Abstract
The pattern of somatic mutations observed at diagnosis of acute myeloid leukemia (AML) has been well-characterized. However, the premalignant mutational landscape of AML and its impact on risk and time to diagnosis is unknown. Here we identified 212 women from the Women’s Health Initiative who were healthy at study baseline, but eventually developed AML during follow-up (median time: 9.6 years). Deep sequencing was performed on peripheral blood DNA of these cases and compared to age-matched controls that did not develop AML. We discovered that mutations in IDH1, IDH2, TP53, DNMT3A, TET2 and spliceosome genes significantly increased the odds of developing AML. All subjects with TP53 mutations (n = 21 out of 21 patients) and IDH1 and IDH2 (n = 15 out of 15 patients) mutations eventually developed AML in our study. The presence of detectable mutations years before diagnosis suggests that there is a period of latency that precedes AML during which early detection, monitoring and interventional studies should be considered.
The pathogenesis of AML is characterized by serial acquisition of somatic mutations and several genes are recurrently mutated in AML1–3. However, it is not known when such mutations appear prior to the development of overt disease, how they evolve and the specific risk associated with each one. Furthermore, the acquisition of AML-associated mutations has also been found in normal aging, and approximately 10% of people older than 65 years of age4–7 have clonal hematopoiesis. The presence of clonal hematopoiesis is associated with an increased risk of hematologic malignancies and cardiovascular disease. However, studies of clonal hematopoiesis to date have included very few subjects who subsequently developed AML. Whole-exome sequencing on peripheral blood samples from 17,182 individuals was preformed in a previous study5. Mutations in genes that are associated with hematologic cancers were found in 5.6% of subjects aged 60–69 years and in up to 18.4% of those aged >90 years. Only 16 hematologic cancers were reported in this group, of which 6 were cases with AML. In another study4, DNA sequencing was performed on peripheral blood from 12,380 Swedish individuals (mean age, 55 years), with clonal mutations detectable most commonly in DNMT3A, ASXL1 and TET2. Clonal hematopoiesis was strongly associated with increased risk of hematologic cancer (hazard ratio, 12.9; 95% confidence interval, 5.8–28.7), but there were only 12 cases with AML reported in the study4. The objective of the present work was to establish a genomically defined premalignant state of AML and ascertain the premalignant mutational landscape of AML years before the diagnosis of the disease. We further sought to investigate whether specific mutations, allele burdens or patterns of coexisting mutations would affect the risk and time-to-diagnosis of AML. To this end, we performed deep sequencing of serially collected peripheral blood samples obtained from 212 women a median of 9.6 years before their diagnosis of AML, along with 212 age-matched controls.
Results
Study population and samples.
The Women’s Health Initiative (WHI) was a large, prospective clinical trial and observational study of more than 160,000 women initiated by the US National Institutes of Health (NIH) in 1991 and consisted of three clinical trials and an observational cohort designed to assess the impact of hormonal therapy on postmenopausal health issues in women with an average follow-up of 10.8 years (s.d., 3.3 years)8,9. Detailed clinical data regarding medical history, medications and complete blood counts were available at baseline assessment. New diagnoses were updated in follow-up, with central confirmation of all new diagnoses of cancer. The participants in the clinical trials were followed at baseline and three, six and nine years after study entry, whereas the participants of the observational cohort were followed at baseline, one and three years after study entry. In addition, all participants of the observational cohort filled out yearly questionnaires for the first eight years of follow-up, updating any new medical diagnoses during follow-up.
Identification of cases.
In the WHI cohort, 212 study participants eventually developed pathologically confirmed AML. Of these, baseline peripheral blood DNA at study entry was available and passed quality control in 188 participants; these were identified as cases to be included in the final analyses. Additional follow-up samples for 130 cases were available at one and/or three years after baseline, all prior to the diagnosis of AML. Exclusion criteria for cases included known diagnosis of any hematological disorder (myeloid and lymphoid), including AML, prior to WHI baseline evaluation.
Controls.
Age-matched controls (n = 212) were selected from participants who were confirmed not to have a diagnosis of AML while being followed during the WHI study. Exclusion criteria included concurrent or history of prior hematologic disorder (myeloid and lymphoid), including AML, at WHI baseline. Controls were matched to cases by age at baseline within two years, history of non-hematologic cancers at baseline and type and timing of any cancers that occurred in cases after WHI baseline, but before the diagnosis of AML as well as the exact timing of blood sample collection and follow up in the WHI. Matching was done in a time forward manner to ensure that each control had as much control time as its matched case10. Of the 212 controls, peripheral blood was available and passed quality standard in 181 controls at WHI baseline and these were included in the analyses. Additional follow up samples for 126 controls were available at one and/or three years after baseline.
Landscape of mutations in blood before AML diagnosis
With a median of 9.6 years before diagnosis of AML, cases were more likely to harbor mutations than controls (odds ratio, 4.86; 95% confidence interval, 3.07–7.77; P = 3.8 × 10−13). The most common mutations identified above 1% variant allele fraction (VAF) included DNMT3A (36.7% of cases compared to 18.8% of controls), TET2 (25.0% of cases compared to 5.5% of controls), TP53 (11.2% of cases compared to 0% of controls), SRSF2 (6.9% of cases compared to 0% of controls), IDH2 (6.4% of cases compared to 0% controls), SF3B1 (5.9% of cases compared to 1.1% of controls), JAK2 (5.3% of cases compared to 0.6% of controls) and ASXL1 (3.2% of cases compared to 3.3% of controls; Fig. 1a, Supplementary Fig. 2 and Supplementary Table 2). Spliceosome mutations in SF3B1, SRSF2 and U2AF1 were identified as a group in 13.8% of cases (n = 26 out of 188) compared to 1.1% of controls (n = 2 out of 181). Similarly, IDH mutations as a group (both IDH1 and IDH2) were identified in 8% of cases (n = 15 out of 188) compared to 0% of controls. There was no association between the presence of any mutation and abnormal hemoglobin, white-blood cell count and/or platelet level (Supplementary Table 1). We were not able to evaluate the relationship between red cell distribution width (RDW), absolute neutrophil count and mean corpuscular volume (MCV) with mutations as the WHI did not collect these data.
