Abstract
Preparation of dry starters for alcohol production is an age-old traditional technology in the Eastern Himalayan regions of east Nepal, the Darjeeling hills, Sikkim, and Arunachal Pradesh in India, and Bhutan. We studied the bacterial diversity in 35 samples of traditionally prepared dry starters, represented by marcha of Nepal, Sikkim, the Darjeeling hills, and Bhutan, phab of Bhutan, and paa, pee, and phut of Arunachal Pradesh, respectively. Populations of bacteria in these starters were 105 to 108 cfu/g. A total of 201 bacterial strains were isolated from starter samples, phenotypically characterized, and their identities confirmed by the 16S rRNA sanger sequencing method. The dominant phylum was Firmicutes (85%), followed by Proteobacteria (9%), and Actinobacteria (6%). Lactic acid bacteria (LAB) (59%) formed the most abundant group, followed by non-LAB (32%) and Gram-negative bacteria (9%). Based on the 16S rRNA gene sequencing result, we identified LAB: Enterococcus durans, E. faecium, E. fecalis, E. hirae, E. lactis, Pediococcus acidilactici, P. pentosaceus, Lactobacillus plantarum subsp. plantarum, Lb. pentosus, Leuconostoc mesenteroides, and Weissella cibaria; non-LAB: Bacillus subtilis subsp. inaquosorum, B. circulans, B. albus, B. cereus, B. nakamurai, B. nitratireducens, B. pseudomycoides, B. zhangzhouensis, Kocuria rosea, Staphylococcus hominis subsp. hominis, S. warneri, S. gallinarum, S. sciuri, Lysinibacillus boronitolerans, Brevibacterium frigoritolerans, and Micrococcus yunnanensis; Gram-negative bacteria: Pseudomonas putida, Klebsiella pneumoniae, Enterobacter hormaechei subsp. xiangfangensis, E. hormaechei subsp. steigerwaltii, and Stenotrophomonas maltophilia. We characterized diversity indexes of the bacterial community present in traditionally prepared dry starters. This is the first report on the bacterial diversity of traditionally dry starters of the Eastern Himalayas by sanger sequencing.
Keywords: Eastern Himalayas, starters, 16S rRNA sequencing, bacterial diversity, lactic acid bacteria
Introduction
The Himalayas, well known for high mountains with natural beauty and rich biological resources, extend from peak Nanga Parbat in Pakistan to peak Namcha Barwa across India, Nepal, and Bhutan (Le Fort, 1975). Based on geo-morphology and demography, the Himalayas are divided into three regions, the Western, Central, and Eastern Himalayas (Nandy et al., 2006). The geographical location of the Eastern Himalayas extends from eastern Nepal, North East India (Darjeeling hills, Sikkim, and Arunachal Pradesh), Bhutan, and Tibet Autonomous Regions in China (Saha, 2013). Agrarian and pastoral types of mountain farming dominate the agriculture and animal husbandry systems in the Eastern Himalayas, and these are practiced by diverse ethnic communities (Sharma et al., 2007; Bhasin, 2013). Many major and rare types of ethnic fermented foods and beverages are traditionally produced from locally available plant and animal resources and are made into a wide variety of flavorsome cuisine that is consumed as staple diets, side-dishes, curries, soups, condiments, and alcoholic drinks by ethnic people of the Eastern Himalayas (Tamang, 2010; Tamang et al., 2012). The majority of ethnic Himalayan people drink home-made traditional alcoholic beverages and distilled liquor prepared from cereals (rice, finger millets, and maize) as per socio-compulsion but also for enjoyment. Vinification, malting, and brewing processes for alcohol production are completely unknown in the food culture of the Himalayan people; instead, rice or finger millets are fermented into mildly alcoholic (∼4%) beverages (Thapa and Tamang, 2004) by using dry starters, which are unique to these regions.
The Himalayan people have been practicing the art of starter-making using indigenous technology for centuries by using overnight-soaked and pounded rice flours mixed with wild herbs, spices, and 1–2% of previously prepared dry starters in powder form to make doughs. Doughs mixtures with desirable shapes and sizes are placed in fresh fern leaves and allowed to ferment for 2–3 days at room temperature, and the freshly fermented doughs are then sun dried for 2–3 days to get dry starters (Thakur et al., 2015; Anupma et al., 2018). Every ethnic community in the Western, Central, and Eastern Himalayas prepare amylase and alcohol-producing starters with slight variation in the use of substrates, such as rice or wheat, and wrapping materials, such as fern fronds, paddy straw, or plant leaves. In local languages, these are termed marcha in Nepal, the Darjeeling hills, and Sikkim in India (Shrivastava et al., 2012; Thakur et al., 2015; Anupma et al., 2018), mana and manapu in Nepal (Nikkuni et al., 1996), phab in Bhutan (Tamang, 2010), chowan in Tripura, dawdim in Mizoram, humao, modor pitha in Assam, hamei in Manipur, khekhrii in Nagaland, and phut in Arunachal Pradesh (Anupma et al., 2018) in India. Similar types of alcohol-producing starters are also prepared in South East Asia by ethnic Asian communities, such as the Vietnamese benh (Dung et al., 2007), Korean nuruk (Jung et al., 2012), Indonesian ragi (Surono, 2016), Philippine bubod (Kozaki and Uchimura, 1990), Chinese daque or chiu or chu (Chen et al., 2014), Thai loogpang (Limtong et al., 2002), and Cambodian dombea (Ly et al., 2018). The most remarkable advent in the traditional preparation of starter cultures is the practice of the “back-slopping method” (terminology in modern food microbiology) used by ethnic Asians irrespective of their geographical locations for sub-culturing the desirable and essential microbiota.
Traditionally prepared dry starters show coexistence of mixed microbiota represented by different genera and species of filamentous molds (Hesseltine et al., 1988; Tamang et al., 1988; Sha et al., 2019), yeasts (Hesseltine and Kurtzman, 1990; Jeyaram et al., 2008, 2011; Sha et al., 2017, 2018, 2019), and bacteria (Hesseltine and Ray, 1988; Tamang et al., 2007; Sha et al., 2017) for saccharification (Lee and Lee, 2002; Thapa and Tamang, 2004), liquefaction (Pervez et al., 2014), and ethanol production (Tsuyoshi et al., 2005; Zheng et al., 2011) to produce traditional alcoholic beverages and distilled liquor in many South East Asian countries, including Nepal, India, and Bhutan in the Himalayas. Filamentous molds (species of Rhizopus, Mucor, Aspergillus), and yeasts (species of Saccharomyces, Pichia, Sacharomycopsis, Candida) are involved in saccharification and liquefaction; they produce amylolytic enzymes for degrading starch into sugars, and the main alcohol-producing yeasts are Saccharomyces for alcohol production (Nout and Aidoo, 2002; Thapa and Tamang, 2004; Li et al., 2012; Nile, 2015). Besides the saccharifying and alcohol-producing ability of mycelia molds and yeasts, some bacterial species present in starters also contribute by imparting flavor, antagonism, and acidification onto the fermenting substrates (Tamang et al., 2007; Huang et al., 2017). Extensive profiling of the diversity of yeasts and mycelial molds in various traditionally prepared dry starters collected from different places of North East India have been reported earlier (Tamang et al., 1988; Tamang and Sarkar, 1995; Tsuyoshi et al., 2005; Jeyaram et al., 2008, 2011; Bora et al., 2016; Sha et al., 2017, 2018, 2019). Samples of marcha collected from the Darjeeling hills and Sikkim were analyzed earlier and reported few species of bacteria: Pediococcus pentosaceus (Tamang and Sarkar, 1995), Pediococcus pentosaceus and Lb. brevis (Tamang et al., 2007), Acetobacter, Fructobacillus, Lactococcus, Lactobacillus, Leuconostoc, Burkholderia, and Gluconacetobacter (Sha et al., 2017). However, no published reports on bacterial diversity associated with marcha in Nepal and Bhutan, phab in Bhutan, and paa, pee, and phut in Arunachal Pradesh are available to date. Marcha (Figures 1A–D) is a dry rice-based starter, prepared by the Gorkha/Nepali community in the Darjeeling hills and Sikkim in India, east Nepal, and south Bhutan, to ferment boiled finger-millets into a sweet-sour, mildly alcoholic beverage called kodo ko jaanr or chyang (Tamang et al., 1996). Marcha is prepared from soaked and pounded rice flours mixed with some wild herbs, few spices, 1–2% of previously prepared powdered marcha by the back-slopping method to make doughs that are placed in fresh ferns leaves, are allowed to ferment for 2–3 days, and are then sun dried for 2–3 days to get dry starters. Phab or pho (Figure 1E) is a dark brown, flattened, cake-like starter prepared from powdered maize by the Drukpa community in Bhutan to produce a home-made distilled alcoholic drink called ara from barley and finger millets (Anupma et al., 2018). Paa (Figure 1F), pee (Figure 1G), and phut (Figure 1H) are dry starters prepared from rice by the Nyshing, Apatani, and Mongpa communities of Arunachal Pradesh, respectively (Anupma et al., 2018). Pee is used to ferment rice into a mildly alcoholic beverage called opo by the Nyshing tribes, a mildly alcoholic drink called apong by the Apatani, and phut is used to prepare a sweet-sour, mildly alcoholic beverage called themsing by the Mongpa tribes of Arunachal Pradesh (Shrivastava et al., 2012). Preparation of marcha, phab, paa, pee, and phut is more or less similar except for some variation in the use of substrates, such as rice in the case of marcha, phut, paa, and pee and maize-rice husk in phab, and wrapping materials, of which fern leaves are used for fermenting rice flour during marcha preparation, dry paddy straws are used for phab preparation, and locally available plant leaves are used for the preparation of paa, pee, and phut. We collected dry samples of marcha, pee, paa, phut, and phab from different places in the Eastern Himalayan regions of Nepal, India, and Bhutan to profile the bacterial diversity as information on yeasts and the mycelial molds community is already available (Sha et al., 2017, 2018, 2019). The present study aimed to profile bacterial diversity isolated from marcha, pee, paa, phut, and phab based on phenotypic and biochemical tests that use the 16S rRNA gene sequencing method.
FIGURE 1.
Different types of dry starters from the Eastern Himalayas: (A) Marcha from Nepal, (B) Marcha from Darjeeling, (C) Marcha from Sikkim, (D) Marcha from Bhutan, (E) Phab from Bhutan, (F) Paa from Arunachal Pradesh, (G) Pee from Arunachal Pradesh, and (H) Phut from Arunachal Pradesh.
Materials and Methods
Samples
A total of 35 samples of traditionally prepared dry starters were collected in pre-sterile poly bags from different places located in the Eastern Himalayas viz marcha (8 samples) from Nepal, marcha (5) from the Darjeeling hills, marcha (8) from Sikkim, marcha (5) from Bhutan, paa (2), pee (3), and phut (2) from Arunachal Pradesh, and phab (2) from Bhutan (Table 1). Collected samples were transported and kept in a desiccator at room temperature since traditionally sun-dried starters are stored in a dry place for more than a year (Tamang et al., 1996).
