Abstract
Alzheimer’s disease (AD) is the most common cause of dementia and cannot be cured. The etiology and pathogenesis of AD is still not fully understood, the genetics is considered to be one of the most important factors for AD onset, and the identified susceptible genes could provide clues to the AD mechanism and also be the potential targets. MicroRNA-146a-5p (miR-146a) is well known in the regulation of the inflammatory response, and the functional SNP of miR-146a was associated with AD risk. In this study, using a noninvasive nasal administration, we discovered that a miR-146a agomir (M146AG) rescued cognitive impairment in the APP/PS1 transgenic mouse and alleviated the overall pathological process in the AD mouse model, including neuroinflammation, glia activation, Aβ deposit, and tau phosphorylation in hippocampi. Furthermore, the transcriptional analysis revealed that besides the effect of neuroinflammation, M146AG may serve as a multi-potency target for intervention in AD. In addition, Srsf6 was identified as a target of miR-146a, which may play a role in AD progression. In conclusion, our study supports that the nasal-to-brain pathway is efficient and operable for the brain administration of microRNAs (miRNAs), and that miR-146a may be a new potential target for AD treatment.
Keywords: miR-146a-5p, Alzheimer’s disease, intranasal administration, srsf6, neuroinflammation
Introduction
Alzheimer’s disease (AD), a progressive neurodegenerative disease that occurs in elderly and pre-senile individuals, is the most common form of dementia in the elderly population. Although the etiology and pathogenesis of AD are not fully understood, the disease is thought to result from a combination of genetic, aging, and environmental factors.1 The pathological hallmarks of AD are the presence of senile plaques and neurofibrillary tangles in the brain, but targeted drugs for AD treatment in clinical trials are unsatisfactory. The current clinical drugs, such as cholinesterase inhibitors and glutamate receptor antagonists, can only ameliorate the symptoms and do not cure the disease; effective therapeutic strategies for AD are still lacking and need to be developed.2 Genetic factors are important risk factors for the development of AD, and the mutations with presenilin 1 (PS1), presenilin 2 (PS2), and amyloid precursor protein (APP) are associated with early-onset AD (EOAD). Increasing numbers of susceptible genes and SNPs have been identified in different populations that contribute to the onset of late-onset AD (LOAD), which accounts for more than 90% of AD patients, and have been identified in different populations.3,4 Analysis of susceptibility genes helps us to elucidate the pathogenesis of LOAD and the AD mechanism and to identify potential therapeutic targets for AD treatment.
MicroRNAs (miRNAs) are often thought to be determinants of cell fate and are increasingly recognized as critical regulators of various brain pathologies, ranging from neurodevelopmental disorders to brain tumors to neurodegenerative diseases, indicating that miRNAs are very promising targets for the treatment of diseases.2,5 miRNA-146a-5p (miR-146a) is a well-known miRNA associated with inflammation, and the related SNPs of miR-146a were also associated with several inflammatory diseases.6 Increasing evidence suggests that miR-146a is associated with AD and may play an important role in the progression of AD in recent years. Upregulation of miR-146a was observed in brain regions affected by AD pathology, including the hippocampus and temporal cortex, while its expression was unchanged in unaffected areas.7 Studies have shown that miR-146a targets include complement factor H (CFH), interleukin (IL)-1 receptor-associated kinase 1 (IRAK1), and tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), among others, which are associated with the innate immunity of AD.8 In addition, current studies have indicated that miR-146a is not only involved in the regulation of innate immunity but also affects other biological functions, such as hematopoietic function and cell differentiation, suggesting extensive functions in physiological and disease processes.9,10 We previously reported that a functional SNP located in the promoter region of miR-146a could influence miR-146a expression and is associated with the risk of AD and rate of cognitive decline in AD patients, strongly suggesting that miR-146a plays an important role in the pathogenesis of AD and may be a potential new target for its treatment.11
In this study, we aimed to assess whether the upregulation of miR-146a is beneficial to inhibit the progression of AD. A miR-146a agomir (named M146AG) was used to evaluate the development of an AD mouse model by a nasal-to-brain pathway. Furthermore, the mechanism of miR-146a in rescuing the course of AD was also explored.
Results
Nasal Administration of M146AG Was an Effective Delivery Route to the Hippocampi of Mice
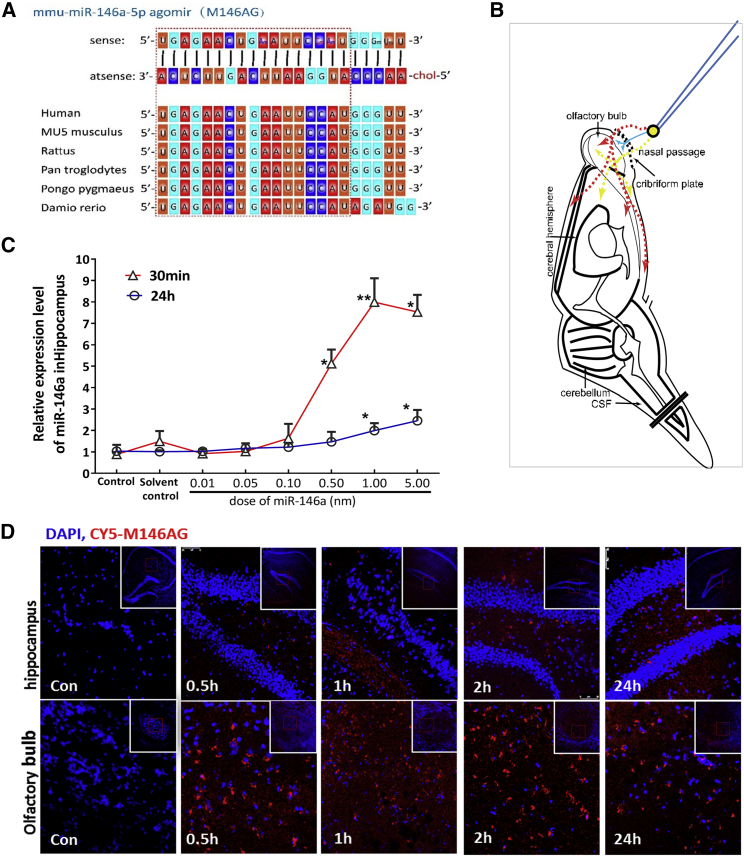
miRNAs are small molecules with a short half-life and are easily degraded. M146AG is a chemically modified miR-146a mimic that is more stable and has a higher cell membrane affinity than does the endogenous miRNA (Figure 1A). For AD drug therapy, methods to effectively reach the brain through or bypassing the blood-brain barrier (BBB) are the difficulties and key to achieving targeted interventions. Therefore, in this study, a nose-to-brain drug delivery approach was used to evaluate the efficiency of miRNA mimics as hippocampal targets. First, we evaluated the feasibility of this administration of miRNAs through preliminary experiments. M146AG was delivered via the nose-to-brain route to 12-month-old C57BL/6 mice (Figure 1B), and the levels of miR-146a in the hippocampi were evaluated at different time points (Figure 1C). As shown in Figure 1, M146AG could be intranasally delivered to the hippocampi, reaching an 8.0-fold peak at 30 min and a still 1.75-fold peak at 24 h compared with that of the vehicle control. Furthermore, to further verify that the exogenous M146AG reached the hippocampi to increase the miR-146a level, 1 nmol of CY5-labeled M146AG was administered to 12-month-old C57BL/6 mice by nasal instillation, and the results showed that the fluorophores were observed in the olfactory bulb and hippocampi of the treated mice from 30 min to 1 h, and that the fluorescence intensity weakened with time, and the fluorescence was still observed in the hippocampi and olfactory bulb 24 h after administration (Figure 1D), suggesting that the nasal administration of M146AG effectively delivered this molecule to the hippocampi. In addition, we obtained similar results with administration of another control miRNA agomir through the same nose-to-brain drug delivery approach (Figure S1), suggesting that this method is feasible for the miRNA drug administration to the brain.
Figure 1.
Nasal Administration of M146AG Effectively Delivered to the Hippocampi
(A) Construct of mmu-miR-146a-5p agomir and the homology structure of miR-146a in multiple species. In the sense chain, Am, Um, Cm, and Gm mean that A, U, C, and G were modified by 2′-ome, respectively. The antisense chain was modified with cholesterol (chol). (B) Schematic diagram of the possible pathways of nose-to-brain drug delivery to bypass the BBB. Intranasally delivered drugs are rapidly transported into the CNS tissue by the peripheral olfactory system (red), by small-molecular-weight solutes that may rapidly enter cerebrospinal fluid (CSF) after nasal administration (blue), and by the neural pathway connecting the nasal passage to the olfactory bulb/rostral brain (yellow). (C) Expression levels of miR-146a in the hippocampi of 12-month-old C57BL/6 mice at different time points detected by qRT-PCR analysis. (D) Olfactory bulb and hippocampi images of CY5-labeled M146AG treatment of 12-month-old C57BL/6 mice were taken at ×40 magnification. Nuclei are stained blue, and CY5-labeled M146AG displays red fluorescence. All data are presented as the mean ± SEM. At least three independent experiments were performed (n = 5 mice per group). *p < 0.05; **p < 0.01.
