Abstract
NTNG2 encodes netrin-G2, a membrane-anchored protein implicated in the molecular organization of neuronal circuitry and synaptic organization and diversification in vertebrates. In this study, through a combination of exome sequencing and autozygosity mapping, we have identified 16 individuals (from seven unrelated families) with ultra-rare homozygous missense variants in NTNG2; these individuals present with shared features of a neurodevelopmental disorder consisting of global developmental delay, severe to profound intellectual disability, muscle weakness and abnormal tone, autistic features, behavioral abnormalities, and variable dysmorphisms. The variants disrupt highly conserved residues across the protein. Functional experiments, including in silico analysis of the protein structure, in vitro assessment of cell surface expression, and in vitro knockdown, revealed potential mechanisms of pathogenicity of the variants, including loss of protein function and decreased neurite outgrowth. Our data indicate that appropriate expression of NTNG2 plays an important role in neurotypical development.
Keywords: developmental delay, intellectual disability, autism, neurodevelopmental disorder, NTNG2
Main Text
Based on studies in invertebrates and chicken and mouse, netrins are considered to be the paradigmatic axon guidance molecules, yet the essential role of this family of proteins in humans remains unclear. Members of the classical netrin family are secreted proteins that include UNC6 (uncoordinated-6) in C. elegans and netrins NTN1–4 in vertebrates.1, 2 Netrin-G proteins (NTNG1 and NTNG2) are distinct from classical netrins in that they are vertebrate-specific, membrane-bound proteins tethered to the plasma membrane by glycosyl phosphatidylinositol (GPI) anchors.3 NTNG1 (MIM: 608818) and NTNG2 are predominantly expressed in a non-overlapping and complementary pattern in specific neuronal subsets of the developing and mature central nervous system.4, 5, 6 The proteins interact with the extracellular region of their specific netrin-G ligand receptors NGL-1/LRRC4C (MIM: 608817) and NGL-2/LRRC4, respectively.7 Selectivity in binding between netrin-G molecules and their cognate receptors is mediated by the interactions of three loops of the laminin domain, and the extracellular leucine rich repeats (LRR) domain of NGLs results in a molecular hand-clasp interaction of high affinity.8
NTNG2 encodes netrin-G2, a vertebrate-specific protein that is part of a distinct functional sub-class of the highly conserved netrin family. The netrin family provides axonal guidance cues during central nervous system development.9 NTNG2 is located on 9q34.13, and the canonical transcript consists of eight exons including seven coding exons; it encodes a 530-amino-acid protein. NTNG2 demonstrates evidence of missense constraint in the ExAC database, with a Z score of 4.34, and review of population-based (gnomAD) and ethnically diverse in-house databases reveals an absence of homozygous damaging and/or deleterious variants. Despite this constraint, its potential role in human genetic disease is not clear. Here we show that bi-allelic missense variants in NTNG2 cause a distinctive neurological and behavioral disorder that highlights the importance of this family of genes in human nervous system development.
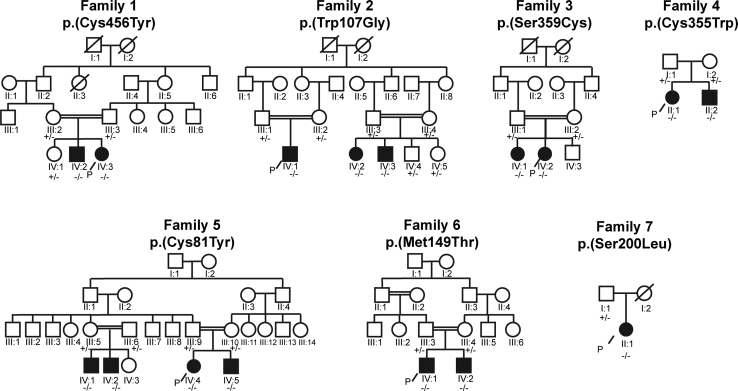
We have identified 16 individuals (from seven unrelated families) who have ultra-rare bi-allelic variants in NTNG2 and who present with shared clinical features of a neurodevelopmental disorder. Consent for clinical data and biological material collection, use, and storage was obtained from all participating families after written informed consent was provided, and studies for each family were approved by their respective institutional review boards (see Supplemental Data for further details). Following genomic DNA extraction from blood, exome sequencing, and homozygosity mapping, we identified 16 individuals in seven unrelated families from different parts of Iran (Families 1, 2, 3), Mexico (Family 4), Turkey (Family 5), Egypt (Family 6), and Bangladesh (Family 7) who have a similar clinical phenotype and have homozygous missense variants in NTNG2 (Figure S1,Table S1). Researchers and physicians for all families were connected using the GeneMatcher/Matchmaker exchange.10, 11 All families except for Families 4 and 7 had a known history of consanguinity, and all of the variants were segregated in the affected families in accordance with Mendelian expectations for a recessive disease trait (Figure 1). Autozygosity mapping for Families 4 and 7 revealed distant relatedness, and parents of the proband in Family 7 come from the same village (Figure S2, Table S2). There was no evidence of neuropsychiatric disorders in the heterozygous family members presented here.
