Abstract
Background
The maintenance of genomic integrity is the responsibility of a complex network, denominated the DNA damage response (DDR), which controls the lesion detection and DNA repair. The main repair pathways are base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), homologous recombination repair (HR) and non-homologous end joining repair (NHEJ). They correct double-strand breaks (DSB), single-strand breaks, mismatches and others, or when the damage is quite extensive and repair insufficient, apoptosis is activated.
Methods
In this study we used the BLAST reciprocal best-hit methodology to search for DDR orthologs proteins in Aedes aegypti. We also provided a comparison between Ae. aegypti, D. melanogaster and human DDR network.
Results
Our analysis revealed the presence of ATR and ATM signaling, including the H2AX ortholog, in Ae. aegypti. Key DDR proteins (orthologs to RAD51, Ku and MRN complexes, XP-components, MutS and MutL) were also identified in this insect. Other proteins were not identified in both Ae. aegypti and D. melanogaster, including BRCA1 and its partners from BRCA1-A complex, TP53BP1, PALB2, POLk, CSA, CSB and POLβ. In humans, their absence affects DSB signaling, HR and sub-pathways of NER and BER. Seven orthologs not known in D. melanogaster were found in Ae. aegypti (RNF168, RIF1, WRN, RAD54B, RMI1, DNAPKcs, ARTEMIS).
Conclusions
The presence of key DDR proteins in Ae. aegypti suggests that the main DDR pathways are functional in this insect, and the identification of proteins not known in D. melanogaster can help fill gaps in the DDR network. The mapping of the DDR network in Ae. aegypti can support mosquito biology studies and inform genetic manipulation approaches applied to this vector.
Keywords: Aedes aegypti, DDR, DNA damage response, DNA repair
Background
Aedes aegypti is one of the most important insect vectors due to its ability to transmit dengue, Zika, chikungunya, and yellow fever [1]. The disease vector capacity of this mosquito is related to its blood-feeding habits. In a single meal Ae. aegypti females can ingest an amount of blood up to three times their body weight [2, 3]. Hemoglobin, which is about 60% of the blood protein fraction, releases its prosthetic group heme when digested in mosquito gut. In the insect midgut, heme accumulation and hydrolysis by heme oxygenase lead to iron release that catalyzes the formation of reactive oxygen species (ROS) via Fenton reaction [2]. In larval stages, water pollutants, heavy metals and plant metabolites present in breeding sites, as well as UV exposure, contribute to ROS formation and can alter insect physiology and insecticide tolerance [4, 5].
Low levels of ROS are important for many biological processes such as signal transduction, and insect immunity [6, 7]. However, high levels of ROS can induce lipid peroxidation, protein and DNA oxidation, generate DNA single-strand breaks (SSBs) and double-strand breaks (DSBs) [8–10].
To repair DNA damage and maintain genome integrity, organisms rely on a complex system denominated the DNA damage response (DDR). The DDR includes signaling and repair pathways, as base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), homologous recombination repair (HR) and non-homologous end joining repair (NHEJ) [11–13]. In addition, when the damage is quite extensive and repair insufficient, apoptosis is activated [14].
The DDR network has been extensively studied in model organisms such as Drosophila melanogaster, that encodes many key DDR proteins [15], but little is known about these pathways in insect vectors [16]. In mosquitoes the repair of DSB has been initially studied to improve genomic manipulation and generation of transgenic insects [17–20]. Thus, the wide identification of the DDR players in Ae. aegypti can support the genomic manipulation tools development, and the resistance and transmission-blockage studies.
In this study we used bioinformatics tools to search for and annotate proteins from and related to DDR pathways in Ae. aegypti. We show here that key genes coding for DDR proteins are present, suggesting that the main DDR pathways are functional in this organism. Additionally, Ae. aegypti, like other dipterans, lacks important DDR proteins such as BRCA1, TP53BP1 and XRCC4, raising questions about how they deal with the lack of these DDR components.
Databases
The DDR signaling and repair pathways analyzed were as follows: ATR signaling; double-strand break repair (DSB); homologous recombination repair (HR); non-homologous end joining repair (NHEJ); mismatch repair (MMR); base excision repair (BER); and nucleotide excision repair (NER).
The following databases were used: (i) Uniprot-Swissprot release May 2018 (http://ftp.ebi.ac.uk); (ii) one custom DDR database compiled by us containing DDR proteins from Homo sapiens, Apis mellifera and Drosophila melanogaster. The H. sapiens DDR proteins listed in Reactome pathways “base excision repair”, “nucleotide excision repair”, “mismatch repair”, “DNA double-strand break response”, “HDR through homologous recombination (HRR) or single-strand annealing”, “nonhomologous end-joining (NHEJ)”, “HDR through MMEJ (alt-NHEJ) and “DNA damage reversal” plus A. mellifera and D. melanogaster DDR proteins listed in literature (Arcas et al. [25]) were obtained from database (i); (iii) Ae. aegypti proteins version 5.1 from VectorBase; (iv) KEGG eukaryotes (KE) proteins, release 5 June 2017; (v) Gene Ontology (GO) proteins, release August 2018 (http://archive.geneontology.org/); and (vi) Conserved Domain Database (CDD), version 3.16 from NCBI (ftp://ftp.ncbi.nlm.nih.gov).
Reciprocal best-hit methodology [21] was used to manually identify and annotate Ae. aegypti orthologs for the proteins present in the DDR database (Additional file 1: Figure S1). The first BLASTP used proteins from DDR database (ii) as queries and Ae. aegypti database (iii) as subject. The blastp e-value cut-off (10−15) was determined experimentally by our group to restrict the BLAST results, lowering the potentially false positive hits. The top 5 hits were considered if the e-value was smaller than 10−15. These Ae. aegypti hit proteins were compared by BLASTP with the databases db(i), db(iv), db(v) and db(vi). The top 2 back-hits were considered if the e-value was smaller than 10−15. The orthology was assumed to the Ae. aegypti protein that have (i) both top 2 back-hits (for databases db(i), db(iv) and db(v)) with the same annotation as the initial query; and (ii) the same typical conserved domains (database db(vi) result) as those present in the initial query.
Multiple sequence alignment was carried out using the Clustal Omega web server with standard parameters [22]. Kinase-specific and unspecific phosphorylation sites prediction was made using NetPhos 3.1 web prediction server [23] also with standard parameters.
Signal transduction
The DDR signaling pathway consists of DNA damage sensors, signal transducers and effectors proteins, and at core of this machinery are the transducer kinases ATM (ataxia telangiectasia mutated), ATR (ATM and Rad3-related) DNAPKcs (DNA-dependent protein kinase, catalytic subunit). These three phosphoinositide 3-kinase related kinases (PIKKs) are responsible to phosphorylate the effector proteins, which participate in cell cycle control, DNA repair pathways and apoptosis. ATM and DNAPKcs are involved in repair of double-strand breaks (DSBs) while ATR is activated by a variety of damages, being important in signaling of UV lesions, stalled replication forks and in damage surveillance during DNA replication [24]. ATM seems to have emerged in plants, while DNAPKcs ATR appears in early eukaryotes [25]. These kinases are present in Ae. aegypti, whereas D. melanogaster encodes orthologs only for ATM and ATR [25]. The role of ATR, ATM and DNAPKcs will be discussed in the next sections.
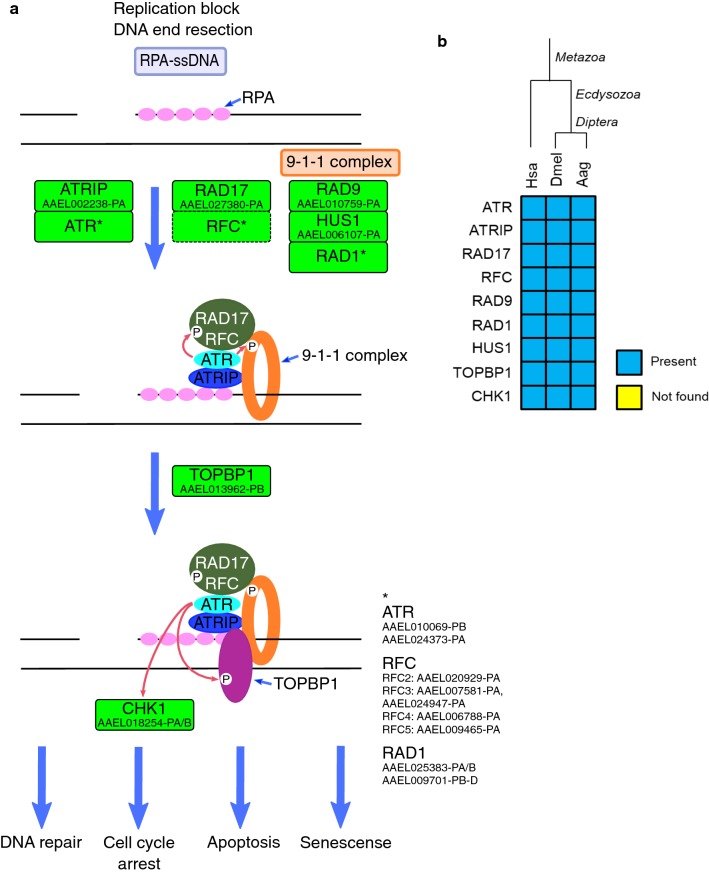
ATR is recruited in response to single-stranded DNA (ssDNA), through the binding of ATR-interacting protein (ATRIP) to RPA, that coats ssDNA structure [26]. RPA-ssDNA also recruits Rad17-RFC clamp loader to ssDNA/dsDNA junction, which loads RAD9-RAD1-HUS1 (9-1-1) clamp onto double-strand DNA (dsDNA) [27]. 9-9-1 promotes the recruitment of TOPBP1 that fully activates ATR, which phosphorylates effector proteins such as checkpoint kinase 1 (Chk1) involved in arrest of cell cycle progression [28, 29]. The proteins of the ATR network seem to have appeared in early eukaryotes, with exception of ATRIP that emerged in plants, and CHK1 that appeared before fungi and animals split [25]. Due to the early origin of these pathways, all proteins were identified in Ae. aegypti (Fig. 1). The complete list of Ae. aegypti ATR signaling proteins is provided in Additional file 2: Table S1.
