Abstract
A recent bioinformatic analysis of well-characterized classes of riboswitches uncovered subgroups unable to bind to the regulatory molecule of the parental class. Within the guanine/adenine class, seven groups of RNAs were identified that deviate from the consensus sequence at one or more of three positions directly involved purine nucleobase recognition, one of which was validated as a second class of 2′-deoxyguanosine riboswitch (called 2′-dG-II). To understand how 2′-dG-II riboswitches recognize their cognate ligand and how they differ from a previously identified class of 2′-deoxyguanosine binding riboswitches, we have solved the crystal structure of a 2′-dG-II aptamer domain bound to 2′-deoxyguanosine. This structure reveals a global architecture similar to other members of the purine riboswitch family, but contains key differences within the ligand binding core. Defining the 2′-dG-II riboswitches is a two-nucleotide insertion in the three-way junction that promotes novel base-base interactions. Unlike 2′-dG-I riboswitches, the 2′-dG-II class only requires local changes to the ligand binding pocket of the guanine/adenine class to achieve a change in ligand preference. Notably, members of the 2′-dG-II family have variable ability to discriminate between 2′-deoxyguanosine and riboguanosine, suggesting that a subset of 2′-dG-II riboswitches may bind either molecule to regulate gene expression.
INTRODUCTION
It is becoming increasingly realized that within bioinformatically-defined classes of riboswitches there exists variation in both the cognate metabolite that regulates gene expression and the ability to discriminate between chemically related metabolites (1). Diversity in ligand recognition within a defined class is exemplified by the ykkC family of riboswitches. Originally identified as a sequence upstream of genes encoding multidrug resistance efflux pumps and other transporters (2), these riboswitches remained orphan until it was determined that some members respond to guanidine (3). Further examination of this class revealed multiple distinct classes sharing a secondary structure, but containing sequence variation within some of the most conserved regions. Distinct classes of the ykkC family of riboswitches have most recently been verified to bind the purine biosynthetic precursor phosphoribosyl pyrophosphate (PRPP) (4), the bacterial alarmone ppGpp (5), and nucleoside diphosphates (6). Other examples of ligand recognition diversity have been observed within class-II cobalamin riboswitches that bind adenosyl- and methylcobalamin with a spectrum of selectivities (7), a subset of GEMM-I riboswitches that bind cyclic AMP-GMP rather than cyclic di-GMP (8) and rare FMN riboswitches that bind an alternative but unknown compound (9).
Despite the realization that effector ligand diversity might be a widespread phenomenon within bioinformatically-defined riboswitch classes, identifying functional variation is challenging. Based on current knowledge, most sequence variation within a class does not lead to a change in ligand selectivity (1). To identify functional variants, the Breaker laboratory recently developed a new computational approach to examine established riboswitch classes for detecting unrecognized ligand binding diversity. This was achieved by using crystal structures of riboswitch aptamer–ligand complexes to identify the first shell of ligand-interacting nucleotides and survey the Rfam sequence alignment of that class to identify members that vary from consensus at these positions. This approach resulted in identification of variant groups within the guanine, c-di-GMP, glycine and FMN classes (1).
One of the variant classes of guanine riboswitches discovered was predicted and validated as a second class of 2′-deoxyguanosine (2′-dG) binders–referred to as the 2′-dG-II class (originally annotated as the ‘UCC variant group’) (1). These RNAs contain a first shell of ligand-interacting nucleotides whose identity is U22, C51, C74, similar to the equivalent nucleotides in the Mesoplasma florum (Mfl) I-A 2′-dG-I riboswitches (C31, C58, C80) (red nucleotides, Figure 1) (10). The identity of the pyrimidine at position 51/58, which directly interacts with the N3/N9 face (sugar edge) of the purine ligand, is critical for establishing selectivity for nucleobase versus nucleoside binding across the purine riboswitch family (11,12) (Figure 2A, B). However, there are further differences between the two classes of 2′-dG binders around the ligand binding pocket that distinguish them. First, the 2′-dG-II riboswitches have an insertion element within J3/1 that is unique amongst the purine riboswitches (1). A second notable change is that the junction-proximal base pair in P1 is a G–C pair whereas in all other purine riboswitch classes it is a nearly invariant A–U pair (1). These, along with other differences further away from the ligand binding pocket such as in L2 and L3, may be important for the ability of this new class of riboswitches to bind 2′-dG with high affinity and selectivity.
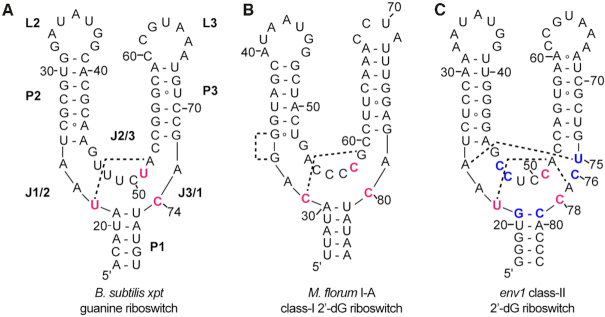
Figure 1.
Secondary structure of representatives of the three classes of riboswitches within the purine family. Sequences are based upon those used for determination of the crystal structure of (A) the B. subtilis xpt guanine riboswitch aptamer domain (PDB ID: 4FE5), (B) the M. florum I-A class-I 2′-dG riboswitch aptamer domain (PDB ID: 3SKI), and (C) the env1 class-II 2′-dG riboswitch (this study). The secondary structural elements which are common to all three classes, are labeled as P (paired), J (joining) and L (loop). Nucleotides highlighted in red directly interact with the ligand nucleobase and nucleotides in blue within the env1 sequence represent key differences in the ligand binding three-way junction between the xpt and env1 RNAs. The dashed lines represent two sets of base-base interactions in the junction that distinguish the three classes of riboswitches.
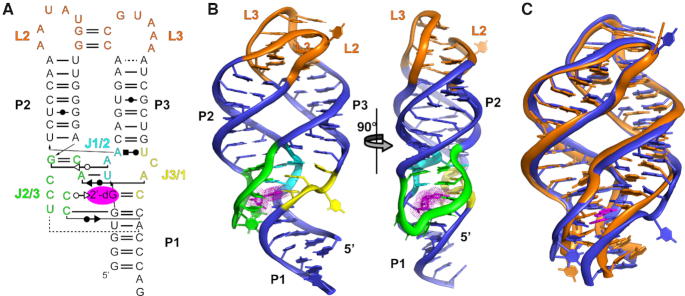
Figure 2.
