Abstract
CSB, a member of the SWI2/SNF2 superfamily, has been implicated in evicting histones to promote the DSB pathway choice towards homologous recombination (HR) repair. However, how CSB promotes HR repair remains poorly characterized. Here we demonstrate that CSB interacts with both MRE11/RAD50/NBS1 (MRN) and BRCA1 in a cell cycle regulated manner, with the former requiring its WHD and occurring predominantly in early S phase. CSB interacts with the BRCT domain of BRCA1 and this interaction is regulated by CDK-dependent phosphorylation of CSB on S1276. The CSB–BRCA1 interaction, which peaks in late S/G2 phase, is responsible for mediating the interaction of CSB with the BRCA1-C complex consisting of BRCA1, MRN and CtIP. While dispensable for histone eviction at DSBs, CSB phosphorylation on S1276 is necessary to promote efficient MRN- and CtIP-mediated DNA end resection, thereby restricting NHEJ and enforcing the DSB repair pathway choice to HR. CSB phosphorylation on S1276 is also necessary to support cell survival in response to DNA damage-inducing agents. These results altogether suggest that CSB interacts with BRCA1 to promote DNA end resection for HR repair and that although prerequisite, CSB-mediated histone eviction alone is insufficient to promote the pathway choice towards HR.
INTRODUCTION
DNA double-strand breaks (DSBs), one of the most lethal forms of DNA damage, can threaten genomic integrity and promote tumorigenesis or premature aging if not repaired properly. In S and G2 cells, repair of DSBs is highly regulated to promote the choice of homologous recombination (HR) repair over nonhomologous end joining (NHEJ) (1,2). While NHEJ can ligate two broken ends in the absence of sequence homology and is often associated with small deletions/insertions, HR uses a sister chromatid in S/G2 as a template to repair broken DNA and thus is considered to be error free. HR requires resection of DNA at DSBs to produce 3′ single-stranded DNA, which is first protected and bound by RPA and subsequently replaced by RAD51 to form RAD51–ssDNA nucleoprotein filament responsible for homology search and strand invasion (3).
DNA end resection is initiated by the MRE11/RAD50/NBS1 complex, also known as MRN, along with its cofactor CtIP (4–6). Both MRN and CtIP interact with BRCA1 in S/G2, forming the BRCA1/MRN/CtIP complex (4), also referred to as the BRCA1-C complex, which is implicated in DNA end resection. BRCA1 binds phosphorylated CtIP via its BRCT domain and BRCA1-CtIP is reported to promote DNA end resection (6,7), perhaps by enhancing the nuclease activity of MRN required for effective end resection. BRCA1 is also reported to stimulate the speed of CtIP-mediated DNA end resection (8) although other studies have suggested that CtIP-mediated end resection can operate independently of BRCA1 (9).
CSB, a member of the SWI2/SNF2 family, is a a multifunctional protein that participates in a wide range of cellular processes. First described for its role in transcription-coupled nucleotide excision repair of UV-induced DNA damage (10), CSB is also implicated in regulating the choice of DSB repair pathways (11,12). CSB contains a N-terminal region, a central ATPase domain as well as a C-terminal region. A winged helix domain (WHD), which was first predicted by computational modeling (11) and subsequently confirmed via crystallography (13), resides within the last 76 amino acids of CSB and this domain mediates recruitment of CSB to DSBs in S phase (11). It has been reported that at DSBs, CSB removes histones from damaged chromatin, restricts RIF1-mediated NHEJ and promotes BRCA1-mediated HR (11). However, whether CSB might play a direct role in BRCA1-mediated HR has not been characterized.
Here, we report that CSB interacts with MRN and BRCA1 independently of each other in a cell cycle regulated manner, with the CSB–MRN interaction in early S phase and the CSB–BRCA1 interaction predominantly in late S/G2 phase. We demonstrate that CSB is phosphorylated by CDK activity on S1276, located outside of the WHD. This phosphorylation mediates the CSB–BRCA1 interaction but not the CSB–MRN interaction, the latter of which requires the WHD. While dispensable for histone eviction at DSBs, CSB phosphorylation on S1276 promotes efficient BRCA1 recruitment to DSBs, which facilitates efficient MRN- and CtIP-mediated DNA end resection. Taken together, these results suggest that eviction of histones by CSB alone is insufficient to support BRCA1 recruitment and that CSB actively recruits BRCA1 to promote MRN- and CtIP-mediated DNA end resection.
MATERIALS AND METHODS
Plasmids, siRNA and antibodies
Retroviral expression constructs of Myc-tagged CSB full length and CSB truncation alleles have been described (11,12). Wild type CSB was used as a template to generate, via site-directed mutagenesis, CSB mutants S1276A and S1276D, which were cloned into the retroviral expression vector pLPC-N-Myc (14) or mammalian expression vector mCherry-LacR-NLS (11). The expression construct pDEST-mCherry-LacR-NLS-MRE11 was generated via Gateway (Invitrogen) using MRE11 cDNA (a kind gift from John Petrini, Sloan Kettering Institute) as a template. The expression constructs pDEST-mCherry-LacR-NLS-BRCA1 (Addgene #71115), pDEST-Flag-BRCA1 (Addgene #71117) and pcDNA5-GFP-BRCA1 (Addgene #71116) were previously described (15). The plasmid pcDNA5-GFP-BRCA1 was used as a template for deletion-PCR to generate BRCA1-ΔBRCT (Δ1582–1863). The pcDNA5-GFP-BRCA1–BRCT (amino acids from 1582 to 1863) and pcDNA5-GFP-BRCA1-RING (amino acids from 1 to 102) were generated by amplifying the regions of interest by PCR and the resulting fragments were cloned into the EcoRI and BamHI sites of the pcDNA5-GFP vector. The BRCA1–BRCT domain was also cloned into mammalian expression pLPC-N-Flag as well as bacterial expression pGST-parallel-2 as described (16). The primers used to generate these constructs are available upon request.
siRNAs used were from Dharmacon: non-targeting siRNA (siControl; D-001206-14-05); siBRCA1 (D-003461-05); siCtIP (GCUAAAACAGGAACGAAUC) (17); siNBS1 (CCAACUAAAUUGCCAAGUA).
The rabbit polyclonal anti-pS1276 antibody was developed by LifeTein against a CSB peptide containing phosphorylated serine 1276 (KHDAIMDGA-pS-PDYVLVE) (LifeTein). Other antibodies used include MRE11/RAD50/NBS1 (18) (kind gifts from John Petrini, Sloan Kettering Institute); BRCA1 (MS110, Abcam); BRCA1 (07-434, Millipore); CtIP (ab70163, Abcam); RAP80 (A300-763A, Bethyl Laboratories); CSB (A301-354A, Bethyl Laboratories); CSB (553C5a, Fitzgerald); γ-H2AX (Milipore); RIF1 (sc55979, Santa Cruz); Myc (9E10, Calbiochem); Flag (F3165, Sigma); H2A (ab18255, Abcam); H2B (ab1790, Abcam); mCherry (NBP2-25157, Novus Biologicals); γ-tubulin (GTU88, Sigma).
Cell culture, transfection, retroviral infection and treatment
All cells were grown in DMEM medium with 10% fetal bovine serum supplemented with non-essential amino acids, l-glutamine, 100 U/ml penicilin and 0.1 mg/ml streptomycin. Cell lines used: hTERT-RPE parental and CSB knockout (12), Phoenix (14), U2OS (19) (ATCC), U2OS-265 (20) (a kind gift from Roger Greenberg, University of Pennsylvania), U2OS-265-CSB knockout (KO) (11), 293T (ATCC), HCT116 (Life Technology), HCT116-CSB-KO (11), AID-DIvA-U2OS (21) (a generous gift from Gaëlle Legube, University of Toulouse). Parental cells were examined for mycoplasma contamination. Retroviral gene delivery was carried out as described (22,23) to generate stable cell lines. DNA and siRNA transfections were carried out with respective JetPrime® transfection reagent (Polyplus) and Lipofectamine RNAiMax (Invitrogen) according to their respective manufacturer's instructions.
To induce expression of FokI, U2OS-265 or U2OS-265-CSB-KO cells were treated with both 1 μM Shield-1 (CheminPharm) and 1 μM 4-hydroxytamoxifen (4-OHT, Abcam) for 4 h.
Generation of AID-DIvA-U2OS CSB knockout (KO) cells CRISPR/Cas9 genome editing of CSB
CRISPR/Cas9-mediated gene targeting to generate two independent AID-DIvA-U2OS CSB KO clones (KO-1 and KO-2) was done as described (11).
Cell synchronization, immunoprecipitation and immunoblotting
Cell synchronization was done as described (11). Immunoprecipitation (IP) was carried out as described (11). Immunoblotting was done as described (18,19).
In vivo DNA end resection assays
AsiSI-induced DNA end resection assays were carried out essentially as described (24) with minor modifications. AID-DIvA-U2OS WT, CSB-KO and CSB-KO complemented with various CSB alleles were seeded in 24-well plates in triplicate. Twenty-four hours later, cells were treated with 1 μM 4-hydroxytamoxifen (4-OHT, Sigma) for 4 h or indicated hours to induce DSBs. Subsequently genomic DNA was isolated using the Gentra Puregene Cell Kit (Qiagen) according to the manufacturer's protocol. Genomic DNA was then digested with 20 units of BsrG1 (NEB), BamHI (NEB) or HindIII (NEB) overnight at 37°C. Real-time PCR (qPCR) was performed in duplicate for each sample using primers and probes as described (24). The percentage of ssDNA (ssDNA%) was calculated based on the following equation: ssDNA% = 1/(2dCt-1 + 0.5) × 100. The value for dCt was calculated by subtracting the Ct value of the mock-digested sample from the Ct value of the digested sample.
