Highlights
-
•
Genetic screening of SCA10 in large cohort of Indian SCA patients.
-
•
Estimation of at-risk haplotype using population genetics approach in south Asians.
-
•
Suggestive rarity of SCA10 in the Indian Population.
Abbreviations: SCA, Spinocerebellar ataxia; SNPs, Single Nucleotide Polymorphisms; RP-PCR, Repeat-primed PCR; EUR, European population; EAS, East Asian population; AMR, American population; AFR, African population; SAS, South Asian population; KGP, 1000 Genomes Project
Keywords: Spinocerebellar ataxia, SCA10, Autosomal dominant cerebellar ataxia, ATXN10, Pentanucleotide repeat expansion, Haplotype markers
Abstract
Spinocerebellar ataxia type 10 (SCA10) is a rare autosomal dominant cerebellar ataxia caused by nucleotide ATTCT expansion in ATXN10 gene. SCA10 has been reported in patients of cerebellar ataxia from Amerindian/Latin America and in East Asian ancestry. A common founder has been ascribed to the origin of ATTCT repeat expansion mutation in both the population. Here we present our investigation of the SCA10 pentanucleotide repeat expansion in 461 SCA patients of the Indian population. The analysis of multi-ethnic at-risk haplotype C-(ATTCT)n-GGC was performed using genotype data of various ethnic population included in the 1000 Genomes Project (KGP) to infer the prevalence of at-risk haplotype in the Indian populations. Unsurprisingly, none of the patient’s DNA samples with (ATTCT)n expansion was observed in pathological range, however, the observed normal range of (ATTCT)n was 8–22 repeats, suggesting very rare or absence of the occurrence of SCA10 in Indian SCA patients. The at-risk haplotype, CGGC was found to be the most prevalent haplotype across different populations and no segregation of CGGC haplotype with large normal or small normal ATTCT repeats length was observed. However, on extended haplotype analysis, some lineage of CGGC with a flanking divergence at 5′ end was observed specifically in the American or East Asian population but not in other population in KGP dataset. Together, these evidence points towards the absence of SCA10 in Indian population and haplotype-based analysis also suggests its occurrence to be rare in South Asian, European and African population. Further investigations are required to establish the present finding.
Significance
The implications of the findings of this study are 1.) For the diagnostic work-up of SCAs in the Indian population and to decide upon inclusion of SCA10 in panel based genetic investigations even for Indians living abroad. 2.) The haplotype based inference of its presumptive prevalence through the estimation of at-risk haplotype using population genetics approach (South-Asians as the background) allowed us to estimate the possible absence of SCA10 in Indian population. SCA10 is a rare autosomal dominant cerebellar ataxia mostly reported among SCA patients from Latin America and recently described in East Asia population. The genetic study of SCA10 performed in the unrelated Indian spinocerebellar ataxia patients with heterogeneous ethnicity confirmed its absence from the Indian population and that conforms to population genetic based inference of its rarity or absence. 3.) This approach may be adopted for the screening of other subtypes of SCAs, i.e. other rare SCAs e.g. SCA31, SCA36, and SCA37.
1. Introduction
Spinocerebellar ataxia type 10 (SCA10) is a rare autosomal dominant cerebellar ataxia caused by pentanucleotide (ATTCT) repeat expansion in intron 9 of the ATXN10 gene which maps to chr22q13.3 region [[1], [2], [3]]. The number of (ATTCT)n repeats commonly found among different populations ranged from 10 to 29, whereas the pathological expanded alleles ranged from 800 to 4500 repeats in affected patients. Furthermore, the intermediate size alleles of repeats 280 to 850 exhibited reduced penetrance [2,4,5]. SCA10 is characterized by progressive cerebellar dysfunctions together with epilepsy and peripheral neuropathy. The first identification of SCA10 in two Mexican families [1,6] revealed the characteristic phenotype of pure cerebellar ataxia associated with seizures. Subsequently, general features of ataxia including gait ataxia, dysarthria, variable limb ataxia, and ocular movement abnormalities have been witnessed in SCA10 positive patients [5,7,8]. Further characterization of new SCA10 families unveiled association with diverse phenotypes like polyneuropathy, pyramidal signs, cognitive impairment and neuropsychiatric disturbance, and even manifestation of ataxia without seizures [5,9]. The observed variability in the clinical spectrum of patients could be linked to the ethnicity of the reported cases [5]. In the reported cases, the age of onset of SCA10 varied from 12 to 48 years, however, progressive earlier disease onset has been anticipated in successive generations in affected members of SCA10 families [1,2,9].
Initially, SCA 10 was widely found in regions of Latin America mostly in Mexico and Brazil, where it was the second most common SCA after SCA3 (Brazil) and SCA2 (Mexico) [10]. An extensive haplotype analysis by Almeida et al., demonstrated ancestral origin of SCA10, presumed to have arisen in the Amerindian population. These findings relatively combined with the sequential dispersion of SCA10 all over Latin American countries, provide evidence for a common founder mutation [11,12]. SCA10 cases reported in patients from the USA, Mexico, Argentina, Venezuela, Colombia, Bolivia, Brazil, Peru, China and recently in Japan shared common haplotype found in the Amerindian population [5,7,[13], [14], [15], [16], [17]].
