Abstract
We have previously shown that autophagy is required for post meiotic anther development including programmed cell death-mediated degradation of the tapetum and pollen maturation in rice. However, the spatiotemporal dynamics of autophagy in the tapetum remain poorly understood. We here established an in vivo imaging technique to analyze the dynamics of autophagy in rice tapetum cells by expressing green fluorescent protein-tagged AtATG8, a marker for autophagosomes. 3D-imaging analysis revealed that the number of autophagosomes/autophagy-related structures is extremely low at the tetrad stage (stage 8), and autophagy is dramatically induced at the uninucleate stages (stage 9–10) throughout the tapetal cells during anther development. The present monitoring system for autophagy offers a powerful tool to analyze the regulation of autophagy in rice tapetal cells during pollen maturation.
Keywords: autophagy, imaging, pollen maturation, rice tapetal programmed cell death
Introduction
Rice (Oryza sativa L.) is the staple food for more than half of the world’s population, making it the world’s most important food crop. Due to the importance of this plant in agriculture and scientific research, elucidating the mechanism of pollen development in rice is of considerable significance.
Reproductive development is accompanied by drastic changes in metabolism for the plentiful supply of nutrients and thus requires its appropriate regulation. In flowering plants, anthers exhibit four-layered structure composed of epidermis, endothecium, middle layer, and tapetum. Of these layers, the tapetum provides metabolites and nutrients to pollen grains, microspores, and pollen coat during their development (Ariizumi and Toriyama 2011). The tapetum includes triacylglycerol (TAG)-containing lipid bodies, which supply essential lipid components during pollen maturation (Li-Beisson et al. 2010; Murphy 2012).
As pollens develop, the tapetum is degraded to provide nutrients, metabolites, and sporopollenin precursors to the developing microspores from stage 7 to 11. Defects in tapetal degradation have been suggested to result in the development of abnormal pollen coats and grains, leading to severe male sterility (Ariizumi and Toriyama 2011; Ku et al. 2003; Li et al. 2006; Zhang et al. 2008). Tapetal degradation is tightly regulated, and characteristic features of programmed cell death (PCD) such as chromatin condensation, cell shrinkage, endoplasmic reticulum (ER) swelling, mitochondrial persistence have been reported (Rogers et al. 2005).
Autophagy is an evolutionarily conserved system for degradation of intracellular components through the vacuoles/lysosomes and has been shown to play essential roles in growth, development, and survival of eukaryotic cells (Mizushima et al. 2010). Intracellular components are enveloped by the autophagosomal membrane and fuse with the vacuoles/lysosomes, where they are broken down by lytic enzymes. These changes are referred to as autophagic flux (Kurusu et al. 2016; Yoshii and Mizushima 2017).
More than 30 known autophagy-related genes (ATG) have been identified in yeast, many of which are conserved in most eukaryotes, including animals and plants (Yoshimoto and Ohsumi 2018). The formation of autophagosomes requires two ubiquitin conjugation-like reactions with ATG12 and ATG8. The C-terminal glycine-residue of autophagy-related protein 8 (ATG8) is essential for conjugation reactions and the formation of autophagosomes (Mizushima et al. 2011). The green fluorescent protein (GFP)–ATG8 fusion protein has been used as a marker for monitoring the entire process of autophagy in animals and fungi (Klionsky et al. 2007) as well as in plant cells (Contento et al. 2005; Hanamata et al. 2013; Thompson et al. 2005; Toyooka et al. 2006; Yoshimoto et al. 2004). Autophagosomes were detected as ring-shaped and punctate structures containing GFP–ATG8 in the cytosol, which increased under nutrient starvation and stress conditions (Toyooka et al. 2006; Yoshimoto et al. 2009).
