Abstract
Cell-free protein synthesis (CFPS) is an established biotechnology tool that has shown great utility in many applications such as prototyping proteins, building genetic circuits, designing biosensors, and expressing cytotoxic proteins. Although CFPS has been widely deployed, the many, varied methods presented in the literature can be challenging for new users to adopt. From our experience and others who newly enter the field, one of the most frustrating aspects of applying CFPS as a laboratory can be the large levels of variability that are present within experimental replicates. Herein we provide a retrospective summary of CFPS methods that reduce variability significantly. These methods include optimized extract preparation, fully solubilizing the master mix components, and careful mixing of the reaction. These have reduced our coefficient of variation from 97.3% to 1.2%. Moreover, these methods allow complete novices (e.g. semester rotation undergraduate students) to provide data that is comparable to experienced users, thus allowing broader participation in this exciting research area.
Keywords: Cell-free protein synthesis, CFPS, Cell extract, In vitro protein synthesis, In vitro transcription-translation, Cell-free synthetic biology
1. Introduction
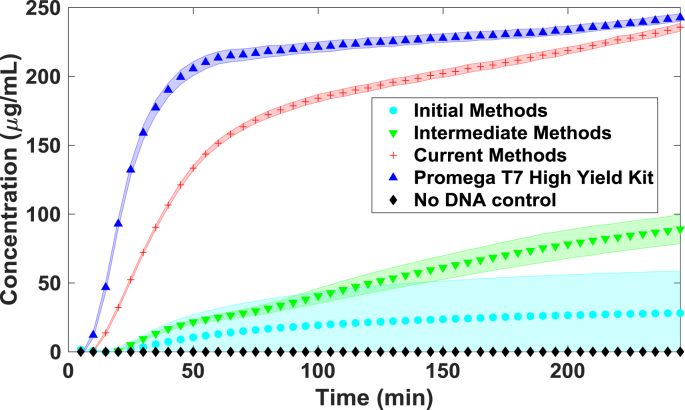
Cell-free protein synthesis (CFPS) is an established biotechnology tool, used over 50 years ago to decipher the relationship between mRNA codons and amino acid sequences [1]. It has had a recent renaissance due to many new applications such as producing protein therapies [[2], [3], [4]], prototyping proteins [5], engineering metabolic networks [6,7], and biosensors [[8], [9], [10]]. Additionally, the composition of cell extract from different strains has been analyzed including TB3, BL21 Star™ (DE3), A19, and BL21 Rosetta2 [[11], [12], [13], [14]]. Many of the components needed for CFPS (cell extract, supplements and additives) are now commercially available. This has led many groups, including our own, to adopt CFPS as a convenient tool for in-house expression of custom proteins. This adoption is evident in a rapid increase in reviewed papers that use E. coli based CFPS [15]. There are many good reviews on CFPS to orient new users on basic methods, current uses, and potential future applications [5,[16], [17], [18], [19], [20]]. Despite this rapid adoption and widespread information, CFPS can still be difficult to implement, especially for new users, due to variability that stems from the small reaction volumes and sensitive reagents. Multiple recent papers have pointed out this issue of variability [[21], [22], [23], [24]]. There has also been some work aimed at identifying and quantifying interlaboratory variability [22]. In fact, the National Institute of Standards and Technology (NIST) recently held a workshop to address the sources of variability in CFPS experiments [25]. Commercial kits (such as Promega S30 T7 High-Yield, PURExpress) that standardize reagent production can improve variation [26], but careful methods are also necessary to reduce variability. In this methods paper, we summarize the evolution of best practices that have improved the variability in our CFPS reactions such that the variability is comparable with a commercial kit (Promega S30 T7 High-Yield - Fig. 1).
Fig. 1.
Reporter protein, sfGFP, expression kinetics obtained by our group showing progression of methods to reduce variability. ‘Initial Methods’ were our first attempts at expression using a PANOx-SP system [27], ‘intermediate methods’ refer to our implementation of Sutro's modified Cytomim system [4], ‘current methods’ refer to improved techniques (detailed herein) using the PANOx-SP system, and the Promega S30 T7 High-Yield Protein Expression kit is a commercial kit used for benchmarking against these methods. The shaded area represents 1 standard deviation (n = 4).
2. Materials and methods
All chemical reagents were purchased from Sigma Aldrich unless otherwise noted.
2.1. Cell growth and extract preparation
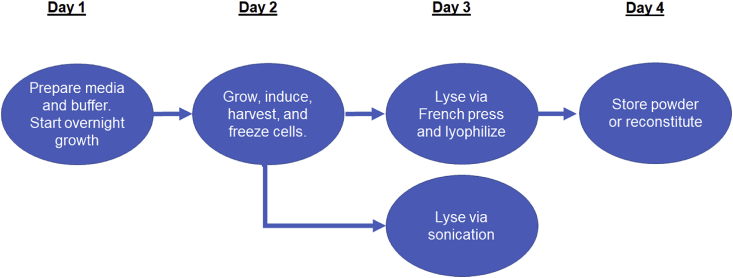
Our process to develop scalable methods to improve cell extract with reduced variability is published in detail [26] and outlined here (Fig. 2). The E. coli strain BL21 Star™ (DE3) (Invitrogen) is used due to its rapid doubling time, T7 transcription machinery, and ability to express proteins from linear DNA due to a mutation in the RNaseE gene (rne131) [28,29]. This strain of E. coli is induced with IPTG during growth to produce T7 RNA polymerase (T7RNAP) for the cell-free reaction. This is useful when the genetic template utilizes a T7 promoter as it eliminates the need to supplement T7RNAP to the extract post growth, which can be another source of variance. Optimal conditions for IPTG exposure and total growth time were determined and reported in terms of the extent of the cell growth curve (as optical density metrics are not absolute and can change based on instrument and vessel used). If a new cell line is used, a designed experiment (DoE) should be used to determine optimal growth conditions (induction and harvest times) as these can have a large impact on protein expression efficiency of the extract [26,30]. However, it may be of interest to note that observed variability in CFPS performance is not significantly impacted by extract preparation as long as protocols are followed faithfully [22]. The combined cell growth and extract preparation process is by far the most laborious and time intensive; it is presented here as daily steps.
Fig. 2.
High level overview of the cell growth and extract preparation process. Steps in the process are grouped into categories based on which day the steps are performed.
2.1.1. Day 1 (media and buffer preparation)
-
1.