Previous reports have suggested the absence ATXN10 pathogenic expansion in certain Asian populations like Japanese, Chinese, and Indian [14,15]. Contrary to this definition/belief, recently SCA10 positive families have been identified in the East Asian population of China and Japan. This intriguing finding prompted us to investigate the presence of SCA10 in the Indian population. Henceforth, the aim of the present study was to investigate SCA10 (ATTCT)n repeat expansion in a reasonable cohort of 461 patients of Indian origin presenting with ADCA phenotype which were essentially negative for primary genetic screening panel for Ataxia. The distribution of ATXN10 normal alleles was also determined in a cohort of control samples. Furthermore, the analysis and distribution of known intragenic haplotype CGGC and extended haplotype lineages flanking the pentanucleotide repeat of ATXN10 was performed utilizing the dataset from 1000 Genome Project to support our findings through population genetics approach [18].
2. Methods
2.1. Patient recruitment
A total of 461 unrelated patients enrolled in the CSIR-GOMED project [an initiative taken for investigation of neurogenetic and other prevalent genetic disorders in the Indian population (http://gomed.igib.in)] during 2015–2018 were included in the study. These patient samples had essentially come for genetic screening for SCA and was found negative for the primary screening panel inclusive of SCA1, SCA2, SCA3, SCA6, SCA7, SCA12, and SCA17.
We also included 117 neurologically healthy subjects as controls in this study. The study was approved by the ethics committee CSIR-IGIB. Written informed consent was obtained from all patients and controls for molecular genetic testing and research.
2.2. Molecular testing for ATXN10 repeat expansion analysis
Blood samples was collected from all patients and controls included in the study. Genomic DNA was extracted from peripheral blood lymphocytes using modified salting out method ((Miller et al., 1988).
2.2.1. PCR and fragment analysis
Flanking PCR was performed using fluorescently labelled ATXN10 locus-specific primers to assess (ATTCT)n repeats in all patients and controls included in the study. The sequence of primers used were as follows: SCA10-FP (5´–6-FAM-CAGATGGCAGAATGATAAACTCAA–3´) primer and SCA10-RP (5´–AGCCTGGGCAACATAGAGAGA–3´). PCR was performed in a 10-μl reaction containing 100ոg of genomic DNA, 0.3 pmol of each primer, 2.5 mM of each dNTP, 15 mM MgCl2, 5 M betaine and 1U of AmpliTaq Gold polymerase in the 10X buffer (Applied Biosystems). The reaction conditions were as follows: initial denaturation at 96 °C for 5 min; 35 cycles of denaturation at 96 °C for 45 s, annealing at 60 °C for 1 min, and extension at 72 °C for 3 min (all steps were completed using ABI thermal cycler). PCR products were separated on ABI 3500XL Genetic Analyzer (Applied Biosystems) and evaluated using the GeneMapper v4.1 software with ROX-500 as an internal standard marker (Applied Biosystems).
Repeat-primed (RP) PCR protocol was followed for the detection of expanded ATXN10 allele for all samples which showed homozygous peak in the flanking PCR [19,20]. It was carried out using three primers including one FAM labelled forward primer (FP) (SCA10-5′-6-FAM- GAAGACAAATAGAAAACAGATGGCACA), a first reverse primer RP-R1 (5′- TACGCATCCCAGTTTGAGACG GAATAGAATAGAATAGAATAGAATAGAATAGAATAGAATAG) and a second reverse RP-R2 anchor tail (5′- TACGCATCCCAGTTTGAGACG). RP-PCR was carried out as follows: 100 ng of genomic DNA, 0.8 μM FP, 0.08 μM of RP-R1 primer and 0.8 μM of the RP-R2 primer, with FAILSAFE I reagent (epicentre), and 1U of AmpliTaq Gold polymerase (Applied Biosystems). Amplified fragments were separated and analyzed on ABI 3500XL Genetic Analyzer (Applied Biosystems) and evaluated using the GeneMapper v4.1 software (Applied Biosystems).
2.3. SCA10 haplotype analysis
SCA10 at-risk haplotype (CGGC) analysis was carried out using reported marker SNPs C(rs5764850), G(rs72556348), G(rs72556349), C(rs72556350) which are considered as stable haplotype markers in SCA10 cases flanking the repeat expansion region [12]. The study of risk haplotype variants was performed using the 1000 Genomes population data which provides information about genetic variants found in European (EUR), East Asian (EAS), American (AMR), African (AFR) and South Asian (SAS) population [18]. The genetic variants of five different populations distributed across the world were reconstructed for estimating haplotype frequency with most likely haplotype pairing among individuals of different population using PHASE v2.1 [21]. Similarly, the normal count and distribution of ATTCT repeat in five different populations with varying SNP lineages was studied using the 1000 Genomes Project and PHASE v2.1. For extended haplotype analysis in (ATTCT)n repeat flanking region, SNPs genotype variant data was obtained for ATXN10 region covering 5118 SNPs. From these, SNPs with allele frequency >0.05 were chosen for further haplotype analysis. A total of 56 SNPs (spanning ∼26kbp region) remained for the haplotype analysis and were analyzed using PHASE v2.1.
