Highlights
-
•
First-year hippocampal atrophy in stroke is more accelerated ipsi-lesionally.
-
•
Volume estimation is not impacted by hemisphere side, study group, or scan timepoint.
-
•
Segmentation method-hippocampal size interaction determines volume estimation.
-
•
FreeSurfer/Subfields and fsl/FIRST segmentations agreed best with manual tracing.
Keywords: Hippocampal atrophy, Freesurfer, Linear mixed-effect model, Magnetic resonance imaging, Stroke
Abstract
We assessed first-year hippocampal atrophy in stroke patients and healthy controls using manual and automated segmentations: AdaBoost, FIRST (fsl/v5.0.8), FreeSurfer/v5.3 and v6.0, and Subfields (in FreeSurfer/v6.0). We estimated hippocampal volumes in 39 healthy controls and 124 stroke participants at three months, and 38 controls and 113 stroke participants at one year. We used intra-class correlation, concordance, and reduced major axis regression to assess agreement between automated and ‘Manual’ estimations. A linear mixed-effect model was used to characterize hippocampal atrophy.
Overall, hippocampal volumes were reduced by 3.9% in first-ever stroke and 9.2% in recurrent stroke at three months post-stroke, with comparable ipsi-and contra-lesional reductions in first-ever stroke. Mean atrophy rates between time points were 0.5% for controls and 1.0% for stroke patients (0.6% contra-lesionally, 1.4% ipsi-lesionally). Atrophy rates in left and right-hemisphere strokes were comparable. All methods revealed significant volume change in first-ever and ipsi-lesional stroke (p < 0.001).
Hippocampal volume estimation was not impacted by hemisphere, study group, or scan time point, but rather, by the interaction between the automated segmentation method and hippocampal size. Compared to Manual, Subfields and FIRST recorded the lowest bias. FreeSurfer/v5.3 overestimated volumes the most for large hippocampi, while FIRST was the most accurate in estimating small volumes. AdaBoost performance was average.
Our findings suggest that first-year ipsi-lesional hippocampal atrophy rate especially in first-ever stroke, is greater than atrophy rates in healthy controls and contra-lesional stroke. Subfields and FIRST can complementarily be effective in characterizing the hippocampal atrophy in healthy and stroke cohorts.
1. Introduction
Stroke is a major global cause of disability and death (Strong et al., 2007; Maier et al., 2015; Aerts et al., 2016). People with stroke have a higher incidence of dementia (Makin et al., 2013), and up to one-third of stroke survivors develop post-stroke dementia (PSD) in the years following the initial stroke incident (Mok et al., 2017). Understanding the trajectory of cognitive decline and associated brain changes over the first year after stroke is crucial for developing treatments to delay the onset of PSD and modify its course.
Magnetic resonance imaging (MRI) has been successfully used for the detection of structural changes in PSD (Mijajlović et al., 2017). Combined with neuropsychological testing, quantitative MRI methods show promise as important diagnostic tools (Ystad et al., 2009). In many quantitative MRI studies of diseased populations (Frodl et al., 2006, M Meisenzahl et al., 2009, Woon et al., 2010, Apfel et al., 2011), including subcortical ischemic vascular dementia (Fein et al., 2000), smaller hippocampal volumes have been reported. In major stroke survivors, hippocampal atrophy has been associated with cognitive decline (Gemmell et al., 2012; Kliper et al., 2013).
The segmentation of structures characterized by morphological complexity – like the hippocampi – is challenging. Despite serious efforts to automate hippocampal segmentation, this is still commonly performed by human experts (Maglietta et al., 2016). However, manual segmentation is restricted by time and is costly for large datasets, has lower reproducibility unless protocols are strictly adhered to, and its results may be influenced by rater bias (Colon-Perez et al., 2016). The high reproducibility of automated methods reduces bias and facilitates the replication of findings between studies, in addition to allowing for faster processing. However, most longitudinal validation studies of automated segmentation have focused on healthy (Morey et al., 2009; Perlaki et al., 2017) and Alzheimer's disease (AD) cohorts (Morra et al., 2010; Mulder et al., 2014; Cover et al., 2016; Maglietta et al., 2016; Cover et al., 2018). Automated segmentation has not been validated for the characterization of hippocampal atrophy in stroke.
Assessing longitudinal brain atrophy in conditions with destructive brain lesions, such as stroke, can be particularly challenging given the dynamic structural alterations before and after the stroke. The timing and sample size of MRI datasets pose challenges to conclusions about structural atrophy. There is also the challenge posed by the inclusion of patients with recurrent (i.e., prior) stroke who may already have suffered significant regional brain atrophy including in the hippocampus. While we have looked at brain atrophy in the first three months post-onset (Brodtmann et al., 2012; Brodtmann et al., 2013; Li et al., 2015), relatively longer term trajectories of hippocampal volume changes have not been examined. Given that most functional recovery occurs in the first three months after stroke (Lee et al., 2015), the three-month time point may serve as a better baseline for assessing longitudinal brain atrophy.
Researchers have examined whole-brain (Seghier et al., 2014) and hippocampal (Schaapsmeerders et al., 2015) atrophy in young and old patients many years after stroke. They reported lower volumes ipsi-lesionally in the whole-brain and in the hippocampus. However, the profile and rates of hippocampal atrophy over the first year remain largely unknown.
Previously (Khlif et al., 2018), we assessed the agreement between a number of automated segmentation methods and manual tracing in estimating hippocampal volumes in healthy participants and stroke patients at three months post-stroke. The top performers (AdaBoost, FIRST, and Subfields) in that study, in addition to FreeSurfer/v5.3 and v6.0, were used in the current study. The latter two methods were included in order to validate the improvement in hippocampal volume estimation sought with the evolution of FreeSurfer algorithms. We expected the algorithms’ performance at 12 months to be consistent with their previous performance at three months. Moreover, we needed to evaluate their performance in assessing hippocampal atrophy between three and 12 months to better inform future studies. Accordingly, our aims were to:
-
(1)
Quantify hippocampal volume change in healthy and ischemic stroke participants in the period between three and 12 months after stroke (based on volumes estimated using manual and automated segmentation methods).
-
(2)
Quantify hippocampal volume change in left-hemisphere stroke compared to right-hemisphere stroke, in first-ever stroke compared to recurrent stroke, and in ipsi-lesional hippocampus compared to contra-lesional hippocampus.
-
(3)
Evaluate the agreement between manual tracing and automated segmentation in estimating hippocampal volumes at 12 months post-stroke.
-
(4)
Assess the sensitivity of automated algorithms in detecting longitudinal hippocampal volume change between three and 12 months post-stroke.
2. Materials and methods
2.1. Participants
Participants were sampled from the Cognition And Neocortical Volume after Stroke (CANVAS) study (Brodtmann et al., 2014). Briefly, participants were recruited from three sites in Melbourne (Austin Health, Eastern Health, and Melbourne Health), with all MRI scans held at The Florey Institute of Neuroscience and Mental Health, Austin Hospital campus. Ethical approval was granted by each hospital's Human Research Ethics Committee and all participants provided informed consent. Participants completed an interview (to collect basic demographic and medical history information), 3T MRI scans and neuropsychological assessments, at five time points: baseline (within six weeks of index stroke), three months, one year, three years, and five years post-stroke (post-baseline for controls). Patients diagnosed with primary hemorrhagic stroke, transient ischemic attack (TIA), venous infarction, or significant medical comorbidities were excluded from participation. Age-matched healthy controls with no history of stroke or TIA were also recruited. All participants had no pre-existing dementia, neurodegenerative disorders, major psychiatric illnesses or substance abuse problems.
The findings presented in this paper were based on CANVAS data from 163 participants at three months (39 control and 124 stroke) and 151 participants at one year (38 control and 113 stroke). Data for 38 control and 110 stroke participants were available at both time points.
2.2. MRI acquisition
Whole brain images were acquired on a 3T Siemens Tim Trio Scanner (Siemens, Erlangen, Germany). The MR images were obtained using a T1-weighted 3D magnetization-prepared rapid gradient sequence: 160 coronal 1 mm-thick slices, 1900 ms repetition time, 2.6 ms echo time, 900 ms inversion time, 9° flip angle, 256 × 256 field-of-view, and 1 × 1 mm2 in-plane resolution.
