Abstract
O-GlcNAc is an abundant post-translational modification found on nuclear and cytoplasmic proteins in all metazoans. This modification regulates a wide variety of cellular processes, and elevated O-GlcNAc levels have been implicated in cancer progression. A single essential enzyme, O-GlcNAc transferase (OGT), is responsible for all nucleocytoplasmic O-GlcNAcylation. Understanding how this enzyme chooses its substrates is critical for understanding, and potentially manipulating, its functions. Here we use protein microarray technology and proteome-wide glycosylation profiling to show that conserved aspartate residues in the tetratricopeptide repeat (TPR) lumen of OGT drive substrate selection. Changing these residues to alanines alters substrate selectivity and unexpectedly increases rates of protein glycosylation. Our findings support a model where sites of glycosylation for many OGT substrates are determined by TPR domain contacts to substrate side chains five to fifteen residues C-terminal to the glycosite. In addition to guiding design of inhibitors that target OGT’s TPR domain, this information will inform efforts to engineer substrates to explore biological functions.
Graphical Abstract
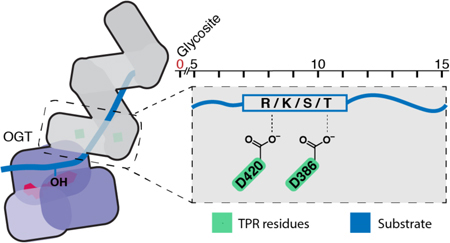
O-GlcNAc transferase (OGT), a protein found in all metazoans, is a nutrient- and stress-responsive glycosyltransferase that regulates the functions of nuclear and cytoplasmic proteins by catalyzing the transfer of N-acetylglucosamine (GlcNAc) to serine and threonine side chains.1 O-GlcNAc modifications can alter protein localization, activity, stability, and protein-protein interactions.2 OGT activity is required to maintain cellular homeostasis, but chronically elevated protein O-GlcNAc levels have been linked to insulin resistance, diabetic complications, and cancer.3 To better understand OGT’s function and potentially develop inhibitors that selectively disrupt subsets of OGT-substrate interactions, it is critical to know how OGT chooses its substrates.
In addition to its catalytic glycosyltransferase domain, OGT has a tetratricopeptide repeat (TPR) domain that is necessary for protein glycosylation.1,4 It has been speculated that adaptor proteins that bind to the TPR domain drive OGT substrate selection.5 However, information on how changes to the TPRs affect substrate selectivity is surprisingly limited. We previously obtained a structure of human OGT complexed with a peptide substrate that binds in the TPR lumen.6 The structure showed that this substrate is anchored in the lumen through bidentate contacts from the side chains of a highly conserved ladder of asparagines that extends the length of the TPR domain (Figure 1A). We asked whether these asparagines were important for substrate binding and found that mutating them led to decreased glycosylation of most OGT substrates even through the OGT active site was fully functional.7 These studies identified a shared mode of substrate binding but did not provide insight into how selectivity is achieved because the asparagines only make amide backbone contacts. Here we report the first functional evidence that residues in the TPR lumen drive OGT substrate selectivity.
Figure 1.
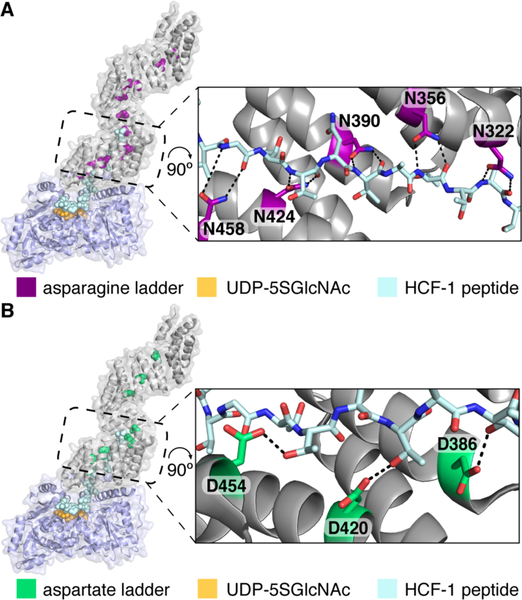
Two conserved amino acid ladders line the OGT TPR lumen. A) Composite structure of human OGT complexed with a 26 residue peptide (light blue) was built by aligning overlapping residues from two structures (PDB codes 4N3B and 1W3B). Asparagine residues form a ladder, and the expanded view shows that five sequential asparagines closest to the active site make bidentate contacts to the bound peptide backbone. B) Composite structure as in A, but with TPR aspartates highlighted. Three sequential aspartates contact threonine sides chains of the bound peptide.