3. Results
3.1. Brief clinical details of the study cohort
The patients included in the study was essentially diagnosed with cerebellar ataxia with variable phenotype. It included a clinically heterogeneous cohort with mild to severe apparent signs of cerebellar ataxia, dysarthria and other neurologic signs and symptoms of varying durations. Of the total samples taken for the study, 324 patients were male and 137 were female. The patients were further categorized as Early onset cerebellar ataxia (6–25 years; mean age:18.6; n = 72), adult onset cerebellar ataxia (26–45 years; mean age: 35.4 years; n = 156) and late onset cerebellar ataxia (46–80 years; mean age: 58.8 years; n = 207) with major phenotype of ADCA/SCA cases (for 6% of cases age information were not available with the referral documents).
3.2. ATTCT repeats in patients and control population
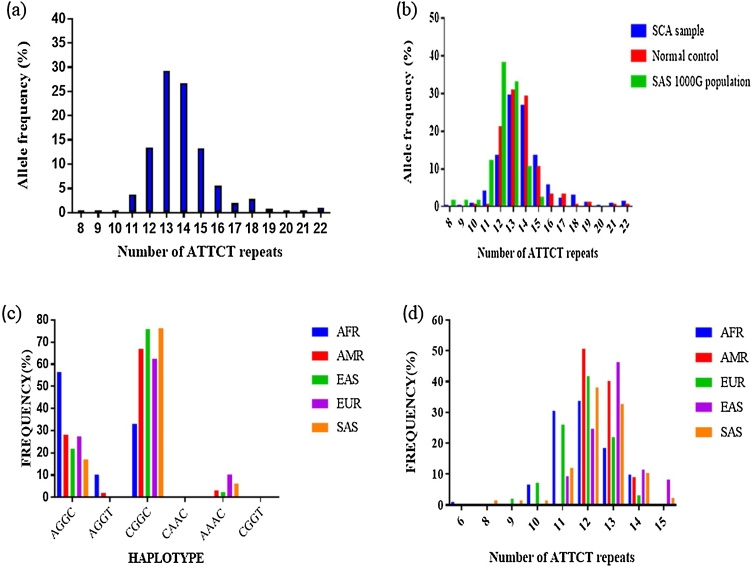
Pathogenic repeat expansion for SCA10 was not observed in any of the patients included in the study. Moreover, genetic analysis of ATTCT repeat region in ATXN 10 gene (allele count n = 922) revealed that 78.8% of patients were heterozygous with repeats ranging from 8 to 22. The most common ATTCT repeat was 13 found in 29.27% of patients followed by 14 repeats in 26.67% of cases (Fig. 1a). Further, repeat primed PCR analysis confirmed that the remaining 21.2% of patients were homozygous with allele ranging from 11 to 19 repeats. Also, in the homozygous state, allele 13/13 ATTCT repeats were the most recurrent with the frequency of 45% followed by 14/14 repeats.
Fig. 1.
Distribution of (ATTCT)n repeat and haplotype (a) Distribution of (ATTCT)n repeat in SCA patient samples (b) Comparative distribution of (ATTCT)n repeat in SCA samples, normal control and SAS reference samples (c) Reconstructed haplotype in five populations with estimated frequency (d) CGGC haplotype with repeat number in different population.
In the 117 control individuals, the ATTCT repeat ranged from 10 to 22, with 82.1% heterozygosity, and the remaining 17.9% samples were homozygous. The control group showed 13 ATTCT repeats as most frequent with an estimated frequency of 30.8% followed by 14/14 repeat. We also compared the results with the reference samples present in the 1000 Genomes Project SAS population showing 12 ATTCT repeats as the most common repeat in the cohort of 86 reference samples.
3.3. Haplotype distribution and its inference in Indian population
To assess the frequency distribution of at-risk haplotype for ATTCT expansion ‘C-(ATTCT)n-GGC’, the genotype variants of SNPs rs5764850, rs72556348, rs72556349, and rs72556350 along with the ATTCT repeats from 1000 Genomes Population database were used. Haplotype construction with most likely haplotype pairing in European, East Asian, American, African and South Asian population using PHASE v2.1 revealed six possible haplotype pairing. The possible pair with common SNPs found was CGGC, AGGC, AGGT, CAAC, AAAC, and CGGT (Table 1). The CGGC (spanning over 2 kbp) haplotype was most common among all the populations followed by AGGC whereas another haplotype variant was found in specific population only as shown in (Fig. 1c). Amongst five different populations, estimated frequency of CGGC haplotype was found to be 33% in AFR, 67% in AMR, 75.7% in EAS, 62.3% in EUR and 76.13% in SAS population. The CGGC haplotype was not found to be segregating with either large normal or small normal ATTCT repeat length. The distribution of ATTCT repeat number with CGGC haplotype in five populations demonstrated that 12 repeats of ATTCT was most frequently associated with CGGC haplotype across all populations including the SAS population (Fig. 1d). As the at-risk haplotype lineage was found to be quite frequent, we investigated the differences in at-risk haplotype sub lineage, spanning the repeat region of the different populations. We studied the genetic variants in an extended region surrounding ATTCT repeats. The extended haplotype analysis showed that the neighbouring haplotype arm surrounding the at-risk haplotype CGGC’ majorly comprised of haplotype-A, 1-CGGC-A (55%) (Table 2) and haplotype-B, 2-CGGC-B (32%) and were present across all five sub-population of KGP dataset. As SCA10 is known to occur only in people of Amerindian/Latin America and East Asian Ancestry (China and Japan), we looked across any diverged haplotype lineage with ATTCT > 13 repeats specific to AMR and EAS population. On further analysing the 5′ neighbouring divergent arm, we observed haplotype lineages (comprising of different SNPs) in either AMR or EAS populations. Furthermore, none of the haplotype lineages had shown presence in both the populations together, suggesting that these haplotype lineages are population specific as well as they were absent in other KGP (EUR, SAS and AFR) populations.
