Abstract
Sphagnum mosses dominate peatlands by employing harsh ecosystem tactics to prevent vascular plant growth and microbial degradation of these large carbon stores. Knowledge about Sphagnum‐produced metabolites, their structure and their function, is important to better understand the mechanisms, underlying this carbon sequestration phenomenon in the face of climate variability. It is currently unclear which compounds are responsible for inhibition of organic matter decomposition and the mechanisms by which this inhibition occurs. Metabolite profiling of Sphagnum fallax was performed using two types of mass spectrometry (MS) systems and 1H nuclear magnetic resonance spectroscopy (1H NMR). Lipidome profiling was performed using LC‐MS/MS. A total of 655 metabolites, including one hundred fifty‐two lipids, were detected by NMR and LC‐MS/MS—329 of which were novel metabolites (31 unknown lipids). Sphagum fallax metabolite profile was composed mainly of acid‐like and flavonoid glycoside compounds, that could be acting as potent antimicrobial compounds, allowing Sphagnum to control its environment. Sphagnum fallax metabolite composition comparison against previously known antimicrobial plant metabolites confirmed this trend, with seventeen antimicrobial compounds discovered to be present in Sphagnum fallax, the majority of which were acids and glycosides. Biological activity of these compounds needs to be further tested to confirm antimicrobial qualities. Three fungal metabolites were identified providing insights into fungal colonization that may benefit Sphagnum. Characterizing the metabolite profile of Sphagnum fallax provided a baseline to understand the mechanisms in which Sphagnum fallax acts on its environment, its relation to carbon sequestration in peatlands, and provide key biomarkers to predict peatland C store changes (sequestration, emissions) as climate shifts.
Keywords: antimicrobial metabolites, fungal metabolites, lipidomics, metabolomics, Sphagnum fallax
Significance.
This work is significant because it illustrates using a comprehensive metabolomics approach, the potential mechanisms by which Sphagnum fallax inhibits microbial activity in peatlands and hence litter decomposition, leading to C sequestration. The identified metabolites provide a baseline to determine key biomarkers to predict peatland C store accumulation, sequestration, and emissions as climate shifts.
1. INTRODUCTION
Peatlands cover over 4 million km2 worldwide and are the most efficient terrestrial ecosystems in storing carbon (Gorham, 1991). The total amount of carbon stored in these systems is approximately 250–450 Pg C, equivalent to half of what is stored in the atmosphere as CO2 (750 Pg C; Frolking et al., 2011; Gorham, 1991). Northern peatlands are formed of bogs and fens, the former being a significant driver of organic matter accumulation, total amount of carbon stored, and total storage capacity (Armentano & Menges, 1986; Sjors, 1981). The bog arcotelm, defined as the peat thickness above the summer water table (Clymo, 1978), varies largely in the hummock/hollow microtopography, with a thicker layer of peat and correlative minimal C losses characteristic to hummocks (Wallén, 1986). This low organic carbon (OC) consumption and decomposition in peat bog hummocks is by and large due to the most important peat‐forming plants, the bryophytic moss Sphagnum (van Breemen, 1995; Verhoeven & Liefveld, 1997).
Sphagnum mosses, an ancient early‐branching lineage of land plants, are keystone species that shape their habitat through unique biochemical and morphological adaptations. Since these plants lack roots and sophisticated shoot systems for resource accumulation and distribution, Sphagnum mosses rely on employing hostile ecosystem tactics to ensure light, nutrient, and water availability (Kostka et al., 2016; Turetsky, 2018; van Breemen, 1995; Verhoeven & Liefveld, 1997). Sphagnum mosses have high capillarity and create permanently wet and water‐logged soil conditions, with resultant anoxia and relatively low temperatures. The characteristic cell‐wall structure, made of polysaccharides and highly reactive carbonyl groups, has been linked to a high‐cation exchange capacity which simultaneously traps nutrients and acidifies the environment (Ca concentration <2 mg/L and pH < 4.2; Clymo, 1978; van Breemen, 1995). As such, these adaptations wage environmental warfare on vascular plants and microbial degrading counterparts, which promotes Sphagnum growth, suppresses microbial and vascular plant growth, leads to subsequent slow litter (organic matter) decomposition, and ultimately results in the high carbon storage capacity observed in peat bogs.
Climate is one of the most important determinants of peatlands’ character, distribution, and biodiversity, yet peatland‐specific processes that control C decomposition, accumulation, and subsequent feedback to climate, despite their significance, are not included in current climate models (Limpens et al., 2008). Variability in primary productivity of peatlands is projected with increasing temperature, where some areas will benefit from lengthened growing seasons and concurrent warmer temperatures, and other areas will see a decrease in primary productivity due to evapotranspiration (Hufnagel & Garamvölgyi, 2014). A plausible shift from Sphagnum moss dominance to vascular plant species (i.e., sedges) is generally agreed upon in climate models, and because Sphagnum currently dictates and limits organic matter decomposition, in part by its restriction on microbial decomposer growth (and related organic matter degradation/greenhouse gas by‐products from microbial metabolism), these climates impacted shifts could affect the mobility of organic matter within peatland soils. While the understanding of individual biotic and abiotic drivers of C cycling in peatlands is developing (Chanton et al., 2008; Chasar, Chanton, Glaser, Siegel, & Rivers, 2000; Fenner & Freeman, 2011; Freeman, Evans, Monteith, Reynolds, & Fenner, 2001; Freeman, Ostle, & Kang, 2002), the plant–soil–microbe interactions in the context of rising temperatures remain poorly understood. This uncertainty in peatland response to climate change has therefore led to increase interest and urgency in better understanding the decomposition pathways of these large and important carbon stocks, and more specifically, the Sphagnum moss–microbial OM degradation interactions that underlay it.
While it is widely agreed upon that decomposition of peat carbon is partially inhibited by antimicrobial properties of Sphagnum, that is, by deterring the main soil decomposers, it is currently unclear which compounds are responsible for inhibition of organic matter decomposition and the mechanisms by which this inhibition occurs. Some researchers have suggested that the amorphous phenolic polymer that covers the surface of the Sphagnum cell wall, whose structure and function is similar to lignin, forms a physical barrier against decomposition (Freeman et al., 2001; Hájek, Ballance, Limpens, Zijlstra, & Verhoeven, 2011; Talbot & Treseder, 2012; Turetsky, Crow, Evans, Vitt, & Wieder, 2008; Verhoeven & Liefveld, 1997). However, Mellegård, Stalheim, Hormazabal, Granum, & Hardy, 2009 were unable to determine any antibacterial effects of phenolic compounds from moss extracts. Others have suggested that sphagnan, a pectin‐like compound within the cell‐wall holocellulose, is slowly released into the environment when the cell wall undergoes autogenic acid hydrolysis, liberating uronic acid for high‐cation exchange and acidification (Børsheim, Christensen, & Painter, 2001; Hájek et al., 2011; Painter, 1991; Stalheim, Ballance, Christensen, & Granum, 2009). The ambiguity in compounds and related function of various Sphagnum compounds on ecosystem variables elucidates an important research gap to gauge how these mosses control their ecosystems. Furthermore, this gap is widened by limitations in testing methodology, where previous studies have used singular analytical techniques which cannot identify a full suite of compounds (i.e., gas chromatography‐mass spectrometry, pyrolysis molecular beam mass spectrometry, 13C NMR; Black, Cornhill, & Woodward, 1955; Limpens, Bohlin, & Nilsson, 2017; Turetsky et al., 2008; Verhoeven & Liefveld, 1997), have focused only on specific compounds or classes of compounds (Ballance, Børsheim, Inngjerdingen, Paulsen, & Christensen, 2007; Klavina, Springe, Steinberga, Mezaka, & Ievinsh, 2018), or have concentrated on applied function rather than identifying structures (Bengtsson, Granath, & Rydin, 2016; Bengtsson, Rydin, & Hájek, 2018; Hájek et al., 2011). While all of these are crucial to understanding Sphagnum and its ecosystem roles, a full‐suite chemical characterization of Sphagnum fallax metabolites is currently missing from the literature.
Due to the importance of Sphagnum and the chemical structures underlying its dominance of peatland ecosystems, as well as the little available information of these structures and the general composition of Sphagnum fallax, this research aims to characterize Sphagnum fallax metabolite and lipid profile. We implemented a full‐analytical suite of mass spectrometry (MS) and 1H liquid state nuclear magnetic resonance spectroscopy (NMR) characterization techniques in order to identify known and unknown compounds of Sphagnum fallax to expand on and improve current databases. Our research further aims to understand the antimicrobial activity of Sphagnum and better understand its microbial inhibition mechanisms by using Fourier‐transform ion cyclotron resonance mass spectrometry (FTICR‐MS) to compare metabolomes and identify antimicrobial compounds in Sphagnum against the metabolomes of nine medicinal plants with previously known antimicrobial and medicinal properties (Ahmed et al., 2016; Cowan, 1999; Oliveira‐Alves et al., 2017; Ortega‐Ramirez et al., 2014; Riaz, Zia‐Ul‐Haq, & Jaafar, 2013; Singh, Khanam, Misra, & Srivastava, 2011; Surjushe, Vasani, & Saple, 2009; Yang et al., 2012). This research will provide fundamental chemical and structural data to further bridge the knowledge gap of how Sphagnum fallax acts on its environment, with the peatland microbiome, and the role it plays in the vulnerability or resilience of peatland carbon stores to changing environmental conditions. This information can be further extrapolated upon to improve climate models by identifying environmental biomarkers for understanding and predicting carbon changes in peatlands.
2. MATERIALS AND METHODS
2.1. Sample site—Sphagnum fallax
The experimental site was the S1 bog in the Marcell Experimental Forest (MEF; N 47°30.4760; W93°27.1620), approximately 40 km north of Grand Rapids, Minnesota, USA. The bog surface is characterized by hummock and hollow microtopography with a relief of 10–30 cm between the tops of the hollows and the hummocks. For more detailed information about this site refer to Kolka (2011) and Tfaily et al. (2014). S1 bog vegetation consists of two dominant tree species, black spruce (Picea mariana) and larch (Larix laricina), with both hollow and hummock features. The bryophytes in hollows are mainly colonized by Sphagnum fallax. Three biological replicates of individual plants of Sphagnum fallax were collected during August 2017. Only green living plants were sampled: samples focused on the capitulum plus about 2–3 cm of green living stem. Sphagnum stems (phyllosphere) were cleaned from unrelated plant debris and frozen immediately on dry ice and shipped overnight to the laboratory.
2.2. Sample collection and preparation
The 14 plant species analyzed in the antimicrobial portion of this study are included in Table 1. The Sphagnum collection site and collection procedure are described above. Medicinal plants were obtained as dry, loose leaf from the Tucson Herb Store; seed from Food Conspiracy Coop; dry, loose leaf from Hippie Gypsy; and live, dry, or powder from distributors on Amazon (JMBamboo, Bixa Botanical, Kart It). Creosote and controls (i.e., plants that were randomly collected and may not specifically have antimicrobial compounds) were collected from five locations on the University of Arizona campus in 15 ml sterile tubes. In preparation for organic matter metabolite extraction, triplicates of all plant samples were lyophilized using a Labconco FreeZone, Benchtop freeze dryer for 24 hr to remove moisture from samples and ensure moisture homogeneity between plant species for starting weight and extraction.
Table 1.
