here highlights that NUDT15 hydrolyzes 6-thio-dGTP and 6-thio-GTP more efficiently than the canonical guanosine triphosphates, GTP and dGTP, due to higher substrate affinity (lower KM). To explain the observed substrate preferences, we solved the first crystal structure of NUDT15 with 6-thio-GMP bound to the active site. Surprisingly, the NUDT15 R139C mutant hydrolyzed these substrates at similar efficiencies as NUDT15 WT, indicating that sensitivity in patients may be independent of enzymatic capability. When expressed in cells, R139C was rapidly degraded, suggesting that structural abnormalities could be a factor. Thermal instability of the R139C mutant was confirmed in vitro, and analysis of a variety of Arg139 mutants indicated that the loss of arginine and disruption of the ionic bonding network destabilizes the protein. Our attempts to crystalize NUDT15 R139C have been unsuccessful so far, which is likely an additional consequence of decreased stability. Since NUDT15 R139C was rapidly degraded in cells, as presumably is the case in patients carrying this variant, we wanted to mimic this situation with NUDT15 knockdown experiments. Depletion of NUDT15 clearly sensitized cells to 6-thioguanine, similar to the response of patients carrying the R139C mutation. This sensitization was enhanced by functional mismatch repair. Taken together, the data confirm the importance of NUDT15 in thiopurine metabolism and efficacy (Fig. 6).
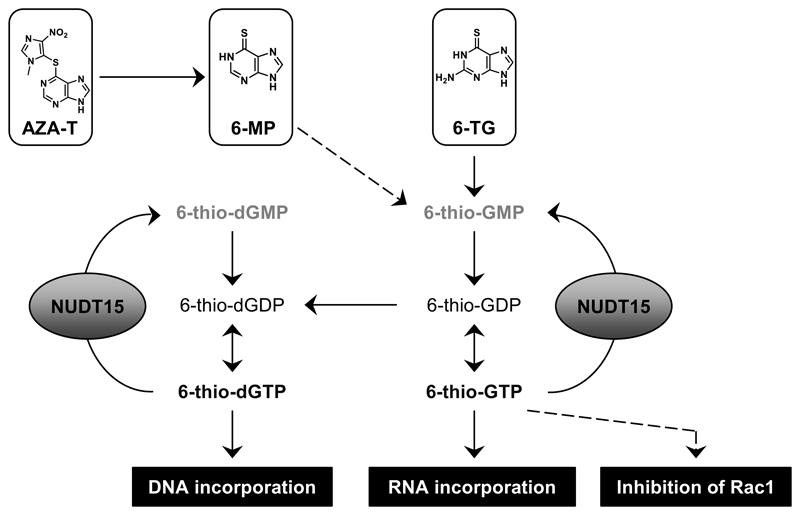
Figure 6. Schematic overview of how NUDT15 is involved in thiopurine metabolism.
The azathioprine (AZA-T), 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG) pro-drugs undergo a series of enzymatic reactions that result in the active species, 6-thio-GTP and 6-thio-dGTP, which are then incorporated into DNA/RNA or can inhibit Rac1 GTPase activity (6-thio-GTP). NUDT15 hydrolyzes 6-thio-dGTP and 6-thio-GTP to the corresponding monophosphates and thereby acts as a barrier to the maximum efficacy of thiopurines in cells.
These data support a recent study that demonstrated a role for NUDT15 in thiopurine metabolism and explored the biological ramifications of missense polymorphisms (43). Our results extend the implications of this study through further mechanistic and biochemical characterizations, including a high-resolution structure of NUDT15 with the 6-thio-GTP hydrolysis product, 6-thio-GMP, and analysis of the structural contribution of Arg139 to protein stability. Furthermore, we show that NUDT15 depletion sensitizes both MMR-proficient and -deficient cells to thioguanine, which is indicative of increased DNA incorporation, as well as contribution of other MMR-independent toxicity mechanisms. For example, defective RNA production (11), influence on GTPases, like RAC1 (44), and interference in purine biosynthesis have been suggested as additional sources of activity and potential toxicity of thiopurines (4, 45).
