Summary
It is of great significance to understand CO2 fixation in the oceans. Using single cell Raman spectra (SCRS) as biochemical profiles, Raman activated cell ejection (RACE) was able to link phenotypes and genotypes of cells. Here, we show that mini‐metagenomic sequences from RACE can be used as a reference to reconstruct nearly complete genomes of key functional bacteria by binning shotgun metagenomic sequencing data. By applying this approach to 13C bicarbonate spiked seawater from euphotic zone of the Yellow Sea of China, the dominant bacteria Synechococcus spp. and Pelagibacter spp. were revealed and both of them contain carotenoid and were able to incorporate 13C into the cells at the same time. Genetic analysis of the reconstructed genomes suggests that both Synechococcus spp. and Pelagibacter spp. contained all genes necessary for carotenoid synthesis, light energy harvesting and CO2 fixation. Interestingly, the reconstructed genome indicates that Pelagibacter spp. harbored intact sets of genes for β‐carotene (precursor of retional), proteorhodopsin synthesis and anaplerotic CO2 fixation. This novel approach shines light on the role of marine ‘microbial dark matter’ in global carbon cycling, by linking yet‐to‐be‐cultured Synechococcus spp. and Pelagibacter spp. to carbon fixation and flow activities in situ.
Introduction
Microorganisms in the oceans are responsible for approximately half of the carbon dioxide fixation on Earth (Sabine et al., 2004), yet we know little about their ecological and functional roles in situ (Amann et al., 1995; Whitman et al., 1998; Dumont and Murrell, 2005). It is hypothesized that significant populations of uncultivated microorganisms play a pivotal role in carbon fixation in oceans. As it is a challenge to establish a direct link between the phenotype (functions) and the genotype (genomics) of cells within a complex microbial community, the actual identity and activity of these contributors in the oceans remain elusive (Weller et al., 1992; Zehr et al., 2008; Tripp et al., 2010).
This hurdle is mainly caused by the dominant and not‐yet‐cultured marine microbes (Venter et al., 2004). More importantly, studies based on pure cultures are insufficient to reveal bacterial ecological function in their natural and biological context (Huang et al., 2009). As a result, many ecologically important but uncultured microbes were first discovered by advanced molecular tools, in the absence of direct validation of in situ functional activities. For example, the introduction of metagenomic approaches has enabled discovery of SAR11 bacteria (Pelagibacterales) as one of the most abundant bacteria in global oceans in 1990 (Giovannoni et al., 1990; Giovannoni, 2017), although it remained uncultured until 2002 (Rappe et al., 2002). In recent years, metagenomic analyses, which circumvent cultivation procedures and directly sequence DNA extracted from the environment (Handelsman, 2004), have already revealed massive amounts of diversity and novel genotypes from microbial ecosystems (Nayfach and Pollard, 2016). However, linking such genotypes (e.g., species identity and genes) to the phenotypes (e.g., biochemical profile and functions) in situ remains difficult. The ability to distinguish or validate ecological functions at the single cells is usually completely lost during the metagenomic sequencing process where all the cells in microbial community were extracted for DNA as a mixture, regardless of their specific functions (Huang et al., 2015; Nayfach and Pollard, 2016).
Single‐cell Raman spectra (SCRS) reflect phenotypic and intrinsic biochemical fingerprints of individual cells (Huang et al., 2004; Xu et al., 2017). Certain Raman bands in SCRS would shift when cells incorporate stable isotopes (e.g., 13C, 15N, and 2H), indicating metabolism of a specific isotopic substrate or general metabolic activity (Huang et al., 2004; Li et al., 2012b; Wang et al., 2016, 2013a; Berry et al., 2015; Tao et al., 2017). Raman‐stable isotope probing (Raman‐SIP) has been employed to probe bacterial function at the single cell level (Huang et al., 2009; Bankevich et al., 2012; Li et al., 2012b; Wang et al., 2016a). To link the function to its underlying genotypes, we have developed a series of Raman‐Activated Cell Sorting (RACS) techniques, such as Raman‐activated cell ejection (RACE) (Wang et al., 2017) and Raman‐activated microfluidic sorting (RAMS) (Zhang et al., 2015b; McIlvenna et al., 2016; Song et al., 2017), to isolate individual cells based on their SCRS characteristics. After RACS, genomic DNA from single cells was amplified by multiple displacement amplification (MDA) and subsequently processed for sequencing (Song et al., 2017).
