Abstract
The complex between Trm7 and Trm734 (Trm7–Trm734) from Saccharomyces cerevisiae catalyzes 2′-O-methylation at position 34 in tRNA. We report biochemical and structural studies of the Trm7–Trm734 complex. Purified recombinant Trm7–Trm734 preferentially methylates tRNAPhe transcript variants possessing two of three factors (Cm32, m1G37 and pyrimidine34). Therefore, tRNAPhe, tRNATrp and tRNALeu are specifically methylated by Trm7–Trm734. We have solved the crystal structures of the apo and S-adenosyl-L-methionine bound forms of Trm7–Trm734. Small angle X-ray scattering reveals that Trm7–Trm734 exists as a hetero-dimer in solution. Trm7 possesses a Rossmann-fold catalytic domain, while Trm734 consists of three WD40 β-propeller domains (termed BPA, BPB and BPC). BPA and BPC form a unique V-shaped cleft, which docks to Trm7. The C-terminal region of Trm7 is required for binding to Trm734. The D-arm of substrate tRNA is required for methylation by Trm7–Trm734. If the D-arm in tRNAPhe is docked onto the positively charged area of BPB in Trm734, the anticodon-loop is located near the catalytic pocket of Trm7. This model suggests that Trm734 is required for correct positioning of tRNA for methylation. Additionally, a point-mutation in Trm7, which is observed in FTSJ1 (human Trm7 ortholog) of nosyndromic X-linked intellectual disability patients, decreases the methylation activity.
INTRODUCTION
Transfer RNA is as an adapter molecule that interprets the codons in mRNA to corresponding amino acids. Primary tRNA transcripts must undergo maturation by processing, splicing and post-transcriptional modification for their biological function. Post-transcriptional modifications in tRNA are found in three domains of life (1), and contribute to structural integrity and stability of tRNA as well as decoding fidelity and efficiency in protein synthesis (2,3).
To date, more than 100 modified nucleosides have been found in tRNA (http://mods.rna.albany.edu/mods/) (1). Methylated nucleosides are frequently found in tRNA, and 2′-O-methylation of ribose is most abundant (4). Given that the methyl-group in 2′-O-methylated nucleoside shifts the equilibrium of ribose-puckering from the C2′-endo form to the C3′-endo form, 2′-O-methylation stabilizes a local structure in tRNA (5). The first position (position 34) of anticodon in tRNA is often modified to 2′-O-methylated nucleoside (6), and it is considered to reinforce the codon-anticodon interaction during protein synthesis. Furthermore, because 2′-O-methylation protects RNA from hydrolysis, the 2′-O-methylation in the anticodon-loop may prolong the half-life of tRNA.
S-adenosyl-L-methionine (SAM)-dependent methyltransferases are divided into more than five classes on the basis of the structure of their catalytic domain (4,7). Most methyltransferases have a Rossmann fold catalytic domain and are classified as class I enzymes. Members of SpoU-TrmD (SPOUT) RNA methyltransferase superfamily are classified as class IV enzymes (7,8). While 2′-O-methyation in anticodons is generally widespread, completely different types of tRNA methyltransferases function in the modification in the three domains of life. For example, in Escherichia coli, 2′-O-methylation at position 34 in the formation of 2′-O-methylcytidine (Cm34) and 5-carboxymethylaminomethyl-2′-O-methyluridine (cmnm5Um34) is catalyzed by TrmL (9), a homo-dimeric enzyme of the SPOUT RNA methyltransferase superfamily (10). In contrast, 2′-O-methylation in tRNATrp from Methanocaldococcus jannashii, a hyper-thermophilic archaeon, is mediated by a complex of multiple proteins and RNA (L7Ae, Nop5, archaeal fibrillarin and box C/D guide RNA system) (11), in which archaeal fibrillarin is a class I methyltransferase (12,13). In the case of Saccharomyces cerevisiae, a complex between Trm7 and Trm734 (Trm7–Trm734) catalyzes the 2′-O-methylation at position 34 (Nm34) in tRNA (14,15). The amino acid sequence of Trm7 strongly suggests that Trm7, the catalytic subunit, belongs to the class I enzyme superfamily (16). Thus, it is clear that methyltransferases responsible for the Nm34 modifications in tRNA differ among the three domains of life.
Analysis of the trm7 gene deletion strain of S. cerevisiae reveals that Trm7 is responsible for all Nm34 modifications [2′-O-methylguanosine (Gm34) in tRNAPhe, Cm34 in tRNATrp and 5-carbamoylmethyl-2′-O-methyluridine (ncm5Um34) in tRNALeu] (14,16). The modification site targeted by Trm7 switches dependent on its partner subunit: a complex of Trm7 and Trm732 (Trm7–Trm732) catalyzes 2′-O-methylation at position 32 in tRNA (14,16). Furthermore, it has been reported that the formation of wyosine at position 37 requires Gm34 in tRNAPhe (17). Moreover, during the course of study, it was reported that trm7-gene deletion mutant strains of S. cerevisiae and Schizosaccharomyces pombe caused constitutive activation of general amino acid control response (18). In addition, it has been reported that mutations in human FTSJ1 (ortholog of Trm7) cause non-syndromic X-linked intellectual disability (19).
However, there remain important questions. For example, why does Trm7–Trm734 act only on specific tRNAs? Furthermore, why is a partner subunit, Trm734, required for methylation on position 34 in tRNA? To address these issues, we report biochemical and structural studies of Trm7–Trm734.
MATERIALS AND METHODS
Materials and yeast strains
[Methyl-14C]-SAM (1.95 GBq/mmol) was purchased from PerkinElmer. Non-radioisotope-labeled SAM and sinefungin were obtained from Sigma. DNA oligomers were obtained from Thermo Fisher Scientific. T7 RNA polymerase was purchased from Toyobo. All other chemical reagents were of analytical grade.
Construction of expression vectors
Saccharomyces cerevisiae BY4741 cells were grown in YPD medium at 30°C for 48 h. The genomic DNA was extracted from the cells with a yeast genomic DNA extraction and purification high recovery kit (TAKARA, Japan). All the primers used for the construction of cloning and expression vectors of trm7, trm732 and trm734 genes are listed in Supplementary Table S1. The details of construction of Trm7–Trm734 and Trm7–Trm732 co-expression vectors are described in Supplementary File.
Expression of Trm7–Trm734 and Trm7–Trm732 in E. coli
Transformation of the E. coli BL21 (DE3) Rosetta 2 strain was performed with the plasmids. The transformats were cultivated in 100 ml of LB liquid medium containing 100 μg/ml of ampicillin at 37°C for 12 h. The cells were sub-cultured in 1 l of the same medium at 37°C. When the optical density reached ∼0.8, isopropyl-β-D-thiogalactopyranoside was added into the growth medium to a final concentration of 0.5 mM with subsequent cultivation at 20°C for 24 h. The cultured cells were harvested by centrifugation (4320 × g at 20°C for 20 min).
Purification of Trm7–Trm734 and Trm7–Trm732
Trm7–Trm734 and Trm7–Trm732 were purified using the same method as described below. 3.0 g of cells were suspended in buffer A [50 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 5 mM imidazole, 200 mM KCl and 5% glycerol] supplemented with Halt protease inhibitor single-use cocktail (Thermo Fisher Scientific), and then disrupted with an ultrasonic disruptor (model VCX-500, Sonics & Materials. Inc, USA). The supernatant was collected by centrifugation at 38 900 × g at 4°C for 20 min, and then loaded onto a Ni-NTA Super-flow column (QIAGEN) equilibrated with buffer A. After the unbound proteins were washed with buffer A, the bound proteins were eluted stepwise using buffer A containing 500 mM imidazole. The concentration of KCl in the eluted fraction was adjusted to final concentration of 100 mM by the addition of buffer C [50 mM Tris–HCl (pH 8.0), 5 mM 2-mercaptoethanol, 50mM KCl and 5% glycerol]. The diluted fraction was loaded onto a HiTrap Heparin-Sepharose column (GE Healthcare) pre-equilibrated with buffer C, and then the bound proteins eluted with a linear gradient from 50 mM to 1 M KCl in buffer C. The eluted fraction was concentrated, and then loaded onto a Superdex-200 gel-filtration column (GE Healthcare) equilibrated with buffer D [50 mM Tris–HCl (pH 8.0), 5 mM 2-mercaptoethanol, 400 mM KCl and 5% glycerol]. The eluted fractions were pooled, and then concentrated. The purity of Trm7–Trm734 and Trm7–Trm732 were confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of TrmD
Aquifex aeolicus TrmD, a tRNA m1G37 methyltransferase, was purified as reported in our previous study (20).