Table 1.
The core SCA10 haplotype and its frequency distribution with ATTCT repeats.
| (ATTCT)n | 6 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|
| AFR | |||||||||
| AGGC | 3.2 | 1.3 | 1.3 | 8.3 | 30.6 | 35.0 | 16.6 | 3.8 | 0.0 |
| AGGT | 0 | 0 | 0 | 0 | 46.43 | 46.43 | 7.14 | 0 | 0 |
| CGGC | 1.1 | 0.0 | 0.0 | 6.5 | 30.4 | 33.7 | 18.5 | 9.8 | 0.0 |
| CAAC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 |
| AAAC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| CGGT | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| EAS | |||||||||
| AGGC | 0.0 | 0.0 | 0.0 | 0.0 | 14.7 | 17.6 | 29.4 | 14.7 | 23.5 |
| AGGT | 0.0 | 0.0 | 0.0 | 0.0 | 9.3 | 24.7 | 46.4 | 11.3 | 8.2 |
| CGGC | 0.0 | 0.0 | 0.0 | 0.0 | 10.1 | 27.0 | 50.6 | 12.4 | 0.0 |
| CAAC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AAAC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 33.3 | 33.3 | 33.3 | 0.0 |
| CGGT | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| AMR | |||||||||
| AGGC | 7.1 | 0.0 | 0.0 | 0.0 | 7.1 | 32.1 | 46.4 | 7.1 | 0.0 |
| AGGT | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 50.0 | 50.0 | 0.0 | 0.0 |
| CGGC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 50.7 | 40.3 | 9.0 | 0.0 |
| CAAC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| AAAC | 0.0 | 0.0 | 0.0 | 0.0 | 66.7 | 33.3 | 0.0 | 0.0 | 0.0 |
| CGGT | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| EUR | |||||||||
| AGGC | 0.0 | 0.0 | 0.0 | 9.5 | 23.8 | 28.6 | 31.0 | 4.8 | 2.4 |
| AGGT | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| CGGC | 0.0 | 0.0 | 2.0 | 7.1 | 25.5 | 40.8 | 21.4 | 3.1 | 0.0 |
| CAAC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| AAAC | 0.0 | 0.0 | 0.0 | 0.0 | 25.0 | 37.5 | 31.3 | 0.0 | 6.3 |
| CGGT | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| SAS | |||||||||
| AGGC | 0.0 | 0.0 | 0.0 | 0.0 | 33.3 | 50.0 | 6.7 | 6.7 | 3.3 |
| AGGT | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| CGGC | 0.0 | 1.5 | 1.5 | 1.5 | 11.9 | 38.1 | 32.8 | 10.4 | 2.2 |
| CAAC | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| AAAC | 0.0 | 0.0 | 0.0 | 0.0 | 18.2 | 0.0 | 54.5 | 27.3 | 0.0 |
| CGGT | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 |
According to KGP database and Phase v2.1. EUR: European; EAS: East Asian; AMR: American; AFR: African; SAS: South Asian.
Table 2.
Extended Haplotype flanking CGGC-SCA10 core haplotype structure. The text in italics represents haplotype lineage specifically observed in AMR or EAS population. The other haplotype in Boxed manner represents prevalent haplotype 1-CGGC-A and 2-CGGC-B.