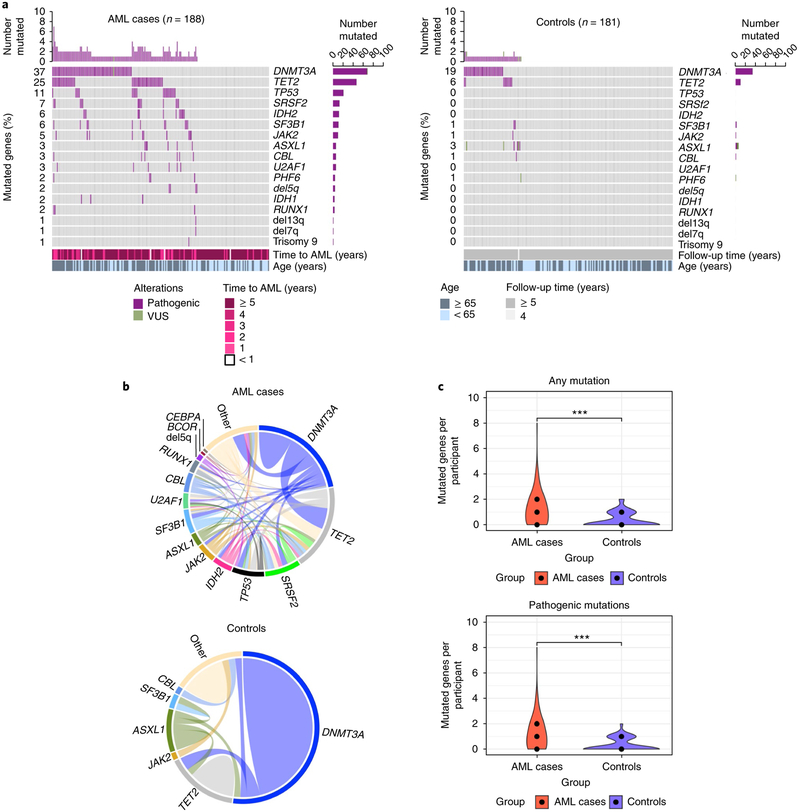
Fig. 1 |. Spectrum of mutations seen at baseline years prior to the diagnosis of AML alongside matched controls.
a, Mutated genes in cases with AML (n = 188; left) compared to controls (n = 181; right). For each gene (rows), pathogenic mutations (purple) and variants of unknown significance (VUS; green) are indicated. Side bar plots indicate the number of participants in whom the gene is mutated in each group. Top bar plots indicate the number of mutated genes per participant. In cases with AML, time to AML and age are shown. For controls, the time of follow-up and age are shown. b, Comutations between genes in cases with AML (n = 129) and controls (n = 56). ‘Other’ indicates other genes. c, Violin plot indicating the number of mutated genes per participant in cases with AML (red) compared to controls (blue). Median number of mutated genes, first and third quantile are shown for each group (middle, lower and upper black dots, respectively). Left plot indicates the number of mutated genes per participant in cases with AML (median, 1; range, 0–8; n = 129 out 188 mutated) versus controls (median, 0; first quantile, 0; range 0–2; n = 56 out of 181 mutated) (P < 2.2 × 10−16, two-tailed Mann–Whitney U test). The right plot indicates the number of pathogenic mutations in cases with AML (median, 1; range, 0–8; n = 127 out of 188 mutated) versus controls (median, 0; first quantile, 0; range, 0–2; n = 53 out of 181 mutated). P < 2.2 × 10−16, two-tailed Mann–Whitney U test). ***P < 0.001, Mann–Whitney U test.
Overall, cases with AML demonstrated greater clonal complexity than controls, and 46.8% of the cases with AML showed comutations, compared to 5.5% of controls (odds ratio, 9.01; 95% confidence interval, 4.1–21.4; P = 9.7 × 10−10). As shown in Fig. 1b, the most common comutations present in cases with AML were DNMT3A with TET2, DNMT3A with SRSF2, TET2 with SRSF2, and IDH2 with SRSF2. Among controls, mutations were generally present individually. When tested for mutual exclusivity and co-occurrence patterns, mutations in DNMT3A, TP53, TET2 and ASXL1 had a tendency toward mutual exclusivity. Conversely, co-occurrence was noted for IDH2 and SRSF2 as well as RUNX1 and PHF6. Among controls, no common co-occurrence pattern was identified and DNMT3A with TET2 was found to be mutually exclusive (Supplementary Fig. 3). Overall, cases with AML had a median of one mutated gene (median, 1; range, 0–8), whereas controls had median of no mutated genes (median, 0; range, 0–2) (P < 2.2 × 10−16). Older individuals (≥65 years) had more mutated genes in both cases and controls. The same held true for mutations categorized as pathogenic versus variant of unknown significance (Fig. 1c). We also found that the incremental increase in the number of mutations increased age-adjusted odds of AML per mutation (odds ratio, 3.27; 95% confidence interval, 2.47–4.45).