However, genome coverage from RACS was frequently low when sequencing one cell at a time (< 20%), which was presumably due to severe non‐specific and biased amplification when starting from a very low amount of DNA template. Recently mini‐metagenomics, which starts from a small number of cells rather than one single cell, has been reported to improve the genome coverage and assembly integrity (McLean et al., 2013; Yao et al., 2017; Yu et al., 2017). Moreover, deep sequencing and advanced binning algorithms can facilitate informative mining from the assembled genomic sequences (Albertsen et al., 2013; Langille et al., 2013; Nielsen et al., 2014; Magnusdottir et al., 2017). We proposed that, mini‐metagenome from RACE sorted cells can serve as reference to reconstruct genomes encoding enzymes for complete metabolic pathways by recruiting and binning deep metagenomic sequencing data, thus establish a direct link between the phenotype (in situ functions) and genotype (genomes) of bacteria.
Nearly all photosynthetic cells contain carotenoids, which generate a strong resonance Raman signal. The Raman shifts of carotenoid bands caused by 13C incorporation in SCRS can indicate CO2‐fixing activity in photosynthetic cells (Li et al., 2012b). By application of RACE and subsequent DNA sequencing to marine bacteria, we reconstructed near‐complete genomes encoding metabolic pathways of two key bacteria responsible for CO2 fixation in the Yellow Sea of China. First, the CO2‐fixing bacterial cells were detected by Raman‐SIP after spiking 13C bicarbonate to the seawater. Second, RACE was applied to isolate bacterial cells that showed 13C shift in SCRS, and subsequently sequenced. Third, genes encoding metabolic pathways from the mini‐metagenomes were identified according to characterisation for both carotenoid synthesis and CO2‐fixing functions that were revealed by SCRS. Finally, the mini‐metagenomes were employed as reference to guide reconstruction of functional single‐cell genomes from deep metagenomic sequencing. It reveals a significant role of CO2 fixation of Synechococcus spp. and Pelagibacter spp. in the seawater.
Results
An ‘all‐in‐one’ device that integrates Raman‐activated cell sorting and sequencing
The coupling of Raman Activated Cell Ejection (RACE) and subsequent single‐cell sequencing is achieved via a specially designed device (Fig. 1 and Supporting Information Fig. S1). In our previous design, the slide with cell sample had to be manually flipped over for cell ejection (Song et al., 2017). This procedure increases the risk of contamination for single‐cell DNA amplification and slows down the sorting process and represents a hurdle for automation.
Figure 1.
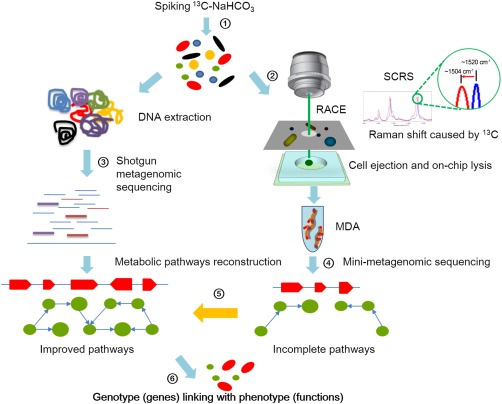
Pipeline of metagenomics aided Raman‐activated cell ejection (RACE) that links genotype to phenotype.
