Abstract
Immunotherapy has become a promising new approach for cancer treatment due to the immune system’s ability to remove tumors in a safe and specific manner. Many tumors express anti-inflammatory factors that deactivate the local immune response or recruit peripheral macrophages into pro-tumor roles. Because of this, effective and specific ways of activating macrophages into anti-tumor phenotypes is highly desirable for immunotherapy purposes. Here, the use of a small molecule TLR agonist as a macrophage activator for anti-cancer therapy is reported. This compound, referred to as PBI1, demonstrated unique activation characteristics and expression patterns compared to treatment with LPS, through activation of TLR4. Furthermore, PBI1 treatment resulted in anti-tumor immune behavior, enhancing macrophage phagocytic efficiency five-fold versus non-treated macrophages. Additive effects were observed via use of a complementary strategy (anti-CD47 antibody), resulting in ~10-fold enhancement of phagocytosis, suggesting this small molecule approach could be used in conjunction with other therapeutics.
Keywords: Inflammatory, Immunotherapy, Macrophage, Adjuvant, Cancer, Phagocytosis
1. Introduction
Cancer is one of the leading causes of death around the world, and fighting it is a focus of research programs globally [1]. However, most cancer therapy regimens continue to rely on non-specific chemotherapy and radiotherapy to eliminate tumors, which have severe side effects. They are also ineffective against many cancer types and can increase disease recurrence [1]. As a result, alternative strategies are sought after, such as immunotherapy [2]. Cancer immunotherapy involves utilizing the immune system to eliminate the disease, and is attractive on account of the immune system’s specificity and biocompatibility compared to traditional cancer therapy. Several immunotherapies have been approved clinically, and others have reached the clinical trial stage [3].
While immunotherapy is alluring, developing immune-based strategies is challenging. Normally, the immune system detects and eliminates pre-oncogenic cells [4], however, cancer cells can generate cytokines and receptors for immune evasion and reprogramming [5]. In this manner, tumorigenic cells are able to escape detection and disable pro-inflammatory behaviors. An example of the former is over-expression of “don’t eat me” surface marker CD47 by cancer cells, preventing phagocytosis [6], while in the latter the tumor releases chemo-attractants and anti-inflammatory signals, such as IL-4, IL-10, CS1, CSF1R, and MFG-E8, to reprogram immune cells to perform protumorigenic roles [4,7]. These include facilitation of angiogenesis, epithelial to mesenchymal transition (EMT), and microenvironment remodeling [8]. Once this point is reached, the likelihood of patient survival decreases sharply [9].
Because of the immune system’s role in cancer progression, there is great interest in the re-education of immune elements into anti-cancer entities. Polarization of macrophages into M1 (classically activated) phenotypes is important toward refocusing the immune system for eliminating cancer [10]. In this immune-stimulating phenotype, macrophages attack and phagocytose tumor cells. This facilitates a larger overall attack by the immune system, resulting in tumor elimination [10]. Macrophages also generate reactive oxygen species (ROS) and present tumor antigens, which recruit T cells and B cells to the tumor site [11]. In contrast, immune-suppressing M2 (alternatively activated) phenotypes, or tumor-associated macrophages (TAMs), contribute to tumor progression, joining the tumor mass and microenvironment [10]. TAMs release pro-tumor growth factors, such as vascular endothelial growth factor (VEGF), promote vascularization, remodel the microenvironment, and silence the immune response [12–14]. Overall, reprogramming macrophages into M1-like states and away from M2/TAM phenotypes has great potential as an anti-cancer immunotherapeutic approach [15].
The polarization of macrophages toward M1 phenotypes is a well-studied phenomenon, with known pathways identified. Specifically, the interferon gamma (IFN-γ) receptor (IFNGR) and the toll-like receptor (TLR) class are understood to activate macrophages into pro-inflammatory roles [16]. The most common strategies for in vitro M1 macrophage polarization involve treatment with IFN-γ (ligand for IFNGR) and lipopolysaccharide (LPS, a TLR4 agonist originating from bacterial cell walls) [16]. While these agonists are useful as research tools, both IFN-γ and LPS have drawbacks that make them non-viable therapeutically. IFN-γ is a small protein, difficult to consistently modify and incorporate into delivery vehicles [17], while LPS is a bacterial cell wall component consisting of a mixture of structures and may be contaminated with other bacterial components, resulting in off-target immune effects [18]. Systemic administration of these agents results in immune overstimulation, leading to negative outcomes including septic shock, cytokine storms, and death [18].
Because of these issues, there is a need to identify alternative macrophage activators. Additionally, therapeutic candidates should be amenable to chemical modifications and association with targeted delivery vehicles. To date, there are few immune-modulating compounds approved clinically. One example is the TLR7 activator imiquimod, which is approved for the topical treatment of genital warts and basal cell carcinoma [19]. However, in terms of general anti-cancer agents, systemically delivered drugs are desirable to provide access to a range of tumor locations and facilitate immune cell recruitment. There have been examples of anti-inflammatory antibody blockades utilized to reprogram macrophages for cancer therapy [7,20], but in terms of modifications, antibodies are more difficult to alter than small organic molecules, and are also linked to uncontrollable immune-based toxicity [21].