We contend that sensitivity in patients with the R139C mutation is due to protein instability that results in NUDT15 R139C degradation (Fig. 3, 4). This implies that sensitivity results from limited NUDT15 availability and is not a direct consequence of enzymatic capacity (Table 1). Notably, we found that, when overexpressed, R139C protein levels were significantly lower than WT protein levels despite having essentially the same mRNA expression. Treatment with proteasome inhibitor was able to restore this difference, suggesting the ubiquitin-proteasome system flags the protein for destruction (38), and subsequent evidence supported that loss of structural integrity is the likely cause.
Pharmacogenetics studies have emphasized the importance of screening patients for NUDT15 missense variants before administering thiopurines, as the tolerated dose can be <10% of that for unaffected patients (17, 19, 43). Screening for mutations to thiopurine methyl transferase (TPMT), an enzyme that methylates thiopurines and prevents their DNA incorporation, is a common routine prior to initiating thiopurine treatments. However, TPMT variations are less common in the Asian population compared to Europeans (17, 46), and only a quarter of European IBD patients suffering from thiopurine-induced leukopenia carry mutated TPMT (17, 47, 48). Other enzymes, such as ITPase (49, 50) and NT5C2 (51, 52) have also been suggested to influence sensitivity to thiopurines. Thus, thiopurine sensitivity and resistance factors are multifaceted.
Currently, the physiological role of NUDT15 remains unclear – as thiopurines are not endogenous metabolites. Since cellular concentrations of dGTP have been reported to be around 5 μM (36, 37), we measured time-dependent catalysis at this concentration (Supplementary Fig. S2) and performed a thorough kinetic analysis (Figure 1B and C). No detectable NUDT15-catalyzed hydrolysis of dGTP at 5 μM would suggest that it is not a relevant substrate in the cell; however, the prospect that NUDT15 may have a role in dGTP metabolism under certain situational contexts cannot be excluded at this time. The metabolism of dGTP is regulated by multiple enzymes with substantial overlap, which further complicates evaluating the role for NUDT15 in these processes. Nonetheless, depletion of NUDT15 alone does not have a discernable effect on cell cycle progression, DNA damage or cell viability (23), nor has the R139C mutation been identified as a risk factor for disease or sensitivity to other therapeutics in patients.
In the presence of thiopurines, however, NUDT15 appears to be a key mediator of efficacy, as our and other’s results clearly suggest (43). Intriguingly, we found NUDT15 mRNA expression to be upregulated in clinical ALL samples (53) (Supplementary Fig. S8), which raises the enticing possibility of modulating thiopurine dosing by specifically targeting NUDT15.
Supplementary Material
Acknowledgements
We thank Prof. Astrid Gräslund and Axel Abelein for help and advice regarding the circular dichroism analysis. We thank the beamline scientists at BESSY, Berlin, ESRF, Grenoble for their support in data collection, PSF for protein purification and Biostruct-X for support. We also thank Kristina Edfeldt and Sabina Eriksson for their support in the Helleday Lab, Elisée Wiita for help with DSF experiments and Saeed Eshtad for providing us with the non-targeting shRNA construct. This project is supported by the Knut and Alice Wallenberg Foundation (T.H. and P.S.), the Swedish Research Council [2014-5667], the Wenner-Gren Foundation, Åke Wibergs Foundation and the Swedish Cancer Society (P.S.).
Financial support: This project is primarily supported by The Knut and Alice Wallenberg Foundation (T. Helleday, P. Stenmark). Further support was received from the Swedish Research Council (T. Helleday, P. Stenmark), the European Research Council (T. Helleday), Göran Gustafsson Foundation (T. Helleday), Swedish Cancer Society (T. Helleday, P. Stenmark), the Swedish Children’s Cancer Foundation (N.G. Sheppard, T. Helleday), the Swedish Pain Relief Foundation (T. Helleday), the Wenner-Gren Foundation (P. Herr, P. Stenmark), the Åke Wiberg Foundation (P. Stenmark), the Torsten and Ragnar Söderberg Foundation (T. Helleday), the Canadian Institutes of Health Research and the Breast Cancer Society of Canada (B.D.G. Page), and the David and Astrid Hagelén Foundation (B.D.G. Page).