① Feeding of 13C substrate to probe the metabolism of microbial community, for example, from a seawater sample; ② cells were split into the following two processes: DNA extraction and cell sorting according to SCRS, which exhibits shifts caused by 13C incorporation; ③ the extracted DNA is processed for metagenomic sequencing; ④ the sorted cells are lysed and the mini‐metagenomes were amplified by MDA; ⑤ incomplete pathways from mini‐metagenomic sequencing serve as a reference of metagenomic data to reconstruct nearly complete metabolic pathways and ⑥ metabolic functions are validated by the reconstructed metabolic pathways and genes, and a link between genotype (genes) and phenotype (functions) is established at the single cell level. [Colour figure can be viewed at http://wileyonlinelibrary.com]
To ensure precise yet efficient workflow and prevent environmental contamination, an ‘all‐in‐one’ device was designed and fabricated, which consists of three components, assembled in a sandwich‐like manner, that couples SCRS measurement (the sampling chip), cell ejection (the transparent ejection slide), and on‐chip DNA extraction (the collection chip) in a closed format (Fig. 1 and Supporting Information Fig. S1). The process starts with adding samples onto the wells of the sampling slide (Fig. 1 and Supporting Information Fig. S1B). Then, at the SCRS measurement, the cells were not directly exposed to the Raman laser; instead, the Raman incident laser went through the quartz slide before it reached the cells. Raman signal was acquired after passing through the same quartz slide and coating layer (Supporting Information Fig. S1). As a result, the new design avoids the need to physically flip the ejection chip between SCRS acquisition and cell ejection, reducing the risk of DNA contamination and increasing the throughput of RACE (Fig. 1).
Supplementation of 12C/13C NaHCO3 had a minimal effect on abundance and diversity of microbial community in seawater samples
The seawater samples were collected from a typical epipelagic (euphotic) zone in the Yellow Sea of China (Supporting Information Fig. S2; physiochemical parameters in Supporting Information Table S1). Pre‐experiment showed that the marine bacteria with 13C incorporation were detectable using Raman micro‐spectroscopy after 5‐day incubation with 13C NaHCO3 (Supporting Information Figs S3 and S4). To assess the influence of supplemented biocarbonate (e.g., 12C NaHCO3 and 13C NaHCO3) on microbiota structure, 16S rRNA amplicon sequencing results were compared among the original seawater (Primary), 5‐day seawater spiked with 13C NaHCO3 (13C), 5‐day seawater with 12C NaHCO3 (12C) and the 5‐day seawater control without any treatment (C_free). For each of these conditions, triplicate samples were taken, with 12 samples in total. For each sample, averaged 11.4 million 16S rDNA read pairs were sequenced (Supporting Information Table S2). After read trimming, screening and removal of chimeras and singletons, averagely ∼ 96,399 high‐quality reads per sample were obtained (Supporting Information Table S2), revealing a total of 4466 OTUs from the 12 samples.
The rarefaction curves suggested that the sequencing depth was sufficient to capture the diversity of the microbial community (Supporting Information Table S2). Comparison of α diversity indexes (including Observed_OTUs, Simpson, Chao1 and Shannon; Supporting Information Table S2) revealed no significant difference between the four conditions in species richness and bacterial diversity (Kruskal–Wallis test or Wilcoxon test for pairwise comparison; all p > 0.05; Supporting Information Fig. S5). Abundance distribution at the phylum and family levels suggested that supplementation of carbon source 12C/13C NaHCO3 had introduced minor influence on the structure of microbial community (Fig. 2). At the phylum level, Cyanobacteria, Bacteroidetes, and Proteobacteria were the most abundant taxa, contributing to 91.58%–93.26% of the bacterial communities in each sample (Fig. 2A). Classification at the family level detected no significant difference either in microbiota structure among the four conditions of seawater (Fig. 2B). Apart from the (bacterio)chlorophyll‐based phototrophs of Synechococcaceae, Balneolaceae and Rhodobacteraceae, proteorhodopsin‐based Pelagonacteraceae (SAR11) were amongst the most dominant bacteria within the community (Fig. 2B).