2.3. Hippocampal volumetry
Left and right hippocampal volumes were estimated for each participant using the segmentation algorithms described below. Volumes were expressed in mm3.
2.3.1. AdaBoost
AdaBoost is a machine learning-based segmentation method. It produced accurate, high-quality automated segmentations in 400 participants from the Alzheimer's disease Neuroimaging Initiative (Morra et al., 2008). The AdaBoost-based method classifies voxels as belonging to the hippocampus based on thousands of features, which are learned from a training set of manually delineated data. The implementation of AdaBoost used in the current study, applied by one of the authors (A.R.) using the neuGRID platform (Redolfi et al., 2013), was trained on the EADC-ADNI harmonized protocol for hippocampal segmentation. Adaboost corrects images through the N3 bias field correction algorithm and then registers the brain to the standard ICBM-152 template through affine registration using the fsl/FLIRT algorithm.
2.3.2. FIRST
FIRST, (fsl/5.0.8) (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki), is a model-based segmentation tool (Patenaude et al., 2011). The shape and appearance models used in FIRST are constructed from manually segmented images provided by the Center for Morphometric Analysis, Boston MA, United States. The manual labels are parameterized as surface meshes and modelled as a point distribution model. Deformable surfaces are used to automatically parameterize the volumetric labels in terms of meshes. The deformable surfaces are constrained to preserve vertex correspondence across the training data. Furthermore, normalized intensities along the surface normals are sampled and modeled. The shape and appearance model is based on multivariate Gaussian assumptions. FIRST searches through linear combinations of shape modes of variation for the most probable shape instance given the observed intensities in a T1-weighted image.
T1-weighted volumes were RAS-oriented and centred using the alignment script ‘acpcdetect_v2.0′ (https://www.nitrc.org/frs/?group_id=90&release_id=3772). Centring was based on a midpoint between anterior and posterior commissures. The images were then registered using ‘first_flirt’ which runs two-stage affine registration to MNI152 space at 1 mm resolution. The first stage is a standard 12 degrees of freedom registration to the template. The second stage applies 12 degrees of freedom registration using an MNI152 sub-cortical mask to exclude voxels outside the sub-cortical regions.
2.3.3. FreeSurfer
FreeSurfer (http://surfer.nmr.mgh.harvard.edu/) segmentation includes 1) motion correction and averaging (Reuter et al., 2010) of multiple images when available, 2) removal of non-brain tissue (Segonne et al., 2004), 3) Talairach transformation (affine transform to the MNI305 atlas), 4) segmentation of subcortical white matter and deep gray matter structures, including hippocampus, amygdala, caudate, putamen, ventricles (Fischl et al., 2002; Fischl et al., 2004), 5) intensity normalization (Sled et al., 1998), 6) tessellation of the gray matter-white matter boundary, 7) topology correction (Fischl et al., 2001; Segonne et al., 2007), and 8) surface deformation following intensity gradients (Dale et al., 1999; Fischl and Dale 2000).
Segmentation was performed using the longitudinal processing stream (recon-all -long) in FreeSurfer (Han et al., 2006; Reuter et al., 2012). We used both v.5.3 and v.6.0 as we expected differences between the two versions of FreeSurfer. In particular, a bug for time point addition present in longitudinal processing of FreeSurfer/v5.3 was fixed in FreeSurfer/v6.0. In addition, there were changes in the computation of the cerebellum which affected the estimation of total intracranial volume (TIV). This was particularly of concern given that TIV was included as a covariate in this study.
2.3.4. Subregional segmentation in FreeSurfer
We used the longitudinal ‘Subfields’ function in FreeSurfer/v6.0 (longHippoSubfieldsT1.sh) which performs automated subregional segmentation of the hippocampus based on a statistical atlas built primarily upon ultra-high resolution (∼0.1 mm isotropic) ex vivo MRI data (Iglesias et al., 2015). This atlas has addressed a number of limitations of the in vivo atlas distributed with FreeSurfer/v5.3. The Subfields segmentation requires a completion of the standard segmentation beforehand (recon-all -long). We used the sum of all subregions’ volumes to compute the (whole) hippocampal Subfields volume.
2.3.5. Manual tracing
Hippocampi were manually traced following guidelines described in the EADC-ADNI harmonized protocol for manual hippocampal segmentation (Boccardi et al., 2015; Frisoni et al., 2015). Special attention was paid to the determination of the most rostral and caudal slices, exclusion of the choroid plexus, inclusion of the alveus/fimbria, and exclusion of internal CSF pools within the hippocampus. Coronal slices were used to trace the hippocampi on 3D Slicer 4.5.0.1 (Fedorov et al., 2012) available from http://www.slicer.org. Images in sagittal views were consulted to confirm hippocampal boundaries.
2.3.6. Quality control
The registered images were visually checked to confirm that the orientation and size of the subject brain corresponded with that of the template and to verify that the subcortical structures were appropriately situated. The segmented images were manually inspected to verify that the segmentation of the hippocampus did not overlap with that of another adjacent structure, such as the amygdala, and that no part of the hippocampus was left out during segmentation.
2.4. Agreement between segmentation methods
Agreement between manual tracing and each of the automated segmentation methods was quantified using the intra-class correlation coefficient (ICC) (Shrout and Fleiss 1979), and Lin's concordance correlation Coefficient (CCC) (Lin 1989). In the computation of ICC, no generalization to a larger population of raters was assumed. Since we were interested in comparing specific segmentations at hand, the methods were assumed fixed. If assumed random, ICC would be similar to CCC.
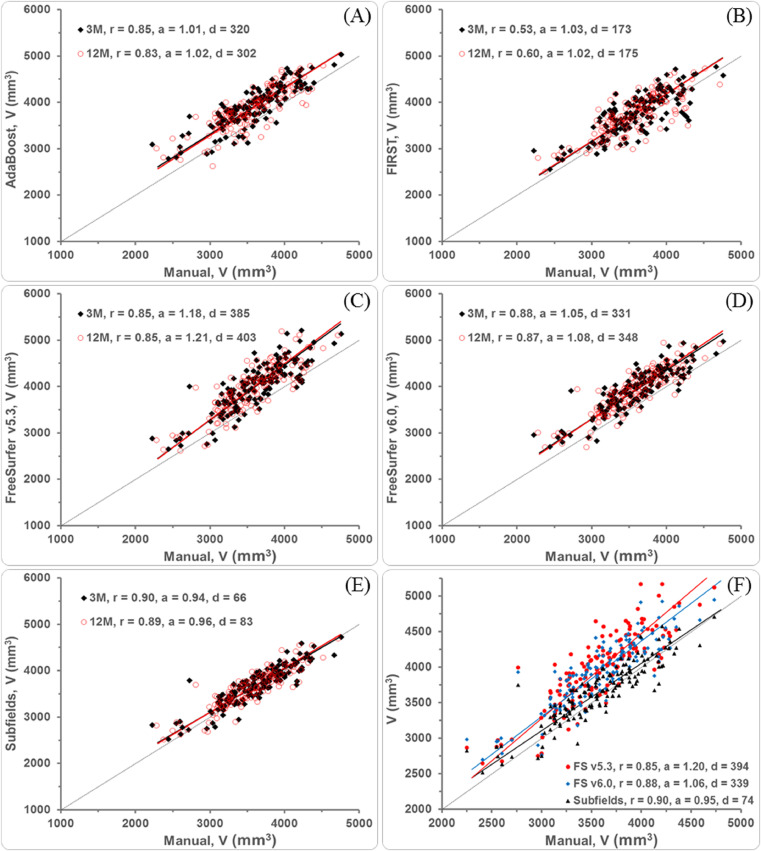
In the correlation plots (Fig. 1), we graphically included ‘best-fit’ lines with slopes and intercepts determined by a reduced major axis regression (RMAR) analysis, where neither method is assumed ‘gold-standard’. For a perfect fit line (slope, a = 1), the intercept serves to indicate the amount of fixed bias in the compared method. For slopes larger or smaller than 1, the bias is proportional. Thus, we only reported the bias at mean hippocampal volume (volume difference, d).
