Abstract
Ciliogenesis associated kinase 1 (CILK1) was previously known as intestinal cell kinase (ICK) because it was cloned from that origin. However, CILK1 is now recognized as a widely expressed and highly conserved serine/threonine protein kinase. Mutations in the human CILK1 gene have been associated with ciliopathies, a group of human genetic disorders with defects in the primary cilium. In mice, both Cilk1 knock-out and Cilk1 knock-in mutations have recapitulated human ciliopathies. Thus, CILK1 has a fundamental role in the function of the cilium. Several candidate substrates have been proposed for CILK1 and the challenge is to relate these to the mutant phenotypes. In this review, we summarize what is known about CILK1 functions and targets, and discuss gaps in current knowledge that motivate further experimentation to fully understand the role of CILK1 in organ development in humans.
Keywords: primary cilia, ciliopathy, ciliogenesis, intraflagellar transport, kinesin family member 3A, Hedgehog signalling, epilepsy, stereocilia, autophagy
1. Introduction
The large CMGC (CDK/MAPK/GSK/CLK) branch of the human kinome includes the subgroup called RCK (v-ros cross-hybridizing kinase) (1). These RCK protein kinases consist of ICK/MRK (intestinal cell kinase/MAK-related kinase) (2, 3), MAK (male germ cell-associated kinase) (1, 4), and MOK (MAPK/MAK/MRK-overlapping kinase) (5). In the course of searching for their biological functions and signalling mechanisms, ICK was established as the prototype for this group of kinases that have similarity to both MAPKs (mitogen-activated protein kinases) and CDKs (cyclin-dependent protein kinases). The name of ICK is misleading because, while initially cloned from intestinal cells, ICK is broadly expressed in tissues and cells, and more importantly this name has no implication on the key character or function of the gene. The HUGO gene nomenclature committee recently approved the change of the gene name and symbol from intestinal cell kinase (ICK) to ciliogenesis associated kinase 1 (CILK1), to convey its evolutionarily conserved function in ciliogenesis. In this review and hereafter, ICK is referred to as CILK1.
1.1. Regulation of CILK1 by phosphorylation
RCKs are “unusual remote cousins” of MAPKs in that they share significant homology with MAPKs in the catalytic domain and contain a MAPK-like TXY motif in the activation loop that is dually phosphorylated for kinase activation (6). Unlike classic MAPKs, RCKs are not acutely activated by growth factors through the canonical dual-specificity MEKs (3, 7, 8). Instead, both CILK1 and MAK are activated by phosphorylation of Thr157 in the TDY motif by CDK20 (cyclin-dependent kinase 20), also known as CCRK (cell cycle-related kinase) (8, 9). In contrast to CDK20 as a “yang” positive regulator, PP5 (protein phosphatase 5) was identified from a yeast two-hybrid screen as a “yin” negative regulator that dephosphorylates Thr157 and inactivates CILK1 in response to oxidative stress (8). The tyrosine in the TDY motif is thought to undergo auto-phosphorylation for full activation of CILK1 (8). Both CILK1 and MAK also possess the CDK-like regulatory sites T14Y15 near their N-terminus, but lack the PSTAIRE cyclin binding motif found in most CDKs (6). CILK1 can be partially inactivated by fibroblast growth factor (FGF) signalling-mediated phosphorylation of the conserved Tyr15 (10). Little else is known about the mechanism and the effect of the T14Y15 motif phosphorylation in CILK1 and MAK.
1.2. Intrinsically disordered but functionally critical non-catalytic domain
Although CILK1 is very similar to MAPK in the N-terminal catalytic domain, it diverges significantly in both the length and sequence of the C-terminal non-catalytic domain (CTD). No recognized structural domain has been found in the CTD (285–632 aa) of CILK1 using NCBI Conserved Domain Architecture Retrieval Tool (CDART) (11). The predicted secondary structure for this CTD is entirely random coil (12), suggesting that the CTD is an intrinsically disordered protein (IDP) region (13). Although IDPs lack well-defined three-dimensional structures, they are known to rely on short linear motifs (SLiMs) to interact with folded protein domains and facilitate key protein functions (13, 14). Indeed, we have recently reported that the intrinsically unstructured CTD is critical for CILK1 functions by determining substrate binding and subcellular localization (15). Better understanding of the basis for CTD functions merits further investigation.
1.3. CILK1 substrate phosphorylation consensus
The CILK1 substrate phosphorylation consensus sequence has been determined by a positional scanning peptide array as [R]-[P]-[X]-[S/T]-[P/A/S/T], with the strongest selection for arginine at −3 and proline at −2 positions (8). One distinction from MAPKs and CDKs is that CILK1 does not absolutely require proline at +1 position or a basic residue (K/R) at +3 position in substrates, respectively (16, 17). The phosphorylation consensus of CILK1 is similar to that (R-P-X-S/T-P) of DYRKs (dual-specificity tyrosine phosphorylation-regulated kinases) (18), except in the stringency for requirement of proline at +1 position. It is worth pointing out that the current CILK1 substrate consensus sequence was determined using only the N-terminal kinase domain. Since the CTD of CILK1 has been subsequently shown as indispensable for substrate recognition and phosphorylation (15), we speculate that CILK1 substrate specificity may be influenced by the MAPK-like “docking” interactions through its CTD. This would probably not affect interactions at the CILK1 active site that contribute to specificity with peptide substrates. The CILK1 substrate consensus sequence may be of some use in identification of physiological substrates.
1.4. Discovery of CILK1 functions and disease relevance
CILK1 relevance to human disease remained obscure for more than a decade after its discovery in the late 1990’s. The major breakthrough came in 2009, first from the report of human ECO (endocrine-cerebro-osteodysplasia) syndrome, where CILK1 was implicated in development of the human central nervous, skeletal, and endocrine systems (19). Later, a knockdown study in human colon cancer cells showed that suppression of CILK1 reduced cell proliferation and G1 cell cycle progression (20). The essential role of CILK1 in development was further substantiated by both Cilk1 knock-out (KO) (21, 22) and Cilk1 mutation knock-in (KI) mouse models (23, 24). Mechanistic analysis of phenotypes in CILK1-deficient mouse models revealed a role for CILK1 in the primary cilium, as described below.
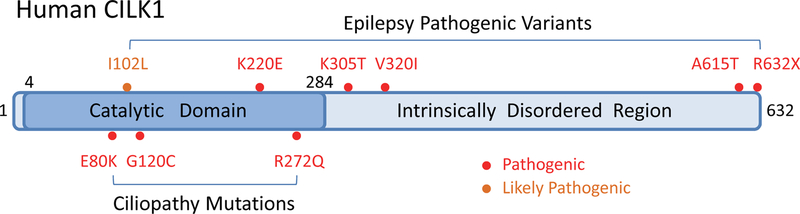
Most mammalian cells have a primary cilium protruding from their surface that senses environmental cues and transduces extracellular signals to regulate intracellular processes (25). Human diseases called ciliopathies are attributed to mutations in the proteins and enzymes supporting the function of this organelle (26). Ciliopathies manifest as a constellation of clinical features in nearly every major body organ, highlighting the essential role of the primary cilium in development and homeostasis. To date, there are still major gaps in our knowledge about how primary cilia are formed, maintained and function in signal transduction, and the mechanisms associated with ciliary dysfunction are yet to be fully elucidated. Homologues of the RCK family in Caenorhabditis elegans (DYF5, dye-filling defective 5), Chalmydomonas reinhardtii (LF4, long flagella protein 4), Tetrahymena thermophila (LF4A, long flagella protein 4A), and Leishmania mexicana (LmxMPK9, Leishmania mexicana MAP kinase 9) are negative regulators of cilia and flagella length (27–30), because mutations in these genes result in abnormal extension of the cilium/flagellum. So far, three inactivating mutations in human CILK1 gene have been identified in human ciliopathies (19, 31, 32), and pathogenic variants were linked to Juvenile Myoclonic Epilepsy (33) (Fig. 1). Candidate substrates for CILK1 emerged from these revelations of its biological functions (Fig. 2).
