Abstract
A spectroscopic study of the interactions of Λ- and Δ-[Ru(phen)2(dppz)]2+ with i-motif DNA containing thymine loops of various lengths. In the presence of i-motifs, the luminescence of the Λ enantiomer was enhanced much more than the Δ. Despite this, the effect of each enantiomer on i-motif thermal stability was comparable. The sequences most affected by [Ru(phen)2(dppz)]2+ were those with long thymine loops; this suggests that long-looped i-motifs are attractive targets for potential transition metal complex drugs and should be explored further in drug design.
Keywords: i-motif, DNA, ruthenium, melting, stabilization, luminescence
Introduction
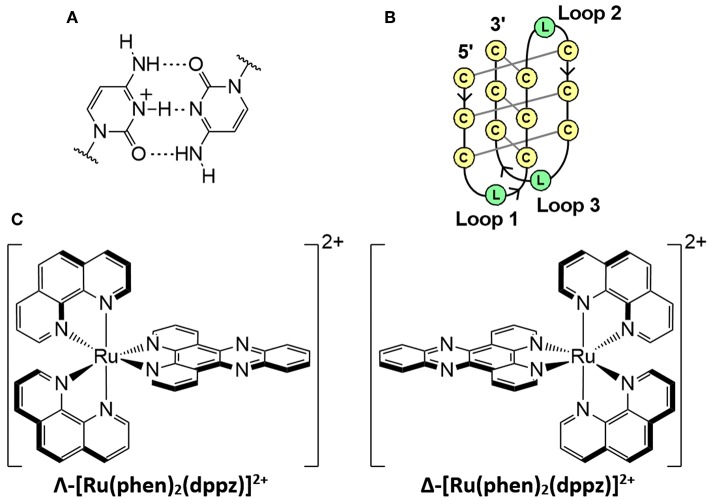
The intercalated motif (i-motif) is a DNA structure containing intercalated cytosine+-cytosine base pairs between four strands (Figures 1A,B) (Gehring et al., 1993; Školáková et al., 2019). I-motifs were originally thought to only form in acidic pH due to the protonation of cytosine required; however, stable i-motif formation has been reported at alkaline (Zhou et al., 2010) and neutral pH (Day et al., 2013; Fujii and Sugimoto, 2015; Wright et al., 2017), as well as conditions mimicking physiological molecular crowding (Rajendran et al., 2010). Moreover, recent demonstrations of the presence of i-motifs in the nuclei of human cells (Dzatko et al., 2018; Zeraati et al., 2018), and their ability to inhibit DNA polymerase (Takahashi et al., 2017) has increased interest in their biological function (Abou assi et al., 2018). Sequences that are complementary to those that form G-quadruplexes have been shown to form i-motif structures in the promoter regions of several cancer genes (Brooks et al., 2010; Brazier et al., 2012; Li et al., 2016). These include the transcription factors that code the cellular myelocytomatosis (c-Myc) (Mathur et al., 2004) and B-cell lymphoma-2 (Bcl-2) oncogenes, the latter of which is overexpressed in some cancers and may be underexpressed in some neurodegenerative diseases (Bar-Am et al., 2005; Knight et al., 2019). The transcription factor hnRNP LL reportedly binds to i-motifs, which suggests that the i-motif acts as a recognition site for the activation of transcription of Bcl-2(Kang et al., 2014).
Figure 1.
(A) Hemi-protonated cytosine+-cytosine base pair. (B) Schematic diagram of an intramolecular i-motif where C is cytosine and L represents the loop regions with any DNA base. (C) Enantiomers of the polypyridyl ruthenium complex; [Ru(phen)2(dppz)]2+.
The unique structure and potential biological roles of the i-motif makes it an attractive binding target for small molecules, particularly if the binding results in stabilization. For example, the porphyrin TMPyP4 reportedly binds to i-motifs similar in structure to the human telomeric sequence [5′-(C3TAA)3C3-3′] (Fedoroff et al., 2000). A variety of other i-motif-binding ligands have been reported, including carbon nanotubes (Li et al., 2006), bis-acridines (Alberti et al., 2001), mitoxantrone (Wright et al., 2016), crystal violet (Ma et al., 2011), and derivatives of thiazole orange (Sheng et al., 2017) and coumarin (Satpathi et al., 2019). An under-researched family of ligands in the field of i-motifs is transition metal complexes, of which i-motif binding studies are limited (Shi et al., 2010a,b; Lu et al., 2015). Metal complexes have been studied as DNA binders for decades due to their uses as therapeutic and diagnostic agents (Metcalfe and Thomas, 2003; Pages et al., 2015; Deo et al., 2016). In particular, polypyridyl ruthenium complexes have been extensively researched for medicinal purposes due to their high stability, affinity for DNA, and luminescence properties (Gill and Thomas, 2012; Deo et al., 2016; Poynton et al., 2017). The complex [Ru(phen)2(dppz)]2+ (Figure 1C, where phen = 1,10-phenanthroline; dppz = dipyrido[3,2-a:2′3′-c]phenazine) and derivatives demonstrate a DNA “light switch” effect, in which luminescence is greatly enhanced when bound to DNA (Friedman et al., 1990; Cardin et al., 2017). The equilibrium between dark and emissive states is influenced by both changes in solvent environment around the ancillary phen ligands and changes in the hydrogen bonding of solvent molecules with the pyrazine nitrogen atoms of the dppz ligand (Chantzis et al., 2013; Véry et al., 2014). The emissive state occurs during intercalative binding of [Ru(phen)2(dppz)]2+ with DNA (Hartshorn and Barton, 1992; Olofsson et al., 2004). Solution and crystallographic studies have revealed that this complex and its derivatives bind to duplex and G-quadruplex DNA (Wilson et al., 2013; Hall et al., 2016; McQuaid et al., 2019a), and can inhibit telomerase activity (Yu et al., 2012). Preliminary work has demonstrated non-specific binding between i-motifs and rac-[Ru(phen)2(dppz)]2+ (rac-Ru) (Shi et al., 2010a,b).
