Abstract
Histone deacetylase 3 (Hdac3) is a target of the FDA approved HDAC inhibitors, which are used for the treatment of lymphoid malignancies. Here, we used Cd19-Cre to conditionally delete Hdac3 to define its role in germinal center B cells, which represent the cell of origin for many B cell malignancies. Cd19-Cre-Hdac3−/− mice showed impaired germinal center formation along with a defect in plasmablast production. Analysis of Hdac3−/− germinal centers revealed a reduction in dark zone centroblasts and accumulation of light zone centrocytes. RNA-seq revealed a significant correlation between genes up-regulated upon Hdac3 loss and those up-regulated in Foxo1-deleted germinal center B cells, even though Foxo1 typically activates transcription. Therefore, to determine whether gene expression changes observed in Hdac3−/− germinal centers were a result of direct effects of Hdac3 deacetylase activity, we used an HDAC3 selective inhibitor and examined nascent transcription in germinal center-derived cell lines. Transcriptional changes upon HDAC3 inhibition were enriched for light zone gene signatures as observed in germinal centers. Further comparison of PRO-seq data with ChIP-seq/exo data for BCL6, SMRT, FOXO1 and H3K27ac identified direct targets of HDAC3 function including CD86, CD83 and CXCR5 that are likely responsible for driving the light zone phenotype observed in vivo.
INTRODUCTION
The ability of our adaptive immune system to generate antibody diversity requires multiple steps to alter the germline-encoded antibody loci. B cell immunologic diversity is first created through VDJ recombination to assemble a wide-ranging antibody repertoire. Upon activation in the periphery, B cells expressing these relatively low affinity antibodies can form germinal centers (GCs) where they undergo affinity maturation to generate high affinity antibodies (1). Within the germinal centers, B cells undergo bursts of rapid proliferation and somatic hypermutation in a specialized compartment known as the dark zone (DZ) before transit to the light zone (LZ), where they receive selection signals through interactions with follicular dendritic cells (FDCs) and T follicular helper (TFH) cells (2–4). Importantly, germinal center B cells may cycle between the dark and light zones, potentially undergoing multiple rounds of mutation and selection, to generate high affinity antibodies (5,6). Successful affinity maturation ultimately culminates in germinal center B cell differentiation into either antibody-secreting plasma cells or memory B cells.
Dramatic changes in transcriptional programs are required for the establishment of germinal center B cell identity and for subsequent differentiation of these cells into plasma or memory B cells (7,8). Transcriptional networks composed of key DNA binding transcription factors and chromatin modifying enzymes coordinate these programs. The transcription factor, BCL6, plays a prominent role in the establishment of germinal center B cell identity where it recruits co-repressor complexes to genes associated with cell cycle checkpoints and DNA damage responses (e.g. Tp53, Atr, Chek1), which would normally be invoked by somatic hypermutation, as well as genes associated with plasma cell differentiation (e.g. Prdm1, Irf4) (9–13). In fact, deletion of Bcl6 in mice prevented germinal center formation upon antigen exposure, suggesting that BCL6 is a master regulator of the germinal center (14–16). A second transcription factor, FOXO1, cooperates with BCL6 at a number of gene targets (17,18). However, unlike BCL6, FOXO1 is not required for germinal center formation. Rather, it plays a critical role in the establishment of the dark zone and successful affinity maturation (17,18). Thus, the collaboration between BCL6 and FOXO1 regulates gene expression to promote the generation of high affinity antibodies and plasma cell differentiation.
BCL6 represses transcription in part, through associations between the BCL6-BTB domain with either BCOR or NCOR1/SMRT co-repressor complexes (19–21). Specifically, in germinal center-derived lymphomas, BCL6 recruits NCOR1/SMRT to maintain enhancers in ‘poised’ rather than active states (9). Furthermore, disruption of the interaction of BCOR and NCOR/SMRT complexes with the BTB domain of BCL6 effectively kills BCL6-dependent lymphomas (12,22,23), and mutation of the BCL6 BTB domain prevents germinal center formation (24). Thus, co-repressor interactions mediated by the BCL6 BTB domain are critical for BCL6 function both in normal and malignant B cells.
HDAC3 is the catalytic component of the NCOR1/SMRT co-repressor complexes. Mouse models demonstrated that Hdac3 is absolutely required for early lymphocyte development and biology, as deletion of Hdac3 in hematopoietic stem cells caused multi-lineage dysplasia with the loss of the earliest lymphoid-primed multipotent progenitor cells (25). Moreover, deletion of Hdac3 in early B-progenitor cells caused a defect in VDJ recombination and failure in B cell development, which precluded the analysis of Hdac3 functions in the later stages of B cell maturation (26). Similar defects were observed when Hdac3 was deleted during T cell development, which caused a failure in positive selection (27–29). Given that the SMRT/HDAC3 complex is recruited by Bcl6 during the germinal center reaction and that disruption of this association affected the growth and survival of BCL6-dependent diffuse large B cell lymphoma, we sought to determine the requirement of Hdac3 for germinal center formation and function.
Using CD19-cre to drive recombination at the Hdac3 locus, we found that deletion of Hdac3 caused only a minor defect in overall numbers of germinal centers formed, suggesting that Hdac3 was not necessary for the full complement of Bcl6 functions. However, germinal center B cell numbers were reduced upon Hdac3 deletion, and those germinal centers that formed were characterized by an accumulation of light zone centrocytes. This phenotype bore a striking similarity to that caused by Foxo1 deletion from GC B cells (17,18). Indeed, we found significant overlap between genes up-regulated in Foxo1-deleted germinal centers and those up-regulated upon Hdac3 loss, yet Foxo1 predominantly activates transcription, indicating that these changes in gene expression could be indirect (17,18). Therefore, we used short treatments with an Hdac3 selective inhibitor in germinal center-derived lymphoma cell lines with wild type or mutant FOXO1 to try to determine if Hdac3 directly regulates the expression of FOXO1-regulated germinal center-associated transcripts. Consistent with in vivo phenotypes, precision nuclear run-on transcription sequencing showed that HDAC3 repressed the expression of a light zone gene expression signature. ChIP-seq data indicated that these genes were more highly correlated with BCL6 rather than FOXO1 binding. In particular, HDAC3 regulated the expression of targets critical for germinal center polarization including CD86, CD83 and CXCR5, likely explaining the accumulation of light zone centrocytes observed in mouse germinal centers in the absence of Hdac3.
MATERIALS AND METHODS
Mice
Mice harboring a conditional Hdac3 allele (30) were crossed to CD19-Cre knock in mice (the Jackson Laboratory), with mice heterozygous for CD19-Cre being used for experiments. Mice were maintained on a C57BL/6J background. Experiments were performed at 6–10 weeks of age. Animals were housed under specific pathogen-free conditions in accordance with guidelines set forth by Vanderbilt University. All experiments were conducted according to an Institutional Animal Care and Use Committee-approved protocol.
Flow cytometry
A 70 μM cell strainer was used to make single cell suspensions of mouse splenocytes. Mouse bone marrow was flushed from the femur and tibia. Following erythrocyte lysis (Buffer EL, Qiagen), cells were stained with combinations of fluorophore-conjugated antibodies for 20 min at 4°C, washed and analyzed on a 5-laser BD LSRII. Flow cytometry to detect Prdm1 was carried out using the Transcription factor buffer set (BD; 56725) according to manufacturer's protocol. Antibodies- from eBioscience- IgM(ll/41), B220(RA3-6B2), CD23(B3B4), CD21(eBio4E3), IgD (11–25,28), Fas(15A7), CD43 (eBioR2/60); from BD Biosciences- CXCR4(2B11), CD86(GL1), CD138(281-2), Prdm1/Blimp-1(5E7). Data analysis performed using FlowJo software (Tree Star). For phospho-flow, 1 × 106 isolated B cells were rested for 30 min at 37°C prior to BCR crosslinking with 10 μg/ml of α-IgM (Jackson ImmunoResearch Laboratories). Stimulated B cells were fixed with paraformaldehyde prior to staining with antibodies to phospho-PLCγ2 (BD Biosciences). Phospho-flow data was analyzed using Cytobank (31).
RNA-Seq
10 days post-immunization with SRBCs, total RNA was isolated from sorted B220+GL7+ and B220+GL7− spleen cells by Trizol (Ambion). Library construction was performed by the Vanderbilt VANTAGE Shared Resource following enrichment of poly-adenylated RNAs from biological replicates. Samples were sequenced using an Illumina HiSeq 2500 instrument on an SR-50 run generating approximately 30M reads/sample.
RNA-seq analysis
Pre-processed reads were aligned to the mouse transcriptome (mm10, downloaded from UCSC) using TopHat and differential gene expression was determined by Cuffdiff as previously described (32). Gene set enrichment analysis (GSEA) was performed using software available from the Broad Institute and compared the list of expressed genes pre-ranked by the log2-fold change in expression between Hdac3−/− compared to wild-type FPKM and gene signatures identified in the denoted publication using an FDR of 0.05 as the cutoff for significance.
ChIP-Exo
ChIP-exo for FOXO1 (CST; C29H4) and H3K27ac (abcam; ab4729) was performed on OCI-LY1 cells. Cells were cross-linked with 1% formaldehyde for 8 min prior to quenching with 125 mM glycine. Following cell lysis, chromatin was sonicated for 10 cycles; 30 s ON 30 s OFF with a Bioruptor (diagenode) followed by immunoprecipitation for 3 h using antibody plus Protein A:G beads. ChIP-exo library construction was carried out as previously described (33). Peaks were called using MACS2 (version 2.0.10; q = 0.005) (34) and BCL6 and SMRT ChIP-seq datasets from OCI-LY1 cells (9). For FOXO1 ChIP-exo, adapter sequences were trimmed with Trimomatic-0.32 (35) prior to alignment to the hg19 genome using Bowtie (36).