Plant species tested in antimicrobial comparison study
| Testing group | Plant | Plant material type |
|---|---|---|
| Sphagnum sp. | Sphagnum fallax | Stems, capitulum |
| Medicinal plants | Chamomile (Matricaria recutita) | Flower, stems |
| Senna (Cassia angustifolia) | Leaves | |
| Mullein (Verbascum thapsus) | Leaves, stems | |
| Chia (Salvia hispanica) | Seeds | |
| Rosemary (Rosemarinus officinacis) | Leaves | |
| Cinnamon (Cinnamomum cassia) | Bark | |
| Aloe (Aloe vera) | Stems | |
| Creosote (Larrea tridentate) | Leaves, stems | |
| Cannabidiol (CBD – Hemp cannabis) | Flower, stems, seeds | |
| Controls | Virgin Palm (Dioon edule) | Leaves, stems |
| Arizona Cypress (Cupressus arizonica) | Leaves, stems | |
| Fountain Grass (Cenchrus setaceus) | Leaves | |
| Mexican Flame (Anisacanthus quadrifidus) | Leaves, stems |
2.3. Organic matter metabolite extraction
Extracts were prepared using an adjusted Folch extraction (Folch, Lees, & Sloane Stanley, 1957). Folch extractions are used to extract labile (polar and semipolar compounds), and therefore, the detection of terpenes from these extracts can be limited. Lyophilized triplicate samples were ground using a standard electronic coffee grinder, weighed into three equal weights (about 0.1–0.05 g), and each triplicate per sample was wetted with 200 µl of milliQ water. Extraction was performed on each sample by sequentially adding 2 ml MeOH, followed by a 5 s vortex; 4 ml CHCl3, followed by a 5 s vortex; sonication at 25°C for 1 hr (CPX3800 Ultrasonic Bath, Fisherbrand); addition of 1.25 ml of H2O, followed by a slight mix to achieve bi‐layer separation; and incubated at 4°C overnight. The top, aqueous layer (metabolite—polar) was pipetted off into 1 ml glass vials and stored at −80°C until FTICR‐MS (all samples) and 1H NMR (Sphagnum only) analysis. The bottom, chloroform layer (Sphagnum only—nonpolar) was dried down and stored in 50:50 methanol:chloroform until lipidomics analysis.
2.4. Metabolomic analysis by FTICR‐MS
A Bruker 9.4‐Tesla FTICR spectrometer located at University of Arizona was used to collect high‐resolution mass spectra of the organic matter in the aqueous extract by direct injection. A standard Bruker electrospray ionization (ESI) source was used to generate negatively charged molecular ions. Samples were then introduced directly to the ESI source. The instrument settings were optimized by tuning on a Suwannee River fulvic acid (SRFA) standard, purchased from International Humic Substances Society (IHCC). Blanks (HPLC grade methanol) were analyzed at the beginning and end of the day to monitor potential carry over from one sample to another. The instrument was flushed between samples using a mixture of water and methanol. The ion accumulation time (IAT) was varied to account for differences in C concentration between samples and varied between 0.03 and 0.05 s. One hundred and forty‐four individual scans were averaged for each sample and internally calibrated using an organic matter homologous series separated by 14 Da (CH2 groups). The mass measurement accuracy was <1 ppm for singly charged ions across a broad m/z range (100–1,200 m/z). The mass resolution was ~240 K at 341 m/z. The transient was 0.8 s. Data Analysis software (BrukerDaltonik version 4.2) was used to convert raw spectra to a list of m/z values applying FTICR‐MS peak picker module with a signal‐to‐noise ratio (S/N) threshold set to 7 and absolute intensity threshold to the default value of 100. Putative chemical formulae were then assigned using Formularity (Tolić et al., 2017) software, as previously described in Tfaily, Hess, Koyama, and Evans (2018). Chemical formulae were assigned based on the following criteria: S/N > 7 and mass measurement error < 1 ppm, taking into consideration the presence of C, H, O, N, S, and P and excluding other elements. To ensure consistent formula assignment, we aligned all sample peak lists for the entire dataset to each other in order to facilitate consistent peak assignments and eliminate possible mass shifts that would impact formula assignment. We implemented the following rules to further ensure consistent formula assignment: (a) we consistently picked the formula with the lowest error between predicted and observed m/z and with the lowest number of heteroatoms, and (b) the assignment of one phosphorus atom requires the presence of at least four oxygen atoms. The chemical character of thousands of peaks in each sample's ESI FTICR‐MS spectrum was evaluated using van Krevelen diagrams. Compounds were plotted on the van Krevelen diagram based on their molar H:C ratios (y‐axis) and molar O:C ratios (x‐axis; Kim, Kramer, & Hatcher, 2003). Van Krevelen diagrams provide a means to visualize and compare the average properties of organic compounds and assign compounds to the major biochemical classes (e.g., lipid‐, protein‐, lignin‐, carbohydrate‐, and condensed aromatic‐like). In this study, biochemical compound classes are reported as relative abundance values based on counts of C, H, and O for the following H:C and O:C ranges: lipids (0 < O:C ≤ 0.3 and 1.5 ≤ H:C ≤ 2.5), unsaturated hydrocarbons (0 ≤ O:C ≤ 0.125 and 0.8 ≤ H:C < 2.5), proteins (0.3 < O:C ≤ 0.55 and 1.5 ≤ H:C ≤ 2.3), amino sugars (0.55 < O:C ≤ 0.7 and 1.5 ≤ H:C ≤ 2.2), lignin (0.125 < O:C ≤ 0.65 and 0.8 ≤ H:C < 1.5), tannins (0.65 < O:C ≤ 1.1 and 0.8 ≤ H:C < 1.5), and condensed hydrocarbons (aromatics; 0 ≤ 200 O:C ≤ 0.95 and 0.2 ≤ H:C < 0.8; Tfaily et al., 2015, 2017). It is important to note that FTICR‐MS by direct injection does not differentiate between isomers (i.e., compounds with same m/z, same molecular formula but different structure).
A 21 Tesla Agilent FTICR mass spectrometer (MS) equipped with a Waters ultra‐performance liquid chromatography (UP‐LC) system, was used to collect tandem mass spectrometry (LC‐MS/MS) spectra of triplicate Sphagnum organic matter extracts in negative and positive ion modes. Sphagnum extracts were separated using a Zorbax C18 column (0.5 mm × 150 mm × 5 µm particle size). Samples were injected in a 10 μL volume on column and eluted with solvent A (5 mM aqueous ammonium formate) and solvent B (5 mM ammonium formate in mass spectrometry grade methanol) with a 60 min linear gradient from 5% to 95% B, followed by isocratic elution at 95% B for 10 min at a flow rate of 0.2 ml/min. The flow from the LC was coupled to the 21 T FTICR‐MS using a heated ESI source set to a capillary voltage of 3,500 V; sheath, auxiliary, and sweep gas flow rates were 12, 6, and 2 (arbitrary units); and ion transfer tube and vaporizer temperatures were 300°C and 75°C, respectively. Mass spectra were collected with 1–2‐s transients. MS/MS fragmentation spectra were collected by collision‐induced dissociation (CID) of the major features using a collision energy of 40. Data were processed using Compound Discoverer 3.0 Software (Thermo Fisher Scientific—US) a qualitative data‐processing application that uses accurate mass data, isotope pattern matching, and mass spectral library searches for the structural identification of small molecules. The complete workflow is summarized in Figure S1. Briefly, we performed spectral library and similarity searches to identify compounds, searched chemical databases for putative candidates, and annotated spectra with predicted fragmentation. During the process, spectra were first aligned followed by peak picking. Predicted elemental composition using the TrueCompositionTM algorithm was generated for all unknowns using: exact mass, isotopic pattern, fine isotopic pattern, and MS/MS data using the built in HighChem Fragmentation Library of reference fragmentation mechanisms. Only compounds with MS/MS data were used for the analysis (i.e., compounds with relatively high intensity needed to generate MS/MS profiles). Metabolite identification was then performed using both spectral libraries and compound databases: mzCloud (ddMS2; online spectral library with >2 million spectra) that searches the online mzCloud database of fragmentation scans by using the MW or predicted formulas when available, and ChemSpider (exact mass or formula—chemical structure database with >480 data sources, 59 million structures) that searches selected databases of MS1 scans by using the MW or predicted formulas when available. For predicted composition, identification is dependent on mass error, number of matched isotopes, number of missed isotopes, number of matched fragments, spectral similarity score between theoretical and measured isotope pattern, matched intensity percentage of the theoretical pattern, matched intensity percentage of the relevant portion of the MS, and finally matched intensity percentage of the MS/MS scan. The mass tolerance was set to 5 ppm. After metabolite identification, data were grouped into two groups: group 1 included metabolites that were confirmed by the online database mzCloud and either predicted composition and ChemSpider or both, whereas group 2 included compounds that were validated by predicted composition and/or ChemSpider only. It is important to note that metabolites that were identified by Chemspider are only putative identifications since it is based on mass alone. Only metabolites that were identified in the three biological replicates were reported.
2.5. Metabolomic analysis by 1H NMR
One of the methanol extracted samples prepared as described above was used for metabolite profiling and quantitation by 1H NMR analysis to characterize common metabolites involved in primary metabolism and get an estimate of their abundance in living plants. The one‐dimensional (1D) 1H NMR spectrum of the Sphagnum sample was collected in accordance with standard Chenomx sample preparation and data collection guidelines (Weljie, Newton, Mercier, Carlson, & Slupsky, 2006). The sample was vacuum dried then reconstituted in deionized H2O. Even though vacuum drying would remove the majority of methanol in the sample, traces of methanol might remain in the sample after drying. Thus, our reported methanol concentration might be an overestimation of the true concentration of methanol in the plant extract. water extract was then diluted by 10% (v/v) with 5 mM 2,2‐dimethyl‐2‐silapentane‐5‐sulfonate‐d6 (DSS) as an internal standard, and the dilutions were accounted for in the normalized concentrations reported. NMR data were collected using a Varian Direct Drive 600 MHz NMR spectrometer equipped with a 5‐mm triple resonance salt‐tolerant cold probe and a cold‐carbon pre‐amplifier. The 1D 1H NMR spectra of all samples were processed, assigned, and analyzed using Chenomx NMR Suite 8.3 that combines chemistry and advanced algorithms to identify and quantify metabolites in NMR spectra with an extensive library of spectral models of metabolites (https://www.chenomx.com). In contrast to other analytical methods, only one internal standard is necessary for NMR analysis. Here, quantification was achieved through the use of an internal standard 4,4‐dimethyl‐4‐silapentane‐1‐sulfonic acid (DSS) that was used as chemical shift references for the calibration of the NMR data (at δ 0.0 ppm) as well as an internal standard for quantitation where spectral intensities of metabolites were quantified relative to the DSS internal standard. Candidate metabolites present in each of the complex mixtures were determined by matching the chemical shift, J‐coupling, and intensity information of experimental NMR signals screened against the NMR signals of standard metabolites in the Chenomx library (along with HMDB and some custom metabolite additions totaling ~1,000 metabolites). A list of the average chemical shifts per identified metabolite is included in Table S1. NMR spectra were collected as described previously (Dalcin Martins et al., 2017), and the detection limit of the method was ~1 µM. Only metabolites above 1 µM were included in this study. All concentrations were limited to three significant figures only.
2.6. Lipidomics analysis by LC‐MS/MS
Lipids were analyzed by LC‐MS/MS in both positive and negative ESI modes on the triplicate Sphagnum samples using a linear trap quadropole (LTQ) Orbitrap Velos mass spectrometer (Thermo Fisher Scientific), as described in detail previously (Kyle et al., 2017). Lipid species were identified using the LIQUID tool (Kyle et al., 2017) followed by manual data inspection. Confidently identified lipid species were quantified using MZmine 2 (Pluskal, Castillo, Villar‐Briones, & Orešič, 2010), and the peak intensities were normalized by linear regression and central tendency (i.e., identifying a central or typical value for a probability distribution) using InfernoRDN. Lipids that were identified in all three biological replicates were reported.
