Highlights
-
•
Computational neuroimaging captures focal brainstem pathology in motor neuron diseases in contrast to both healthy- and disease controls.
-
•
ALS patients exhibit progressive medulla oblongata, pontine and mesencephalic volume loss over time.
-
•
Brainstem atrophy in ALS and PLS is dominated by medulla oblongata volume reductions.
-
•
Vertex analyses of ALS patients reveal flattening of the medullary pyramids bilaterally.
-
•
Morphometric analyses in ALS detect density reductions in the mesencephalic crura consistent with corticospinal tract degeneration.
Keywords: Motor neuron disease, Amyotrophic lateral sclerosis, Primary lateral sclerosis, Longitudinal study, Brainstem, Medulla, Pons, Mesencephalon, Neuroimaging
Glossary: AD, axial diffusivity; ALS, amyotrophic lateral sclerosis; ALS T1, ALS cohort at the first timepoint; ALS T2, ALS cohort at the second timepoint; ANCOVA, analysis of covariance; BG, basal ganglia; BRS, brainstem; C9orf72, chromosome 9 open reading frame 72; CN, cranial nerve; CNN, cranial nerve nuclei; CST, corticospinal tract; Cr, creatine‐phosphocreatine; DTI, diffusion tensor imaging; EPI, echo-planar imaging; FA, fractional anisotropy; FDR, false discovery rate; FTD, frontotemporal dementia; FOV, field-of-view; FSL, FMRIB Software Library; FWE, familywise error; GM, grey matter; HARDI, high angular resolution diffusion imaging; HC, healthy control; HSP, hereditary spastic paraplegia; IR-SPGR, Inversion Recovery prepared Spoiled Gradient Recalled echo; LMN, lower motor neuron; M, mean; MB, midbrain; MEM, mixed-effect model; MD, mean diffusivity; MND, motor neuron disease; MNI152, Montreal Neurological Institute 152 standard space; MR, magnetic resonance; MRS, magnetic resonance spectroscopy; Myo, myoinositol; NAA, N-acetylaspartate; NODDI, Neurite orientation dispersion and density imaging; PBA, pseudobulbar affect; PCL, pathological crying and laughing; PLS, primary lateral sclerosis; PMC, primary motor cortex; pTDP-43, phosphorylated 43 kDa TAR DNA-binding protein; QBI, q-ball imaging; RE, repeat expansion; RD, radial diffusivity; SC, spinal cord; SD, standard deviation; SBMA, spinal and bulbar muscular atrophy / Kennedy's disease; SOD1, superoxide dismutase 1; T1, Timepoint 1; T1W, T1-weighted imaging; T2, Timepoint 2; TBSS, tract-based spatial statistics; TE, echo time; TFCE, threshold-free cluster enhancement; TI, inversion time; TIV, total intracranial volume; TR, repetition time; UMN, upper motor neuron; WM, white matter
Abstract
Background
Brainstem pathology is a hallmark feature of ALS, yet most imaging studies focus on cortical grey matter alterations and internal capsule white matter pathology. Brainstem imaging in ALS provides a unique opportunity to appraise descending motor tract degeneration and bulbar lower motor neuron involvement.
Methods
A prospective longitudinal imaging study has been undertaken with 100 patients with ALS, 33 patients with PLS, 30 patients with FTD and 100 healthy controls. Volumetric, vertex and morphometric analyses were conducted correcting for demographic factors to characterise disease-specific patterns of brainstem pathology. Using a Bayesian segmentation algorithm, the brainstem was segmented into the medulla, pons and mesencephalon to measure regional volume reductions, shape analyses were performed to ascertain the atrophy profile of each study group and region-of-interest morphometry was used to evaluate focal density alterations.
Results
ALS and PLS patients exhibit considerable brainstem atrophy compared to both disease- and healthy controls. Volume reductions in ALS and PLS are dominated by medulla oblongata pathology, but pontine atrophy can also be detected. In ALS, vertex analyses confirm the flattening of the medullary pyramids bilaterally in comparison to healthy controls and widespread pontine shape deformations in contrast to PLS. The ALS cohort exhibit bilateral density reductions in the mesencephalic crura in contrast to healthy controls, central pontine atrophy compared to disease controls, peri-aqueduct mesencephalic and posterior pontine changes in comparison to PLS patients.
Conclus
ions: Computational brainstem imaging captures the degeneration of both white and grey matter components in ALS. Our longitudinal data indicate progressive brainstem atrophy over time, underlining the biomarker potential of quantitative brainstem measures in ALS. At a time when a multitude of clinical trials are underway worldwide, there is an unprecedented need for accurate biomarkers to monitor disease progression and detect response to therapy. Brainstem imaging is a promising addition to candidate biomarkers of ALS and PLS.
1. Introduction
Motor neuron diseases (MND) encompass a range of clinically heterogeneous conditions which typically affect young adults, are relentlessly progressive and have no effective disease-modifying therapies (Chipika et al., 2019). Some of the barriers to the development of effective pharmacological interventions include the considerable clinical heterogeneity of the condition, lack of validated biomarkers and late recruitment into pharmacological trials (Mitsumoto et al., 2014; Schuster et al., 2015; Hardiman et al., 2016). The majority of imaging studies in MND focus on cortical grey matter atrophy (Omer et al., 2017; Bede et al., 2013a), corticospinal tract degeneration (Schuster et al., 2016a), corpus callosum pathology (Filippini et al., 2010; Bede et al., 2015) and spinal cord degeneration (Bede et al., 2012; El Mendili et al., 2019; Lebouteux et al., 2014). With the recognition of extra-motor deficits in MND (Burke, 2017; Elamin et al., 2017; Burke et al., 2016; Christidi et al., 2018; Finegan et al., 2019a) the frontotemporal (Nasseroleslami et al., 2019; Iyer, 2017), basal ganglia (Feron et al., 2018; Christidi et al., 2019) and cerebellar (Abidi et al., 2019) profiles of MNDs have also been gradually characterised (Bede et al., 2018a), but there remains a striking lack of dedicated brainstem studies. Brainstem pathology is a hallmark feature of the disease and has been associated with the condition since its earliest descriptions (Clarke and Jackson, 1867). In one of the first pathologically supported reports, Lockhart Clarke eloquently describes progressive bulbar involvement in a patient with ALS: “September 1865; Her voice changed; she did not pronounce words as usual…Her deglutition now became difficult… The tongue is protruded badly; about a quarter of an inch beyond the lower teeth. It is atrophied on each side, and in folds, reminding one of cerebral convolutions. Her talking is nearly unintelligible; to me it is generally quite unintelligible, but the nurse can usually manage to make out what the patient says.” This moving description from over 150 years ago illustrates the bulbar symptoms in ALS which continue to affect patients today (Yunusova et al., 2019). Following his clinical description, Clarke comments on post mortem brainstem changes; “The medulla oblongata was below the average size. It was not, however, softer than usual, nor was it anywhere damaged by disintegration of tissue; but many of its nerve-cells, particularly about the floor of the fourth ventricle and calamus scriptorius, were in different stages of degeneration.” Despite historical descriptions of brainstem atrophy, it remains surprisingly understudied in vivo, in spite of considerable advances in ALS neuroimaging (Bede and Hardiman, 2014). Moreover, brainstem pathology is regarded as ‘stage 1’ of a recently proposed four-stage pathological staging system based on pathological TDP-43 burden patterns (Brettschneider et al., 2013). Furthermore, brainstem pathology is not unique to ALS, it is a unifying feature of most motor neuron diseases (ALS, PLS, SBMA) affecting the descending pyramidal tracts, brainstem nuclei or both, depending on the phenotype (Brettschneider et al., 2013; Querin et al., 2018a; Li et al., 1998; Finegan et al., 2019b; Geser et al., 2011). So, while brainstem degeneration is a pathognomonic feature of most MNDs, it is seldom evaluated specifically in dedicated imaging studies. This is a missed opportunity at a time when wet and dry biomarkers are urgently sought to inform clinical trial outcomes (Devos et al., 2019; Blasco et al., 2018). Accordingly, our main objective is the systematic characterisation of brainstem involvement in ALS and PLS, using both healthy- and disease controls in a prospective longitudinal study design. Based on the extensive post mortem literature of the disease (Brettschneider et al., 2013; Geser et al., 2011), we hypothesise that brainstem pathology can not only be detected in vivo, but it exhibits progressive changes over time.