Fig. 1.
Correlations of automated segmentation methods to manual tracing (plots A-E) at three (3 M) and 12-month (12 M) post-stroke time points (a = slope of RMAR fit line, d = mean hippocampal volume difference for compared methods, r = Pearson's correlation coefficient). Plot (F): Correlations of the FreeSurfer (FS) methods to manual tracing with 3 M and 12 M volumes combined.
2.5. Statistical analysis–group comparison
Statistical analyses were completed in MATLAB (Statistics Toolbox, Release 2015a, The MathWorks Inc., Massachusetts, USA). All analyses used an alpha threshold of 0.05 to determine statistical significance.
2.5.1. Group demographics
Differences between groups on demographics were examined using two-sample t tests (for age and TIV), the Fisher Exact test (for sex), and the Wilcoxon rank sum test (for years of education).
2.5.2. Cross-sectional and longitudinal group analysis of hippocampal volume and volume change
We compared hippocampal volumes in the control and stroke groups cross-sectionally and longitudinally between the three-month (3 M) and 12-month (12 M) time points. For this, we used a linear mixed-effect (lme) model where the estimated volumes were taken as the dependent variable and age, sex, years of education, and TIV were included as covariates. Age was included to correct for brain volume loss due to normal aging, sex to account for sexual dimorphism, education to adjust for its protective effect on cerebral structures, and TIV to control for head size variations. The covariates were checked for non-collinearity (the maximum variance inflation factor was 1.3) and residuals from model fitting were checked for normality and heteroscedasticity.
The lme model allowed for repeated measures and accommodated missing values in our dataset. For better generalization of our findings, we designed our mixed-effect model based on a maximal random effects structure. Time (a categorical variable with 3 M and 12 M as input) was included as a fixed effect, in addition to group (categorical variable with control and stroke, or stroke subclasses as input), sex (categorical variable), age, years of education, and TIV. The inclusion of first-order interactions between fixed effects was determined by comparing models (with and without the interaction term) using the simulated likelihood ratio test (500 replications, 95% confidence level). Among all interactions, only the group-time interaction was found to be significant. For fitting the data, maximum likelihood was used to estimate the model parameters.
A random intercept and a random (time) slope varied by subject were also included in the lme model to fulfil the non-dependency requirement for within-subject and to allow for heterogeneity in estimated measures between subjects. The random intercept and slope were modelled as uncorrelated based on comparisons between models (with and without the correlation assumption). The likelihood ratio test showed the correlation between the random intercept and random slope to be non-significant.
A model (lme.1) was first used to fit hippocampal volumes averaged across hemispheres. The MATLAB ‘fitlme’ function was employed to fit the model using the ‘reference’ method of coding dummy variables (I[.]). Here, the response (i.e., dependent variable) at three months for male control participants was used as the reference. Accordingly, the lme.1 model was formulated as:
| (1) |
where Yijm represents hippocampal volume, i is the observation index, j = 2 corresponds to the 12 M time point, m is the participant index, and β[.] are the fixed-effect coefficients. The random effects and observation error had the following prior distributions:
After fitting the lme model, we tested the residual errors for normality using the Kolmogorov-Smirnov test. We excluded influential participants (maximum of three) as needed to meet the normality test for residuals. Residuals from a sample model fitting are shown in Supplemental Fig. 1 where they appear random regardless of group or hippocampal size and where their distribution is shown normal.
In a second model (lme.2), we fitted responses after grouping the stroke volumes based on hemisphere side in which stroke had occurred. Hippocampal volumes were grouped ipsi-lesionally and contra-lesionally, where both the ipsi-lesional and contra-lesional groups contained a mixture of left and right hemisphere volumes. Since the ipsi-lesional and contra-lesional groups were not independent, the random effects were further grouped by a subject-group interaction. For the control participants, we used volumes averaged across hemispheres as in lme.1. The three control, ipsi-lesional, and contra-lesional groups were then compared using the following extended model:
| (2) |
where k = 2, 3 is the group index.
Models lme.1 and lme.2 were also used to fit stroke volumes grouped as left vs. right, as well as first-ever stroke vs. recurrent stroke. There were 18 cases of recurrent stroke with an average of 6.5 years since last stroke. The MATLAB function ‘predict (lme)’ was used to compute fitted volumes subsequently used for estimating the percentage rate of volume change between time points per the following:
where Y3M and Y12M are respectively the fitted hippocampal volumes at three and 12 months post-stroke. The MATLAB script for implementing the linear mixed-effect models is shown in Appendix A.
3. Results
Hippocampal volumes were estimated using manual tracing and three publicly available segmentation tools, including AdaBoost, FIRST, and longitudinal FreeSurfer (v5.3, v6.0, and Subfields).
3.1. Agreement between manual and automated segmentation methods
The correlation plots showing Pearson's correlation coefficient and RMAR analysis are presented in Fig. 1 where the behaviour of each method is shown similar at three and 12-month time points. RMAR analysis showed all automated methods having a positive bias (d > 0), with Subfields and FreeSurfer/v5.3 showing lowest and highest bias, respectively. The bias shown by AdaBoost and FIRST is the least proportional to hippocampal volume (a ≈ 1). On the other hand, FreeSurfer/v5.3 overestimated large hippocampal volumes, while Subfields slightly overestimated smaller volumes.
Hippocampal volumes estimated by Subfields correlated best to manually traced volumes at both time points (r ≥ 0.89), while FIRST volumes correlated the least (r ≤ 0.6). In terms of intra-class correlation, Adaboost performed slightly better than FreeSurfer/v5.3 but slightly worse than FreeSurfer/v6.0. For absolute agreement based on Lin's CCC, Subfields scored highest; followed by FIRST (see Table 1). Side-by-side comparison showed improvement of the FreeSurfer segmentation with the introduction of v6.0 and Subfields (see Fig. 1, plot F).
Table 1.
Agreement scores between automated segmentation and manual tracing at the three and 12-month post-stroke time points.
| Method | 3-month time point | 12-month time point | ||
|---|---|---|---|---|
| CCC | ICC | CCC | ICC | |
| AdaBoost | 0.68 | 0.85 | 0.66 | 0.83 |
| FIRST | 0.75 | 0.80 | 0.74 | 0.79 |
| FreeSurfer/v5.3 | 0.64 | 0.84 | 0.62 | 0.83 |
| FreeSurfer/v6.0 | 0.69 | 0.87 | 0.68 | 0.87 |
| Subfields | 0.89 | 0.89 | 0.88 | 0.89 |
3.2. Group comparison per segmentation method
3.2.1. Demographic comparison
There was no difference in age, sex, or TIV between stroke and control participants, but the healthy controls were better educated (p < 0.05), see Table 2.
Table 2.
Demographic characteristics of the stroke and control groups at each time point.
| Variable | 3-month time point | 12-month time point | ||||
|---|---|---|---|---|---|---|
| Control | Stroke | p | Control | Stroke | p | |
| Number | 39 | 124 | 38 | 112 | ||
| Age, years | 68.5 (6.7) | 67.0 (11.7) | 0.77a | 69.3 (6.8) | 67.9 (12.3) | 0.49a |
| Mean (SD) | ||||||
| Men | 24 (61.5) | 86 (69.4) | 0.36b | 23 (60.5) | 79 (70.5) | 0.25b |
| Number (%) | ||||||
| Education, years | 17 (11, 18) | 12 (10, 15) | 0.0006c | 17 (11, 18) | 12 (10, 15) | 0.0003c |
| Median (Q1, Q3) | ||||||
| TIV (106 mm3) | 1.53 (0.12) | 1.51 (0.17) | 0.47a | 1.53 (0.12) | 1.51 (0.17) | 0.62a |
| Mean (SD) | ||||||
Q1, Q3 = 25th, 75th percentiles;.
Two-sample t-test;.
Fisher exact test;.
Wilcoxon Rank Sum test, SD = standard deviation; TIV = total intracranial volume.