Overall structure of the env1 2′-dG-II riboswitch. (A) Secondary structure of the environmental sequence 1 (env1) 2′-dG-II riboswitch annotated with nucleobase-nucleobase interactions. Base interaction notation is that of Leontis and Westhof (57). (B) Cartoon representation of the crystal structure of the env1 riboswitch aptamer domain in complex with 2′-deoxyguanosine (magenta). Watson–Crick helices (P1 – P3) are denoted in blue, the two terminal loops (L2 and L3) in orange and the three joining regions of the three-way junction (J1/2, J2/3 and J3/1) are shown in cyan, green and yellow, consistent with panel (A). (C) Superimposition of the B. subtilis xpt guanine riboswitch aptamer domain (orange) with env1 2′-dG riboswitch aptamer domain (blue). In each RNA, the P1 helix was truncated to the three Watson–Crick base pairs proximal to the three way junction. The RNAs align with an r.m.s. deviation of 1.95 Å using PyMOL.
In this work, we present a structural and biochemical analysis of the 2′-dG-II class of riboswitches that explains its selectivity for 2′-dG over other purine nucleobases and nucleosides. While the global architecture of the 2′-dG-II riboswitch aptamer domain is highly similar to the other classes within the purine riboswitch family, there are significant differences around the ligand binding pocket between the two 2′-dG classes. Notably, the two-nucleotide insertion element in J3/1 enables base-base interactions not observed in the other purine classes. Analysis of a series of mutants indicates that while the J3/1 insertion element and the identity of the junction-proximal base pair in P1 play a central role in promoting 2′-dG affinity, other nucleotides within the junction also contribute towards achieving high affinity binding. Binding analysis of a subset of aptamers from the 2′-dG-II family revealed a spectrum of selectivities for 2′-dG versus 2′-rG, suggesting that some members may respond to both nucleosides, further highlighting potential hidden variation within bioinformatically-defined riboswitch classes.
MATERIALS AND METHODS
RNA preparation
The env1 2′-dG aptamer domain RNA and derivatives were in vitro transcribed by T7 RNA polymerase from a PCR-generated DNA template using previously described methods (13). Sequences of all RNAs used in this study are given in Supplementary Table S1. After incubating for 3 h at 37°C, the transcription reaction was buffer exchanged into Milli-Q water and purified using denaturing polyacrylamide gel electrophoresis. Product was extracted using the crush and soak technique at 4°C into 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0) followed by buffer exchange with 0.5× TE buffer (5 mM Tris, 0.5 mM EDTA, pH 8.0) and concentrated to a volume of 500 μl using a 10 kDa Amicon centrifugal microconcentrator. RNA concentrations were determined using a calculated molar extinction coefficient at 260 nm wavelength; RNA was stored at –20°C until use. For crystallization and ITC, RNA was refolded by heat/cooling at 95°C for 3 min and incubating on ice for 5 min in TE buffer. Native gel electrophoresis was used to assess whether the refolded RNA adopted a single, monomeric conformation (14).
RNA crystallization
The env1 2′-dG aptamer domain RNA was crystallized at 30°C using the hanging drop diffusion method. 4 μl drops composed of a 1:1 ratio of RNA-ligand mixture to precipitant mixture were suspended above 500 μl of 35% 2-methyl-2,4-pentanediol (MPD). The RNA-ligand mixture contained 300 μM RNA and 600 μM 2′-deoxyguanosine while the precipitant mixture contained 40 mM sodium cacodylate pH 6.0, 23% v/v MPD, 18 mM cobalt hexammine, 80 mM potassium chloride, and 12 mM sodium chloride. Hanging drops were incubated for 2–3 days to allow crystals to reach maximum size. To cryoprotect, crystals were soaked in a precipitant mixture containing 35% MPD for 5 min, and flash frozen in liquid nitrogen. Data were collected on a home-source Rigaku MicroMax-007 HF X-ray source with a Dectris Pilatus 3R 200K-A detector and processed with HKL3000. Screening for crystals that diffract X-rays to high resolution and data collection methods were using standard practice for X-ray crystallography (15,16). Crystallographic data collection statistics are given in Supplementary Table S2.
Structure determination, refinement and analysis
An electron density map was calculated by molecular replacement using Phaser (17) in PHENIX (18) using the three helices of the B. subtilis xpt guanine riboswitch (GR) RNA (P1 nucleotides (nt) 15–21 and 75–81, P2 nt 25–31 and 39–45, P3 nt 54–59 and 67–72) as a starting model (PDB ID: 4FE5), excluding all non-helical linker regions (J1/2, L2, J2/3, L3, J3/1). Model refinement was initiated by replacing the sequence of the GR helices with that of env1 using Coot (19), followed by refinement in PHENIX. Further rounds of iterative model building and refinement were performed in which the rest of the RNA model was built, the ligand placed within the binding pocket and finally solvent molecules were placed. Standard approaches to reduce model bias in molecular replacement were employed throughout electron density map calculation and model building (20,21). The final model statistics are provided in Supplementary Table S2.
Isothermal titration calorimetry (ITC)
RNA-ligand binding affinities were quantified by ITC using methods described previously (22). RNA was dialyzed overnight in ITC buffer containing 50 mM K-HEPES pH 7.5, 20 mM MgCl2, and 100 mM KCl at 4°C, similar to previously published conditions (10–12). Ligand was dissolved in ITC buffer at 65°C and centrifuged to remove undissolved particles. Experiments were performed in triplicate using a Microcal ITC-200 calorimeter. Titrations were performed at 30°C with a reference power of 9 μcal/s. RNA was loaded into the sample cell at concentrations of 5–50 μM, after which ligand was titrated into the cell using the syringe in 19, 2.0 μl injections at concentrations between 50–500 μM. c values were between 0.5 and 1120, with all but the weakest binders >1 and all but the tightest binder below 500. Data were fitted to a single-site binding model with Origin 7.0 ITC software (Microcal Software) to determine the apparent dissociation constant, KD,app; representative data and fits are shown in Supplementary Figure S1. Data and statistical analysis was performed in accordance to best practice as described (23,24).
