Abstract
Understanding variation in physiological traits across taxa is a central question in evolutionary biology that has wide-ranging implications in biomedicine, disease ecology, and environmental protection. Sialic acid (Sia), and in particular, 5-N-acetylneuraminic acid (Neu5Ac), is chemically bound to galactose and the underlying glycan via α2–3 or α2–6 glycosidic linkage (i.e., Siaα2–3Galactose or Siaα2–6Galactose), conferring two different cell surface structures that affects cell to cell communication and interactions with foreign agents including microparasites and toxins. As an initial step towards understanding variation of Sia across the class Aves, we collected red blood cells (RBCs or erythrocytes) and measured Sia quantity in 76 species and 340 individuals using HPLC-MS/MS and glycosidic linkage type in 24 species and 105 individuals using hemagglutination assay. Although Sia quantity did not, α2–6 glycosidic linkage did exhibit a discernable phylogenetic pattern as evaluated by a phylogenetic signal (λ) value of 0.7. Sia quantity appeared to be higher in after hatch year birds than hatch year birds (P<0.05); moreover, ~80% of the measured Sia across all individuals or species was expressed by ~20% of the individuals or species. Lastly, as expected, we detected a minimal presence of 5-N-glycolylneuraminic acid in the avian RBCs tested. These data provide novel insights and a large baseline dataset for further study on the variability of Sia in the class Aves which might be useful for understanding Sia dependent processes in birds.
Keywords: Birds, Phylogenetic, Sialic Acid, Avian Influenza, Plasmodium, Receptor
1. Introduction
Understanding variation in physiological traits across taxa is a central question in evolutionary biology that has wide-ranging implications from biomedicine and disease ecology to environmental protection. Susceptibility to stressors can vary greatly across species. The same agent can be tolerated in one species while causing disease or mortality in another species. One stark example of this phenomenon important to animal and public health is that the same H5N1 avian influenza strain can be highly pathogenic in chickens (Gallus gallus), yet produces minimal clinical signs of disease in mallard ducks (Anas platyrhynchos) (Perkins and Swayne, 2003). These major differences in susceptibility, and therefore the levels of surrogacy of one species for another may depend on variation in receptor sites in host species, immunological response differences, or a combination of known and unknown factors.
Extending the application of surrogacy further, the selection of a model species for the development of a pharmaceutical is informed by knowledge of how the organism is similar and different to the species for which the drug is being developed. Additionally, establishing environmental quality standards aimed at protecting humans and wildlife also relies on an understanding of interspecies variation in susceptibility to ensure that the most sensitive groups are protected by the standard. For example, variation in protein structure across taxa can predict susceptibility to a toxicant (LaLone et al., 2016). Additionally, the presence of phylogenetic signal (Blomberg et al., 2003) in fish toxicity data for metals (Buchwalter et al., 2008) and pesticides (Hylton et al., 2018) provides more recent examples in support of the idea that the correlation between physiology and phylogeny (Garland et al., 2005) can be used to address practical problems in various fields.
One physiological factor known to play an important role in the biology of disease, such as influenza, in birds and other vertebrate taxa is sialic acid. Sialic acid is a nine-carbon negatively charged sugar present on the cell membrane of all vertebrate and a few higher invertebrate species (Varki, 1992). Sialic acids are involved in cell to cell and cell to environment interactions. Sialic acid (Sia), and in particular, 5-N-acetylneuraminic acid (Neu5Ac), is chemically bound to galactose and the underlying glycan via α2–3 or α2–6 glycosidic linkage (i.e., Siaα2–3Galactose or Siaα2–6Galactose), conferring two different cell surface structures that affects cell to cell communication and interactions with foreign agents including microparasites and toxins (Varki, 2007).
The receptor site for influenza A viruses (IAVs) are sialic acids bound to conserved amino acids surrounded by antibody binding sites (Weis et al., 1988). IAVs emerging from birds tend to exhibit a higher affinity to cells expressing α2–3 linked Sias whereas IAVs from humans are often specific to the α2–6 conformation (Suzuki et al., 2000). In addition, the invasion of erythrocytes by Plasmodium falciparum, the causative agent of malaria in humans, is dependent on a high affinity recognition of sialic acid on cell surface receptors (Malpede et al., 2013). Sialic acid is known to serve as a red blood cell (RBC)-bound receptor for P. falciparum in humans (Duraisingh et al., 2003; Orlandi et al., 1992), with the glycolated form of Sia (5-N-glycolylneuraminic acid, Neu5Gc) potentially conferring resistance to P. reichenowi in humans who do not express it, but not chimpanzees that express it (Martin et al., 2005) . From this information, while it has not been observed experimentally, it may be postulated that Sia is a receptor site for Plasmodium species causing avian malaria in birds. Other associations with Sias include organism age (Kumar and Rizvi, 2013; Mehdi et al., 2012) and exposure to environmental stressors affecting inflammatory status such as bacterial pathogens (Lehmann et al., 2006), which can change the quantity of Sia expressed on the cell surface. Lastly, of relevance for the present study, sialic acids afford protection against oxidative stress, which may be highly important for RBCs (Mehdi et al., 2012), whose primary function is to transport oxygen.
Because Sias are known to be important in zoonotic agents, a deeper understanding of Sia expression across avian taxonomic groups is important. For example, if phylogenetic position can be used as a proxy for α2–6 expression in respiratory or gastrointestinal tissues, it may be possible to predict taxonomic groups with a higher potential for hosting IAVs that may affect human populations. With data about Sia expression across taxa, an improved capacity to understand organismal level susceptibility to specific pathogenic agents can be scaled to higher performing epidemiologic models.
However, cataloging phylogenetic or other patterns in physiological traits (susceptibility to IAV, e.g.) is a daunting endeavor. The difficulty stems from issues such as the sheer number of potential organisms to test to the often unknown taxonomic applicability of a method to measure a biomolecule (Brock et al., 2014; Fassbinder-Orth et al., 2016), to the unknown extent to which the sampling and handling of an organism affects its physiological state (Wingfield et al., 1995). Added challenge comes from the “moving goalpost problem” or the “red queen effect” (Varki, 2006); that is, evolutionary processes change organisms over time in response to local environmental factors. A study of the variability of glycosidic linkage and Sia quantity across taxa is feasible because species-specific reagents are not required and capture and sampling is not known to alter Sia in the rapid timeframe of handling.
The influence of evolutionary relationships on species physiology is well recognized (Blomberg et al., 2003; Garland et al., 2005). Groups of closely related individuals inhabiting a common location with similar life histories tend to interact with similar environmental factors to which their physiological systems respond and confer competitive survival and reproductive rates. Consequently, closely related species tend to express similar physiological traits as compared to more distantly related species, which is measured as degree of phylogenetic signal.
We hypothesized the presence of phylogenetic signal in Siaα2–3Gal and Siaα2–6Gal glycosidic linkages, but not Neu5Ac quantity on RBC membranes. Linkage is a modification of Sia that requires the presence of specific inherited biosynthetic machinery (i.e., genes) while variation about current Sia quantity is a result of up or down regulation of existing biosynthetic machinery in response to the current presence of a Sia ligand (Angata and Varki, 2002) and factors such as RBC age and redox status (Mehdi et al., 2012). More broadly, the current work fits within an aim to discern phylogenetic relationships in Sia in birds that could be useful in the understanding of host-pathogen systems such as influenza and Plasmodium.
2. Materials and Methods
2.1. Summary of dataset
Samples for the measurement of Sia quantity were obtained from 76 avian species across 65 genera, 29 families, and 16 orders for a total of 340 individuals. Glycosidic linkage measurements were made in 24 species across 21 genera, 13 families, and 5 orders for a total of 105 individuals. Sample sets were not the same for each analysis because sample requirements and access to samples differed for each assay as described below. Given the underlying questions for the work, samples were acquired in a way so as to maximize phylogenetic distance within analysis type in favor of strictly matching taxonomic groups.
2.2. Sample collection
Both captive and wild birds were sampled for this study and multiple potential covariates were collected at the time of sampling including bird mass, sex, date, and location. Birds were handled according to the Guidelines for the Care and Use of Wild Birds in Research (Fair et al., 2010) and IACUC protocol (#10–60) at Los Alamos National Laboratory to ensure the humane treatment of all experimental subjects. All wild birds captured as part of this study were approved under applicable State and Federal permits for scientific collecting and banding permits. No more than 1% of a bird’s blood volume was collected by venipuncture into lithium heparin tubes. For quantification of Neu5Ac, sample was placed on ice (4°C) until it was centrifuged at 1000 × g for 10 minutes within 3 hours of collection. The plasma components and RBCs were placed into separate tubes and the buffy coat (white blood cells) was discarded. RBC samples were then washed 2x in PBS (pH 7.6) and all samples were frozen at −80°C until further processing. For analysis of Sia-Gal glycosidic linkage type via hemagglutination assay (as described below), samples were collected as above, and maintained at 4°C for no more than one week before being processed as fresh whole blood.
Red blood cell plasma membranes were isolated from cytoplasmic and nuclear components by osmotic disruption and serial centrifugation as modified from Georgatos and Blobel (1987). Lysis buffer (5 mM NaPO4, 2 mM MgCl2, 2 mM β-mercaptoethanol, 1 mM phenylmethanesulfonyl fluoride, pH 7.6) was added 5x sample volume followed by vortexing and incubation and gentle agitation of the sample for 15 mins on wet ice. The sample was then centrifuged (16K × g, 1 min) and the middle layer of RBC ghosts were collected and resuspended in lysis buffer. Centrifugation and wash steps were repeated 3–5x to ensure the removal of all traces of hemoglobin. These steps allowed for the separation of nuclear components and any remaining leukocytes and fibrin from the plasma membranes. After final wash, the samples were suspended in a PBS/protease inhibitor cocktail (HALT™, Thermo Cat No. 78430). One aliquot was saved for total protein measurements by Bradford assay and the other for sialic acid quantitation by mass spectrometry. Samples were stored at −80C until analysis by mass spectrometry.