M146AG Attenuated Memory Deficits in APP/PS1 Transgenic Mice
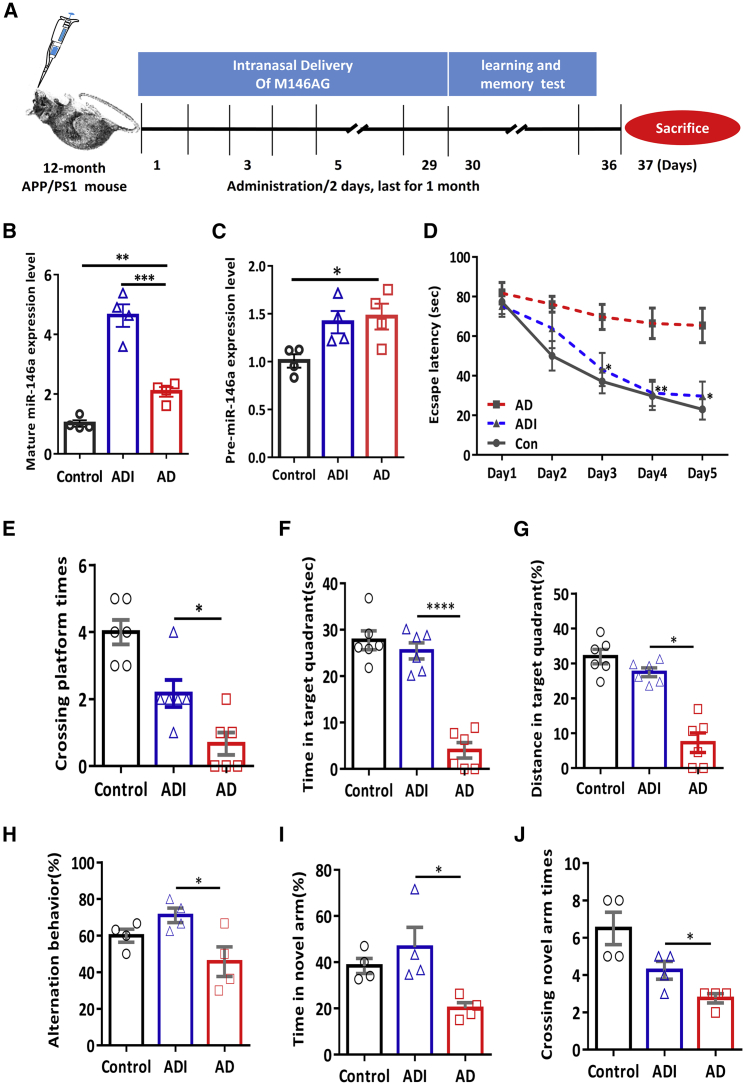
Then, M146AG was administered to 12-month-old APP/PS1 transgenic mice via the nose-to-brain route to evaluate the potential therapeutic effects on AD (Figure 2A). After 1 month of M146AG administration of the AD mouse model (ADI groups), the levels of mature miR-146a were evaluated in hippocampi of ADI groups than those in the C57 control group (Figure 2B), and the levels of mature miR-146a and precursor miR-146a (pre-miR-146a) in the AD group were found to be higher than those in the control group (Figures 2B and 2C). Meanwhile, the pre-miR-146a level in the ADI group was similar to that in the AD group (Figure 2C), which suggested that the administration of M146AG increased the level of mature miR-146a in hippocampi. Moreover, our results showed that scrambled M146AG administration showed no significant difference in either mature miR-146a or pre-miR-146a with the solvent administration by the same way of nasal delivery, in both C57 mice group and the APP/PS1 transgenic mice groups (Figure S2), suggesting that the elevated miR-146a level in the hippocampus was caused by the administration of exogenous M146AG.
Figure 2.
M146AG Attenuated the Memory Deficits in APP/PS1 Transgenic Mice
(A) Graphic procedure of drug delivery, point in time of behavioral test, and sacrifice in the subsequent experiments. (B and C) The expression levels of mature miR-146a (B) and pre-miR-146a (C) in the hippocampus of control mice (C57 mice), AD mice (APP/PS1 transgenic mice), and ADI mice (M146AG-APP/PS1 mice) at 12 months of age were detected by qRT-PCR. U6 was used as the control of mature miR-146a; GAPDH was used as the control of pre-miR-146a. (D) The escape latencies and pathway diagram of AD mice, ADI mice, and C57 mice groups were tested in the MWM for 5 consecutive days. (E–G) Probe trials performed at day 6: the crossing times in the platform site (E), the time spent in the target quadrant (F), and the more swimming distances in the target quadrant (G). (H–J) Y-maze tests were performed at day 6 after the MWM test, and the spontaneous alternation (H), the time in the novel arm (I), and the number of crossings in the novel arm (J) were measured during a 5-min session. All data are presented as the mean ± SEM. At least three independent experiments were performed (n = 4–6 mice per group). *p < 0.05; **p < 0.01;***p < 0.001; ****p < 0.001.
The memory deficits in APP/PS1 transgenic mice were evaluated by a Morris water maze (MWM) test and Y-maze test. As shown in Figure 2D, the ADI groups displayed significantly reduced escape latencies compared with those of the 12-month-old APP/PS1 transgenic mice (AD groups) (Figure 2D). In the spatial probe test, the ADI groups crossed the platform more often and spent more time and had a greater swimming distance in the target quadrant than did the control mice (Figures 2E–2G). In addition, consistent conclusions were observed in the Y-maze test, as the ADI groups had better alternation behavior (Figure 2H), more time in the novel arm (Figure 2I), and had more novel arm crossings (Figure 2J) than did the AD groups. Furthermore, we also examined whether the modification of mimics, rather than the sequence of miR-146a, might affect the cognitive degradation of AD, and we compared the cognitive status of scrambled M146AG and normal solvents in both APP/PS1 transgenic mice and C57 mice. The results showed that scrambled M146AG administration showed no significant difference in the MWM test with the solvent administration by the same nasal delivery in either C57 mice or APP/PS1 transgenic mice (Figure S3), suggesting that the sequence of miR-146a is the core to influence the course of AD in the mouse model. Collectively, these results suggested that treatment with M146AG attenuates learning and memory cognitive deficits in APP/PS1 transgenic mice.
M146AG Decreased Glial Cell Activation in the APP/PS1 Mice Hippocampi
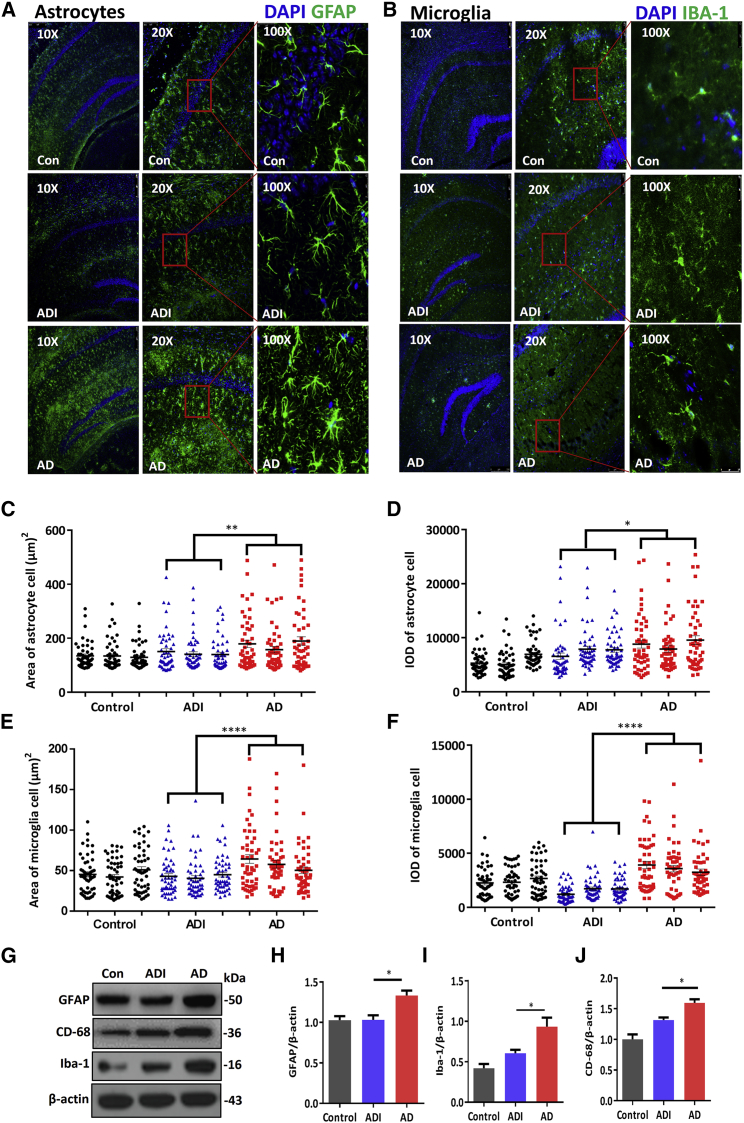
Astrocytes and microglia are involved in the pathogenesis of AD.4,12 To determine whether M146AG treatment alleviates glial cell activation in APP/PS1 mice hippocampi, we examined brain sections to observe hippocampal GFAP and Iba-1 fluorescence, which denoted astrocytes and microglia, respectively. As shown in Figure 3, hypertrophy of astrocytes was observed in the hippocampi of 12-month-old APP/PS1 mice, and the density of astrocytes, visualized by anti-GFAP, showed that the astrocyte cell area and integrated optical density (IOD) were increased significantly; these parameters were dramatically reduced in M146AG-treated 12-month-old APP/PS1 mice (Figures 3C and 3D). Furthermore, we observed that compared with the AD group, the GFAP protein expression in the hippocampus showed the lower trend in the ADI group (Figures 3G and 3H) and was similar in the AD + negative control (NC) group (Figures S4A and S4B), suggesting that astrocytes in the hippocampi of 12-month-old APP/PS1 mice were significantly activated compared with those in the hippocampi of C57 mice. Similarly, microglia in the hippocampi of 12-month-old APP/PS1 mice appeared amoeboid macrophage-like with short, thick processes expanding as lamellipodia, and the microglia cell area and IOD were increased significantly; these parameters were reduced in M146AG-treated 12-month-old APP/PS1 mice, as visualized by anti-Iba-1 (Figures 3E and 3F). The microglia showed a circular shape, denoting the morphology of inactive microglia, and the Iba-1 and CD68 levels were significantly decreased in the M146AG-treated 12-month-old APP/PS1 mice compared with those in the 12-month-old APP/PS1 mice (Figures 3G, 3I, and 3J); meanwhile, no significance different was observed in the AD+NC group (Figures S4A and S4C). Taken together, these results indicated that M146AG treatment inhibited the activation of astrocytes and microglia in the hippocampi in the AD mice model.