We have previously demonstrated that the stability of i-motifs is influenced by loop length, with longer loops resulting in lower overall stability and vice-versa; this is likely due to the differences in flexibility of the loop regions (Gurung et al., 2015). We speculate that these regions are more desirable binding sites as they aren't as tightly packed as the intercalated cytosine core. In fact, the nature of the lateral loops of the Bcl-2 i-motif was determined to be vital to the binding potency of IMC-48 (Kang et al., 2014). Here we report the first study of interactions between enantiomerically resolved ruthenium complexes and i-motifs with various loop lengths. We have used synchrotron radiation circular dichroism (SRCD), UV, and luminescence spectroscopy to probe the binding of Λ-[Ru(phen)2(dppz)]2+ (Λ-Ru) and Δ-[Ru(phen)2(dppz)]2+ (Δ-Ru) to a series of i-motif sequences. These consisted of a block of three paired cytosines and combinations of long and short thymine loops. Loop lengths were either uniform (C3TX, e.g., C3T4) or combinations of 3 and 8 thymines (C3TXXX e.g., C3T383), with C3T3/C3T333, and C3T8/C3T888 being in both “groups” (Table 1).
Table 1.
DNA melting temperatures (TM) of the C3TX and C3TXXX sequences (1 μM ss) with and without 1 equiv. Λ-Ru or Δ-Ru.
| Label | Sequence 5′ → 3′ | Native | Λ-Ru | Δ-Ru | ||
|---|---|---|---|---|---|---|
| TM | TM | ΔTM | TM | ΔTM | ||
| C3T3/333 | (CCCTTT)3CCC | 60.0 | 60.3 | +0.3 | 60.3 | +0.3 |
| C3T4 | (CCCTTTT)3CCC | 59.6 | 58.9 | −0.7 | 58.7 | −0.9 |
| C3T5 | (CCCTTTTT)3CCC | 51.7 | 51.4 | −0.3 | 51.4 | −0.3 |
| C3T6 | (CCCTTTTTT)3CCC | 47.0 | 48.4 | +1.4 | 48.2 | +1.2 |
| C3T7 | (CCCTTTTTTT)3CCC | 41.9 | 45.8 | +3.9 | 45.7 | +3.8 |
| C3T8/888 | (CCCTTTTTTTT)3CCC | 37.8 | 44.1 | +6.3 | 43.2 | +5.4 |
| C3T338 | CCCTTTCCCTTTCCCTTTTTTTTCCC | 49.1 | 50.8 | +1.7 | 50.5 | +1.4 |
| C3T383 | CCCTTTCCCTTTTTTTTCCCTTTCCC | 55.3 | 55.2 | −0.1 | 55.0 | −0.3 |
| C3T388 | CCCTTTCCCTTTTTTTTCCCTTTTTTTTCCC | 46.1 | 48.1 | +2.0 | 47.2 | +1.1 |
| C3T833 | CCCTTTTTTTTCCCTTTCCCTTTCCC | 49.5 | 51.0 | +1.5 | 50.8 | +1.3 |
| C3T838 | CCCTTTTTTTTCCCTTTCCCTTTTTTTTCCC | 39.8 | 45.7 | +6.0 | 45.6 | +5.9 |
| C3T883 | CCCTTTTTTTTCCCTTTTTTTTCCCTTTCCC | 46.6 | 48.8 | +2.2 | 47.8 | +1.2 |
Values are in degrees Celsius with a standard deviation of ±~0.1–0.6°C. Standard deviation for individual measurements are shown in Table S3.1.