3.2.2. Control vs. stroke participants: based on averaged left and right hippocampal volumes
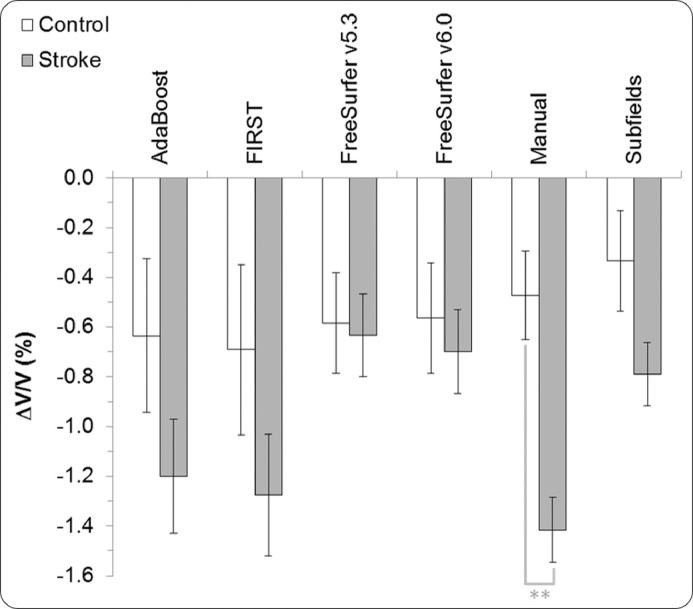
Cross-sectional comparison at three months revealed lower mean hippocampal volume for the stroke group compared to the control group, regardless of segmentation method. This difference was larger at 12 months due to the accelerated rate of hippocampal atrophy in the stroke group relative to controls (see Table 3).
Table 3.
Control and stroke hippocampal volume (V) per segmentation method at three and 12 months and relative volume change (ΔV/V) between time points (3 M = three-month time point, 12 M = 12-month time point).
| V (mm3) | Control | Stroke | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | 3M | 12M | ΔV/V (%) | 3M | 12M | ΔV/V (%) |
| AdaBoost | 4013 (348) | 3987 (341) | −0.64 (1.96) | 3863 (450) | 3816 (459) | −1.20 (2.67) |
| FIRST | 3877 (371) | 3851 (374) | −0.70 (2.16) | 3687 (461) | 3641 (457) | −1.26 (2.83) |
| FreeSurfer/v5.3 | 4143 (383) | 4119 (387) | −0.58 (1.28) | 3894 (553) | 3871 (556) | −0.63 (1.92) |
| FreeSurfer/v6.0 | 4040 (340) | 4019 (349) | −0.56 (1.40) | 3841 (496) | 3816 (505) | −0.70 (1.97) |
| Manual | 3682 (365) | 3665 (359) | −0.46 (1.13) | 3531 (460) | 3484 (453) | −1.34 (1.54) |
| Subfields | 3751 (314) | 3739 (321) | −0.33 (1.28) | 3591 (441) | 3563 (444) | −0.79 (1.46) |
At three months, a significant difference between control and stroke mean hippocampal volumes was found using FreeSurfer/v5.3 (p = 0.011) and FreeSurfer/v6.0 (p = 0.027) methods, but not with Manual. This finding was replicated at the 12-month time point: this was a result of the directly proportional bias shown by the FreeSurfer algorithms, especially in version 5.3 (see Fig. 1, plot F). Between three and 12 months, the mean hippocampal volume for controls had decreased between 0.33% (with Subfields) and 0.7% (with FIRST) compared to a volume reduction in stroke between 0.63% (with FreeSurfer/v5.3) and 1.34% (with Manual; see Fig. 2). Volume reduction in stroke was significant in all methods (p < 0.001). In particular, volume reduction in right-hemisphere strokes was significant in all methods (p ≤ 0.003). Left-hemisphere stroke volume reduction was significant in most methods (p ≤ 0.002), except for FreeSurfer/v5.3 and v6.0 (p ≥ 0.08).
Fig. 2.
Averaged hippocampal volume change per segmentation method (ΔV/V) between three and 12 months post-stroke (error bars = standard error, ** p < 0.01).
3.2.3. Control vs. stroke participants: based on lesion side
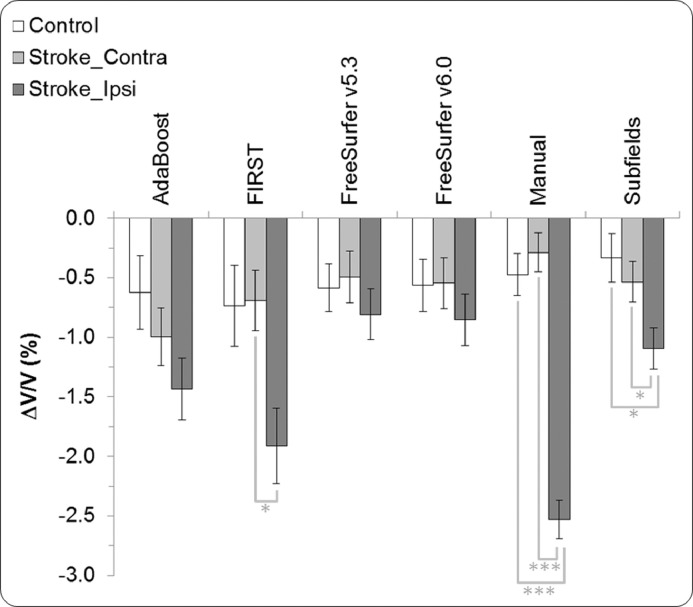
In this analysis, we compared three hippocampal volume classifications using lme.2: healthy control, ipsi-lesional stroke, and contra-lesional stroke. Hippocampal volume and atrophy for controls, estimated using this model, were very similar to those estimated using lme.1 (see Table 3). Cross-sectional analysis again revealed lower hippocampal volumes in stroke patients (both ipsi- and contra-lesionally) compared to controls at three months, with further reductions at 12 months as shown by all methods (see Table 4). Compared to the other segmentation methods, FreeSurfer/v5.3 showed that contra-lesional volumes in stroke were significantly different from control volumes at both three months (p = 0.023) and 12 months (p = 0.031). FIRST, FreeSurfer/v5.3, and FreeSurfer/v6.0 showed that the ipsi-lesional stroke volumes were significantly lower (p < 0.05) than controls at both time points, while AdaBoost, Manual, and Subfields did not show a significant difference at either time point.
Table 4.
Ipsi-lesional and contra-lesional hippocampal volume (V) per segmentation method at three (3 M) and 12 months (12 M) and relative volume change (ΔV/V) between time points.
| V (mm3) | Stroke – Contra-lesional volumes | Stroke – Ipsi-lesional volumes | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | 3M | 12M | ΔV/V (%) | 3M | 12M | ΔV/V (%) |
| AdaBoost | 3863 (487) | 3827 (499) | −0.99 (2.80) | 3860 (526) | 3803 (547) | −1.43 (3.04) |
| FIRST | 3718 (495) | 3692 (491) | −0.69 (2.97) | 3654 (497) | 3587 (504) | −1.91 (3.65) |
| FreeSurfer/v5.3 | 3889 (587) | 3872 (596) | −0.49 (2.54) | 3900 (581) | 3871 (587) | −0.81 (2.49) |
| FreeSurfer/v6.0 | 3853 (513) | 3834 (523) | −0.55 (2.50) | 3830 (523) | 3800 (535) | −0.85 (2.53) |
| Manual | 3510 (487) | 3502 (488) | −0.24 (1.90) | 3558 (482) | 3473 (480) | −2.47 (1.84) |
| Subfields | 3592 (442) | 3574 (447) | −0.54 (1.99) | 3594 (476) | 3557 (485) | −1.09 (2.00) |
In the longitudinal analysis, all segmentation methods revealed non-significant hippocampal volume change for the control group and significant volume reduction (p < 0.001) for the ipsi-lesional stroke group. No method found a statistical difference between control and contra-lesional atrophy rates. On the other hand, only Subfields (p = 0.038) agreed with Manual (p < 0.001) in showing a statistically significant difference in hippocampal volume reductions between control and the ipsi-lesional group.