Preparation of tRNA transcripts
The wild-type and mutant tRNAPhe transcripts were prepared by in vitro T7 RNA polymerase transcription as reported previously (21) using the primers listed in Supplementary Table S2. The transcripts were purified by Q-Sepharose chromatography. Methylated tRNAPhe transcripts were prepared as follows. For Cm32 formation by Trm7–Trm732, 0.1 μM Trm7–Trm732, 8.5 μM transcript and 50 μM SAM in 200 μl of buffer E [50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 6 mM 2-mercaptoethanol, and 50 mM KCl] were incubated at 30°C for 2 h. For m1G37 formation, 1.25 μM TrmD, 8.5 μM transcript, 50 μM SAM in 200 μl of buffer E were incubated at 55°C for 2 h. The methylated tRNAs were recovered by phenol–chloroform extraction and ethanol precipitation.
tRNA methylation assay
The enzymatic activity of Trm7–Trm734 was measured by incorporation of 14C-methyl groups from [methyl-14C]-SAM into tRNA transcripts; 0.22 μM Trm7–Trm734, 11 μM transcript and 13 μM [methyl-14C]-SAM in 180 μl of buffer E were incubated for various times (0, 2, 5, 15, 30 and 60 min) at 30°C. An aliquot (20 μl) was then used for the filter assay. In order to detect Gm formation in tRNAPhe transcript by Trm7–Trm734, we employed two-dimensional thin-layer chromatography (22). The 14C-methylated tRNAPhe transcript was dissolved in 5 μl of 50 mM ammonium acetate (pH 5.0) and digested with 2.5 units of nuclease P1, and then 2 μl of standard nucleotides (0.05 A260 units each of pA, pG, pC and pU) was added. A total of 2 μl of the sample was spotted onto a thin layer plate (Merck code number 1.05565, cellulose F) and separated. Incorporation of 14C-methyl groups was monitored with a with a FLA-2000 (GE Healthcare) imaging analyzer. Standard nucleotides were marked by UV 260 nm irradiation. To visualize the methylation activity, we employed 10% PAGE (7 M urea) and an imaging analyzer system. Briefly, tRNA (0.2 A260 units) sample was incubated with 0.8 μM Trm7–Trm734 and 19.5 μM [methyl-14C]-SAM for 30 min at 30°C in 50 μl of buffer E, and then loaded onto a 10% polyacrylamide gel (7 M urea). The gel was stained with methylene blue and dried. The incorporation of 14C-methyl groups into tRNA was monitored with the imaging analyzer as described above.
Crystallization
A concentrated solution of ∼3 mg/ml of Trm7–Trm734 was used for crystallization using the hanging drop vapor diffusion technique. Initial crystallization screening was performed using Crystal screening kits 1 and 2, and VDX48 plates with sealant (Hampton Research). The drop solution was equilibrated against 200 μl of reservoir solution at 20°C. A few small single crystals appeared after 5 days in the drop in Crystal screening kit 1 #39. The crystallization conditions were further optimized by using Additive screening kit (Hampton Research). A single crystal with rectangular shape (size μm, 200 × 100 × 50) was obtained under the crystallization conditions [0.1 M HEPES-NaOH (pH 7.5), 2 M (NH4)2SO4, 2% PEG400, 2 mM phenol and 3% ethylene-glycol] at 20°C for 5 days. The crystal of Trm7–Trm734 in complex with SAM was prepared as follow. The crystal of Trm7–Trm734 was soaked in the mother liquor containing 1 mM SAM at 20°C for 1h. For the experimental phase determination using single-wavelength anomalous dispersion (SAD) method, the crystal was soaked in crystallization mother liquor supplemented with 0.2 mM K2OsO4 at 20°C for 1 h and then soaked in the mother liquor at 20°C for 1 h to remove unbound Os atoms. Cryo-protection was achieved by stepwise transfer to the respective artificial mother liquor containing 25% ethylene-glycol. The crystals were then flash-frozen in liquid nitrogen.
Structural determination
X-ray diffraction datasets from the crystals of apo-Trm7–Trm734 and Trm7–Trm734–SAM complex (λ = 1.0000) and SAD datasets from the Os-induced crystal (λ = 1.13986) were collected at 100 K on the BL26B1 and BL38B1 beamlines at SPring-8 (Hyogo, Japan). All datasets were processed, merged and scaled using the HKL2000 program (23). Using the deduced Os-SAD dataset, all 10 Os positions were identified and refined in the monoclinic space group C2, and the initial phase was calculated by using AutoSol in PHENIX (24), followed by automated model building using RESOLVE (25). The resulting map and partial model were used for manually building the model using COOT (26). The model was further refined by using PHENIX (20). Using the apo-Trm7–Trm734 and Trm7–Trm734–SAM complex datasets with the refined model as a search coordinate, the structures were determined by molecular replacement with the Phaser program (27). The models were further manually built with COOT (22) and refined with PHENIX (24). The structure of apo-Trm7–Trm734 was refined to Rwork/Rfree of 19.4%/23.9% at 2.70 Å resolution, and the structure of Trm7–Trm734–SAM complex was refined to Rwork/Rfree of 20.3%/23.5% at 2.32 Å resolution, respectively (Table 1). The space group of the crystals belonged to C2, where two Trm7–Trm734 molecules are present in an asymmetric unit and were almost identical one another. The final model of apo-Trm7–Trm734 contained residues [Trm7 (chain B and D), 9–212 and 233–259; Trm734 (chain A and C), 1–547, 555–758, 763–992, 997–1013], 310 water molecules, four SO42− molecules and two HEPES molecules, and the Trm7–Trm734–SAM complex contained residues [Trm7 (chain B and D), 9–212 and 233–259; Trm734 (chain A and C), 1–547, 555–758, 763–992, 997–1013], 511 water molecules, 2 SAM molecules, 10 SO42− molecules and 2 HEPES molecules. The final models were further checked using PROCHECK (28), showing the quality of the refined models. Ramachandran plots (%) of their structures are tabulated in Table 1. The structure factors and coordinates of apo-Trm7–Trm734 and Trm7–Trm734–SAM complex have been deposited in the Protein Data Bank (PDB codes: 6JP6 and 6JPL). All structural figures were generated by PyMOL (DeLano Scientific, Palo Alto, CA, USA).
Table 1.
Data collection and refinement statistics
| Trm7–Trm734 | Trm7–Trm734-Os-derivative | Trm7–Trm734–SAM | |
|---|---|---|---|
| Data Collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a (Å) | 253 643 | 258 179 | 253 558 |
| b (Å) | 110 351 | 109 479 | 110 671 |
| c (Å) | 133 503 | 131 164 | 133 559 |
| α, β, γ (°) | 90, 90.02, 90 | 90, 89.98, 90 | 90, 90.01, 90 |
| Resolution (Å) | 50–2.70 (2.75–2.70) | 50–3.20 (3.26–3.20) | 50–2.32 (2.36–2.32) |
| Completeness (%) | 99.9 (99.0) | 100 (100) | 99.9 (99.5) |
| Rmerge | 8.7 (46.5) | 12.6 (67.9) | 7.0 (68.7) |
| I/σIa | 22.2 (3.7) | 30.2 (4.1) | 29.7 (2.7) |
| Redundancy | 4.2 (4.2) | 7.6 (7.6) | 4.6 (4.2) |
| Refinement | |||
| Resolution (Å) | 36.43–2.70 | 47.42–2.32 | |
| No. Reflections | 100 973 | 159 317 | |
| No. of non-hydrogen atoms | 19 867 | 20 155 | |
| Protein atoms | 19 560 | 19 560 | |
| Water molecules | 257 | 461 | |
| No. ions | SO42− × 4, HEPES × 2 | SO42− × 10, HEPES × 2 | |
| No. Ligands | SAM × 2 | ||
| R work b | 19.4 | 20.32 | |
| R free c | 23.87 | 23.51 | |
| Bond (Å) | 0.01 | 0.01 | |
| Angle (°) | 1.23 | 1.25 | |
| Average B-factor (Å2) | 46.6 | 50.4 | |
| Ramachandran plot (%) | |||
| Most favored | 94.41 | 93.18 | |
| Additional allowed | 5.05 | 6.12 | |
| Generously allowed | 0.53 | 0.7 | |
| Disallowed | 0 | 0 |
The value in the parentheses is for the highest resolution shell.
a R merge = ΣΣj|<I(h)> – I(h)j|/ΣΣj|<I(h)>|, where <I(h)> is the mean intensity of symmetry-equivalent reflections.
b R work = Σ (IIFp(obs) – Fp(calc)II)/ΣIFp(obs)I.
c R free = R factor for a selected subset (5%) of reflections that was not included in earlier refinement calculations.
Small angle X-ray scattering (SAXS) data collection and processing
Small angle X-ray scattering (SAXS) data for apo Trm7–Trm734 and Trm7–Trm734 in complex with tRNAPhe and sinefungin were recorded at 20°C on the BL-10C and BL45XU beamlines at PF (Tsukuba, Japan) and SPring-8 (Hyogo, Japan), respectively (29). The details of SAXS measurement are described in the Supplementary File. The structures are submitted to small angle scattering biological data bank (SASBDB; https://www.sasbdb.org/aboutSASBDB/) (30). Deposited SASBDB ID of Trm7–Trm734 is SASDDR3.
Site-directed mutagenesis and purification of mutant proteins
Site-directed mutagenesis was performed using the QuichChange site directed mutagenesis kit (Stratagene) with the primers listed in Supplementary Table S3. The expression vectors for deletion mutants of Trm7 (Δ233-310 Trm7 and Δ260-310 Trm7) were constructed by reverse polymerase chain reaction with the two primers listed in Supplementary Table S3. These mutations were verified by DNA sequencing. All the mutants were expressed and purified using the same method for the wild-type Trm7–Trm734. The purity of mutants was confirmed by SDS-PAGE.