Figure 1.
Pedigrees of All Families with Affected Individuals and Variants and Segregation Findings
+ indicates wild-type allele, - indicates variant allele, P indicates proband.
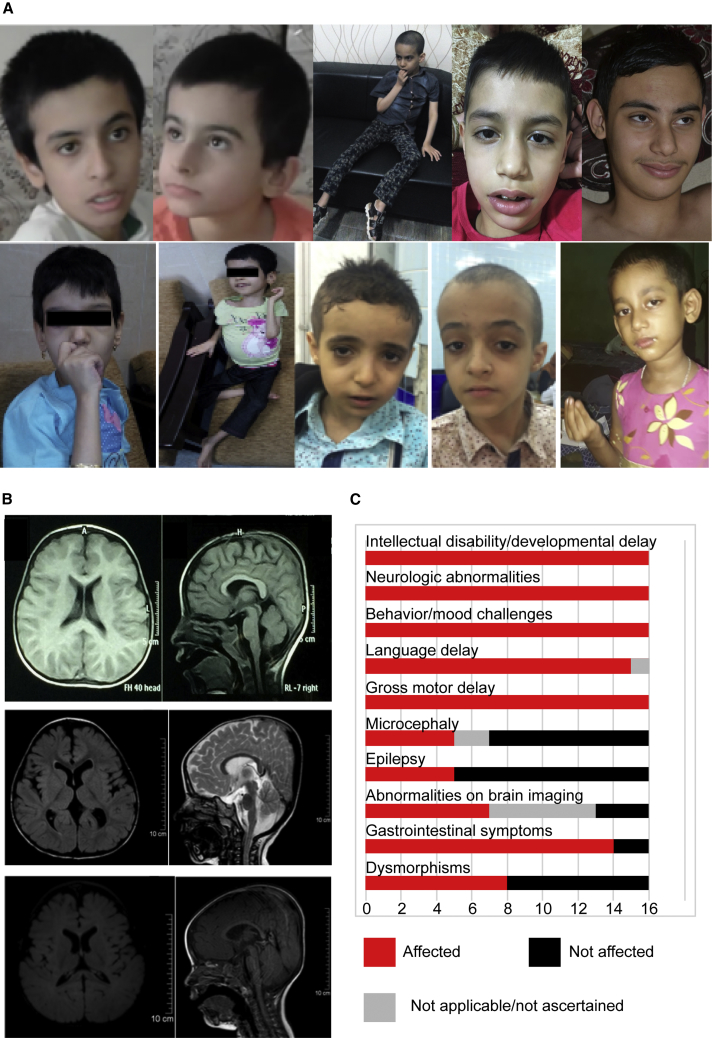
Clinical features of affected individuals are presented in Table 1. Affected individuals presented with global developmental delay with severe to profound intellectual disability; the majority were non-verbal and non-ambulatory. Most individuals also had features of autism and all were noted to have mood and/or behavioral challenges, many of which were similar to those seen in Rett syndrome, such as hand stereotypy, episodes of laughing and/or screaming, and bruxism, and in Angelman syndrome (Videos S1–S4). Gastrointestinal symptoms, including constipation and bloating, were also common. Growth parameters were below average, and four individuals had documented failure to thrive. Secondary microcephaly was also observed. Dysmorphic features were variable and included low-set ears, hypotelorism, and frontal bossing (Figure 2A). Neurologically, hypotonia in infancy and muscle weakness and/or atrophy were common findings. Five individuals had early-onset seizures, and four were noted to have ocular findings of esotropia, nystagmus, or strabismus. Brain imaging, conducted in both infancy and childhood, demonstrated findings ranging from normal to mild brain atrophy with white matter abnormalities (Figure 2B). In summary, affected individuals display a neurodevelopmental disorder of severe-to-profound intellectual disability with marked motor involvement and mood and behavioral challenges including autistic features, as well as poor growth and facial dysmorphisms. Detailed phenotypic descriptions are provided in Table 1, Table S1, Figure 2, and the Supplemental Note: Case Reports in Supplemental Data.
Table 1.