2. Methods
2.1. Ethics statement
The study protocol, recruitment, and data management procedures were approved by our institutional ethics committee (Beaumont Hospital, Dublin, Ireland), in accordance with the 1964 Helsinki declaration and its later amendments. All participants provided informed consent and data was handled in accordance with the relevant EU regulations (GDPR).
2.2. Participants
A total of 100 ALS patients, 33 patients with PLS, 30 patients with FTD, and 100 healthy controls were included. Exclusion criteria included prior traumatic brain injury, prior cerebrovascular events, space-occupying intracranial findings, and inability to undergo MRI scanning due to implanted metallic devices (baclofen pumps, pacemakers), claustrophobia, or orthopnoea. Participating ALS patients had ‘probable’ or ‘definite’ ALS according to the El Escorial criteria (Brooks et al., 2000), PLS patients were diagnosed based on the Gordon criteria (Gordon et al., 2006) and FTD patients diagnosed in accordance with the Rascovsky criteria (Rascovsky et al., 2011). ALS patients were scanned twice with a follow-up interval of 4 months. Healthy controls had no known neurological or psychiatric conditions, previous head injuries or established vascular risk factors and no family history of neurodegenerative conditions.
2.3. Neuroimaging methods
T1-weighted images were acquired on a 3 Tesla Philips Achieva system using an 8-channel receive-only head coil using a 3D Inversion Recovery prepared Spoiled Gradient Recalled echo (IR-SPGR) sequence with the following settings; spatial resolution: 1 mm3, field-of-view (FOV): 256 × 256 × 160 mm, TR/TE = 8.5/3.9 ms, TI = =1060 ms, flip angle = 8°, SENSE factor = 1.5, acquisition time: 7 min 30 s. A multimodal approach was undertaken to comprehensively characterise brainstem degeneration in MND. First, total intracranial volumes (TIV) were estimated and a model-based approach utilised to estimate the volume of the entire brainstem. Subsequently, a Bayesian segmentation algorithm was used to estimate the volumes of the medulla oblongata, pons, and midbrain. Brainstem atrophy patterns in ALS were evaluated with reference to healthy controls, disease controls and PLS patients using vertex analyses. Finally, region of interest morphometry analyses were carried out to evaluate focal brain stem alterations in the study groups.
2.4. TIV and total brainstem volumes
Total intracranial volume (TIV) was calculated for each study participant to be used as a covariate for subsequent volumetric comparisons. The FMRIB Software Library (FSL) was used for TIV estimations. Each subject's skull-stripped brain was linearly aligned to the standard MNI152 brain image, the inverse of the determinant of the affine registration matrix was calculated and multiplied by the size of the template. FSL-FLIRT was used for registration to template (Jenkinson and Smith, 2001), and tissue type segmentation was performed using FSL-FAST (Zhang et al., 2001). For the estimation of total brain stem volume the model-based approach of FSL-FIRST was implemented. As described previously (Bede et al., 2018b), FSL-FIRST (Patenaude et al., 2011) uses a two-stage affine registration approach to register raw T1 data sets to the Montreal Neurological Institute 152 (MNI152) standard space first and then a model-based method is implemented for the segmentation of subcortical structures.
2.5. Brainstem segmentation
A Bayesian segmentation algorithm was used for the segmentation of the brainstem into the medulla oblongata, pons and midbrain. The protocol relies on a probabilistic atlas of the brainstem and its neighbouring anatomical structures generated based on 49 scans (Iglesias et al., 2015). The FreeSurfer image analysis suite (Fischl, 2012) was used for the pre-processing of T1-wieghted data which included the removal of non-brain tissue, segmentation of the subcortical white matter and deep grey matter structures, intensity normalization, tessellation of the grey matter-white matter boundary, and automated topology correction. Subsequent to pre-processing and brainstem segmentation, raw volumetric values were retrieved for each study participant at each timepoint for statistical interpretation with the appropriate covariates.
2.6. Vertex analyses
In order to characterise anatomical patterns of degenerative change in the brainstem beyond overall volume reductions, shape analyses were performed between the relevant study groups. FMRIB's subcortical segmentation and registration tool FIRST (Patenaude et al., 2011) was used for vertex analyses to generate quantitative, surface-projected information about focal brainstem involvement. Vertex locations of individual participants were projected on the surface of an average template shape as scalar values, positive value being outside the surface and negative values inside. Permutation based non-parametric statistics were used for group comparisons, design matrices included age, gender and education (Winkler et al., 2014), probability maps were corrected for multiple comparisons, and a statistical threshold of p < .05 FWE was considered significant.
2.7. Region of interest morphometry
In order to detect focal density alterations within the brain stem, region-of-interest morphometry was performed to complement the vertex analyses. The FMRIB Software Library (FSL) was used for brain extraction and tissue-type segmentation, following which grey-matter partial volume images were aligned to the MNI152 standard space using affine registration. Subsequently, a study specific template was created, to which the grey matter images from each subject were non-linearly coregistered. A voxelwise generalized linear model and permutation-based non-parametric testing was used to highlight density alterations in a merged brainstem mask accounting for age, gender and education (Winkler et al., 2014; Nichols and Holmes, 2002). The labels of the Harvard-Oxford probabilistic structural atlas (Desikan et al., 2006) and the labels of the Talairach probabilistic (Lancaster et al., 2000) atlas were used to generate a merged brainstem mask incorporating the medulla oblongata, pons and midbrain.
2.8. Genetics
Of the 100 ALS patients, C9orf72 repeat expansion status was determined in 97 patients (Byrne et al., 2012). DNA samples from patients were tested for the presence of the pathogenic GGGGCC hexanucleotide repeat expansion in C9orf72 by repeat-primed PCR using the Applied Biosystems (Foster City, CA, USA) 3130xl Genetic Analyser and visualised using GeneMapper version 4.0 as described previously (Byrne et al., 2012; Bede et al., 2013b). Patients carrying more than 30 hexanucleotide repeats were considered positive for the expansion. Whole-genome sequence data were available for 44 ALS patients (Project Min, 2018) and targeted DNA sequence data for a further 27 ALS patients (Kenna et al., 2013). Whole exome sequence data were available for 29 of the 33 PLS patients (Finegan et al., 2019a). Putative variants were defined as protein altering variants in the exons and splice sites of 33 genes linked to ALS on the ALS online database (Abel et al., 2013) and 70 genes linked to HSP (Klebe et al., 2015). C9orf72 hexanucleotide repeat expansion status in the PLS cohort was also determined using repeat-primed polymerase chain reaction (PCR).
2.9. Statistics
Analyses of covariance (ANCOVA) were performed to evaluate intergroup differences in brainstem volumes. Assumptions of normality, linearity and homogeneity of variances were verified. Brainstem volumes were included as dependent variables, and study group allocation as the categorical independent variable. TIV, age, education, and gender were included as covariates (Bede et al., 2013c). Summary tables were generated to present the estimated marginal means of brainstem volumes, standard error, between-group ANCOVA significance and p-values for Bonferroni-corrected post hoc testing. Estimated marginal means of volumes were plotted with confidence intervals to illustrate the volumetric profile of each study group. To demonstrate the divergent subcortical profiles of ALS, PLS and FTD percentage volume reductions were calculated with reference to the estimated marginal means of healthy controls and plotted on a radar (spider) chart. Differences in longitudinal change between C9orf72 hexanucleotide carriers and those without the repeat expansions were evaluated with mixed effect models.