RESULTS
Design and crystallization of 2′-dG-II aptamer domains
To validate their identities as 2′-dG riboswitches, we first verified the 2′-deoxyguanosine binding ability of several 2′-dG-II class RNAs using isothermal titration calorimetry (ITC). The env23 sequence (RNA nomenclature adopted from Weinberg et al. (1)) used to determine the ligand binding specificity of this class was observed to bind 2′-dG with a KD,app of 290 ± 50 nM (Table 1), consistent with in-line probing values (1). In addition, we investigated a sequence (env1) from the sequence alignment of this class of riboswitches that most closely resembled the B. subtilis xpt guanine riboswitch, particularly with respect to the terminal loop-loop (L2–L3) interaction (25). Prior structural studies using the xpt guanine aptamer to host alternative ligand binding pockets in the three-way junction revealed that these RNAs, like the parental sequence, were readily crystallizable (11,26–28). 2′-dG binding by env1 was determined by ITC to be 34 ± 1 nM (Table 1), less than an order of magnitude weaker than the ∼4 nM binding affinity of xpt guanine riboswitch for guanine (29).
Table 1.
ITC analysis of 2′-dG and rG binding to purine family riboswitches
| Riboswitch | Class | K D,2′-dG (nM) | K D,rG (nM) | K rel (rG/2′-dG) |
|---|---|---|---|---|
| env1 | 2′-dG-II | 34 ± 1 | 89 ± 10 | 2.6 |
| B. subtilis xpt | G/A | 7700 ± 100 | 36 000 ± 1000 | 2.6 |
| env30 | 2′-dG-II | 2400 ± 900 | 36 000 ± 1000 | 15 |
| M. florum I-A | 2′-dG-I | 190 ± 10 | 5000 ± 600a | 27 |
| xpt GdG 5B a | synthetic | 350 ± 100 | 10 000 ± 100 | 29 |
| env6 | 2′-dG-II | 88 ± 3 | 3000 ± 200 | 35 |
| env23 | 2′-dG-II | 290 ± 50 | 51 000 ± 4000 | 180 |
aData from reference (11).
The env23 and env1 RNAs in complex with 2′-dG were screened for diffraction quality crystals using a strategy similar to that of the xpt aptamer-hypoxanthine complex (30). For each, a series of RNAs was created whose P1 helix lengths varied between 5 and 8 bp, with all variants having a single 3′-adenosine overhang. Initial screening yielded crystals of the env1 RNA, but were found to be irreproducible. Analysis of the length of RNA in the crystals and that remaining in solution using denaturing acrylamide gel electrophoresis revealed that RNA in crystals were slightly longer than the non-crystallizing RNA remaining in the mother liquor, suggesting a nontemplated 3′-nucleotide was added by T7 RNA polymerase (31,32). We subsequently screened an additional set of single nucleotide additions to the 3′-end of env1 using transcription templates that have two 2′-O-methoxy groups at the 3′-end to prevent nontemplated nucleotide addition (33,34) and found that a guanosine added to the 3′-end resulted in reproducible crystals, while addition of an adenosine or uridine did not. Subsequent analysis of the lattice contacts in the env1 crystal structure revealed that the Watson–Crick face of the 3′ guanosine forms hydrogen bonding interactions with the β-phosphate of the 5′-residue of a neighboring molecule. It should be noted that a similar strategy was used to obtain reproducible crystals of the SAM-I/IV riboswitch aptamer domain (35).
Crystals of the aptamer domain of the env1 2′-dG-II class riboswitch in complex with 2′-deoxyguanosine were obtained in the P4122 space group and diffracted using a home source rotating copper anode x-rays to 2.8 Å resolution (crystallographic data and model refinement statistics provided in Supplementary Table S2). The structure was determined by molecular replacement (17) using the three helices of the xpt guanine riboswitch RNA as an initial search model (PDB ID: 4FE5, (36)), as described in the methods. The initial electron density map (Supplementary Figure S2) was sufficiently clear to build the remaining RNA and the model was further built and refined using PHENIX (18). The final model had Rwork = 24.4% and Rfree = 27.6% values with overall good geometry as determined using MolProbity (37,38).
Global structure of the env1 2′-dG-II aptamer domain
The secondary structure and global architecture of the env1 2′-dG-II riboswitch aptamer domain (Figure 2A, B) adopts the same three-way junction supported by a distal tertiary interaction observed in all structures of purine family riboswitches (12,39–41). Superimposition of the B. subtilis xpt guanine and env1 2′-dG-II aptamers reveals that the three helices are organized around the central three-way junction in an identical fashion (Figure 2C). The main architectural difference between the two RNAs is the presence of an additional base pair in the env1 P3 helix (seven versus six base pairs in xpt)—similar to the M. florum 2′-dG-I aptamer. To preserve critical tertiary loop contacts between L2 and L3, this additional base pair shifts the P3 helix further into the binding core relative to xpt in both the 2′-dG-I (12) and 2′-dG-II RNAs. However, it should be noted that a seven base pair P3 helix is common within the guanine class of purine family riboswitches as well (36), so this feature is not unique to 2′-deoxyguanosine binding.
The L2–L3 interaction of 2′-dG-II is nearly identical to the tertiary interactions observed in the B. subtilis xpt guanine (39,41), B. subtilis pbuE adenine (40), and V. vulnificus add adenine (41) riboswitches (Figure 3). However, env1 exhibits three differences from xpt in the tertiary loop region: (i) an adenosine versus guanosine at the first position of L2, (ii) an A•A mismatch as the closing pair at the top of the P3 helix in env1, which is an A–U base pair in xpt, and (iii) a different Watson–Crick pair closing P2. To determine if these differences impact ligand recognition, we substituted the L2-L3 loop-loop region of env1 and tested affinity for 2′-dG. Substitution of the entire env1 L2-L3 loop and proximal two base pairs of P2 and P3 with the corresponding xpt sequence had no effect on 2′-dG binding (KD = 30 ± 2 nM, Table 2), indicating that the env1 tertiary loop region is interchangeable with that of xpt and likely plays no role in determining ligand selectivity in the guanine and 2′-dG-II classes of purine riboswitches. Conversely, because of significant differences between L2 of M. florum and env1 beyond the central two Watson–Crick G–C base pairs (Figure 3A, B), the two RNAs superimpose poorly in this region (Figure 3D).
Figure 3.

L2-L3 interactions in the purine riboswitch family. (A) Map of long-range interactions between L2 and L3 in the env1 2′-dG-II riboswitch aptamer domain. Notation is that of Leontis and Westhof (57). (B) Stereoscopic image of long-range interactions between L2 and L3 in the M. florum I-A 2′-dG-I riboswitch aptamer domain. Dashed lines represent single hydrogen bonding interactions. (C) Wall-eyed stereo view of the alignment of the L2-L3 interaction of the xpt aptamer domain (orange) and env1 aptamer domain (blue). R.m.s. deviation between the substructures is 0.50 Å. (D) Wall-eyed stereo view of the alignment of the L2–L3 interaction of the M. florum I-A riboswitch aptamer domain (green) and env1 aptamer domain (blue).