Materials and Methods
Materials
Λ- and Δ-[Ru(phen)2dppz]2+ were synthesized and resolved through our previously reported methods; full details are included in the Supporting Information (Ortmans et al., 2004; McQuaid et al., 2019a). Unless otherwise stated, all materials and chemicals were sourced from Sigma-Aldrich (Merck) or Honeywell research chemicals. Oligonucleotides were obtained from Eurogentec (RP-HPLC purified) and used without further purification. All solvents were obtained at HPLC grade and used without further purification.
Solution Preparation and Annealing
Initial stock solutions of the ruthenium complexes and oligonucleotides were made in water and checked for concentration using the extinction coefficient of 20 000 M−1 cm−1 at 440 nm for the former, and for the latter, the Eurogentec-provided extinction coefficients at 260 nm, calculated using the nearest-neighbor model. The stocks were then diluted with buffer and combined to form solutions with either a 1:0 or 1:1 ratio of DNA strand to ruthenium complex. Annealing of the oligonucleotides, both with and without ruthenium complex present, was achieved by incubating the buffered solution at 90°C for 5 min, and then allowing it to cool to room temperature overnight. Preliminary experiments showed that adding ruthenium complex before or after i-motif annealing did not affect the melting temperature (Figure S3.1), and so all ruthenium additions in this study were done pre-annealing.
Synchrotron Radiation Circular Dichroism
Samples consisted of the oligonucleotide [100 μM single stranded (ss)] and ruthenium complex (100 μM) in 20 mM sodium cacodylate buffer at pH 5. The concentration of buffer was lowered relative to other experiments to obtain the lowest data resolution cut-off. CD spectra were recorded at 20 °C between 195 and 350 nm with a 1 nm increment. Experiments were performed in a 0.01 cm pathlength cuvette, on beamline B23 at Diamond Light Source Ltd.
UV Melting
UV melting experiments were carried out using Agilent Cary 100 with a temperature controlled six-cell changer. Samples consisted of the oligonucleotide (1 μM ss) and either 0 or 1 molar equivalent of the ruthenium complex. The buffer consisted of 50 mM sodium cacodylate at a pH of either 5 or 8. Absorption was recorded at 260 and 295 nm at 1 °C intervals between 20–90°C, with a temperature change rate of 0.5 °C/min in a 1 cm pathlength quartz cuvette. Melting curves were generated from this data. To determine the melting temperature, the curves were fitted with a sigmoidal function: where the constants are H, St, S and TM: the height, steepness, starting point and inflexion point (melting temperature) of the function, respectively, and the variable is T, the temperature. This function was generated for each curve through minimization of the sum of residuals between the raw data and model. The fit was applied to the region that best represented a sigmoid. Experiments were performed in triplicate and the results averaged.
Luminescence Spectroscopy
Luminescence spectroscopy measurements were performed using a 1 cm pathlength quartz cell at room temperature. Samples consisted of oligonucleotides and ruthenium complex with a final concentration of 20 μM each (ss DNA), dissolved in 50 mM sodium cacodylate buffer (pH 5 or 8). Emission spectra were measured between 550 and 875 nm with an excitation wavelength of 440 nm.
Results and Discussion
Synchrotron Radiation Circular Dichroism
SRCD spectra of the C3TX sequences were obtained with and without the presence of rac-Ru at pH 5 (Figure S2.1). Rac-Ru had to be used here as the CD signal of Λ-Ru and Δ-Ru would obscure that of the i-motif. Only minor changes in CD spectra were observed with addition of rac-Ru, demonstrating that the i-motif structure still forms in the presence of ruthenium complexes. This is supported by the UV melting profiles of both native and ruthenium-bound C3TX and C3TXXX, which show characteristic hyperchromicity at 260 nm and hypochromicity at 295 nm (Figures S3.2–5) (Phan and Mergny, 2002).
UV Melting
UV melting experiments revealed that for C3TX sequences with loops of 6 thymines or less, there was no appreciable difference (<1.5°C) in melting temperature (TM) with one equivalent of Λ-Ru or Δ-Ru, while stabilization of ~4°C occurred for C3T7 and ~6°C for C3T8 (Table 1, Figure 2A, and Figure S3.7). This result suggests that the longer loops have cavities that allow binding of ruthenium complexes, resulting in stabilization. For C3TXXX, sequences with longer loops 1 and 3 experienced the most stabilization with ruthenium, while a long loop 2 did not affect stabilization (Figure 2B, Table 1). For example, the ΔTM of C3T838 was ~6°C, while C3T383 was not stabilized. Interestingly, the sequences with the lowest native stability were those that were stabilized by Λ-Ru and Δ-Ru the most. There was little difference in the effect of Λ-Ru and Δ-Ru on sequence stability, although Λ-Ru did occasionally produce higher ΔTM values. These experiments were repeated at pH 8 for C3T3 and C3T8; a melting curve was not observed, implying an absence of i-motif structure (Figure S3.6).
Figure 2.