Clinical Features of Affected Individuals
| Family | 1 | 1 | 2 | 2 | 2 | 3 | 3 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | IV:2 | IV:3 | IV:1 | IV:2 | IV:3 | IV:1 | IV:2 | II:1 | II:2 | IV:1 | IV:2 | IV:4 | IV:5 | IV:1 | IV:2 | II:1 |
| Age (years) | 18 | 10 | 11 | 16 | 9 | 15 | 11 | 11 | 21 | 11 | 1.25 | 9 | 5 | 11 | 8 | 3 |
| Sex | M | F | M | M | F | F | F | F | M | M | M | F | M | M | M | F |
| ID/GDD | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Motor delay | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Language delay | + | + | + | + | + | + | + | + | + | + | NA | + | + | + | + | + |
| Autistic features/stereotypy | + | + | + | + | + | + | + | + | - | + | NA | + | + | + | - | + |
| Hyperactivity | - | + | + | + | + | - | - | - | + | - | - | - | - | + | + | - |
| Screaming/laughing spells | + | + | + | + | + | + | + | + | + | + | NA | + | - | + | - | + |
| Self-injury/hand-biting | + | + | + | + | + | - | - | + | + | - | - | - | - | - | - | - |
| Bruxism | - | + | + | + | + | - | - | - | - | - | NA | + | + | + | - | - |
| Hypotonia in infancy | + | + | + | + | + | + | + | + | + | + | + | + | + | - | - | + |
| Nonambulatory | + | + | + | + | + | + | + | + | + | + | NA | + | + | - | - | - |
| Brain imaging abnormalities | - | NA | + | NA | NA | - | + | - | NA | NA | NA | + | + | + | + | + |
| Seizures | - | - | + | - | - | - | - | + | + | - | - | - | - | + | - | + |
| Microcephaly | - | + | - | + | + | - | - | + | NA | NA | - | + | - | - | - | - |
| Secondary Microcephaly | + | NA | + | NA | NA | |||||||||||
| Dysmorphic features | - | - | + | - | - | + | + | - | - | NA | + | + | + | + | + | - |
| Ophthalmologic features | - | - | - | - | - | + | + | - | - | - | - | + | + | - | - | - |
| GI symptoms | + | + | + | + | + | + | + | + | + | + | + | + | + | - | - | + |
ID/GDD, intellectual disability/global developmental delay; GI, gastrointestinal; -, absent; +, present; NA, not ascertained/not applicable
Figure 2.
Clinical Features of Affected Individuals
(A) Representative photographs demonstrating clinical features of affected individuals; these features include facial features, muscular atrophy, and hand stereotypy. Top row from left to right: Family 2 IV:2, IV:3, IV:1; Family 1 IV:3, IV:2. Bottom row: Family 3 IV:1, IV:2; Family 6 IV:2, IV:1; Family 7 II:1.
(B) Representative MRIs of affected individuals, demonstrating decreased brain volume. From top to bottom: Family 6 IV:2; Family 5 IV:4; Family 5 IV:5.
(C) Bar graph summarizing proportions of various clinical findings affecting individuals.
Movements are similar to those observed in Angelman syndrome.
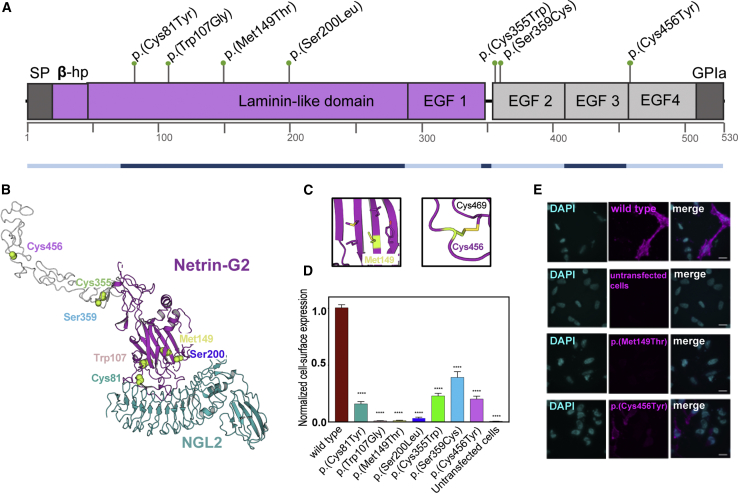
The ultra-rare variants we identified from family based genomic studies in the above individuals were notable for several reasons (Table S2). All variants were absent from both local ethnically diverse in-house databases, as well as large population databases. Because most of the NTNG2 missense variants observed are rare to their specific “clan,” they may reflect variants that arose recently and according to the clan genomics hypothesis are therefore expected to have a larger influence on disease.12 All variants were predicted by a majority of prediction tools (FATHMM, MutationAssessor, MutationTaster, PolyPhen-2, SIFT, PROVEAN, and CADD) to be likely damaging to protein function, and genomic evolutionary rate profiling (GERP) indicated that these sites may be under evolutionary constraint (Table S2). In fact, all variants impact residues conserved in NTNG1, a result that gives further evidence for the argument that they are pathogenic. Annotation of the variant locations on the protein domains of NTNG2 revealed that they are not confined to one domain, but they fall within the laminin and EGF domains and are predicted to disrupt structural motifs within NTNG2 (Figure 3A). No other alternative candidate variants common to the families were identified (Table S3).