PRO-Seq
Nuclear run-on and PRO-Seq library construction was performed according to methods described previously (37). Briefly, cells were treated in duplicate with 10 μM RGFP966 (Selleckchem) or DMSO control for 3 h prior to nuclei isolation. 20 × 106 nuclei were used per run-on. Nuclear run-ons were carried out in the presence of 375 μM GTP, ATP, UTP and biotin-11-CTP and 0.5% Sarkosyl at 30°C for 3 min. Total cellular RNA was isolated by Trizol extraction, hydrolyzed in 0.2N NaOH, and nascent RNAs incorporating biotin-CTP were isolated by Streptavidin bead binding (Invitrogen). Following 3′ adapter ligation, 5′ cap removal and triphosphate repair, 5′ hydroxyl repair, and 5′ adapter ligation, RNA was reverse transcribed and amplified. Amplified cDNA libraries were PAGE purified and provided to Vanderbilt VANTAGE Core Shared Resource for sequencing. Adapter trimming was conducted with Trimommatic-0.32 (35) prior to alignment to hg19 human genome build using Bowtie 2 (38). Analysis of PRO-seq data was carried out using the Nascent RNA Sequencing Analysis (NRSA) pipeline (39).
RESULTS
CD19-cre-mediated deletion of Hdac3 bypasses its requirement for B cell development
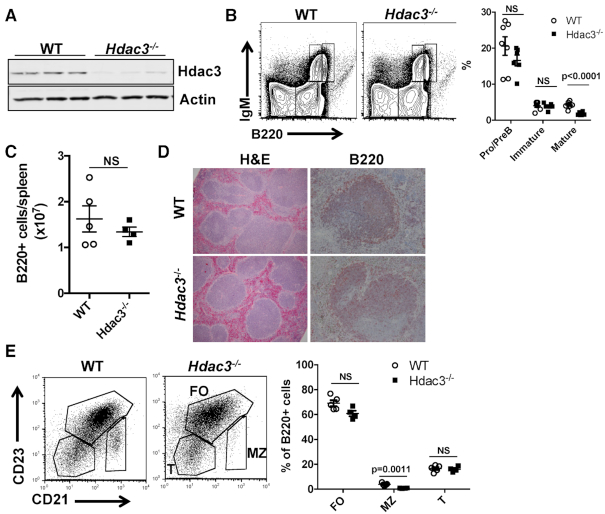
CD19-Cre was used to drive recombination of a conditional Hdac3 allele and genotyping of B220+ cells from the spleens of CD19-cre-Hdac3flox/- heterozygous mice showed that Hdac3 was efficiently deleted (Supplementary Figure S2F). This resulted in the efficient loss of Hdac3 protein from B220+ splenocytes (Figure 1A; note the small amount of residual Hdac3 could be due to its long half-life or contaminating non-B cells). While previous studies demonstrated that deletion of Hdac3 from early B cell progenitors halted B cell development due to defective VDJ recombination (26), Hdac3 deletion later in development by CD19-Cre did not impair B cell development within the bone marrow except for a reduction in mature recirculating B cell percentages and absolute numbers corresponding to Hardy fraction F (Figure 1B and Supplementary Figure S1A–C). However, CD19-Cre-mediated deletion of Hdac3 had no effect on splenic B cell numbers (Figure 1C) or splenic architecture (Figure 1D). Further classification of splenic B cell populations based on CD23 and CD21 expression revealed a reduction in marginal zone B cells, while transitional and follicular B cell populations were unchanged (Figure 1E).
Figure 1.
B cell development is unaffected by CD19-cre driven deletion of Hdac3. (A) Loss of Hdac3 protein was determined by western blot analyses of proteins isolated from B220+ splenocytes of Hdac3+/+CD19cre+/−(WT) controls or Hdac3F/−CD19cre+/−(Hdac3−/−) animals. (B) Flow cytometric analysis utilizing IgM and B220 antibodies characterizes B cell development within the bone marrow of control and Hdac3-deleted mice. Graphical representation of B cell populations as a percentage of the total bone marrow from at least 6 mice per group is presented at the right. (C) Flow cytometry was used to determine the number of B220+ cells per spleen from multiple mice. (D) In order to characterize the structure of splenic follicles, formalin-fixed spleens were subject to H&E staining and immunohistochemistry with α-B220. (E) Flow cytometric characterization of splenic B cell populations was carried out. B220+ splenocytes were further analyzed based on surface CD23 and CD21 expression. Graph on right depicts five WT mice and four Hdac3−/− mice.
Loss of Hdac3 impaired LPS-induced differentiation
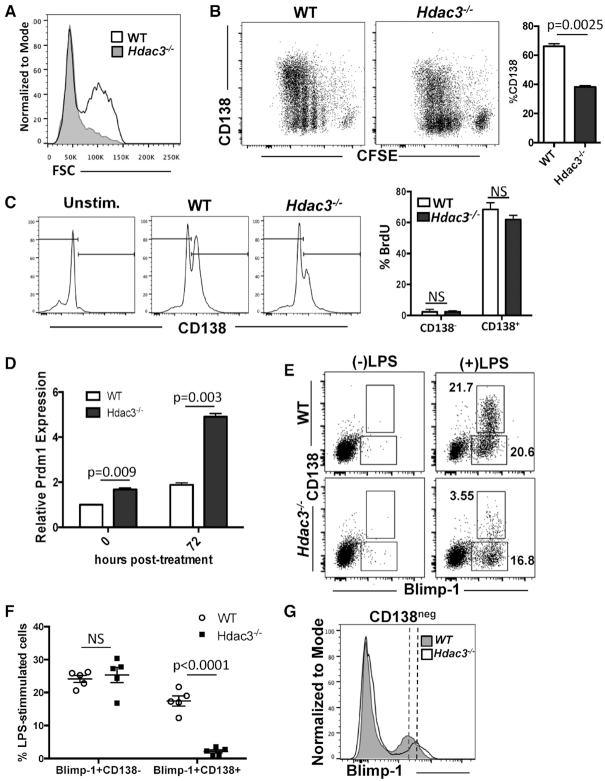
Previous conditional knockout of Hdac3 in stem cells, early B and early T cells all showed signs of replicative stress (25,26) that might be missed under steady state conditions in the CD19-Cre model. Therefore, we isolated splenic B cells and cultured them ex vivo in the presence of LPS to stimulate cell division and plasmablast differentiation. As splenic B cells become plasmablasts they increase in size and 72 h post-stimulation, an FSChi differentiated cell population had emerged in the wild-type B cell cultures, while the FSChi population was markedly reduced in the absence of Hdac3 (Figure 2A). We labeled cells with carboxyfluorescein succinimidyl ester (CSFE) to monitor cell division and 72 h following LPS stimulation, cells were harvested and stained for surface CD138 expression and relative levels of CD138 and CFSE were determined by flow cytometry. In wild-type cultures, LPS stimulation yielded a synchronous, stepwise reduction in CFSE that correlated with the emergence of CD138+ plasmablasts (Figure 2B, left). In the absence of Hdac3, LPS stimulation readily induced proliferation as demonstrated by a reduction in CFSE intensity; however, cell division resulted in the emergence of significantly fewer CD138+ cells (Figure 2B, right), suggesting a defect in plasmablast differentiation. This defect is unlikely due to a reduction in marginal zone B cells, as similar results were obtained in B cell cultures isolated from lymph nodes (Supplementary Figure S2A–C) that are devoid of marginal zone B cells (40,41). Interestingly, while Hdac3−/− cultures showed a marked reduction in CD138+ cells (Figure 2C, left), those cells that did differentiate incorporated BrdU at a rate similar to wild-type CD138+ cells (Figure 2C, right), consistent with the finding that there is no intrinsic proliferation defect in Hdac3-deleted B cells.
Figure 2.
Activated Hdac3−/− B cells show only minor defects in proliferation but show impaired differentiation. (A) B220+ splenocytes were stimulated ex vivo with LPS. Seventy-two hours post-stimulation, flow cytometry was performed to detect FSChi plasmablasts. (B) B cells were labeled with CFSE and stimulated with LPS for 72 h, at which time cells were stained with α-CD138 and flow acquired. The graph on the right depicts the percentage of CD138+ cells present in culture following 72-hour stimulation. (C) Seventy-two hours post-LPS stimulation, B cells were pulsed with BrdU and stained for surface CD138. Cells were then fixed and stained for BrdU incorporation. Flow cytometry analysis of CD138 showed reduced CD138 staining in Hdac3−/− cultures, while BrdU quantification (right) showed that while CD138+ cells proliferated at a much higher rate than CD138− cells, the loss of Hdac3 did not alter proliferation rates in either population. (D) Prdm1 transcript levels were determined by q-RT-PCR at time 0 and 72 h post-stimulation with LPS. (E) ex vivo B cell cultures were stimulated with LPS for 72 h and were then stained for surface CD138 and intracellular Blimp-1 (encoded by Prdm1). (F) Quantification of the experiment depicted in (E) carried out from cells isolated from three mice per genotype, with technical replicates carried out for two mice per genotype. (G) Overlay of Blimp-1 staining from CD138− B cells after LPS stimulation revealed an increase in Blimp-1 MFI in Hdac3-deleted cultures.
In hematopoietic stem and progenitor cells, defects in cell cycle progression were attributed to a defect in DNA replication fork velocity, likely secondary to chromatin conformation changes (25). However, Hdac3−/− B cells showed only a subtle slowing of DNA replication forks by DNA fiber labeling assays (Supplementary Figure S2D). Furthermore, Hdac3−/− splenic B cells exhibited relatively normal micrococcal nuclease sensitivity (Supplementary Figure S2E). Thus, the relatively mild effects of Hdac3 loss on chromatin structure, DNA replication and proliferation in B cell cultures are unlikely to account for the overall impairment of plasmablast formation in the absence of Hdac3. Furthermore, genotyping of B cell cultures before and after LPS stimulation revealed that the few CD138+ plasmablasts that were able to form in Hdac3−/− cultures, were not simply derived from non-deleted B cells (Supplementary Figure S2F). Thus, while LPS-stimulated B cells proliferated in response to LPS-stimulation, they failed to efficiently differentiate into CD138+ plasmablasts.