Rice mutants defective in autophagy, Osatg7-1, Osatg7-2, and Osatg9, show sporophytic severe male sterility in normal growth conditions, suggesting that autophagy is crucial for sexual reproductive development as well as phytohormone metabolism of rice (Hanamata et al. 2014; Kurusu et al. 2014, 2017). Pollens from the Osatg7-1 mutant are premature due to significant defects in the anther during pollen maturation. Of note, autophagosomes as well as autophagy-related structures including multilamellar bodies, presumably intermediate structures of autophagosomes, and dense globular bodies enclosed within the vacuoles emerge in the tapetum in the wild-type but not in the Osatg7-1 mutant, as determined by transmission electron microscopy (TEM) analysis (Hanamata et al. 2014; Kurusu et al. 2014), suggesting that autophagy contribute to tapetal degradation and PCD in rice (Kurusu and Kuchitsu 2017). Little is known, however, on the molecular links and the timing between the spatiotemporal dynamics of autophagy and tapetal PCD. Moreover, TEM is not suitable for quantification, but quantitative assessment techniques of autophagy in the tapetum have not yet been available.
We here established an in vivo imaging technique to analyze the dynamics of autophagy in rice tapetum cells by expressing GFP–ATG8 fusion protein, a marker of autophagosomes, and revealed that autophagy is dramatically induced at stage 9–10 throughout the tapetal cells during pollen maturation.
Materials and methods
Establishment of transgenic rice plants stably expressing GFP–ATG8 specifically in the tapetum
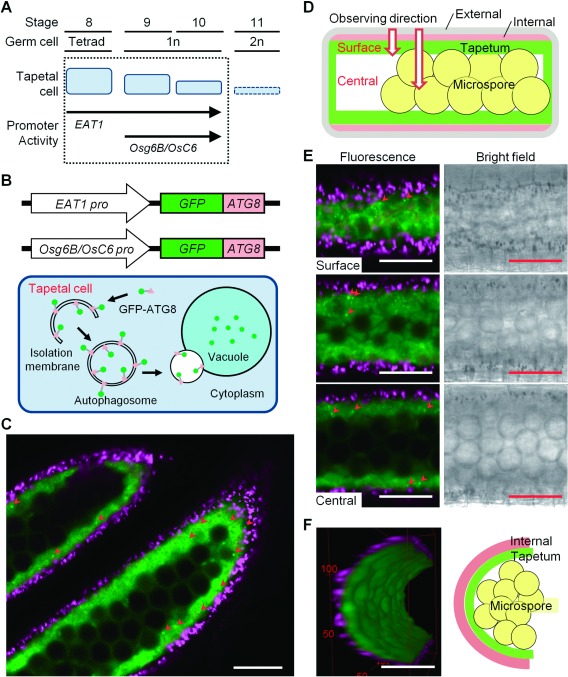
To visualize the dynamics of autophagy in rice tapetal cells at various developmental stages, we generated transgenic rice lines expressing GFP–AtATG8a (GFP–ATG8) fusion protein (Yoshimoto et al. 2004) under the control of ETERNAL TAPETUM1 (EAT1) and ANTHER SPECIFIC PROTEIN 6 (Osg6B/OsC6) promoters, which were mainly expressed in the tapetum during anther development stages (Ono et al. 2018; Yokoi et al. 1997; Zhang et al. 2010) (Figure 1A). The GFP–AtATG8a fusion construct (pBI121 binary vector) was kindly provided by Dr. Koki Yoshimoto (Meiji University, Kawasaki, Japan).
Figure 1. Imaging and quantitative characterization of autophagy based on GFP–ATG8 protein in rice tapetum during anther development and pollen maturation. (A) A schematic diagram of developmental stages of rice anthers. Tapetal cells are gradually degraded from stage 8 until 11 by programmed cell death. Stages when the activity of the two tapetum-specific promoters are active are shown. (B) A schematic diagram of visualization of the autophagy using GFP–ATG8 fusion proteins under the control of tapetum-specific promoters, EAT1 and Osg6B/OsC6. ATG8 is incorporated into the isolation membrane and the autophagosome. Autophagosomes are fused with the vacuole, where ATG8 is degraded. (C, E, F) Visualization of the dynamics of autophagosomes/autophagy-related structures in rice tapetum using transgenic plants stably expressing GFP–ATG8 under the control of EAT1 promoter. Confocal fluorescence images were obtained using a CLSM. Green and magenta fluorescence indicate GFP and autofluorescence of chlorophyll, respectively. Arrowheads indicate punctate signals of GFP–ATG8 (autophagosomes/autophagy-related structures). Scale bars: 50 µm. Similar images were also observed in plants stably expressing GFP–ATG8 under the control of Osg6B/OsC6 promoter (data not shown). (C) Cross-section of rice anthers containing immature microspores. Data are representative of three experiments. (D) A schematic diagram of a cross section of rice anther. (E) Z-Stack images of anthers from the surface to the central area. (F) 3-Dimensional reconstruction of the tapetum undergoing autophagy at stage 10 in rice anther based on the Z-Stack images. Figures were generated by rotating the cell 360° horizontally. Refer to the Supplementary Movie S1 for 3-dimensional visualization. Green signal represents GFP, red signal represents autofluorescence of chlorophyll in the endothecium.