Here, we utilize a small molecule TLR4 activator to induce the M1 phenotype and enhance anti-oncogenic properties in macrophages (Fig. 1). This molecule, a pyrimido[5,4-b]indole referred to as PBI1, was previously identified among a series of compounds that activate TLR4 [22]. This molecule was shown to bind to TLR4 and induce expression of various pro-inflammatory cytokines in dendritic cells [22]. Preclinical studies have shown that structurally similar molecules are effective immune adjuvants for influenza therapy [23], however, these compounds have not been evaluated in terms of macrophage activation and anti-cancer activity. Our hypothesis is that this small molecule TLR4 agonist can activate and polarize macrophages into an anti-cancer phenotype nearly as well as naturally occurring cytokines or existing adjuvants. We demonstrate that PBI1 upregulates pro-inflammatory genes in macrophages and induces M1-associated phenotypic changes and cytokine production. Macrophages treated with PBI1 demonstrate enhanced anti-tumor activity toward B-cell lymphoma cells as determined by phagocytosis assays. We also show that treatment of M2-like macrophages with PBI1 results in their re-education of macrophages toward an M1-like phenotype.
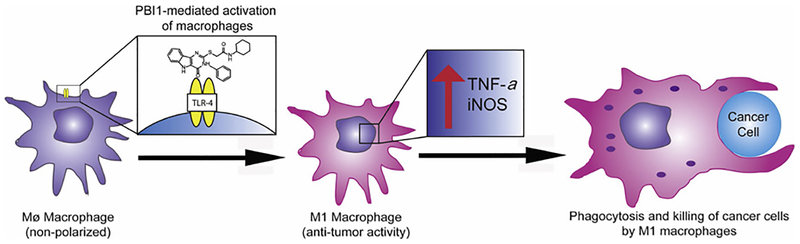
Fig. 1.
Illustration of pyrimido[5,4-b]indole (PBI1)-mediated macrophage activation. Following TLR4 stimulation, resulting M1 macrophages have enhanced levels of TNF-α and iNOS, increased ROS generation, and phagocytosis efficiency.
2. Materials and methods
All reagents were purchased from Thermo-Fisher Scientific except where otherwise noted. All DMSO utilized was cell culture grade (Sigma). Confocal microscopy images were obtained on an Eclipse Ti-E microscope (Nikon, Tokyo, Japan) using a 63X or 20X objective at room temperature. Images were acquired and processed using NIS-Elements and ImageJ. Flow cytometry analysis was performed on a BD LSRFortessa 5 L flow cytometer equipped with FACSDiva (BD Sciences, USA) at the Flow Cytometry Core Facility at the University of Massachusetts Amherst. RT-PCR data was generated using a CFX Connect Real-Time PCR Detection System (Biorad, Hercules, CA). For assays requiring absorbance measurements, a SpectraMax M2 plate reader was used (Molecular Devices, San Jose, CA). Student’s t-test was used to analyze statistical significance between control groups.
2.1. Cell culture
RAW 264.7 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). Primary immortalized macrophages were a gift from Prof. Susan Carpenter at the University of California, Santa Cruz. Both types of cells were cultured at 37 °C under a humidified atmosphere containing 5% CO2. Standard growth media consisted of high glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (100 μg/ml penicillin and 100 μg/ml streptomycin). Daudi cells were a gift from Prof. Vincent Rotello at the University of Massachusetts Amherst and were cultured in Roswell Park Memorial Institute media (RPMI 1640) supplemented with 10% fetal bovine serum and 1% antibiotics (100 μg/ml penicillin and 100 μg/ml streptomycin). Under the above culture conditions, the cells were sub-cultured approximately once every four days and only cells between passages 7 and 15 were used for all experiments.
2.2. Synthesis
The synthesis of PBI1 was largely performed as previously described [22]. A complete description of procedures and analytical data may be found in the supporting information.
2.3. RT-PCR preparation
RAW 264.7 cells were plated in 24 well plates at a density of 100,000 cells/well. Media was aspirated and PBI1-treated cells were dosed with 20 μg/mL compound in fresh media for 24 h. M1 treatment group cells were treated with 50 ng/mL IFN-γ for 24 h after which LPS was added to a final concentration of 50 ng/mL for an additional 24 h. M2 group cells were treated with 50 ng/mL IL-4 for 48 h. For the M2 + PBI1 group, the cells were treated with 50 ng/mL IL-4 for 24 h. 24 h after IL-4 dosing, the media was removed and replaced with fresh media containing 20 μg/mL PBI1 for 24 h. Each experiment included three biological replicates per treatment condition. 20 μg/mL of PBI1 was used for this assay and several others as PBI1 demonstrated no toxicity at this concentration. Following treatments, RNA was extracted following the procedure below.