Figure 2.
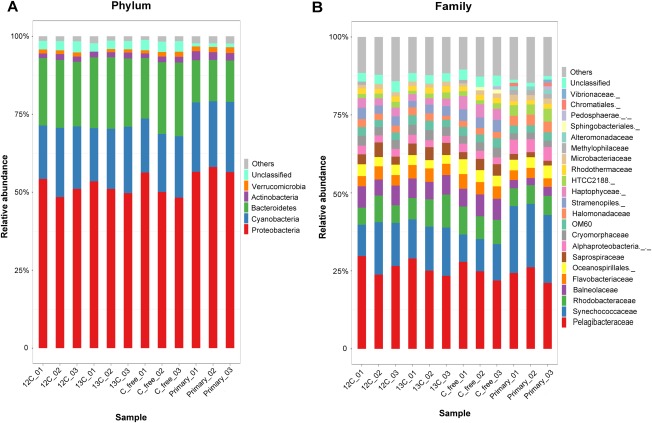
Taxonomical structure and relative abundance of each sample at the phylum level (A) and at the family level (B).
‘Primary’ represents the original seawater, ‘13C’ represents the 5‐day seawater spiked with 13C NaHCO3, ‘12C’ represents the 5‐day seawater with 12C‐NaHCO3 and ‘C_free’ represents the 5‐day seawater control without any treatment. For each of the conditions, triplicate samples were taken marked as _01, _02 and _03. Those accounting for < 1% of the total OTUs are included in ‘Others’, which are shown in gray color at the top of each bar. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Raman shift biomarkers indicated CO2 fixing activities
Single‐cell Raman spectroscopy coupled with stable‐isotope probing (SIP) has been used to reveal specific metabolic functions or to screen for the active species in a microbial community, which is based on the fact that the integration of stable isotope into a cell causes significant shifts of Raman bands in SCRS (Li et al., 2012b; Wang et al., 2016a). Carotenoid vibrations are dominant Raman signals in SCRS of photosynthetic cells, and Raman shifts on carotenoid bands indicate 13C incorporation into the whole cells, which provides unambiguous indication of CO2 fixation (Li et al., 2012b). The characteristic Raman spectra of carotenoid‐containing cells are in the ranges of 1500–1550, 1150–1170 and 1000–1020 cm−1, which are caused by in‐phase C=C (v1), C–C stretching (v2) vibrations of the polyene chain and in‐plane rocking mode of CH3 groups attached to the polyene chain (v3) (Schulz and Baranska, 2007; Robert, 2009; Li et al., 2012a). Although v1, v2 and v3 positions in SCRS of photosynthetic cells might vary, they would all shift to lower wavenumbers when cells have incorporated 13C (Li et al., 2012a). After analyzing many different photosynthetic cells with 13C incorporation (Li et al., 2012b), it is found that 13C in cells usually cause the v2 and v3 bands shifting to the wavenumbers lower than 1150 and 1000 cm−1 respectively. For example, after Synechococcus sp. PCC7002 grown in 13C NaHCO3, its v2 and v3 bands shift to 1147 and 999 cm−1 from original 1159 and 1008 cm−1 in SCRS of 12C cells (Supporting Information Fig. S3C). Hence, in this study, we used the simultaneous shifts in v2 and v3 as criteria, to identify and sort photosynthetic cells that are actively taking in 13C NaHCO3 (Fig. 3 and Supporting Information Fig. S3C).
Figure 3.
