Abstract
NKAP is a ubiquitously expressed nucleoplasmic protein that is currently known as a transcriptional regulatory molecule via its interaction with HDAC3 and spliceosomal proteins. Here, we report a disorder of transcriptional regulation due to missense mutations in the X chromosome gene, NKAP. These mutations are clustered in the C-terminal region of NKAP where NKAP interacts with HDAC3 and post-catalytic spliceosomal complex proteins. Consistent with a role for the C-terminal region of NKAP in embryogenesis, nkap mutant zebrafish with a C-terminally truncated NKAP demonstrate severe developmental defects. The clinical features of affected individuals are highly conserved and include developmental delay, hypotonia, joint contractures, behavioral abnormalities, Marfanoid habitus, and scoliosis. In affected cases, transcriptome analysis revealed the presence of a unique transcriptome signature, which is characterized by the downregulation of long genes with higher exon numbers. These observations indicate the critical role of NKAP in transcriptional regulation and demonstrate that perturbations of the C-terminal region lead to developmental defects in both humans and zebrafish.
Keywords: spliceosome, transcriptome, P-complex, splicing
Main Text
We have identified a cohort of ten affected males from eight families who carry missense mutations in the X chromosome gene NKAP (MIM: 300766). Two of these individuals, subjects 7 and 9, were previously reported with minimal clinical detail, one in a cohort of individuals with a potential Lujan-Fryns syndrome (MIM: 309520),1 and one in a cohort of individuals with intellectual disability.2 The remaining eight individuals with NKAP mutations were identified independently by exome sequencing and recruited through GeneMatcher3 and collaborating clinicians. All individuals were enrolled in the research study under an institutional review board-protocol. The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional review boards), and informed consent was obtained from the parents/guardians of the affected individuals.
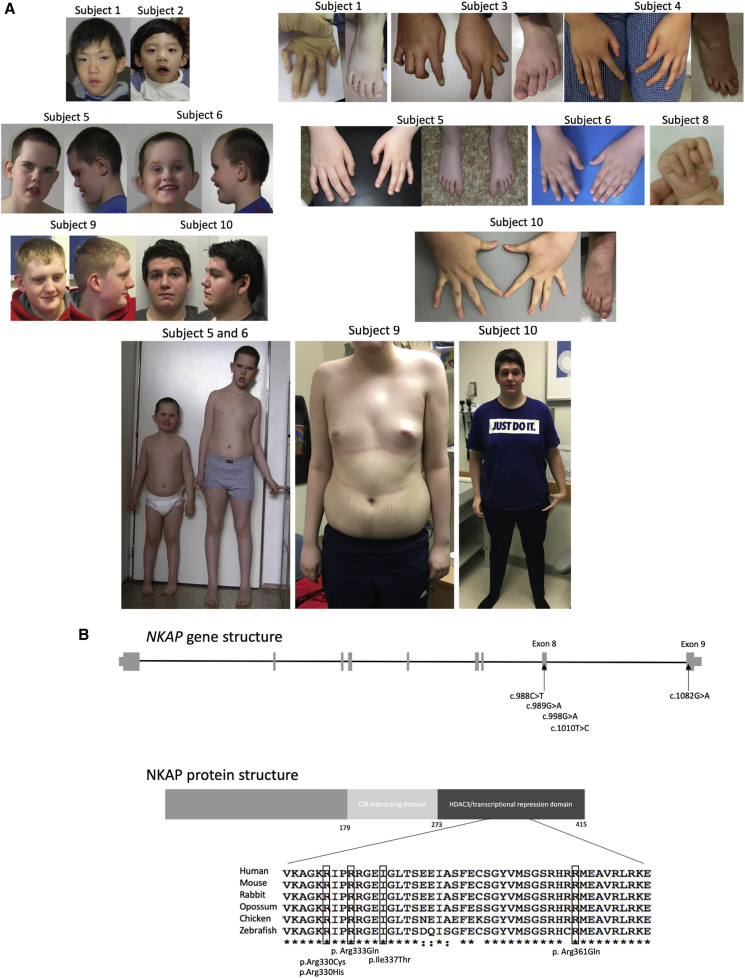
The clinical features of these ten affected males were highly conserved. All subjects had developmental delay/intellectual disability (ID), tall stature with Marfanoid habitus, and hypotonia. Shared facial dysmorphisms included: a long face (8/10), open-mouth appearance (7/10), midface hypoplasia (7/10), prominent or large ears (8/10), and a short philtrum (7/10). Musculoskeletal findings were common, with 5 of 10 reported to have pectus carinatum or excavatum, 6 of 10 with scoliosis, and 8 of 10 with arachnodactyly. Talipes equinovarus was identified in one subject. Other less common features included: genitourinary anomalies such as cryptorchidism (4/10), central obesity (5/10), and behavioral abnormalities that included attention deficit hyperactivity disorder (ADHD) or aggressive behaviors (5/10; Figure 1A, Tables 1 and S1, and Supplemental Note). Cardiac manifestations included mitral valve regurgitation (3/10), atrial septal defect (1/10), ventricular septal defect (1/10), small patent ductus arteriosus (1/10), and aortic dilatation (1/10). Although developmental delay or learning disability and tall stature/Marfanoid habitus were observed in all subjects with NKAP mutations, the other features, as noted above, were more variable. Mothers carrying NKAP mutations were unaffected or much less severely affected. While NKAP has been shown to play a role in chromosome alignment during mitosis,4 no chromosomal abnormalities were seen in the subjects with NKAP mutations. Similarly, although NKAP has been demonstrated to be a key developmental regulator of the hematopoietic and immune system,5 none of the subjects with NKAP mutations were known to have hematological or immunological manifestations.