Figure 3.
M146AG Decreased the Activation of Glia in the Hippocampi of APP/PS1 Mice
(A, C, D) Representative images showing the activation of astrocytes and the fluorescence intensity by immunofluorescence staining for GFAP (green) (A). Quantification of the area (C) and IOD (D) of astrocytes are also shown (three sections per group with 50 astrocytes randomly selected from each section). (B, E, F) Representative images showing the activation of microglia and the fluorescence intensity by immunofluorescence staining for Iba-1 (green) (B). Quantification of the area (E) and IOD (F) of microglia cells are also shown (three sections per group with 50 microglia randomly selected from each section). (G–J) The expression levels of GFAP (H), CD68 (I), and Iba-1 (J) were detected by western blotting (G); β-actin was used as the control. All data are presented as the mean ± SEM. At least three independent experiments were performed. AD represents the hippocampi of 12-month-old APP/PS1 mice; ADI represents the hippocampi of 12-month-old APP/PS1 mice with M146AG administration; control represents the hippocampi of 12-month-old C57 mice (n = 3–4 mice per group). *p < 0.05; **p < 0.01; ****p < 0.0001.
M146AG Relieved Neuroinflammation in the APP/PS1 Mice Hippocampi
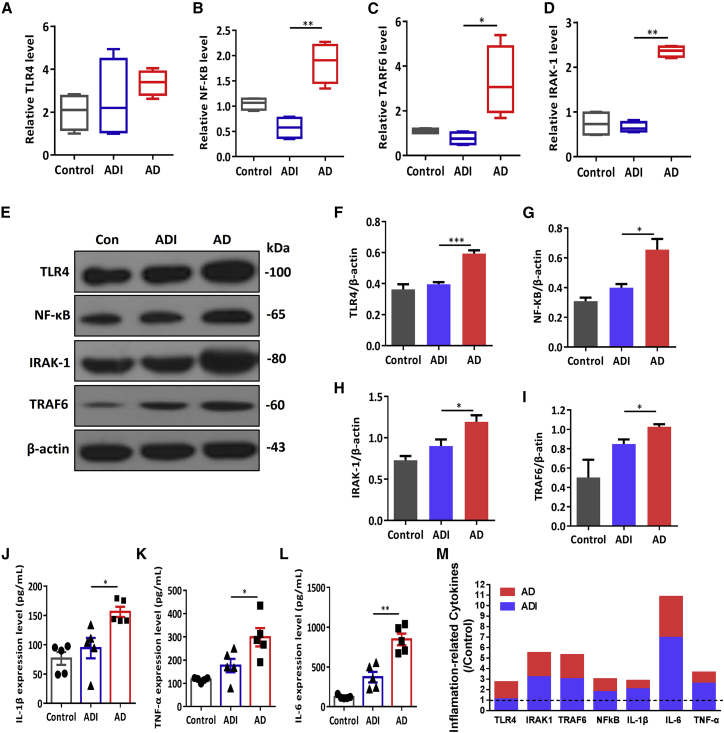
Chronic inflammation enhances hippocampal neuron damage and aggravates the pathophysiological process of AD,13 and miR-146a is reportedly an endogenous regulatory factor in the Toll-like receptor 4 (TLR4) pathway involved in the development of inflammatory diseases.14 Therefore, we further evaluated whether M146AG reduced neuroinflammation in the progression of AD. As shown in Figure 4, the mRNA levels of IRAK1, nuclear factor κB (NF-κB), TRAF6, and TLR4 showed the reduced trend in the hippocampi of M146AG-treated mice compare with the AD groups (Figures 4A–4D). Subsequently, TLR4, IRAK1, TRAF6, and NF-κB protein levels were decreased in hippocampi of the ADI group compared with the AD group (Figure 4E–4I). Meanwhile, the NF-κB protein level was not significantly altered in hippocampi of the AD+NC group (Figures S4A and S4D). Moreover, as shown in Figures 4J–4L, IL-1β, TNF-α, and IL-6 levels were also decreased in hippocampi of the ADI group. In summary, these results indicated that M146AG treatment inhibited the expression of the TLR4 signaling pathway and its related inflammatory genes IRAK1, TRAF6, and NF-κB, reducing the release of inflammatory factors, IL-1β, TNF-α, and IL-6 levels, ultimately relieving the progression of AD neuroinflammation (Figure 4M).
Figure 4.
M146AG Relieved the Neuronflammation in Hippocampi of 12-Month-Old APP/PS1 Mice
(A–D) The relative mRNA levels of TLR4 (A), NF-κB (B), TRAF6 (C), and IRAK1 (D) were detected in hippocampi of AD, ADI, and control groups by the qRT-PCR; GAPDH was used as the control. (E–I) The protein levels of TLR4 (F), IRAK1 (G), NF-κB (H), and TRAF6 (I) in hippocampi of AD, ADI, and control groups were detected by western blotting analysis (E); β-actin was used as the control. (J–L) The expression of IL-1β (J), TNF-α (K), and IL-6 (L) in hippocampi of AD, ADI, and control groups was detected by ELISAs. (M) Graphic shows the changes of inflammatory-related factors in the hippocampi of AD mice under the interference of M146AG. All data are presented as the mean ± SEM. At least three independent experiments were performed (n = 5 mice per group). *p < 0.05; **p < 0.01; ***p < 0.001.
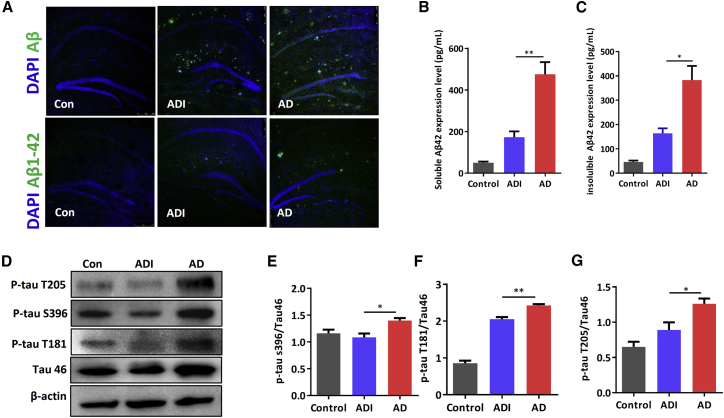
M146AG Reduced Aβ Levels and Tau Hyperphosphorylation in APP/PS1 Mice
The classical pathological features of AD, Aβ, and Tau phosphorylation were also investigated to evaluate the effect of M146AG administration on the pathogenetic pathway of AD. As shown in Figure 5A, the total Aβ and Aβ42 levels were lower in the hippocampi of ADI groups than those of the AD groups. Additionally, the levels of soluble Aβ42 and insoluble Aβ42 were significantly decreased in ADI groups compared with those in the AD groups (Figures 5B and 5C). With regard to Tau, the levels of P-tau S396, P-tau T181, and P-tau T205 were significantly decreased in ADI groups compared with the 12-month-old APP/PS1 mice (Figures 5D–5G). These data demonstrated that M146AG intervention alleviated the formation and deposition of Aβ and the hyperphosphorylation of Tau in APP/PS1 mice, which revealed a significant alleviation of pathological progression in APP/PS1 mice.
Figure 5.
M146AG Reduced Aβ Level and Tau Phosphorylation in 12-Month-Old APP/PS1 Mice
(A) Representative images of total Aβ and Aβ42 deposition levels in hippocampi of 12-month-old mice were determined from Aβ and Aβ42 immunostaining (in green); the images were taken at ×10 magnification. (B) The expression level of soluble Aβ42 was detected in hippocampi of AD, ADI, and control groups by ELISAs. (C) The expression level of insoluble Aβ42 was detected in hippocampi of AD, ADI, and control groups by ELISAs. (D–G) The expression levels of P-tau S396 (E), P-tau T181 (F), and P-tau T205 (G) in hippocampi of AD, ADI, and control groups were detected by western blotting (D); β-actin was used as the control. All data are presented as the mean ± SEM. At least three independent experiments were performed (n = 4 mice per group). *p < 0.05; **p < 0.01.
M146AG Alleviated the Progression of AD through Multiple Pathways
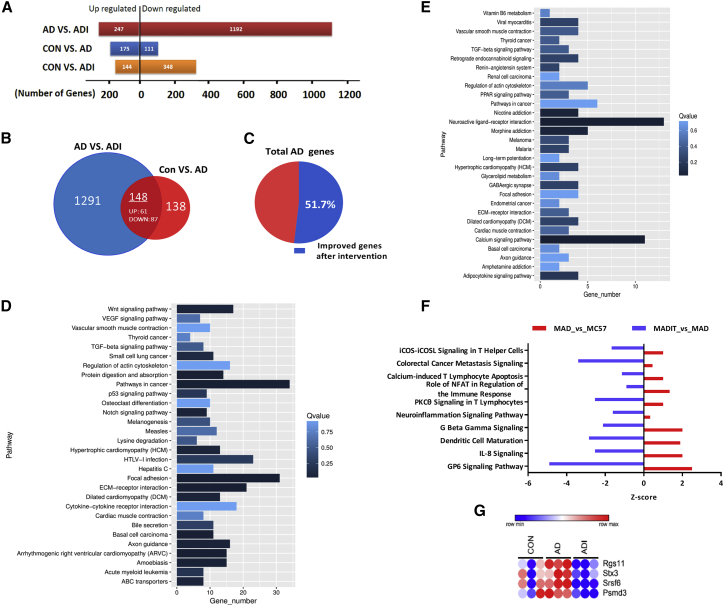
Based on the above results, we concluded that administration of miR-146a alleviated the cognitive deterioration and pathological process of AD and potentially by affecting multiple pathogenic pathways of AD. To further comprehensively elucidate the possible gene-regulated pathways involved in the protective effect of miR-146a on the development of AD, we performed a genome-wide transcriptional microarray analysis of the hippocampi of miR-146a-administered APP/PS mice (ADI), APP/PS1 mice (AD), and C57 (control) mice groups. As shown in Figure 6A, 247 upregulated genes and 1,192 downregulated genes were identified in hippocampi of ADI groups compared to the AD groups, suggesting that M146AG significantly influenced these genes at the transcriptional level. Notably, the number of genes changed at the transcriptional level in the miR-146a group was greater than that in the AD group, especially for the downregulated genes, which suggested a negative role in the extensive regulation of miR-146a (Figure 6A).