RESULTS
Requirements of Cm32, m1G37 and pyrimidine34 in tRNA for methylation by Trm7–Trm734
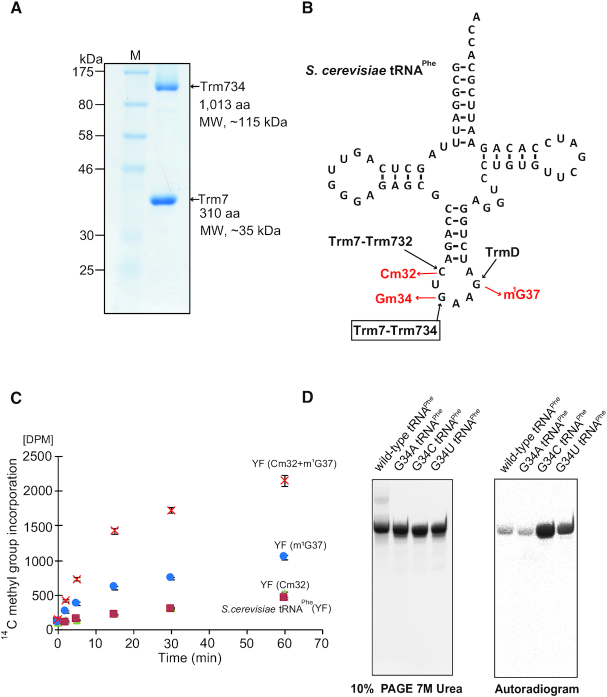
In this study, we have constructed E. coli expression systems for Trm7–Trm734 and Trm7–Trm732. Trm7–Trm734 and Trm7–Trm732 were successfully purified as shown in Figure 1A and Supplementary Figure S1.
Figure 1.
Characterization of the recombinant Trm7–Trm734 (A) SDS-PAGE (12.5%) of purified recombinant Trm7–Trm734. The gel was stained with Coomassie Brilliant Blue. (B) Cloverleaf structure of Saccharomyces cerevisiae tRNAPhe transcript. Three SAM-dependent tRNA methyltransferases, Trm7–Trm732, Trm7–Trm734 and TrmD, catalyze formation of Cm32, Gm34 and m1G37, respectively. (C) Time-dependent methyl transfer activity of Trm7–Trm734 using S. cerevisiae tRNAPhe transcript (YF, green triangles), YF with Cm32 (red squares), YF with m1G37 (cyan circles) and YF with both Cm32 and m1G37 (orange cross marks). Error-bars indicate the standard deviation calculated from the results of three independent experiments. (D) Methyl-group acceptance activities of mutant tRNAPhe transcripts were measured. G34 in the wild-type transcript was replaced by A, U or C. Left, the transcripts were analyzed by 10% PAGE (7 M urea). The gel was stained with methylene blue. Right, autoradiogram of the same gel.
Because Trm7, Trm732 and Trm734 are localized to cytoplasm in S. cerevisiae (31), the events through nuclear membrane are not required for the methylation by Trm7–Trm734. Furthermore, precursor tRNAPhe contains an intron, which is removed in cytoplasm (32–35). Moreover, precursor tRNAs are transported between nuclear and cytoplasm repeatedly during their maturation in S. cerevisiae (36–38). Thus, the 2′-O-methylations in the anticodon-loop of tRNAPhe are expected to be relatively late events in the maturation process (37). Therefore, we considered whether other modified nucleosides may be involved or required for the methylation by Trm7–Trm734. To investigate this idea, methylated tRNAPhe transcripts were prepared (Figure 1B). When the Cm32 modification was introduced into tRNAPhe transcript by Trm7–Trm732, the methyl-group acceptance activity was not increased (Figure 1C). In contrast, when m1G37 was introduced, the methyl-group acceptance activity clearly increased. Furthermore, when both the Cm32 and m1G37 modifications were introduced, the initial velocity of methyl-transfer reaction by Trm7–Trm734 was very fast (Figure 1C). Thus, the full activity of Trm7–Trm734 requires other modifications (Cm32 and m1G37) in tRNA. The formation of Gm in the tRNA transcript by Trm7–Trm734 was confirmed as shown in Supplementary Figure S2A and B. In addition, we substituted the guanine base at position 34 in tRNAPhe transcript for A, C and U (Figure 1D). Although these mutant tRNAPhe transcripts were methylated by Trm7–Trm734, the substitution of G34 by C or U (G34C or G34U) strongly enhanced the methyl-group acceptance activity (Figure 1D). Thus, Trm7–Trm734 preferentially methylates pyrimidine nucleoside at position 34 (pyrimidine34) rather than purine nucleoside. These results answer the question, ‘Why does Trm7–Trm734 act only on specific tRNAs (tRNAPhe, tRNATrp and tRNALeu)?’ Of the 43 tRNA molecular species in S. cerevisiae, only tRNAPhe, tRNATrp and tRNALeu possess the combination of Cm32, m1G37 and/or pyrimidine34 in the normal anticodon-loop, which is required for the effective methylation by Trm7–Trm734.
Overall structure
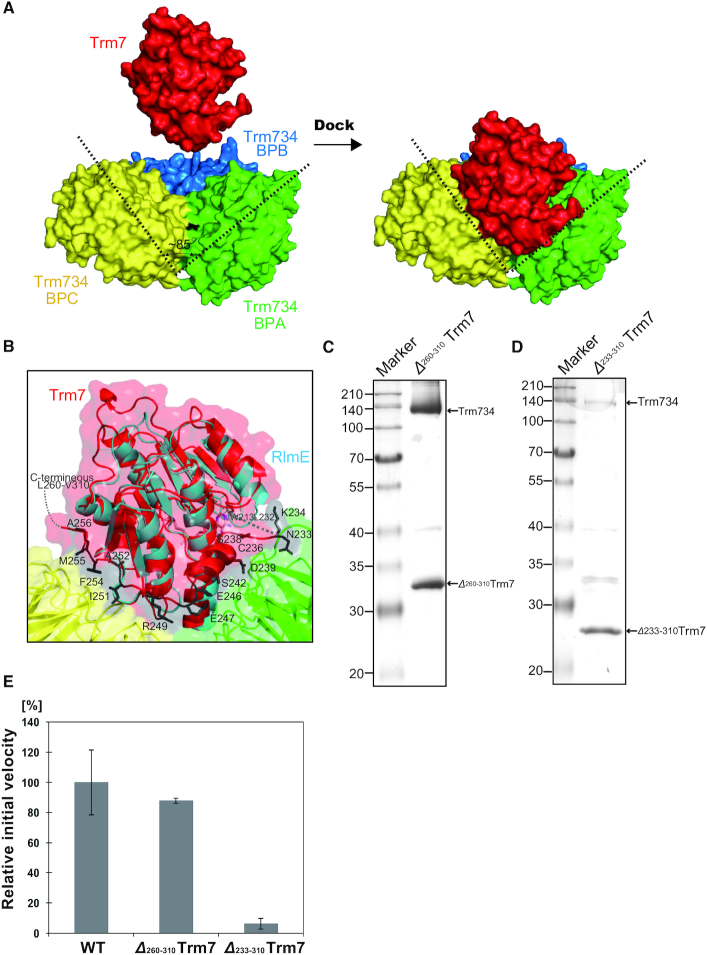
We determined the X-ray structure of Trm7–Trm734 at 2.70 Å resolution (Figure 2A). Trm7 forms a heterodimer with Trm734 with an overall size of ∼100 × 60 × 80 Å3. Furthermore, the X-ray structure of Trm7–Trm734 in complex with SAM was determined at 2.32 Å resolution and shown to be almost identical to the structure of apo state (Supplementary Figure S3A). Two heterodimers are found in asymmetric units of the crystal which belongs to the monoclinic space group C2. One HEPES and five SO42− molecules from the reagents used for crystallization are bound to each heterodimer (Supplementary Figure S3B). A structural model of two C-terminal regions of Trm7, W213-L232 and L260-V310, could not be built as these regions were not visible in the electron density (Figure 2B). The C-terminal region of L260-V310 was not degraded by contaminating proteases during the crystallization because we confirmed by SDS-PAGE analysis that Trm7 in the crystals retained its full length (data not shown). Therefore, the C-terminal region of Trm7 is disordered and likely to reside in the large space (Supplementary Figure S4). Trm734 consists of three WD-40 β-propeller domains (hereafter abbreviated as BPA, BPB and BPC), and each domain is connected by three linker regions (Figure 2A and B; black). Only the BPA and BPC domains directly interact with Trm7. A detailed description of the bipartite interaction between Trm7 and Trm734 is provided later. We next performed SAXS analysis to confirm the Trm7–Trm734 heterodimeric complex structure in solution. As shown in Figure 2C, the structural model of Trm7–Trm734 with randomized loops (L260-V310) in the C-terminal Trm7 is superimposed well onto the envelope shape of the solution model generated from SAXS analysis, with good χ2 and normalized spatial discrepancy (NSD) values (2.18 and 1.92, Supplementary Figure S5 and Table S4). Thus, these results confirm the heterodimer of the Trm7–Trm734 complex structure in solution as well as in the crystal and suggest the presence of a disordered C-terminal region (L260-V310) of Trm7.
Figure 2.