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
References
- 1.Hitchings GH, Elion GB, Falco EA, Russell PB, Vanderwerff H. Studies on analogs of purines and pyrimidines. Annals of the New York Academy of Sciences. 1950;52(8):1318–35. doi: 10.1111/j.1749-6632.1950.tb54032.x. Epub 1950/07/07. [DOI] [PubMed] [Google Scholar]
- 2.Schmiegelow K, Nielsen SN, Frandsen TL, Nersting J. Mercaptopurine/Methotrexate Maintenance Therapy of Childhood Acute Lymphoblastic Leukemia: Clinical Facts and Fiction. Journal of Pediatric Hematology/Oncology. 2014;36(7):503–17. doi: 10.1097/MPH.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford K, Shih DQ. Optimizing 6-mercaptopurine and azathioprine therapy in the management of inflammatory bowel disease. World journal of gastroenterology. 2011;17(37):4166–73. doi: 10.3748/wjg.v17.i37.4166. Epub 2011/11/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karran P. Thiopurines, DNA damage, DNA repair and therapy-related cancer. Br Med Bull. 2006;79–80:153–70. doi: 10.1093/bmb/ldl020. [DOI] [PubMed] [Google Scholar]
- 5.Nelson JA, Carpenter JW, Rose LM, Adamson DJ. Mechanisms of Action of 6-Thioguanine, 6-Mercaptopurine, and 8-Azaguanine. Cancer Research. 1975;35(10):2872–8. [PubMed] [Google Scholar]
- 6.Ling YH, Nelson JA, Cheng YC, Anderson RS, Beattie KL. 2'-Deoxy-6-thioguanosine 5'-triphosphate as a substrate for purified human DNA polymerases and calf thymus terminal deoxynucleotidyltransferase in vitro. Mol Pharmacol. 1991;40(4):508–14. [PubMed] [Google Scholar]
- 7.Warren DJ, Andersen A, Slordal L. Quantitation of 6-thioguanine residues in peripheral blood leukocyte DNA obtained from patients receiving 6-mercaptopurine-based maintenance therapy. Cancer Res. 1995;55(8):1670–4. [PubMed] [Google Scholar]
- 8.Cuffari C, Li DY, Mahoney J, Barnes Y, Bayless TM. Peripheral blood mononuclear cell DNA 6-thioguanine metabolite levels correlate with decreased interferon-gamma production in patients with Crohn's disease on AZA therapy. Dig Dis Sci. 2004;49(1):133–7. doi: 10.1023/b:ddas.0000011614.88494.ee. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida S, Yamada M, Masaki S, Saneyoshi M. Utilization of 2'-deoxy-6-thioguanosine 5'-triphosphate in DNA synthesis in vitro by DNA polymerase alpha from calf thymus. Cancer Res. 1979;39(10):3955–8. [PubMed] [Google Scholar]
- 10.Karran P, Attard N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nat Rev Cancer. 2008;8(1):24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 11.You C, Dai X, Yuan B, Wang Y. Effects of 6-thioguanine and S6-methylthioguanine on transcription in vitro and in human cells. The Journal of biological chemistry. 2012;287(49):40915–23. doi: 10.1074/jbc.M112.418681. Epub 2012/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swann PF, Waters TR, Moulton DC, Xu YZ, Zheng Q, Edwards M, et al. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273(5278):1109–11. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 13.Aquilina G, Giammarioli AM, Zijno A, Di Muccio A, Dogliotti E, Bignami M. Tolerance to O6-methylguanine and 6-thioguanine cytotoxic effects: a cross-resistant phenotype in N-methylnitrosourea-resistant Chinese hamster ovary cells. Cancer Res. 1990;50(14):4248–53. Epub 1990/07/15. [PubMed] [Google Scholar]
- 14.Yan T, Berry SE, Desai AB, Kinsella TJ. DNA mismatch repair (MMR) mediates 6-thioguanine genotoxicity by introducing single-strand breaks to signal a G2-M arrest in MMR-proficient RKO cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(6):2327–34. Epub 2003/06/11. [PubMed] [Google Scholar]