TABLE 1.
Bacterial load of dry starters from the Eastern Himalayas.
| Sample | Altitude | Moisture | cfu/g | |||||
| Sample | Region | Collection Site | (Meter) | Latitude | Longitude | content (%) | pH | (×107) |
| Marcha | Nepal | Dharan | 371 | 26°48′ N | 87°17′ E | 12.5 (9.6−17.0) | 5.6 (5.5−5.9) | 2.1 (1.1−2.9) |
| Dhankuta | 1154 | 26°53′ N | 87°8 ′ E | |||||
| Hiley | 857 | 27°02′ N | 87°24′ E | |||||
| Hathikharka | 1394 | 27°01′ N | 87°32′ E | |||||
| Marcha | Darjeeling hills | Darjeeling | 2059 | 27°04′ N | 88°26′ E | 13.1 (12.9−13.3) | 5.4 (5.2−5.6) | 15.3 (11.0−19.6) |
| Kalimpong | 1176 | 27°07′ N | 88°47′ E | |||||
| Marcha | Sikkim | Pakyong | 1341 | 27°24′ N | 88°59′ E | 11.8 (10.0−13.4) | 5.7 (5.6−5.9) | 18.5 (10.2−26.5) |
| Gangtok | 1637 | 27°32′ N | 88°61′E | |||||
| Recab | 1072 | 27°21′ N | 88°50′E | |||||
| Basilakha | 906 | 27°22′ N | 88°60′ E | |||||
| Marcha | Bhutan | Gedumari | 1045 | 26°90′ N | 89°39′E | 13.76 (11.8−15.72) | 5.7 (5.5−5.9) | 0.01 (0.01−0.02) |
| Thimphu | 2401 | 27°47′ N | 89°62′ E | |||||
| Paa | Arunachal Pradesh | Lower Subansiri | 661 | 27°8′ N | 93°6′E | 11.7 (11−12) | 5.1 (5−5.2) | 2.3 (2.0−2.6) |
| Pee | Ziro valley | 1576 | 27°53′ N | 93°81′E | 12.1 (11−13) | 5.5 (5.2−5.8) | 17.6 (16.8−18.4) | |
| Phut | Upper Subansiri | 1816 | 28°3′ N | 94°E | 11.6 (11.4−11.8) | 5.2 (5.1−5.3) | 11.5 (9.8−13.2) | |
| Phab | Bhutan | Dhonakha | 2311 | 27°66′ N | 89°70′ E | 6.17 (6.13−6.2) | 5.2 (5.0−5.4) | 0.03 (0.02−0.04) |
Analysis of Moisture and pH
The moisture content of the samples was estimated by a moisture analyzer (OHAUS/MB-45, United States). The pH of the samples was determined by homogenizing 1 g of sample in 10 mL of distilled water, and the readings were taken using a digital pH-meter (Orion 910003, Thermo Fisher Scientific, United States).
Microbiological Analysis
Dry starter samples were taken from a desiccator, coarsely crushed by a sterile spatula, and 10 g of the powered sample was then homogenized with 90 mL of 0.85% physiological saline in a stomacher lab blender 40 (Seward, United Kingdom) for 2 min. The homogenized samples were serially diluted in the same diluents, and 1 mL of appropriate diluents was then plated using specific media by the pour plate method. Nutrient agar (MM102, HiMedia, Mumbai, India) for aerobic mesophilic bacterial count, MRS (Man-Rogosa-Sharpe) agar (M641, HiMedia, Mumbai, India) and M17 Agar Base (M929, HiMedia, Mumbai, India) for lactic acid bacteria (LAB), and VRBGA (violet red bile glucose agar) (M581, HiMedia, Mumbai, India) for Gram-negative bacteria were used for the enumeration of bacteria in respective plates. Nutrient agar plates and VRBGA plates were incubated at 37°C for 24 h, and MRS plates and M17 plates were incubated at 30°C for 24–48 h aerobically. The number of colonies was counted as colony forming unit cfu/g. The purity of colonies was maintained by re-streaking them into fresh medium, and this was further confirmed by microscopic examination. The pure colonies were then preserved in 50% glycerol at −20°C for further identification and analysis.
Phenotypic and Biochemical Characterization
Bacterial isolates were phenotypically characterized for their presumptive identification, and groupings were done on the basis of cell morphology, Gram’s reaction, colony morphology, catalase test, sporulation tests, gas production from glucose, and ammonia production from arginine (Holt et al., 1994). The physiological tests including growth at different pHs, temperatures, and salt tolerance were performed (Tamang et al., 2007). Biochemical characterization of isolates such as sugar fermentation tests, IMViC (Indole, Methyl red; Voges-Proskauer and Citrate) tests specifically for Gram-negative isolates, nitrate reduction tests, and urease tests were also performed using the method of Hammes and Hertel (2003).
Genotypic Characterization
Genomic DNA Extraction
The genomic DNA of each bacterial isolate was extracted by the standard phenol/chloroform method of Cheng and Jiang (2006) with slight modifications. A total of 1 ml of culture grown overnight in MRS broth (M369, HiMedia, Mumbai, India) at 30°C was centrifuged at 8,000 rpm for 10 min. The pellets were centrifuged at 3,000 rpm, suspended in 40 μl 1× TE buffer, and freshly prepared 15 μl lysozyme and 15 μl RNAse enzyme were added to the pellets and incubated at 37°C for 3 h. After incubation, 15 μl of 20% SDS (sodium dodecyl sulfate) and 15 μl of proteinase-K were added and further incubated at 55°C for 3 h. An equal volume of phenol-chloroform solution (49:48) was added to the above mixture, centrifuged at 10,000 rpm for 15 min, and the aqueous upper layer formed was transferred to a fresh vial containing chloroform-isoamyl solution (48:1). It was centrifuged again at 10,000 rpm for 15 min, and the upper aqueous layer formed was transferred to a fresh vial containing 15 μl of 3M sodium acetate and 400 μl of cold absolute alcohol and kept at −20°C for 1 h. The mixture was again centrifuged at 10,000 rpm for 30 min, and the pellets were washed with 70% ethanol and further centrifuged at 10,000 rpm for 30 min. The pellets were then collected, air dried, and suspended in 30 μl 1× TE buffer and stored at −20°C for further analysis. The quality of the genomic DNA was checked by electrophoresis in 0.8% agarose gel and quantified using a NanoDrop spectrometer (ND-1000 spectrometer, NanoDrop technologies, Willington, CT, United States) (Kumbhare et al., 2015).
PCR Amplification
The PCR of the 16S rRNA gene from the isolated genomic DNA was amplified using a universal oligonucleotide primer pair 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) (Lane, 1991) in a Thermal cycler (Applied Biosystems-2720, United States). The reaction mixture, conditions, and protocol for the polymerase chain reaction amplification were performed following the method of Chagnaud et al. (2001). PCR amplification was performed in a mixture containing a final volume of 50 μl of Go green Taq master mix (1×) (NEB), 10 μM of F primer, 10 μM of R primer, and nuclease-free water (NEB). The PCR reaction program was set under the following PCR conditions: 94°C for 10 min; 94°C for 1 min, 65°C for 1 min, 72°C for 30 s for 35 cycles, and 72°C for 7 min. PCR products were detected by electrophoresis using 1% agarose, and the bands were stained with 7 μl/100 mL of ethidium bromide (RM813, HiMedia, Mumbai, India) and visualized in UV source Gel-Doc 1000 (Bio-Rad, 97-0186-02, United States). A standard 100 base pair DNA ladder (HiMedia, Mumbai, India) was used for the verification of amplicon size.
Purification of the PCR Amplicons
The amplified PCR products were then purified using PEG (polyethylene glycol)-NaCl (sodium chloride) precipitation (20% w/v of PEG, 2.5 M NaCl) precipitation method with little modifications of method described by Schmitz and Riesner (2006). About 0.6 volume of 20% PEG-NaCl was added to final volume of PCR products and incubated at 37°C for 30 min. After centrifugation at 12,000 rpm for 30 min, the aqueous solution was discarded, the pellet was washed twice with freshly prepared ethanol (70%) by centrifugation at 12,000 rpm for 30 min. The collected pellet was then air-dried overnight and 20 μl of nuclease-free water was added, and the final purified product was loaded in 1% agarose gel.
16S rRNA Gene Sequencing
PCR products were set up in 5 μl volume for single primer amplification with the same universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) (Lane, 1991) for separate reactions for each primer. PCR reaction was set as follows: denaturation for (96°C, 10 s), annealing (50°C, 5 s), and elongation (60°C, 2 min) with a stop reaction at 4°C. The amplicons were then precipitated with 1 μl sodium acetate (3M, pH 5.2) and 24 μl of absolute alcohol, mixed briefly in vortex and incubated at room temperature for 15 min, spun at 12,000 rpm for 20 min, further washed with 70% ethanol, air-dried, and suspended in 10 μl formamide. Sequencing of the amplicons was performed by the Sanger Sequencing method or the Chain-termination DNA (Sanger et al., 1977), the automation of a modified Sanger method that is commonly used to check the sequence of the templates (Heather and Chain, 2016), was carried out in an automated DNA Analyzer (ABI 3730XL Capillary Sequencers, Applied Biosystems, Foster City, CA, United States).
Bioinformatics
The sequence quality was checked by Sequence Scanner v.1.0 (Applied Biosystems, Foster City, CA, United States). After checking the sequence quality, the sequences were assembled using a ChromasPro 1.5 (McCarty, 1998). The orientation of the assembled sequences was checked using an orientation checker v.1.0. The identity of bacterial isolates was assigned by comparing their DNA sequences with those available in the GenBank NCBI (National Center for Biotechnology Information) database using a BLAST (basic local alignment search tool) 2.0 program (Altschul et al., 1990). The sequences were then aligned by pairwise alignment using clustalW, and the phylogenetic tree was constructed using MEGA7.0 software by the neighbor joining method (Gascuel and Steel, 2006; Kumar et al., 2016). Diversity indices were calculated using a PAST (PAleontological STatistics) v.3.25, which is a comprehensive statistics package used in many fields of life sciences, economics, earth science, engineering, and paleontology (Hammer et al., 2001). The Chao 1 value for species richness was calculated following the method of Chao and Chiu (2016).
Data Availability
The sequences retrieved from the 16S rRNA sequencing were deposited at GenBank-NCBI under the nucleotide accession number: MK748250-MK748278, MK202997-MK203032, and MK752675-MK752677.
Results
Microbial Population
Populations of bacteria in 35 samples of traditionally prepared dry starters collected from different regions of the Eastern Himalayas were 1.0 × 105 to 2.7 × 108 cfu/g (Table 1). The moisture contents of all samples analyzed were 10%–17% except for phab of Bhutan in which the moisture content was comparatively low (<6%). Average pH of all samples was 5.5 (Table 1).