Fig. 1.
a ATR signaling in Ae. aegypti. Rectangles: green, identified; solid line, protein; dashed, protein complex. Replication fork proteins were omitted to improve clarity. b Heatmap of H. sapiens (Hsa); D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S1
Double-strand break repair
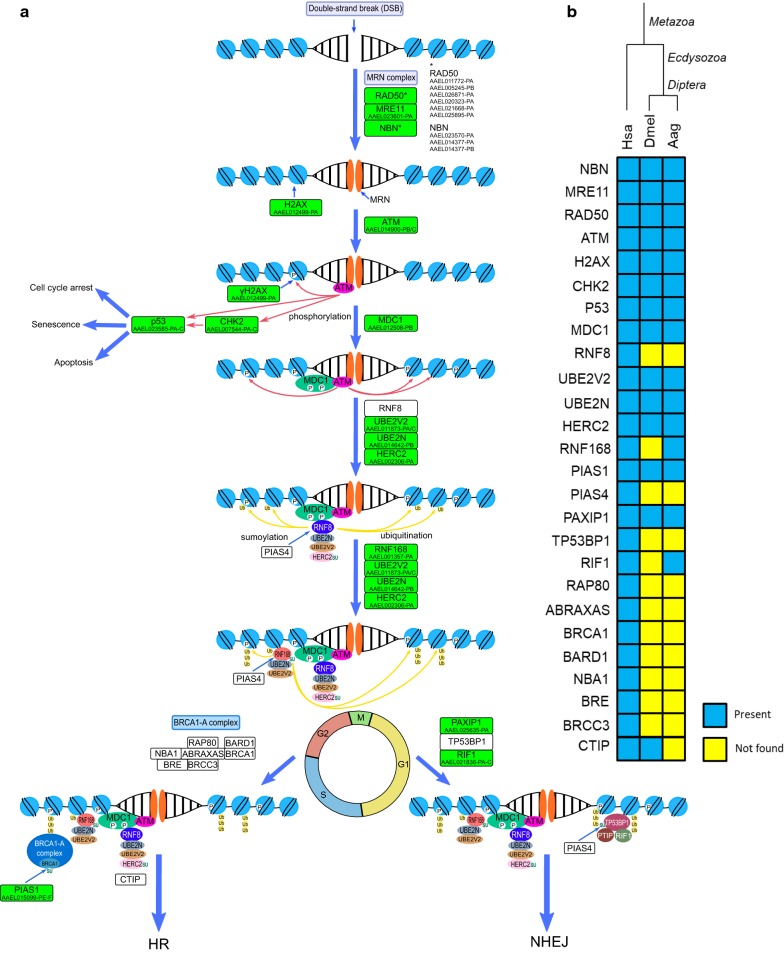
Double-strand breaks (DSBs) are potential harmful lesions that can be repaired by homologous recombination (HR) and by non-homologous end joining (NHEJ) [30]. The MRN complex, composed of RAD50, NBN (also known as NBS1 and XRS2 in yeast) and MRE11, is the sensor that recognizes a DSB and recruits ATM to damage site [31]. Activated ATM phosphorylates CHK2 and p53, regulating cell cycle arrest, senescence and apoptosis in human cells [11]. ATM also phosphorylates the adjacent histones H2A/H2AX, producing gamma-H2A (γH2A) and gamma-H2AX (γH2AX), which is relevant for the foci formation and the recruitment of the mediator of DNA damage checkpoint 1 (MDC1) [32]. MDC1 promotes ATM signaling amplification and recruits E3 ubiquitin ligase RNF8 that is responsible for the initial ubiquitination of the histones H2A/H2AX followed by poly ubiquitination by E3 ubiquitin ligase RNF168 [33–36]. This ubiquitination process is necessary for the recruitment of BRCA1-A complex, composed of BRCA1-BARD1 heterodimer, RAP80, ABRAXAS, BRCC3, BRE, BABAM1 [35, 37, 38].
The choice of DSB repair pathway depends of cell cycle stage. In the S and G2, CtIP associates with BRCA1 and MRN complex to stimulate DSB end resection promoting homologous recombination (HR) [39, 40], whereas in G1 the association of TP53BP1 with RIF1 and PTIP inhibits the DSB end resection leading to NHEJ [41].
The genes encoding proteins at the first steps of the pathway, damage recognition and initial response, possess an early origin. MRE11, RAD50, CHK2 and p53 all emerged in early eukaryotes, and NBN appeared in plants [25]. In Ae. aegypti, the MRN complex (NBN: AAEL023570-PA, AAEL014377-PA/B; MRE11: AAEL023601-PA; RAD50: AAEL011772-PA, AAEL005245-PB, AAEL026871-PA, AAEL020323-PA, AAEL021668-PA, AAEL025895-PA), ATM (AAEL014900-PB/C), CHK2 (AAEL007544-PA-C) and p53 (AAEL023585-PA-C) are all present (Fig. 2).
Fig. 2.
a Double-strand break (DSB) signaling in Ae. aegypti. Rectangles: green, identified; white, not identified. b Heatmap of H. sapiens (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S2
Arcas et al. [25], suggested that MDC1 appeared only in vertebrates; however, in our analysis we were able to identify an ortholog of this protein in Ae. aegypti (AAEL012508-PB), which is also present in D. melanogaster [15] (Fig. 2).
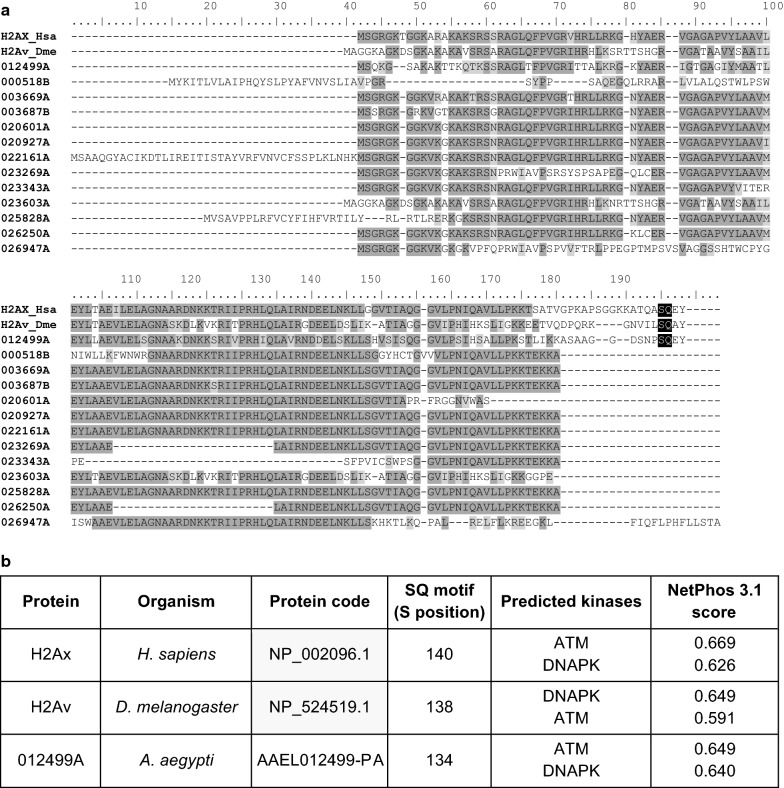
The histone H2A is highly conserved through the evolution, being identified in early eukaryotes [25]. Some of H2A histone variants include the SQ motif, located at the C-terminal region, which is required for ATM phosphorylation. The variant H2AX, in humans, possesses the SQ motif and is phosphorylated in Ser139. In D. melanogaster, the H2Av is the functional ortholog of human H2AX, being phosphorylated at Ser138, in response to DSB [42]. We identified 94 histones H2A in Ae. aegypti and, to search for the SQ motif, we made an alignment between human H2Ax, D. melanogaster H2Av, and H2A identified in Ae. aegypti (Fig. 3a). The C-terminal region of AAEL012499-PA was the only one among all the Ae. aegypti histones that showed the SQ motif. Furthermore, it aligned correctly with the SQ motifs from the human H2AX and D. melanogaster H2Av. Phosphorylation prediction analyses, made with NetPhos 3.1 web prediction server, also showed the same result to AAEL012499-PA Ser134, to H2AX Ser140 (NCBI and Uniprot records to human H2AX (NP_002096.1) point Ser140 as the phosphorylation position but the literature [42] points Ser139, despite the numbering differences both refer to the same serine) and to H2Av Ser138, indicating the SQ motif is phosphorylated by ATM (Fig. 3b). Taken together, these data suggest that AAEL012499-PA is the Ae. aegypti functional ortholog of the human H2AX and D. melanogaster H2Av. The H2A identified in Ae. aegypti are provided in Additional file 2: Table S3.
Fig. 3.
a Multiple sequence alignment of H2Ax histones. Identical proteins were clustered and codes were shortened for brevity. Background: light grey, low amino acid conservation; dark, high conservation; black, SQ-motif. b SQ-motif phosphorylation prediction. Only predictions above NetPhos cut-off score (0.5) were considered
The ubiquitin-protein ligase RNF8 was detected in early eukaryotes, while the RNF168 was identified only in Chordata. Both are absent in D. melanogaster and in the model species C. elegans and S. cerevisiae [25]. Aedes aegypti lacks the RNF8 ubiquitin ligase, but, curiously, encodes an ortholog for the RNF168 (AAEL001357-PA) (Fig. 2).
The SUMO E3 ligase PIAS4 is absent in Ae. aegypti, but PIAS1 (AAEL015099-PE/F) is present (Fig. 2). In vertebrates both proteins are involved in sumoylation of DSB response/repair proteins, such as HERC2, RNF168, BRCA1 and TP53BP1 [43–45]. While PIAS4 is present only in vertebrates, PIAS1 appeared earlier, before division of plants [25], suggesting that PIAS1 should plays the role of PIAS4, not only in Ae. aegypti, but also in other species that lack this protein, such as D. melanogaster.
The CtIP and the BRCA1-A complex, including BRCA1, were not identified in Ae. aegypti (Fig. 2). CtIP emerged in Bilateria and an ortholog have already been found in D. melanogaster [15, 25]. Most BRCA1-A complex proteins possess an early origin; BRCA1, BRE and BRCC3, originated in early eukaryotes, while NBA1 and BARD1 in the common ancestor of plants and animals. Only RAP80 appeared in vertebrates. However, this complex is also absent in D. melanogaster and seems to have been lost in Diptera [25]. It has already been reported that the components of DSB response have originated in different periods of time, suggesting that this pathway may have assembled in a modular way during evolution [25]. The lack of BRCA1-A complex proteins together with the fact that HR is functional [46–48] suggest that dipterans should have rewired the HR pathway activation.
