Abstract
Variants in genes encoding ribosomal proteins have thus far been associated with Diamond-Blackfan anemia, a rare inherited bone marrow failure, and isolated congenital asplenia. Here, we report one de novo missense variant and three de novo splice variants in RPL13, which encodes ribosomal protein RPL13 (also called eL13), in four unrelated individuals with a rare bone dysplasia causing severe short stature. The three splice variants (c.477+1G>T, c.477+1G>A, and c.477+2 T>C) result in partial intron retention, which leads to an 18-amino acid insertion. In contrast to observations from Diamond-Blackfan anemia, we detected no evidence of significant pre-rRNA processing disturbance in cells derived from two affected individuals. Consistently, we showed that the insertion-containing protein is stably expressed and incorporated into 60S subunits similar to the wild-type protein. Erythroid proliferation in culture and ribosome profile on sucrose gradient are modified, suggesting a change in translation dynamics. We also provide evidence that RPL13 is present at high levels in chondrocytes and osteoblasts in mouse growth plates. Taken together, we show that the identified RPL13 variants cause a human ribosomopathy defined by a rare skeletal dysplasia, and we highlight the role of this ribosomal protein in bone development.
Keywords: RPL13, chondrodysplasia, bone dysplasia, ribosome, short stature, Spondyloepimetaphyseal dysplasia
Main Text
Because ribosomes translate mRNAs, they are universally responsible for the quality and quantity of proteins in all cells. Their production is highly regulated by many cellular processes, including growth, proliferation, and differentiation.1 Assembly of the 80 ribosomal proteins (RPs) and four ribosomal RNAs (rRNAs) that form the two ribosomal subunits involve hundreds of ribosome biogenesis factors (RBFs). Variants in genes encoding RPs or RBFs have been linked to a growing class of genetic conditions called ribosomopathies. These conditions often include inherited bone marrow failure (IMBF), but various other tissues are affected, such as bones, spleen, pancreas, and heart.2 Variants in genes encoding RPs have been described in a restricted set of congenital pathologies: Diamond-Blackfan anemia (DBA; >20 different RP genes [MIM: 105650]), isolated congenital asplenia (ICA [MIM: 271400], RPSA),3 an autism syndrome (MIM: 617412) (RPS23),4 and predisposition to hereditary nonpolyposis colorectal carcinoma (MIM: 120435) (RPS20).5 DBA is described as a pure red-cell aplasia, while ICA is characterized by absence of the spleen without any other developmental defect. How variants in ubiquitously expressed genes can yield such tissue-specific phenotypes remains an open question.
We report four unrelated individuals affected by a severe spondyloepimetaphyseal dysplasia (SEMD) which was first described in 2013.6 The clinical and radiological abnormalities observed in these individuals are summarized in Table S1. Individuals 1 and 2 were reported as having a rare form of SEMD6 which is similar to SEMD matrilin-3 related (MIM: 608728) and SEMD Strudwick type (MIM: 184250). All individuals shared common major features: normal birth length, early postnatal growth deficiency, severe short stature, and genu varum. Skeletal radiographies showed platyspondyly and severe epiphyseal and metaphyseal changes in lower limbs. None of the individuals presented any episodes of anemia, and they all had a normal blood count at last follow-up.
Through trio exome sequencing of individuals P1 and P2 and their parents, we identified two de novo intronic single-nucleotide variants in RPL13 (NM_000977.3, [MIM: 113703]): c.477+1G>T in individual P1 and c.477+2T>C in individual P2 (Table 1). These variants are absent in public variant databases (dbSNP138, 1000 Genomes, NHLBI GO Exome Sequencing Project, gnomAD). Through GeneMatcher,7 we subsequently identified two additional individuals (P3 and P4) with RPL13 variants (c.477+1G>A and c.548G>C [p.Arg183Pro]); we also identified these variants through the use of trio-based exome sequencing. Both individuals were followed for short stature with SEMD (Figure 1A).
Table 1.