Figure 3.
Structural Mapping and Cell Surface Expression
(A) Netrin-G2 domain overview. The positions of altered residues relative to the protein domains are indicated. Domain nomenclature is: SP, signal peptide; β-hp, N-terminal β-hairpin domain; EGF, epidermal-growth factor like; GPIa, GPI anchor. Corresponding exons are represented underneath the domain organization in blue.
(B) Full-length model of netrin-G2 based on the crystal structure of the Laminin-like domain and EGF18 (purple) and on homology models of EGF2-4 (gray), in complex with its ligand NGL2 (cyan). The residues that are mutated in the presented variants are indicated as green spheres.
(C) Close-up views of Met149 and Cys456 residues as found in the structural model shown in panel B. For close-up views of the other mutated residues, see Figure S3.
(D) The quantification of the cell surface expression levels of wild type (WT) and mutant netrin-G2 constructs (see panel E) is shown (mean ± SEM). The variants show significantly reduced cell-surface expression compared to the WT (∗∗∗∗p < 0.0001).
(E) Netrin-G2 constructs were expressed in HeLa cells with an N-terminal flag tag. Flag-tagged protein was detected via cell-surface immunostaining (magenta). DAPI (blue) highlights cell nuclei. Representative images are shown for WT netrin-G2, untransfected cells (negative control), p.Met149Thr and p.Cys456Tyr variants. Representative images of other variants are shown in Figure S3. Scale bar is 15 μm.
The available NGL2/netrin-G2 crystal structure contains a model of the netrin-G2 N terminus up to the first EGF domain. We used MODELER13 to create a homology for the EGF domains 2–4, which were not included in that crystal structure. Using these models, we found that the variants are located in the laminin, EGF2, or EGF4 domain (Figure 3B–3C, Figure S3). In addition to possible effects on specific protein-to-protein interaction sites, this suggests a more global mechanism of functional disruption. Strikingly, we found that four of the seven, i.e., 57% of the variants, involve the loss or addition of cysteine residues (GenBank: NM_032536.3: c.242G>A [p.Cys81Tyr], c.1065C>G [p.Cys355Trp], c.1076C>G [p.Ser359Cys], c.1367G>A [p.Cys456Tyr]). Given that the cysteine content of NTNG2 is only 7.9% in humans, the enrichment for cysteine variants in this cohort suggests a mechanism of pathogenicity. Due to the oxidizing environment in the endoplasmic reticulum (ER) and extracellular space, cysteine residues found in extracellular proteins typically appear in pairs and form disulfide bridges. Such bridges can stabilize a protein by reducing the entropy of the unfolded state and/or they can facilitate the path to the native state if they link parts of a protein that must come into contact early during a folding reaction.14 Unpaired exposed cysteines are detected by the ER quality control machinery and targeted for refolding or degradation.14 We hypothesize that the NTNG2 variants involving cysteine could have a negative effect on protein stability and cell surface expression.
Three out of the seven variants do not involve cysteines: c.599C>T (p.Ser200Leu), c.319T>G (p.Trp107Gly), and c.446T>C, (p.Met149Thr). For both p.Trp107Gly and p.Met149Thr, a large hydrophobic residue (Trp or Met) is changed to either one lacking a side chain (Gly) or one bearing a small polar side chain (Thr). Both of these residues form part of the hydrophobic core that stabilizes the folding of the netrin-G2 laminin domain. Disruption likely causes protein misfolding and lack of expression at the cell surface. Thus the consequence of both types of variants, cysteine-dependent or hydrophobic core disruptive, is potentially a similar reduction in protein stability and expression at the cell surface. The p.Ser200Leu variant does not fit into either of the above categories, with Ser200 located at the periphery of the laminin domain. It is also located ∼0.9 nm away from the surface of NGL2 as found in the crystal structure (Figure S3). With typical hydrogen bonds about 0.25 nm in length, Ser200 is not interacting directly with NGL2, although allosteric effects could still influence the netrin-G2 binding loops.