Figure 1.
Clinical Phenotype of the Subjects with NKAP Mutations
(A) Clinical pictures of the subjects. Written permission to publish the photographs was obtained from the parents of the subjects.
(B) Location of NKAP mutations. Identified mutations are located within exon 8 and exon 9 of NKAP. The missense mutations alter highly conserved amino acids. CIR and HDAC3 binding domains of NKAP are depicted.5
Table 1.
Clinical Features of Individuals with NKAP Mutations
|
Subject 1 |
Subject 2 |
Subject 3 |
Subject 4 |
Subject 5 |
Subject 6 |
Subject 7 |
Subject 8 |
Subject 9 |
Subject 10 |
|
|---|---|---|---|---|---|---|---|---|---|---|
| NKAP Mutation |
p.Arg330Cys |
p.Arg330His |
p.Arg333Gln |
p.Arg333Gln |
p.Arg333Gln |
p.Arg333Gln |
p.Ile337Thr |
p.Arg361Gln |
||
|
c.988C>T |
c.989G>A |
c.998G>A |
c.998G>A |
c.998G>A |
c.998G>A |
c.998G>A |
c.998G>A |
c.1010T>C |
c.1082G>A |
|
| de novo | de novo | apparently de novo | maternally inherited | maternally inherited | unk | de novo | presumably de novo | |||
| Neurological | ||||||||||
| Developmental delay/intellectual disability | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| ADHD/aggressive behavior | no | no | no | yes | yes | yes | no | no | yes | yes |
| Hypotonia | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Dysmorphisms | ||||||||||
| Long face | yes | yes | yes | yes | yes | no | yes | unk | yes | yes |
| Short philtrum | yes | yes | yes | yes | no | yes | yes | unk | yes | no |
| Open mouth appearance | yes | yes | yes | yes | yes | yes | yes | unk | no | no |
| Midface hypoplasia | no | yes | yes | yes | yes | yes | yes | unk | yes | no |
| Prominent/large ears | yes | yes | no | yes | yes | yes | yes | unk | yes | yes |
| Cardiac | ||||||||||
| Pathology (age at the last evaluation) | atrial septal defect (10 yo) | small patent ductal arteriosus (6 yo) | mild mitral valve regurgitation (18 yo) | mitral valve prolapse, mitral regurgitation and aortic root dilatation (10 yo) | no (11 yo) | no (5 yo) | mitral valves prolapse with the minimal mitral regurgitation (21 yo) | history of ventricular septal defect (7 yo) | no (10 yo) | no (16 yo) |
| Skeletal | ||||||||||
| Tall stature | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Scoliosis | yes | yes | yes | yes | yes | no | yes | no | no | no |
| Pectus | yes | no | no | no | carinatum | carinatum | carinatum | no | excavatum | no |
| Slender limbs | yes | yes | yes | yes | yes | no | yes | yes | yes | yes |
| Joint laxity | yes | yes | yes | yes | yes | no | yes | yes | yes | yes |
| Camptodactyly | yes | no | yes | no | no | no | yes | yes | no | no |
| Arachnodactyly | yes | yes | yes | yes | no | no | yes | yes | yes | yes |
Unk, unknown. Full table can be found as Table S1.
Four affected males had apparently de novo NKAP variants, three had maternally inherited variants (subjects 5 and 6 and the unrelated subject 7), and inheritance for one subject (subject 8) was unknown. Two brothers (subject 3 and 4) apparently had de novo inheritance of the variant, consistent with maternal germline mosaicism. We identified five different missense mutations in the ten subjects (c.988C>T [p.Arg330Cys], 1 male; c.989G>A [p.Arg330His], 1 male; c.G998A [p.Arg333Gln], 6 males from 4 families; c.1010T>C [p.Ile337Thr], 1 male; and c.1082G>A [p.Arg361Gln], 1 male). All variants were located in exons 8 and 9 of NKAP (GenBank: NM_024528) encoding the C-terminal part of the protein, and all altered highly conserved amino acids (Table 1, Figure 1B).