Clinical and Radiological Observation in Affected Individuals
| - | P1 | P2 | P3 | P4 |
|---|---|---|---|---|
| RPL13 variant (NM_000977.3) | c.477+1G>T | c.477+2T>C | c.477+1G>A | c.548G>C |
| DNA (RefSeq accession number: NC_000016.9, ref GRCh37.p13) | g.89628800 G>T | g.89628801 T>C | g.89628800 G>A | g.89629362 G>C |
| Protein (RefSeq: NP_000968.2) | p.Asn159_Val160ins18 | p.Asn159_Val160ins18 | p.Asn159_Val160ins18 | p.Arg183Pro |
| ExAC/gnomAD frequency | absent | absent | absent | absent |
| Mode of inheritance | de novo | de novo | de novo | de novo |
| Method of variant detection | ES trio | ES trio | ES trio | ES trio |
| Gender | M | M | M | F |
| Skeletal Findings | ||||
| Prenatal length < 2SD | - | - | - | - |
| Short stature | +++ | +++ | +++ | + |
| Genu varum | + | + | + | + |
| Scoliosis | - | + | - | - |
| Hyperlaxity | - | - | - | - |
| Extraskeletal Findings | ||||
| Cone-rod dystrophy | - | - | - | - |
| Myopia | - | - | - | - |
| Deafness | - | - | - | - |
| Anemia | - | - | - | - |
| Radiological Findings | ||||
| Platyspondyly | + | + | + | + |
| Bowed femora | + | + | ||
| Shortened long bones | + | + | + | + |
| Metaphyseal involvement | + | + | + | + |
| Lower limbs epiphyseal changes | + | + | + | + |
| Upper limbs epiphyseal changes | - | - | + | - |
| Pelvis | ||||
| Coxa vara | + | + | + | + |
| Hands | ||||
| Short phalanges | - | - | - | - |
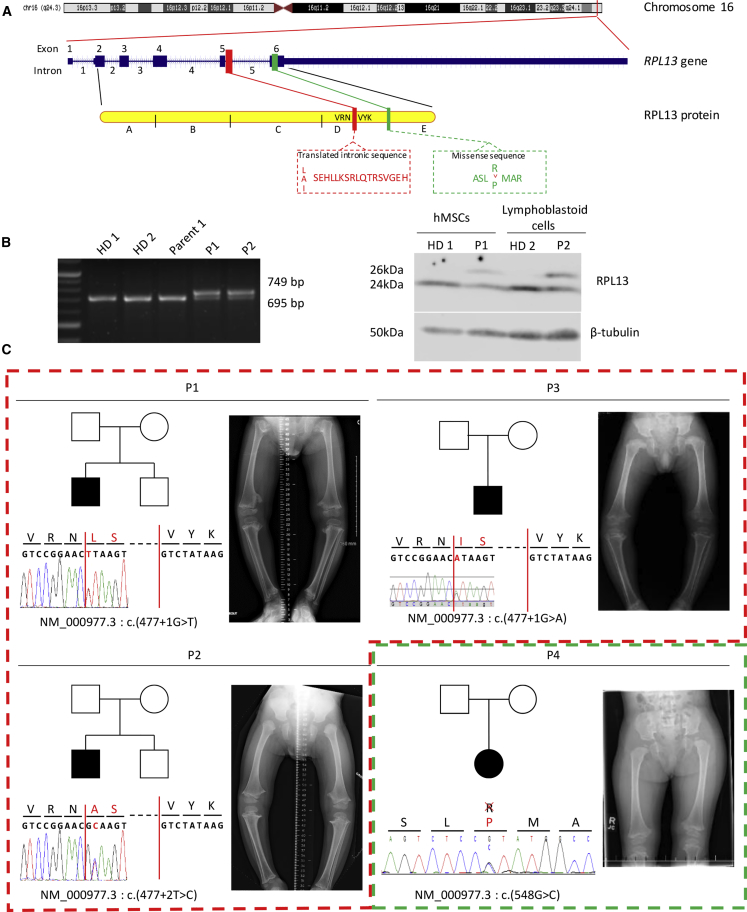
Figure 1.