3. Results
Study participants were matched for age, gender, handedness and education. Despite the comparable demographic profile of the study groups, each statistical model of our imaging analyses included age, gender, education and total intracranial volumes as covariates Table 1.
Table 1.
The clinical and demographic profile of the study cohorts.
| ALS T1 n = 100 | ALS T2 n = 100 | PLS n = 33 | FTD n = 30 | HC n = 100 | p value | |
|---|---|---|---|---|---|---|
| Age | 59.83 (11.155) | 60.21 (11.235) | 60.48 (10.488) | 63.97 (14.571) | 59.65 (11.876) | 0.484 |
| Gender (male) | 62 (62%) | 62 (62%) | 19 (58%) | 13 (43%) | 54 (54%) | 0.332 |
| Education (years) | 13.45 (3.176) | 13.45 (3.176) | 12.88 (3.380) | 12.80 (3.210) | 12.47 (3.383) | 0.171 |
| Handedness (right) | 90 (90%) | 90 (90%) | 29 (88%) | 27 (90%) | 94 (94%) | .784 |
| ALSFRS-r | 36.62 (7.470) | 35.40 (7.335) | 34.36 (5.337) | N/a | N/a | 0.23 |
3.1. Volumetric analyses
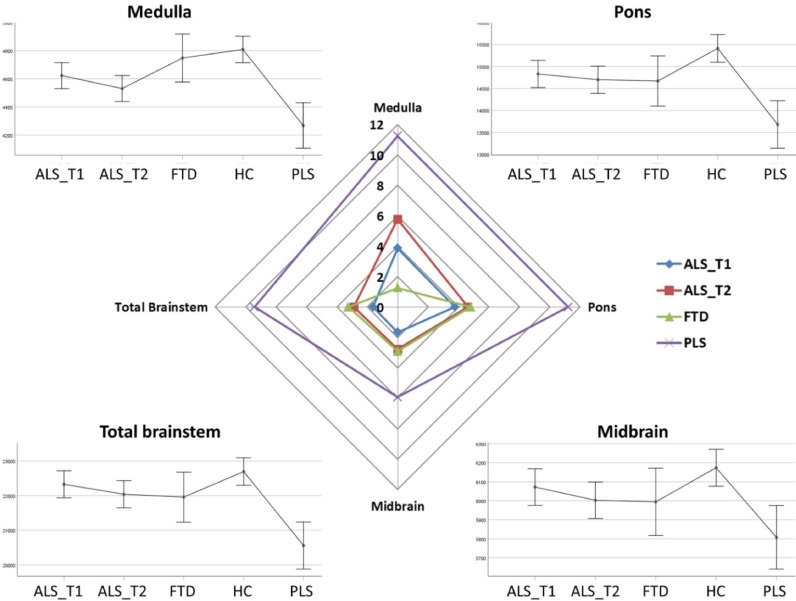
The volumetric analyses of brainstem regions revealed considerable multisegmental brainstem atrophy in both ALS and PLS with medullar predominance. ALS patients at their second timepoint exhibited statistically significant volume reductions in both the medulla and pons compared to healthy controls. Total brainstem volume reductions in ALS did not reach statistical significance with reference to controls, suggestive of selective medullar and pontine involvement. In the PLS cohort, brainstem atrophy was detectable in all three regions and medullar atrophy was not only apparent in comparison to healthy-, but also with reference to disease controls. PLS patients also had significantly lower medullar, pontine and mesencephalic volumes compared to ALS patients. Across the two time points ALS patients exhibited progressive volume loss in all three brainstem segments but longitudinal changes did not reach statistical significance. ALS patients showed a trend of medullar atrophy compared to healthy controls at their first time point which has reached significance by their second timepoint. Similarly, while pontine differences were not significant between healthy controls and ALS patients at the first time point, due to progressive pontine volume loss, ALS patients at the their second time point exhibited statistically significant pontine changes (Table 2 and Fig. 1).
Table 2.
Brainstem volumes in healthy controls (HC), patients with frontotemporal dementia (FTD), patients with amyotrophic lateral sclerosis (ALS) and primary lateral sclerosis (PLS). Estimated marginal means and standard error are adjusted for age, gender, education and total intracranial volume (TIV) (Covariates appearing in the model are evaluated at the following values: Age = 59.59, Gender = 1.43, Edu = 13.63, TIV = 1435355.28) Pairwise post hoc comparisons are Bonferroni corrected. Significant intergroup differences are flagged with asterisks ‘*’. Statistical trends are flagged with superscript ‘t’. ALS_T1: ALS patients first time point, ALS_T2: ALS patients second time point, FTD: patients with frontotemporal dementia, HC: healthy controls, PLS: patients with primary lateral sclerosis.
| Structure | Study group | Estimated marginal means(adjusted for age, gender, education and TIV) | Standard error | ANCOVA Sig. (p) | Post-hoc comparisons (Bonferroni-corrected) |
|---|---|---|---|---|---|
| Medulla | <0.001 |
ALS T1 vs HCp = .061t ALS T1 vs PLSp = .002* ALS T2 vs HCp < .001* ALS T2 vs PLSp= = .057 t FTD vs PLSp = .001* HC vs PLSp < .001* |
|||
| HC | 4808.641 | 4714.924 | |||
| ALS T1 | 4623.070 | 47.012 | |||
| ALS T2 | 4530.937 | 47.023 | |||
| FTD | 4747.986 | 86.610 | |||
| PLS | 4268.433 | 81.904 | |||
| Pons | <0.001 |
ALS T1 vs PLSp = .003* ALS T2 vs HCp = .017* ALS T2 vs PLSp = .014* HC vs PLSp < .001* |
|||
| HC | 15410.591 | 159.537 | |||
| ALS T1 | 14829.294 | 157.394 | |||
| ALS T2 | 14699.411 | 157.433 | |||
| FTD | 14670.462 | 289.969 | |||
| PLS | 13682.707 | 274.212 | |||
| Midbrain | 0.004 |
ALS T1 vs PLSp = .073 t HC vs PLSp = .02* |
|||
| HC | 6173.757 | 49.524 | |||
| ALS T1 | 6071.860 | 48.859 | |||
| ALS T2 | 6002.527 | 48.871 | |||
| FTD | 5994.261 | 90.014 | |||
| PLS | 5807.431 | 85.122 | |||
| Whole Brainstem | < 0.001 |
ALS T1 vs PLSp < .001* ALS T2 vs PLSp = .002* FTD vs PLSp = .058 t HC vs PLSp < .001* |
|||
| HC | 22694.377 | 202.067 | |||
| ALS T1 | 22326.919 | 199.353 | |||
| ALS T2 | 22038.915 | 199.402 | |||
| FTD | 21956.655 | 367.272 | |||
| PLS | 20556.064 | 347.314 |
Fig. 1.
The volumetric profile of the brainstem in ALS, PLS, disease and healthy controls. Corners: Estimate marginal means of the medulla, pons, midbrain are plotted for each study group adjusted for age, gender, education and total intracranial volumes. Centre: Percentage volumetric change is plotted with reference to healthy controls. ALS_T1: ALS patients first time point, ALS_T2: ALS patients second time point, FTD: patients with frontotemporal dementia, HC: healthy controls, PLS: patients with primary lateral sclerosis.