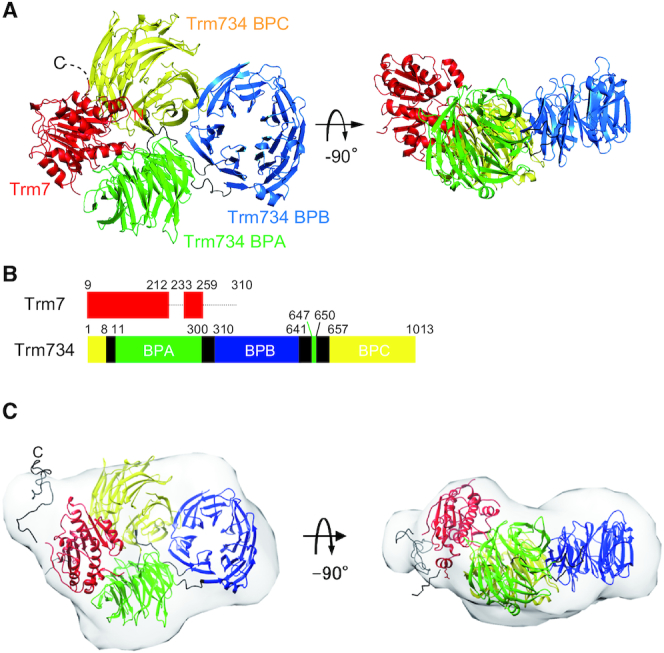
Structure of Trm7–Trm734. (A) Ribbon diagram of the X-ray structure of Trm7–Trm734. Trm7 is colored red. The three domains of Trm734 (BPA, BPB and BPC) and linker regions are colored green, blue, yellow and black, respectively. The structures on the right are rotated through −90° along the horizontal axis. (B) Schematic representation of Trm7 and Trm734 with domain boundaries. The dotted lines show the regions of Trm7 which are invisible. (C) Overlay of the envelope shape of Trm7–Trm734 calculated with DAMMIN using the SAXS data and the CORAL model of Trm7–Trm734 based on its X-ray structure.
Figure 3A shows the structure of Trm7 in the Trm7–Trm734 complex. Trm7 is composed of one 310 helix, seven α helices and seven β strands (Supplementary Figure S6). All the α helices surround one parallel/antiparallel mixed β-sheet that is formed by the seven β strands (Supplementary Figure S6A), indicating that the structural motif of Trm7 is a Rossmann fold, as found in the class I methyltransferases. The structure of S. cerevisiae Trm7 is considered similar to that of S. pombe Trm7 and Homo sapiens FTSJ1, given that there is a striking similarity of amino acid residues (Supplementary Figure S7). In the Trm734 structure (Figure 3B), BPA, BPB and BPC are WD40 domains consisting of a seven-bladed β-propeller fold (numbered from 1 to 7 in Supplementary Figure S6B) with each of the WD40 blades being composed of a four-stranded anti-parallel β-sheet (hereafter referred to as a, b, c and d, from the innermost to the outermost strands, Supplementary Figure S6B). The N-terminus of Trm734 starts from the β strand (d7) of BPC and transfers to the β strand (a1) of BPA via one linker. After the formation of six WD40 blades in BPA, the β strand (c7) of BPA also transfers to the β strand (d1) of BPB via one linker with BPA being incomplete because of one missing β strand (d7) required for the formation of the seventh WD40 blade. The β strand (c1) of complete BPB transfers to the β strand (d7) of BPA, thereby completing the formation of BPA. At last, d7 in BPA transfers to the β strand (a1) of BPC via one linker and BPC is formed. In the WD40 blades, insertions of the following secondary structure elements were found: two α helices (α1 and α2) are inserted between the two β strands [c2-d2 (BPA) and d1-a2 (BPB)], and four 310 helices are also inserted between two β strands [d2-a3 (BPB), a3-b3 (BPB), a1-b1 (BPC) and c4-d4 (BPC)]. Three β strands (β1-β3) are further inserted between two β strands [c4-d4 (BPC)]. As shown in Supplementary Figure S8, there is little amino acid sequence homology between S. cerevisiae Trm734, S. pombe Trm734 and human WD8 (a homolog of Trm734) other than His-Gly (HG) and Trp-Asp (WD) sequences, which are conserved in the WD40 repeats (39). The structures of BPA and BPC are the most similar of the three domains of Trm734, while the structure of BPB differs slightly from that of BPA and BPC (Supplementary Figure S9). A search for structural homology to Trm734 with the Dali server (40) has suggested that BPA and BPC have structural homology with the WD40 repeat-containing protein 61 involved in human transcription (i.e. the structure with the highest Z-score: 2.2 Å r.m.s.d for 268 aligned residues; Protein Data Bank (PDB) code: 3OW8-A), and that BPB is homologous to S. cerevisiae Sec12 which has the fold of the WD40 repeat and which is a guanine nucleotide exchange factor responsible for activating the small G protein (i.e. the structure with the highest Z-score: 3.3 Å r.m.s.d for 278 aligned residues; PDB code: 4H5J-B). Therefore, Trm734 consists of three domains of classical WD40 domains conserved in other proteins, especially those that interact with nucleic acids.
Figure 3.
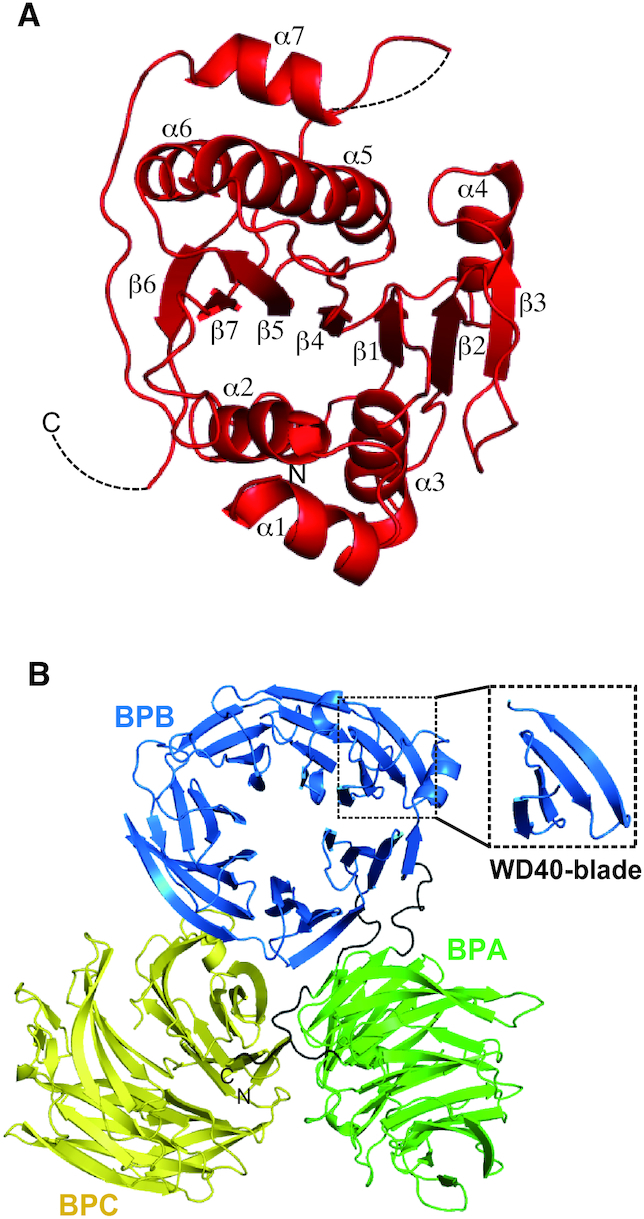
Structures of Trm7 and Trm734 subunits in the Trm7–Trm734 complex. (A) Ribbon diagram of the Trm7 structure. (B) Ribbon diagram of Trm734. Three WD40 domains—BPA, BPB and BPC—are colored, green, blue and yellow, respectively. The linker regions are colored black. Close-up view enclosed in a dotted square indicates one WD40 blade formed by four antiparallel β strands.
Active site of Trm7
In the structure of Trm7–Trm734 in complex with SAM (Figure 4A and Supplementary Figure S3), SAM is bound to the pocket of Trm7. As shown in Figure 4B and Supplementary Figure S10A, 14 residues form the SAM binding pocket: 12 residues (S25, C51, A52, A53, P54, S56, L84, A98, D99, I100, G125 and A126) bind to SAM by hydrophobic interactions and van der Waals contacts, and two residues (D83 and D124) form three hydrogen bonds with SAM (D83 Oδ1-SAM ribose O2′, D83 Oδ2-SAM ribose O3′ and D124 Oδ1-SAM carboxyl OXT). All these residues are conserved in Trm7 and RlmE proteins (Supplementary Figure S7). RlmE is a eubacterial 23S rRNA methyltransferase, which catalyzes 2′-O-methylation of U2552 of the 23S rRNA (41–43). The structural characteristics of these residues are well conserved around the SAM binding pocket (Supplementary Figure S10B). The overall structure of Trm7 is very similar to that of the E. coli RlmE (44). RlmE is reported to use four catalytic residues (K38, D124, K164 and E199; the number of each amino acid residue is based on the sequence of E. coli RlmE.) for the transfer of the methyl group from SAM (44). These resides of E. coli RlmE are conserved in Trm7 proteins (Supplementary Figure S7). Furthermore, there is a striking conformational similarity of the four residues and SAM between S. cerevisiae Trm7 and E. coli RlmE (Supplementary Figure S10C). Therefore, these observations strongly suggest that the methylation mechanism of Trm7 is very similar to that of RlmE. However, the long C-terminal region is distinct in Trm7 and FTSJ1 proteins (Supplementary Figure S7).
Figure 4.