We observed that the TPR domain of OGT contains a ladder of conserved aspartates that, like the asparagine ladder, extends the full length of the superhelix (Figure 1B, Table S1). In the OGT:peptide structure, three aspartates proximal to the active site, D386, D420, and D454, contact threonine side chains in the peptide (Figure 1B), suggesting they play a role in substrate selectivity.
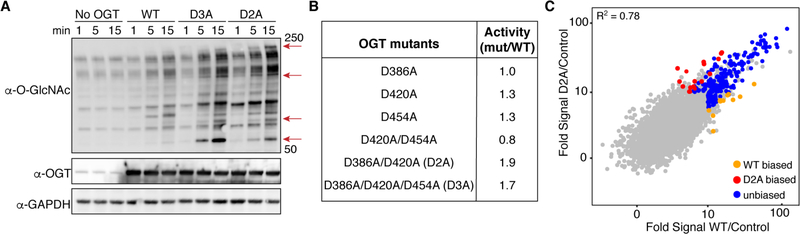
To test the importance of these aspartates, we made mutants in which some or all were changed to alanine (Figure 2). Kinetic analysis of the two mutants showed that these changes did not affect glycosylation of a model peptide that only binds in the OGT active site (Figure S2A). Therefore, OGT’s catalytic machinery was unaffected by the TPR mutations. We next evaluated the activity of each mutant using HeLa cell extracts, which allowed us to assess how the mutations affected protein glycosylation on a proteome-wide scale (Figure 2, S2, S3). Adding OGTWT to the extracts resulted in a time-dependent increase in O-GlcNAcylation (Figure 2A). Most of the mutants showed similar glycosylation activity to OGTWT (Figure 2B). However, the triple mutant and the D386A/D420A mutant (called hereafter D2A) showed increased glycosylation activity (Figure 2A, S4). Moreover, the appearance of new O-GlcNAc bands suggested altered selectivity. Taken together, these experiments showed that the aspartates in the TPR lumen of OGT influence substrate recognition.
Figure 2.
Aspartate residues in the TPR lumen affect glycosylation profiles. A) Glycosylation of HeLa extracts by recombinant OGT variants shows increased O-GlcNAcylation by D2A and D3A. Red arrows highlight bands prominent only for D2A and D3A. B) Table of aspartate to alanine mutants showing relative glycosylation activity in HeLa cell extracts compared to OGTWT. See Methods. C) Scatterplot showing glycosylation signal for microarray proteins treated with OGTWT or OGTD2A. Glycosylation hits had > 10-fold signal above control at < 5% FDR for at least one OGT variant (Figure S5). Hits with ≥ 1.8-fold difference between enzyme variants at a p-value < 0.05 are considered “biased” for WT (orange) or D2A (red) (Figure S6). Data points are the average of three microarrays per condition (no enzyme, WT, D2A).
Commercially available protein microarrays containing thousands of diverse human proteins, including kinases, phosphatases, GPCRs, nuclear receptors, and proteases, provide an alternative platform to cell extracts for globally profiling substrate preferences of OGT and its variants.7,8 An advantage of using protein microarrays is that substrates can be readily identified by location on the array and the amount of glycosylation can be quantified. We evaluated microarray glycosylation profiles for OGTWT and OGTD2A and found that they were highly correlated and shared a substantial number (116) of substrates (Figure 2C, S5, Table S2). However, some substrates were preferentially glycosylated by either OGTWT or OGTD2A (Figure 2C, S6). Therefore, two orthogonal strategies for profiling global protein glycosylation demonstrated that changing two aspartates in the TPR lumen to alanine affects OGT’s substrate preferences.
