Abstract
Background
The DMD gene is one of the largest human genes, being composed of 79 exons. Dystrophin Dp116 expressed from the promoter in intron 55 is a Schwann cell-specific isoform. The pathophysiological roles of Dp116 are largely unknown, because of its limited expression. This study assessed the expression of Dp116 in glioblastoma cells and evaluated the splicing patterns of the DMD gene in these cells.
Methods
Full-length Dp116 cDNA was PCR amplified from U-251 glioblastoma cells. Dp116 protein was analyzed by Western blotting.
Results
Full-length Dp116 cDNA, extending from exon S1 to exon 79, was PCR amplified to avoid confusion with other DMD isoforms. The full-length Dp116 transcript was amplified as nearly 3 kb in size. Western blotting of U-251 cell lysates revealed a signal at a position corresponding to vector-expressed Dp116 protein, indicating that Dp116 is expressed in glioblastoma cells. Sequencing of the amplified product revealed five splice variants, all skipping exon 78. The most abundant transcript lacked only exon 78 (Dp116b), whereas the second most abundant transcript lacked both exons 71 and 78 (Dp116ab). A third transcript lacking exons 71–74 and 78 was also identified (Dp116bc). Two novel splicing patterns were also observed, one with a deletion of exons 68 and 69 (Dp116bΔ68-69) and the other with a 100 bp deletion in the 5’ terminal end of exon 75 (75s), which was produced by the activation of a cryptic splice acceptor site (Dp116b75s). However, the splicing patterns in glioblastoma cells of DMD exons in Dp116 and Dp71 showed no significant differences.
Conclusions
Dp116 is expressed in glioblastoma cells as five splicing variants, with Dp116b being the most abundant. Two novel splicing patterns of DMD exons were observed.
Keywords: Dp116, DMD gene, Dystrophin, Glioblastoma, Splicing, Splice variants
Highlights
-
•
Dp116 is a Schwann cell-specific dystrophin isoform.
-
•
Dp116 was shown to be expressed in glioblastoma, a lethal cerebral malignancy.
-
•
Skipping of exon 78 was the default pathway.
-
•
Of the five alternatively spliced variants detected, Dp116b was the most abundant.
-
•
DMD exons showed two novel splicing patterns, one with cryptic splice activation.
1. Introduction
The DMD gene is one of the largest genes in the human genome, encoding a 14-kb long transcript consisting of 79 exons spread over more than 2.4 Mb on the X chromosome [1]. The gene exhibits a highly complex arrangement, with eight alternative promoters scattered in its introns driving the expression of four full-length and four short dystrophin isoforms in a tissue- or development-specific manner [2,3]. Dp427 m is a full-length muscle-specific isoform, a lack of which causes Duchenne muscular dystrophy (DMD) (OMIM310200), a fatal progressive muscle wasting disease [4]. Four alternative promoter-first exon regions are embedded in downstream introns and produce the Dp260, Dp140, Dp116 and Dp71 isoforms. Dp71, the shortest isoform, is transcribed from a promoter in intron 62 of the DMD gene, with the Dp71 transcript consisting of Dp71-specific exon G1 and DMD exons 63–79 [5]. Thus, DMD exons 63–79 are incorporated into the mRNA of not only Dp71 but also all other isoforms.
Dp116, the second shortest isoform of the DMD gene, is transcribed from the Dp116 promoter in intron 55. Dp116 transcript is 5.2 kb long and consists of the Dp116-specific exon S1 joined to DMD exons 56–79 [6,7]. The Dp116 promoter is characterized by its very specific activation in Schwann cells [2]. Due to its limited expression and large size, the pathophysiological roles of Dp116 are largely unknown [7]. We recently reported that Dp116 plays a role in the development of cardiac dysfunction in DMD patients [8], suggesting that Dp116 expression is not limited to Schwann cells. However, exact mechanism of Dp116 to enhance cardiomyopathy remains unknown. It is necessary to understand physiological roles of Dp116 well, since cardiomyopathy is a leading cause of early death in DMD [9].
Alternative splicing is a mechanism that enables cells to generate various diverse proteins from a limited number of genes [10], as well as having important physiological functions in different developmental processes in humans [11]. The most frequent type of alternative splicing of the DMD gene consists of the skipping of exons [12]. Though most skipping of DMD exons occurs in-frame exons, skipping of exon 78 shifts the reading frame to produce a large dystrophin with an elongated C-terminal amino acid sequence [13,14]. Analysis of Dp71 transcripts has identified multiple exon skipping patterns in the region from exon 71 to exon 78 [5,15,16].