2.7. Network analysis
Network analysis was used to identify potential antimicrobial compounds in FTICR‐MS data based on specific chemical transformations between the pairs of m/z values, that is, m/z values of compounds of known antimicrobial effect and those m/z of unknown compounds. We used Cytoscape (Shannon et al., 2003) with the MetaNetter 2.0 plug‐in (Burgess, Borutzki, Rankin, Daly, & Jourdan, 2017) to conduct the network analysis. The list of m/z values from the Sphagnum sample was imported into Cytoscape, and each m/z value was defined as a node as described previously in Longnecker and Kujawinski (2016). In these networks, nodes are connected to each other by edges, where the edges are defined as the mass difference between two m/z values resulting from a chemical transformation. For example, the gain or loss of CO2 (Δm = 43.9898) between two compounds would be represented as two “nodes,” or the corresponding smaller and larger m/z values, connected by an edge named CO2. For each chemical transformation, there may be a series of connected m/z values and one m/z value may be connected to other m/z values by more than one transformation. In this study, we used a pre‐defined list of m/z values for use as edges adopted from Breitling, Ritchie, Goodenowe, Stewart, and Barrett (2006).
3. RESULTS
3.1. LC‐FTICR‐MS/MS and 1H NMR analysis of the Sphagnum methanol layer (metabolomics)
Primary and secondary metabolites were analyzed by liquid state 1H NMR and LC‐FTICR‐MS/MS. A total of 29 metabolites were identified by 1H NMR (Table 2), representing metabolites commonly found in plants and soils. These metabolites were broadly categorized into eight groups: amino acids, acids, sugars, amines, polyols, choline, alcohols, and aromatic compounds. Sugars and acids were the most abundant metabolites and represented the highest concentration of total measured metabolites, with ~3,360 µM and ~1,010 µM in total per 50 mg of Sphagnum in 1 ml of DI H2O (extraction efficiency ~ 15%, Tfaily et al. 2015, 2017), respectively. Fructose was the most abundant measured metabolite present in Sphagnum (sugar; 2,490 µM), followed by glucose (sugar; 796 µM), malate (acid; 555 µM), and glycerol (polyol; 254 µM). Alcohols and aromatics represented the lowest overall concentration in Sphagnum (7.6 µM and 9.3 µM in total), with methylamine (amine; 3.3 µM), and propylene glycol (polyol; 2.2 µM) measured at the lowest concentrations of present metabolites. The majority of these compounds are of known biological activity, as inferred from the KEGG database (https://www.genome.jp/), signifying metabolites that could act as biomarkers of Sphagnum metabolic pathways (Table S2).
Table 2.
Primary metabolites (with KEGG compound ID) found in Sphagnum fallax detected by 1H NMR
| Sphagnum | KEGG compound ID | Formula | Compound concentrationa | Compound class |
|---|---|---|---|---|
| Dimethylamine | C00543 | C2H7N | 10.2 | Amines |
| 4‐Aminobutyrate | C00334 | C4H9NO2 | 142 | |
| Methylamine | C00218 | CH5N | 3.3 | |
| Propylene glycol | C00583 | C3H8O2 | 2.2 | Polyols |
| Glycerol | C00116 | C3H8O3 | 254 | |
| O‐Phosphocholine | C00588 | C5H15NO4P | 3.8 | Cholines |
| Choline | C00114 | C5H14NO | 43.0 | |
| Quinone | C00472 | C6H4O2 | 9.3 | Quinones |
| Methanol | C00132 | CH4O | 7.6 | Alcohol |
| Pyruvate | C00022 | C3H4O3 | 6.0 | Acids |
| Lactate | C00186 | C3H6O3 | 12.0 | |
| 3‐Hydroxybutyrate | C01089 | C4H8O3 | 12.1 | |
| Acetate | C00033 | C2H4O2 | 236 | |
| Fumarate | C00122 | C4H4O4 | 13.2 | |
| Malate | C00711 | C4H6O5 | 555 | |
| Formate | C00058 | CH2O2 | 175 | |
| Isoleucine | C00407 | C6H13NO2 | 27.5 | Amino Acids |
| Phenylalanine | C00079 | C9H11NO2 | 30.7 | |
| Uridine | C00299 | C9H12N2O6 | 30.0 | |
| Leucine | C00123 | C6H13NO2 | 45.2 | |
| Valine | C00183 | C5H11NO2 | 58.4 | |
| Alanine | C00041 | C3H7NO2 | 160 | |
| Threonine | C00188 | C4H9NO3 | 41.6 | |
| Asparagine | C00152 | C4H8N2O3 | 188 | |
| Aspartate | C00049 | C4H7NO4 | 210 | |
| Glucose | C00031 | C6H12O6 | 796 | Sugars |
| Fructose | C00095 | C6H12O6 | 2,480 | |
| Sucrose | C00089 | C12H22O11 | 26.5 | |
| myo‐Inositol | C00137 | C6H12O6 | 48.9 |
µM per 50 mg of Sphagnum in 1 ml of DI H2O (extraction efficiency 15%).
LC‐FTICR‐MS/MS was performed to further identify and characterize secondary metabolites against TrueComposition TM algorithm (Predicted Composition—only compounds with MS2 data), mzCloud (online fragmentation spectral library with >2 million spectra), and ChemSpider (exact mass or formula—chemical structure database with >480 data sources, 59 million structures). Due to the high‐resolution of the instrument, this technique allows for high sensitivity detection and greater ability to detect previously uncharacterized compounds. Furthermore, all LC‐FTICR‐MS/MS and 1H NMR data were cross‐checked against the Plant Metabolic Pathway Network (PMN—https://www.plantcyc.org) database of documented Sphagnum fallax chemical compounds to determine which detected metabolites were novel or known metabolites in Sphagnum fallax. Positive mode LC‐FTICR‐MS/MS identified a total of 234 compounds of which 54 were validated by Predicted Composition and ChemSpider, and 20 were validated by Predicted Compositions, and mzCloud, and/or ChemSpider (Table 3; Table S3). Acids and sugars (in particular flavonoid glycosides) dominated the chemical classifications of the validated compounds, in addition to triols, amides, cholines, antioxidants, and flavones/oids. Riboflavin, luteolin, and adenosine (and their derivatives) were the only metabolites of the 54 named compounds that were recorded in the PMN database, signifying 51 identified compounds previously not known to be present in Sphagnum. Out of the 20 compounds, 10 were associated with a KEGG ID and involved in diverse pathways including flavone and flavonol biosynthesis, flavonoid biosynthesis, biosynthesis of secondary metabolites, purine metabolism, sphingolipid metabolism, alpha‐linolenic acid metabolism, biosynthesis of antibiotics, biosynthesis of unsaturated fatty acids, aflatoxin biosynthesis, and glutathione metabolism, all of which play important roles in plant metabolism and in its control and integration and defense against pathogens. The remaining 160 compounds were not found using mzCloud, Predicted Compositions, ChemSpider, or the PMN database, and signify novel metabolites detected in Sphagnum not identified or characterized in previous literature or databases.
Table 3.
Secondary metabolites identified by LC‐FTICR‐MS/MS (positive ion mode) with KEGG ID and validated by mzCloud and predicted composition and/or ChemSpider (in triplicate samples)
| Class | Name | Formula |
mzCloud Search |
mzCloud score | Predicted Compositions | Chemspider | Molecular Weight | KEGG ID |
|---|---|---|---|---|---|---|---|---|
| Triol | 2‐Amino‐1,3,4‐octadecanetriol | C18H39NO3 | Full match | 99.7 | Full match | Full match | 317.2 | C12144 |
| Amide | α‐Linolenoyl ethanolamide | C20H35NO2 | Full match | 75.5 | Full match | Not the top hit | 321.2 | |
| Acid | 9S,13R‐12‐Oxophytodienoic acid | C18H28O3 | Full match | 98.3 | Full match | Not the top hit | 292.2 | |
| Acid | 3‐(4‐Hydroxy‐5‐oxo‐3‐phenyl‐2,5‐dihydro‐2‐furanyl)propanoic acid | C13H12O5 | Full match | 67.2 | Full match | Partial match | 248 | |
| Vitamin | Riboflavin | C17H20N4O6 | Full match | 98.8 | Full match | 376.1 | D00050 | |
| Sugar | 2‐(3,4‐Dihydroxyphenyl)‐5,7‐dihydroxy‐4‐oxo‐4H‐chromen‐3‐yl 6‐O‐β‐D‐xylopyranosyl‐β‐D‐glucopyranoside | C26H28O16 | Full match | 69.6 | Full match | 596.1 | ||
| Antioxidants in plants | L‐Glutathione oxidized | C20H32N6O12S2 | Full match | 99.3 | Full match | 612.1 | C00127 | |
| Sugar | 2‐(3,4‐Dihydroxyphenyl)‐5,7‐dihydroxy‐4‐oxo‐4H‐chromen‐3‐yl 6‐O‐β‐D‐xylopyranosyl‐β‐D‐glucopyranoside | C26H28O16 | Full match | 69.6 | Full match | 596.1 | ||
| Uronic acid | 6‐O‐Methylscutellarin | C22H20O12 | Full match | 99.3 | 476 | |||
| Sugar | (2S,2'S,3R,3'R)‐7'‐(β‐D‐Glucopyranosyloxy)‐5,5'‐dihydroxy‐2,2'‐bis(4‐hydroxyphenyl)‐4,4'‐dioxo‐3,3',4,4'‐tetrahydro‐2H,2'H‐3,3'‐bichromen‐7‐yl β‐D‐glucopyranoside | C42H42O20 | Full match | 64.7 | 866.2 | |||
| Sugar | (1ξ)‐1,5‐Anhydro‐1‐[2‐(3,4‐dihydroxyphenyl)‐5,7‐dihydroxy‐4‐oxo‐4H‐chromen‐8‐yl]‐D‐galactitol | C21H20O11 | Full match | 91.1 | Not the top hit | 448.1 | ||
| Mold metabolite | Aflatoxin B2 | C17H14O6 | Full match | 61.3 | Partial match | 314 | C16753 | |
| flavonoid, antioxidant | Luteolin | C15H10O6 | Full match | 66.1 | Partial match | 286 | C01514 | |
| Sugar | (1S)‐1,5‐Anhydro‐2‐O‐(6‐deoxy‐α‐L‐mannopyranosyl)‐1‐[5,7‐dihydroxy‐2‐(4‐hydroxyphenyl)‐4‐oxo‐4H‐chromen‐6‐yl]‐D‐glucitol | C27H30O14 | Full match | 93.8 | Partial match | 578.1 | ||
| Sugar | 4‐(3,4‐Dihydroxyphenyl)‐7‐methoxy‐2‐oxo‐2H‐chromen‐5‐yl 6‐O‐[3,4‐dihydroxy‐4‐(hydroxymethyl)tetrahydro‐2‐furanyl]hexopyranoside | C27H30O15 | Full match | 83.5 | Partial match | 594.1 | ||
| Pigenin flavone glucoside | Vitexin | C21H20O10 | Full match | 92.6 | Partial match | 432.1 | C01460 | |
| Nucleoside | Adenosine | C10H13N5O4 | Full match | 99.7 | Not the top hit | Full match | 267 | C00212 |
| Flavone | Hispidulin | C16H12O6 | Full match | 99.1 | Not the top hit | Full match | 300 | C10058 |
| Acid | α‐Linolenic acida | C18H30O2 | Full match | 88.9 | Not the top hit | Full match | 278.2 | C06427 |
| Acid | 9S,13R‐12‐Oxophytodienoic acid | C18H28O3 | Full match | 99.5 | Not the top hit | Not the top hit | 292.2 | C12144 |
Possible lipid isomer (fatty acid).