2.3. 5-N-acetylneuraminic acid quantification
Sialic acid (Neu5Ac) was quantified by stable isotope addition and analysis by high-performance liquid chromatography-mass spectrometry as in (Allevi et al., 2008). This technique relies on the addition of a known quantity of N-acetyl-D-neuraminic acid-1,2,3- 13C3 (13C3-Neu5Ac, molecular weight, 312.25 g/mol, Sigma-Aldrich, Cat No. 649694) to each sample prior to processing in order to estimate the quantity of native Neu5Ac by an equivalency calculation (see Eq. 1, 2 below). Thus, quantitation of native Neu5Ac is based on the fact that any sample processing associated loss of analyte occurs to both native and stable isotope labeled Neu5Ac.
| Eq. 1 |
μg Neu5Ac is the only unknown variable, so it is solved for by using Eq. 2
| Eq. 2 |
TIC = total ion counts from HPLC-MS/MS analysis
Once RBCs were processed as above, total protein was measured as in Bradford (1976) using Coomassie Plus assay kit (Pierce, cat. no. 23236) in order to (1) estimate the amount of 13C3-Neu5Ac to add to the sample, and (2) for normalization of sialic acid quantity. For best quantitation, we aimed for a ratio of 1 mol Neu5Ac:1 mol 13C3-Neu5Ac and achieved this via the estimate of 100 pmol Neu5Ac/μg protein in chicken (Gallus gallus) and rock dove (Columbia livia) RBCs based on preliminary studies that produced a standard curve (a 1:1 molar ratio produces an approximately 1:1 TIC ratio). Samples (500 μls) treated with stable isotope enriched 13C3-Neu5Ac were then processed to cleave the glycosidic linkage between Sia and underlying glycans by the addition of 2 M acetic acid and heating at 80°C for 3 hours to completely release all Sias (Varki and Diaz, 1984). Following acid hydrolysis, samples were cleaned by removing all compounds over 10 kDa by filtration (Untracel, Cat #UFC801008, Millipore Sigma, Billerica, MA, USA) and then lyophilized and stored at −20°C until HPLC-MS/MS analysis.
For quantification of free Neu5Ac in prepared samples, the general method of van der Ham et al. (2007) was used with the following modifications. A Thermo Scientific LTQ XL 2D linear quadrupole ion trap mass spectrometer interfaced with an electrospray ionization source and equipped with a Surveyor LC system was used for detection. Separations were performed using a GlycoSep R HPLC column (4.6 mm × 150 mm) (ProZyme, California, USA, Cat No. GKI-4728). The injection volume was 10 μL. A 3.5-minute linear gradient (acetonitrile 5–10%, 0.1% formic acid in water 88–83%, methanol 7%) was used at a flow rate of 500 μL/min. Retention times for both Neu5Ac and isotope enriched 13C3-Neu5Ac were 2.9–3.3 minutes. After 3.5 minutes, the gradient changed to 78% acetonitrile (over 0.5 minutes) to wash the column for an additional 1 minute, followed by a 2-minute hold at original concentrations to re-equilibrate the column. Blank water runs were performed between each sample to ensure complete washing of the column. Each sample was tested three times for consistency. The MS was operated in positive single ion mode (SIM), negative SIM, as well as ms2 for m/z 310, 313, and 326 (Neu5GC) for comparative analysis. Total Ion Counts (TIC) for the M+1 peaks (and M-1 peaks) and major fragments of Neu5Ac, 13C3-Neu5Ac, and Neu5Gc were compared for each sample. The ratio of the two peaks, the known concentration of the isotope enriched Neu5Ac, and volume corrections were used to calculate the concentration of native Neu5Ac for each sample. Duplicate blanks for each batch of samples were generated by treating sample diluent (water) just as samples were treated above. The TIC from the duplicate blanks was averaged and this value was subtracted from a sample’s TIC to correct for diluent effects. We found that using the TIC from the positive ms2 mode, using the molecular ion major fragments m/z 292 (Neu5Ac), 295 (13C3-Neu5Ac), and 308 (Neu5Gc), corresponding to loss of H2O, gave the most precise and consistent results based on known standards. Results from the other modes of analysis were in general quite internally consistent.
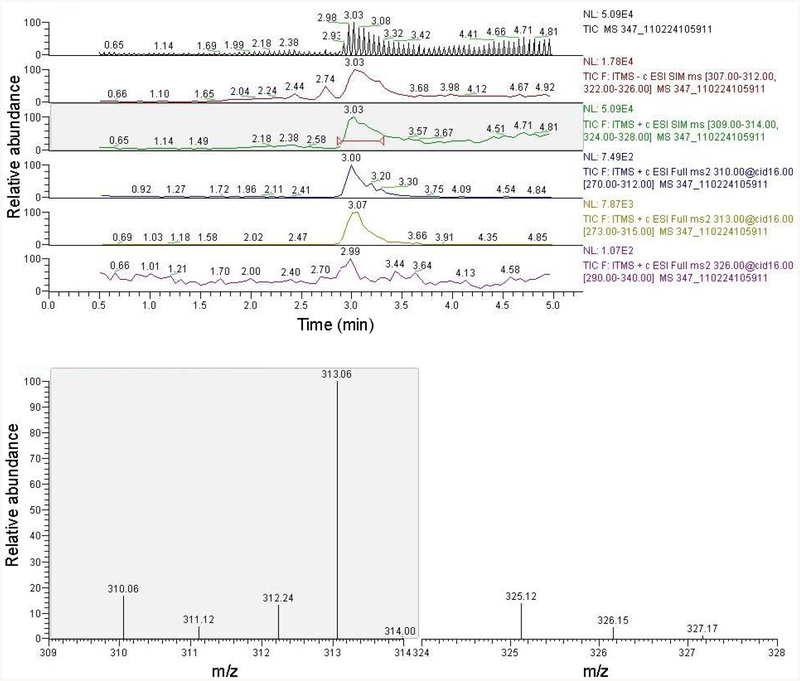
An example LC/MS run is shown in Figure 1. The top cell shows 5 separate but simultaneous TIC chromatograms (the top is a combination of all) of each mode of analysis. The positive SIM chromatogram (green trace) was chosen for demonstration purposes. The red inclusion arrow represents the time period over which continuous spectra were averaged, with the results displayed in the lower cell. The m/z peak at 313 (13C3-Neu5Ac) dominates over 310 (native Neu5Ac) and 326 (Neu5Gc).
Figure 1.
Each liquid chromatographic (LC) trace shows the relative abundance of signal intensity in the mode and region of interest over the time period of the LC run. The bottom mass spectrometer (MS) spectra depict the average signal between 2.9 and 3.4 minutes for positive mode, from mass (M)=309.00 to 314.00 and from M=324.00 to 328.00 (averaged from the green LCMS trace). Data for multiple modes and regions were collected simultaneously over the course of the LCMS run for each.
High (125 μg) and low (12.5 μg) quantity bovine fetuin standards were used to check for assay repeatability by testing for Neu5Ac as with samples. Bovine fetuin is known to contain both Neu5Ac and Neu5Gc forms of sialic acid in known quantities (Spiro, 1960) and therefore is an accepted means for quality assurance (e.g. repeatability) in studies of sialic acid (Allevi et al., 2008; Shaw et al., 2001). The interassay coefficient of variation for high and low quantity fetuin standards were 13.1% and 9.5%, respectively. Mean Neu5Ac quantity for fetuin standards was 454.9 pmol/μg total protein. Neu5Gc content analysis was qualitative, as there was no commercial source at the time of analysis, nor was there 13C enriched Neu5Gc commercially available. This situation limited our ability to detect Neu5Gc as well as substantiate its presence through MS2 fragmentation patterns. Thus, qualitative analysis was done on the main M+1 Neu5Gc peak averaged over the portion of the LC run with the highest TIC.
2.4. Sia-Gal Glycosidic Linkage Determination
Hemagglutination assay was used to determine the presence of α2–3 and α2–6 Sia-Gal glycosidic linkage on the membrane surface of RBCs. Hemagglutination and hemagglutination inhibition assays have been conventionally used for detection of influenza viruses (Ito et al., 1997; Pawar et al., 2012) and antibody response (Pedersen, 2014; Wegmann and Smithies, 1966); but, instead of influenza virus, a specific lectin for a specific Sia-Gal glycosidic linkage was used in the current study. The idea of the assay is that the magnitude (titer) of RBC agglutination is correlated with the quantity of either Sia-Gal glycosidic linkage type in the presence of a linkage specific lectin. To determine the abundance of each glycosidic linkage type, a titered quantity of α2–3 (Maackia amurensis II extract, Vector Laboratories, Burlingame, CA, Cat. No. L-1260–2) or α2–6 (Sambucus nigra extract, Vector Laboratories, Burlingame, CA, Cat No. L-1300) lectin was added to a standard volume of RBCs. As the quantity of lectin increased, so too did the level of agglutination. The titer of the most dilute lectin concentration that produced a RBC “button” was recorded for each RBC sample and lectin type and used in subsequent statistical and phylogenetic analyses. Serum was heat inactivated in a water bath at 56°C for 30 minutes. Lectin dilution consisted of 50 ml phosphate buffer solution (ph 7.3) with 2.5 μl Tween 80.
2.5. Statistical analysis
Sialic acid quantity data were checked for their adherence to the assumptions of parametric statistical methods and were often found to be normal after natural log transformation (Shapiro-Wilk W test, P > 0.05); multiple linear regression (MLR) models selected by Akaike’s information criterion (AIC) (Burnham and Anderson, 2003) was used in these cases. Statistically significant parameters within the final model are reported. The dataset used for model selection work was a subset of the complete dataset because of sample to sample variation in missing age, sex, mass, or other covariates. Thus, to enhance the sample size in the examination of important specific factors on Sia quantity as determined by AIC, all available data were used for each statistical test conducted and the resultant sample size is noted. This process yielded multivariate (MLR) and univariate (ANOVA) results with samples sizes noted in the results section.
Non-parametric methods were used to examine the statistical significance of several factors on glycosidic linkage type, as linkage type did not adhere to assumptions of parametric tests. The above analyses were conducted in R (R Core Team, 2016) using the base stats package. The lm and step functions were used for the MLR and model selection work, respectively.