Glioblastoma is an aggressive brain tumor highly resistant to treatment [17]. Molecular characterization of glioblastoma may help in designing effective therapies. One method of treatment may be the manipulation of RNA processing of tumor drivers [18]. The DMD gene is regarded as a tumor suppressor gene [19], with Dp71 shown to have tumor suppressive activity [20,21]. We previously showed that glioblastoma cells express Dp71, with this Dp71 composed of six splice variants [16], suggesting that glioblastoma provides a specific environment for regulating the splicing of DMD exons.
During the study on Dp71, we have obtained a signature that indicates the expression of Dp116 in U-251 cells. This study was designed to characterize Dp116 and the alternative splicing of DMD exons in glioblastoma cells. Dp116 mRNA and protein were shown to be expressed in glioblastoma cells. Furthermore, five splice variants and two novel splicing patterns of the DMD gene were identified. Splicing patterns of DMD exons were compared in Dp116 and Dp71. Identification of Dp116 in U-251 cells facilitates studies on the roles of Dp116.
2. Methods
2.1. Cell lines
The U-251 glioblastoma cell line was purchased from the Japanese Collection of Research Bioresources (JCRB; Osaka, Japan) within the past year. Cells were cultured in minimum essential medium (MEM; Gibco Life Technologies, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone, GE Healthcare, Chicago, IL, USA) at 37 °C in a 5% CO2 humidified incubator. The HEK293 embryonic kidney was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) within the last 2 years. HEK293 was cultured in Eagle's Minimum Essential Medium (Wako Pure Chemical Industries Ltd., Osaka, Japan). These media were supplemented with 10% FBS (Gibco Life Technologies, Grand Island, NY, USA) and 1% antibiotic-antimycotic solution (Gibco Life Technologies) at 37 °C in a 5% CO2 humidified incubator.
2.2. Transcript analysis
Cultured cells were rinsed twice with phosphate buffered saline (PBS; Sigma-Aldrich Co., St. Louis, MO, USA) and collected using the lysis/binding buffer from High Pure RNA isolation kits (Roche Diagnostics, Basel, Switzerland). RNA was extracted from these cells using High Pure RNA isolation kits (Roche Diagnostics). Human total RNA from skeletal muscle was obtained from a human total RNA Master Panel II (Clontech Laboratories, Inc., Mountain View, CA, USA). cDNA was synthesized from 0.5 μg of each total RNA using random primers, as described [22]. The 5′ end fragment of Dp116 transcript was PCR amplified as described [23]. The integrity and concentration of these cDNA preparations were assessed by amplifying the mRNA of the house keeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), as described [24]. Full-length Dp116 transcript was PCR amplified using a set of primers on exon S1 (Dp116F2; 5′-GGGTTTTCTCAGGATTGCTAT-3′) and exon 79 (5F; 5′-ATCATCTGCCATGTGGAAAAG-3′).
PCR amplification was performed in a total volume of 10 μl, containing 1 μl of cDNA, 1 μl of 10 × ExTaq buffer (Takara Bio, Inc., Shiga, Japan), 0.25 U of ExTaq polymerase (Takara Bio, Inc.), 500 nM of each primer, and 250 μM dNTPs (Takara Bio, Inc.). Amplification was performed on a Mastercycler Gradient PCR machine (Eppendorf, Hamburg, Germany), with each consisting of an initial denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 0.5 min, annealing at 60 °C for 0.5 min, and extension at 72 °C for 1.5 min. Amplified PCR products were electrophoresed on 2% agarose gels. The amplification was done at least two times.
To examine the exon structure of the full-length transcript, the amplified product was subcloned into the pT7 blue T vector (Novagen, Inc., San Diego, CA, USA) and sequenced as described before [16].
2.3. Overexpression of Dp116
A Dp116-expressing plasmid was constructed by inserting the Dp116 coding sequence, consisting of DMD exons S1 and 56–79, into the plasmid pcDNA3, a mammalian expression vector with a CMV promoter (Invitrogen, Thermo Fisher Scientific Inc., Carlsbad, CA, USA). This construct was synthesized by FASMAC Co., Ltd. (Atsugi, Japan), and its sequence was confirmed by sequencing. HEK293 cells grown to 80% confluence on six-well culture dishes were transfected with 2 μg of synthesized plasmid in 4 μL lipofectamine2000 (Thermo Fischer Scientific, Waltham, MA, USA). After culturing for 24 h, the cells were harvested.