Mapping and Localization of the RPL13 (eL13) Variants Observed in Four Individuals
(A) Schematic representation of the gene RPL13 located in 16q24.3 region and its protein with the site of variants in the intron 5.
(B) Agarose gel electrophoresis of RT-PCR products of RPL13 mRNA from two healthy donors, one parent, P1, and P2 (left panel). Western blot analysis of cell lysates from MSCs or lymphoblastoid cells from healthy donors (HD), P1, and P2 illustrating the presence of the variant protein RPL13, 2kDa larger than the wild type protein. β-tubulin was used as loading control (right panel).
(C) Pedigree drawings, intragenic variants of RPL13 revealed by Sanger and X-Rays photographs of individuals at the ages (respectively) of 4 years and 4 months (P1), 3 years and 4 months (P2), 4 years (P3), and 2 years and 10 months (P4) showing shortened long bones, bowed femora with metaphyseal enlargement, and irregularities.
To confirm the pathological effect of the c.477+1G>T and c.477+2T>C variants, we performed RT-PCR on RNA isolated from peripheral blood samples from individuals P1 and P2. cDNA was used as a template for PCR using primers (Table 1). Agarose gel electrophoresis using cDNA showed an aberrant longer fragment amplified in both individuals, in addition to the normal 695 bp amplification product observed with control cDNA (Figure 1B, left panel).
Sanger sequencing of the aberrant amplified products revealed the presence of the first 54 bp of intron 5 (Figure 1C). These results confirmed that the intronic missense variants abolish the natural donor splice-site of intron 5 and demonstrated that a cryptic new donor splice located in intron 5 of RPL13 was utilized in individuals P1 and P2. This leads to an abnormal mRNA containing 54 bp of intron 5. Translation of this abnormal mRNA is expected to encode a 229-amino-acid protein containing the 18 supplementary amino acids. The fourth c.548G>C variant was confirmed by Sanger sequencing and was de novo. It leads to the substitution of arginine 183 by a proline, which is expected to break the long alpha-helix present at the C terminus of the protein. The arginine 183 is highly conserved throughout species (PhastCons score 1, Mutation Taster8). This variant is also absent from all genomic databases and is predicted to be pathogenic (score 0.966 [PolyPhen9], PHRED score 26.0 [CADD10], score 106 [Mutation Taster]).
At the protein level, western blot analysis carried out on protein lysates from mesenchymal stem cells (MSCs, individual P1) and from lymphoblastoid cells (individual P2) confirmed RT-PCR data. Two bands (24kDa and 26kDa) were detected for the two individuals, and only one band was detected in control cells (Figure 1B, right panel). The two bands differed by 2kDa, corresponding to an 18-residue peptide coded by the first 54 bp of intron 5.
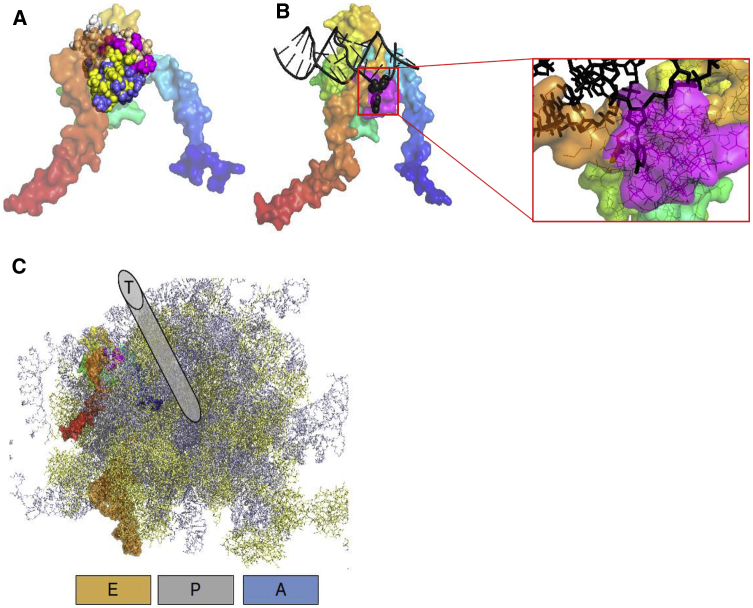