Phylogenetic signal was estimated for sialic acid quantity and glycosidic linkage titer data using the phylogenetic generalized least squares model (PGLS) implemented in the package caper version 0.5.2 (Orme, 2013) for the R computing environment (R Core Team, 2016). As the input for PGLS, we used previously published avian phylogenetic trees (Jetz et al., 2012). Briefly, Jetz et al. (2012) generated sets of 10,000 trees for all bird species by combining separate phylogenetic analyses of individual avian clades with published backbone phylogenies from Hackett et al. (2008) or Ericson et al. (2006) using a relaxed molecular clock approach. For PGLS, we computed a single consensus tree as an average across all 10,000 trees for each backbone avian phylogeny in which all polytomies were broken randomly.
Phylogenetic signal in PGLS is quantified through the parameter λ, which can vary between 0 and 1 (Freckleton et al., 2002; Pagel, 1997). A λ value of 0 indicates no phylogenetic signal (independent evolution) and species can be considered statistically independent, whereas a value of 1 indicates very strong phylogenetic signal conforming to a Brownian motion model of evolution (i.e., similarity among species is proportional to the time of shared evolution). Because λ can take any value between 0 and 1, caper evaluates whether the estimated maximum likelihood λ value for the data differs from the extremes using a likelihood ratio test (Freckleton et al., 2002; Pagel, 1997). In a PGLS linear model with independent explanatory variables, λ is estimated on the model’s residuals (Freckleton et al., 2002; Pagel, 1997). λ was estimated in PGLS with the following dependent variables: log10-transformed sialic acid data, log10-transformed 2:3 and 2:6 glycosydic linkage titer data, and the ratio of 2:3 to 2:6 linkage data. Since we hypothesized that body size or field metabolic rate (FMR) could influence RBC sialic acid quantity in particular, we ran PGLS with and without these as explanatory variables.
3. Results
3.1. Individual scale variation of sialic acid
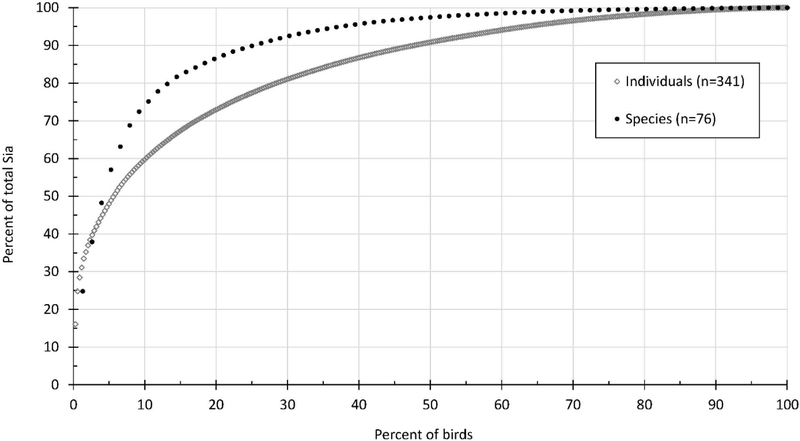
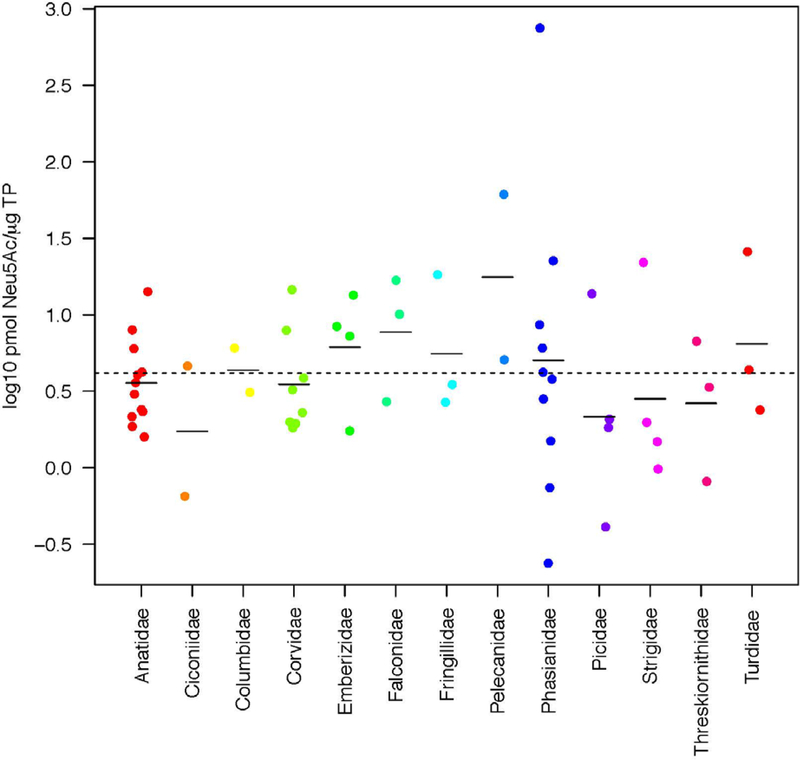
Sialic acid quantity across 76 species ranged by a factor of approximately 10,000; however, for 99% of the data, Sia ranged by a factor of approximately 1,600 after removing a very high Sia concentration from Attwater’s prairie chicken (Tympanuchus cupido) (1014.57 pmol/ug TP) (Table 1). Note sample volumes from this species were very small and so may be less reliable than samples with a higher total protein content for Neu5Ac normalization. Geometric mean and median Sia quantity was 7.08 and 6.63 pmol/ug TP, respectively, while 80% of the data was below 18.06 pmol/ug TP. The full range of the data can be found in Table 2. It was also determined that 20% of the 76 species or 340 of the individuals accounted for approximately 86% or 73%, respectively, of the total Sia measured across the entire dataset (Figure 2). The bird species with the lowest geometric mean Sia (0.24 pmol/ug TP) was Indian peafowl (Pavo cristatus) and the species with the highest (after Attwater’s prairie chicken and with more than one sample) mean Sia was Western bluebird (Sialia mexicana; 25.80 pmol/μg TP) (Table 2). Family level Sia varies somewhat uniformly around the mean family value (Figure 3).
Table 1.
Summary of species level RBC Sia quantity as geometric (Geo) mean ± geometric standard deviation (SD) pmol Neu5Ac/μg total protein (TP), with percent male and hatch year (HY) birds.
| Species | Family | Order | GeoMean ± GeoSD | N | %Male | %HY |
|---|---|---|---|---|---|---|
| Aix sponsa | Anatidae | Anseriformes | 2.15±3.78 | 2 | 0.0 | 100.0 |
| Anas acuta | Anatidae | Anseriformes | 4.22±1.87 | 6 | 50.0 | 100.0 |
| Anas americana | Anatidae | Anseriformes | 14.16±7.58 | 2 | 0.0 | 0.0 |
| Anas crecca | Anatidae | Anseriformes | 7.95±3.07 | 2 | 50.0 | 0.0 |
| Anas platyrhynchos | Anatidae | Anseriformes | 3.03±1.62 | 12 | ND | 100.0 |
| Anser anser | Anatidae | Anseriformes | 1.86±0 | 1 | 100.0 | ND |
| Asarcornis scutulata | Anatidae | Anseriformes | 2.32±0 | 1 | 0.0 | 0.0 |
| Branta sanduicpasis | Anatidae | Anseriformes | 6.01±0 | 1 | 100.0 | 0.0 |
| Cygnus bucccinator | Anatidae | Anseriformes | 1.59±0 | 1 | 0.0 | 0.0 |
| Dendrocygna viduata | Anatidae | Anseriformes | 4.04±0 | 1 | 100.0 | 0.0 |
| Mergus cucullatus | Anatidae | Anseriformes | 3.60±1.32 | 2 | 50.0 | 100.0 |
| Tadorna tadorna | Anatidae | Anseriformes | 2.40±1.72 | 4 | ND | 0.0 |
| Bubulcus ibis | Ardeidae | Ciconiiformes | 6.63±0 | 1 | 100.0 | 0.0 |
| Ephippiorhynchus senegalensis | Ciconiidae | Ciconiiformes | 0.65±0 | 1 | ND | 0.0 |
| Leptoptilos crumeniferus | Ciconiidae | Ciconiiformes | 4.62±0 | 1 | 100.0 | 0.0 |
| Columba livia | Columbidae | Columbiformes | 6.06±5.02 | 16 | ND | 33.3 |
| Zenaida macroura | Columbidae | Columbiformes | 3.12±2.17 | 6 | 80.0 | 0.0 |
| Falco peregrinus | Falconidae | Falconiformes | 2.70±0 | 1 | 100.0 | 100.0 |
| Falco sparverius | Falconidae | Falconiformes | 10.06±2.09 | 26 | 43.8 | 61.5 |
| Fulica americana | Falconidae | Falconiformes | 16.79±1.34 | 7 | ND | ND |
| Alectoris chukar | Phasianidae | Galliformes | 4.21±1.42 | 6 | 83.3 | 100.0 |
| Centrocercus urophasianus | Phasianidae | Galliformes | 22.54±2.34 | 22 | 45.5 | 72.7 |
| Chrysolophus pictus | Phasianidae | Galliformes | 0.74±1.00 | 2 | 50.0 | ND |
| Colinus virginianus | Odontophoridae | Galliformes | 16.07±1.42 | 10 | 50.0 | 0.0 |
| Gallus gallus | Phasianidae | Galliformes | 8.60±1.91 | 11 | 0.0 | 27.3 |
| Lophophorus impejanus | Phasianidae | Galliformes | 2.81±0 | 1 | 0.0 | ND |
| Meleagris gallopavo | Phasianidae | Galliformes | 1.49±2.38 | 7 | 75.0 | 0.0 |
| Numida meleagris | Numididae | Galliformes | 6.00±1.44 | 11 | 36.4 | 9.1 |
| Pavo cristatus | Phasianidae | Galliformes | 0.24±2.65 | 4 | 50.0 | 100.0 |
| Phasianus colchicus | Phasianidae | Galliformes | 6.05±1.37 | 7 | 28.6 | 100.0 |