Negative mode LC‐FTICR‐MS/MS identified a total of 240 compounds of which 71 compounds were validated by Predicted Composition and ChemSpider, and 31 were compounds validated by mzCloud, Predicted Composition, and/or ChemSpider (Table 4; Table S4). Acids dominated the compounds characterized by negative mode, followed by sugars (flavonoid glycosides), lipids, esters (one fatty acid ester), and an antioxidant. Prostaglandin E2 and pheophorbide b (and their derivatives) were the only metabolites of the 71 named compounds that were recorded in the PMN database, signifying 69 identified compounds previously not known to be present in Sphagnum. Eleven of the compounds (with 3 isomers) were identified in KEGG database involved in diverse pathways including as follows: Arachidonic acid metabolism (an elicitor of the plant defense response to phytopathogens), biosynthesis of unsaturated fatty acids, linoleic acid metabolism (common in plant leaves), biosynthesis of secondary metabolites, fatty acid biosynthesis, diterpenoid biosynthesis, cutin, suberine and wax biosynthesis, and glutathione metabolism (plays an important role in plant metabolism and cell function). The remaining 138 compounds with partial or no matches signify novel metabolites present in Sphagnum not identified or characterized in previous literature or databases.
Table 4.
Secondary metabolites identified by LC‐FTICR‐MS/MS (negative ion mode) with KEGG ID and validated by mzCloud and predicted composition and/or ChemSpider (in triplicate samples)
| Class | Name | Formula | mzCloud Search | mzCloud score | Predicted Compositions | ChemSpider | Molecular Weight | KEGG ID |
|---|---|---|---|---|---|---|---|---|
| Sugar | 3‐Hydroxy‐3,5,5‐trimethyl‐4‐(3‐oxo‐1‐buten‐1‐ylidene)cyclohexyl β‐D‐glucopyranoside | C19H30O8 | Full match | 52.7 | Full match | 432.19971 | ||
| Acid | (2α,3β,19α)‐2,3,19‐Trihydroxyolean‐12‐en‐28‐oic acid | C30H48O5 | Full match | 54.5 | Partial match | 488.35047 | ||
| Acid | (2β,5ξ,8α,9β,10α,13α,15α)‐15‐Hydroxy‐2‐{[2‐O‐(3‐methylbutanoyl)‐β‐D‐glucopyranosyl]oxy}kaur‐16‐ene‐18,19‐dioic acid | C31H46O12 | Full match | 63 | Full match | 610.32098 | ||
| Lipid | Prostaglandin E2a | C20H32O5 | Full match | 75.8 | Full match | 352.2251 | C00584 | |
| Acid | Neochlorogenic acid | C16H18O9 | Full match | 82.1 | Full match | 354.09513 | ||
| Sugar | (1S)‐1,5‐Anhydro‐2‐O‐(6‐deoxy‐α‐L‐mannopyranosyl)‐1‐[5,7‐dihydroxy‐2‐(4‐hydroxyphenyl)‐4‐oxo‐4H‐chromen‐6‐yl]‐D‐glucitol | C27H30O14 | Full match | 89.3 | Partial match | 578.16412 | ||
| Acid | 8(S)‐Hydroxy‐(5Z,9E,11Z,14Z)‐eicosatetraenoic acid | C20H32O3 | Full match | 90.9 | Partial match | 320.2352 | C14748 | |
| Acid | (15Z)‐9,12,13‐Trihydroxy‐15‐octadecenoic acid | C18H34O5 | Full match | 92.7 | Not the top hit | 330.24069 | ||
| Sugar | (1ξ)‐1,5‐Anhydro‐1‐[2‐(3,4‐dihydroxyphenyl)‐5,7‐dihydroxy‐4‐oxo‐4H‐chromen‐8‐yl]‐D‐galactitol | C21H20O11 | Full match | 97.6 | Not the top hit | 448.10082 | ||
| Acid | Pinolenic acid | C18H30O2 | Full match | 99.3 | Not the top hit | 278.2247 | ||
| Acid/fatty acid | Arachidonic acidb | C20H32O2 | Full match | 99.8 | Full match | 304.24041 | C00219 | |
| Acid | (2α,3β,19α)‐2,3,19‐Trihydroxyolean‐12‐en‐28‐oic acid | C30H48O5 | Full match | 54.5 | Not the top hit | Partial match | 488.35047 | |
| Lipid autacoids | 6α‐Prostaglandin I1a | C20H34O5 | Full match | 70.8 | Full match | Partial match | 354.24071 | |
| Acid | 8(S)‐Hydroxy‐(5Z,9E,11Z,14Z)‐eicosatetraenoic acida | C20H32O3 | Full match | 77.5 | Full match | Partial match | 320.2352 | C14748 |
| Acid | 14(Z)‐Eicosenoic acidb | C20H38O2 | Full match | 89.7 | Full match | Partial match | 310.28723 | |
| Acid | Corchorifatty acid F (plant metabolite, an antioxidant, and an antifungal agent.) | C18H32O5 | Full match | 93.5 | Full match | Not the top hit | 328.22505 | |
| Acid | 16‐Hydroxyhexadecanoic acidb | C16H32O3 | Full match | 94.5 | Full match | Full match | 272.23518 | C18218 |
| Acid | 11,12‐Epoxy‐(5Z,8Z,11Z)‐icosatrienoic acida | C20H32O3 | Full match | 94.9 | Full match | Not the top hit | 320.2352 | C14748 |
| Acid | Oleic acidb | C18H34O2 | Full match | 95.1 | Full match | Full match | 282.25602 | C01712 |
| Acid | 8Z,11Z,14Z‐Eicosatrienoic acidb | C20H34O2 | Full match | 98.3 | Full match | Partial match | 306.25593 | C03242 |
| Acid | cis‐7‐Hexadecenoic acidb | C16H30O2 | Full match | 98.7 | Full match | Partial match | 254.22466 | C08362 |
| Fatty acid ethyl ester | Ethyl myristate | C16H32O2 | Full match | 98.7 | Full match | Not the top hit | 256.24036 | |
| Antioxidant in plants | L‐Glutathione oxidized | C20H32N6 O12S2 | Full match | 99.1 | Full match | Full match | 612.15245 | C00127 |
| Acid | Eicosapentaenoic acidb | C20H30O2 | Full match | 99.5 | Full match | Partial match | 302.22472 | C06087 |
| Acid | 9(Z),11(E)‐Conjugated linoleic acidb | C18H32O2 | Full match | 99.7 | Full match | Not the top hit | 280.24036 | |
| Acid | 5‐(3,5‐Di‐sec‐butyl‐1‐cyclopenten‐1‐yl)‐5‐hydroxy‐3‐oxopentanoic acid | C18H30O4 | Partial match | Full match | Not the top hit | 310.21445 | ||
| Ester | 2‐methyl‐1,1'‐(1,10‐decanediyl) ester | C18H30O4 | Not the top hit | Full match | Full match | 310.21454 | ||
| Acid | (E,Z)‐9‐Hydroperoxy‐10,12‐octadecadienoic acida | C18H32O4 | Partial match | Full match | Full match | 312.23018 | ||
| Acid | 5‐(3,5‐Di‐sec‐butyl‐1‐cyclopenten‐1‐yl)‐5‐hydroxy‐3‐oxopentanoic acid | C18H30O4 | Not the top hit | Full match | Not the top hit | 310.21445 |
Possible oxidative product/derivative of lipid.
Possible lipid isomer (fatty acid).
3.2. LC‐MS/MS analysis of the chloroform layer (lipidomics)
One hundred and fifty‐two lipids were detected in Sphagnum, the majority of which were glycerophospholipid species, as well as 34 previously unidentified lipids (Figure 1, Table 5, Table S5). Polyunsaturated long‐chain fatty acids and glycerolipids have been broadly shown to be a naturally occurring group in bryophytic mosses (Mikami & Hartmann, 2004), and our data confirm their presence (Table 5). Previous studies on lipid composition of Sphagnum sp. have shown primarily alkyl compounds, sterols, triterpenoids, fatty acids, fatty alcohols, hydrocarbons, glycolipids, phospholipids, and some glycerophospholipids (Baas, Pancost, Geel, & Sinninghe Damsté, 2000; Klavina et al., 2018; Koskimies & Simola, 1980; Koskimies‐Soininen & Nyberg, 1987; Moore et al., 2015; Karunen & Ekman 1982). Our data confirmed the presence of sterols, fatty acids, prenyl derivative—ubiquinone, and glycerophospholipids, yet also demonstrated an abundance of sphingolipids and glycerolipids. Carotenoids and terpenes are also generally found in many plants (Tao, 2007) but were not detected in the lipidomics analysis of Sphagnum fallax.
Figure 1.
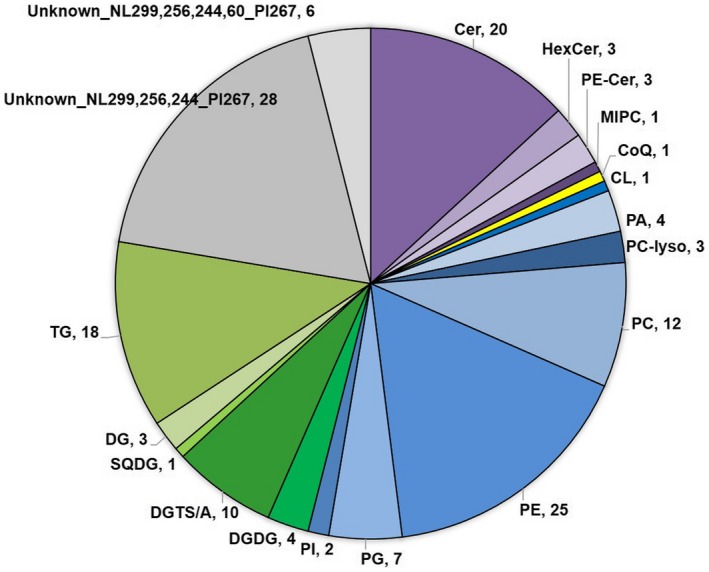
Distribution of identified lipids in Sphagnum fallax by subclass. Cer = ceramide, HexCer = galactose or glucose ceramide, PE‐Cer = ceramide phosphoethanolamine, MIPC = ceramide phosphoinositol, CoQ = coenzyme Q, CL = cardiolipin, PA = Diacylglycerophosphate, PC‐lyso = monoacylglcerophosphocholine, PC = diaoacylglcerophosphocholine, PE = diacylglcerophosphoethanolamine, PG = diacylglycerophosphoglycerol, PI = diacylglycerophosphoinositols, DGDG = digalactosyldiacylglycerol, DGTS/A = diacylglyceryl‐trimethyl‐homoserine or diacylglyceryl‐hydroxymethyl‐trimethyl‐alanine, SQDG = sulfoquinovosyldiacylglycerol, DG = diacylglycerol, TG = triacylglycerol. The number next to the lipid class abbreviation signifies the number of lipid species identified in that lipid class
Table 5.