Figure 1:
Human CILK1 protein domain structure and the pathogenic variants identified in human ciliopathies and epilepsy. Human CILK1 has two basic structural domains: the N-terminal catalytic domain (4–284 aa) and the C-terminal non-catalytic domain (285–632 aa), which is an intrinsically disordered protein region with critical functions. Three pathogenic mutations (E80K, G120C, and R272Q) in the catalytic domain are associated with ciliopathies. Six pathogenic or likely pathogenic variants located in both the catalytic (I102L and K220E) and the non-catalytic (K305T, V320I, A615T, and R632) domains are associated with epilepsy.
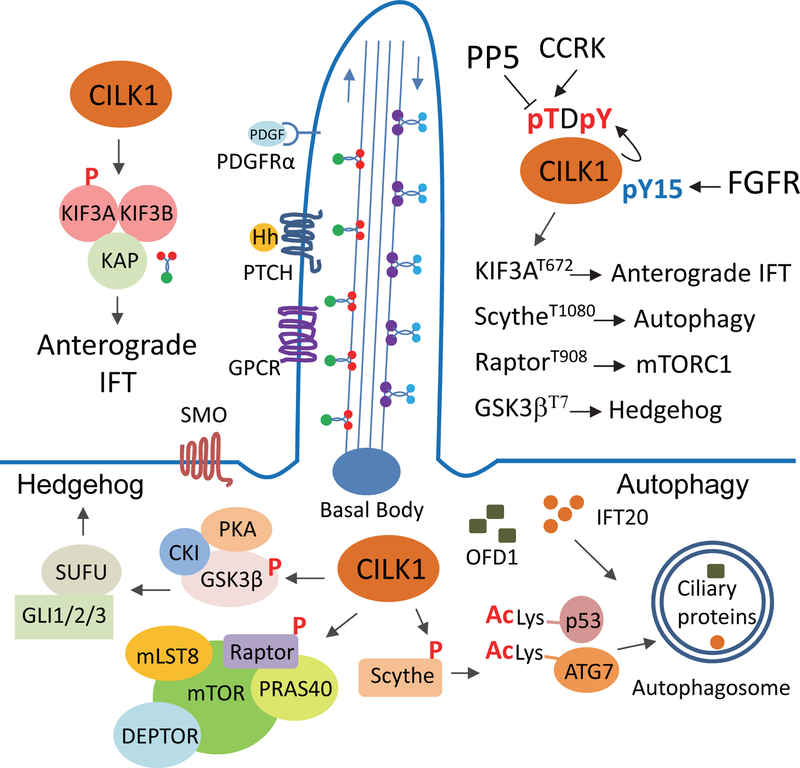
Figure 2:
A working model for CILK1 signalling. The activity of CILK1 can be regulated by phosphorylation of the TDY motif and the conserved N-terminal regulatory site Tyr15. CILK1 is localized to the primary cilium, which provides a unique cellular environment with higher concentration of second messengers such as calcium and cyclic AMP, and many signalling outputs such as GPCR, Hedgehog, PDGFRα, TGFβ, and WNT. How CILK1 is up and down regulated in this unique signalling environment is largely unknown. Four candidate substrates (KIF3A, Scythe, Raptor, and GSK3β) for CILK1 have been identified that can potentially mediate CILK1 effect on cilia structure, signalling, and function. (1) KIF3A is in the kinesin II motor complex that mediates anterograde IFT (intraflagellar transport), which is essential for cilia formation and maintenance. (2) Scythe is a co-chaperone protein that has a critical role in autophagy by modulating the acetylation of p53 and ATG7. Autophagy regulates ciliogenesis by controlling the levels of ciliary proteins IFT20 and OFD1 through autophagic degradation. (3) Raptor is a key regulatory protein for mTOR in mTORC1, which can regulate cilia length or ciliogenesis through an unknown mechanism. (4) GSK3β is a positive regulator of ciliogenesis and ciliary Hedgehog signalling pathway. How these substrates mediate CILK1 effects on cilia morphology and function is still poorly defined.
In this review, we will highlight recent advances in our knowledge about CILK1 functions in multi-organ systems and putative substrates in various signalling pathways, as well as discuss perspectives on future studies that may provide mechanistic insights into the role of CILK1 in human development and disease.
2. CILK1 functions in multi-organ systems
2.1. CILK1 in the intestinal epithelium
CILK1 is broadly expressed in tissues and highly abundant in the intestine (2, 3, 34, 35). In the mouse small intestine, CILK1 mRNA is specifically enriched in the proliferative compartment, the intestinal crypts, where stem/progenitor cells and transient amplifying epithelial cells reside (3, 34). The restricted localization in the crypt raised an early speculation that CILK1 may function in the regulation of intestinal epithelial cell proliferation and intestinal stem or progenitor cell activities (3). Using lentiviral short hairpin RNA (shRNA) interference, we tested this hypothesis by knockdown of CILK1 expression in cultured colorectal carcinoma and intestinal epithelial cell lines. Indeed, the knockdown of CILK1 significantly reduced the proliferation of these cell lines. The results provided the first in vitro evidence that CILK1 promotes cell proliferation and cell cycle progression (20). To further address the functions of CILK1 in vivo, we generated an intestine specific KO mouse model. It was puzzling that there was no obvious phenotype in CILK1-deficient intestinal epithelium during normal development and homeostatic maintenance (unpublished data). But, in a radiation injury mouse model, we observed up-regulation of CILK1 expression in regenerating intestinal epithelium that coincides with elevated expression of intestinal stem cell markers. The CILK1 deficiency stunted villi length in the regenerating epithelium, suggesting a supportive role of CILK1 in regenerative growth of intestinal epithelium after radiation injury (unpublished data). CILK1 expression in the mouse intestinal epithelium is also significantly elevated during the intestinal response to protein malnutrition (36), implicating that CILK1 signaling may contribute to compensatory mucosal growth under nutritional stress. Further studies are required to find out how CILK1 supports “catch-up” growth of intestinal epithelium in response to nutrient deficiency and impacts the pathophysiological outcomes induced by the vicious cycle of malnutrition and infection.
2.2. CILK1 in brain development and epilepsy
Homozygous CILK1 R272Q was identified as the causative mutation for human ECO syndrome (19). In a SwissModel structure of CILK1 catalytic domain, the conserved R272 in the PKKRP motif is in L15 of subdomain XI and forms ion pairs with conserved E169 and W184; these interactions are predicted to stabilize the interface of αh-L15-αi and help to create an active conformation for CILK1 (7). Mutation of R272 to either alanine or glutamine inactivates CILK1 as a kinase (7, 19). In ECO syndrome, inactivation of CILK1 caused extensive neuropathological defects in embryonic development, including holoprosencephaly, corpus callosum hypoplasia, and cerebral cortex malformation (19). In a CILK1-deficient mouse model, embryos displayed similar abnormalities, such as enlarged cerebral cortexes and hydrocephalus (21). These embryonic phenotypes in the developing brain are associated with reduced number and reduced activity of neuronal progenitor cells that show aberrant ciliogenesis and sonic hedgehog signaling (21). Furthermore, conditional deletion of CILK1 resulted in growth defects in postnatal brains in the cerebellum and hippocampus dentate gyrus, which are likely caused by compromised postnatal neurogenesis (21). Surprisingly, loss of CILK1 function in brain affects ciliogenesis only in neural progenitor cells, not in mature neurons, suggesting a neuronal cell type-specific effect of CILK1 in ciliogenesis.