2.4. Western blotting
Dp116 was analyzed by Western blotting, as described [25]. Blots were incubated overnight with a 1:1000 dilution of rabbit polyclonal antibody directed against the C-terminal domain of human dystrophin (ab154168; Abcam, Cambridge, UK), followed by incubation with anti-rabbit IgG secondary antibody (GE Healthcare). As a loading control, membranes were incubated with a 1:4000 dilution of GAPDH antibody (2118S; Cell Signaling Technology Inc.), followed by incubation with anti-mouse IgG secondary antibody (GE Healthcare). Immunoreactive bands were detected with Immobilon Forte Western HRP Substrate (Merck Millipore, MA, USA).
2.5. Sequence analysis
Splicing acceptor sites and branch points were assessed in nucleotide sequences using the Human Splicing Finder - Version 3.1 bioinformatic tool (http://www.umd.be/HSF3/). Searches for homology of amino acid sequences were performed using the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
3. Results
3.1. Dp116 transcript in U-251 glioblastoma cells
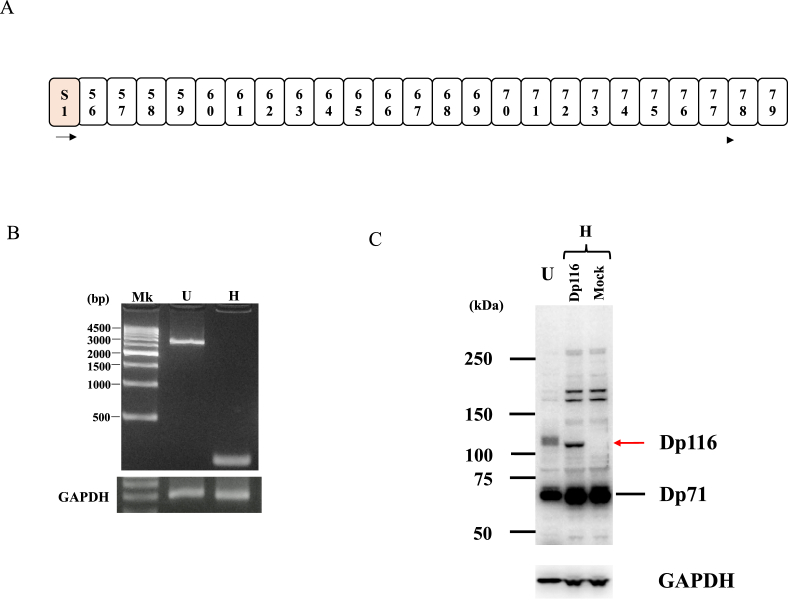
Although Dp116 is expressed in a Schwann cell-specific manner [7], its expression was analyzed in U-251 glioblastoma cells. To assess its expression, it was necessary to amplify full-length Dp116 cDNA as a single product. Conventional PCR amplification of full-length Dp116 cDNA produced a single band of expected size (2.9 kb) on an electropherogram (Fig. 1B). Expression of full-length Dp116 transcript was subsequently analyzed in HEK293 cells. However, no PCR amplified product was observed (Fig. 1B).
Fig. 1.
Identification of Dp116 transcripts. A. Schematic illustration of the exon structures of Dp116 and Dp71 In Dp116, the specific exon S1 was joined to DMD exons 56–79. The open and shaded boxes indicate DMD exons and promoter specific exons, respectively. The number in each box indicates the exon number. Arrows indicate the location and direction of primers used for PCR amplification. B. Amplification of full-length Dp116 cDNA. The full-length Dp116 cDNA was amplified using a forward primer on exon S1 and a reverse primer on exon 79. Electropherograms of PCR amplified products are shown. The amplification revealed a single band near the 3000 bp marker in U-251 cells (U), but not in HEK293 cells (H). As a control, GAPDH was also amplified (GAPDH). Mk refers to a size marker. C. Identification of Dp116 protein by Western blotting Protein extracts were prepared from U-251 cells and from HEK293 cells transfected with a Dp116 expressing or an empty vector. Following electrophoresis and transfer to PVDF membranes, the membranes were incubated with a rabbit antibody against the C-terminal region of dystrophin. A band slightly above 100 kDa was observed in HEK293 cells (H) transfected with a Dp116 expressing vector (Dp116), but not in mock-transfected cells (Mock). In U-251 cells (U), a broad band was revealed at the position corresponding to Dp116. As a control, the blots were incubated with an antibody against GAPDH (bottom). Size markers are shown at the left.
3.2. Dp116 protein in U-251 cells
The presence of Dp116 protein in glioblastoma cell lysates was examined by Western blotting, using an antibody against C-terminal region of dystrophin (Fig. 1C). To confirm the ability of this antibody to detect Dp116, it was tested against HEK293 cells transfected with a vector expressing Dp116. Dp116 expressed in HEK293 cells was present as a band just above the 100kDa size marker, but no band was detected in empty vector-transfected cells. A signal corresponding to the artificially-expressed Dp116 in HEK293 cells was detected in the lysates of glioblastoma cells, confirming that glioblastoma cells expressed Dp116 protein, as well as Dp116 mRNA.