3.2. Vertex analyses
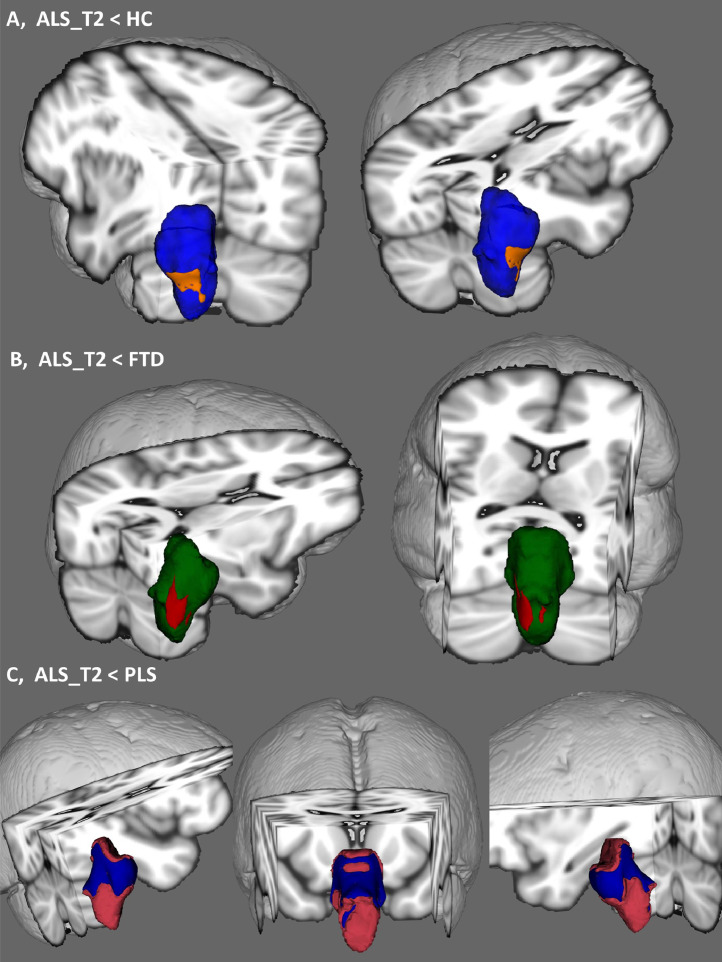
Focal shape deformations were detected at the second timepoint of ALS patients compared to both disease- and healthy controls following the appropriate statistical corrections. Considerable symmetric medullary pyramid atrophy was detected in comparison to healthy controls. In contrast to disease controls, an asymmetric pattern of anterolateral medullary atrophy was detected which centred on the right pyramid but incorporated the right olivary nuclei. In contrast to PLS, ALS patients exhibited widespread anterior and posterior pontine shape deformations including atrophy of the floor of the fourth ventricle.
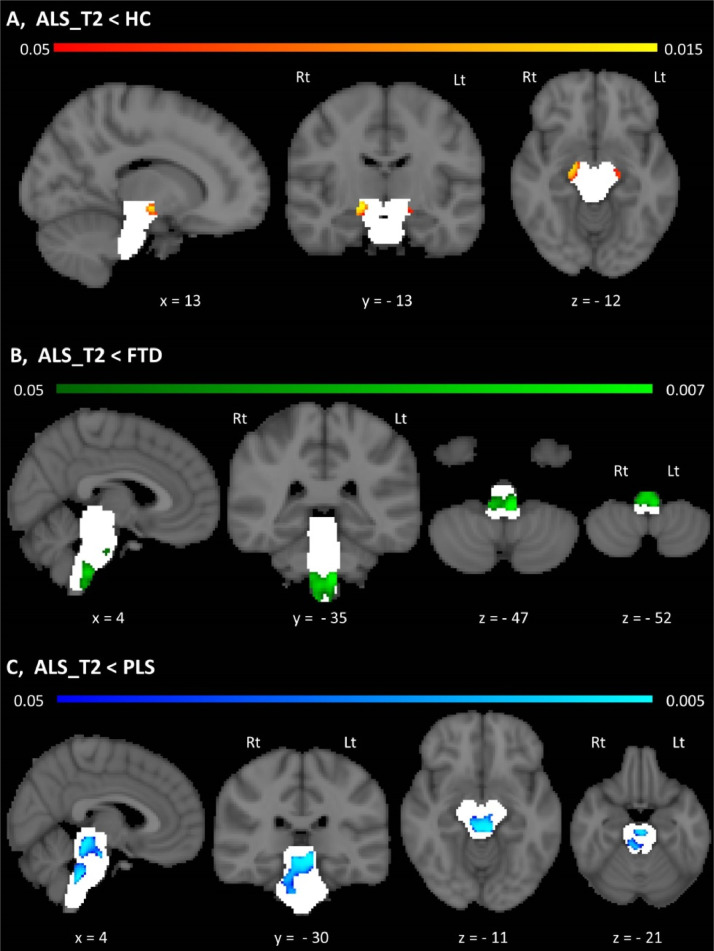
3.3. Brainstem morphometry
Morphometric analyses revealed density reductions in the ALS cohort compared to healthy controls in the mesencephalic crura. The ALS FTD contrast highlighted anterior medullar and central pontine atrophy at the second timepoint of ALS patients compared to disease controls. Compared to PLS patients, the ALS cohort exhibited peri-aqueduct mesencephalic, central pontine and posterior pontine density reductions. No statistically significant morphometric changes were detected between the two timepoints following the appropriate corrections in the ALS cohort.
3.4. Genetics
Eleven ALS patients and none of the PLS patients carried the GGGGCC hexanucleotide repeat expansion in C9orf72. ALS patients tested negative for other genes previously implicated in ALS. No PLS patients carried ALS or HSP associated mutations. In the ALS cohort, no brainstem differences were identified between hexanucleotide repeat carriers and those without the C9orf72 mutation at baseline or on follow-up. Moreover, patients with repeat expansions have not shown increased volume loss over time compared to C9orf72 negative ALS patients.
4. Discussion
Our study provides evidence of focal and progressive brainstem pathology in ALS and highlights that brainstem alterations can be characterised in vivo using routine MRI sequences. While volumetric analyses provide an overall indication of atrophy, the characterisation of shape deformations and focal density reduction enables the identification of the most vulnerable brainstem regions.
Our data indicate that total brainstem volume reductions are primarily driven by medulla oblongata pathology, but pontine changes also contribute to brainstem atrophy. PLS which is an upper motor neuron disorder (Finegan et al., 2019b) exhibits marked medullar, pontine and mesencephalic atrophy compared to ALS, suggesting that volume reductions are primarily caused by upper motor neuron degeneration. Furthermore, the medullar atrophy observed in PLS was not only confirmed with reference to healthy controls but also in contrast to disease controls. It is also noteworthy that using only a 4-month follow-up interval progressive volume loss can be detected in all three brainstem segments. Our morphometric analyses revealed focal density reductions in ALS compared to healthy controls in the bilateral mesencephalic crura where the descending corticospinal and corticonuclear tracts run. This further supports the notion the UMN degeneration contributes to brainstem volume reductions. This is echoed by the ALS FTD contrast which revealed focal changes in the medullar pyramids. The ALS PLS morphometric comparison is particularly interesting as both conditions are associated with widespread UMN degeneration. This contrast revealed widespread peri-aqueduct mesencephalic, central and posterior pontine grey matter changes.