TP53BP1 originated in Metazoa, whereas PAXIP1 and RIF1 emerged in early eukaryotes [25]. Although TP53BP1 is absent in Ae. aegypti, both PAXIP1 and RIF1 are present. Regarding the TP53BP1 absence, it has already been shown that PAXIP1 could also associate with ARTEMIS (a known NHEJ factor detailed below), induced by DNA damage. This association happens downstream of TP53BP1 and leads to the trimming of the DNA ends to facilitate NHEJ and avoid the extensive resection necessary for HR [49]. As ARTEMIS is a conserved nuclease that is present in Ae. aegypti, it is a possibility that in this insect the interaction between ARTEMIS and PAXIP bypass TP53BP1 signaling and promotes NHEJ activation. The complete list of Ae. aegypti double-strand break repair proteins is provided in Additional file 2: Table S2.
Homologous recombination
DSB repair by homologous recombination (HR) occurs during S and G2 phases of the cell cycle and uses the sister chromatid as a template for repair [50].
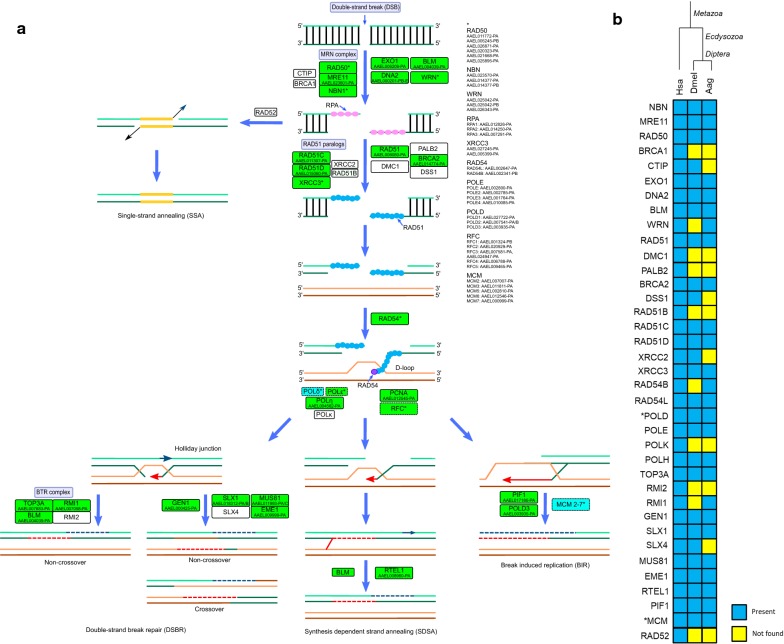
To initiate, HR requires the resection of the DSB, which is initially carried out by the association of the MRN complex, CtIP and BRCA1, followed by the exonuclease EXO1 or the endonuclease DNA2, in a process facilitated by the helicases BLM and WRN [40, 51–54]. As discussed above, orthologs of human CtIP and BRCA1 were not found in Ae. aegypti, being still unclear how this organism carries out this step. The helicase BRIP1, that interacts with BRCA1 and is supposed to participate in the recruitment of RPA and RAD51 [55], is also absent. The MRN complex as well as the three RPA subunits were identified in Ae. aegypti (discussed above). EXO1, DNA2 and WRN emerged in early eukaryotes, and BLM is an ancient protein found in prokaryotes. All these proteins are present in Ae. aegypti (Fig. 4).
Fig. 4.
a Homologous recombination (HR) repair in Ae. aegypti. Rectangles: solid line, protein; dashed, protein complex; green, identified; cyan, partially identified; white, not identified. b Heatmap of H. sapiens (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S4. *Subunits not identified: Aag-MCM4, Aag-POLD4 and Dmel-POLD4
The formation of invasive RAD51 nucleoprotein filaments occurs by the replacement of RPA from ssDNA by RAD51, mediated by BRCA2 and PALB2 [56, 57]. The filaments are stabilized by RAD51 paralogs and invade the sister chromatid in cooperation with RAD54 [58, 59]. After invasion, the replicative DNA polymerases (POL δ, ε) or translesion DNA polymerases (POL η, κ) extend the DNA strand generating a D-loop [60]. The RAD51 allow the HR during meiosis and mitosis. It is one of the two eukaryotic functional homologs of the strand exchange bacterial RecA [61]. This protein is present in early eukaryotes [25] and an ortholog was also found in Ae. aegypti (AAEL006080-PA). The other eukaryotic RecA functional homolog is DMC1, which acts only in meiosis [61]. An ortholog of this protein was not identified in Ae. aegypti and is also lacking in D. melanogaster. Humans encode five RAD51 paralogs: RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3 (Fig. 4). In Ae. aegypti RAD51B was not identified, as expected since it is lacking in Ecdysozoa [15]. Surprisingly, XRCC2 that is present in D. melanogaster [15] and in other mosquitoes such as An. gambiae and Cx. quinquefasciatus (KEGG orthology group K10879) was not found. Otherwise, RAD51C (AAEL011307-PA), RAD51D (AAEL015060-PA) and XRCC3 (AAEL027245-PA, AAEL005399-PA) were all identified in Ae. aegypti. BRCA2 emerged in early eukaryotes [25] and was identified, but PALB2 (Fig. 4), which seems to have appeared in vertebrates [25], was not. Two orthologs of RAD54 were identified in Ae. aegypti, RAD54L and RAD54B, otherwise one of them (RAD54B) have been lost in many insects, including D. melanogaster [15]. The replicative DNA polymerases POL δ and ε are complexes both formed by four subunits. It was not found in, Ae. aegypti, the subunit 4 of POL δ but the catalytic subunit was (POLD1 - AAEL027722-PA; POLD2 - AAEL007541-PB, AAEL007541-PA; POLD3 - AAEL003935-PA). All the subunits of POL ε (POLE - AAEL002800-PA; POLE2 - AAEL002785-PA; POLE3 - AAEL001764-PA; POLE4 - AAEL010085-PA) were found. The translesion DNA polymerase POL η (AAEL004562-PA) was identified in Ae. aegypti, but the POL κ was not, it is also absent in D. melanogaster and in several insects [15] (Fig. 4).
D-loop structures can be solved by three pathways: double-strand break repair (DSBR), synthesis-dependent strand annealing (SDSA) or break induced replication (BIR) [50]. In the DSBR the D-loop is processed by the formation of the Holliday junction that can be dissolved (by the BTR complex generating non-crossover products) or resolved (by the nucleases SLX1-SLX4 and MUS81-EME1 or GEN1 forming both crossover and non-crossover products) [62–65]. The BTR complex proteins TOP3A and BLM are ancient proteins, being found even in prokaryotes, whereas RMI1 and RMI2 emerged in plants and animals, respectively. Both MUS81 and SLX1, as well as GEN1, seems to have originated in early eukaryotes, EME1 and SLX4 emerged later in animals [25]. Of these proteins, only RMI2 and SLX4 were not identified in Ae. aegypti. RMI2 is also lacking in D. melanogaster and in most insects [15]. Although it was proposed that dipterans have lost RMI1 [15], we identified an ortholog of this protein in Ae. aegypti (Fig. 4).
In the SDSA, the invading strand dissociates from the sister chromatid and anneals with the complementary strand of the broken DNA end, which results in non-crossover products [50]. This pathway is carried out by the helicases RTEL1 or BLM [66, 67], which were both identified in Ae. aegypti (RTEL1 - AAEL008960-PA; BLM - AAEL004039-PA) (Fig. 4).
In BIR a replication fork is assembled after D-loop formation and the entire chromosome arm is synthesized [68]. BIR was extensively studied in yeasts and is carried out by Pol 32 (POLD3 ortholog) and the helicases PIF1 and MCM2-7 [69, 70]. All these proteins, except for MCM4, are found in Ae. aegypti (POLD3; PIF1 - AAEL017186-PA; MCM2 - AAEL007007-PA; MCM3 - AAEL011811-PA; MCM5 - AAEL002810-PA; MCM6 - AAEL012546-PA; MCM7 - AAEL000999-PA).
The DSB can also be repaired by the RAD51 independent pathway, denominated single-strand annealing (SSA), which anneals complementary DSB ends generated by the extensive resection, in a process mediated by RAD52 [71], that is absent in Ae. aegypti (Fig. 4). In D. melanogaster SSA occurs normally although the absence of RAD52. The lack of RAD52 and BRCA1 or PALB2 is lethal to human cells [72], questions about how dipterans deal with the loss of these proteins, have already been raised, but the answer is still unknown [15]. The complete list of Ae. aegypti HR proteins is provided in Additional file 2: Table S4.
Non-homologous end joining (NHEJ)
NHEJ is responsible for re-joining DNA broken ends and is the main DSB repair pathway in eukaryotes, occurring in G1 phase [73].