| AFR | AMR | EAS | EUR | SAS | ATTCTn Allele | ATTCTn length | Haplo-5’ | 5'ARM | SCA10 core haplotype | Haplo-3’ | 3’arm |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 0 | 0 | 8 | 14 | TTGGGCCTTGTGTCAGAAAAAAAAGGG | 16 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 1 | 0 | 0 | 0 | 7 | 13 | TTGAACTATGCGTCTCAAAAAAAAGAG | 13 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 1 | 0 | 0 | 8 | 14 | TTGGACCTTGTGTCAGAAAAAAAAGAG | 4 | CGGC | TCACGTGTGAAGAGTAGCCGAACC | C |
| 0 | 0 | 2 | 0 | 0 | 7 | 13 | TTGGGCCTCGTGTCAGAAAGAAAAGAG | 8 | CGGC | TCACATCTGAAGATTGGCCGAACC | D |
| 0 | 0 | 1 | 0 | 0 | 7 | 13 | TTGGGCCATGCGTCAGAAAGGGAGGGG | 18 | CGGC | TCACGTGCGCGGAGTAGCCCAACC | H |
| 0 | 0 | 1 | 0 | 0 | 7 | 13 | TTGGGCCATGCGTCAGAAAGGGAGGGG | 18 | CGGC | TCACGTGTGCGGAGTAGCCCAACC | L |
| 1 | 0 | 0 | 0 | 0 | 1 | 6 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 0 | 0 | 0 | 1 | 2 | 8 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 0 | 0 | 2 | 1 | 3 | 9 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 4 | 0 | 0 | 6 | 1 | 4 | 10 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 24 | 10 | 1 | 17 | 10 | 5 | 11 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 28 | 24 | 7 | 23 | 33 | 6 | 12 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 11 | 20 | 15 | 6 | 16 | 7 | 13 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 3 | 3 | 3 | 2 | 7 | 8 | 14 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 0 | 2 | 0 | 1 | 9 | 15 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 2 | 0 | 0 | 0 | 0 | 5 | 11 | TTGGGCCATGTGTCAGAAAGAAAAGAG | 5 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 1 | 0 | 1 | 0 | 5 | 11 | TTGGGCCATGTGTCAGAAAAAAAAGAG | 6 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 1 | 0 | 0 | 1 | 0 | 6 | 12 | TTGGGCCATGTGTCAGAAAAAAAAGAG | 6 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 1 | 0 | 0 | 0 | 0 | 7 | 13 | TTGGGCCATGTGTCAGAAAAAAAAGAG | 6 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 5 | 0 | 0 | 0 | 0 | 8 | 14 | TTGGGCCATGTGTCAGAAAAAAAAGAG | 6 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 2 | 0 | 0 | 0 | 0 | 6 | 12 | TTGGGCCTTGTGTCAGAAAGAAAAGAG | 11 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 1 | 0 | 0 | 0 | 0 | 8 | 14 | TTGGGCCTTGTGTCAGAAAGAAAAGAG | 11 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 0 | 1 | 0 | 0 | 6 | 12 | TTGGGCCTCGTGTCAGAAAAAAAAGAG | 14 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 0 | 0 | 1 | 0 | 6 | 12 | TTGGGCCTTGTGTCAGAAAAAAAAGGG | 16 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 1 | 0 | 2 | 0 | 7 | 13 | TTGGGCCTTGTGTCAGAAAAAAAAGGG | 16 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 0 | 0 | 0 | 2 | 8 | 14 | TTGGGCCTTATGTCAGAAAAAAAAGAG | 20 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 1 | 0 | 0 | 0 | 0 | 7 | 13 | TTGGGCCTTGTGCCAGAAAAAAAAGAG | 22 | CGGC | TCACGTGTGAAGAGTAGCCCAACC | A |
| 0 | 0 | 0 | 0 | 1 | 2 | 8 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 0 | 0 | 1 | 3 | 9 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 1 | 0 | 0 | 1 | 0 | 4 | 10 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 1 | 6 | 7 | 5 | 4 | 5 | 11 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 10 | 15 | 12 | 11 | 6 | 12 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 2 | 6 | 24 | 12 | 23 | 7 | 13 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 2 | 7 | 1 | 5 | 8 | 14 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 6 | 0 | 2 | 9 | 15 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 1 | 0 | 0 | 0 | 0 | 4 | 10 | TTGGGCTATGTGTCAGAAAAAAAAGAG | 3 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 1 | 0 | 0 | 0 | 5 | 11 | TTGGGCTTTGTGTCAGAAAAAAAAGAG | 7 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 1 | 0 | 0 | 0 | 0 | 6 | 12 | TTGGGCTTTGTGTCAGAAAAAAAAGAG | 7 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 0 | 2 | 0 | 5 | 11 | TTGGGCTTTATGTCAGAAAAAAAAGAG | 9 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 0 | 1 | 0 | 6 | 12 | TTGGGCTTTATGTCAGAAAAAAAAGAG | 9 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 1 | 0 | 1 | 0 | 6 | 12 | TTGAACTATGCGTCTCAAAAAAAAGAG | 13 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 1 | 0 | 0 | 6 | 12 | TTGGACTATATGTCAGAAAAAAAAGAG | 15 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 1 | 1 | 0 | 7 | 13 | TTGGACTATATGTCAGAAAAAAAAGAG | 15 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 0 | 0 | 1 | 6 | 12 | TTGGGCCTTATGTCAGAAAAAAAAGAG | 20 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 0 | 0 | 1 | 6 | 12 | TTGGACTTTATGTCAGAAAAAAAAGGG | 21 | CGGC | TCACGTGTGAAGATTAGCCGAACC | B |
| 0 | 0 | 0 | 0 | 1 | 6 | 12 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTGTGAAGAGTAGCCGAACC | C |