Our vertex analyses also confirmed the symmetric involvement of the medullary pyramids in ALS. In contrast to PLS, ALS patients also exhibited widespread posterior pontine shape deformations including atrophy of the floor of the fourth ventricle as historically described by Clarke in 1867 (Clarke and Jackson, 1867). The widespread pontine shape deformations detected in ALS in contrast to PLS raises important questions about central brainstem involvement in ALS. As vertex analyses are essentially surface-projected representations of atrophy, the circumferential, anterior-posterior-lateral deformation detected in ALS compared to PLS (Fig. 2(C)) is likely to be driven by central volume reductions i.e. grey matter degeneration. This would not only be consistent with the differential grey matter (LMN / CNN) involvement in ALS compared to PLS clinically, but also consistent with our morphometric findings, where central pontine and mesencephalic density reductions were also detected in ALS compared to PLS (Fig. 3(C)). The integrative interpretation of the shape and morphometric alterations in ALS compared to PLS, which is a pure UMN condition, suggest that brainstem grey matter degeneration can be detected in vivo. In summary, while overall volume reductions are more prominent in PLS (Fig. 1) vertex and voxelwise analyses capture central brainstem pathology in ALS (Figs. 2(C) and 3(C)).
Fig. 2.
Anatomical patterns of brainstem pathology based on vertex analyses correcting for age, gender, education and total intracranial volumes. Shape deformation in ALS are shown compared to healthy controls (A), disease controls (B) and PLS patients (C). 3D brainstem mesh is presented in blue (A), green (B) and pink (C). Patterns of atrophy is illustrated in orange with respect to healthy controls (A), in red with reference to disease controls (B) and blue in comparison to PLS patients (C). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Morphometric changes in the brainstem A, Morphometric brainstem alterations in ALS patients compared to healthy controls at p < .05 TFCE FWE corrected for age, gender, education and TIV. B, Morphometric brainstem alterations in ALS patients compared to disease controls (FTD) at p < .05 TFCE FWE corrected for age, gender, education and TIV. C, Morphometric brainstem alterations in ALS patients compared to PLS patients at p < .05 TFCE FWE corrected for age, gender, education and TIV. Statistical maps are presented in MNI space, MNI coordinates are provided under each view, and the brainstem mask is presented as a white underlay. Radiological convention is used; Lt – Left, Rt – Right.
Surprisingly few human imaging studies have specifically evaluated brain stem structures in ALS, but some whole brains studies have commented on infratentorial changes. Diffusion tensor imaging studies have been inconsistent in detecting pyramidal tract degeneration at medullar or pontine levels. Some studies readily detected inferior brain stem diffusivity changes (Schuster et al., 2016a; Bede et al., 2015; Iwata et al., 2008) while others only captured mesencephalic pyramidal tract alterations (Yin et al., 2004). Brainstem FA reductions based on pooled multisite data have been linked to stage 2 of a recently proposed pathological staging system (Muller et al., 2016). The findings of functional studies in ALS are even more conflicting (Proudfoot et al., 2018; Bede, 2017). Several PET studies have shown increased pontine metabolism (Cistaro et al., 2012) which was interpreted as reactive astrocytosis (Johansson et al., 2007). Hypermetabolism in the brainstem has been linked to bulbar onset (Cistaro et al., 2012) and C9orf72 hexanucleotide repeats (Cistaro et al., 2014; Floeter and Gendron, 2018). Some spectroscopy studies however have detected decreased Naa/Cr ratios in the brainstem (Pioro et al., 1999; Bradley et al., 1999) while other did not (Sivak et al., 2010; Kalra, 2019). Animal imaging studies enable the histological interpretation of radiological changes (Evans et al., 2012; Bede, 2019) and high-field SOD1 transgenic mouse models showed T2 weighted hyperintensities in the nucleus ambiguus, facial nucleus, trigeminal motor nucleus, rostroventrolateral reticular nucleus, lateral paragigantocellular nucleus and the substantia nigra with associated neuronal loss of histological evaluation (Zang et al., 2004). Our morphometric and vertex analyses captured central pontine and inferior mesencephalic changes in ALS compared to PLS. Few imaging studies have evaluated lower motor neuron pathology to date in ALS, and without exception, these were advanced, quantitative spinal cord imaging studies, where the accurate segmentation of white and grey matter components is possible (Querin et al., 2018b,2019a).
The benefit of using standard T1-weighted sequences and morphometric analyses is that high resolution 3D T1W sequences are typically already part of clinical protocols and are readily available on any imaging platform irrespective of manufacturer or field strength. Morphometric brain stem measures could therefore be easily integrated in clinical trial protocols and data acquisition could be harmonised across multiple centres (Muller et al., 2016; Turner et al., 2012). The brainstem measures derived from T1-weighted data are therefore relatively attractive candidate biomarkers as they can be acquired non-invasively without special sequences, the acquisition time is typically short (<10 min), the measures discriminate patients from disease controls, and the derived metrics exhibit progressive change over time.
The identification of imaging measures which readily discriminate ALS patients from healthy and disease controls is hugely relevant for emerging machine learning applications (Bede et al., 2018a) and survival prediction (Schuster et al., 2017). ALS is associated with strikingly focal anatomical vulnerability profiles (Bede et al., 2016) which enable disease-specific feature selection for classification algorithms (Grollemund et al., 2019; Schuster et al., 2016b). Classification models enable the categorisation of blinded datasets into diagnostic, phenotypic or prognostic categories and the incorporation of the discriminatory brain regions such as the brainstem is likely to increase the accuracy of such models further (Querin et al., 2018b; Bede et al., 2017).
Longitudinal imaging in ALS is typically limited by high attrition rates (Chipika et al., 2019) and relatively few large presymptomatic studies have been published to date (Schuster et al., 2015; Eisen et al., 2014; Querin et al., 2019b). Despite the challenges of longitudinal imaging, a number of multi-timepoint studies have now been published (Menke et al., 2018; Bede and Hardiman, 2018) which suggest early white matter degeneration with a ceiling effect and progressive grey mater degeneration. From a biomarker point of view, the ability to track progressive changes in the later stages of the disease is essential; as indices exhibiting early ceiling effects are not suitable for monitoring.
Despite a number of pioneering studies, relatively little is known of the imaging profile of PLS and existing studies suffer from considerable sample size limitations. Furthermore, most PLS studies report overlapping imaging patterns with ALS (Müller et al., 2018; Van Weehaeghe et al., 2016), highlighting motor cortex (Butman and Floeter, 2007; Schuster et al., 2013), pyramidal tract (Müller et al., 2018; Iwata et al., 2011), and corpus callosum pathology (Unrath et al., 2010; Agosta et al., 2014) and very few PLS studies highlight distinctive imaging features (Finegan et al., 2019a, b; Clark et al., 2018). Our data indicate that brainstem atrophy is particularly marked in PLS and the differences between the ALS and PLS cohort are statistically significant. The description of features which discriminate PLS and ALS may have implications for the categorisation of UMN predominant patients and ‘suspected PLS’ patients who don't fulfil current diagnostic criteria (Gordon et al., 2006,2009).
The presented study has several limitations. No post-mortem data are available to validate our imaging findings which would be of particular interest in the PLS where patterns of pTDP-43 burden are poorly characterised. The longitudinal arm of the study is a two-time point design which precludes commenting on linear versus exponential longitudinal changes. While we intentionally used T1-weighted data alone in the various analysis streams, novel diffusion sequences such as HARDI, Q-ball imaging or NODDI (Barritt et al., 2018; Broad et al., 2018) may have been particularly useful in characterising crossing fibre integrity in the brainstem. Notwithstanding these limitations, our study provides evidence that standard T1-weighted data can be used to detect global and focal brainstem pathology in motor neuron diseases in contrast to both healthy and disease controls.
5. Conclusions
Progressive brainstem pathology can be detected in ALS in vivo and brainstem measures may have a biomarker role in diagnostic, monitoring and prognostic applications.
Declaration of Competing Interest
Peter Bede is a member of the UK Motor Neuron Disease Association (MNDA) Research Advisory Panel, the steering committee of Neuroimaging Society in ALS (NiSALS) and the medical patron of the Irish Motor Neuron Disease Association (IMNDA). These affiliations had no impact on the design, analyses, and presentation of this work.