Hdac3 −/− B cells fail to differentiate upon up-regulation of Prdm1
Prdm1 encodes Blimp-1, a key transcription factor that drives plasma cell differentiation, as forced expression of Prdm1 induced differentiation and immunoglobulin secretion in mature B cells (42,43). Stimulation of wild-type B cells with LPS resulted in an approximately 2-fold increase in Prdm1 mRNA as determined by qRT-PCR. In contrast, there was a nearly 2-fold increase in Prdm1 mRNA expressed by Hdac3−/− B cells prior to LPS stimulation, which became even more pronounced following LPS activation (Figure 2D), which is consistent with Bach2 repressing Prdm1 by recruiting HDAC3 (44). Flow cytometry comparing surface CD138 expression with intracellular Blimp-1 protein levels revealed a similar up-regulation of Blimp-1 in both wild-type and Hdac3-deleted cultures prior to differentiation to a CD138+ plasmablast. However, even in the presence of increased Blimp-1, Hdac3-deficient cells failed to efficiently differentiate into CD138+ cells (Figure 2E, F). Moreover, overlay of Blimp-1 staining from CD138− cells revealed an increase in Blimp-1 protein in Hdac3−/− cells compared with wild-type controls, consistent with the idea that Prdm1 was up-regulated in the absence of Hdac3 (Figure 2G). Blimp-1 recruits Hdac3 to repress transcription in germ cells (45) and we confirmed that Blimp-1 can associate with Hdac3 (Supplementary Figure S3A). In addition, a selective Hdac3 inhibitor was able to up-regulate Blimp-1-regulated genes (46) in cells that express high levels of Blimp-1 (Supplementary Figure S3B). Therefore, the failure of Blimp-1 to drive differentiation to a CD138+ plasmablast in Hdac3-deleted cells could be due to impaired Blimp-1/Hdac3-mediated repression.
Hdac3−/− B cells show impaired plasma cell differentiation
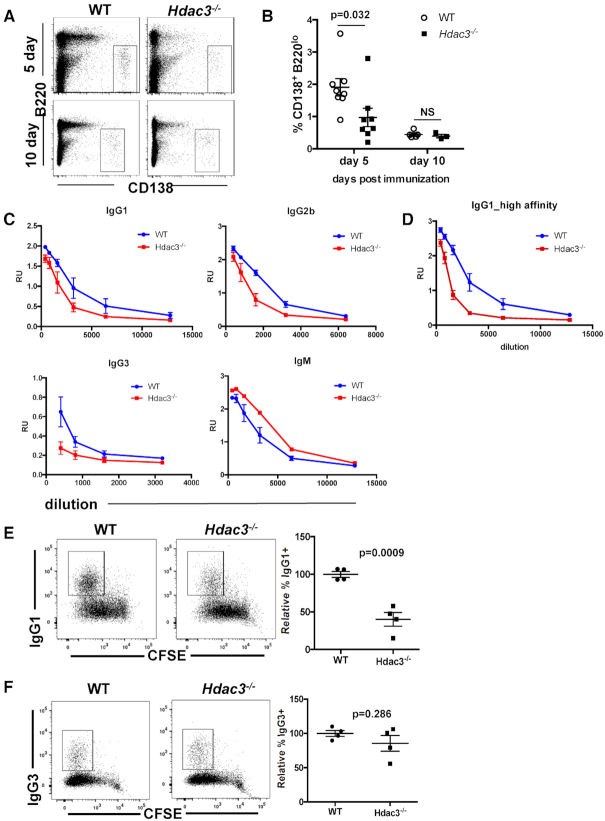
We next sought to determine if Hdac3 loss resulted in a similar reduction in extrafollicular plasmablast formation in vivo. We immunized mice with sheep red blood cells and examined CD138+ plasmablast numbers. In the absence of Hdac3, there was a significant reduction in CD138+ plasma cells 5 days following SRBC immunization (Figure 3A, B). Thus, Hdac3 was required for terminal B cell differentiation into plasmablasts in both in vivo and ex vivo settings. Consistent with the reduction in CD138+ plasma cells, serum levels of IgG1, IgG2b, and IgG3 were reduced while serum IgM levels were elevated in Hdac3−/− animals 8 days post-immunization (Figure 3C). In addition, levels of high affinity IgG1 were reduced in the serum of Hdac3−/− animals, suggesting reduced antibody affinity in the absence of Hdac3 (Figure 3D). The reduction in serum IgG levels in combination with elevated serum IgM may also indicate an impairment of class switch recombination. Therefore, we tested the impact of Hdac3 loss on class-switch recombination using ex vivo assays.
Figure 3.
Extrafollicular plasma cell differentiation is impaired in the absence of Hdac3. (A) 5 or 10 days post-immunization with SRBCs, spleens were dissociated and analyzed for the presence of CD138+B220lo plasma cells. (B) Graph depicts quantitation of the cohort from A. (C) Mice were immunized with NP-CGG. Eight days post-immunization, serum was harvested and serial serum dilutions were used in ELISAs detecting the indicated immunoglobulin using NP(31)-BSA coated plates. (D) Serum from C was used to detect IgG1 binding to NP(7)-BSA coated plates. (E) Splenic B cells were isolated and stimulated ex vivo with αCD40 and IL-4 for 4 days to induce immunoglobulin class switching to IgG1. Right panel shows quantification of populations identified from multiple mice/genotype. (F) Splenic B cells were isolated and stimulated ex vivo with LPS for 4 days to induce immunoglobulin class switching to IgG3. Right panel shows quantification of populations identified in from multiple mice/genotype.
B cell cultures were either stimulated to switch to IgG1 following treatment with αCD40 and IL-4, or stimulated to switch to IgG3 following treatment with LPS (Figure 3E–F, Supplementary Figure S3C–E). Loss of Hdac3 resulted in a significant reduction in IgG1-switched B cells (Figure 3E) that corresponded to lower levels of γ1 post-switch transcripts (Supplementary Figure S3E). In contrast, there were no significant effects on IgG3 class-switching or on post-switch γ3 expression (Figure 3F, Supplementary Figure S3E). RT-PCR detected a similar induction of Aicda expression in both wild-type and Hdac3-deleted B cell cultures following stimulation with both LPS and αCD40/IL-4, thus a failure to activate AID expression cannot account for reduced IgG1 class-switching (Supplementary Figure S3C, D). Finally, germline transcript analysis (47) revealed an induction of only γ1 expression in wild-type cultures following αCD40/IL-4 stimulation, while in Hdac3-deleted cultures, both γ1 and γ3 RNAs were induced by αCD40/IL-4. Thus, it is possible that in the absence of Hdac3, γ3 transcription is not efficiently repressed downstream of IL-4 (48). Alternatively, γ3 is more proximal to μ than γ1 (49), so loss of Hdac3 could affect general chromatin structure at this locus to reduce the efficiency of distal recombination events, similar to the effect of Hdac3 deletion on VDJ recombination in early B cell progenitor cells (26). Thus, Hdac3 plays a nuanced, context-specific role in facilitating class-switch recombination.
Altered Germinal center formation in the absence of Hdac3
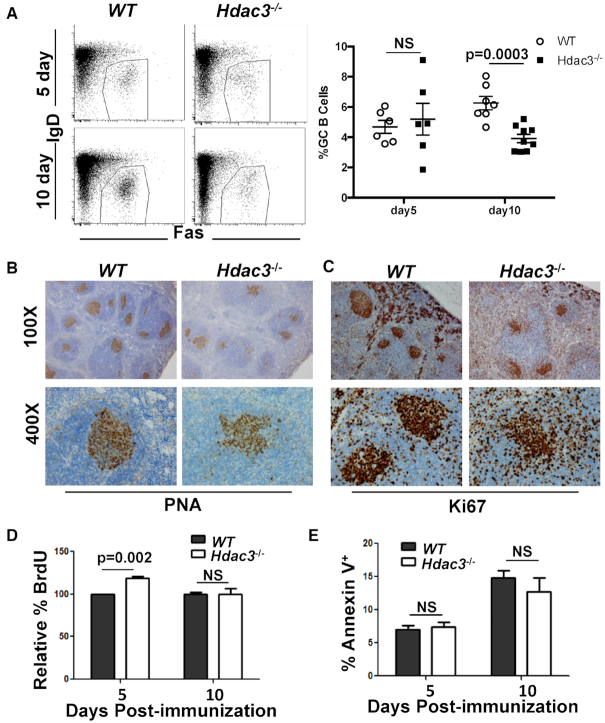
Plasma cells generated 5 days after immunization are likely derived from extrafollicular plasmablasts and are generated in a Bcl6-independent manner. Similarly, LPS-driven B cell differentiation (Figure 2) also occurs in a Bcl6-independent manner (14,50). However, given that Bcl6 recruits Hdac3-containing repressor complexes, we sought to directly address the role of Hdac3 in Bcl6-dependent B cell biology. Mice were immunized with sheep red blood cells (SRBCs) and the germinal center response analyzed 5- and 10-days post-immunization (dpi). At 5 dpi, Hdac3−/− animals contained similar numbers of B220+Fas+IgD− germinal center B cells. However, by 10 dpi, when the germinal center is fully developed (7), there was a reduction in GC B cells in the absence of Hdac3 (Figure 4A). Detection of germinal centers by staining with peanut agglutinin (PNA; Figure 4B) or Ki67 (Figure 4C) 10 days after immunization revealed a disorganized and often dispersed germinal center appearance.
Figure 4.