Prepare and autoclave 500 mL of LB media (5 g tryptone, 2.5 g yeast extract, 5 g sodium chloride, and ddH2O to 500 mL)
-
2.
Prepare 900 mL of 2x YTP media (16 g tryptone, 10 g yeast extract, 5 g sodium chloride, 7 g potassium phosphate dibasic, 3 g potassium phosphate monobasic, and ddH2O to 900 mL). Titrate the 2x YTP to pH 7.2 using 5 M KOH and autoclave.
-
3.
Significant: Prepare a 500 mL solution of 1 M glucose and filter using a Nalgene™ Rapid-Flow™ sterile disposable filter unit with PES membrane (ThermoFisher #166-0045). Glucose will eventually be added to the 2x YTP media in order to produce 2x YTPG. We choose to filter the glucose to avoid a Maillard reaction which has been shown to impair media performance [31]. This can also be avoided by autoclaving 2xYTP and glucose in separate containers.
-
4.
Prepare and filter a 500 mL solution of buffer A (10 mM tris-acetate pH 8.2, 14 mM magnesium acetate, and 60 mM potassium acetate).
-
5.
Once the LB media has cooled to room temperature, aliquot 20 mL into a 50 mL conical tube and add a cell pick of BL21 Star™ (DE3). Allow this starter culture to grow overnight (14–18 h) at 37 °C and 230 rpm in a MaxQ 4000 shaker (Thermo Scientific). We have found that the quantity of the cell pick and this timing do not have to be exact as the overnight cultures double to the point of resource exhaustion and thus enter the cell growth in Day 2 with a similar concentration of cells (little difference in growth curves observed). Allow the 2x YTP to set at room temperature overnight.
2.1.2. Day 2 (cell growth and harvest)
-
6.
Add 100 mL 1 M glucose to the 900 mL 2x YTP to make 1L of 2x YTPG and warm in the shaker incubator for 30 min at 37 °C.
-
7.
If an optimum growth time has already been determined, inoculate the 2x YTPG with the 20 mL starter culture (step 5 above). Otherwise, generate a new growth curve using OD600 measurements taken at a regular time interval (15–30 min). First, remove 20 mL of the 2x YTPG to use as a baseline reference in the spectrophotometer. Then, inoculate the growth media with the 20 mL starter culture.
-
8.
Significant: Once the desired point of induction has been reached (acquired via an earlier designed experiment campaign [26]) add 1 mL of 1 M IPTG. For the BL21 Star™ we have previously determined this to be 3 h 20 min for 1 L growths in 2.5L Tunair flasks [26] shaken at 270 rpm.
Note: Optimizing cell growth in this way is meant to improve yield while also reducing the number of measurements taken by the researcher. In contrast, recent literature shows that optimization may not be necessary when using cell-free autoinduction (CFAI) media. This offers another viable option for those who wish to reduce variability caused by manual steps as much as possible [32].
-
9.
Harvest at the desired overall growth time. We have determined this to be 4 h 15 min for 1 L growths in 2.5 L Tunair flasks [26]. Distribute the growth evenly among centrifuge tubes and weigh each. Ensure the mass of all tubes is identical by adding ddH2O.
-
10.
Centrifuge the cell suspension at 5000 xg and 4 °C for 15 min to pellet the cells.
-
11.
Decant the supernatant and collect the cell pellets in a 50 mL conical tube using a metal spatula. Keep on ice
-
12.
Resuspend the cell pellet in cold buffer A via vortexing.
-
13.
Centrifuge the cell pellet at 5000 xg and 4 °C for 10 min to pellet the cells.
-
14.
Discard the supernatant and wipe any residual liquid from inside the tube using a Kimwipe.
-
15.
Significant: Record the mass of the cells. This is referred to as the wet cell mass. Under these conditions, a wet cell mass of 5–7 g is typically obtained from each liter.
Optional: Freeze the cell pellet and store at −80 °C for later use. The pellet can be flash frozen in liquid N2 or frozen in the freezer with no noticeable detrimental effects downstream. This step is optional because the cells can be lysed immediately after harvesting, if desired.
2.1.3. Day 3 (cell lysis and extract lyophilization)
There are two methods we commonly use to disrupt the cell membrane: sonication and French press homogenization. When lysing small amounts of cells (<25 g wet cell mass) it is more efficient to use a sonication protocol. However, large amounts (≥25 g wet cell mass) are best handled with homogenization [26].
2.1.3.1. Option 1: French press homogenization (preferred)
-
17.
Add 1 mL buffer A and 1 mL ddH2O per gram wet cell mass to the pellet. Resuspend the pellet via vortexing. The buffer and the extra water lower the viscosity of this mixture enough to eliminate homogenizer clogging and maintain a more consistent pressure swing of the homogenizer piston. The excess water will be driven off later through lyophilization.
-
18.
Turn on the vacuum pump and the homogenizer (Avestin EmulsiFlex C3). Clean the homogenizer by flushing it 3 times with water and a 70% (v/v) ethanol solution. Repeat 3 times with ddH2O to ensure removal of all ethanol.
-
19.
Prime the homogenizer with ddH2O and put on standby so it doesn't run dry. Only keep a small amount of water in the hopper so as not to dilute the cells any more than necessary.
-
20.
Significant: Place the metal heat exchanger coil located in the outlet tube in a beaker of ice to facilitate cooling.
-
20.
Pour the cell suspension into the hopper and start the machine.
-
21.
Ensure there is a clean beaker to collect the resulting crude lysate.
-
22.
Maintain a pressure swing of 25,000–30,000 psi by continuously adjusting the pressure knob (again this can only be done if the cells are diluted, if the mixture is too thick the swing is inconsistent and the homogenizer will stall).
Note: The above conditions have been optimized for use with the homogenizer in our lab. By implementing the same method (i.e. using a designed experiment), the optimum for any membrane disruption method can be determined.
-
23.
Upon completion, collect the crude lysate in 5 mL microcentrifuge tubes.
-
24.
Centrifuge the crude lysate at 12,000 xg and 4 °C for 10 min.
-
25.
Decant the supernatant into a 50 mL conical tube and place on ice.
Optional: It is historically common to perform a run-off reaction at this point to improve protein yields by digesting any remaining nucleic acids with endogenous enzymes. This is typically done in small volumes (~500 μL) around 37 °C and 250 rpm for an optimized amount of time [16,33]. However, certain E. coli strains such as BL21 Star™ (DE3) produce a robust extract that does not require a runoff reaction [23,34]. To optimize protein yield, it may be necessary to optimize new strains or promoter systems [34,35]. Because of this, we do not outline them in this protocol. The extract will then need to be centrifuged and collected as in steps 24 and 25.