Lipid profile distribution of Sphagnum fallax by class and subclass as identified by LC‐MS/MS (positive and negative ion modes, in triplicate samples)
| Lipid category | Subclass | # identifications |
|---|---|---|
| Sphingolipid | Cer | 20 |
| HexCer | 3 | |
| PE‐Cer | 3 | |
| MIPC | 1 | |
| Prenyl lipid | CoQ | 1 |
| Glycerophospholipid | CL | 1 |
| PA | 4 | |
| PC‐lyso | 3 | |
| PC | 12 | |
| PE | 25 | |
| PG | 7 | |
| PI | 2 | |
| Glycerolipid | DGDG | 4 |
| DGTS/A | 10 | |
| SQDG | 1 | |
| DG | 3 | |
| TG | 18 | |
| Unknown | Unknown_NL299,256,244_PI267 | 28 |
| Unknown_NL299,256,244,60_PI267 | 6 | |
| Total identifications | 152 |
3.3. Fungal metabolites and the presence of amine and nitrogen‐containing compounds
Three fungal metabolites were identified through positive mode LC‐FTICR‐MS/MS and confirmed using predicted compositions (matched by number and intensity of isotopes, and number and intensity of fragments) and ChemSpider (matched by mass): (8beta)‐6‐Allyl‐N‐[3‐(dimethylamino)propyl]ergoline‐8‐carboxamide, dehydroergosterol, and ergosterol (Table S6). Ergosterol is the predominant sterol of many fungi (ascomycetes and basidiomycetes) and is not naturally occurring in plants. Its concentration in plant samples is indicative of fungal growth and used to estimate fungal biomass associated with plant litter (Frainer & McKie, 2015; Gulis, Kuehn, & Suberkropp, 2009). Additionally, MIPC, a sphingolipid identified in the lipidomics data, is a fungal lipid commonly found in yeast (Kohlwein, 2017).
Thirty‐two nitrogen‐containing compounds were detected using LC‐FTICR‐MS/MS positive mode, which is greater than 60% of the identified compounds in this mode. Negative mode LC‐FTICR‐MS/MS detected 21 additional nitrogen‐containing compounds. 1H NMR analysis measured 4‐aminobutyrate (an amide) at a concentration of 142 µM, and while methylamine and dimethylamine were all also detected in 50 mg of Sphagnum per 1 ml of DI H2O by 1H NMR. Typically, boreal forests are nitrogen‐limited (Rousk, Jones, & DeLuca, 2013); however, this abundance of nitrogen could signify a nutrient pool available for nitrogen fixation in these ecosystems.
3.4. Acids and flavonoid sugars dominate the metabolome of Sphagnum fallax
LC‐FTICR‐MS/MS negative mode data show a surplus of acids present in Sphagnum, with 22 of the 31 validated compounds confirmed as acids, and seven more partial matches (Table 4; Table S4). Seven acid structures were confirmed via NMR, with acidic compounds present at the second highest total concentration in Sphagnum (1,010 µM/50 mg Sphagnum) following sugars (3,360 µM/50 mg Sphagnum; Table 2). Fittingly, Sphagnum moss is known to create an acidic environment in peatlands, which is suggested to be linked, in part, to Sphagnum's antimicrobial nature. Many sugar‐containing compounds (simple sugars and glucosides/glycosides), which can be “probiotic” and aid in microbial consumption and growth, were not only identified at highest concentrations in the NMR data but also confirmed in both LC‐FTICR‐MS/MS negative and positive mode. Fructose and glucose were measured at the highest concentrations of all identified compounds in the NMR characterization of Sphagnum, and 10 compounds were verified as sugar derivatives (flavonoid glycosides) via LC‐FTICR‐MS/MS.
3.5. Comparison of Sphagnum to medicinal plants using principal component analysis
We used direct inject (DI) FTICR‐MS to compare Sphagnum's metabolite signature to the metabolite composition of different plant species with known antimicrobial effects (i.e., medicinal plants). Similarities and differences of chemical composition between medicinal, control, and Sphagnum were demonstrated using principal component analysis (Figure 2). Sphagnum showed greatest similarities across both components to mullein, creosote, CBD, and Arizona cypress. Lignin, amino sugars, and proteins were drivers of similarities between Sphagnum and like plants stated above. It should be noted that Sphagnum fallax is still driven by unique features, demonstrated by the separate clusters of each Sphagnum replicate plotted away from many of the other plant clusters, which signify there are likely distinctive metabolites that could serve as environmental biomarkers of Sphagnum.
Figure 2.
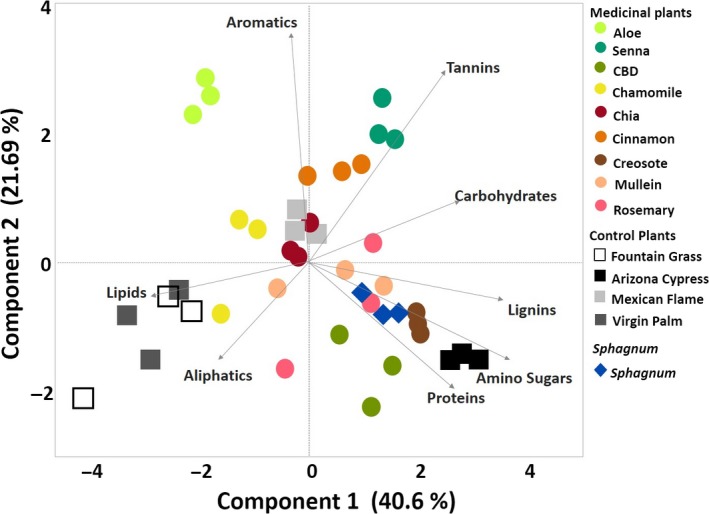
Principal component analysis plot derived from FTICR‐MS data (negative ion mode by direct injection), showing the distribution of the relative abundances of the different biogeochemical classes of compounds observed in each sample. For each plant, three biological replicates were analyzed
3.6. Seventeen putative antimicrobial substances detected by FTICR‐MS in Sphagnum
To assess the antimicrobial similarities of chemical compounds between Sphagnum and medicinal plants, all m/z values from Sphagnum fallax were cross‐checked against a database of antimicrobial compounds confirmed in the FTICR‐MS analysis of the tested medicinal plants. The m/z of 17 known antimicrobial chemical compounds (Abedini et al., 2013; Akroum, Bendjeddou, Satta, & Lalaoui, 2009; Appendino et al., 2008; Bai et al., 2018; Chiruvella, Mohammed, Dampuri, Ghanta, & Raghavan, 2007; del Valle et al., 2016; Fiamegos et al., 2011; Huang, George, & Ebersole, 2010; Kandakumar & Manju, 2017; Lou et al., 2012; Mazimba, 2017; Nakatsuji et al., 2009; Narasimhan, Ohlan, Ohlan, Judge, & Narang, 2009; Sati, Dhyani, Bhatt, & Pandey, 2019; Shi et al., 2016; Singh, Sharma, Rani, Mishra, & Sharma, 2011) were confirmed to be present within Sphagnum. Two compounds had an associated isomer (Table 6); therefore, while 17 compounds were associated with antimicrobial traits medicinal, only 15 m/z values were detected. Of the 15 m/z values, five were detected in LC‐FTICR‐MS/MS positive and negative mode, including O‐glycoside‐luteolin and isomer kaempferol, O‐glycoside‐kaempferol and isomer luteolin glucoside, and caffeoylquinic acid. The absence of the 12 other compounds in the LC‐FTICR‐MS/MS data is likely that these compounds are present in low abundance, and hence, no MS/MS spectra were available during analysis of these ions in Sphagnum. Sphagnum showed the greatest similarities in antimicrobial qualities with chia, followed by chamomile and CBD. Of these antimicrobial compounds, nine of the seventeen are acids, which correlate with previous literature on Sphagnum's antimicrobial nature. Seven of these structures are sugar derivatives (e.g., flavonoid glycosides), which could be inhibiting microbial activity due to its antimicrobial properties. The presence of these putative antimicrobial compounds should be further tested via microbiological assays using S. fallax dominating strains to confirm the biological (antimicrobial) activity of these metabolites.
Table 6.
Known antimicrobial compounds shared between Sphagnum fallax and the medicinal plants used in this study
| Compound | Molecular formula | Plant associated with and confirmed |
|---|---|---|
| O‐glycoside‐aglycones apigenin | C15H10O5 | CBD |
| O‐glycoside‐luteolina, c | C15H10O6 | |
| Tetrahydrocannabinolic acid | C22H30O4 | |
| O‐glycoside‐kaempferola, d | C21H20O11 | |
| Umbelliferone | C9H6O3 | Chamomile |
| Isorhamnetin | C16H12O7 | |
| Luteolin glucosidea, d | C21H20O11 | |
| Quercetin glycoside | C21H20O12 | |
| Coumaric acid | C9H8O3 | Chia |
| Quinic acid | C7H12O6 | |
| Syringic acid‐glucoside | C15H18O9 | |
| Caffeoylquinic acida | C16H18O9 | |
| Methyl rosmarinic acid‐glucoside | C18H16O8 | |
| Dodecanoic acidb | C12H24O2 | Cinnamon |
| Palmitic acid ethyl ester | C18H36O2 | Creosote |
| Veratric acid | C9H10O4 | Mullein |
| Kaempferola, c | C15H10O6 | Senna |
Compounds confirmed by LC‐FTICR‐MS/MS analysis.
Possible lipid isomer—fatty acid 12:0.
Isomers.
Isomers.
3.7. Novel and known compounds identified by network analysis suggest potential pool of antimicrobial compounds present in Sphagnum fallax
Network analysis was performed on the potential antimicrobial substances present in Sphagnum to further identify known and unknown compounds that could be linked to Sphagnum's antimicrobial ability. In order to do so, all m/z values detected by DI‐FTICR‐MS in Sphagnum were analyzed and plotted by their differences in chemical transformations, and then, compounds were selected for that were first neighbors of (one transformation different from) the 17 putative antimicrobial metabolites. Compounds were submitted to Formularity software to generate formulas based on specific formula assignment rules, specifically assigning C, H, O, N, S, and P. Chemical classes were then determined by specific O:C and H:C ratios. Two hundred and thirty‐two masses were identified to be first neighbors of the 17 antimicrobial compounds, with 211 compounds that were assigned formulas and chemical classes (Table S7). For 21 of these compounds, no molecular formula was generated either because they contained elements outside of formula assignment rules or because they did not follow the rules used by Formularity. Therefore, these compounds are considered unknown. Polyphenol (lignin‐like) species dominate the antimicrobial and first neighbor compounds, followed closely by carbohydrate and aromatic compounds (Figure 3a). Two subnetworks are outlined in Figure 3b,c. Tetrahydrocannabinolic acid is one carbon/dehydrogenation from a previously unidentified species C22H31O4, which could suggest an undiscovered antimicrobial compound. Antimicrobial substances palmitic acid ethyl ester and dodecanoic acid are only three ethyl additions different, signifying the compounds C16H32O2 and C14H28O2 could also have similar antimicrobial effects within the environment. It should be noted that both C16H32O2 and C14H28O2 could be fatty acids 16:0 and 14:0, respectively, and each has 29 and 26 isomers. Further research should be done to structurally characterize these compounds and investigate their antimicrobial activity.
Figure 3.