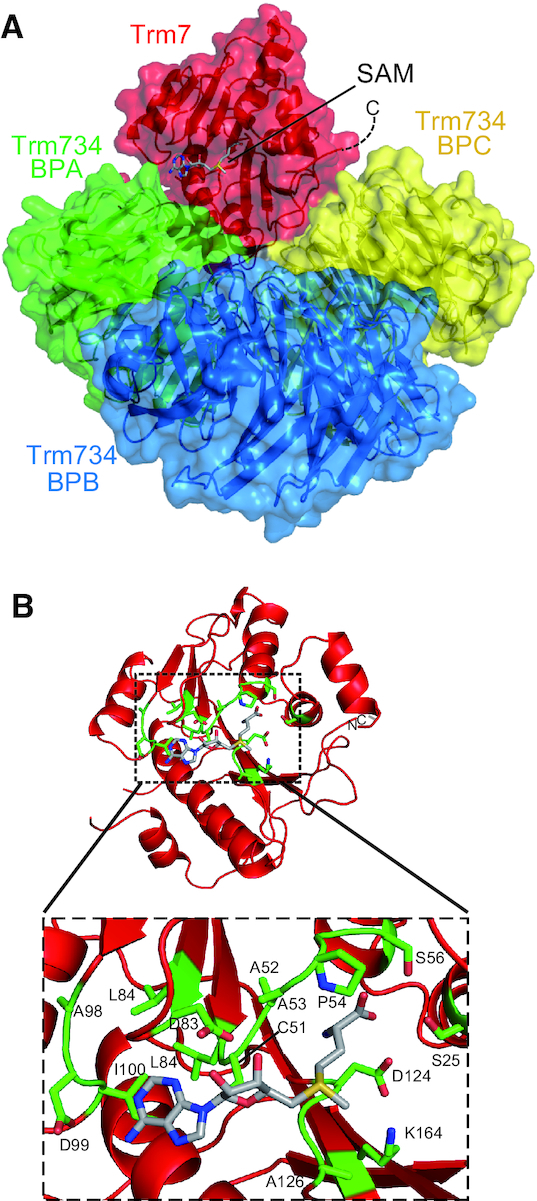
SAM bound form of Trm7–Trm734. (A) Ribbon and surface schematic of the overall structure of Trm7–Trm734 in complex with SAM. Trm7, and BPA, BPB and BPC of Trm734 are colored red, and green, blue, and yellow, respectively. SAM is depicted as a stick model. (B) SAM binding pocket in Trm7. Close-up view of SAM binding pocket is shown in a dotted square. The amino acid residues responsible for the interactions with SAM are highlighted as stick models (green).
Bipartite interaction between Trm7 and Trm734
Trm7 directly interacts with the two WD40 domains, BPA and BPC, in Trm734. In the interaction between Trm7 and BPA, seven hydrogen bonds and three salt bridges play a major role (Supplementary Figure S11A and Table 2A). In contrast, seven hydrogen bonds and one salt bridge are observed in the interaction between Trm7 and BPC (Supplementary Figure S11B and Table 2B). In addition to these hydrogen bonds and salt brides, many residues play a significant role in hydrophobic interaction and van der Waals contacts (Supplementary Tables S5 and 6). The buried surface area (BSA) of the interface between Trm7 and Trm734 is estimated to be 1728.7 Å2 in Trm7 (45), whereas the estimated BSA values of Trm7 for BPA and BPC have been found to be 994.4 and 734.3 Å2, respectively. As shown in Figure 5A, BPA and BPC cooperatively form a unique V-shaped cleft, which has not been reported in other WD40 proteins. The cleft is ∼85° wide and is adjusted for docking to Trm7. Most residues of BPA and BPC involved in the interaction with Trm7 are not conserved in other Trm734 homologs (Supplementary Figure S8). Figure 5B shows the superimposition of the RlmE structure onto Trm7 in complex with Trm734 with the conformation of RlmE being well superimposed with that of Trm7 with the exception of the C-terminal region of Trm7 (N233-A256) absent in RlmE. Most of the residues in the C-terminal region of Trm7 interact with BPA and BPC in Trm734, with an approximate BSA of 374.7 Å2 (21.7%, the estimated occupancy for the total BSA of Trm7), indicating that the C-terminal region of Trm7 significantly contributes to the interaction between Trm7 and Trm734. Thus, the C-terminal region (N233-A256) of Trm7 might determine the specificity of binding to Trm734. The role of the disordered region (L260-V310) in Trm7 remains unclear because the region is invisible.
Table 2.
Hydrogen bonds and salt bridges between Trm7 and Trm734
| A. Hydrogen bonds and salt bridges between Trm7 and Trm734 BPA. | ||
|---|---|---|
| Trm7 | Trm734 BPA | length (Å) |
| Q142 NE2 | Y112 OH | 3.0 |
| R169 NH1 | N48 OD1 | 3.2 |
| R169 NH2 | N48 OD1 | 3.2 |
| V129 O | R223 NH1 | 3.0 |
| D134 OD2 | H51 NE2 | 2.7 |
| N233 N | G132 O | 3.4 |
| N233 OD1 | K114 NZ | 2.5 |
| C236 SG | R134 NH1 | 2.8 |
| E246 OE2 | R67 NH2 | 3.4 |
| E247 OE1 | R67 NH2 | 2.5 |
| B. Hydrogen bonds and salt bridges between Trm7 and Trm734 BPC. | ||
| Trm7 | Trm734 BPC | length (Å) |
| R167 NH1 | T938 OG1 | 3.1 |
| R167 NH1 | D906 OD1 | 3.6 |
| R167 NH2 | N908 OD1 | 2.9 |
| R167 NH2 | D906 O | 2.9 |
| R167 O | S937 OG | 2.6 |
| D172 OD2 | S936 OG | 3.3 |
| T196 OG1 | R658 NH2 | 3.2 |
| Y254 O | Q882 NE2 | 2.7 |
Figure 5.
The V-shaped cleft of Trm734 docks to Trm7. (A) Docking of Trm7 onto Trm734 through the V-shaped cleft. The V-shaped cleft is opened to 85° (dotted line). (B) Close-up view of the ribbon diagram of the interaction of Trm7 (red) with BPA (green) and BPC (yellow) of Trm734. The stick models (dark-gray) show the C-terminal region (N233-A256) of Trm7 essential for the interaction between Trm7 and Trm734. The structure of Escherichia coli RlmE (cyan) is superimposed onto that of Trm7 (red). (C) 12% SDS-PAGE of purified Δ260–310 Trm7 mutant. The gel was stained with Coomassie Brilliant Blue. (D) About 12% SDS-PAGE of purified Δ233–310 Trm7 mutant. (E) Relative methyl-transfer activities of the wild-type Trm7–Trm734 (WT) and deletion mutants (Δ233–310 Trm7 and Δ260–310 Trm7). The initial velocity of the WT for tRNAPhe transcript is expressed as 100.0%.
To examine the role of C-terminal region, we performed mutational analysis using two mutants (Δ233-310 Trm7 and Δ260-310 Trm7), in which the two C-terminal regions (N233-V310 and L260-V310, respectively) were deleted. As shown in Figure 5C, the complex of Δ260-310 Trm7 and Trm734 could be purified. In contrast, only a small amount of the complex between Δ233-310 Trm7 and Trm734 could be purified by Ni-NTA column chromatography (Figure 5D). Instead, the majority of purified Δ233-310 Trm7 was obtained in the Trm734-unbound form. As shown in Figure 5E, the complex of Δ260-310 Trm7 and Trm734 retained methyl-transfer activity. In contrast, the activity of Δ233-310 Trm7 and Trm734 fraction was barely detectable. Thus, these results suggest that the C-terminal region (N260-V310) is not required for the methylation activity of Trm7–Trm734, and that another region (N233-A256) is important for the interaction between Trm7 and Trm734. Thus, it is clear that overall the C-terminal region of Trm7 is required for the interaction with Trm734.
Substrate tRNA recognition by Trm7–Trm734
Prior to this study, the intriguing question of how substrate tRNA bound to Trm7–Trm734 remained. To determine the tRNA-binding mode of Trm7–Trm734, we created an electrostatic potential surface using the Trm7–Trm734 structure (Figure 6). As shown in Figure 6A, many positively charged residues are distributed on the surface of BPB in Trm734 and are thus considered to be the recognition sites of phosphate groups in the tRNA substrate. To date, only one structure of complex between WD40 repeat protein and nucleic acid, namely the DNA bound form of human DDB1 and DDB2 complex, has been reported (46,47). The human DDB1 and DDB2 complex can bind to DNA and DDB1 is composed of three WD40 domains (Supplementary Figure S12A and B). However, the interaction between Trm7–Trm734 and tRNA could not be predicted from this structure because the conformational location of each WD40 domain in DDB1 is different from that in Trm734 (Supplementary Figure S12B) and the WD40 domains in DDB1 do not contact the DNA (Supplementary Figure S12A).
Figure 6.