Presence of mutations at baseline increases risk of AML
Having a mutation at baseline assessment was associated with statistically increased odds of developing AML (odds ratio, 4.86; 95% confidence interval, 3.07–7.77; P = 3.8 × 10−13) (Table 1). This finding was independent of age: odds ratio of 4.39 (2.08–9.61) for cases aged <65 years; odds ratio of 6.19 (3.25–12.14) for cases aged ≥65 years. Overall, 68.62% (n = 129) of cases and only 30.9% of controls (n = 56) were found to have mutations. Mutations were found in 53.75% of cases and 20.78% of controls <65 years old, and in 79.63% of cases and 38.46% of controls who were ≥65 years old. This rate of clonal hematopoiesis in the control group is generally higher than previous reports4,5,11, which were performed using low-coverage whole-exome sequencing and a higher VAF cutoff. We therefore also determined the clonal hematopoiesis rate using previous variant classification and VAF cutoff criteria (Supplementary Fig. 4 and Supplementary Table 4). When performing this adjustment, the observed rate of clonal hematopoiesis in controls was more in line with previous studies at around 7% (Supplementary Fig. 4 and Supplementary Table 4).
Table 1 |.
Mutation frequencies and odds ratios of AML
| Cases with AML (n = 188) | Controls (n = 181) | Odds ratio | ||||
|---|---|---|---|---|---|---|
| Age | Number of cases harboring any mutations (%) | Number of non-mutated cases (%) | Number of controls harboring any mutations (%) | Number of controls with no mutations (%) | Odds ratio (95% confidence interval) | P value |
| <65 | 43 (53.75) | 37 (46.25) | 16 (20.78) | 61 (79.22) | 4.39 (2.08–9.61) | 3.1 × 10−5 |
| ≥65 | 86 (79.63) | 22 (20.37) | 40 (38.46) | 64 (61.54) | 6.19 (3.25–12.14) | 1.0 × 10−9 |
| Total | 129 (68.62) | 59 (31.38) | 56 (30.94) | 125 (69.06) | 4.86 (3.07–7.77) | 3.8 × 10−13 |
Number and frequency of mutations in cases with AML versus controls overall and for participants younger than 65 or at least 65 years of age.
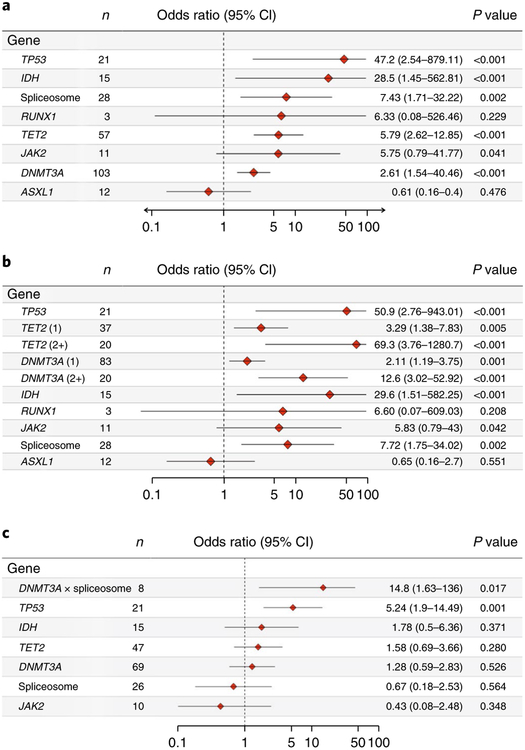
Among the recurrently mutated genes, some mutations demonstrated increased specificity and penetrance for the development of AML. All participants with mutations in TP53 (n = 21 out of 21), IDH1 or IDH2 (n = 15 out of 15), or RUNX1 with PHF6 (n = 3 out of 3) in our cohort eventually developed AML. Multivariable analysis was performed to evaluate potential associations between individual mutated genes and the development of AML, adjusting for confounders including comutated genes and age (Fig. 2a). This analysis found that TP53 (odds ratio, 47.2; 95% confidence interval, 2.5–879.1), IDH (including IDH1 and IDH2) (odds ratio, 28.5; 95% confidence interval, 1.4–562.8), spliceosome genes (including SF3B1, SRSF2 and U2AF1) (odds ratio, 7.4; 95% confidence interval, 1.7–32.2), TET2 (odds ratio, 5.8; 95% confidence intreval, 2.6–12.9) and DNMT3A (odds ratio, 2.6; 95% confidence interval, 1.5–4.5) were associated with significantly increased odds of developing AML relative to controls and will be referred to as ‘high-risk genes’ hereafter. Similar results were obtained when considering only those variants flagged as ‘likely pathogenic’ (Supplementary Fig. 16). Study participants who had more than one variant in DNMT3A or TET2 demonstrated significantly increased odds of AML (Fig. 2b): two or more DNMT3A variants (odds ratio, 12.6; 95% confidence interval, 3.0–52.9), one DNMT3A variant (odds ratio, 2.1; 95% confidence interval, 1.2–3.8); two or more TET2 variants (odds ratio, 69.3; 95% confidence interval, 3.8–1,280.7), one TET2 variant (odds ratio, 3.29; 95% confidence interval, 1.4–7.8). Because single mutations in DNMT3A and TET2 were the most commonly found mutations in controls, this finding distinguishes the pattern of DNMT3A and TET2 mutations in cases compared to controls. Mutations in IDH1 and IDH2 were exclusively found in the known recurring hotspots in Arg132 and Arg1402,12. Similarly, SRSF2 mutations were confined to the well-known Pro95 hotspot13,14. TP53 mutations were primarily in the DNA-binding, transactivation and oligomerization domains15,16. The distribution of mutations within functional protein domains, as well as the type of mutations found at each location, is shown in the Supplementary Information (Supplementary Figs. 5–8, 13–15). We also identified relatively few non-canonical mutations, such as non-RING domain CBL mutations, non-kinase JAK2 mutations, as well as mutations in FLT1 and CARD11. These mutations were almost always associated with the presence of probable driver comutations in DNMT3A and/or TET2 and were unique to particular study subjects. Notably, these mutations typically demonstrated persistence in the study participant when serial samples were available (Supplementary Fig. 11). Although these mutations are not likely to drive the observed clonal expansions, they are likely passenger events that are symptomatic of driver mutations and thus accurately reflect the premalignant state in cases with AML and clonal hematopoiesis when present in the controls.