Heterozygous variants in human CILK1 gene were associated with juvenile myoclonic epilepsy in 7% of the patients in a recent study (33). When plasmids encoding the pathogenic CILK1 variants (K220E, K305T, A615T, and R632X in epilepsy and R272Q in ECO syndrome) were electroporated into the mouse neocortex they impaired radial migration of neural progenitor cells such that fewer neurons reached the cortical plate. Thus, introduction of the inactive kinase inhibited cell migration, possibly through a dominant negative action. Cortical progenitor cells expressing pathogenic CILK1 variants showed a lower index of mitosis and cell-cycle exit but higher index of apoptosis (except A615T) when compared with cells expressing wild-type CILK1 (33). Heterozygous Cilk1 null mice experienced tonic-clonic convulsions more frequently than wild-type mice during 2% isoflurane induced light sleep (33). However, there is striking variation in epilepsy phenotypes both within and among families, which strongly suggests that CILK1 is pleiotropic, and the phenotypic effect of CILK1 variants in epilepsy depends on the interaction with modifier genes in epistatic loci (33).
In the central nervous system, the neural primary cilium is emerging as a non-synaptic signaling mechanism through which environmental signals shape and refine interneuronal connectivity (37, 38). An important question yet to be addressed is whether the mechanism by which heterozygous CILK1 variants trigger epilepsy is related to its highly conserved role in the primary cilium. Surprisingly, epilepsy phenotype can be induced in heterozygous KO mice whose brain structures and neural cilia morphology appear to be indistinguishable from that of wild-type mice (21, 33). If one functional allele can produce enough active CILK1 protein to fulfill its role in the formation and maintenance of primary cilia, as suggested by both Cilk1 KO and Cilk1 R272Q KI mouse models (21–23), the epilepsy phenotype of CILK1 variants is unlikely the direct consequence of neural ciliary defects. However, since CILK1 is pleiotropic in epilepsy (33), the genetic background of mouse models may play a critical role in determining the epilepsy phenotype of variants, which should be taken into consideration when designing further studies to elucidate the mechanism underlying the epileptic effect of CILK1 variants.
2.3. CILK1 in lung morphogenesis and primitive alveoli formation
ECO syndrome is an autosomal recessive, neonatal-lethal developmental disorder (19). Both homozygous Cilk1 KO and Cilk1 R272Q KI mice died around birth due to respiratory distress (21, 23). Autopsy revealed obvious lung hypoplasia, among other ECO clinical features. R272Q homozygous lungs had the normal shape, number and arrangement of lobes but were hypoplastic and severely deficient in airspace (23). These observations support the idea that CILK1 is required for cell proliferation and tissue development in vivo. Morphometric analyses further indicated a significant reduction in lung saccular area and a marked increase in interstitial mesenchymal thickness, suggesting that the formation of primitive alveoli was severely disrupted by CILK1 dysfunction (23).
During branching morphogenesis at the pseudoglandular stage, the number of lung buds (branching points) per area was reduced by about 50% in CILK1 mutant lungs as compared with normal littermate controls, indicating that loss of CILK1 function severely compromised lung branching morphogenesis (23). The bronchial tree develops through extensive proliferation of distal epithelium and surrounding mesenchyme. In CILK1 mutant lungs, reduced proliferation of mesenchymal but not epithelial cells impaired lung branching (23).
During alveolar development at the saccular stage (E16.5–18.5), two major morphogenetic events occur and are required for the formation of air saccules: differentiation of the two main specialized epithelial cell types of the future alveolus, and the thinning and remodeling of lung interstitium. In the developing lung, CILK1 deficiency did not alter the morphology and functions of alveolar type 1 and type 2 epithelial cells but caused hypercellularity in the interstitium (23). These results suggest that CILK1 function is dispensable for differentiation of alveolar epithelial cells but is essential for the thinning and remodeling of interstitial mesenchyme. Further analysis indicated that a deficiency in apoptosis is unlikely a major cause of the excessive cellularity in the interstitium; instead abnormal mesenchymal differentiation in the interstitium may result in the hypercellularity that blocks the thinning of interstitium and the expansion of airspace (23). Furthermore, CILK1 dysfunction induced elongation of primary cilia and perturbation of ciliary Hedgehog signaling during lung sacculation (23).
In the developing lung, loss of CILK1 function perturbed not only ciliary structure and Hedgehog signaling but also autophagy, a highly conserved intracellular process in the maintenance of cell homeostasis (23). Autophagy is intrinsically linked to ciliogenesis (39–41) and is implicated for a critical role of facilitating the thinning of the alveolar septa that is necessary for effective gas exchange during the transition to air breathing at birth (42). The mechanism by which CILK1 regulates lung morphogenesis and sacculation through ciliary signaling and autophagy is still unclear and awaits future investigation.
2.4. CILK1 in skeletal development
Homozygosity for mutations in CILK1 including c.1305G-A (R272Q), c.358G-T (G120C), and c.238G-A (E80K), produced ECO and ECO-like syndromes that present profound and wide-ranging skeletal abnormalities such as polydactyly, short ribs, bowed limbs, and shortened and hypoplastic long bones (19, 31, 43). Cilk1 null and R272Q mutant mouse models showed marked disruption of growth plate architecture, with a shortened proliferative zone and poorly formed hypertrophic zone, indicating compromised proliferation and faulty differentiation of chondrocytes (24, 43). Cilk1 R272Q KI mice showed a deformed spine with defective intervertebral disc (24). Analysis of the spinal column and long bones of R272Q mutants revealed reduced proliferation zones and reduced prevalence of type X collagen-expressing hypertrophic chondrocytes (24). Because analysis of apoptotic cells showed no significant difference between normal and CILK1 mutant growth plates, CILK1 is implicated in the regulation of chondrocyte proliferation and maturation. Although the mechanisms underlying these skeletal phenotypes are not fully understood, studies from patient-derived fibroblasts (E80K) and CILK1 null chondrocytes have suggested a link between Hedgehog and ERK signaling pathways and ciliary dysfunction (43).
2.5. CILK1 in inner ear development and auditory function
The cochlea in the inner ear is the hearing organ. The planar cell polarity (PCP) signalling pathway plays a critical role in the establishment of cellular asymmetry within the plane of a sheet of inner ear sensory hair cells. PCP in hair cells in the cochlea refers to the asymmetric structure consisting of staircase-like stereocilia bundles and one kinocilium on the apical surface of the cell body. CILK1 deletion caused PCP defects in the cochlea, including misorientation of stereocilia and aberrant position of the kinocilium, leading to auditory dysfunction (44). Furthermore, disruption of CILK1 caused accumulation of ciliary protein Ift88 at the tip of cilia and cilia elongation in the cochlea, suggesting IFT dysfunction is closely related to cilia elongation in CILK1-deficient cochlea (44). This result is consistent with prior observations in cultured cells that CILK1 knockdown induces cilia elongation and increases the anterograde IFT velocity (45). Overall, these results demonstrate that CILK1 is essential for auditory function by regulating PCP formation in inner ear hair cells. The mechanism by which CILK1 controls cilia morphology and function and the establishment of PCP in the cochlea remains to be elucidated.