| Tragopan temminckii | Phasianidae | Galliformes | 3.79±0 | 1 | 0.0 | ND |
| Tympanuchus cupido | Phasianidae | Galliformes | 748.50±0 | 1 | ND | ND |
| Gavia immer | Gaviidae | Gaviiformes | 6.54±1.67 | 10 | 50.0 | 0.0 |
| Cariama cristata | Cariamidae | Gruiformes | 15.30±0 | 1 | 0.0 | 0.0 |
| Grus canadensis tabida | Gruidae | Gruiformes | 17.24±1.61 | 12 | 50.0 | 0.0 |
| Agelaius phoeniceus | Icteridae | Passeriformes | 3.00±2.19 | 7 | 85.7 | 14.3 |
| Aphelocoma californica | Corvidae | Passeriformes | 3.23±0 | 1 | ND | ND |
| Aphelocoma insularis | Corvidae | Passeriformes | 2.29±1.12 | 3 | ND | 50.0 |
| Carduelis pinus | Fringillidae | Passeriformes | 3.50±0 | 1 | 100.0 | 0.0 |
| Carduelis tristis | Fringillidae | Passeriformes | 2.68±1.25 | 3 | 100.0 | 0.0 |
| Carpodacus mexicanus | Fringillidae | Passeriformes | 18.27±1.73 | 4 | 100.0 | 25.0 |
| Catharus guttatus | Turdidae | Passeriformes | 2.38±0 | 1 | ND | 0.0 |
| Corvus brachyrhynchus | Corvidae | Passeriformes | 1.82±0 | 1 | ND | 0.0 |
| Corvus corax | Corvidae | Passeriformes | 3.86±0 | 1 | ND | ND |
| Corvus cryptoleucus | Corvidae | Passeriformes | 14.55±3.07 | 31 | 50.0 | 0.0 |
| Guiraca caerulea | Cardinalidae | Passeriformes | 3.24±2.73 | 3 | 100.0 | 33.3 |
| Gymnorhinus cyanocephalus | Corvidae | Passeriformes | 1.94±1.48 | 5 | ND | 20.0 |
| Junco hyemalis | Emberizidae | Passeriformes | 1.74±1.24 | 5 | ND | 0.0 |
| Nucifraga columbiana | Corvidae | Passeriformes | 7.92±1.35 | 2 | ND | 0.0 |
| Pica hudsonia | Corvidae | Passeriformes | 1.99±2.55 | 3 | ND | 0.0 |
| Pipilo maculatus | Emberizidae | Passeriformes | 8.37±1.33 | 2 | ND | ND |
| Sialia mexicana | Turdidae | Passeriformes | 25.80±2.99 | 11 | 36.4 | 10.0 |
| Spizella passerina | Emberizidae | Passeriformes | 7.24±1.81 | 9 | 100.0 | 0.0 |
| Trichoglossus haematodus | Loriinae | Passeriformes | 21.33±1.80 | 2 | 50.0 | 0.0 |
| Turdus migratorius | Turdidae | Passeriformes | 4.36±1.35 | 5 | 0.0 | 75.0 |
| Vermivora virginiae | Parulidae | Passeriformes | 2.64±0 | 1 | ND | 100.0 |
| Zonotrichia leucophryus | Emberizidae | Passeriformes | 13.41±0 | 3 | 66.7 | 0.0 |
| Eudocimus albus | Threskiornithidae | Pelecaniformes | 0.81±1.21 | 2 | 50.0 | 100.0 |
| Pelecanus erthrorhynchos | Pelecanidae | Pelecaniformes | 5.07±1.32 | 2 | 0.0 | ND |
| Pelecanus occidentalis | Pelecanidae | Pelecaniformes | 61.15±0 | 1 | ND | 0.0 |
| Platalea ajaja | Threskiornithidae | Pelecaniformes | 6.70±0 | 1 | 100.0 | ND |
| Threskiornis aethiopicus | Threskiornithidae | Pelecaniformes | 3.36±0 | 1 | ND | 0.0 |
| Phoenicopterus ruber | Phoenicopteridae | Phoenicopteriformes | 10.95±1.36 | 3 | 0.0 | 0.0 |
| Melanerpes formicivorus | Picidae | Piciformes | 0.41±0 | 1 | 100.0 | 0.0 |
| Picoides pubescens | Picidae | Piciformes | 2.07±0 | 1 | ND | ND |
| Picoides scalaris | Picidae | Piciformes | 1.83±0 | 1 | 100.0 | 0.0 |
| Sphyrapicus thyroideus | Picidae | Piciformes | 13.70±4.41 | 2 | 0.0 | 0.0 |
| Melopsittacus undulatus | Psitticidae | Psittaciformes | 29.30±0 | 1 | 100.0 | 0.0 |
| Spheniscus humboldti | Spheniscidae | Sphenisciformes | 2.19±2.57 | 4 | 25.0 | 75.0 |
| Asio otus | Strigidae | Strigiformes | 1.48±0 | 1 | ND | 0.0 |
| Athene cunicularia | Strigidae | Strigiformes | 1.98±0 | 1 | ND | 0.0 |
| Bubo virginianus | Strigidae | Strigiformes | 22.00±1.79 | 2 | 100.0 | 0.0 |
| Megascops asio | Strigidae | Strigiformes | 0.98±0 | 1 | ND | 0.0 |
| Dromaius novaehollandiae | Casuariidae | Struthioniformes | 6.96±0 | 1 | 100.0 | 0.0 |
| Struthio camelus | Struthionidae | Struthioniformes | 12.25±0 | 1 | 100.0 | 0.0 |
| Phalacrocorax auritus | Phalacrocoracidae | Suliformes | 16.84±1.72 | 9 | ND | ND |
| All | All | All | 7.08±3.46 | 340 | 51.7 | 32.3 |
Table 2.
Sialic acid quantity and Sia-Gal glycosidic linkage descriptive statistics
| Statistic | Neu5Ac pmol/ug TP |
α2–6 titer | α2–3 titer | α2–6:α2–3 ratio |
|---|---|---|---|---|
| Minimum value | 0.10 | 2.00 | 2.00 | 0.33 |
| 10th centile value | 1.62 | 2.40 | 2.00 | 0.76 |
| 25th centile value | 3.21 | 8.00 | 2.00 | 1.00 |
| Median value | 6.63 | 24.00 | 4.00 | 1.67 |
| 75th centile | 14.64 | 93.00 | 8.00 | 3.00 |
| 90th centile value | 32.70 | 192.00 | 16.00 | 4.55 |
| Maximum value | 1014.57 | 1280.00 | 192.00 | 8.00 |
Figure 2.
Contribution of sialic acid (Neu5Ac) by each of 76 avian species or 340 individuals to the total sialic acid pool in the dataset. Each species or individual symbol represents the sialic acid condition of either one species or one individual, respectively.
Figure 3.
Sialic Acid (Neu5Ac) quantity by avian family. Dotted line represents cross-species mean log Neu5Ac concentration across families. Solid lines represent species mean log Neu5Ac concentration within each family.
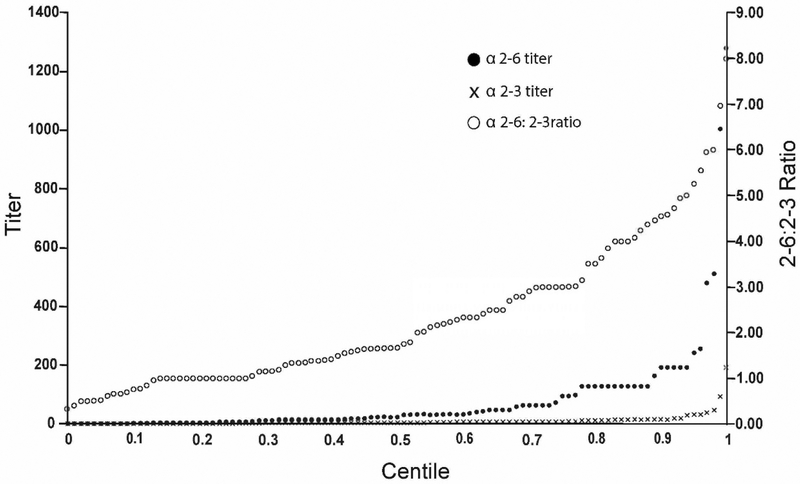
The sialic acid-galactose glycosidic linkage was measured in 24 species. Most (22 of 24) species exhibited a higher α2–6 titer than α2–3 titer (Table 3), indicating that most birds expressed a higher abundance of α2–6 than α2–3 Sia-Gal linkages on the surface of their RBCs. The 90th centile of the α2–6 titer, α2–3 titer, and α2–6:α2–3 titer ratio was 192.00, 16.00, and 4.55, respectively (Table 2 and Figure 4). The mean & median α2–6 titer, α2–3 titer, and α2–6:α2–3 titer ratio was 81.00 (SD=175.00) & 24.00, 10.00 (SD=21.00) & 4.00, and 12.58 (SD=22.00) & 1.67, respectively. The only species with a higher α2–3 than α2–6 titer was (Pica hudsonia) (mean α2–6:α2–3 ratio 0.60, SD=0.25, n=6). Wild turkey (Meleagris gallopavo) RBCs yielded a α2–6:α2–3 ratio of 1.00, but only from one sample. The species with the highest α2–6:α2–3 ratio was red-naped sapsucker (Spyrapicus nuchalis) (64.00, n=1), or with n>1, the white-breasted nuthatch (Sitta carolinensis) (mean ratio 38.59, SD=40.49, n=3). The range of glycosidic linkage titer data can be found in Table 2 and viewed as cumulative distribution functions (Figure 3).
Table 3.