Table 2.
ITC analysis of 2′-dG binding to 2′-dG-II riboswitch mutants
| RNA | K D,dG (nM) | K rel (KD,mut/KD,wt) |
|---|---|---|
| env1 (wild type) | 34 ± 1 | 1 |
| env1 with xpt L2-L3 | 30 ± 2 | 0.88 |
| env1 U75A | 1600 ± 300 | 46 |
| env1 C76G | 100 ± 40 | 2.9 |
| env1 Δ(U75,C76) | 240 ± 50 | 7.1 |
| env1 A77U | 250 ± 10 | 7.4 |
| env1 C78A | 41 000 ± 9000 | 1200 |
| env1 C51U | 620 ± 180 | 18 |
| env1 G21A,C79U | 570 ± 30 | 17 |
| env1 C47,48U | 110 ± 10 | 3.2 |
| env1 C48A | 7.1 ± 1.4 | 0.21 |
Local architecture of the three-way junction
Within the three-way junction of the 2′-dG-II aptamer domain is a set of nucleobase-mediated interactions that are universally conserved within the purine riboswitch family (grey box, Figure 4). These interactions are primarily mediated by highly conserved nucleotides and/or nucleobase interactions in the joining strands (J1/2, J2/3 and J3/1) of the three-way junction along with the two junction-proximal base pairs of the P1 helix. Most distal to the P1 helix is a universally conserved base triple formed by a Watson–Crick pair between the first and last nucleotides of J2/3 (G46–C53) and a Watson–Crick/sugar edge interaction between A23 of J1/2 and G46. Adjacent to this triple is a Watson–Crick pair between A52 and U22. While in the guanine/adenine and 2′-dG-II classes this is a U–A pair, it is a C–G pair in 2′-dG-I (12); mutation of the U–A pair in the B. subtilis xpt guanine riboswitch from to a C–G pair imparts only a modest loss in binding affinity (42) in support of the general requirement for a Y22–R52 base pair. The third layer in the junction is a base triple involving C51, C78 and 2′-dG. This triple is universal within the purine family, although the mode of base-base interactions differs amongst the classes (vide infra). C50 of J2/3 engages the junction-proximal Watson–Crick pair (G21–C79) of the P1 helix through a two hydrogen bonding interaction that is conserved throughout the purine family, despite the differing G–C nucleotide identity of the junction-proximal base pair of P1 in 2′-dG-II compared to the A–U pair found in the other classes. The one variable structural aspect of the core is how the second junction proximal base pair in P1 (U20–A80) interacts with J2/3. Disruption of this interaction in the B. subtilis xpt guanine riboswitch has a very modest negative affect on ligand binding affinity (42).
Figure 4.
Architecture of the three-way junction. (A) Schematic of the base interactions between the three strands of the junction (coloring consistent with Figure 2) and the P1 helix. Grey shading represents base-base interactions universal to the purine riboswitch family. Red interactions denote those unique to the 2′-dG-II aptamer domain. Notation is that of Leontis and Westhof (57). (B) Wall-eyed stereo view of the architecture of the three-way junction. (C) Wall-eyed stereo view of the three-way junction rotated 90° clockwise relative to the perspective in panel B.
A two-nucleotide insertion in J3/1 is unique to 2′-dG-II purine riboswitches
The most significant difference between the 2′-dG-II and other purine riboswitches within the three-way junction is a two nucleotide insertion in J3/1 that is absent in both the guanine/adenine and 2′-dG-I classes of purine riboswitches (Figure 5A, B). This insertion is present in all members of the 2′-dG-II class, with the first position being an invariant uridine nucleotide while the second nucleotide is variable. In the crystal structure of env1(2′-dG-II), this insertion element promotes new base-base interactions within the three-way junction. U75 forms a base pair with the Hoogsteen face of A24 (Figure 5C). In the xpt guanine riboswitch this nucleotide is unpaired and stacks with the bottom of the P3 helix while the equivalent nucleotide in Mfl-IA(2′-dG-I) forms a base triple with the junction-proximal base pair in P2. The second nucleotide of the insertion, C76, is flipped away from the junction such that it does not make contacts with other bases in the RNA. Supporting this observation, mutation of C76 to a guanosine has minimal effect on 2′-dG binding affinity, reducing it only 2.9-fold (Table 2).
Figure 5.
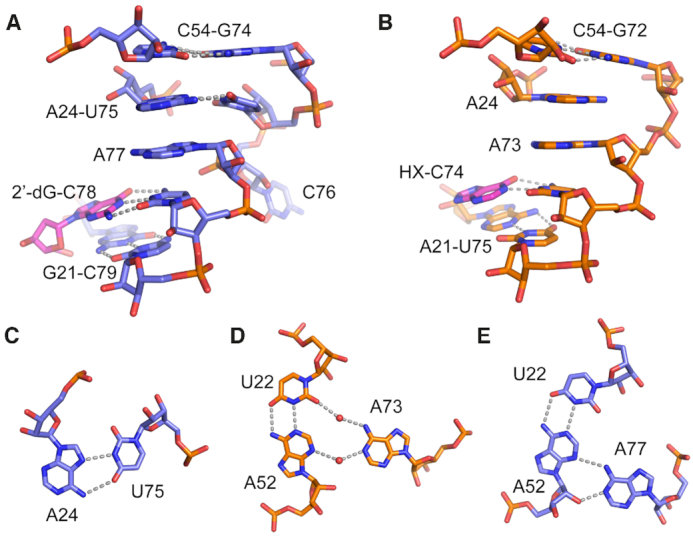
Interactions mediated by the U75-C76 J3/1 insertion element. (A) Local structure of the J3/1 element (nucleotides U75–C78) in the env1 2′-dG-II riboswitch aptamer domain and its interaction with ligand. Base-base interactions are denoted by grey dashed lines. (B) Local structure of J3/1 (nucleotides A73–C74) and its interactions with ligand (hypoxanthine, HX) in the xpt guanine riboswitch aptamer domain. (C) Base pairing interaction between U75 of the J3/1 insertion element and A24. (D) Indirect water-mediated interaction between A73 and the U22–A52 base pair in the xpt riboswitch (4FE5). Note that these highly ordered waters are consistently observed across multiple high-resolution crystal structures of the xpt riboswitch. (E) Direct interaction between A77 and the U22–A52 base pair in the junction.