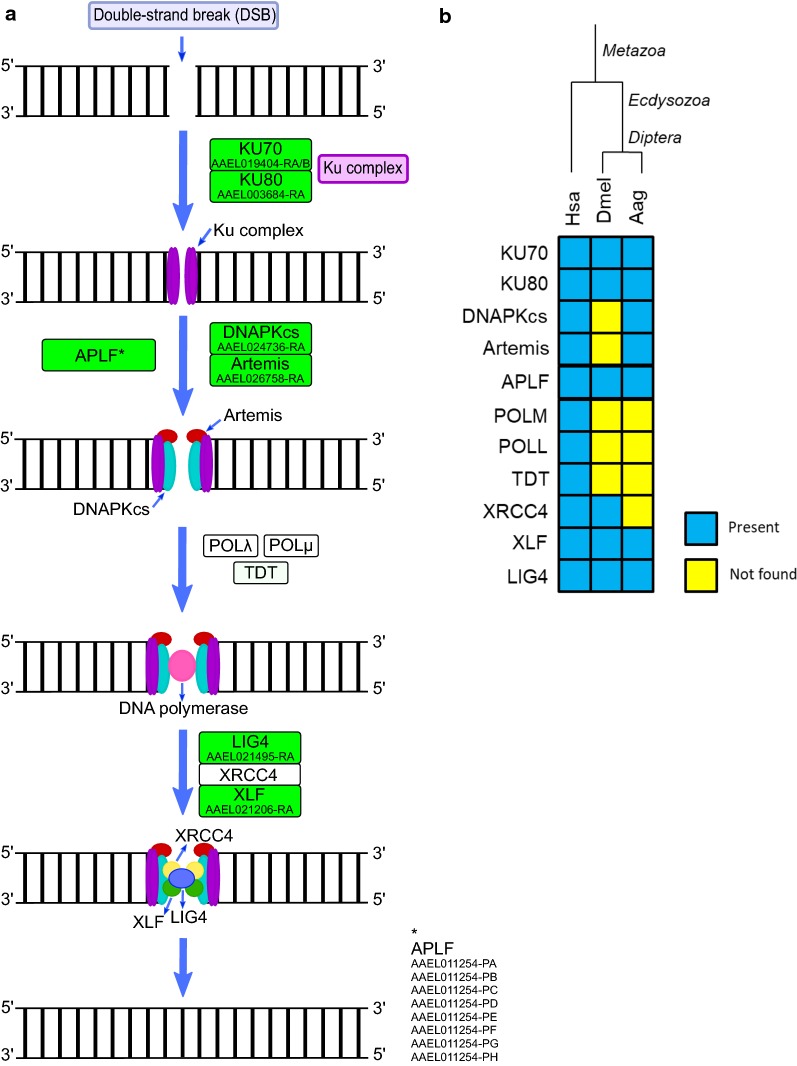
The first step of NHEJ is the binding of Ku complex (KU70 and KU80) to DSBs, which prevents the DNA ends resection and recruits the DNA-dependent protein kinase catalytic subunit (DNAPKcs), forming a multiprotein complex in both DSB ends that interact and aligns the broken ends [74–77]. KU70, KU80 and DNAPKcs were originated in early eukaryotes, being identified in Ae. aegypti. Curiously, DNAPKcs is lacking in D. melanogaster and in many insects [15] (Fig. 5).
Fig. 5.
a Non-homologous end joining (NHEJ) repair in Ae. aegypti. Rectangles: green, identified; white, not identified. b Heatmap of H. sapiens (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S5
Subsequently, when the overhangs are not complementary, DNAPKcs activates ARTEMIS, an endonuclease that processes the broken ends to find cohesive nucleotides [78]. ARTEMIS is lacking in D. melanogaster and is suggested to be lost in dipterans [15]. However, we could identify an ortholog of this protein (AAEL026758-PA) in Ae. aegypti (Fig. 5). Although ARTEMIS is the major nuclease in NHEJ, there are others proteins that might be involved in DSBs end resection such as the PNKP-like factor (APLF), polynucleotide kinase/phosphatase (PNKP), aprataxin (APTX), tyrosyl DNA phosphodiesterase 1 (TDP1) and tyrosyl DNA phosphodiesterase 2 (TDP2) [79–81]. All these proteins were found in Ae. aegypti (APLF - AAEL011254-PA-H; APTX - AAEL014945-PB-D; PNKP - AAEL025882-PA; TDP1 - AAEL011629-PB-C), except for TDP2, which is also lacking in D. melanogaster and A. mellifera [25].
The processing of DNA ends continues with the Pol X family polymerases that fill small single-strand gaps in DSB ends. The POL μ and POL λ, are members of this family, and both can incorporate nucleotides in a template-dependent and independent manner [79, 82–84]. Aedes aegypti lacks the polymerases from Pol X family (Fig. 5) suggesting that this step occurs without the dNTPs insertion; however, other polymerases can incorporate dNTPs in a template-dependent manner during NHEJ [79]. The Pol X family polymerases are also absent in most insects, including D. melanogaster [15].
In the last step, the non-homologous end-joining factor 1 (XLF) interacts with the XRCC4-LIG4 complex (X-ray repair cross-complementing protein 4 - DNA ligase 4), to catalyze the DSB ligation [85, 86]. Both LIG4 (AAEL021495-PA) and XLF (AAEL021206-PA) were found in Ae. aegypti (Fig. 5). XRCC4 was not found in this insect (Fig. 5), although it is present in some Diptera, such as D. melanogaster, and its emergence reported before plants [15, 25, 87]. Further investigation is necessary to know if (and how) NHEJ works in mosquitoes without XRCC4 as it seems to be very important. The knockout of XRCC4 in mouse cells results in 20-fold reduction of NHEJ, increasing the ends degradation and ends joining by microhomology [88]. The complete list of Ae. aegypti NHEJ proteins is provided in Additional file 2: Table S5.
Microhomology-mediated end joining (MMEJ)
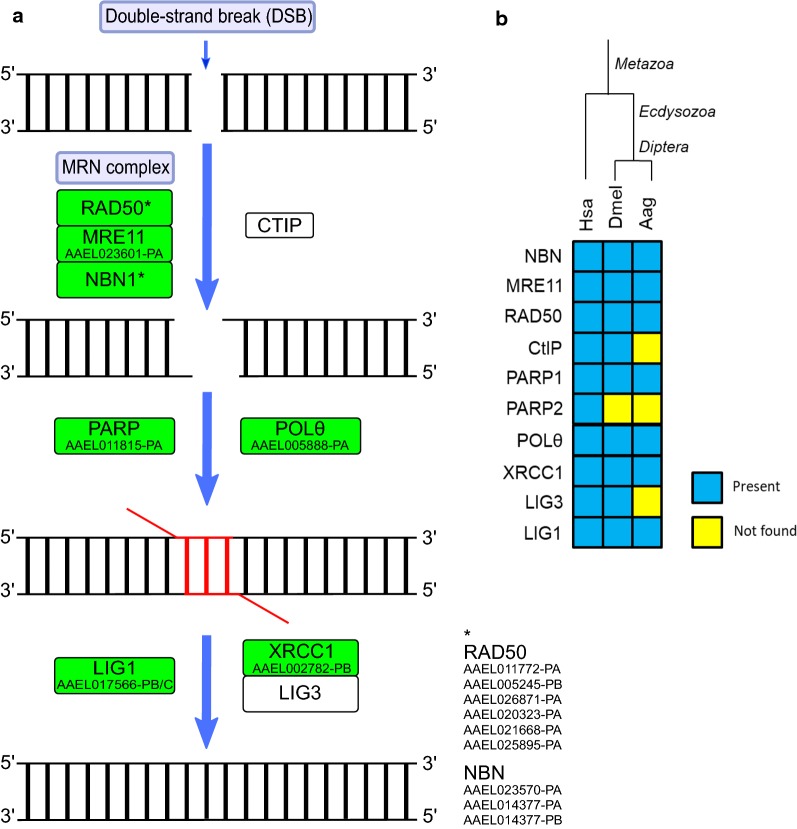
The DSB end joining can also occur by microhomology-mediated end joining (MMEJ), also known as alternative NHEJ (alt-NHEJ), which does not require ATM activation or classical NHEJ components such as the Ku complex and XRCC4-LIG4 [89]. To initiate, MMEJ needs a limited end resection that, as in HR, is mediated by MRN complex and CtIP [90]. The ssDNA overhangs recruit PARP1 or PARP2, POLθ and 5-flap endonuclease (FEN1) involved in the microhomology events, with POLθ responsible to promote the 3′-ssDNA overhangs annealing [91, 92]. Finally, the MRN complex recruits XRCC1 and LIG3 to catalyze DNA ends ligate [93].
As already discussed, CtIP was not identified in Ae. aegypti, although D. melanogaster encodes a functional ortholog of this protein. Orthologs for PARP1, FEN1 and XRCC1 were found in Ae. aegypti, but LIG3 is absent in this insect. As these proteins participate in BER, they will be discussed in more detail later in this paper. Furthermore, an ortholog of POLθ (AAEL005888-RA) was found in Ae. aegypti and is also present in D. melanogaster (Fig. 6). In fact, the role of POLθ in MMEJ was first identified in this fly, during a P-element transposition experiment [94]. The presence of the proteins involved in MMEJ, especially POLθ, in Ae. aegypti suggests that this pathway is functional in this mosquito. The absence of LIG3 may not affect MMEJ in Ae. aegypti due to the possible role of LIG1 in this process [95]. The complete list of Ae. aegypti MMEJ proteins is provided in Additional file 2: Table S6.
Fig. 6.
a Microhomology-mediated end joining (MMEJ) repair in Ae. aegypti. Rectangles: green, identified; white, not identified. b Heatmap of H. sapiens (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S6
Mismatch repair
DNA mismatch repair (MMR) is a highly conserved pathway responsible for recognizing and correcting mismatched base pairs and insertion/deletion loops (IDLs), that occur mostly during the replication process [96].
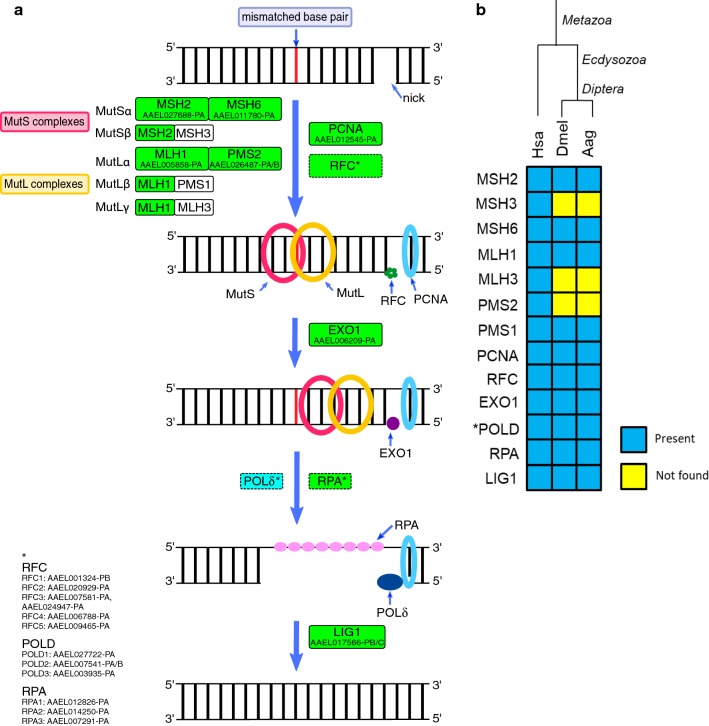
In prokaryotes, the MutL and MutS proteins are the central players of MMR [97]. In eukaryotes, MutS homologs form the functional heterodimers MutSα (MSH2-MSH6) and MutSβ (MSH2-MSH3), that repair mismatches and IDLs of up to two bases and of more than two bases, respectively [98]. Eukaryote MutL homologs form three functional heterodimers: MutLα (MLH1-PMS2), MutLβ (MLH1-MLH3) and MutLγ (MLH1-PMS1); however, MutLα is the one most involved in MMR [99–101].