NKAP encodes a ubiquitously expressed, nuclear speckle protein, initially reported as a mediator of NF-κB signaling.6 Subsequently, diverse biological roles of NKAP have been reported, including transcriptional regulation and chromosome alignment.4, 5 NKAP was suggested to be involved in mRNA splicing,7 and very recently, NKAP was shown to be a part of post-catalytic P-complex, a large splicing complex responsible for neighboring exonic ligation and processed mRNA release.8
To analyze the effect of identified NKAP mutations, we measured NKAP mRNA and protein levels in lymphoblastoid cell line (LCL) samples from subjects with NKAP mutations compared to control samples and did not note any differences (Figure S1). Immunofluorescence showed no differences in intracellular distribution of NKAP between control and NKAP mutant samples (Figure S1). We could confirm previous findings that NKAP localizes in a nuclear speckled pattern, which contain nucleoplasmic granules enriched in mRNA processing proteins such as splicing factors.7
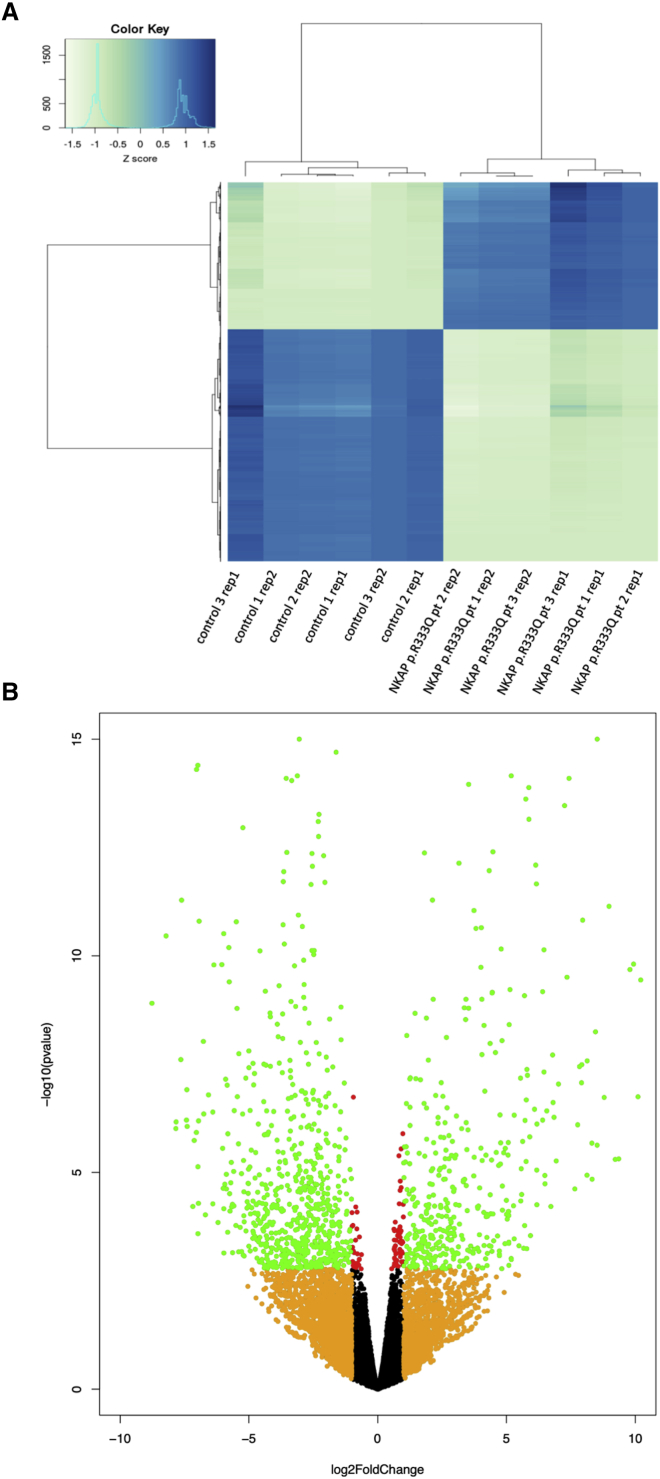
Given the previously reported functions of NKAP, we hypothesized that these NKAP mutations alter transcription. Transcriptome analysis was performed using RNA sequencing (RNA-seq) of LCLs from case and control subjects. RNA-seq was initially performed using three LCLs with the p.Arg333Gln substitution and three control LCLs. Clustering analysis revealed that subjects with NKAP mutations exhibited a consistent unique, disrupted transcriptome profile, with 455 upregulated genes and 721 downregulated genes compared to control lines (FDR < 0.05) (Table S2). This suggests that the majority of differentially expressed genes (DEGs) are downregulated in NKAP mutant samples (Figures 2A and 2B). DEGs include HES1 and JAG1, Notch signaling pathway genes. The RNA-seq results were validated by qRT-PCR of two DEGs (HES1 and APP) (Figure S2). RNA-seq was also performed using LCL with the p.Arg361Gln substitution, and it demonstrated a similar DEG profile (Figure S3). Pathway analysis for the DEGs identified in subjects with NKAP mutations was performed with DAVID Functional Annotation Bioinformatics Microarray Analysis (Table S3). Terms such as “extracellular matrix” were enriched in the NKAP mutant DEGs. Because individuals with NKAP mutations demonstrated features associated with connective tissue disorder, misexpression of these genes may be related to their clinical features. Previously, the Notch signaling pathway was shown to be a transcriptional target of NKAP and mediates the various signaling pathways, such as TGF-β, involved in regulating the extracellular matrix gene expression.5, 9, 10 Therefore, alterations in the Notch signaling pathway may be involved in the mechanism of this transcriptome profile.
Figure 2.
Transcriptome Profile of the Genetic Disorder due to NKAP Mutations
RNA sequencing reveals 455 upregulated and 721 downregulated genes in subjects with NKAP mutations. RNA sequencing was performed using 3 subject with NKAP p.Arg333Gln substitution and control LCL samples. Biological duplicates were included for each sample.
(A) Hierarchical clustering and heatmap demonstrated the presence of a unique transcriptome profile seen in the subjects with NKAP mutations. Differentially expressed genes in the LCLs with NKAP mutations defined by FDR < 0.05 are depicted. Dark blue color indicates higher gene expression, and light green color indicates lower gene expression. The distances between two clusters are depicted on the top and left of the heatmap.