To assess the putative consequences of RPL13 variants on the ribosome structure, the 18-residue-long (individuals P1, P2, and P3) peptide was inserted into the RPL13 sequence and submitted to I-TASSER for structure prediction. The first model obtained had a TM-Score of 0.44,11 which is indicative of a high confidence in the models. All five predicted models are very similar, with a root-mean-square deviation (RMSD) between them less than 2 Å to the native RPL13 fold. The 18-residue-long polypeptide addition is modeled as a compact structure conserving the U-shaped RPL architecture (Figure 2A). In the human ribosome structure, RPL13 is in contact with Expansion Segment 7L (ES7L) of the 28S RNA around positions 500–530 (complementary strand: 630–660). A direct interaction between adenine 509 and RPL13 tyrosine 161 is clearly observed in the deposited 4V6X structure, with the adenine extruded from the double-stranded DNA. The additional polypeptide insertion, as modeled in our study, would introduce steric clashes with the DNA at that position, thus preventing this specific interaction (Figures 2B and 2C). This polypeptide patch seems nevertheless compatible with a merely intact ribosome structure because RPL13 presents an extended contact zone at the surface of the ribosome, close to the tRNA exit site (Figure 2D).
Figure 2.
Structural Consequences of the Polypeptide Insertion on RPL13 and Ribosomal Architecture
(A) The U-shaped RPL13 structure in surface representation is colored from the N-terminal in blue to the C-terminal in red. The polypeptide insertion introduce a compact patch displayed in sphere representation (model 1, magenta; model 2, yellow; model 3, tan; model 4, white; model 5, blue).
(B) The insertion for model 1 (magenta surface) would induce a steric clash with Adenine 509 (in sphere representation) of the 28S RNA (black) in its ES7L region.
(C) General organization of the ribosome including the variant RPL13 protein. The 40S subunit is removed for clarity, 60S proteins are indicated in yellow, 60S RNAs are indicated in gray. Letters A, P, and E allow approximatively location of the aminacyl, the peptidyl, and the exit sites. The tRNA found in the deposited structure at the exit site is displayed in orange surface. The polypeptidic exit tunnel (T) is indicated above the picture although its exact location is more buried in the ribosome structure.
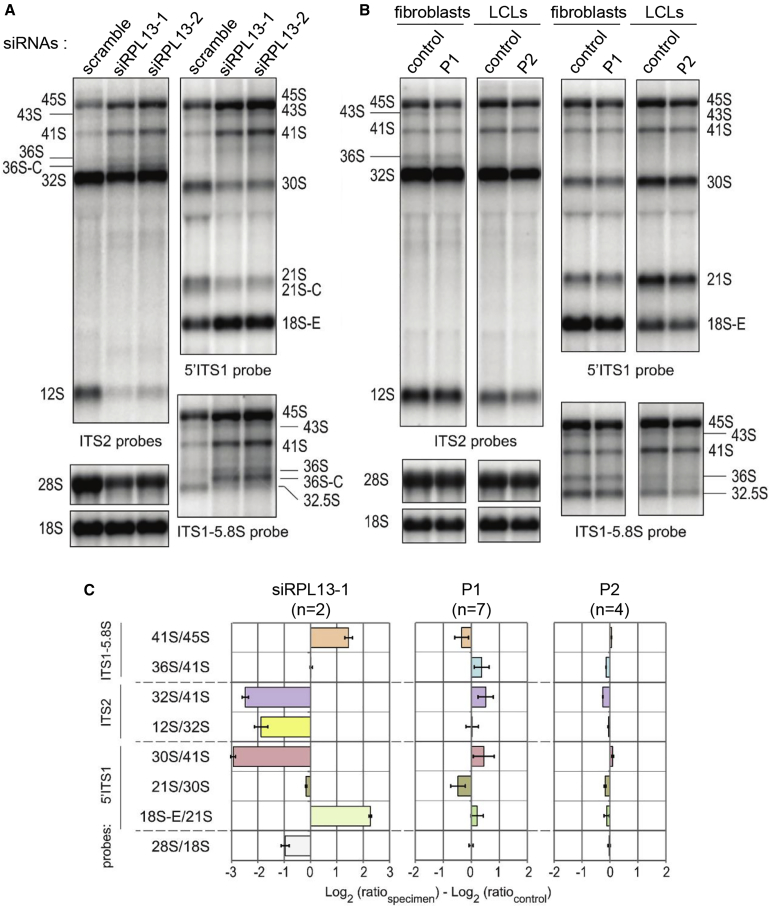
To investigate the functional consequences of RPL13 variants on ribosome biogenesis, we first assessed their impact on pre-rRNA processing by comparison with cells depleted of RPL13. As shown in Figure 3A, knockdown of RPL13 with siRNAs in HeLa cells severely affected the pre-rRNA pattern detected by northern blot. The most conspicuous effect was a defect in pre-rRNA cleavage at site 2 in ITS1, as revealed by the accumulation of 41S pre-rRNA relative to the 30S and 21S pre-rRNAs, and the high levels of 36S and 36S-C pre-rRNAs. We also noticed a drop of the levels of 12S pre-rRNA