Abstract
Background
Climate change is expected to have a negative impact on food availability. While most efforts have been directed to reducing greenhouse gas emissions, complementary strategies are necessary to control the detrimental effects of climate change on farm animal performance. The objective of this study was to develop novel animal resilience phenotypes using reaction norm slopes, and examine their genetic and genomic parameters. A closely monitored dairy goat population was used for this purpose.
Results
Individual animals differed in their response to changing atmospheric temperature and a temperature-humidity index. Significant genetic variance and heritability estimates were derived for these animal resilience phenotypes. Furthermore, some resilience traits had a significant unfavourable genetic correlation with animal performance. Genome-wide association analyses identified several candidate genes related to animal resilience to environment change.
Conclusions
Heritable variation exists among dairy goats in their production response to fluctuating weather variables. Results may inform future breeding programmes aimed to ensure efficient animal performance under changing climatic conditions.
Keywords: Animal resilience, Climate change, Selective breeding, Heritability, Candidate genes
Background
According to the United Nations Intergovernmental Panel on Climate Change, human activities since the pre-industrial times have had a strong impact on climate. Agriculture is believed to contribute to climate change mostly due to greenhouse gas emissions through the use of fertilisers, methane production by livestock and nitrous oxide emissions from soils [1]. Furthermore, indirect consequences of the agricultural industrialisation such as deforestation [2], intensive monoculture leading to a reduction in variation [3], and the improper use of water irrigation and industrial machinery have also contributed to climate change. The substantial rise in global atmospheric temperature has been particularly steep over the past few decades (0.17 °C/decade), and has been largely noticeable in the northern hemisphere during spring and winter and more uniform throughout the year in the southern hemisphere. In Europe, in addition to the gradual increase in temperature, climate change has also been manifesting in alterations in intra-seasonal and inter-annual variability, with decreasing variability of winter mean temperatures and increased variability of summer mean temperatures [4]. An increase in temperature variability is also predicted for tropical countries [5]. Additional modifications in precipitation and humidity patterns are also evident, with increased and decreased annual precipitation in northern and southern Europe, respectively, and fluctuations in precipitation in central Europe [6].
With regards to agriculture and livestock farming, the main focus to-date has been on mitigating the effects of methane and other greenhouse gas emissions [7, 8]. At the same time, there is a growing concern that climate change may adversely affect the quality and quantity of both plant [9] and livestock [10] products leading to reduced food availability as well as increased frequency and severity of disease [11]. Therefore, there is a recognised need to address the current detrimental effects of environmental degradation on animal and plant production, and to develop additional strategies to mitigate the problem [10, 12, 13].
Selective breeding for enhanced animal resilience to environmental variation may offer a novel strategy to address the impact of climate change on livestock production [14, 15]. The few genetic studies conducted to-date have focussed on extreme directional climate challenges such as heat stress from very high temperatures [16–19]. While these considerations are appropriate in specific geomorphological areas, climate change leading to increased seasonal variability in weather conditions may also affect animal performance [10, 12], even within the moderate temperature range.
Animal resilience must be properly defined in order to derive appropriate phenotypes across the range of prevailing and expected environmental conditions [20–22]. These phenotypes could be included in selective breeding programmes aiming at sustainable animal production levels in presence of environmental (climate) perturbations.
Different theoretical frameworks have been used to model resilience to environmental changes and its effects on animal performance. Recent studies have shown the potential use of genotype by environment interaction (GxE) to estimate resilience phenotypes for animal production traits [23–25]. In this context, individual phenotypes can be described as a continuous function of an environmental variable using random regression model approaches [26]. Reaction norm functions can then be used to express resilience as a phenotypic response of animal performance to changing environment (for example, weather).
The objectives of the present study were to (i) derive novel animal resilience phenotypes based on milk production changes in response to weather variability and (ii) investigate the genetic and genomic architecture of these newly derived animal phenotypes.
We deployed reaction norm functions to derive resilience phenotypes, mixed models for statistical analyses to estimate relevant genetic parameters, and genome-wide association studies to detect molecular markers and candidate genes controlling resilience. We used data from a well-monitored dairy goat population but our approach is applicable to any livestock species.
Results
Animal performance records and weather measurements
Descriptive statistics of animal performance and weather records are presented in Table 1. Daily milk yield, temperature and a temperature-humidity index (THI) reflected averages of a 10-day period.
Table 1.
Descriptive statistics of animal performance and weather records
| DMY (kg) | T (°C) | RH (%) | THI | |
|---|---|---|---|---|
| Mean | 3.58 | 9.89 | 83.05 | 50.48 |
| SD | 1.51 | 4.62 | 5.35 | 7.62 |
| Minimum | 0.20 | −3.27 | 65.65 | 29.13 |
| Quantile 25 | 2.50 | 6.04 | 79.38 | 44.41 |
| Median | 3.50 | 9.99 | 83.13 | 50.95 |
| Quantile 75 | 4.60 | 14.14 | 86.73 | 57.52 |
| Maximum | 12.60 | 18.92 | 97.27 | 65.25 |
Daily milk yield (DMY), daily temperature (T), daily relative humidity (RH) and daily temperature-humidity index (THI)
The prevailing weather conditions in the geographic region during the time of the study are illustrated in Additional file 1: Figure S1. These conditions are concordant with other weather reports in the UK [27], with average temperatures of 17-20 °C in July–August and 3-4 °C in January–February.
Individual animal resilience phenotypes
Descriptive statistics of animal resilience phenotypes are shown in Table 2. These phenotypes reflect the change of individual animal daily performance (milk yield) in response to changing weather (temperature and THI). Values of individual phenotypes were both positive, suggestive of increased milk production at higher values of the weather measurement, and negative, reflecting decreased animal performance at higher values of the weather measurement. An additional phenotype was the absolute value of these records indicating stable (values close to zero) versus volatile milk production response to weather change.
Table 2.
