Abstract
In this work, we statistically improved culture media for rPOXA 1B laccase production, expressed in Pichia pastoris containing pGAPZαA-LaccPost-Stop construct and assayed at 10 L bioreactor production scale (6 L effective work volume). The concentrated enzyme was evaluated for temperature and pH stability and kinetic parameter, characterized by monitoring oxidation of different ABTS [2, 20-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)] substrate concentrations. Plackett–Burman experimental design (PBED) implementation improved previous work results by 3.05-fold, obtaining a laccase activity of 1373.72 ± 0.37 U L−1 at 168 h of culture in a 500 mL shake flask. In contrast, one factor experimental design (OFED) applied after PBED improved by threefold the previous study, additionally increasing the C/N ratio. Employing OFED media at 10 L bioreactor scale was capable of producing 3159.93 ± 498.90 U L−1 at 192 h, representing a 2.4-fold increase. rPOXA 1B concentrate remained stable between 10 and 50 °C and retained over 70% residual enzymatic activity at 60 °C and 50% at 70 °C. Concerning pH stability, the enzyme was stable at pH 4.0 ± 0.2 with a residual activity greater than 90%. The lowest residual activity (60%) was obtained at pH 10.0 ± 0.2. Furthermore, the apparent kinetic parameters were Vmax of 3.163 × 10−2 mM min−1 and Km of 1.716 mM. Collectively, regarding enzyme stability our data provide possibilities for applications involving a wide range of pH and temperatures.
Keywords: Plackett–Burman experimental design, One-factor experimental design, Pichia pastoris, Recombinant laccase, Enzyme stability, Enzyme kinetics
Introduction
Laccases (EC 1.10.3.2) also known as p-diphenol oxidases are blue multicopper oxidases (MCOs) with the ability to catalyze the oxidation of organic aromatic compounds, with concomitant reduction of molecular oxygen to water. Laccases have an extraordinary range of natural substrate degradation activity, such as phenols, polyphenols, anilines, arylamines, methoxyphenols, hydroxyindoles, benzenethiols, organic and inorganic compounds (Gelo-Pujic et al. 1999; Brown et al. 2002) in the presence of a mediator substance, such as ABTS [2, 20-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)], (O’Callaghan et al. 2002; Taha et al. 2013).
Laccases have been proposed for a great number of potential industrial and biotechnological applications, such as textile, food processing, paper and pulp, pharmaceutical, nanobiotechnology, and cosmetic industry. In addition, laccases have also been used in bioremediation. Due to its activity, laccases can decolorize synthetic dyes, such as anthraquinone, azo compounds (Hou et al. 2004; Zeng et al. 2012), triarylmethane, indigo carmine (Abadulla et al. 2000; Colao et al. 2006), triphenylmethane (Qing Yang et al. 2009; Morales-Álvarez et al. 2016; Morales-Álvarez et al. 2017b), neolane (Zouari-Mechichi et al. 2006), and other dyes such as remazol bright blue in the presence of organic mediators (Soares et al. 2001) from textile effluents. Laccase effectiveness has been demonstrated in pulping black liquor (PBL) from the paper industry (Rivera-Hoyos et al. 2018). Additionally, laccase can aid in low density polyethylene (LDPE) sequential physical–chemical–biologic biotransformation (Gómez-Méndez et al. 2018) and in bioconversion of lignocellulosic and oxodegradable polyethylene residues (Moreno-Bayona et al. 2019). Majeau et al. (2010) found that some phenolic or xenobiotic compounds could be eliminated from industrial wastewaters using laccases (Majeau et al. 2010). García et al. (2011) employed laccases to remove pharmaceuticals and personal care products from conventional wastewater (Garcia et al. 2011). Another important laccase industrial application is pulp bleaching for paper production (Camarero et al. 2004).
At present, laccases have great potential as biological catalysts and are of considerable importance for the biotechnological industry; however, laccase secreted from natural sources are generally not adequate for high-scale production, usually due to low production yield and high preparation and purification costs. Nonetheless, heterologous expression allows obtaining high yields and producing laccase in greater quantities for industrial applications.
Laccases have been expressed in methylotrophic yeast such as P. pastoris, using different promoters with interesting results. From our perspective as described in previous work by our group (Rivera-Hoyos et al. 2015), pAOX1 is not compatible with the environment. Enzyme production by using pAOX1 requires the addition of methanol to culture media. Hence, the enzyme must be isolated from the media to avoid methanol disposal into the effluent to be treated. Additionally, methanol concentration in culture media could result in a problem when scaling up the process, mainly because methanol is highly flammable (Brooks and Crowl 2007), fire hazardous, toxic (Çelik and Çalık 2012), and with elevated costs associated with storage and handling (Cos et al. 2006). However, currently few studies have been devoted to laccase recombinant heterologous expression in P. pastoris using glyceraldehyde-3-phosphate dehydrogenase promoter (pGAP). Nevertheless, it has been demonstrated that glucose is an adequate carbon source for constitutive laccase production, contrary to glycerol or maltose (Bohlin et al. 2006a).
The objective of this work was to increase P. ostreatus recombinant POXA 1B (rPOXA 1B) laccase activity at low-scale production by improving culture media, taking into account nitrogen concentration and source type (organic and inorganic), carbon source concentration (glucose), copper concentration, media volume (attempting to impact oxygen transfer), and inoculum percentage to assay improved media at 10 L bioreactor scale with 6 L effective work volume (EWV). In addition, we characterized concentrated enzyme kinetics in terms of maximum reaction rate Vmax (mM min−1) and Michaelis constant KM (mM), by using ABTS as a substrate.
Materials and methods
Strain
Pichia pastoris X33 containing the expression vector pGAPZαA-LaccPost-Stop (Clone 1) with previously optimized POXA 1B synthetic gene codifying for Pleurotus ostreatus laccase was used. This strain was previously conserved in YPG media [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose] supplemented with 20% (w/v) glycerol and stored at − 80 °C (Rivera-Hoyos et al. 2015).
Inoculum preparation
Vials from Master Cell Bank (MCB) (Poutou et al. 1994) clone 1 of the P. pastoris X33/pGAPZαA-LaccPost-Stop strain were used. Once thawed, 5 mL sterile YPG media supplemented with 40 µg mL−1 Zeocin (Z) were inoculated and incubated at 30 °C overnight at 180 r.p.m. 500 mL Erlenmeyer flasks with 100 mL fresh YPG-Z (EWV) were inoculated and cultured under the same conditions for 12 h. The number of Erlenmeyer flasks depended on the assay as defined by factorial design at shake flask or 10 L bioreactor scale (6 L EWV). To dismiss the presence of any contaminant morphology by Gram stain, the remaining culture was evaluated and used as inoculum.
Plackett–Burman experimental design
Seven factors with two levels were evaluated to implement this design as follows: media volume (150 and 300 mL), CuSO4 concentration (0.1 and 1.0 mM), inoculum percentage [2 and 10% (v/v)], glucose concentration (10 and 30 g L−1), (NH4)2SO4 concentration (5 and 20 mM), peptone concentration (10 and 20 g L−1), and yeast extract concentration (5 and 10 g L−1). In addition, the design included a center point that was evaluated in triplicate, assayed values were 225 mL media volume, 0.55 mM CuSO4 concentration, 6% (v/v) inoculum, 20 g L−1glucose concentration, 12.50 mM (NH4)2SO4 concentration, 15 g L−1 peptone concentration, and 7.5 g L−1 yeast extract concentration (Morales-Álvarez et al. 2017a).
One factor design
Results obtained from the Plackett–Burman experimental design were taken into account for this design as a function of the significant factor and its contribution percentage on culture enzyme activity (Sarria-Alfonso et al. 2013).
The following variables were evaluated in time: enzyme activity (U L−1), protein concentration (mg mL−1), glucose concentration (g L−1), specific enzyme activity (U mg −1), and productivity (U L−1 h−1) based on “enzyme activity” (Rivera-Hoyos et al. 2015; Morales-Álvarez et al. 2017a). Enzyme activity (U L−1) was used for statistical analysis as the response variable. All assays for statistical improvement were performed in 500 mL Erlenmeyer flasks at 30 °C and 180 r.p.m., always using the same shaker for 168 h of culture with a pH variable, starting at 7.0 ± 0.2. Design Expert (V9.0) software (Stat-Ease, Inc. Minneapolis, MN USA) was used to design the experiments as well as to perform data analysis.