MutS and MutL homologs possess an early origin, since MSH3 and MSH6 and MLH1 are ancient proteins, being present in prokaryotes, and MSH2, PMS2 and MLH3 (KEGG orthology group K08739) emerged in early eukaryotes [25]. Only PMS1 appears later in Metazoa (KEGG orthology group K10864). The MSH2 (AAEL027688-PA), MSH6 (AAEL011780-PA), MLH1 (AAEL005858-PA) and PMS2 (AAEL026487-PA/B) were found in Ae. aegypti, while the MSH3, MLH3 and PMS1 were not (Fig. 7). They seem to have been lost in the species of Diptera [15]. The absence of these proteins suggests that dipterans should not be able to form the MutSβ, MutLβ and MutLγ complexes, at least as vertebrates do, and raises questions about how they deal with mismatches and if MutSα and MutLα are enough to do this task.
Fig. 7.
a Mismatch repair (MMR) repair in Ae. aegypti. Rectangles: solid line, protein; dashed, protein complex; green, identified; cyan, partially identified; white not identified. b Heatmap of H. sapiens (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S7. *Subunits not identified: Aag-POLD4 and Dmel-POLD4
Before excision, the 5′-ends of Okazaki fragments and PCNA help discriminate between the leader and the lagging strand [102, 103]. Subsequently, the excision is orchestrated by EXO1 in cooperation with PCNA [104, 105], POL δ synthesizes a new fragment and DNA ligase I (LIG1) catalyzes strand ligation [106, 107].
The PCNA and LIG1 proteins, present in prokaryotes and eukaryotes, were both found in Ae. aegypti. As discussed above, RPA, EXO1 and POL δ (expect subunit 4) were all identified in this mosquito (Fig. 7). The complete list of Ae. aegypti MMR proteins is provided in Additional file 2: Table S7.
Base excision repair (BER) and single-strand break repair (SSBR)
Base excision repair (BER) is responsible to handle with endogenous small base lesions as oxidation, alkylation, deamination and depurination. This pathway also repairs abasic sites (AP sites) and single-strand breaks (SSBs) [108, 109].
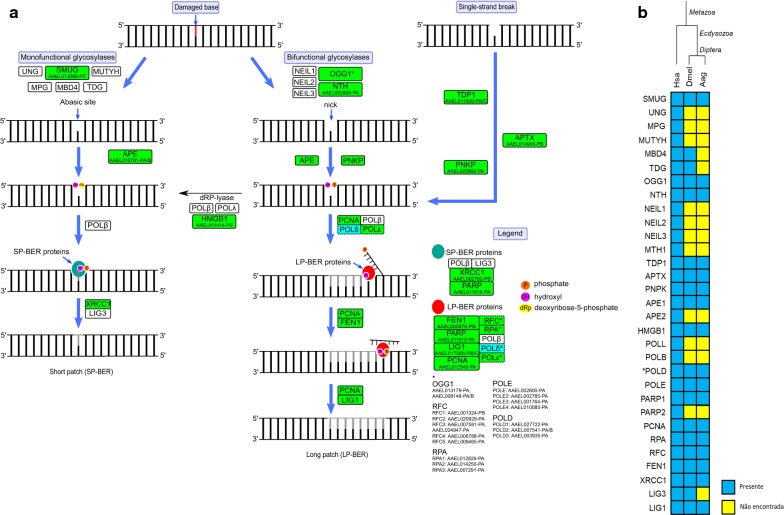
The mechanism of BER involves five major steps, and starts with the recognition and excision of the damaged base by a DNA glycosylase, that can be mono- or bifunctional and cleaves the N-glycosidic bond generating an AP site [73, 110, 111]. Humans possess eleven glycosylases: uracil DNA N-glycosylase (UNG); thymine DNA glycosylase (TDG); single-strand-selective monofunctional uracil DNA glycosylase (SMUG); methyl-CpG-binding domain 4 (MBD4); 3-methylpurine glycosylase (MPG); 8-oxoguanine DNA glycosylase (OGG); MutY homolog DNA glycosylase (MUTY); endonuclease III-like (NTH); endonuclease VIII-like 1 (NEIL1); endonuclease VIII-like 2 (NEIL2); and endonuclease VIII-3 (NEIL3). In Ae. aegypti there are only three DNA glycosylases: the monofunctional SMUG (AAEL013286-PC); the bifunctional OGG (AAEL013179-PA, AAEL008148-PA/B); and NTH (AAEL003906-PA) (Fig. 8). Comparing with other organisms, the monofunctional glycosylases UNG, MUTY, MPG are absent in the Diptera while MBD4 is present only in some species of this group such as D. melanogaster; and TDG is absent in mosquitoes [15]. The absence of UNG (the major uracil DNA glycosylase) in D melanogaster, and the downregulation of deoxyuracil triphosphatase (dUTPase) has already been correlated with high levels of uracil incorporation in larvae DNA. The higher levels of uracil-containing DNA are well tolerated in larval stages but corrected during development [15, 112]. In fact, D. melanogaster encodes a protein, denominated uracil-DNA degrading factor (UDE), present in holometabolous insects, which can degrade uracil-containing DNA [113]. The UDE protein is also found in Ae. aegypti (AAEL003864-PA), indicating that this mosquito may deal with uracil-containing DNA in the same way as D. melanogaster. The glycosylases OGG1, MUTY and the hydrolase MTH1 (named MutT in bacteria) are involved in the repair of the major oxidative lesion 7,8-dihydro-8-oxoguanine (8-oxoG) [114]. MTH1 catalyzes the hydrolysis of 8-oxo-dGTP to avoid its incorporation in DNA [115]. OGG1 is responsible for the removal of 8-oxoG residues from the DNA initiating the repair that will restore the G:C base pair [116]. If the 8oxoG:C bypass the excision by OGG1 and the DNA replication occurs, one of the DNA copies will have 8-oxoG:A pair. It is recognized by MUTY, that also removes the mismatched adenine [117]. In Ae. aegypti only an ortholog for OGG1 was identified. Otherwise, the 8-oxoG:A generated during DNA replication can be repaired by the MMR pathway [118] which, as discussed above, seems to be functional in this insect.
Fig. 8.
a Base excision repair (BER) repair in Ae. aegypti. Rectangles: solid line, protein; dashed, protein complex; green, identified; cyan, partially identified; white not identified. b Heatmap of H. sapiens (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S8. *Subunits not identified: Aag-POLD4 and Dmel-POLD4
The second step is the action of the AP endonuclease (APE), which cleaves the DNA backbone at the 5′-end removing the remaining sugar-phosphate structure. When the damaged base is removed by a bifunctional glycosylase its lyase activity cleaves AP-site leaving an 3′α,β-unsaturated aldehyde (3′-PUA) or a phosphate group (3′-P), that are removed by APE and polynucleotide kinase 3′-phosphatase (PNKP), respectively [108, 119]. In the case of SSBs the DNA ends can be processed, to generate the necessary 3′- and 5′-termini, by aprataxin (APTX), tyrosyl-DNA phosphodiesterase 1 (TDP1) and PNKP [8]. Humans possess two APE, APE1 and APE2, but Ae. aegypti encodes only APE1 (AAEL010781-PA, AAEL010781-PB) and is lacking the APE2 ortholog that is also lacking in all dipterans [15]. Otherwise, PNKP (AAEL025882-PA), APTX (AAEL014945-PB-D) and TDP1 (AAEL011629-PB-C) are all present in Ae. aegypti (Fig. 8).
The next steps of BER can occur via two different pathways: short-patch (SP-BER) and long-patch (LP-BER). SP-BER proceeds when 3′-OH and 5′-dRP termini are present, in which DNA polymerase β (POL β) removes 5′dRP and inserts a new nucleotide, filling the gap. Then the complex of x-ray repair cross-complementing 1 (XRCC1) and DNA ligase 3 (LIG3) seals the nick [120]. The LP-BER occurs when 5′-terminal is not a Pol β substrate. In this pathway, between 2 and 10 nucleotides of the 3′-termini are displaced and removed from the DNA backbone and a new nucleotide chain is synthetized by any of the POL (β, δ or ε) complexed with PCNA and flap endonuclease 1 (Fen1). The final ligation step is performed by LIG1 [117].
As indicated above, POLβ and POLλ are members of Pol X family and both are lacking in Ae. aegypti and in the Diptera. It was already suggested that dipterans use only LP-BER due to the lack of POLβ [15]. Moreover, Ae. aegypti seems to have lost LIG3 (Fig. 8) while it is present in many dipterans (KEGG orthology group K10776), reinforcing the hypothesis that this specie uses only LP-BER pathway. The LP-BER polymerases POL δ and POL ε were found in Ae. aegypti (Fig. 8), but POL δ lacks the subunit 4 (discussed above). Humans possess two PARPs that participate in BER, PARP1 and PARP2. In Ae. aegypti only one ortholog of PARP (AAEL011815-PA) was identified (Fig. 8), which is more similar to PARP1. Although PARP1 and PARP2 have specific functions, both possess overlapping roles [121, 122]. Considering that PARP2 is absent is arthropods [25], it is possible that PARP1 is performing the functions of PARP2. The complete list of Ae. aegypti BER proteins is provided in Additional file 2: Table S8.