The EAT1 promoter and terminator linked to GFP–AtATG8a fragment was constructed as follows. The pPZP-EAT1pro-GFP-NOSter plasmid was constructed from pPZP-EAT1pro-EAT1-GFP (Ono et al. 2018) by inverse PCR using primers 5′-CTT GCT CAC CAT GGA TCC TTT GGC AAA ACA GTG CTA GGC-3′ and 5′-CTG TTT TGC CAA AGG ATC CAT GGT GAG CAA GGG CG-3′, and by self-circularization using an In-Fusion HD Cloning Kit (TaKaRa, Japan). The AtATG8a fragment was amplified from a construct containing GFP-AtATG8a with the following primers: 5′-GCT GCC CGA CAA CCA CTA CCT G-3′ and 5′-TAC TTG TAC AGA GCA ACG GTA AGA GAT CCA AAAG-3′ (BsrGI site is underlined). The AtATG8a PCR product was digested with BsrGI and subcloned into the BsrGI site of pPZP-EAT1pro-GFP-NOSter (Figure 1B). The binary vector pSMA35H2-NG was kindly provided by Dr. Hiroaki Ichikawa (NARO, Tsukuba, Japan).
The Osg6B/OsC6 promoter (Accession No. GenBank: D21160.1) linked to GFP–AtATG8a fragment was constructed using PCR-based cloning. Briefly, the Osg6B/OsC6 promoter fragment was amplified by PCR from rice genome DNA (cv. Nipponbare) with the following primers: 5′-CCG GAT ATC GAA TTC TTT TTT TTA CAC AGT TCA AAG TG-3′ and 5′-acagctcctcgcccttgctcaccatGCT AGC TTA ATT AGC TTT GG-3′ for Osg6B/OsC6 containing the overlapping region of GFP-AtATG8a (EcoRV site is underlined, and lowercase indicates the overlapping region). The GFP-fused AtATG8a fragment was amplified from a construct containing GFP-AtATG8a with the following primers: 5′-attaaccaaagctaattaagctagcATG GTG AGC AAG GGC GAG GA-3′ and 5′-CGGAGCTCTCA AGC AAC GGT AAG AGA TCC AAA AGT GTT C-3′ for GFP-AtATG8a containing the overlapping region of Osg6B/OsC6 (SacI site is underlined, and lowercase indicates the overlapping region). The PCR products were linked by PCR using the following primers: 5′-CCGGATATCGAA TTC TTT TTT TTA CAC AGT TCA AAG TG-3′ and 5′-CGGAGCTCTCA AGC AAC GGT AAG AGA TCC AAA AGT GTT C-3′. The resulting product (Osg6B/OsC6-GFP-AtATG8a) was digested with EcoRV and SacI and then cloned into the binary vector pSMA35H2-NG (Chen et al. 2016) using the SacI site and a HindIII site blunted with T4 DNA polymerase.
Each construct was introduced into rice calli derived from OsATG7 (+/−) hetero seeds as well as Nipponbare using Agrobacterium-mediated transformation. Transformed calli were screened by hygromycin selection (50 µg ml−1) and transgenic plants were then regenerated. Transgenic cell lines derived from T2 plants were genotyped and used for various analyses.
Plant materials and growth conditions
Surface-sterilized seeds of transgenic rice lines (Oryza sativa L. cv. Nipponbare) were germinated on MS medium (Murashige and Skoog 1962) containing 0.8% agar and grown for 10 days in a growth chamber under long-day conditions (16-h light/8-h darkness, 28°C). Seedlings were transplanted into soil and grown in the greenhouse.