Optional: The extract can be dialyzed to remove metabolic byproducts. This is typically performed with using 10K MWCO cutoff cassette multiple times with chilled buffer A. This step has been popular in the past but has become less common [23,26,34]. Dialysis may also be necessary to optimize new strains and promoter systems [34,35]. The extract will then need to be centrifuged and collected as in steps 24 and 25.
-
26.
Prepare the dilute extract for lyophilization by placing it in a container that will provide a large surface area and good heat transfer (a metal baking pan works well). Cover the pan with a large Kimwipe and tape it to the pan.
Note: Lyophilization, traditionally, is an optional step that may or may not meet the needs of the user. In this case, the extract we produce using a French press is dilute, so we perform lyophilization to remove the excess water. In this way, we can reconstitute our extract to match the protein concentration achieved using other lysing methods (i.e. sonication) or other desired concentrations.
-
27.
Freeze the diluted extract at −80 °C for 30 min.
-
28.
Significant: Prepare the 35 L VirTis Genesis Pilot-scale Lyophilizer (SP Scientific) by setting the shelf temperature to 15 °C. A smaller, bench top lyophilization unit that sublimates in vessels at room temperature can also be used, but we have found better extract performance from extract that has been lyophilized at a chilled shelf temperature.
-
29.
Place the baking pan in the lyophilizer and turn on the vacuum. The vacuum should be set to 200 mTorr.
-
28.
Turn on the condenser and allow it to lyophilize overnight. Optional: Depending on the volume being lyophilized, the necessary water loss can occur in as little as 4 h [26].
2.1.4. Day 4 (extract reconstitution and storage)
-
29.
Follow the lyophilization shutdown procedure by turning off the condenser, then turn off the vacuum, then turn on the vacuum release.
-
30.
Once the internal pressure has equilibrated, transfer the contents of the baking pan to a pre-weighed 50 mL conical tube and record the mass. The resulting powder is fluffy and has a charge, so it is best to crush the powder with a spatula before transferring to the tube.
Optional: Store the lyophilized extract for later use. Lyophilized extract can remain active at elevated temperatures (−20 °C or 4 °C) for at least a year [36].
-
31.
Add water to the lyophilized extract according to the following equation ddH2O (mL) = (mass of lyophilized extract (g))(1.5 mL/0.094 g) and allow to sit at room temperature for 5 min before resuspension. Resuspend by pipetting up and down.
-
32.
Significant: Use PCR tubes to store the extract in 45 μL aliquots at −80 °C. While this takes a large amount of time, it saves reagent and avoids multiple freeze thaw cycles. The aliquots will depend on personal preferences but 45 μL works well since each 15 μL reaction has 3.6 μL. This is enough extract to reliably support 2 sets of 5 replicates.
2.1.5. Day 3 (alternative method)
2.1.5.1. Option 2: sonication
Sonication is suitable for small amounts of cell lysis (<25g wet cell mass). When developing a sonication protocol, designed experiments can test parameters such as amplitude, energy, and sample volume. This allows for the possibility of a more controlled lysing environment for extract since settings on piston-based continuous homogenizers cannot be as rigorously controlled. However, the throughput of the continuous homogenizer will always be greater when large volumes of extract need to be processed [23,26,34].
Optional: Turn on the cooling system for the sonicator. The cooling system helps prevent sample to sample variation by keeping a more consistent sample temperature than a typical ice bath.
-
17.
Add 1 mL buffer A per gram wet cell mass to the pellet. Resuspend the pellet via vortexing.
-
18.
Transfer the suspension to 1.5 mL microcentrifuge tubes in 1 mL aliquots and keep on ice.
-
19.
Set the sonicator (Qsonica Q125 with a 2 mm (5/64 in) tip) to 50% amplitude and 10 s on/off pulse.
-
20.
Significant: Sonicate the samples, one by one, on ice (if not using a cooling system) until an energy input of 532 J has been reached. This number was determined by previously optimized methods [23]. This takes about 5 min per sample. Place each sample on ice post sonication.
-
21.
Centrifuge the crude lysate at 12,000 xg and 4 °C for 10 min.
-
22.
Using a 200 μL pipette carefully transfer the supernatant to a 50 mL conical tube. Gently homogenize the clarified extract to ensure any batch to batch variability is eliminated.
Optional: Run-off reactions, dialysis, and lyophilization steps after this are the same as detailed in the homogenizer section above.
-
23.
Significant: Use PCR tubes to store the extract in 45 μL aliquots at −80 °C. While this takes a large amount of time, it saves reagent and avoids multiple freeze thaw cycles.
2.3. DNA amplification
One of the advantages of CFPS is that it can use both plasmids and polymerase chain reaction (PCR) products directly as expression templates [5,17,37]. We have found both to work equally well and can be made in house. Plasmids are useful once desired sequences are settled on as they can be transformed into cells and then harvested to create large stocks of DNA template. PCR products are much more useful in the prototyping phase, as the work stream is much shorter (no transformation of cells) but they are more expensive to produce and the DNA yield is orders of magnitude smaller than by cells. We have recently demonstrated how rolling circle amplification (RCA) can be used to mitigate this problem by amplifying a small amount of mail-order gene fragment to large amounts of DNA for protein expression [5].
2.3.1. Plasmids
The pJL1-sfGFP plasmid was used for all data in this manuscript [23]. Plasmids are amplified using the following steps.
-
1.
After cloning the gene of interest into the plasmid and transformation into E. coli, colonies containing the plasmid are grown and stored in a glycerol stock.
-
2.
This glycerol stock is then used for subsequent growth harvest for DNA purification.
-
3.
The cells are grown, lysed, and the DNA is purified using ZymoPURE II MaxiPrep Kit (Zymo Research); this kit is selected for purification due to the column design. The column has a narrow neck which allows for smaller volume elution and higher concentration purifications, if desired. The instructions provided with the kit are followed exactly except a different elution buffer that does not contain EDTA (such as Qiagen EB #19086) must be used since EDTA is known to chelate magnesium.
-
4.
DNA is then quantified using a NanoPhotometer® N80 (Implen).