- 15.Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, et al. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes & development. 2004;18(11):1331–44. doi: 10.1101/gad.294404. Epub 2004/06/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.York SJ, Modrich P. Mismatch repair-dependent iterative excision at irreparable O6-methylguanine lesions in human nuclear extracts. The Journal of biological chemistry. 2006;281(32):22674–83. doi: 10.1074/jbc.M603667200. Epub 2006/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang SK, Hong M, Baek J, Choi H, Zhao W, Jung Y, et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet. 2014;46(9):1017–20. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakuta Y, Naito T, Onodera M, Kuroha M, Kimura T, Shiga H, et al. NUDT15 R139C causes thiopurine-induced early severe hair loss and leukopenia in Japanese patients with IBD. Pharmacogenomics J. 2015 doi: 10.1038/tpj.2015.43. [DOI] [PubMed] [Google Scholar]
- 19.Yang JJ, Landier W, Yang W, Liu C, Hageman L, Cheng C, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol. 2015;33(11):1235–42. doi: 10.1200/JCO.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Y, Kato M, Hasegawa D, Urayama KY, Nakadate H, Kondoh K, et al. Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br J Haematol. 2015;171(1):109–15. doi: 10.1111/bjh.13518. [DOI] [PubMed] [Google Scholar]
- 21.Chiengthong K, Ittiwut C, Muensri S, Sophonphan J, Sosothikul D, Seksan P, et al. NUDT15 c.415C>T increases risk of 6-mercaptopurine induced myelosuppression during maintenance therapy in children with acute lymphoblastic leukemia. Haematologica. 2015 doi: 10.3324/haematol.2015.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai JP, Ishibashi T, Takagi Y, Hayakawa H, Sekiguchi M. Mouse MTH2 protein which prevents mutations caused by 8-oxoguanine nucleotides. Biochem Biophys Res Commun. 2003;305(4):1073–7. doi: 10.1016/s0006-291x(03)00864-7. [DOI] [PubMed] [Google Scholar]
- 23.Carter M, Jemth AS, Hagenkort A, Page BD, Gustafsson R, Griese JJ, et al. Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2. Nat Commun. 2015;6 doi: 10.1038/ncomms8871. 7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC biotechnology. 2011;11:92. doi: 10.1186/1472-6750-11-92. Epub 2011/10/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie AGW, Powell HR. Processing diffraction data with MOSFLM. Nato Sci Ser Ii Math. 2007;245:41–51. [Google Scholar]
- 26.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallographica Section D, Biological Crystallography. 2013;69(Pt 7):1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica Section D, Biological crystallography. 1997;53(Pt 3):240–55. doi: 10.1107/S0907444996012255. Epub 1997/05/01. [DOI] [PubMed] [Google Scholar]
- 30.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Mol Biol. 1999;6(5):458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. Epub 2010/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. Epub 2010/01/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gad H, Koolmeister T, Jemth A-S, Eshtad S, Jacques SA, Strom CE, et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508(7495):215–21. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 34.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 35.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. The Journal of biological chemistry. 2005;280(7):5516–26. doi: 10.1074/jbc.M412105200. Epub 2004/12/22. [DOI] [PubMed] [Google Scholar]
- 36.Traut TW. Physiological concentrations of purines and pyrimidines. Molecular and cellular biochemistry. 1994;140(1):1–22. doi: 10.1007/BF00928361. Epub 1994/11/09. [DOI] [PubMed] [Google Scholar]
- 37.Wilson PM, Labonte MJ, Russell J, Louie S, Ghobrial AA, Ladner RD. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic acids research. 2011;39(17):e112. doi: 10.1093/nar/gkr350. Epub 2011/05/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–32. doi: 10.1038/nature10317. Epub 2011/07/22. [DOI] [PubMed] [Google Scholar]