2.4. RNA extraction and cDNA conversion
Approximately 1.5 μg RNA was directly harvested from cells using the PureLink RNA Mini Kit (Ambion) following the manufacturer’s instructions. SuperScript IV Reverse Transcriptase, RNaseOut, 10 mM dNTPs, and 50 μM Random Hexamers were used for the conversion of approximately 150 ng of RNA to cDNA (ThermoFisher, Pittsburgh, PA), also following the manufacturer’s instructions with a sample volume of 20 μL. Briefly, primers were annealed to RNA at 65 °C for 5 min. Then the annealed RNA was combined with the reaction mixture (containing 500 units of reverse transcriptase per sample) and amplified at 53 °C for 10 min and melted at 80 °C for 10 min. The cDNA was frozen at −20 °C and then used for RT-PCR within 1 week. RNA and cDNA were quantified using a NanoDrop 2000 (ThermoFisher). RNA and cDNA contamination, inhibition, and integrity were assessed by analyzing the A260/A280 ratio, where 1.8 for DNA and 2.0 for RNA was considered “pure.”
2.5. Quantitative RT-PCR
RT-PCR was performed on cDNA as prepared above using a CFX Connect real-time system (Biorad) with iTaq Universal SYBR Green Supermix (Biorad, Hercules, CA). All DNA primers were purchased from Integrated DNA Technologies (Caralville, Iowa). The following primer sequences (as determined by NCBI primer-BLAST) were used: β-actin (ACTB, accession number: NM_007393, amplicon length: 86 pairs, exon6) (forward) GATCAGCAAGCAGGAGTACGA, (reverse) AAAACGCAGC GCAGTAACAGT; iNOS (NOS2, accession number: NM_010927.4, amplicon length: 127 pairs, exon 2,3, splice variants: 1–3) (forward) GTTCTCAGCCCAACAATACAAGA, (reverse) GTGGACGGGTCGATGT CAC; TNF-α (TNF, accession number: NM_013693.3, amplicon length: 145 pairs, exon 3,4) (forward) CCTGTAGCCCACGTCGTAG, (reverse) GGGAGTCAAGGTACAACCC. 200 nM of each primer was mixed with 1 μl (100 ng) of cDNA, 10 μL SYBR green supermix and H2O to a final volume of 20 μL. Analyses were performed as follows: the samples were first activated at 50 °C for 2 min, then 95 °C for 2 min. Then denaturing occurred at 95 °C for 30 s followed by annealing at 57 °C; the denature/ anneal process was repeated over 40 cycles. Relative gene expression was determined by comparing the Ct value of the gene of interest to that of the β-actin housekeeping gene, by the 2ΔΔCt method [24]. Three biological replicates were performed for each treatment condition and three technical replicates were used for each biological replicate. There was no amplification for the no-template control (NTC). Data was analyzed using the CFX Manager 3.1 software. CQ values were generated by using the point at which the sample fluorescence value exceeded the software’s default threshold value. Each sample was normalized to the untreated control. β-actin was used as a reference gene since it is commonly used and none of the treatments were expected to affect its expression.
2.6. Phagocytosis assay with flow cytometry
RAW 264.7 and Daudi cells were separately plated in 24 well plates at a density of 2 × 105 cells/well each. The same day, designated RAW 264.7 wells were treated with PBI1 to reach a final concentration of 5 μg/mL in 0.5% DMSO. A slightly lower concentration was utilized in this experiment to demonstrate that profound changes in cell phenotype are possible at low concentrations of PBI1. 24 h following treatment, RAW cells were trypsinized using TrypLE (ThermoFisher), washed twice in PBS, and labelled with 1 μg/mL PE-F4/80 antibody (BD Biosciences, cat. No: 565410, Clone:T45–2342) for 30 min at 4 °C. All RAW cells were resuspended in culture media and 10 μM cytochalasin D was added to the designated samples. Simultaneously, Daudi cells were removed from the 24 well plate, washed twice with LCIS (ThermoFisher) and labelled with pHRhodo Green according to the manufacturer’s instructions (ThermoFisher). These cells were washed with PBS and then labelled with 10 μg/mL anti-CD47 antibody (Bio Xcell, cat. No: BE0019–1, Clone: B6.H12) for 30 min at 4 °C. All of the Daudi cells (approximately 4 × 105) were resuspended in culture media and combined with the RAW 264.7 cells for 2 h at 37 °C. The approximate ratio of RAW:Daudi was 1:1. The samples were then washed with PBS, resuspended in FACS buffer (1% FBS in PBS) and transferred to flow cytometry tubes. The samples were analyzed on a LSRFortessa 5 L flow cytometer (BD Biosciences) using 488 nm and 561 nm lasers, counting 30,000 events, at the University of Massachusetts Amherst Flow Cytometry Facility. Three samples per treatment group were evaluated. Phagocytotic index was calculated using the following equation: [F4/80+pHRhodoGreen+ events]/[F4/80+pHRhodoGreen− events], normalized to the untreated control group.