Figure 6.
Transcriptional Signatures in hippocampi of miR-146a-Administered APP/PS Mice, APP/PS1 Mice, and C57 Mice
(A) Number of mice significantly upregulated or downregulated in all three pairwise comparisons. (B) Venn diagram represents the numbers of differentially expressed genes from miR-146a-administered APP/PS mice (ADI) versus control APP/PS mice (AD) and from APP/PS mice (AD) versus control C57 mice (Con), and their overlap. (C) Pie chart represents the percentage of alleviated genes after M146AG administration to the number of all AD pathogenic genes. (D and E) Statistics of pathway enrichment in all three pairwise comparisons: (D) pathway enrichment upregulated in AD mice and downregulated in ADI mice; (E) pathway enrichment downregulated in hippocampi of AD mice and upregulated in hippocampi of ADI mice. (F) The significant enrichment of the differentially expressed genes in the IPA-based classical pathway was compared among the three groups in pairs. (G) Heatmap of the miR-146a potential targets (n = 3 mice per group). One-way ANOVA, p < 0.05.
Next, we analyzed the pathways of altered genes between the AD group and the M146AG intervention group to determine which major pathophysiological pathways of AD were improved by M146AG. In a comparison of the differential pathways in the hippocampal area between the 12-month-old APP/PS1 mice and control mice, we observed that besides the inflammation pathways, more than 22 pathways were significantly reversed or weakened under the M146AG intervention (Figure S6), suggesting that the therapeutic effect of M146AG on AD was a multichannel effect. To further identify which genes play important roles in the M146AG intervention in AD, we analyzed the genes that were both related to the progression of AD and reversed after M146AG administration in the hippocampi of each group. Finally, 148 genes with 61 upregulated genes and 87 downregulated genes were identified (Figure 6B; Figure S7). The number of alleviated genes after M146AG administration accounted for 51.7% of the number of all AD pathogenic genes (Figure 6C). Moreover, we analyzed the potential pathways based on the screened gene and found that these gene-related pathways play different roles in the pathogenic pathways of AD. The top five biological pathways with the downregulated genes were extracellular matrix (ECM)-receptor interaction, focal adhesion, axon guidance, the notch signaling pathway, and the wnt signaling pathway (Figure 6D), and the top five biological pathways with the upregulated genes were the calcium signaling pathway, neuroactive ligand-receptor interaction, nicotine addiction, morphine addiction, and the adipocytokine signaling pathway (Figure 6E). Furthermore, Ingenuity Pathway Analysis (IPA) was also used to label the signal pathway under the M146AG intervention in AD, and the results showed that 24 pathways in the pathogenic pathway of AD were reversed or alleviated under the M146AG intervention (Figure S8), including the top 5 neurological pathways, namely the GP6 signaling pathway, the neuroinflammation signaling pathway, the neuroprotective role of thimet oligopeptidase 1 (THOP1) in AD, dendritic cell maturation, and axonal guidance signaling (Figure 6F). Combined with the above analysis, these results indicate that M146AG may delay the development of AD through diverse pathways, and the effect of miR-146a on the pathological pathway may involve multiple targets.
To further identify genes playing important roles in the M146AG intervention of AD, we further compared the downregulated genes after M146AG administration and the predicted database of the miR-146a potential targets. Finally, four genes, Srsf6, Stx3, Psmd3, and Rgs11, were identified (Figure 6G); these genes both have potential binding sites in the 3′ UTR for miR-146a and were also downregulated in the hippocampi of the AD mouse model after M146AG administration in vivo. Thus, these genes may be potential target genes and new pathological factors contributing to AD.
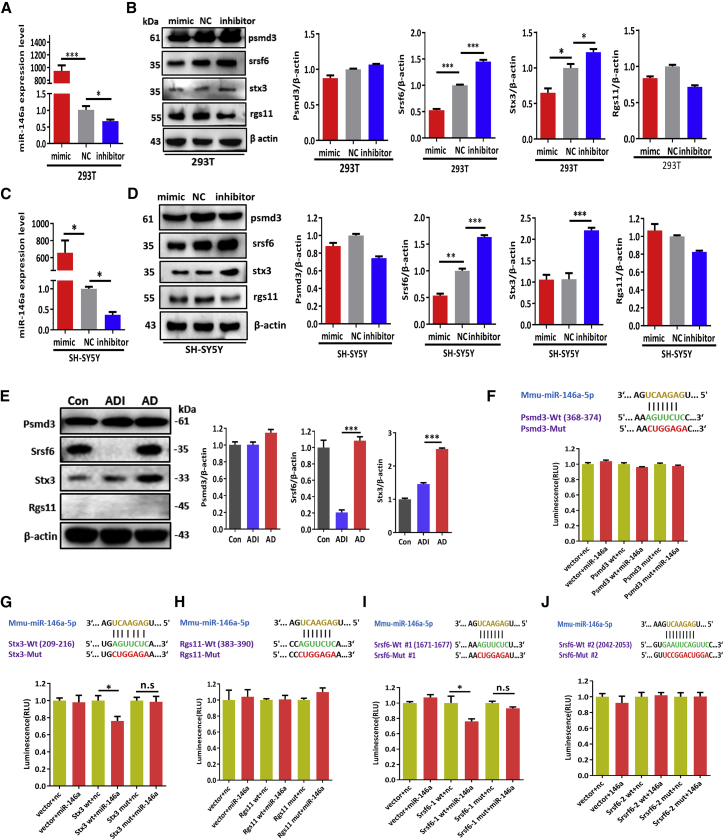
Srsf6 Was a Target of miR-146a and May Played a Potential Role in the Pathophysiological Process of AD
To determine whether the screened genes are targets of miR-146a, the protein levels of these four genes by the miR-146a intervention were further validated in 293T cell and SH-SY5Y cell lines. The results showed that Srsf6 protein levels were negatively regulated by miR-146a in both 293T cells and SH-SY5Y cells, whereas Stx3, Psmd3, and Rgs11 protein levels were not well correlated with miR-146a in a negative manner in the both cell lines in vitro (Figures 7A–7D). We further validated the protein levels of these four genes in the hippocampi of the AD, ADI, and control groups. As shown in Figure 7E, the protein levels of Srsf6 and Stx3 were decreased in the ADI group compared with the AD group, although no significant difference was observed in the Psmd3 protein level, and the protein level of Rgs11 was not detected by western blotting analysis. Furthermore, Dual-Luciferase reporter assays were performed to confirm whether the screened genes are the miR-146a targets. The results showed that the fluorescence intensity was reduced with the mutation of the “1671–1677” binding region of Srsf6 and the “209–216” binding region of Stx3 under the miR-146a mimics administration in 293T cell lines, respectively, suggesting that miR-146a directly binding to the 3′ UTRs of the Srsf6 gene and Stx3 gene (Figures 7G and 7I), while the 3′ UTRs of Psmd3, Rgs11, and Srsf6-2 were not significantly altered (Figures 7F, 7H, and 7J). Collectively, these results suggest that Srsf6 is the direct target of miR-146a with the binding sites 209–216, significantly reduced in mRNA and protein level in hippocampi of the ADI group, which may thus contribute to the pathophysiological process of AD.
Figure 7.
Srsf6 Was Identified as the Direct Target of miR-146a
(A) 293T cells were transfected with miR-146a mimics, scrambled miRNA (NC), or inhibitor for 24 h, and the relative levels of miR-146a were detected by qRT-PCR. (B) Then the protein levels of Psmd3, Srsf6, Stx3, and Rgs11 in 293T cells were also detected by western blotting analysis; β-actin was used as the control. (C) SH-SY5Y cells were transfected with miR-146a mimics, scrambled miRNA, or inhibitor for 24 h, and the relative levels of miR-146a were detected by qRT-PCR. (D) Then the protein levels of Psmd3, Srsf6, Stx3, and Rgs11 in SH-SY5Y cells were also detected by western blot analysis; β-actin was used as the control. (E) The protein levels of Psmd3, Srsf6, Stx3, and Rgs11 in hippocampi of AD, ADI, and control groups were detected by western blotting analysis; β-actin was used as the control (n = 4). (F–J) Luciferase reporter assays were used to detect the interaction between the putative Psmd3 (F), Stx3 (G), Rgs11 (H), Srsf6-1 (I), and Srsf6-2 (J) binding sites with miR-146a. Relative activity of luciferase constructs bearing either wild-type (wt) or mutant (mut) Psmd3, Srsf6-1, Srsf6-2, or Stx3, Rgs11 3′ UTRs in cells co-transfected with miR-146a mimic is presented as Renilla/firefly luminescence units (RLUs). All data are presented as the mean ± SEM. At least three independent experiments were performed. *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
In the present study, we reported the potential feasibility of using miRNAs administered via the nasal-to-brain pathway as targets in AD treatment. The chemically synthesized miRNA mimics were effectively delivered to the hippocampi through nasal administration. Moreover, our study demonstrated that the intranasal delivery of miR-146a agomir improved behavioral and cognitive dysfunction and relieved the pathophysiological process in the APP/PS1 mice model, suggesting the potentially important role of miR-146a as a new target for AD.
miRNAs, important epigenetic regulators that function as pleiotropic modulators due to their ability to target multiple genes, are involved in a wide range of biological processes and are thus considered to be closely related to diseases.15, 16, 17 miR-146a is well known for its role as an anti-inflammatory mediator of immune cell activation. miR-146a targets various immune inflammation-related genes, including IRAK1, TRAF6, CXCR4, CFH, COX2, ROCK1, EGFR, Notch1, and STAT1,18,19 and it participates in a wide range of physiological processes or diseases related to the immune inflammation response.9,14,20,21 Recently, elevated evidence suggests that the intervention results by targeting miR-146a showed that it was unquestionably a potent inhibitor of neuroinflammation.22, 23, 24, 25, 26 Nevertheless, the understanding of the molecular mechanisms through which miR-146a mediates its various biological activities remains poor. With regard to AD, the function and mechanism of miR-146a in the pathogenesis of AD remain unclear. Our previous study showed that a functional SNP of miR-146a could influence the miR-146a level and was associated with AD risk,11 stimulating our interest to explore the function of miR-146a in AD. In the present study, we further confirmed that the high level of miR-146a in the brain could protect against the neuroinflammation and cognitive impairment caused by AD through diverse complex pathways, making it a potential new miRNA target for AD treatment.