Nucleotide excision repair
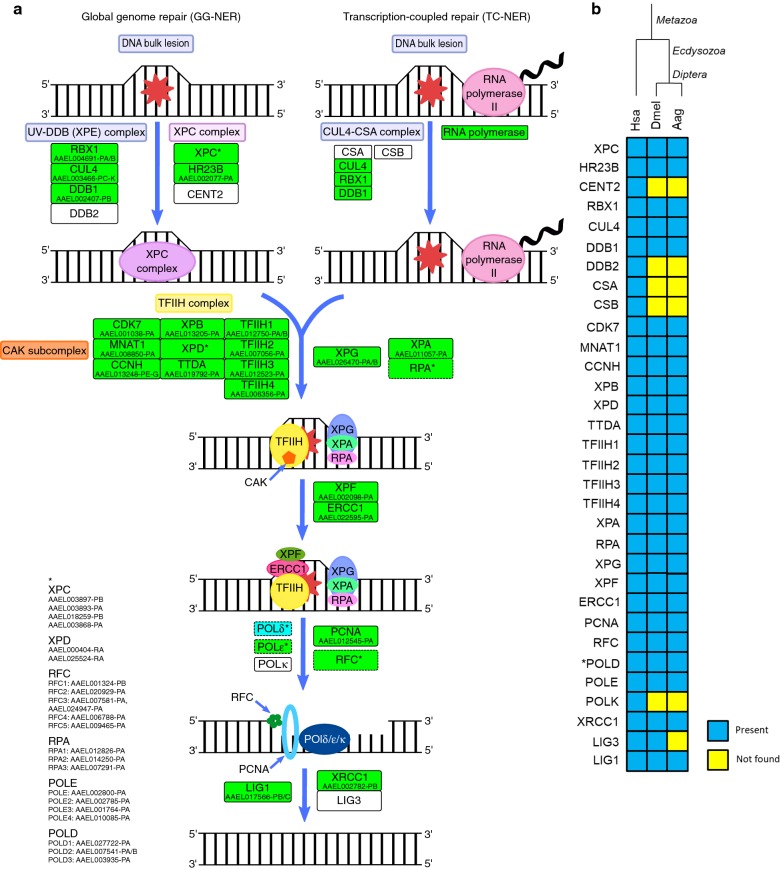
Nucleotide excision repair (NER) is a versatile pathway that repairs helix-distorting DNA lesions such as intra- and interstrand crosslinks and ultraviolet (UV) damages [123].
NER starts with damage recognition, which can occur via two sub-pathways: global genome repair (GG-NER) that detects lesions in all over the genome, and transcription-coupled repair (TC-NER) which recognizes damages in transcribed strand of active genes [124]. In GG-NER, detection occurs with the binding of the XPC complex (XPC, HR23B and CENT2) to the non-damaged strand, a process that is enhanced by the UV-DDB (XPE) complex (DDB1, DDB2 CUL4 and RBX1) [125].
The two initial NER complexes emerged in early eukaryotes [25], except for the DDB2 that appears only in plants (KEGG orthology group K10140). The XPC complex (XPC - AAEL003897-PA/B, AAEL018259-PB, AAEL003868-PA; HR23B - AAEL002077-PA) is almost complete in Ae. aegypti, only CENT2 was not found (Fig. 9). Although CENT2 enhances the DNA-binding activity of XPC-HR23B, it is not essential for NER [126], thus the absence of this protein in Ae. aegypti may not interfere in GG-NER. Furthermore, the UV-DDB complex (DDB1 - AAEL002407-PB; CUL4A - AAEL003466-PC-K) was partially identified, lacking the DDB2 protein (Fig. 9). However, the XPC complex can recognize the damage in the absence of the UV-DDB complex, which indeed is necessary to keep the repair proteins around the lesion site [127]; so the lacking DDB2 probably will not avoid GG-NER but can decrease its efficiency (Fig. 9).
Fig. 9.
a Nucleotide excision repair (NER) repair in Ae. aegypti. Rectangles: solid line, protein; dashed, protein complex; green, identified; cyan, partially identified; white not identified. b Heatmap of H. sapiens (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins. Protein codes are provided in Additional file 2: Table S9. *Subunits not identified: Aag-POLD4 and Dmel-POLD4
TC-NER repairs helix-distorting DNA lesions that block the RNA polymerase II such as inter- and intra-strand crosslinks generated by chemotherapeutics such as cisplatin, and UV damages such as cyclobutane pyrimidine dimers (CPDs) [128]. In this sub-pathway, the blockage of RNA polymerase II is the signal to recruit cockayne syndrome group A (CSA) and cockayne syndrome group B (CSB) proteins [123]. Both proteins were not identified in Ae. aegypti (Fig. 9), and seem to have been lost in all of the Diptera, otherwise the literature [15] states their origin in early eukaryotes. In humans, mutations in CSA and CSB proteins lead to cockayne syndrome, an autosomal recessive disease, characterized by microcephaly, photosensitivity, premature aging, short stature, learning and developmental delay [129]. Although mutations in these genes lead to a severe syndrome in humans, the TC-NER could not be identified in D. melanogaster [15] and is possible that DNA photolyases (discussed below) play a role in the repair of these UV lesions in these insects.
After damaged site recognition, both pathways require the same enzymatic machinery, and the following step is the recruitment of the ten subunits transcriptional factor IIH complex (TFIIH) [130].
The TFIIH helicases subunits XPB and XPD unwind DNA around the damage (~30 bp) generating a bubble, where the single strands are stabilized by XPA and RPA [73, 130, 131]. The following step is the dual incision around the damage, which is catalyzed by Xeroderma pigmentosum complementation group F (XPF)-DNA excision repair protein ERCC-1 (ERCC1) (5′) and XPG endonuclease (3′) [132]. The result is the removal of ~30 nucleotides, generating a gap that is filled by POL (δ, ε or κ), in cooperation with PCNA and RFC [133], and the remaining nick is sealed by XRCC1-LIG3 or LIG1 [134].
Arcas et al. [25] have already shown that NER central players originated in early eukaryotes, so is not surprising that these proteins are present in Ae. aegypti, except for LIG3 and POL δ, that lacks the subunit 4 (discussed above) (Fig. 9). The complete list of Ae. aegypti NER proteins is provided in Additional file 2: Table S9.
Direct repair
In addition to the DNA repair pathways, discussed in this paper, the organisms also possess mechanisms that directly reverse the DNA damage. The DNA photolyases, the α-ketoglutarate-dependent dioxygenases (AlkB) and the O-6-methylguanine DNA methyltransferase (MGMT) are the main proteins involved in this type of DNA repair [135]. The complete list of Ae. aegypti direct repair proteins is provided in Additional file 2: Table S10.
Photolyase repair
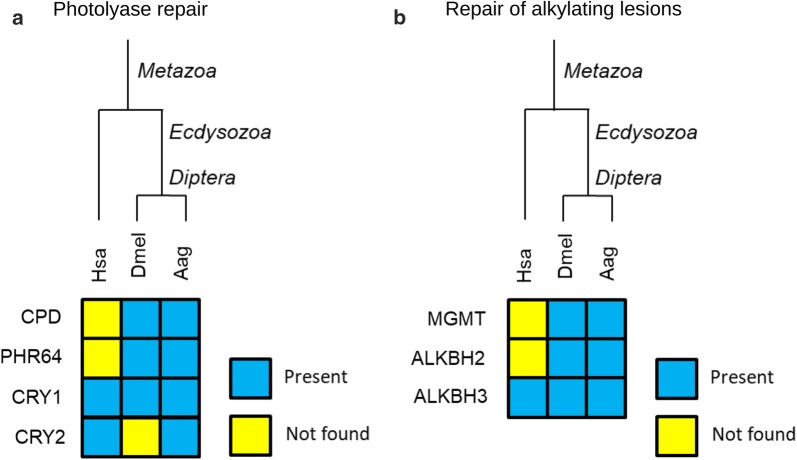
The photolyases are ancient flavoproteins activated by blue light that repair UV-induced DNA damages as cyclobutane pyrimidine dimer (CPD) and pyrimidine-pyrimidone (6-4) photoproduct [136]. Based on the substrate affinity, the photolyases are classified as CPD photolyase and (6-4) photolyase (PHR6-4). Photolyases are also ancestors of cryptochromes (CRY), a flavoprotein involved in circadian clock [137]. Although both are similar in sequence and crystal structure, CRY lacks the ability to repair UV-induced damage [138]. While humans only encode CRY, D. melanogaster possesses orthologs of the two types of photolyases plus CRY [139]. In Ae. aegypti it was possible to identify orthologs for CPD (AAEL001787-PA-E) and PHR6-4 (AAEL001175-PA), and two orthologs of CRY, CRY1 (AAEL004146-PA) and CRY2 (AAEL011967-PA) (Fig. 10a).
Fig. 10.
a Direct repair in Ae. aegypti. Heatmaps of human (Hsa), D. melanogaster (Dmel) and Ae. aegypti (Aag) proteins for photolyase repair and b alkylating lesions repair. Protein codes are provided in Additional file 2: Table S10
Repair of alkylating lesions
Endogenous and exogenous alkylating agents can damage the genomic DNA by the generation of mutagenic and cytotoxic adducts. To deal with these lesions the cell encodes mechanisms to remove the alkylated base, such as DNA glycosylases and the direct repair enzymes MGMT and AlkB.
The MGMT repairs O-6-methylguanine, which is one of the most cytotoxic and mutagenic DNA lesions, due to the ability to pair with C and T during DNA replication [140]. The MGMT transfers the O-6-methyl group from guanine to its cysteine 145 [139]. The covalent bond between Cys145 and methyl group inactivates the MGMT, which is then degraded in the ubiquitin/proteasome pathway [141]. Although MGMT is a conserved protein that is also present in D. melanogaster [142, 143], an ortholog was not identified in Ae. aegypti (Fig. 10b), keeping unanswered how this mosquito deals with this alkylating lesion.
The DNA repair function of AlkB family dioxygenases was initially identified in E. coli AlkB, which removes the 1-methyladenine and 3-methylcytosine through an oxidative dealkylation reaction. Among the nine human AlkB orthologs only two (ALKBH2 and ALKBH3) possess the repair activity [144]. In Ae. aegypti orthologs for both ALKBH2 and ALKBH3 were not identified, which is also lacking in D. melanogaster (Fig. 10b).
Alkylating lesions can also be repaired by the DNA glycosylases from BER pathway, such as TDG, MBD4, MPG and SMUG [145]. Otherwise, Ae. aegypti only encodes ortholog for SMUG (Fig. 8). The absence of MGMT, ALKBH2, ALKBH3 and the DNA glycosylases raise questions about how this mosquito deals with alkylating DNA lesions.