Fig. 2 |. Multivariable analysis of the risk to develop AML associated with the presence of mutated genes.
a, Forest plot indicating the odds ratio of mutations in each gene occurring in the cases with AML compared to controls. Genes or gene categories significantly associated with AML include TP53 (P = 5.5 × 10−6), IDH (P = 3.0 × 10−4), spliceosome, TET2 (P = 2.4 × 10 −6) and DNMT3A (P = 3.4 × 10−4). b, Forest plot indicating odds ratio of mutations in each gene including number of mutations in DNMT3A and TET2 occurring in the cases with AML compared to controls when one mutation in each gene is present per participant (1) compared to two or more mutations in present in each gene per participant (2+). P < 0.001: TP53 (P = 3.2 × 10−6); TET2 (2) (P = 1.8 × 10 −7); DNMT3A (2) (P = 1.9 × 10−5) and IDH (P = 2.8 × 10−4). c, Mutations in TP53 and DNMT3A are significantly associated with rapid development of AML. Odds ratios per gene were adjusted by age (years) as a continuous variable. IDH category includes IDH1 and IDH2. The spliceosome category includes SRSF2, SF3B1 and U2AF1. Interaction between DNMT3A and spliceosome is indicated (DNMT3A × spliceosome). CI, confidence interval; N, number of affected cases. P values are shown for penalized likelihood multivariable logistic regression.
We next sought to derive population-based estimates of the impact of mutations in high-risk genes on AML development using a pseudolikelihood-based approach17. Mutation-free participants in the high-risk genes (DNMT3A, TET2, TP53, IDH1 or IDH2, spliceosome (SRSF2, SF3B1 or U2AF1)) were estimated to develop AML with an incidence of 2.7 per 100,000 per year. Mutations increased this risk at varying rates depending on the gene: TP53, 13.9 per 100,000 per year; IDH, 12.6 per 100,000 per year; spliceosome, 9.2 per 100,000 per year; DNMT3A, 4.6 per 100,000 per year; and TET2, 5.5 per 100,000 per year (Supplementary Fig. 12).
Mutations are associated with an accelerated time to AML presentation
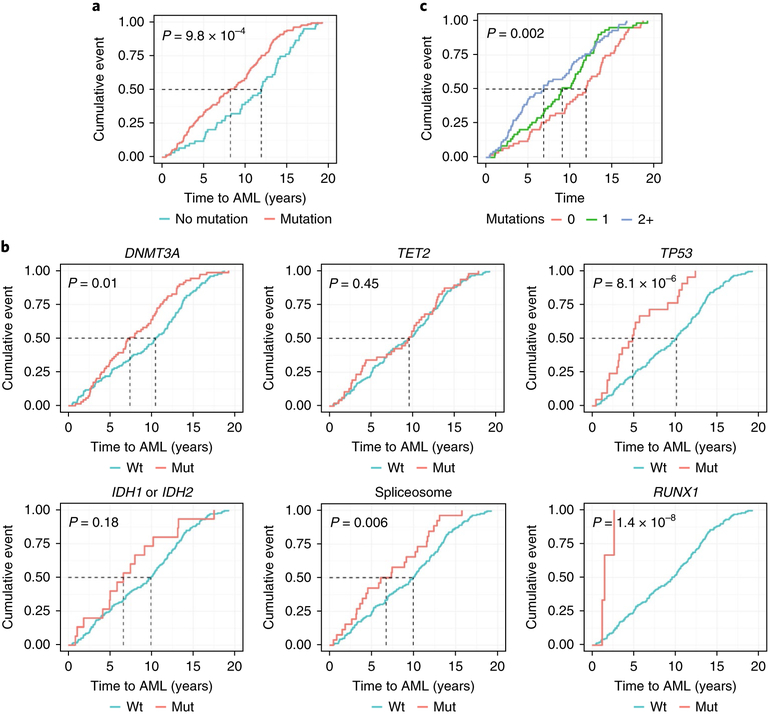
Cases with AML who had mutations at baseline experienced significantly shorter latency of disease than cases without baseline mutations. The presence of any mutation shifted the median time to AML diagnosis from 11.9 years (no mutations) to a median of 8.2 years after baseline assessment (P = 9.8 × 10−4, log-rank test; Fig. 3a). Univariable analysis of mutated genes demonstrated that median time to AML varied according to mutated gene (Fig. 3b): DNMT3A (7.4 compared to 10.5 years), TP53 (4.9 compared to 10.1 years), spliceosome genes (6.7 compared to 9.9 years) and RUNX1 (1.5 compared to 9.6 years). The presence of RUNX1 mutations seemed to be associated with especially rapid development of AML within two years, but the number of participants in this subgroup was small (n = 3). Clonal complexity also affected the latency of disease as patients with one mutation in the high-risk genes developed AML in 9.1 years, but those with two or more mutations in the high-risk genes developed AML in only 6.9 years (Fig. 3c).