2.3.2. Rolling circle amplification
As our lab relies on new, custom proteins for downstream applications, we use the aforementioned RCA method to quickly create genetic templates to prototype the proteins. We have previously published the development process, advantages, and next steps for this method [5]. Here we outline the methods:
-
1.
A minimal genetic template is designed to contain primer binding sites, restriction enzyme sites, a T7 promoter, a ribosome binding site, a start codon, and a T7 terminator. The desired amino acid is codon optimized using an online tool (IDT Codon Optimization Tool) [38,39] and is encoded between the start and end codon.
-
2.
The gene fragment can then be ordered from a vendor of choice (e.g. IDT, Twist).
-
3.
Once the gene fragment has been received, it is suspended and undergoes traditional linear amplification. We use both GoTaq (Promega) and OneTaq (New England BioLabs) with no observed differences. If there are concerns about template length (>1.5 kb) a product like Phusion (New England BioLabs) is more precise in amplification. Protocols supplied by the vendor are followed in this step. Promega and New England BioLabs (NEB) both have online melting temperature (Tm) calculators that are dependent on the product and the primers used for amplification.
-
4.
The resulting linear template (LET) is then purified (using Zymo Kit #D4003) and eluted in at least 50 μL of buffer (Qiagen EB #19086). If desired, the LET can be quantified, but 45 μL must be reserved for the restriction digest.
-
5.
Restriction digest is performed by adding 5 μL of Cut Smart buffer (NEB) and 1 μL (20 units) restriction enzyme (HindIII-HF from NEB #R3104S) to the 45 μL of LET. Run the digestion for 1 h at 37 °C and then inactivate the enzyme at 80 °C for 20 min.
-
6.
After digestion, the “sticky ends” from the digest are ligated together to form a small circle template using T4 (or T7) ligase (New England Biolabs). This is done by adding 5 μL T4 DNA Ligase buffer (NEB #M0202S) and 2 μL T4 (800 units) DNA ligase to the digested product and run at room temperature for 1 h.
-
7.
The minimal circular expression template (CET) is then purified (using Zymo Kit #D4003) and eluted in 45 μL of buffer (Qiagen EB #19086).
-
8.
Rolling circle amplification can work on DNA concentrations in the pg/μL range, but it is not necessary to dilute the DNA to this low of a concentration. Typically, we add 1–2 μL of DNA from the purified CET stock to 100 μL of double distilled water as our dilute stock for isothermal RCA.
NOTE: The dilute CET is only meant for a one-time use. Do not store it for extended periods (>1 week at −20 °C) as we find its performance suffers in later CFPS experiments.
-
9.
Rolling circle amplification (RCA) is performed using a TempliPhi™ kit (GE Healthcare). The kit comes with 3 solutions: a lysis buffer (red cap), amplification buffer (blue cap), and phi29 (Φ29) polymerase (yellow cap). It is best to aliquot master mixes of this for future amplifications to reduce the number of freeze cycles. For one group of RCA reactions, mix 20 μL lysis buffer, 20 μL reaction buffer, and 0.8 μL Φ29 polymerase. Add 4 μL of dilute CET (from step 8 above) and distribute evenly among 4 PCR tubes (11.2 μL per tube). According to the protocol from the manufacturer, the reaction is exhausted after 4 h at 30 °C. To parallelize lab work, we tend to let this reaction run overnight and purify the following morning.
-
10.
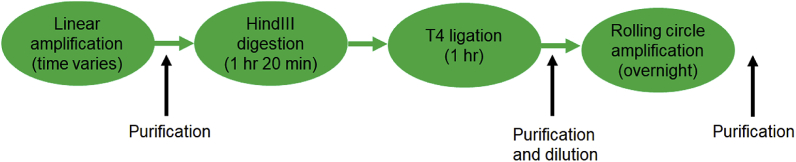
The resulting product is diluted with 14 μL of double distilled water, purified (using Zymo Kit #D4003), eluted in at least 25 μL of buffer (Qiagen EB #19086), quantified, and stored at −20 °C for later use in CFPS. A high-level overview of the process is shown in Fig. 3.
Fig. 3.
Simplified overview of the rolling circle amplification from minimal template process. The entire process takes less than 24 h and results in a large amount of genetic temple that is suitable for CFPS.
2.4. Master mix preparation
There are many recipes of supplements to add to the CFPS reaction. We have previously used the modified Cytomim system outlined by Sutro Biopharma but found the low yield to be troublesome for small-scale research applications [4,26]. One that we have found to be most productive and least variable is the PANOx-SP recipe [28]. Our variation of the PANOx-SP “master mix” includes: HEPES, ATP, GMP, CMP, UMP, phosphoenolpyruvate (PEP), folinic acid, E. coli tRNA, 20 amino acids (purchased from Formedium, United Kingdom), NAD, CoA, potassium glutamate, ammonium glutamate (MP Biomedicals), magnesium glutamate, potassium oxalate, putrescine, and spermidine. Since we induce the production of T7RNAP during cell growth, there is no need to add to the master mix. The best practices for the development of a master mix are as follows:
-
1.
Significant: Create concentrated stock mixes of each of these components which can then be combined to form the master mix solution. Creating a stock solution for most of these components is relatively straightforward since most of them are soluble in water. It is important to note that some are only partially soluble in water; these take some additional steps.
-
2.
Significant: To prepare a 1 M PEP solution, one must use a combination of water and KOH until PEP is completely dissolved. Titrate the solution to a pH of 7 with KOH.
-
3.
Significant: The most difficult stock to make is the amino acid solution. Our stock solution is 50 mM of 20 amino acids and not all are readily soluble at neutral pH. Tyrosine, in particular, will not dissolve even when heated. While it has been reported that a non-homogenous amino acid suspension is not detrimental to CFPS [4,27], we prefer to completely solubilize our amino acid mixture using previously reported methods [40]. This reduces the chance of having insoluble fragments of the amino acids present in reactions. The amino acids are dissolved in a mixture of water and 5 M KOH until a pH of 12 is achieved. The pH of the solution can be adjusted by adding acetic acid. It is important to note that amino acids will start to drop out of solution if the pH drops below 12. The amino acids should also be added in order of most hydrophobic to least hydrophobic, and completely solubilized before adding the next amino acid. While a stock amino acid solution pH of 12 may seem high, the buffering capacity of the master mix is maintained when all components are combined. We find the final pH of the master mix is 7.0–7.3.
-
4.