Phenotypic Characterization
We isolated 201 total bacterial isolates from 35 different samples of traditionally prepared starters collected from the Eastern Himalayas, which were represented by 139 isolates from marcha (Sikkim 49; Darjeeling 38; Nepal 34, Bhutan 18), 12 isolates from paa (Arunachal Pradesh), 17 isolates from pee (Arunachal Pradesh), 11 isolates from phut (Arunachal Pradesh), and 22 isolates from phab (Bhutan). All 201 bacterial isolates were phenotypically characterized based on various biochemical and physiological parameters (Table 2). A total of nine different bacterial genera including unidentified group were presumptively identified based on phenotypic results following Bergey’s manual of bacteriological classification (Holt et al., 1994), which were mostly represented by Gram-positive bacteria (Pediococcus, Lactobacillus, Enterococcus, Leuconostoc, Bacillus, and Staphylococcus) and two Gram-negative bacteria (Enterobacter and Citrobacter). We randomly grouped 201 isolates into 68 representative bacterial strains based on phenotypic characterization results (data not shown).
TABLE 2.
Phenotypic characterization of bacterial isolates from dry starters from the Eastern Himalayas.
| Presumptive Identification (Total number of isolates) | Tolerance | IMViC test | |||||||||||||||||||||||||
| Sugar fermentation | NaCl | pH | Temperature(°C) | Indole | MR | VP | Citrate | Urease | Nitrate | ||||||||||||||||||
| Cellobiose | Raffinose | Sorbitol | Arabinose | Mellibiose | Xylose | Lactose | Ribose | Melizitose | Glucose | Sucrose | Mannitol | Rhamnose | 5% | 10% | 3.6 | 9.6 | 10.6 | 15 | 10 | 45 | |||||||
| Leuconostoc (15) | +(6) −(9) | +(7) −(8) | +(5) −(10) | +(4) −(11) | +(9) −(6) | −(10) +(5) | +(9) −(5) v(1) | −(6) +(8) v(1) | +(10) −(5) | + | +(8) −(6) v(1) | +(12) −(3) | +(4) −(10) v(1) | + | +(1) −(14) | + | +(5) −(10) | +(9) −(6) | +(12) −(3) | IMViC test was not determined for Gram-positive bacteria. | |||||||
| Enterococcus (41) | +(18) −(23) | +(29) −(12) | +(23) −(18) | +(14) −(27) | +(34) −(7) | +(18) −(24) | +(34) −(6) v(1) | +(34) −(7) | +(33) −(3) v(5) | + | −(7) +(34) | +(7) −(34) | +(23) −(17) v(1) | +(36) −(5) | +(6) −(30) v(5) | +(6) −(35) | +(35) −(6) | +(13) −(28) | +(32) −(4) v(5) | +(35) −(6) | |||||||
| Pediococcus (57) | +(19) −(38) | +(27) −(30) | +(19) −(38) | +(23) −(26) v(8) | +(28) −(29) | +(23) −(29) v(5) | +(36) −(12) v(9) | +(49) −(5) v(3) | +(32) −(24) v(1) | + | +(33) −(24) | +(13) −(44) | +(38) −(18) v(1) | +(48) v(9) | +(4) −(51) v(2) | +(3) −(51) v(3) | +(40) −(16) v(1) | +(5) −(52) | +(7) −(44) v(6) | +(38) −(13) v(6) | +(46) −(9) v(2) | ||||||
| Lactobacillus (15) | +(12) −(3) | +(5) −(10) | +(10) −(5) | +(5) −(10) | +(10) −(5) | +(5) −(10) | + | +(12) −(3) | +(4) −(11) | + | +(12) −(3) | +(10) −(2) v(3) | +(4) −(11) | + | +(7) −(8) | +(10) −(5) | +(3) −(6) v(6) | + | +(9) −(6) | +(5) −(5) v(5) | |||||||
| Bacillus (21) | +(17) −(4) | +(3) −(18) | +(3) −(18) | +(3) −(18) | +(3) −(18) | +(6) −(15) | +(15) −(6) | +(17) −(4) | +(9) −(12) | +(18) −(3) | −(11) +(10) | +(15) −(6) | + | +(7) −(14) | +(6) −(15) | +(17) v(4) | + | +(19) v(2) | + | ||||||||
| Staphylococcus (13) | +(8 −(5) | +(6) −(5) v(2) | +(5) −(8) | +(4) −(9) | +(6) −(7) | +(5) −(6) v(2) | +(9) −(2) v(2) | +(5) −(8) | +(3) −(10) | + | +(8) −(4) v(1) | +(8) −(5) | +(8) −(3) v(2) | +(6) −(2) v(5) | +(2) −(9) v(2) | +(2) −(9) v(2) | +(5) −(7) v(1) | +(2) −(10) v(1) | +(5) −(7) v(1) | +(9) −(3) v(1) | |||||||
| Unidentified (21) | +(12) −(9) | +(7) −(14) | +(4) −(17) | +(11) −(8) v(2) | +(8) −(13) | +(9) −(10) v(2) | +(12) −(6) v(3) | +(18) −(3) | +(12) −(9) | + | +(11) −(10) | +(3) −(18) | +(12) −(9) | + | +(2) −(19) | +(1) −(19) v(1) | +(14) −(6) v(1) | +(3) −(17) v(1) | +(11) −(8) v(2) | +(15) −(3) v(3) | +(15) −(4) v(2) | ||||||
| Enterobacter (8) | + | − | +(2) −(6) | + | − | + | − | +(6) −(2) | − | + | + | + | + | Physiological tests were not done for Gram–negative bacteria | − | − | + | + | + | + | |||||||
| Citrobacter (10) | +(5) −(5) | - | − | +(5) −(5) | +(8) −(2) | +(7) −(3) | +(8) −(2) | −(7) +(3) | −(5) v(5) | −(7) +(3) | − | − | − | +(6) −(4) | −(6) +(4) | +(6) −(4) | |||||||||||
+,Positive; −,Negative; v, Variable; Numbers in parenthesis indicates number of isolates; All strains of Gram-negative bacteria were tested for IMViC either +/−. +,Positive; −,Negative; v, Variable; Numbers in parenthesis indicates number of isolates; All strains of Gram-negative bacteria were tested for IMViC either ±.
Molecular Identification of Bacterial Isolates
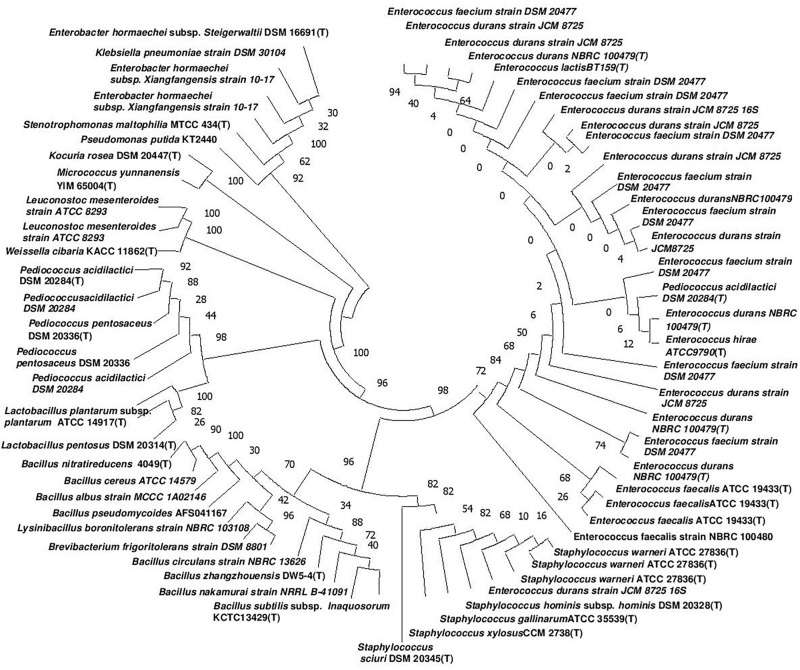
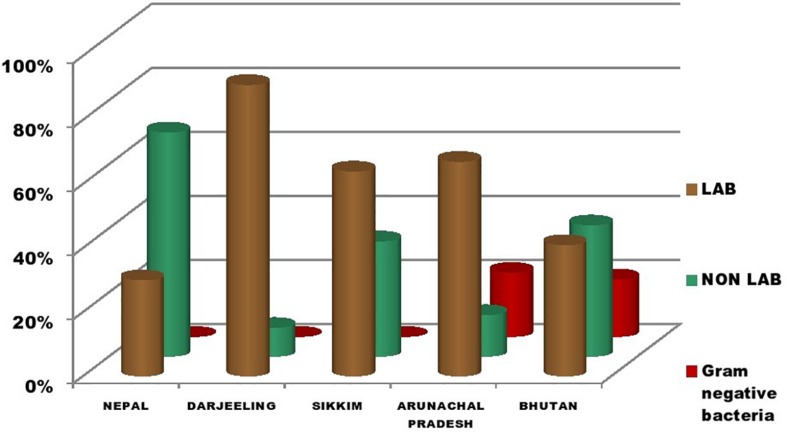
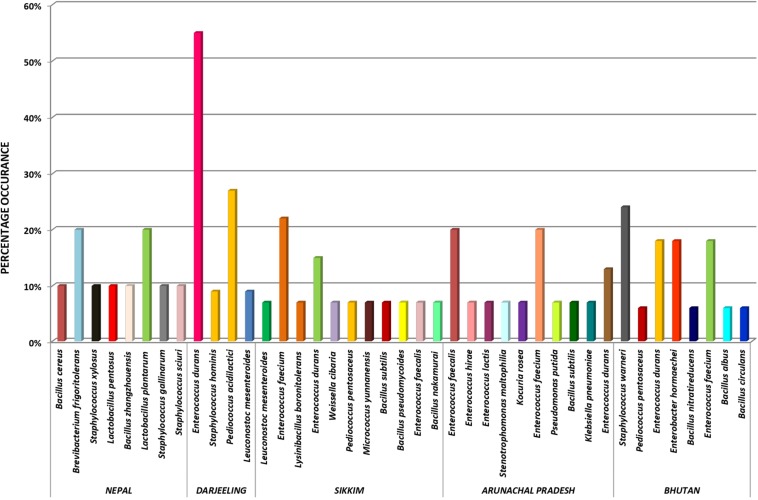
The genomic DNA of each isolate of all 68 representative bacteria strains was extracted and PCR products were prepared for identification by 16S rRNA gene sequence using the Sanger method. DNA sequences of bacterial isolates were assigned by comparing them with those available in the GenBank NCBI database using a BLAST 2.0 program (Altschul et al., 1990) for identification. The phylogenetic trees of the nucleotide sequences of 68 bacteria isolates from samples of marcha, paa, pee, phut, and phab were constructed using the Neighbor-joining method with 1,000 bootstrap value replicates (Figure 2). The 16S rRNA sequencing results showed three bacterial phyla represented by Firmicutes (85%), Proteobacteria (9%), and Actinobacteria (6%). The phylum distribution of the marcha samples from Nepal showed Firmicutes (80%) followed by Actinobacteria (20%); Darjeeling showed Firmicutes (100%); Sikkim showed Firmicutes (92%), and Actinobacteria (8%); Bhutan showed Firmicutes (100%). In starters from Arunachal Pradesh the variable distribution pattern in phyla level was observed. Samples of paa showed Firmicutes (80%), and Proteobacteria (20%), pee showed Firmicutes (67%), Proteobacteria (16%), and Actinobacteria (17%), and phut showed Firmicutes (75%), and Proteobacteria (25%). Similarly, phylum distribution in phab from Bhutan showed Firmicutes (57%) and Proteobacteria (43%). Based on results of the 16S rRNA gene sequencing, 15 different genera viz. Leuconostoc, Enterococcus, Bacillus, Staphylococcus, Lactobacillus, Enterobacter, Klebsiella, Pseudomonas, Pediococcus, Stenotrophomonas, Kocuria, Brevibacterium, Lysinibacillus, Weissella, and Micrococcus with 32 species from starters of the Eastern Himalayas were identified (Tables 3, 4). A wide diversity of bacteria (mainly LAB) was reported for the first time in traditionally prepared dry starters of the Eastern Himalayas (Table 5). The dominance of species of LAB was observed with 59% of total isolates in samples over non-LAB isolates (31%) (Figure 3). Enterococcus durans, E. faecium, E. fecalis, E. hirae, E. lactis, Pediococcus acidilactici, P. pentosaceus, Lactobacillus plantarum subsp. plantarum, Lb. pentosus, Leuconostoc mesenteroides, and Weissella cibaria were lactic acid bacterial species found in starter samples. Enterococcus durans (54.5%) was the most dominant species present in marcha samples from India (Darjeeling), whereas Pediococcus pentocaseus (5.8%) showed the lowest prevalence in marcha samples from Bhutan (Figure 4). LAB were found in all samples with highest occurrence in marcha samples of Darjeeling (91%). Non-LAB species were also recovered in many samples of starters, which were represented by Bacillus subtilis subsp. inaquosorum, B. circulans, B. albus, B. cereus, B. nakamurai, B. nitratireducens, B. pseudomycoides, B. zhangzhouensis, Kocuria rosea, Staphylococcus hominis subsp. hominis, S. warneri, S. gallinarum, S. sciuri, Lysinibacillus boronitolerans, Brevibacterium frigoritolerans, and Micrococcus yunnanensis. Interestingly, we detected few Gram-negative bacteria in some of the starter cultures from Arunachal Pradesh such as Stenotrophomonas maltophilia in paa, Klebsiella pneumoniae in pee, Pseudomonas putida in phut, and Enterobacter hormaechei subsp. xiangfangensis, and E. hormaechei subsp. steigerwaltii in some samples of phab from Bhutan (Table 5).