We tested all variants for function by overexpressing wild-type (WT) and variant netrin-G2 constructs in HeLa cells and assessing their presence at the cell surface through the use of indirect immunofluorescence and immunoblotting validation (Figure S4). All variants displayed substantially decreased cell surface expression as compared to WT (Figures 3D–3E). Notably, some of these variants had more cell surface expression compared to others, suggesting that some netrin-G2 may still be localized in these individuals. The variants may nevertheless show deficient ligand-receptor binding or signaling since we did not observe a clear association with cell surface expression levels and clinical phenotype severity.
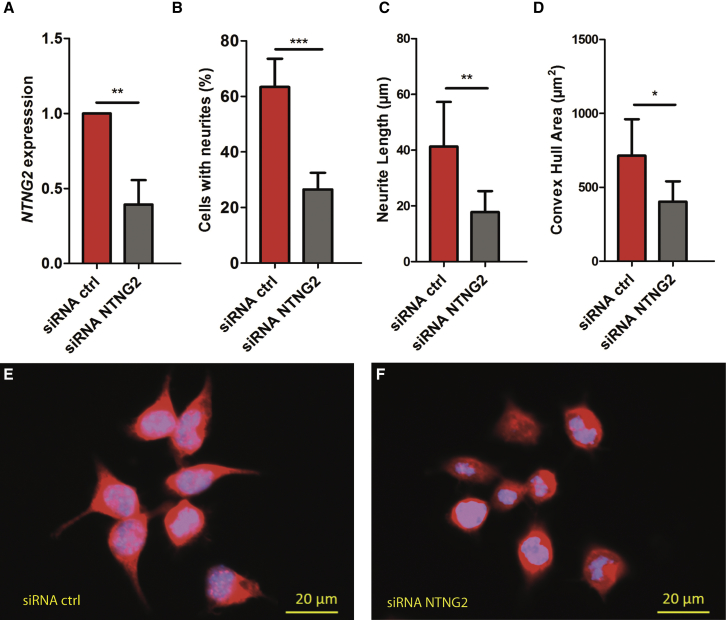
Given the decreased cell surface expression pattern observed in all seven variants, we sought to determine the more global effects of NTNG2 loss of function. Using siRNA to target endogenous Ntng2 expression in mouse N2A cells, we first confirmed that transfection with our Ntng2-specific siRNA led to decreased expression with quantitative polymerase chain reaction (Figure 4A). We next assessed neurite outgrowth and found a significant reduction for all parameters assessed, which included neurite number, neurite length, and convex hull area—a measurement used for measuring dendritic field (Figures 4B–4F). These findings demonstrate a potential mechanism by which the NTNG2 variants may contribute to pathological neurodevelopment.
Figure 4.
Ntng2 Knockdown in N2a Cells
(A) Knockdown of endogenous Ntng2 by Ntng2-specific siRNAs as normalized to control siRNA. Results of quantitative RT-PCR 30 h post-transfection.
(B–D) Effects of Ntng2 knockdown on neurite outgrowth. Data presented as mean and SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Quantification was conducted by counting the absolute number of cells with neurites (B), measuring the neurite length by NeuroLucida tracing (C), and quantification of the Convex hull area (D). All analyses show a significant reduction of neurite number and length as a consequence of Ntng2 knockdown.
(E–F) Example of the N2a appearance at 30 h post-transfection with control (E) and Ntng2-specific siRNA (F). Visualization was done using MAP2 counterstaining (red).