Altered germinal centers in the absence of Hdac3. (A) Mice were immunized with sheep red blood cells (SRBCs). 5 and 10 days post-immunization, spleens were harvested and analyzed by flow cytometry for the presence of B220+IgDloFas+ germinal center B cells. Graphical representation of GC B cell populations as a percentage of total splenic B cells from at least 6 mice per group is presented at the right. (B) Spleens were isolated from SRBC-injected mice ten days following immunization and sections were stained with PNA to identify germinal center B cells. (C) Ki67 staining was performed on 10 dpi spleens to identify proliferating cells. (D) Immunized mice were injected with BrdU 2 h prior to tissue harvest. Germinal center B cells were analyzed by flow cytometry for BrdU incorporation. (E) Splenocytes were labeled with annexin V and analyzed by flow cytometry. The percentage of B220+IgDloFas+ Annexin V+ splenocytes are depicted.
Mechanistically, these defects could be due to deficiencies in cell proliferation, survival and/or the activation potential of Hdac3−/− B cells. BrdU incorporation assays indicated that there was a slight increase in the number of cycling Hdac3−/− germinal center B cells five days post-immunization, yet no impairment of BrdU incorporation in Hdac3-deleted germinal center B cells 10 days after immunization (Figure 4D). In addition, there was no difference in the number of annexin V positive cells at either time point examined (Figure 4E), suggesting that reductions in Hdac3−/− germinal center B cell numbers were not due to reduced proliferation rates or increased cell death in those germinal centers that did form. Finally, B cell receptor (BCR) crosslinking of isolated splenic B cells with α-IgM revealed robust BCR signaling in the absence of Hdac3 (Supplementary Figure S4). This included the effective phosphorylation of PLCγ2 (Supplementary Figure S4A) and subsequent calcium release (Supplementary Figure S4B) as well as efficient phosphorylation of ERK (Supplementary Figure S4C; note that phospho-Erk was consistently slightly higher in the null cells). Thus, signaling events stimulated upon B cell activation were not markedly reduced by Hdac3 loss.
Altered gene expression in Hdac3−/− germinal center B cells disrupts differentiation to plasma cells
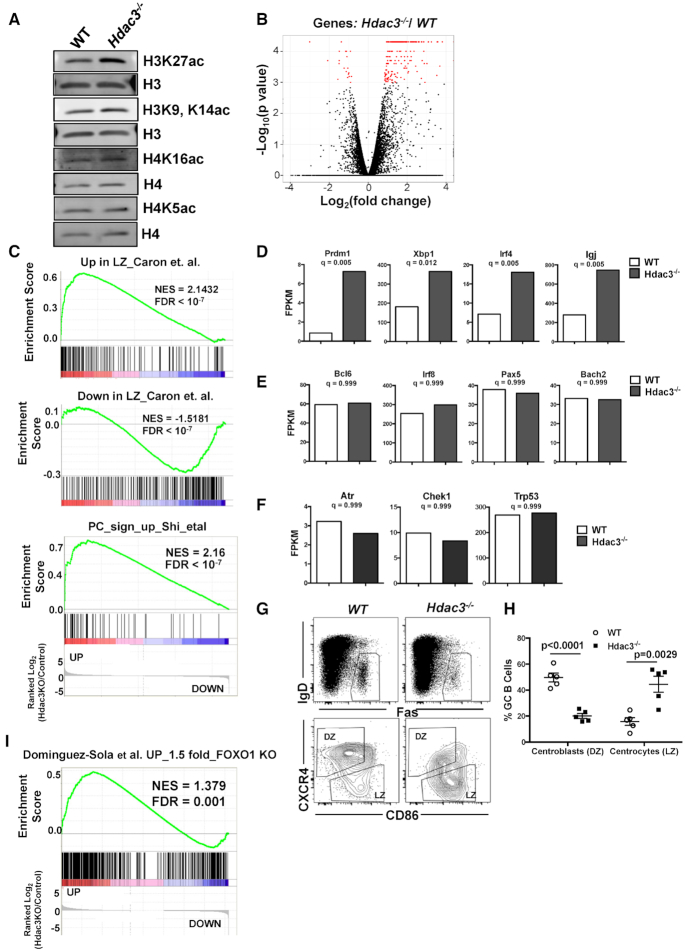
Examination of histone acetylation levels in Hdac3 deleted B cells revealed an increase in H3K27ac with only modest effects on other histone acetylation marks (Figure 5A). The ability of Hdac3 to regulate H3K27ac is consistent with previous reports in B cell lymphoma (9), and accumulation of H3K27ac is associated with activation of promoter and enhancer activity. Therefore, to address the impact of Hdac3 loss on germinal center transcriptional programs, B cell populations were sorted from the spleens of immunized mice and analyzed by RNA-seq. Hdac3−/− germinal center B cells (B220+GL7+) showed increased expression of a relatively low number of genes (just over 200), with very few genes down-regulated (Figure 5B), a result consistent with the role of Hdac3 in transcriptional repression rather than wide-spread chromatin effects. Because so few genes were affected, general gene ontology analysis revealed enrichment of only a few pathways among those genes altered in Hdac3-deleted germinal center B cells (Supplementary Figure S5A). In addition, several B-cell specific pathways were even more highly correlated with Hdac3-regulated gene signatures. First, genes up-regulated in the absence of Hdac3 were highly correlated with those up-regulated in the germinal center light zone compared with the dark zone and genes down-regulated in the light zone were also correlated with genes down-regulated upon Hdac3 loss (Figure 5C, upper and middle). In addition, there was a highly significant correlation between genes up-regulated upon plasma cell differentiation and those up-regulated in the absence of Hdac3 (Figure 5C, lower). This is particularly interesting given the reduction in plasmablast formation observed both in vitro and in vivo, but is consistent with the observation that Prdm1 up-regulation was not sufficient for plasmablast formation in the absence of Hdac3 (Figures 2 and 3).
Figure 5.
Hdac3 loss is associated with a reduction in dark zone centroblasts. (A) Western blot analysis detecting acetylated histone marks from B220+ splenocytes isolated from Hdac3−/− animals or WT controls. (B) Spleens from SRBC-injected mice were sorted for germinal center B cells (B220+GL7hi). The volcano plot depicts genes with significantly altered expression in Hdac3-deleted cell populations in red. (C) GSEA was performed comparing gene expression changes in Hdac3−/− germinal center B cells to a published signature of genes upregulated in light zones compared to dark zones (75) (upper), genes down-regulated in light zones compared with dark zones (75) (middle) and a signature of the 50 most up-regulated transcripts in differentiating plasma cells (76). (D) Transcript levels of key regulators of plasma cell differentiation (Prdm1, Irf4, Xbp1, IgJ) were significantly up-regulated upon Hdac3 loss in germinal center B cells. (E) Transcripts associated with germinal center B cell indentity (Bcl6, Irf8, Pax5, and Bach2) were unaltered in the absence of Hdac3. (F) Canonical Bcl6 targets associated with DNA damage checkpoints (Atr, Chek1 and Trp53) were unchanged in Hdac3−/−germinal center B cells. (G) B220+IgDloFas+ GC B cells (upper) were further classified as dark zone centroblasts or light zone centrocytes on the bases of surface CXCR4 and CD86 expression. (H) Quantification of populations identified in (A) from 5 mice/genotype. (I) GSEA identifies a significant correlation between genes up-regulated in GC B cells upon loss of Hdac3 and genes up-regulated upon loss of Foxo1 (17).
Among the specific genes activated in Hdac3−/− germinal center B cells were Bcl6-regulated genes that function as critical determinants of plasma cell differentiation, including Prdm1 (encoding Blimp-1), Irf4, and Xbp1 (Figure 5D). Not only are these transcription factors critical for plasma cell identity, but they also silence germinal center B cell transcriptional programs to drive terminal differentiation (51). For example, Blimp-1 (Prdm1) repressed the expression of both Bcl6 and Pax5, such that the induction of plasma cell transcriptional networks was tightly coupled to the cessation of germinal center B cell-associated transcriptional programs (52,53). However, in Hdac3−/− cells, Bcl6, Pax5, Irf8 and Bach2 expression remained high, even in the presence of accumulating Prdm1 (Figure 5E). While Bcl6-regulated genes that are associated with plasma cell differentiation were up-regulated in Hdac3−/− germinal centers, other Bcl6-regulated genes that modulate cell cycle checkpoints and the DNA damage response were unaffected by Hdac3 deletion (Figure 5F). Thus, Hdac3 inactivation impaired transcriptional repression of a subset of Bcl6-controlled genes, resulting in the elevation of plasma cell factors within germinal center B cells.
Hdac3-deleted germinal centers exhibit a loss of dark zone centroblasts
The germinal center can be divided into dark zone centroblasts, which are undergoing somatic hypermutation, and light zone centrocytes that are undergoing selection through association with antigen presenting cells and TFH cells. The up-regulation of genes associated with a plasma cell identity begins to occur in the light zone of the germinal center prior to differentiation. Given that we observed an up-regulation of mRNAs associated with light zone cells (Figure 5C), we asked whether Hdac3 was skewing the ratio of dark zone centroblasts to light zone centrocytes within germinal centers. Germinal center B cells that were B220+IgDloFas+ (5G, upper panels) were further classified as either CXCR4hiCD86lo dark zone centroblasts or CXCR4loCD86hi light zone centrocytes (Figure 5G, lower panels). In wild-type germinal centers, the majority of B cells were located in the dark zone compartment (Figure 5G and H). In contrast, Hdac3−/− germinal centers revealed a significant depletion of dark zone centroblasts and an accumulation of light zone centrocytes (Figure 5G and H). Given that most germinal center proliferation occurs in the dark zone it may seem surprising that we did not observe reductions in germinal center B cell proliferation in the absence of Hdac3; however, these results are consistent with other genetic models that found an uncoupling of proliferation from dark zone centroblasts (17). We did note that while an E2F signature was induced in both wild-type and Hdac3−/− germinal centers compared with naïve B cells, it was not induced to the same degree in the absence of Hdac3, suggesting that there may be subtle proliferative differences in these cells (Supplementary Figure S5B).