(B) Volcano plot demonstrated a larger number of downregulated genes compared to upregulated genes. Red dots represent genes with FDR < 0.05. Orange dots represent its log fold expression changes (log2FC) greater than 1. Green dots represent genes whose FDR < 0.05 and log2 FC > 1.
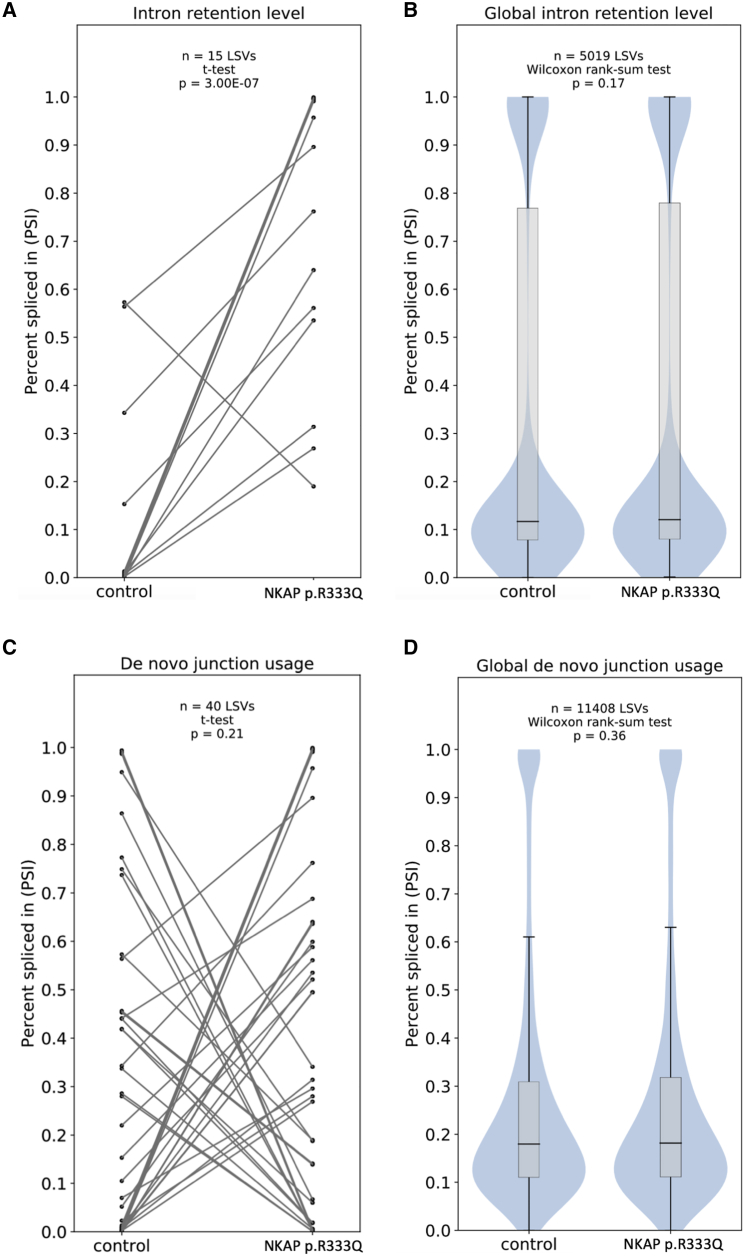
NKAP localizes to the post-catalytic P-complex, whose function is exonic ligation and mRNA release after 5′ and 3′ splice site selection.8 Therefore, although NKAP is a part of the spliceosomal complex, the dysfunction of NKAP may not cause increased aberrant or alternative splicing. Consistent with this notion, splicing analysis with MAJIQ11 did not identify major splicing pattern alterations with little to no alternative splicing, intron retention, or de novo (unannotated) splicing junction usage changes (Figure 3, Table S4).
Figure 3.
NKAP p.Arg333Gln Has Minimal Impact on Splicing Regulation
(A) Level of intron retention is increased among splicing variations with changes > 15 percent spliced in (dPSI) between control and mutant cells.
(B) Intron retention is not affected on a global level by the NKAP mutation.
(C) De novo (unannotated) junctions are not preferentially used in NKAP mutant cells for splicing variations with dPSI > 15 between control and mutant cells.
(D) De novo junction usage is not affected on a global level by the NKAP mutation.
We found only 39 splicing variations with changes in percent spliced in (PSI) of more than 15 and with a probability that dPSI is above 15 (P(|dPSI|> = 0.15)) of more than 0.95 in control versus NKAP mutant LCLs (Table S4). Although 14 out of 15 splicing variations in which a retained intron has a delta percent spliced in (dPSI) of more than 15 display increased intron retention in NKAP mutants (Figure 3A), intron retention levels are not globally affected when looking at all splicing variations detected in the transcriptome (Figure 3B). NKAP mutant cells also show few splicing variations involving de novo junctions (unannotated, putatively involving cryptic splice sites) and no preference in the inclusion of these junctions (Figure 3C) and a global usage of de novo junctions similar to control cells (Figure 3D).