2.6. CILK1 in cardiac development and hypertrophy
No major cardiac phenotype in embryonic development was reported in ECO and ECO-like syndromes, except that an affected patient bearing the G120C mutation in CILK1 presented ventricular septal defect (VSD), which is a common heart defect at birth (31). Heart phenotype was either not detected or understudied in both KO and mutant CILK1 KI mouse models. Abe and colleagues isolated ICK/MRK from a rat heart cDNA library, and they reported CILK1 expression in myocardium of embryonic and adult hearts (2). Interestingly, they observed an increase in the intensity of CILK1 staining and the number of CILK1-positive cardiomyocytes in hypertrophic hearts under experimentally induced stenosis of the abdominal aorta (2). This result implicates that CILK1 expression is inducible in the heart by external stress such as pressure overload. This phenomenon is reminiscent of what we observed in the intestinal epithelium where CILK1 expression can be induced under nutritional stress (36) or radiation injury (unpublished data). Conditional KO mouse models will be needed to interrogate the role of CILK1 in the heart under both normal and pathophysiological conditions.
2.7. CILK1 in kidney and adrenal glands
In ECO patients with the homozygous R272Q mutation, kidney cortex and medulla showed cystically dilated tubules to a variable extent (19). An affected fetus with homozygosity for the G102C mutation showed large and hyperechogenic kidneys (31). Hyperechogenicity is a non-specific finding but a significant one in that it suggests the presence of renal abnormalities. The role of cilia in the pathogenesis of cystic kidney disease has been well established but the exact mechanism by which mutant genes and cilia cause cysts remain poorly defined (46, 47). Since CILK1 has a highly conserved role in regulating cilia function, it will be an intriguing question whether CILK1 dysfunction contributes to the pathogenesis of cyst formation in kidney disease.
ECO and ECO-like syndromes presented remarkable defects in endocrine glands, including absent or hypoplastic adrenal glands and pituitary gland (19, 31). Primary cilia are signalling hubs for Hedgehog and Wnt signalling pathways, both of which play a major role in the development and regeneration of adrenal glands (48, 49). It is thus conceivable that CILK1 may be involved in the development of adrenal glands through regulating various ciliary signalling pathways.
3. CILK1 targets in signalling pathways
3.1. Targeting KIF3A in anterograde IFT
In primary cilia, intraflagellar transport (IFT) is a bi-directional system that consists of kinesin-mediated anterograde and dynein-mediated retrograde movement of cargos. IFT, especially anterograde, is critical for proper cilium formation and maintenance of the structure (50). Knockdown of CILK1 accelerates anterograde IFT in the primary cilium, which may provide a possible explanation for CILK1 deficiency-induced cilium elongation (45). Anterograde IFT is mediated by the heterotrimeric kinesin-2 motor complex (KIF3A/KIF3B/KAP3). KIF3A and KIF3B (kinesin family member 3A and 3B) form a heterodimer that functions as a microtubule-based fast anterograde translocator. Human KIF3A-Thr672 is located within a CILK1 consensus sequence RPRTS that is highly conserved among metazoans (15) and is phosphorylated by CILK1 in vitro and in vivo (15, 21).This CILK1 site is in the C-terminal cargo-binding region of human KIF3A that contains multiple phosphorylation sites, including PKA site Ser687 and CaMKII sites Thr692 and Ser696 (51). Phosphorylation of S687/T692/S696 enhances the cargo-binding and trafficking activities of KIF3A (51). It remains to be determined whether phosphorylation of KIF3A-Thr672 by CILK1 is another mechanism for regulation of KIF3A activity in IFT. Mouse Kif3a mutant protein containing mutation of all 8 phospho-sites at the C-terminal could not rescue the ciliogenesis defects in Kif3a-knockdown cells (21), consistent with the notion that phosphorylation of KIF3A C-terminal cargo-binding domain is critical for IFT and ciliogenesis. Interestingly, mouse Kif3a-T674A mutant protein exhibited a stronger capacity than Kif3a-WT protein to rescue cilia formation in Kif3a-knockdown cells (21). This intriguing observation supports the hypothesis that KIF3A-pThr672 acts as a downstream effector through which CILK1 negatively regulates ciliogenesis. Further studies are required to address how CILK1 phosphorylation of KIF3A-Thr672 affects IFT and ciliogenesis, and whether deregulation of this phosphorylation event is required for the ciliopathy phenotypes caused by CILK1 dysfunction.
3.2. Targeting Raptor in mTORC1 signalling
The serine-threonine protein kinase mTOR (mammalian target of rapamycin) is the core catalytic component of two structurally and functionally distinct protein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which collectively integrate nutrient, hormonal, and energy signal inputs to control cell growth, proliferation and survival (52–54). Suppression of CILK1 expression in intestinal epithelial cells markedly impaired cell proliferation and G1 cell cycle progression (20). Furthermore, CILK1 deficiency led to a significant decrease in mTORC1 activity, concomitant with reduced expression of specific mTORC1 downstream targets cyclinD1 and c-Myc (20). These results suggest that CILK1 may target the mTORC1 signalling pathway to regulate cell proliferation and cell cycle progression.
Raptor, regulatory associated protein of mTOR, plays an important role as a scaffolding protein to recruit substrates to mTOR (55) and also positively regulate mTOR activity by directly interacting with Rag family GTPases to induce mTORC1 re-localization to the lysosomes (56–58). Raptor is a substrate of multiple protein kinases and the complex phosphorylation status of Raptor is tightly associated with the activity of mTORC1. CILK1 physically interacts with mTORC1 and phosphorylates Raptor at Thr908 in cells (59). Although the phospho-deficient mutant, Raptor T908A, did not affect mTORC1 assembly, it did markedly impair the mTORC1 activation by insulin or by the small GTP-binding protein RheB under nutrient starvation (59). These findings point to phosphorylation of Raptor by CILK1 as a requirement for activation of mTORC1.
Cilia function and mTOR activation are linked. Primary cilia regulate mTORC1 activity through the ciliary Lkb1-AMPK-Rheb pathway (60). Cilium formation and length are regulated by the mTOR pathway. Inhibition of mTORC1 by rapamycin resulted in shorter cilia in zebrafish and C. reinhardtii embryos (61). In mammalian cells, however, inactivating mTORC1 by rapamycin or Raptor knockdown promoted ciliogenesis (62). Rapamycin treatment suppressed the effect of CILK1 depletion on cilium length and IFT (45). These results suggest that CILK1 could regulate ciliogenesis and IFT through phosphorylation of Raptor and activation of mTORC1.