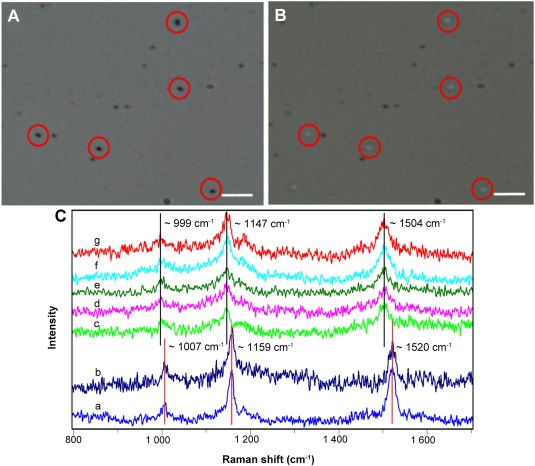
Raman‐activated cell ejection of CO2‐fixing microbes in the sea water sample for mini‐metagenomics.
A. Cells on the ejection slide were identified by Raman measurement.
B. Cells were ejected off the slide.
C. Single‐cell Raman spectra of (a) cells treated with 13C NaHCO3 at time t = 0 days; (b) cells treated with 12C NaHCO3 at time t = 5 days; (c–g) five cells shown in (A) and (B) that were treated with 13C NaHCO3 at time t = 5 days. The scale bar in (A) and (B) is 10 µm. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Isolation of CO2‐fixing cells from seawater samples based on their characteristic SCRS
13C shift SCRS in cells were observed in day five after continuous monitoring seawater reactors. Cells were sorted using RACE illustrated in Fig. 1 and Supporting Information Fig. S3. In the seawater samples spiked with 13C NaHCO3, cells with 13C shifted carotenoid bands in SCRS were sorted (Fig. 3C), based on the Raman sorting criteria (Supporting Information Fig. S3C). One group was taken as negative control with no cells (details showed in Supporting Information Fig. S6). Eight groups were sorted, each contained 30 cells, which all showed significant Raman shifts at v2 and v3 bands in SCRS, similar to those in 13C Synechococcus sp. PCC7002 (Supporting Information Fig. S3C and Fig. 3C).
To link the CO2‐fixing function to genotype, the sorted cells were processed for subsequent mini‐metagenomic analysis. The cell lysis was carried out on the cell collection chip (see Experimental procedures), and the lysate was then transferred to centrifuge tube for genomic DNA amplification. The MDA products were checked for 16S rRNA sequences by PCR to confirm successful amplification (Supporting Information Fig. S6; primers shown in Supporting Information Table S3). Among eight sorted samples, two sample groups that yield positive PCR results, which were sorted independently using identical RACE criteria from a single seawater sample, were designated as TET3 and TET4, and further processed to construct the library for high‐throughput genomic sequencing.
Mini‐metagenomic analysis was consistent with phenotypic properties identified by RACE in terms of carotenoid synthesis and CO2 fixation
Due to the low rate of assembly errors (Bankevich et al., 2012), SPAdes was chosen to assemble the reads into contigs, with total length of 4.72 and 4.49 Mbp for TET3 and TET4, respectively (Supporting Information Table S4). For TET3 and TET4, 4.0% and 7.6% of the contigs (in length) were < 200 bp, 15.6% and 10.3% were > 1500 bp, and 80.4% and 82.1% were ranged from 200 to 1500 bp, respectively (Supporting Information Table S4). These contigs were used as draft mini‐metagenomes for downstream analysis.