Recent studies reported a loss of dark zone centroblasts and defects in IgG1 class-switching upon deletion of the transcription factor, Foxo1, from germinal center B cells (17,18), two phenotypes that bore striking similarity to those observed upon Hdac3 loss (Figures 3E and 5G). Furthermore, Foxo1 co-regulates a subset of Bcl6 target genes, including Prdm1 and CD86 (17). Gene set enrichment analysis (GSEA) identified a significant overlap between genes up-regulated at least 1.5-fold in Foxo1-deleted germinal centers and those up-regulated in the absence of Hdac3 (Figure 5I). Hdac3 is inactive unless bound by NCOR or SMRT, and Foxo1 and SMRT were both reported to bind at Bcl6-regulated targets (9,17), including Prdm1, suggesting a convergence on the regulation of a subset of BCL6 target genes.
HDAC3 regulates a subset of BCL6 repressed genes
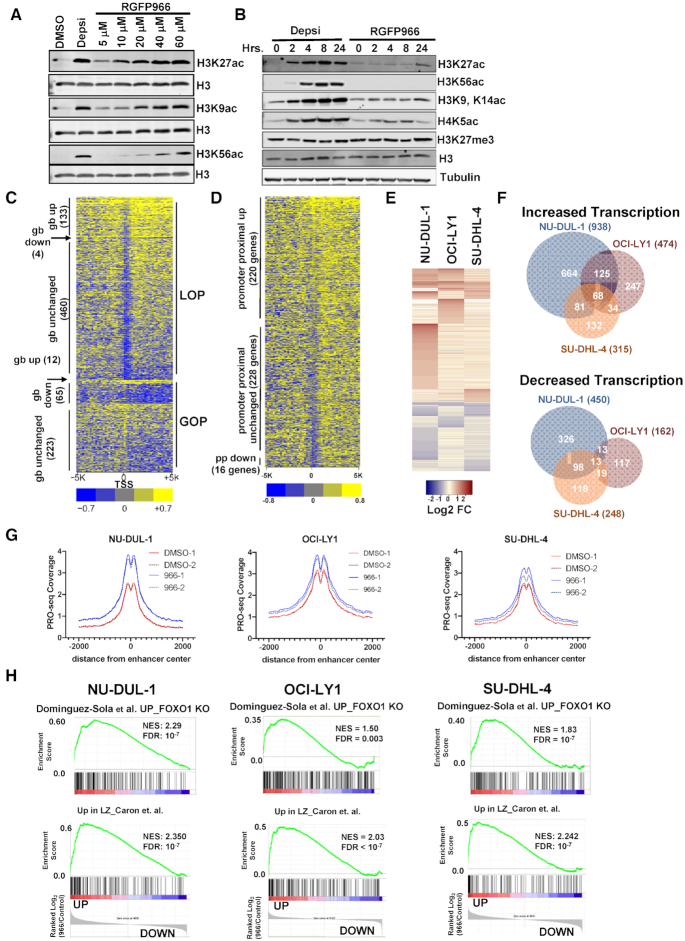
Murine genetic analysis identified critical points in B cell maturation that required the continued presence of Hdac3 (Figures 1-4) (25,26). While RNA-seq from Hdac3-deleted germinal centers identified changes in germinal center transcripts, whether these signatures are directly regulated by Hdac3 or merely correlate with the accumulation of LZ centrocytes in the absence of Hdac3 is hard to distinguish. Therefore, we used a small molecule inhibitor, RGFP966, that is 8-fold more selective for Hdac3 over other histone deacetylase proteins (54–56) and treated OCI-LY1 germinal center-derived lymphoma cells to assess presumably direct gene expression changes at early timepoints. Western blot analysis for HDAC3-regulated histone acetylation marks (Figure 6A) showed that H3K27ac, a mark of active enhancers, was increased at 5 to 10 μM RGFP966 with little to no effect on H3K56ac, which is potently increased by inhibitors of HDAC1 and HDAC2 such as Depsipeptide (Figure 6A and B) (54).
Figure 6.
PRO-seq analysis of lymphoma cells treated with an Hdac3-selective inhibitor. (A) Western blot analysis of acetyl-histone H3K27, H3K9 and H3K56 using anti-Histone H3 as a loading control 24 h after addition of 10 nM Depsipeptide or RGFP966 at the concentration shown at the top of each lane. (B) Western blot shows accumulation of histone acetylation over time in response to either 10 nM Depsipeptide or 10 μM RGFP966. (C) Heat map displaying the relative intensity of active RNA polymerases ±5 kb from the transcriptional start site (TSS) of genes affected by a 3 h treatment of OCI-LY1 cells with 10 uM of RGFP966. LOP, loss of promoter-proximal pausing; GOP, gain of pausing; gb, gene body. (D) Heat map as described in B, but showing genes with increased polymerase in the gene body. (E) Heat map showing the changes up and down for genes affected by a 3 h treatment of RGFP966 of the lymphoma cell lines indicated at the top of each lane. (F) Venn diagrams show the number of genes whose transcription was commonly increased or decreased in PRO-seq data from the three lymphoma cell lines treated with RGFP966 for 3 h in cell lines shown. (G) PRO-seq signal corresponding to eRNA levels around NRSA-called intergenic enhancers following a 3-hour treatment with RGFP966 (H) Gene set enrichment analysis comparing gene changes identified by PRO-seq following HDAC3 inhibition with RGFP66 with the FOXO1 gene signature (upper) or a light zone gene signature (lower) for all three DLBCL cell lines.
In order to simultaneously address changes in both gene transcription and enhancer function following HDAC3 inhibition, LY1 cells were treated with RGFP966 for 3 h prior to harvest for precision nuclear run-on sequencing (PRO-seq). PRO-seq maps transcriptionally engaged RNA polymerase by performing nuclear run-on in the presence of biotinylated NTPs and then purifying nascent transcripts based on the biotin-streptavidin interaction. Using this method, we can detect not only nascent protein-coding transcripts, but also nascent non-coding transcripts including eRNAs, lncRNAs and miRNAs. Furthermore, polymerase mapping by PRO-seq allows detailed characterization of transcription initiation, promoter-proximal pausing and transcription elongation. The sensitivity of this method allows the use of early timepoints following HDAC3 inhibition, which means that, unlike RNA-seq, we can identify genes likely to represent direct transcriptional targets of HDAC3 and avoid characterization of secondary events.
PRO-seq was performed after just a 3-hour treatment of OCI-LY1 cells with 10 μM RGFP966. At this early time point there were only modest changes in global histone acetylation (Figure 6B). The nascent RNA sequence analysis (NRSA) informatics package (39) was used to identify genes with a gain of RNA polymerase pausing near the start site relative to the body of the gene (pausing ratio) or a loss of pausing. This analysis detected nearly 600 genes with a loss in promoter-proximal pausing, but this was associated with only 133 genes with increased numbers of active polymerases in the gene body (Figure 6C shows a heatmap of the region ±5 KBP of the transcription start site). This analysis identified another 12 genes with more polymerase in the gene body but with no change in pausing, and 65 genes with a loss of polymerase in the gene body and a gain in promoter-proximal pausing (Figure 6C).
While pausing ratio is calculated within a single gene, we also normalized these data using DESeq2 (57) to allow us to determine the effect of inhibiting HDAC3 on nascent gene body transcription. Increases in gene body polymerase generally correlate with increases in transcription and decreases in gene body polymerase generally correlate with reduced transcription. This analysis identified 474 genes that showed increased polymerase activity in the gene body following HDAC3 inhibition (Figure 6D). When these 474 up-regulated genes were examined for changes in promoter proximal polymerase pausing, 220 genes showed an increase in polymerase at the pause site, 228 genes showed no change in polymerase at the pause site and only 16 genes showed a reduction in polymerase at the pause site. Note that 10 genes did not have sufficient promoter proximal signal in one or both conditions to determine changes in promoter proximal polymerase and therefore were not plotted (Figure 6D). Thus, these results suggest that the inhibition of HDAC3 induced gene expression by driving transcription initiation without significant effects on pausing or elongation.
OCI-LY1 cells contain the M1L mutation that causes FOXO1 to be translated from the second methionine producing an N-terminal truncation that escapes negative regulation by AKT (58). We expanded our PRO-seq analysis to a second FOXO1 mutant cell line (NU-DUL-1) and SU-DHL-4 that has wild type FOXO1. NU-DUL-1 showed the largest number of changes within the gene body with 968 genes with increased numbers of active RNA polymerases and 450 genes showing loss of transcription (Figure 6E, F). Although OCI-LY1 and SU-DHL-4 showed less dramatic changes, the general trend was the same (Figure 6E, F). While 125 genes were specifically activated in the two FOXO1 mutant cell lines (Figure 6F, Supplementary Figure S6), there were also many genes up-regulated in all three cell lines (Figure 6F, Supplementary Figure S6). In fact, KEGG analysis revealed deregulation of many common pathways across all three cell lines, including Antigen Processing and Presentation, Allograft rejection and Cell Adhesion Molecules (Supplementary Figure S7A).
One advantage of PRO-seq over other quantitative measures of gene expression is that PRO-seq identifies all polymerase activity, including the transcription of small regulatory RNAs such as miRNAs and enhancer RNAs (eRNAs) (39). Importantly, eRNA synthesis is an even better predictor of enhancer activity than changes in histone modifications (59–61). Consistent with the accumulation of H3K27ac following HDAC3 inhibition (Figure 6A, B), RGFP966 treatment increased the eRNA signal around PRO-seq identified intergenic enhancers in all three cell lines (Figure 6G). The most dramatic increase in eRNA levels was observed in the NU-DUL-1 cell line, which also exhibited the largest numbers of gene expression changes (Figure 6G, F). Gene set enrichment analysis showed that inhibition of HDAC3 caused increases in genes up-regulated by deletion of Foxo1 from germinal centers and enriched in light zone centrocytes (Figure 6H). Importantly, these are the same gene signatures observed in Hdac3-deleted germinal centers and suggests that light zone-associated genes are likely direct targets of HDAC3 transcriptional activity.