Acknowledgement
We gratefully acknowledge all the motor neuron disease patients and controls who have participated in this research study. Without their generosity this study would have not been possible. Peter Bede and the computational neuroimaging group are supported by the Health Research Board (HRB EIA-2017-019), the Andrew Lydon scholarship, the Irish Institute of Clinical Neuroscience (IICN), the Iris O'Brien Foundation, the Research Motor Neuron (RMN-Ireland) Foundation and the Irish Motor Neuron Disease Association (IMNDA). Russell L McLaughlin is supported by the Motor Neurone Disease Association (957-799) and Science Foundation Ireland (17/CDA/4737). Mark A Doherty is supported by Science Foundation Ireland (15/SPP/3244). The sponsors of this study had no direct role in the design, analyses, presentation of this work or the decision to submit these findings for publication.
References
- Chipika R.H., Finegan E., Li Hi Shing S. Tracking a fast-moving disease: longitudinal markers, monitoring, and clinical trial endpoints in ALS. Front. Neurol. 2019;10:229. doi: 10.3389/fneur.2019.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsumoto H., Brooks B.R., Silani V. Clinical trials in amyotrophic lateral sclerosis: why so many negative trials and how can trials be improved? Lancet Neurol. 2014;13(11):1127–1138. doi: 10.1016/S1474-4422(14)70129-2. [DOI] [PubMed] [Google Scholar]
- Schuster C., Elamin M., Hardiman O. Presymptomatic and longitudinal neuroimaging in neurodegeneration–from snapshots to motion picture: a systematic review. J. Neurol. Neurosurg. Psychiatry. 2015;86(10):1089–1096. doi: 10.1136/jnnp-2014-309888. [DOI] [PubMed] [Google Scholar]
- Hardiman O., Doherty C.P., Elamin M. Springer International Publishing; Switzerland: 2016. Neurodegenerative Disorders: A Clinical Guide; pp. 1–336. 2016 ed. 2016, Springer Cham Heidelberg New York Dordrecht London©Springer International Publishing. [Google Scholar]
- Omer T., Finegan E., Hutchinson S., Doherty M., Vajda A., McLaughlin R.L., Pender N., Hardiman O., Bede P. Neuroimaging patterns along the ALS-FTD spectrum: a multiparametric imaging study. Amyotroph Lateral Scler Frontotemporal Degener. 2017;18(7-8):611–623. doi: 10.1080/21678421.2017.1332077. Epub 2017 May 31.PMID: 28562080. [DOI] [PubMed] [Google Scholar]
- Bede P., Bokde A., Elamin M. Grey matter correlates of clinical variables in amyotrophic lateral sclerosis (ALS): a neuroimaging study of ALS motor phenotype heterogeneity and cortical focality. J. Neurol. Neurosurg. Psychiatry. 2013;84(7):766–773. doi: 10.1136/jnnp-2012-302674. [DOI] [PubMed] [Google Scholar]
- Schuster C., Elamin M., Hardiman O. The segmental diffusivity profile of amyotrophic lateral sclerosis associated white matter degeneration. Eur. J. Neurol. 2016;23(8):1361–1371. doi: 10.1111/ene.13038. [DOI] [PubMed] [Google Scholar]
- Filippini N., Douaud G., Mackay C.E. Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis. Neurology. 2010;75(18):1645–1652. doi: 10.1212/WNL.0b013e3181fb84d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Elamin M., Byrne S. Patterns of cerebral and cerebellar white matter degeneration in ALS. J. Neurol. Neurosurg. Psychiatry. 2015;86(4):468–470. doi: 10.1136/jnnp-2014-308172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Bokde A.L., Byrne S. Spinal cord markers in ALS: diagnostic and biomarker considerations. Amyotroph. Lateral Scler. 2012;13(5):407–415. doi: 10.3109/17482968.2011.649760. [DOI] [PubMed] [Google Scholar]
- El Mendili M.M., Querin G., Bede P. Spinal cord imaging in amyotrophic lateral sclerosis: historical concepts-novel techniques. Front Neurol. 2019;10:350. doi: 10.3389/fneur.2019.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebouteux M.V., Franques J., Guillevin R. Revisiting the spectrum of lower motor neuron diseases with snake eyes appearance on magnetic resonance imaging. Eur. J. Neurol. 2014;21(9):1233–1241. doi: 10.1111/ene.12465. [DOI] [PubMed] [Google Scholar]
- Elamin M., Pinto-Grau M., Burke T. Identifying behavioural changes in ALS: validation of the beaumont behavioural inventory (BBI) Amyotroph. Lateral Scler. Frontotemporal Degener. 2017;18(1–2):68–73. doi: 10.1080/21678421.2016.1248976. [DOI] [PubMed] [Google Scholar]
- Burke A Cross-sectional population-based investigation into behavioral change in amyotrophic lateral sclerosis: subphenotypes, staging, cognitive predictors, and survival. Ann Clin Transl Neurol. 2017;4:305–317. doi: 10.1002/acn3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T., Elamin M., Bede P., Pinto-Grau M., Lonergan K., Hardiman O., Pender N. Discordant performance on the ‘Reading the Mind in the Eyes’ Test, based on disease onset in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2016;17(7-8):467–472. doi: 10.1080/21678421.2016.1177088. Epub 2016 May 6.PMID: 27152765. [DOI] [PubMed] [Google Scholar]
- Christidi F., Karavasilis E., Rentzos M. Clinical and radiological markers of extra-motor deficits in amyotrophic lateral sclerosis. Front. Neurol. 2018;9:1005. doi: 10.3389/fneur.2018.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegan E., Chipika R.H., Li Hi Shing S. The clinical and radiological profile of primary lateral sclerosis: a population-based study. J. Neurol. 2019;266:2718–2733. doi: 10.1007/s00415-019-09473-z. [DOI] [PubMed] [Google Scholar]
- Nasseroleslami B., Dukic S., Broderick M., Mohr K., Schuster C., Gavin B., McLaughlin R., Heverin M., Vajda A., Iyer P.M., Pender N., Bede P., Lalor E.C., Hardiman O. Characteristic Increases in EEG Connectivity Correlate With Changes of Structural MRI in Amyotrophic Lateral Sclerosis. Cereb Cortex. 2019;29(1):27–41. doi: 10.1093/cercor/bhx301. PMID: 29136131. [DOI] [PubMed] [Google Scholar]
- Feron M., Couillandre A., Mseddi E. Extrapyramidal deficits in ALS: a combined biomechanical and neuroimaging study. J. Neurol. 2018;265:2125–2136. doi: 10.1007/s00415-018-8964-y. [DOI] [PubMed] [Google Scholar]
- Christidi F., Karavasilis E., Rentzos M. Hippocampal pathology in amyotrophic lateral sclerosis: selective vulnerability of subfields and their associated projections. Neurobiol. Aging. 2019;84:178–188. doi: 10.1016/j.neurobiolaging.2019.07.019. [DOI] [PubMed] [Google Scholar]
- Abidi M., de Marco G., Couillandre A. Adaptive functional reorganization in amyotrophic lateral sclerosis: coexisting degenerative and compensatory changes. Eur. J. Neurol. 2019 doi: 10.1111/ene.14042. PMID: 31310452. [DOI] [PubMed] [Google Scholar]
- Bede P., Querin G., Pradat P.F. The changing landscape of motor neuron disease imaging: the transition from descriptive studies to precision clinical tools. Curr. Opin. Neurol. 2018;31(4):431–438. doi: 10.1097/WCO.0000000000000569. [DOI] [PubMed] [Google Scholar]