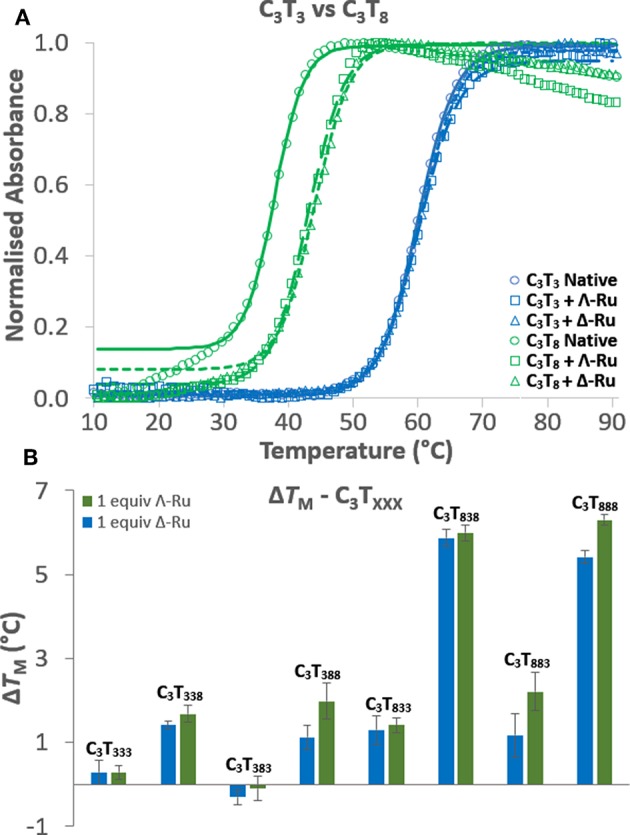
(A) UV melting curves of C3T3 (blue) and C3T8 (green) in their native forms (circles) and in the presence of Λ-Ru (squares) or Δ-Ru (triangles). (B) Comparison of the ΔTM induced by Λ-Ru (green) and Δ-Ru (blue) for C3TXXX.
Luminescence
Enhancement of luminescence was observed for Λ-Ru and Δ-Ru when bound to all i-motif sequences, aside from C3T3, for which little enhancement occurred (Figure 3, Figure S4.1). For non-C3T3 sequences, emission was much higher for Λ-Ru than for Δ-Ru, meaning that the solvent environment and pyrazine nitrogen exposure were more favorable for luminescence of Λ-Ru. Without the presence of DNA, the emission of each complex was very low (Figure 3, Figure S4.1). For C3TX, emission generally increased with loop length, with a large increase for C3T7 and C3T8 (Figure S4.1). For C3TXXX, trends differed between Λ-Ru and Δ-Ru. For Δ-Ru, C3T838 and C3T888 produced the highest emission (>300x Ru alone), C3T333 showed relatively little enhancement (20x Ru alone), and the other sequences showed relatively moderate enhancement (100-140x Ru alone). For Λ-Ru, emission was more varied per sequence and was correlated with loop length (Figure 3). A long loop 2 resulted in more than double the emission of a long loop 1 or 3, as evidenced by the higher emission of C3T383− (600x Ru alone) relative to C3T338, C3T833 (200-250x Ru alone), and C3T838 (550x Ru alone).
Figure 3.
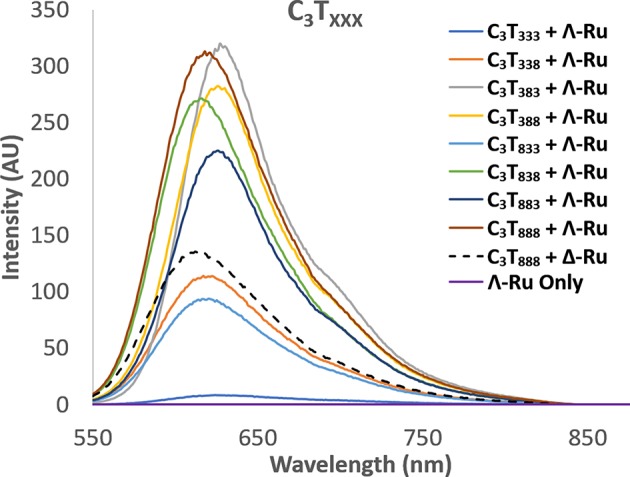
Luminescence spectra of Λ-Ru (20 μM) with C3TXXX (20 μM ss). The spectrum of Δ-Ru with C3T888 is also shown for comparison (broken line).