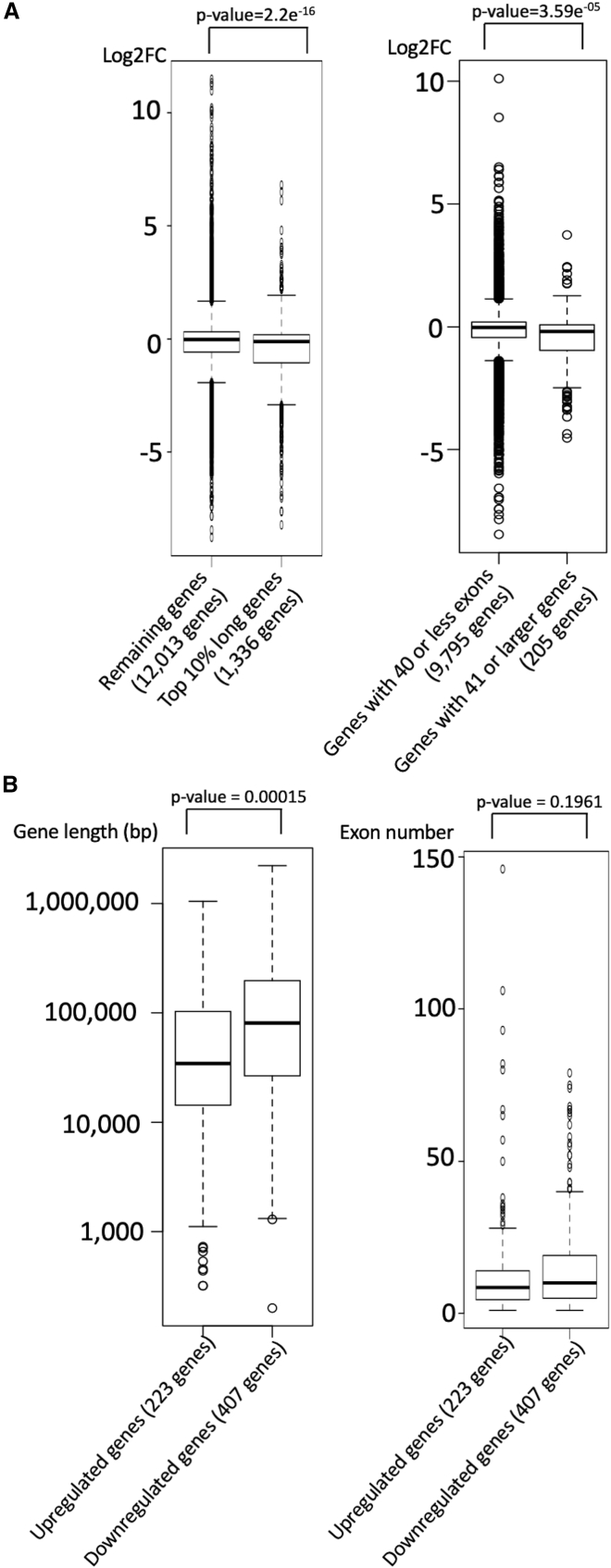
Alternatively, we did note that long genes with a higher number of exons were downregulated. As such, we evaluated the differential expression levels of long genes. Expression levels of the genes whose coding region length is in the top 10 percentile were lower than other genes whose coding region length is below 90th percentile (p value = 2.2e−16) (Figure 4A). The coding region length of downregulated genes was significantly longer than that of upregulated genes (Figure 4B). Finally, we evaluated the differential expression levels of genes with higher exon numbers in the subjects with NKAP mutations. As the majority of human genes contains less than 40 exons,12 we separately evaluated the level of differential gene expression among genes with ≤40 exons and genes with ≥41 exons (top 2 percentile). Expression levels of the genes with ≥41 exons were lower than those of the genes with ≤40 exons (p value = 3.59e−05) (Figure 4A). The exon numbers of downregulated genes were higher than those of upregulated genes, although the difference was not significant (p value = 0.196) (Figure 4B). These data indicate that NKAP dysfunction due to NKAP mutations results in misexpression of longer genes with larger numbers of exons.
Figure 4.
Effects of NKAP Mutations on Long Genes with a Greater Number of Exons
The genes, whose length and exon number were available in the Gene Base 1.1., were considered in this analysis.
(A) Differential gene expression levels of long genes with a greater number of exons. Left: Comparison of differential gene expression levels between long genes (top 10 percentile) and the rest in the subjects with NKAP mutations. Boxplot demonstrated that expression levels of the long genes tend to be downregulated in the subjects with NKAP mutations, compared to remainder of the genes. Right: Comparison of differential gene expression levels between genes with 40 or less exons and genes with 41 or more exons in the subjects with NKAP mutations. Boxplot demonstrated that expression levels of the genes with 41 or more exons tend to be downregulated in the subjects with NKAP mutations, compared to those of the genes with 40 or less exons. The top of each box represents the first quartile, and the bottom of each box represents the third quartile. The top of each whisker indicates the upper extreme, and the bottom of each whisker indicates the lower extreme. p value was calculated by t test.
(B) Comparison of gene length and exon number between upregulated genes and downregulated genes in individuals with NKAP mutations. Boxplot demonstrated that downregulated genes tend to have longer gene length and higher exon numbers. The top of each box represents the first quartile, and the bottom of each box represents the third quartile. The top of each whisker indicates the upper extreme, and the bottom of each whisker indicates the lower extreme. p values were calculated by t test.