The rates of volume reduction are graphically presented in Fig. 3. For the contra-lesional group, hippocampal volume reductions ranged between 0.24% (Manual) and 0.99% (AdaBoost). Ipsi-lesionally, hippocampal volume reductions ranged between 0.81% (FreeSurfer/v5.3) and 2.47% (Manual). The second largest ipsi-lesional atrophy rate was estimated by FIRST at 1.91%. Finally, FIRST (p = 0.026), Manual (p < 0.001), and Subfields (p = 0.033) all showed statistically different hippocampal atrophy rates between contra-lesional and ipsi-lesional stroke.
Fig. 3.
Contra-lesional and ipsi-lesional hippocampal volume change per segmentation method between three and 12 months post stroke (error bars: standard error, * p < 0.05, *** p < 0.001).
3.2.4. Stroke participants: first-ever vs. recurrent
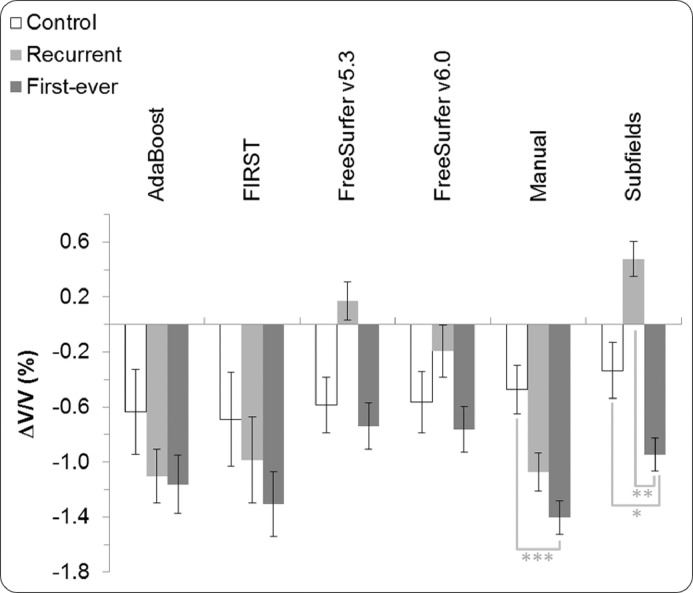
In this analysis, we compared control hippocampal volumes to first-ever stroke and recurrent stroke average volumes using lme.1. Hippocampal volume and atrophy for control participants again remained unchanged. With all methods, cross-sectional analysis revealed recurrent stroke volumes to be lower than first-ever stroke volumes at both three and 12 months (see Table 5). However, none of the methods found first-ever and recurrent stroke volumes to be significantly different at either time point. Both FreeSurfer/v5.3 and v6.0 showed first-ever and recurrent stroke volumes to be significantly lower than control at both time points. In addition, FIRST found recurrent stroke hippocampal volumes to be significantly lower than control hippocampal volumes at three (p = 0.045) and 12 months (p = 0.039). Although smaller recurrent stroke hippocampal volumes were estimated by Manual and Subfields, the latter methods did not find the difference between the two groups to be significant.
Table 5.
Hippocampal volume (V) in first-ever and recurrent stroke patients, per segmentation method, at three and 12 months and relative volume change (ΔV/V) between time points (3 M = three-month, 12 M = 12-month).
| V (mm3) | First-ever stroke | Recurrent stroke | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | 3M | 12M | DV/V (%) | 3M | 12M | DV/V (%) |
| AdaBoost | 3882 (444) | 3836 (454) | −1.16 (1.67) | 3747 (480) | 3695 (487) | −1.10 (1.57) |
| FIRST | 3720 (469) | 3673 (465) | −1.29 (0.39) | 3467 (376) | 3433 (372) | −0.98 (0.41) |
| FreeSurfer/v5.3 | 3930 (552) | 3902 (557) | −0.74 (1.75) | 3676 (527) | 3681 (517) | 0.17 (1.31) |
| FreeSurfer/v6.0 | 3876 (504) | 3849 (515) | −0.76 (1.75) | 3607 (415) | 3599 (404) | −0.19 (1.74) |
| Manual | 3547 (466) | 3499 (458) | −1.37 (1.26) | 3437 (414) | 3400 (417) | −1.08 (1.31) |
| Subfields | 3620 (448) | 3587 (453) | −0.94 (1.25) | 3393 (369) | 3410 (370) | 0.48 (1.16) |
The longitudinal analysis found the hippocampal atrophy in the first-ever stroke group to be statistically significant (p < 0.001) as shown by all methods. For the recurrent stroke group, FIRST (p = 0.046) and Manual (p = 0.005) showed a significant hippocampal volume reduction between three and 12 months. Rates of volume reduction for the control and first-ever stroke groups were significantly different as shown by Manual (p < 0.001) and Subfields (p = 0.023). However, no method showed a significant difference in atrophy rates between control and recurrent stroke groups. Between first-ever and recurrent stroke patients, only Subfields showed a significant difference in hippocampal atrophy rates between three and 12 months, in contradiction to findings based on Manual data. This discrepancy could be attributed to the small sample size of the recurrent stroke group and to relatively smaller hippocampi in this group.
The profiles of hippocampal atrophy estimated by the various segmentation methods are presented in Fig. 4. The atrophy profiles by AdaBoost, FreeSurfer/v6.0, and especially FIRST, resemble the atrophy profile by Manual, while only FreeSurfer/v5.3 and Subfields showed an increase in hippocampal volumes for recurrent stroke between three and 12-month time points. The hippocampal volume reduction for first-ever stroke ranged between 0.74% with FreeSurfer/v5.3 and 1.37% with Manual. For the recurrent stroke group, hippocampal volume change ranged from −1.10% with FIRST to 0.48% with Subfields. Further analysis based on lesion side found that neither contra-lesional nor ipsi-lesional hippocampal volume reductions were statistically different between first-ever and recurrent stroke groups as revealed by all methods, except for Subfields (p = 0.03).
Fig. 4.
Hippocampal volume change for first-ever stroke and recurrent stroke between three and 12 months per segmentation method (error bars: standard error, * p < 0.05, ** p < 0.01, *** p < 0.001).
3.3. Summary of clinical findings across methods
In previous sections, we have seen a general tendency of all automated methods to replicate – with variable degree of accuracy – the group comparisons quantified by manual segmentation (see for instance Fig. 2–4). In this section, we report on the general clinical findings with respect to hippocampal atrophy state and rate in the first year after stroke across all segmentation methods. Overall, the stroke hippocampal volume was found to be reduced at the three-month time point by 4.7% (3.7%–6.0%) compared to controls. In particular, the contra-lesional and ipsi-lesional hippocampal volumes in the stroke group were reduced at this time point by 4.6% (3.7%–6.1%) and 4.7% (3.4%–5.9%) respectively. On the other hand, volumes for first-ever and recurrent stroke groups were respectively reduced by 3.9% (3.3%–5.1%) and 9.2% (6.6%–11.3%) at three months. At the 12-month time point, the segmentation methods showed a widening of the gap between control and stroke average hippocampal volumes; now estimated at 5.1% (4.3%–6.0%); representing an increase of 0.4%. The gaps for the contra-lesional and recurrent stroke groups remained unchanged at 4.6% and 9.2% respectively. The gaps for ipsi-lesional and first-ever stroke groups, however, increased to 5.5% (4.6%–6.0%) and 4.4% (3.8%–5.3%) respectively; representing a reduction of 0.8% in the ipsi-lesional group and of 0.5% in first-ever stroke group.
As for hippocampal volume reduction between the three and 12-month time points, it was estimated across methods to be at 0.5% (0.33%–0.7%) in healthy controls and 1% (0.7%–1.34%) in ischemic stroke patients; 0.6% (0.24%–0.99%) contra-lesionally and 1.4% (0.81%–2.47%) ipsi-lesionally. Over the same period, the hippocampal volume change was estimated at 0.5% (−1.1%–0.48%) in the recurrent stroke group (0% (−1.22%–0.64%) contra-lesionally and 0.9% (−2.41%–0.23%) ipsi-lesionally) and 1% (−1.37% to −0.74%) in the first-ever stroke group (0.6% (−0.96% to −0.32%) contra-lesionally and 1.5% (−2.48% to −0.89%) ipsi-lesionally).