| 0 | 0 | 1 | 0 | 0 | 7 | 13 | TTGGACTTTATGTCAGAAAAAAAAGAG | 2 | CGGC | TCACGTGTGAAGAGTAGCCGAACC | C |
| 0 | 0 | 0 | 0 | 1 | 4 | 10 | TTGGACCTTGTGTCAGAAAAAAAAGAG | 4 | CGGC | TCACGTGTGAAGAGTAGCCGAACC | C |
| 0 | 0 | 1 | 0 | 0 | 5 | 11 | TTGGGCCTCGTGTCAGAAAGAAAAGAG | 8 | CGGC | TCACATCTGAAGATTGGCCGAACC | D |
| 0 | 0 | 0 | 0 | 1 | 5 | 11 | CCGAGCCACGTGTCAGAAAAAAAAGAG | 10 | CGGC | TCACATCTGAAGAGTGTTGCGGTT | E |
| 1 | 0 | 0 | 0 | 0 | 6 | 12 | TTGGACTTTACGTTAGAAAAGGAGGAG | 12 | CGGC | TCACGTGTGAGGATTAGCCGAACC | F |
| 0 | 0 | 0 | 1 | 0 | 6 | 12 | CCGAGCCACGTGTCACAGGGAAAAGAG | 17 | CGGC | TCACGTGTGAAGAGTGGTGCGGTT | G |
| 0 | 0 | 0 | 0 | 1 | 6 | 12 | TTGGGCCATGCGTCAGAAAGGGAGGGG | 18 | CGGC | TCACGTGCGCGGAGTAGCCCAACC | H |
| 0 | 0 | 0 | 0 | 1 | 6 | 12 | TTGGGCCTTGCATCTGAAAAAAAAGAG | 19 | CGGC | GCACGTGTGAAGAGTAGCCCAACC | I |
| 0 | 0 | 0 | 0 | 1 | 7 | 13 | TTGGGCCTTGCATCTGAAAAAAAAGAG | 19 | CGGC | GCACGTGTGAAGAGTAGCCCAACC | I |
| 0 | 0 | 0 | 0 | 1 | 7 | 13 | TTGGACCATATGTCAGAAAAAAAAGAG | 24 | CGGC | GCACGTGTGAAGAGTAGCCCAACC | I |
| 0 | 0 | 0 | 0 | 2 | 6 | 12 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACGTCTGAAGAGTAGCCCAACC | J |
| 1 | 0 | 0 | 0 | 0 | 7 | 13 | TTAGGGCTTGCGTCAGGAAAAAGAAAG | 23 | CGGC | TCAGGTGTGAAGGGCAGCCCAACC | K |
| 0 | 0 | 0 | 0 | 1 | 7 | 13 | TTGGGCCTTGTGTCAGAAAAAAAAGAG | 1 | CGGC | TCACATCTGAAGATTAGCCGAACC | M |
| 0 | 0 | 0 | 0 | 1 | 7 | 13 | TTGGACTTCATGTCAGAAAGAAAAGAG | 25 | CGGC | TCACGTGTGAAGATTGGCCGAACC | N |
4. Discussion
SCA10 is one of the rare ADCA caused by pentanucleotide repeat expansion in the intron 9 of the ATXN10 gene. Till date, SCA10 cases have been reported in several countries including, USA, Mexico, Argentina, Colombia, Bolivia, Brazil and Peru (Table 3) [5,7,13,16,17,[22], [23], [24]]. The reported cases share common SNPs associated with the SCA10 expansion locus proposing that SCA10 in Latin America has a common origin coming from an Amerindian populace. Recently, two SCA10 affected families has been documented in China and Japan having ATTCT expansion sharing common haplotype within the repeat region [14,15].
Table 3.
The comparative studies of SCA10 in different world populations.
| Population | Number of patients | Age of onset (years) | Number of ATTCT repeat Controls | Number of ATTCT repeats In Patients | References |
|---|---|---|---|---|---|
| Brazil | 90 | 35.5 (22-46) | 10 - 29 | 850-1500 | [13,24] |
| Mexico | 19 | 26.7 (14-44) | 10 - 22 | 920 - 4,140 | [7] |
| Argentina | 5 | 35 | 10 - 29 | 1100 | [22] |
| Peru | 1 | 47 | 9 - 32 | 1,760 | [16] |
| Minnesota | 1 | 89 | <32 | 1400 | [29] |
| Bolivia | 2 | 38-60 | ? | 1611 | [23] |
| China | 1 | 45 | 9 - 32 | 852 | [15] |
| Japan | 2 | 34-65 | 1500 | [14] | |
| Cypriot | 58 | 20-50 | 11 - 20 | 10–19 | [26] |
| German | 440 | 19-58 | 10 - 22 | 15-16 | [25] |
| White American, French-Canadian, Italian, Japanese & Spanish | 478 | ? | 10 - 22 | 10-21 | [27] |
| French | 123 families | ? | 11 - 22 | 11 - 22 | [11] |
| India | 461 | 43 (6-80) | 10-22 | 8-22 | Present study |
Genetic testing of SCA10 in a substantial cohort of Indian SCA patients did not show any pathological repeat expansion in the ATXN10 gene. Similar studies on SCA10 have documented the absence of pathological repeat expansion in the Cyprus and German populations [25,26]. In the current study, the ATTCT repeat length was found to fall between 8 to 22 repeats which is the non-pathological or normal range. It may be suggested that the near absence of SCA10 in Indian population could possibly be due to differing ethnicities and might manifest in populations with specific genetic backgrounds [27]. Additionally to strengthen our hypothesis of absence of SCA10 in the Indian population, haplotype analysis flanking the ATTCT expansion region was carried out across various ethnic population in 1000 Genomes data (KGP). The study revealed that the at-risk haplotype is comparatively more frequent in all populations with large normal or small normal ATTCT length. However, extended haplotype analysis showed some diversified lineage at 5′ end of the CGGC haplotype with ATTCT > 13 repeats. This lineage was specifically observed in the American or East Asian population excluding other populations in KGP dataset. As common risk haplotype has been observed across all populations, so we can’t rule down the possible hypothesis that independent expansion events on typically shared haplotype lineage but in different geographical regions may have possibly happened by chance event or by the backflow of human migrants from Beringia to other parts of the world [15,28]. Altogether, these facts point towards the absence of SCA10 in Indian population and haplotype-dependent assessment furthermore suggests its occurrence to be rare in South Asian, European and African population.