- Clarke J.L., Jackson J.H. On a case of muscular atrophy, with disease of the spinal cord and medulla oblongata. Med. Chir. Trans. 1867;50(1):489–498. doi: 10.1177/095952876705000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunusova Y., Plowman E.K., Green J.R. Clinical measures of bulbar dysfunction in ALS. Front. Neurol. 2019;10:106. doi: 10.3389/fneur.2019.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Hardiman O. Lessons of ALS imaging: pitfalls and future directions—a critical review. NeuroImage Clin. 2014;4(0):436–443. doi: 10.1016/j.nicl.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J., Del Tredici K., Toledo J.B. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann. Neurol. 2013;74(1):20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querin G., Bede P., Marchand-Pauvert V. Biomarkers of spinal and bulbar muscle atrophy (SBMA): a comprehensive review. Front. Neurol. 2018;9:844. doi: 10.3389/fneur.2018.00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Nakagomi Y., Kobayashi Y. Nonneural nuclear inclusions of androgen receptor protein in spinal and bulbar muscular atrophy. Am. J. Pathol. 1998;153(3):695–701. doi: 10.1016/S0002-9440(10)65612-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegan E., Chipika R.H., Shing S.L.H., Hardiman O., Bede P. Primary lateral sclerosis: a distinct entity or part of the ALS spectrum? Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(3-4):133–145. doi: 10.1080/21678421.2018.1550518. Epub 2019 Jan 18.PMID: 30654671. [DOI] [PubMed] [Google Scholar]
- Geser F., Prvulovic D., O'Dwyer L. On the development of markers for pathological TDP-43 in amyotrophic lateral sclerosis with and without dementia. Prog. Neurobiol. 2011;95(4):649–662. doi: 10.1016/j.pneurobio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Moreau C., Kyheng M. A ferroptosis-based panel of prognostic biomarkers for amyotrophic lateral sclerosis. Sci. Rep. 2019;9(1):2918. doi: 10.1038/s41598-019-39739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco H., Patin F., Descat A. A pharmaco-metabolomics approach in a clinical trial of ALS: identification of predictive markers of progression. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0198116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Miller R.G., Swash M. El escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron. Disord. 2000;1(5):293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- Gordon P.H., Cheng B., Katz I.B. The natural history of primary lateral sclerosis. Neurology. 2006;66(5):647–653. doi: 10.1212/01.wnl.0000200962.94777.71. [DOI] [PubMed] [Google Scholar]
- Rascovsky K., Hodges J.R., Knopman D. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer Mismatch Negativity as an Indicator of Cognitive Sub-Domain Dysfunction in Amyotrophic Lateral Sclerosis. Front Neurol. 2017;8:395. doi: 10.3389/fneur.2017.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Brady M., Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Bede P., Omer T., Finegan E. Connectivity-based characterisation of subcortical grey matter pathology in frontotemporal dementia and ALS: a multimodal neuroimaging study. Brain Imaging Behav. 2018;12(6):1696–1707. doi: 10.1007/s11682-018-9837-9. [DOI] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N. A bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias J.E., Van Leemput K., Bhatt P. Bayesian segmentation of brainstem structures in MRI. Neuroimage. 2015;113:184–195. doi: 10.1016/j.neuroimage.2015.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler A.M., Ridgway G.R., Webster M.A. Permutation inference for the general linear model. Neuroimage. 2014;92:381–397. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols T.E., Holmes A.P. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum. Brain Mapp. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R.S., Segonne F., Fischl B. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Lancaster J.L., Woldorff M.G., Parsons L.M. Automated talairach atlas labels for functional brain mapping. Hum. Brain Mapp. 2000;10(3):120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S., Elamin M., Bede P. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11(3):232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Bokde A.L., Byrne S. Multiparametric mri study of als stratified for the C9orf72 genotype. Neurology. 2013;81(4):361–369. doi: 10.1212/WNL.0b013e31829c5eee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Project Min, E.A.L.S.S.C. Project mine: study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur. J. Hum. Genet. EJHG. 2018;26(10):1537–1546. doi: 10.1038/s41431-018-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna K.P., McLaughlin R.L., Byrne S. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J. Med. Genet. 2013;50(11):776–783. doi: 10.1136/jmedgenet-2013-101795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel O., Shatunov A., Jones A.R. Development of a smartphone app for a genetics website: the amyotrophic lateral sclerosis online genetics database (ALSoD) JMIR Mhealth Uhealth. 2013;1(2):e18. doi: 10.2196/mhealth.2706. PMID: 25098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe S., Stevanin G., Depienne C. Clinical and genetic heterogeneity in hereditary spastic paraplegias: from SPG1 to SPG72 and still counting. Rev. Neurol. (Paris) 2015;171(6–7):505–530. doi: 10.1016/j.neurol.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Bede P., Elamin M., Byrne S. Sexual dimorphism in ALS: exploring gender-specific neuroimaging signatures. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013 doi: 10.3109/21678421.2013.865749. [DOI] [PubMed] [Google Scholar]
- Iwata N.K., Aoki S., Okabe S. Evaluation of corticospinal tracts in ALS with diffusion tensor MRI and brainstem stimulation. Neurology. 2008;70(7):528–532. doi: 10.1212/01.wnl.0000299186.72374.19. [DOI] [PubMed] [Google Scholar]
- Yin H., Lim C.C., Ma L. Combined MR spectroscopic imaging and diffusion tensor mri visualizes corticospinal tract degeneration in amyotrophic lateral sclerosis. J. Neurol. 2004;251(10):1249–1254. doi: 10.1007/s00415-004-0526-9. [DOI] [PubMed] [Google Scholar]
- Muller H.P., Turner M.R., Grosskreutz J. A large-scale multicentre cerebral diffusion tensor imaging study in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2016;87(6):570–579. doi: 10.1136/jnnp-2015-311952. [DOI] [PubMed] [Google Scholar]
- Proudfoot M., Bede P., Turner M.R. Imaging cerebral activity in amyotrophic lateral sclerosis. Front. Neurol. 2018;9:1148. doi: 10.3389/fneur.2018.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cistaro A., Valentini M.C., Chio A. Brain hypermetabolism in amyotrophic lateral sclerosis: a FDG pet study in ALS of spinal and bulbar onset. Eur. J. Nucl. Med. Mol. Imaging. 2012;39(2):251–259. doi: 10.1007/s00259-011-1979-6. [DOI] [PubMed] [Google Scholar]
- Johansson A., Engler H., Blomquist G. Evidence for astrocytosis in ALS demonstrated by [11C](L)-deprenyl-D2 pet. J. Neurol. Sci. 2007;255(1–2):17–22. doi: 10.1016/j.jns.2007.01.057. [DOI] [PubMed] [Google Scholar]
- Cistaro A., Pagani M., Montuschi A. The metabolic signature of C9ORF72-related ALS: FDG pet comparison with nonmutated patients. Eur. J. Nucl. Med. Mol. Imaging. 2014 doi: 10.1007/s00259-013-2667-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floeter M.K., Gendron T.F. Biomarkers for amyotrophic lateral sclerosis and frontotemporal dementia associated with hexanucleotide expansion mutations in C9orf72. Front. Neurol. 2018;9:1063. doi: 10.3389/fneur.2018.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioro E.P., Majors A.W., Mitsumoto H. 1H-MRS evidence of neurodegeneration and excess glutamate + glutamine in ALS medulla. Neurology. 1999;53(1):71–79. doi: 10.1212/wnl.53.1.71. [DOI] [PubMed] [Google Scholar]