3.3. Targeting Scythe in autophagy
Increasing evidence has shown that ciliogenesis and autophagy are intricately linked and reciprocally regulated (39, 40, 63). Autophagy regulates ciliogenesis by controlling the levels of ciliary proteins that are either essential for ciliogenesis, such as IFT20 (intraflagellar transport 20) (40), or negative regulators of ciliogenesis, such as the centriolar satellite protein OFD1 (oral-facial-digital syndrome 1) (39). Under basal conditions, basal autophagy prevents ciliary growth through degradation of IFT proteins such as IFT20. Early upon nutrient removal, induction of autophagy triggers degradation of the endogenous inhibitor of ciliogenesis OFD1, thus promoting cilium formation. This switch in autophagy cargo upon starvation promotes IFT and ciliogenesis. In ECO ciliopathy mouse lungs, elongation of primary cilia is closely correlated with increased autophagy (23). Furthermore, CILK1 mutant cells displayed a faster autophagy flux and quicker degradative process upon induction of autophagy by starvation. These observations raise the question whether CILK1 loss-of-function increased autophagy for the elongation of primary cilia in ECO ciliopathy.
Scythe was identified as a CILK1 interacting protein and a candidate substrate from a yeast two-hybrid screen (8). In mammalian cells, CILK1 directly interacts with Scythe and specifically phosphorylates Scythe at Thr-1080 (8). Scythe is a co-chaperon protein involved in protein quality control (64) and is essential for basal and starvation-induced autophagy (65). Whether CILK1 controls ciliogenesis in part through regulating autophagy via phosphorylation of Scythe awaits further investigation.
3.4. Targeting GSK3β in Hedgehog signalling
Glycogen synthase kinase 3β (GSK3β) is a positive regulator of ciliogenesis. In green alga Chalmydomonas reinhardtii, flagellar assembly requires GSK3β activity. Adding lithium chloride, an inhibitor of GSK3β, to cells undergoing flagellar assembly results in flagella less than half the normal length (66, 67). In mammalian cells, the inhibitory Ser-9 phosphorylation state of GSK3β correlates with the reduced frequency of primary cilia formation in renal cysts (68). GSK3β promotes assembly of primary cilia after mitosis through the GSK3β-Dzip1-Rab8 signalling cascade (69). In addition, GSK3β is a positive regulator of Hedgehog (Hh) signalling. Down-regulation of GSK3β expression by RNA interference in Hh-responsive cells attenuated Hedgehog signalling (70). GSK3β binds Sufu, a negative regulator of Hh signalling. GSK3β phosphorylation of Sufu at T407 decreases its ability to bind Gli and suppress Gli-mediated transcription (70).
GSK3β Thr7 is a highly conserved CILK1 site. Inhibitory Ser9 phosphorylation of GSK3β by AKT is an important mechanism that negatively regulates GSK3β activity upon insulin stimulation (71). Thr7 and Thr8 are located in the AKT substrate consensus sequence on GSK3β and are essential for insulin-stimulated Ser9 phosphorylation in vivo and GSK3β inactivation (72). CILK1 phosphorylates GSK3β Thr7 in vitro and in vivo. Thr7 phosphorylation enhances Ser9 phosphorylation and promotes phospho-Ser9-dependent autoinhibition of GSK3β (72). These results raise the hypothesis that CILK1 negatively regulates GSK3β activity through inhibitory Thr7 phosphorylation to suppress cilia formation and Hedgehog signalling.
4. Conclusions and Perspectives
CILK1 has an essential role in human development. Both CILK1 and its upstream activating kinase CDK20/CCRK are highly conserved regulators of ciliogenesis and cilia length. CILK1 KO and the ECO mutation (R272Q) KI mouse models revealed a requirement for CILK1 in multiple organ systems. Alteration in cilia number, cilia length, ciliary signalling and autophagy presumably accounts for the pathological phenotypes caused by CILK1 dysfunction. Several candidate substrates for CILK1 have been identified, such as KIF3A, Raptor and Scythe but how they mediate CILK1 effects on cilia morphology and function is still poorly defined. How the phosphorylation of these substrates relates to mutant CILK1 ciliopathies and epilepsy is completely unknown. The link of CILK1 mutations to human ciliopathies and epilepsy has brought CILK1 under the spotlight on the centre stage of human health and disease. Although remarkable progress has been made within the past two decades, many significant questions remain to be addressed in future research.
4.1. Does CILK1 have cellular functions separate from the primary cilium?
If so, what are the substrates and do these functions contribute to the pathogenesis of ciliopathies and epilepsy? Studies using either GFP-tagged CILK1 or antibody staining of endogenous CILK1 have shown that CILK1 is localized in the primary cilium. We and others have focussed on CILK1 regulation of KIF3A and ciliary anterograde transport. But CILK1 is observed in other intracellular compartments, including nucleus and cytoplasm. There are unique sequence requirements for subcellular targeting of CILK1. For example, we have shown that mutation of the conserved R272 within the PKKRP motif in the catalytic domain is sufficient to exclude CILK1 from the nucleus (7). Furthermore, we have shown the non-catalytic CTD of CILK1 is required for ciliary localization (15). It has been shown that CILK1 with different ciliopathy mutations exhibits distinct patterns of subcellular localization (31, 43), which predicts that these ciliopathy mutations may cause their disease phenotypes through different molecular mechanisms. It is likely that CILK1 interacts with distinct pools of substrates and signalling proteins in its various locations within cells. Biochemical isolation and characterization of these mutation-specific CILK1 complexes may expose other, new targets for the diversity of CILK1 signalling.
4.2. What are the environmental stimuli that activate or inactivate CILK1?
Primary cilium offers a unique signalling environment. The primary cilium contains many cell surface receptors, including GPCR, RTK, Hedgehog, PDGFRα, TGFβ, and WNT. The cilium provides highly efficient signal processing, with a remarkably large ratio of sensing surface to internal volume. This restricted intracellular environment favours generation of high concentrations of second messengers such as calcium and cyclic AMP and protein-protein interactions. How CILK1 is regulated in this unique signalling environment is largely unknown. What extracellular signals impact CILK1 activity? Is CILK1 intracellular localization regulated? We need to move beyond establishing a requirement for CILK1 in ciliary-dependent functions and define with more precision what CILK1 does, and how. The answers to these questions will require tools to track CILK1 activity in time and space under different cellular conditions. Plus, we need to understand how ciliopathy-associated mutations affect CILK1 activation and inactivation, as well as association with and phosphorylation of different substrates, and the effects on these targets.
4.3. Do CILK1, MAK, and MOK have similar or distinct functions and mechanisms of action?
RCK family kinases and their homologs, including C. elegans DYF5, Chalmydomonas LF4, Tetrahymena LF4A, and Leishmania LmxMPK9, are negative regulators of cilium and flagellum length (27–30). Based on the sequence similarity in their serine/threonine kinase domain, LF4 is closer to MOK, and DYF5 is closer to CILK1 and MAK. Jansen and colleagues have shown that while both CILK1 and MOK are negative regulators of cilia length, they have distinct effects on intraflagellar transport (45). Loss- or gain-of-function of CILK1 affected IFT velocity, which is similar to the phenotype of its C. elegans homolog DYF5. In contrast, MOK overexpression or knockdown had no effect on IFT speed. MOK homologs LF4 and LF4A have inconsistent IFT phenotypes (28, 30). These results support independent functions for CILK1 and MOK. The effects of CILK1 and MOK on cilium length both require mTORC1 signalling, but identification of their individual downstream targets and effectors require further investigation.