To further understand the role of the J3/1 insertion element in ligand recognition, we examined it in the context of both env1(2′-dG-II) and xpt. Strikingly, deletion of these two nucleotides in env1 only results in a moderate loss of affinity for 2′-dG (7.1-fold, Table 2), while a U75A mutant reduces affinity 46-fold. A simple explanation for this observation is that a more sterically bulky purine at position 75 that still engages with A24 would push the backbone of J3/1 outwards, potentially disrupting a number of neighboring interactions as opposed to the deletion mutant that only ablates the A24–U75 pair. The deletion mutant is more modest than the C51U mutation that results in an 18-fold loss of 2′-dG binding affinity. These data indicate that the two nucleotide insertion element in J3/1 is not essential for promoting 2′-dG binding in this class of riboswitches, although the A24-U75 interaction increases affinity. Insertion of these two nucleotides into the xpt guanine riboswitch aptamer at the equivalent position does not promote 2′-dG binding, as affinity does not change compared to wild type xpt (8.1 ± 0.6 μM and 7.7 ± 0.4 μM, respectively). However, its affinity for hypoxanthine (HX)—one of its cognate ligands—is weakened from 0.73 ± 0.1 μM (39) to 21 ± 2 μM, a 29-fold reduction in affinity. Thus, the J3/1 insertion in the context of the xpt guanine riboswitch changed selectivity from favoring HX by ∼10-fold to favoring 2′-dG by 2.6-fold. Further alteration of xpt by including the U51C mutation that was previously shown to promote 2′-dG binding (xpt(U51C), KD,app = 12 ± 1 μM, (11)) increased the affinity 4.2-fold to 1.6 ± 0.1 μM while nearly abolishing affinity for hypoxanthine (>100 μM). Thus, the identity of position 51 in concert with the two nucleotide insertion in J1/3 promotes both tighter and more selective 2′-dG binding in the context of the xpt aptamer. These data suggest that these nucleotides are the strongest drivers of nucleoside versus nucleobase binding selectivity in purine riboswitches, but other changes to the sequence in the junction are required to achieve the highest affinity for 2′-dG.
The second direct base-base interaction promoted by the insertion element is via adenosine 77. In other purine riboswitch family members, this nucleotide is coplanar with a Y22-R52 Watson–Crick pair above the ligand-binding triple in the three-way junction. In xpt, the U22–A52 base pair forms a water-mediated interaction with A73 (Figure 5D), while in Mfl-IA the equivalent nucleotide is too distant to form a direct or indirect interaction (12,39). However, in the env1(2′-dG-II) structure, A77 forms a direct two hydrogen bond interaction with the U22–A52 base pair, establishing a direct base triple between the three joining strands of the junction (Figure 5E). The role of this interaction in ligand binding affinity is revealed by an A77U mutation, which disrupts the base triple and weakens 2′-dG binding affinity by ∼7-fold. The equivalent A73C mutation in xpt riboswitch aptamer results in only a two-fold decrease in ligand binding affinity (42). Thus, the unique insertion element of the 2′-dG-II class promotes two new base-mediated interactions that further organizes the three-way junction at the base of the P3 helix, thereby increasing ligand binding affinity.
2′-Deoxyguanosine recognition by the env1 riboswitch aptamer domain
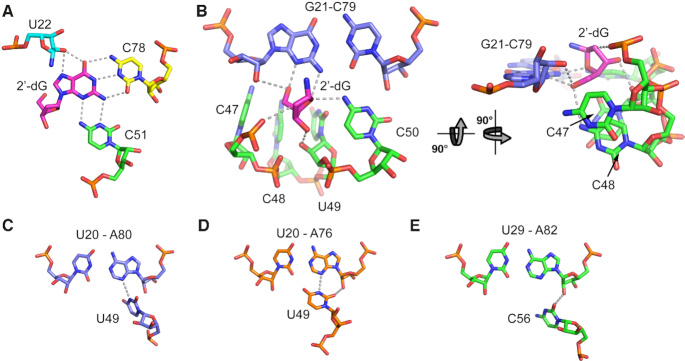
As with other purine family riboswitches, env1(2′-dG-II) recognizes the ligand nucleobase through a base triple involving a pyrimidine residue at position 51 in J2–3 and a pyrimidine at position 78 in J3–1 (the equivalent positions in xpt are 51 and 74, respectively). C78 forms a Watson–Crick base pair with the 2′-deoxyguanosine nucleobase. Nucleotide 78, which is universally conserved, is essential for high affinity ligand binding; its mutation to adenosine results in 1200-fold weaker binding affinity (Table 2). C51 pairs with the ligand's sugar edge, in the same fashion as the equivalent base, C58, in Mfl-IA (11,12) (Figure 6A). Mutation of C51 to uridine results in significant loss of ligand binding affinity (∼18-fold, Table 2), similar to the same mutation in Mfl-IA(2′-dG-I) (83-fold, (12)). Finally, while U47 in xpt directly interacts with U51, the equivalent nucleotide in both classes of 2′-dG riboswitches is positioned within a three base stack and does not play a direct role in ligand binding. Thus, base recognition in 2′-dG-II uses the same set of same contacts as 2′-dG-I and accommodates the 2′-deoxyribose sugar moiety through a shift of C51 towards C74 as compared to the positioning of U51 in guanine and adenine riboswitch aptamers (11,12).
Figure 6.
Ligand recognition by the env1 2′-dG-II riboswitch aptamer domain. (A) Interactions between nucleotides in the three-way junction and the guanine moiety of the ligand. Hydrogen bonding interactions are shown as grey dashes. Note that two hydrogen bonding interactions are drawn between O2′ of U22 and the N7 and O6 of 2′-dG since these two interactions are nearly equidistant (2.9 and 2.7 Å, respectively). (B) Interactions between the 2′-deoxyribose moiety and RNA. The nucleobase moiety of the 2′-dG ligand (magenta) has been omitted for clarity. (C–E) Base triple between J2/3 and the second proximal base pair in P1 in the (C) env1 2′-dG-II riboswitch, (D) xpt guanine riboswitch and (E) M. florum 2′dG-I riboswitch.