4. Discussion
We estimated hippocampal volume, and volume change, in the first year after ischemic stroke using automated hippocampal segmentation methods, in addition to manual tracing performed according to the EADC-ADNI harmonized protocol for hippocampal segmentation.
Various segmentation methods have been used in healthy and diseased populations for the estimation of whole and regional brain volumes and volume change over time (Morey et al., 2009; Morra et al., 2010; Mulder et al., 2014; Yassi et al., 2015; Cover et al., 2016; Maglietta et al., 2016; Perlaki et al., 2017; Rana et al., 2017; Cover et al., 2018). Depending on the targeted brain structure, segmentation methods differ in accuracy and repeatability, and preferences are determined based on comparison to manual delineation, still regarded as the ‘gold-standard’. In one study, hippocampal volumes with FreeSurfer (v4.0.5) were found to be closer to manual volumes than FIRST (fsl/v4.0.1). However, amygdala volumes with FreeSurfer were farther from hand-traced volumes than FIRST (Morey et al., 2009). In a separate study, FreeSurfer (v4.5 and v5.3) and FIRST (fsl/v5.0.7) were also assessed and compared to manual delineation of the putamen and caudate nucleus (Perlaki et al., 2017). FIRST achieved better results than both versions of FreeSurfer for putamen segmentation when compared to manual segmentation. FreeSurfer overestimated putamen volume, while FIRST did not significantly differ from manual tracing. However, for caudate segmentation, FreeSurfer and FIRST had similar performance (Perlaki et al., 2017).
In the current study, hippocampal volumes with Subfields were the closest (d3M = 66 mm3, Fig. 1) to manually traced volumes, while volumes with FreeSurfer/v5.3 were the farthest (d3M = 385 mm3). The bias with FIRST (d3M = 173 mm3) was nearly half that with FreeSurfer/v6.0 and AdaBoost. The recorded bias with FIRST and AdaBoost was nearly fixed (a ≈ 1). FreeSurfer/v6.0 (a3M = 1.05) and Subfields (a3M = 0.94) had similar but slightly higher and reversed proportional bias (i.e., increasing bias for increasing hippocampal volume with FreeSurfer/v6.0 and for decreasing volume with Subfields). In terms of correlation (r), intra-class correlation (ICC) and absolute agreement (CCC), Subfields scored the highest, while FIRST scored second for absolute agreement. FreeSurfer/v6.0 had second highest Pearson's and intra-class correlation coefficients.
We note that the performance of the presented automated segmentation methods for the three-month time point remained unchanged for the 12-month time point. We have also shown that such performance was influenced neither by study population (e.g., healthy vs. stroke) nor by hemisphere (i.e., left vs. right), but was mainly dictated by a method−hippocampal size interaction (see Supplemental Fig. 2). The slope of best-fit line (see Fig. 1) determined by RMAR is a manifestation of this interaction where methods exhibiting a bias directly proportional to the size of the hippocampus were found to overestimate the cross-sectional difference between the healthy and disease groups and to underestimate the difference in their longitudinal change. The artificial inflation and deflation of effect sizes shown in our cross-sectional and longitudinal analyses have also been reported in other studies (Xie et al., 2019). Finally, the presence of ischemic infarcts did not negatively impact on the performance of automatic segmentation. On the contrary, the automated methods performed worse when the stroke lesions were accounted for (Khlif et al., 2018).
The overall results presented in Section 3.3 indicated that, at three months, contra-lesional and ipsi-lesional stroke hippocampal volumes were nearly equally reduced compared to control participants (4.6% contra-lesionally vs. 4.7% ipsi-lesionally). A similar pattern emerged in the first-ever stroke group (3.8% contra-lesional vs. 4.0% ipsi-lesional reduction compared to control). Moreover, the small difference between contra-lesional and ipsi-lesional reductions at three months accounted for the faster atrophy rate on the ipsi-lesional side, which we postulate is accelerated by the stroke. This translated to a difference of 0.8% in atrophy rate over nine months. The higher atrophy rate between the three and 12-month time points in the stroke group, compared to controls, was driven by greater volume loss in first-ever stroke compared to recurrent stroke; though the difference in their atrophy rates was shown to be non-significant by most methods, especially Manual. This may be related to the baseline degree of atrophy being higher in recurrent stroke patients and may reflect a limitation in atrophy rate once brain volumes are already reduced.
Individually and collectively, the segmentation methods showed that the average hippocampal volume is larger in control than stroke participants at three months and even more so at 12 months. The statistical difference between these two groups depended on the way the segmentation method modulated the hippocampal volume. In particular, FreeSurfer/v5.3 overestimated hippocampal volumes the most, but more importantly, this overestimation increased with hippocampal size. Our findings showed a significant improvement of the FreeSurfer algorithm from versions 5.3 to 6.0 and eventually to the implementation of an ex vivo atlas with Subfields.
In Section 3.3, we noted that at three months, the first-ever and recurrent stroke group hippocampal volumes were reduced by 3.9% and 9.2%, respectively. Based on an average of 6.5 years between prior and index stroke in the recurrent stroke group, we estimated this group's annualised atrophy rate (prior to recent stroke) to be around 0.8% on average. On the other hand, the annualised atrophy rate for first-ever stroke patients was estimated to be 1.3% in the first year after stroke. We predict that hippocampal atrophy in our first-ever stroke group will slow over subsequent years, and plan to test this hypothesis in the CANVAS cohort.
Accelerated hippocampal volume loss has been described in many neurological and psychiatric disorders, such as AD and temporal lobe epilepsy. The rate of hippocampal atrophy is a strong predictor of progression to AD (Henneman et al., 2009). Hippocampal atrophy has also been associated with cognitive decline in stroke survivors (Gemmel et al., 2012, Kliper et al., 2013). Barnes et al. (2009) performed a meta-analysis of longitudinal hippocampal atrophy in 595 AD participants and 212 healthy controls (Barnes et al., 2009). The mean (95% CIs) annualised hippocampal atrophy rates were found to be around 4.7% (3.9, 5.4) in AD and 1.4% (0.5, 2.3) in healthy controls. The ratio of atrophy rates between these two cohorts was calculated to be 3.4 times higher for AD. In our study, we report a non-significant average hippocampal volume reduction of 0.7% (annualised) for the healthy control group (from 0.4% with Subfields to 0.9% with FIRST) compared to a significant annualised 1.3% volume reduction in the stroke group (from 0.8% with FreeSurfer/v5.3 to 1.8% with Manual). Ipsi-lesionally, the average annualised hippocampal volume reduction was 1.9%: ranging from 1.1% with FreeSurfer/v5.3 to 3.3% with Manual. Thus, the ratio of ipsi-lesional to control atrophy was at 2.6 on average (from 1.4 with FreeSurfer/v5.3 to 5.4 with Manual). The ratio of ipsi-lesional to control atrophy found in the current study trends similar to the ratio of AD atrophy to control atrophy reported in Barnes et al. (2009). The ratio of atrophy of ipsi-lesional stroke to control revealed by Manual in our study even surpassed the average ratio of atrophy in AD compared to healthy controls in Barnes et al. (2009).
Finally, the linear mixed-effect model used in this work has effectively captured the variability in the data given the normally distributed low magnitude residuals obtained. We chose maximal models for better generalization and we have provided the MATLAB script (Appendix A) to facilitate result reproducibility. However, given the small number of recurrent stroke participants in our study, findings regarding this group should be interpreted with caution in the context of this sample size limitation. We also note that the performance of the segmentation methods is governed by the type of data we used. A different MRI scanner and/or acquisition protocol may lead to different comparisons between the segmentation methods. Still, the FreeSurfer algorithm used for subregion segmentation (i.e., Subfields) can adapt to MRI variations from different acquisition hardware or pulse sequences. This has been demonstrated on three datasets with different types of MRI contrast and resolution (Iglesias et al., 2015).
5. Conclusion
Improvements in volumetric estimation methods have a significant impact on findings from MRI studies. We validated improvements in hippocampal volume estimation with the introduction of subregional segmentation in FreeSurfer based on an ex vivo atlas. FIRST also achieved good performance, and together with Subfields, they replicated the hippocampal atrophy profile defined by manual tracing for healthy control and stroke populations.