relative to its precursor, the 32S pre-rRNA (Figures 3A). A similar phenotype was observed in a control lymphoblastoid cell line treated with these siRNAs (Figure S1). When analyzed through the use of northern blot, total RNAs from cells established from individuals P1 and P2 did not show such an abnormal pre-rRNA pattern, indicating that the RPL13 variants do not affect pre-rRNA maturation like a null mutant does (Figures 3B and 3C). Consistent with the lack of impact on pre-rRNA processing, a ribosome profile on a sucrose gradient in lymphoblastoid cells from individual P2 showed a normal ratio of free 40S to 60S subunits compared to the control (Figures 4A and 4B). Importantly, we found that the RPL13 variant protein distributed in the gradient exactly as the wild-type protein. The variant is therefore incorporated into 60S subunits that form ribosomes competent for translation. We noticed, however, a decrease of the 80S and polysome peaks in the individual sample, which may indicate a change in translation dynamics (Figure 4C).
Figure 3.
Consequence of Mutant RPL13 on Pre-rRNA Processing
(A) Northern blot analysis of pre-RNAs in HeLa cells treated with RPL13 siRNAs. The 5′ITS1 and the ITS2 probes detect the precursors to the 40S and 60S subunit RNAs, respectively. The ITS1-5.8S probe evidences accumulation of 36S and 36S-C precursors, indicative of a defect in cleavage at site 2.
(B) Northern blot analysis of pre-rRNAs from fibroblasts of individual P1, lymphoblastoid cells of individual P2, and healthy donors with the same probes as in (A).
(C) Quantification of changes in the pre-rRNA pattern in (A) and (B) by ratio analysis of multiple precursors (RAMP)(27), expressed as variations relative to the respective controls (number of replicate are stated in the figure). The analysis shows no significant variation in individual cells, unlike in RPL13 siRNA-treated cells.
Figure 4.
Incorporation of Variant RPL13 in Large Ribosomal Subunits
(A and B) Polysome profiles of lymphoblastoid cells from a healthy control (A) and from individual P2 (B) displaying free small ribosomal subunits (40S peak) and free large ribosomal subunits (60S peak). Monosomes (80S peak) and polysomes correspond to mRNAs bearing single ribosomes or several ribosomes, respectively. Western blot analyses probed with anti-RPL13 and anti-RPS19 antibodies are displayed below the corresponding gradient fractions.
(C) Overlay of the two polysome profiles shown in (A) and (B) for lymphoblastoid cells from a healthy control (ctl; dotted gray line) and from individual P2 (P2; solid black line).
CD34+ cells isolated from peripheral blood were cultured in liquid culture over 15 days to evaluate erythroid proliferation. Only the late phase of the terminal erythroid differentiation was affected in the individual with the variant of RPL13 (Figure S2A). Cell viability during the complete erythroid differentiation time course has been confirmed (Figure S2B).