10 L bioreactor scale production containing 6 L of effective work volume
The media resulting from OFED evaluation was employed. Production was performed in 10 L bioreactor (TECFERM) with 60% (6 L) effective work volume (EWV). Production conditions were 30 °C, 1 v.v.m., (air volume/media volume), and 180 r.p.m., for 192 h. Samples for kinetic study were collected every 2 h during the first 12 h and every 24 h up to 192 h. Three lots were produced and were kinetically characterized as a function of dried biomass (g L−1), extracellular protein concentration (mg mL−1), residual glucose concentration (g L−1), enzyme activity (U L−1), specific enzyme activity (U mg−1), and pH. The following kinetic parameters were calculated: specific growth rate µx (h−1), duplication time Td (h), biomass/substrate yield Y(x/s) (g g−1), biomass productivity P(x) (g L−1 h−1), enzyme/substrate yield “Y(p/s)”, (U L−1 g−1), and enzyme productivity “P(p)” (U L−1 h−1) (quotation marks “” used for values based on enzyme activity).
Sigma Plot (V11.0), (Systat Software, Inc. San Jose, CA USA) was used for all culture kinetic graphs.
Dry biomass determination
Dry biomass concentration (g L−1) was calculated by using a calibration curve (Landázuri et al. 2009) (Eq. 1).
| 1 |
where X is the g L−1 dry biomass and Absλ600nm is the absorbance at 600 nm. Later, X (g L−1) was transformed to Ln (X/X0).
Supernatant concentration
The supernatant obtained from 10 L bioreactor was centrifuged at 8000×g and filtered through Whatman No. 3 paper, followed by filtration through Sartopore®2 (pore size: 0.45/0.2 µm heterogeneous PES double layer, Sartorius). The filtrate was ultrafiltered using Vivaflow-200, 100,000, and 10,000 MWCO PES cross flow cassettes (Sartorius), and the concentrate rPOXA 1B was stored at 4 °C until use.
Before and after each step, the following were measured: volume (mL), pH, enzyme activity (U L−1), and protein concentration (mg mL−1). Specific activity (U mg−1) was also calculated. A fraction of the enzyme concentrate was used for pH and temperature stability, and to determine KM and Vmax enzyme kinetic parameters, as well as zymogram functional activity.
Functional identification of the enzyme
The zymogram was carried out in 12% (w/v) native-PAGE under non-denaturing conditions. Enzyme activity or functionality was visualized in the gel by 0.5 M ABTS stain. A control laccase Lac® (Sigma-Aldrich, St. Louis, MO USA) was used for functional enzyme detection (Morales-Álvarez et al. 2017a).
Temperature stability
Concentrated enzyme stability was evaluated by incubating for 1 h at the following temperatures: 10, 20, 30, 40, 50, 60 and 70 °C, followed by residual laccase activity evaluation. All measurements were carried out at least three times (Morales-Álvarez et al. 2017a).
pH stability
To determine pH stability, the concentrated enzyme was previously incubated for 1 h at 25 °C in the absence of substrate, using Britton–Robinson buffer (Reynolds et al. 2013), with pH values in the ranges of 2.0 to 10.0 ± 0.2, then residual laccase activity was determined under standard assay conditions (Morales-Álvarez et al. 2017a). All determinations were carried out at least in triplicate.
Enzyme kinetic constants
Concentrated enzyme kinetic constants were evaluated using ABTS as a substrate in a concentration ranging from 0.1 to 3 mM, in 600 mM sodium acetate buffer, pH 4.5 ± 0.2. For all assays, 800 µl concentrated enzyme with an activity of 10.6 U L−1 was used. Assays were performed at 25 °C (Morales-Álvarez et al. 2017a). After adjusting the hyperbola using Michaelis–Menten equation, apparent KM and Vmax were calculated following Lineweaver–Burk linearization method (Nelson and Cox 2005), by employing the SIMFIT (V7.4.6) software (Burquillo et al. 2003). All kinetic assays were performed in triplicate.
Determination of total residual reducing sugar concentration
Total residual reducing sugar concentrations were determined for each sample in triplicate. To this end, 3,5-dinitrosalicylic acid was used (Miller 1959). A standard curve was prepared with values ranging from 0.1 to 5 g L−1 d-glucose and values were calculated according to Eq. 2.
| 2 |
Total extracellular protein concentration
For each sample residual protein was determined by measuring direct absorbance (Abs) at 280 nm, using UV–Vis NanoDrop spectrophotometer (Thermo Scientific).
Enzyme activity assay
Laccase enzyme activity was monitored by a change in absorbance at 436 nm (ε436 = 29,300 M−1 cm−1) due to ABTS oxidation in a 60 mM sodium acetate buffer (pH 4.5 ± 0.2). 800 µL sample was added to 100 µL 600 mM sodium acetate buffer and 100 µL 0.5 mM ABTS at room temperature (25 °C). Green radical formation was evaluated spectrophotometrically for 3 min. A unit of activity is defined as the quantity of enzyme required for oxidizing 1 µmol of ABTS in 1 min. Blanc solution contained 800 µL distilled water, 100 µL 600 mM sodium acetate buffer solution, and 100 µL 5 mM ABTS. Enzyme activity was expressed as U L−1 (Tinoco et al. 2001).
Specific activity was calculated by dividing enzymatic activity for each hour of culture by total extracellular protein concentration (Eq. 3).
| 3 |
where Act. Enz. = U L−1, Prot. Conc. = mg mL−1
Productivity, enzyme activity production per hour (UL−1 h−1), was calculated by using Eq. 4:
| 4 |
Results
Plackett–Burman experimental design
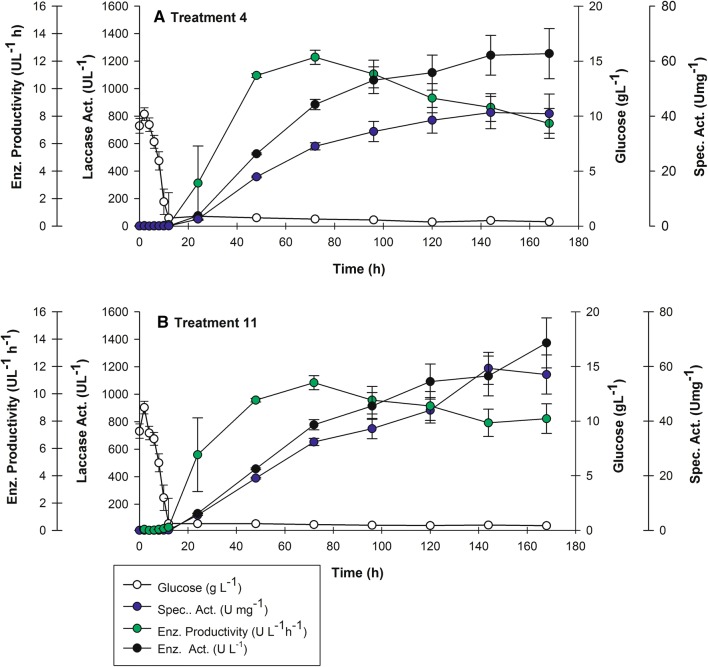
Plackett–Burman experimental design was implemented to evaluate the primary effect of the seven factors assayed on extracellular enzyme activity. Treatments (T4 and T11) were distinctive with enzyme activity greater than 1200 U L−1 (Fig. 1).
Fig. 1.
Plackett-Burman Experimental Design (PBED). a T4 kinetic performance. b T4 kinetic performance (Table 2)
From 72 h of culture and onward, laccase enzyme activity for treatments T4 and T11 exceeded 451.08 ± 6.46 U L−1: results obtained in a previous work (Rivera-Hoyos et al. 2015), (Fig. 1). The tendency was similar for specific activity. Statistical results for T11 at 168 h (best treatment) of culture are shown in Table 1; parameters include central point information (curvature) and regression model (unadjusted).
Table 1.
Laccase activity for T11 treatment at 168 h of culture
| Statistical and kinetic parameters | Unadj. |
|---|---|
| Model analysis of ANOVA | |
| Time of culture (h) | 168 |
| Model p value | 0.0158* |
| Model F value | 5.8000 |
| Lack of fit p value | 0.4022 |
| Lack of fit F value | 1.5300 |
| R2 | 0.4718 |
| Adj. R2 | 0.3905 |
| Pred. R2 | 0.2325 |
| Adeq. precision | 6.4240 |
| R2–adj. R2 | 0.0813 |
| Factor | Effect | p value | Contribution percent |
|---|---|---|---|
| A—culture media volume | 116.33 | 0.4697** | 3.540 |
| B—CuSO4 | − 14.79 | 0.9255 | 0.057 |
| C—inoculum | 304.04 | 0.0300* | 24.170 |
| D—glucose | − 11.95 | 0.9398 | 0.037 |
| E—(NH4)2SO4 | 19.62 | 0.9013 | 0.100 |
| F—peptone | 18.20 | 0.9084 | 0.087 |
| G—yeast extract | 297.50 | 0.0331* | 23.140 |
In the PBED, an ANOVA was performed to determine the effect of every variable on laccase activity. In addition, effect and contribution percent were also estimated
*Significant at 97%; **significant at 53%; other factors had less than 10% of significance
Table 1 depicts the influence of different factors on enzyme activity after applying a hierarchical model. This resulted in the removal of factors A, B, D, E, and F from the model. The polynomial equation representing laccase activity was (Eq. 5):
| 5 |
A comparison among observed enzyme activity results and model predicted enzyme activities are detailed in Table 2.