Netrin signaling has been implicated in neurologic and psychiatric disorders. For example, conditional Ntng1 knockout in distinct neuronal subtypes is associated with alterations in fear and anxiety-like behaviors in rodent models, and abnormal expression of NTNG2 has been found in the human brain in refractory epilepsy.15, 16 Studies of Ntng2 and Ngl2 knockout mice have shown that both types of mutant mice have an identical phenotype of lack of behavioral startle in response to acoustic stimulus, with no structural abnormalities noted in the inner ear.17 Single-nucleotide polymorphisms (SNPs) and differential expression patterns of NTNG1 and NTNG2 have been associated with schizophrenia and bipolar disorder in humans.18, 19, 20 A de novo genomic rearrangement involving NTNG1 was proposed to potentially cause features of Rett syndrome in an isolated individual.21 Additionally, de novo missense variants in NTNG1 were reported in two individuals with autism spectrum disorder.22 An in vitro study of variants in the histone demethylase KDM5C (Lysine demethylase 5C [MIM: 314690]), which is known to cause intellectual disability, showed that NTNG2 seemed to be important in mediating effects on neurite growth and length; these results are consistent with our findings here.23 In fact, several clinical features, in addition to intellectual disability, are shared between these two disorders, including variable neurologic, behavioral, and dysmorphic features. Extensive behavioral battery on Ntng2 knockout mice demonstrated marked deficits in learning, memory, and visual and motor functioning.24 Although NTNG2 does not appear to be necessary for axon guidance, it has been shown to be important in the laminar distribution of its receptors and synaptic plasticity.25, 26 A homozygous founder frameshift variant in NTNG2 has recently been identified in eight individuals from four families with a similar clinical phenotype, and this further strengthens the evidence supporting the pathogenicity of the variants presented here.27
Other genes involved in netrin signaling have also been implicated in neurodevelopmental disorders in isolated case reports, some of which have involved examples of de novo variation.28 Specifically, variations in LRRC4C and LRRC4 have both been associated with intellectual disability and autism.29, 30 Furthermore, functional work in mice has shown that LRRC4 expression regulates N-methyl-D-aspartate receptor (NMDAR)-dependent synaptic plasticity and prevents autistic-like behaviors.31 LRRC4C and LRRC4 have both been shown to be important in hippocampal synapse formation and function.32, 33 The marked findings of severe intellectual disability and autistic features in our cohort are particularly intriguing given the unique role of NTNG2 in vertebrates. As we previously mentioned, netrin-g family members express in distinct, non-overlapping, and complementary neuronal circuits, suggesting a role in establishing appropriate neuronal patterning. This neuronal compartmentalization parallels distinct behavioral compartmentalization, as in mouse knockout models, Ntng2 knockouts demonstrated sensorimotor, spatial memory, working memory, procedural learning, and attentional deficits, while Ntng1 knockouts demonstrated distinct learning and fear conditioning deficits.24 Our findings here, in conjunction with the known role of NTNG2 in the control of synaptic plasticity and postsynaptic membrane organization, illustrate the clinical relevance of these neuronal functions to higher cognitive processes. In fact, given the profound finding of intellectual disability in the individuals presented here, it is intriguing that NTNG2 expression is enriched in the human claustrum, an enigmatic brain region posited to play a role in the integration of conscious perception.34
Our work provides the groundwork for establishing a genotype-to-phenotype relationship with NTNG2 variants, and establishes an initial description of the clinical spectrum. NTNG2 should be considered in the clinical evaluation of children with severe intellectual disability and neuropsychiatric symptoms. In addition to identification by exome sequencing, it will be important to add NTNG2 to clinical gene-panel tests for intellectual disability given the marked yet variable clinical phenotype. In summary, our results implicate rare bi-allelic missense NTNG2 variants in the pathobiology of a neurodevelopmental disorder consisting of severe intellectual disability, autistic features, and motor impairment. Our findings provide strong clinical and functional evidence for the importance of the appropriate expression of NTNG2 in neurodevelopment.
Declaration of Interests
Baylor College of Medicine (BCM) and Miraca Holdings have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), which performs clinical microarray analysis and clinical exome sequencing. J.R.L. serves on the Scientific Advisory Board of BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The other authors declare no competing interests.
Acknowledgments
This work was supported by the United States National Institutes of Health: U54HG003067 (E.L.), UM1HG008900 (D.M., H.R.), U54HG006504 (R.L., M.G.), R35 NS105078 (J.R.L.), UM1HG006542 (J.R.L.), K08HG008986 (J.E.P.), R01NS048453 (J.G.G.), R01NS052455 (J.G.G.), T32 NS043124-17 (D.P.), R01NS035129 (C.A.W.), T32 MH112510 (C.M.D.). Additional support includes the Muscular Dystrophy Association (#512848[J.R.L]., H.H.); the Clinical Research Training Scholarship in Neuromuscular Disease partnered by American Brain Foundation and Muscle Study Group (D.P.); Howard Hughes Medical Institute (C.A.W., J.G.G.); Wellcome Trust (202827/Z/16/Z, [E.S.], Wellcome Trust DPhil. Cellular Structural Biology [C.Z.]; SYNaPS Study Group and Synaptopathies Strategic Award 165908, WT093205 MA, WT104033AIA [H.H].); National Institute for Health Research University College London Hospitals Biomedical Research Centre (H.H.); Rosetree Trust (H.H.); Ataxia UK (H.H.); MSA Trust (H.H.); Brain Research UK (H.H.); Muscular Dystrophy UK (H.H.); Higher Education Commission of Pakistan (H.H.); The MRC (MR/S01165X/1, MR/S005021/1, G0601943 [H.H.]); European Union Seventh Framework Program (Gencodys: 241995 [H.vB.]); CAPES Fellowship, Brazil (99999.013311/2013-01 [D.L.P.]); Uehara Memorial Foundation (T.M.); the Broad Institute of MIT; Yale Center for Mendelian Disorders; Harvard Center for Mendelian Disorders; and Queen Square Genomics group at University College London. Genome sequencing was performed through an in-kind donation of sequencing from Human Longevity Inc., and from the Rady Children’s Institute of Genomic Medicine.