2.7. Griess assay
RAW 264.7 cells were plated in 24 well plates at a density of 1.5 × 105 cells/well 24 h prior to the experiment. The following day, the culture media was removed and replaced with serum free Opti-Mem media for 2 h. Cells designated for TLR4 inhibition were pre-treated with 7.2 μg/mL TAK242 (Cayman Chemical) 1 h before additional treatment. Then, media was replaced again with 250 μL phenol red-free DMEM culture media containing either LPS or PBI1 at the indicated concentrations, and cells were incubated for an additional 48 h. Cell supernatant was collected from wells and centrifuged at 5000 rpm for 5 min. Then, Griess reagent (Thermofisher) was prepared according to the manufacturer’s instructions and 60 μl Griess reagent was combined with 60 μL cell supernatant in a clear 96 well plate. After 15 min in the dark, absorbance was read using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA) at 548 nm. Three biological replicates per treatment condition were used. Actual NO2− concentrations were determined by comparing absorbance values to a standard curve generated using pure NaNO2− solutions.
2.8. Confocal microscopy
For acquisition of representative cell morphology images, RAW 264.7 cells were plated in a 4 chamber Lab-Tek II chambered cover-glass system (Nunc, NY) at a density of 5 × 104/well and allowed to adhere overnight. Media was aspirated and cells were then polarized with 20 μg/mL PBI1 in 0.2% DMSO in fresh media for 48 h. After polarization, cells were imaged using an Eclipse Ti-E microscope at 20x magnification. M1 cell images were acquired similarly, using polarization conditions as outlined in the RT-PCR Preparation section above.
For acquisition of representative phagocytosis images, 5 × 104 RAW 264.7 cells were plated in a 4 chamber Lab-Tek II chambered cover-glass system. Designated wells were directly treated with PBI1 to a final concentration of 4.5 μg/mL in 0.4% DMSO for 24 h. The RAW cells were labeled with Cell Tracker Blue according to the manufacturer’s instructions. 1 × 105 Daudi cells were counted, labeled with phRhodo Green and added to the wells containing RAW cells. After 2 h incubation, wells were imaged using an Eclipse Ti-E microscope at 63x magnification. The final ratio of RAW to Daudi cells was approximately 2:1.
2.9. ELISA
RAW 264.7 cells were plated in 24 well plates at a density of 60,000 cells/well and allowed to adhere overnight. Media was removed and PBI1-treated cells were dosed with 5, 10, and 20 μg/mL compound in0.4% DMSO in fresh media for 48 h. M1 treatment group cells were treated with 50 ng/mL IFN-γ for 24 h after which LPS was added to a final concentration of 50 ng/mL for an additional 24 h. M2 group cells were treated with 50 ng/mL IL-4 for 48 h. For the M2 + PBI1 group, the cells were treated with 50 ng/mL IL-4 for 24 h. 24 h after IL-4 dosing, the cells were treated with 20 μg/mL PBI1 for 24 h. Each experiment included four biological replicates per treatment condition. After treatment, supernatant was collected from the wells and then analyzed using a Mouse IL-6 ELISA set and a Mouse TNF-α ELISA set (BD Biosciences, Bedford, MA) according to the manufacturer’s instructions.
3. Results
3.1. Confocal microscopy
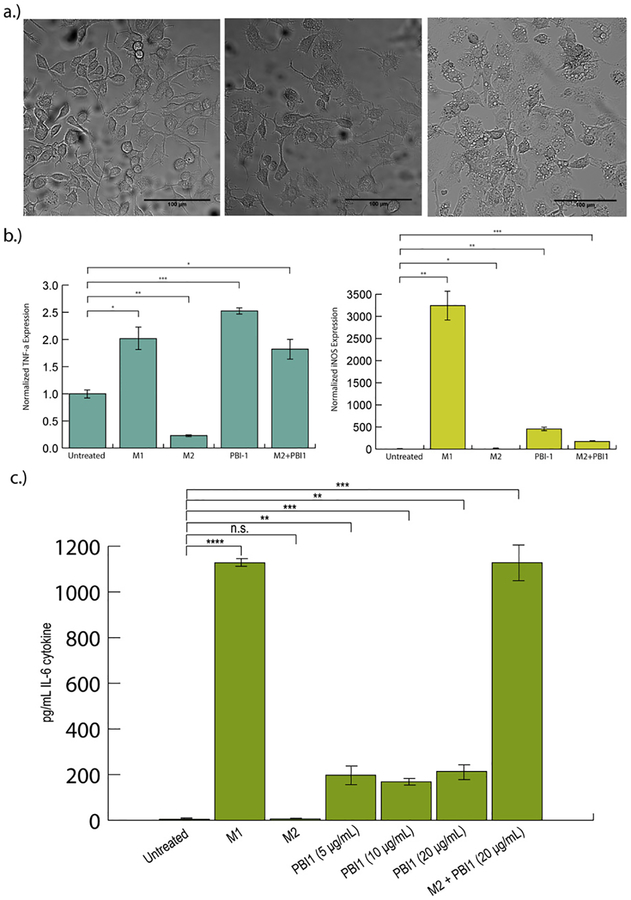
Following synthesis and characterization of PBI1 (Fig. S1–S8), RAW 264.7 murine macrophages were dosed with the compound, and changes in morphology and gene expression were assessed using confocal microscopy and RT-PCR, respectively (Fig. 2). Confocal microscopy revealed significant changes in macrophage morphology after 24 h. Treated macrophages acquired a phenotype that is associated with M1-polarized macrophages (Fig. 2a): the cells became flatter and produced longer pseudopodia [16] compared to non-treated cells.