Table 2.
PBED matrix comparison between T4 and T11 for the observed and predicted results
| Treatment | Factor type | Volume of culture media (mL) | CuSO4 (mM) | Inoculum (% v/v) | Glucose (g L−1) | (NH4)2SO4 (mM) | Peptone (g L−1) | Yeast extract (g L−1) | Observed Enz Act. (U L−1) 168 h | Predicted Enz Act. (U L−1) 168 h |
|---|---|---|---|---|---|---|---|---|---|---|
| T4 | Factorial | 150 | 1 | 10 | 10 | 20 | 20 | 10 | 1254.270 | 1276.130 |
| T11 | Factorial | 300 | 1 | 10 | 10 | 5 | 10 | 10 | 1373.720 | 1276.130 |
| Center point | 225 | 0.55 | 6 | 20 | 12.5 | 15 | 7.5 | 1151.880 | 975.363 | |
| Center point | 225 | 0.55 | 6 | 20 | 12.5 | 15 | 7.5 | 1275.600 | 975.363 | |
| Center point | 225 | 0.55 | 6 | 20 | 12.5 | 15 | 7.5 | 1339.590 | 975.363 | |
Defined center points as consequence of factors influencing laccase activities. Factors at two levels in addition to a center point, as described in “Materials and methods”
One-factor experimental design
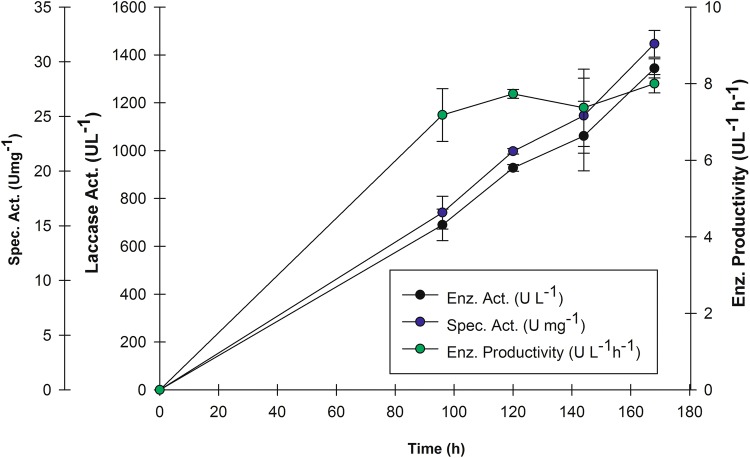
Results employing PBED allowed to formulate OFED (Table 1). After 168 h of culture, laccase enzyme activity from assays performed at level (− 1) had an activity of 961.43 ± 72.40 U L−1 with 10 g L−1 yeast extract. The highest enzyme activity observed (1343.52 ± 40.30 U L−1) was for level (0) with 15 g L−1 yeast extract. Last, level (+ 1) had an enzyme activity of 1251.37 ± 30.65 U L−1 with 20 g L−1 yeast extract. Figure 2 presents the kinetics at level (0) with 15 g L−1 yeast extract. It is noteworthy that the initial glucose concentration (10 g L−1) was depleted for the three experiments during the first 12 h of culture, as was observed in PBED.
Fig. 2.
One Factor experimental design (OFED). Level (0) kinetic results (only determined at 0, 96, 120, 144, and 168 h of culture) [OFED-15 g L−1 de yeast extract: 500 mL Erlenmeyer flask (300 mL EWV of culture media, 10 % (v/v) inoculum, 1 mM CuSO4, 10 g L−1 glucose, 20 mM (NH4)2SO4, 20 g L−1 peptone, 15 g L−1 yeast extract), Enzyme activity 1,343.52 ± 40.30 U L−1 at 168 h of culture]
One-factor experimental design results are shown in Table 3, and as observed the quadratic model was significant.
Table 3.
Effect of yeast extract concentration on laccase activity as determined by OFED (ANOVA)
| Source | Sum of squares | df | Mean square | F value | p value |
|---|---|---|---|---|---|
| Prob > F | |||||
| Model | 1.59E+08 | 2 | 79512.89 | 30.56 | 0.0101 |
| A—yeast extract | 90185.85 | 1 | 90185.85 | 34.66 | 0.0098 |
| A2 | 74965.36 | 1 | 74965.36 | 28.81 | 0.0127 |
| Pure error | 7805.45 | 3 | 2601.82 | ||
| Cor total | 1.67E+08 | 5 | |||
| R2 | 0.9532 | ||||
| Adj. R2 | 0.9220 | ||||
| Pred. R2 | 0.8129 | ||||
| Adeq. precision | 10.5930 |
Significant terms are presented in bold
Enzyme activity of 1365.67 U L−1 with desirability of 0.9860 could be obtained maintaining all factors and increasing yeast extract to 16.53 g L−1 (an additional 1.53 g), as predicted by the OFED. In comparison to the experimental activity observed, this would imply an increase in only 22.15 U L−1. Statistical results for both PBED and OFED are illustrated in Table 4.
Table 4.
PBED and OFED result summary
| Factor | Previous result | Experimental design | |
|---|---|---|---|
| PBED | OFED | ||
| EWV (mL) | 100 | 300 | 300 |
| Ratio media vol/flask vol. | 1/5 | 3/5 | 3/5 |
| Inoculum % (v/v) | 2.0 | 10.0 | 10.0 |
| CuSO4 (mM) | 0.0 | 1.0 | 1.0 |
| Glucose (g L−1) | 20.0 | 10.0 | 10.0 |
| (NH4)2SO4 (mM) | 0.0 | 5.0 | 20.0 |
| Peptone (g L−1) | 20.0 | 10.0 | 20.0 |
| Yeast extract (g L−1) | 10.0 | 10.0 | 15.0 |
| Time of culture (h) | 144 | 168 | 168 |
| Enzyme activity (U L−1) | 451.08 ± 6.46 | 1373.72 ± 0.37 | 1343.52 ± 40.30 |
| Reference | (Rivera-Hoyos et al. 2015) | Present study | |
Factors evaluated for both experimental designs are presented and compared with a previous study (Rivera-Hoyos et al. 2015)
Production at 10 L bioreactor scale containing 6 L of effective work volume
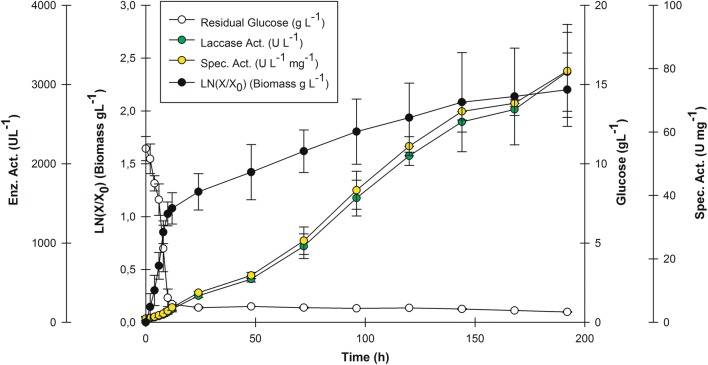
The media obtained from OFED (Table 4) was used to perform rPOXA 1B kinetics at 10 L bioreactor scale (triplicate). Figure 3 illustrates the average kinetics of three production lots.
Fig. 3.
rPOXA 1B kinetics at 10 L bioreactor scale (6 L EWV). Residual glucose, biomass’ Ln (g L−1), laccase activity (U L−1), and enzyme specific activity (U mg−1)
As observed for the PBED results, the highest glucose consumption was detected at 12 h (Fig. 5). In contrast, biomass cellular growth (g L−1) expressed as Ln (X/X0) displayed two exponential growth phases. A specific growth rate (μx = 0.10 h−1) and a duplication time Td of 7 h characterized the first phase (0–12 h of culture). The second exponential phase started once residual glucose was depleted (12–192 h) with a specific growth rate of μx = 0.006 h−1 and Td = 110 h. However, laccase activity and specific activity increased exponentially from 0 to 192 h, with values of 3159.93 ± 498.90 UL−1 and 79.28 ± 14.70 U mg−1, respectively. At this scale of production, once glucose was depleted, the highest enzyme activity and biomass were produced.