Descriptive statistics of resilience phenotypes expressed as milk production change (kg) per unit increase in weather variables
| Phenotype | Mean | SD | Min | Max | K | S |
|---|---|---|---|---|---|---|
| T | 0.029 | 0.048 | −0.170 | 0.372 | 1.31 | 0.23 |
| abs (T) | 0.044 | 0.035 | 2.28E-6 | 0.372 | 3.03 | 1.38 |
| THI | 0.018 | 0.029 | −0.102 | 0.224 | 1.31 | 0.22 |
| abs (THI) | 0.026 | 0.021 | 5.44E-7 | 0.224 | 3.02 | 1.38 |
T Performance change per unit of temperature (°C) change, THI Performance change per unit of temperature-humidity index change, abs Absolute value of corresponding performance change, SD Standard deviation, K Kurtosis, S Skewness
Genetic parameters of resilience phenotypes
Variance components and genetic parameter estimates for animal resilience phenotypes are shown in Table 3. All estimates were significantly greater than zero (P < 0.01). Genetic correlation of total lifetime milk yield with the resilience phenotypes related to absolute slopes (volatility phenotypes) were also significantly positive (P < 0.01). The latter implies an unfavourable correlation where animals with high milk yield potential are also more likely to have their milk production affected by change in changing weather.
Table 3.
Genetic parameters of resilience phenotypes expressed as milk production change per unit increase in weather variables
| Phenotype | VP | VA | h2 | rA |
|---|---|---|---|---|
| T | 2.06E-3 ± 2.27E-5* | 2.18E-4 ± 2.79E-5* | 0.11 ± 0.01* | 0.05 ± 0.07 |
| abs (T) | 6.70E-3 ± 7.30E-5* | 6.13E-4 ± 8.68E-5* | 0.09 ± 0.01* | 0.46 ± 0.07* |
| THI | 7.51E-4 ± 8.27E-6* | 7.92E-5 ± 1.01E-5* | 0.11 ± 0.01* | 0.04 ± 0.07 |
| abs (THI) | 4.03E-3 ± 4.39E-5* | 3.63E-4 ± 5.20E-5* | 0.09 ± 0.01* | 0.46 ± 0.07* |
T Performance change per unit of temperature (°C) change, THI Performance change per unit of temperature-humidity index change, abs Absolute value of corresponding performance change (square root transformed), VP Phenotypic variance, VA Additive variance, h2 Heritability, rA Genetic correlation with total lifetime milk yield (square root transformed)
Genomic association analysis
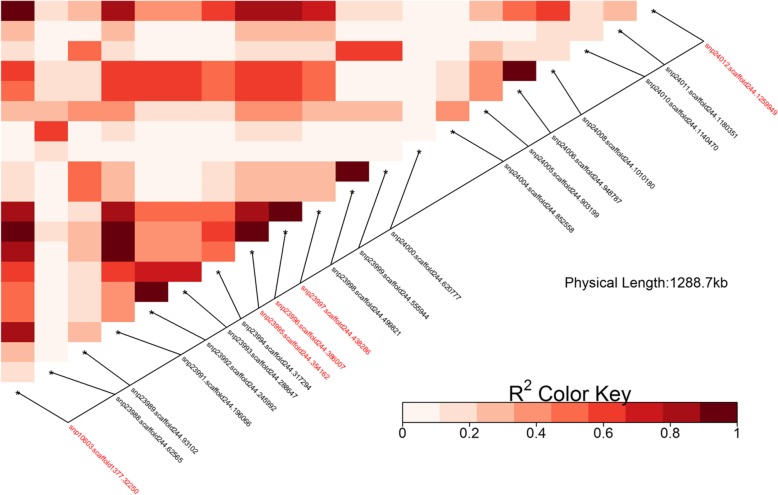
Population structure was not detected and values of the inflation factor λ ranged from 0.996 to 1.001 for all analysed phenotypes. Several single nucleotide polymorphisms (SNPs) were found to be significantly associated with total lifetime milk yield and resilience phenotypes either at genome- or chromosome-wide levels (Table 4, Additional file 2: Figure S2). Table 4 summarises these results and includes annotated genes found in the respective genomic regions. One genome-wide significant SNP was detected on chromosome 19 for total lifetime milk yield and resilience phenotypes based on absolute slopes, which was also significant at chromosome-wide level for all the other resilience phenotypes. Another two genome-wide significant SNPs were detected for total lifetime milk yield on chromosomes 8 and 13, with no effect on resilience traits. Other chromosome-wide associations were detected on chromosomes 3, 4, 13, 14, 19, 20 and 24. All significant SNPs on chromosome 19 span a region of 1289 kb, representing a relatively high linkage disequilibrium block (Fig. 1).
Table 4.
Single Nucleotide Polymorphisms significantly associated with goat milk yield and resilience phenotypes at genome-* and chromosome-wide level
| Chr | Position (bp) | Trait | Beta effect ± SE | -log10(P) | pve (%) | Nearest genes |
|---|---|---|---|---|---|---|
| 3 | 2,389,800 | TMY | 0.6580 ± 0.1501 | 4.93 | 0.18 | HDAC4 |
| 4 | 42,384,455 | TMY | 0.7455 ± 0.1646 | 5.22 | 0.19 | HECW1 |
| 8 | 39,875,253 | TMY | 1.1943 ± 0.1863 | 9.82* | 0.37 | SLC1A1; GLIS3 |
| 13 | 58,370,939 | TMY | 0.7424 ± 0.1484 | 6.24* | 0.24 | BMP7; TFAP2C |
| 13 | 66,542,955 | T | −0.0041 ± 0.0010 | 4.59 | 0.17 | RPRD1B; LOC108637373 |
| THI | −0.0025 ± 0.0006 | 4.53 | 0.16 | |||
| 14 | 69,473,160 | TMY | −0.7232 ± 0.1597 | 5.22 | 0.19 | LOC102186225 |
| 19 | 26,192,068 | T | 0.0040 ± 0.0009 | 4.64 | 0.17 | TRNAS-UGA; KIF1C |
| 19 | 26,578,715 | T | 0.0040 ± 0.0010 | 4.52 | 0.16 | LOC102178853; ALOX12 |
| 19 | 26,610,550 | T | 0.0042 ± 0.0010 | 4.90 | 0.18 | RNASEK |
| abs (T) | 0.0156 ± 0.0015 | 24.55* | 0.91 | |||
| THI | 0.0024 ± 0.0006 | 4.60 | 0.17 | |||
| abs (THI) | 0.0120 ± 0.0012 | 24.12* | 0.89 | |||
| TMY | 3.6668 ± 0.1916 | 79.53* | 2.78 | |||
| 19 | 26,662,221 | T | 0.0039 ± 0.0010 | 4.50 | 0.16 | ASGR2 |
| 19 | 27,480,733 | T | 0.0043 ± 0.0009 | 5.40 | 0.20 | ALOXE3 |
| THI | 0.0025 ± 0.0006 | 5.13 | 0.19 | |||
| 20 | 3,621,302 | TMY | −0.7984 ± 0.1828 | 4.90 | 0.18 | FGF18; SMIM23 |
| 20 | 10,173,528 | TMY | 0.8545 ± 0.1882 | 5.25 | 0.19 | NAIP |
| 24 | 52,818,318 | abs (T) | −0.0074 ± 0.0017 | 4.74 | 0.17 | LOC108633777; LOC108633778 |
| abs (THI) | −0.0058 ± 0.0013 | 4.66 | 0.17 |
TMY Total lifetime milk yield, T Performance change per unit of temperature (°C) change, THI Performance change per unit of temperature-humidity index change, abs Absolute value of corresponding performance change, Chr Chromosome, pve Percentage of explained phenotypic variance. Genome-wide significances are indicated with an asterisk *
Fig. 1.