To quantify the amount of AsiSI-induced DSB1 and DSB2 sites on chromosome 1, qPCR was performed with the mock-digested sample and primers flanking these two sites as described (24). The Ct values from AsiSI sites were then normalized to the Ct values from a HindIII site (also designated as NO DSB) on chromosome 22 using the ddCt method, giving rise to the percentage of AsiSI-induced DSBs.
In vivo assays of histone occupancy at a I-PpoI-induced DSB
ChIP and I-PpoI-induced DSB assays were carried out as described (11). Primers for real-time PCR were as described (11). Each qPCR product of the I-PpoI locus on chromosome 1 was first normalized to that from input DNA as internal control and then normalized to the corresponding ChIP efficiency. The y-axis in figures displaying ChIP results represents the relative occupancy normalized to the untreated control.
Immunofluorescence and quantification of protein recruitment to FokI-induced DSBs
Immunofluorescence (IF) was performed as described (12,14). All cell images were recorded on a Zeiss Axioplan 2 microscope with a Hammamatsu C4742–95 camera and processed in Open Lab.
To quantify recruitment of RAP80, BRCA1, MRE11, RAD50, NBS1, CtIP or RIF1 to FokI-induced DSBs, fixed cells were co-immunostained with respective primary antibody in conjunction with γH2AX. The γH2AX signal was used to mark the area of FokI-induced damage and the intensity of respective aforementioned proteins within the marked area was measured. Their intensity marked by γH2AX was normalized respectively to their intensity of the same size area but away from the FokI-induced damage site in the same nucleus, giving rise to normalized signal intensity. All images for a given experiment were captured on the same day with the same exposure time. All analyses were carried out on unmodified images with ImageJ software (NIH). Data were represented as scatter plot graphs with the mean indicated. P values were derived using a two-tailed Mann-Whitney test.
Recombinant BRCA1–BRCT protein and in vitro pulldown assays
Production of GST-fused BRCA1–BRCT domain carrying amino acids from 1582–1863 was carried out essentially as described (16,25) with minor modifications. Induction of GST–BRCA1–BRCT was carried out overnight with 1 mM isopropylthiogalactoside at room temperature. The cell pellet was resuspended in lysis buffer [25 mM Tris–HCl pH7.4, 1 M NaCl, 1 mM dithiothreitol and 1 mM EDTA] and lysed by sonication. Triton X-100 was then added to 1% and the lysate was shaken at 4°C for 30 min. Following centrifugation, the supernatant was incubated with a 50% slurry of glutathione-Sepharose 4B (GE Life Sciences) overnight at 4°C. Bound protein was eluted with 10 mM reduced glutathione in buffer containing 50 mM Tris–HCl (pH 8.0) and 1 mM dithiothreitol, subsequently dialyzed against a buffer containing 500 mM KCl, 20 mM HEPES pH 7.9, 20% glycerol and 3 mM MgCl2, followed by flash-freezing and storage in aliquots at –80°C.
For in vitro pulldown assays, 100 μl dry Strep-Tactin Sepharose beads (iba, Cat# 2-1201) was conjugated to 25 μg CSB peptide carrying either S1276 (Biotin-KHDAIMDGASPDYVLV, LifeTein) or phosphorylated S1276 (Biotin-KHDAIMDGApSPDYVLV, LifeTein) in 900 μl PBS/0.02% Tween for 2 h. Bead were washed four times with 1 ml PBS/0.02% Tween and resuspended in 100 μl PBS. Pull down assays were performed with 20 μl dry beads with no peptide, nonphosphorylated or phosphorylated CSB peptide and 1 μg recombinant GST–BRCA1–BRCT protein in 500 μl of PBS/0.02% Tween for 1 h at 4°C. Beads were washed four times with 1 ml of PBS/0.02% Tween and resuspended in 40 μl of SDS sample buffer. GST–BRCA1–BRCT was visualized by western blotting with a homemade anti-GST antibody.
Clonogenic survival assays
Clonogenic survival assays were done as described (12).
Statistical analysis
A Student's two-tailed unpaired t test was used to derive all P values except for where specified.
RESULTS
CSB supports efficient recruitment of the BRCA1-C complex to DSBs
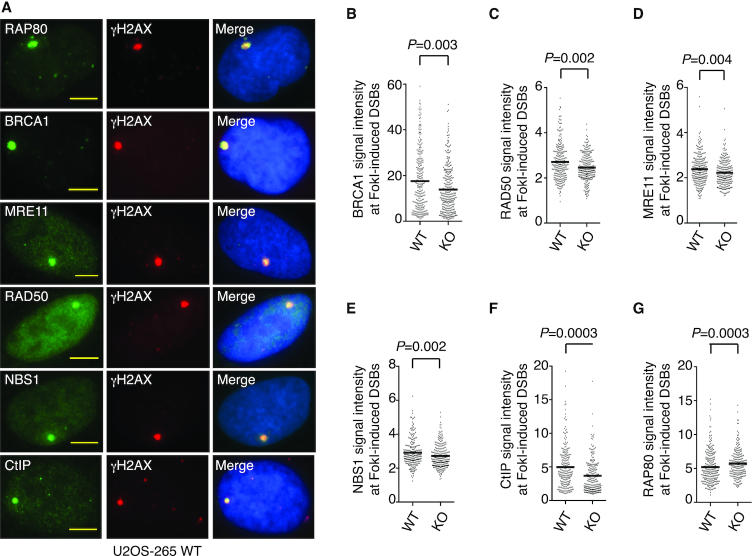
RAP80 of the BRCA1-A complex and MRN of the BRCA1-C complex are implicated in recruiting BRCA1 to DSBs (26–29). To investigate if CSB might regulate recruitment of RAP80 and MRN to DSBs, we first employed a pair of reporter cell lines U2OS-265 wild type (WT) and CSB knockout (KO), which allow measurement of recruitment of proteins to FokI-induced DSBs. Loss of CSB not only abrogated its own recruitment (as expected) but also impaired BRCA1 recruitment to FokI-induced DSBs (Figure 1A and 1B, Supplementary Figure S1A–S1C), in agreement with previous findings (11,12). Loss of CSB also impaired recruitment of MRE11/RAD50/NBS1 and CtIP to FokI-induced DSBs (Figure 1A, 1C–1F, Supplementary Figure S1C and S1D). On the other hand, RAP80 recruitment to FokI-induced DSBs was not compromised but instead increased in U2OS-265 CSB-KO cells (Figure 1A and 1G). To further substantiate the role of CSB in regulating MRN to DSBs, we examined IR-induced damage foci of MRE11 and NBS1 in two isogenic cell lines hTERT-RPE WT and CSB-KO (12). Loss of CSB impaired IR-induced MRE11 and NBS1 foci formation (Supplementary Figure S1E and S1F). We have previously reported that loss of CSB impairs IR-induced BRCA1 foci formation (12). These results altogether suggest that CSB promotes efficient recruitment of the BRCA1-C complex to DSBs.
Figure 1.
Loss of CSB impairs recruitment of BRCA1, MRE11/RAD50/NBS1 and CtIP but not RAP80 to DSBs. (A) Representative images of U2OS-265 WT cells with induction of FokI expression. Fixed cells were stained with an anti-γH2AX antibody in conjunction with an antibody against various endogenous proteins as indicated. Nuclei were stained with DAPI in blue in this and following figures. Scale bars in this and subsequent figures: 5 μm. (B) Quantification of the intensity of BRCA1 signal at the site of FokI-induced DSBs. The respective numbers of cells analyzed for U2OS-265 WT and CSB-KO were 277 and 309. Data are represented as scatter plot graphs from single experiments with the mean indicated in this and subsequent panels. The P value was determined using non-parametric Mann-Whitney rank-sum t-test in this and subsequent panels. (C) Quantification of the intensity of RAD50 signal at the site of FokI-induced DSBs. The respective numbers of cells analyzed for U2OS-265 WT and CSB-KO were 265 and 235. (D) Quantification of the intensity of MRE11 signal at the site of FokI-induced DSBs. The respective numbers of cells analyzed for U2OS-265 WT and CSB-KO were 267 and 254. (E) Quantification of the intensity of NBS1 signal at the site of FokI-induced DSBs. The respective numbers of cells analyzed for U2OS-265 WT and CSB-KO were 248 and 267. (F) Quantification of the intensity of CtIP signal at the site of FokI-induced DSBs. The respective numbers of cells analyzed for U2OS-265 WT and CSB-KO were 249 and 249. (G) Quantification of the intensity of RAP80 signal at the site of FokI-induced DSBs. The respective numbers of cells analyzed for U2OS-265 WT and CSB-KO were 309 and 356.