An unusual feature of the 2′-dG-II class relative to other classes in the purine family is the presence of a G21–C79 Watson–Crick base pair in P1 proximal to the three-way junction rather than an A–U pair. While an A–U pair is nearly universally conserved at this position, a G–C pair is observed in a few guanine and adenine riboswitches (43), and an A–U to G–C mutation is well tolerated by the xpt riboswitch (42). In env1, this base pair is directly involved in ligand recognition through interactions between N2 of G21 and N4 of C50 hydrogen bonding with the deoxyribose furan oxygen (O4′) (Figure 6B). This is an additional hydrogen bonding interaction over that observed in Mfl-IA(2′-dG-I), in which only the exocyclic amine of C57 (equivalent of C50 in env1) interacts with O4′ of 2′-deoxyguanosine. This additional interaction substantially contributes to env1′s higher affinity for 2′-dG than Mfl-IA since a G21A–C79U mutation in env1 binds 2′-dG with 16.9 times lower affinity than wild type (Table 2).
The env1(2′-dG-II) aptamer domain accommodates the 2′-deoxyribose sugar through a reorganization of five nucleotides in J2/3 (nt 47–51) relative to the guanine/adenine class in a fashion similar to the 2′-dG-I class. The first key difference is the shift of C51 towards C78, which maintains a two hydrogen bond interaction between C51 and 2′-dG (Figure 6A). This shift was first documented in a designed 2′-dG hybrid variant of xpt and Mfl-IA and was proposed to be essential for 2′-dG recognition (11). The second difference between env1(2′-dG-II) and guanine/adenine aptamers is the stacking of C47, C48 and U49 to create a pocket for the deoxyribose sugar, which is again similar to the configuration both in a designed 2′-dG hybrid variant of the xpt aptamer as well as Mfl-IA. The last nucleotide of the stack (U49) interacts differently with the universally conserved U–A pair in P1 between the three classes (Figure 6C–E). U49 forms a single hydrogen bond with U20–A80 pair while the equivalent uridine residue in guanine/adenine riboswitches forms a two hydrogen bond interaction with the U–A pair. In contrast, the equivalent residue in Mfl-IA(2′-dG-I) is a cytosine that does not interact with the U–A pair, but rather interacts with two hydroxyl groups in the backbone. Overall, the env1(2′-dG-II) and Mfl-IA(2′-dG-I) aptamers use a similar strategy to create a binding pocket for the deoxyribose sugar.
The 2′-deoxyguanosine sugar is recognized by extensive interactions with the pocket formed by J2/3 and P1. As described above, the furan oxygen (O4′) forms two hydrogen bonding interactions with N2 of G21 and N4 of C50 (Figure 6B). The 3′-OH of the ligand is recognized by the 2′-OH of U49, similar to what is observed in Mfl-IA(2′-dG-I). Finally, the 5′-OH of the ligand hydrogen bonds with N3 of G21 and likely N4 of C48. This latter hydrogen bonding interaction is slightly longer at 3.6 Å, but can still be productive. Within the 2′-dG-II class, residue 48 is often adenosine which could also position a hydrogen bond donating amine adjacent to the 5′-OH of 2′-dG. This substitution in env1(C48A) results in a 4.8-fold increase in binding affinity, strongly suggesting that this interaction contributes to binding affinity in other members of the 2′-dG-II class. Remarkably, the binding affinity for env1(C48A) is nearly identical that of the B. subtilis xpt riboswitch aptamer for guanine (7 and 5 nM, respectively) and displays a >25-fold higher affinity for 2′-dG than Mfl-IA(2′-dG-I) (Table 1). These data indicate that a subset of class II 2′-dG riboswitches have an additional set of interactions that result in significantly higher affinity for their cognate ligand as compared to their 2′-dG-I counterparts.
env1 displays modest discrimination between 2′-dG and rG
In the cellular context, 2′-dG riboswitches are challenged by chemically similar compounds to their cognate ligand, the most similar being riboguanosine (rG). In rapidly growing E. coli, the intracellular concentration of rG is 1.6 μM as compared to a 2′-dG concentration of 0.57 μM (44). Measurement of the affinity of env1 for rG revealed that the aptamer binds this metabolite with only a 2.6-fold lower affinity than 2′-dG (Table 1). This contrasts with Mfl-IA, which binds 2′-dG with ∼30-fold (Table 1) to 50-fold (12) higher affinity than rG. To determine whether other class-II riboswitches exhibit reduced selectivity for 2′-dG over rG, we examined three other 2′-dG-II riboswitches with varying sequences within the three-way junction (Table 1). Overall, class II 2′-dG riboswitches vary broadly in their ability to discriminate between 2′-dG and rG, with env1 being the least selective while the env23 sequence has a far greater selectivity for 2′-dG than Mfl-IA. This range in selectivity is accompanied by a broad range in affinities, varying almost 2 orders of magnitude in 2′-dG binding affinity and approaching 3 orders of magnitude in rG binding affinity. This broad spectrum of binding affinities for the cognate ligand is similar to that observed in other riboswitches, such as the SAM-I riboswitch (45,46).
env1 shows decreased discrimination against antiretroviral 2′-dG derivatives compared to M. florum
In addition to high affinity rG binding, env1 also recognizes more structurally divergent 2′-dG derivatives. Compared to M. florum 2′-dG-I, env1 is more tolerant toward chemically related compounds as demonstrated by its higher binding affinities and weaker discrimination against the antiviral drugs acyclovir and pencyclovir. Env1 binds acyclovir and pencyclovir more tightly (KD = 560 ± 10 nM and 2.0 ± 0.1 μM, respectively) than M. florum (KD = 4.8 ± 1.3 and 22 ± 1.1 μM, respectively), and displays a lowered ability to discriminate between the antiviral and 2′-dG (Krel,env1(lig/2′-dG) = 16 for acyclovir and 59 for pencyclovir; Krel,M. florum(lig/2′-dG) = 28 for acyclovir and 132 for pencyclovir). Thus, while the env1 riboswitch achieves higher affinity binding for 2′-dG than other riboswitches, it appears that its ability to discriminate between closely related metabolites and RNA-binding drugs is significantly diminished—a noteworthy example that higher affinity binding does not necessarily correlate with higher specificity.
DISCUSSION
The structure and associated mutational analysis of a member of the 2′-dG-II class of purine riboswitches in complex with 2′-deoxyguanosine has yielded new insights into how modification of similar RNA sequences can potentially alter selectivity to bind alternative small molecules. The 2′-dG-II riboswitch aptamer domain shares significant sequence and structural homology with other members of the purine riboswitch family in regions critical for establishing global 3D architecture and ligand recognition. Compared to the guanine/adenine class, both classes of 2′-dG binding riboswitches use similar changes in sequence and architecture in J2/3 to accommodate the 2′-deoxyribose sugar: a shift of the pyrimidine residue that contacts the sugar edge of the ligand nucleobase in all purine riboswitches and flipping out of three nucleotides from J2/3. This five nucleotide element (xpt and env1 nucleotides 47–51) is critical for conferring 2′-dG selectivity. A single point mutation, U51C, in the xpt guanine riboswitch is sufficient to switch the ligand preference from guanine to 2′-dG, and additional changes within this element further enhance 2′-dG selectivity and affinity.