Linkage Disequilibrium structure on chromosome 19 spanning region between significant SNP for resilience phenotypes: significant SNP are marked in red
Discussion
Climate change is expected to affect future livestock performance due to not only directional changes such as rising atmospheric temperature but also an increased volatility in weather conditions. Selective breeding for enhanced animal resilience to weather changes may contribute to the mitigation of the problem, leading to stable animal performance that is unaffected by weather variability. The present study set out to identify novel phenotypes of animal resilience and address their potential use in breeding schemes by estimating genetic parameters and identifying potential candidate genes. Results would determine how amenable animal resilience might be to improvement through genetic selection and how to inform relevant breeding programmes.
The use of linear slopes derived from reaction norm functions fitted to random regression models provided an assessment of the response of individual animal performance to changing weather including atmospheric temperature and THI. While the assumption of linearity was valid for the available range of weather measurements in the present study, other weather measurements and/or different value ranges of the same measurements in other geomorphological regions of the world might warrant investigation of non-linear models. In the latter case, the methodology presented here would still be relevant, as slopes at specific ranges in the weather measurement trajectory may be derived and used as distinct resilience phenotypes. For example, in areas where climate change is expected to lead to increased temperatures beyond the heat stress threshold (around 35°C for dairy goats [28]), slopes of performance traits below and above this threshold could be treated as separate phenotypes in a multi-trait breeding index.
When considering a range of temperatures below the heat stress threshold, as was the case in the present study, low temperatures are associated with lower average animal performance. Under cold stress (temperatures below 10 °C), animal feed intake is mostly directed towards maintaining their thermal balance requirements at the cost of producing less milk. Under higher temperatures, but still below the heat stress threshold, thermal balance requirements will be reduced, leading to better animal performance. Indeed, population curves from the reaction norm in the present study revealed a favourable impact of rising temperature and THI on performance manifested as increased daily milk production. The effect of THI almost mirrored that of temperature, partly because of the formula used to estimate THI [29] and partly because a relatively wider range of temperature values was observed in our data compared to humidity.
The individual animal resilience phenotypes derived in the present study exhibited significant phenotypic variation. Thus, the same weather change invoked positive or negative responses in different individuals while, for others, production was not affected at all (zero slopes). The latter individuals could be considered as the most resilient to climate change.
Furthermore, a significant proportion of the observed phenotypic variation among animals was genetic and heritable. Heritability estimates for resilience phenotypes ranged from 0.09 to 0.11, which is within the range of estimates for other fitness-related traits previously reported in goats [27], cattle [28] and sheep [29]. Although relatively low, these estimates are significantly greater than zero meaning that animal resilience to weather change is amenable to improvement via selective breeding. Since the outcome of selective breeding is cumulative, it is recommended that relevant programmes be put in place immediately in order to gradually and systematically enhance animal resilience to weather conditions as climate change becomes more pronounced.
When resilience was defined as the absolute value of the slope, reflecting volatility of animal performance with changing weather, a significant antagonistic genetic correlation with the actual level of milk production was estimated. This correlation suggests that animals with the genetic capacity for high milk yield will also be genetically predisposed to less stable milk production when challenged with changing weather. Although our range of temperatures is outside the heat stress interval, this result is also in agreement with previous studies on heat stress, where high merit animals were found to be more susceptible to environmental change [30, 31]. Therefore, careful consideration of resilience phenotypes should be made when including these traits in the breeding goal, in order to properly account for potentially unfavourable correlations with other traits of interest. Selection index theory can be used to effectively combine genetically antagonistic traits leading to overall genetic improvement in livestock [32–35]. Furthermore, index weights will need to be re-estimated every few generations in order to account for potentially new genetic correlations between traits emanating from changes in linkage disequilibrium due to selection.
Our genome-wide analyses identified several genomic markers associated with resilience phenotypes, particularly on chromosome 19. Although the significant SNPs identified on this chromosome were in mid to high linkage disequilibrium, only three of them, positioned within less than 0.1 Mb of each other, defined an actual haplotype with an overall squared correlation greater than 0.8 [36]. In this haplotype, one genome-wide significant SNP was detected affecting milk production level and relevant resilience phenotypes based on absolute slopes, which was also significant at chromosome-wide level for all the other resilience phenotypes. A previous study in goats [37] has shown genome-wide significant association of milk yield with another SNP in the same region located within 32 kb from the SNP identified here. Our SNP was found in exon 3 of the RNASEK gene, which encodes ribonuclease K protein. While the particular function of the latter is unknown, other ribonuclease pathways have been previously shown to be related to milk production [38] as well as host defence tissues and secretions in cattle [39]. Furthermore, ribonucleases are often involved in detention of protein synthesis to conserve energy under stress conditions [40]. Other chromosome-wide significant SNP associations for slopes on temperature and THI were also detected in this haplotype on chromosome 19, close to genes ALOX12 and ASGR2. Gene ALOX12 encodes the arachidonate 12-lipoxygenase, previously linked to goat milk and protein yield [37, 41], and the development and maintenance of the skin barrier [42]. Gene ASGR2 encodes a subunit of the asialoglycoprotein receptor, associated with udder attachment in goats and cattle [37, 43].