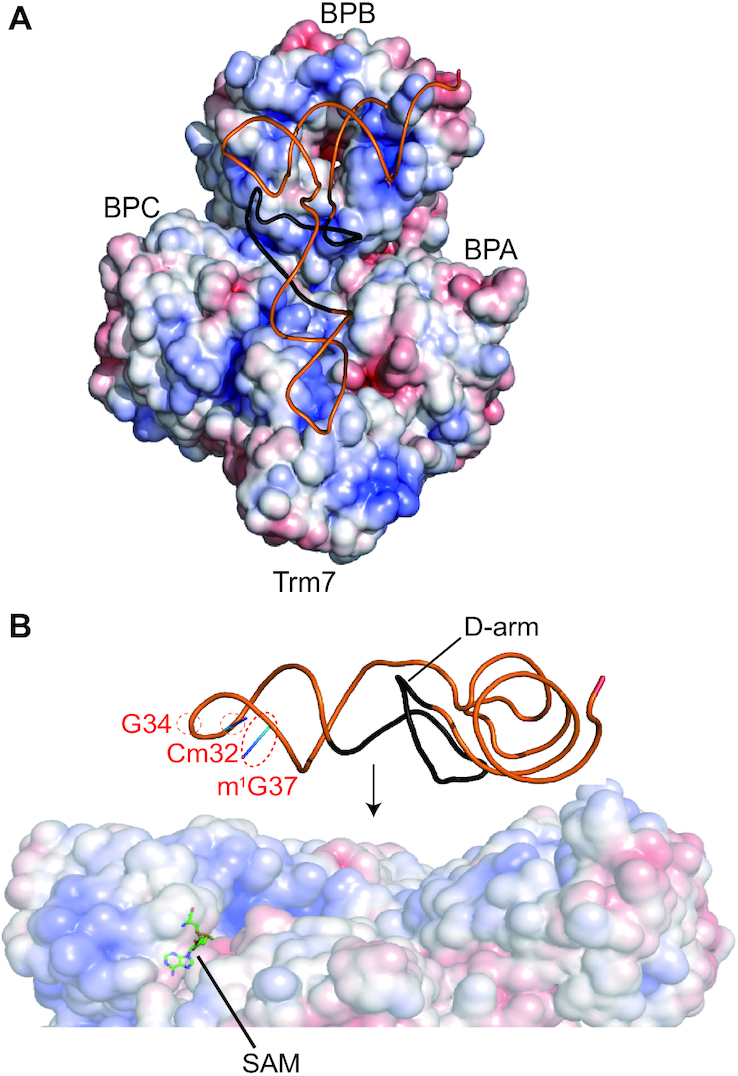
Search for the RNA-binding mode of Trm7–Trm734. (A) Surface model of Trm7–Trm734 colored according to the electrostatic potential represented as a gradient from negative (red) to positive (blue). Saccharomyces cerevisiae tRNAPhe is placed along the distribution of positively charged surface of Trm7–Trm734–SAM. A line diagram of S. cerevisiae tRNAPhe is colored orange. The D-arm of S. cerevisiae tRNAPhe is colored black. (B) Close-up view of the placement in panel A. The D-arm of S. cerevisiae tRNAPhe is colored black. Cm32, G34, m1G37 and SAM are indicated by the red dotted circles and black line with labels, respectively.
To examine what parts of the tRNA are responsible for recognition by Trm7–Trm734, we measured the methyl group acceptance activity of four truncated mutant transcripts of S. cerevisiae tRNAPhe, in which the D-arm, T-arm, anticodon-arm and aminoacyl-stem were individually deleted (Supplementary Figure S13A). Methyl group acceptance activity was clearly detected for the wild-type and the Δ T-arm and Δ aminoacyl-stem mutants, but could not be observed in the Δ D-arm and Δ anticodon-arm mutants (Supplementary Figure S13B). These results demonstrate the importance of D-arm of S. cerevisiae tRNAPhe for recognition by Trm7–Trm734. On the basis of these results, we manually placed the structure of S. cerevisiae tRNAPhe on Trm7–Trm734 with SAM (Figure 6). The L-shaped structure of tRNA is positioned along the distribution of positively charged residues in Trm7–Trm734. In this model, BPB in Trm734 can be placed near the aminoacyl-stem and D-arm of S. cerevisiae tRNAPhe, and Trm7 can be placed making contact with the anticodon loop of tRNA. BPC of Trm734 seems to contact with a part of anticodon loop. As shown in Figure 6B, the methylation site (G34) and positive determinants (Cm32, m1G37 and D-arm) for Trm7–Trm734 are able to be placed on the same side of the tRNA. In this model, Trm734 regulates the angle and distance from the aminoacyl-stem and D-arm to the catalytic pocket in Trm7, which captures the ribose of G34 of the tRNA. Thus, this model suggests that Trm734 is required for the positioning of tRNA.
However, a docking model could not be built without clashes between the L-shaped tRNA and Trm7–Trm734, suggesting that the structures of tRNA and Trm7–Trm734 change for the methyl-transfer reaction to proceed. The methyl-transfer reactions mediated by tRNA methyltransferases often require structural changes of both the tRNA and enzyme itself (induced-fit process) (48–51). To investigate how the Trm7–Trm734 changes structurally upon tRNA binding, we performed SAXS analysis of Trm7–Trm734 in complex with sinefungin and tRNA. Compared to the envelope shape of apo Trm7–Trm734, the structure of the complex of Trm7–Trm734–tRNA is significantly changed, showing a shifted radius of gyration (Rg) of 40.98 ± 0.11 Å and maximum size parameter (Dmax) of 155 Å in P(r) analysis (Supplementary Table S4). These values are a remarkable increase relative to those of the SAXS envelop of Trm7–Trm734 which has values for Rg of 37.67 ± 0.11 Å and Dmax of 130 Å (Supplementary Table S4). Therefore, these findings suggest that tRNA binding induces a conformational change of Trm7–Trm734, although the precise tRNA binding mode remains unclear because the structural model of tRNA in Trm7–Trm734–tRNA complex is not fitted to the SAXS envelop.
NSXLID
Saccharomyces cerevisiae Trm7 is highly homologous to human FTSJ1, with a sequence identity of 49%, and A26 and its adjacent sequences in both enzymes are identical (Supplementary Figure S7). In FTSJ1, the replacement of A26 with proline causes NSXLID in humans (19). As shown in Figure 7A, A26 of S. cerevisiae Trm7 is in the second position of the α helix (α2). Furthermore, A26 is adjacent to the catalytic residue K28 in E. coli RlmE. We prepared a recombinant A26P mutant, in which A26 in Trm7 was replaced with proline. We purified the A26P mutant in complex with Trm734 as judged by the band intensities of proteins by SDS-PAGE analysis (Figure 7B). This indicates that the A26P mutation does not affect the interaction with Trm734. We further found that the activity of the A26P mutant was significantly decreased compared with the wild-type (Figure 7C). Therefore, these results suggest that the A26P mutant might prevent the proper folding of the α2 helix, in which one of the important residues for the catalytic activity, K28 is sited. It has been reported that tRNAPhe molecules from a NSXLID patient with FTSJ1 A26P mutation and a S. cerevisiae Trm7 A26P mutant strain lack the Gm34 modification (19). Our in vitro experiments using the purified Trm7-A26P and Trm734 complex support these in vivo observations and show that the lack of Gm34 in tRNAs from human and S. cerevisiae mutant cells is explainable without the need to consider the presence of other protein factors.
Figure 7.
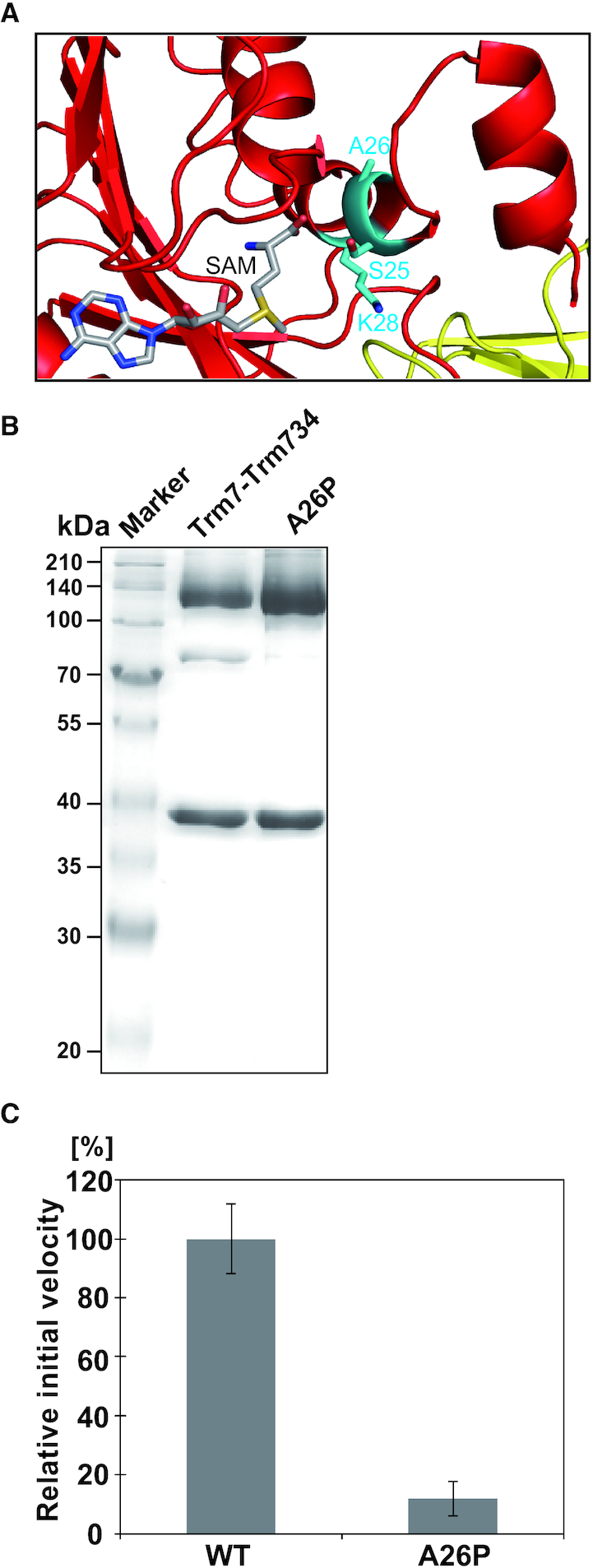
Activity of the A26P mutant. (A) Close-up view of the peripheral structure of α helix (α2) and SAM in Trm7. The residues, S25 A26 and K28 are shown as stick models (cyan). SAM is illustrated as a stick model. (B) About 12% SDS-PAGE of purified A26P mutant. The gel was stained with Coomassie Brilliant Blue. (C) Relative methyl-transfer activities of the wild-type Trm7–Trm734 (WT) and A26P mutant. The initial velocity of the WT for tRNAPhe transcript is expressed as 100.0%.