Tos17-insertional rice Osatg7-1 mutant (OsATG7−/−), wild-type (OsATG7+/+), and heterozygous (OsATG7+/−) plants were selected in seed pools obtained from heterozygous lines expressing GFP–AtATG8 by genomic PCR using the following primer mixture: OsATG7 forward primer 5′-CAT ACT ACC ACC TCA GCT TGC TAG-3,′ Tos-17 forward primer 5′-ACT ATT GTT AGG TTG CAA GTT AGT TAA GA-3,′ and OsATG7 reverse primer 5′-GCA TTC AGG AAA ACC TCG TAT CG-3′ for OsATG7. HPTII forward 5′-GAT GTA GGA GGG CGT GGA TAT GTC-3′ and HPTII reverse 5′-CTT CTA CAC AGC CAT CGG TCC AGA-3′ were used for hygromycin phosphotransferase II detection.
In vivo imaging of autophagy
Imaging analysis of rice anther was performed by a modification of the method described by (Ono et al. 2018). Rice spikelets were fixed in phosphate buffered saline without calcium and magnesium (PBS (−)) buffer containing 4% paraformaldehyde (PFA) for 3 h and washed 6 times in PBS (−) buffer for 2 h. Anther samples of different developmental stages were separated based on the length and color of the anthers (Table 1) (Zhang et al. 2011). To visualize the fluorescence of GFP, whole anthers were mounted on slides in PBS (−) buffer, and images of autophagosomes/autophagy-related structures were observed using an LSM5 EXCITER confocal fluorescence microscope with a 40× objective lens (Carl Zeiss, Germany). For all experiments, the laser intensity was adjusted to the lowest level that retained a significant signal-to-noise ratio. Images were processed using the Zeiss LSM Image Browser (Carl Zeiss, Germany) and the ImageJ software. 3D-image processing was carried out using the 3D viewer of ImageJ software. Z-Stack images obtained by CLSM were reconstructed into a 3-dimensional model.
Table 1. Anther groups defined for imaging analyses.
| Sample stages | Stage 8 | Stage 9 | Stage 10 | Stage 11 |
|---|---|---|---|---|
| Anther length (mm) | 0.8–0.9 | 0.9–1.0 | 1.1–1.5 | 1.6–1.8 |
| Colors | Transparent | Transparent | Transparent/Whitish yellow | Yellow |
Results and discussion
To visualize the dynamics of autophagy in rice tapetal cells during pollen maturation, we established transgenic rice plants stably expressing the GFP–ATG8 under the control of tapetal specific promoters, EAT1 or Osg6B/OsC6 (Figure 1A, B). As shown in Figure 1C, GFP fluorescence was detected specifically within the cytosol of tapetal cells stably expressing GFP–ATG8 under the control of EAT1 promoter. Similar images were also observed in plants stably expressing GFP–ATG under the control of Osg6B/OsC6 promoter (data not shown). We also performed Z-stack-imaging analysis to determine the spatiotemporal expression of GFP–ATG8 in rice anthers (Figure 1D). As shown in Figure 1E and F, GFP signals were detected in the inner layer of the chlorophyll autofluorescence, and autophagosomes/autophagy-related structures were clearly observed throughout the tapetum at stage 10 (Supplementary Movie S1), indicating that the GFP–ATG8 protein was expressed specifically in the tapetum in anthers.