Conclusions
The bioinformatics analysis of this study helped identify orthologs of many key DDR proteins in Ae. aegypti, such as RAD51, RAD50, MRE11, NBN, KU80, KU70, LIG4, XLF, XPA, XPC, XPB, XPD, XPE, XPF, XPG, MSH2, MSH6, PMS2, MLH1 MutS, MutL, SMUG, OGG and NTH. Our analysis also identified a functional ortholog of human H2Ax (Drosophila H2Av) histone in Ae. aegypti. These findings indicate that the ATR and ATM signaling, DSB, HR, NHEJ, MMR, LP-BER and GG-NER repair pathways should be functional in this mosquito. Both insects showed similarities regarding the proteins not identified in Ae. aegypti (BRCA1 and its partners from the BRCA1-A complex, TP53BP1, PALB2, POLk, CSA, CSB and POLβ). It is relevant to stress that some unidentified proteins can be a result from real gene absences but also can represent a very divergent ortholog or a functional ortholog. In humans, almost all of them are essential and their lack affects DSB signaling, HR, GG-NER and SP-BER, raising questions about how these insects deal with DSB repair pathway choice and suggesting that both GG-NER and SP-BER could have been rewired or be absent. The differences between Ae. aegypti and D. melanogaster included seven proteins not reported in D. melanogaster that were found in Ae. aegypti (RNF168, RIF1, WRN, RAD54B, RMI1, DNAPKcs and ARTEMIS) and also other known six proteins in Drosophila that were not identified in Ae. aegypti (CTIP, DSS1, XRCC2, SLX4, XRCC4 and LIG3). Despite the lack of XRCC4 (important for NHEJ ligation step), NHEJ is functional in Ae. aegypti, since it was already used in the generation of genetically modified mosquitoes [18], suggesting a rewire of this pathway. This review provides an initial overview of DDR in Ae. aegypti. Understanding this system, especially the DSBs repair pathways, may help improve genomic manipulation and the establishment of transgenic mosquitoes.
Supplementary information
Additional file 1: Figure S1. Reciprocal-blast based methodology workflow. Arrows labelled “a–e” represent BLASTp analysis where the queries are the arrow-base group proteins and the subject database is at the arrowhead. Forward BLASTp (arrow “a”) top 5 hits were considered if they have e-value < 10−15, forming “Hits” group. Reverse blasts (arrows “b–e”) top 2 hits were considered if they have e-value < 10−15.
Additional file 2: Table S1. ATR signaling protein codes. Table S2. DSB protein codes. Table S3. H2A codes. Table S4. HR protein codes. Table S5. NHEJ protein codes. Table S6. MMEJ protein codes. Table S7. MMR protein codes. Table S8. BER protein codes. Table S9. NER protein codes. Table S10. Direct repair protein codes.
Acknowledgements
We thank André Luiz Quintanilha Torres, Eloy da Silva Seabra Junior and Thayany Ferreira da Costa for helpful suggestions.
Abbreviations
- DDR
DNA damage response, BER: base excision repair, NER: nucleotide excision repair, MMR: mismatch repair, HR: homologous recombination, NHEJ: non-homologous end joining, DSB: double-strand break
Authors’ contributions
MM, AM, MC and RM designed the study. MM performed the bioinformatics analysis. MM and RM interpreted the data. MM, AM, MC and RM reviewed and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq): INCT - Entomologia molecular Consortium and MM PhD fellowship.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maria Beatriz S. Mota, Email: mbsmota@gmail.com
Marcelo Alex Carvalho, Email: marcelo.carvalho@inca.gov.br.
Alvaro N. A. Monteiro, Email: alvaro.monteiro@moffitt.org
Rafael D. Mesquita, Email: rdmesquita@iq.ufrj.br
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-019-3792-1.
References
- 1.Powell JR. Perspective piece mosquito-borne human viral diseases: why Aedes aegypti? Am J Trop Med Hyg. 2018;98:1563–1565. doi: 10.4269/ajtmh.17-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lima VL, Dias F, Nunes RD, Pereira LO, Santos TSR, Chiarini LB, et al. The antioxidant role of xanthurenic acid in the Aedes aegypti midgut during digestion of a blood meal. PLoS ONE. 2012;7:e38349. doi: 10.1371/journal.pone.0038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliveira JHM, Gonçalves RLS, Lara FA, Dias FA, Gandara ACP, Menna-Barreto RFS, et al. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 2011;7:e1001320. doi: 10.1371/journal.ppat.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poupardin R, Reynaud S, Strode C, Ranson H, Vontas J, David JP. Cross-induction of detoxification genes by environmental xenobiotics and insecticides in the mosquito Aedes aegypti: impact on larval tolerance to chemical insecticides. Insect Biochem Mol Biol. 2008;38:540–551. doi: 10.1016/j.ibmb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Tetreau G, Chandor-Proust A, Faucon F, Stalinski R, Akhouayri I, Prud’homme SM, et al. UV light and urban pollution: bad cocktail for mosquitoes? Aquat Toxicol. 2014;146:52–60. doi: 10.1016/j.aquatox.2013.10.031. [DOI] [PubMed] [Google Scholar]
- 6.Diaz-Albiter H, Sant’Anna MRV, Genta FA, Dillon RJ. Reactive oxygen species-mediated immunity against Leishmania mexicana and Serratia marcescens in the phlebotomine sand fly Lutzomyia longipalpis. J Biol Chem. 2012;287:23995–24003. doi: 10.1074/jbc.M112.376095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina-Cruz A, DeJong RJ, Charles B, Gupta L, Kumar S, Jaramillo-Gutierrez G, et al. Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium. J Biol Chem. 2008;283:3217–3223. doi: 10.1074/jbc.M705873200. [DOI] [PubMed] [Google Scholar]
- 8.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 9.Graça-Souza AV, Maya-Monteiro C, Paiva-Silva GO, Braz GRC, Paes MC, Sorgine MHF, et al. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem Mol Biol. 2006;36:322–335. doi: 10.1016/j.ibmb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciccia A, Elledge SJ. The DNA Damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giglia-Mari G, Zotter A, Vermeulen W. DNA damage response. Cold Spring Harb Perspect Biol. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Sekelsky J. DNA repair in Drosophila: mutagens, models, and lacking genes. Genetics. 2017;205:471–490. doi: 10.1534/genetics.116.186759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overcash JM, Aryan A, Myles KM, Adelman ZN. Understanding the DNA damage response in order to achieve desired gene editing outcomes in mosquitoes. Chromosom Res. 2015;23:31–42. doi: 10.1007/s10577-014-9450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aryan A, Myles KM, Adelman ZN. Targeted genome editing in Aedes aegypti using TALENs. Methods. 2014;69:38–45. doi: 10.1016/j.ymeth.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aryan A, Anderson MAE, Myles KM, Adelman ZN. Germline excision of transgenes in Aedes aegypti by homing endonucleases. Sci Rep. 2013;3:1603. doi: 10.1038/srep01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu S, Aryan A, Overcash JM, Samuel GH, Anderson MAE, Dahlem TJ, et al. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc Natl Acad Sci USA. 2015;112:4038–4043. doi: 10.1073/pnas.1502370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11:51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chojnacki S, Cowley A, Lee J, Foix A, Lopez R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 2017;45:550–553. doi: 10.1093/nar/gkx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 24.Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arcas A, Fernández-Capetillo O, Cases I, Rojas AM. Emergence and evolutionary analysis of the human DDR network: implications in comparative genomics and downstream analyses. Mol Biol Evol. 2014;31:940–961. doi: 10.1093/molbev/msu046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 27.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 29.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodgers K, Mcvey M. Error-prone repair of DNA double-strand breaks. J Cell Physiol. 2016;231:15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 32.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 33.Coster G, Goldberg M. The cellular response to DNA damage: a focus on MDC1 and its interacting proteins. Nucleus. 2010;1:166–178. doi: 10.4161/nucl.11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 36.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brcal A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JTF, Tkáč J, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 42.Madigan JP. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002;30:3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 44.Danielsen JR, Povlsen LK, Villumsen BH, Streicher W, Nilsson J, Wikström M, et al. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol. 2012;197:179–187. doi: 10.1083/jcb.201106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rong YS, Golic KG. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics. 2003;165:1831–1842. doi: 10.1093/genetics/165.4.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staeva-Vieira E, Yoo S, Lehmann R. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 2003;22:5863–5874. doi: 10.1093/emboj/cdg564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo S, McKee BD. Functional analysis of the Drosophila Rad5l gene (spn-A) in repair of DNA damage and meiotic chromosome segregation. DNA Repair (Amst). 2005;4:231–242. doi: 10.1016/j.dnarep.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Aroumougame A, Lobrich M, Li Y, Chen D, Chenj J, et al. PTIP associates with artemis to dictate DNA repair pathway choice. Genes Dev. 2014;28:2693–2698. doi: 10.1101/gad.252478.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nimonkar AV, Özsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, et al. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J Biol Chem. 2014;289:27314–27326. doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Nievera CJ, Lee AYL, Wu X. Cell cycle-dependent complex formation of BRCA1·CtIP·MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 55.Xie J, Peng M, Guillemette S, Quan S, Maniatis S, Wu Y, et al. FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS Genet. 2012;8:e1002786. doi: 10.1371/journal.pgen.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buisson R, Dion-Côté AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goyal N, Rossi MJ, Mazina OM, Chi Y, Moritz RL, Clurman BE, et al. RAD54 N-terminal domain is a DNA sensor that couples ATP hydrolysis with branch migration of Holliday junctions. Nat Commun. 2018;9:34. doi: 10.1038/s41467-017-02497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiianitsa K, Solinger JA, Heyer WD. Terminal association of Rad54 protein with the Rad51-dsDNA filament. Proc Natl Acad Sci USA. 2006;103:9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sebesta M, Burkovics P, Juhasz S, Zhang S, Szabo JE, Lee MYWT, et al. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Repair (Amst). 2013;12:691–698. doi: 10.1016/j.dnarep.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borgogno MV, Monti MR, Zhao W, Sung P, Argarana CE, Pezza RJ. Tolerance of DNA mismatches in Dmc1 recombinase-mediated DNA strand exchange. J Biol Chem. 2016;291:4928–4938. doi: 10.1074/jbc.M115.704718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu L, Hickson IO. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 63.Matos J, West SC. Holliday junction resolution: regulation in space and time. DNA Repair (Amst). 2014;19:176–181. doi: 10.1016/j.dnarep.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West SC, Blanco MG, Chan YW, Matos J, Sarbajna S, Wyatt HDM. Resolution of recombination intermediates: mechanisms and regulation. Cold Spring Harb Symp Quant Biol. 2016;80:103–109. doi: 10.1101/sqb.2015.80.027649. [DOI] [PubMed] [Google Scholar]