Summary of species level Sia-Gal glycosidic linkage type (as hemagglutination titer).
| Species | Mean α2–3 titer |
SD α2–3 titer |
Mean α2–6 titer |
SD α2–6 titer |
Mean α2–6:α2–3 ratio |
SD α2–6:α2–3 ratio |
N |
|---|---|---|---|---|---|---|---|
| Pica hudsonia | 7.33 | 4.50 | 3.67 | 0.82 | 0.60 | 0.25 | 6 |
| Meleagris gallopavo | 4.00 | NA | 4.00 | NA | 1.00 | NA | 1 |
| Gallus gallus | 3.50 | 2.28 | 3.92 | 4.19 | 1.23 | 0.99 | 12 |
| Aphelocoma californica | 8.00 | 5.66 | 14.00 | 2.83 | 2.50 | 2.12 | 2 |
| Turdus migratorius | 20.00 | 16.97 | 24.00 | 22.63 | 2.63 | 3.36 | 2 |
| Anas platyrhynchos | 11.69 | 6.83 | 26.31 | 34.79 | 2.74 | 3.88 | 16 |
| Passerina amoena | 3.00 | NA | 12.00 | NA | 4.00 | NA | 1 |
| Carpodacus mexicanus | 10.40 | 7.27 | 36.40 | 26.32 | 5.42 | 7.36 | 5 |
| Sphyrapicus thyroideus | 58.67 | 33.31 | 277.33 | 205.73 | 5.56 | 4.54 | 3 |
| Setophaga auduboni | 16.00 | NA | 96.00 | NA | 6.00 | NA | 1 |
| Carduelis psaltria | 3.00 | 1.41 | 22.00 | 14.14 | 7.00 | 1.41 | 2 |
| Contopus sordidulus | 7.33 | 7.57 | 32.00 | 28.84 | 7.17 | 7.75 | 3 |
| Sialia mexicana | 9.63 | 6.28 | 72.75 | 75.27 | 7.77 | 5.52 | 8 |
| Catharus guttatus | 8.00 | NA | 93.00 | NA | 11.63 | NA | 1 |
| Pheucticus melanocephalus | 97.50 | 133.64 | 672.00 | 859.84 | 14.00 | 10.37 | 2 |
| Pipilo chlorurus | 2.00 | NA | 32.00 | NA | 16.00 | NA | 1 |
| Vermivora virginiae | 2.00 | 0.00 | 35.33 | 15.53 | 17.67 | 7.77 | 3 |
| Columba livia | 3.73 | 2.49 | 65.87 | 59.57 | 18.80 | 16.93 | 15 |
| Guiraca caerulea | 8.00 | 14.14 | 114.86 | 186.63 | 24.97 | 45.52 | 7 |
| Toxostoma crissale | 4.00 | NA | 128.00 | NA | 32.00 | NA | 1 |
| Pipilo maculatus | 6.20 | 3.90 | 102.40 | 94.39 | 34.19 | 53.16 | 5 |
| Carduelis pinus | 3.75 | 0.96 | 128.00 | 52.26 | 37.07 | 21.07 | 4 |
| Sitta carolinensis | 9.67 | 2.08 | 427.33 | 516.73 | 38.59 | 40.49 | 3 |
| Sphyrapicus nuchalis | 4.00 | NA | 256.00 | NA | 64.00 | NA | 1 |
Figure 4.
Cumulative distribution of RBC hemagglutination titers with lectins specific for Siaα2–3Gal or Siaα2–6Gal; the ratio of these two titers (i.e., α2–6:α2–3) is also shown. N = 24 species and 105 individuals.
The best statistical model for sialic acid quantity on RBCs included the date of the sample, the species from which it was collected, and the age of the individual. An individual’s sex and mass were not maintained in the final selected model (ΔAIC = 3.10, F(36,82) = 6.12, R2 = 0.729, P < 0.0001). Species and sample date but not age were significant effects in the final model (Table 4). Univariate statistical analysis for the effect of age and sex on Sia quantity revealed no difference by sex, but after hatch year birds ( = 15.80 pmol Neu5Ac/μg TP) exhibited a higher Sia concentration than hatch year birds ( = 12.00 pmol Neu5Ac/μg TP) (F(1,277) = 5.56, P = 0.0190) (Table 5). In contrast to Sia quantity results, no statistically significant effect of an individual scale variable (age and sex) on the type of glycosidic linkage was detected (Table 6).
Table 4.
Model selection results for the effect of the noted variables on log Sia quantity (pmol Neu5Ac/μg TP). Statistically significant (P < 0.05) variables are bolded with direction (+ or −) of effect noted in final model. The dataset for this analysis was a subset (N = 119) of the full dataset (N = 340) due to missing values within variables.
| Model | AIC | ΔAIC | DF | F | R2 | P |
|---|---|---|---|---|---|---|
| log(Neu5Ac) ~ date+mass+sex+species+age | −330.69 | 0 | 38,80 | 5.718 | 0.7309 | <0.0001 |
| log(Neu5Ac) ~ date+mass+species+age | −332.54 | 1.85 | 37,81 | 5.936 | 0.7306 | <0.0001 |
| log(Neu5Ac) ~ date(+)+species+age | −333.79 | 3.10 | 36,82 | 6.123 | 0.7289 | <0.0001 |
Table 5.
Effect of individual scale variables on Sia quantity. This analysis utilized the full dataset (N = 340), but ignored missing values as noted by the N for each subclassifier entry.
| Classifier | Subclassifier | N | Mean (pmol Neu5Ac/μg TP) |
SD | Statistic |
|---|---|---|---|---|---|
| Age | Hatch Year | 90 | 12.00 | 20.00 | F(1,277)=5.56 (P = 0.0190) |
| After Hatch Year | 189 | 15.80 | 26.08 | ||
| Sex | Female | 98 | 14.74 | 28.43 | F(1,201)=0.186 (P = 0.6671) |
| Male | 105 | 12.21 | 13.58 |
Table 6.
Effect of individual scale variables on glycosidic linkage. No statistical differences were detected between groups.
| Classifier | Subclassifier | N | Mean (SD) | ||
|---|---|---|---|---|---|
| α2–3 titer | α2–6 titer | α2–6:α2–3 Ratio | |||
| Age | Hatch Year | 30 | 7.0 (6.5) | 64.4 (100.9) | 17.4 (27.0) |
| After Hatch Year | 32 | 15.8 (34.2) | 141.7 (284.1) | 13.8 (18.8) | |
| Sex | Female | 16 | 25.1 (47.0) | 278.6 (375.7) | 27.4 (35.2) |
| Male | 11 | 5.2 (4.2) | 72 (75.9) | 17.1 (19.7) | |
3.2. Comparative phylogenetic analysis
Sialic acid quantity data did not exhibit significant phylogenetic signal (λ ≠ 0, p=1; Table 7). The cross-species mean of sialic acid quantity fell within the range of species variation within each avian family except for Pelicanidae (Figure 3). We examined the relationship between species body size and FMR on Sia quantity and found no relationship (R2 < 0.001, P > 0.05). Significant phylogenetic signal was exhibited by a2:6 titer data (λ ≠ 0 and λ ≠ 1, p<0.05), but not a2:3 titer data (λ ≠ 0, p=1) or the ratio of 2:6-to-2:3 titer data (λ ≠ 0, p=0.49). The pattern of statistical significance in phylogenetic signal for sialic acid or glycosidic linkage data did not change based on which backbone phylogenetic tree used (i.e., (Hackett et al., 2008) or (Ericson et al., 2006); Table 7).
Table 7. Phylogenetic Signal in Avian Sialic Acid Quantity and Glycosidic Linkage Data.
Phylogenetic signal (λ) estimates were generated using phylogenetic consensus trees from Jetz et al. (2012) based on avian backbone phylogenies from Hackett et al. (Hackett et al., 2008) and Ericson et al. (Ericson et al., 2006).
| Data Type | Species (N) | Phylogenetic Signal (λ) | ||
|---|---|---|---|---|
| Hackett et al. (2008) | Ericson et al. (2006) | |||
| Sia quantity (pmol Neu5Ac/μg TP) | 76 | 0i | 0i | |
| Sia glycosidic linkage | log α2–3 | 24 | 0i | 0i |
| log α2–6 | 0.724i,ii | 0.700i,ii | ||
| α2–3:α2–6 ratio | 0.242i | 0.240i | ||
, Lambda significantly different from 1 (P < 0.05)
, Lambda significantly different from 0 (P < 0.05)
4. Discussion
Sialic acid is involved with numerous processes including immunity, pathogen recognition, and oxidative stress (Varki, 2007). Sia provides an important receptor site for influenza viruses (Suzuki et al., 2000), malaria parasites, Mycoplasma gallisepticum (Glasgow and Hill, 1980) and many other agents (see review by Lehmann et al. (2006)). Although there is a large literature reporting on the quantity, structure, and linkage of Sia to underlying cell-surface glycans, much of it has been focused on mammals and on understanding pathogen interactions in a small number of species known to be important for a particular disease, as above. Consequently, a large knowledge gap exists with respect to baseline variation of Sia in the Class Aves. For example, it is not known whether phylogenetic position is associated with one or more aspects of Sia structure. Although the RBC is not directly involved with disease processes caused by most of the above agents, the RBC can be obtained non-lethally and thus provides a sampling matrix to enable large comparative studies. Our study provides findings on this matter in that we report on the quantity of Sia across 76 avian species and the linkage of Sia to the underlying glycan across 24 avian species in a non-lethally obtained matrix, the RBC. We found that avian RBC Sia varied along two scales: individual and phylogenetic. RBC Sia quantity was higher in after hatch year birds than hatch year birds. Additionally, approximately 20% of the birds or species expressed approximately 80% of the Sia expressed in the group, respectively. And, most avian species expressed more α2–6 than α2–3 linkages on RBCs. While Sia quantity did not, Siaα2–6Gal glycosidic linkage did exhibit phylogenetic signal. This means that closely related avian species exhibited more similar Siaα2–6Gal glycosidic linkage titer values as compare to more distantly related species.