Published: October 24, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.025.
Accession Numbers
The accession numbers for the variants reported in this paper are ClinVar SCV000994967.1, SCV000994968.1, SCV000994969.1, SCV000994970.1, SCV000994971.1, SCV000994972.1, and SCV000994973.1.
LOVD variant identification numbers are 597120–597126.
Web Resources
FATHMM web server, http://fathmm.biocompute.org.uk/
GeneMatcher, https://genematcher.org/
GERP, http://mendel.stanford.edu/sidowlab/downloads/gerp/index.html
MutationAssessor web server, http://mutationassessor.org/r3/
MutationTaster, http://www.mutationtaster.org/
Online Mendelian Inheritance in Man, https://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org/index.php
Supplemental Data
References
- 1.Serafini T., Kennedy T.E., Galko M.J., Mirzayan C., Jessell T.M., Tessier-Lavigne M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell. 1994;78:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 2.Dickson B.J. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- 3.Cirulli V., Yebra M. Netrins: beyond the brain. Nat. Rev. Mol. Cell Biol. 2007;8:296–306. doi: 10.1038/nrm2142. [DOI] [PubMed] [Google Scholar]
- 4.Nakashiba T., Nishimura S., Ikeda T., Itohara S. Complementary expression and neurite outgrowth activity of netrin-G subfamily members. Mech. Dev. 2002;111:47–60. doi: 10.1016/s0925-4773(01)00600-1. [DOI] [PubMed] [Google Scholar]
- 5.Meerabux J.M., Ohba H., Fukasawa M., Suto Y., Aoki-Suzuki M., Nakashiba T., Nishimura S., Itohara S., Yoshikawa T. Human netrin-G1 isoforms show evidence of differential expression. Genomics. 2005;86:112–116. doi: 10.1016/j.ygeno.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Lin J.C., Ho W.H., Gurney A., Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat. Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- 7.Woo J., Kwon S.K., Kim E. The NGL family of leucine-rich repeat-containing synaptic adhesion molecules. Mol. Cell. Neurosci. 2009;42:1–10. doi: 10.1016/j.mcn.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Seiradake E., Coles C.H., Perestenko P.V., Harlos K., McIlhinney R.A., Aricescu A.R., Jones E.Y. Structural basis for cell surface patterning through NetrinG-NGL interactions. EMBO J. 2011;30:4479–4488. doi: 10.1038/emboj.2011.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakashiba T., Ikeda T., Nishimura S., Tashiro K., Honjo T., Culotti J.G., Itohara S. Netrin-G1: a novel glycosyl phosphatidylinositol-linked mammalian netrin that is functionally divergent from classical netrins. J. Neurosci. 2000;20:6540–6550. doi: 10.1523/JNEUROSCI.20-17-06540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics. 2016;15:5.6.1–5.6.30. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feige M.J., Hendershot L.M. Disulfide bonds in ER protein folding and homeostasis. Curr. Opin. Cell Biol. 2011;23:167–175. doi: 10.1016/j.ceb.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Y., Liu G., Fang M., Shen L., Wang L., Han Y., Shen D., Wang X. Abnormal expression of netrin-G2 in temporal lobe epilepsy neurons in humans and a rat model. Exp. Neurol. 2010;224:340–346. doi: 10.1016/j.expneurol.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q., Sano C., Masuda A., Ando R., Tanaka M., Itohara S. Netrin-G1 regulates fear-like and anxiety-like behaviors in dissociable neural circuits. Sci. Rep. 2016;6:28750. doi: 10.1038/srep28750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W., Rajan I., Savelieva K.V., Wang C.Y., Vogel P., Kelly M., Xu N., Hasson B., Jarman W., Lanthorn T.H. Netrin-G2 and netrin-G2 ligand are both required for normal auditory responsiveness. Genes Brain Behav. 2008;7:385–392. doi: 10.1111/j.1601-183X.2007.00361.x. [DOI] [PubMed] [Google Scholar]