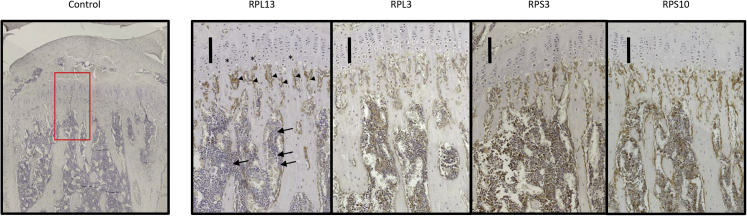
To assess the localization pattern of RPL13 in bone growth plate, cross sections of growth plate from 8-week-old healthy male mice were analyzed through the use of immunohistochemistry. Immunostaining revealed that RPL13 is present in cells from the hypertrophic zone and the remodelling zone. A weaker immunostaining was observed in cells located in the bone marrow compartment (Figure 5). Subsequently, we also evaluated three different RPs from the large and the small subunits of the ribosomes. RPL3 (MIM: 604163), RPS3 (MIM: 600454), and RPS10 (MIM: 603632) were immunohistochemically detected in cells from the growth plate, and they displayed the same localization pattern as did RPL13. RPL3, RPS3, and RPS10 were also found in the bone marrow compartment (Figure 5).
Figure 5.
Immunochemistry of Four Riboproteins in Mouse Growth Plate
Micrographs of longitudinal sections of tibiae from 8-week-old mice immunohistochemically stained for the expression of RPL13, RPL3, RPS3, and RPS10. RPL13 is expressed in cells from the hypertrophic zone (black stars) and the remodeling zone (arrowheads) with a weaker staining in cells from the bone marrow compartment (solid arrows). Expression of RPL3, RPS3, and RPS10 was detected with the same pattern in the three compartments. Scale bars correspond to 100μm.
In this study, we report four unrelated individuals with a severe bone dysplasia sharing common clinical and radiological features. The abnormalities shared by the four individuals confirm a rare form of SEMD previously described for two of them.6 All individuals presented with short stature, genu varum, epimetaphyseal anomalies, and platyspondyly but did not show any other symptoms, nor did they show any hematological abnormality on blood counts. The decreased erythroid cell proliferation reported at the terminal orthochromatophilic erythroblast stage should not be sufficient to generate an erythroid blockade and a aregenerative anemia as seen in DBA.
All individuals harbored a de novo RPL13 splice-site or missense variant. Based on the modeling of the adverse structural effects of the splice-site variants from two individuals, the mutant proteins are expected to be incorporated in pre-60S particles. Indeed, pre-rRNA processing does not seem to be affected in analyzed cells. We also showed that for variants c.477+1G>T and c.477+2 T>C, the variant protein is incorporated into the ribosomes to the same extent as is wild-type RPL13 in analyzed cells.
These data provide evidence that these de novo variants in RPL13 cause a rare form of bone dysplasia with severe short stature, responsible for a human ribosomopathy which specifically affects bone tissue. Affected individuals do not show any other symptoms such as anemia, bone marrow failure, or cranio-facial abnormalities, as can be observed in other ribosomopathies such as DBA, Shwachman-Diamond syndrome (SDS [MIM: 617941]) and cartilage hair hypoplasia (CHH-AD [MIM: 250250–607095]).1
RPL13, also known as eL13,12 is a 211-amino-acid-long protein. RPL13 is known to be essential to ribosomal assembly because its depletion leads to maturation arrest at an early ribosomal assembly stage in yeast.13 Consistently, we show that knockdown of RPL13 by siRNA in HeLa cells leads to an early defect in pre-rRNA processing, a defect which is characterized by an increase of the 45S and 41S precursors. However, our data suggest that ribosome biogenesis is not significantly affected by the variants reported here: we did not observe similar abnormality in pre-rRNA processing in the cells of individuals P1 and P2 (Figure 4B). We cannot exclude the possibility that a mild defect in ribosome biogenesis occurs in highly proliferative tissues like the growth plate, but could not be unambiguously detected here. The 60S subunits containing the RPL13 variants were detected in the same proportion as were the normal 60S subunits, including in the polysome fractions, which further indicates that incorporation of a variant RPL13 into a nascent 60S particle does not compromise its maturation, nor does it affect its stability in the cytoplasm. This is a fundamental