CSB interacts with the BRCA1-C complex
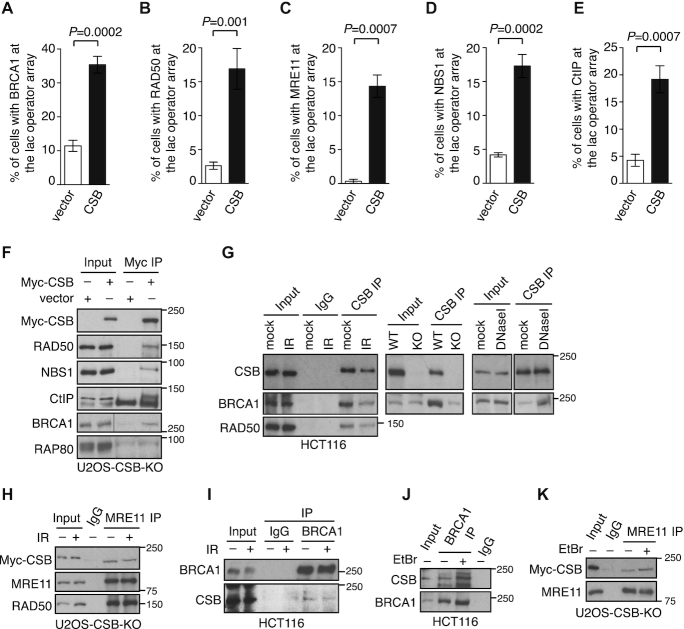
To investigate if CSB might interact with the BRCA1-C complex, we returned to the reporter U2OS-265 CSB-KO cell line, which also allows for analysis of protein-protein interactions of endogenous proteins with CSB fused to mCherry-LacR at the lac operator array. The use of U2OS-265 CSB-KO cells was meant to minimize any interference in analysis of protein-protein interactions from endogenous CSB. Overexpression of mCherry-LacR-CSB in U2OS-265 CSB-KO cells led to an accumulation of endogenous BRCA1, MRE11/RAD50/NBS1 and CtIP but not RAP80 at the lac operator array (Figure 2A–2E, Supplementary Figure S2A and S2B), indicative of an interaction between CSB and the BRCA1-C complex. To investigate if the chromatin remodeling activity of CSB might drive the accumulation of the BRCA1-C complex at the lac operator array, we turned to two CSB mutants, one being CSB carrying a ATPase-dead W851R mutation and the other being CSB lacking the first 30 amino acids (CSB-ΔΝ30), both of which have been reported to be defective in chromatin remodeling (11). While mCherry-LacR-CSB-W851R failed to recruit BRCA1 to the lac operator array, mCherry-LacR-CSB-ΔΝ30 was fully competent in doing so (Supplementary Figure S2C). These results suggest that a defect in chromatin remodeling activity per se is unlikely the sole reason behind the inability of mCherry-LacR-CSB-W851R to recruit BRCA1 to the lac operator array. We have previously reported that the ATPase domain of CSB is engaged in intramolecular interactions with both of its N- and C-terminal regions (30). Perhaps the W851R mutation alters the conformation of CSB, resulting in its inability to interact with BRCA1.
Figure 2.
CSB interacts with the BRCA1-C complex. (A–E) CSB colocalizes with the BRCA1-C complex at the lac operator array. Quantification of vector- and mCherry-LacR-CSB-expressing U2OS-265 CSB-KO cells exhibiting an accumulation of BRCA1 (A), RAD50 (B), MRE11 (C), NBS1 (D), CtIP (E) at the lac operator array. At least 100 cells positive for mCherry staining were scored per condition in a blind manner. Standard deviations from three independent experiments are indicated. (F) CoIPs with anti-Myc antibody in U2OS CSB-KO cells expressing the vector alone or Myc-CSB. Immunoblotting was performed with antibodies against various proteins as indicated in this and subsequent Figures. (G) Anti-CSB coIPs done with HCT116 cells treated with or without 10 Gy IR (the left panel), with HCT116 WT or CSB-KO cells (the middle panel) and with HCT116 cells treated with or without 100 units/ml DNAase I (the right panel). (H) Anti-MRE11 coIPs of Myc-CSB-expressing U2OS CSB-KO cells treated with or without 10 Gy IR. (I) Anti-BRCA1 coIPs of HCT116 cells treated with or without 10 Gy IR. (J) Anti-BRCA1 coIPs of HCT116 cells treated with or without 100 ng/ml ethidium bromide (EtBr). (K) Anti-MRE11 coIPs of Myc-CSB-expressing U2OS CSB-KO cells treated with or without 100 ng/ml EtBr.
To further substantiate the interaction of CSB with the BRCA1 complex, we performed a series of coimmunoprecipitation (coIP) experiments. CoIPs revealed that exogenously-expressed Myc-CSB brought down endogenous BRCA1, RAD50, NBS1 and CtIP but not RAP80 in U2OS-CSB-KO cells (Figure 2F). BRCA1 and RAD50 were also brought down by an antibody against endogenous CSB with or without 10 Gy IR (Figure 2G, the left panel). Although loss of CSB did not affect BRCA1 expression, coIPs with an anti-CSB antibody highly enriched BRCA1 from HCT116 WT cells compared to HCT116 CSB-KO cells (Figure 2G, the middle panel), indicating that the CSB–BRCA1 interaction is specific. The CSB–BRCA1 interaction was not affected by treatment with DNase I, suggesting that it is not mediated by DNA (Figure 2G, the right panel). Reverse IPs with either anti-MRE11 or anti-BRCA1 antibodies brought down CSB with or without 10 Gy IR (Figure 2H and 2I). The addition of ethidium bromide did not affect the interaction of CSB with BRCA1 and MRE11 (Figure 2J and 2K), supporting the notion that their interaction is not mediated by DNA. Taken together, these results reveal that CSB interacts with the BRCA1-C complex and that this interaction is independent of the presence of DNA damage.
CSB interacts with MRN and BRCA1 in a cell cycle regulated manner
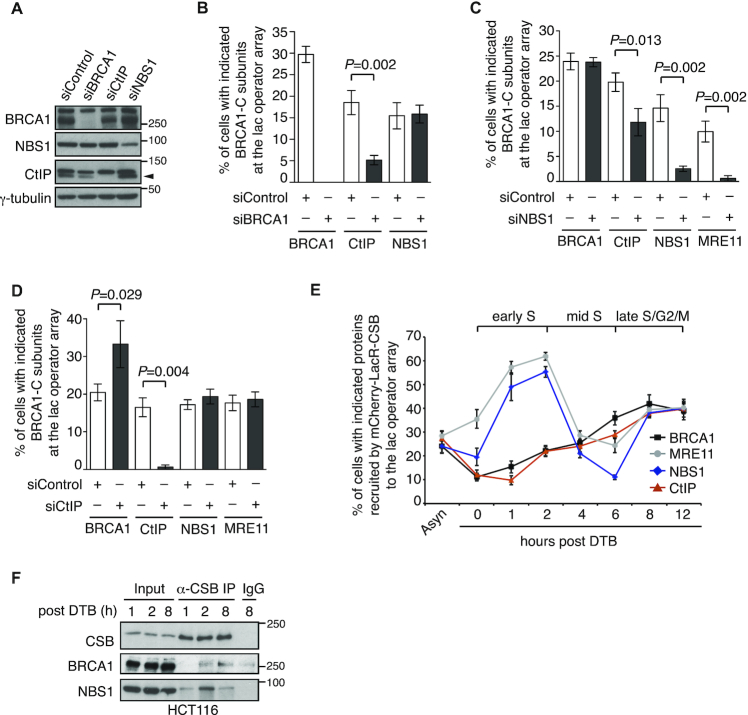
To further investigate the interaction of CSB with the BRCA1-C complex, we knocked down BRCA1, NBS1 and CtIP one at a time in U2OS-265 CSB-KO cells expressing mCherry-LacR-CSB (Figure 3A). IF analysis revealed that knockdown of BRCA1 abrogated its own accumulation, drastically reduced CtIP accumulation but had little effect on NBS1 accumulation at the lac operatory array in U2OS-265 CSB-KO cells expressing mCherry-LacR-CSB (Figure 3B and Supplementary Figure S3A–S3C). Knockdown of NBS1 severely reduced not only its own but also MRE11 accumulation, moderately compromised CtIP accumulation but had little impact on BRCA1 accumulation at the lac operatory array in U2OS-265 CSB-KO cells expressing mCherry-LacR-CSB (Figure 3C and Supplementary Figure S3A-S3D). Knockdown of CtIP did not affect accumulation of MRE11 and NBS1 but led to an increase in BRCA1 accumulation at the lac operator array in U2OS-265 CSB-KO cells expressing mCherry-LacR-CSB (Figure 3D and Supplementary Figure S3A–D), the latter of which was unlikely due to any effect of CtIP depletion on the level of BRCA1 expression (Figure 3A). BRCA1 is known to bind CtIP. Perhaps depletion of CtIP might have freed up additional BRCA1 that interacted with CSB at the lac operator array, contributing to the observed increase in BRCA1 recruitment to the lac operator array in CtIP-depleted cells. Experiments described in Figure 3B-3D were not done at the same time, which might have contributed to the observed variations in the percentage of siControl-expressing cells exhibiting BRCA1, CtIP and MRN at the lac operator array. Taken together, these results suggest that CSB interacts with BRCA1 and MRN, either of which may mediate the interaction of CSB with the BRCA1-C complex.