Several lines of in vivo evidence previously showed miR-146a to be upregulated in brains of AD patients and mice.5,7,8,27 However, the expression level of miR-146a was not consistent with the different pathological processes of diseases or different diseases. For example, the level of miR-146a was downregulated in patients with postpartum psychosis, type 2 diabetes, and acute myeloid leukemia, as well as in patients exhibiting an acute inflammatory response to monosodium urate crystals;28, 29, 30, 31 conversely, it was also upregulated in diseases such as inflammatory arthritis, atherosclerosis, myocardial damage,32 and chronic obstructive pulmonary disease (COPD).32, 33, 34, 35 Similarly, in the nervous system, the upregulation of miR-146a was reported in neurological diseases such as AD,5,7,8,27 epilepsy,36 and multiple sclerosis37, while the downregulation of miR-146a was observed in patients with acute ischemic stroke38 and Parkinson’s disease.39 These irregular results reflect the flexible regulatory role of endogenous miR-146a in different stages of disease or in different diseases. However, the current consistent evidence supports that exogenous intervention of miR-146a or gene knockout/knockdown of this miRNA plays a role in accelerating the inflammatory response and seems to exert no benefits on the pathophysiological process or biological process of dieaseas.40, 41, 42 Therefore, despite that previous evidence showed miR-146a to be upregulated in AD, we infer that the upregulation of miR-146a plays a protective role in the process of AD, so we attempted to evaluate the therapeutic effect on AD by increasing miR-146a expression, and the results confirm our hypothesis that increasing the miR-146a level alleviates AD disease progression.
Notably, in this study, a chemosynthetic miR-146a agomir was administered via the intranasal route to AD model mice as a targeted intervention treatment. Methods allowing effective drug bypass of the BBB have always been a challenge in the field of brain-related disorders.43 The in vivo half-lives of miRNA mimics are limited, and high doses are often required to cross the BBB and reach the brain to obtain a specific concentration.16 The current AD treatment drugs are administered mainly via oral and intravenous routes, and the main disadvantages are low drug concentrations, poor therapeutic efficacy, and major peripheral side effects. Other common strategies, such as surgery and intraventricular injections, are obviously highly invasive and painful for patients. In contrast, the intranasal route provides a direct and noninvasive pathway to bypass the BBB, with increased intracerebral drug concentrations, decreased quantities, and few side effects observed.44 Several proteins and polypeptides, such as BDNF and CNTF, have been reported that can efficiently enter the CNS target location through the intranasal route in a mouse model,45 and it has also been demonstrated that the intranasal administration route could increase the delivery of low-permeable small molecules to the brain in both rats and monkeys,46 and also to be effective in EcoHIV-infected mice and voluntary AD patients.47,48 Although carrier and intrusive issues still need to be addressed, direct nose-to-brain drug delivery offers a potentially efficient strategy for AD treatment.44 Agomir is a specially labeled and chemically modified double-stranded small RNA that modulates the biological function of the target gene by mimicking endogenous miRNA, has a higher affinity for the cell membrane, and is more enriched than the unmodified miRNA mimic, making it particularly suitable for in vivo interference experiments.49 As expected, our data showed that exogenous miRNAs can effectively reach the hippocampi and maintain an effective concentration when administered via nose-to-brain delivery. Moreover, the dosage administered was also much lower than the dose of miRNA agomir administered via the blood.50,51
Chronic neuroinflammation is generally considered to be a downstream factor in the pathology of AD and is closely related to the activation of glial cells.52 However, the clinical and experimental evidence in studies targeting the inflammation-related pathway as a potential therapy for AD remains controversial.53, 54, 55 Whether miR-146a, as an inflammatory response “braker,” has an effect on AD mainly through inflammation-related pathways or through another mechanism should be explored. Our results first confirmed that M146AG inhibited the activation of glia and neuroinflammation in the AD mice model, and the further results of transcriptome analysis revealed that inflammation-related pathways were clearly one of the significant pathways in the alleviated AD under the M146AG intervention. Notably, our evaluation of miR-146a administration in AD treatment showed that miR-146a intervention alleviated the overall pathological process of AD, including neuroinflammation, activation of glial cells, Aβ level, and Tau hyperphosphorylation, and this synthetic effect was also associated with the conclusion drawn from our transcriptome analysis, suggesting that the effect of miR-146a on AD may be complex and involve multiple targets. In the subsequent search for downstream targets of miR-146a, we confirmed that one gene, Srsf6, was targeted by miR-146a, and protein levels were downregulated in the hippocampi of the AD mice model after M146AG administration in vivo, suggesting that Srsf6 may be a new potential target or pathogenic factor in AD treatment. Alternative precursor mRNA splicing is a key mechanism for regulating gene expression in mammals, and Srsf6 has been reported to act as a splicing factor that mainly accumulates in nuclear inclusion bodies and cytoplasm.56 The splicing factor Srsf6 was shown to be altered in the brains of Huntington’s disease patients and mouse models.57 However, to the best of our knowledge, no direct evidence of the association between Srsf6 and AD has been reported, and the functions of Srsf6 in AD are still vague and needed to be elucidated in the future.
Some concerns should be addressed. In this study, although miR-146a has been reported that was elevated in the brain of AD, our data showed that the miR-146a agomir slowed down the pathological process of AD in an AD mice model; however, our results did not infer that the inhibition of miRNA-146a was not beneficial to AD. There may be different intervention methods for administration, the time point of intervention, dosage, and different animal models, which may all lead to different results; besides, the mechanism of miR-146a in AD is far from clear. Thus, only when we truly understand the dynamic change and the molecular mechanism of miRNA-146a in the AD process can we better promote its potential as a target, and more in-depth research is needed in the future. Notably, the number of downregulated genes following miR-146a intervention was much higher than that of upregulated genes in the pathological process of AD compared with those of AD mice, which indicated the possible negative regulatory mechanism of miR-146a and also the potential side effects of miR-146a. In addition, through transcriptome screening, bioinformatics analysis, and in vitro validation, we showed that the miR-146a target gene Srsf6 may be a potential target or that it played a role in AD progression. However, the functions of Srsf6 in AD are still vague and need to be elucidated, and these results do not suggest that the effects of miR-146a on AD are facilitated by only or mainly this target. The mechanism by which miR-146a functions in AD needs to be further explored.
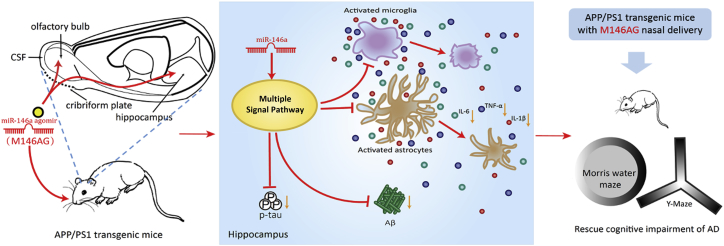
In conclusion, our study demonstrated that the intranasal delivery of miR-146a agomir improves behavioral and cognitive dysfunction in an AD mice model and provides a profile of miR-146a interventions in the AD pathological pathway, identifying one gene, Srsf6, which may play a role in the progression of AD, and our data also support that the nasal-to-brain pathway is efficient and operable for the administration of miRNAs to treat brain diseases (Figure 8). These results provide some new clues for the treatment of AD with new target and future drug delivery.
Figure 8.
M146AG Rescued the Pathological Process and Cognitive Impairment through Nasal Administration in an AD Mouse Model
Materials and Methods
Animals
Wild-type (WT) C57BL/6J and B6C3-Tg (APPswe, PSEN1dE9) male mice were purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China). The mice were housed under specific pathogen-free conditions in a controlled animal facility (23 ± 2°C, 60 ± 5% relative humidity) with a standard 12-h light/12-h dark cycle and allowed free access to rodent chow and water. APP/PS1 mice were housed in individual cages, and WT C57BL/6 mice were housed as four or five mice per cage. These included 12-month-old animals, and each group had four or more animals. All studies involving mice were performed in accordance with guidelines established by the Institutional Animal Care and Use Committee of Guangdong Medical University, which approved all protocols used in this study.
Dynamic Changes in Relative miR-146a Levels after Intranasal Delivery of M146AG
In total, 70 C57BL/6J mice at 12 months of age were randomly selected and equally divided into 14 groups, 6 of which were used as 30-min groups, and 6 others that were used as 24-h groups; the remaining 2 groups served as controls. One or 2 weeks before the formal experiment, the back of the mouse was grasped with the left hand every day, and its head and abdomen were turned upward. The mouse was fixed in this position for 30 s to 1 min and then administered the treatments in 4-μL drops by pipette.58 Six groups of mice at each time point were administered different doses of M146AG (0.01, 0.05, 0.1, 0.5, 1, and 5 nmol) by nasal instillation (IGE Biotech, Guangzhou, China). According to the manufacturer’s instructions, M146AG was dissolved in 24 μL of RNase-free water and administered by pipette in 4-μL drops (total of six fractions), alternating between each nostril every 2–3 min. Control mice received an equal volume of vehicle. All mice were sacrificed individually for collection of the hippocampi. Then, real-time qPCR was used to quantitate the relative miR-146a levels in the hippocampi, and the dynamic changes in the relative miR-146a levels after intranasal delivery of M146AG were further analyzed and compared with those of the vehicle control.