CILK1 is most closely related to MAK in sequence and domain structure. Pathogenic MAK variants are associated with retinitis pigmentosa, a retinal ciliopathy (73, 74). MAK plays a key role as a negative regulator of cilium length in retinal photoreceptor survival (75). Furukawa and colleagues proposed that MAK regulates microtubule stability and controls cilium length through phosphorylation of microtubule-associated proteins, such as retinitis pigmentosa 1 (RP1) (75). A recent study in C. elegans further indicated that the CDK20-MAK kinase cascade controls cilia shape and structure by regulating axonemal microtubule dynamics (76). Indeed, CILK1, MAK and their closest homolog DYF5 affect the motility of kinesin motors and movement of IFT particles, as well as docking and undocking of IFT cargos in primary cilia (27, 45, 77). Although CILK1 and MAK are almost identical in the sequence of the N-terminal catalytic domain, they differ significantly in the sequence of the C-terminal non-catalytic domain (CTD). We recently demonstrated that the intrinsically disordered CTD of CILK1 is required for its ciliary targeting, substrate binding and phosphorylation. This suggests that the divergent CTDs of CILK1 and MAK may determine their individual functions by specifying substrate recognition and subcellular compartmentalization. A key question that remains to be addressed in our future study is whether the ciliary functions of CILK1 and MAK are distinct or redundant, and whether these kinases utilize different targets and pathways to regulate ciliary length and ciliary transport machinery.
Table 1:
CILK1 dysfunction phenotypes in multi-organ systems
| Organ | Major Clinical Abnormalities | Developmental and Cellular Defects | Ref |
|---|---|---|---|
| Brain | holoprosencephaly, hydrocephalus, corpus callosum hypoplasia, abnormal cerebral cortex | reduced number and reduced activity of neuronal progenitor cells, impaired radial migration of neural progenitor cells, impaired postnatal neurogenesis | (19) (21) (33) |
| Lung | hypoplasia respiratory distress airspace deficiency | abnormal primitive alveoli with reduced saccular area, reduced branching and mesenchymal proliferation, hypercellular interstitium and abnormal mesenchymal differentiation | (19) (21) (23) |
| Skeletal | polydactyly, short ribs, bowed limbs, shortened and hypoplastic long bones, deformed spine with defective intervertebral disc | disruption of growth plate architecture, shortened proliferative zone and poorly formed hypertrophic zone, reduced mineralization and reduced number of type X collagen-expressing hypertrophic chondrocytes, impaired chondrocytes differentiation and maturation | (19) (24) (31) (43) |
| Cochlea Inner Ear | compromised auditory function | disrupted planar cell polarity in hair cells, misorientation of stereocilia, and aberrant position of the kinocilium | (44) |
| Heart | ventricular septal defect (only one case) | no description, understudied | (31) |
| Kidney | large and hyperechogenic kidneys | cystically dilated tubules to a variable extent in cortex and medulla | (19) (31) |
| Endocrine Glands | absent or hypoplastic adrenal glands and pituitary gland | no description, understudied | (19) |
Acknowledgments
This work was supported by the National Institute of Health grant GM127690 to Z.F. and the F. Palmer Weber endowed professorship to D.L.B. We thank our colleagues Thomas Sturgill, Michael Weber, Thurl Harris, Li Jin, and Mark Beenhakker for constructive comments and valuable advice.
Abbreviations:
- CILK1
ciliogenesis associated kinase 1
- ICK
intestinal cell kinase
- RCK
v-ros cross-hybridizing kinase
- MAK
male germ cell-associated kinase
- MOK
MAPK/MAK/MRK-overlapping kinase
- MRK
MAK-related kinase
- LF4
long flagella protein 4
- DYF5
dye-filling defective 5
- LmxMPK9
Leishmania mexicana MAP kinase 9
- MAPK
mitogen-activated protein kinase
- CDK
cyclin-dependent kinase
- CDK20
cyclin-dependent kinase 20
- CCRK
cell cycle-related kinase
- PP5
protein phosphatase 5
- FGF
fibroblast growth factor
- IDP
intrinsically disordered protein
- SLiMs
short linear motifs
- DYRKs
dual-specificity tyrosine phosphorylation-regulated kinases
- ECO
endocrine-cerebro-osteodysplasia
- KO
knock-out
- KI
knock-in
- PCP
planar cell polarity
- IFT
intraflagellar transport
- KIF3A
kinesin family member 3A
- mTOR
mammalian target of rapamycin
- Raptor
regulatory associated protein of mTOR
- IFT20
intraflagellar transport 20
- OFD1
oral-facial-digital syndrome 1
- RP1
retinitis pigmentosa 1
References
- 1.Bladt F, and Birchmeier C (1993) Characterization and expression analysis of the murine rck gene: a protein kinase with a potential function in sensory cells. Differentiation 53, 115–122 [DOI] [PubMed] [Google Scholar]
- 2.Abe S, Yagi T, Ishiyama S, Hiroe M, Marumo F, and Ikawa Y (1995) Molecular cloning of a novel serine/threonine kinase, MRK, possibly involved in cardiac development. Oncogene 11, 2187–2195 [PubMed] [Google Scholar]
- 3.Togawa K, Yan YX, Inomoto T, Slaugenhaupt S, and Rustgi AK (2000) Intestinal cell kinase (ICK) localizes to the crypt region and requires a dual phosphorylation site found in map kinases. J Cell Physiol 183, 129–139 [DOI] [PubMed] [Google Scholar]
- 4.Matsushime H, Jinno A, Takagi N, and Shibuya M (1990) A novel mammalian protein kinase gene (mak) is highly expressed in testicular germ cells at and after meiosis. Mol Cell Biol 10, 2261–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyata Y, Akashi M, and Nishida E (1999) Molecular cloning and characterization of a novel member of the MAP kinase superfamily. Genes Cells 4, 299–309 [DOI] [PubMed] [Google Scholar]
- 6.Miyata Y, and Nishida E (1999) Distantly related cousins of MAP kinase: biochemical properties and possible physiological functions. Biochem Biophys Res Commun 266, 291–295 [DOI] [PubMed] [Google Scholar]
- 7.Fu Z, Schroeder MJ, Shabanowitz J, Kaldis P, Togawa K, Rustgi AK, Hunt DF, and Sturgill TW (2005) Activation of a nuclear Cdc2-related kinase within a mitogen-activated protein kinase-like TDY motif by autophosphorylation and cyclin-dependent protein kinase-activating kinase. Mol Cell Biol 25, 6047–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Z, Larson KA, Chitta RK, Parker SA, Turk BE, Lawrence MW, Kaldis P, Galaktionov K, Cohn SM, Shabanowitz J, Hunt DF, and Sturgill TW (2006) Identification of yin-yang regulators and a phosphorylation consensus for male germ cell-associated kinase (MAK)-related kinase. Mol Cell Biol 26, 8639–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang LY, and Kung HJ (2012) Male germ cell-associated kinase is overexpressed in prostate cancer cells and causes mitotic defects via deregulation of APC/CCDH1. Oncogene 31, 2907–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunova Bosakova M, Nita A, Gregor T, Varecha M, Gudernova I, Fafilek B, Barta T, Basheer N, Abraham SP, Balek L, Tomanova M, Fialova Kucerova J, Bosak J, Potesil D, Zieba J, Song J, Konik P, Park S, Duran I, Zdrahal Z, Smajs D, Jansen G, Fu Z, Ko HW, Hampl A, Trantirek L, Krakow D, and Krejci P (2019) Fibroblast growth factor receptor influences primary cilium length through an interaction with intestinal cell kinase. Proc Natl Acad Sci U S A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geer LY, Domrachev M, Lipman DJ, and Bryant SH (2002) CDART: Protein homology by domain architecture. Genome Research 12, 1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollastri G, and McLysaght A (2005) Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics 21, 1719–1720 [DOI] [PubMed] [Google Scholar]