difference compared to DBA, in which haploinsufficiency of RPs impairs synthesis of ribosomes and thereby decreases their production rate. The ribosomal profile on sucrose gradient in cells expressing the RPL13 variants is mildly changed, which suggests that translation is affected in individuals’ cells. We assume that RPL13 could play a specific role in the translation of particular mRNAs in chondrocytes or/and osteoblasts of the growth plate. In support of this hypothesis, loss of function of RPs has already been linked to very specific developmental defects in mouse models in the absence of global translation defects.14, 15
In DBA, mutations affecting RPs primarily affect erythropoiesis, but they also affect multiple other organs with variable penetrance. Skeletal abnormalities (e.g., abnormal thumbs, radial hypoplasia, short stature) have been reported in ribosomopathies such as DBA, but these conditions are not classified as skeletal dysplasias. In contrast, CHH-AD is a metaphyseal chondrodysplasia caused by RMRP (MIM: 157660) pathogenic variants and is considered to be a ribosomopathy.16 RMRP is the RNA component of RNase MRP, a nucleolar ribonucleoprotein particle which catalyzes the endonucleolytic cleavage of the pre-rRNA at site 2.17, 18 RMRP is a key factor for the regulation of hypertrophic chondrocytes. It has been shown that the knockdown of RMRP leads to a defect of chondrocyte differentiation.19 Our data show that RPL13, together with other RPs, is highly expressed in the growth plate and in cells lining the newly primary bone trabeculae, similar to RNase MRP. These high expression levels further stress the importance of ribosome biogenesis and translation in the growth plate. Whether the pathophysiological mechanisms underlying these two skeletal dysplasias share some common grounds remains to be explored.
Interestingly, a direct interaction between adenine 509 of the 28S rRNA and tyrosine 161 of RPL13 has been shown, with the adenine extruded from the double-stranded DNA.20 Here, molecular modeling suggested that the 18-amino-acid insertion localized in this region might induce the removal of this interaction. A primary consequence of this insertion might be the blockade of a conserved 28S RNA binding on ES7L. Consistent with the production and stability of the 60S subunit containing the variants, the polypeptide insertion in RPL13 does not seem to influence the overall ribosome architecture. The p.Arg183Pro missense variant affects an amino acid which interacts with the 28S rRNA and is predicted to break the C-terminal alpha helix of RPL13, an alpha helix which extensively contacts the 28S rRNA. Hence, both splice and missense variants may destabilize the interaction of RPL13 C terminus with the 28S rRNA and compromise its function in translation.
In summary, our data demonstrate that RPL13 variants cause a rare skeletal dysplasia with short stature. The identified RPL13 variants do not modify overall ribosome biogenesis, even when the resulting variant RPL13 protein is integrated to the mature large subunit. This ribosomopathy might affect bone development by affecting the translation level of specific mRNAs in cells from the growth plate, as shown in the erythroid lineage. Future studies will attempt to unravel the specific impact of these RPL13 variants on translation of key factors of the growth plate.
Declaration of Interests
K.McW. is an employee of GeneDx. A.P. is an employee of Center for Genomics and Transcriptomics and Praxis für Humangenetik Tübingen. The other authors declare no competing interests.
Acknowledgments
We would like to thank all families for participating in this study. P.E.G. and M.F.O’D. were funded by the Agence Nationale de la Recherche (ANR) grants ANR-15-CE12-0001 and ANR-16-CE11-0029 and by the European Research Area Network (ERA NET) E-RARE project and ANR grant ANR-15-RAR3-0007-04 (European Diamond-Blackfan Anemia Consortium [EuroDBA]).
Published: October 17, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.024.
Contributor Information
Marc Baud’huin, Email: marc.baudhuin@univ-nantes.fr.
Bertrand Isidor, Email: bertrand.isidor@chu-nantes.fr.