3.3. Alternative splicing of Dp116 transcripts in glioblastoma cells
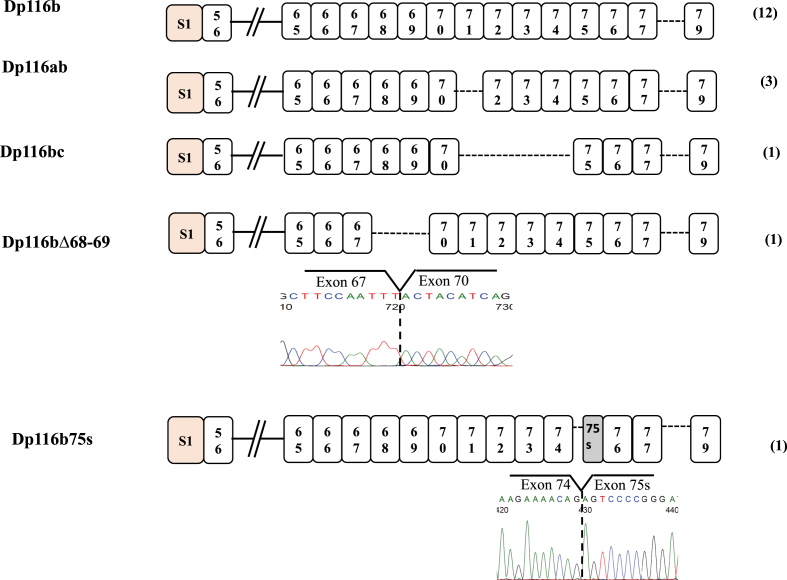
To assess whether DMD exons 71–78 of Dp116 are alternatively spliced, as in Dp71, the PCR amplified products of 20 clones of full-length Dp116 were sequenced. Initially, all clones were sequenced using a primer on exon S1, revealing a nearly 1000 nucleotide-long sequence. Two clones were identified as no-specific, whereas the other 18 had completely normal exon structures, from exon S1 to exon 61. Subsequently, these 18 clones were subjected to sequencing starting from exon 61. Seventeen clones had normal sequences from exons 61 to 70, whereas one clone showed the direct joining of exon 67 to exon 70, deleting both exons 68 and 69 (Fig. 2). Finally, exons 71–79 were sequenced in these 18 clones. All clones lacked exon 78, suggesting that omission of exon 78 was a default splicing pathway in glioblastoma. Some clones also showed skipping of exons 71–74 and both exons 71 and 78 (Fig. 2). Alternative splicing, omitting exons 68 and 69, had never been reported in any DMD isoforms. Because these two exons were removed from the Dp116 sequence, skipping of these exons was likely considered a novel alternative splicing of the DMD gene. One clone had the same exon structure as Dp116b, but exon 75 lacked 100 bp at its 5′end (exon 75s).
Fig. 2.
Dp116 splice variants. Sequencing of the amplified product in 18 clones revealed Dp116b in 12 clones, Dp116ab in three, and Dp116bc, Dp116bΔ68–69 and Dp116b75s in one each. Their exon structures are illustrated schematically. Boxes represent exons and the numbers in the boxes represent exon numbers. The number of clones are shown on the right (parenthesis). Partial nucleotide sequences at the junctions between exons 67 and 70 and between exon 74 and exon 75s are shown under the boxes.