Fig. 2.
Effects of PBI1 treatment on cell morphology and M1 marker expression. a) Confocal microscopy images (20x) depicting changes in cell morphology following treatment with 20 μg/ml PBI1. PBI1-treated macrophages (center) resembled those polarized to an M1 phenotype (right), with a “flatter” appearance and more pseudopodia, in comparison with non-treated cells (left). b) RT-PCR analysis of TNF-α (left) and iNOS (right) expression in PBI1 treated cells revealed significantly greater expression of both relative to non-treated controls. M2 + PBI1 refers to cells polarized to the M2 phenotype and subsequently treated with PBI1. c) ELISA analysis of IL-6 secretion following treatment. Unpaired, two-tailed student T-tests with equal variance were used to determine significance; *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.00005. n.s. = non-significant.
3.2. RTPCR analysis
RT-PCR analysis of the mRNA expression of two M1-related markers, tumor necrosis factor-α (TNF-α) and inducible nitric oxide synthase (iNOS) [11], revealed that PBI1-treated macrophages express significantly higher levels of both genes compared to non-treated cells, in a similar trend as IFN-γ/LPS treatment (Fig. 2b). This effect was also observed in an immortalized primary macrophage cell line (Fig. S9). Furthermore, PBI1 treatment was also able to ‘re-educate’ macrophages that had been polarized toward the anti-inflammatory M2 phenotype, resulting in enhanced levels of iNOS and TNF-α. Alamar blue assays indicated that no significant toxicity occurred as a result of compound treatment (Fig. S10).
3.3. ELISA analysis
ELISA analysis of the protein expression of the M1-related cytokines, interleukin 6 (IL-6) and TNF-α, revealed that cells secreted both cytokines to greater extents when treated with PBI1 or the combination of IFN-γ/LPS compared to untreated cells (Fig. 2c, S11). PBI1 treatment following M2 polarization also resulted in secretion of both cytokines.
3.4. Griess assay
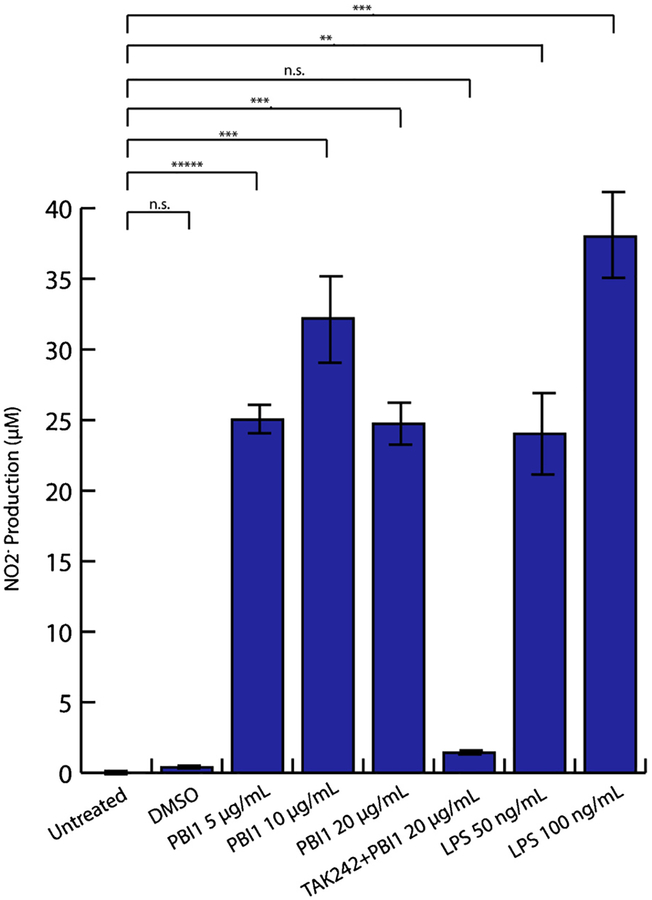
Generation and release of reactive oxygen and nitrogen species (ROS and RNS, respectively) are important processes of M1 macrophage anti-tumor and pathogen responses [25]. RNS are generally derived from nitric oxide (NO). To evaluate relative NO production by PBI1-treated cells, a Griess assay was performed [26]. This method indirectly measures NO via evaluation of nitrite (NO2−), one of its two primary stable and non-volatile breakdown products. The determined NO2− concentrations show that PBI1 effectively induced production of NO2−, achieving significantly higher levels than non-treated samples, and within 2-fold of LPS-treated cells (Fig. 3). Additionally, pretreatment with the TLR4 specific inhibitor TAK242 completely eliminated the induction of NO by PBI1 (Fig. 3) [27].