Immunofluorescence Was Used to Detect the Effectiveness of M146AG to Bypass the BBB
Twenty 12-month-old C57BL/6J mice were selected and randomly divided into four groups to receive 1 nmol of CY5-labeled M146AG (IGE Biotech, Guangzhou, China). The four groups were sacrificed individually at 30 min and 1, 2, and 24 h after intranasal delivery, and another group of five mice were sacrificed immediately after vehicle administration (0 min) as a control. Their brains were removed, incubated in 4°C paraformaldehyde overnight, transferred to 20% sucrose for 24 h, soaked in 30% sucrose for 24 h, embedded with O.C.T compound, and stored at −80°C. Ten-microliter frozen sections in the coronal plane were taken to observe the olfactory bulb and hippocampal fluorescence expression (Leica TCS SP5 confocal microscope, Leica Microsystems, USA).
Treatment of APP/PS1 Mice with M146AG
APP/PS1 mice were randomly selected, and each group had at least eight mice. We administered 1 nmol of M146AG (sequence: 5′-UGAGAACUGAAUUCCAUGGGUU-3′) (IGE Biotech, Guangzhou, China) by pipette in 4-μL drops (total of six fractions), alternating between each nostril every 2–3 min. Control APP/PS1 mice and C57BL/6J mice received an equal volume of vehicle or M146AG NC (sequence: 5′-UCACAACCUCCUAGAAAGAGUAGA-3′) (IGE Biotech, Guangzhou, China). Treatment was performed every other day for a total of 30 days. Then, the mice were trained and tested on the MWM and Y-maze. Finally, all mice were sacrificed individually, and brain tissue samples were taken for subsequent experiments.
MWM Test
The MWM test, which has been widely utilized in the study of AD models, was used to test spatial sense of location and sense of direction (spatial positioning) of the experimental animals as well as their learning and memory abilities. The maze consisted of a circular pool (120 cm in diameter, 50 cm high) that was filled with opaque water (22 ± 3°C), and a circular antholeucin platform (10 cm in diameter, 28 cm high) was placed in one quadrant that was submerged 1 cm below the surface of the water. Each mouse was subjected to four tests on each day of the 5-day place navigation test. In each test, the mouse was placed in the water from the midpoint of a different quadrant edge, allowed to swim for a maximum of 90 s to find the hidden platform, and then allowed to have a 10-s rest period on the platform. When mice failed to find the hidden platform within the allotted time period, they were guided to the platform and were placed on it for 10 s, and escape latency was recorded as 90 s. On the final day of the spatial probe test, the platform was removed, and the swimming time and distance in the quadrant in which the platform had previously been located and the platform-crossing times were recorded for 90 s. The experimental process was recorded and analyzed by the data acquisition system.
Y-Maze
The Y-maze was made of a medical organic board painted black. The maze had three arms that were 30 cm long, 15 cm high, and 8 cm wide and positioned at equal angles (120°). There was a removable partition in the center of each arm of the labyrinth that displayed different geometric shapes as visual markers. The three arms were randomly set as the novel arm, start arm, and other arm. The novel arm was blocked by a partition in the first phase of the experiment and opened in the second phase. The start arm was the arm entered by the mouse where the maze was located. During the entire experiment, the start arm and other arm were open all of the time, and the mice could freely access these arms. Wood chips were laid in the maze, and after each training or test, the wood chips in each arm were mixed to prevent residual odor. Mice were placed at the end of the start arm and allowed to move freely through the maze for 5 min. Arm entry was considered to be completed when the hind paws of the mouse were completely placed in the arm. The camera was placed 1.5 m above the maze, and the entire process was recorded.
Sample Preparation
After the behavioral tests, the mice were anaesthetized with 10% chloral hydrate (0.35 mL/100 g body weight) by intraperitoneal injection. The right heart auricle was cut with small scissors, and the left ventricle was rapidly perfused with saline precooled at 4°C. When the liver turned white, it was replaced with 4°C precooled 4% paraformaldehyde, and the animals were then sacrificed by decapitation. Their brains were removed and divided into two parts along the sagittal plane. One hippocampus was quickly isolated from the hemisphere and stored at −80°C for subsequent biochemical analysis. The other hemisphere was placed in a 4% paraformaldehyde-PBS solution prechilled at 4°C (pH 7.4) overnight to prepare the frozen slices.
Immunohistochemistry
Mouse hemispheres were placed in a 4% paraformaldehyde-PBS solution prechilled at 4°C (pH 7.4) overnight, transferred to 20% sucrose for 24 h, soaked in 30% sucrose for 24 h, embedded in O.C.T compound (Tissue-Tek), and stored at −80°C. Then, the tissues were cut with a Leica cryostat to obtain 25-μm-thick floating coronal sections. After removing the sections from the −80°C freezer, they were placed on a rack and then in a 37°C incubator for 30 min and rinsed three times at room temperature for 5 min each with 1× PBS (pH 7.2–7.4). The sections were blocked by incubation with 0.1% Triton X-100/10% bovine serum for 30 min. The blocking solution covered all brain tissues and prevented the tissue from drying out. Then, the samples were incubated with primary antibodies overnight at 4°C. NC sections without primary antibodies were processed at the same time. The primary antibodies included rabbit anti-GFAP (1:200, CST), rabbit anti-IBA-1 (1:100, Abcam), rabbit anti-β-amyloid (1:100, CST), and rabbit anti-β-amyloid (1-42 specific, 1:1,600, CST). The next morning, the sections were washed in PBS, incubated for 1 h at room temperature with DyLight 488-conjugated goat anti-rabbit IgG (1:200, Abcam), stained with DAPI for 3 min, washed with PBS, and coverslipped. Fluorescence images were captured using a Leica TCS SP5 confocal microscope. For at least three sections per group, 50 astrocytes or microglia were randomly selected from each section, and the cell area and IOD value were measured. Image analysis was performed using Image-Pro Plus 6.0 software (Media Cybernetics, Bethesda, USA).
Cell Culture and Transfection
293T cells were obtained in our laboratory and cultured in DMEM (HyClone, Logan, UT). Human neuroblastoma SH-SY5Y cells were purchased from Shanghai Genechem. These cells were trypsinized and plated in 6-well plates or 12-well plates (Nest Biotechnology). 293T cells and SH-SY5Y cells were cultured in DMEM (HyClone). The media were supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY), 100 U/mL penicillin (HyClone), and 100 mg/mL streptomycin (HyClone). The cells (8.3 × 104 cells/cm2) were maintained in a humidified incubator with 5% CO2 at 37°C, cultured for 24 h until reaching 50%–70% confluence, and then incubated with miR-146a mimics or scrambled miRNA (50 nM final concentration). In addition, the concentration of the locked nucleic acid (LNA) inhibitor was 50 nM. Finally, these cells were incubated with Lipofectamine 2000 diluted in Opti-MEM (Invitrogen). After 6 h of transfection, the medium was removed and replaced with fresh medium, which was composed of DMEM supplemented with 10% FBS and 100 U/mL penicillin/streptomycin. All cells were cultured in a humidified incubator at 5% CO2 and 37°C. After 24 h, the cells were subjected to total RNA extraction for qRT-PCR or were lysed after 48 h to obtain protein samples for western blot analysis.
ELISAs
The levels of TNF-α, IL-1β, IL-6, and Aβ42 in the hippocampal homogenates were individually determined using ELISA kits, including a TNF-α ELISA kit (catalog no. C708, GenStar, Beijing, China), an IL-1β ELISA kit (catalog no. C701, GenStar, Beijing, China), an IL-6 ELISA kit (catalog no. C704, GenStar, Beijing, China), and an Aβ1-42 ELISA kit (catalog no. KMB3441, Invitrogen, USA). All ELISAs were performed according to the manufacturers’ instructions, and the insoluble Aβ42 was treated with hexafluoroethane before the assay. Briefly, the hippocampal homogenates were added to a 96-well ELISA plate and then reacted with appropriate primary antibodies and horseradish peroxidase-conjugated secondary antibodies; 3,3′,5,5′-tetramethylbenzidine (TMB) was used as the substrate, and the results were quantified using an ELISA reader (Bio-Rad, Hercules, USA) at 450 nm. The detectable dose limits of the TNF-α, IL-1β, IL-6, and Aβ42 ELISA kits were 8, 5, 2, and 3 pg/mL, respectively. Averages from two replicate wells were used for each sample. All quantitative analyses were performed by the external standard method, producing a standard curve with a correlation coefficient >0.99.
qRT-PCR Assay
Total RNA was extracted from the hippocampi or cultures by using TRIzol (Invitrogen, CA, USA), according to the manufacturer’s protocol in an RNase-free environment. Equal amounts of total RNA were treated with DNase I and then followed by reverse transcription using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions. qRT-PCR was performed by using the SYBR Green method on a LightCycler 480 sequence detector system (Roche Applied Science, Penzberg, Germany). Each cDNA sample was repeatedly tested with three replicates. The transcription levels of U6 or GAPDH were used as an internal control. The specific primers are listed in Table S1. The relative expression levels were calculated using the 2−ΔΔCT method.