- 13.Oldfield CJ, and Dunker AK (2014) Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 83, 553–584 [DOI] [PubMed] [Google Scholar]
- 14.Tompa P (2012) Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci 37, 509–516 [DOI] [PubMed] [Google Scholar]
- 15.Oh YS, Wang EJ, Gailey CD, Brautigan DL, Allen BL, and Fu Z (2019) Ciliopathy-Associated Protein Kinase ICK Requires Its Non-Catalytic Carboxyl-Terminal Domain for Regulation of Ciliogenesis. Cells 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, and Cantley LC (1994) Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol 4, 973–982 [DOI] [PubMed] [Google Scholar]
- 17.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, and Turk BE (2004) A rapid method for determining protein kinase phosphorylation specificity. Nat Methods 1, 27–29 [DOI] [PubMed] [Google Scholar]
- 18.Himpel S, Tegge W, Frank R, Leder S, Joost HG, and Becker W (2000) Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem 275, 2431–2438 [DOI] [PubMed] [Google Scholar]
- 19.Lahiry P, Wang J, Robinson JF, Turowec JP, Litchfield DW, Lanktree MB, Gloor GB, Puffenberger EG, Strauss KA, Martens MB, Ramsay DA, Rupar CA, Siu V, and Hegele RA (2009) A multiplex human syndrome implicates a key role for intestinal cell kinase in development of central nervous, skeletal, and endocrine systems. Am J Hum Genet 84, 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Z, Kim J, Vidrich A, Sturgill TW, and Cohn SM (2009) Intestinal cell kinase, a MAP kinase-related kinase, regulates proliferation and G1 cell cycle progression of intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 297, G632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaya T, Omori Y, Kuwahara R, and Furukawa T (2014) ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J 33, 1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon H, Song J, Shin JO, Lee H, Kim HK, Eggenschwiller JT, Bok J, and Ko HW (2014) Intestinal cell kinase, a protein associated with endocrine-cerebro-osteodysplasia syndrome, is a key regulator of cilia length and Hedgehog signaling. Proc Natl Acad Sci U S A 111, 8541–8546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong Y, Park SH, Wu D, Xu W, Guillot SJ, Jin L, Li X, Wang Y, Lin CS, and Fu Z (2017) An essential role of intestinal cell kinase in lung development is linked to the perinatal lethality of human ECO syndrome. FEBS Lett [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding M, Jin L, Xie L, Park SH, Tong Y, Wu D, Chhabra AB, Fu Z, and Li X (2018) A Murine Model for Human ECO Syndrome Reveals a Critical Role of Intestinal Cell Kinase in Skeletal Development. Calcif Tissue Int 102, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malicki JJ, and Johnson CA (2017) The Cilium: Cellular Antenna and Central Processing Unit. Trends Cell Biol 27, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiter JF, and Leroux MR (2017) Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Bio 18, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burghoorn J, Dekkers MP, Rademakers S, de Jong T, Willemsen R, and Jansen G (2007) Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci U S A 104, 7157–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman SA, Wilson NF, Haas NA, and Lefebvre PA (2003) A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol 13, 1145–1149 [DOI] [PubMed] [Google Scholar]
- 29.Bengs F, Scholz A, Kuhn D, and Wiese M (2005) LmxMPK9, a mitogen-activated protein kinase homologue affects flagellar length in Leishmania mexicana. Mol Microbiol 55, 1606–1615 [DOI] [PubMed] [Google Scholar]
- 30.Jiang YY, Maier W, Baumeister R, Minevich G, Joachimiak E, Wloga D, Ruan Z, Kannan N, Bocarro S, Bahraini A, Vasudevan KK, Lechtreck K, Orias E, and Gaertig J (2019) LF4/MOK and a CDK-related kinase regulate the number and length of cilia in Tetrahymena. Plos Genet 15, e1008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oud MM, Bonnard C, Mans DA, Altunoglu U, Tohari S, Ng AY, Eskin A, Lee H, Rupar CA, de Wagenaar NP, Wu KM, Lahiry P, Pazour GJ, Nelson SF, Hegele RA, Roepman R, Kayserili H, Venkatesh B, Siu VM, Reversade B, and Arts HH (2016) A novel ICK mutation causes ciliary disruption and lethal endocrine-cerebro-osteodysplasia syndrome. Cilia 5, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paige Taylor S, Kunova Bosakova M, Varecha M, Balek L, Barta T, Trantirek L, Jelinkova I, Duran I, Vesela I, Forlenza KN, Martin JH, Hampl A, Bamshad M, Nickerson D, Jaworski ML, Song J, Ko HW, Cohn DH, Krakow D, and Krejci P (2016) An inactivating mutation in intestinal cell kinase, ICK, impairs hedgehog signalling and causes short rib-polydactyly syndrome. Hum Mol Genet 25, 3998–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey JN, de Nijs L, Bai D, Suzuki T, Miyamoto H, Tanaka M, Patterson C, Lin YC, Medina MT, Alonso ME, Serratosa JM, Duron RM, Nguyen VH, Wight JE, Martinez-Juarez IE, Ochoa A, Jara-Prado A, Guilhoto L, Molina Y, Yacubian EM, Lopez-Ruiz M, Inoue Y, Kaneko S, Hirose S, Osawa M, Oguni H, Fujimoto S, Grisar TM, Stern JM, Yamakawa K, Lakaye B, and Delgado-Escueta AV (2018) Variant Intestinal-Cell Kinase in Juvenile Myoclonic Epilepsy. N Engl J Med 378, 1018–1028 [DOI] [PubMed] [Google Scholar]
- 34.Tsutsumi R, Chaya T, and Furukawa T (2018) Enriched expression of the ciliopathy gene Ick in cell proliferating regions of adult mice. Gene Expr Patterns 29, 18–23 [DOI] [PubMed] [Google Scholar]
- 35.Chen T, Wu D, Moskaluk CA, and Fu Z (2013) Distinct expression patterns of ICK/MAK/MOK protein kinases in the intestine implicate functional diversity. PLoS One 8, e79359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolick DT, Chen T, LA OA, Tong Y, Wu D, Joyner LT 2nd, Oria RB, Guerrant RL, and Fu Z (2014) Intestinal cell kinase is a novel participant in intestinal cell signaling responses to protein malnutrition. PLoS One 9, e106902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J, Otis JM, Higginbotham H, Monckton C, Cheng J, Asokan A, Mykytyn K, Caspary T, Stuber GD, and Anton ES (2017) Primary Cilia Signaling Shapes the Development of Interneuronal Connectivity. Dev Cell 42, 286–300 e284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirschen GW, and Xiong Q (2017) Primary cilia as a novel horizon between neuron and environment. Neural Regen Res 12, 1225–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, and Zhong Q (2013) Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 502, 254–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, and Cuervo AM (2013) Functional interaction between autophagy and ciliogenesis. Nature 502, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pampliega O, and Cuervo AM (2016) Autophagy and primary cilia: dual interplay. Curr Opin Cell Biol 39, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheong H, Wu J, Gonzales LK, Guttentag SH, Thompson CB, and Lindsten T (2014) Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy 10, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paige Taylor S, Kunova Bosakova M, Varecha M, Balek L, Barta T, Trantirek L, Jelinkova I, Duran I, Vesela I, Forlenza KN, Martin JH, Hampl A, Bamshad M, Nickerson D, Jaworski ML, Song J, Ko HW, Cohn DH, Krakow D, and Krejci P (2016) An inactivating mutation in intestinal cell kinase, ICK, impairs hedgehog signalling and causes short rib-polydactyly syndrome. Hum Mol Genet [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okamoto S, Chaya T, Omori Y, Kuwahara R, Kubo S, Sakaguchi H, and Furukawa T (2017) Ick Ciliary Kinase Is Essential for Planar Cell Polarity Formation in Inner Ear Hair Cells and Hearing Function. J Neurosci 37, 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broekhuis JR, Verhey KJ, and Jansen G (2014) Regulation of cilium length and intraflagellar transport by the RCK-kinases ICK and MOK in renal epithelial cells. PLoS One 9, e108470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma M, Gallagher AR, and Somlo S (2017) Ciliary Mechanisms of Cyst Formation in Polycystic Kidney Disease. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avasthi P, Maser RL, and Tran PV (2017) Primary Cilia in Cystic Kidney Disease. Results Probl Cell Differ 60, 281–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes DC, Leal LF, Mermejo LM, Scrideli CA, Martinelli CE Jr., Fragoso MC, Latronico AC, Tone LG, Tucci S, Yunes JA, Cardinalli IA, Mastellaro MJ, Brandalise SR, Ramalho F, Moreira AC, Ramalho LN, de Castro M, and Antonini SR (2014) Sonic hedgehog signaling is active in human adrenal cortex development and deregulated in adrenocortical tumors. J Clin Endocrinol Metab 99, E1209–1216 [DOI] [PubMed] [Google Scholar]
- 49.Finco I, Lerario AM, and Hammer GD (2018) Sonic Hedgehog and WNT Signaling Promote Adrenal Gland Regeneration in Male Mice. Endocrinology 159, 579–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelke MF, Waas B, Kearns SE, Suber A, Boss A, Allen BL, and Verhey KJ (2019) Acute Inhibition of Heterotrimeric Kinesin-2 Function Reveals Mechanisms of Intraflagellar Transport in Mammalian Cilia. Curr Biol 29, 1137–1148 e1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichinose S, Ogawa T, and Hirokawa N (2015) Mechanism of Activity-Dependent Cargo Loading via the Phosphorylation of KIF3A by PKA and CaMKIIa. Neuron 87, 1022–1035 [DOI] [PubMed] [Google Scholar]
- 52.Hall MN (2008) mTOR-what does it do? Transplant Proc 40, S5–8 [DOI] [PubMed] [Google Scholar]
- 53.Bhaskar PT, and Hay N (2007) The two TORCs and Akt. Dev Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 54.Laplante M, and Sabatini DM (2009) mTOR signaling at a glance. J Cell Sci 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, and Yonezawa K (2003) The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem 278, 15461–15464 [DOI] [PubMed] [Google Scholar]
- 56.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, and Guan KL (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanrahan J, and Blenis J (2006) Rheb activation of mTOR and S6K1 signaling. Methods Enzymol 407, 542–555 [DOI] [PubMed] [Google Scholar]
- 59.Wu D, Chapman JR, Wang L, Harris TE, Shabanowitz J, Hunt DF, and Fu Z (2012) Intestinal cell kinase (ICK) promotes activation of mTOR complex 1 (mTORC1) through phosphorylation of Raptor Thr-908. J Biol Chem 287, 12510–12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boehlke C, Kotsis F, Patel V, Braeg S, Voelker H, Bredt S, Beyer T, Janusch H, Hamann C, Godel M, Muller K, Herbst M, Hornung M, Doerken M, Kottgen M, Nitschke R, Igarashi P, Walz G, and Kuehn EW (2010) Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol 12, 1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan S, Li J, Diener DR, Choma MA, Rosenbaum JL, and Sun Z (2012) Target-of-rapamycin complex 1 (Torc1) signaling modulates cilia size and function through protein synthesis regulation. Proc Natl Acad Sci U S A 109, 2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi K, Nagai T, Chiba S, Nakayama K, and Mizuno K (2018) Glucose deprivation induces primary cilium formation through mTORC1 inactivation. J Cell Sci 131 [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Livingston MJ, Su Y, and Dong Z (2015) Reciprocal regulation of cilia and autophagy via the MTOR and proteasome pathways. Autophagy 11, 607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JG, and Ye Y (2013) Bag6/Bat3/Scythe: a novel chaperone activity with diverse regulatory functions in protein biogenesis and degradation. Bioessays 35, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sebti S, Prebois C, Perez-Gracia E, Bauvy C, Desmots F, Pirot N, Gongora C, Bach AS, Hubberstey AV, Palissot V, Berchem G, Codogno P, Linares LK, Liaudet-Coopman E, and Pattingre S (2014) BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc Natl Acad Sci U S A 111, 4115–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams R, Ryves WJ, Dalton EC, Eickholt B, Shaltiel G, Agam G, and Harwood AJ (2004) A molecular cell biology of lithium. Biochem Soc Trans 32, 799–802 [DOI] [PubMed] [Google Scholar]
- 67.Wilson NF, and Lefebvre PA (2004) Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot Cell 3, 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thoma CR, Frew IJ, Hoerner CR, Montani M, Moch H, and Krek W (2007) pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat Cell Biol 9, 588–595 [DOI] [PubMed] [Google Scholar]
- 69.Zhang B, Zhang T, Wang G, Chi W, Jiang Q, and Zhang C (2015) GSK3beta-Dzip1-Rab8 cascade regulates ciliogenesis after mitosis. PLoS Biol 13, e1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takenaka K, Kise Y, and Miki H (2007) GSK3beta positively regulates Hedgehog signaling through Sufu in mammalian cells. Biochem Biophys Res Commun 353, 501–508 [DOI] [PubMed] [Google Scholar]
- 71.Cross DA, Alessi DR, Cohen P, Andjelkovich M, and Hemmings BA (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 72.Tong Y, Park S, Wu D, Harris TE, Moskaluk CA, Brautigan DL, and Fu Z (2018) Modulation of GSK3beta autoinhibition by Thr-7 and Thr-8. FEBS Lett 592, 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozgul RK, Siemiatkowska AM, Yucel D, Myers CA, Collin RW, Zonneveld MN, Beryozkin A, Banin E, Hoyng CB, van den Born LI, Bose R, Shen W, Sharon D, Cremers FP, Klevering BJ, den Hollander AI, and Corbo JC (2011) Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet 89, 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tucker BA, Scheetz TE, Mullins RF, DeLuca AP, Hoffmann JM, Johnston RM, Jacobson SG, Sheffield VC, and Stone EM (2011) Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci U S A 108, E569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Omori Y, Chaya T, Katoh K, Kajimura N, Sato S, Muraoka K, Ueno S, Koyasu T, Kondo M, and Furukawa T (2010) Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc Natl Acad Sci U S A 107, 22671–22676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maurya AK, Rogers T, and Sengupta P (2019) A CCRK and a MAK Kinase Modulate Cilia Branching and Length via Regulation of Axonemal Microtubule Dynamics in Caenorhabditis elegans. Curr Biol 29, 1286–1300 e1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yi P, Xie C, and Ou G (2018) The kinases male germ cell-associated kinase and cell cycle-related kinase regulate kinesin-2 motility in Caenorhabditis elegans neuronal cilia. Traffic 19, 522–535 [DOI] [PubMed] [Google Scholar]