Fig. 3.
Supernatant NO2− levels are increased following LPS or PBI1 treatment as measured by Griess assay. Pretreatment with TAK242 inhibited TLR4 activity prior to PBI1 addition (PBI1 + TAK242). Unpaired, two-tailed student T-tests with equal variance were used to determine significance; **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.00005. n.s. = non-significant.
3.5. Phagocytosis
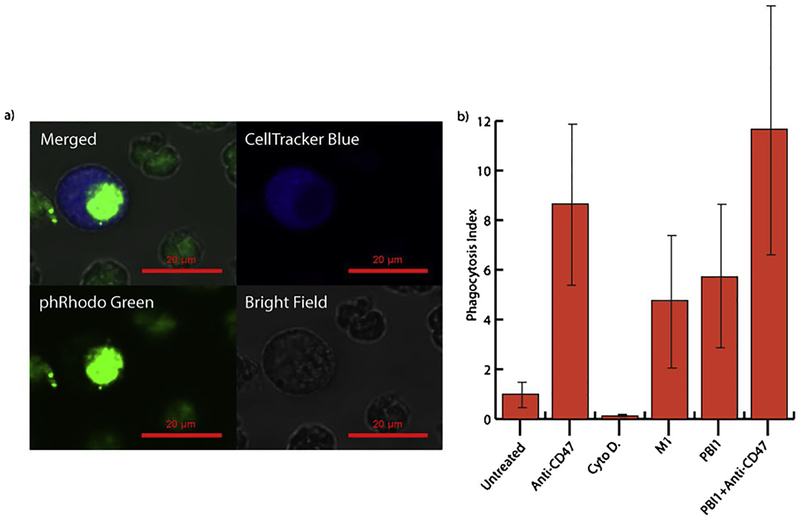
A key aspect of the macrophage antitumor response is the identification and phagocytosis, or engulfment, of cancerous cells [6]. To assess the phagocytic efficacy of PBI1 treated macrophage cells, a fluorescent flow cytometry assay was conducted using Daudi (B cell lymphoma) as the target cancer cell line (Fig. 4, S12). RAW 264.7 cells treated with PBI1 had a nearly 5-fold greater average phagocytic efficiency versus non-treated macrophages. This was increased further when macrophage PBI1 treatment was combined with antibody-mediated blocking of Daudi cell CD47, the ‘don’t eat me’ signal involved in phagocytosis inhibition signaling. Cytochalasin D, a potent phagocytosis inhibitor, was used as a negative control [28].
Fig. 4.
Effects of PBI1 on phagocytosis of Daudi (B-cell lymphoma) cells. a) Representative confocal microscopy images illustrating RAW 264.7 macrophage (CellTracker Blue) phagocytosis of Daudi cells (phRodo Green). b) Phagocytosis indices generated from flow cytometry data of RAW 264.7 macrophages phagocytosing Daudi cells following various treatments.
4. Discussion
In summary, PBI1 was demonstrated to polarize macrophages toward an anti-cancer phenotype. RAW 264.7 macrophages treated with PBI1 adopted an “M1-like” pro-inflammatory morphology, as cells became flattened and produced extended pseudopodia. RT-PCR and ELISA results also corroborated this phenotype via an increase in the expression of M1 inflammatory genes. PBI1 treatment increased the expression of TNF-α, IL-6 and iNOS approximately 2.5, 200 and 500-fold, respectively. Dosing of PBI1 also resulted in the re-education of M2-polarized macrophages, whereupon following treatment, cells expressed higher levels of inflammatory cytokines. Notably, PBI1 treatment after M2 polarization increased TNF-α, IL-6 and iNOS to levels nearly identical to those of M1 polarized macrophages. This is particularly relevant for the conversion of tumor-associated macrophages into tumor-killing macrophages within cancer microenvironments.