Additional SNPs affecting resilience phenotypes were found on chromosome 19 outside the defined haplotype. Of particular interest is the association close to ALOXE3, which encodes the arachidonate lipoxygenase 3, a protein implicated in skin differentiation. In humans, mutations of this gene cause congenital ichthyosis, a skin disease with several symptoms including intolerance to heat and humidity [44]. Furthermore, this protein is also involved in the development of fat cells, and was previously linked to udder depth in goats [37] and to the pathway of arachidonic acid, a polyunsaturated fatty acid present in mammals’ milk [45].
Furthermore, several SNPs were detected significantly affecting total lifetime milk yield in the present study without any significant association with resilience. Among these SNPs, a genome-wide significant SNP was detected on chromosome 8, previously associated with goat milk production [37] and close to genes SLC1A1 and GLIS3. Another genome-wide significant SNP was detected on chromosome 13, close to genes BMP7 and TFAP2C, with the latter (transcription factor AP-2 gamma) having been previously associated to mammary development and several milk traits in sheep [46]. Other chromosome-wide significant SNPs for milk yield were detected on chromosomes 3, 4, 13, 14 and 20. The region on chromosome 4, in particular, is between exons 1 and 2 of gene HECW1, previously associated with vitamin B-12 content in cow milk [47].
Significant SNPs detected in the present study were generally located close to genes encoding proteins related to lipid metabolism, skin differentiation and stress response. Particular genetic variants segregating in the studied population could be related to variation in tolerance to temperature and humidity through a combination of direct effects on metabolism and indirect effects on stress and discomfort, even within the range of thermoneutral temperatures. However, identified SNPs explained only a small proportion of the trait variances, thus potentially indicating a polygenic architecture underlying resilience of animal milk production to weather change. Therefore, these results support the hypothesis of a complex underlying genetic link between animal production and environmental comfort involving multiple biological networks.
Additional considerations are warranted when addressing the impact of climate change on breeding schemes. Of particular importance are the strength and direction of the expected changes. In the case of the UK, temperatures are expected to rise by about 2 °C by 2100, with a potential 4.2 °C increase in summer temperatures in southern England by 2080 [48]. Under the same scenario, winters will become wetter by up to 23% by 2080 and summers drier by up to a 24%, with more frequent and severe droughts [48]. These changes will bring higher weather instability and impose a threat to animal performance if animal resilience is not considered in breeding schemes. Therefore, selective breeding schemes should include resilience phenotypes based on absolute values of slopes in order to select animals that are more resilient to short- and medium-term changes.
However, in other cases, the directionality of the individual animal production change might be more important than the effect of the increased variability. Resilience phenotypes based on actual slopes rather than their absolute values might then prove more useful, allowing to select animals that have an increased performance in the direction of the expected climate change. Example scenarios where these schemes would be useful are countries where the changes in weather will potentially lead to seasonal values in a particular direction. Selective breeding schemes could then be informed by multiple resilience phenotypes measured at different values of weather measurements (for example temperature), thus creating an animal index based on a combination of the directional increase in animal performance up to the inflection (stress) threshold and stability of performance thereafter.
Furthermore, an economic assessment of the reaction norm as a novel animal phenotype is important, particularly when combining multiple traits in selection indices. Previous studies [49, 50] have shown that the economic values of phenotypes derived with reaction norms depend on the trait whose stability in different conditions is measured as well as the diversity of environments where progeny of the selected animals will be raised. However, this consideration was out of the scope of this study, and further research needs to be conducted within the context of particular breeding schemes.
Finally, the use of reaction norms to derive resilience phenotypes can be applied not only to production traits, as shown in the present study, but also to other animal traits related with health and reproduction. While previous studies of fitness traits have not detected large genotype by environment interactions [51, 52] in dairy cattle, studies in beef cattle have revealed a significant impact of such interactions on animal reproduction [53]. Therefore, it is important to consider the possibility of a genetic basis of resilience in all biological functions of interest and the potential inclusion in selection indices for breeding schemes.
Conclusions
The present study has demonstrated the applicability of reaction norms to obtain resilience phenotypes for animal performance to weather variability. Phenotypes obtained exhibited significant heritable genetic variance and can be used to underpin selective breeding schemes aiming to enhance animal performance and production stability in varying weather conditions. Candidate genes were detected for several resilience phenotypes, including genes related to stress response, lipid metabolism and skin development. These results can be used to further improve the accuracy of selective breeding. Non-linear models and a more extensive range of environments should be considered in future studies to account for variation outside the range studied here.
Methods
Data
Daily milk production records of individual animals were obtained from two UK dairy goat farms located at latitude 53° and 54° north. Strong genetic connectedness existed between the two farms as a result of inter-farm breeding program [54]. Animals in these farms are kept in an environment consisting of sheds without climate-controlled conditions. Because of specific management practices in these farms, daily animal milking records obtained were actually the average over 10 consecutive days.
Only records in the first 720 days of lactation were kept for the present study. Data were limited to goats that kidded from 2007 onwards, at a kidding age between 9 and 89 months and with at least three valid milk records. In addition, animal records with a lifetime estimate of the average daily milk yield outside three standard deviations from the mean were removed. The final dataset consisted of 980,689 milk records for 20,546 goats.
Animal pedigree was extracted from the farm database and comprised 46,825 animals spanning 19 generations, including 524 sires and 20,205 dams.
Weather data were obtained from the nearest weather station (less than 20 miles from the farms) and included average daily temperature and humidity. A temperature-humidity index (THI) was then calculated using the National Research Council formula [29]:
| 1 |
where THI = temperature-humidity index; T = average daily temperature (°C) and RH = average daily humidity (%). In consistence with the definition of animal performance, weather measurements used in the study represented averages of the same 10-day periods corresponding to each milk production record.