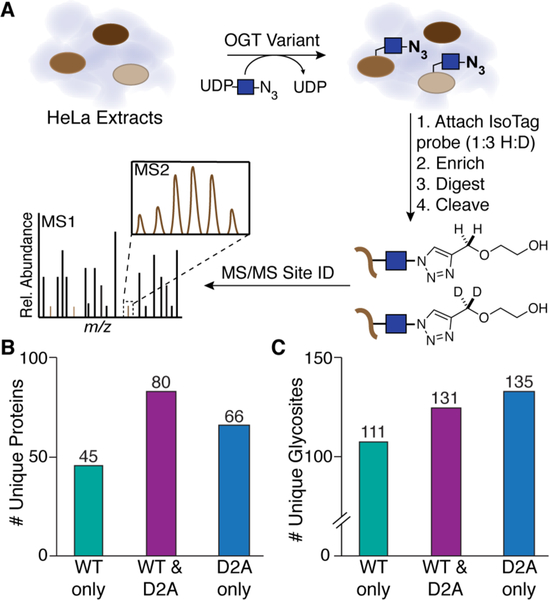
We sought to obtain information on the specific sites glycosylated by OGTWT or OGTD2A to better understand the role of the TPR aspartates in substrate selection. We adapted a previously developed glycosite mapping strategy to identify sites of new protein glycosylation in HeLa extracts after addition of recombinant OGTWT or OGTD2A (Figure 3A).9 After isotope tagging, enrichment, digestion, and probe cleavage, glycopeptides were analyzed using mass spectrometry. Consistent with the microarray results, we found that a substantial number of protein substrates were preferentially glycosylated by either OGTD2A or OGTWT (Figure 3B). From 191 total glycosylated proteins, we identified 111 and 135 unique glycosites for OGTWT and OGTD2A, respectively (Figure 3C, Table S5). For the 80 proteins that were glycosylated by both OGTWT and OGTD2A, we also observed multiple instances in which the sites of glycosylation were different (Figure S8, Table S6). These results show that substrate preferences change substantially when the TPR aspartates are replaced with small, nonpolar alanines.
Figure 3.
Glycosite mapping shows that changing aspartates in the TPR lumen to alanine alters substrate preferences. A) Schematic of glycosite mapping strategy. HeLa cell extracts were treated with OGT and UDP-GlcNAz10, an azide-containing analog, to enable IsoTag probe attachment. Following work-up, the samples were analyzed by MS/MS to identify glycosite sequences. Only unambiguously localized O-glycosites were used in subsequent analyses (see Methods). Glycosite mapping was performed in duplicate for each OGT variant. B) Number of unique proteins glycosylated by OGTWT and OGTD2A. C) Number of unique glycosites identified for OGTWT and OGTD2A.
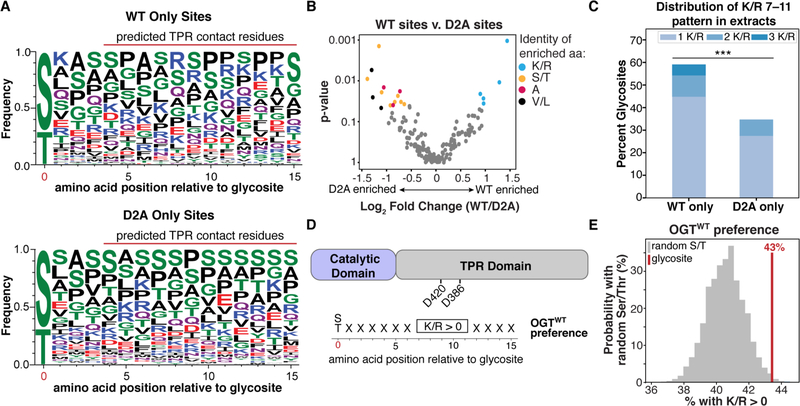
We used the glycosite mapping data to determine if there are amino acid preferences in substrates glycosylated by either OGTWT or OGTD2A. Sequence frequency plots for a 16 amino acid region that includes the glycosite and 15 C-terminal amino acids, are shown for peptides glycosylated by only OGTWT or OGTD2A (Figure 4A; S9).11 We chose this window to capture sequence preferences of residues that could bind in the TPR domain proximal to the active site. For OGTWT-specific substrates, visual inspection revealed an apparent preference for positively charged amino acids. To better quantify the amino acids that are selectively enriched for OGTWT and OGTD2A, we counted the number of occurrences of each amino acid within a sliding three amino acid window at all positions between 1 and 15 amino acids C-terminal to the glycosite (Figure S10). Relative to OGTD2A, substrates selectively glycosylated by OGTWT were enriched in lysines and arginines within multiple three amino acid windows (Figure 4B, Table S7). In contrast, alanine, large hydrophobic residues, and Ser/Thr were enriched for OGTD2A(Figure 4B, Table S7). Testing alternative window sizes revealed that OGTWT-specific glycosites were most strongly enriched for arginine and lysine at amino acid positions 7 through 11 (Figure 4C, Table S8), which is the region of the substrate that would engage the D420 and D386 aspartates in the TPR lumen (Figure 4D).