We also evaluated the mechanism by which PBI1 promoted macrophage activation. The most common pathways for macrophage stimulation include the TLR family of receptors that recognize a wide variety of substrates [8]. To confirm that TLR4 is the target of PBI1 for macrophage activation, a competition experiment was performed. Cells were treated with a highly specific TLR4 inhibitor, TAK242, and activation by PBI1 was determined by Griess assay. This experiment revealed that macrophages treated with either PBI1 or LPS released significant levels of nitric oxide, another confirmation of the activation potential of PBI1. However, with pre-treatment of cells with TAK242, PBI1 did not induce any detectable level of nitric oxide, confirming that TLR4 is the target of PBI1 and is crucial to resulting macrophage activation.
Having evaluated the pro-inflammatory responses of macrophages to PBI1 treatment, the ability of these cells to subsequently phagocytose cancers cells was investigated. It has been previously shown that activation of local macrophages into an M1 inflammatory phenotype can result in significant anti-tumor macrophage activity, which is of great interest for the generation of cancer immunotherapies [25]. One of the major anti-cancer macrophage mechanisms involves phagocytosis of cancer cells. Inflammatory macrophages can invade tumor tissue, engulf resident tumor cells, and release immune signaling factors that drive further immune responses against the tumor site [25]. A phagocytosis assay revealed that PBI1-treated macrophages engulfed targeted cancer cells with a 5-fold higher efficacy than untreated macrophages, revealing that inflammatory activation by PBI1 does in fact increase anti-tumor activity. It was also demonstrated that pre-treatment of the targeted cancer cells with a CD47 blocking antibody, which blocks the SIRPα-CD47 phagocytosis inhibitory pathway, further increases the efficacy of PBI1-induced phagocytosis. This effect occurred independently of PBI1-activation as well and could be used to increase the anti-cancer efficacy of PBI1 treatment.
5. Conclusion
The immune system and cancer progression have a complex relationship. While effective immune-based strategies are of interest in enervating and eliminating cancer progression, their development can be challenging due to varying macrophage-activating signals [29,30]. PBI1 has significant macrophage activation capability. As a potential immune adjuvant for cancer therapy, PBI1 is relatively non-toxic and effectively increases both phagocytic and oxidative burst mechanisms of the macrophage anti-tumor response. While PBI1 has shown promise as a therapeutic on its own, it will likely show the greatest efficacy if used in conjugation with drug delivery vehicles or targeting elements. This is especially relevant in trying to avoid broad immune-activation and runaway immune responses. As a small molecule, it should be fairly straight-forward to attach to a variety of carriers, including nano-particles, proteins, and cells; further studies will explore these options. Incorporation of PBI1 with more complex therapeutics, as opposed to its use independently, could result in robust combinatorial anti-cancer strategies. Additional studies will evaluate the anti-cancer efficacy of PBI1 in vivo, including in concert with delivery vehicles, and as an adjuvant with other chemotherapeutics. In the future, we will also seek to identify other potential therapeutic targets of PBI1-activated macrophages.
Supplementary Material
Funding sources
This research was supported by a SEED grant from the University of Massachusetts Institute for Applied Life Sciences. JH was supported by the NIH (EB014277) and an NIH National Research Service Award (T32 GM008515). JAM-R was supported by the UMass Amherst NIH Postbaccalaureate Research Program (GM086264–08), a Northeast Alliance for Graduate Education and the Professoriate (NEAGEP) fellowship from the STEM Diversity Institute at UMass Amherst, and an NIH National Research Service Award (T32 GM008515).
Abbreviations:
- CD47
cluster of differentiation 47
- IL-4
interleukin 4
- CS1
CCND3 subset 1
- CSF1R
colony stimulating factor receptor 1
- MFEG8
milk fat globule-EGF factor 8 protein
- VEGF
vascular endothelial growth factor
- SEAP
secreted embryonic alkaline phosphatase
- LPS
lipopolysaccharide
- IFN-γ
interferon-γ
Footnotes
Declaration of Competing Interest
None.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.phrs.2019.104452.