Individual resilience phenotypes
A theoretical random regression model including a reaction norm function is:
| 2 |
where yij corresponds to the performance record of individual animal i, at a given environment j, X corresponds to a set of fixed effects describing all environments, f(β, Xj) corresponds to a function (population reaction norm) describing the relationship between average animal performance and environment j, fi(ai, Xj) corresponds to a function (individual animal reaction norm) describing the relationship between individual animal i and environment j (expressed as a deviation from the population reaction norm) and eij corresponds to the residual.
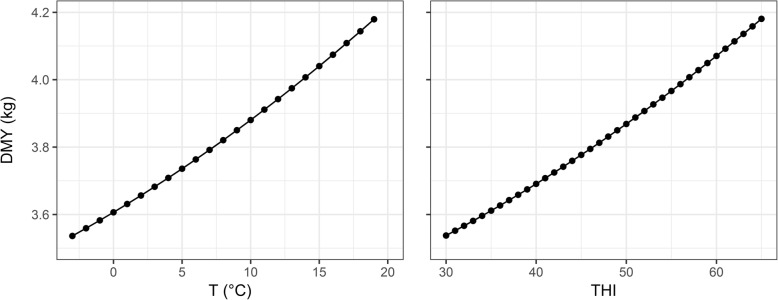
This model was fitted to milk yield records and corresponding temperature and THI values using second degree Legendre polynomials for the reaction norm function and the BLUPF90 suite of programs [55]. Initial exploration revealed a relatively linear behaviour for both weather measurements and animal performance (Fig. 2). Therefore, further analyses were conducted using the following simplified model with first degree Legendre polynomials [23]:
| 3 |
where μ corresponded to the population average intercept, μi corresponded to the animal i intercept deviated from the population intercept, and s and si corresponded to the population and individual i (as deviation) slopes on the fixed effect (environment); all other terms were as in model (2). The population reaction norm then was μ + sXij and the individual reaction norm μi + siXij, expressed as deviations from the population reaction norm.
Fig. 2.
Population reaction norms: Daily milk yield evolution in response to temperature (T) and temperature-humidity index (THI) variability. Reaction norms were modelled with second degree Legendre polynomials
Pedigree information was not included in model (3). Fixed effects included in this model were farm, interaction of calendar year and season of kidding, age at most recent kidding prior to milking date, number of days in milk, interaction between farm and date of record, and lactation (milking period) number.
Subsequently, individual reaction norms were computed per animal by adding the overall population norm to the corresponding individual animal deviation. Slopes of these individual reaction norms were estimated using derivatives, indicating the change in animal performance (milk yield) in response to weather fluctuations. These slopes were considered as the animal resilience phenotypes. Furthermore, absolute values of the estimated individual slopes were considered as additional resilience phenotypes, reflecting the stability/volatility of animal performance in relation to weather change, with values closer to zero representing more stable (resilient) animals.
Genetic parameters of resilience phenotypes
Variance components and heritability estimates of all animal resilience phenotypes were derived from mixed models including the available pedigree information, using the ASReml software [56]. The distribution of resilience phenotypes based on the absolute value of slopes was normalised by applying a square root transformation. Fixed effects in the mixed models included total number of milking days, farm, total number of lactations, age at first kidding (onset of productive life) and interaction between calendar year and season of first kidding.
Univariate analyses were conducted for each resilience phenotype separately to estimate its additive genetic variance and heritability. Bivariate analyses of resilience with total milk produced throughout the animal’s productive life (square root transformed to normalise) were also conducted to estimate genetic correlations.
Genomic association analysis of resilience phenotypes
A total of 10,620 animals with resilience phenotypes had been genotyped with the Illumina Caprine 50 K BeadChip containing 53,347 Single Nucleotide Polymorphisms (SNPs). Marker quality assurance removed SNPs on the sex chromosomes and those autosomal SNPs with Illumina GC score < 0.6, call rate < 95%, minor allele frequency < 0.05 and deviations from Hardy-Weinberg equilibrium (Bonferroni corrected threshold of 10− 7). Sample quality was assessed, and samples with call rates >90% were kept. These quality assurance edits resulted in a final set of 10,620 animals and 44,280 SNPs across all 29 autosomes with positions based on the most recent goat genome assembly ARS1 [57].
Association analyses were performed using the multi-locus mixed model algorithm [58] implemented in Golden Helix SNP & Variation Suite v8.8.3. The following model was used:
| 4 |
where y was the vector of animal phenotypes for the analysed trait; β was a vector of coefficients for the SNP effects and other fixed effects (same as described for the estimation of genetic parameters); a was the vector of random animal polygenic effects; e was the vector of random residual effects; and X and Z were incidence matrices relating observations to fixed and random animal effects, respectively.
The vector of random animal effects a and residual effects e in model (3) were assumed to follow normal distributions with a ~ N ) and e ~ N ), where G corresponds to the genomic relatedness matrix, I corresponds to the identity matrix and and correspond to the genetic and residual variances, respectively. Covariance between a and e was assumed to be zero.
The genomic relationship matrix G was calculated following VanRaden [59].
where S is a centred incidence matrix of SNP genotypes, N is the number of SNP markers, and pi is allele frequency of marker i.
Statistical significance of SNPs was assessed using Wald tests. Following a forward-backward stepwise regression [58], once the algorithm performed an initial scan testing each marker, additional genome scans were performed adjusting the model to account for the most significant SNPs on the initial scan. Significance thresholds were set at both genome- and chromosome-wide levels using Bonferroni corrections for multiple marker testing with a significance level of P < 0.05. This resulted in a genome-wide significance threshold of -log10(P) = 5.95. For significant markers, the proportion of explained phenotypic variance (pve) was estimated as:
where mrssh0 is the Mahalanobis root sum of squares for the null hypothesis and mrssk is the Mahalanobis root sum of squares for marker k.
Supplementary information
Additional file 1: Figure S1. Monthly average of weather measurements: daily temperature (T) and temperature-humidity index (THI). Standard deviations are shown as bars.