3.4. Novel exon 75s created by cryptic splice acceptor site activation within exon 75
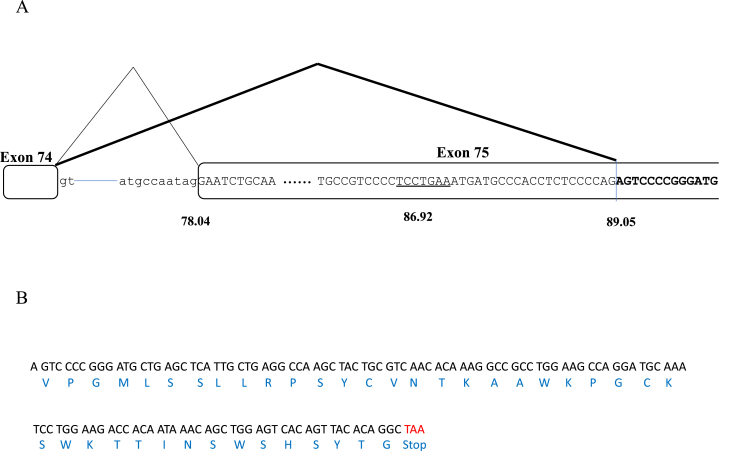
Because exon 75s was an unexpected product but was inserted into mRNA, its nucleotide sequence was examined in detail. As expected, AG dinucleotides that usually comprise splice acceptor sites were identified upstream of truncated exon 75 (Fig. 3). Human Splicing Finder analysis showed that this acceptor site had a higher probability score than the conventional splice acceptor site of exon 75 (Fig. 3). In addition, hepta-nucleotides 22 bp upstream of the cryptic splice site were found to have a high probability score as a branch point (Fig. 3). The sequence between the acceptor site and branch point were rich in pyrimidine nucleotides (C and T), forming a polypyrimidine tract that is necessary for proper splicing [26]. These findings indicated that exon 75s was an alternative splicing product formed using a cryptic splice site within exon 75. The deletion of 100 bp shifted the original dystrophin reading frame and was expected to encode new peptides. Exon 75s encoded 41 amino acids, with codon 42 being a stop codon (Fig. 3). A homology search of its amino acid sequence failed to identify any highly homologous protein.
Fig. 3.
Exon 75s created by cryptic splice site activation. A. Schematic illustration of part of the genomic structure of the DMD gene. Authentic splicing proceeds from exon 74 to exon 75 (diagonal line), whereas novel alternative splicing proceeds from exon 74 to the cryptic splice acceptor site (bold diagonal line). Boxes and lines indicate exons and introns, respectively. Partial nucleotide sequences of exons and introns are shown as upper- and lower-case letters, respectively. The branch point sequence is underlined. Numbers under the schema indicate probability scores for splice acceptor sites and branch points. B. Nucleotide sequence of exon 75s (top) and its translated amino acid sequence (bottom). Codon 42 of exon 75s is a stop codon.
3.5. Isoforms of Dp116
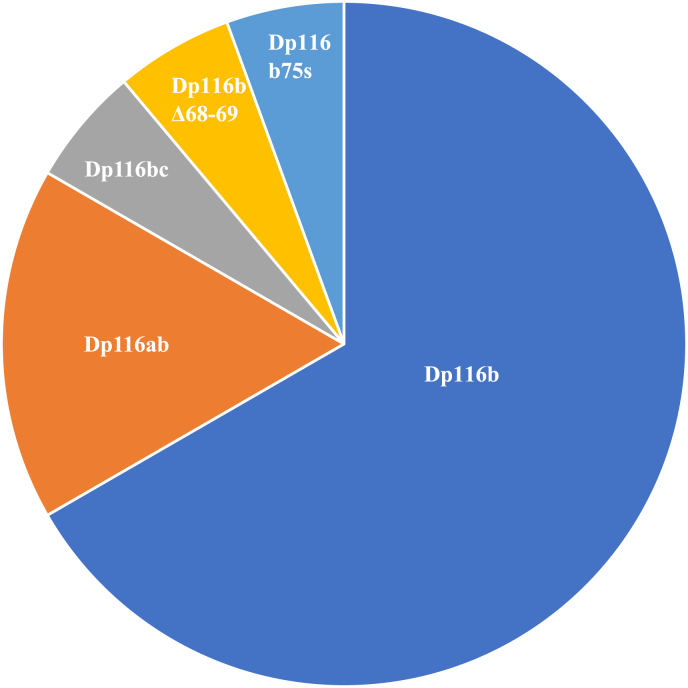
Of the 18 clones, the most abundant (12 clones, 66.6 %) lacked only exon 78. This variant was named Dp116b (Fig. 4), based on the naming of Dp71 transcripts [5]. Three clones (16.6%) lacked both exons 71 and 78, with this variant named Dp116ab. Each of the three remaining clones had different splicing patterns. One lacked exons 71–74 and exon 78 (Dp116bc), one lacked exons 68, 69 and 78 (Dp116bΔ68–69). The deletion of exons 68 and 69 maintained the reading frame of the protein, with Dp116bΔ68–69 expected to lack 93 amino acid residues. Skipping of exon 78 was the default condition in Dp116, indicating specific splicing regulatory mechanisms for Dp116.
Fig. 4.
Isoforms of Dp116. Frequency of isoforms of Dp116 is shown. The percentages of isoforms of Dp116 in U-251 cells were calculated from colony numbers. The most abundant isoform was Dp116b.