In addition to luminescence enhancement, the emission maxima shifted depending on the sequence. For Δ-Ru, λmax was generally blue-shifted with increasing loop length (Table S4.1, Figure S4.3). This correlated positively with emission intensity, which has been observed for ruthenium complexes that undergo stacking with G-quadruplex DNA (Wilson et al., 2010). However, for Λ-Ru, there are few trends relating λmax to loop length, or to intensity (Figure S4.3); this overall suggests while Δ-Ru is likely to undergo base stacking with i-motif loops, Λ-Ru may also interact through different modes. The luminescence of Λ-Ru and Δ-Ru with C3T3 and C3T8 was also recorded at pH 8; interestingly, emission enhancement still occurred despite the lack of i-motif structure. Relative to their i-motif forms, C3T3 and C3T8 produced higher and lower enhancement, respectively (Figure S4.2). Luminescence enhancement of [Ru(phen)2(dppz)]2+ due to interactions with single stranded sequences without tertiary structure has been previously reported (Coates et al., 2001), and could explain this phenomenon.
Melting vs. Luminescence
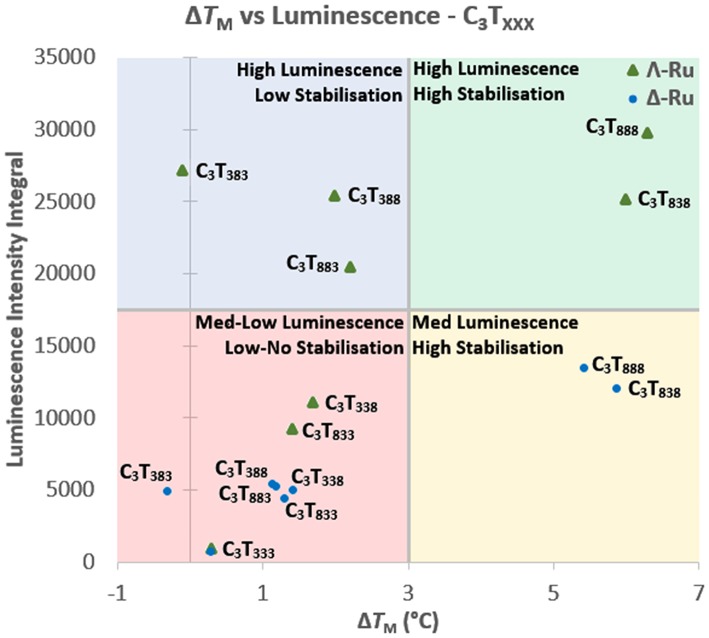
When comparing the UV melting and luminescence data for these complexes, some trends do emerge. For C3TX, the binding of Λ-Ru and Δ-Ru to longer-looped sequences resulted in both higher ΔTM and higher ruthenium emission (Figure S5.1). For shorter loops, luminescence generally increased with increasing length, but melting stabilization did not. For the C3TXXX series, there are some trends relating emission, TM and the position of the long loops (Figure 4). The luminescence of Λ-Ru indicates that it binds to longer loops irrespective of where they are positioned, but the TM data only shows stabilization when bound to loops 1 or 3. Λ-Ru demonstrated high luminescence with C3T383, but no stabilization, whereas interaction with C3T838 and C3T888 resulted in high luminescence and stabilization. C3T333 was not stabilized and did not enhance luminescence. The other sequences were slightly stabilized and produced medium-low luminescence enhancement (Figure 4). Overall, the presence of Λ-Ru with longer loops 1 and 3 resulted in increased stability and luminescence, while only luminescence was increased with a long loop 2. Δ-Ru did not follow the same C3TXXX trends as Λ-Ru, aside from low luminescence and ΔTM for C3T333 (Figure 4). Luminescence and ΔTM were only notable for C3T888 and C3T838, while these values were medium-low for the remaining sequences. Unlike Λ-Ru, the interactions of Δ-Ru with a long loop 2 did not increase luminescence relative to loops 1 or 3, despite resulting in approximately the same thermal stabilization as Λ-Ru.
Figure 4.
Comparison of the integrated luminescence intensity and ΔTM of Λ-Ru (green triangles) and Δ-Ru (blue circles) when bound with C3TXXX. The graph is quartered into sections based upon the relative luminescence enhancement and thermal stabilization for each i-motif-ruthenium combination.
Conclusion
In this study we have demonstrated that the length and position of i-motif thymine loops not only impacts the native structure, but also the degree of stabilization by Λ-Ru and Δ-Ru. These complexes do not stabilize short-looped sequences, but do stabilize the relatively less stable, long-looped i-motifs. It is possible that the longer loops are more flexible, and that they may form T-T hairpins; we recently reported binding of Λ-[Ru(tetraazaphenanthrene)2(dppz)]2+ to mismatched T-T base pairs and speculate something similar may be occurring here in the longer i-motif loops (McQuaid et al., 2019b). The luminescence blue-shifting implies that base stacking and perhaps other modes may contribute to ruthenium-loop interactions, but further spectroscopic and crystallographic experiments are required to elucidate the true binding behavior. Overall, these results demonstrate that i-motif sequences with longer loops are potential transition metal drug targets for therapeutic and diagnostic purposes.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
BP led the writing of the manuscript and performed UV and luminescence experiments. SG performed the SRCD experiments, supported by JH. BP and SG each were responsible for analysis of their collected data. KM synthesized and resolved the ruthenium enantiomers. CC and JB conceived the project and supervised the work. All authors contributed to the writing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the University of Reading for access to instruments in the Chemical Analysis Facility. BP was supported by the BBSRC grant BB/P021328/1. SG was supported by a joint Ph.D. studentship between the University of Reading and Diamond Light Source. KM was supported by a joint Ph.D. studentship between the Engineering and Physical Sciences Research Council and Diamond Light Source.