4.1. Individual scale variation—Sia quantity
We used HPLC-MS/MS in conjunction with an isotope spiking protocol to quantify RBC Sia as pmol of 5-N-acetylneuraminic acid per μg protein of RBC membranes. Isotope dilution liquid chromatography-tandem mass spectrometry is a well-known technique that has been used to quantify a variety of analytes in various matrices, and is considered a “gold standard” technique by both the National Institute for Standards and Technology (e.g., Bunk and Lowenthal (2012)) as well as the Center for Disease Control (e.g., Pantazides et al. (2015)) to certify analyte concentrations. Much of the mammalian RBC Sia data (Jancik and Schauer, 1974; Kumar and Rizvi, 2013; Mazzanti et al., 1997; Mehdi et al., 2012) have been generated using chromogenic procedures involving periodate with thiobarbituric acid (Aminoff, 1961; Jancik and Schauer, 1974; Warren, 1959), or resorcinol based reactions (Jourdian et al., 1971; Spyridaki and Siskos, 1996; Svennerholm, 1957) to quantify free, bound, or total Sia as 5-N-acetylneuraminic acid. The focused measurement of Neu5Ac is justified because although there are some 50 known forms of Sia (Angata and Varki, 2002), it is the primary Sia on vertebrate cell membranes (Angata and Varki, 2002; Schauer, 2000). Although we quantified Sia differently from others (i.e., as pmol Neu5Ac/μg protein vs. μg Neu5Ac/mg protein), our data are qualitatively analogous to these works and so we can draw at least qualitative comparisons on observed Sia expression patterns, such as changes in Sia abundance with the age of the organism.
Age is known to influence sialic acid expression in mammalian RBCs (Kumar and Rizvi, 2013; Mazzanti et al., 1997; Mehdi et al., 2012), and our data suggest age may be a factor in bird RBCs as well, but in an opposite manner to mammals. However, age was only a statistically significant (P < 0.05) factor on Sia quantity when analyzed on a univariate basis across all individuals. In mammals, RBC Sia quantity has been shown to decline with age whereas our avian data show the opposite trend in that it was higher in older after hatch year than in younger (<1-year-old) hatch year birds. The discrepancy between RBC Sia quantity trends in the avian data versus available mammalian data may be a result of factors associated with differences in the assays used to quantify Sia as described above, the breadth and depth of the datasets, or may reflect real taxonomic (Class) differences between patterns in RBC Sia quantity.
Our dataset consisted of 76 avian species with only two age categories (i.e., ‘hatch year’ and ‘after hatch year’) while Kumar and Rizvi (2013) present rat data across six age categories (1 to 24 months) and Mehdi et al. (2012) present data in a wide age range (21–90 years) of adult humans. If, as our data suggest, after hatch year (older) birds do exhibit higher Sia quantity than hatch year (younger) birds, it may be that age-related patterns in RBC Sia quantity differ between birds and mammals; the reasons for which are not yet clear, suggesting more specific studies assessing age versus Sia quantity are warranted in birds and mammals. If like in rats and humans, oxidative stress increases as birds age, then it is not clear why RBC Sia would be higher in older birds. Contrary to what is suggested in mammals in which RBC bound Sia is rapidly degraded in older animals due to a higher abundance of free radicals (Mehdi et al., 2012), it may be hypothesized that elevated Sia quantity in birds is a compensatory response to higher levels of oxidative stress in older birds. Or, it may also be that RBC-bound Sia provides a different function in birds than in mammals. To this point, we examined the relationship between a species FMR and its RBC Sia quantity and found no relationship. That is, our data did not support the hypothesis that a higher metabolic rate (vis-à-vis higher oxygen consumption) and thus a higher free radical production rate would result in a higher RBC Sia concentration in order to mitigate oxidative stress on these oxygen-carrying cells. As avian RBCs are much shorter-lived than mammalian RBCs (~25% as long), perhaps they are not buffered by the antioxidative functions of Sia as in mammals; there are known differences in antioxidative actions between birds versus mammals (El-Mekawi et al., 1993). Our data do not provide the final analysis on this issue because although we found higher Sia in after hatch year birds across all species, age was not a significant effect in the final multilinear regression model. Given this and a lack of association between FMR and Sia quantity, RBC Sia may primarily serve other functions including pathogen binding (Baseman et al., 1982; Klotz et al., 1992; Useh et al., 2006), provide cellular negative electrostatic charge (Eylar et al., 1962) to improve RBC mobility, and a signal for RBC removal for older RBCs with reduced Sia levels (Durocher et al., 1975).. Clearly, many questions regarding RBC Sia quantity in birds remain unaddressed.
4.2. Individual scale variation—Sia-Gal Linkage
As the use of the HA assay for our research question is novel, there are no published titer data against which we can compare our results, so we discuss patterns of linkage type on a qualitative basis. Although the HA assay has not previously been used to assess Sia-Gal linkage patterns across taxa, the procedure is well-suited for the assessment of relative lectin-Sia binding across samples. We used the same lectin reagents that are used for Sia receptor research in lectin histochemistry assays (Cohen et al., 2012). That is, SNA binds Sia bound to Gal via α2–6 linkage while MAL II binds to α2–3 linkages (Cohen et al., 2012). In the HA assay, the capacity for an RBC sample to bind a specific lectin is indirectly measured by the inverse of the lowest lectin dilution that results in visible RBC agglutination (i.e., the titer). Agglutination is a result of multiple RBCs linking together through RBC Sia-Lectin-RBC Sia chains that form a visible precipitation at the base of a cell culture well. An analogous version of the HA assay is used to determine the Sia-receptor binding affinity of an influenza virus by using RBCs with known linkage type such as sheep or pig (Ito et al., 1997). In Ito et al. (1997), the question was the Sia binding affinity of the lectin (the virus) rather than the Sia receptor on the RBC, as in our study. That is, hemagglutination to examine Sia binding characteristics is an often-used and relied-upon approach.
The majority of avian species (22 of 24) tested exhibited a higher α2–6 titer than α2–3 titer. Although the RBC is not a recognized target cell for avian influenza infection, one might predict that birds express a higher α2–3 than α2–6 titer, as most avian influenza viruses bind to α2–3 receptors (Suzuki et al., 2000). Perhaps a higher abundance of α2–6 receptors than α2–3 receptors serves to mitigate RBC infection with the most common IAVs confronting birds. However, because IAVs do not typically circulate in the bloodstream, we do not suspect this function to be in operation. Keeping in mind the purported functions of Sia on RBCs as mentioned above, it is not immediately clear why birds would express a higher α2–6 than α2–3 titer. An assessment of titer patterns across species has the potential to provide some insight, however.
4.3. Species scale variation—Sia Quantity
The lack of phylogenetic signal in sialic acid quantity data is not surprising given the large variation in values within each family and the similarity in mean values within families and across the entire avian dataset. Such variation may be because a cell’s quantity of Sia is a labile property that reflects a cell’s condition (e.g., its age, redox status) the organism’s physiologic condition (e.g., its age or health status), and environmental factors (e.g., bacterial infection) (Schauer, 2000; Varki, 2007). Although individuals within a species tend to exist in similar habitats and environmental envelope, suggesting species-related patterns in Sia quantity, our data do not support such a process. Other studies in four common peridomestic avian species would also lend to such a conclusion (Useh et al., 2006). Therefore, discussing Sia quantity on the species scale is not currently a fruitful endeavor. In contrast, our Sia-Gal linkage data did exhibit some species level variation.
4.4. Species scale variation—Sia-Gal Linkage
The finding of phylogenetic signal in α2–6 glycosidic linkage, but not in α2–3 linkage or α2–3:α2–6 linkage ratio, may indicate that the latter two metrics are driven more by species-specific environmental and/or evolutionary factors. However, the phylogenetic signal may be driven in part by the high similarity in α2–6 titer values between the two phasianid and two picid species. Further investigation with wider sampling within each avian family or order is needed to make definitive conclusions about the ecological and evolutionary drivers of glycosidic linkage variation. It is possible that the particular selection of species affected our results, but that concern is not unique to the current study and we have no information to contradict our results. A λ of 0 (P > 0.05 for statistical difference from λ = 1, but not from λ = 0) for Siaα2–3Gal and 0.724 (P < 0.05 for statistical difference from λ = 0 or 1) for Siaα2–6Gal indicates a relatively large contrast in our results and a low chance for type I error (α, false positive) in the 2–6 linkage data and type II error (β, false negative) in the 2–3 linkage data. These findings were achieved despite the small samples size (N=24). Moreover, the wide phylogenetic spread of species in the Siaα2–3Gal and Siaα2–6Gal data including passeriformes, piciformes, and galliformes suggests that it is unlikely that adding or changing species would change the overall outcome of the analysis, although it could change the Siaα2–6Gal lambda value or level of significance.
A range of biological explanations for phylogenetic signal in α2–6 data are plausible. IAVs infect many species of birds (Dusek et al., 2009; Fuller et al., 2010; Munster et al., 2007; Olsen et al., 2006) and avian influenza viruses are thought to primarily bind to α2–3 receptors (Connor et al., 1994). As it is thought that birds typically interact with α2–3 binding influenza viruses more than α2–6 binding viruses (Suzuki et al., 2000), it may be conjectured that avian species have been exposed to a large range of α2–3 binding viruses leading to a lack of phylogenetic signal in this linkage type. Alpha2–6 linkages on the other hand may exhibit phylogenetic signal as a result of less environmental pressure on linkage expression because α2–6 binding viruses generally affect humans or peridomestic species rather than wild birds (Connor et al., 1994; Suzuki et al., 2000). As a consequence of this hypothesized limited environmental selective pressure, the distribution of α2–6 receptors across avian taxa may have developed through a Brownian motion evolutionary pattern resulting in the presence of phylogenetic signal in our dataset (Garland et al., 2005). Birds with the highest α2–6 titer (and high linkage ratios) were both Passerine species, whereas birds with lower α2–6 titers (and low linkage ratios) included the domestic species mallard duck and chicken. Perhaps species that interact with human societies more than other species express a lower RBC α2–6 titer as a result of a possible higher overall exposure to α2–6 binding viruses than more wild species. A lower titer may reduce virus-RBC binding and hemagglutination in such species. This idea is highly speculative and further research is warranted.