4. Discussion
Schwann cell-specific Dp116 was found to be expressed in U-251 glioblastoma cells, the third type of human non-Schwann cell found to express Dp116. Expression of Dp116 in skin fibroblast cells was shown to be lower in DMD patients than in controls [27], and PCR amplification showed that Dp116 was expressed in cardiac muscle [8]. Although SH-SY5Y cells expressed brain specific Dp427c, they did not express Dp116 [23], indicating that Dp116 expression is not universal in neuronal cells. The ability of glioblastoma cells to express Dp116 suggests that they have characteristics of Schwann cells. Future studies, however, are required for confirmation.
Glioblastoma is a highly malignant brain tumor. Despite the development of novel therapeutic modalities, median overall patient survival is only 15 months [17]. Further molecular characterization of glioblastomas may reveal new therapeutic targets [18]. Dp116 has not been included in studies of the molecular characteristics of U-251 glioblastoma [28,29]. In our study, conventional PCR successfully amplified full-length Dp116, suggesting that Dp116 expression is upregulated in U-251 cells and may be a molecular biomarker in glioblastoma. The involvement of Dp116 in G-protein signaling pathways important in glioblastoma tumorigenesis suggests that Dp116 may play a role in the molecular pathogenesis of glioblastoma [27,30]. Additional molecular studies are required to test this hypothesis.
Five splice variants of Dp116 were shown to be present in U-251 glioblastoma cells, with skipping of exon 78 found to be a default splicing pathway. Skipping of this exon has also been reported to occur in brain DMD transcripts [13]. Of the five splice variants, four were produced by alternative splicing of authentic exons and were expected to produce truncated Dp116 protein isoforms. In contrast, the fifth splice variant (Dp116b75s) was produced by activating a cryptic splice site, creating exon 75s. Although RNA-Seq technology showed that cryptic splice sites in four DMD exons were activated in adult human skeletal muscle, exon 75s had never been identified [12]. Dp116b75s is expected to produce a C-terminal truncated Dp116, as Dp40 encoded in a transcript containing part of a DMD intron has been reported functional [31,32].
Cancer cells have been reported to show many alternative splicing events, with these events associated with cancer progression and metastasis, and with the deregulation of splicing factors [33]. Although the DMD gene has been reported to be a tumor suppressor gene, few studies have assessed alternative splicing of the DMD gene in cancer [25]. Results to date cannot determine whether identified novel splicing patterns were cancer- or glioblastoma-specific. If cancer-specific, elucidation of their physiological roles would suggest novel cancer treatments, especially for glioblastoma.
Identification of Dp116 in cultured cells is expected to facilitate studies on physiological roles of Dp116. Thereby, understanding of pathophysiology of cardiomyopathy of DMD, a leading cause of early death of DMD patients, would be promoted.
Funding
This work was supported in part by an Intramural Research Grant (28-6) for Neurological and Psychiatric Disorders of NCNP and JSPS KAKENHI Grant Number 18K07861.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100703.
Contributor Information
Abdul Qawee Mahyoob Rani, Email: rani@reha.kobegakuin.ac.jp.
Kazuhiro Maeta, Email: maeta@reha.kobegakuin.ac.jp.
Tatsuya Kawaguchi, Email: kawaguchi.tatsuya.sg@rdn.daiichisankyo.co.jp.
Hiroyuki Awano, Email: awahiro@med.kobe-u.ac.jp.
Masashi Nagai, Email: natsu@med.kobe-u.ac.jp.
Hisahide Nishio, Email: nishio@reha.kobegakuin.ac.jp.