Spermidine may come as a liquid. If this is the case, do not make a stock, just use the specific gravity to determine how much should be added to the final master mix. This is done in the spreadsheet provided in the supplement.
-
5.
Determine the final concentration for each component in the master mix. This is typically done by setting up a spreadsheet and working backwards from needed final concentrations (see example sheet in Supplement 1). Set the cell extract and genetic template concentrations, then determine what volume of the final reaction is reserved for the master mix. For instance, our CFPS reactions are typically 15 μL total volume with 5 μL reserved for the master mix. In the provided spreadsheet, the variables typically manipulated (DNA concentration, number of replicates, etc.) are highlighted in green.
-
6.
A supplement mix can be stored as separate solutions (salts, energy, PEP, etc.) [27,33] or all components can be stored together into one master mix. We combine all components into a single tube, vortex, and store in 60 μL aliquots at −80 °C. Stock solutions are thawed on ice but can also be thawed more quickly on a thermo block at 30 °C if necessary. Upon thawing, the solutions should be vortexed for homogeneity.
The variability in our experiments was drastically reduced when we started using completely soluble master mix. To illustrate, the PANOx-SP (final) sample in Fig. 1 is a fully soluble master mix that is on par with the master mix provided by Promega's S30 T7 High-Yield CFPS kit (#L1110). The initial sample in Fig. 1 were not completely solubilized and the increased variance is apparent. Despite this, there is an advantage to splitting the master mix into multiple mixtures in order to calibrate magnesium ion concentration. Each batch of cell extract is different and requires a different magnesium concentration in the final reaction to optimize protein production [16,27,33,35]. This step of creating a custom master mix, we have found, is not critical but should be done with large batches of extract that will be used for long periods of time. It should also be noted that observed variability is heavily influenced by the preparation of the master mix [22]. As such, we believe this is the most critical step in CFPS.
2.5. Cell-free reaction
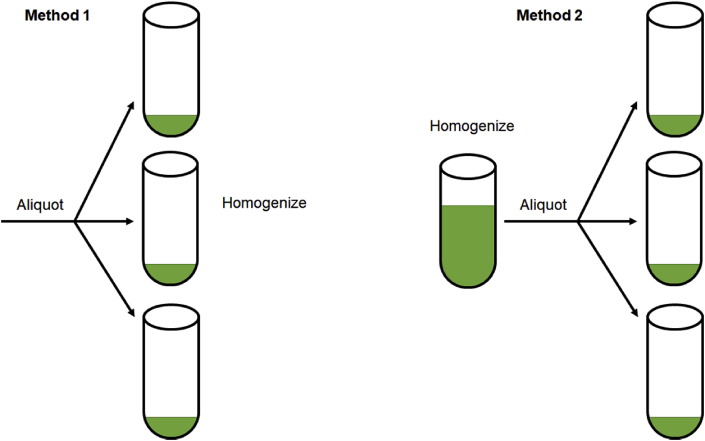
Finally, in setting up the cell-free reactions there are also key steps to reduce variability. A CFPS reaction requires only 4 components: cell extract, DNA template, master mix, and water to bring the solution to final reaction volume. Final reaction volumes are typically small (<20 μL) and there are two ways to create sample replicates (Fig. 4). Method 1 involves aliquoting each small volume into separate wells, or tubes, and homogenizing each reaction individually. These experimental replicates include the error introduced by pipetting small sample volumes. Method 2 involves adding all the components into a single mixture tube, homogenizing, and then aliquoting the replicates from this mixture into separate wells. This method eliminates the error introduced by small volume pipetting errors as each are derived from the same mixture and the combined volumes are larger and easier to manage by standard pipettes. In this manner the end variability measured is caused solely by differences in CFPS expression variation and not pipetting. One drawback to this method is that it requires slightly more reagent than desired. For example, in order to conduct 5 replicates, we typically mix the necessary volume for 6 replicates due to loss in pipetting and wetting of plastic surfaces. We have also noticed that the reduction of pipetting error from Method 2 allows CFPS novices to perform experiments with reduced variance (Fig. 5). The master mix used to generate Fig. 5 replaces ATP with AMP; which is used to reduce cost in training, but also results in >3x reduction in expression efficiency (comparing Figs. 1–5).
Fig. 4.
Two methods of preparing sample replicates. In Method 1, components for each reaction are directly aliquoted to the individual reactors, then subsequently homogenized. In Method 2, all components are homogenized first in a mixture tank and then aliquoted as complete reaction replicates to single containers.
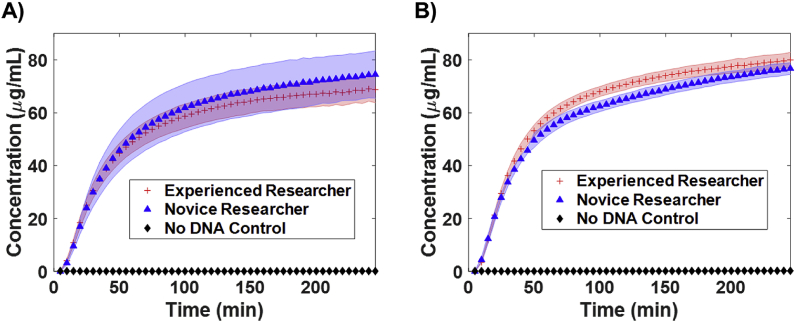
Fig. 5.
Comparison of data obtained from the two replicate generating methods mentioned in Fig. 4, where A) is obtained from mixing all reaction components individually in wells (includes pipetting error) and B) involves mixing all reactants in a single mixture tube first, then aliquoting the mixture replicates to independent wells (n = 5). Shaded error region is for one standard deviation. Note: the master mix used in these training experiments uses AMP instead of ATP to further reduce cost [41], but comes at a loss of yield (compare to Fig. 1 which used a modified ATP based master mix).