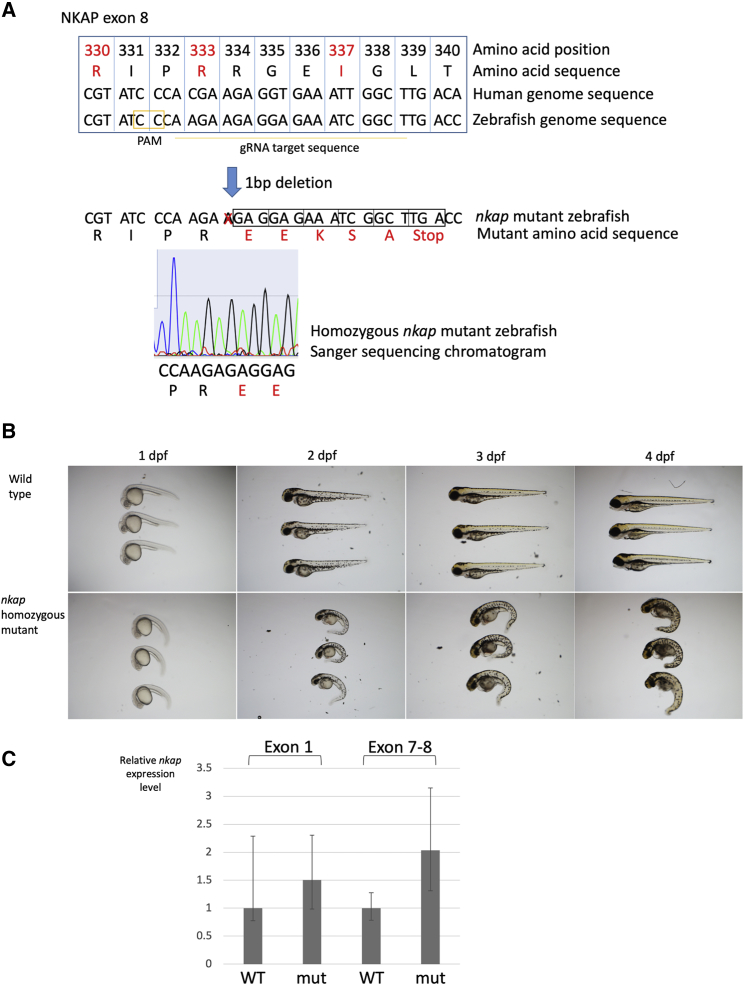
To evaluate the effects of NKAP mutation during embryogenesis, we established an nkap mutant zebrafish model using CRISPR/Cas9. Reciprocal BLAST of the human NKAP cDNA sequence (GenBank: NM_024528) against the zebrafish genome identified only one ortholog, nkap (GenBank: NM_001003414, with 68% identity to human). In fish, nkap is located on chromosome 14, with two copies of nkap present. We identified a gRNA targeting a region where human NKAP missense mutations reside (Figures 5A and S5) and have established F0 fish had a germline heterozygous 1 bp deletion in nkap (c.859delA; GenBank: NM_001003414), resulting in a frameshift mutation (p.Arg298GlufsTer6; GenBank: NP_001003414) (Figures 5A and S5). These heterozygous F0 1 bp deletion fish were then outcrossed to a wild-type fish, and 40 F1 heterozygous fish with the 1 bp deletion in nkap were identified (Figure S5). These heterozygous nkap mutant F1 zebrafish did not show a striking early developmental phenotype. Heterozygous nkap mutant F1 fish were incrossed to obtain an F2 generation that contained offspring with a homozygous 1 bp deletion in nkap (Figure 5A). Transmission of nkap 1 bp deletion followed a Mendelian inheritance pattern, with approximately 25% of F2 fish showing an obvious phenotype due to homozygous nkap 1 bp deletion. These homozygous nkap mutant fish consistently demonstrated striking early developmental defects, characterized by edema in the eye, intestinal tract, and heart with noticeable curvature of the notochord (Figure 5B). CNS necrosis was noticeable from 2 dpf. All homozygous nkap mutant fish lacked a heartbeat at 4 dpf and began to decompose; none survived beyond 4 dpf.
Figure 5.
Zebrafish Model of nkap Mutation
(A) CRISPR/Cas9 genome editing of zebrafish nkap. gRNA target sequence of zebrafish nkap is indicated. Amino acids mutated in human subjects with NKAP mutations are highlighted by red. Sanger sequence chromatogram demonstrating 1 bp deletion is depicted at the bottom. 1 bp deletion results in frameshift mutation.
(B) Developmental defects of homozygous 1 bp nkap deletion zebrafish. Top images are wild-type zebrafish, and bottom images are nkap mutant zebrafish. Morphological differences are noticeable at 1 dpf. 2× magnification images.
(C) Nkap expression level in the homozygous 1 bp nkap deletion fry. Relative expression levels of mutant nkap to wild-type zebrafish are depicted. Two pairs of nkap qPCR primer were used and listed in the Table S5. Both demonstrated comparable results. Expression level was normalized against actb2 expression. qPCR analyses were performed in sextuplicate (technical replicates). Three biological replicates demonstrated consistent results. Error bars: 2 SD.