Western Blotting
Hippocampal tissue and cultures were homogenized in radioimmunoprecipitation assay (RIPA) buffer (Solarbio, Beijing, China) and embodied protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). Homogenates were centrifuged at 12,000 × g for 10 min at 4°C. Supernatants were collected, and equal amounts of extracted protein were separated by 6%–12% SDS-PAGE, then transferred onto polyvinylidene fluoride (PVDF) membranes. Nonspecific binding sites were blocked in PBS containing 5% nonfat dried milk for 1 h, and then the membranes were probed with the following primary antibodies diluted in the blocking medium overnight at 4°C: rabbit anti-NF-κB (1:2,000, CST), anti-IRAK1 (1:500, CST), anti-TRAF6 (1:400, Santa Cruz), anti-GFAP (1:2,000, Abcam), anti-Iba-1 (1:500, Abcam), anti-CD68 (1:300, Proteintech), anti-p-tau T205 (1:1,000, Abcam), anti-tau 46 (1:500, CST), anti-p-tau S396 (1:500, Abcam), anti-p-tau T181 (1:500, CST), anti-Srsf6 (1:400, Sigma), anti-Psmd3 (1:1,000, Abcam), anti-Stx3 (1:500, Sigma), anti-Rgs11 (1:100, Santa Cruz), and anti-β-actin (1:5,000, EarthOx). The membranes were washed with PBS containing 0.1% Tween 20, followed by incubation with the appropriate species-specific HRP-conjugated secondary antibody at 1:5,000 for 1 h at room temperature. Images were visualized on an Azure Biosystems C600 imaging system, and relative band intensity was normalized relative to β-actin using ImageJ software (NIH).
Transcriptome Assay
Total RNAs purified from 12-month-old mice were analyzed by spectrophotometry. An optical density (OD)260/OD280 ratio between 1.8 and 2.0 was standard, indicating that the RNA purity was high and met the experimental requirements. The concentration of RNA was greater than 100 ng/μL, and the total volume was greater than 30 μL. The RNA samples were placed into a small enzyme-free 1.5-mL Eppendorf (EP) tube, embedded in dry ice, and sent to Nanjing Vazyme Biotech. RNA sequencing was performed by following the standard protocols provided in the commercial kits, as stated elsewhere. We performed expression analysis to identify single genes, but also to discover differentially activated key cell functions. Differential gene expression was compared using the DESeq2 database (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html). The KEGG pathway database (https://www.kegg.jp) was used for pathway analysis. IPA analysis was conducted using the QIAGEN database (https://www.qiagen.com/cn/shop/analytics-software/ingenuity-pathway-analysis/, version 45868156). An adjusted p value less than 0.05 was considered significant. The TargetScan (http://www.targetscan.org) and miRDB search public database (http://www.mirdb.org) were used for the prediction of the 3′ UTR binding sites of genes with miR-146a.
Validation of miR-146a Targets by Luciferase Reporter Assays
The predicted target sequences of Srsf6-1, Srsf6-2, Rgs11, Psmd3, and Stx3 were cloned into the psiCHECK2 plasmid downstream of Renilla luciferase using XhoI and NotI (Shanghai Genechem). Briefly, 293T cells were split 1 day before the transfection to achieve 60%–70% confluence on the day of transfection. One thousand nanograms of each constructs was cotransfected with either miRNA mimics (100 nM final concentration) or scrambled miRNA (100 nM). Then, these cells were treated with Lipofectamine 2000 diluted in Opti-MEM (Invitrogen). After 6 h of transfection, the medium was removed and replaced with fresh medium, which was composed of DMEM (HyClone) supplemented with 10% FBS and 100 U/ml penicillin/streptomycin. All cells were cultured in a humidified incubator at 5% CO2 and 37°C. At 24 h after transfection, cell lysates were processed for Dual-Luciferase assays using the Dual-Glo luciferase assay kit (Promega, USA) and detected with a Mithras LB 940 multilabel reader (Berthold Technologies, Bad Wildbad, Germany).
Statistical Analysis
All experiments were performed with side-by-side controls and in random order and were replicated at least three times. All data are presented as mean SEM. A one-way ANOVA and t test were used to analyze the differences between the values of the samples in each group. The experimental results were all statistically analyzed and plotted using GraphPad Prism 6.0 software. When p < 0.05, the difference between the two groups was considered statistically significant. In the figures, p values are indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
Author Contributions
L.C., B.Z., and C.W conceived and designed the experiments. H.M., W.F., Y.W., Y.C., X.L, X.C, and F.C. performed the experiments. H.M., W.F., F.C., J.Y., and P.T. analyzed data and contributed to the bioinformatics analysis. L.C., H.M., and W.F. contributed to transcriptome analysis. H.M., L.C., W.F., and Y.W. wrote the original draft. T.Z., H.C., X.C, and T.H. provided methodology aid and project administration. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81671181 and 81771161), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2017), and Guangdong Province: Special Support Plan for High-Level Talents.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.002.
Contributor Information
Conghua Wan, Email: wanchh1964@qq.com.
Bin Zhao, Email: binzhaoe@163.com.
Lili Cui, Email: cuilili@gdmu.edu.cn.
Supplemental Information
References
- 1.Huang Y., Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miya Shaik M., Tamargo I.A., Abubakar M.B., Kamal M.A., Greig N.H., Gan S.H. The Role of microRNAs in Alzheimer’s disease and their therapeutic potentials. Genes (Basel) 2018;9:E174. doi: 10.3390/genes9040174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulland T.K., Colonna M. TREM2—a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 2018;14:667–675. doi: 10.1038/s41582-018-0072-1. [DOI] [PubMed] [Google Scholar]
- 4.Thei L., Imm J., Kaisis E., Dallas M.L., Kerrigan T.L. Microglia in Alzheimer’s disease: a role for ion channels. Front. Neurosci. 2018;12:676. doi: 10.3389/fnins.2018.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta P., Bhattacharjee S., Sharma A.R., Sharma G., Lee S.S., Chakraborty C. miRNAs in Alzheimer disease—a therapeutic perspective. Curr. Alzheimer Res. 2017;14:1198–1206. doi: 10.2174/1567205014666170829101016. [DOI] [PubMed] [Google Scholar]
- 6.Rusca N., Monticelli S. miR-146a in immunity and disease. Mol. Biol. Int. 2011;2011:437301. doi: 10.4061/2011/437301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayers D., Scerri C. Non-coding RNA influences in dementia. Noncoding RNA Res. 2018;3:188–194. doi: 10.1016/j.ncrna.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethi P., Lukiw W.J. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 9.Magilnick N., Reyes E.Y., Wang W.L., Vonderfecht S.L., Gohda J., Inoue J.I., Boldin M.P. miR-146a-Traf6 regulatory axis controls autoimmunity and myelopoiesis, but is dispensable for hematopoietic stem cell homeostasis and tumor suppression. Proc. Natl. Acad. Sci. USA. 2017;114:E7140–E7149. doi: 10.1073/pnas.1706833114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labbaye C., Testa U. The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J. Hematol. Oncol. 2012;5:13. doi: 10.1186/1756-8722-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui L., Li Y., Ma G., Wang Y., Cai Y., Liu S., Chen Y., Li J., Xie Y., Liu G. A functional polymorphism in the promoter region of microRNA-146a is associated with the risk of Alzheimer disease and the rate of cognitive decline in patients. PLoS ONE. 2014;9:e89019. doi: 10.1371/journal.pone.0089019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter S.F., Herholz K., Rosa-Neto P., Pellerin L., Nordberg A., Zimmer E.R. Astrocyte biomarkers in Alzheimer’s disease. Trends Mol. Med. 2019;25:77–95. doi: 10.1016/j.molmed.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paterson M.R., Kriegel A.J. miR-146a/b: a family with shared seeds and different roots. Physiol. Genomics. 2017;49:243–252. doi: 10.1152/physiolgenomics.00133.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maciotta S., Meregalli M., Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front. Cell. Neurosci. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olde Loohuis N.F., Kos A., Martens G.J., Van Bokhoven H., Nadif Kasri N., Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell. Mol. Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 18.Mastroianni J., Stickel N., Andrlova H., Hanke K., Melchinger W., Duquesne S., Schmidt D., Falk M., Andrieux G., Pfeifer D. miR-146a controls immune response in the melanoma microenvironment. Cancer Res. 2019;79:183–195. doi: 10.1158/0008-5472.CAN-18-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L.L., Huang Y., Wang G., Chen S.D. The potential role of microRNA-146 in Alzheimer’s disease: biomarker or therapeutic target? Med. Hypotheses. 2012;78:398–401. doi: 10.1016/j.mehy.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Mann M., Mehta A., Zhao J.L., Lee K., Marinov G.K., Garcia-Flores Y., Lu L.F., Rudensky A.Y., Baltimore D. An NF-κB-microRNA regulatory network tunes macrophage inflammatory responses. Nat. Commun. 2017;8:851. doi: 10.1038/s41467-017-00972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su W., Aloi M.S., Garden G.A. MicroRNAs mediating CNS inflammation: small regulators with powerful potential. Brain Behav. Immun. 2016;52:1–8. doi: 10.1016/j.bbi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Z.F., Sun Z.W., Zhang Y., Wang S., Yu Q.G., Wu Z.B. Regulatory effects of miR-146a/b on the function of endothelial progenitor cells in acute ischemic stroke in mice. Kaohsiung J. Med. Sci. 2017;33:369–378. doi: 10.1016/j.kjms.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Zhang Z.G., Lu M., Zhang Y., Shang X., Chopp M. miR-146a promotes oligodendrocyte progenitor cell differentiation and enhances remyelination in a model of experimental autoimmune encephalomyelitis. Neurobiol. Dis. 2019;125:154–162. doi: 10.1016/j.nbd.