Acknowledgments
This work was supported by the National Health and Medical Research Council project grant (APP1020526), the Brain Foundation, Wicking Trust, Collie Trust, and Sidney and Fiona Myer Family Foundation. The Florey Institute of Neuroscience and Mental Health acknowledges the strong support from the Victorian Government and in particular the funding from the Operational Infrastructure Support Grant. The authors acknowledge the facilities, and the scientific and technical assistance of the National Imaging Facility at the Florey Node. The authors would also like to thank the Victorian Life Sciences Computation Initiative in the University of Melbourne (http://www.vlsci.org.au/) for support of data supercomputing in SGI Altix XE Cluster. N.E. was supported by the Australian Research Council (DE180100893).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nicl.2019.102008.
Appendix A: Linear mixed-effect model–MATLAB script

Appendix B. Supplementary materials
References
- Aerts H., Fias W., Caeyenberghs K., Marinazzo D. "Brain networks under attack: robustness properties and the impact of lesions. Brain. 2016;139:3063–3083. doi: 10.1093/brain/aww194. (Pt 12) [DOI] [PubMed] [Google Scholar]
- Apfel B.A., Ross J., Hlavin J., Meyerhoff D.J., Metzler T.J., Marmar C.R., Weiner M.W., Schuff N., Neylan T.C. "Hippocampal volume differences in gulf war veterans with current versus lifetime posttraumatic stress disorder symptoms. Biol. Psychiatry. 2011;69(6):541–548. doi: 10.1016/j.biopsych.2010.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J. A meta-analysis of hippocampal atrophy rates in Alzheimer's disease. Neurobiol Aging. 2009;30(11):1711–1723. doi: 10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccardi M., Bocchetta M., Apostolova L.G., Barnes J., Bartzokis G., Corbetta G., DeCarli C., deToledo-Morrell L., Firbank M., Ganzola R., Gerritsen L., Henneman W., Killiany R.J., Malykhin N., Pasqualetti P., Pruessner J.C., Redolfi A., Robitaille N., Soininen H., Tolomeo D., Wang L., Watson C., Wolf H., Duvernoy H., Duchesne S., Jack C.R., Frisoni G.B. Delphi definition of the EADC-ADNI harmonized protocol for hippocampal segmentation on magnetic resonance. Alzheimers Dement. 2015;11(2):126–138. doi: 10.1016/j.jalz.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodtmann A., Pardoe H., Li Q., Lichter R., Ostergaard L., Cumming T. Changes in regional brain volume three months after stroke. J. Neurol. Sci. 2012;322(1–2):122–128. doi: 10.1016/j.jns.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Brodtmann A., Pardoe H., Li Q., Werden E., Raffelt A., Cumming T. "Early ipsilesional hippocampal atrophy occurs after both anterior and posterior circulation strokes (P06.052) Neurology. 2013;80(7 Supplement) [Google Scholar]
- Brodtmann A., Werden E., Pardoe H., Li Q., Jackson G., Donnan G., Cowie T., Bradshaw J., Darby D., Cumming T. "Charting cognitive and volumetric trajectories after stroke: protocol for the cognition and neocortical volume after stroke (CANVAS) study. Int. J. Stroke. 2014;9(6):824–828. doi: 10.1111/ijs.12301. [DOI] [PubMed] [Google Scholar]
- Colon-Perez L.M., Triplett W., Bohsali A., Corti M., Nguyen P.T., Patten C., Mareci T.H., Price C.C. A majority rule approach for region-of-interest-guided streamline fiber tractography. Brain Imaging Behav. 2016;10(4):1137–1147. doi: 10.1007/s11682-015-9474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover K.S., van Schijndel R.A., Bosco P., Damangir S., Redolfi A. Can measuring hippocampal atrophy with a fully automatic method be substantially less noisy than manual segmentation over both 1 and 3 years? Psychiatry Res. Neuroimaging. 2018;280:39–47. doi: 10.1016/j.pscychresns.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Cover K.S., van Schijndel R.A., Versteeg A., Leung K.K., Mulder E.R., Jong R.A., Visser P.J., Redolfi A., Revillard J., Grenier B., Manset D., Damangir S., Bosco P., Vrenken H., van Dijk B.W., Frisoni G.B., Barkhof F. Reproducibility of hippocampal atrophy rates measured with manual, FreeSurfer, Adaboost, FSL/first and the Maps-HBSI methods in Alzheimer's disease. Psychiatry Res. 2016;252:26–35. doi: 10.1016/j.pscychresns.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J.C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., Buatti J., Aylward S., Miller J.V., Pieper S., Kikinis R. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein G., Di Sclafani V., Tanabe J., Cardenas V., Weiner M.W., Jagust W.J., Reed B.R., Norman D., Schuff N., Kusdra L., Greenfield T., Chui H. Hippocampal and cortical atrophy predict dementia in subcortical ischemic vascular disease. Neurology. 2000;55(11):1626–1635. doi: 10.1212/wnl.55.11.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Dale A.M. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. USA. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Liu A., Dale A.M. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans. Med. Imaging. 2001;20(1):70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C., van der Kouwe A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., Dale A.M. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., van der Kouwe A.J., Makris N., Segonne F., Quinn B.T., Dale A.M. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23(Suppl 1):S69–84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Frisoni G.B., Jack C.R., Jr., Bocchetta M., Bauer C., Frederiksen K.S., Liu Y., Preboske G., Swihart T., Blair M., Cavedo E., Grothe M.J., Lanfredi M., Martinez O., Nishikawa M., Portegies M., Stoub T., Ward C., Apostolova L.G., Ganzola R., Wolf D., Barkhof F., Bartzokis G., DeCarli C., Csernansky J.G., deToledo-Morrell L., Geerlings M.I., Kaye J., Killiany R.J., Lehericy S., Matsuda H., O'Brien J., Silbert L.C., Scheltens P., Soininen H., Teipel S., Waldemar G., Fellgiebel A., Barnes J., Firbank M., Gerritsen L., Henneman W., Malykhin N., Pruessner J.C., Wang L., Watson C., Wolf H., deLeon M., Pantel J., Ferrari C., Bosco P., Pasqualetti P., Duchesne S., Duvernoy H., Boccardi M. The EADC-ADNI harmonized protocol for manual hippocampal segmentation on magnetic resonance: evidence of validity. Alzheimers Dement. 2015;11(2):111–125. doi: 10.1016/j.jalz.2014.05.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T., Schaub A., Banac S., Charypar M., Jager M., Kummler P., Bottlender R., Zetzsche T., Born C., Leinsinger G., Reiser M., Moller H.J., Meisenzahl E.M. Reduced hippocampal volume correlates with executive dysfunctioning in major depression. J. Psychiatry Neurosci. 2006;31(5):316–323. [PMC free article] [PubMed] [Google Scholar]
- Gemmell E., Bosomworth H., Allan L., Hall R., Khundakar A., Oakley A.E., Deramecourt V., Polvikoski T.M., O'Brien J.T., Kalaria R.N. Hippocampal neuronal atrophy and cognitive function in delayed poststroke and aging-related dementias. Stroke. 2012;43(3):808–814. doi: 10.1161/STROKEAHA.111.636498. [DOI] [PubMed] [Google Scholar]
- Han X., Jovicich J., Salat D., van der Kouwe A., Quinn B., Czanner S., Busa E., Pacheco J., Albert M., Killiany R., Maguire P., Rosas D., Makris N., Dale A., Dickerson B., Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage. 2006;32(1):180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Iglesias J.E., Augustinack J.C., Nguyen K., Player C.M., Player A., Wright M., Roy N., Frosch M.P., McKee A.C., Wald L.L., Fischl B., Van Leemput K. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage. 2015;115:117–137. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlif M.S., Egorova N., Werden E., Redolfi A., Boccardi M., DeCarli C.S., Fletcher E., Singh B., Li Q., Bird L., Brodtmann A. A comparison of automated segmentation and manual tracing in estimating hippocampal volume in ischemic stroke and healthy control participants. Neuroimage Clin. 2019;21 doi: 10.1016/j.nicl.2018.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliper E., Bashat D.B., Bornstein N.M., Shenhar-Tsarfaty S., Hallevi H., Auriel E., Shopin L., Bloch S., Berliner S., Giladi N., Goldbourt U., Shapira I., Korczyn A.D., Assayag E.B. Cognitive decline after stroke: relation to inflammatory biomarkers and hippocampal volume. Stroke. 2013;44(5):1433–1435. doi: 10.1161/STROKEAHA.111.000536. [DOI] [PubMed] [Google Scholar]