Fig. 5.
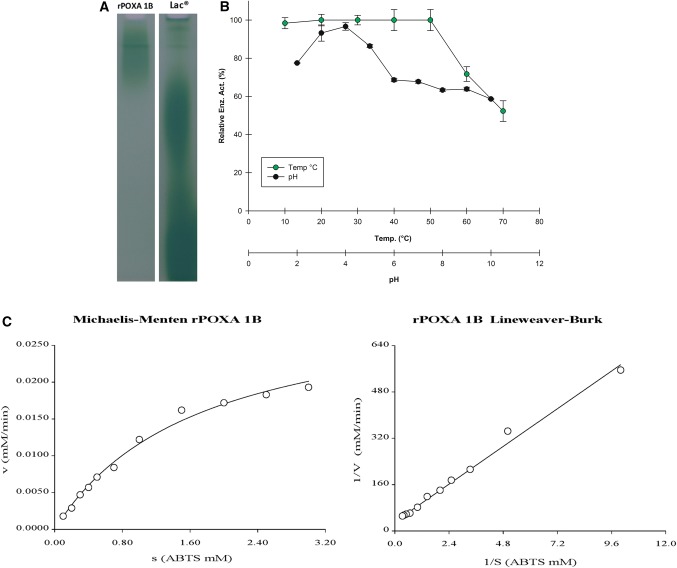
rPOXA 1B concentrate characterization. a Zymogram (native-PAGE 12% w/v) identification of functional rPOXA 1B, developed with 0.5 M ABTS. Samples: rPOXA 1B concentrate, Lac®: Positive control Laccase Sigma-Aldrich®. b Temperature and pH effect on relative residual enzyme activity, all assays were performed in triplicate. Relative enzyme activity relates to the enzyme activity at the assay's starting point. c Concentrate of rPOXA 1B enzyme kinetic results, left. Michaelis-Menten graph and right. Lineweaver-Burk plot
Supernatant concentration
Figure 4 demonstrates the average concentration of three rPOXA 1B produced lots in 10 L bioreactor, using OFED improved media.
Fig. 4.
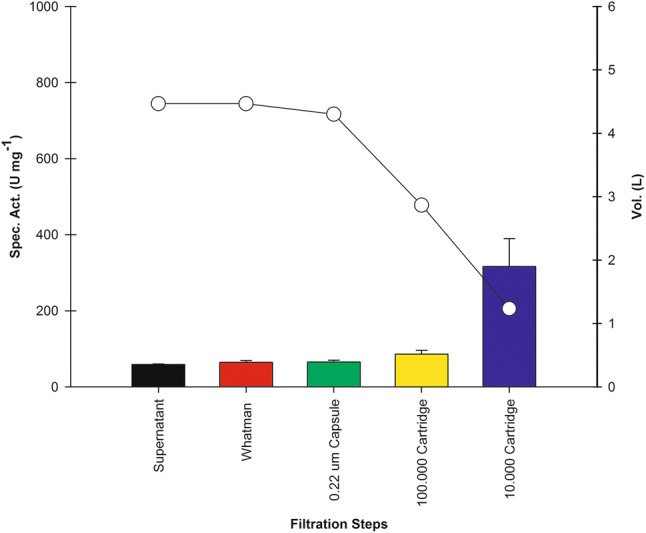
Average specific activity obtained from concentrate of three lots at 10 L bioreactor scale. Color bars depict average values of specific activity, at the end of culture in a 10 L bioreactor (supernatant, black bar), followed by different concentration steps. For each step, changes in volume are illustrated (line with open circles). Since the enzyme has an approximate weight of 54.18090 kDa, the filtrate was collected (0.22) for the Whatman, capsule, and 100.000 Dalton steps, whereas for the 10,000 Dalton cassette, the concentrate was collected
Functional enzyme identification
P. pastoris rPOXA 1B laccase functional identification was performed by zymogram (native PAGE) using ABTS as a developing agent (Fig. 5a).
Temperature and pH stability
Concentrated rPOX 1B laccase was exposed for 1 h to different temperatures and pH values to evaluate residual enzyme stability (Fig. 5b).
Kinetic constants
Last, enzyme kinetics were characterized using ABTS as a substrate as shown in Fig. 5c, Table 5, Eq. 6. Moreover, assay conditions were used to calculate maximum velocity values (Vmax) and substrate affinity or Michaelis constant (KM) for enzyme concentrates obtained from the supernatant:
| 6 |
where V0 is the initial velocity, Vmax is the maximum reaction rate, KM is the Michaelis constant, and [S] is the substrate concentration.
Table 5.
Apparent kinetic parameters. Data for rPOXA 1B using ABTS as substrate
| Kinetic parameters | Standard error | Confidence limit 95% | p | ||
|---|---|---|---|---|---|
| Vmax (mM min−1) | 3.163 × 10−2 | 0.001 | 0.029 | 0.034 | 0.000 |
| KM (mM) | 1.716 | 0.121 | 1.440 | 1.990 | 0.000 |
Discussion
Plackett–Burman experimental design (PBED)
In comparison with previous work, all treatments exceeded 451.08 ± 6.46 U L−1 enzyme activity (Rivera-Hoyos et al. 2015). The highest laccase activities were 1,254.27 ± 0.37 U L−1 and 1,373.72 ± 0.37 U L−1 for the most promising treatments T4 and T11, respectively (Fig. 1, Table 3), representing a 3.1-fold increase in comparison with our previous report (Rivera-Hoyos et al. 2015). However, productivity (based on enzyme activity) for the selected treatments T4 and T11 reached their highest values at 72 h of culture with 12.28 (U L−1 h−1) and 10.81 (U L−1 h−1), respectively (Fig. 1).
ANOVA for PBED statistical design can be presented in two ways, as an adjusted or unadjusted model. For an adjusted model, the central point information (curvature) is separated from the regression model. For the unadjusted model, the central points (curvature) are included within the regression model. We selected an unadjusted model to calculate standard deviation from the curvature (central points) and to decrease the number of replicas in treatments, assuming for all treatments the SDs of the curvature (central points).
For an unadjusted model, if the curvature is significant, the prediction of factorial points would be partially higher or lower depending on additional information given by the central points. If so, because the quadratic coefficients required to model the curvature are indistinguishable among themselves, the curvature could not be modeled, and the sum of squares (SS) of the curvature would be included in the SS lack of fit. Last, if the curvature is significant, this model would not be appropriate to perform a prediction.
In our case, the hierarchical model was significant for all analyzed hours for the unadjusted model curvature (Table 1). However, at 72 h, despite productivity “based on enzyme activity” being the highest (Fig. 1), the lack of fit p value was significant, thus indicating discontinuing the analysis. A comparison of the remaining hours of production suggested that the most significant p value observed was at 168 h, where lack of fit was not significant. At 168 h difference between R2 and Adj. R2 was the lowest comparing the difference among different hours of culture. Therefore, 168 h was selected as an adequate time point to analyze the main effect all factors would have on enzyme activity (Table 1).
For unadjusted model, an F value of 5.8 suggested that the model was significant (Table 1) and there would only be a 1.58% possibility for a greater F value due to noise in the experiment. A lack of fit F value of 1.53 implied it was not significant in relation to pure error. In addition, there would be a 40.22% probability that such large F value lack of fit could occur due to noise; therefore, a non-significant lack of fit is good for the model.
On the other hand, a predicted R2 of 0.2325 was in agreement with the Adj. R2 of 0.3905, since the difference between them was less than 0.2 (Table 1). Moreover, Adeq. Precision indicates the ratio signal/noise greater than 4 is desirable. For our case, estimated Adeq. Precision was 6.424; thus, this model could be used to navigate the design space.
To further continue improving the culture media and taking into account the obtained results (Table 1), the following factors were maintained: A at 300 mL, B at 0.1 mM, D at 10 g L−1, E at 20 mM, and F at 20 g L−1. Factors C had a positive contribution effect of 24.17% and G of 23.14%, suggesting that in a future statistical design they should be assayed at higher concentrations. However, for factor C, 10% (v/v) inoculum has already been tested, representing 30 mL for the next experimental design. Implementing such an increase in volume would not be practical; thus, we decided to continue with factor C at 10% (v/v), despite its significance.
Prediction for T11 did not exceed the obtained result. In contrast, T4 prediction slightly exceeded observed enzyme activity, but was only 7% less than the observed value for T11 (Table 2). The only remaining significant factor was G (yeast extract), with a percentage contribution of 23.14%. It was decided to perform a three-level one-factor statistical design with (− 1, 0, + 1) at 10, 15, and 20 g L−1 with a duplicate to improve enzyme activity.