Footnotes
Funding. This work was supported by the Biotechnology and Biological Sciences Research Council grant BB/M004635/1 and BB/P021328/1 (to JH, CC, and JB). The authors are also grateful to Diamond Light Source for provision of beamtime on beamline B23 (SM19962 and SM16002).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2019.00744/full#supplementary-material
References
- Abou assi H., Garavís M., González C., Damha M. J. (2018). i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 46, 8038–8056. 10.1093/nar/gky735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti P., Ren J., Teulade-Fichou M. P., Guittat L., Riou J.-F., Chaires J. B., et al. (2001). Interaction of an acridine dimer with DNA quadruplex structures. J. Biomol. Struct. Dyn. 19, 505–513. 10.1080/07391102.2001.10506758 [DOI] [PubMed] [Google Scholar]
- Bar-Am O., Weinreb O., Amit T., Youdim M. B. H. (2005). Regulation of Bcl-2 family proteins, neurotrophic factors, and APP processing in the neurorescue activity of propargylamine. FASEB J. 19, 1899–1901. 10.1096/fj.05-3794fje [DOI] [PubMed] [Google Scholar]
- Brazier J. A., Shah A., Brown G. D. (2012). I-Motif formation in gene promoters: unusually stable formation in sequences complementary to known G-quadruplexes. Chem. Commun. 48, 10739–10741. 10.1039/C2CC30863K [DOI] [PubMed] [Google Scholar]
- Brooks T. A., Kendrick S., Hurley L. (2010). Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 277, 3459–3469. 10.1111/j.1742-4658.2010.07759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin C. J., Kelly J. M., Quinn S. J. (2017). Photochemically active DNA-intercalating ruthenium and related complexes – insights by combining crystallography and transient spectroscopy. Chem. Sci. 8, 4705–4723. 10.1039/c7sc01070b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantzis A., Very T., Daniel C., Monari A., Assfeld X. (2013). Theoretical evidence of photo-induced charge transfer from DNA to intercalated ruthenium (II) organometallic complexes. Chem. Phys. Lett. 578, 133–137. 10.1016/j.cplett.2013.05.068 [DOI] [Google Scholar]
- Coates C. G., Mcgarvey J. J., Callaghan P. L., Coletti M., Hamilton J. G. (2001). Probing the Interaction of [Ru(phen)2(dppz)]2+ with single-stranded DNA what degree of protection is required for operation of the “Light-Switch Effect”? J. Phys. Chem. B. 105, 730–735. 10.1021/jp002856w [DOI] [Google Scholar]
- Day H. A., Huguin C., Waller Z. A. E. (2013). Silver cations fold i-motif at neutral pH. Chem. Commun. 49, 7696–7698. 10.1039/C3CC43495H [DOI] [PubMed] [Google Scholar]
- Deo K. M., Pages B. J., Ang D. L., Gordon C. P., Aldrich-Wright J. R. (2016). Transition metal intercalators as anticancer agents—recent advances. Int. J. Mol. Sci. 17, 1818. 10.3390/ijms17111818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzatko S., Krafcikova M., Hänsel-Hertsch R., Fessl T., Fiala R., Loja T., et al. (2018). Evaluation of the stability of DNA i-motifs in the nuclei of living mammalian cells. Angew. Chem. Int. Ed. 57, 2165–2169. 10.1002/anie.201712284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff O. Y., Rangan A., Chemeris V. V., Hurley L. H. (2000). Cationic porphyrins promote the formation of i-Motif DNA and bind peripherally by a nonintercalative mechanism. Biochemistry 39, 15083–15090. 10.1021/bi001528j [DOI] [PubMed] [Google Scholar]
- Friedman A. E., Chambron J. C., Sauvage J. P., Turro N. J., Barton J. K. (1990). A molecular light switch for DNA: Ru(bpy)2(dppz)2+. J. Am. Chem. Soc. 112, 4960–4962. 10.1021/ja00168a052 [DOI] [Google Scholar]
- Fujii T., Sugimoto N. (2015). Loop nucleotides impact the stability of intrastrand i-motif structures at neutral pH. Phys. Chem. Chem. Phys. 17, 16719–16722. 10.1039/C5CP02794B [DOI] [PubMed] [Google Scholar]
- Gehring K., Leroy J.-L., Guéron M. (1993). A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 363, 561–565. 10.1038/363561a0 [DOI] [PubMed] [Google Scholar]
- Gill M. R., Thomas J. A. (2012). Ruthenium(ii) polypyridyl complexes and DNA-from structural probes to cellular imaging and therapeutics. Chem. Soc. Rev. 41, 3179–3192. 10.1039/C2CS15299A [DOI] [PubMed] [Google Scholar]