Figure 3.
CSB interacts with MRN and BRCA1 in a cell cycle regulated manner. (A) Western analysis of U2OS-265 CSB-KO cells expressing various siRNAs as indicated. The γ-tubulin blot was used as a loading control in this and subsequent figures. The arrowhead indicates the CtIP protein band. (B) Quantification of the percentage of siControl- and siBRCA1-expressing cells exhibiting BRCA1, CtIP and NBS1 at the lac operator array. At least 100 mCherry-positive cells was scored per condition in a blind manner in this panel, 3C and 3D. Standard deviations from three independent experiments are indicated in this and subsequent panels. (C) Quantification of the percentage of siControl- and siNBS1-cells exhibiting BRCA1, CtIP, NBS1 and MRE11 at the lac operator array. (D) Quantification of the percentage of siControl- and siCtIP-cells exhibiting BRCA1, CtIP, NBS1 and MRE11 at the lac operator array. (E) Quantification of the percentage of synchronized mCherry-LacR-CSB-expressing U2OS-265 cells exhibiting indicated proteins at the lac operator array. At least 200 mCherry-positive cells were scored per condition in a blind manner. Asyn: asynchronous; DTB: double thymidine block. (F) Anti-CSB coIPs of synchronized HCT116 cells.
To gain further insights into the interaction of CSB with the BRCA1-C complex, we examined the dynamics of recruitment of endogenous BRCA1, MRE11, NBS1 and CtIP to the lac operator array in synchronized U2OS-265 cells overexpressing mCherry-LacR-CSB. The interaction of CSB with MRE11 and NBS1 went through a rise-fall-rise cycle at the lac operator array. Recruitment of both MRE11 and NBS1 by mCherry-LacR-CSB to the lac operator array started as synchronized cells were released into S phase (Figure 3E). This recruitment reached the first peak at 2 h post release from a double thymidine block and then declined drastically (Figure 3E). After bottoming out at 6 h post release, the recruitment of both MRE11 and NBS1 by mCherry-LacR-CSB recovered quickly and reached the second peak 8 hr post release (Figure 3E). On the other hand, the interaction of CSB with BRCA1 did not go through such a rise-fall-rise cycle. Instead, recruitment of BRCA1 by mCherry-LacR-CSB to the lac operator array started as synchronized cells were released into S phase, continued to rise gradually throughout the S/G2 phase and peaked at 8 h post release, a time point when both MRE11 and NBS1 were observed to reach the second highest accumulation at the lac operator (Figure 3E). The recruitment of CtIP to the lac operator array followed a similar dynamics to that of BRCA1 (Figure 3E). These results altogether suggest that CSB interacts with MRN and BRCA1 in a cell cycle regulated manner.
This notion was further supported by coIP analysis with synchronized cells collected 1, 2 and 8 h post release. The CSB-NBS1 interaction was visible at 1 h post release, substantially enriched at 2 h post release and then decreased at 8 h post release (Figure 3F). The CSB–BRCA1 interaction was not detectable at 1 h post release (Figure 3F). Compared to the IgG control, this interaction was barely enriched at 2 h post release but visibly enriched 8 h post release (Figure 3F). These results suggest that the CSB–MRN interaction in early S cells occurs independently of the CSB–BRCA1 interaction in late S/G2 cells. These results further imply that the CSB–BRCA1 interaction is responsible for the interaction of CSB with the BRCA1-C complex in late S/G2 cells.
CSB interacts with MRN and BRCA1 via distinct regions, with the former requiring the WHD
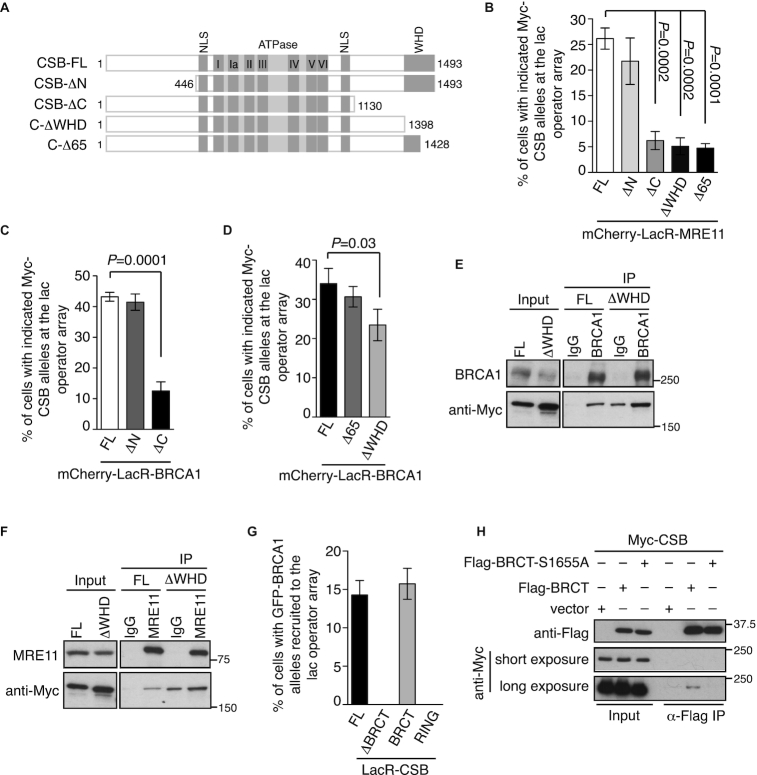
To investigate if a particular domain of CSB might be engaged in its interaction with MRN and BRCA1, we separately fused MRE11 and BRCA1 to mCherry-LacR and examined their ability to recruit Myc-CSB carrying either a N-terminal deletion (CSB-ΔN) or a C-terminal deletion (CSB-ΔC) (Figure 4A) to the lac operator array. While the N-terminal domain of CSB was dispensable for its interaction with either MRE11 or BRCA1, the C-terminal region of CSB was necessary for its interaction with MRE11 and BRCA1 (Figure 4B and 4C, Supplementary Figure S4A and S4B). Either a partial or a complete deletion of the WHD domain severely impaired CSB interaction with MRE11 but it had no or mild effect on the interaction of CSB with BRCA1 (Figure 4B and 4D, Supplementary Figure S4A and S4C). We also examined the requirement of the WHD for the interaction of CSB with BRCA1 and MRE11 via coIPs. For unknown reasons, Myc-CSB-ΔWHD exhibited a higher level of nonspecific binding to IgG than Myc-CSB (Figure 4E and 4F). Nevertheless, compared to the IgG control, coIPs with an anti-BRCA1 antibody but not an anti-MRE11 antibody substantially enriched Myc-CSB-ΔWHD (Figure 4E and 4F). Taken together, these results suggest that CSB interacts, via two separate regions of its C-terminal region, with MRN and BRCA1 and that the WHD mediates the CSB–MRN interaction.
Figure 4.
The WHD mediates the CSB–MRN interaction whereas BRCA1 interacts with CSB via its BRCT domain. (A) Schematic diagram of CSB full length and CSB truncation mutants. NLS: nuclear localization signal; WHD: winged helix domain. (B) Quantification of the percentage of mCherry-LacR-MRE11-expressing cells exhibiting various Myc-CSB alleles at the lac operator array. At least 250 cells positive for expression of Myc-CSB alleles were scored per condition in a blind manner in this and subsequent panels. Standard deviations from three independent experiments are indicated in this and subsequent panels. (C, D) Quantification of the percentage of mCherry-LacR-BRCA1-expressing cells exhibiting various Myc-CSB alleles as indicated at the lac operator array. (E) Anti-BRCA1 coIPs of U2OS-CSB-KO cells stably expressing Myc-CSB-FL or Myc-CSB-ΔWHD. (F) Anti-MRE11 coIPs of U2OS-CSB-KO cells stably expressing Myc-CSB-FL or Myc-CSB-ΔWHD. (G) Quantification of mCherry-LacR-CSB-expressing cells exhibiting various GFP-BRCA1 alleles as indicated at the lac operator array. (H) CoIPs with anti-Flag antibody in 293T cells expressing Myc-CSB in conjunction with the vector alone, Flag-BRCA1–BRCT or Flag-BRCA1–BRCT-S1655A.
BRCA1 interacts with CSB via its BRCT domain
To investigate if a particular domain of BRCA1 might mediate its interaction with CSB, we co-expressed mCherry-LacR-CSB with various GFP-fused BRCA1 alleles either being full length (FL), lacking the BRCT domain (ΔBRCT), carrying the BRCT domain alone or the RING domain alone in U2OS-265 CSB-KO cells. While both GFP-BRCA1-FL and GFP-BRCA1–BRCT were recruited to the lac operator array, neither GFP-BRCA1-ΔBRCT nor GFP-BRCA1-RING was recruited to the lac operator array (Figure 4G and Supplementary Figure S5A), suggesting that BRCA1 interacts with CSB via its BRCT domain. The BRCT domain of BRCA1 is known to directly bind phosphorylated serine (pSer) (31,32). S1655 is a key residue in the BRCA1–BRCT domain that bonds with pSer and thus we examined if S1655 might mediate the interaction of BRCA1 with CSB. IF analysis revealed that unlike Flag-BRCA1, Flag-BRCA1 carrying a S1655A mutation failed to interact with mCherry-LacR-CSB at the lac operator array in U2OS-265 CSB-KO cells (Supplementary Figure S5B and S5C). In addition, coIP analysis revealed that the S1655A mutation abrogated the ability of the BRCA1–BRCT domain to interact with CSB (Figure 4H), indicative of a role of phosphorylation in mediating the CSB–BRCA1 interaction. These results altogether demonstrate that the BRCT domain of BRCA1 is necessary and sufficient for its interaction with CSB.