After decontamination, contigs from the de novo assembled mini‐metagenome of RACE‐sorted cells were assigned to particular taxa based on their sequence similarity (Table 1). They belonged to seven phyla: Proteobacteria, Bacteroidetes, Chlorophyta, Fusobacteria, Firmicutes, Actinobacteria and Cynaobacteria (Table 1). Contigs > 1000 bp were clustered and visualized using t‐SNE via their 4‐mer signatures, which represented the major clusters of genomes in each sample (Fig. 4 and Supporting Information Table S4). As expected, a significant portion of the contigs was not assigned to any known bacteria. On the other hand, contigs from Chroococcales (an order of Cyanobacteria) were dominant in TET3 (Fig. 4A) while those from Pelagibacterales (SAR11) were dominant in TET4 (Fig. 4B). Consistent with this, in terms of percentages of reads in mini‐metagenomic data, the most abundant bacteria were Chroococcales (an order of cyanobacteria; 66.17%) in TET3 and Pelagibacterales (SAR11, an order of Alphaproteobacteria composed of oligotrophic photochemotroph bacteria; 54.99%) in TET4 (Scanlan et al., 2009; Giovannoni, 2017). TET3 and TET4 shared other three phyla (Proteobacteria, Cynaobacteria and Firmicutes) and six 16S rRNA sequences in TET3 had the same taxonomy identity as TET4 (Supporting Information Table 1). Interestingly, a proteorhodopsin (PR) gene was detected in TET3 (Fig. 4A), suggesting the presence of PR‐containing bacteria in our sorted samples.
Table 1.
Predicted genome completeness and 16S rRNA genes of RACE‐sorted bacteria.
| Metagenome | Mini‐metagenome | |||||
|---|---|---|---|---|---|---|
| RACE‐sorted samples | Class * | Estimated genome completeness (%) | Estimated genome completeness (%) | Identity 16S rRNA (%) | Percentage of reads ** (%) | References supporting presence of carotenoids |
| TET3 | Proteobacteria | 100 | 7.29 | 27.92 | ||
| Proteobacteria‐undefined | 97.41 | 4.17 | – | 2.90 | Shindo and Misawa (2014) | |
| Vibrionales | 50.10 | 3.89 | 99.35 | 0.04 | Meziti et al. (2015) | |
| → Pelagibacterales | 100 | 0.17 | – | 16.06 | Haro‐Moreno et al. (2017) | |
| Xanthomonadales | 30.57 | 1.72 | – | 0.00 | Paret et al. (2012) | |
| Pseudomonadales | 10.42 | 0.00 | 99.36/100 | 0.33 | Reddy and Garcia‐Pichel (2015) | |
| Cyanobacteria | 100 | 6.55 | 66.17 | |||
| → Chroococcales | 100 | 6.55 | 100 | 66.17 | Wang et al. (2016c) | |
| Actinobacteria | 100 | 0.86 | 0.01 | |||
| Micrococcales | 87.72 | 0.86 | – | 0.00 | Trujillo (2016) | |
| Firmicutes | 100 | 0.00 | 0.01 | |||
| Bacillales | 89.34 | 0.00 | 100 | 0.01 | Steiger et al. (2015) | |
| Undefined sequences | – | – | 5.89 | – | ||
| TET4 | Proteobacteria | 100 | 42.40 | 56.84 | ||
| → Pelagibacterales | 100 | 13.66 | – | 54.99 | Haro‐Moreno et al. (2017) | |
| Vibrionales | 50.10 | 4.75 | 99.67/100 | 0.01 | Meziti et al. (2015) | |
| Pseudomonadales | 10.42 | 2.98 | 98.61/96.31 | 0.48 | Reddy and Garcia‐Pichel (2015) | |
| Xanthomonadales | 30.57 | 0.16 | – | 0.00 | Paret et al. (2012) | |
| Caulobacterales | 9.98 | 0.16 | – | 0.00 | Fukui et al. (2013) | |
| Bacteroidetes | 100 | 1.88 | – | 9.87 | ||
| Flavobacteriales | 100 | 1.88 | – | 9.87 | Shahina et al. (2013) | |
| Chlorophyta (Eukarya) | 77.59 | 0.94 | – | 0.00 | ||
| Sphaeropleales | 1.33 | 0.94 | – | 0.00 | Lee et al. (2016) | |
| Fusobacteria | 11.05 | 0.51 | – | 0.01 | ||
| Fusobacteriales | 11.68 | 0.51 | – | 0.01 | – | |
| Firmicutes | 100 | 0.00 | – | 4.66 | ||
| Bacillales | 89.34 | 0.00 | 100 | 0.83 | Steiger et al. (2015) | |
| Cyanobacteria | 100 | 0.00 | – | 0.02 | ||
| → Chroococcales | 100 | 0.00 | 100/100 | 0.02 | Wang et al. (2016c) | |
| Undefined sequences | – | – | 28.60 | – | ||
*In this column, phylum names are in italics bold, and order names are only in italics; ‘→’ represent the bacteria that are numerically abundant, that is, the focus of this study.