Fig. 3 |. Time to AML diagnosis is influenced by mutation status.
Cumulative incidence of AML diagnoses (cumulative event; y axis) as a function of time (years to AML diagnosis) is shown. a, Participants include cases with AML only at baseline (n = 188) with any mutated gene (n = 129 out of 188) versus no mutations (n = 59 out of 188). b, Participants with mutations in genes associated with AML versus participants with no mutations in these genes (DNMT3A, n = 69 out of 188; TET2, n = 47 out of 188; TP53, n = 21 out of 188; IDH1 or IDH2, n = 15 out of 188; spliceosome, n = 26 out of 188) in addition to RUNX1 (n = 3 out of 188). Data on RUNX1 are provided because all participants with a RUNX1 (n = 3) mutation rapidly developed AML (< 2 years) although significance was not achieved due to the few participants who had mutations in RUNX1 within the cohort. c, Cases with AML who had no mutated genes (n = 86 out of 188), one mutated gene (n = 56 out of 188), or two or more mutated genes (2+) (n = 46 out of 188) in significant high-risk genes associated with development of AML. Two-sided P values are shown for the log-rank test.
Multivariable analyses produced similar findings (Fig. 2c). Mutations in TP53 were independently associated with AML occurring within five years of baseline (odds ratio, 5.2; 95% confidence interval, 1.9–14.5; P = 0.001) as well as an increased annual rate of AML develoment (hazard ratio, 3.01; 95% confidence interval, 1.8–5.0; P =2.8 × 10−5) when adjusted for confounding comutations and age. DNMT3A mutations when present with spliceosome mutations were also associated with increased odds of AML development within five years (odds ratio, 14.8; 95% confidence interval, 1.6–136; P = 0.02). None of the other genes were associated with increased odds of earlier AML development when present alone or with other comutations. For TP53 and DNMT3A mutations, there was no appreciable difference in time to development of AML by mutation subtype, for example, in Arg882 in DNMT3A. Finally, whereas all participants (15 out of 15) with IDH mutations were eventually diagnosed with AML, there was no increased risk of AML earlier than five years after baseline assessment, suggesting the possibility that further downstream mutational events are required for AML development in these cases.
Mutations at any VAF are associated with increased AML risk
When we considered only mutations in the genes that are associated with development of AML (high-risk genes), as shown in Fig. 4a, mutations present at VAF > 10% were found more commonly among cases compared to controls (odds ratio, 14.8; 95% confidence interval, 5.8–48.8; P = 1.5 × 10−13). As demonstrated in the histograms, mutations in DNMT3A and TET2 at lower VAFs were notably more distributed among cases and controls. By contrast, mutations present at higher VAFs in DNMT3A and TET2 were almost exclusively seen in cases with AML, suggesting that mutations in DNMT3A and TET2 are less specific to cases with AML at lower VAFs. In a multivariable model evaluating high (≥10%) and low VAFs (<10%) in the high-risk genes and adjusting for age and other comutations, high VAFs in DNMT3A (high VAF: odds ratio, 4.8; 95% confidence interval, 1.6–14.8; P = 0.004; low VAF: odds ratio, 2.5; 95% confidence interval, 1.4–4.4; P = 0.002), TET2 (high VAF: odds ratio, 20.4; 95% confidence interval, 3.5–120.1; P = 0.002; low VAF: odds ratio, 3.6; 95% confidence interval, 1.5–8.9; P = 0.004) and spliceosome genes (high VAF: odds ratio, 14.1; 95% confidence interval, 0.8–260.9; P = 0.019, low VAF: odds ratio, 3.4; 95% confidence interval, 0.7–16.8; P = 0.105) were associated with higher odds of developing AML (Supplementary Fig. 22). For TP53 and IDH mutations, the odds of AML development did not significantly change between lower and higher VAFs as all of these subjects developed AML. Next, we further examined the specificity of mutations in genes that were significantly associated with cases with AML (Fig. 1a) by determining their frequency in controls. The true-positive and false-positive rate of mutations at varying VAF cutoffs was visualized using receiver operating characteristic analysis (Fig. 4b). Individually, mutations in TP53, SRSF2, U2AF1, SF3B1 and IDH2 produced a <1% rate of false-positive detection at VAF cutoffs ranging from 1 to 2%. Although not exactly replicating the false-positive rate in the general population, these data provide a starting point for future studies to assess the false-positive rate in the general population. Tabulated results are available in Supplementary Fig. 9. For the subset of participants for whom serial samples were available, changes in VAF were assessed from baseline to one or three or years. We evaluated whether the time to AML development was influenced by the fold increase in VAF in participants who had demonstrated a statistically significant increase in VAF upon serial testing (Fig. 4c). The rate of increase in VAF for IDH2 mutations (R2 = 0.67; slope, −3.3; 95% confidence interval, −21.5 to −3.7; P = 0.02) and TP53 mutations (R2 = 0.87; slope, −3.0; 95% confidence interval, −14.0 to −5.5; P = 0.009) was significantly associated with a shorter time to AML development in a linear regression analysis. Although this relationship was similar for mutations in DNMT3A, it did not achieve statistical significance. Overall, >95% of mutations persisted when evaluated longitudinally at one year or three years after baseline. However, there were fewer changes from baseline allelic fraction across all genes in year 1 compared to year 3 (Supplementary Figs. 10, 11).