Masafumi Matsuo, Email: matsuo@kobe-u.ac.jp.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ahn A.H., Kunkel L.M. The structural and functional diversity of dystrophin. Nat. Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. ([doi]) [DOI] [PubMed] [Google Scholar]
- 2.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 3.Massourides E., Polentes J., Mangeot P.E., Mournetas V., Nectoux J., Deburgrave N., Nusbaum P., Leturcq F., Popplewell L., Dickson G., Wein N., Flanigan K.M., Peschanski M., Chelly J., Pinset C. Dp412e: a novel human embryonic dystrophin isoform induced by BMP4 in early differentiated cells. Skelet. Muscle. 2015;5:40. doi: 10.1186/s13395-015-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnkrant D.J., Bushby K., Bann C.M., Apkon S.D., Blackwell A., Brumbaugh D., Case L.E., Clemens P.R., Hadjiyannakis S., Pandya S., Street N., Tomezsko J., Wagner K.R., Ward L.M., Weber D.R. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and neuromuscular, rehabilitation, endocrine, and gastrointestinal and nutritional management. Lancet Neurol. 2018;17:251–267. doi: 10.1016/S1474-4422(18)30024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tadayoni R., Rendon A., Soria-Jasso L.E., Cisneros B. Dystrophin Dp71: the smallest but multifunctional product of the Duchenne muscular dystrophy gene. Mol. Neurobiol. 2012;45:43–60. doi: 10.1007/s12035-011-8218-9. ([doi]) [DOI] [PubMed] [Google Scholar]
- 6.Byers T.J., Lidov H.G., Kunkel L.M. An alternative dystrophin transcript specific to peripheral nerve. Nat. Genet. 1993;4:77–81. doi: 10.1038/ng0593-77. [DOI] [PubMed] [Google Scholar]
- 7.Matsuo M., Awano H., Matsumoto M., Nagai M., Kawaguchi T., Zhang Z., Nishio H. Dystrophin Dp116: a yet to Be investigated product of the Duchenne muscular dystrophy gene. Genes. 2017;8 doi: 10.3390/genes8100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto T., Awano H., Zhan Z., Enomoto-Sakuma M., Kitaaki S., Matsumoto M., Nagai M., Sato I., Imanishi T., Hayashi N., Matsuo M., Iijima K., Jun S. Cardiac dysfunction in Duchenne muscular dystrophy is less frequent in patients with mutations in the dystrophin Dp116 coding region than in other regions. Circ Genome Precis Med. 2018;11 doi: 10.1161/CIRCGEN.117.001782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamdar F., Garry D.J. Dystrophin-deficient cardiomyopathy. J. Am. Coll. Cardiol. 2016;67:2533–2546. doi: 10.1016/j.jacc.2016.02.081. [DOI] [PubMed] [Google Scholar]
- 10.Park E., Pan Z., Zhang Z., Lin L., Xing Y.I. The expanding landscape of alternative splicing variation in human populations. Am. J. Hum. Genet. 2018;102:11–26. doi: 10.1016/j.ajhg.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baralle F., Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017;18:437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouge A.L., Murauer E., Beyne E., Miro J., Varilh J., Taulan M., Koenig M., Claustres M., Tuffery-Giraud S. Targeted RNA-Seq profiling of splicing pattern in the DMD gene: exons are mostly constitutively spliced in human skeletal muscle. Sci. Rep. 2017;7:39094. doi: 10.1038/srep39094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feener C.A., Koenig M., Kunkel L.M. Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus. Nature. 1989;338:509–511. doi: 10.1038/338509a0. [DOI] [PubMed] [Google Scholar]
- 14.Rau F., Laine J., Ramanoudjame L., Ferry A., Arandel L., Delalande O., Jollet A., Dingli F., Lee K.Y., Peccate C., Lorain S., Kabashi E., Athanasopoulos T., Koo T., Loew D., Swanson M.S., Le Rumeur E., Dickson G., Allamand V., Marie J., Furling D. Abnormal splicing switch of DMD's penultimate exon compromises muscle fibre maintenance in myotonic dystrophy. Nat. Commun. 2015;6:7205. doi: 10.1038/ncomms8205. ncomms8205 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aragon J., Gonzalez-Reyes M., Romo-Yanez J., Vacca O., Aguilar-Gonzalez G., Rendon A., Vaillend C., Montanez C. Dystrophin Dp71 isoforms are differentially expressed in the mouse brain and retina: report of new alternative splicing and a novel nomenclature for Dp71 isoforms. Mol. Neurobiol. 2018;55:1376–1386. doi: 10.1007/s12035-017-0405-x. [DOI] [PubMed] [Google Scholar]
- 16.Rani A.Q.M., Farea M., Maeta K., Kawaguchi T., Awano H., Nagai M., Nishio H., Matsuo M. Identification of the shortest splice variant of Dp71, together with five known variants, in glioblastoma cells. Biochem. Biophys. Res. Commun. 2019;508:640–645. doi: 10.1016/j.bbrc.2018.11.168. [DOI] [PubMed] [Google Scholar]
- 17.Kawano H., Hirano H., Yonezawa H., Yunoue S., Yatsushiro K., Ogita M., Hiraki Y., Uchida H., Habu M., Fujio S., Oyoshi T., Bakhtiar Y., Sugata S., Yamahata H., Hanaya R., Tokimura H., Arita K. Improvement in treatment results of glioblastoma over the last three decades and beneficial factors. Br. J. Neurosurg. 2015;29:206–212. doi: 10.3109/02688697.2014.967750. [DOI] [PubMed] [Google Scholar]