The composition of our typical cell-free reaction are as follows: 57 mM HEPES (pH 7), 1.2 mM ATP, 0.85 mM GMP, 0.85 mM CMP, 0.85 mM UMP, 33 mM PEP, 34 μg/mL folinic acid, 171 μg/mL E. coli tRNA, 2 mM 20 amino acids, 0.33 mM NAD, 0.26 mM CoA, 175 mM potassium glutamate, 10 mM ammonium glutamate, 16 mM magnesium glutamate, 2.7 mM potassium oxalate, 1 mM putrescine, 1.5 mM spermidine, 13.3 ng/μL pJL1-sfGFP plasmid DNA, and 24% (v/v) cell extract. The components were added to a 1.5 mL tube (DNA added last) and homogenized before being aliquoted to wells in 15 μL aliquots. Due to the variability of small CFPS reactions, ours are always carried out in n = 4 or 5 samples in order to obtain better population sampling. When using the Promega kit, the ratio of master mix to total reaction volume in the manufacturer's protocol is maintained when scaling down the reaction. To keep results comparable, we use previously optimized extract [26]. The reactions are typically carried out at 37 °C in a 384 black-walled, flat-bottom well plate (Greiner #781906) and covered with a colorless film (Axygen). Fluorescence measurements are taken with a Synergy Neo2 HTS Multi-Mode Microplate Reader (BioTek) for at least 4 h. In cases where expression longer than 4 h is desired, the temperature is reduced to 30 °C. The fluorescence of sfGFP is measured at an excitation of 485 nm and emission of 528 nm using a ±20 bandpass window and 61 gain setting. The reaction is also stirred in orbital motion at 237 cpm; although it is disputed whether mixing is needed for such small volumes, we find it does improve sample homogeneity [28]. To summarize the reaction procedure:
-
1.
Determine the amount of extract, DNA, master mix, and ddH2O needed for six 15 μL reactions.
-
2.
Thaw, on ice, the amount of reagent needed to run all samples and controls.
-
3.
In a 1.5 mL microcentrifuge tube, add in the following order: ddH2O, extract, master mix, and DNA. While we see no observed significance in the order of addition, DNA should always be added last since it starts the reaction.
-
4.
Homogenize the reaction mix by pipetting up and down at least 10 times. The total volume should be 90 μL so the pipette should be set to 45 μL.
-
5.
Pipette the reaction mix into 5 individual wells in 15 μL aliquots.
-
6.
Cover the plate with film and run under desired reaction conditions.
We use a plate reader to measure sfGFP since we believe activity is the most objective measurement of correctly folded protein. Other proteins (enzymes, antibodies, etc.) may require more specific assays (binding, kinetics, toxicity, etc.) to determine their quality. In these cases, it may be best to assess titer through purification. In this way, activity per concentration can be used to objectively assess the quality of the process.
3. Results and discussion
The data presented in Figs. 1 and 5 show how CFPS results can have reduced variability with improved methods. The initial trials (Fig. 1) used the PANOx-SP system [23] but without concern for completely dissolving all the master mix components and using replicate Method 1 in Fig. 4. The final trials (Fig. 1) also used PANOx-SP master mix but following the methods described in this work. Since the average signal of each sample between these experiments is different, it's best to compare the variability between these samples using the coefficient of variation (CV) or relative standard deviation, which is computed by dividing the standard deviation by the mean value and expressed as a percentage. We find the CV of the initial attempt to be 97.3% and was improved to 1.2% using these methods. This is comparable to the commercial kit (Promega #L1110) CV of 1.14%. When comparing the data obtained from an experienced user to a novice (undergrad researcher with limited experience in CFPS), method 1 of replicate preparation results in CVs of 6.8% and 11.0%. When method 2 of replicate preparation is used, there is a noticeable decrease in variance from both experimentalists, with CVs of 3.5% and 3.0% for experienced and novice respectively. This also achieves some level of standardization so results between team members can be compared.
We believe the small difference in performance and variation between the Promega master mix and our own (Fig. 1) is likely due to makeup of the commercial master mix. The use of nucleoside triphosphates (GTP, CTP, UTP) may provide a performance advantage over the less expensive monophosphates (GMP, CMP, UMP) used in our master mix [27]. It could also be due to differences in reagent concentration. The Promega documentation does not state the concentrations of their master mix components, therefore a true comparison is difficult to make. Despite the lack of information, we do observe that the variability using both these mixes is comparable when following the remainder of the methods presented in this work.
While it is impossible to completely eliminate all sources of variation, following the procedures outlined here, we have found, reduces experimental error. Machine errors such as unequal heating, shaking and measurement error will remain. Other sources can be attributed to the stochastic nature of biological processes. It is also important to note that the methods outlined in this paper are for the expression of sfGFP but the methods used may need to be further optimized for other proteins of interest. For example, expressing various immunoglobulins required different temperatures for optimum expression [42]. Therefore, the process should be optimized for the protein(s) of interest, if maximum expression is desired.
If CFPS is being used for larger batch production (such as in distributed manufacturing scenarios [43]) or large scale production (such as for cytotoxic therapies [2]), many of these sources of variability (e.g. pipetting methods) are no longer a concern. The 15 μL reactions used for prototyping discussed herein are very different from the large reactors found in a pilot plant. At that scale, sources of variability would include mixing and heating methods in various reactors. Also, at the production scale the master mix composition and cell lysis method would be driven by cost factors. Thus, the methods outlined herein are targeted at the researcher using CFPS for fast protein prototyping where small yields (μg-mg) are sufficient.
4. Conclusions
In conclusion, the variability that can be observed in CFPS experiments, especially in runs performed by new users, can be reduced significantly by adopting the methods presented in this paper. Of the method steps we highlight in this paper, the most significant are 1) completely dissolving all master-mix components for their individual stock solutions, 2) making enough master mix to last the entire experimental campaign, and 3) homogenizing all reactants before aliquoting replicates to reduce the small-volume pipetting error (Fig. 6). In this manner the researcher can obtain more statistically significant results and those new to the field of in vitro expression can more reliably contribute to CFPS experimental campaigns. This is of importance as CFPS moves from a niche tool to a technique used by larger teams for biomaterial, metabolic network, and sensor discovery and design.