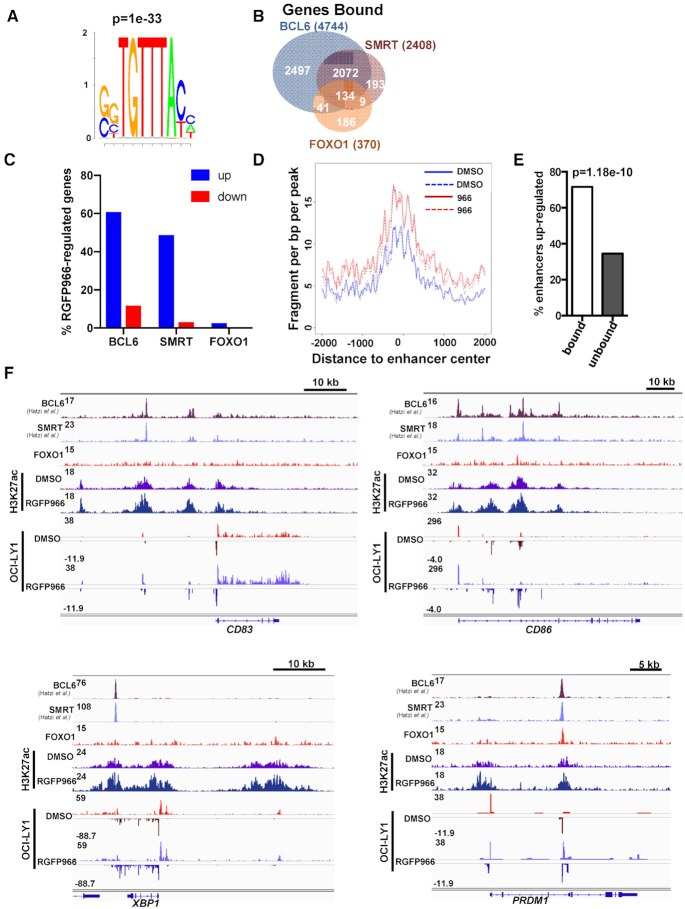
While it is clear that Hdac3 and Foxo1 deletion from germinal centers yields similar phenotypes and gene expression changes (17,18), it remained to be determined if HDAC3 and FOXO1 directly coordinate gene expression at common targets. Therefore, we sought to determine if endogenous HDAC3 and FOXO1 co-localize on chromatin. We utilized the human germinal center-derived cell line, OCI-LY1, to perform ChIP-exo for FOXO1. The FOXO1 peaks identified by ChIP-exo were highly enriched for the canonical Forkhead DNA binding motif (Figure 7A). In addition, we mined published ChIP-seq datasets for BCL6 and SMRT binding (9) to compare the peak location of BCL6 and SMRT with our data for FOXO1. While the antibodies to HDAC3 do not yield a good ChIP signal, likely because SMRT is much larger and obscures HDAC3, SMRT is the regulatory component of the HDAC3 complex, so ChIP-seq for SMRT identifies active HDAC3 complexes (62,63). SMRT binding was detected at about a third of FOXO1-bound genes (Figure 7B), but most of these genes were also regulated by BCL6 (Figure 7B). Conversely, FOXO1 was only detected at just over 5% of the SMRT-bound genes, which suggests that Hdac3 inactivation is unlikely to mirror the transcriptional effects of Foxo1 inactivation. Similar effects were observed when individual peaks were compared within these datasets (Supplementary Figure S7B, C). In contrast, there was nearly a 50% overlap between BCL6 and SMRT. When genes bound by BCL6/SMRT/FOXO1 were compared with genes identified as deregulated upon HDAC3 inhibition by PRO-seq, there was a high correlation with increased expression and BCL6/SMRT binding. However, very few up-regulated genes were also bound by FOXO1. At the same time the correlation with genes that are bound by BCL6/SMRT and those down-regulated upon HDAC3 inhibition were quite low (Figure 7C, Supplementary Figure S7B). Thus, BCL6/SMRT binding correlated selectively with HDAC3-mediated repression. Consistently, enhancers bound by BCL6/SMRT showed increases in the bi-directional transcription that is a hallmark of the enhancer center (Figure 7D). In fact, up-regulation of eRNAs observed upon HDAC3 inhibition was more likely to occur at BCL6/SMRT bound enhancers than unbound enhancers (Figure 7E).
Figure 7.
Hdac3 associates with and regulates a subset of BCL6/Foxo1 gene targets. (A) ChIP-exo was performed from OCI-LY1 cells using α-FOXO1. Motif analysis of called FOXO1 peaks revealed a highly significant enrichment for the forkhead DNA binding motif. (B) FOXO1 peaks were annotated to nearest gene and compared with BCL6- and SMRT-bound genes. FOXO1 peaks obtained were compared with published ChIP-seq data for BCL6 and SMRT from the same cell line (9). (C) BCL6, SMRT or FOXO1 bound genes were compared with genes identified as either up-regulated or down-regulated by PRO-seq following HDAC3 inhibition. The percentage of genes bound by the respective transcription factor is shown. (D) The histogram depicts changes in the activity at enhancers located within 100 bp of a BCL6:SMRT binding site following treatment with RGFP966. (E) Graph shows enhancers bound by BCL6:SMRT complexes were more likely to be up-regulated upon HDAC3 inhibition than non-bound enhancers (P value determined by Fisher's exact test). (F) Loci associated with Light Zone centrocytes (CD83, CD86) and plasma cell differentiation (XBP1, PRDM1) are displayed. Tracks depict BCL6, SMRT and FOXO1 ChIP signal as well as ChIP for H3K27ac ± 10 μM RGFP966 for 3 h and PRO-seq signal +/− 10μM RGFP966 for 3 h.
Among the BCL6/SMRT bound genes were those critical for light zone establishment and maintenance including CD83 and CD86 (Figure 7F, upper). Moreover, inhibition of HDAC3 by RGFP966 caused an increase in H3K27ac and eRNA transcription around BCL6/SMRT binding sites at these loci, which further correlated with an increase in gene body transcription. Similarly, BCL6/SMRT binding was also detected at the XBP1 and PRDM1 loci, two genes also up-regulated in Hdac3-deleted germinal centers (Figure 5) and critical for plasma cell differentiation. HDAC3 inhibition was associated with an increase in H3K27ac and an increase in gene body transcription at XBP1 and PRDM1 as well (Figure 7F, lower). Interestingly, while FOXO1 did not appear to bind CD83 or XBP1, it did bind in close proximity to BCL6 and SMRT at the PRDM1 locus. Thus, while HDAC3 activity is more highly correlated with BCL6 binding, it may still coordinate with FOXO1 at specific loci. These data suggest that Hdac3 is required for germinal center dark zone maintenance, possibly by allowing BCL6 to suppress light zone gene expression during B cell development and is critical for repressing targets associated with terminal plasma cell differentiation.
DISCUSSION
HDAC3 is the enzymatic component of the nuclear hormone co-repressor (NCOR/SMRT:HDAC3) complex. These complexes are recruited by BCL6, a transcription factor critical for cell cycle checkpoint inactivation, formation of germinal centers, and repression of plasma cell differentiation within the germinal center (11,50,64,65). Disruption of NCOR/SMRT:HDAC3 recruitment by peptides or small molecules that target the BCL6 BTB domain suggested that HDAC3 would be a good therapeutic target in DLBCL (23,66). However, the physiological role of Hdac3 in the germinal center remained unexplored, because MB1-Cre-mediated deletion of Hdac3 blocked early B cell development (26). CD19-Cre is expressed late enough in B cell development to allow VDJ recombination and the production of normal B cell numbers in the absence of Hdac3 (Figure 1), allowing us to assess the role of Hdac3 during the germinal center reaction (Figures 4 and 5). While the phenotypes observed exhibit some hallmarks of impaired BCL6-mediated repression, primarily with regards to gene expression, germinal centers still formed in the absence of Hdac3 whereas they failed to form in the absence of Bcl6 (14–16). In fact, the phenotype more closely resembled deletion of the transcription factor, Foxo1 (17,18).
Foxo1 is required for the establishment and/or maintenance of the germinal center dark zone and efficient affinity maturation (17,18,67). By comparison, loss of Hdac3 resulted in a similar reduction of the germinal center dark zone and accumulation of light zone centrocytes (Figure 5). Foxo1 has primarily been characterized in the context of transcriptional activation; however, it has been proposed to repress specific gene targets (e.g. Prdm1) in the context of germinal centers (17). In fact, cDNA microarray data in conjunction with ChIP-seq identified a subset of Bcl6 target genes that were co-regulated by Foxo1 (17). Thus, to determine if HDAC3 was directly regulating FOXO1 function, we turned to lymphoma cell lines that allowed genomic analysis following HDAC3 inhibition. We used a brief treatment of three germinal center-derived lymphoma cell lines (two with activating FOXO1 mutations) (58) with a slow on, slow off, selective HDAC3 inhibitor, RGFP966 (55), to enrich for direct targets of HDAC3 activity. PRO-seq provides a direct readout of transcription as opposed to RNA-seq, which monitors steady state RNA levels subject to multiple levels of regulation. Consistent with a role for HDAC3 in transcriptional repression, more genes were activated than repressed upon RGFP966 treatment (Figure 6). Surprisingly, all three cell lines showed an up-regulation of the germinal center light zone signature. This was the same signature enriched upon Hdac3 deletion from germinal centers and suggests that HDAC3 directly represses genes associated with the light zone.