- 65.Wyatt HDM, Sarbajna S, Matos J, West SC. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for holliday junction resolution in human cells. Mol Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 66.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MIR, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uringa EJ, Lisaingo K, Pickett HA, Brind’Amour J, Rohde JH, Zelensky A, et al. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol Biol Cell. 2012;23:2782–2792. doi: 10.1091/mbc.e12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malkova A. Break-induced replication: the where, the why, and the how. Trends Genet. 2018;34:518–531. doi: 10.1016/j.tig.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakofsky CJ, Malkova A. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit Rev Biochem Mol Biol. 2017;52:395–413. doi: 10.1080/10409238.2017.1314444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, et al. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010;38:2917–2930. doi: 10.1093/nar/gkp1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–3558. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iyama T, Wilson DM. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst). 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson E. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun J, Lee KJ, Davis AJ, Chen DJ. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J Biol Chem. 2012;287:4936–4945. doi: 10.1074/jbc.M111.306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to dna and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 77.Yoo S. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moscariello M, Wieloch R, Kurosawa A, Li F, Adachi N, Mladenov E, et al. Role for Artemis nuclease in the repair of radiation-induced DNA double strand breaks by alternative end joining. DNA Repair (Amst). 2015;31:29–40. doi: 10.1016/j.dnarep.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 79.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gómez-Herreros F, Romero-Granados R, Zeng Z, Álvarez-Quilón A, Quintero C, Ju L, et al. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013;9:e1003226. doi: 10.1371/journal.pgen.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heo J, Li J, Summerlin M, Hays A, Katyal S, McKinnon PJ, et al. TDP1 promotes assembly of non-homologous end joining protein complexes on DNA. DNA Repair (Amst). 2015;30:28–37. doi: 10.1016/j.dnarep.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan W, Wu X. DNA polymerase λ can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem Biophys Res Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 83.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, et al. Implication of DNA polymerase λ in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 84.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and Ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 86.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku Recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/MCB.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gorski MM, Eeken JCJ, De Jong AWM, Klink I, Loos M, Romeijn RJ, et al. The Drosophila melanogaster DNA ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-dtrand breaks and acts synergistically with Rad54. Genetics. 2003;165:1929–1941. doi: 10.1093/genetics/165.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mateos-Gomez PA, Kent T, Deng SK, McDevitt S, Kashkina E, Hoang TM, et al. The helicase domain of Poltheta counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol. 2017;24:1116–1123. doi: 10.1038/nsmb.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015;6:e1697. doi: 10.1038/cddis.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Della-Maria J, Zhou Y, Tsai M-S, Kuhnlein J, Carney JP, Paull TT, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:1–16. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sfeir A, Symington LS. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu D, Keijzers G, Rasmussen LJ. DNA mismatch repair and its many roles in eukaryotic cells. Mutat Res. 2017;773:174–187. doi: 10.1016/j.mrrev.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 97.Fukui K. DNA mismatch repair in eukaryotes and bacteria. J Nucleic Acids. 2010;2010:1–16. doi: 10.4061/2010/260512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cannavo E, Marra G, Sabates-Bellver J, Menigatti M, Lipkin SM, Fischer F, et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 100.Erie DA, Weninger KR. Single molecule studies of DNA mismatch repair. DNA Repair (Amst). 2014;20:71–81. doi: 10.1016/j.dnarep.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Räschle M, Marra G, Nyström-Lahti M, Schär P, Jiricny J. Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- 102.Nick McElhinny S, Kissling GE, Kunkel T. Differential correction of lagging-strand replication errors made by DNA polymerases alpha and {delta} Proc Natl Acad Sci USA. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov F, Modrich P. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/S1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 105.Liberti SE, Andersen SD, Wang J, May A, Miron S, Perderiset M, et al. Bi-directional routing of DNA mismatch repair protein human exonuclease 1 to replication foci and DNA double strand breaks. DNA Repair (Amst). 2011;10:73–86. doi: 10.1016/j.dnarep.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 108.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst). 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Markkanen E. Not breathing is not an option: how to deal with oxidative DNA damage. DNA Repair (Amst). 2017;59:82–105. doi: 10.1016/j.dnarep.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 110.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Muha V, Horváth A, Békési A, Pukáncsik M, Hodoscsek B, Merényi G, et al. Uracil-containing DNA in Drosophila: stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 2012;8:e1002738. doi: 10.1371/journal.pgen.1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Békési A, Pukáncsik M, Muha V, Zagyva I, Leveles I, Hunyadi-Gulyás É, et al. A novel fruitfly protein under developmental control degrades uracil-DNA. Biochem Biophys Res Commun. 2007;355:643–648. doi: 10.1016/j.bbrc.2007.01.196. [DOI] [PubMed] [Google Scholar]
- 114.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cadet J, Davies KJA. Oxidative DNA damage & repair: an introduction. Free Radic Biol Med. 2017;107:2–12. doi: 10.1016/j.freeradbiomed.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase 1: beyond repair of the oxidatively modified base lesions. Redox Biol. 2018;14:669–678. doi: 10.1016/j.redox.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst). 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Z, Pearlman AH, Hsieh P. DNA mismatch repair and the DNA damage response. DNA Repair (Amst). 2016;38:94–101. doi: 10.1016/j.dnarep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu G, Herzig M, Rotrekl V, Walter CA. Base excision repair, aging and health span. Mech Ageing Dev. 2008;129:366–382. doi: 10.1016/j.mad.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst). 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 121.Hanzlikova H, Gittens W, Krejcikova K, Zeng Z, Caldecott KW. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017;45:2546–2557. doi: 10.1093/nar/gkw1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: new players in tumour development. Am J Cancer Res. 2011;1:328–346. [PMC free article] [PubMed] [Google Scholar]
- 123.Marteijn J, Lans H, Vermeulen W, Hoeijmakers JHJ. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 124.Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mu H, Geacintov NE, Broyde S, Yeo JE, Schärer OD. Molecular basis for damage recognition and verification by XPC-RAD23B and TFIIH in nucleotide excision repair. DNA Repair (Amst). 2018;71:33–42. doi: 10.1016/j.dnarep.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, et al. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol Cell Biol. 2005;25:5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oh KS, Imoto K, Emmert S, Tamura D, Digiovanna JJ, Kraemer KH. Nucleotide excision repair proteins rapidly accumulate but fail to persist in human XP-E (DDB2 mutant) cells. Photochem Photobiol. 2011;87:729–733. doi: 10.1111/j.1751-1097.2011.00909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Geijer ME, Marteijn JA. What happens at the lesion does not stay at the lesion: transcription-coupled nucleotide excision repair and the effects of DNA damage on transcription in cis and trans. DNA Repair (Amst). 2018;71:56–68. doi: 10.1016/j.dnarep.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 129.Karikkineth AC, Scheibye-Knudsen M, Fivenson E, Croteau DL, Bohr VA. Cockayne syndrome: clinical features, model systems and pathways. Ageing Res Rev. 2017;33:3–17. doi: 10.1016/j.arr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jeppesen DK, Bohr VA, Stevnsner T. DNA repair deficiency in neurodegeneration. Prog Neurobiol. 2011;94:166–200. doi: 10.1016/j.pneurobio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Diderich K, Alanazi M, Hoeijmakers JHJ. Premature aging and cancer in nucleotide excision repair-disorders. DNA Repair (Amst). 2011;10:772–780. doi: 10.1016/j.dnarep.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 134.Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LHF, Fousteri MI. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 135.Yi C, He C. DNA repair by reversal of DNA damage. Cold Spring Harb Perspect Biol. 2013;5:a012575. doi: 10.1101/cshperspect.a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang M, Wang L, Zhong D. Photolyase: dynamics and mechanisms of repair of sun-induced DNA damage. Photochem Photobiol. 2017;93:78–92. doi: 10.1111/php.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mei Q, Dvornyk V. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS One. 2015;10:e0135940. doi: 10.1371/journal.pone.0135940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang J, Du X, Pan W, Wang X, Wu W. Photoactivation of the cryptochrome/photolyase superfamily. J Photochem Photobiol C Photochem Rev. 2015;22:84–102. doi: 10.1016/j.jphotochemrev.2014.12.001. [DOI] [Google Scholar]
- 139.Selby CP, Sancar A. The second chromophore in Drosophila photolyase/cryptochrome family photoreceptors. Biochemistry. 2012;51:167–171. doi: 10.1021/bi201536w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Soll JM, Sobol RW, Mosammaparast N. Regulation of DNA alkylation damage repair: lessons and therapeutic opportunities. Trends Biochem Sci. 2017;42:206–218. doi: 10.1016/j.tibs.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mostofa A, Punganuru SR, Madala HR, Srivenugopal KS. S-phase specific downregulation of human O(6)-methylguanine DNA methyltransferase (MGMT) and its serendipitous interactions with PCNA and p21(cip1) proteins in glioma cells. Neoplasia. 2018;20:305–323. doi: 10.1016/j.neo.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 143.Kooistra R, Zonneveld JB, Watson AJ, Margison GP, Lohman PH, Pastink A. Identification and characterisation of the Drosophila melanogaster O6-alkylguanine-DNA alkyltransferase cDNA. Nucleic Acids Res. 1999;27:1795–1801. doi: 10.1093/nar/27.8.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fu D, Samson LD, Hubscher U, van Loon B. The interaction between ALKBH2 DNA repair enzyme and PCNA is direct, mediated by the hydrophobic pocket of PCNA and perturbed in naturally-occurring ALKBH2 variants. DNA Repair (Amst). 2015;35:13–18. doi: 10.1016/j.dnarep.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zdzalik D, Domanska A, Prorok P, Kosicki K, van den Born E, Falnes PO, et al. Differential repair of etheno-DNA adducts by bacterial and human AlkB proteins. DNA Repair (Amst). 2015;30:1–10. doi: 10.1016/j.dnarep.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Reciprocal-blast based methodology workflow. Arrows labelled “a–e” represent BLASTp analysis where the queries are the arrow-base group proteins and the subject database is at the arrowhead. Forward BLASTp (arrow “a”) top 5 hits were considered if they have e-value < 10−15, forming “Hits” group. Reverse blasts (arrows “b–e”) top 2 hits were considered if they have e-value < 10−15.
Additional file 2: Table S1. ATR signaling protein codes. Table S2. DSB protein codes. Table S3. H2A codes. Table S4. HR protein codes. Table S5. NHEJ protein codes. Table S6. MMEJ protein codes. Table S7. MMR protein codes. Table S8. BER protein codes. Table S9. NER protein codes. Table S10. Direct repair protein codes.
Data Availability Statement
Not applicable.