FIGURE 2.
Phylogenetic tree of the nucleotide sequences of 68 bacteria isolates from 35 different samples of dry starter from the Eastern Himalayas based on 16S rRNA sequencing. The tree was constructed by using the Neighbor-joining method (Gascuel and Steel, 2006) with bootstrap values for 1,000 replicates shown at the nodes of the tree using MEGA-7 (Kumar et al., 2016). The optimal tree with the sum of branch length = 0.98855936 is shown. The evolutionary distances were computed by the Maximum Composite Likelihood method (Varin et al., 2011) and are expressed in the units of the number of nucleotide substitutions per site. All positions containing gaps and missing data were eliminated. There were 308 total positions in the final dataset.
TABLE 3.
Identification of LAB isolates from dry starters from the Eastern Himalayas based on 16S rRNA gene sequencing.
| Isolate code | Sample (Place) | Identity | Type species (% similarity) | GenBank Accession No. | Size (base pair) |
| AKB6 | Marcha (Darjeeling) | Leuconostoc mesenteroides | Leuconostoc mesenteroides ATCC 8293 (99.54%) | MK748250 | 1315 |
| BPB18 | Marcha (Bhutan) | Enterococcus durans | Enterococcus durans JCM 8725 16S (99.52%) | MK748251 | 1254 |
| DMB4 | Marcha (Darjeeling) | Enterococcus durans | Enterococcus durans JCM 8725 (99.55%) | MK748252 | 1325 |
| SMB13 | Marcha (Sikkim) | Leuconostoc mesenteroides | Leuconostoc mesenteroides ATCC 8293 (99.55%) | MK748253 | 1339 |
| AKB3 | Marcha (Darjeeling) | Pediococcus acidilactici | Pediococcus acidilactici DSM 20284 (99.62%) | MK748254 | 833 |
| AOB14 | Pee (Arunachal Pradesh) | Enterococcus durans | Enterococcus durans JCM 8725 (99.62%) | MK748255 | 1333 |
| AOB15 | Pee (Arunachal Pradesh) | Enterococcus faecium | Enterococcus faecium DSM 20477 (98.11%) | MK748256 | 1430 |
| AOB25 | Phut (Arunachal Pradesh) | Enterococcus faecium | Enterococcus faecium DSM 20477 (99.71%) | MK748258 | 1406 |
| AOB4 | Paa (Arunachal Pradesh) | Enterococcus faecium | Enterococcus faecium DSM 20477 (99.38%) | MK748259 | 1460 |
| BPB11 | Marcha (Bhutan) | Enterococcus faecium | Enterococcus faecium DSM 20477 (98.91%) | MK748260 | 1476 |
| BPB31 | Phab (Bhutan) | Enterococcus faecium | Enterococcus faecium DSM 20477 (99.28%) | MK748264 | 1390 |
| BPB33 | Phab (Bhutan) | Enterococcus faecium | Enterococcus faecium DSM 20477 (99.51%) | MK748265 | 1432 |
| DMB11 | Marcha (Darjeeling) | Enterococcus durans | Enterococcus durans JCM 8725 (99.78%) | MK748267 | 1342 |
| DMB12 | Marcha (Darjeeling) | Pediococcus acidilactici | Pediococcus acidilactici DSM 20284 (99.59%) | MK748268 | 1462 |
| DMB6 | Marcha (Darjeeling) | Enterococcus durans | Enterococcus durans JCM 8725 (99.38%) | MK748269 | 1443 |
| MBV14 | Pee (Arunachal Pradesh) | Enterococcus durans | Enterococcus durans JCM 8725 (99.86%) | MK748270 | 1436 |
| SMB15 | Marcha (Sikkim) | Enterococcus faecium | Enterococcus faecium DSM 20477 (99.86%) | MK748274 | 1447 |
| SMB21 | Marcha (Sikkim) | Enterococcus faecium | Enterococcus faecium DSM 20477 (99.64%) | MK748276 | 1400 |
| SMB5 | Marcha (Sikkim) | Enterococcus faecium | Enterococcus faecium DSM 20477 (99.78%) | MK748277 | 1391 |
| SMB7 | Marcha (Sikkim) | Enterococcus durans | Enterococcus durans JCM 8725 (98.71%) | MK748278 | 1158 |
| AOB5 | Paa (Arunachal Pradesh) | Enterococcus faecalis | Enterococcus faecalis ATCC 19433(T) (99.86%) | MK202997 | 1421 |
| BPB13 | Marcha (Bhutan) | Pediococcus pentosaceus | Pediococcus pentosaceus DSM 20336(T) (99.73%) | MK203008 | 1456 |
| BPB21 | Phab (Bhutan) | Enterococcus durans | Enterococcus durans NBRC 100479(T) (99.79%) | MK203010 | 1430 |
| BPB4 | Marcha (Bhutan) | Enterococcus durans | Enterococcus durans NBRC 100479(T) (99.65%) | MK203013 | 1437 |
| DMB3 | Marcha (Darjeeling) | Enterococcus durans | Enterococcus durans NBRC 100479(T) (99.72%) | MK203015 | 1441 |
| AOB24 | Phut (Arunachal Pradesh) | Enterococcus hirae | Enterococcus hirae ATCC 9790(T) (99.86%.) | MK202998 | 1411 |
| DMB13 | Marcha (Darjeeling) | Enterococcus durans | Enterococcus durans NBRC 100479(T) (99.58%) | MK203017 | 1433 |
| DMB14 | Marcha (Darjeeling) | Pediococcus acidilactici | Pediococcus acidilactici DSM 20284(T) (99.52%) | MK203018 | 1461 |
| DMB11 | Marcha (Darjeeling) | Pediococcus acidilactici | Pediococcus acidilactici DSM 20284(T) (99.66%) | MK203019 | 1456 |
| DMB15 | Marcha (Darjeeling) | Enterococcus durans | Enterococcus durans NBRC 100479(T) (99.72%) | MK203020 | 1437 |
| NMB3 | Marcha (Nepal) | Lactobacillus pentosus | Lactobacillus pentosus DSM 20314(T) (97.44%) | MK203022 | 1276 |
| NMB8 | Marcha (Nepal) | Lactobacillus plantarum subsp. plantarum | Lactobacillus plantarum subsp. plantarum ATCC 14917(T) (99.65%) | MK203024 | 1441 |
| AOB26 | Phut (Arunachal Pradesh) | Enterococcus lactis | Enterococcus lactis BT159 (T) (98.0%) | MK202999 | 1398 |
| NMB7 | Marcha (Nepal) | Lactobacillus plantarum subsp. plantarum | Lactobacillus plantarum subsp. plantarum ATCC 14917(T) (100%) | MK203027 | 1435 |
| SMB9 | Marcha (Sikkim) | Weissella cibaria | Weissella cibaria KACC 11862(T) (99.66%) | MK203028 | 1455 |
| SMB13 | Marcha (Sikkim) | Pediococcus pentosaceus | Pediococcus pentosaceus DSM 20336(T) (99.79%) | MK203029 | 1433 |
| AOB2 | Paa (Arunachal Pradesh) | Enterococcus faecalis | Enterococcus faecalis ATCC 19433(T) (99.79%). | MK203002 | 1420 |
| AOB11 | Paa (Arunachal Pradesh) | Enterococcus faecalis | Enterococcus faecalis ATCC 19433(T) (99.58%). | MK203003 | 1421 |
| SMB11 | Marcha (Sikkim) | Enterococcus durans | Enterococcus durans JCM 8725 (96.44%) | MK752677 | 1432 |
| SMB3 | Marcha (Sikkim) | Enterococcus faecalis | Enterococcus faecalis NBRC 100480 (97.42%) | MK752675 | 1123 |
ATCC, American Type Cell Culture; JCM, Japan Collection of Microorganisms; DSM, Deutsche Sammlung von Mikroorganismen; MCCC, Microbial Culture Collection; NBRC, Biological Resource Centre, NITE; CCM, Czech collection of microorganisms; KACC, Korean Agricultural Culture Collection; YIM, Yunnan Institute of Microbiology; KCTC, Korean Collection for Type Cultures; NRRL, Agricultural Research Service Culture Collection.
TABLE 4.