4.5. Minimal N-glycolylneuraminic acid on avian RBC membranes
As a result of the nature of our HPLC-MS/MS based assay for Sia quantity, we were able to investigate the presence of a form of Sia (N-glycolylneuraminic acid (Neu5Gc)) that is considered to be mostly absent in avian taxa (Schauer et al., 2009). In assessing mass spectra for each sample, we discovered a detectable but limited abundance of Neu5Gc across all species tested. Total ion counts were greater than background for some samples, but signal to noise ratios suggested minimal quantity. Any detected Neu5Gc may have been a result of the consumption of food items that contained Neu5Gc (Tangvoranuntakul et al., 2003). It is possible that birds with detected Neu5Gc express the gene for endogenous production of Neu5Gc, CMP-Neu5Ac hydroxylase (CMAH), but we have no way of knowing this from our data and would expect a higher quantity if Neu5Gc was endogenously produced and expressed on RBC membranes. Therefore, our data support the current thinking (Schauer et al., 2009) that birds do not endogenously produce Neu5Gc.
4.6. Future Directions
Although Sia is not the sole cellular receptor for IAVs (reviewed in Ge and Wang (2011)), a significant application of continued similar work may be with influenza surveillance programs. If the RBC Siaα2–6:α2–3Gal linkage ratio reliably corresponds to the linkage ratio in key intestinal or respiratory tissues important for avian influenza infection (Franҫa and Brown, 2014), this work may help to increase our understanding influenza infection and transmission dynamics. That is, if intestinal or respiratory Siaα2–6:α2–3Gal linkage ratio also exhibited phylogenetic signal, a phylogenetic guide map could be built and assist in constructing zoonotic disease surveillance programs targeted at species most likely to carry a virus of concern. Although evolutionary distance from duck to human may be too far for a direct jump by an avian influenza virus, some avian species, such as chickens that share virus-receptor binding characteristics with ducks and humans (Kim et al., 2005; Kuchipudi et al., 2009), may act as a transmission “bridge” between these species given their role in agriculture. Other species that may also share receptor binding characteristics include the quail and pheasants (Campitelli et al., 2004; Gambaryan et al., 2005; Neumann and Kawaoka, 2006) which can be both wild or domestic, suggesting they may also serve as a bridge between avian and human influenza transmission. A phylogenetic guide map for Sia-Gal linkage type in birds may enhance our capacity to make similar predictions a priori, given any influenza virus of concern, provided the virus’ Sia receptor affinity is known. Although capacity to be infected with a given virus is somewhat dependent on Sia type, tissue tropism and Sia glycosidic linkage did not appear to be directly correlated in a study involving 37 species and 11 bird orders (Franca et al., 2013).
Our findings are potentially informative for avian malaria as well. While it has been shown that complement receptor 1 (CR1) is a sialic acid-independent receptor for the invasion of erythrocytes by P. falciparum (Spadafora et al. 2010), it was shown earlier that P. falciparum depends heavily on sialic acid present on glycophorins to invade erythrocytes (Miller et al 1977). Mycoplasmosis in house finches and poultry, influenza viruses, and Plasmodium parasites both cause disease in birds that have led to largescale mortality events (Chen et al., 2005) and in the case of avian malaria, local extinction events (Warner 1968, van Riper 1986). Each of these pathogens displays inter- and intraspecific differences in host susceptibility, suggesting the importance of investigating sources for this variation. Sialic acid abundance and the way in which Sia is chemically bound to cell surface glycans is associated with host susceptibility for influenza (Suzuki et al., 2000) and perhaps many other agents (Lehmann et al., 2006). Uncovering broad patterns of Sia expression stimulates additional hypothesis driven studies as well providing insights useful for the development of disease monitoring and mitigation strategies.
Our observation that approximately 20% of the individuals or species contributed to approximately 80% of the total RBC sialic acid of the group may stimulate future investigations to understand which individuals or species may be more predisposed to infection with certain disease agents. For example, those with a higher quantity of sialic acid on respiratory or intestinal cell surfaces may be more likely to become infected with influenza. If that were the case, perhaps that would help to explain why 20% of the birds shed approximately 80% of the influenza virus as reported in Jankowski et al. (2013). The variability of Sia quantity between individuals within species is somewhat low, with mean Sia quantity > the standard deviation of Sia quantity for most species (41 of 46 species with N > 1) despite a mean N of just 4.5 individuals per species. Therefore, it does appear that there were a subset of individuals and species that contributed more to overall total group Sia quantity than others. Understanding the inequality in the expression of a physiological trait such as Sia quantity is certainly worth further investigation.
There are approximately 10,000 extant avian species, so our research provides an initial step towards characterizing avian RBC Sia more broadly. Larger, phylogenetically guided surveys of avian RBC Sia, coupled with examinations of its functions and relationships to Sia in other tissues and across age classes will help to place our results in a more nuanced context. Our study is the first of its kind to provide RBC sialic acid (Neu5Ac) quantity and Sia-Gal linkage type across a high number of avian taxa. Our primary findings were the identification of phylogenetic signal in α2–6 linkage, an effect of age on Sia quantity that may be different from the pattern in mammals, and no detection of Neu5Gc in avian RBC membranes. These data may stimulate future studies to explore the range of functions provided by RBC-bound Sia and decipher the environmental and evolutionary drivers most responsible for the observed variation.
Acknowledgements
We are grateful to the many organizations and individuals that offered avian blood samples for this project. In particular, we would like to thank Dr. Mike Murray and the Monterey Bay Aquarium, the Aquarium of the Pacific, Chicago Zoological Society, St. Louis Zoo, and the Wildlife Conservation Society. We also thank C. Hathcock, David Keller, Kassidy Burnett, Sherri Sherwood, and Rhonda Robinson for assistance in the field.
Funding Source
This work was completed under the Laboratory Directed Research and Development program at Los Alamos National Laboratory (LANL) operated by Los Alamos National Security for the U.S. Department of Energy contract no. W-7405-ENG-36. Current operation of LANL is by Triad National Security, LLC under Contract No. 89233218CNA000001 with the U.S. Department of Energy.
Footnotes
Declarations of interest: none
References
- Allevi P, Femia EA, Costa ML, Cazzola R, Anastasia M, 2008. Quantification of N-acetyl- and N-glycolylneuraminic acids by a stable isotope dilution assay using high-performance liquid chromatography-tandem mass spectrometry. Journal of Chromatography A 1212, 98–105. [DOI] [PubMed] [Google Scholar]
- Aminoff D, 1961. Methods for the quantitative estimation of N-acetylneuraminic acid and their application to hydrolysates of sialomucoids. Biochemical Journal 81, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Varki A, 2002. Chemical diversity in the sialic acids and related α-keto acids: an evolutionary perspective. Chemical reviews 102, 439–470. [DOI] [PubMed] [Google Scholar]
- Baseman J, Banai M, Kahane I, 1982. Sialic acid residues mediate Mycoplasma pneumoniae attachment to human and sheep erythrocytes. Infection and immunity 38, 389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg SP, Garland T Jr, Ives AR, 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. [DOI] [PubMed] [Google Scholar]
- Bradford MM, 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brock PM, Murdock CC, Martin LB, 2014. The history of ecoimmunology and its integration with disease ecology. The Society for Integrative and Comparative Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter DB, Cain DJ, Martin CA, Xie L, Luoma SN, Garland T, 2008. Aquatic insect ecophysiological traits reveal phylogenetically based differences in dissolved cadmium susceptibility. Proceedings of the National Academy of Sciences 105, 8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunk DM, Lowenthal MS, 2012. Isotope dilution liquid chromatography-tandem mass spectrometry for quantitative amino acid analysis, Amino Acid Analysis. Springer, 29–38. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR, 2003. Model selection and multimodel inference: a practical information-theoretic approach. Springer Science & Business Media. [Google Scholar]
- Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, 2004. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology 323, 24–36. [DOI] [PubMed] [Google Scholar]
- Chen H, Smith G, Zhang S, Qin K, Wang J, Li K, Webster R, Peiris J, Guan Y, 2005. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436, 191. [DOI] [PubMed] [Google Scholar]
- Cohen M, Varki NM, Jankowski MD, Gagneux P, 2012. Using unfixed, frozen tissues to study natural mucin distribution. Journal of visualized experiments: JoVE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC, 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205, 17–23. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Maier AG, Triglia T, Cowman AF, 2003. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and-independent pathways. Proceedings of the National Academy of Sciences 100, 4796–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher JR, Payne RC, Conrad ME, 1975. Role of sialic acid in erythrocyte survival. Blood 45, 11–20. [PubMed] [Google Scholar]
- Dusek RJ, Bortner JB, DeLiberto TJ, Hoskins J, Franson JC, Bales BD, Yparraguirre D, Swafford SR, Ip HS, 2009. Surveillance for high pathogenicity avian influenza virus in wild birds in the Pacific Flyway of the United States, 2006–2007. Avian Diseases 53, 222–230. [DOI] [PubMed] [Google Scholar]
- El-Mekawi S, Yagil R, Meyerstein N, 1993. Effect of oxidative stress on avian erythrocytes. Journal of basic and clinical physiology and pharmacology 4, 199–212. [DOI] [PubMed] [Google Scholar]
- Ericson PGP, Zuccon D, Ohlson JI, Johansson US, Alvarenga H, Prum RO, 2006. Higher-level phylogeny and morphological evolution of tyrant flycatchers, cotingas, manakins, and their allies (Aves: Tyrannida). Molecular Phylogenetics and Evolution 40, 471–483. [DOI] [PubMed] [Google Scholar]
- Eylar EH, Madoff MA, Brody OV, Oncley JL, 1962. The contribution of sialic acid to the surface charge of the erythrocyte. Journal of Biological Chemistry 237, 1992–2000. [PubMed] [Google Scholar]
- Fair JM, Paul E, Jones J, Eds., 2010. Guidelines to the use of wild birds in research. Ornithological Council, Washington, D.C. [Google Scholar]