Accession Numbers
The ClinVar accession numbers for the newly reported sequence variants are VCV000689800, VCV000689801, VCV000689802, and VCV000689803 (GenBank: NM_000977.3).
Web Resources
1000 Genomes, http://www.1000genomes.org/
Combined Annotation Dependent Depletion, https://cadd.gs.washington.edu
Decipher, https://decipher.sanger.ac.uk/
ExAC Browser, http://exac.broadinstitute.org/
Genematcher, https://genematcher.org/
The Human Protein Atlas, http://www.proteinatlas.org/
MutationTaster, http://www.mutationtaster.org
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, https://evs.gs.washington.edu/EVS/
OMIM, https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Trainor P.A., Merrill A.E. Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochim. Biophys. Acta. 2014;1842:769–778. doi: 10.1016/j.bbadis.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert M., O’Donohue M.F., Lebaron S., Gleizes P.E. Pre-Ribosomal RNA Processing in Human Cells: From Mechanisms to Congenital Diseases. Biomolecules. 2018;8:E123. doi: 10.3390/biom8040123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolze A., Boisson B., Bosch B., Antipenko A., Bouaziz M., Sackstein P., Chaker-Margot M., Barlogis V., Briggs T., Colino E. Incomplete penetrance for isolated congenital asplenia in humans with mutations in translated and untranslated RPSA exons. Proc. Natl. Acad. Sci. USA. 2018;115:E8007–E8016. doi: 10.1073/pnas.1805437115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paolini N.A., Attwood M., Sondalle S.B., Vieira C.M.D.S., van Adrichem A.M., di Summa F.M., O’Donohue M.F., Gleizes P.E., Rachuri S., Briggs J.W. A Ribosomopathy Reveals Decoding Defective Ribosomes Driving Human Dysmorphism. Am. J. Hum. Genet. 2017;100:506–522. doi: 10.1016/j.ajhg.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieminen T.T., O’Donohue M.F., Wu Y., Lohi H., Scherer S.W., Paterson A.D., Ellonen P., Abdel-Rahman W.M., Valo S., Mecklin J.P. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology. 2014;147:595–598.e5. doi: 10.1053/j.gastro.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isidor B., Geffroy L., de Courtivron B., Le Caignec C., Thiel C.T., Mortier G., Cormier-Daire V., David A., Toutain A. A new form of severe spondyloepimetaphyseal dysplasia: clinical and radiological characterization. Am. J. Med. Genet. A. 2013;161A:2645–2651. doi: 10.1002/ajmg.a.36132. [DOI] [PubMed] [Google Scholar]
- 7.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 9.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J., Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L., Lindahl L., Liljas A., Lipton J.M. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamalinda M., Ohmayer U., Jakovljevic J., Kumcuoglu B., Woolford J., Mbom B., Lin L., Woolford J.L., Jr. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes & development. 2014;28:198–210. doi: 10.1101/gad.228825.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondrashov N., Pusic A., Stumpf C.R., Shimizu K., Hsieh A.C., Ishijima J., Shiroishi T., Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson S.J., Lauritsen J.P., Hartman M.G., Foushee A.M., Lefebvre J.M., Shinton S.A., Gerhardt B., Hardy R.R., Oravecz T., Wiest D.L. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Mattijssen S., Welting T.J., Pruijn G.J. RNase MRP and disease. Wiley Interdiscip. Rev. RNA. 2010;1:102–116. doi: 10.1002/wrna.9. [DOI] [PubMed] [Google Scholar]
- 17.Gill T., Cai T., Aulds J., Wierzbicki S., Schmitt M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldfarb K.C., Cech T.R. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes & development. 2017;31:59–71. doi: 10.1101/gad.286963.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinbusch M.M.F., Caron M.M.J., Surtel D.A.M., Friedrich F., Lausch E., Pruijn G.J.M., Verhesen W., Schroen B.L.M., van Rhijn L.W., Zabel B., Welting T.J.M. Expression of RMRP RNA is regulated in chondrocyte hypertrophy and determines chondrogenic differentiation. Sci. Rep. 2017;7:6440. doi: 10.1038/s41598-017-06809-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.