Identification of non-LAB and Gram-negative bacteria from dry starters from the Eastern Himalayas based on 16S rRNA gene sequencing.
| Isolate code | Sample (Place) | Identity | Type species (% similarity) | GenBank Accession No. | Size (base pair) |
| AOB48 | Phut (Arunachal Pradesh) | Pseudomonas putida | Pseudomonas putida KT2440 (99.85%) | MK203004 | 1379 |
| AOB18 | Pee (Arunachal Pradesh) | Klebsiella pneumoniae | Klebsiella pneumoniae DSM 30104 (99.3%) | MK748257 | 1439 |
| BPB23 | Phab (Bhutan) | Enterobacter hormaechei subsp. xiangfangensis | Enterobacter hormaechei subsp. Xiangfangensis 10−17 (99.58%) | MK748261 | 1431 |
| BPB27 | Phab (Bhutan) | Enterobacter hormaechei subsp. Xiangfangensis | Enterobacter hormaechei subsp. Xiangfangensis 10−17 (98.88%) | MK748263 | 1446 |
| BPB26 | Phab (Bhutan) | Enterobacter hormaechei subsp. steigerwaltii | Enterobacter hormaechei subsp. Steigerwaltii DSM 16691(T) (99.23%) | MK203011 | 1422 |
| AOB9 | Paa (Arunachal Pradesh | Stenotrophomonas maltophilia | Stenotrophomonas maltophilia MTCC 434(T) (99.79%) | MK203000 | 1416 |
| NMB10 | Marcha (Nepal) | Bacillus zhangzhouensis | Bacillus zhangzhouensis DW5−4(T) (99.58%) | MK203023 | 1432 |
| NMB23 | Marcha (Nepal) | Staphylococcus xylosus | Staphylococcus xylosus CCM 2738(T) (99.86%) | MK203021 | 1426 |
| BPB24 | Phab (Bhutan) | Bacillus albus | Bacillus albus MCCC 1A02146 (99.02%) | MK748262 | 1437 |
| BPB8 | Marcha (Bhutan) | Bacillus circulans | Bacillus circulans NBRC 13626 (98.64%) | MK748266 | 1412 |
| NMB11 | Marcha (Nepal) | Bacillus cereus | Bacillus cereus ATCC 14579 (100%) | MK748271 | 1460 |
| NMB12 | Marcha (Nepal) | Brevibacterium frigoritolerans | Brevibacterium frigoritolerans DSM 8801 (99.72%) | MK748272 | 1426 |
| NMB13 | Marcha (Nepal) | Brevibacterium frigoritolerans | Brevibacterium frigoritolerans DSM 8801 (100%) | MK748273 | 1388 |
| SMB19 | Marcha (Sikkim) | Lysinibacillus boronitolerans | Lysinibacillus boronitolerans NBRC 103108 (99.59%) | MK748275 | 1220 |
| BPB1 | Marcha (Bhutan) | Staphylococcus warneri | Staphylococcus warneri ATCC 27836(T) (99.72%) | MK203006 | 1437 |
| BPB10 | Marcha (Bhutan) | Staphylococcus warneri | Staphylococcus warneri ATCC 27836(T) (99.79%) | MK203007 | 1432 |
| BPB17 | Marcha (Bhutan) | Staphylococcus warneri | Staphylococcus warneri ATCC 27836(T) (99.72%) | MK203009 | 1429 |
| BPB3 | Marcha (Bhutan) | Staphylococcus warneri | Staphylococcus warneri ATCC 27836(T) (98.92%) | MK203012 | 1490 |
| BPB7 | Marcha (Bhutan) | Bacillus nitratireducens | Bacillus nitratireducens 4049(T) (99.36%) | MK203014 | 1404 |
| DMB5 | Marcha (Darjeeling) | Staphylococcus hominis subsp. hominis | Staphylococcus hominis subsp. hominis DSM 20328(T) (99.93%) | MK203016 | 1425 |
| NMB20 | Marcha (Nepal) | Staphylococcus gallinarum | Staphylococcus gallinarum ATCC 35539(T) (99.86%) | MK203025 | 1437 |
| NMB22 | Marcha (Nepal) | Staphylococcus sciuri | Staphylococcus sciuri DSM 20345(T) (99.65%) | MK203026 | 1439 |
| SMB22 | Marcha (Sikkim) | Micrococcus yunnanensis | Micrococcus yunnanensis YIM 65004(T) (99.64%) | MK203030 | 1379 |
| SMB1 | Marcha (Sikkim) | Bacillus subtilis subsp. inaquosorum | Bacillus subtilis subsp. Inaquosorum KCTC 13429(T) (99.65%) | MK203031 | 1425 |
| SMB8 | Marcha (Sikkim) | Bacillus pseudomycoides | Bacillus pseudomycoides AFS041167 (99.93%) | MK203032 | 1407 |
| AOB19 | Pee (Arunachal Pradesh) | Kocuria rosea | Kocuria rosea DSM 20447(T) (99.79%.) | MK203001 | 1399 |
| AOB20 | Pee (Arunachal Pradesh) | Bacillus subtilis subsp. inaquosorum | Bacillus subtilis subsp. Inaquosorum KCTC 13429(T) (99.79%) | MK203005 | 1431 |
| SMB14 | Marcha (Sikkim) | Bacillus nakamurai | Bacillus nakamurai NRRL B-41091 (96.65%) | MK752676 | 1103 |
TABLE 5.
Bacterial diversity in dry starters from the Eastern Himalayas.
| Country/place | Starter | Bacterial species | |
| Nepal | Marcha | LAB: | Lactobacillus pentosus, Lb. plantarum subsp. plantarum |
| Non-LAB: | Bacillus cereus, B. zhangzhouensis, Brevibacterium frigoritolerans, Staphylococcus xylosus, S. gallinarum, S. sciuri | ||
| Gram-ve bacteria: | NR | ||
| India (Darjeeling hills) | Marcha | LAB: | Enterococcus durans, Pediococcus acidilactici, Leuconostoc mesenteroides |
| Non-LAB: | Staphylococcus hominis subsp. hominis | ||
| Gram-ve bacteria: | NR | ||
| India (Sikkim) | Marcha | LAB: | Pediococcus pentosaceus, Leuconostoc mesenteroides, Enterococcus faecium, E. faecalis, E. durans, Weissella cibaria |
| Non-LAB: | Lysinibacillus boronitolerans, Micrococcus yunnanensis, Bacillus subtilis subsp. inaquosorum, B. pseudomycoides, B. nakamurai | ||
| Gram-ve bacteria: | NR | ||
| India (Arunachal Pradesh) | Paa | LAB: | Enterococcus faecalis, E. faecium |
| Non-LAB: | NR | ||
| Gram-ve bacteria: | Stenotrophomonas maltophilia | ||
| Pee | LAB: | Enterococcus faecalis, E. durans, E. faecium | |
| Non-LAB: | Kocuria rosea, Bacillus subtilis subsp. inaquosorum | ||
| Gram-ve bacteria: | Klebsiella pneumoniae | ||
| Phut | LAB: | Enterococcus hirae, E. lactis, E. faecium | |
| Non-LAB: | NR | ||
| Gram-ve bacteria: | Pseudomonas putida | ||
| Bhutan | Marcha | LAB: | Pediococcus pentosaceus, Enterococcus durans, E. faecium |
| Non-LAB: | Staphylococcus warneri, Bacillus nitratireducens, B. circulans | ||
| Gram-ve bacteria: | NR | ||
| Phab | LAB: | Enterococcus durans, E. faecium | |
| Non-LAB: | Bacillus albus | ||
| Gram-ve bacteria: | Enterobacter hormaechei subsp. xiangfangensis, Enterobacter hormaechei subsp. steigerwaltii | ||
LAB, lactic acid bacteria; NR, not recovered.
FIGURE 3.
Distribution of LAB, non-LAB, and Gram-negative bacteria in dry starters from the Eastern Himalayas.
FIGURE 4.
Distribution of bacterial species in dry starters from the Eastern Himalayas.
Diversity indexes of bacterial communities of different starter cultures were characterized by the Shannon diversity index H, the Simpson’s index, and the Dominance and Chao1 index (Table 6). The Shannon diversity index H for evaluating bacterial diversity recorded highest in marcha from Sikkim (H:2.305) and lowest in marcha from Darjeeling (H:1.121). Simpson’s diversity index (1-D) values were 0.8878, 0.8711, 0.86, and 0.8374 for starters from Sikkim, Arunachal Pradesh, Nepal, and Bhutan, respectively. An estimation of species richness based on abundance was shown by the Chao 1 index. The dominance D-values were recorded as being highest for marcha samples from Darjeeling and lowest for marcha samples from Sikkim.
TABLE 6.
Diversity indices of different dry starters from the Eastern Himalayas.
| Country/Region | Diversity indices | |||
| Simpson’s index (1-D) | Shannon’s index (H) | Dominance (D) | Chao-1 | |
| Nepal | 0.86 | 2.025 | 0.14 | 13 |
| India (Darjeeling hills) | 0.6116 | 1.121 | 0.3884 | 5 |
| India (Sikkim) | 0.8878 | 2.305 | 0.1122 | 29 |
| India (Arunachal Pradesh) | 0.8711 | 2.176 | 0.1289 | 20.5 |
| Bhutan | 0.8374 | 1.925 | 0.1626 | 14 |
Discussion
In this study five types of traditionally prepared dry starters (marcha, pha, paa, pee, and phut) were collected from different regions of the Eastern Himalayas, and they were analyzed for microbial load, pH, and moisture. The average bacterial population of all samples was 108 cfu/g, which was not reported earlier except for marcha from the Darjeeling hills and Sikkim (Tamang and Sarkar, 1995; Tsuyoshi et al., 2005; Tamang et al., 2007). The bacterial load of marcha from Sikkim was 106 to 108 cfu/g (Table 1), which was almost the same as that of populations of yeasts and filamentous molds in marcha from Sikkim (Tsuyoshi et al., 2005). This shows that bacterial populations in traditionally prepared starters of the Eastern Himalayas may have co-existed equally with filamentous molds and yeasts (Hesseltine et al., 1988; Zheng et al., 2015). The moisture content of all starters was low due to the sun-drying process that followed immediately after fermentation, the step necessary to maintain the potency of traditionally prepared starters to be able to be stored in a dry place at room temperature for future use. The pH of all samples was mildly acidic, which may be due to the dominance of LAB (∼108 cfu/g) in dry starters (Tamang and Sarkar, 1995).