- 39.Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53(1) Cancer Res. 1996;56(11):2649–54. Epub 1996/06/01. [PubMed] [Google Scholar]
- 40.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, et al. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N'-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54(16):4308–12. Epub 1994/08/15. [PubMed] [Google Scholar]
- 41.Hawn MT, Umar A, Carethers JM, Marra G, Kunkel TA, Boland CR, et al. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995;55(17):3721–5. Epub 1995/09/01. [PubMed] [Google Scholar]
- 42.Yan T, Desai AB, Jacobberger JW, Sramkoski RM, Loh T, Kinsella TJ. CHK1 and CHK2 are differentially involved in mismatch repair–mediated 6-thioguanine-induced cell cycle checkpoint responses. Molecular Cancer Therapeutics. 2004;3(9):1147–57. [PubMed] [Google Scholar]
- 43.Moriyama T, Nishii R, Perez-Andreu V, Yang W, Klussmann FA, Zhao X, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet. 2016 doi: 10.1038/ng.3508. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts RL, Barclay ML. Update on thiopurine pharmacogenetics in inflammatory bowel disease. Pharmacogenomics. 2015;16(8):891–903. doi: 10.2217/pgs.15.29. [DOI] [PubMed] [Google Scholar]
- 45.Tay BS, Lilley RM, Murray AW, Atkinson MR. Inhibition of phosphoribosyl pyrophosphate amidotransferase from Ehrlich ascites-tumour cells by thiopurine nucleotides. Biochemical pharmacology. 1969;18(4):936–8. doi: 10.1016/0006-2952(69)90069-0. Epub 1969/04/01. [DOI] [PubMed] [Google Scholar]
- 46.Cooper SC, Ford LT, Berg JD, Lewis MJ. Ethnic variation of thiopurine S-methyltransferase activity: a large, prospective population study. Pharmacogenomics. 2008;9(3):303–9. doi: 10.2217/14622416.9.3.303. [DOI] [PubMed] [Google Scholar]
- 47.Colombel JF, Ferrari N, Debuysere H, Marteau P, Gendre JP, Bonaz B, et al. Genotypic analysis of thiopurine S-methyltransferase in patients with Crohn's disease and severe myelosuppression during azathioprine therapy. Gastroenterology. 2000;118(6):1025–30. doi: 10.1016/s0016-5085(00)70354-4. [DOI] [PubMed] [Google Scholar]
- 48.Booth RA, Ansari MT, Tricco AC, Loit E, Weeks L, Doucette S, et al. Assessment of thiopurine methyltransferase activity in patients prescribed azathioprine or other thiopurine-based drugs. Evid Rep Technol Assess (Full Rep) 2010;(196):1–282. [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka Y, Manabe A, Nakadate H, Kondoh K, Nakamura K, Koh K, et al. The activity of the inosine triphosphate pyrophosphatase affects toxicity of 6-mercaptopurine during maintenance therapy for acute lymphoblastic leukemia in Japanese children. Leuk Res. 2012;36(5):560–4. doi: 10.1016/j.leukres.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 50.Wan Rosalina WR, Teh LK, Mohamad N, Nasir A, Yusoff R, Baba AA, et al. Polymorphism of ITPA 94C>A and risk of adverse effects among patients with acute lymphoblastic leukaemia treated with 6-mercaptopurine. J Clin Pharm Ther. 2012;37(2):237–41. doi: 10.1111/j.1365-2710.2011.01272.x. [DOI] [PubMed] [Google Scholar]
- 51.Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nature Genetics. 2013;45(3):290–4. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19(3):368–71. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Homminga I, Pieters R, Langerak Anton W, de Rooi Johan J, Stubbs A, Verstegen M, et al. Integrated Transcript and Genome Analyses Reveal NKX2-1 and MEF2C as Potential Oncogenes in T Cell Acute Lymphoblastic Leukemia. Cancer Cell. 2011;19(4):484–97. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.