References
- [1].DeSantis C, Ma J, Bryan L, Jemal A, Breast cancer statistics 2013, Cancer J. Clin 64 (2014) 52–62, 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- [2].Blattman JN, Greenberg PD, Cancer immunotherapy: a treatment for the masses, Science 305 (2004) 200–206, 10.1126/science.1100369. [DOI] [PubMed] [Google Scholar]
- [3].Butterfield LH, Dendritic cells in cancer immunotherapy clinical trials: are we making progress? Front. Immunol 4 (2013) 3–9, 10.3389/fimmu.2013.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jinushi M, Chiba S, Yoshiyama H, et al. , Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells, Proc. Natl. Acad. Sci 108 (2011) 12425–12430, 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Solinas G, Germano G, Mantovani A, Allavena P, Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation, J. Leukoc. Biol 86 (2009) 1065–1073, 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- [6].Jaiswal S, Jamieson CH, Pang WW, et al. , CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis, Cell 138 (2009) 271–285, 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu Y, Knolhoff BL, Meyer MA, et al. , CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models, Cancer Res 74 (2014) 5057–5069, 10.1158/0008-5472.CAN-13-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Noy R, Pollard JW, Tumor-associated macrophages: from mechanisms to therapy, Immunity 41 (2014) 49–61, 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Q, Liu L, Gong CY, et al. , Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature, PLoS One 7 (2014) e50946, 10.1371/journal.pone.0050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zheng X, Turkowski K, Mora J, et al. , Redirecting tumor-associated macrophages to become tumoricidal effectors as a novel strategy for cancer therapy, Oncotarget 8 (2015) 48436–48452, 10.18632/oncotarget.17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Murray PJ, Wynn TA, Protective and pathogenic functions of macrophage subsets, Nat. Rev. Immunol 11 (2011) 723–737, 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bingle L, Lewis CE, Corke KP, Reed M, Brown NJ, Macrophages promote angiogenesis in human breast tumour spheroids in vivo, Br. J. Cancer 94 (2006) 101–107, 10.1038/sj.bjc.6602901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu CY, Xu JY, Shi XY, et al. , M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway, Lab. Invest 93 (2013) 844–854, 10.1038/labinvest.2013.69. [DOI] [PubMed] [Google Scholar]
- [14].Liu X, Pu Y, Cron K, et al. , CD47 blockade triggers T cell-mediated destruction of immunogenic tumors, Nat. Med 21 (2015) 1209–1215, 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ray M, Lee YW, Hardie J, et al. , CRISPRed macrophages for cell-based cancer immunotherapy, Bioconjug. Chem 29 (2018) 445–450, 10.1021/acs.bioconjchem.7b00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mosser DM, Edwards JP, Exploring the full spectrum of macrophage activation, Nat. Rev. Immunol 8 (2008) 958–969, 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fu A, Tang R, Hardie J, Farkas ME, Rotello VM, Promises and pitfalls of intracellular delivery of proteins, Bioconjug. Chem 25 (2014) 1602–1608, 10.1021/bc500320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dasu MR, Devaraj S, Du Clos TW, Jialal I, The biological effects of CRP are not attributable to endotoxin contamination: evidence from TLR4 knockdown human aortic endothelial cells, J. Lipid Res 48 (2007) 509–512, 10.1194/jlr.C600020-JLR200. [DOI] [PubMed] [Google Scholar]
- [19].van der Fits L, Mourits S, Voerman JS, et al. , Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis, J. Immunol 182 (2009) 5836–5845, 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- [20].Georgoudaki AM, Prokopec KE, Boura VF, et al. , Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis, Cell Rep 15 (2016) 2000–2011, 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- [21].Pardoll DM, The blockade of immune checkpoints in cancer immunotherapy, Nat. Rev. Cancer 12 (2012) 252–264, 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chan M, Hayashi T, Matthewson RD, et al. , Identification of substituted pyrimido [5,4- b]indoles as selective Toll-Like receptor 4 ligands, J. Med. Chem 56 (2013) 4206–4223, 10.1021/jm301694x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goff PH, Hayashi T, Martinez-Gil L, et al. , Synthetic Toll-like receptor 4 (TLR4) and TLR7 ligands as influenza virus vaccine adjuvants induce rapid, sustained, and broadly protective responses, J. Virol 89 (2015) 3221–3235, 10.1128/JVI.03337-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative C(T) method, Nat. Protoc 3 (2008) 1101–1108, 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- [25].Sinha P, Clements VK, Ostrand-Rosenberg S, Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease, J. Immunol 174 (2005) 636–645, 10.4049/jimmunol.174.2.636. [DOI] [PubMed] [Google Scholar]
- [26].Misko TP, Schilling RJ, Salvemini D, Moore WM, Currie MG, A fluorometric assay for the measurement of nitrite in biological samples, Anal. Biochem 214 (1993) 11–16, 10.1006/abio.1993.1449. [DOI] [PubMed] [Google Scholar]
- [27].Kawamoto T, Ii M, Kitazaki T, Iizawa Y, Kimura H, TAK-242 selectively suppresses Toll-like receptor 4-signaling mediated by the intracellular domain, Eur. J. Pharmacol 584 (2008) 40–48, 10.1016/j.ejphar.2008.01.026. [DOI] [PubMed] [Google Scholar]
- [28].Schrijvers DM, De Meyer GRY, Herman AG, Martinet W, Phagocytosis in atherosclerosis: molecular mechanisms and implications for plaque progression and stability, Cardiovasc. Res 73 (2007) 470–480, 10.1016/j.cardiores.2006.09.005. [DOI] [PubMed] [Google Scholar]
- [29].Liu Y, Hardie J, Zhang X, Rotello VM, Effects of engineered nanoparticles on the innate immune system, Semin. Immunol 34 (2017) 25–32, 10.1016/j.smim.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Joshi BP, Hardie J, Farkas ME, Harnessing biology to deliver therapeutic and imaging entities via cell-based methods, Chem. Eur. J 24 (2018) 8717–8726, 10.1002/chem.201706180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.