Foxo1 −/− mice also showed a germinal center light zone signature, but when we compared PRO-seq data with ChIPseq data for FOXO1, BCL6 (9), SMRT (9), and H3K27ac, HDAC3 function was more tightly coupled to BCL6 than FOXO1, as genes up-regulated following RGFP966 treatment were bound by BCL6 and/or SMRT much more frequently than by FOXO1 (Figure 7C). Moreover, an increase in RNA polymerase activity was observed at enhancers located near BCL6:SMRT binding sites and these BCL6:SMRT-bound enhancers were more likely to be up-regulated than non-bound enhancers upon HDAC3 inhibition (Figure 7E). Thus, it seems likely that the similar germinal center phenotypes observed in Hdac3- and Foxo1-deleted animals are unlikely to be directly related. For example, approximately 25% of the genes up-regulated upon Hdac3 loss from germinal centers were up-regulated upon HDAC3 inhibition in at least one of the DLBCL cell lines, suggesting that many of the genes up-regulated in Hdac3-deleted germinal center B cells are in fact direct targets of Hdac3 activity. By contrast, we have used inducible degradation of the endogenous FOXO1 protein and confirmed that FOXO1 primarily activates transcription in OCI-LY1 cells (Stengel and Hiebert, unpublished results). Thus, while genes up-regulated following Foxo1 deletion were highly enriched for genes up-regulated upon Hdac3 deletion, many of these changes, at least in the case of Foxo1−/− mice, are likely indirect and may reflect loss of the germinal center dark zone cells. Conversely, there were a handful of genes co-bound by BCL6:SMRT:FOXO1 (see PRDM1, Figure 7F), so it is possible that specific loci co-bound by BCL6:SMRT:FOXO1 may be particularly important for HDAC3 phenotypes. It is also possible that differences in the active transcriptional networks between germinal centers and DLBCL cell lines have obscured some of the links between FOXO1 and HDAC3.
Combining PRO-seq with ChIP-seq datasets for SMRT and H3K27ac after RGFP966 treatment allowed even more confident identification of direct HDAC3 targets that are required for B cell development and plasma cell differentiation (Figure 7). Gene targets that are associated with plasma cell differentiation (PRDM1, XBP1) were identified as direct targets of HDAC3 regulation and were up-regulated (Figure 5 and 7). However, in spite of the elevated expression of plasma cell-associated transcripts, plasma cell formation was reduced in Hdac3-deleted animals, particularly at early time points after antigen challenge, which is reflective of extrafollicular immune responses (Figures 2 and 3) (68). In addition, LPS-stimulated plasmablast differentiation ex vivo was compromised in Hdac3-deleted B cell cultures in spite efficient up-regulation of Prdm1 both at the transcript and protein levels. Given that previous studies reported that enforced expression of Prdm1 was sufficient to drive plasma cell differentiation (42), our data suggest that Prdm1 is not sufficient to drive plasma cell development in the absence of Hdac3. Prdm1 is a well-established transcriptional repressor and one possible explanation for this is that Hdac3 mediates the repression function of Prdm1 in B cells at some loci. Consistent with this hypothesis are reports that the repressive function of Blimp-1 (Prdm1) involves the recruitment of deacetylase activity (69) and that Blimp-1 recruits Hdac3 to mediate gene repression and germ cell fate determination (45). Here we confirmed the association between BLIMP-1 and HDAC3, and our data from myeloma cells further supports the idea that HDAC3 is also involved in BLIMP-1-mediated repression in the B cell lineage.
Additional direct targets of HDAC3 included CD86 and CD83 (up in all three cell lines, Figure 7) and CXCR5 (up in two of three cell lines). The geographic segmentation of the germinal center into the dark and light zones is dependent on specific interactions between B cells and other cells within the germinal center including T helper and follicular dendritic cells present in the light zone and reticular cells present in the dark zone (70). For instance, CXCR4 not only marks dark zone centroblasts, but also facilitates the interaction with CXCL12 on reticular cells resulting in their localization in the dark zone (71,72). Similarly, CXCR5-expressing germinal center B cells interact with CXCL13 expressed by follicular dendritic cells resulting in their localization to the germinal center light zone (72). Elevated CD86 expression on the surface of centrocytes mediates interactions with T helper cells in the light zone that promote proliferation and class-switching (73). CD83, another marker of light zone centrocytes, also plays functional roles in the polarization of the germinal center, as B cell-specific deletion of CD83 resulted in the accumulation of germinal center B cells in the dark zone (74). Thus, direct repression of CXCR5, CD83 and CD86 by HDAC3 likely maintains normal dark zone/light zone polarization of germinal center B cells by maintaining appropriate contacts with supportive cells present within the germinal center.
DATA AVAILABILITY
Data have been deposited in GEO: GSE130502, GSE130516, GSE77113.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of Hiebert lab for helpful discussions, reagents and advice. We thank the Flow Cytometry, Translational Pathology and Genome Sciences/VANTAGE shared resources for services and support.
Author contributions: K.S. designed and performed experiments, analyzed data and wrote the manuscript, S.B. designed and performed some experiments and wrote the manuscript, J.W. and Q.L. analyzed data, SH designed experiments, analyzed data and wrote the manuscript, S.S. and J.E. performed experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
T.J. Martell Foundation; Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation; National Institutes of Health [RO1-CA109355, RO1-CA164605, R01-CA64140 to SWH]; Vanderbilt Digestive Disease Research [NIDDK P30DK58404]; Vanderbilt Institute for Clinical and Translational Research; Vanderbilt-Ingram Cancer Center [NCI P30CA68485]; K.S. was supported by 5 T32 CA009582-31 and a postdoctoral fellowship [PF-13-303-01-DMC] from the American Cancer Society. Funding for open access charge: National Institutes of Health [R01 CA164605].
Conflict of interest statement. The authors have declared that no relevant conflict of interest exists, but S.H. receives research funding from Incyte Inc. for work not related to this study.
REFERENCES
- 1. DeFranco A.L. The germinal center antibody response in health and disease. F1000Res. 2016; 5:F1000 Faculty Rev-999. [Google Scholar]
- 2. Allen C.D., Okada T., Cyster J.G.. Germinal-center organization and cellular dynamics. Immunity. 2007; 27:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Victora G.D., Nussenzweig M.C.. Germinal centers. Annu. Rev. Immunol. 2012; 30:429–457. [DOI] [PubMed] [Google Scholar]
- 4. Mesin L., Ersching J., Victora G.D.. Germinal center B cell dynamics. Immunity. 2016; 45:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., Nussenzweig M.C.. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010; 143:592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gitlin A.D., Shulman Z., Nussenzweig M.C.. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014; 509:637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Recaldin T., Fear D.J.. Transcription factors regulating B cell fate in the germinal centre. Clin. Exp. Immunol. 2016; 183:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cattoretti G., Shaknovich R., Smith P.M., Jack H.M., Murty V.V., Alobeid B.. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J. Immunol. 2006; 177:6930–6939. [DOI] [PubMed] [Google Scholar]
- 9. Hatzi K., Jiang Y., Huang C., Garrett-Bakelman F., Gearhart M.D., Giannopoulou E.G., Zumbo P., Kirouac K., Bhaskara S., Polo J.M. et al.. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013; 4:578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Basso K., Dalla-Favera R.. Roles of BCL6 in normal and transformed germinal center B cells. Immunol. Rev. 2012; 247:172–183. [DOI] [PubMed] [Google Scholar]
- 11. Shaffer A.L., Yu X., He Y., Boldrick J., Chan E.P., Staudt L.M.. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000; 13:199–212. [DOI] [PubMed] [Google Scholar]
- 12. Polo J.M., Dell’Oso T., Ranuncolo S.M., Cerchietti L., Beck D., Da Silva G.F., Prive G.G., Licht J.D., Melnick A.. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 2004; 10:1329–1335. [DOI] [PubMed] [Google Scholar]
- 13. Phan R.T., Dalla-Favera R.. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004; 432:635–639. [DOI] [PubMed] [Google Scholar]
- 14. Fukuda T., Yoshida T., Okada S., Hatano M., Miki T., Ishibashi K., Okabe S., Koseki H., Hirosawa S., Taniguchi M. et al.. Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 1997; 186:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dent A.L., Shaffer A.L., Yu X., Allman D., Staudt L.M.. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997; 276:589–592. [DOI] [PubMed] [Google Scholar]
- 16. Ye B.H., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R.S. et al.. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997; 16:161–170. [DOI] [PubMed] [Google Scholar]
- 17. Dominguez-Sola D., Kung J., Holmes A.B., Wells V.A., Mo T., Basso K., Dalla-Favera R.. The FOXO1 transcription factor instructs the germinal center dark zone program. Immunity. 2015; 43:1064–1074. [DOI] [PubMed] [Google Scholar]
- 18. Sander S., Chu V.T., Yasuda T., Franklin A., Graf R., Calado D.P., Li S., Imami K., Selbach M., Di Virgilio M. et al.. PI3 kinase and FOXO1 transcription factor activity differentially control B cells in the germinal center light and dark zones. Immunity. 2015; 43:1075–1086. [DOI] [PubMed] [Google Scholar]
- 19. Ahmad K.F., Melnick A., Lax S., Bouchard D., Liu J., Kiang C.L., Mayer S., Takahashi S., Licht J.D., Prive G.G.. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell. 2003; 12:1551–1564. [DOI] [PubMed] [Google Scholar]