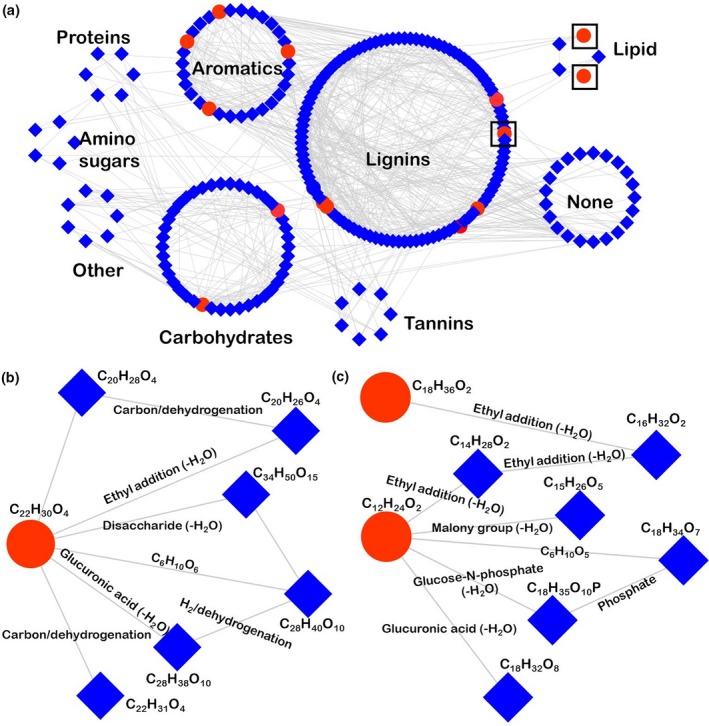
(a) Network analysis derived from FTICR‐MS (negative ion mode by direct injection) of the 17 antimicrobial compounds (dark purple circles) and 232 possible antimicrobial compounds (light purple squares) all detected in Sphagnum fallax. Possible antimicrobial compounds were determined by selecting first neighbors of the 17 identified antimicrobial compounds, that is, compounds that were one chemical reaction different between the pairs of m/z values (masses). Compounds are grouped by identified class, determined by oxygen:carbon and hydrogen:carbon ratios. Black squares represent the subnetworks have shown in b (polyphenols) and c (lipids) (b) Subnetwork of antimicrobial compound tetrahydrocannabinolic acid (circle) and all first neighbor compounds (squares) detected in Sphagnum fallax. (c) Subnetwork of antimicrobial compounds (circles) palmitic acid ethyl ester and dodecanoic acid, and all first neighbor compounds (squares) detected in Sphagnum fallax
4. DISCUSSION
A comprehensive, state‐of‐the‐art approach to Sphagnum fallax metabolite characterization was used in order to further understand the Sphagnum metabolome and highlight the chemical compounds underlying Sphagnum's dominance of peatlands. By using a combination of 1H NMR, LC‐MS/MS, LC‐FTICR‐MS/MS, and DI‐FTICR‐MS to identify known and unknown compounds specific to Sphagnum, we were able to provide an extensive chemical characterization of Sphagnum fallax that is more in depth than previous literature (PMN Plant Cyc, Sphagnum fallax). A total of 655 metabolites, including 152 lipids, were detected by NMR and LC‐MS/MS—329 of which were novel metabolites including 31 unknown lipids. Acids, sugar, and their derivatives (flavonoid glycosides) dominated Sphagnum chemical composition. The lipid composition confirmed previous literature reports (Baas et al., 2000; Klavina et al., 2018; Koskimies & Simola, 1980; Koskimies‐Soininen & Nyberg, 1987; Moore et al., 2015) and also demonstrated a surplus of sphingolipids and glycerolipids. Given the high‐resolution data generated by these analytical techniques, this chemical profile of Sphagnum fallax has implications for identifying biomarkers that can dictate organic matter transformations in peatlands and help understand the mechanisms by which Sphagnum inhibits microbial activity.
The presence and importance of the fungal microbiome of Sphagnum sp. has been outlined in more recent years, with most of these fungal–moss relationships defined as parasitic and pathogenic toward Sphagnum (Kostka et al., 2016). Stenroos et al. (2010), however, showed a possible symbiotic relationship between Sphagnum girgensohnii, nitrogen‐fixing cyanobacteria Nostoc, and Trizodia fungi, where the fungus provided a less hostile environment for the cyanobacteria to reside and perform nitrogen fixation for the moss and fungi benefit. Our data identified three fungal metabolites, membrane lipid ergosterol (used to measure fungal growth), and its derivatives ((8beta)‐6‐Allyl‐N‐[3‐(dimethylamino)propyl]ergoline‐8‐carboxamide and dehydroergosterol), as well as one fungal sphingolipid, MIPC. We also identified 57 nitrogen‐containing compounds. Since nitrogen can be a limiting nutrient in peatlands (Rousk et al., 2013), our data suggest the possibility of Sphagnum metabolites as nitrogen sources and, furthermore, a possible moss–bacterial–fungal nitrogen fixation relationship due to the plethora of N‐containing compounds and ergosterol signifying fungal growth. More research should be conducted to characterize fungal species and the overall microbiome of Sphagnum fallax in this context.
One of the large factors in slow decay and carbon sequestration in peatlands is Sphagnum's ability to create a hostile environment and inhibit decomposer growth/decomposition. It has been suggested that phenolic compounds readily react with nutrients in the environment making them unavailable for decomposers, and the amorphous phenolic polymer that covers the surface of Sphagnum cell wall forms a physical barrier against decomposition (Freeman et al., 2001; Hájek et al., 2011; Talbot & Treseder, 2012; Turetsky et al., 2008; Verhoeven & Liefveld, 1997). Yet of all the compounds detected, our data only classified two compounds as phenols, 2‐{Bis[(4‐phenyl‐1,3‐thiazol‐2‐yl)amino]methyl}phenol and 4‐tert‐Octylphenol monoethoxylate, detected in LC‐MS negative mode (Table S3), and one quinone detected via 1H NMR at a concentration of 8.4 µM (Table 2). While these three could be the players in antimicrobial properties, it is likely these are not fully responsible for decomposer suppression, which corresponds with previous research (Bengtsson et al., 2018; Mellegård et al., 2009).
Acidification and nutrient sequestration is likewise suggested to be a player in the antimicrobial nature of Sphagnum, where highly reactive cation exchange (carbonyl/carboxyl) groups react with nutrient sources in the environment making them unavailable to decomposers and subsequently releasing protons into the environment lowering the pH to <4.2 (Børsheim et al., 2001; Hájek et al., 2011; Painter, 1991; Stalheim et al., 2009). This has mostly been attributed to Sphagnan, Sphagnum acid, and uronic acids in previous literature (Stalheim et al., 2009; Verhoeven & Liefveld, 1997). Our data support this trend, with more than 60% of structures identified by 1H NMR containing carbonyl/carboxyl functional groups, 1,010 µM of Sphagnum contributed to acids, and the discovery of thirty‐six acids via LC‐MS not previously known to be present in Sphagnum. Furthermore, in the comparison of Sphagnum to medicinal plants, greater than half (9 of 17) of the identified potential antimicrobial compounds were acids. Further research is needed to fully understand whether these acids are acting to control pH of peatlands, but it is likely these acidic, high‐cation exchange compounds are acting by suppression of microbial species.
Interestingly, Sphagnum makeup was also dominated by sugar‐like derivatives (glycosidases) and other complex sugars, which can be broken down by microbes into simple sugars (free glucose) and used for metabolism. However, 11 complex sugars were detected by LC‐FTICR‐MS/MS, and of the seventeen potential antimicrobial compounds present in Sphagnum, seven were also complex sugars derivatives. The detection of these complex sugars and their derivatives in Sphagnum are likely attributed to three phenomena. First, it is possible that the presence of acids and therefore low pH inhibits microbial growth and degradation of complex sugars for energy, which would account for both the detection of complex sugars and wealth of acids. Secondly, these complex sugars could be playing a role in inhibiting microbial growth, which has been shown in previous literature for these seven sugar derivatives in other plants (Akroum et al., 2009; Chiruvella et al., 2007; Sati et al., 2019; Shi et al., 2016; Singh, Sharma, et al., 2011; del Valle et al., 2016). Finally, network analysis identified 211 known compounds and 21 unknown compounds that were first neighbors of these 17 antimicrobial compounds, suggesting there are potentially other compounds in the Sphagnum metabolome that contribute to antimicrobial effects. Our data indicate that it is likely all three scenarios contributing to the antimicrobial nature of Sphagnum, which is crucial knowledge to understand biological factors underlying slow decay rates, peatland carbon sequestration, and how the shift from Sphagnum to vascular plants may act on or change this effect. With further research, the presence and intensity of these compounds could be used as biomarkers of antimicrobial activity in peatlands and indicate changes in carbon cycling and greenhouse gas fluxes as climate shifts.
Peatland terrestrial ecosystems are the largest carbon sinks worldwide, driven largely by Sphagnum's dominance over other plant species, control of slow decay rates and water table fluctuations, and subsequent changes in redox chemistry of peat bogs (Tfaily et al., 2014). The processes by which Sphagnum controls C decomposition, accumulation, and transformation are still poorly understood and, while relevant, are not included in current climate models. Once structural biomarkers are defined and linked to carbon specific processes, shifts in organic matter composition, and decomposition changes due to water table fluctuations, we can use these biomarkers to predict and anticipate peatland C transformations. With our high‐resolution mass spectrometry and liquid state NMR approach, we found 298 novel metabolites and few hundreds of compounds previously unknown to be present in Sphagnum fallax, which highlights the gap of knowledge on this species. Our research provides a much‐needed baseline characterization that can be extrapolated upon to understand the chemical structures (biomarkers) and their functions that underlay carbon sequestration and will aid in predicting what will happen in the face of climate shift, vascular plant dominance, and subsequent greenhouse gas fluxes.
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
MMT and JF wrote the manuscript with contributions from all coauthors. MMT, TOM, and HMH designed the experiment. RC, JT, DH, NT, HH, RA, JL, HG, and JEK collected and processed the data. MMT and JF conducted the analysis and interpretation. TM, NH, and MMT provided funding and guidance.
Supporting information
ACKNOWLEDGMENTS
Metabolomics analyses were supported in part by the Microbiomes in Transition (MinT) Initiative as part of the Laboratory Directed Research and Development Program at PNNL. A portion of this research was performed using Environmental Molecular Sciences Laboratory a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at PNNL in Richland Washington. PNNL is a multi‐program national laboratory operated by Battelle for the DOE under contract DE‐AC05‐76RLO 1830. We would like to thank Rachel Wilson and David Weston for sample collection and the rest of SPRUCE team. The SPRUCE experiment is supported by the United States Department of Energy, Office of Science, Biological and Environmental Research under contract DE‐AC05‐00OR22725. Oak Ridge National Laboratory is managed by UT‐Battelle, LLC for the United States Department of Energy. We would also like to thank two anonyms reviewer for their constructive comments.