Fig. 4 |. Mutations pose AML risk irrespective of the variant allele fraction.
a, Maximum allelic fraction for baseline mutations per gene: DNMT3A, n = 69 cases, 34 controls; TET2, n = 47 cases, 10 controls; TP53, n = 21 cases; SRSF2, n = 13 casees; IDH2, n = 12 cases; JAK2, n = 10 cases, 1 control; SF3B1, n = 11 cases, 2 controls; U2AF1, n = 6 cases; IDH1, n = 3 cases; RUNX1, n = 3 cases. Proportion of cases with AML (red) and controls (blue) is shown in each bin (width, 2.5%). b, Receiver operating characteristic curves indicating the percentage of true-positive rates compared to the percentage of false-positive rates for detecting cases with AML. Performance is shown for mutations in any gene significantly associated with cases with AML (left; DNMT3A, TET2, IDH1, IDH2, SRSF2, SF3B1, U2AF1, TP53; n = 164 (118 cases with AML, 44 controls)) or the same set of genes excluding DNMT3A; n = 94 (81 cases with AML, 12 controls) (right). c, Annual rate of VAF change per year influences kinetics of AML diagnosis for TP53 (n = 7) and IDH2 (n = 8). Time to AML (years) is plotted against fold change in VAF at year 1 or year 3 compared to baseline. Regression line is indicated. R2 and P values, linear regression. DNMT3A, n = 16; TET2, n = 13; SRSF2, n = 4; JAK2, n = 7; SF3B1, n = 5; U2AF1, n = 4.
Association between non-AML cancer history and mutations
We examined the mutational patterns of the 21 cases and 19 controls that had a history of malignancy at baseline, including breast, lung, bladder, endometrial, ovarian and colon cancer. Treatment history for the cancer was not known. In total, 42% of cases (n = 9 out of 21) and 26% of controls (n = 5 out of 19) with a prior history of cancer at baseline were found to have mutations (not significant). Although the absolute number of cases and controls with a prior cancer history was low, we did note that 3 out of 10 cases with a prior history of cancer had IDH2 mutations at baseline evaluation and that all three of these cases had prior breast cancer. None of the cases or controls with a prior history of cancer had TP53 gene mutations. Overall, because of the low number of cases and controls with a prior history of cancer, we did not have sufficient power to study this effect.
Progression to AML from baseline mutations
Despite having selected for participants who eventually developed AML, we noted the absence of NPM1 and FLT3 mutations, which are among the two most frequently recurring driver mutations in AML1,18. This finding is consistent with other reports of their absence in clonal hematopoiesis, and suggests that they may have a cooperative role in AML pathogenesis4,5. We identified a single case for whom follow-up at year 1 preceded AML diagnosis by <30 days (Fig. 5, case A) and compared mutations at baseline and year 1 (<30 days prior to AML diagnosis). The depth of sequencing coverage at NPM1 insertion sites was similar at both time points (around 450x). The participant had an IDH2 mutation (8% VAF) at baseline and after one year had acquired a new NPM1 type-A mutation (14% VAF), along with an increased VAF of a mutation in IDH2 (13%). AML was diagnosed less than 30 days later. The rapid development of AML after the acquisition of an NPM1 mutation suggests cooperation with the pre-existing IDH2 clone given the similarity of their allelic fractions.
Fig. 5 |. Clonal evolution towards AML in selected patients.
Clonal composition and evolution are shown for four selected examples of participants who were evaluable serially (cases A, B, C and D). Peripheral blood was sampled at baseline and years 1 or 3. The x axis indicates time (years). The y axis indicates the VAF where the maximum possible VAF is 1 (100%). Mutated genes are shown at each time point as indicated on the line chart. Time of AML diagnosis (AML Dx) relative to baseline is indicated by the red vertical dotted line. a, Case A, an IDH2 mutation (8% VAF) is present at baseline at lower VAF and persists at year 1 at 13% VAF with an acquired NPM1 type-A mutation at 14% VAF. AML diagnosis occurs <30 days after the year 1 sample. b, Case B: a DNMT3A mutation remained stable from baseline to year 1 follow-up. VAFs of JAK2 and SF3B1 increased from 5% to 24% before AML diagnosis at 4.3 years after baseline. c, Case C: clonal expansion of TP53 from 3% to 21% with acquired SRSF2 and CUX1 mutations between baseline and year 3 follow-up. TET2 remains relatively stable. AML diagnosed at 4.4 years after baseline. d, Case D: low VAF mutations in IDH2 and TET1 expand by year 3 along with acquisition of SRSF2 in the presence of a relatively stable DNMT3A mutation. AML diagnosis occurs 6.6 years after baseline.
Other patterns of clonal evolution and expansion are demonstrated in representative cases B, C and D (Fig. 5). Mutations in genes that are typically associated with clonal hematopoiesis, such as DNMT3A and TET2, were shown to generally have stable or minimally increased VAFs in follow-up. Progression to AML was often preceded by the acquisition of new mutations or by the expansion of mutations in other genes, such as RUNX1 or TP53 (Fig. 5). With the exception of case B, who had a large DNMT3A-mutated clone with multiple subclones, comutations in the same cell formally require single cell analysis.