- Fassbinder-Orth CA, Wilcoxen TE, Tran T, Boughton RK, Fair JM, Hofmeister EK, Grindstaff JL, Owen JC, 2016. Immunoglobulin detection in wild birds: effectiveness of three secondary anti-avian IgY antibodies in direct ELISAs in 41 avian species. Methods in ecology and evolution 7, 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca M, Stallknecht D, Howerth E, 2013. Expression and distribution of sialic acid influenza virus receptors in wild birds. Avian pathology 42, 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franҫa MS, Brown JD, 2014. Influenza pathobiology and pathogenesis in avian species, Influenza Pathogenesis and Control-Volume I. Springer, 221–242. [DOI] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M, 2002. Phylogenetic analysis and comparative data: a test and review of evidence. The American Naturalist 160, 712–726. [DOI] [PubMed] [Google Scholar]
- Fuller TL, Saatchi SS, Curd EE, Toffelmier E, Thomassen HA, Buermann W, DeSante DF, Nott MP, Saracco JF, Ralph C, 2010. Mapping the risk of avian influenza in wild birds in the US. BMC infectious diseases 10, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan A, Yamnikova S, Lvov D, Tuzikov A, Chinarev A, Pazynina G, Webster R, Matrosovich M, Bovin N, 2005. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology 334, 276–283. [DOI] [PubMed] [Google Scholar]
- Garland T, Bennett AF, Rezende EL, 2005. Phylogenetic approaches in comparative physiology. Journal of experimental Biology 208, 3015–3035. [DOI] [PubMed] [Google Scholar]
- Georgatos SD, Blobel G, 1987. Two distinct attachment sites for vimentin along the plasma membrane and the nuclear envelope in avian erythrocytes: a basis for a vectorial assembly of intermediate filaments. The Journal of Cell Biology 105, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow LR, Hill RL, 1980. Interaction of Mycoplasma gallisepticum with sialyl glycoproteins. Infection and immunity 30, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han K-L, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T, 2008. A Phylogenomic Study of Birds Reveals Their Evolutionary History. Science 320, 1763–1768. [DOI] [PubMed] [Google Scholar]
- Hylton A, Chiari Y, Capellini I, Barron MG, Glaberman S, 2018. Mixed phylogenetic signal in fish toxicity data across chemical classes. Ecological Applications 28, 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y, 1997. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology 227, 493–499. [DOI] [PubMed] [Google Scholar]
- Jancik J, Schauer R, 1974. Sialic acid—a determinant of the life-time of rabbit erythrocytes. Hoppe-Seyleŕ s Zeitschrift für physiologische Chemie 355, 395–400. [DOI] [PubMed] [Google Scholar]
- Jankowski MD, Williams CJ, Fair JM, Owen JC, 2013. Birds Shed RNA-Viruses According to the Pareto Principle. PloS one 8, e72611–e72611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO, 2012. The global diversity of birds in space and time. Nature 491, 444–448. [DOI] [PubMed] [Google Scholar]
- Jourdian GW, Dean L, Roseman S, 1971. The sialic acids XI. A periodate-resorcinol method for the quantitative estimation of free sialic acids and their glycosides. Journal of Biological Chemistry 246, 430–435. [PubMed] [Google Scholar]
- Kim JA, Ryu SY, Seo SH, 2005. Cells in the respiratory and intestinal tracts of chickens have different proportions of both human and avian influenza virus receptors. The Journal of Microbiology 43, 366–369. [PubMed] [Google Scholar]
- Klotz FW, Orlandi PA, Reuter G, Cohen SJ, Haynes JD, Schauer R, Howard RJ, Palese P, Miller LH, 1992. Binding of Plasmodium falciparum 175-kilodalton erythrocyte binding antigen and invasion of murine erythrocytes requires N-acetylneuraminic acid but not its O-acetylated form. Molecular and biochemical parasitology 51, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchipudi SV, Nelli R, White GA, Bain M, Chang KC, Dunham S, 2009. Differences in influenza virus receptors in chickens and ducks: implications for interspecies transmission. Journal of molecular and genetic medicine: an international journal of biomedical research 3, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Rizvi S, 2013. Erythrocyte membrane bound and plasma sialic acid during aging. Biologia 68, 762–765. [Google Scholar]
- LaLone CA, Villeneuve DL, Lyons D, Helgen HW, Robinson SL, Swintek JA, Saari TW, Ankley GT, 2016. Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS): A web-based tool for addressing the challenges of cross-species extrapolation of chemical toxicity. Toxicological sciences : an official journal of the Society of Toxicology. [DOI] [PubMed] [Google Scholar]
- Lehmann F, Tiralongo E, Tiralongo J, 2006. Sialic acid-specific lectins: occurrence, specificity and function. Cellular and Molecular Life Sciences CMLS 63, 1331–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpede BM, Lin DH, Tolia NH, 2013. Molecular basis for sialic acid-dependent receptor recognition by Plasmodium falciparum erythrocyte binding antigen 140/BAEBL. Journal of Biological Chemistry, jbc. M113. 450643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A, 2005. Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proceedings of the National Academy of Sciences 102, 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzanti L, Rabini RA, Salvolini E, Tesei M, Martarelli D, Venerando B, Curatola G, 1997. Sialic acid, diabetes, and aging: a study on the erythrocyte membrane. Metabolism-Clinical and Experimental 46, 59–61. [DOI] [PubMed] [Google Scholar]
- Mehdi MM, Singh P, Rizvi SI, 2012. Erythrocyte sialic acid content during aging in humans: correlation with markers of oxidative stress. Disease markers 32, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, Osterhaus ADME, Fouchier RAM, 2007. Spatial, Temporal, and Species Variation in Prevalence of Influenza A Viruses in Wild Migratory Birds. PLoS Pathog 3, e61–e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Kawaoka Y, 2006. Host range restriction and pathogenicity in the context of influenza pandemic. Emerging infectious diseases 12, 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM, 2006. Global Patterns of Influenza A Virus in Wild Birds. Science 312, 384–388. [DOI] [PubMed] [Google Scholar]
- Orlandi PA, Klotz FW, Haynes JD, 1992. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac (alpha 2–3) Gal-sequences of glycophorin A. The Journal of cell biology 116, 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D, 2013. The caper package: comparative analysis of phylogenetics and evolution in R.
- Pagel M, 1997. Inferring evolutionary processes from phylogenies. Zoologica Scripta 26, 331–348. [Google Scholar]
- Pantazides BG, Crow BS, Garton JW, Quiñones-González JA, Blake TA, Thomas JD, Johnson RC, 2015. Simplified method for quantifying sulfur mustard adducts to blood proteins by ultrahigh pressure liquid chromatography–isotope dilution tandem mass spectrometry. Chemical research in toxicology 28, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar SD, Parkhi SS, Koratkar SS, Mishra AC, 2012. Receptor specificity and erythrocyte binding preferences of avian influenza viruses isolated from India. Virology journal 9, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JC, 2014. Hemagglutination-inhibition assay for influenza virus subtype identification and the detection and quantitation of serum antibodies to influenza virus, Animal Influenza Virus. Springer, 11–25. [DOI] [PubMed] [Google Scholar]
- Perkins LEL, Swayne DE, 2003. Comparative susceptibility of selected avian and mammalian species to a Hong Kong–origin H5N1 high-pathogenicity avian influenza virus. Avian diseases 47, 956–967. [DOI] [PubMed] [Google Scholar]
- R_Core_Team, 2016. R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Schauer R, 2000. Achievements and challenges of sialic acid research. Glycoconjugate journal 17, 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R, Srinivasan GV, Coddeville B, Zanetta J-P, Guérardel Y, 2009. Low incidence of N-glycolylneuraminic acid in birds and reptiles and its absence in the platypus. Carbohydrate research 344, 1494–1500. [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Chao H, Xiao B, 2001. Determination of sialic acids by liquid chromatography-mass spectrometry. Journal of Chromatography A 913, 365–370. [DOI] [PubMed] [Google Scholar]
- Spiro RG, 1960. Studies on Fetuin, a Glycoprotein of Fetal Serum. Journal of Biological Chemistry 235, 2860–2869. [PubMed] [Google Scholar]
- Spyridaki M-HE, Siskos PA, 1996. An improved spectrophotometric method for the determination of free, bound and total N-acetylneuraminic acid in biological fluids. Analytica chimica acta 327, 277–285. [Google Scholar]
- Suzuki Y, Ito T, Suzuki T, Holland RE, Chambers TM, Kiso M, Ishida H, Kawaoka Y, 2000. Sialic acid species as a determinant of the host range of influenza A viruses. Journal of virology 74, 11825–11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerholm L, 1957. Quantitive estimation of sialic acids: II. A colorimetric resorcinol-hydrochloric acid method. Biochimica et biophysica acta 24, 604–611. [DOI] [PubMed] [Google Scholar]
- Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, Muchmore E, 2003. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U. S. A 100, 12045–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Useh N, Omeiza G, Nok A, Esievo K, 2006. Comparative studies on erythrocyte sialic acid levels in apparently healthy indigenous Nigerian poultry species. Cell Biochemistry and Function: Cellular biochemistry and its modulation by active agents or disease 24, 143–146. [DOI] [PubMed] [Google Scholar]
- van der Ham M, Prinsen BH, Huijmans JG, Abeling NG, Dorland B, Berger R, de Koning TJ, 2007. Quantification of free and total sialic acid excretion by LC–MS/MS. Journal of Chromatography B 848, 251–257. [DOI] [PubMed] [Google Scholar]
- Varki A, 1992. Diversity in the sialic acids. Glycobiology 2, 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, 2006. Nothing in glycobiology makes sense, except in the light of evolution. Cell 126, 841–845. [DOI] [PubMed] [Google Scholar]
- Varki A, 2007. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 446, 1023. [DOI] [PubMed] [Google Scholar]
- Varki A, Diaz S, 1984. The release and purification of sialic acids from glycoconjugates: Methods to minimize the loss and migration of O-acetyl groups. Analytical biochemistry 137, 236–247. [DOI] [PubMed] [Google Scholar]
- Warren L, 1959. The thiobarbituric acid assay of sialic acids. J biol chem 234, 1971–1975. [PubMed] [Google Scholar]
- Wegmann TG, Smithies O, 1966. A simple hemagglutination system requiring small amounts of red cells and antibodies. Transfusion 6, 67–73. [Google Scholar]
- Weis W, Brown J, Cusack S, Paulson J, Skehel J, Wiley D, 1988. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333, 426. [DOI] [PubMed] [Google Scholar]
- Wingfield JC, O’REILLY KM, Astheimer LB, 1995. Modulation of the adrenocortical responses to acute stress in arctic birds: a possible ecological basis. American Zoologist 35, 285–294. [Google Scholar]