Fudyma JD, Lyon J, AminiTabrizi R, et al. Untargeted metabolomic profiling of Sphagnum fallax reveals novel antimicrobial metabolites. Plant Direct. 2019;3:1–17. 10.1002/pld3.179
REFERENCES
- Abedini, A. , Roumy, V. , Mahieux, S. , Biabiany, M. , Standaert‐Vitse, A. , Rivière, C. , … Hennebelle, T. (2013). Rosmarinic acid and its methyl ester as antimicrobial components of the hydromethanolic extract of Hyptis atrorubens Poit. (Lamiaceae). Evidence‐based Complementary and Alternative Medicine, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S. I. , Hayat, M. Q. , Tahir, M. , Mansoor, Q. , Ismail, M. , Keck, K. , & Bates, R. B. (2016). Pharmacologically active flavonoids from the anticancer, antioxidant and antimicrobial extracts of Cassia angustifolia Vahl. BMC Complementary and Alternative Medicine, 16, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akroum, S. , Bendjeddou, D. , Satta, D. , & Lalaoui, K. (2009). Antibacterial activity and acute toxicity effect of flavonoids extracted from Mentha longifolia . American‐Eurasian Journal of Scientific Research, 4, 93–96. [Google Scholar]
- Appendino, G. , Gibbons, S. , Giana, A. , Pagani, A. , Grassi, G. , Stavri, M. , … Rahman, M. M. (2008). Antibacterial cannabinoids from Cannabis sativa: A structure‐activity study. Journal of Natural Products, 71, 1427–1430. [DOI] [PubMed] [Google Scholar]
- Armentano, T. V. , & Menges, E. S. (1986). Patterns of change in the carbon balance of organic soil‐wetlands of the temperate zone. Journal of Ecology, 74, 755 10.2307/2260396 [DOI] [Google Scholar]
- Baas, M. , Pancost, R. , Van Geel, B. , & Sinninghe Damsté, J. S. (2000). A comparative study of lipids in Sphagnum species. Organic Geochemistry, 31, 535–541. 10.1016/S0146-6380(00)00037-1 [DOI] [Google Scholar]
- Bai, J. , Wu, Y. , Zhong, K. , Xiao, K. , Liu, L. , Huang, Y. , … Gao, H. (2018). A comparative study on the effects of quinic acid and shikimic acid on cellular functions of Staphylococcus aureus . Journal of Food Protection, 81, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Ballance, S. , Børsheim, K. Y. , Inngjerdingen, K. , Paulsen, B. S. , & Christensen, B. E. (2007). A re‐examination and partial characterisation of polysaccharides released by mild acid hydrolysis from the chlorite‐treated leaves of Sphagnum papillosum . Carbohydrate Polymers, 67, 104–115. 10.1016/j.carbpol.2006.04.020 [DOI] [Google Scholar]
- Bengtsson, F. , Granath, G. , & Rydin, H. (2016). Photosynthesis, growth, and decay traits in Sphagnum ‐ a multispecies comparison. Ecology and Evolution, 6, 3325–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson, F. , Rydin, H. , & Hájek, T. (2018). Biochemical determinants of litter quality in 15 species of Sphagnum . Plant and Soil, 425, 161–176. 10.1007/s11104-018-3579-8 [DOI] [Google Scholar]
- Black, W. A. P. , Cornhill, W. J. , & Woodward, F. N. (1955). A preliminary investigation on the chemical composition of Sphagnum moss and peat. Journal of Applied Chemistry, 5, 484–493. 10.1002/jctb.5010050907 [DOI] [Google Scholar]
- Børsheim, K. Y. , Christensen, B. E. , & Painter, T. J. (2001). Preservation of fish by embedment in Sphagnum moss, peat or holocellulose: Experimental proof of the oxopolysaccharidic nature of the preservative substance and of its antimicrobial and tanning action. Innovative Food Science and Emerging Technologies, 2, 63–74. [Google Scholar]
- Breitling, R. , Ritchie, S. , Goodenowe, D. , Stewart, M. L. , & Barrett, M. P. (2006). Ab initio prediction of metabolic networks using fourier transform mass spectrometry data. Metabolomics, 2, 155–164. 10.1007/s11306-006-0029-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, K. E. V. , Borutzki, Y. , Rankin, N. , Daly, R. , & Jourdan, F. (2017). MetaNetter 2: A Cytoscape plugin for ab initio network analysis and metabolite feature classification. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 1071, 68–74. 10.1016/j.jchromb.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanton, J. P. , Glaser, P. H. , Chasar, L. S. , Burdige, D. J. , Hines, M. E. , Siegel, D. I. , … Cooper, W. T. (2008). Radiocarbon evidence for the importance of surface vegetation on fermentation and methanogenesis in contrasting types of boreal peatlands. Global Biogeochemical Cycles, 22 10.1029/2008GB003274 [DOI] [Google Scholar]
- Chasar, L. S. , Chanton, J. P. , Glaser, P. H. , Siegel, D. I. , & Rivers, J. S. (2000). Radiocarbon and stable carbon isotopic evidence for transport and transformation of dissolved organic carbon, dissolved inorganic carbon, and CH4 in a northern Minnesota peatland. Global Biogeochemical Cycles, 14, 1095–1108. [Google Scholar]
- Chiruvella, K. K. , Mohammed, A. , Dampuri, G. , Ghanta, R. G. , & Raghavan, S. C. (2007). Phytochemical and antimicrobial studies of methyl angolensate and luteolin‐7‐O‐glucoside isolated from callus cultures of Soymida febrifuga . International Journal of Biomedical Science, 3, 269–278. [PMC free article] [PubMed] [Google Scholar]
- Clymo, R. S. (1978). A model of peat bog growthIn Heal O. W. & Perkins D. F. (Eds.), Production ecology of British moors and montane grasslands. Ecological studies (analysis and synthesis) (pp. 187–223). Berlin, Heidelberg: Springer. [Google Scholar]
- Cowan, M. M. (1999). Plant products as antimicrobial agents. Clinical Microbiology Reviews, 12, 564–582. 10.1128/CMR.12.4.564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalcin Martins, P. , Hoyt, D. W. , Bansal, S. , Mills, C. T. , Tfaily, M. , Tangen, B. A. , … Wilkins, M. J. (2017). Abundant carbon substrates drive extremely high sulfate reduction rates and methane fluxes in prairie pothole wetlands. Global Change Biology, 23, 3107–3120. 10.1111/gcb.13633 [DOI] [PubMed] [Google Scholar]
- del Valle, P. , García‐Armesto, M. R. , de Arriaga, D. , González‐Donquiles, C. , Rodríquez‐Fernández, P. , & Rúa, J. (2016). Antimicrobial activity of kaempferol and resveratrol in binary combinations with parabens or propyl gallate against Enterococcus faecalis . Food Control, 61, 213–220. 10.1016/j.foodcont.2015.10.001 [DOI] [Google Scholar]
- Fenner, N. , & Freeman, C. (2011). Drought‐induced carbon loss in peatlands. Nature Geoscience, 4, 895–900. 10.1038/ngeo1323 [DOI] [Google Scholar]
- Fiamegos, Y. C. , Kastritis, P. L. , Exarchou, V. , Han, H. , Bonvin, A. M. J. J. , Vervoort, J. , … Tegos, G. P. (2011). Antimicrobial and efflux pump inhibitory activity of caffeoylquinic acids from Artemisia absinthium against gram‐positive pathogenic bacteria. PLoS ONE, 6(4), e18127 10.1371/journal.pone.0018127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch, J. , Lees, M. , & Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. The Journal of Biological Chemistry, 226, 497–509. [PubMed] [Google Scholar]
- Frainer, A. , & McKie, B. G. (2015). Advances in ecological research In Pawar S., Woodward G., & Dell A. I. (Eds.), Trait‐based ecology – From structure to function (pp. 169–200). London, UK: Academic Press. [Google Scholar]
- Freeman, C. , Evans, C. D. , Monteith, D. T. , Reynolds, B. , & Fenner, N. (2001). Export of organic carbon from peat soils. Nature, 412, 785–785. 10.1038/35090628 [DOI] [PubMed] [Google Scholar]
- Freeman, C. , Ostle, N. , & Kang, H. (2002). An enzymic ‘latch’ on a global carbon store. Nature, 409, 149–149. 10.1038/35051650 [DOI] [PubMed] [Google Scholar]
- Frolking, S. , Talbot, J. , Jones, M. C. , Treat, C. C. , Kauffman, J. B. , Tuittila, E.‐S. , & Roulet, N. (2011). Peatlands in the Earth’s 21st century climate system. Environmental Reviews, 19, 371–396. 10.1139/a11-014 [DOI] [Google Scholar]
- Gorham, E. (1991). Northern peatlands: Role in the carbon cycle and probable responses to climatic warming. Ecological Applications, 1, 182–195. 10.2307/1941811 [DOI] [PubMed] [Google Scholar]
- Gulis, V. , Kuehn, K. A. , & Suberkropp, K. (2009). Fungi. Encyclopedia of Inland Waters, 233–243. [Google Scholar]
- Hájek, T. , Ballance, S. , Limpens, J. , Zijlstra, M. , & Verhoeven, J. T. A. (2011). Cell‐wall polysaccharides play an important role in decay resistance of Sphagnum and actively depressed decomposition in vitro. Biogeochemistry, 103, 45–57. 10.1007/s10533-010-9444-3 [DOI] [Google Scholar]
- Huang, C. B. , George, B. , & Ebersole, J. L. (2010). Antimicrobial activity of n‐6, n‐7 and n‐9 fatty acids and their esters for oral microorganisms. Archives of Oral Biology, 55, 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel, L. , & Garamvölgyi, Á. (2014). Impacts of climate change on vegetation distribution No. 2: Climate change induced vegetation shifts in the new world. Applied Ecology and Environmental Research, 12, 355–422. 10.15666/aeer/1202_355422 [DOI] [Google Scholar]
- Kandakumar, S. , & Manju, D. V. (2017). Pharmacological applications of isorhamnetin: A short review. International Journal of Trend in Scientific Research and Development, 1, 672–676. 10.31142/ijtsrd2202 [DOI] [Google Scholar]
- Karunen, P. , & Ekman, R. (1982). Age‐dependent content of polymerized lipids in Sphagnum fuscum . Physiologia Plantarum, 54, 162–166. 10.1111/j.1399-3054.1982.tb06320.x [DOI] [Google Scholar]
- Kim, S. , Kramer, R. W. , & Hatcher, P. G. (2003). Graphical method for analysis of ultrahigh‐resolution broadband mass spectra of natural organic matter, the Van Krevelen diagram. Analytical Chemistry, 75, 5336–5344. [DOI] [PubMed] [Google Scholar]
- Klavina, L. , Springe, G. , Steinberga, I. , Mezaka, A. , & Ievinsh, G. (2018). Seasonal changes of chemical composition in boreonemoral moss species. Environmental and Experimental Biology, 16, 9–19. [Google Scholar]
- Kohlwein, S. D. (2017). Analyzing and understanding lipids of yeast: A challenging endeavor. Cold Spring Harbor Protocols, , 2017(5). 10.1101/pdb.top078956 [DOI] [PubMed] [Google Scholar]
- Kolka, R. (2011). Peatland Biogeochemistry and Watershed Hydrology at the Marcell Experimental Forest. Boca Raton, FL: CRC Press. [Google Scholar]
- Koskimies, K. , & Simola, L. K. (1980). The fatty acid composition of some Sphagnum species. Canadian Journal of Botany, 58, 259–263. [Google Scholar]
- Koskimies‐Soininen, K. , & Nyberg, H. (1987). Effects of temperature and light on the lipids of Sphagnum magellanicum . Phytochemistry, 26, 2213–2221. 10.1016/S0031-9422(00)84686-5 [DOI] [Google Scholar]