Discussion
Peripheral blood samples collected years before AML diagnosis enabled a unique and unprecedented opportunity to establish the existence of a genomically defined premalignant state of AML years before the development of overt disease and to determine the specific risk and time to AML development that are associated with various mutations, comutations and clone sizes. We found that study participants who eventually developed AML had higher mutational complexity at WHI baseline evaluation than age-matched controls. The presence of a mutation at baseline was associated with increased odds of developing AML. Individually, the most-significant mutations associated with increased odds of AML included those in TP53, IDH1 and IDH2, spliceosome (SRSF2, SF3B1 and U2AF1), TET2 and DNMT3A. As mutations in DNMT3A and TET2 were common in controls as well, it is important to note that both higher VAFs (>10%) and the presence of a higher number of variants (two or more) in these genes were associated with higher risk of AML. The median time to development of AML was shorter in subjects with mutations present at baseline WHI evaluation and those participants with baseline TP53 mutations and DNMT3A comutated with spliceosome genes were more likely to develop AML within the following five years. Participants with mutations in the RUNX1 gene all developed AML within two years after baseline, but the number of cases was too low to make statistical inferences. The time to develop AML was inversely correlated with an increasing VAF for somatic mutations in TP53 and IDH2. Although having a mutation in any gene from our panel at baseline increased the risk of developing AML, we identified a set of genes with high AML penetrance, including TP53, IDH1 and IDH2, SRSF2, SF3B1 and U2AF1. Any detectable VAF in the high-penetrance genes at a sensitivity cutoff of 1% was associated with increased risk of AML. When serial samples were available, we found that a more rapid rise in the allelic fraction of IDH2 or TP53 mutations was significantly associated with a shorter time to AML development. As expected, serial samples also revealed the stepwise acquisition of mutations leading to AML. Mutations in genes commonly associated with clonal hematopoiesis, such as DNMT3A and TET2, were maintained over time, while new dominant subclones arose in genes such as NPM1, TP53, and SRSF2 preceding the development of AML. We note that the absolute rate of clonal hematopoiesis in our study was on the higher end of the reported range for controls4–6,19,20, likely in part because our study consisted of participants older than 50 years of age and was performed at relatively high non-duplicate sequencing depth with a lower 1% allelic fraction cutoff. Additionally, while there is increasing consensus around the biological definition of clonal hematopoiesis, there is no universally accepted technical definition. In general, studies have varied with respect to both the demographics of the study population and technical factors, among which are the sequencing techniques used (whole-genome, whole-exome or targeted sequencing), analytical approaches, in-solution capture versus amplicon sequencing, sequencer technology, depth of coverage and allelic fraction cutoff. For example, early seminal studies that helped to establish our general understanding of the prevalence of clonal hematopoiesis mutations were performed with relatively low-coverage whole-exome sequencing with lesser sensitivity for mutations present at low allelic fractions, as well as suboptimal coverage of genes recurrently mutated in clonal hematopoiesis (for example, mutations in TET2 for which there was essentially no coverage of coding exons outside of exon 3)4,5. Our study finds that >70% of TET2 mutations in both our case and control groups outside exon 3, suggesting possibly >3-fold underestimation of clonal hematopoiesis mutations in TET2 by previous work. Moreover, recent data20 using high-depth error-corrected sequencing demonstrated nearly ubiquitous clonal hematopoiesis dominated by mutations in DNMT3A and TET2. Irrespective of these differences, we observe in our study significantly increased mutations in cases with AML relative to clonal hematopoiesis in the control population, a significantly shorter latency period to AML development when mutations were detected, and mutations in genes of known significance among the cases with AML, such as TP53 and IDH2, that are not found among the clonal hematopoiesis controls.
A major strength of our study is having a cohort of normal controls that was reliably followed over 10 years for outcomes, including detailed baseline demographics, medical history and laboratory data, thus providing a solid scientific base for a matched case–control analyses at the population level. A limitation to our study is that we could not perform sequencing studies of blood or bone marrow taken at the time of AML diagnosis. However, for participants with serial samples, we noted that the vast majority of mutations persist at one or three years after baseline, indicating their stability and often increasing clone size (Supplementary Fig. 11). Another limitation is that because our study included only women, there is also likely to be underestimation of mutations in X-linked genes with male predominance, such as PHF6 and ZRSR221,22, as well as possible overrepresentation of mutations in DNMT3A, as has been reported previously4. Several commonly mutated genes in AML were absent from our study, notably FLT3 and NPM1, suggesting that the acquisition of these mutations may be later events in AML ontogeny. Although pre-existing mutations in TP53 and spliceosome genes were strongly associated with the development of AML in this study, it is well-known that these mutations are not exclusive to AML and are also associated with other hematological malignancies and as a rare clonal hematopoiesis event in normal populations23. However, these data provide a strong rationale for following participants with TP53 mutations. By contrast, IDH2 Arg140 mutations are relatively more specific to AML. While it is possible that some of the participants in our study developed myelodysplastic syndrome after baseline and prior to developing AML, it should be noted that all of our cases and controls had normal baseline hematological parameters. Thus, further prospective monitoring studies will be required to confirm the specificity, or lack thereof, of these mutations in predicting AML versus other hematological disorders. Within our study, we determined that for specific genes (that is, TP53, IDH2, SF3B1, SRSF2 and U2AF1), a VAF cutoff correlating with a falsepositive fraction of less than 1% could be achieved. These estimates would be a starting point for future studies to estimate the real falsepositive fraction in a general population.