**This column lists the fraction of reads mapped to the specific organisms among the reads mapped to all the annotated contigs.
Figure 4.
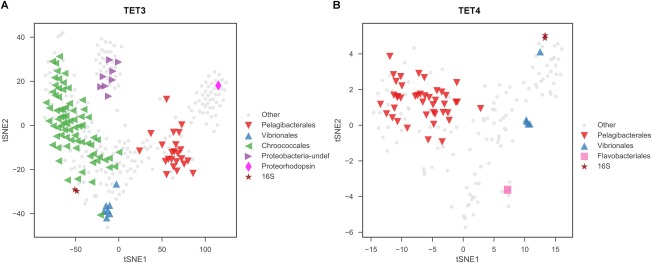
t‐SNE embeddings of the 4‐mer signatures of assembled mini‐metagenomic contigs.
Plots represent samples TET3 (A) and TET4 (B), and only those contigs longer than 1000 bp were considered. Contigs were colored and shaped based on taxonomic annotations listed in Table 1. This result suggests that Pelagibacterales (including Proteobacteria‐undef) and Chroococcales were dominant in sample TET3, while Pelagibacterales was dominant in sample TET4. [Colour figure can be viewed at http://wileyonlinelibrary.com]
RACE sorted cells should contain carotenoids and utilize 13C NaHCO3. According to literature, all bacteria identified in TET3 and TET4 were potentially able to synthesize carotenoids (Table 1), which was in good agreement with the sorting criteria. Two dominant types of cells in TET3 and TET4 were Cyanobacteria and Pelagibacterales (SAR11), both of which are able to use sunlight as energy source, although cyanobacteria use chlorophyll whilst Pelagibacterales (SAR11) employ PR to harvest sunlight energy (Finkel et al., 2013). It is expected that cyanobacteria were detected, as they are dominant photoautotrophic bacteria in the ocean, which usually contain carotenoids and use 13C NaHCO3. Interestingly, the sequencing data show that Pelagibacterales (SAR11) was found in TET3 (16.06%) and TET4 (54.99%), which should be 13C‐labelled and contain carotenoids.
Mini‐metagenome from RACE and metagenome from whole community DNA are coupled to reconstruct genomes of dominant CO2‐fixing bacteria in the seawater
The 16S rRNA metagenomic sequencing also indicated that Synechococcus spp. (Cyanobacteria) and Pelagibacter spp. (Pelagibacterales) were the two dominant bacteria in the seawater (Supporting Information Fig. S7). To reconstruct their genomes, deep metagenomic sequencing of the seawater sample was performed. Over 117 million paired end clean reads were produced, which were assembled into 1,990,773 contigs (totally 1.67 Gb; Supporting Information Table S4). Among them, totally 47,274 contigs were annotated as either Synechococcus spp. (24,834 contigs, total length of 36.2 Mb, average GC content of 60%) or Pelagibacter spp. (Candidatus Pelagibacter and Pelagibacteraceae‐undef; 22,440 contigs, combined length of 20.5 Mb, average GC content of 31%). GC distribution plots of the re‐mapped sequences were smooth (Supporting Information Fig. S8), which was consistent with the absence of significant contamination. The overall GC contents of the binned genomes (represented by curve's central peak; Supporting Information Fig. S8) were close to those of known sequenced genomes of corresponding taxa, respectively (Candidatus Pelagibacter ubique HTCC1062 with RefSeq accession NC_007205.1: GC content 29.7%; Synechococcus spp. WH 7803 with RefSeq accession NC_009481.1: GC content 60.2%). Thus, these two sets of contigs should represent, to a certain degree, the collective genetic complements of CO2‐fixing Synechococcus spp. and Pelagibacter spp., respectively, in this particular ecosystem of seawater. In fact, if taken as a whole, respectively, each of them made nearly complete genomes (100% completeness; as estimated by CheckM) of Synechococcus spp. (Cyanobacteria) and Pelagibacter spp. (Pelagibacterales) (Table 1).