Although the rarity of SCA10 has already been demonstrated previously, we attempted to investigate a large cohort of Ataxia patients for SCA10 keeping in view the recently identified cases from China and Japan. Accordingly, this study does not target SCA patients manifesting the core features of SCA10. However, we adopted a large sample size to reduce the false negatives in view of phenotypic heterogeneity to overcome the limitation. We acknowledge that patients specifically exhibiting SCA10 features may give the correct scenario of its presence in the Indian population. Another major limitation may be stated as the study being hospital based rather than a population level survey to document the exact absence of SCA10 from India. However, our being the largest single centre in this context, catering to major tertiary care centres which are accessible to patients far and wide. Yet, SCA10 cases with precise clinical phenotype may be missed or go undiagnosed keeping in view the diverse ethnicity and geographical background prevailing in India.
5. Conclusion
This study establishes the rarity of SCA10 in the unrelated Indian spinocerebellar ataxia patients with heterogeneous ethnicity. It is further confirmed by population genetics based inferences suggesting it be rare or absent and might manifest in populations with specific genetic background from where it originated. However, this study provides future references while considering the diagnostic work-up of SCAs in the Indian population and to decide upon inclusion of SCA10 in panel based genetic investigations even for Indians living abroad. This approach may also be adopted for the screening of other subtypes of SCAs, i.e. other rare SCAs e.g. SCA31, SCA36, and SCA37.
Financial disclosure
Nothing to report for any author
Funding source for study
Financial support was received from Council of Scientific and Industrial Research funded GOMED:MLP1601 and GOMED-Tech:MLP1802 projects.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgment
Financial support was received from CSIR funded GOMED (MLP1601) and GOMED-Tech (MLP1802). We thank all the patients and their family members for their participation.
References
- 1.Zu L., Figueroa K.P., Grewal R., Pulst S. Mapping of a new autosomal dominant spinocerebellar Ataxia to chromosome 22. Am. J. Hum. Genet. 1999;64:594–599. doi: 10.1086/302247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuura T., Yamagata T., Burgess D.L., Rasmussen A., Grewal R.P., Watase K., Khajavi M., McCall A.E., Davis C.F., Zu L., Achari M., Pulst S.M., Alonso E., Noebels J.L., Nelson D.L., Zoghbi H.Y., Ashizawa T. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- 3.Pennisi E. ENCODE project writes eulogy for junk DNA. Science. 2012;337 doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 4.Alonso I., Jardim L.B., Artigalas O., Saraiva-Pereira M.L., Matsuura T., Ashizawa T., Sequeiros J., Silveira I. Reduced penetrance of intermediate size alleles in spinocerebellar ataxia type 10. Neurology. 2006;66:1602–1604. doi: 10.1212/01.wnl.0000216266.30177.bb. [DOI] [PubMed] [Google Scholar]
- 5.Teive H.A.G., Munhoz R.P., Arruda W.O., Raskin S., Werneck L.C., Ashizawa T. Spinocerebellar ataxia type 10 - A review. Park. Relat. Disord. 2011;17:655–661. doi: 10.1016/j.parkreldis.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Matsuura T., Achari M., Khajavi M., Bachinski L.L., Zoghbi H.Y., Ashizawa T. Mapping of the gene for a novel spinocerebellar ataxia with pure cerebellar signs and epilepsy. Ann. Neurol. 1999;45:407–411. doi: 10.1002/1531-8249(199903)45:3<407::aid-ana21>3.0.co;2-d. AID-ANA21>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen A., Matsuura T., Ruano L., Yescas P., Ochoa A., Ashizawa T., Alonso E. Clinical and genetic analysis of four Mexican families with spinocerebellar ataxia type 10. Ann. Neurol. 2001;50:234–239. doi: 10.1002/ana.1081. [DOI] [PubMed] [Google Scholar]
- 8.Teive H.A.G., Ashizawa T. Spinocerebellar ataxia type 10: from amerindians to latin americans. Curr. Neurol. Neurosci. Rep. 2013;13:9–11. doi: 10.1007/s11910-013-0393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin X., Ashizawa T. Recent progress in spinocerebellar ataxia type-10 (SCA10) Cerebellum. 2005;4:37–42. doi: 10.1080/14734220510007897. [DOI] [PubMed] [Google Scholar]
- 10.Teive H.A.G., Roa B.B., Raskin S., Fang P., Arruda W.O., Neto Y.C., Gao R., Werneck L.C., Ashizawa T. Clinical phenotype of Brazilian families with spinocerebellar ataxia 10. Neurology. 2004;63:1509–1512. doi: 10.1212/01.wnl.0000142109.62056.57. [DOI] [PubMed] [Google Scholar]
- 11.Jackson A.C., Melanson M., Rossiter J.P. 2002. Familial Herpes Simplex Encephalitis; pp. 406–409. [DOI] [PubMed] [Google Scholar]