Additional file 2: Figure S2. Manhattan plots and QQ-plots: Total milk yield (A), performance change due to daily temperature change (B), performance change due to temperature-humidity index change (C), absolute value of performance change due to daily temperature change (D), absolute value of performance change due to THI change (E).
Acknowledgements
Data were kindly made available from the Yorkshire Dairy Goats company. The authors are thankful to Dr. Clara Díaz, Dr. Maria Jesús Carabaño and Dr. Manuel Ramon Fernández of Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA, Spain) for helpful feedback on the methodology and results.
Abbreviations
- SNP
Single Nucleotide Polymorphism
- THI
Temperature-Humidity Index
- UK
United Kingdom
Authors’ contributions
ESM, VK, JI, SD and SM prepared and performed the data analysis. Reaction norm phenotypes and analyses were performed by ESM, VK and JJI. Genetic analyses were performed by VK, ESM and SD. Genomic analysis was performed by SM. ESM, GB, JC and SM interpreted the results. ESM drafted the manuscript and all co-authors provided comments. GB and JC were responsible for the inception, funding, study design and implementation of the project. All authors have read and approved the final version of this manuscript.
Funding
Funding for this study was provided mainly from the Horizon2020 project iSAGE (grant 679302), which covered design and implementation of the work. Additionally, animal genotyping, pertinent data collection and contributions of SM and JC were supported by UK Biotechnology and Biological Sciences Research Council (BBSRC) and Agritech Strategy BB/M02833X/1 project 102093 (2015–2018); BBSRC/Agritech Strategy BB/M027570/1 project 102129 (2015–2018); and Technology Strategy Board (TSB) Project 101072 (2011–2015). The contribution of ESM was supported by the BBSRC ISP3 (BBS/E/D/30002275). The contribution of GB was partly supported by the Rural & Environment Science & Analytical Services Division of the Scottish Government.
Availability of data and materials
The animal genotypes are commercially sensitive. For more information, please contact GB.
Ethics approval and consent to participate
All authors declare that animal samples were obtained in compliance with local/national laws in force at the time of sampling. Data exchange was in accordance with national and international regulations, and approved by the owners.
Consent for publication
All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Vanessa V. Kapsona and Joanna J. Ilska contributed equally to this work.
Contributor Information
Enrique Sánchez-Molano, Email: Enrique.Sanchez-Molano@roslin.ed.ac.uk.
Vanessa V. Kapsona, Email: Vanessa.Kapsona@sruc.ac.uk
Joanna J. Ilska, Email: Joanna.Ilska@ed.ac.uk
Suzanne Desire, Email: sdesire@exseed.ed.ac.uk.
Joanne Conington, Email: Joanne.Conington@sruc.ac.uk.
Sebastian Mucha, Email: Sebastian.Mucha@sruc.ac.uk.
Georgios Banos, Email: Georgios.Banos@sruc.ac.uk.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12863-019-0787-z.
References
- 1.Agovino M, Casaccia M, Ciommi M, Ferrara M, Marchesano K. Agriculture, climate change and sustainability: the case of EU-28. Ecol Indic. 2018;105:525. doi: 10.1016/j.ecolind.2018.04.064. [DOI] [Google Scholar]
- 2.Lawrence D, Vandecar K. Effects of tropical deforestation on climate and agriculture. Nat Clim Chang. 2014;5:27. doi: 10.1038/nclimate2430. [DOI] [Google Scholar]
- 3.Lin BB. Resilience in agriculture through crop diversification: adaptive Management for Environmental Change. BioScience. 2011;61(3):183–193. doi: 10.1525/bio.2011.61.3.4. [DOI] [Google Scholar]
- 4.Rowell DP. A scenario of European climate change for the late twenty-first century: seasonal means and interannual variability. Clim Dyn. 2005;25(7):837–849. doi: 10.1007/s00382-005-0068-6. [DOI] [Google Scholar]
- 5.Bathiany S, Dakos V, Scheffer M, Lenton TM. Climate models predict increasing temperature variability in poor countries. Sci Adv. 2018;4(5):eaar5809. doi: 10.1126/sciadv.aar5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruosteenoja K, Räisänen P. Seasonal changes in solar radiation and relative humidity in Europe in response to global warming. J Clim. 2013;26(8):2467–2481. doi: 10.1175/JCLI-D-12-00007.1. [DOI] [Google Scholar]
- 7.Moran D, MacLeod M, Wall E, Eory V, McVittie A, Barnes A, Rees RM, Topp CFE, Pajot G, Matthews R, et al. Developing carbon budgets for UK agriculture, land-use, land-use change and forestry out to 2022. Clim Chang. 2011;105(3):529–553. doi: 10.1007/s10584-010-9898-2. [DOI] [Google Scholar]
- 8.Ross SA, Chagunda MGG, Topp CFE, Ennos R. Effect of cattle genotype and feeding regime on greenhouse gas emissions intensity in high producing dairy cows. Livest Sci. 2014;170:158–171. doi: 10.1016/j.livsci.2014.09.011. [DOI] [Google Scholar]
- 9.Lobell DB, Schlenker W, Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333(6042):616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- 10.Thornton PK, van de Steeg J, Notenbaert A, Herrero M. The impacts of climate change on livestock and livestock systems in developing countries: a review of what we know and what we need to know. Agric Syst. 2009;101(3):113–127. doi: 10.1016/j.agsy.2009.05.002. [DOI] [Google Scholar]
- 11.Chowdhury FR, Ibrahim QSU, Bari MS, Alam MMJ, Dunachie SJ, Rodriguez-Morales AJ, Patwary MI. The association between temperature, rainfall and humidity with common climate-sensitive infectious diseases in Bangladesh. PLoS One. 2018;13(6):e0199579. doi: 10.1371/journal.pone.0199579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rojas-Downing MM, Nejadhashemi AP, Harrigan T, Woznicki SA. Climate change and livestock: impacts, adaptation, and mitigation. Clim Risk Manag. 2017;16:145–163. doi: 10.1016/j.crm.2017.02.001. [DOI] [Google Scholar]