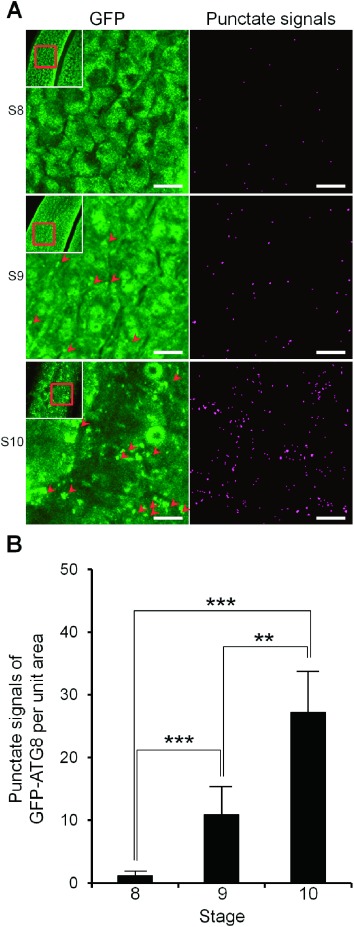
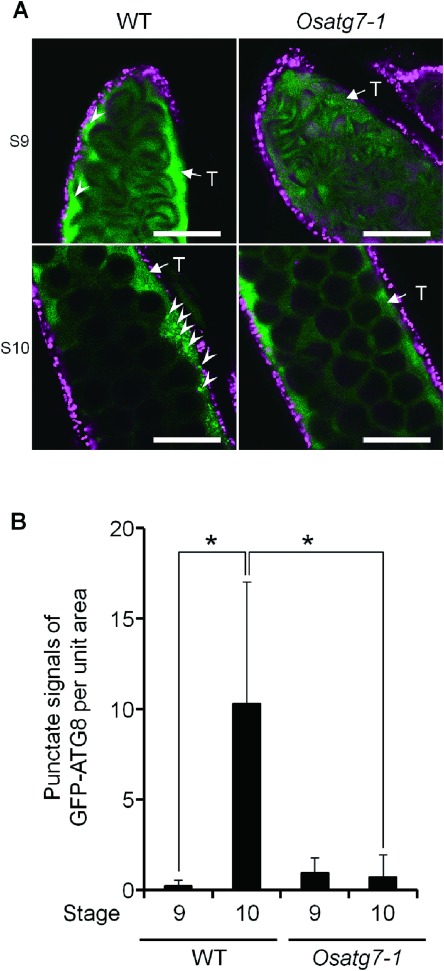
We applied this technique to analyze the spatiotemporal dynamics of autophagy in the tapetum during tapetal PCD in the wild type plants as well as the autophagy deficient mutant Osatg7-1 (Kurusu et al. 2014). ATG7 is specific to autophagy and possesses E1-like activity in the ATG12 conjugation system that is essential for autophagosome biogenesis (Li and Vierstra 2012). A few punctate signals of GFP–ATG8 were observed at the tetrad stage (stage 8), and an increase in punctate signals was observed from early uninucleate stage (stage 9) to the late uninucleate stage (stage 10) (Figure 2). Increase in the number of autophagosomes/autophagy-related structures of tapetum at stage 10 was also confirmed by the transgenic plants stably expressing GFP–ATG8 under the control of Osg6B/OsC6 promoter (Figure 3; wild-type). In contrast, no increase in punctate signals was detected in the Osatg7-1 mutant at stage 10 (Figure 3; Osatg7-1), indicating that punctate signals of GFP–ATG8 were derived from autophagy-related structures such as the autophagosomes.
Figure 2. Quantitative monitoring of autophagosomes/autophagy-related structures in the tapetal cells in each developmental stage during anther development and pollen maturation. (A) Visualization of the dynamics of autophagosomes/autophagy-related structures in rice tapetum during anther development using a transgenic plant stably expressing GFP–AtATG8a under the control of EAT1 promoter. Pictures were obtained by the maximum intensity projection (Left panels). Extraction of the punctate signals of GFP–ATG8 from left panels was performed by rolling ball background subtraction algorithm of the ImageJ software, then thresholds of all images were adjusted using Yen algorithm (Right panels). Arrowheads indicate punctate signals of GFP–AtATG8a. Scale bars: 10 µm. (B) Levels of autophagosomes/autophagy-related structures from the tetrad (stage 8) to the uninucleate stages (stage 9 and 10) in the tapetum. To quantify the levels of autophagosomes/autophagy-related structures, the number of GFP punctate signals (arrowheads) per unit area were counted at the indicated stages using analyze particles plugin of the ImageJ software. Data are means±SD; n=5 independent samples. ** p<0.005, *** p<0.0005; significantly different from each indicated stage data.