- Lee K.B., Lim S.H., Kim K.H., Kim K.J., Kim Y.R., Chang W.N., Yeom J.W., Kim Y.D., Hwang B.Y. Six-month functional recovery of stroke patients: a multi-time-point study. Int. J. Rehabil. Res. 2015;38(2):173–180. doi: 10.1097/MRR.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Pardoe H., Lichter R., Werden E., Raffelt A., Cumming T., Brodtmann A. Cortical thickness estimation in longitudinal stroke studies: a comparison of 3 measurement methods. Neuroimage Clin. 2015;8:526–535. doi: 10.1016/j.nicl.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.I. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45(1):255–268. [PubMed] [Google Scholar]
- Meisenzahl M., Seifert E.D., Bottlender R., Teipel S., Zetzsche T., Jäger M., Koutsouleris N., Schmitt G., Scheuerecker J., Burgermeister B., Hampel H., Rupprecht T., Born C., Reiser M., Möller H.-J., Frodl T. Differences in hippocampal volume between major depression and schizophrenia: a comparative neuroimaging study. Eur. Arch. Psychiatry Clin. Neurosci. 2010;260(2):127–137. doi: 10.1007/s00406-009-0023-3. [DOI] [PubMed] [Google Scholar]
- Maglietta R., Amoroso N., Boccardi M., Bruno S., Chincarini A., Frisoni G.B., Inglese P., Redolfi A., Tangaro S., Tateo A., Bellotti R. Automated hippocampal segmentation in 3D MRI using random undersampling with boosting algorithm. Pattern Anal. Appl. 2016;19(2):579–591. doi: 10.1007/s10044-015-0492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier O., Schroder C., Forkert N.D., Martinetz T., Handels H. Classifiers for ischemic stroke lesion segmentation: a comparison study. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makin S.D., Turpin S., Dennis M.S., Wardlaw J.M. Cognitive impairment after lacunar stroke: systematic review and meta-analysis of incidence, prevalence and comparison with other stroke subtypes. J. Neurol. Neurosurg. Psychiatry. 2013;84(8):893–900. doi: 10.1136/jnnp-2012-303645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijajlović M.D., Pavlović A., Brainin M., Heiss W.-.D., Quinn T.J., Ihle-Hansen H.B., Hermann D.M., Assayag E.B., Richard E., Thiel A., Kliper E., Shin Y.-.I., Kim Y.-.H., Choi S., Jung S., Lee Y.-.B., Sinanović O., Levine D.A., Schlesinger I., Mead G., Milošević V., Leys D., Hagberg G., Ursin M.H., Teuschl Y., Prokopenko S., Mozheyko E., Bezdenezhnykh A., Matz K., Aleksić V., Muresanu D., Korczyn A.D., Bornstein N.M. Post-stroke dementia–a comprehensive review. BMC Med. 2017;15(1):11. doi: 10.1186/s12916-017-0779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok V.C.T., Lam B.Y.K., Wong A., Ko H., Markus H.S., Wong L.K.S. Early-onset and delayed-onset poststroke dementia [mdash] revisiting the mechanisms. Nat. Rev. Neurol. 2017;13(3):148–159. doi: 10.1038/nrneurol.2017.16. [DOI] [PubMed] [Google Scholar]
- Morey R.A., Petty C.M., Xu Y., Hayes J.P., Wagner H.R., Lewis D.V, LaBar K.S., Styner M., McCarthy G. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45(3):855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Avedissian C., Madsen S.K., Parikshak N., Hua X., Toga A.W., Jack C.R., Weiner M.W., Thompson P.M. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43(1):59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Toga A.W., Thompson P.M. "Comparison of Adaboost and support vector machines for detecting Alzheimer's disease through automated hippocampal segmentation. IEEE Trans. Med. Imaging. 2010;29(1):30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E.R., de Jong R.A., Knol D.L., van Schijndel R.A., Cover K.S., Visser P.J., Barkhof F., Vrenken H. "Hippocampal volume change measurement: quantitative assessment of the reproducibility of expert manual outlining and the automated methods FreeSurfer and first. Neuroimage. 2014;92:169–181. doi: 10.1016/j.neuroimage.2014.01.058. [DOI] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlaki G., Horvath R., Nagy S.A., Bogner P., Doczi T., Janszky J., Orsi G. Comparison of accuracy between FSL's First and Freesurfer for caudate nucleus and putamen segmentation. Sci. Rep. 2017;7(1):2418. doi: 10.1038/s41598-017-02584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A.K., Sandu A.L., Robertson K.L., McNeil C.J., Whalley L.J., Staff R.T., Murray A.D. A comparison of measurement methods of hippocampal atrophy rate for predicting Alzheimer's dementia in the Aberdeen birth Cohort of 1936. Alzheimers Dement. 2017;6:31–39. doi: 10.1016/j.dadm.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redolfi A., Bosco P., Manset D., Frisoni G.B., c. neu G. Brain investigation and brain conceptualization. Funct. Neurol. 2013;28(3):175–190. [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Rosas H.D., Fischl B. Highly accurate inverse consistent registration: a robust approach. Neuroimage. 2010;53(4):1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Schmansky N.J., Rosas H.D., Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61(4):1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaapsmeerders P., Tuladhar A.M., Maaijwee N.A.M., Rutten-Jacobs L.C.A., Arntz R.M., Schoonderwaldt H.C., Dorresteijn L.D.A., van Dijk E.J., Kessels R.P.C., de Leeuw F.-E. Lower ipsilateral hippocampal integrity after ischemic stroke in young adults: a long-term follow-up study. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0139772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghier M.L., Ramsden S., Lim L., Leff A.P., Price C.J. Gradual lesion expansion and brain shrinkage years after stroke. Stroke. 2014;45(3):877–879. doi: 10.1161/STROKEAHA.113.003587. [DOI] [PubMed] [Google Scholar]
- Segonne F., Dale A.M., Busa E., Glessner M., Salat D., Hahn H.K., Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22(3):1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Segonne F., Pacheco J., Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans. Med. Imaging. 2007;26(4):518–529. doi: 10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- Shrout P.E., Fleiss J.L. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Strong K., Mathers C., Bonita R. Preventing stroke: saving lives around the world. Lancet Neurol. 2007;6(2):182–187. doi: 10.1016/S1474-4422(07)70031-5. [DOI] [PubMed] [Google Scholar]
- Woon F.L., Sood S., Hedges D.W. Hippocampal volume deficits associated with exposure to psychological trauma and posttraumatic stress disorder in adults: a meta-analysis. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2010;34(7):1181–1188. doi: 10.1016/j.pnpbp.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Xie L., Wisse L.E.M., Pluta J., de Flores R., Piskin V., Manjon J.V., Wang H., Das S.R., Ding S.L., Wolk D.A., Yushkevich P.A. Automated segmentation of medial temporal lobe subregions on in vivo T1-weighted MRI in early stages of Alzheimer's disease. Hum. Brain Mapp. 2019;40(12):3431–3451. doi: 10.1002/hbm.24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassi N., Campbell B.C., Moffat B.A., Steward C., Churilov L., Parsons M.W., Desmond P.M., Davis S.M., Bivard A. Know your tools–concordance of different methods for measuring brain volume change after ischemic stroke. Neuroradiology. 2015;57(7):685–695. doi: 10.1007/s00234-015-1522-8. [DOI] [PubMed] [Google Scholar]
- Ystad M.A., Lundervold A.J., Wehling E., Espeseth T., Rootwelt H., Westlye L.T., Andersson M., Adolfsdottir S., Geitung J.T., Fjell A.M., Reinvang I., Lundervold A. Hippocampal volumes are important predictors for memory function in elderly women. BMC Med. Imaging. 2009;9(1):17. doi: 10.1186/1471-2342-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.