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 24.Wu D., Cerutti C., Lopez-Ramirez M.A., Pryce G., King-Robson J., Simpson J.E., van der Pol S.M., Hirst M.C., de Vries H.E., Sharrack B. Brain endothelial miR-146a negatively modulates T-cell adhesion through repressing multiple targets to inhibit NF-κB activation. J. Cereb. Blood Flow Metab. 2015;35:412–423. doi: 10.1038/jcbfm.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao H., Zhao J., Liu T., Cai Y., Zhou X., Xing H., Wang Y., Yin M., Zhong W., Liu Z. Intranasal delivery of miR-146a mimics delayed seizure onset in the lithium-pilocarpine mouse model. Mediators Inflamm. 2017;2017:6512620. doi: 10.1155/2017/6512620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L., Dong R., Lu Y., Zhou Y., Li K., Zhang Z., Peng M. MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain Behav. Immun. 2019;78:188–201. doi: 10.1016/j.bbi.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Müller M., Kuiperij H.B., Claassen J.A., Küsters B., Verbeek M.M. MicroRNAs in Alzheimer’s disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol. Aging. 2014;35:152–158. doi: 10.1016/j.neurobiolaging.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Weigelt K., Bergink V., Burgerhout K.M., Pescatori M., Wijkhuijs A., Drexhage H.A. Down-regulation of inflammation-protective microRNAs 146a and 212 in monocytes of patients with postpartum psychosis. Brain Behav. Immun. 2013;29:147–155. doi: 10.1016/j.bbi.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 29.Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M., Mayr A., Weger S., Oberhollenzer F., Bonora E. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 30.Mohr S., Doebele C., Comoglio F., Berg T., Beck J., Bohnenberger H., Alexe G., Corso J., Ströbel P., Wachter A. Hoxa9 and Meis1 cooperatively induce addiction to Syk signaling by suppressing miR-146a in acute myeloid leukemia. Cancer Cell. 2017;31:549–562.e11. doi: 10.1016/j.ccell.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalbeth N., Pool B., Shaw O.M., Harper J.L., Tan P., Franklin C., House M.E., Cornish J., Naot D. Role of miR-146a in regulation of the acute inflammatory response to monosodium urate crystals. Ann. Rheum. Dis. 2015;74:786–790. doi: 10.1136/annrheumdis-2014-205409. [DOI] [PubMed] [Google Scholar]
- 32.Shu L., Zhang W., Huang G., Huang C., Zhu X., Su G., Xu J. Troxerutin attenuates myocardial cell apoptosis following myocardial ischemia-reperfusion injury through inhibition of miR-146a-5p expression. J. Cell. Physiol. 2019;234:9274–9282. doi: 10.1002/jcp.27607. [DOI] [PubMed] [Google Scholar]
- 33.Ammari M., Presumey J., Ponsolles C., Roussignol G., Roubert C., Escriou V., Toupet K., Mausset-Bonnefont A.L., Cren M., Robin M. Delivery of miR-146a to Ly6Chigh monocytes inhibits pathogenic bone erosion in inflammatory arthritis. Theranostics. 2018;8:5972–5985. doi: 10.7150/thno.29313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raitoharju E., Lyytikäinen L.P., Levula M., Oksala N., Mennander A., Tarkka M., Klopp N., Illig T., Kähönen M., Karhunen P.J. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219:211–217. doi: 10.1016/j.atherosclerosis.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 35.Tsai M.-J., Tsai Y.-C., Chang W.-A., Lin Y.-S., Tsai P.-H., Sheu C.-C., Kuo P.L., Hsu Y.L. Deducting microRNA-mediated changes common in bronchial epithelial cells of asthma and chronic obstructive pulmonary disease—a next-generation sequencing-guided bioinformatic approach. Int. J. Mol. Sci. 2019;20:E553. doi: 10.3390/ijms20030553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omran A., Peng J., Zhang C., Xiang Q.L., Xue J., Gan N., Kong H., Yin F. Interleukin-1β and microRNA-146a in an immature rat model and children with mesial temporal lobe epilepsy. Epilepsia. 2012;53:1215–1224. doi: 10.1111/j.1528-1167.2012.03540.x. [DOI] [PubMed] [Google Scholar]
- 37.Ma X., Zhou J., Zhong Y., Jiang L., Mu P., Li Y., Singh N., Nagarkatti M., Nagarkatti P. Expression, regulation and function of microRNAs in multiple sclerosis. Int. J. Med. Sci. 2014;11:810–818. doi: 10.7150/ijms.8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S.H., Chen L., Pang X.M., Su S.Y., Zhou X., Chen C.Y., Huang L.G., Li J.P., Liu J.L. Decreased miR-146a expression in acute ischemic stroke directly targets the Fbxl10 mRNA and is involved in modulating apoptosis. Neurochem. Int. 2017;107:156–167. doi: 10.1016/j.neuint.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Caggiu E., Paulus K., Mameli G., Arru G., Sechi G.P., Sechi L.A. Differential expression of miRNA 155 and miRNA 146a in Parkinson’s disease patients. eNeurologicalSci. 2018;13:1–4. doi: 10.1016/j.ensci.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan Y.J., Li J., Yang X., Du S., Ding J., Gao Y., Zhang Y., Yang K., Chen Q. Evidence that miR-146a attenuates aging- and trauma-induced osteoarthritis by inhibiting Notch1, IL-6, and IL-1 mediated catabolism. Aging Cell. 2018;17:e12752. doi: 10.1111/acel.12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B., Wang X., Choi I.Y., Wang Y.C., Liu S., Pham A.T., Moon H., Smith D.J., Rao D.S., Boldin M.P., Yang L. miR-146a modulates autoreactive Th17 cell differentiation and regulates organ-specific autoimmunity. J. Clin. Invest. 2017;127:3702–3716. doi: 10.1172/JCI94012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatt K., Lanting L.L., Jia Y., Yadav S., Reddy M.A., Magilnick N., Boldin M., Natarajan R. Anti-inflammatory role of microRNA-146a in the pathogenesis of diabetic nephropathy. J. Am. Soc. Nephrol. 2016;27:2277–2288. doi: 10.1681/ASN.2015010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gump J.M., Dowdy S.F. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol. Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Alexander A., Saraf S. Nose-to-brain drug delivery approach: a key to easily accessing the brain for the treatment of Alzheimer’s disease. Neural Regen. Res. 2018;13:2102–2104. doi: 10.4103/1673-5374.241458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boudreau R.L., Rodríguez-Lebrón E., Davidson B.L. RNAi medicine for the brain: progresses and challenges. Hum. Mol. Genet. 2011;20(R1):R21–R27. doi: 10.1093/hmg/ddr137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwasaki S., Yamamoto S., Sano N., Tohyama K., Kosugi Y., Furuta A., Hamada T., Igari T., Fujioka Y., Hirabayashi H., Amano N. Direct drug delivery of low-permeable compounds to the central nervous system via intranasal administration in rats and monkeys. Pharm. Res. 2019;36:76. doi: 10.1007/s11095-019-2613-8. [DOI] [PubMed] [Google Scholar]
- 47.Kim B.H., Kelschenbach J., Borjabad A., Hadas E., He H., Potash M.J., Nedelcovych M.T., Rais R., Haughey N.J., McArthur J.C. Intranasal insulin therapy reverses hippocampal dendritic injury and cognitive impairment in a model of HIV-associated neurocognitive disorders in EcoHIV-infected mice. AIDS. 2019;33:973–984. doi: 10.1097/QAD.0000000000002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Craft S., Baker L.D., Montine T.J., Minoshima S., Watson G.S., Claxton A., Arbuckle M., Callaghan M., Tsai E., Plymate S.R. Intranasal insulin therapy for Alzheimer disease and amnestic mild cognitive impairment: a pilot clinical trial. Arch. Neurol. 2012;69:29–38. doi: 10.1001/archneurol.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X., Gu S., Chen B.F., Shen W.L., Yin Z., Xu G.W., Hu J.J., Zhu T., Li G., Wan C. Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway. Biomaterials. 2015;53:239–250. doi: 10.1016/j.biomaterials.2015.02.071. [DOI] [PubMed] [Google Scholar]
- 50.Kim J., Im H.I., Moon C. Intravenous morphine self-administration alters accumbal microRNA profiles in the mouse brain. Neural Regen. Res. 2018;13:77–85. doi: 10.4103/1673-5374.224374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urgard E., Lorents A., Klaas M., Padari K., Viil J., Runnel T., Langel K., Kingo K., Tkaczyk E., Langel Ü. Pre-administration of PepFect6-microRNA-146a nanocomplexes inhibits inflammatory responses in keratinocytes and in a mouse model of irritant contact dermatitis. J. Control. Release. 2016;235:195–204. doi: 10.1016/j.jconrel.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Nordengen K., Kirsebom B.E., Henjum K., Selnes P., Gísladóttir B., Wettergreen M., Torsetnes S.B., Grøntvedt G.R., Waterloo K.K., Aarsland D. Glial activation and inflammation along the Alzheimer’s disease continuum. J. Neuroinflammation. 2019;16:46. doi: 10.1186/s12974-019-1399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nizami S., Hall-Roberts H., Warrier S., Cowley S.A., Di Daniel E. Microglial inflammation and phagocytosis in Alzheimer’s disease: potential therapeutic targets. Br. J. Pharmacol. 2019;176:3515–3532. doi: 10.1111/bph.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McQuade A., Blurton-Jones M. Microglia in Alzheimer’s disease: exploring how genetics and phenotype influence risk. J. Mol. Biol. 2019;431:1805–1817. doi: 10.1016/j.jmb.2019.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaur D., Sharma V., Deshmukh R. Activation of microglia and astrocytes: a roadway to neuroinflammation and Alzheimer’s disease. Inflammopharmacology. 2019;27:663–677. doi: 10.1007/s10787-019-00580-x. [DOI] [PubMed] [Google Scholar]
- 56.Fernández-Nogales M., Santos-Galindo M., Hernández I.H., Cabrera J.R., Lucas J.J. Faulty splicing and cytoskeleton abnormalities in Huntington’s disease. Brain Pathol. 2016;26:772–778. doi: 10.1111/bpa.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernández-Nogales M., Cabrera J.R., Santos-Galindo M., Hoozemans J.J., Ferrer I., Rozemuller A.J., Hernández F., Avila J., Lucas J.J. Huntington’s disease is a four-repeat tauopathy with tau nuclear rods. Nat. Med. 2014;20:881–885. doi: 10.1038/nm.3617. [DOI] [PubMed] [Google Scholar]
- 58.Hanson L.R., Fine J.M., Svitak A.L., Faltesek K.A. Intranasal administration of CNS therapeutics to awake mice. J. Vis. Exp. 2013;(74):e4440. doi: 10.3791/4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.