The 1 bp deletion in this nkap mutant line is expected to cause a frameshift mutation (Figure 5A). It is in exon 8, the penultimate exon, leaving the possibility that the mutant nkap mRNA is escaping nonsense-mediated decay. To examine this possibility, we evaluated nkap expression levels in the F2 homozygous mutant zebrafish at 2 dpf using qRT-PCR. The RNA expression levels of nkap were not significantly altered at either the 5′ or 3′ regions of nkap in the homozygous mutant zebrafish compared to wild-type, indicating that the mutant nkap transcripts likely escape nonsense-mediated decay (Figure 5C). Therefore, as a consequence of the deletion, this nkap mutant zebrafish likely produced a C-terminal-truncated NKAP. The striking developmental phenotype of these zebrafish supports a critical role of the C-terminal region of NKAP during embryogenesis. Interestingly, the ZFIN database reports a homozygous transgenic zebrafish that has an insertion at the nkap start codon (nkaphi1477Tg).13, 14 Its developmental defects closely resemble those of homozygous 1 bp nkap deletion zebrafish embryos, including CNS necrosis, curvature of the notochord, and pericardial edema with most dying between 2 dpf and 5 dpf. Because the C-terminal truncation causes effects comparable to those of start codon elimination, the C-terminal NKAP region is likely essential for developmental regulation governed by NKAP.
Both the highly conserved clinical phenotype characterized by developmental delay and Marfanoid habitus identified in this study and the unique transcriptome profile identified in subject-derived LCLs establish a distinct clinical entity. Germline truncating mutations in NKAP have never been observed in the public database, gnomAD, among healthy individuals, indicating that loss-of-function NKAP mutations are likely detrimental to human health and development.15
Consistent with an X-linked recessive condition, NKAP mutations resulted in symptoms only in males, and females with heterozygous NKAP mutations appeared to be unaffected or significantly less affected. In fact, NKAP undergoes X-inactivation.16 Similar to human NKAP, the mouse Nkap also resides on the X chromosome, and hematopoietic lineage-specific hemizygous Nkap-null mutant male mice show embryonic lethality.17 In zebrafish, nkap is located on chromosome 14. We observed developmental defects only in homozygous nkap mutant zebrafish, while defects were not obvious in heterozygous animals. These observations suggest that elimination of non-mutated NKAP is required to impair development.
The clinical findings of individuals with NKAP mutations demonstrate essential roles of NKAP in neural development as well as in bone and connective tissue. This notion is supported by the striking developmental defects seen in nkap mutant zebrafish, which include spine deformities, a common finding also in human subjects with NKAP mutations. In addition, our gene ontology analysis of NKAP mutant cells revealed enrichment of extracellular matrix genes. Although the transcriptional target genes of NKAP may differ between LCLs and connective tissues, misexpression of extracellular matrix genes may result in defective connective tissue development in subjects with NKAP mutations.
This connective tissue role is supported by the clinical phenotype of this disorder which overlaps with connective tissue disorders, such as Marfan syndrome (MIM: 154700)18, 19 and Beals syndrome (MIM: 121050),20 with the presence of tall stature and joint contractures. In addition, Lujan-Fryns syndrome,1 characterized by Marfanoid habitus, tall stature, and intellectual disability was considered in one individual. Thus, NKAP alterations should be considered for individuals with suspected Marfan, Beals, or Lujan-Fryns syndromes.
The missense mutations identified in subjects were strictly clustered toward the C-terminal end of NKAP. This C-terminal end of NKAP is highly conserved, and missense mutations in the region between Pro296 and Glu394 (exon 7 to exon 9) are also entirely absent in gnomAD,15 underscoring the critical importance of this region (Figure S4). In addition, the lethal mutations caused by C-terminal truncations in zebrafish also support that this region of NKAP is essential in embryogenesis. The C-terminal end of NKAP (aa 273–415), where the subjects have missense mutations, harbors the HDAC3-interacting domain.5 Since HDAC3 is an important transcriptional regulatory molecule, NKAP mutations may affect the HDAC3-mediated transcriptional regulatory role of NKAP, although we did not detect any changes in HDAC3 dosage or intracellular distribution in the subjects (Figure S1).
Very recently, NKAP was shown to be a part of the post-catalytic P-complex, which is responsible for the last steps of splicing, and a crystal structural analysis revealed that the C-terminal region of NKAP (aa 329–358) interacts with SLU7 and PRP8, subunits of P-complex.8 Therefore, the disruption of exon ligation or processed mRNA release is another candidate mechanism explaining transcriptional alteration seen in this diagnosis. Consistent with the specific role of NKAP in the P-complex, we did not identify an increase of aberrant or alternative splicing events in the LCL samples with NKAP mutations.
RNA-seq analysis of LCL samples with NKAP mutations revealed that longer genes with more exons are more often downregulated. This suggests that genes which require more exonic ligation/mRNA release steps before the complete processing of the entire transcript may be more sensitive to NKAP dysfunction. Downregulation of long gene expression may be of key importance for normal neurodevelopment, as a large number of neuronal lineage-specific genes are characterized by long coding regions, and several neurological disorders, such as Rett syndrome (MIM: 312750) and autism, have been shown to be associated with the dysregulation of long genes.21, 22 Although intriguing, further work is needed to determine the role of NKAP in transcriptional regulation of long genes, as long genes are more sensitive to many factors including splicing, transcription efficiency, rate of transit of the transcriptional machinery, and mRNA stabilization.