In conclusion, our study establishes the premalignant landscape of mutations preceding overt AML that is present in peripheral blood at a median of 9.6 years prior to the diagnosis of AML. In our study, all participants with TP53 and IDH mutations eventually developed AML. In comparison to the mutational landscape of de novo AML24,25, our premalignant AML ‘case’ group demonstrated a similar frequency of mutations in genes that have high AML penetrance, especially IDH2 and TP53. DNMT3A and TET2 are accordingly overrepresented in our case cohort compared to de novo AML since most instances of these mutations do not lead to AML on a population level. Our results showing an association of high allelic fractions in DNMT3A and TET2 along with clonal complexity with a higher risk of AML is important in discriminating who would likely progress to AML. The ability to detect and identify high-risk mutations suggests that monitoring strategies for patients, as well as clinical trials of potentially preventative or disease-intercepting interventions should be considered. Indeed, molecularly targeted therapy is already available for IDH2 mutations26 and is under development for mutations in other candidate genes found in this study including IDH1, TP53 and spliceosome genes27–29. Moreover, preclinical data suggest that vitamin C, in clinically achievable doses, may reverse leukemogenesis mediated by TET2 deficiency30. Thus, as the cost of detecting mutations declines and its use in cancer surveillance increases, the feasibility of monitoring becomes more reasonable, especially when associated with a low false-positive rate and the potential availability of therapies with favorable toxicity profiles. We propose that patients at increased risk of developing AML should be followed longitudinally in long-term monitoring studies using next-generation sequencing to further evaluate the prognostic and predictive importance of a premalignant state in this disease. Data from these studies will provide a robust rationale for clinical trials of preventative intervention strategies in populations at high risk of developing AML.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41591-018-0081-z.
Methods
De-identified samples and data were obtained from the WHI and collection of these samples and data was performed with approval from the WHI Ancillary Committee and the WHI Extension Study Steering Committee. Informed consent from participants was obtained at the onset of the WHI trial conducted by participating centers (IRB approval reference number 3467-EXT). All ethical regulations were followed. Genomic DNA was provided by WHI in a blinded manner, in which case–control status and clinical covariates were revealed only after variant calling was completed. Library generation and amplification were performed using a low-error rate Hi-Fi DNA polymerase according to the Kapa HyperPrep protocol (Kapa Biosystems). Dual sample indexing, rather than single indexing, of libraries was performed to minimize signal spread errors arising from misidentification of multiplexed samples31. Targeted sequencing of genes recurrently mutated in hematological malignancies was performed using custom capture probes (Nimblegen) to a median coverage of 2,000× for both cases with AML and controls (Supplementary Fig. 1). Reads were aligned to the 1000 Genomes Phase 2 human reference genome and decoy contigs (hs37d5) using BWA MEM32. Variants were detected using VarDictJava33 using a 1% VAF cutoff and filtered for artifacts as described previously34. Additional method details can be found in the Supplementary Information.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We acknowledge all our donors to Leukemia Fighters, without whom this work would not have been possible; the women who generously participated in the WHI study. We thank J. Z. Xiang and the Weill Cornell Genomics Core Facility as well as J. Catalano of the Englander Institute for Precision Medicine for assistance in sequencing; L.-B. Yan for technical assistance. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts N01WH22110, 24152, 32100–2, 32105–6, 32108–9, 32111–13, 32115, 32118–32119, 32122, 42107–26, 42129–32, and 44221, and the Cancer Center Support Grant NIH:NCI P30CA022453. We acknowledge the dedicated efforts of investigators and staff at the WHI clinical centers, the WHI Clinical Coordinating Center, and the National Heart, Lung and Blood program office (listing available at http://www.whi.org). We are additionally grateful for funding from the Sandra and Edward Meyer Cancer Center, which partially supported this study (D.C.H.).
Footnotes
Competing interests
The authors declares no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-018-0081-z.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mardis ER et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med 361, 1058–1066 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley TJ et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med 363, 2424–2433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding L et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genovese G et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie M et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coombs CC et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials 19, 61–109 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Anderson GL et al. Implementation of the Women’s Health Initiative study design. Ann. Epidemiol 13, S5–S17 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Bergstralh EJ, Kosanke JL & Jacobsen SJ Software for optimal matching in observational studies. Epidemiology 7, 331–332 (1996). [PubMed] [Google Scholar]
- 11.Bowman RL, Busque L & Levine RL Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward PS et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makishima H et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 119, 3203–3210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang SJ et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 119, 4480–4485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivier M, Hollstein M & Hainaut P TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol 2, a001008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadia TM et al. TP53 mutations in newly diagnosed acute myeloid leukemia: clinicomolecular characteristics, response to therapy, and outcomes. Cancer 15, 3484–3491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuelsen SO A psudolikelihood approach to analysis of nested case–control studies. Biometrika 84, 379–394 (1997). [Google Scholar]
- 18.Palmisano M et al. NPM1 mutations are more stable than FLT3 mutations during the course of disease in patients with acute myeloid leukemia. Haematologica 92, 1268–1269 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Zink F et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young AL, Challen GA, Birmann BM & Druley TE Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun 7, 12484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SS et al. Loss-of-function mutations in the splicing factor ZRSR2 are common in blastic plasmacytoid dendritic cell neoplasm and have male predominance. Blood 122, 741 (2013). [Google Scholar]
- 22.Mori T et al. Somatic PHF6 mutations in1760 cases with various myeloid neoplasms. Leukemia 30, 2270–2273 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Lawrence MS et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Cancer Genome Atlas Research Network.. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papaemmanuil E et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enasidenib approved for AML, but best uses unclear. Cancer Discov 7, OF4 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Lee SC & Abdel-Wahab O Therapeutic targeting of splicing in cancer. Nat. Med 22, 976–986 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montalban-Bravo G, Garcia-Manero G & Jabbour E Therapeutic choices after hypomethylating agent resistance for myelodysplastic syndromes. Curr. Opin. Hematol 25, 146–153 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Sabapathy K & Lane DP Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol 15, 13–30 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Cimmino L et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170, 1079–1095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kircher M, Sawyer S & Meyer M Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
- 33.Lai Z et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 44, e108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.