- Bradley W.G., Bowen B.C., Pattany P.M. 1H-magnetic resonance spectroscopy in amyotrophic lateral sclerosis. J. Neurol. Sci. 1999;169(1–2):84–86. doi: 10.1016/s0022-510x(99)00221-x. [DOI] [PubMed] [Google Scholar]
- Sivak S., Bittsansky M., Kurca E. Proton magnetic resonance spectroscopy in patients with early stages of amyotrophic lateral sclerosis. Neuroradiology. 2010;52(12):1079–1085. doi: 10.1007/s00234-010-0685-6. [DOI] [PubMed] [Google Scholar]
- Kalra S. Magnetic resonance spectroscopy in ALS. Front. Neurol. 2019;10:482. doi: 10.3389/fneur.2019.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.C., Modo M., Talbot K. Magnetic resonance imaging of pathological processes in rodent models of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2012;13(3):288–301. doi: 10.3109/17482968.2011.623300. [DOI] [PubMed] [Google Scholar]
- Bede Deciphering neurodegeneration: A paradigm shift from focality to connectivity. Neurology. 2017;89:1758–1759. doi: 10.1212/WNL.0000000000004582. [DOI] [PubMed] [Google Scholar]
- Bede P. The histological correlates of imaging metrics: postmortem validation of in vivo findings. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(7-8):457–460. doi: 10.1080/21678421.2019.1639195. Epub 2019 Jul 11. No abstract available. PMID: 31293187. [DOI] [PubMed] [Google Scholar]
- Zang D.W., Yang Q., Wang H.X. Magnetic resonance imaging reveals neuronal degeneration in the brainstem of the superoxide dismutase 1 transgenic mouse model of amyotrophic lateral sclerosis. Eur. J. Neurosci. 2004;20(7):1745–1751. doi: 10.1111/j.1460-9568.2004.03648.x. [DOI] [PubMed] [Google Scholar]
- Querin G., El Mendili M.M., Bede P. Multimodal spinal cord MRI offers accurate diagnostic classification in ALS. J. Neurol. Neurosurg. Psychiatry. 2018;89:1220–1221. doi: 10.1136/jnnp-2017-317214. [DOI] [PubMed] [Google Scholar]
- Querin G., El Mendili M.M., Lenglet T. The spinal and cerebral profile of adult spinal-muscular atrophy: a multimodal imaging study. Neuroimage Clin. 2019;21 doi: 10.1016/j.nicl.2018.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M.R., Agosta F., Bede P. Neuroimaging in amyotrophic lateral sclerosis. Biomark Med. 2012;6(3):319–337. doi: 10.2217/bmm.12.26. [DOI] [PubMed] [Google Scholar]
- Schuster C., Hardiman O., Bede P. Survival prediction in amyotrophic lateral sclerosis based on MRI measures and clinical characteristics. BMC Neurol. 2017;17(1):73. doi: 10.1186/s12883-017-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Iyer P.M., Schuster C. The selective anatomical vulnerability of ALS: 'disease-defining' and 'disease-defying' brain regions. Amyotroph. Lateral Scler. Frontotemporal Degener. 2016;17(7–8):561–570. doi: 10.3109/21678421.2016.1173702. [DOI] [PubMed] [Google Scholar]
- Grollemund V., Pradat P.F., Querin G. Machine learning in amyotrophic lateral sclerosis: achievements, pitfalls, and future directions. Front. Neurosci. 2019;13:135. doi: 10.3389/fnins.2019.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C., Hardiman O., Bede P. Development of an automated MRI-based diagnostic protocol for amyotrophic lateral sclerosis using disease-specific pathognomonic features: a quantitative disease-state classification study. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0167331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Iyer P.M., Finegan E. Virtual brain biopsies in amyotrophic lateral sclerosis: diagnostic classification based on in vivo pathological patterns. Neuroimage Clin. 2017;15:653–658. doi: 10.1016/j.nicl.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A., Kiernan M., Mitsumoto H. Amyotrophic lateral sclerosis: a long preclinical period? J. Neurol. Neurosurg. Psychiatry. 2014 doi: 10.1136/jnnp-2013-307135. [DOI] [PubMed] [Google Scholar]
- Querin G., Bede P., El Mendili M.M. Presymptomatic spinal cord pathology in c9orf72 mutation carriers: a longitudinal neuroimaging study. Ann. Neurol. 2019;86(2):158–167. doi: 10.1002/ana.25520. [DOI] [PubMed] [Google Scholar]
- Menke R.A.L., Proudfoot M., Talbot K. The two-year progression of structural and functional cerebral mri in amyotrophic lateral sclerosis. NeuroImage Clin. 2018;17:953–961. doi: 10.1016/j.nicl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P., Hardiman O. Longitudinal structural changes in ALS: a three time-point imaging study of white and gray matter degeneration. Amyotroph. Lateral Scler. Frontotemporal Degener. 2018;19(3–4):232–241. doi: 10.1080/21678421.2017.1407795. [DOI] [PubMed] [Google Scholar]
- Müller H.P., Gorges M., Kassubek R. Identical patterns of cortico-efferent tract involvement in primary lateral sclerosis and amyotrophic lateral sclerosis: a tract of interest-based MRI study. NeuroImage Clin. 2018;18:762–769. doi: 10.1016/j.nicl.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Weehaeghe D., Ceccarini J., Delva A. Prospective validation of 18F-FDG brain pet discriminant analysis methods in the diagnosis of amyotrophic lateral sclerosis. J. Nucl. Med. 2016;57(8):1238–1243. doi: 10.2967/jnumed.115.166272. [DOI] [PubMed] [Google Scholar]
- Butman J.A., Floeter M.K. Decreased thickness of primary motor cortex in primary lateral sclerosis. Am. J. Neuroradiol. 2007;28(1):87–91. [PMC free article] [PubMed] [Google Scholar]
- Schuster C., Kasper E., Machts J. Focal thinning of the motor cortex mirrors clinical features of amyotrophic lateral sclerosis and their phenotypes: a neuroimaging study. J. Neurol. 2013;260(11):2856–2864. doi: 10.1007/s00415-013-7083-z. [DOI] [PubMed] [Google Scholar]
- Iwata N.K., Kwan J.Y., Danielian L.E. White matter alterations differ in primary lateral sclerosis and amyotrophic lateral sclerosis. Brain. 2011;134(9):2642–2655. doi: 10.1093/brain/awr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unrath A., Muller H.-.P., Riecker A. Whole brain-based analysis of regional white matter tract alterations in rare motor neuron diseases by diffusion tensor imaging. Hum. Brain Mapp. 2010;31(11):1727–1740. doi: 10.1002/hbm.20971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F., Galantucci S., Riva N. Intrahemispheric and interhemispheric structural network abnormalities in PLS and ALS. Hum. Brain Mapp. 2014;35(4):1710–1722. doi: 10.1002/hbm.22286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.G., Smallwood Shoukry R., Huang C.J. Loss of functional connectivity is an early imaging marker in primary lateral sclerosis. Amyotroph. Lateral Scler. Frontotemporal Degener. 2018;19(7–8):562–569. doi: 10.1080/21678421.2018.1517180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon P.H., Cheng B., Katz I.B. Clinical features that distinguish PLS, upper motor neuron-dominant ALS, and typical ALS. Neurology. 2009;72(22):1948–1952. doi: 10.1212/WNL.0b013e3181a8269b. [DOI] [PubMed] [Google Scholar]
- Barritt A.W., Gabel M.C., Cercignani M. Emerging magnetic resonance imaging techniques and analysis methods in amyotrophic lateral sclerosis. Front. Neurol. 2018;9:1065. doi: 10.3389/fneur.2018.01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad R.J., Gabel M.C., Dowell N.G. Neurite orientation and dispersion density imaging (NODDI) detects cortical and corticospinal tract degeneration in ALS. J. Neurol. Neurosurg. Psychiatry. 2018;90:404–411. doi: 10.1136/jnnp-2018-318830. [DOI] [PMC free article] [PubMed] [Google Scholar]