- 13.Department for Environment Food and Rural Affairs (DEFRA): UK Climate Change Risk Assessment 2017 . In: Synthesis report: priorities for the next five years. UCoC C, editor. 2017. [Google Scholar]
- 14.Weindl I, Lotze-Campen H, Popp A, Müller C, Havlík P, Herrero M, Schmitz C, Rolinski S. Livestock in a changing climate: production system transitions as an adaptation strategy for agriculture. Environ Res Lett. 2015;10(9):094021. doi: 10.1088/1748-9326/10/9/094021. [DOI] [Google Scholar]
- 15.Shields S, Orme-Evans G. The impacts of climate change mitigation strategies on animal welfare. Animals. 2015;5(2):361–394. doi: 10.3390/ani5020361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen TTT, Bowman PJ, Haile-Mariam M, Pryce JE, Hayes BJ. Genomic selection for tolerance to heat stress in Australian dairy cattle. J Dairy Sci. 2016;99(4):2849–2862. doi: 10.3168/jds.2015-9685. [DOI] [PubMed] [Google Scholar]
- 17.Boonkum W, Duangjinda M. Estimation of genetic parameters for heat stress, including dominance gene effects, on milk yield in Thai Holstein dairy cattle. Anim Sci J. 2015;86(3):245–250. doi: 10.1111/asj.12276. [DOI] [PubMed] [Google Scholar]
- 18.Carabaño MJ. The challenge of genetic selection for heat tolerance: the dairy cattle example. Adv Anim Biosci. 2016;7(2):218–222. doi: 10.1017/S2040470016000169. [DOI] [Google Scholar]
- 19.Ravagnolo O, Misztal I. Effect of heat stress on nonreturn rate in Holstein cows: genetic analyses. J Dairy Sci. 2002;85(11):3092–3100. doi: 10.3168/jds.S0022-0302(02)74396-8. [DOI] [PubMed] [Google Scholar]
- 20.Pryce Jennie E., de Haas Yvette. Burleigh Dodds Series in Agricultural Science. 2017. Genetic selection for dairy cow welfare and resilience to climate change; pp. 81–102. [Google Scholar]
- 21.Colditz IG, Hine BC. Resilience in farm animals: biology, management, breeding and implications for animal welfare. Anim Prod Sci. 2016;56(12):1961–1983. doi: 10.1071/AN15297. [DOI] [Google Scholar]
- 22.Berghof TVL, Poppe M, Mulder HA. Opportunities to Improve Resilience in Animal Breeding Programs. Front Genet. 2019;9:692. doi: 10.3389/fgene.2018.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryant J, López-Villalobos N, Holmes C, Pryce J. Simulation modelling of dairy cattle performance based on knowledge of genotype, environment and genotype by environment interactions: current status. Agric Syst. 2005;86(2):121–143. doi: 10.1016/j.agsy.2004.09.004. [DOI] [Google Scholar]
- 24.Mulder HA. Genomic selection improves response to selection in resilience by exploiting genotype by environment interactions. Front Genet. 2016;7:178. doi: 10.3389/fgene.2016.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes BJ, Carrick M, Bowman P, Goddard ME. Genotype×environment interaction for Milk production of daughters of Australian dairy sires from test-day records. J Dairy Sci. 2003;86(11):3736–3744. doi: 10.3168/jds.S0022-0302(03)73980-0. [DOI] [PubMed] [Google Scholar]
- 26.Martin JGA, Nussey DH, Wilson AJ, Réale D. Measuring individual differences in reaction norms in field and experimental studies: a power analysis of random regression models. Methods Ecol Evol. 2011;2(4):362–374. doi: 10.1111/j.2041-210X.2010.00084.x. [DOI] [Google Scholar]
- 27.UK Department for Business Energy and Industrial Strategy: Monthly average daily temperatures in the United Kingdom (UK) from 2013 to 2018 (in degrees Celsius). https://www.statista.com/statistics/322658/monthly-average-daily-temperatures-in-the-united-kingdom-uk. Accessed 15 Jan 2018.
- 28.Salama AAK, Caja G, Hamzaoui S, Badaoui B, Castro-Costa A, Façanha DAE, Guilhermino MM, Bozzi R. Different levels of response to heat stress in dairy goats. Small Rumin Res. 2014;121(1):73–79. doi: 10.1016/j.smallrumres.2013.11.021. [DOI] [Google Scholar]
- 29.National Research Council (NRC) A guide to environmental research on animals. Washington, DC: National Academy of Science; 1971. [Google Scholar]
- 30.West JW. Effects of heat-stress on production in dairy cattle. J Dairy Sci. 2003;86(6):2131–2144. doi: 10.3168/jds.S0022-0302(03)73803-X. [DOI] [PubMed] [Google Scholar]
- 31.Das R, Sailo L, Verma N, Bharti P, Saikia J, Imtiwati Kumar R. Impact of heat stress on health and performance of dairy animals: a review. Vet World. 2016;9(3):260–268. doi: 10.14202/vetworld.2016.260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacNeil MD. Genetic evaluation of an index of birth weight and yearling weight to improve efficiency of beef production1,2. J Anim Sci. 2003;81(10):2425–2433. doi: 10.2527/2003.81102425x. [DOI] [PubMed] [Google Scholar]
- 33.Dekkers JCM. Prediction of response to marker-assisted and genomic selection using selection index theory. J Anim Breed Genet. 2007;124:331–341. doi: 10.1111/j.1439-0388.2007.00701.x. [DOI] [PubMed] [Google Scholar]
- 34.van der Werf J. Genomic Selection in Animal Breeding Programs. In: Gondro C, van der Werf J, Hayes B, editors. Genome-Wide Association Studies and Genomic Prediction. Totowa, NJ: Humana Press; 2013. pp. 543–561. [Google Scholar]
- 35.Dekkers JCM, Van der Werf JH. Breeding Goals and Phenotyping Programs for Multi-Trait Improvement in the Genomics Era. In: Proceedings of the World Congress on Genetics Applied to Livestock Production. Vancouver: WCGALP; 2014. p. 8
- 36.Gu S, Pakstis AJ, Li H, Speed WC, Kidd JR, Kidd KK. Significant variation in haplotype block structure but conservation in tagSNP patterns among global populations. Eur J Hum Genet. 2007;15:302. doi: 10.1038/sj.ejhg.5201751. [DOI] [PubMed] [Google Scholar]