The two 2′-dG binding classes diverge in how they fully establish both high affinity and selectivity for 2′-dG over guanine beyond differences in J2/3. Structural and mutational analysis of the Mfl-IA(2′-dG-I) RNA revealed that both local and distal differences in the RNA are required to achieve high affinity, selective binding for 2′-dG over guanine (11,12). These differences include an altered conformation of the 2′-dG-I A33 (A24 equivalent) nucleotide in the three way junction to form a triple at the base of P2 that is not observed in the guanine/adenine and 2′-dG-II classes, as well as an L2-L3 interaction that preserves the G–C Watson–Crick pairs between the loops but differs substantially otherwise. In contrast, the 2′-dG-II class of RNAs appears to exploit only local differences around the ligand binding pocket to achieve high affinity and selective binding of 2′-dG, which is driven in part by a unique two-nucleotide insertion in J3/1. The ability of classes of riboswitches to exploit local changes around the binding pocket to yield differences in ligand binding selectivity has also been observed within the ykkC family of riboswitches. This family contains classes that recognize guanidine, PRPP or ppGpp that are distinguished by sequence and structural differences around and within the junctional binding pocket that include a point substitution and an insertion element (the P1 helix) (47–50).
Another distinctive feature of the 2′-dG-II class of riboswitches is a broad range of selectivites for 2′-dG over the chemically related compound riboguanosine. While the env1 variant binds 2′-dG with high affinity as compared to other members of the 2′-dG-I and -II classes, it has very low ability to discriminate between these two compounds. Since the concentrations of these two compounds are similar in rapidly dividing E. coli, this raises the distinct possibility that a subset of members of the 2′-dG-II class of riboswitches are responsive to either compound to regulate gene expression. This property was also observed in the 2′-dG-I riboswitches, which was divided into two ‘types’ based upon experimental analysis of ligand selectivity (10). The first, exemplified by the Mfl-IA riboswitch, selectively binds 2′-dG over 3′-dG and guanosine, but the type II sequences bind these compounds with similar affinities. Thus, the ability to respond to a larger pool of guanosine metabolites is found within both classes of 2′-dG riboswitches and likely reflects a regulatory need by a subset of genes whose expression is controlled by these riboswitches.
The ability of a riboswitch to respond to multiple chemically similar compounds may be fairly common. Examples of this behavior include the glmS riboswitch that can interact with both activating and inhibitory metabolites (51), the class-II cobalamin riboswitches, some of which bind the biologically active forms of B12 with near equal affinities (7) and THF riboswitches that bind to a spectrum of reduced folates (52,53). This behavior may enable these riboswitches to respond to metabolic and physiological states of the cell that cannot be readily sensed by interacting with a single compound within a pool of related metabolites.
Insights into the structural and ligand binding properties of env1 and other class-II 2′-dG riboswitches potentially provide new avenues for harnessing these RNAs for synthetic biological applications. Synthetic riboswitches derived from natural purine riboswitches have been implemented in diverse bacteria including industrially important strains of cyanobacteria (54) and thermophiles (55) to regulate gene expression. Furthermore, it has been shown that the guanine riboswitch can be reprogrammed to recognize pyrimidine (56) and pterin (27) compounds with only a limited set of mutations, further diversifying the utility of this riboswitch platform. More recently, it was shown that the purine riboswitch aptamer can be used as an architectural ‘scaffold’ to evolve radically new binding pockets within the three-way junction for compounds dissimilar to purines such as serotonin and dopamine (26). The current study suggests how small insertion elements in the purine riboswitch may be leveraged to engineer RNAs responsive to purine-like compounds and proposes potential enhancement of binding affinities by expanding hydrogen-bonding networks in the binding core. As the demand for a diverse set of robust small molecule responsive regulatory systems increases, the purine riboswitch continues to present new opportunities to meet these needs in synthetic biology.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure has been deposited in the RCSB PDB (www.rcsb.org) under the accession number 6P2H. Other data is available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the technical support and assistance of Dr Annette Erbse in diffraction and calorimetry experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 GM073850 to R.T.B. and T32 GM065103 to M.M.M.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Weinberg Z., Nelson J.W., Lunse C.E., Sherlock M.E., Breaker R.R.. Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2077–E2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I. et al.. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nelson J.W., Atilho R.M., Sherlock M.E., Stockbridge R.B., Breaker R.R.. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol. Cell. 2017; 65:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sherlock M.E., Sudarsan N., Stav S., Breaker R.R.. Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria. Elife. 2018; 7:e33908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sherlock M.E., Sudarsan N., Breaker R.R.. Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:6057–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sherlock M.E., Sadeeshkumar H., Breaker R.R.. Variant bacterial riboswitches associated with nucleotide hydrolase genes sense nucleoside diphosphates. Biochemistry. 2018; 58:401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Polaski J.T., Webster S.M., Johnson J.E. Jr, Batey R.T.. Cobalamin riboswitches exhibit a broad range of ability to discriminate between methylcobalamin and adenosylcobalamin. J. Biol. Chem. 2017; 292:11650–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kellenberger C.A., Wilson S.C., Sales-Lee J., Hammond M.C.. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013; 135:4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atilho R.M., Perkins K.R., Breaker R.R.. Rare variants of the FMN riboswitch class in Clostridium difficile and other bacteria exhibit altered ligand specificity. RNA. 2019; 25:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J.N., Roth A., Breaker R.R.. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:16092–16097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edwards A.L., Batey R.T.. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J. Mol. Biol. 2009; 385:938–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pikovskaya O., Polonskaia A., Patel D.J., Serganov A.. Structural principles of nucleoside selectivity in a 2′-deoxyguanosine riboswitch. Nat. Chem. Biol. 2011; 7:748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edwards A.L., Garst A.D., Batey R.T.. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol. Biol. 2009; 535:135–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eschbach S.H., Lafontaine D.A.. RNA conformational changes analyzed by comparative gel electrophoresis. Methods Mol. Biol. 2014; 1086:255–264. [DOI] [PubMed] [Google Scholar]