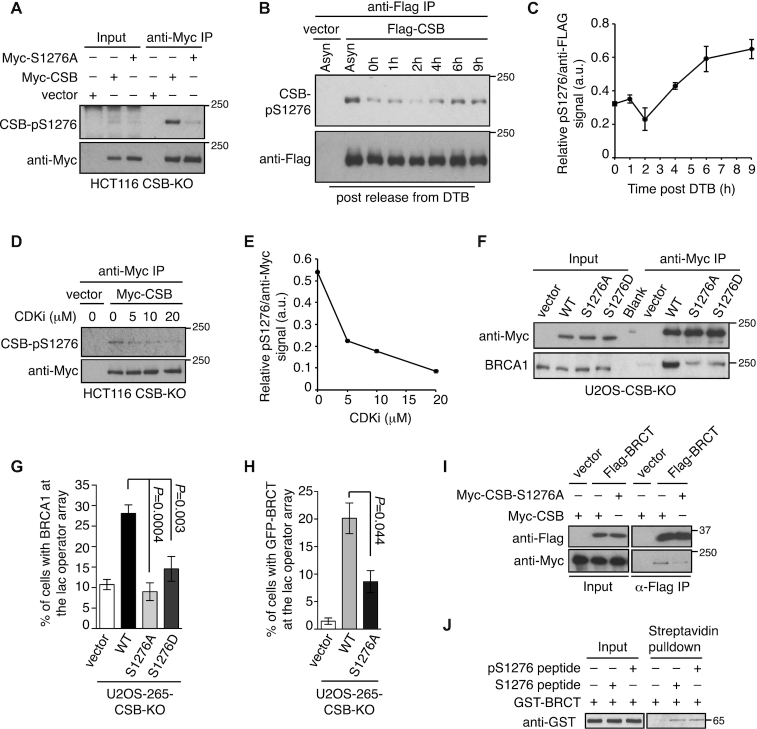
CSB phosphorylation on S1276 mediates its interaction with the BRCT domain of BRCA1
Two quantitative phosphoproteomics studies have identified that CSB is phosphorylated on serine 1276 (S1276P) (33,34), which matches the consensus motif pS/TP for CDKs. In addition, S1276 also resides in the region of CSB required for its interaction with BRCA1. Therefore we investigated if S1276 phosphorylation might regulate the CSB–BRCA1 interaction. Western analysis with a phospho-S1276 (pS1276) antibody detected Myc-CSB but not Myc-CSB carrying a S1276A mutation immunoprecipitated from HCT116 CSB-KO cell lysates (Figure 5A), confirming that CSB is phosphorylated on S1276 in vivo. Western analysis of Flag-CSB immunoprecipitated from synchronized 293T cell lysate revealed an enrichment in the level of S1276 phosphorylation in cells 6 h and 9 h post release from a double thymidine block (Figure 5B and 5C). Furthermore, CSB phosphorylation on S1276 was also sensitive to CDK1 inhibitor CGP74514A (Figure 5D and 5E). These results suggest that CDK activity regulates S1276 phosphorylation of CSB.
Figure 5.
CSB phosphorylation on S1276 mediates its interaction with BRCA1. (A) CoIPs with anti-Myc antibody in HCT116 CSB-KO cells expressing various Myc-CSB alleles as indicated. (B) CoIPs with anti-Flag antibody in synchronized 293T cells expressing the vector alone or Flag-CSB. (C) Quantification of the CSB-pS1276 signal relative to the signal of Flag-CSB immunoprecipitated from synchronized 293T cells expressing Flag-CSB from (B). Quantification was done with ImageJ. Standard deviations from two independent experiments are indicated. (D) Western analysis of Myc-CSB immunoprecipitated from Myc-CSB-expressing HCT116 CSB-KO cells treated with no or various amount of CDK1 inhibitor CGP74514A (CDKi) for 2 h. (E) Quantification of the CSB-pS1276 signal relative to the signal of immunoprecipitated Myc-CSB from (D). Quantification was done with ImageJ. (F) CoIPs with anti-Myc antibody in U2OS CSB-KO stably expressing various Myc-CSB alleles as indicated. (G) Quantification of vector- and various indicated mCherry-LacR-CSB allele-expressing U2OS-265-CSB-KO cells exhibiting BRCA1 at the lac operator array. At least 250 cells positive for expression of mCherry were scored per condition in a blind manner. Standard deviations from three independent experiments are indicated in this panel and 5H. (H) Quantification of cells exhibiting GFP-BRCT at the lac operator array. U2OS-265 CSB-KO cells were co-transfected with GFP-BRCT along with either the vector alone or various mCherry-LacR-CSB alleles. Scoring was done as in 5G. (I) CoIPs with anti-Flag antibody in 293T cells transfected with indicated Myc-CSB alleles in conjunction with the vector alone or Flag-BRCT. (J) Strepavidin bead pulldown assays with GST–BRCA1–BRCT and biotinylated CSB peptide with or without pS1276.
CoIPs revealed that while Myc-CSB interacted with BRCA1, Myc-CSB carrying a non-phosphorylatable S1276A mutation was severely defective in bringing down BRCA1 (Figure 5F). The S1276A mutation also impaired the ability of mCherry-LacR-CSB to recruit BRCA1 to the lac operator array in U2OS-265-CSB-KO cells (Figure 5G and Supplementary Figure S5D). These results suggest that S1276 phosphorylation mediates the CSB–BRCA1 interaction. CSB carrying a phosphomimic S1276D mutation was defective in interacting with BRCA1 in both coIP and IF analysis (Figure 5F and 5G), suggesting that S1276D does not recapitulate S1276 phosphorylation for mediating the CSB–BRCA1 interaction.
We also investigated if S1276 phosphorylation might mediate the interaction of CSB with the BRCA1–BRCT domain. IF analysis revealed that while mCherry-LacR-CSB readily recruited GFP-BRCA1–BRCT to the lac operator array in U2OS-265-CSB-KO cells, mCherry-LacR-CSB-S1276A was defective in doing so (Figure 5H and Supplementary Figure S5E). CoIPs further confirmed that the S1276A mutation impaired the interaction of Myc-CSB with Flag-BRCA1–BRCT (Figure 5I), suggesting that CSB phosphorylation on S1276 mediates its interaction with the BRCA1–BRCT domain.
To further investigate if the BRCA1–BRCT domain might bind directly to phosphorylated S1276 of CSB, we incubated bacterially-expressed recombinant GST-fused BRCA1–BRCT and biotinylated CSB peptide with or without chemically-phosphorylated S1276. Although we detected an interaction between GST–BRCA1–BRCT with biotinylated CSB-pS1276 peptide, this interaction was not enriched compared to that of GST–BRCA1–BRCT with biotinylated CSB peptide without S1276 phosphorylation (Figure 5J). Analysis of surface plasmon resonance (SPR) also did not reveal any enrichment in binding of GST–BRCA1–BRCT with pS1276-containing CSB peptide (Y. Coulombe and J.Y. Masson, unpublished data). These results suggest that although S1276 phosphorylation mediates the CSB–BRCA1 interaction, pS1276 of CSB is not a direct binding target of BRCA1–BRCT.
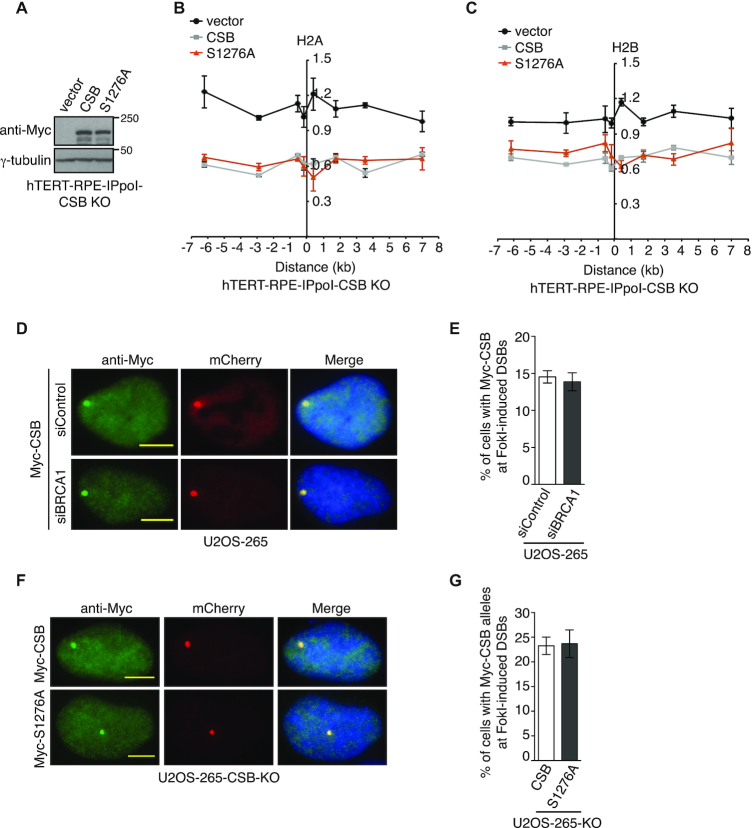
CSB phosphorylation on S1276 is dispensable for its chromatin remodeling activity at DSBs
CSB has been previously reported to evict histones from chromatin flanking DSBs to promote HR (11). To investigate if CSB phosphorylation on S1276A might regulate its chromatin remodeling activity at DSBs, we generated inducible ddI-PpoI-expressing hTERT-RPE CSB KO cells stably expressing the vector alone, Myc-CSB wild type and Myc-CSB carrying a S1276A mutation (Figure 6A). Analysis of histone occupancy revealed that Myc-CSB-S1276A behaved like Myc-CSB, fully restoring I-PpoI-induced loss of H2A and H2B from the I-PpoI cleavage site on chromosome 1 in CSB KO cells, (Figure 6B and 6C), suggesting that CSB phosphorylation on S1276 is dispensable for its chromatin remodeling activity at DSBs.