First, we phenotypically characterized all 201 bacterial strains isolated from samples of marcha, paa, pee, phut, and phab and presumptively identified four genera of LAB- Enterococcus, Pediococcus, Leuconostoc, and Lactobacillus, two genera of non-LAB-Bacillus and Staphylococcus, and two Gram-negative bacterial genera, Enterobacter and Citrobacter. We grouped 201 isolates into 68 representative bacterial strains on the basis of phenotypic and biochemical tests for confirmation of their identity and assigned the taxonomical nomenclature by using 16S rRNA gene sequencing. In our study, we found a dominance of phylum Firmicutes (85%) over Proteobacteria (9%) and Actinobacteria (6%) in starters from the Eastern Himalayas. Firmicutes was also reported as the major abundant phylum in daqu, a starter for Chinese strongly flavored liquor (Zou et al., 2018; He et al., 2019), and in nuruk, a starter from Korean used to produce makgeolli, a Korean alcoholic beverage (Jung et al., 2012). The sequence data based on a constructed phylogenetic tree revealed a dominance of LAB (59%) with five different genera and 11 species represented by Enterococcus durans, E. faecium, E. fecalis, E. hirae, E. lactis, Pediococcus acidilactici, P. pentosaceus, Lactobacillus plantarum subsp. plantarum, Lb. pentosus, Leuconostoc mesenteroides, and Weissella cibaria. Only two genera of LAB represented by Pediococcus pentosaceus and Lactobacillus brevis were reported earlier from marcha samples from Sikkim and the Darjeeling hills (Tamang and Sarkar, 1995; Tamang et al., 2007). However, in this study we found a wide diversity of LAB in samples of marcha collected from the Darjeeling hills and Sikkim in India, which included Pediococcus pentosaceus, P. acidilactici, Enterococcus faecium, E. durans, E. faecalis, Leuconostoc mesenteroides, and Weissella cibaria, whereas, Lactobacillus pentosus and Lb. plantarum subsp. plantarum were found only in marcha samples from Nepal. Variations in altitude and other geographical factors may affect the composition of microbiota in dry starters (Jeyaram et al., 2011; Lv et al., 2012). Traditional methods of preparation of marcha, phab, paa, pee, and phut are more or less similar except for some variations that were observed in the use of substrates, such as rice for marcha, phut, paa, and pee, and maize-rice husk for phab from Bhutan, and also wrapping materials for fermenting substrates such as fern leaves (Glaphylopteriolopsis erubeseens) for marcha preparation, dry paddy straws for phab, and locally available plant leaves for the preparation of paa, pee, and phut. Bacterial diversity in dry starters from the Eastern Himalayas may be influenced by hygienic conditions, quality of cereal substrates, wrapping materials, and sources of natural or tap water during traditional methods of preparation (Peter-Ikechukwu et al., 2016; Gonelimali et al., 2018; Sha et al., 2019).
The bacterial profile of marcha from Nepal and Bhutan, paa, pee, and phut of Arunachal Pradesh, and phab from Bhutan has been reported for the first time in our study. A similar type of dry starter for Assam in North East India called xaj-pitha also contained several species of LAB such as Lactobacillus plantarum, Lb. brevis, Weissella cibaria, W. paramesenteroides, W. confusa, Lactococcus lactis, Lactobacillus casei group, Leuconostoc lactis, Leuconostoc pseudomesenteroides, Pediococcus pentosaceus, Lactococcus garvieae, and Enterococcus sp. (Bora et al., 2016). Thanh et al. (2008) reported many species of LAB in Vietnamese banh men, which included Pediococcus pentosaceus, Lactobacillus plantarum, Lb. brevis, Lb. fermentum, Lb. agilis, W. confusa, W. paramesenteroides, and Lactococcus lactis. Enterococcus faecium, Lactobacillus plantarum, Leuconostoc mesenteroides, Pediococcus acidilactici, P. pentosaceus, Weissella paramesenteroides, and W. cibaria, were reported in nuruk from Korea (Hoon et al., 2013). Several species of LAB in Cambodian dombea were also reported: Weissella cibaria, Lactobacillus plantarum, Lactococcus lactis, Pediococcus pentosaceus, and Enterococcus durans (Ly et al., 2018). This indicates that species of LAB predominate the microbial composition of traditionally prepared dry starters in Asia, including the Eastern Himalayas. LAB have been considered as favorable bacteria in cereal-based beverages due their ability to improve protein digestibility, enhance organoleptic quality, and increase nutritional bioavailability (Luana et al., 2014). Species of Weissella, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, and Enterococcus are known for flavor development, the production of organic acids, and antimicrobial activities in Chinese daqu used for liquor production (Gou et al., 2015). Enterococcus sp. has been reported to produce enterocins, which play a major role in preventing the growth of foodborne and spoilage-causing pathogens (Javed et al., 2011).
Non-LAB species formed the next abundant group (32%) in starters from the Eastern Himalayas with the dominance of Bacillus spp. The abundance of Bacillus sp. may be due to its ability to survive in environments with low moisture and high temperature (Nuding et al., 2017). Also, the Bacillus species are important sources of amylase and protease enzymes, which are involved in saccharification and flavor production (Beaumont, 2002). A dominance of Bacillus sp. was also reported in daqu from China (Wang et al., 2008; Zheng et al., 2012) and banh men from Vietnam (Thanh et al., 2008). The next most abundant bacterium was Staphylococcus spp., found in the Himalayan starters, which secretes amylase (Li et al., 2014) and protease in Chinese daqu (Yang et al., 2017) and also produces lipases for the production of esters for flavor (Talon et al., 1996); thus, this group of bacteria probably plays a major role in the flavor enhancement of the final product. The prevalence of phylum Actinobacteria in some starters of the Eastern Himalayas was only 6%, represented by Kocuria rosea, Micrococcus yunnanensis, and Brevibacterium frigoritolerans. The presence of Actinobacteria has been reported in Chinese daqu (Zou et al., 2018) and Indian marcha and thiat (Sha et al., 2017).
Few species of opportunist pathogens and environmental contaminants such as Micrococcus, Stenotrophomonas, Enterobacter, Klebsiella, and Pseudomonas were detected, and they were found only in samples of paa, pee, and phut from Arunachal Pradesh, and phab from Bhutan. However, both the prevalence and populations of these contaminants were low and it is presumed that these organisms might have contaminated the samples during the traditional method of preparation from substrates, herbs, water, utensils, wrapping materials, etc., Gram-negative bacteria were not detected in any samples of marcha collected from Nepal, India, or Bhutan. In our previous study on marcha, no Gram-negative bacteria were found at the genus level, and this was discovered through an analysis using a high-throughput sequencing method (Sha et al., 2019). Although most of these bacteria are opportunists and probable foodborne pathogens, some of them, such as Enterobacter sp., are involved in the production of amylases and lipases and also the formation of flavor in daqu (Li et al., 2015). The presence of LAB inhibits the growth of pathogenic and spoilage microorganisms in foods (Cizeikiene et al., 2013; Castellano et al., 2017) and produces flavor compounds (Mukisa et al., 2017).
A diversity index, or phylogenetic metric, is a quantitative measure to show phylogenetic relations within different species in a community (Birtel et al., 2015). We characterized diversity indexes of the bacterial community present in starters from the Eastern Himalayas by using the Shannon diversity index H, Simpson’s index, and Dominance and Chao1 index (Table 6). The Shannon diversity index H for evaluating bacterial diversity was recorded as being highest in marcha from Sikkim (H:2.305) and lowest in marcha from Darjeeling (H:1.121), indicating a higher bacterial diversity in marcha from Sikkim as compared to other starters. The Simpson’s diversity index (1-D) index, which considers both the number of species as well as the relative abundance of each species for evaluating diversity, showed the highest values for marcha from Sikkim. The dominance D-values were recorded as being highest for marcha samples from Darjeeling and lowest for marcha samples from Sikkim, which supports the above inference regarding bacterial diversity. The dominance D-value ranged between 0–1, where the value 0 indicated that all taxa were equally present and value 1 indicated the dominance of one taxon over the whole community (Wagner et al., 2018). Thus, the values near zero indicate a highly diverse ecosystem and values near 1 indicate a less diverse or homogenous ecosystem (Lv et al., 2012). Hence, the phylogenetic matric of the bacterial community present in dry starters from the Eastern Himalayas showed high diversity within the community. The Eastern Himalayas are known for their rich floral and faunal diversity within a wide ecosystem (Chettri et al., 2010). Our findings thus highlight the richness of microbial diversity in the food ecosystem of the Eastern Himalayas.
The microbial communities and their interactions in starters are extremely important for proper fermentation, which may determine the productivity and flavor quality of the final alcoholic beverage (Cai et al., 2018). There has been an increasing amount of concern regarding the safety of fermented beverages due to the presence of ethyl carbamate, which is considered to be carcinogenic (Ryu et al., 2015), biogenic amines (Liu et al., 2016), mycotoxin (Sivamaruthi et al., 2018), and contamination by opportunistic microbial pathogens (Hong et al., 2016). All these considerations mandate a deep understanding of the microbial community in starters. Also, the profile of native microbiota in these starters opens a possibility of finding novel strain(s) with functional properties for industrial purposes. This study also records the bacterial diversity of phab from Bhutan, which is found to be produced rarely by a few ethnic people of Bhutan. This is probably due to their preference for commercial marcha, similar to phab, which is sold in local markets. Bacteria present in traditionally prepared dry starters have no amylolytic activities (Thapa and Tamang, 2004); however, they may contribute to the acidification of fermenting substrates and impart flavor with a mildly acidic and sour taste to traditional alcoholic beverages (kodo ko jaanr, opo, apong, and themsing) preferred by the Himalayan people (Thapa and Tamang, 2006; Tamang et al., 2007).
Conclusion
Information on the microbial composition of traditionally prepared dry starters of the Eastern Himalayan regions of India, Nepal, and Bhutan viz. phab, paa, pee, and phut, was unknown except for marcha from Sikkim in India. These traditional starters are used by the Himalayan people to ferment cereals into various alcoholic beverages for home consumption. The main objective of this study was to profile and assign the taxonomical identity of bacteria isolated from these traditional starters of the Eastern Himalayas based on 16S rRNA sequencing. Firmicutes was the most dominant phylum in all starters and was represented by several genera and species of LAB and also by some non-LAB. Interestingly our study showed high diversity within the bacterial community in traditionally prepared starters of the Eastern Himalayas, which may supplement the richness of microbial conservation in the food ecosystem of the regions. Besides diversity, some bacteria isolated from these traditional starters may have commercial and industrial importance. This is the first report on the bacterial diversity of dry starters of the Eastern Himalayas by Sanger sequencing.
Data Availability Statement
The datasets generated for this study can be found in the sequences retrieved from the 16S rRNA sequencing were deposited at GenBank-NCBI under the nucleotide accession number: MK748250-MK748278, MK202997-MK203032, and MK752675-MK752677.
Author Contributions
PP performed the majority of the experiments. JT supervised the experiments and finalized the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The authors are grateful to the Department of Biotechnology (DBT), Government of India, for financial support. PP is grateful to DBT for the award of Traineeship in DBT-funded Bioinformatics Centre of Sikkim University sanctioned to JT.