Figure 4.
Comparative sequence analysis of glycosites reveals that the TPR aspartates select for sequences containing Arg/Lys. A) Amino acid probability logos for OGTWT or OGTD2A-specific glycosites from the glycosite mapping data. Amino acids are ordered top to bottom by frequency, with height proportional to frequency. B) Comparison of amino acid preferences for OGTWT versus OGTD2A. Each dot represents the enrichment of a single type of amino acid in a three amino acid window between 1 and 15 amino acids C-terminal of the glycosite. See Methods. C) Percentages of glycosites in HeLa extracts having one or more Arg/Lys at positions 7–11 from the glycosite for each OGT variant. p<0.001 = ***. D) Schematic of Arg/Lys preference identified for OGT. The enrichment is found in the 7–11 region, which would contact the indicated aspartates. E) 43% of 1817 glycosites from the PhosphoSitePlus database have at least one Arg/Lys at positions 7–11 (red line). Randomly drawn Ser/Thr residues from the same proteins are far less likely to have this preference. The histogram (grey bars) summarizes results of n=100,000 sets of 1817 randomly drawn Ser/Thr residues.
We next asked whether the Arg/Lys preference identified for OGTWT in HeLa extracts was also observed in cells. To do this, we used data from the PhosphoSitePlus database, which includes a curated list of 1,817 O-GlcNAc glycosites.12 We reasoned that if the Arg/Lys preference was truly important in glycosite selection, then authentic glycosites should be far more likely than randomly drawn sets of Ser/Thr residues in the same protein to match the preference. Indeed, we found this to be the case for the OGT glycosites in the database (Figure 4E). Therefore, the Arg/Lys sequence preference we have identified for OGT substrates from the HeLa extracts experiments is also observed for cellular substrates.
Our study shows that residues in OGT’s TPR lumen control proteome-wide substrate recognition. Mutations of only two aspartate residues to alanine increase overall glycosylation activity and result in glycosylation of proteins that are not substrates of OGTWT. At the same time, some substrates that are glycosylated by OGTWT are no longer recognized by the double aspartate mutant. We used a pattern searching approach to show that substrates only glycosylated by OGTWT tend to have positively charged residues within a window spanning residues 7–11 C-terminal to the site of glycosylation. This window is in the region of the substrate that would contact D386 and D420, leading us to conclude that many substrates directly interact with the TPR lumen. Supporting this conclusion, we observed the same Arg/Lys substrate bias in annotated cellular glycosites. Taken together with the previous study showing that asparagine ladder residues are globally important for protein glycosylation,7 our findings indicate that a large fraction of OGT substrates span from the catalytic domain into the TPR lumen. While the asparagines anchor the backbone of substrates, aspartate ladder residues influence substrate preferences and determine glycosite positioning.
The finding that a substantial fraction of OGT substrates bind in the TPR lumen implies that one can use the curated glycosite data to predict substrate preferences. Using a pattern searching approach based on the assumption that residues C-terminal to the glycosite bind in the TPR lumen, we have identified a positive selection not only for Arg/Lys, but for Ser/Thr (Figure S12, Table S9). The latter preference has been observed in the reported structures.6,13 We also observed a striking selection against negatively charged residues that is consistent with our conclusion that the luminal aspartates control substrate specificity.
In closing, it is worth noting that mutations in the TPR domain of OGT have been linked to human diseases. Mutations that distort the TPR helix are found in X-linked intellectual disability.14 Moreover, mutations at D386, D420, and D454, the three residues investigated here, have been found in different cancers.15 The results presented here show that these positions are important for OGT specificity, suggesting that altering OGT specificity may contribute to disease pathogenesis. Further work on the role of the TPR lumen in driving substrate selection may help explain OGT’s role in human disease.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH (GM094263 to S.W. and U01 CA242098–01 to C.M.W.), Burroughs Wellcome Fund (C.M.W.), and the Sloan Foundation (C.M.W). We thank Martha Bulyk and the National Center for Functional Glycomics for aid in scanning microarrays. Glycoproteomics data was collected at the Harvard University Mass Spectrometry and Proteomics Resource Laboratory.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
Experimental protocols, detailed description of data analysis, supplemental figures, and tables are available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b06061.