- Kostka, J. E. , Weston, D. J. , Glass, J. B. , Lilleskov, E. A. , Shaw, A. J. , & Turetsky, M. R. (2016). The Sphagnum microbiome: New insights from an ancient plant lineage. New Phytologist, 211, 57–64. [DOI] [PubMed] [Google Scholar]
- Kyle, J. E. , Crowell, K. L. , Casey, C. P. , Fujimoto, G. M. , Kim, S. , Dautel, S. E. , … Metz, T. O. (2017). LIQUID: An‐open source software for identifying lipids in LC‐MS/MS‐based lipidomics data. Bioinformatics, 33, 1744–1746. 10.1093/bioinformatics/btx046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens, J. , Berendse, F. , Blodau, C. , Canadell, J. G. , Freeman, C. , Holden, J. , … Schaepman‐Strub, G. (2008). Peatlands and the carbon cycle: From local processes to global implications ‐ A synthesis. Biogeosciences, 5, 1475–1739. 10.5194/bg-5-1475-2008 [DOI] [Google Scholar]
- Limpens, J. , Bohlin, E. , & Nilsson, M. B. (2017). Phylogenetic or environmental control on the elemental and organo‐chemical composition of Sphagnum mosses? Plant and Soil, 417, 69–85. 10.1007/s11104-017-3239-4 [DOI] [Google Scholar]
- Longnecker, K. , & Kujawinski, E. B. (2016). Using network analysis to discern compositional patterns in ultrahigh‐resolution mass spectrometry data of dissolved organic matter. Rapid Communications in Mass Spectrometry, 30, 2388–2394. 10.1002/rcm.7719 [DOI] [PubMed] [Google Scholar]
- Lou, Z. , Wang, H. , Rao, S. , Sun, J. , Ma, C. , & Li, J. (2012). P‐coumaric acid kills bacteria through dual damage mechanisms. Food Control, 25, 550–554. 10.1016/j.foodcont.2011.11.022 [DOI] [Google Scholar]
- Mazimba, O. (2017). Umbelliferone: Sources, chemistry and bioactivities review. Bulletin of Faculty of Pharmacy, Cairo University, 55, 223–232. [Google Scholar]
- Mellegård, H. , Stalheim, T. , Hormazabal, V. , Granum, P. E. , & Hardy, S. P. (2009). Antibacterial activity of sphagnum acid and other phenolic compounds found in Sphagnum papillosum against food‐borne bacteria. Letters in Applied Microbiology, 49, 85–90. [DOI] [PubMed] [Google Scholar]
- Mikami, K. , & Hartmann, E. (2004). Lipid metabolism in mosses In Wood A.J., Oliver M.J., & Cove D.J. (Eds.),New frontiers in bryology (pp. 133–155). Norwell, MA: Kluwer Academic Publishers; [Google Scholar]
- Moore, E. K. , Villanueva, L. , Hopmans, E. C. , Rijpstra, W. I. C. , Mets, A. , Dedysh, S. N. , & Sinninghe Damsté, J. S. (2015). Abundant trimethylornithine lipids and specific gene sequences are indicative of planctomycete importance at the oxic/anoxic interface in Sphagnum‐dominated northern wetlands. Applied and Environmental Microbiology, 81, 6333–6344. 10.1128/AEM.00324-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji, T. , Kao, M. C. , Fang, J. Y. , Zouboulis, C. C. , Zhang, L. , Gallo, R. L. , & Huang, C. M. (2009). Antimicrobial property of lauric acid against propionibacterium acnes: Its therapeutic potential for inflammatory acne vulgaris. Journal of Investigative Dermatology, 129, 2480–2488. 10.1038/jid.2009.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan, B. , Ohlan, S. , Ohlan, R. , Judge, V. , & Narang, R. (2009). Hansch analysis of veratric acid derivatives as antimicrobial agents. European Journal of Medicinal Chemistry, 44, 689–700. 10.1016/j.ejmech.2008.05.008 [DOI] [PubMed] [Google Scholar]
- Oliveira‐Alves, S. C. , Vendramini‐Costa, D. B. , Betim Cazarin, C. B. , Maróstica Júnior, M. R. , Borges Ferreira, J. P. , Silva, A. B. , … Bronze, M. R. (2017). Characterization of phenolic compounds in chia (Salvia hispanica L.) seeds, fiber flour and oil. Food Chemistry, 232, 295–305. [DOI] [PubMed] [Google Scholar]
- Ortega‐Ramirez, L. A. , Rodriguez‐Garcia, I. , Leyva, J. M. , Cruz‐Valenzuela, M. R. , Silva‐Espinoza, B. A. , Gonzalez‐Aguilar, G. A. , … Ayala‐Zavala, J. F. (2014). Potential of medicinal plants as antimicrobial and antioxidant agents in food industry: A hypothesis. Journal of Food Science, 79 10.1111/1750-3841.12341 [DOI] [PubMed] [Google Scholar]
- Painter, T. J. (1991). Lindow man, tollund man and other peat‐bog bodies: The preservative and antimicrobial action of sphagnan, a reactive glycuronoglycan with tanning and sequestering properties. Carbohydrate Polymers, 15, 123–142. [Google Scholar]
- Pluskal, T. , Castillo, S. , Villar‐Briones, A. , & Orešič, M. (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry‐based molecular profile data. BMC Bioinformatics, 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz, M. , Zia‐Ul‐Haq, M. , & Jaafar, H. Z. E. (2013). Common mullein, pharmacological and chemical aspects. Brazilian Journal of Pharmacognosy, 23. [Google Scholar]
- Rousk, K. , Jones, D. L. , & DeLuca, T. H. (2013). Moss‐cyanobacteria associations as biogenic sources of nitrogen in boreal forest ecosystems. Frontiers in Microbiology, 4, 10.3389/fmicb.2013.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sati, P. , Dhyani, P. , Bhatt, I. D. , & Pandey, A. (2019). Ginkgo biloba flavonoid glycosides in antimicrobial perspective with reference to extraction method. Journal of Traditional and Complementary Medicine, 9, 15–23. 10.1016/j.jtcme.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N. S. , Wang, J. T. , Ramage, D. , … Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, C. , Sun, Y. I. , Zheng, Z. , Zhang, X. , Song, K. , Jia, Z. , … Xia, X. (2016). Antimicrobial activity of syringic acid against Cronobacter sakazakii and its effect on cell membrane. Food Chemistry, 197, 100–106. 10.1016/j.foodchem.2015.10.100 [DOI] [PubMed] [Google Scholar]
- Singh, D. , Sharma, S. K. , Rani, R. , Mishra, S. , & Sharma, R. A. (2011). Kaempferol‐7‐o‐glucoside and their antimicrobial screening isolate from cassia renigera wall. International Journal of Pharmaceutical and Clinical Research, 3, 30–34. [Google Scholar]
- Singh, O. , Khanam, Z. , Misra, N. , & Srivastava, M. (2011). Chamomile (Matricaria chamomilla L.): An overview. Pharmacognosy Reviews, 5, 82–95. 10.4103/0973-7847.79103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjors, H. (1981). The zonation of northern peatlands and their importance for the carbon balance of the atmosphere. International Journal of Ecology and Environmental Sciences, 7, 11–14. [Google Scholar]
- Stalheim, T. , Ballance, S. , Christensen, B. E. , & Granum, P. E. (2009). Sphagnan ‐ a pectin‐like polymer isolated from Sphagnum moss can inhibit the growth of some typical food spoilage and food poisoning bacteria by lowering the pH. Journal of Applied Microbiology, 106, 967–976. [DOI] [PubMed] [Google Scholar]
- Stenroos, S. , Laukka, T. , Huhtinen, S. , Döbbeler, P. , Myllys, L. , Syrjänen, K. , & Hyvönen, J. (2010). Multiple origins of symbioses between ascomycetes and bryophytes suggested by a five‐gene phylogeny. Cladistics, 26, 281–300. 10.1111/j.1096-0031.2009.00284.x [DOI] [PubMed] [Google Scholar]
- Surjushe, A. , Vasani, R. , & Saple, D. (2009). Aloe vera: A short review. Indian Journal of Dermatology, 53, 163–166. 10.4103/0019-5154.44785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, J. M. , & Treseder, K. K. (2012). Interactions among lignin, cellulose, and nitrogen drive litter chemistry‐decay relationships. Ecology, 93, 345–354. [DOI] [PubMed] [Google Scholar]
- Tao, B.Y. , (2007). Industrial applications for plant oils and lipids In Yang S.T. (Ed.), Bioprocessing for value-added products from renewable resources (pp. 611–627). Amsterdam: Elsevier. [Google Scholar]
- Tfaily, M. M. , Chu, R. K. , Tolić, N. , Roscioli, K. M. , Anderton, C. R. , Paša‐Tolić, L. , … Hess, N. J. (2015). Advanced solvent based methods for molecular characterization of soil organic matter by high‐resolution mass spectrometry. Analytical Chemistry, 87(10), 5206–5215. 10.1021/acs.analchem.5b00116 [DOI] [PubMed] [Google Scholar]
- Tfaily, M. M. , Chu, R. K. , Toyoda, J. , Tolić, N. , Robinson, E. W. , Paša‐Tolić, L. , & Hess, N. J. (2017). Sequential extraction protocol for organic matter from soils and sediments using high resolution mass spectrometry. Analytica Chimica Acta, 972, 54–61. 10.1016/j.aca.2017.03.031 [DOI] [PubMed] [Google Scholar]
- Tfaily, M. M. , Cooper, W. T. , Kostka, J. E. , Chanton, P. R. , Schadt, C. W. , Hanson, P. J. , … Chanton, J. P. (2014). Organic matter transformation in the peat column at Marcell Experimental Forest: Humification and vertical stratification. Journal of Geophysical Research: Biogeosciences, 119(4), 661–675. 10.1002/2013JG002492 [DOI] [Google Scholar]
- Tfaily, M. M. , Hess, N. J. , Koyama, A. , & Evans, R. D. (2018). Elevated [CO2] changes soil organic matter composition and substrate diversity in an arid ecosystem. Geoderma, 330, 1–8. 10.1016/j.geoderma.2018.05.025 [DOI] [Google Scholar]
- Tolić, N. , Liu, Y. , Liyu, A. , Shen, Y. , Tfaily, M. M. , Kujawinski, E. B. , … Hess, N. J. (2017). Formularity: Software for automated formula assignment of natural and other organic matter from ultrahigh‐resolution mass spectra. Analytical Chemistry, 89, 12659–12665. 10.1021/acs.analchem.7b03318 [DOI] [PubMed] [Google Scholar]
- Turetsky, M. R. (2018). The role of bryophytes in carbon and nitrogen cycling. The Bryologist, 106, 395–409. 10.1639/05 [DOI] [Google Scholar]
- Turetsky, M. R. , Crow, S. E. , Evans, R. J. , Vitt, D. H. , & Wieder, R. K. (2008). Trade‐offs in resource allocation among moss species control decomposition in boreal peatlands. Journal of Ecology, 96, 1297–1305. 10.1111/j.1365-2745.2008.01438.x [DOI] [Google Scholar]
- van Breemen, N. (1995). How Sphagnum bogs down other plants. Trends in Ecology & Evolution, 10, 270–275. 10.1016/0169-5347(95)90007-1 [DOI] [PubMed] [Google Scholar]
- Verhoeven, J. T. A. , & Liefveld, W. M. (1997). The ecological significance of organochemical compounds in Sphagnum . Acta Botanica Neerlandica, 46, 117–130. [Google Scholar]
- Wallén, B. O. , & Wallen, B. O. (1986). Above and below ground dry mass of the three main vascular plants on hummocks on a subarctic peat bog. Oikos, 46, 51–56. 10.2307/3565379 [DOI] [Google Scholar]
- Weljie, A. M. , Newton, J. , Mercier, P. , Carlson, E. , & Slupsky, C. M. (2006). Targeted pofiling: Quantitative analysis of 1H NMR metabolomics data. Analytical Chemistry, 78, 4430–4442. [DOI] [PubMed] [Google Scholar]
- Yang, C. H. , Yang, C.‐S. , Hwang, M.‐L. , Chang, C.‐C. , Li, R. X. , & Chuang, L. Y. (2012). Antimicrobial activity of various parts of Cinnamomum cassia extracted with different extraction methods. Journal of Food Biochemistry, 36, 690–698. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