DISCUSSION
For a long time, it has been an enigma that of 43 tRNA molecular species in S. cerevisiae, specific tRNAs, namely tRNAPhe, tRNATrp and tRNALeu contain 2′-O-methylations at position 34. Our biochemical study reveals that the responsible tRNA methyltransferase, Trm7–Trm734, requires Cm32 and m1G37 modifications for efficient activity toward the tRNAPhe transcript and that Trm7–Trm734 preferentially acts on pyrimidine34. In the case of tRNAPhe, although position 34 is G, Cm32 and m1G37 modifications are present. In the case of tRNATrp, although position37 is not m1G but rather is an A, Cm32 and pyrimidine34 are present. In the case of tRNALeu, all three factors, Cm32, pyrimidine34 and m1G37 are present. Therefore, these tRNAs can act as substrates for Trm7–Trm734.
Of the other tRNAs in S. cerevisiae, two tRNAPro molecular species possess pyrimidine34 and m1G37 but not Cm32 and 2′-O-methylation at position 34 is not found in these tRNAs. Instead of Cm32 and purine38, these tRNAPro contain the Ψ32 and Ψ38 modifications where the Ψ32 and Ψ38 modifications are conferred by Pus8 (52) and Pus3 (53), respectively. A base pair between Ψ32 and Ψ38 is probably formed in tRNAPro and enhances stacking effect with the anticodon-stem (2,54,55). The structure of ribose-phosphate backbone of anticodon-loop in these tRNAPro seems to be different to the other tRNAs. This different ribose-phosphate backbone structure of the anticodon-loop in tRNAPro may disturb the introduction of ribose at position 34 into the catalytic pocket or change the distance and angle of anticodon-loop from the D-arm, which are important for the positioning of tRNA.
The degree of some of the modifications in tRNAs from S. cerevisiae may be regulated by the balance between substrate tRNAs and modification enzymes. During the course of this study, it was reported that over-expression of Trm10 [tRNA m1G9 methyltransferase (56)] causes the methylation of tRNAs, which are not methylated by Trm10 in S. cerevisiae wild-type cells (57). Because Trm7–Trm734 can act on unmodified tRNAPhe transcript slowly, over-expression of Trm7–Trm734 in S. cerevisiae cells may cause incorrect methylation of tRNAs, which do not contain Cm32, pyrimidine34 and/or m1G37. However, even in such a situation, the distance and angle of anticodon-loop from the D-arm seem to be important for the methylation by Trm7–Trm734. Furthermore, because one of key modifications, Cm32, is conferred by Trm7–Trm732, it is clear that the activity of Trm7–Trm734 is regulated by the quantitative balance between Trm732 and Trm734.
Trm7 has the class I-type Rossmann fold, frequently found in SAM-dependent RNA methyltransferases, and probably employs a 2′-O-methylation mechanism similar to that of E. coli RlmE, given that their overall structures are similar and that catalytic residues and SAM-binding mode are highly conserved. Hence, these findings suggest that both enzymes share a common ancestor. In the interaction between Trm7 and Trm734, Trm7 docked to the unique V-shaped cleft formed by BPA and BPC in Trm734. The C-terminal region of Trm7, which is lacking in E. coli RlmE, is a major contributor to the interaction with the cleft of Trm734. Thus it is likely that Trm7 acquired the C-terminal region (N233-A256) in the evolutionary process, to provide the ability to interact with Trm734 to allow strict recognition of the 2′-O-methylation site at position 34 in specific tRNA species. In contrast, there was no significant difference in the methylation activity between the wild-type Trm7 and Δ260–310 Trm7 mutant.
Trm734 is comprised by three WD40 repeat domains (BPA, BPB and BPC), and the V-shaped cleft formed by BPA and BPC in Trm734 captures Trm7. Trm734 is reported to be involved in other functions (58,59), but Trm734 structural information alone makes it difficult to speculate what parts of Trm734 are involved in these additional functions. Besides Trm734, Trm82 [a partner subunit in the Trm8 and Trm82 complex (Trm8–Trm82); tRNA m7G46 methyltransferase from S. cerevisiae (60,61)] is known to consist of one WD40 domain and to form a heterodimer with the catalytic domain of Trm8, which belongs to the class I-type SAM-dependent RNA methyltransferases (62). The interaction of Trm8 with Trm82 rearranges the N-terminal conformation of Trm8. Furthermore, Trm82 is expected to be the RNA-binding site. However, the substrate tRNA recognition mechanism of Trm8–Trm82 differs completely from that of Trm7–Trm734: the interaction between the D-arm and T-arm of tRNA is an essential element for recognition by Trm8–Trm82 (63) whereas this is not the case for Trm7–Trm734. WD40 domains are also present in other RNA-binding proteins. For example, Germin5, which is responsible for the biosynthesis of snRNA and can directly bind to snRNA, contains two WD40 domains (64,65). Although our model strongly suggests that Trm734 is required for the positioning of tRNA onto Trm7–Trm734, the L-shaped tRNA cannot be simply placed on the surface of Trm7–Trm734. Thus, dynamic conformational changes of Trm7–Trm734 and tRNAs are likely to be necessary for tRNA recognition and 2′-O-methylation. To address these issues, the structural determination of Trm7–Trm734 in complex with tRNAPhe in future studies is needed.
Although Trm732 has been predicted to contain a DUF2428 domain and Armadillo repeats based on the amino acid sequence (14), the precise structure of Trm732 and the interaction between Trm7 and Trm732 are unknown.
Over the past decades, defects in the modification of human tRNA species have been reported to cause certain diseases (66–68). Indeed, a recent study has shown that a mutation in WDR4 (the human Trm82 homolog) impairs the formation of the m7G46 modification resulting a distinct form of microcephalic primordial dwarfism (69). Furthermore, on the basis of the structural information from S. cerevisiae Trm8–Trm82 (62), the mutated residue in WDR4 is expected to play a significant role in interaction with the catalytic subunit. Most of the structures of tRNA modification enzymes involved in human disease are not yet determined. Herein, our structure-guided mutagenesis led us to suggest that the A26P mutation of human FTSJ1 may induce the conformational change in the α2-helix. We present a novel bipartite interaction in Trm7–Trm734 and molecular hypothesis for the methylation activity of this enzyme.