Culture media volume (factor A) in PBED was significant at 53% with a 3.54% positive effect on enzyme activity (U L−1), (Table 1). In light of Table 4 results, and taking into account PBED is a screening method and does not value interactions among factors, it is necessary to expand the model’s navigation area by increasing Factor A (culture media volume), since it might have an association with the surface for oxygen transfer, as well as interactions with other assayed factors.
It is noteworthy to highlight PBED improved the media in comparison with previous experiments (Rivera-Hoyos et al. 2015), as CuSO4 (important for enzyme activity) was increased, as well as (NH4)2SO4 (a valuable inorganic nitrogen source). In contrast, peptone and yeast extract were decreased (organic nitrogen sources). Additionally, volume within the shake flask was also increased, which could be associated with decreased surface for oxygen transfer.
One-factor experimental design
ANOVA for one-factor design (Table 3) demonstrated with an F value of 30.56 the model was significant and there would only be a 1.01% possibility of a greater F value due to noise. On the other hand, Pred. R2 of 0.8129 was in agreement with an Adj. R2 of 0.9220, since the difference between them was less than 0.2. Estimated Adeq. Precision was 10.5930, which is an adequate statistical signal (Table 3).
One-factor experimental design quadratic model statistical adjustment, reliability, and significance were dependable for its prediction. However, when results were analyzed we decided not to assay the proposed prediction modification and to utilize 16.53 g L−1 of yeast extract instead of 15.00 g L−1, since the model’s predicted enzyme activity was (1365.67 U L−1), within the SD range from the experimentally observed enzyme activity (1343.52 ± 40.30 U L−1).
When comparing enzyme activity results with the three OFED yeast extract levels, it was clear that 15 g L−1 concentration was the most favorable (Fig. 2). Despite that, enzyme activity results obtained with 15 and 20 g L−1 were similar to the results obtained for PBED treatments T4 and T11, as well as the prediction for the same model. In this study, no considerable differences were observed between the PBED and OFED for enzyme activity (Table 4), since both values were found within the same standard deviation range. Nevertheless, taking into account OFED positive effect, 53% significance, as well as PBED volume of culture contribution (Table 1), and final decrease in C/N ratio (Table 4), we decided to use the media composition obtained with OFED for 10 L bioreactor production.
Production at 10 L bioreactor scale containing 6 L of effective work volume
Changing production geometry from 500 mL Erlenmeyer flask to a 10 L bioreactor with 60% EWV and using OFED obtained media (Table 4, Fig. 3), it was feasible to increase laccase activity from 1343.52 ± 40.30 (Fig. 2) to 2684.85 ± 78.44 U L−1 at 168 h, and even further increase to 3159.93 ± 498.90 U L−1 after 192 h of culture (Fig. 3), representing a 2.0- and 2.4-fold increase, respectively. Moreover, specific activity was also increased with geometry change from 31.64 ± 1.22 U mg−1 in agitated flask to 69.08 ± 1.10 U mg−1 at 168 h culture and 79.28 ± 14.690 U mg−1 at 192 h in 10 L bioreactor culture with 60% EWV (Fig. 3).
At both, shake flask and bioreactor scale, two exponential phases were observed (more evident in Fig. 3), the first one depending on glucose consumption and the second one after residual glucose depletion. Thus, various aspects are noteworthy as follows:
-
(i)
Under our assay conditions, improved media required less carbon concentration source, which is peculiar since more carbon would generate more biomass, clearly related to C/N ratio.
-
(ii)
Under our assay conditions, improved media required an increase in media volume, interpreted as a reduction in surface required for oxygen transfer (observed at shake flask scale).
-
(iii)
All of the above contrast with the fact that a constitutive pGAP governs the optimized synthetic laccase POXA 1B gene expression (Rivera-Hoyos et al. 2015), suggesting enzyme should be produced during cell growth phase, ending in the exponential phase (Figs. 1 and 3). Hence, our results agree with those reported in the literature. It has been proposed that genes under pGAP expression are not entirely constitutive and could be regulated under certain conditions. Kern et al. (2007) studied the expression of an oxidase fused to green fluorescent protein (GFP) under pGAP. Among the results discussed, they mentioned GFP fluorescence markedly increased after media glucose was depleted (Kern et al. 2007). Moreover, transiently small quantities of ethanol were produced, a phenomenon suggested by other authors (Baumann et al. 2008; Vogl and Glieder 2013). In the past, some authors have discussed that alternate oxidase activity does not exert a direct influence on hexose transporters. They suggest a more reasonable plausibility is that an alternate cell energy state can induce them to an increase in glucose uptake, responsible for cell growth (Kern et al. 2007). For our particular case, it is likely that a reduced surface for oxygen transfer, associated with an increase in culture media volume is responsible for an alternate energy state in the cell, increasing in this manner glucose uptake.
However, it is important to note that yeast’s observed metabolic regulation after glucose depletion was due to gluconeogenesis induction, which produces catabolites for growth and cell integrity. On the other hand, under low glucose concentration, biomass generation depends on media composition. Because there was the presence of a nitrogen source, such as peptone or yeast extract, biomass formation was favored in the present work with increased peptone and yeast extract (Table 4). Moreover, ammonium salts were also increased in the present work, an additional nitrogen source. Furthermore, when glucose is limited it has been described that Snf4 activates Snf1 kinase, a protein that phosphorylates proteins and transcriptional regulators in genes implicated in alternative carbon source use, such as gluconeogenesis, respiration, transport, meiosis, and response to stress (Lin et al. 2001; Hedbacker and Carlson 2009).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH enzyme, E.C. 1.2.1.12) catalyzes the reversible phosphorylation of glyceraldehyde-3-phosphate into 1,3-biphosphoglycerate, essential for energy metabolism, thus controlling pGAP expression. Giardina and Chiang (2013) evaluated in S. cerevisiae different enzymes expressed under limited glucose conditions, specifically GAP, participating in glycolytic and gluconeogenic pathways. They evidenced high enzyme activity, demonstrating the importance of this gene under glucose-limited conditions.
Table 6 presents fungal laccase activities expressed in P. pastoris from different origins (publications dated from 1997 to 2017), highlighting volume in which production was performed and activity. As can be observed, pAOX1 resulted in the highest activities. In our opinion, it presents a higher risk and is environmentally unfavorable, since it requires methanol addition. Furthermore, it can increase production costs, since it must be guaranteed methanol will not be employed for contaminated effluent treatment. We highlight Fomes lignosus laccase expression using pGAP, with elevated activities (production volume was not described). Additionally, Botrytis aclada laccase activity was also elevated using pGAP in a 2.5 L volume. On the other hand, laccase activity is influenced by the substrate employed (ABTS, guaiacol, etc.) and the reaction buffer (acetate, citrate, etc.). In this sense, our results are promising, as they are within the range of most of the previously published data.
Table 6.