Figure 6.
CSB phosphorylation on S1276 is dispensable for its own recruitment and histone eviction at DSBs. (A) Western analysis of ddI-PpoI-expressing hTERT-RPE CSB-KO cells stably expressing various Myc-CSB alleles as indicated. (B) Relative occupancy of histone H2A in ddI-PpoI-expressing hTERT-RPE CSB-KO cells stably expressing various Myc-CSB alleles as indicated. The x-axis represents the distance in kb upstream and downstream from the I-PpoI-induced DSB on chromosome 1, which was set as 0. The y-axis represents the relative occupancy of H2A of cells 2 h post induction of I-PpoI relative to uninduced cells. Standard error of the mean (SEM) from three independent experiments are indicated in this panel and 6C. (C) Relative occupancy of histone H2B in ddI-PpoI-expressing hTERT-RPE CSB-KO cells stably expressing the vector alone, Myc-CSB or Myc-CSB-S1276A. Both x-axis and y-axis are as described in 6B. (D) Representative images of cells with induction of FokI expression. U2OS-265 cells were transfected with siControl or siBRCA1 and 24 h later with Myc-CSB. (E) Quantification of cells exhibiting Myc-CSB at FokI-induced DSBs from (D). At total of 250 cells positive for Myc-CSB expression were scored per condition in a blind manner. Standard deviations from three independent experiments are indicated in this panel and 6F. (F) Representative images of Myc-CSB and Myc-CSB-S1276A-expressing U2OS-265-CSB-KO cells with induction of FokI expression. (G) Quantification of cells exhibiting Myc-CSB alleles at FokI-induced DSBs from (F). Scoring was done as in 6E.
CSB phosphorylation on S1276 promotes efficient recruitment of the BRCA1-C complex to DSBs
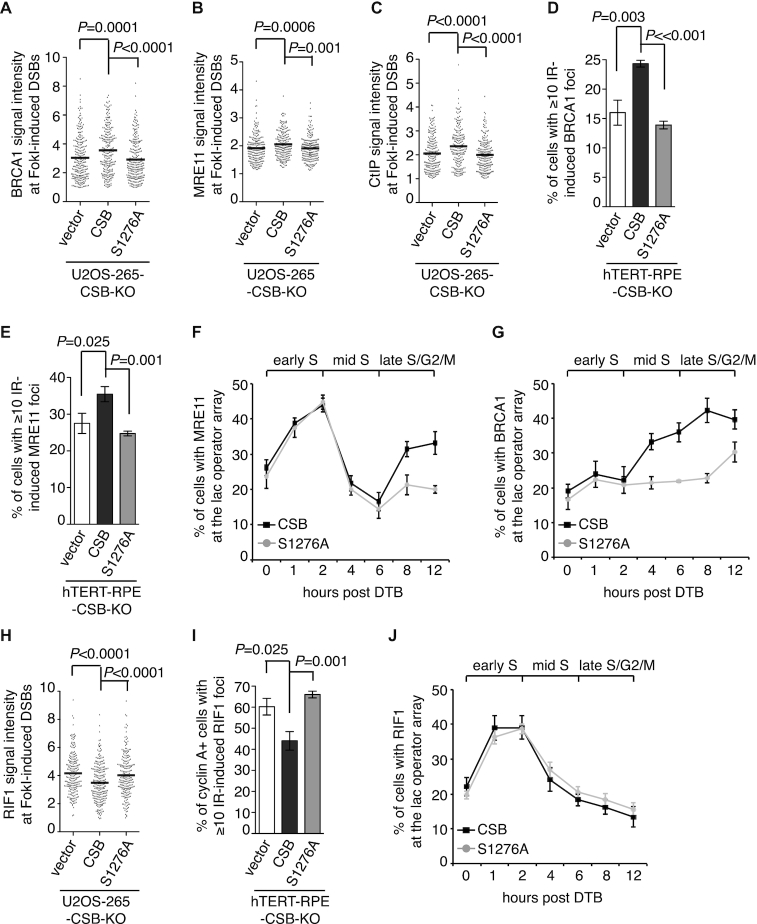
We observed that depletion of BRCA1 did not affect Myc-CSB recruitment to FokI-induced DSBs in U2OS-265 cells (Figure 6D and 6E), suggesting that CSB functions upstream of BRCA1. In agreement with this notion, a S1276A mutation did not affect Myc-CSB recruitment to FokI-induced DSBs (Figure 6F and 6G). To investigate if CSB phosphorylation on S1276 might mediate BRCA1-C recruitment to DSBs, we transfected U2OS-265-CSB-KO cells with the vector alone, Myc-CSB or Myc-CSB-S1276A. While overexpression of Myc-CSB rescued recruitment of BRCA1, MRE11 and CtIP to FokI-induced DSBs, Myc-CSB-S1276A failed to do so (Figure 7A-7C and Supplementary Figure S6A-S6C). We also examined IR-induced damage foci of BRCA1 and MRE11 in hTERT-RPE-CSB-KO cells complemented with either the vector alone, Myc-CSB or Myc-CSB-S1276A. While Myc-CSB rescued IR-induced foci formation of BRCA1 and MRE11, Myc-CSB-S1276A failed to do so (Figure 7D and 7E). These results suggest that CSB phosphorylation on S1276 is necessary to promote efficient recruitment of BRCA1-C to DSBs.
Figure 7.
CSB phosphorylation on S1276 promotes efficient recruitment of BRCA1-C to DSBs. (A) Quantification of the intensity of BRCA1 signal at the site of FokI-induced DSBs in U2OS-265-CSB-KO cells expressing various Myc-CSB alleles as indicated. The respective numbers of cells analyzed for the vector alone, Myc-CSB WT and Myc-CSB-S1267A were 247, 239 and 249. Data are represented as scatter plot graphs from single experiments with the mean indicated in this panel, B, C and H. The P value was determined using non-parametric Mann-Whitney rank-sum t-test in this panel, B, C and H. (B) Quantification of the intensity of MRE11 signal at the site of FokI-induced DSBs in U2OS-265 CSB-KO cells expressing various Myc-CSB alleles as indicated. The respective numbers of cells analyzed for the vector alone, Myc-CSB WT and Myc-CSB-S1267A were 221, 215 and 217. (C) Quantification of the intensity of CtIP signal at the site of FokI-induced DSBs in U2OS-265 CSB-KO cells expressing various Myc-CSB alleles as indicated. The respective numbers of cells analyzed for the vector alone, Myc-CSB WT and Myc-CSB-S1267A were 213, 212 and 232. (D) Quantification of cells with ≥ 10 IR-induced BRCA1 foci. hTERT-RPE-CSB-KO cells expressing various Myc-CSB alleles as indicated were treated with 2 Gy IR and fixed 1 h later in this panel, 7E and 7I. At least 500 cells were scored per condition in a blind manner in this panel, 7E and 7I. Standard deviations from three independent experiments are indicated in this panel, 7E-7G, 7I and 7J. (E) Quantification of cells with ≥ 10 IR-induced MRE11 foci. (F) Quantification of the percentage of synchronized mCherry-LacR-CSB- or mCherry-LacR-CSB-S1276A-expressing U2OS-265 cells with MRE11 at the lac operator array. At least 200 mCherry-positive cells were scored per condition in a blind manner in this panel, 7G and 7J. DTB: double thymidine block. (G) Quantification of the percentage of synchronized mCherry-LacR-CSB- or mCherry-LacR-CSB-S1276A-expressing U2OS-265 cells with BRCA1 at the lac operator array. (H) Quantification of the intensity of RIF1 signal at the site of FokI-induced DSBs in U2OS-265 CSB KO cells expressing various Myc-CSB alleles as indicated. The respective numbers of cells analyzed for the vector alone, Myc-CSB WT and Myc-CSB-S1267A were 218, 245 and 232. (I) Quantification of cyclin A-positive cells with ≥10 IR-induced RIF1 foci. (J) Quantification of the percentage of synchronized mCherry-LacR-CSB- or mCherry-LacR-CSB-S1276A-expressing U2OS-265 cells with RIF1 at the lac operator array.