- 15. Sawyer L., Turner M.. Crystallization of Nucleic Acids And Proteins. A Practical Approach. 1992; Oxford: Oxford University Press; 267–274. [Google Scholar]
- 16. Diederichs K. Crystallographic data and model quality. Nucleic Acid Crystallogr.: Methods Protoc. 2016; 1320:147–173. [DOI] [PubMed] [Google Scholar]
- 17. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 20. Adams P.D., Pannu N.S., Read R.J., Brunger A.T.. Extending the limits of molecular replacement through combined simulated annealing and maximum-likelihood refinement. Acta Crystallogr. D Biol. Crystallogr. 1999; 55:181–190. [DOI] [PubMed] [Google Scholar]
- 21. Terwilliger T.C. Advancing Methods for Biomolecular Crystallography. 2013; Springer; 193–203. [Google Scholar]
- 22. Gilbert S.D., Reyes F.E., Edwards A.L., Batey R.T.. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009; 17:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones C.P., Piszczek G., Ferre-D’Amare A.R.. Isothermal Titration Calorimetry Measurements of Riboswitch-Ligand Interactions. Methods Mol. Biol. 2019; 1964:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liberman J.A., Bogue J.T., Jenkins J.L., Salim M., Wedekind J.E.. ITC Analysis of ligand binding to PreQ(1) riboswitches. Method Enzymol. 2014; 549:435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mandal M., Boese B., Barrick J.E., Winkler W.C., Breaker R.R.. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003; 113:577–586. [DOI] [PubMed] [Google Scholar]
- 26. Porter E.B., Polaski J.T., Morck M.M., Batey R.T.. Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nat. Chem. Biol. 2017; 13:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson C.J., Vincent H.A., Wu M.C., Lowe P.T., Dunstan M.S., Leys D., Micklefield J.. Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species. J. Am. Chem. Soc. 2014; 136:10615–10624. [DOI] [PubMed] [Google Scholar]
- 28. Solem A.C., Halvorsen M., Ramos S.B., Laederach A.. The potential of the riboSNitch in personalized medicine. Wiley Interdiscip. Rev. RNA. 2015; 6:517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilbert S.D., Stoddard C.D., Wise S.J., Batey R.T.. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006; 359:754–768. [DOI] [PubMed] [Google Scholar]
- 30. Gilbert S.D., Montange R.K., Stoddard C.D., Batey R.T.. Structural studies of the purine and SAM binding riboswitches. Cold Spring Harb. Symp. Quant. Biol. 2006; 71:259–268. [DOI] [PubMed] [Google Scholar]
- 31. Krupp G. RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene. 1988; 72:75–89. [DOI] [PubMed] [Google Scholar]
- 32. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kao C., Rüdisser S., Zheng M.. A simple and efficient method to transcribe RNAs with reduced 3′ heterogeneity. Methods. 2001; 23:201–205. [DOI] [PubMed] [Google Scholar]
- 34. Kao C., Zheng M., Rüdisser S.. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999; 5:1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trausch J.J., Xu Z., Edwards A.L., Reyes F.E., Ross P.E., Knight R., Batey R.T.. Structural basis for diversity in the SAM clan of riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6624–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stoddard C.D., Widmann J., Trausch J.J., Marcano-Velazquez J.G., Knight R., Batey R.T.. Nucleotides adjacent to the ligand-binding pocket are linked to activity tuning in the purine riboswitch. J. Mol. Biol. 2013; 425:1596–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B. 3rd, Snoeyink J., Richardson J.S. et al.. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007; 35:W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davis I.W., Murray L.W., Richardson J.S., Richardson D.C.. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004; 32:W615–W619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Batey R.T., Gilbert S.D., Montange R.K.. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004; 432:411–415. [DOI] [PubMed] [Google Scholar]
- 40. Delfosse V., Bouchard P., Bonneau E., Dagenais P., Lemay J.F., Lafontaine D.A., Legault P.. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 2010; 38:2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Serganov A., Yuan Y.R., Pikovskaya O., Polonskaia A., Malinina L., Phan A.T., Hobartner C., Micura R., Breaker R.R., Patel D.J.. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004; 11:1729–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gilbert S.D., Love C.E., Edwards A.L., Batey R.T.. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007; 46:13297–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCown P.J., Corbino K.A., Stav S., Sherlock M.E., Breaker R.R.. Riboswitch diversity and distribution. RNA. 2017; 23:995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D.. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009; 5:593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tomsic J., McDaniel B.A., Grundy F.J., Henkin T.M.. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J. Bacteriol. 2008; 190:823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winkler W.C., Nahvi A., Sudarsan N., Barrick J.E., Breaker R.R.. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003; 10:701–707. [DOI] [PubMed] [Google Scholar]
- 47. Battaglia R.A., Price I.R., Ke A.. Structural basis for guanidine sensing by the ykkC family of riboswitches. RNA. 2017; 23:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Knappenberger A.J., Reiss C.W., Strobel S.A.. Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA. Elife. 2018; 7:e36381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peselis A., Serganov A.. ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands. Nat. Chem. Biol. 2018; 14:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reiss C.W., Xiong Y., Strobel S.A.. Structural basis for ligand binding to the guanidine-I riboswitch. Structure. 2017; 25:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watson P.Y., Fedor M.J.. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat. Struct. Mol. Biol. 2011; 18:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ames T.D., Rodionov D.A., Weinberg Z., Breaker R.R.. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem. Biol. 2010; 17:681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trausch J.J., Ceres P., Reyes F.E., Batey R.T.. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011; 19:1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Higo A., Isu A., Fukaya Y., Hisabori T.. Spatio-temporal gene induction systems in the heterocyst-forming multicellular cyanobacterium Anabaena sp. PCC 7120. Plant Cell Physiol. 2017; 59:82–89. [DOI] [PubMed] [Google Scholar]
- 55. Marcano-Velazquez J.G., Lo J., Nag A., Maness P.C., Chou K.J.. Developing riboswitch-Mediated gene regulatory controls in thermophilic bacteria. ACS Synth. Biol. 2019; 8:633–640. [DOI] [PubMed] [Google Scholar]
- 56. Dixon N., Duncan J.N., Geerlings T., Dunstan M.S., McCarthy J.E., Leys D., Micklefield J.. Reengineering orthogonally selective riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Leontis N.B., Westhof E.. Geometric nomenclature and classification of RNA base pairs. RNA. 2001; 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure has been deposited in the RCSB PDB (www.rcsb.org) under the accession number 6P2H. Other data is available upon request.