P. pastoris (batch culture) fungal laccase activities from different origins, dated from 1997 to 2017
| Origin | Laccase | P. pastoris strain | Enz. Act. (UL−1) | Culture volume (L) | Time of culture (days) | Promoter | Refs |
|---|---|---|---|---|---|---|---|
| Pleurotus ostreatus | POXA1B | X33 | 1373.72 | 0.30 | 7 | GAP | Present work |
| Pleurotus ostreatus | POXA1B | X33 | 3159.93 | 6 | 8 | GAP | Present work |
| Ganoderma lucidum | GlLCC1 | X33 | 4.69 | 0.15 | 7 | GAP | (Morales-Álvarez 2017a) |
| Coriolopsis gallica | LcCg | X33 | 250 | 0.50 | 12 | AOX | (Avelar et al. 2017) |
| Pleurotus ostreatus | POXA1B | X33 | 451.08 | 0.10 | 6.5 | GAP | (Rivera-Hoyos et al. 2015) |
| Ganoderma lucidum | GlLCC1 | X33 | 0.13 | 0.10 | 6.5 | GAP | (Rivera-Hoyos et al. 2015) |
| Trametes trogiiBAFC 463 | lcc3 | GS115 | 5740 | Shake flask | – | AOX | (Campos et al. 2016) |
| Trametes versicolor | lccA | X33 | 11,972 | 5 | 16 | AOX | (Li et al. 2014) |
| Coprinus comatus | lac3/lac4 | KM71H | 689/1465 | 0.05 | 15 | AOX | (Gu et al. 2014) |
| Trametes versicolor | LAC5 | X33 | 14 | 0.04 | 5 | AOX | (Nishibori et al. 2013) |
| Gaeumannomyces graminis | LAC2 | X33 | 100 | 0.04 | 5 | AOX | (Nishibori et al. 2013) |
| Botrytis aclada | BaLac | X33 | 53,300 | 2.50 | 3 | GAP | (Kittl et al. 2012) |
| Botrytis aclada | BaLac | X33 | 51,000 | 0.40 | 4 | AOX | (Kittl et al. 2012) |
| Trametes sp. | LacB | GS115 | 32,000 | – | 13 | AOX | (Li et al. 2007) |
| Trametes sp. 420 | lacD | GS115 | 83,000 | 0.03 | – | AOX | (Hong et al. 2007) |
| Trametes versicolor | Lcc1/Lcc2 | SMD1168 | 1.30 | – | 7 | GAP | (Bohlin et al. 2006b) |
| Trametes trogii | LccI | GS115 | 2520 | 2 | 8 | AOX | (Colao et al. 2006) |
| Trametes sp. | rLacA | GS115 | 5470 | 0.03 | 13 | AOX | (Hong et al. 2006) |
| Fomes lignosus | lcc | GS115 | 9030 | – | 2 | GAP | (Liu et al. 2003) |
| Trametes versicolor | LccIV | GS115 | 1500 | 5 | 4 | AOX | (Brown et al. 2002) |
| Trametes versicolor | lcc1 | SMD1168 | 140,000 | 2.50 | 9 | AOX | (Hong et al. 2002) |
| Pleurotus sajor-caju | lac4 | GS115 and KM71 | 10,200 | 0.015 | 5 | AOX | (Soden et al. 2002) |
| Pycnoporus cinnabarinus | lac1 | X33 | 20 | 0.050 | 10 | AOX | (Otterbein et al. 2000) |
Enzyme concentration
Concentration steps allowed to increase rPOXA 1B specific activity by 5.3-fold, from 147.11 ± 7.24 U mg−1 (average obtained from the centrifuged supernatant) to 782.48 ± 90.20 U mg−1 (average obtained in final concentrate), with a 3.6-fold volume reduction (Fig. 4).
Functional identification of the enzyme
The zymogram allowed functional identification of the enzyme when it oxidized ABTS, generating a green stain (Fig. 5a). Additionally, for rPOXA 1B enzyme only one band was observed, with no additional bands whether larger or smaller; additional bands were observed for the concentrated enzyme, the native fungus or control laccase, suggesting the enzyme retained its monomeric nature as reported by Giardina et al. (1999).
Temperature and pH stability
Stability studies demonstrated P. ostreatus rPOXA 1B concentrated enzyme expressed in P. pastoris X33 was stable between 10 and 50 °C and retained more than 70% of its residual enzyme activity at 60 °C and 50% at 70 °C. Regarding pH stability, rPOXA 1B was more stable at pH 4.0 ± 0.2 with a residual enzyme activity greater than 90%, while at pH 3.0 ± 0.2 or less and greater than pH 5.0 ± 0.2 it was less stable under assay conditions. However, the lowest residual enzyme activity observed was approximately 60% at pH 10.0 ± 0.2. Our results agree with those reported by Giardina et al., (1999). In their study, they worked with purified laccase isoenzyme from P. ostreatus. Their highest activity described was at acid pH 3.0 ± 0.2 and temperatures ranging between 20 and 50 °C (Giardina et al. 1999), in agreement with our results. Taking into consideration temperature and pH stability, it is relevant to point out rPOXA 1B was more stable in comparison with other P. ostreatus isoenzymes (POXA 1 W, POXA2, POXC), characterized in previous studies (Giardina et al. 1999).
Kinetic constants
Whether of fungal or bacterial origin, different laccase KM values present a broad spectrum and can differ even with the same substrate. Nevertheless, for the most part a higher affinity toward ABTS has been reported in comparison with substrates such as syringaldazine or guaiacol, among others. These substrates can be oxidized at a lower velocity with higher Michaelis constants (Manavalan et al. 2013; Patel et al. 2014; Liu et al. 2015). For laccases using ABTS as a substrate, a Vmax of 3.163 × 10−2 mM min−1, KM of 1.716 mM was calculated and found within the values previously reported by others (Manavalan et al. 2013; Gu et al. 2014; Li et al. 2014; You et al. 2014; Liu et al. 2015; Park et al. 2015).
Major and minor differences among other laccase kinetic parameters and ours could be accounted by different factors, where the efficiency of the catalytic process could not only be affected by the enzyme’s intrinsic characteristics, but also by reaction-specific factors, such as complex reactions with various steps involved in the process (Nelson and Cox 2005). Even a minimum variation in the sequence could play an important role in substrate–enzyme interaction and its oxidation, thus highlighting the importance of molecular characterization for the enzyme of interest. Considerable amount of data is necessary to comparatively determine if the difference present among similar laccases is directly related to gene sequences, or if they obey posttranslational modifications, gathered for instance from culture or purification conditions.
The importance of this work resides in statistically significant improved conditions for culture media production. Furthermore, the obtained concentrated enzyme was demonstrated to be stable in a wide range of pH values and temperatures, even though laccase origin is from a mesophilic organism. Therefore, establishing a recombinant enzyme can be done under extreme conditions.
Conclusion
In this work, we improved culture media for rPOXA 1B laccase production by a 2.4-fold increase at 10 L bioreactor in 192 h of culture. Furthermore, POXA 1B was functionally identified by zymography. Moreover, pH stability analysis revealed that the enzyme was more stable at pH 4.0 ± 0.2, with a relative enzyme activity maintained over 60% in a wide range of pH. Regarding temperature stability, it displayed a wide range between 10 and 70 °C. Kinetic parameters obtained for rPOXA 1B showed the enzyme’s affinity for ABTS substrate, which is in the range reported for other laccases. rPOXA 1B expressed in P. pastoris when using improved culture media, under our experimental conditions, is definitively a promising enzyme for several applications involving a wide range of temperatures and pH values.
Acknowledgements
This research was funded by the following grants: Grant ID: 00005575 (Correlación entre la expresión constitutiva la concentración de proteína y la actividad biológica de las lacasas recombinantes POXA1 B de Pleurotus ostreatus y GLlac 1 de Ganoderma lucidum en Pichia pastoris); Grant ID: 00006337 (Optimización del medio de cultivo para producción de la lacasa recombinante POXA 1B de Pleurotus ostreatus en Pichia pastoris) from Pontificia Universidad Javeriana; Grant ID: 00006169 (Joven Investigador Colciencias 2014), Bogotá, D.C. Colombia; Grant ID: 00007885 (Estudio de la estabilidad a tiempo real del concentrado de la lacasa rPOXA 1B de Pleurotus ostreatus producida en Pichia pastoris) from Pontificia Universidad Javeriana. The financing entity had no role in the study design, data collection, or analysis, decision to publish, or preparation of the manuscript. The authors thank Fiona Raikes and María Lucía Gutiérrez, Ph.D., for English editing.
Abbreviations
- MCOs
Multicopper oxidases
- ABTS
2, 20-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)
- PBL
Pulping black liquor
- LDPE
Low density polyethylene
- pAOX1
Alcohol oxidase promoter
- pGAP
Glyceraldehyde-3-phosphate dehydrogenase promoter
- POXA 1B
Laccase from P. ostreatus
- rPOXA 1B
Recombinant laccase from P. ostreatus
- Vmax
Maximum reaction rate
- KM
Michaelis constant
- YPG
Yeast extract, peptone and glucose culture media
- MCB
Master cell bank
- YPG-Z
Yeast extract, peptone, glucose-zeocin culture media
- EWV
Effective work volume
- PBED
Plackett–Burman experimental design
- OFED
One-factor experimental design
- v.v.m.
Air volume per media volume
- µx
Specific growth rate
- Td
Duplication time
- r.p.m.
Revolutions per minute
- Y(x/s)
Biomass/substrate yield
- Y(p/s)
Enzyme/substrate yield
- P(x)
Biomass productivity
- P(p)
Enzyme productivity
- SD
Standard deviation
- GFP
Green fluorescent protein
- Snf4
5′-AMP-activated protein kinase subunit gamma
- Snf1
Carbon catabolite-derepressing protein kinase
- GAPDH
Enzyme glyceraldehyde-3-phosphate dehydrogenase
Compliance with ethical standards
Conflict of interest
The authors declare they have no competing interests.
Footnotes
Leidy D. Ardila-Leal∇, Diego A. Albarracín-Pardo and Claudia M. Rivera-Hoyos contributed equally to this work.