Figure 3. Numbers of autophagosomes/autophagy-related structures in the tapetal cells of the wild-type and the Osatg7-1 mutant. (A) Fluorescence images of the autophagosomes/autophagy-related structures in rice tapetum using a transgenic rice plant stably expressing GFP–AtATG8a under the control of Osg6B/OsC6 promoter. Green and magenta fluorescence indicate GFP and autofluorescence of chlorophyll, respectively. These fluorescence images show one slice containing the tapetal cells obtained by CLSM. Arrowheads indicate punctate signals of GFP–AtATG8a. Scale bars: 50 µm. T, tapetum. (B) Levels of autophagosomes/autophagy-related structures at the uninucleate stages (stage 9 and 10) in the tapetum. To quantify the levels of autophagosome formation, the number of GFP punctate signals (arrowheads) per unit area were counted at the indicated stages. Data are the means±SE of three independent experiments. * p<0.05; significantly different from the wild-type.
In the Osatg7-1 mutant in which tapetal PCD is delayed in comparison with the wild-type, intracellular organelles such as plastids and mitochondria, which could be degraded by autophagy, are clearly observed in the cytoplasm even at the bicellular stage (stage 11) (Kurusu et al. 2014). Moreover, some multilamellar bodies, presumably intermediate structures of autophagosomes (Hariri et al. 2000), were observed in the cytoplasm of the wild-type (Kurusu et al. 2014), indicating that autophagy is activated simultaneously in the tapetum from early uninucleate stage (stage 9), which may be involved in the degradation of intracellular components such as plastids and mitochondria during pollen maturation.
Appropriate temporal regulation of tapetal PCD is vital for normal pollen development. The signal initiating tapetal PCD has been suggested to be first produced during the tetrad stage (Kawanabe et al. 2006). The proper timing of tapetal PCD is tightly controlled by an evolutionarily conserved transcriptional network mediated by several key transcription factors in Arabidopsis and rice (Niu et al. 2013; Ono et al. 2018; Phan et al. 2011). Recently, the expression pattern of ATG genes imply their unique roles in some critical stages of sexual plant reproduction, such as gametophyte development, gametogenesis, and embryogenesis (Zhou et al. 2015). The proper timing of tapetal PCD may be tightly controlled by the specific expression pattern of ATGs. The potential role of autophagy in the regulation of the tapetal transcriptional network is an important topic for future research.
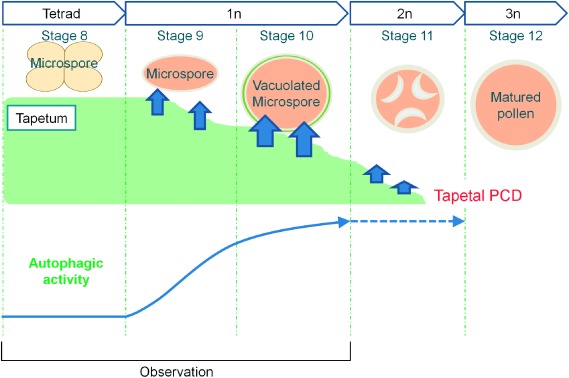
In the present study, we have discovered that the number of autophagosomes/autophagy-related structures is extremely low during early PCD/tetrad stage (stage 8), and autophagy is dramatically induced at stage 9 throughout the tapetal cells during pollen maturation (Figure 4). Since, the Osatg7-1 mutant exhibits a dense thin layer of tapetal tissue that came into contact with the orbicules, which is not observed in the wild-type even at the flowering stage (Hanamata et al. 2014), the activation of autophagy in tapetal cells may play a key role in complete tapetal breakdown in rice. Further imaging analyses of the tapetum using the system established here expressing GFP–ATG8 under the control of various tapetum-specific promoters in other developmental stages along with genetic analyses and characterization of the molecules associated with the autophagy-related structures may reveal novel functions of autophagy and their physiological significance during anther development in rice.
Figure 4. Schematic diagrams of the induction of autophagy at specific stages for rice tapetal degradation. The tapetum, the innermost of the four sporophytic layers of the anther wall directly contacts with the developing gametophytes and acts as a nutritional source for the development of microspore by undergoing degeneration triggered by PCD from stage 8 to 11. Unbroken and broken arrows indicate established and hypothetical links, respectively.