Earlier we have shown that the CSB–MRN interaction in early S phase occurs independently of the CSB–BRCA1 interaction (Figure 3E and 3F). To investigate if the defect in the ability of Myc-CSB-S1276A to rescue MRE11 recruitment to DSBs might have arisen from a defect in its interaction with MRN in early S phase, we examined the effect of the S1276A mutation on the interaction of mCherry-LacR-CSB with MRE11 at the lac operator array in synchronized U2OS-265-CSB-KO cells. IF analysis revealed that although defective in recruiting MRE11 in late S/G2 cells, mCherry-LacR-CSB-S1276A was fully competent in recruiting MRE11 to the lac operator array in early S phase (Figure 7F). On the other hand, the S1276A mutation impaired the ability of mCherry-LacR-CSB to recruit BRCA1 in S/G2 cells (Figure 7G). Taken together, these results suggest that BRCA1 mediates the interaction of CSB with the BRCA1-C complex in late S/G2 cells. These results further imply that a defect in the CSB–BRCA1 interaction is responsible for the inability of Myc-CSB-S1276A to promote efficient recruitment of BRCA1-C complex to DSBs.
CSB phosphorylation on S1276 limits RIF1 at DSBs in S/G2 cells
We observed that Myc-CSB-S1276A failed to suppress NHEJ-promoting factor RIF1 accumulation at FokI-induced DSBs in U2OS-265-CSB-KO cells (Figure 7H and Supplementary Figure S6D). To further investigate the role of S1276 phosphorylation in limiting RIF1, we examined IR-induced RIF1 foci formation in hTERT-RPE-CSB-KO cells expressing the vector alone, Myc-CSB or Myc-CSB-S1276A. The S1276A mutation abrogated the ability of Myc-CSB to suppress IR-induced RIF1 foci formation in cyclin A-positive hTERT-RPE-CSB-KO cells (Figure 7I), suggesting that CSB phosphorylation on S1276 restricts RIF1 at DSBs in S/G2 cells. Previously we have reported that CSB interacts with RIF1 in early S phase (11). Formally it was possible that the inability of CSB-S1276A to limit RIF1 at DSBs might have resulted from a gain in its interaction with RIF1 in late S/G2 phase. Therefore we investigated the effect of a S1276A mutation on the interaction of mCherry-LacR-CSB with RIF1 at the lac operator array in synchronized U2OS-265-CSB-KO cells. mCherry-LacR-CSB-S1276A behaved indistinguishably from mCherry-LacR-CSB in interacting with RIF1 at the lac operator array, predominantly in early S phase and drastically diminished in late S/G2 cells (Figure 7J). These results suggest that it is unlikely that CSB phosphorylation on S1276 limits RIF1 at DSBs in S/G2 through controlling the CSB-RIF1 interaction.
CSB phosphorylation on S1276 promotes DNA end resection and cell survival in response to treatment with olaparib and CPT
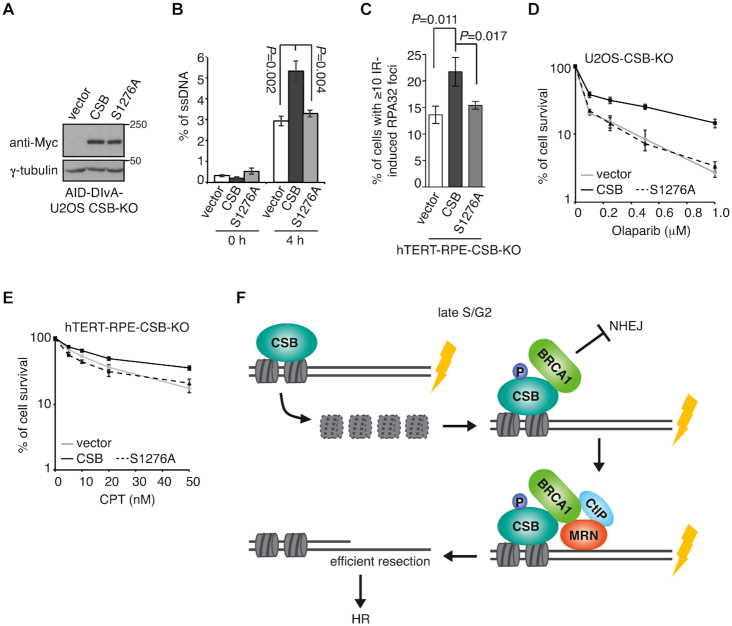
AID-DIvA-U2OS, a well-established cell line with inducible expression of a restriction enzyme AsiSI that cuts the human genome (21), has been used to directly quantify ssDNA generated by 5’ end resection at two AsiSI-induced DSBs on chromosome 1 (21,24) (DSB1 and DSB2, Supplementary Figure S7A). We knocked out CSB via CRISPR-Cas9 in this cell line and generated two independent CSB knockout clones (CSB-KO-1 and CSB-KO-2) (Supplementary Figure S7B). Knockout of CSB did not affect the efficiency of AsiSI-induced DNA cleavage at DSB1 and DSB2 (Supplementary Figure S7C and S7D). Analysis of qPCR assays with three different sets of primers on digested genomic DNA revealed that the generation of ssDNA in AID-DIvA-U2OS wild type cells was evident at respective positions 335 nt and 364 nt from DSB1 and DSB2 but less so at a greater distance away from the cleavage sites, starting to appear 1 h post induction of AsiSI and continuing to rise throughout a period of 4 h we monitored (Supplementary Figure S8A and S8B), in agreement with previous finding (24). At 4 h post induction of AsiSI, the amount of ssDNA generated at positions 335 nt and 364 nt from DSB1 and DSB2 was respectively reduced by ∼25% and ∼26% in AID-DIvA-U2OS CSB-KO-1 cells and ∼42% and ∼30% in AID-DIvA-U2OS CSB-KO-2 cells (Supplementary Figure S8A and S8B). No accumulation of ssDNA was observed in the absence of AsiSI induction (Supplementary Figure S8C). These results demonstrate that CSB promotes efficient DNA end resection.
To investigate the role of S1276 phosphorylation in DNA end resection, we generated AID-DIvA-U2OS CSB-KO-1 cells stably expressing the vector alone, Myc-CSB or Myc-CSB-S1276A. The expression of Myc-CSB-S1276A was comparable to that Myc-CSB (Figure 8A). While overexpression of Myc-CSB fully rescued the amount of ssDNA generated at positions 335 nt from DSB1 in AID-DIvA-U2OS CSB-KO-1 cells, Myc-CSB-S1276A failed to do so (Figure 8B), suggesting that CSB phosphorylation on S1276 promotes efficient MRN- and CtIP-mediated DNA end resection, a key step in HR repair. In support of this notion, Myc-CSB-S1276A also failed to rescue IR-induced foci formation of RPA32 (Figure 8C), a readout for ssDNA. In addition, clonogenic survival assays revealed that overexpression of Myc-CSB-S1276A failed to suppress either the sensitivity of U2OS-CSB-KO cells to the PARP inhibitor olaparib, known to be toxic to cells defective in HR, or the sensitivity of hTERT-RPE-CSB-KO cells to campothecin (CPT), a topoisomerase I inhibitor (Figure 8D and 8E). These results altogether suggest that CSB phosphorylation on S1276 is necessary to support HR repair.
Figure 8.
CSB phosphorylation on S1276 promotes efficient DNA end resection and cell survival in response to DNA damage. (A) Western analysis of AID-DIvA-U2OS CSB-KO cells expressing various Myc-CSB alleles as indicated. (B) Quantification of the amount of ssDNA. AID-DIvA-U2OS CSB-KO cells stably expressing various indicated Myc-CSB alleles were treated with no 4-OHT or 4-OHT for 4 h. The amount of ssDNA generated at position 335 nt from AsiSI-induced DSB1 was measured as described in ‘Methods’. Standard deviations from three independent experiments are indicated. (C) Quantification of cells with ≥ 10 IR-induced RPA32 foci. hTERT-RPE-CSB-KO cells expressing various Myc-CSB alleles as indicated were treated with 10 Gy IR and fixed 1 h later. Standard deviations from three independent experiments are indicated in this and subsequent panels. (D) Clonogenic survival assays of U2OS CSB-KO cells stably expressing various Myc-CSB alleles as indicated. (E) Clonogenic survival assays of hTERT-RPE-CSB-KO cells stably expressing various Myc-CSB alleles as indicated. (F) Model for the role of the CSB–BRCA1 interaction in promoting MRN- and CtIP-mediated DNA end resection. See the text for details.
DISCUSSION
We have previously reported that CSB evicts histones from chromatin surrounding DSBs, restricting RIF1 but promoting BRCA1 at DSBs (11). However, whether eviction of histones by CSB alone might be sufficient for channelling BRCA1-mediated HR repair of DSBs has not been characterized. The work presented here has revealed that CSB-mediated histone eviction and BRCA1 recruitment represent two distinct steps in CSB-regulated DSB repair (Figure 8F) and that CSB-mediated histone eviction alone is insufficient to promote HR repair of DSBs. We have shown that CSB interacts with BRCA1 in late S/G2 and that this interaction is dependent upon the BRCT domain of BRCA1 and S1276 phosphorylation of CSB. Our data suggest that the CSB–BRCA1 interaction actively promotes recruitment of the BRCA1-C complex to DSBs for DNA end resection. Our finding that disrupting the CSB–BRCA1 interaction sensitizes cells to olaparib and camptothecin further highlights the importance of this interaction in HR repair.