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anupma A., Pradhan P., Sha S. P., Tamang J. P. (2018). Traditional skill of ethnic people of the Eastern Himalayas and North East India in preserving microbiota as dry amylolytic starters. Indian J. Tradit. Knowl. 17 184–190. [Google Scholar]
- Beaumont M. (2002). Flavouring composition prepared by fermentation with Bacillus spp. Int. J. Food Microbiol. 75 189–196. 10.1016/S0168-1605(01)00706-1 [DOI] [PubMed] [Google Scholar]
- Bhasin V. (2013). Pastoralists of Himalayas. J. Biodiver. 4 83–113. 10.1080/09766901.2013.11884746 8690004 [DOI] [Google Scholar]
- Birtel J., Walser J. C., Pichon S., Bürgmann H., Matthews B. (2015). Estimating bacterial diversity for ecological studies: methods, metrics, and assumptions. PLoS One 10:e0125356. 10.1371/journal.pone.0125356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora S. S., Keot J., Das S., Sarma K., Barooah M. (2016). Metagenomics analysis of microbial communities associated with a traditional rice wine starter culture (Xaj-pitha) of Assam, India. 3 Biotech. 6:153. 10.1007/s13205-016-0471-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Zhang T., Zhang Q., Luo J., Cai C., Mao J. (2018). Microbial diversity and chemical analysis of the starters used in traditional Chinese sweet rice wine. Food Microbiol. 73 319–326. 10.1016/j.fm.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Castellano P., Pérez Ibarreche M., Blanco Massani M., Fontana C., Vignolo G. M. (2017). Strategies for pathogen biocontrol using lactic acid bacteria and their metabolites: a focus on meat ecosystems and industrial environments. Microorganisms 5:38. 10.3390/microorganisms5030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnaud P., Machinis K., Coutte L. A., Marecat A., Mercenier A. (2001). Rapid PCR –based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. J. Microbiol. Methods 44 139–148. 10.1016/S0167-7012(00)00244-X [DOI] [PubMed] [Google Scholar]
- Chao A., Chiu C. (2016). Nonparametric Estimation and Comparison of Species Richness. Chichester: John Wiley & Sons, 10.1002/9780470015902.a0026329 [DOI] [Google Scholar]
- Chen B., Wu Q., Xu Y. (2014). Filamentous fungal diversity and community structure associated with the solid state fermentation of Chinese Maotai-flavor liquor. Int. J. Food Microbiol. 179 80–84. 10.1016/j.ijfoodmicro.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Cheng H. R., Jiang N. (2006). Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Lett. 28 55–59. 10.1007/s10529-005-4688-z [DOI] [PubMed] [Google Scholar]
- Chettri N., Sharma E., Shakya B., Thapa R., Bajracharya B., Uddin K., et al. (2010). Biodiversity in the Eastern Himalayas: Status, Trends and Vulnerability to Climate Change. Technical Report 2 Kathmandu: ICIMOD. [Google Scholar]
- Cizeikiene D., Gražina J., Paškevičius A., Bartkiene E. (2013). Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control 31 539–545. 10.1016/j.foodcont.2012.12.004 [DOI] [Google Scholar]
- Dung N. T. P., Rombouts F. M., Nout M. J. R. (2007). Characteristics of some traditional Vietnamese starch-based rice wine fermentation starters (men). LWT Food Sci. Technol. 40 130–135. 10.1016/J.LWT.2005.08.004 [DOI] [Google Scholar]
- Gascuel O., Steel M. (2006). Neighbor-Joining revealed. Mol. Biol. Evol. 23 1997–2000. 10.1093/molbev/msl072 [DOI] [PubMed] [Google Scholar]
- Gonelimali F. D., Lin J., Miao W., Xuan J., Charles F., Chen M., et al. (2018). Antimicrobial properties and mechanism of action of some plant extracts against food pathogens and spoilage microorganisms. Front. Microbiol. 9:1639. 10.3389/fmicb.2018.01639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M., Wang H., Yuan H., Zhang W., Tang Y., Kida K. (2015). Characterization of the microbial community in three types of fermentation starters used for Chinese liquor production. J. Inst. Brew. 121 620–627. 10.1002/jib.272 [DOI] [Google Scholar]
- Hammer Ø., Harper D. A. T., Ryan P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9. [Google Scholar]
- Hammes W. P., Hertel C. (2003). “The genus Lactobacillus,” in The Prokaryotes an Electronic Resource for the Microbiological Community, eds Dworkin M., Flakow S., Rosenberg E., Schleifer K. H., Stackbrandt E. (New York, NY:: Springer-Verlag; ). [Google Scholar]
- He G., Huang J., Zhou R., Wu C., Jin Y. (2019). Effect of fortified daqu on the microbial community and flavor in Chinese strong-flavor liquor brewing process. Front. Microbiol. 10:56. 10.3389/fmicb.2019.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heather J. M., Chain B. (2016). The sequence of sequencers: the history of sequencing DNA. Genomics 107 1–8. 10.1016/j.ygeno.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesseltine C. W., Kurtzman C. P. (1990). Yeasts in amylolytic food starters. Anal. Inst. Biol. Univ. Nac. Autón. México, Ser. Bot. 60 1–7. 10.1016/B978-0-444-52149-1.00187-7 [DOI] [Google Scholar]
- Hesseltine C. W., Ray M. L. (1988). Lactic acid bacteria in murcha and ragi. J. Appl. Microbiol. 64 395–401. 10.1111/j.1365-2672.1988.tb05096.x [DOI] [Google Scholar]
- Hesseltine C. W., Rogers R., Winarno F. G. (1988). Microbiological studies on amylolytic Oriental fermentation starters. Mycopathology 101 141–155. 10.1002/jsfa.2740440410 [DOI] [Google Scholar]
- Holt J. G., Krieg N. R., Sneath P. H. A., Staley J. T., Williams S. T. (1994). Bergey’s Manual of Determinative Bacteriology, 9th Edn Baltimore, MD: Williams & Wilkins, 965–1599. [Google Scholar]
- Hong X. T., Chen J., Liu L., Wu H., Tan H. Q., Xie G. F., et al. (2016). Metagenomic sequencing reveals the relationship between microbiota composition and quality of Chinese rice wine. Sci. Rep. 6:26621. 10.1038/srep26621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon S. S., Lee C., Lee S., Park J. M., Lee H. J., Bai D. H., et al. (2013). Analysis of microflora profile in Korean traditional nuruk. J. Microbiol. Biotechnol. 23 40–46. 10.4014/jmb.1210.10001 [DOI] [PubMed] [Google Scholar]
- Huang Y., Yi Z., Jin Y., Zhao Y., He K., Liu D., et al. (2017). New microbial resource: microbial diversity, function and dynamics in Chinese liquor starter. Sci. Rep. 7:14577. 10.1038/s41598-017-14968-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A., Masud T., Ain Q. U., Imran M., Maqsood S. (2011). Enterocins of Enterococcus faecium, emerging natural food preservatives. Ann. Microbiol. 61 699–708. 10.1007/s13213-011-0223-8 [DOI] [Google Scholar]
- Jeyaram K., Singh W., Capece A., Romano P. (2008). Molecular identification of yeast species associated with “Hamei” — A traditional starter used for rice wine production in Manipur. Indian Int. J. Food Microbiol. 124 115–125. 10.1016/j.ijfoodmicro.2008.02.029 [DOI] [PubMed] [Google Scholar]
- Jeyaram K., Tamang J. P., Capece A., Romano P. (2011). Geographical markers for Saccharomyces cerevisiae strains with similar technological origins domesticated for rice-based ethnic fermented beverages production in North East India. Antonie Van Leeuwen. 100 569–578. 10.1007/s10482-011-9612-z [DOI] [PubMed] [Google Scholar]
- Jung M. J., Nam Y. D., Roh S. W., Bae J. W. (2012). Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 30 112–123. 10.1016/j.fm.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Kozaki M., Uchimura T. (1990). Microorganisms in Chinese starter ‘bubod’ and rice wine ‘tapuy’ in the Philippines. J. Brewing Soc. Japan 85 818–824. 10.6013/jbrewsocjapan1988.85.818 [DOI] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhare S. V., Dhotre D. P., Dhar S. K., Jani K., Apte D. A., Shouche Y. S., et al. (2015). Insights into diversity and imputed metabolic potential of bacterial communities in the continental shelf of Agatti Island. PLoS One 10:e0129864. 10.1371/journal.pone.0129864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. (1991). “16S/23S rRNA Sequencing,” in Nucleic Acid Techniques in Bacterial Systematic, eds Stackebrandt E., Goodfellow M. (New York, NY: John Wiley and Sons; ), 115–175. [Google Scholar]
- Le Fort P. (1975). Himalayas: the collided range. Present knowledge of the continental arc. Am. J. Sci. 275A 1–44. [Google Scholar]
- Lee C. H., Lee S. S. (2002). Cereal fermentation by fungi. Appl. Mycol. Biotechnol. 2 151–170. 10.1016/S1874-5334(02)80009-0 [DOI] [Google Scholar]
- Li H., Jiao A., Xu X., Wu C., Wei B., Hu X., et al. (2012). Simultaneous saccharification and fermentation of broken rice: an enzymatic extrusion liquefaction pretreatment for Chinese rice wine production. Bioprocess Biosyst. Eng. 36 1141–1148. 10.1007/s00449-012-0868-0 [DOI] [PubMed] [Google Scholar]
- Li P., Aflakpui F. W. K., Yu H., Luo L., Lin W. (2015). Characterization of activity and microbial diversity of typical types of Daqu for traditional Chinese vinegar. Ann. Microbiol. 65 2019–2027. 10.1007/s13213-015-1040-2 [DOI] [Google Scholar]
- Li Z., Bai Z., Wang D., Zhang W., Zhang M., Lin F., et al. (2014). Cultivable bacterial diversity and amylase production in three typical Daqus of Chinese spirits. Int. J. Food Sci. Technol. 49 776–786. 10.1111/ijfs.12365 [DOI] [Google Scholar]
- Limtong S., Sintara S., Suwannarit P. (2002). Yeast diversity in Thai traditional alcoholic starter. Kasetsart J. Natural Sci. 36 149–158. [Google Scholar]
- Liu S. P., Yu J. X., Wei X. L., Ji Z. W., Zhou Z. L., Meng X. Y., et al. (2016). Sequencing-based screening of functional microorganism to decrease the formation of biogenic amines in Chinese rice wine. Food Control 64 98–104. 10.1016/j.foodcont.2015.12.013 [DOI] [Google Scholar]
- Luana N., Rossana C., Curiel J. A., Kaisa P., Marco G., Rizzello C. G. (2014). Manufacture and characterization of a yogurt-like beverage made with oat flakes fermented by selected lactic acid bacteria. Int. J. Food Microbiol. 185 17–26. 10.1016/j.ijfoodmicro.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Lv X. C., Weng X., Zhang W., Rao P. F., Ni L. (2012). Microbial diversity of traditional fermentation starters for Hong Qu glutinous rice wine as determined by PCR-mediated DGGE. Food Control 28 426–434. 10.1016/j.foodcont.2012.05.025 [DOI] [Google Scholar]