- Gurung S. P., Schwarz C., Hall J. P., Cardin C. J., Brazier J. A. (2015). The importance of loop length on the stability of i-motif structures. Chem. Commun. 51, 5630–5632. 10.1039/C4CC07279K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. P., Keane P. M., Beer H., Buchner K., Winter G., Sorensen T. L., et al. (2016). Delta chirality ruthenium ‘light-switch' complexes can bind in the minor groove of DNA with five different binding modes. Nucleic Acids Res. 44, 9472–9482. 10.1093/nar/gkw753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorn R. M., Barton J. K. (1992). Novel dipyridophenazine complexes of ruthenium(II): exploring luminescent reporters of DNA. J. Am. Chem. Soc. 114, 5919–5925. 10.1021/ja00041a002 [DOI] [Google Scholar]
- Kang H.-J., Kendrick S., Hecht S. M., Hurley L. H. (2014). The transcriptional complex between the BCL2 i-Motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 136, 4172–4185. 10.1021/ja4109352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight T., Luedtke D., Edwards H., Taub J. W., Ge Y. (2019). A delicate balance – The BCL-2 family and its role in apoptosis, oncogenesis, and cancer therapeutics. Biochem. Pharmacol. 162, 250–261. 10.1016/j.bcp.2019.01.015 [DOI] [PubMed] [Google Scholar]
- Li H., Hai J., Zhou J., Yuan G. (2016). The formation and characteristics of the i-motif structure within the promoter of the c-myb proto-oncogene. J. Photochem. Photobiol., B 162, 625–632. 10.1016/j.jphotobiol.2016.07.035 [DOI] [PubMed] [Google Scholar]
- Li X., Peng Y., Ren J., Qu X. (2006). Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Nat. Acad. Sci. U.S.A. 103:19658. 10.1073/pnas.0607245103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Wang M., Liu L.-J., Wong C.-Y., Leung C.-H., Ma D.-L. (2015). A luminescence switch-on probe for terminal deoxynucleotidyl transferase (TdT) activity detection by using an iridium(iii)-based i-motif probe. Chem. Commun. 51, 9953–9956. 10.1039/C5CC02790J [DOI] [PubMed] [Google Scholar]
- Ma D.-L., Kwan M. H.-T., Chan D. S.-H., Lee P., Yang H., Ma V. P.-Y., et al. (2011). Crystal violet as a fluorescent switch-on probe for i-motif: label-free DNA-based logic gate. Analyst 136, 2692–2696. 10.1039/C1AN15091J [DOI] [PubMed] [Google Scholar]
- Mathur V., Verma A., Maiti S., Chowdhury S. (2004). Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Biochem. Biophys. Res. Commun. 320, 1220–1227. 10.1016/j.bbrc.2004.06.074 [DOI] [PubMed] [Google Scholar]
- McQuaid K., Abell H., Gurung S. P., Allan D. R., Winter G., Sorensen T., et al. (2019a). Structural studies reveal enantiospecific recognition of a DNA G-quadruplex by a ruthenium polypyridyl complex. Angew. Chem. Int. Ed. 58, 9881–9885. 10.1002/anie.201814502 [DOI] [PubMed] [Google Scholar]
- McQuaid K., Hall J. P., Baumgaertner L., Cardin D. J., Cardin C. J. (2019b). Three thymine/adenine binding modes of the ruthenium complex Λ-[Ru(TAP)2(dppz)]2+ to the G-quadruplex forming sequence d(TAGGGTT) shown by X-ray crystallography. Chem. Commun. 55, 9116–9119. 10.1039/C9CC04316K [DOI] [PubMed] [Google Scholar]
- Metcalfe C., Thomas J. A. (2003). Kinetically inert transition metal complexes that reversibly bind to DNA. Chem. Soc. Rev. 32, 215–224. 10.1039/B201945K [DOI] [PubMed] [Google Scholar]
- Olofsson J., Önfelt B., Lincoln P. (2004). Three-State Light Switch of [Ru(phen)2dppz]2+: Distinct Excited-State Species with Two, One, or No Hydrogen Bonds from Solvent. J. Phys. Chem. A 108, 4391–4398. 10.1021/jp037967k [DOI] [Google Scholar]
- Ortmans I., Elias B., Kelly J. M., Moucheron C., Kirsch-Demesmaeker A. (2004). [Ru(TAP)2(dppz)]2+: a DNA intercalating complex, which luminesces strongly in water and undergoes photo-induced proton-coupled electron transfer with guanosine-5′-monophosphate. Dalton Trans. 2004, 668–676. 10.1039/B313213G [DOI] [PubMed] [Google Scholar]
- Pages B. J., Ang D. L., Wright E. P., Aldrich-Wright J. R. (2015). Metal complex interactions with DNA. Dalton Trans. 44, 3505–3526. 10.1039/C4DT02700K [DOI] [PubMed] [Google Scholar]