- 18.Aoki-Suzuki M., Yamada K., Meerabux J., Iwayama-Shigeno Y., Ohba H., Iwamoto K., Takao H., Toyota T., Suto Y., Nakatani N. A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia. Biol. Psychiatry. 2005;57:382–393. doi: 10.1016/j.biopsych.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Eastwood S.L., Harrison P.J. Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2008;33:933–945. doi: 10.1038/sj.npp.1301457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eastwood S.L., Harrison P.J. Markers of glutamate synaptic transmission and plasticity are increased in the anterior cingulate cortex in bipolar disorder. Biol. Psychiatry. 2010;67:1010–1016. doi: 10.1016/j.biopsych.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borg I., Freude K., Kübart S., Hoffmann K., Menzel C., Laccone F., Firth H., Ferguson-Smith M.A., Tommerup N., Ropers H.H. Disruption of Netrin G1 by a balanced chromosome translocation in a girl with Rett syndrome. Eur. J. Hum. Genet. 2005;13:921–927. doi: 10.1038/sj.ejhg.5201429. [DOI] [PubMed] [Google Scholar]
- 22.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei G., Deng X., Agarwal S., Iwase S., Disteche C., Xu J. Patient Mutations of the Intellectual Disability Gene KDM5C Downregulate Netrin G2 and Suppress Neurite Growth in Neuro2a Cells. J. Mol. Neurosci. 2016;60:33–45. doi: 10.1007/s12031-016-0770-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q., Goto H., Akiyoshi-Nishimura S., Prosselkov P., Sano C., Matsukawa H., Yaguchi K., Nakashiba T., Itohara S. Diversification of behavior and postsynaptic properties by netrin-G presynaptic adhesion family proteins. Mol. Brain. 2016;9:6. doi: 10.1186/s13041-016-0187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimura-Akiyoshi S., Niimi K., Nakashiba T., Itohara S. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc. Natl. Acad. Sci. USA. 2007;104:14801–14806. doi: 10.1073/pnas.0706919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsukawa H., Akiyoshi-Nishimura S., Zhang Q., Luján R., Yamaguchi K., Goto H., Yaguchi K., Hashikawa T., Sano C., Shigemoto R. Netrin-G/NGL complexes encode functional synaptic diversification. J. Neurosci. 2014;34:15779–15792. doi: 10.1523/JNEUROSCI.1141-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu-Libdeh B., Ashhab M., Shahrour M., Daana M., Dudin A., Elpeleg O., Edvardson S., Harel T. Homozygous frameshift variant in NTNG2, encoding a synaptic cell adhesion molecule, in individuals with developmental delay, hypotonia, and autistic features. Neurogenetics. 2019;20:209–213. doi: 10.1007/s10048-019-00583-4. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 28.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019;104:1182–1201. doi: 10.1016/j.ajhg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maussion G., Cruceanu C., Rosenfeld J.A., Bell S.C., Jollant F., Szatkiewicz J., Collins R.L., Hanscom C., Kolobova I., de Champfleur N.M. Implication of LRRC4C and DPP6 in neurodevelopmental disorders. Am. J. Med. Genet. A. 2017;173:395–406. doi: 10.1002/ajmg.a.38021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangu N., Shimojima K., Takahashi Y., Ohashi T., Tohyama J., Yamamoto T. A 7q31.33q32.1 microdeletion including LRRC4 and GRM8 is associated with severe intellectual disability and characteristics of autism. Hum. Genome Var. 2017;4:17001. doi: 10.1038/hgv.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Um S.M., Ha S., Lee H., Kim J., Kim K., Shin W., Cho Y.S., Roh J.D., Kang J., Yoo T. NGL-2 Deletion Leads to Autistic-like Behaviors Responsive to NMDAR Modulation. Cell Rep. 2018;23:3839–3851. doi: 10.1016/j.celrep.2018.05.087. [DOI] [PubMed] [Google Scholar]
- 32.Choi Y., Park H., Jung H., Kweon H., Kim S., Lee S.Y., Han H., Cho Y., Kim S., Sim W.S. NGL-1/LRRC4C Deletion Moderately Suppresses Hippocampal Excitatory Synapse Development and Function in an Input-Independent Manner. Front. Mol. Neurosci. 2019;12:119. doi: 10.3389/fnmol.2019.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeNardo L.A., de Wit J., Otto-Hitt S., Ghosh A. NGL-2 regulates input-specific synapse development in CA1 pyramidal neurons. Neuron. 2012;76:762–775. doi: 10.1016/j.neuron.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirone A., Cozzi B., Edelstein L., Peruffo A., Lenzi C., Quilici F., Antonini R., Castagna M. Topography of Gng2- and NetrinG2-expression suggests an insular origin of the human claustrum. PLoS ONE. 2012;7:e44745. doi: 10.1371/journal.pone.0044745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movements are similar to those observed in Angelman syndrome.