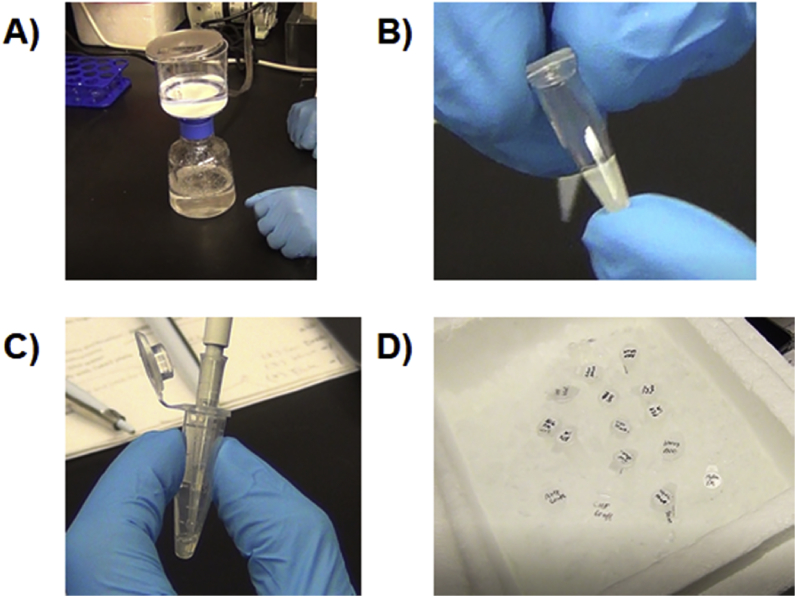
Fig. 6.
Significant method steps in the preparation of a cell-free reaction that can reduce variability and increase efficiency. A) keeping a filtered stock of 1 M glucose for cell growth, B) storing cell extract in 45 μL aliquots for single use, C) homogenizing all reactants in a tube before aliquoting into wells, and D) creating a fully solubilized master mix.
Acknowledgements
We would like to acknowledge Bradley Bundy, Michael Jewett, James Swartz, Vincent Noireaux, and Yongchan Kwon for their assistance in understanding and discussing these protocols. NFR acknowledges funding from the Black & Veatch Building a World of Difference Faculty Fellowship in Engineering and Iowa State University Startup Funds.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2019.10.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Matthaei J.H., Nirenberg M.W. Characteristics and stabilization of DNAase-sensitive protein synthesis in E. coli extracts. Proc Natl Acad Sci. 1961;47:1580–1588. doi: 10.1073/pnas.47.10.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zawada J.F., Yin G., Steiner A.R., Yang J., Naresh A., Roy S.M., Gold D.S., Heinsohn H.G., Murray C.J. Microscale to manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin G., Garces E.D., Yang J., Zhang J., Tran C., Steiner A.R., Roos C., Bajad S., Hudak S., Penta K., Zawada J., Pollitt S., Murray C.J. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. mAbs. 2012;4:217–225. doi: 10.4161/mabs.4.2.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Q., Hanson J.A., Steiner A.R., Tran C., Masikat M.R., Chen R., Zawada J.F., Sato A.K., Hallam T.J., Yin G. A simplified and robust protocol for immunoglobulin expression in Escherichia coli cell-free protein synthesis systems. Biotechnol Prog. 2015;31:823–831. doi: 10.1002/btpr.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dopp J.L., Rothstein S.M., Mansell T.J., Reuel N.F. Rapid prototyping of proteins: mail order gene fragments to assayable proteins within 24 hours. Biotechnol Bioeng. 2019;116:667–676. doi: 10.1002/bit.26912. [DOI] [PubMed] [Google Scholar]
- 6.Jaroentomeechai T., Stark J.C., Natarajan A., Glasscock C.J., Yates L.E., Hsu K.J., Mrksich M., Jewett M.C., DeLisa M.P. Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat Commun. 2018;9:2686. doi: 10.1038/s41467-018-05110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley Q.M., Nash C.J., Jewett M.C. Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synth Biol. 2019;4 doi: 10.1093/synbio/ysz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M., Daringer N.M., Bosch I., Dudley D.M., O'Connor D.H., Gehrke L., Collins J.J. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 9.Salehi A.S.M., Shakalli Tang M.J., Smith M.T., Hunt J.M., Law R.A., Wood D.W., Bundy B.C. Cell-free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal Chem. 2017;89:3395–3401. doi: 10.1021/acs.analchem.6b04034. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi M.K., Tan X., Dy A.J., Braff D., Akana R.T., Furuta Y., Donghia N., Ananthakrishnan A., Collins J.J. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat Commun. 2018;9:3347. doi: 10.1038/s41467-018-05864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurst G.B., Asano K.G., Doktycz C.J., Consoli E.J., Doktycz W.L., Foster C.M., Morrell-Falvey J.L., Standaert R.F., Doktycz M.J. Proteomics-based tools for evaluation of cell-free protein synthesis. Anal Chem. 2017;89:11443–11451. doi: 10.1021/acs.analchem.7b02555. [DOI] [PubMed] [Google Scholar]
- 12.Failmezger J., Rauter M., Nitschel R., Kraml M., Siemann-Herzberg M. Cell-free protein synthesis from non-growing, stressed Escherichia coli. Sci Rep. 2017;7 doi: 10.1038/s41598-017-16767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foshag D., Henrich E., Hiller E., Schäfer M., Kerger C., Burger-Kentischer A., Diaz-Moreno I., García-Mauriño S.M., Dötsch V., Rupp S., Bernhard F. The E. coli S30 lysate proteome: a prototype for cell-free protein production. New Biotechnol. 2018;40:245–260. doi: 10.1016/j.nbt.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Garenne D., Beisel C.L., Noireaux V. Characterization of the all-E. coli transcription-translation system myTXTL by mass spectrometry. Rapid Commun Mass Spectrom. 2019;33:1036–1048. doi: 10.1002/rcm.8438. [DOI] [PubMed] [Google Scholar]
- 15.Gregorio N.E., Levine M.Z., Oza J.P., Gregorio N.E., Levine M.Z., Oza J.P. A user's guide to cell-free protein synthesis. Methods Protoc. 2019;2:24. doi: 10.3390/mps2010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine M.Z., Gregorio N.E., Jewett M.C., Watts K.R., Oza J.P. Escherichia coli-Based Cell-Free Protein Synthesis: protocols for a robust, flexible, and accessible platform technology. J Vis Exp. 2019 doi: 10.3791/58882. [DOI] [PubMed] [Google Scholar]
- 17.Lim H.J., Kim D.-M. Cell-free metabolic engineering: recent developments and future prospects. Methods Protoc. 2019;2:33. doi: 10.3390/mps2020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karig D.K. Cell-free synthetic biology for environmental sensing and remediation. Curr Opin Biotechnol. 2017;45:69–75. doi: 10.1016/j.copbio.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Ogonah O.W., Polizzi K.M., Bracewell D.G. Cell free protein synthesis: a viable option for stratified medicines manufacturing? A brief history of cell free synthesis systems. Curr Opin Chem Eng. 2017;18:77–83. [Google Scholar]