In conclusion, we report a neurodevelopmental disorder with musculoskeletal features caused by NKAP missense mutations that result in transcriptional disruption. Our study supports pleiotropic roles for NKAP in the development of several organ systems and underscores the essential role for NKAP in vertebrate embryogenesis.
Declaration of Interests
The authors declare no competing interests.
Published: October 3, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.009.
Web Resources
DAVID Functional Annotation Bioinformatics Microarray Analysis, https://david.ncifcrf.gov/
DECIPHER, https://decipher.sanger.ac.uk
GeneBase 1.1, http://apollo11.isto.unibo.it/software/
GeneMatcher, https://www.genematcher.org/
OMIM, https://www.omim.org/
ZFIN, https://zfin.org/
Supplemental Data
References
- 1.Hackmann K., Rump A., Haas S.A., Lemke J.R., Fryns J.P., Tzschach A., Wieczorek D., Albrecht B., Kuechler A., Ripperger T. Tentative clinical diagnosis of Lujan-Fryns syndrome--A conglomeration of different genetic entities? Am. J. Med. Genet. A. 2016;170A:94–102. doi: 10.1002/ajmg.a.37378. [DOI] [PubMed] [Google Scholar]
- 2.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li T., Chen L., Cheng J., Dai J., Huang Y., Zhang J., Liu Z., Li A., Li N., Wang H. SUMOylated NKAP is essential for chromosome alignment by anchoring CENP-E to kinetochores. Nat. Commun. 2016;7:12969. doi: 10.1038/ncomms12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pajerowski A.G., Nguyen C., Aghajanian H., Shapiro M.J., Shapiro V.S. NKAP is a transcriptional repressor of notch signaling and is required for T cell development. Immunity. 2009;30:696–707. doi: 10.1016/j.immuni.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D., Li Z., Yang Q., Zhang J., Zhai Z., Shu H.B. Identification of a nuclear protein that promotes NF-kappaB activation. Biochem. Biophys. Res. Commun. 2003;310:720–724. doi: 10.1016/j.bbrc.2003.09.074. [DOI] [PubMed] [Google Scholar]
- 7.Burgute B.D., Peche V.S., Steckelberg A.L., Glöckner G., Gaßen B., Gehring N.H., Noegel A.A. NKAP is a novel RS-related protein that interacts with RNA and RNA binding proteins. Nucleic Acids Res. 2014;42:3177–3193. doi: 10.1093/nar/gkt1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fica S.M., Oubridge C., Wilkinson M.E., Newman A.J., Nagai K. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science. 2019;363:710–714. doi: 10.1126/science.aaw5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaFoya B., Munroe J.A., Mia M.M., Detweiler M.A., Crow J.J., Wood T., Roth S., Sharma B., Albig A.R. Notch: A multi-functional integrating system of microenvironmental signals. Dev. Biol. 2016;418:227–241. doi: 10.1016/j.ydbio.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verrecchia F., Mauviel A. Transforming growth factor-beta signaling through the Smad pathway: role in extracellular matrix gene expression and regulation. J. Invest. Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
- 11.Vaquero-Garcia J., Barrera A., Gazzara M.R., González-Vallinas J., Lahens N.F., Hogenesch J.B., Lynch K.W., Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife. 2016;5:e11752. doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakharkar M.K., Perumal B.S., Sakharkar K.R., Kangueane P. An analysis on gene architecture in human and mouse genomes. In Silico Biol. (Gedrukt) 2005;5:347–365. [PubMed] [Google Scholar]
- 13.Amsterdam A., Nissen R.M., Sun Z., Swindell E.C., Farrington S., Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golling G., Amsterdam A., Sun Z., Antonelli M., Maldonado E., Chen W., Burgess S., Haldi M., Artzt K., Farrington S. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 15.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tukiainen T., Villani A.C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. [Google Scholar]
- 17.Pajerowski A.G., Shapiro M.J., Gwin K., Sundsbak R., Nelson-Holte M., Medina K., Shapiro V.S. Adult hematopoietic stem cells require NKAP for maintenance and survival. Blood. 2010;116:2684–2693. doi: 10.1182/blood-2010-02-268391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietz H.C., Cutting G.R., Pyeritz R.E., Maslen C.L., Sakai L.Y., Corson G.M., Puffenberger E.G., Hamosh A., Nanthakumar E.J., Curristin S.M. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 19.Neptune E.R., Frischmeyer P.A., Arking D.E., Myers L., Bunton T.E., Gayraud B., Ramirez F., Sakai L.Y., Dietz H.C. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 20.Putnam E.A., Zhang H., Ramirez F., Milewicz D.M. Fibrillin-2 (FBN2) mutations result in the Marfan-like disorder, congenital contractural arachnodactyly. Nat. Genet. 1995;11:456–458. doi: 10.1038/ng1295-456. [DOI] [PubMed] [Google Scholar]
- 21.Gabel H.W., Kinde B., Stroud H., Gilbert C.S., Harmin D.A., Kastan N.R., Hemberg M., Ebert D.H., Greenberg M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King I.F., Yandava C.N., Mabb A.M., Hsiao J.S., Huang H.S., Pearson B.L., Calabrese J.M., Starmer J., Parker J.S., Magnuson T. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.