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE16 partner repository with the dataset identifier PXD014731.
The authors declare no competing financial interest.
REFERENCES
- (1).(a) Levine ZG; Walker S. The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells? Annu. Rev. Biochem 2016, 85, 631. [DOI] [PubMed] [Google Scholar]; (b) Janetzko J; Walker S. The making of a sweet modification: structure and function of O-GlcNAc transferase. J. Biol. Chem 2014, 289, 34424. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bond MR; Hanover JA A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell. Biol 2015, 208, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Martinez MR; Dias TB; Natov PS; Zachara NE Stress-induced O-GlcNAcylation: an adaptive process of injured cells. Biochem. Soc. Trans 2017, 45, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).(a) Yang X; Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol 2017, 18, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hart GW; Housley MP; Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007, 446, 1017. [DOI] [PubMed] [Google Scholar]; (c) Zachara NE; Hart GW O-GlcNAc modification: a nutritional sensor that modulates proteasome function. Trends Cell Biol. 2004, 14, 218. [DOI] [PubMed] [Google Scholar]
- (3).(a) Ma J; Hart GW Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics 2013, 10, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Peterson SB; Hart GW New insights: A role for O-GlcNAcylation in diabetic complications. Crit. Rev. Biochem. Mol. Biol 2016, 51, 150. [DOI] [PubMed] [Google Scholar]; (c) Issad T; Masson E; Pagesy P. O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 2010, 36, 423. [DOI] [PubMed] [Google Scholar]; (d) Vaidyanathan K; Wells L. Multiple tissue-specific roles for the O-GlcNAc post-translational dodification in the induction of and complications arising from type II diabetes. J. Biol. Chem 2014, 289, 34466. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Slawson C; Hart GW O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 2011, 11, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Yi W; Clark PM; Mason DE; Keenan MC; Hill C; Goddard WA; Peters EC; Driggers EM; Hsieh-Wilson LC Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).(a) Lazarus MB; Nam Y; Jiang J; Sliz P; Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 2011, 469, 564. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lazarus MB; Jiang J; Gloster TM; Zandber WF; Whitworth GE; Vocadlo DJ; Walker S. Structural snapshot of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol 2012, 8, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Joiner CM; Li H; Jiang J; Walker S. Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms. Curr. Opin. Struct. Biol 2019, 56, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Iyer SPN; Hart GW Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem 2003, 278, 24608. [DOI] [PubMed] [Google Scholar]; (e) Lubas WA; Hanover JA Functional expression of O-linked glcNAc transferase: domain structure and substrate specificity. J. Biol. Chem 2000, 275, 10983. [DOI] [PubMed] [Google Scholar]; (f) Jinek M; Rehwinkel J; Lazarus BD; Izaurralde E; Hanover JA; Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin [alpha]. Nat. Struct. Mol. Biol 2004, 11, 1001. [DOI] [PubMed] [Google Scholar]
- (5).(a) Cheung WD; Hart GW AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose peprivation. J. Biol. Chem 2008, 283, 13009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cheung WD; Sakabe K; Housley MP; Dias WB; Hart GW O-Linked β-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem 2008, 283, 33935. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Deplus R; Delatte B; Schwinn MK; Defrance M; Méndez J; Murphy N; Dawson MA; Volkmar M; Putmans P; Calonne E; Shih AH; Levine RL; Bernard O; Mercher T; Solary E; Urh M; Daniels DL; Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO Journal 2013, 32, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Yang X; Zhang F; Kudlow JE Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 2002, 110, 69. [DOI] [PubMed] [Google Scholar]