- Phan A. T., Mergny J. L. (2002). Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 30, 4618–4625. 10.1093/nar/gkf597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynton F. E., Bright S. A., Blasco S., Williams D. C., Kelly J. M., Gunnlaugsson T. (2017). The development of ruthenium(ii) polypyridyl complexes and conjugates for in vitro cellular and in vivo applications. Chem. Soc. Rev. 46, 7706–7756. 10.1039/C7CS00680B [DOI] [PubMed] [Google Scholar]
- Rajendran A., Nakano S.-I., Sugimoto N. (2010). Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 46, 1299–1301. 10.1039/B922050J [DOI] [PubMed] [Google Scholar]
- Satpathi S., Sappati S., Das K., Hazra P. (2019). Structural characteristics requisite for the ligand-based selective detection of i-motif DNA. Org. Biomol. Chem. 17, 5392–5399. 10.1039/C9OB01020C [DOI] [PubMed] [Google Scholar]
- Sheng Q., Neaverson J. C., Mahmoud T., Stevenson C. E. M., Matthews S. E., Waller Z. A. E. (2017). Identification of new DNA i-motif binding ligands through a fluorescent intercalator displacement assay. Org. Biomol. Chem. 15, 5669–5673. 10.1039/C7OB00710H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Geng X., Zhao J., Yao T., Wang C., Yang D., et al. (2010a). Interaction of [Ru(bpy)2(dppz)]2+ with human telomeric DNA: Preferential binding to G-quadruplexes over i-motif. Biochim. 92, 370–377. 10.1016/j.biochi.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Shi S., Zhao J., Geng X., Yao T., Huang H., Liu T., et al. (2010b). Molecular “light switch” for G-quadruplexes and i-motif of human telomeric DNA: [Ru(phen)2(dppz)]2+. Dalton Trans. 39, 2490–2493. 10.1039/B916094A [DOI] [PubMed] [Google Scholar]
- Školáková P., Renčiuk D., Palacký J., Krafčík D., Dvoráková Z., Kejnovská I., et al. (2019). Systematic investigation of sequence requirements for DNA i-motif formation. Nucleic Acids Res. 47, 2177–2189. 10.1093/nar/gkz046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Brazier J. A., Sugimoto N. (2017). Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc. Nat. Acad. Sci. 114, 9605–9610. 10.1073/pnas.1704258114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry T., Ambrosek D., Otsuka M., Gourlaouen C., Assfeld X., Monari A., et al. (2014). Photophysical properties of ruthenium(II) Polypyridyl DNA intercalators: effects of the molecular surroundings investigated by theory. Chem. Eur. J. 20, 12901–12909. 10.1002/chem.201402963 [DOI] [PubMed] [Google Scholar]
- Wilson T., Costa P. J., Félix V., Williamson M. P., Thomas J. A. (2013). Structural Studies on Dinuclear ruthenium(II) complexes that bind diastereoselectively to an antiparallel folded human telomere sequence. J. Med. Chem. 56, 8674–8683. 10.1021/jm401119b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T., Williamson M. P., Thomas J. A. (2010). Differentiating quadruplexes: binding preferences of a luminescent dinuclear ruthenium(ii) complex with four-stranded DNA structures. Org. Biomol. Chem. 8, 2617–2621. 10.1039/B924263E [DOI] [PubMed] [Google Scholar]
- Wright E. P., Day H. A., Ibrahim A. M., Kumar J., Boswell L. J. E., Huguin C., et al. (2016). Mitoxantrone and analogues bind and stabilize i-Motif forming DNA sequences. Sci. Rep. 6:39456. 10.1038/srep39456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. P., Huppert J. L., Waller Z. A. E. (2017). Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 45, 2951–2959. 10.1093/nar/gkx090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Liu Y., Wang C., Sun D., Yang X., Liu Y., et al. (2012). Chiral Ruthenium(II) polypyridyl complexes: stabilization of G-Quadruplex DNA, inhibition of telomerase activity and cellular uptake. PLoS ONE 7:e50902. 10.1371/journal.pone.0050902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeraati M., Langley D. B., Schofield P., Moye A. L., Rouet R., Hughes W.E., et al. (2018). I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 10, 631–637. 10.1038/s41557-018-0046-3 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wei C., Jia G., Wang X., Feng Z., Li C. (2010). Formation of i-motif structure at neutral and slightly alkaline pH. Mol. Biosyst. 6, 580–586. 10.1039/B919600E [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.