We have shown that the WHD of CSB interacts with MRN in early S phase. The significance of the CSB–MRN interaction in DSB repair is unknown, but is likely distinct from the BRCA1-mediated CSB–MRN interaction in late S/G2. Previously it has been reported that the WHD of CSB also mediates its interaction with RIF1 and that this interaction is required for CSB recruitment to DSBs in early S phase (11). We have observed that depletion of NBS1 impaired CSB recruitment to FokI-induced DSBs (N.L. Batenburg and X.-D. Zhu, unpublished data). Future studies would be required to investigate how MRN and RIF1 might coordinate to recruit CSB to DSBs.
We have shown that pS1276 of CSB is not a direct binding target of BRCA1–BRCT. S1655, located in the BRCA1–BRCT domain, is implicated in binding pSer (35). Our finding that a S1655A mutation abolishes the interaction of BRCA1–BRCT with CSB suggests that BRCA1–BRCT might bind a yet-to-be identified pSer of CSB. Alternatively, the interaction of CSB with BRCA1 might be mediated by a yet-to-be identified bridging factor.
Previously we have reported that CSB is phosphorylated by CDK on S158 in late S/G2 and that this phosphorylation is required for histone eviction at DSBs (11). While a S158A mutation has been reported to impair BRCA1 recruitment to DSBs and DNA end resection (11), a S158A mutation was not observed to affect the interaction of CSB with BRCA1 (N.L. Batenburg and X.-D. Zhu, unpublished data), which agrees with our finding that the N-terminal region of CSB is dispensable for its interaction with BRCA1 (Figure 4C), suggesting that CSB-mediated histone eviction is a prerequisite for CSB-directed recruitment of BRCA1-C to DSBs for DNA end resection. The work presented here has revealed S1276 of CSB as a second target site of CDK. Our current finding that a S1276A mutation does not affect CSB-mediated histone eviction but impairs its interaction with BRCA1 of the BRCA1-C complex suggests that although prerequisite, CSB-mediated histone eviction is not sufficient to support efficient recruitment of BRCA1-C to DSBs. Taken together, these results underscore a crucial role of CDK activity in fine-tuning CSB to coordinate and to ensure these two activities of CSB at the correct time for HR.
We envision a model in which histone eviction by CSB precedes active recruitment of BRCA1 by CSB to DSBs albeit the timing of these two steps are tightly linked via CDK activity in late S/G2 (Figure 8F). CDK-dependent phosphorylation of S1276 mediates the CSB–BRCA1 interaction, which promotes recruitment of the BRCA1-C complex to DSBs (Figure 8F). Subsequent initiation of DNA end resection by MRN and CtIP enforces HR as the pathway choice of DNA DSB repair. Previously, we have reported that the CSB-S158A mutant is defective not only in evicting histones but also in suppressing RIF1 accumulation at DSBs in S/G2 cells (11). In this report, we have shown that a S1276A mutation of CSB does not affect histone eviction but is defective in suppressing RIF1 accumulation at DSBs in S/G2 cells. These findings suggest that both CSB-mediated histone eviction and CSB-directed BRCA1 recruitment are essential to limit RIF1-mediated NHEJ at DSBs in late S/G2 cells and that disrupting one of them leads to misregulation in DSB repair pathway choice.
DATA AVAILABILITY
All the data used in this study are available within the article, Supplementary files or available from the authors upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to John Petrini for MRE11/RAD50/NBS1 antibodies, Roger Greenberg for U2OS-265 cells, Gaëlle Legube for AID-DIvA-U2OS cells and Daniel Durocher for BRCA1 constructs. Franciele Busatto and Daryl Ronato are thanked for technical help.
Author contributions: N.L.B. performed the majority of the experiments. J.R.W. generated expression constructs for CSB mutant alleles, MRE11, Flag- and GST–BRCA1–BRCT, produced recombinant GST–BRCA1–BRCT as well as assisted with IR experiments. A.S. generated the GFP-BRCA1 truncation alleles. Y.C. performed SPR and streptavidin bead pulldown assays. All authors contributed to data analysis and interpretation. X.D.Z. designed the project and wrote the paper with N.L.B., J.R.W. and with input from other authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) [FDN388879 to J.-Y.M., PJT159793 to X.-D.Z.]; J.-Y.M. is a FRQS Chair in Genome Stability; A.S. receives a doctoral award from CIHR. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chapman J.R., Taylor M.R., Boulton S.J.. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012; 47:497–510. [DOI] [PubMed] [Google Scholar]
- 2. Hustedt N., Durocher D.. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2016; 19:1–9. [DOI] [PubMed] [Google Scholar]
- 3. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Ann. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 4. Chen L., Nievera C.J., Lee A.Y., Wu X.. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 5. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 6. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yun M.H., Hiom K.. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009; 459:460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cruz-Garcia A., Lopez-Saavedra A., Huertas P.. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014; 9:451–459. [DOI] [PubMed] [Google Scholar]
- 9. Polato F., Callen E., Wong N., Faryabi R., Bunting S., Chen H.T., Kozak M., Kruhlak M.J., Reczek C.R., Lee W.H. et al.. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 2014; 211:1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J.H.. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992; 71:939–953. [DOI] [PubMed] [Google Scholar]
- 11. Batenburg N.L., Walker J.R., Noordermeer S.M., Moatti N., Durocher D., Zhu X.D.. ATM and CDK2 control chromatin remodeler CSB to inhibit RIF1 in DSB repair pathway choice. Nat. Commun. 2017; 8:1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Batenburg N.L., Thompson E.L., Hendrickson E.A., Zhu X.D.. Cockayne syndrome group B protein regulates DNA double-strand break repair and checkpoint activation. EMBO J. 2015; 34:1399–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takahashi T.S., Sato Y., Yamagata A., Goto-Ito S., Saijo M., Fukai S.. Structural basis of ubiquitin recognition by the winged-helix domain of Cockayne syndrome group B protein. Nucleic Acids Res. 2019; 47:3784–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Batenburg N.L., Mitchell T.R., Leach D.M., Rainbow A.J., Zhu X.D.. Cockayne Syndrome group B protein interacts with TRF2 and regulates telomere length and stability. Nucleic Acids Res. 2012; 40:9661–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G. et al.. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015; 528:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Mitchell T.R., Glenfield K., Jeyanthan K., Zhu X.D.. Arginine methylation regulates telomere length and stability. Mol. Cell Biol. 2009; 29:4918–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. et al.. A cell cycle-dependent regulatory circuit composed of 53BP1-Rif1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 18. Zhu X.D., Kuster B., Mann M., Petrini J.H., Lange T.. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 2000; 25:347–352. [DOI] [PubMed] [Google Scholar]
- 19. Wilson F.R., Ho A., Walker J.R., Zhu X.D.. Cdk-dependent phosphorylation regulates TRF1 recruitment to PML bodies and promotes C-circle production in ALT cells. J. Cell Sci. 2016; 129:2559–2572. [DOI] [PubMed] [Google Scholar]
- 20. Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A.. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010; 141:970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P. et al.. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014; 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu Y., Xiao S., Zhu X.D.. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 2007; 14:832–840. [DOI] [PubMed] [Google Scholar]
- 23. Wu Y., Mitchell T.R., Zhu X.D.. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech. Ageing Dev. 2008; 129:602–610. [DOI] [PubMed] [Google Scholar]
- 24. Zhou Y., Caron P., Legube G., Paull T.T.. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014; 42:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKerlie M., Zhu X.D.. Cyclin B-dependent kinase 1 regulates human TRF1 to modulate the resolution of sister telomeres. Nat. Commun. 2011; 2:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A.. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007; 316:1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J.. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007; 316:1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan J., Kim Y.S., Yang X.P., Li L.P., Liao G., Xia F., Jetten A.M.. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007; 67:6647–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carney J.P., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R. 3rd, Hays L., Morgan W.F., Petrini J.H.. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998; 93:477–486. [DOI] [PubMed] [Google Scholar]
- 30. Batenburg N.L., Qin J., Walker J.R., Zhu X.D.. Efficient UV repair requires disengagement of the CSB winged helix domain from the CSB ATPase domain. DNA Repair. 2018; 68:58–67. [DOI] [PubMed] [Google Scholar]
- 31. Yu X., Chini C.C., He M., Mer G., Chen J.. The BRCT domain is a phospho-protein binding domain. Science. 2003; 302:639–642. [DOI] [PubMed] [Google Scholar]
- 32. Manke I.A., Lowery D.M., Nguyen A., Yaffe M.B.. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003; 302:636–639. [DOI] [PubMed] [Google Scholar]
- 33. Kettenbach A.N., Schweppe D.K., Faherty B.K., Pechenick D., Pletnev A.A., Gerber S.A.. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 2011; 4:rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma K., D'Souza R.C., Tyanova S., Schaab C., Wisniewski J.R., Cox J., Mann M.. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014; 8:1583–1594. [DOI] [PubMed] [Google Scholar]
- 35. Williams R.S., Green R., Glover J.N.. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat. Struct. Biol. 2001; 8:838–842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this study are available within the article, Supplementary files or available from the authors upon request.