- (6).Lazarus MB; Jiang J; Kapuria V; Bhuiyan T; Janetzko J; Zandberg WF; Vocadlo DJ; Herr W; Walker S. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 2013, 342, 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Levine ZG; Fan C; Melicher MS; Orman M; Benjamin T; Walker S. O-GlcNAc transferase recognizes protein substrates using an asparagine ladder in the tetratricopeptide repeat (TPR) superhelix. J. Am. Chem. Soc 2018, 140, 3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).(a) Sutandy FXR; Qian J; Chen C; Zhu H. Overview of protein microarrays. Curr. Proto. Protein Sci 2013, 72, 27.1 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Macbeath G; Schreiber SL Printing proteins as microarrays for high-throughput function determination. Science 2000, 289, 1760. [DOI] [PubMed] [Google Scholar]; (c) Berrade L; Garcia AE; Camarero JA Protein microarrays: novel developments and applications. Pharm Res 2011, 28, 1480. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Ortiz-Meoz RF; Merbl Y; Kirschner MW; Walker S. Microarray discovery of new OGT substrates: the medulloblastoma oncogene OTX2 is O-GlcNAcylated. J. Am. Chem. Soc 2014, 136, 4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Woo CM; Iavarone AT; Spiciarich DR; Palaniappan KK; Bertozzi CR Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 2015, 12, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Woo CM; Felix A; Byrd WE; Zuegel DK; Ishihara M; Azadi P; Iavarone AT; Pitteri SJ; Bertozzi CR Development of IsoTaG, a chemical glycoproteomics technique for profiling intact N- and O-glycopeptides from whole cell proteomes. J. Proteome Res 2017, 16, 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Woo CM; Lund PJ; Huang AC; Davis MM; Bertozzi CR; Pitteri S. Mapping and quantification of over 2,000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (IsoTaG). Mol. Cell. Proteomics 2018, 17, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).(a) Vocadlo DJ; Hang HC; Kim E; Hanover JA; Bertozzi CR A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 9116. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Leavy TM; Bertozzi CR A high-throughput assay for O-GlcNAc transferase detects primary sequence preferences in peptide substrates. Bioorg. Med. Chem. Lett 2007, 17, 3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Crooks GE; Hon G; Chandonia JM; Breener SE WebLogo: a sequence logo generator. Genome Res 2004, 14, 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hornbeck PV; Zhang B; Murray B; Kornhauser JM; Latham V; Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 2015, 43, D512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rafie K; Raimi O; Ferenbach AT; Borodkin VS; Kapuria V; van Aalten DMF Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats. Open Biol. 2017, 7, 170078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).(a) Vaidyanathan K; Niranjan T; Selvan N; Teo CF; May M; Patel S; Weatherly B; Skinner C; Opitz J; Carey J; Viskochil D; Gecz J; Shaw M; Peng Y; Alexov E; Wang T; Schwartz C; Wells L. Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem 2017, 292, 8948. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Selvan N; George S; Serajee FJ; Shaw M; Hobson L; Kalscheuer V; Prasad N; Levy SE; Taylor J; Aftimos S; Schwartz CE; Hug AM; Gecz J; Wells L. O-GlcNAc transferase missense mutations linked to X-linked intellectual disability deregulate genes involved in cell fate determination and signaling. J. Biol. Chem 2018, 293, 10810. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Willems AP; Gungodu M; Kempers MJE; Giltay JC; Prundt R; Elgerink M; Loza BF; Fuijkschot J; Ferenbach AT; van Gassen KLI; van Aalten DMF; Lefeber DJ Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J. Biol. Chem 2017, 292, 12621. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gundogdu M; Llabres S; Gorelik A; Ferenbach AT; Zachariae U; van Aalten DMF The O-GlcNAc transferase intellectual disability mutation L254F distorts the TPR helix. Cell Chemical Biology 2018, 25, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).(a) Kamburov A; Lawrence MS; Polak P; Leshchiner I; Lage K; Golub TR; Lander ES; Getz G. Comprehensiv assessment of cancer missense mutations clustering in protein structures. Proc. Natl. Acad. Sci. U S A 2015, 112, E5486. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tate JG; Bamford S; Jubb HC; Sondka Z; Beare DM; Bindal N; Boutselakis H; Cole CG; Creatore C; Dawson E; Fish P; Harsha B; Hathaway C; Jupe SC; Kok CY; Noble K; Ponting L; Ramshaw CC; Rye CE; Speedy HE; Stefancsik R; Thompson SL; Wang S; Ward S; Campbell PJ; Forbes SA COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Research 2019, 47, D941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Terment T; Brazma A; Vizcaino JA The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019, 47, D442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.