- 20. Ghetu A.F., Corcoran C.M., Cerchietti L., Bardwell V.J., Melnick A., Prive G.G.. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell. 2008; 29:384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dhordain P., Albagli O., Lin R.J., Ansieau S., Quief S., Leutz A., Kerckaert J.P., Evans R.M., Leprince D.. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cardenas M.G., Yu W., Beguelin W., Teater M.R., Geng H., Goldstein R.L., Oswald E., Hatzi K., Yang S.N., Cohen J. et al.. Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J. Clin. Invest. 2016; 126:3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cerchietti L.C., Ghetu A.F., Zhu X., Da Silva G.F., Zhong S., Matthews M., Bunting K.L., Polo J.M., Fares C., Arrowsmith C.H. et al.. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010; 17:400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang C., Hatzi K., Melnick A.. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat. Immunol. 2013; 14:380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Summers A.R., Fischer M.A., Stengel K.R., Zhao Y., Kaiser J.F., Wells C.E., Hunt A., Bhaskara S., Luzwick J.W., Sampathi S. et al.. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J. Clin. Invest. 2013; 123:3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stengel K.R., Barnett K.R., Wang J., Liu Q., Hodges E., Hiebert S.W., Bhaskara S.. Deacetylase activity of histone deacetylase 3 is required for productive VDJ recombination and B-cell development. PNAS. 2017; 114:8608–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stengel K.R., Zhao Y., Klus N.J., Kaiser J.F., Gordy L.E., Joyce S., Hiebert S.W., Summers A.R.. Histone Deacetylase 3 is required for efficient T cell development. Mol. Cell. Biol. 2015; 35:3854–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsu F.C., Belmonte P.J., Constans M.M., Chen M.W., McWilliams D.C., Hiebert S.W., Shapiro V.S.. Histone deacetylase 3 is required for T cell maturation. J. Immunol. 2015; 195:1578–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Philips R.L., Chen M.W., McWilliams D.C., Belmonte P.J., Constans M.M., Shapiro V.S.. HDAC3 is required for the downregulation of RORgammat during thymocyte positive selection. J. Immunol. 2016; 197:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhaskara S., Chyla B.J., Amann J.M., Knutson S.K., Cortez D., Sun Z.W., Hiebert S.W.. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol. Cell. 2008; 30:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kotecha N., Krutzik P.O., Irish J.M.. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010; doi:10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhee H.S., Pugh B.F.. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr. Protoc. Mol. Biol. 2012; doi:10.1002/0471142727.mb2124s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwak H., Fuda N.J., Core L.J., Lis J.T.. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013; 339:950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang J., Zhao Y., Zhou X., Hiebert S.W., Liu Q., Shyr Y.. Nascent RNA sequencing analysis provides insights into enhancer-mediated gene regulation. BMC Genomics. 2018; 19:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin F., Kearney J.F.. Marginal-zone B cells. Nat. Rev. Immunol. 2002; 2:323–335. [DOI] [PubMed] [Google Scholar]
- 41. Cerutti A., Cols M., Puga I.. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol. 2013; 13:118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Turner C.A. Jr, Mack D.H., Davis M.M.. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994; 77:297–306. [DOI] [PubMed] [Google Scholar]
- 43. Lin Y., Wong K., Calame K.. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997; 276:596–599. [DOI] [PubMed] [Google Scholar]
- 44. Tanaka H., Muto A., Shima H., Katoh Y., Sax N., Tajima S., Brydun A., Ikura T., Yoshizawa N., Masai H. et al.. Epigenetic regulation of the Blimp-1 gene (Prdm1) in B cells involves Bach2 and histone deacetylase 3. J. Biol. Chem. 2016; 291:6316–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mochizuki K., Hayashi Y., Sekinaka T., Otsuka K., Ito-Matsuoka Y., Kobayashi H., Oki S., Takehara A., Kono T., Osumi N. et al.. Repression of somatic genes by selective recruitment of HDAC3 by BLIMP1 is essential for mouse primordial germ cell fate determination. Cell Rep. 2018; 24:2682–2693. [DOI] [PubMed] [Google Scholar]
- 46. Minnich M., Tagoh H., Bonelt P., Axelsson E., Fischer M., Cebolla B., Tarakhovsky A., Nutt S.L., Jaritz M., Busslinger M.. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 2016; 17:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T.. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000; 102:553–563. [DOI] [PubMed] [Google Scholar]
- 48. Stavnezer J., Guikema J.E., Schrader C.E.. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008; 26:261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu Z., Zan H., Pone E.J., Mai T., Casali P.. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 2012; 12:517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tunyaplin C., Shaffer A.L., Angelin-Duclos C.D., Yu X., Staudt L.M., Calame K.L.. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 2004; 173:1158–1165. [DOI] [PubMed] [Google Scholar]
- 51. Shapiro-Shelef M., Calame K.. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005; 5:230–242. [DOI] [PubMed] [Google Scholar]
- 52. Lin K.I., Angelin-Duclos C., Kuo T.C., Calame K.. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol. Cell Biol. 2002; 22:4771–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K. et al.. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002; 17:51–62. [DOI] [PubMed] [Google Scholar]
- 54. Wells C.E., Bhaskara S., Stengel K.R., Zhao Y., Sirbu B., Chagot B., Cortez D., Khabele D., Chazin W.J., Cooper A. et al.. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PLoS One. 2013; 8:e68915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Malvaez M., McQuown S.C., Rogge G.A., Astarabadi M., Jacques V., Carreiro S., Rusche J.R., Wood M.A.. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. PNAS. 2013; 110:2647–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leus N.G., van der Wouden P.E., van den Bosch T., Hooghiemstra W.T., Ourailidou M.E., Kistemaker L.E., Bischoff R., Gosens R., Haisma H.J., Dekker F.J.. HDAC 3-selective inhibitor RGFP966 demonstrates anti-inflammatory properties in RAW 264.7 macrophages and mouse precision-cut lung slices by attenuating NF-kappaB p65 transcriptional activity. Biochem. Pharmacol. 2016; 108:58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trinh D.L., Scott D.W., Morin R.D., Mendez-Lago M., An J., Jones S.J., Mungall A.J., Zhao Y., Schein J., Steidl C. et al.. Analysis of FOXO1 mutations in diffuse large B-cell lymphoma. Blood. 2013; 121:3666–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hah N., Murakami S., Nagari A., Danko C.G., Kraus W.L.. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013; 23:1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Henriques T., Scruggs B.S., Inouye M.O., Muse G.W., Williams L.H., Burkholder A.B., Lavender C.A., Fargo D.C., Adelman K.. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018; 32:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dorighi K.M., Swigut T., Henriques T., Bhanu N.V., Scruggs B.S., Nady N., Still C.D. 2nd, Garcia B.A., Adelman K., Wysocka J.. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 Monomethylation. Mol. Cell. 2017; 66:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guenther M.G., Barak O., Lazar M.A.. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell Biol. 2001; 21:6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Watson P.J., Fairall L., Santos G.M., Schwabe J.W.. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012; 481:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ranuncolo S.M., Wang L., Polo J.M., Dell’Oso T., Dierov J., Gaymes T.J., Rassool F., Carroll M., Melnick A.. BCL6-mediated attenuation of DNA damage sensing triggers growth arrest and senescence through a p53-dependent pathway in a cell context-dependent manner. J. Biol. Chem. 2008; 283:22565–22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ranuncolo S.M., Polo J.M., Dierov J., Singer M., Kuo T., Greally J., Green R., Carroll M., Melnick A.. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat. Immunol. 2007; 8:705–714. [DOI] [PubMed] [Google Scholar]
- 66. Cerchietti L.C., Yang S.N., Shaknovich R., Hatzi K., Polo J.M., Chadburn A., Dowdy S.F., Melnick A.. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009; 113:3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Inoue T., Shinnakasu R., Ise W., Kawai C., Egawa T., Kurosaki T.. The transcription factor Foxo1 controls germinal center B cell proliferation in response to T cell help. J. Exp. Med. 2017; 214:1181–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. MacLennan I.C., Toellner K.M., Cunningham A.F., Serre K., Sze D.M., Zuniga E., Cook M.C., Vinuesa C.G.. Extrafollicular antibody responses. Immunol. Rev. 2003; 194:8–18. [DOI] [PubMed] [Google Scholar]
- 69. Yu J., Angelin-Duclos C., Greenwood J., Liao J., Calame K.. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell Biol. 2000; 20:2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Haberman A.M., Gonzalez D.G., Wong P., Zhang T.T., Kerfoot S.M.. Germinal center B cell initiation, GC maturation, and the coevolution of its stromal cell niches. Immunol. Rev. 2019; 288:10–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rodda L.B., Bannard O., Ludewig B., Nagasawa T., Cyster J.G.. Phenotypic and morphological properties of germinal center dark zone Cxcl12-Expressing reticular cells. J. Immunol. 2015; 195:4781–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Allen C.D., Ansel K.M., Low C., Lesley R., Tamamura H., Fujii N., Cyster J.G.. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 2004; 5:943–952. [DOI] [PubMed] [Google Scholar]
- 73. Suvas S., Singh V., Sahdev S., Vohra H., Agrewala J.N.. Distinct role of CD80 and CD86 in the regulation of the activation of B cell and B cell lymphoma. J. Biol. Chem. 2002; 277:7766–7775. [DOI] [PubMed] [Google Scholar]
- 74. Krzyzak L., Seitz C., Urbat A., Hutzler S., Ostalecki C., Glasner J., Hiergeist A., Gessner A., Winkler T.H., Steinkasserer A. et al.. CD83 modulates B cell activation and germinal center responses. J. Immunol. 2016; 196:3581–3594. [DOI] [PubMed] [Google Scholar]
- 75. Caron G., Le Gallou S., Lamy T., Tarte K., Fest T.. CXCR4 expression functionally discriminates centroblasts versus centrocytes within human germinal center B cells. J. Immunol. 2009; 182:7595–7602. [DOI] [PubMed] [Google Scholar]
- 76. Shi W., Liao Y., Willis S.N., Taubenheim N., Inouye M., Tarlinton D.M., Smyth G.K., Hodgkin P.D., Nutt S.L., Corcoran L.M.. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 2015; 16:663–673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in GEO: GSE130502, GSE130516, GSE77113.