References
- Abadulla E, Tzanov T, Costa S, Robra K-H, Cavaco-Paulo A, GuBitz GM. Decolorization and detoxification of textile dyes with a laccase from Trametes hirsuta. Appl Environ Microbiol. 2000;66(8):3357–3362. doi: 10.1128/AEM.66.8.3357-3362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelar M, Olvera C, Aceves-Zamudio D, Luis Folch J, Ayala M. Recombinant expression of a laccase from Coriolopsis gallica in Pichia pastoris using a modified a-factor preproleader. Protein Expr Purif. 2017;136:14–19. doi: 10.1016/j.pep.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Baumann K, Maurer M, Dragosits M, Cos O, Ferrer P, Mattanovich D. Hypoxic fed-batch cultivation of Pichia pastoris increases specific and volumetric productivity of recombinant proteins. Biotechnol Bioeng. 2008;100(1):177–183. doi: 10.1002/bit.21763. [DOI] [PubMed] [Google Scholar]
- Bohlin C, Johnson LJ, Roth R, Van Zyl WH. Heterologous expression of Trametes versicolor laccase in Pichia pastoris and Aspergillus niger. Appl Biochem Biotechnol. 2006;129–132:195–214. doi: 10.1007/978-1-59745-268-7_15. [DOI] [PubMed] [Google Scholar]
- Bohlin C, Jonsson LJ, Roth R, Van Zyl WH. Heterologous expression of Trametes versicolor laccase in Pichia pastoris and Aspergillus niger. Appl Biochem Biotechnol. 2006;129–132:195–214. doi: 10.1385/ABAB:129:1:195. [DOI] [PubMed] [Google Scholar]
- Brooks MR, Crowl DA. Flammability envelopes for methanol, ethanol, acetonitrile and toluene. J Loss Prev Process Ind. 2007;20:144–150. doi: 10.1016/j.jlp.2007.01.001. [DOI] [Google Scholar]
- Brown MA, Zhao Z, Mauk AG. Expression and characterization of a recombinant multi-copper oxidase: laccase IV from Trametes versicolor. Inorg Chim Acta. 2002;331:232–238. doi: 10.1016/S0020-1693(01)00814-3. [DOI] [Google Scholar]
- Burquillo FJ, Holgado M, Bardsley WG. Uso del paquete estadístico SIMFIT en la enseñanza del análisis de datos en ciencias experimentales. J Sci Educ. 2003;4(1):8–14. [Google Scholar]
- Camarero S, García O, Vidal T, Colom J, del Río JC, Gutiérrez A, Gras JM, Monje R, Martínez MJ, Martínez ÁT. Efficient bleaching of non-wood high-quality paper pulp using laccase-mediator system. Enzyme Microb Technol. 2004;35:113–120. doi: 10.1016/j.enzmictec.2003.10.019. [DOI] [Google Scholar]
- Campos PA, Levin LN, Wirth SA. Heterologous production, characterization and dye decolorization ability of a novel thermostable laccase isoenzyme from Trametes trogii BAFC 463. Process Biochem. 2016;51:895–903. doi: 10.1016/j.procbio.2016.03.015. [DOI] [Google Scholar]
- Çelik E, Çalık P. Production of recombinant proteins by yeast cells. Biotechnol Adv. 2012;30:1108–1118. doi: 10.1016/j.biotechadv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Colao MC, Lupino S, Garzillo AM, Buonocore V, Ruzzi M. Heterologous expression of lcc1 gene from Trametes trogii in Pichia pastoris and characterization of the recombinant enzyme. Microb Cell Fact. 2006;5:1–11. doi: 10.1186/1475-2859-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cos C, Ramón R, Montesinos JL, Valero F. Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microb Cell Fact. 2006;5:17. doi: 10.1186/1475-2859-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GaMB Soares, Pessoa de Amorim MT, Costa-Ferreira M. Use of laccase together with redox mediators to decolourize Remazol Brilliant Blue R. J Biotechnol. 2001;89:123–129. doi: 10.1016/S0168-1656(01)00302-9. [DOI] [PubMed] [Google Scholar]
- Garcia HA, Hoffman CM, Kinney KA, Lawler DF. Laccase-catalyzed oxidation of oxybenzone in municipal wastewater primary effluent. Water Res. 2011;45:1921–1932. doi: 10.1016/j.watres.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Gelo-Pujic M, Hyung-Han K, Butlin NG, Palmore GTR. Electrochemical studies of a truncated laccase produced in Pichia pastoris. Appl Environ Microbiol. 1999;65:5515–5521. doi: 10.1128/aem.65.12.5515-5521.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina P, Palmieri G, Scaloni A, Fontanella B, Faraco V, Cennamo G, Sannia G. Protein and gene structure of a blue laccase from Pleurotus ostreatus. Biochem J. 1999;341:655–663. doi: 10.1042/bj3410655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Méndez LD, Moreno-Bayona DA, Poutou-Piñales RA, Salcedo-Reyes JC, Pedroza-Rodríguez AM, Vargas A, Bogoya JM. Biodeterioration of plasma pretreated LDPE sheets by Pleurotus ostreatus. PLoS One. 2018;13(9):e0203786. doi: 10.1371/journal.pone.0203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Zheng F, Long L, Wang J, Ding S. Engineering the expression and characterization of two novel laccase isoenzymes from Coprinus comatus in Pichia pastoris by fusing an additional ten amino acids tag at N-terminus. PLoS One. 2014;9(4):e93912. doi: 10.1371/journal.pone.0093912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2009;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Meinander NQ, Jönsson LJ. Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris. Biotechnol Bioeng. 2002;79(4):438–449. doi: 10.1002/bit.10297. [DOI] [PubMed] [Google Scholar]
- Hong Y, Xiao Y, Zhou H, Fang W, Zhang M, Wang J, Wu L, Yu Z. Expression of a laccase cDNA from Trametes sp. AH28-2 in Pichia pastoris and mutagenesis of transformants by nitrogen ion implantation. FEMS Microbiol Lett. 2006;258:96–101. doi: 10.1111/j.1574-6968.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- Hong Y-z, Zhou H-m, Tu X-m, Li J-f, Xiao Y-z. Cloning of a laccase gene from a novel Basidiomycete Trametes sp. 420 and its heterologous expression in Pichia pastoris. Curr Microbiol. 2007;54:260–265. doi: 10.1007/s00284-006-0068-8. [DOI] [PubMed] [Google Scholar]
- Hou H, Zhou J, Wang J, Du C, Yan B. Enhancement of laccase production by Pleurotus ostreatus and its use for the decolorization of anthraquinone dye. Process Biochem. 2004;39:1415–1419. doi: 10.1016/S0032-9592(03)00267-X. [DOI] [Google Scholar]
- Kern A, Hartner FS, Freigassner M, Spielhofer J, Rumpf C, Leitner L, Frohlich KU, Glieder A. Pichia pastoris “just in time” alternative respiration. Microbiology. 2007;153(4):1250–1260. doi: 10.1099/mic.0.2006/001404-0. [DOI] [PubMed] [Google Scholar]
- Kittl R, Mueangtoom K, Gonaus C, Khazaneh ST, Sygmund C, Haltrich D, Ludwig R. A chloride tolerant laccase from the plant pathogen ascomycete Botrytis aclada expressed at high levels in Pichia pastoris. J Biotechnol. 2012;157:304–314. doi: 10.1016/j.jbiotec.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Landázuri P, Poutou-Piñales RA, Acero-Godoy J, Córdoba-Ruiz HA, Echeverri-Peña OY, Sáenz H, Delgado-Boada JM, Barrera-Avellaneda LA. Cloning and shake flask expression of hrIDS-Like in Pichia pastoris. Afr J Biotech. 2009;8(12):2871–2877. [Google Scholar]
- Li JF, Hong YZ, Xiao YZ, Xu YH, Fang W. High production of laccase B from Trametes sp. in Pichia pastoris. World J Microbiol Biotechnol. 2007;23:741–745. doi: 10.1007/s11274-006-9286-2. [DOI] [Google Scholar]
- Li Q, Pei J, Zhao L, Xie J, Cao F, Wang G. Overexpression and characterization of laccase from Trametes versicolor in Pichia pastoris. Appl Biochem Microbiol. 2014;50(2):140–147. doi: 10.1134/S0003683814020124. [DOI] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J Biol Chem. 2001;276(38):36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- Liu W, Chao Y, Liu S, Bao H, Qian S. Molecular cloning and characterization of a laccase gene from the basidiomycete Fame lignosus and expression in Pichia pastoris. App Microbiol Biochem. 2003;63(2):174–181. doi: 10.1007/s00253-003-1398-0. [DOI] [PubMed] [Google Scholar]