- Ly S., Mith H., Tarayre C., Taminiau B., Daube G., Fauconnier M. L., et al. (2018). Impact of microbial composition of Cambodian traditional dried starters (dombea) on flavor compounds of rice wine: combining amplicon sequencing with HP-SPME-GCMS. Front. Microbiol. 9:894. 10.3389/fmicb.2018.00894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty C. (1998). CHROMASPRO 1.34: Free Program. Available at: http://www.technelysium.com.au/chromas.html [Google Scholar]
- Mukisa I. M., Byaruhanga Y. B., Muyanja C. M. B. K., Langsrud T., Narvhus J. A. (2017). Production of organic flavor compounds by dominant lactic acid bacteria and yeasts from Obushera, a traditional sorghum malt fermented beverage. Food Sci. Nutr. 5 702–712. 10.1002/fsn3.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy S. N., Dhyani P. P., Sanal P. K. (2006). Resources information database of the Indian Himalaya. ENVIS Monogr. 3 1–95. [Google Scholar]
- Nikkuni S., Karki T. B., Terao T., Suzuki C. (1996). Microflora of mana, a Nepalese rice koji. J. Ferment.Bioeng. 81 168–170. 10.1016/0922-338x(96)87597-0 [DOI] [Google Scholar]
- Nile S. H. (2015). The nutritional, biochemical and health effects of makgeolli – a traditional Korean fermented cereal beverage. J. Inst. Brew. 121 457–463. 10.1002/jib.264 [DOI] [Google Scholar]
- Nout M. J. R., Aidoo K. E. (2002). “Asian Fungal Fermented Food,” in Mycota, a Comprehensive Treatise On Fungi as Experimental Systems and Applied Research, Industrial Applications, Vol. 10 ed. Osiewacz H. D. (Berlin: Springer-Verlag; ), 23–47. 10.1007/978-3-662-10378-4_2 [DOI] [Google Scholar]
- Nuding D. L., Gough R. V., Venkateswaran K. J., Spry J. A., Tolbert M. A. (2017). Laboratory investigations on the survival of Bacillus subtilis spores in deliquescent salt mars analog environments. Astrobiology 17 997–1008. 10.1089/ast.2016.1545 [DOI] [PubMed] [Google Scholar]
- Pervez S., Aman A., Iqbal S., Siddiqui N. N., Ul Qader S. A. (2014). Saccharification and liquefaction of cassava starch: an alternative source for the production of bioethanol using amylolytic enzymes by double fermentation process. BMC Biotechnol. 14:49. 10.1186/1472-6750-14-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter-Ikechukwu A. I., Kabuo N. O., Alagbaoso S. O., Njoku N. E., Eluchie C. N., Momoh W. O. (2016). Effect of wrapping materials on physico-chemical and microbiological qualities of fermented melon seed (Citrullus colocynthis L.) used as condiment. Am. J. Food Sci. Technol. 4 14–19. 10.12691/ajfst-4-1-3 [DOI] [Google Scholar]
- Ryu D., Choi B., Kim E., Park S., Paeng H., Kim C. I., et al. (2015). Determination of ethyl carbamate in alcoholic beverages and fermented foods sold in Korea. Toxicol Res. 31 289–297. 10.5487/TR.2015.31.3.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D. (2013). Lesser Himalayan sequences in Eastern Himalaya and their deformation: implications for Paleoproterozoic tectonic activity along the northern margin of India. Geosci. Front. 4 289–304. 10.1016/j.gsf.2013.01.004 [DOI] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Nati. Acad. Sci. U.S.A. 74 5463–5467. 10.1073/pnas.74.12.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Riesner D. (2006). Purification of nuclei acids by selective precipitation with polyethylene glycol 6000. Anal. Biochem. 354 311–313. 10.1016/j.ab.2006.03.014 [DOI] [PubMed] [Google Scholar]
- Sha S. P., Jani K., Sharma A., Anupma A., Pradhan P., Shouche Y., et al. (2017). Analysis of bacterial and fungal communities in Marcha and Thiat, traditionally prepared amylolytic starters of India. Sci. Rep. 7:10967. 10.1038/s41598-017-11609-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha S. P., Suryavanshi M. S., Tamang J. P. (2019). Mycobiome diversity in traditionally prepared starters for alcoholic beverages in India by high-throughput sequencing method. Front. Microbiol. 10:348. 10.3389/fmicb.2019.003482237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha S. P., Suryavanshi M. V., Jani K., Sharma A., Shouche Y. S., Tamang J. P. (2018). Diversity of yeasts and molds by culture-dependent and culture-independent methods for mycobiome surveillance of traditionally prepared dried starters for the production. Front. Microbiol. 9:2237. 10.3389/fmicb.2018.02237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Jianchu X., Sharma G. (2007). Traditional agroforestry in the eastern Himalayan region: land management system supporting ecosystem services. Tropical Ecol. 48 189–200. [Google Scholar]
- Shrivastava K., Greeshma A. G., Srivastava B. (2012). Biotechnology in tradition – a process technology of alcoholic beverages practiced by different tribes of Arunachal Pradesh, North East India. Indian J. Trad. Knowl. 11 81–89. [Google Scholar]
- Sivamaruthi B. S., Kesika P., Chaiyasut C. (2018). Toxins in fermented foods: prevalence and preventions—a mini review. Toxins 11:E4. 10.3390/toxins11010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surono I. S. (2016). “Ethnic fermented foods and beverages of Indonesia,” in Ethnic Fermented Foods and Alcoholic Beverages of Asia, ed. Tamang J. P. (New Delhi: Springer; ), 341–382. [Google Scholar]
- Talon R., Montel M. C., Berdague J. L. (1996). Production of flavor esters by lipases of Staphylococcus warneri and Staphylococcus xylosus. Enzyme Microb. Technol. 19 620–622. 10.1016/S0141-0229(96)00075-0 [DOI] [Google Scholar]
- Tamang J. P. (2010). Himalayan Fermented Foods: Microbiology, Nutrition, and Ethnic Values. New York, NY: CRC Press, 295. [Google Scholar]
- Tamang J. P., Dewan S., Tamang B., Rai A., Schillinger U., Holzapfel W. H. (2007). Lactic acid bacteria in hamei and marcha of North East India. Indian J. Microbiol. 47 119–125. 10.1007/s12088-007-0024-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang J. P., Sarkar P. K. (1995). Microflora of murcha: an amylolytic fermentation starter. Microbios 81 115–122. [PubMed] [Google Scholar]
- Tamang J. P., Sarkar P. K., Hesseltine C. W. (1988). Traditional fermented foods and beverages of Darjeeling and Sikkim–a review. J. Sci. Food Agric. 44 375–385. 10.1002/jsfa.2740440410 [DOI] [Google Scholar]
- Tamang J. P., Tamang N., Thapa S., Dewan S., Tamang B. M., Yonzan H., et al. (2012). Microorganisms and nutritional value of ethnic fermented foods and alcoholic beverages of North East India. Indian J. Tradit. Knowl. 11 7–25. [Google Scholar]
- Tamang J. P., Thapa S., Tamang N., Rai B. (1996). Indigenous fermented food beverages of Darjeeling hills and Sikkim: a process and product characterization. J. Hill Res. 9 401–411. [Google Scholar]
- Thakur N., Saris P. E., Bhalla T. C. (2015). Microorganisms associated with amylolytic starters and traditional fermented alcoholic beverages of North Western Himalayas in India. Food Biosci. 11 92–96. 10.1016/j.fbio.2015.05.002 [DOI] [Google Scholar]
- Thanh V. N., Mai Le T., Tuan D. A. (2008). Microbial diversity of traditional Vietnamese alcohol fermentation starters (banh men) as determined by PCR-mediated DGGE. Int. J. Food Microbiol. 128 268–273. 10.1016/j.ijfoodmicro.2008.08.020 [DOI] [PubMed] [Google Scholar]
- Thapa S., Tamang J. P. (2004). Product characterization of kodo ko jaanr: fermented finger millet beverage of the Himalayas. Food Microbiol. 21 617–622. 10.1016/j.fm.2004.01.004 [DOI] [Google Scholar]
- Thapa S., Tamang J. P. (2006). Microbiological and physico-chemical changes during fermentation of kodo ko jaanr, a traditional alcoholic beverage of the Darjeeling hills and Sikkim. Indian J. Microbiol. 46 333–341. [Google Scholar]
- Tsuyoshi N., Fudou R., Yamanaka S., Kozaki M., Tamang N., Thapa S., et al. (2005). Identification of yeast strains isolated from marcha in Sikkim, a microbial starter for amylolytic fermentation. Int. J. Food Microbiol. 99 135–146. 10.1016/j.ijfoodmicro.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Varin C., Reid N., Firth D. (2011). An overview of composite likelihood methods. Stat. Sin. 21 5–42. [Google Scholar]
- Wagner B. D., Grunwald G. K., Zerbe G. O., Mikulich-Gilbertson S. K., Robertson C. E., Zemanick E. T., et al. (2018). On the use of diversity measures in longitudinal sequencing studies of microbial communities. Front. Microbiol. 9:1037. 10.3389/fmicb.2018.01037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. L., Shi D. J., Gong G. L. (2008). Microorganisms in Daqu: a starter culture of Chinese Maotai-flavor liquor. World J. Microbiol. Biotechnol. 24 2183–2190. 10.1007/s10295-009-0661-5 [DOI] [PubMed] [Google Scholar]
- Yang J.-G., Dou X., Han P.-J., Bai F.-Y., Zhou J., Zhang S.-Y., et al. (2017). Microbial diversity in Daqu during production of Luzhou-flavored liquor. J. Am. Soc. Brew. Chem. 75 136–144. [Google Scholar]
- Zheng X. W., Tabrizi M. R., Nout M. J. R., Han B. Z. (2011). Daqu-a traditional Chinese liquor fermentation starter. J. Institute Brew. 117 82–90. [Google Scholar]
- Zheng X. W., Yan Z., Han B. Z., Zwietering M. H., Samson R. A., Boekhout T., et al. (2012). Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 31 293–300. 10.1016/j.fm.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Zheng X. W., Yan Z., Nout M. J. R., Boekhout T., Han B. Z., Zwietering M. H., et al. (2015). Characterization of the microbial community in different types of Daqu samples as revealed by 16S rRNA and 26S rRNA gene clone libraries. World J. Microbiol. Biotechnol. 31 199–208. 10.1007/s11274-014-1776-z [DOI] [PubMed] [Google Scholar]
- Zou W., Zhao C., Luo H. (2018). Diversity and function of microbial community in Chinese strong-flavor baijiu ecosystem: a review. Front. Microbiol. 9:671 10.3389/fmicb.2018.00671 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences retrieved from the 16S rRNA sequencing were deposited at GenBank-NCBI under the nucleotide accession number: MK748250-MK748278, MK202997-MK203032, and MK752675-MK752677.
The datasets generated for this study can be found in the sequences retrieved from the 16S rRNA sequencing were deposited at GenBank-NCBI under the nucleotide accession number: MK748250-MK748278, MK202997-MK203032, and MK752675-MK752677.