- 12.Almeida T., Alonso I., Martins S., Ramos E.M., Azevedo L., Ohno K., Amorim A., Saraiva-Pereira M.L., Jardim L.B., Matsuura T., Sequeiros J., Silveira I. Ancestral origin of the ATTCT repeat expansion in spinocerebellar ataxia type 10 (SCA10) PLoS One. 2009;4:1–6. doi: 10.1371/journal.pone.0004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afonso H., Teive G., Moro A., Moscovich M., Arruda W.O., Puppi R., Raskin S., Mary G., Teive G., Dallabrida N., Ashizawa T. 2015. Spinocerebellar Ataxia Type 10 in the South of Brazil : The Amerindian-Belgian Connection. 2014–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naito H., Takahashi T., Kamada M., Morino H., Yoshino H., Hattori N., Maruyama H., Kawakami H., Matsumoto M. First report of a Japanese family with spinocerebellar ataxia type 10: the second report from Asia after a report from China. PLoS One. 2017;12:1–11. doi: 10.1371/journal.pone.0177955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K., Mcfarland K.N., Liu J., Zeng D., Landrian I., Xia G., Hao Y., Jin M., Mulligan C.J., Gu W., Ashizawa T. Spinocerebellar Ataxia type 10 in chinese han. Neurol. Genet. 2015;1:1–3. doi: 10.1212/NXG.0000000000000026. doi:10.1212/ NXG.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonardi L., Marcotulli C., McFarland K.N., Tessa A., DiFabio R., Santorelli F.M., Pierelli F., Ashizawa T., Casali C. Spinocerebellar ataxia type 10 in Peru: the missing link in the Amerindian origin of the disease. J. Neurol. 2014;261(September (9)):1691–1694. doi: 10.1007/s00415-014-7394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bushara K., Bower M., Liu J., McFarland K.N., Landrian I., Hutter D., Teive H.A.G., Rasmussen A., Mulligan C.J., Ashizawa T. Expansion of the spinocerebellar ataxia type 10 (SCA10) repeat in a patient with Sioux Native American ancestry. PLoS One. 2013;8:8–11. doi: 10.1371/journal.pone.0081342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.1000 Genomes Project Consortium, A global reference for human genetic variation. Nature. 2015;526:68. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cagnoli C., Michielotto C., Matsuura T., Ashizawa T., Margolis R.L., Holmes S.E., Gellera C., Migone N., Brusco A. Detection of large pathogenic expansions in FRDA1, SCA10, and SCA12 genes using a simple fluorescent repeat-primed PCR assay. J. Mol. Diagn. 2004;6:96–100. doi: 10.1016/S1525-1578(10)60496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warner J.P., Barron L.H., Goudie D., Kelly K., Dow D., Fitzpatrick D.R., Brock D.J. A general method for the detection of large CAG repeat expansions by fluorescent PCR. J. Med. Genet. 1996;33:1022–1026. doi: 10.1136/jmg.33.12.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatto E.M., Gao R., White M.C., Uribe Roca M.C., Etcheverry J.L., Persi G., Poderoso J.J., Ashizawa T. Ethnic origin and extrapyramidal signs in an argentinean spinocerebellar Ataxia type 10 family. Neurology. 2007;69:216–218. doi: 10.1212/01.wnl.0000265596.72492.89. [DOI] [PubMed] [Google Scholar]
- 23.Baizabal-Carvallo J.F., Xia G., Botros P., Laguna J., Ashizawa T., Jankovic J. Bolivian kindred with combined spinocerebellar ataxia types 2 and 10. Acta Neurol. Scand. 2015;132:139–142. doi: 10.1111/ane.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Carvalho Aguiar P., Fuchs T., Borges V., Lamar K.M., Silva S.M.A., Ferraz H.B., Ozelius L. Screening of Brazilian families with primary dystonia reveals a novel THAP1 mutation and a de novo TOR1A GAG deletion. Mov. Disord. 2010;25:2854–2857. doi: 10.1002/mds.23133. [DOI] [PubMed] [Google Scholar]
- 25.Aydin G., Dekomien G., Hoffjan S., Gerding W.M., Epplen J.T., Arning L. Frequency of SCA8, SCA10, SCA12, SCA36, FXTAS and C9orf72 repeat expansions in SCA patients negative for the most common SCA subtypes. BMC Neurol. 2018;18:1–8. doi: 10.1186/s12883-017-1009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Votsi C., Zamba-papanicolaou E., Georghiou A., Kyriakides T., Papacostas S., Kleopa K.A., Pantzaris M., Christodoulou K. Journal of the Neurological Sciences Investigation of SCA10 in the Cypriot population : further exclusion of SCA dynamic repeat mutations. J. Neurol. Sci. 2012;323:154–157. doi: 10.1016/j.jns.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura T., Ranum L.P.W., Volpini V. 2012. Spinocerebellar Ataxia Type 10 Is Rare in Populations Other Than Mexicans the Online Version of This Article, Along With Updated Information and Services, Is Located on the World Wide Web at : Clinical / Scientific Notes Spinocerebellar Ataxia Type. 10 i. [Google Scholar]
- 28.Mulligan C.J., Kitchen A., Miyamoto M.M. Updated three-stage model for the peopling of the Americas. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003199. doi:0.1371/journal.pone.0003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., McFarland K.N., Landrian I., Wu S.S., Bower M., Hutter D., Bushara K., Teive H.A.G., Ashizawa T. Identifying novel interruption motifs in spinocerebellar ataxia type 10 expansions. Neurol. Clin. Neurosci. 2014;2:38–43. [Google Scholar]