DATA AVAILABILITY
The crystal structure factors and coordinates have been deposited in the Protein Data Bank (PDB codes: 6JP6 and 6JPL).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff members of the beam-line facility at SPring-8 (Hyogo, Japan) and Photon Factory (Tsukuba, Japan) for their assistant with data collection. The synchrotron radiation experiments were performed at the BL26B1, BL38B1 and BL45XU in the SPring-8 with the approval of the Japan Synchrotron Radiation. Research Institute (JASRI) (Proposal No. 2013B1272, 2014A1246, 2014B1063, 2015B2047 and 2016A2547), and at the BL-10C in the Photon Factory with the support of the Platform for Drug Discovery, Informatics, and Structural Life Science (PDIS) from Japan Agency for Medical Research and Development (AMED). Furthermore, the authors thank the Division of Material Science and Applied Protein Research the Advanced Research Support Center, Ehime University for technical assistance in VariMax RAPID/α and Typhoon FLA 7000 system. At last, the authors thank Prof. Nicolas Leulliot (CNRS, Paris University) for his valuable comments in this manuscript and Dr Shingo Izawa (Kyoto Institute of Technology) for providing the S. cerevisiae BY4741 strain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI Grant [JP15K06975, JP18K06088 to A.H.; 23350081, JP16H04763 to H.H.]. Funding for open access charge: JSPS KAKENHI Grant [JP15K06975, JP18K06088 to A.H.; 23350081, JP16H04763 to H.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A. et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lorenz C., Lünse C.E., Mörl M.. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017; 7:E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Väre V.Y., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F.. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017; 7:E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014; 5:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M., Hata T., Iimori T., Watanabe T., Miyazawa T., Yokoyama S.. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992; 43:1040–1046. [DOI] [PubMed] [Google Scholar]
- 6. Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J.. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009; 37:D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schubert H.G., Blumenthal R.M., Cheng X.. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003; 28:329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hori H. Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA. Biomolecules. 2017; 7:E23.28264529 [Google Scholar]
- 9. Benítez-Páez A., Villarroya M., Douthwaite S., Gabaldón T., Armengod M.E.. YibK is the 2′-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA (Leu) isoacceptors. RNA. 2010; 16:2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lim K., Zhang H., Tempczyk A., Krajewski W., Bonander N., Toedt J., Howard A., Eisenstein E., Herzberg O.. Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a cofactor bound at a site formed by a knot. Proteins. 2003; 51:56–67. [DOI] [PubMed] [Google Scholar]
- 11. Singh S.K., Gurha P., Tran E.J., Maxwell E.S., Gupta R.. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J. Biol. Chem. 2004; 279:47661–47671. [DOI] [PubMed] [Google Scholar]
- 12. Aittaleb M., Rashid R., Chen Q., Palmer J.R., Daniels C.J., Li H.. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 2003; 10:256–263. [DOI] [PubMed] [Google Scholar]
- 13. Deng L., Starostina N.G., Liu Z.J., Rose J.P., Terns R.M., Terns M.P., Wang B.C.. Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus. Biochem. Biophys. Res. Commun. 2004; 315:726–732. [DOI] [PubMed] [Google Scholar]
- 14. Guy M.P., Podyma B.M., Preston M.A., Shaheen H.H., Krivos K.L., Limbach P.A., Hopper A.K., Phizicky E.M.. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA. 2012; 18:1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guy M.P., Phizicky E.M.. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014; 11:1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pintard L., Lecointe F., Bujnicki J.M., Bonnerot C., Grosjean H., Lapeyre B.. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002; 21:1811–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guy M.P., Phizicky E.M.. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA. 2015; 21:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han L., Guy M.P., Kon Y., Phizicky E.M.. Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet. 2018; 14:e1007288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guy M.P., Shaw M., Weiner C.L., Hobson L., Stark Z., Rose K., Kalscheuer V.M., Gecz J., Phizicky E.M.. Defects in tRNA anticodon Loop 2′-O-Methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum. Mutat. 2015; 36:1176–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeda H., Toyooka T., Ikeuchi Y., Yokobori S., Okadome K., Takano F., Oshima T., Suzuki T., Endo Y., Hori H.. The substrate specificity of tRNA (m1G37) methyltransferase (TrmD) from Aquifex aeolicus. Genes Cells. 2006; 11:1353–1365. [DOI] [PubMed] [Google Scholar]
- 21. Hori H., Yamazaki N., Matsumoto T., Watanabe Y., Ueda T., Nishikawa K., Kumagai I., Watanabe K.. Substrate recognition of tRNA (Guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem. 1998; 273:25721–25727. [DOI] [PubMed] [Google Scholar]
- 22. Keith G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie. 1995; 77:142–144. [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 24. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Terwilliger T.C. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D Biol. Crystallogr. 2003; 59:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 27. McCoy A., Grosse-Kunstleve R., Adams P., Winn M., Storoni L., Read R.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laskowski R., MacArthur M., Moss D., Thoronton J.. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1992; 26:283–291. [Google Scholar]
- 29. Shimizu N., Mori T., Nagatani Y., Ohta H., Saijo S., Takagi H., Takahashi M., Yatabe K., Kosuge T., Igarashi N.. BL-10C, the small-angle x-ray scattering beamline at the photon factory. AIP Conference Proceedings. 2019; 2054:060041. [Google Scholar]
- 30. Valentini E., Kikhney A.G., Previtali G., Jeffries C.M., Svergun D.I.. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 2015; 43:D357–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Breker M., Gymrek M., Moldavski O., Schuldiner M.. LoQAtE–localization and quantitation ATlas of the yeast proteomE. A new tool for multiparametric dissection of single-protein behavior in response to biological perturbations in yeast. Nucleic Acids Res. 2014; 42:D726–D730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshihisa T., Yunoki-Esaki K., Ohshima C., Tanaka N., Endo T.. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell. 2003; 14:3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshihisa T., Ohshima C., Yunoki-Esaki K., Endo T.. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells. 2007; 12:285–297. [DOI] [PubMed] [Google Scholar]
- 34. Hayashi S., Mori S., Suzuki T., Suzuki T., Yoshihisa T.. Impact of intron removal from tRNA genes on Saccharomyces cerevisiae. Nucleic Acids Res. 2019; 47:5936–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takano A., Endo T., Yoshihisa T.. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005; 309:140–142. [DOI] [PubMed] [Google Scholar]
- 36. Ohira T., Suzuki T.. Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10502–10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohira T., Suzuki T.. Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat. Chem. Biol. 2016; 12:648–655. [DOI] [PubMed] [Google Scholar]
- 38. Chatterjee K., Nostramo R.T., Wan Y., Hopper A.K.. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: location, location, location. Biochim. Biophys. Acta. Gene. Rugul. Mech. 2018; 1861:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stirnimann C.U., Petsalaki E., Russell R.B., Müller C.W.. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010; 35:565–574. [DOI] [PubMed] [Google Scholar]
- 40. Holm L., Rosenström P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caldas T., Binet E., Bouloc P., Costa A., Desgres J., Richarme G.. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 2000; 275:16414–16419. [DOI] [PubMed] [Google Scholar]
- 42. Caldas T., Binet E., Bouloc P., Richarme G.. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 2000; 271:714–718. [DOI] [PubMed] [Google Scholar]
- 43. Bügl H., Fauman E.B., Staker B.L., Zheng F., Kushner S.R., Saper M.A., Bardwell J.C., Jakob U.. RNA methylation under heat shock control. Mol. Cell. 2000; 6:349–360. [DOI] [PubMed] [Google Scholar]
- 44. Hager J., Staker B.L., Bugl H., Jakob U.. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 2002; 277:41978–41986. [DOI] [PubMed] [Google Scholar]
- 45. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 46. Xu C., Min J.. Structure and function of WD40 domain proteins. Protein Cell. 2011; 2:202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scrima A., Konícková R., Czyzewski B.K., Kawasaki Y., Jeffrey P.D., Groisman R., Nakatani Y., Iwai S., Pavletich N.P., Thomä N.H.. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008; 135:1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ochi A., Makabe K., Kuwajima K., Hori H.. Flexible recognition of the tRNA G18 methylation target site by TrmH methyltransferase through first binding and induced fit processes. J. Biol. Chem. 2010; 285:9018–9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ochi A., Makabe K., Yamagami R., Hirata A., Sakaguchi R., Hou Y.M., Watanabe K., Nureki O., Kuwajima K., Hori H.. The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process. J. Biol. Chem. 2013; 288:25562–255574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ito T., Masuda I., Yoshida K., Goto-Ito S., Sekine S., Suh S.W., Hou Y.M., Yokoyama S.. Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4197–E4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krishnamohan A., Dodbele S., Jackman J.E.. Insights into catalytic and tRNA recognition mechanism of the dual-specific tRNA methyltransferase from Thermococcus kodakarensis. Genes. 2019; 10:E100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Behm-Ansmant I., Grosjean H., Massenet S., Motorin Y., Branlant C.. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2004; 279:52998–53006. [DOI] [PubMed] [Google Scholar]
- 53. Hepfer C.E., Arnold-Croop S., Fogell H., Steudel K.G., Moon M., Roff A., Zaikoski S., Rickman A., Komsisky K., Harbaugh D.L. et al.. DEG1, encoding the tRNA:pseudouridine synthase Pus3p, impacts HOT1-stimulated recombination in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2005; 274:528–538. [DOI] [PubMed] [Google Scholar]
- 54. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 55. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jackman J.E., Montange R.K., Malik H.S., Phizicky E.M.. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003; 9:574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Swinehart W.E., Henderson J.C., Jackman J.E.. Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA. 2013; 19:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nyswaner K.M., Checkley M.A., Yi M., Stephens R.M., Garfinkel D.J.. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 2008; 178:197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi Y., Stefan C.J., Rue S.M., Teis D., Emr S.D.. Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Mol. Biol. Cell. 2011; 22:4093–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Alexandrov A., Martzen M.R., Phizicky E.M.. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002; 8:1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tomikawa C. 7-Methylguanosine modifications in transfer RNA (tRNA). Int. J. Mol. Sci. 2018; 19:E4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leulliot N., Chaillet M., Durand D., Ulryck N., Blondeau K., van Tilbeurgh H.. Structure of the yeast tRNA m7G methylation complex. Structure. 2008; 16:52–61. [DOI] [PubMed] [Google Scholar]
- 63. Matsumoto K., Toyooka T., Tomikawa C., Ochi A., Takano Y., Takayanagi N., Endo Y., Hori H.. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex). FEBS Lett. 2007; 581:1599–1604. [DOI] [PubMed] [Google Scholar]
- 64. Jin W., Wang Y., Liu C.P., Yang N., Jin M., Cong Y., Wang M., Xu R.M.. Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev. 2016; 30:2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tang X., Bharath S.R., Piao S., Tan V.Q., Bowler M.W., Song H.. Structural basis for specific recognition of pre-snRNA by Gemin5. Cell Res. 2016; 26:1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bohnsack K.E., Höbartner C., Bohnsack M.T.. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes. 2019; 10:pii: E102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hawer H., Hammermeister A., Ravichandran K.E., Glatt S., Schaffrath R., Klassen R.. Roles of elongator dependent tRNA modification pathways in neurodegeneration and cancer. Genes. 2018; 10:E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pereira M., Francisco S., Varanda A.S., Santos M., Santos M.A.S., Soares A.R.. Impact of tRNA modifications and tRNA-modifying enzymes on proteostasis and human disease. Int. J. Mol. Sci. 2018; 19:E3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shaheen R., Abdel-Salam G.M., Guy M.P., Alomar R., Abdel-Hamid M.S., Afifi H.H., Ismail S.I., Emam B.A., Phizicky E.M., Alkuraya F.S.. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015; 16:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure factors and coordinates have been deposited in the Protein Data Bank (PDB codes: 6JP6 and 6JPL).