- Liu H, Cheng Y, Du B, Tong C, Liang S, Han S, Zheng S, Lin Y. Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in Pichia pastoris and its application in synthetic dye decolorization. PLoS One. 2015;10(3):0119833. doi: 10.1371/journal.pone.0119833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau J-A, Brar SK, Tyagi RD. Laccases for removal of recalcitrant and emerging pollutants. Bioresour Technol. 2010;101(7):2331–2350. doi: 10.1016/j.biortech.2009.10.087. [DOI] [PubMed] [Google Scholar]
- Manavalan T, Manavalan A, Thangavelu KP, Heese K. Characterization of optimized production, purification and application of laccase from Ganoderma lucidum. Biochem Eng J. 2013;70:106–114. doi: 10.1016/j.bej.2012.10.007. [DOI] [Google Scholar]
- Miller G. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Morales-Álvarez ED, Rivera-Hoyos CM, González-Ogliastri N, Rodríguez-Vázquez R, Poutou-Piñales RA, Daza CE, Pedroza-Rodríguez AM. Partial removal and detoxification of Malachite Green and Crystal Violet from laboratory artificially contaminated water by Pleurotus ostreatus. Univ Sci. 2016;21(3):259–285. doi: 10.11144/Javeriana.SC21-3.prad. [DOI] [Google Scholar]
- Morales-Álvarez ED, Rivera-Hoyos CM, Cardozo-Bernal ÁM, Poutou-Piñales RA, Pedroza-Rodríguez AM, Díaz-Rincón DJ, Rodríguez-López A, Alméciga-Díaz CJ, Cuervo-Patiño CL. Plackett-Burman design for rGILCC1 laccase activity enhancement in Pichia pastoris: concentrated enzyme kinetic characterization. Enzyme Res. 2017;2017:5947581. doi: 10.1155/2017/5947581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Álvarez ED, Rivera-Hoyos CM, Chaparro-Núnez LE, Daza CE, Poutou-Piñales RA, Pedroza-Rodríguez AM. Decolorization and detoxification of Malachite Green by Ganoderma lucidum: key operating parameters and adsorption studies. J Environ Eng. 2017;143(4):04016093. doi: 10.1061/(ASCE)EE.1943-7870.0001180. [DOI] [Google Scholar]
- Moreno-Bayona DA, Gómez-Méndez LD, Blanco-Vargas A, Castillo-Toro A, Herrera-Carlosama L, Poutou-Piñales RA, Salcedo-Reyes JC, Díaz-Ariza LA, Castillo-Carvajal LC, Rojas-Higuera NS, Pedroza-Rodríguez AM. Simultaneous bioconversion of lignocellulosic residues and oxodegradable polyethylene by Pleurotus ostreatus for biochar production, enriched with phosphate solubilizing bacteria for agricultural use. PLoS One. 2019;14(5):e0217100. doi: 10.1371/journal.pone.0217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger, principles of biochemistry. 5. New York: W.H. Freeman and Company; 2005. [Google Scholar]
- Nishibori N, Masaki K, Tsuchioka H, Fujii T, Iefuji H. Comparison of laccase production levels in Pichia pastoris and Cryptococcus sp. S-2. J Biosci Bioeng. 2013;115(4):394–399. doi: 10.1016/j.jbiosc.2012.10.025. [DOI] [PubMed] [Google Scholar]
- O’Callaghan J, O’Brien MM, McClean K, Dobson AD. Optimisation of the expression of a Trametes versicolor laccase gene in Pichia pastoris. J Ind Microbiol Biotechnol. 2002;29(2):55–59. doi: 10.1038/sj.jim.7000268. [DOI] [PubMed] [Google Scholar]
- Otterbein L, Record E, Longhi S, Asther M, Moukha S. Molecular cloning of the cDNA encoding laccase from Pycnoporus cinnabarinus I-937 and expression in Pichia pastoris. Eur J Biochem. 2000;267:1619–1625. doi: 10.1046/j.1432-1327.2000.01166.x. [DOI] [PubMed] [Google Scholar]
- Park M, Kim M, Kim S, Ha B, Ro H-S. Differential expression of laccase genes in Pleurotus ostreatus and biochemical characterization of laccase isozymes produced in Pichia pastoris. Mycobiology. 2015;43(3):280–287. doi: 10.5941/MYCO.2015.43.3.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H, Gupte S, Gahlout M, Gupte A. Purification and characterization of an extracellular laccase from solid-state culture of Pleurotus ostreatus HP-1. 3 Biotech. 2014;4(1):77–84. doi: 10.1007/s13205-013-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutou RA, Amador E, Candelario M. Banco de células primario (BCP): caracterización y papel en la producción de proteínas recombinantes. Biotecnología Aplicada. 1994;11(1):55–59. [Google Scholar]
- Qing Yang X, Xia Zhao X, Yun Liu C, Zheng Y, Jun Qian S. Decolorization of azo, triphenylmethane and anthraquinone dyes by a newly isolated Trametes sp. SQ01 and its laccase. Process Biochem. 2009;44:1185–1189. doi: 10.1016/j.procbio.2009.06.015. [DOI] [Google Scholar]
- Reynolds JE, III, Josowicz M, Vegh RB, Solntsev KM. Spectral and redox properties of the GFP synthetic chromophores as a function of pH in buffered media. Chem Commun. 2013;49(71):7788–7790. doi: 10.1039/C3CC44113J. [DOI] [PubMed] [Google Scholar]
- Rivera-Hoyos CM, Morales-Álvarez ED, Poveda-Cuevas SA, Reyes-Guzmán EA, Poutou-Piñales RA, Reyes-Montaño EA, Pedroza-Rodríguez AM, Rodríguez-Vázquez R, Cardozo-Bernal ÁM. Computational analysis and low-scale constitutive expression of laccases synthetic genes GlLCC1 from Ganoderma lucidum and POXA 1B from Pleurotus ostreatus in Pichia pastoris. PLoS One. 2015;10(1):e0116524. doi: 10.1371/journal.pone.0116524.g002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Hoyos CM, Morales-Álvarez ED, Abelló-Esparza J, Buitrago-Pérez DF, Martínez-Aldana N, Salcedo-Reyes JC, Poutou-Piñales RA, Pedroza-Rodríguez AM. Detoxification of pulping black liquor with Pleurotus ostreatus or recombinant Pichia pastoris followed by CuO/TiO2/visible photocatalysis. Sci Rep. 2018;8:3503. doi: 10.1038/s41598-018-21597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarria-Alfonso V, Sánchez-Sierra J, Aguirre-Morales M, Gutiérrez-Rojas I, Moreno-Sarmiento N, Poutou-Piñales RA. Culture media statistical optimization for biomass production of a ligninolytic fungus for future rice straw degradation. Indian J Microbiol. 2013;53(2):199–207. doi: 10.1007/s12088-013-0358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soden DM, O’Callaghan J, Dobson ADW. Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous Pichia pastoris host. Microbiology. 2002;148:4003–4014. doi: 10.1099/00221287-148-12-4003. [DOI] [PubMed] [Google Scholar]
- Taha AA, Shwaish II, Mohammed AH, Haider AJ, Stamatis H. Production of a laccase from Botrytis cinerea (DSMZ 877) and application for textile phenolic dye decolorization. Energy Proc. 2013;36:862–871. doi: 10.1016/j.egypro.2013.07.099. [DOI] [Google Scholar]
- Tinoco R, Pickard MA, Vazquez-Duhalt R. Kinetic differences of purified laccases from six Pleurotus ostreatus strains. Lett Appl Microbiol. 2001;32(5):331–335. doi: 10.1046/j.1472-765X.2001.00913.x. [DOI] [PubMed] [Google Scholar]
- Vogl T, Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnol. 2013;30(4):385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- You L-F, Liu Z-M, Lin J-F, Guo L-Q, Huang X-L, Yang H-X. Molecular cloning of a laccase gene from Ganoderma lucidum and heterologous expression in Pichia pastoris. J Basic Microbiol. 2014;54(S1):S134–S141. doi: 10.1002/jobm.201200808. [DOI] [PubMed] [Google Scholar]
- Zeng X, Cai Y, Liao X, Zeng X, Luo S, Zhang D. Anthraquinone dye assisted the decolorization of azo dyes by a novel Trametes trogii laccase. Process Biochem. 2012;47:160–163. doi: 10.1016/j.procbio.2011.10.019. [DOI] [Google Scholar]
- Zouari-Mechichi H, Mechichi T, Dhouib A, Sayadi S, Martínez AT, Martínez MJ. Laccase purification and characterization from Trametes trogii isolated in Tunisia: decolorization of textile dyes by the purified enzyme. Enzyme Microb Technol. 2006;39:141–148. doi: 10.1016/j.enzmictec.2005.11.027. [DOI] [Google Scholar]