- 37.Mucha S, Mrode R, Coffey M, Kizilaslan M, Desire S, Conington J. Genome-wide association study of conformation and milk yield in mixed-breed dairy goats. J Dairy Sci. 2018;101(3):2213–2225. doi: 10.3168/jds.2017-12919. [DOI] [PubMed] [Google Scholar]
- 38.Raven L-A, Cocks BG, Pryce JE, Cottrell JJ, Hayes BJ. Genes of the RNASE5 pathway contain SNP associated with milk production traits in dairy cattle. Genet Sel Evol. 2013;45(1):25. doi: 10.1186/1297-9686-45-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta SK, Haigh BJ, Wheeler TT. Abundance of RNase4 and RNase5 mRNA and protein in host defence related tissues and secretions in cattle. Biochem Biophys Rep. 2016;8:261–267. doi: 10.1016/j.bbrep.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorrentino S. The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010;584(11):2194–2200. doi: 10.1016/j.febslet.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 41.Martin P, Palhière I, Maroteau C, Bardou P, Canale-Tabet K, Sarry J, Woloszyn F, Bertrand-Michel J, Racke I, Besir H, et al. A genome scan for milk production traits in dairy goats reveals two new mutations in Dgat1 reducing milk fat content. Sci Rep. 2017;7(1):1872. doi: 10.1038/s41598-017-02052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mashima R, Okuyama T. The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biol. 2015;6:297–310. doi: 10.1016/j.redox.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrooten C, Bovenhuis H, Coppieters W, Van Arendonk JAM. Whole genome scan to detect quantitative trait loci for conformation and functional traits in dairy cattle. J Dairy Sci. 2000;83(4):795–806. doi: 10.3168/jds.S0022-0302(00)74942-3. [DOI] [PubMed] [Google Scholar]
- 44.Ullah R, Ansar M, Durrani ZU, Lee K, Santos-Cortez RLP, Muhammad D, Ali M, Zia M, Ayub M, Khan S, et al. Novel mutations in the genes TGM1 and ALOXE3 underlying autosomal recessive congenital ichthyosis. Int J Dermatol. 2016;55(5):524–530. doi: 10.1111/ijd.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Idamokoro EM, Muchenje V, Afolayan AJ, Hugo A. Comparative fatty-acid profile and atherogenicity index of milk from free grazing Nguni, Boer and non-descript goats in South Africa. Pastoralism. 2019;9(1):4. doi: 10.1186/s13570-019-0138-8. [DOI] [Google Scholar]
- 46.Gutierrez-Gil B, Arranz JJ, Pong-Wong R, Garcia-Gamez E, Kijas J, Wiener P. Application of selection mapping to identify genomic regions associated with dairy production in sheep. PLoS One. 2014;9(5):e94623. doi: 10.1371/journal.pone.0094623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutten MJM, Bouwman AC, Sprong RC, van Arendonk JAM, Visker MHPW. Genetic variation in vitamin B-12 content of bovine Milk and its association with SNP along the bovine genome. PLoS One. 2013;8(4):e62382. doi: 10.1371/journal.pone.0062382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.UK Meteorological Office . UK Climate Projections: UKCP09 dataset. 2009. [Google Scholar]
- 49.Hermesch S, Amer P. Deriving Economic Values for Reaction Norms of Growth in Pigs. In: Twentieth Conference of the Association for the Advancement of Animal Breeding and Genetics. Australia: Association for the Advancement of Animal Breeding and Genetics; 2013. p. 475–8.
- 50.Kolmodin R, Bijma P. Response to mass selection when the genotype by environment interaction is modelled as a linear reaction norm. Genet Sel Evol. 2004;36(4):435–454. doi: 10.1186/1297-9686-36-4-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haile-Mariam M, Carrick MJ, Goddard ME. Genotype by environment interaction for fertility, survival, and Milk production traits in Australian dairy cattle. J Dairy Sci. 2008;91(12):4840–4853. doi: 10.3168/jds.2008-1084. [DOI] [PubMed] [Google Scholar]
- 52.Streit M, Reinhardt F, Thaller G, Bennewitz J. Reaction norms and genotype-by-environment interaction in the German Holstein dairy cattle. J Anim Breed Genet. 2012;129(5):380–389. doi: 10.1111/j.1439-0388.2012.00999.x. [DOI] [PubMed] [Google Scholar]
- 53.Morris CA, Baker RL, Hickey SM, Johnson DL, Cullen NG, Wilson JA. Evidence of genotype by environment interaction for reproductive and maternal traits in beef cattle. Anim Sci. 2010;56(1):69–83. doi: 10.1017/S0003356100006176. [DOI] [Google Scholar]
- 54.Mucha S, Mrode R, MacLaren-Lee I, Coffey M, Conington J. Estimation of genomic breeding values for milk yield in UK dairy goats. J Dairy Sci. 2015;98(11):8201–8208. doi: 10.3168/jds.2015-9682. [DOI] [PubMed] [Google Scholar]
- 55.Misztal I, Tsuruta S, Strabel T, Auvray B, Druet T, Lee D. Proceedings of 7th World Congress on Genetics Applied to Livestock Production. 2002. BLUPF90 and related programs. [Google Scholar]
- 56.Gilmour AR, Gogel B, Cullis BR, Thompson R. 2009 ASReml user guide release 3.0. Hemel Hempstead: VSN International Ltd; 2009. [Google Scholar]
- 57.Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017;49:643. doi: 10.1038/ng.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012;44:825. doi: 10.1038/ng.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Monthly average of weather measurements: daily temperature (T) and temperature-humidity index (THI). Standard deviations are shown as bars.
Additional file 2: Figure S2. Manhattan plots and QQ-plots: Total milk yield (A), performance change due to daily temperature change (B), performance change due to temperature-humidity index change (C), absolute value of performance change due to daily temperature change (D), absolute value of performance change due to THI change (E).
Data Availability Statement
The animal genotypes are commercially sensitive. For more information, please contact GB.