- 20.Wilding K.M., Schinn S.M., Long E.A., Bundy B.C. The emerging impact of cell-free chemical biosynthesis. Curr Opin Biotechnol. 2018;53:115–121. doi: 10.1016/j.copbio.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Chizzolini F., Forlin M., Yeh Martín N., Berloffa G., Cecchi D., Mansy S.S. Cell-free translation is more variable than transcription. ACS Synth Biol. 2017;6:638–647. doi: 10.1021/acssynbio.6b00250. [DOI] [PubMed] [Google Scholar]
- 22.Cole S.D., Beabout K., Turner K.B., Smith Z.K., Funk V.L., V Harbaugh S., Liem A.T., Roth P.A., Geier B.A., Emanuel P.A., Walper S.A., Chávez J.L., Lux M.W. Quantification of interlaboratory cell-free protein synthesis variability. ACS Synth Biol. 2019 doi: 10.1021/acssynbio.9b00178. [DOI] [PubMed] [Google Scholar]
- 23.Kwon Y.C., Jewett M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci Rep. 2015;5 doi: 10.1038/srep08663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrestha P., Holland T.M., Bundy B.C. Streamlined extract preparation for Escherichia coli Supplement. Biotechniques. 2012 doi: 10.2144/0000113924. [DOI] [PubMed] [Google Scholar]
- 25.Romantseva E., Strychalski E. 2019. NIST cell-free workshop.https://www.nist.gov/sites/default/files/documents/2019/06/19/nist_cell-free_workshop_report.pdf La Jolla, CA. [Google Scholar]
- 26.Dopp J.L., Reuel N.F. Process optimization for scalable E. coli extract preparation for cell-free protein synthesis. Biochem Eng J. 2018;138:21–28. [Google Scholar]
- 27.Yang W.C., Patel K.G., Wong H.E., Swartz J.R. Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol Prog. 2012;28:413–420. doi: 10.1002/btpr.1509. [DOI] [PubMed] [Google Scholar]
- 28.Dopp J.L., Tamiev D.D., Reuel N.F. Cell-free supplement mixtures: elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol Adv. 2018;37:246–258. doi: 10.1016/j.biotechadv.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Ahn J.H., Chu H.S., Kim T.W., Oh I.S., Choi C.Y., Hahn G.H., Park C.G., Kim D.M. Cell-free synthesis of recombinant proteins from PCR-amplified genes at a comparable productivity to that of plasmid-based reactions. Biochem Biophys Res Commun. 2005;338:1346–1352. doi: 10.1016/j.bbrc.2005.10.094. [DOI] [PubMed] [Google Scholar]
- 30.Wilding K.M., Hunt J.P., Wilkerson J.W., Funk P.J., Swensen R.L., Carver W.C., Christian M.L., Bundy B.C. Endotoxin-free E. Coli-based cell-free protein synthesis: pre-expression endotoxin removal approaches for on-demand cancer therapeutic production. Biotechnol J. 2018 doi: 10.1002/biot.201800271. [DOI] [PubMed] [Google Scholar]
- 31.Helou C., Marier D., Jacolot P., Abdennebi-Najar L., Niquet-Léridon C., Tessier F.J., Gadonna-Widehem P. Amino acids. 2014. Microorganisms and Maillard reaction products: a review of the literature and recent findings; pp. 267–277. [DOI] [PubMed] [Google Scholar]
- 32.Levine M.Z., So B., Mullin A.C., Watts K.R., Oza J.P. 2019. Redesigned upstream processing enables a 24-hour workflow from E. coli cells to cell-free protein synthesis. [Google Scholar]
- 33.Sun Z.Z., Hayes C.A., Shin J., Caschera F., Murray R.M., Noireaux V. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J Vis Exp. 2013 doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Copeland, Padumane Kwon. A crude extract preparation and optimization from a genomically engineered Escherichia coli for the cell-free protein synthesis system: practical laboratory guideline. Methods Protoc. 2019;2:68. doi: 10.3390/mps2030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman A.D., Kelley-Loughnane N., Lucks J.B., Jewett M.C. vol. 7. 2019. p. 36. (Deconstructing cell-free extract preparation for in vitro activation of transcriptional genetic circuitry). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salehi A.S.M., Smith M.T., Bennett A.M., Williams J.B., Pitt W.G., Bundy B.C. Cell-free protein synthesis of a cytotoxic cancer therapeutic: onconase production and a just-add-water cell-free system. Biotechnol J. 2016;11:274–281. doi: 10.1002/biot.201500237. [DOI] [PubMed] [Google Scholar]
- 37.Schinn S.M., Broadbent A., Bradley W.T., Bundy B.C. Protein synthesis directly from PCR: progress and applications of cell-free protein synthesis with linear DNA. New Biotechnol. 2016;33:480–487. doi: 10.1016/j.nbt.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Burgess-Brown N.A., Sharma S., Sobott F., Loenarz C., Oppermann U., Gileadi O. Codon optimization can improve expression of human genes in Escherichia coli: a multi-gene study. Protein Expr Purif. 2008;59:94–102. doi: 10.1016/j.pep.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Maertens B., Spriestersbach A., Von Groll U., Roth U., Kubicek J., Gerrits M., Graf M., Liss M., Daubert D., Wagner R., Schäfer F. Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci. 2010;19:1312–1326. doi: 10.1002/pro.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caschera F., Noireaux V. Preparation of amino acid mixtures for cell-free expression systems. Biotechniques. 2015;58:40–43. doi: 10.2144/000114249. [DOI] [PubMed] [Google Scholar]
- 41.Calhoun K.A., Swartz J.R. An economical method for cell-free protein synthesis using glucose and nucleoside monophosphates. Biotechnol Prog. 2005;21:1146–1153. doi: 10.1021/bp050052y. [DOI] [PubMed] [Google Scholar]
- 42.Murakami S., Matsumoto R., Kanamori T. Constructive approach for synthesis of a functional IgG using a reconstituted cell-free protein synthesis system. Sci Rep. 2019;9:671. doi: 10.1038/s41598-018-36691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pardee K., Slomovic S., Nguyen P.Q., Lee J.W., Donghia N., Burrill D., Ferrante T., McSorley F.R., Furuta Y., Vernet A., Lewandowski M., Boddy C.N., Joshi N.S., Collins J.J. Portable, on-demand biomolecular manufacturing. Cell. 2016;167:248–259. doi: 10.1016/j.cell.2016.09.013. e12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.