Acknowledgments
We would like to thank Ms. Yuri Sera for technical support, Dr. Kenji Hashimoto for constructive advice. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology for challenging Exploratory Research (15K14681 and 17K19274) to KK, for Scientific Research on Innovative Areas (16H01207) to KK, for Scientific Research (C) (18K05562) to TK, by the Asahi Glass Foundation to TK, and by the NIG-JOINT (84A2018) to KK.
Abbreviations
- ATGs
autophagy-related genes
- CLSM
confocal laser scanning microscope
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- PCD
programmed cell death
- PFA
paraformaldehyde
- TAG
triacylglycerol
- TEM
transmission electron microscopy
Supplementary Data
References
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Chen L, Guan L, Toyomoto D, Sugita T, Hamaguchi T, Okabe R (2016) Plant regeneration and its functional analysis within transgenic rice of ASG-1, an apomixis-specific gene isolated from apomictic Guinea Grass. Biotech J Inter 16: 1–13 [Google Scholar]
- Contento AL, Xiong Y, Bassham DC (2005) Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP–AtATG8e fusion protein. Plant J 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Hanamata S, Kurusu T, Kuchitsu K (2014) Roles of autophagy in male reproductive development in plants. Front Plant Sci 5: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamata S, Kurusu T, Okada M, Suda A, Kawamura K, Tsukada E, Kuchitsu K (2013) In vivo imaging and quantitative monitoring of autophagic flux in tobacco BY-2 cells. Plant Signal Behav 8: e22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri M, Millane G, Guimond MP, Guay G, Dennis JW, Nabi IR (2000) Biogenesis of multilamellar bodies via autophagy. Mol Biol Cell 11: 255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanabe T, Ariizumi T, Kawai-Yamada M, Uchimiya H, Toriyama K (2006) Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol 47: 784–787 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO (2007) Methods for monitoring autophagy from yeast to human. Autophagy 3: 181–206 [DOI] [PubMed] [Google Scholar]
- Ku S, Yoon H, Suh HS, Chung YY (2003) Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217: 559–565 [DOI] [PubMed] [Google Scholar]
- Kurusu T, Hanamata S, Kuchitsu K (2016) Quantitative live cell imaging of autophagic flux and roles of autophagy in reproductive development in plants. Bioimages 24: 1–11 [Google Scholar]
- Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, et al. (2014) OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10: 878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu T, Koyano T, Kitahata N, Kojima M, Hanamata S, Sakakibara H, Kuchitsu K (2017) Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal Behav 12: e1365211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu T, Kuchitsu K (2017) Autophagy, programmed cell death and reactive oxygen species in sexual reproduction in plants. J Plant Res 130: 491–499 [DOI] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. Arabidopsis Book 8: e0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murphy DJ (2012) The dynamic roles of intracellular lipid droplets: From archaea to mammals. Protoplasma 249: 541–585 [DOI] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Ono S, Liu H, Tsuda K, Fukai E, Tanaka K, Sasaki T, Nonomura KI (2018) EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet 14: e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LA, Dubos C, Surman C, Willment J, Cullis IF, Mansfield SD, Campbell MM (2005) Comparison of lignin deposition in three ectopic lignification mutants. New Phytol 168: 123–140 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K, Moriyasu Y, Goto Y, Takeuchi M, Fukuda H, Matsuoka K (2006) Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2: 96–106 [DOI] [PubMed] [Google Scholar]
- Yokoi S, Tsuchiya T, Toriyama K, Hinata K (1997) Tapetum-specific expression of the Osg6B promoter-β-glucuronidase gene in transgenic rice. Plant Cell Rep 16: 363–367 [DOI] [PubMed] [Google Scholar]
- Yoshii SR, Mizushima N (2017) Monitoring and measuring autophagy. Int J Mol Sci 18: 1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Ohsumi Y (2018) Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol 59: 1337–1344 [DOI] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant 1: 599–610 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang W, Yen Y, Long H, Deng G, Pan Z, Yu M (2010) A novel herbicide-inducible male sterility system. J Sci Food Agric 90: 2526–2530 [DOI] [PubMed] [Google Scholar]
- Zhou XM, Zhao P, Wang W, Zou J, Cheng TH, Peng XB, Sun MX (2015) A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res 22: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.