- 18.Marcelino Meliso F., Hubert C.G., Favoretto Galante P.A., Penalva L.O. RNA processing as an alternative route to attack glioblastoma. Hum. Genet. 2017;136:1129–1141. doi: 10.1007/s00439-017-1819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Marino-Enriquez A., Bennett R.R., Zhu M., Shen Y., Eilers G., Lee J.C., Henze J., Fletcher B.S., Gu Z., Fox E.A., Antonescu C.R., Fletcher C.D., Guo X., Raut C.P., Demetri G.D., van de Rijn M., Ordog T., Kunkel L.M., Fletcher J.A. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat. Genet. 2014;46:601–606. doi: 10.1038/ng.2974. ng.2974 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan S., Xiao X. Dp71 as a tumor suppressor: combined efforts of nuclear lamin B1 and emerin? J Xiangya Med. 2017;2:25. [Google Scholar]
- 21.Tan S., Tan J., Tan S., Zhao S., Cao X., Chen Z., Weng Q., Zhang H., Wang K.K., Zhou J., Xiao X. Decreased Dp71 expression is associated with gastric adenocarcinoma prognosis. Oncotarget. 2016;7:53702–53711. doi: 10.18632/oncotarget.10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuo M., Masumura T., Nishio H., Nakajima T., Kitoh Y., Takumi T., Koga J., Nakamura H. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy kobe. J. Clin. Investig. 1991;87:2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishida A., Minegishi M., Takeuchi A., Awano H., Niba E.T., Matsuo M. Neuronal SH-SY5Y cells use the C-dystrophin promoter coupled with exon 78 skipping and display multiple patterns of alternative splicing including two intronic insertion events. Hum. Genet. 2015;134:993–1001. doi: 10.1007/s00439-015-1581-2. [DOI] [PubMed] [Google Scholar]
- 24.Thi Tran H.T., Takeshima Y., Surono A., Yagi M., Wada H., Matsuo M. A G-to-A transition at the fifth position of intron 32 of the dystrophin gene inactivates a splice donor site both in vivo and in vitro. Mol. Genet. Metab. 2005;85:213–219. doi: 10.1016/j.ymgme.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Niba E.T.E., Yamanaka R., Rani A.Q.M., Awano H., Matsumoto M., Nishio H., Matsuo M. DMD transcripts in CRL-2061 rhabdomyosarcoma cells show high levels of intron retention by intron-specific PCR amplification. Cancer Cell Int. 2017;17:58. doi: 10.1186/s12935-017-0428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habara Y., Doshita M., Hirozawa S., Yokono Y., Yagi M., Takeshima Y., Matsuo M. A strong exonic splicing enhancer in dystrophin exon 19 achieve proper splicing without an upstream polypyrimidine tract. J. Biochem. 2008;143:303–310. doi: 10.1093/jb/mvm227. [DOI] [PubMed] [Google Scholar]
- 27.Labarque V., Freson K., Thys C., Wittevrongel C., Hoylaerts M.F., De Vos R., Goemans N., Van Geet C. Increased Gs signalling in platelets and impaired collagen activation, due to a defect in the dystrophin gene, result in increased blood loss during spinal surgery. Hum. Mol. Genet. 2008;17:357–366. doi: 10.1093/hmg/ddm312. [DOI] [PubMed] [Google Scholar]
- 28.Wang A., Zhang G. Differential gene expression analysis in glioblastoma cells and normal human brain cells based on GEO database. Oncol Lett. 2017;14:6040–6044. doi: 10.3892/ol.2017.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motaln H., Koren A., Gruden K., Ramsak Z., Schichor C., Lah T.T. Heterogeneous glioblastoma cell cross-talk promotes phenotype alterations and enhanced drug resistance. Oncotarget. 2015;6:40998–41017. doi: 10.18632/oncotarget.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherry Stella. G protein-coupled receptors as oncogenic signals in glioma: emerging therapeutic avenues. Neuroscience. 2014;278:222–236. doi: 10.1016/j.neuroscience.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tozawa T., Itoh K., Yaoi T., Tando S., Umekage M., Dai H., Hosoi H., Fushiki S. The shortest isoform of dystrophin (Dp40) interacts with a group of presynaptic proteins to form a presumptive novel complex in the mouse brain. Mol. Neurobiol. 2012;45:287–297. doi: 10.1007/s12035-012-8233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aragon J., Martinez-Herrera A., Bermudez-Cruz R.M., Bazan M.L., Soid-Raggi G., Ceja V., Coy-Arechavaleta A.S., Aleman V., Depardon F., Montanez C. EF-hand domains are involved in the differential cellular distribution of dystrophin Dp40. Neurosci. Lett. 2015;600:115–120. doi: 10.1016/j.neulet.2015.05.038. S0304-3940(15)00395-X [pii] [DOI] [PubMed] [Google Scholar]
- 33.Oltean S., Bates D.O. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. onc2013533 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.