The seawater was sampled from the pelagic zone and had sufficient sunlight but low organic matter (chemical oxygen demand was 1.05 mg L−1; Supporting Information Table S1). These conditions should have favored bacteria, such as Synechococcus spp. (Cyanobacteria) and Pelagibacter spp. (Pelagibacterales), which are able to harvest sunlight energy and undertake CO2 fixation. Application of RACE successfully linked bacterial ecological function (e.g., CO2 fixation in this case) and mini‐metagenomes, which were then served as references or blueprints to bin the complete genomes of key functional bacteria Synechococcus spp. (Cyanobacteria) and Pelagibacter spp. (Pelagibacterales) with aid of high‐throughput metagenomic sequencing. As a result, RACE‐mediated single‐cell genome is effective at establishing a link between phenotype/function and genotype.
Reconstructed metabolic pathways reveal new ecological functions of RACE‐sorted bacteria
Contigs assembled from mini‐metagenomic data with length longer than 200 bp were collected to predict functional genes in this study (Supporting Information Table S4). The number of predicted genes in these functional mini‐metagenomic samples was 4098 and 3102 for TET3 and TET4 respectively. Other details, such as the predicted CDS numbers and tRNA numbers, are provided in Supporting Information Table S5.
RACE was employed to link bacterial CO2 fixing function and genotype in terms of mini‐metagenome. Although the mini‐metagenome was incomplete (Table 1), it provides sufficient genetic information to reconstruct complete metabolic pathways from high‐throughput and deep metagenomic data (see Experimental procedures). The functional genes derived from mini‐metagenome were highly consistent with the functional genes from shotgun metagenomic data in terms of gene similarities (Supporting Information File S1 and Supporting Information Table S7). To unravel the metabolic potential of these functional mini‐metagenomes, the two bacterial mini‐metagenomic gene sets were mapped to the KEGG pathways, which predicted gene function largely based on sequence homology (Kanehisa et al., 2016). Within these gene sets, eight metabolic pathways were examined, including carbon metabolism (ko01200), carbon fixation in photosynthetic organisms (ko00710) and in prokaryotes (ko00720), photosynthesis (ko00195), antenna protein synthesis (ko00196), porphyrin and chlorophyll metabolism (ko00860), carotenoid (ko00906) and terpenoid biosynthesis (ko00900), which were all detected in TET3 and TET4 samples (Supporting Information Table S6 and Supporting Information File S1). A total of 371 matches in the annotation output of these two samples showed high homology to the photosynthetic carbon‐fixing genes.
According to Raman profiles, the sorted cells should contain carotenoids and fix CO2. Using mini‐metagenome as blueprints, complete pathways for carotenoid synthesis were identified for Synechococcus spp. and Pelagibacter spp. (Fig. 5). Calvin cycle for CO2 fixation pathways in Synechococcus spp. (Cyanobacteria; Fig. 6A) and anaplerotic CO2 fixation in Pelagibacter spp. (Pelagibacterales; Fig. 6B) were also revealed by the mini‐metagenomes and metagenomic bins. These observations suggested that Synechococcus spp. should use Calvin cycle, whilst Pelagibacter spp. (Pelagibacterales) may employ anaplerotic reactions to fix CO2 (Fig. 6 and Supporting Information File S2).
Figure 5.
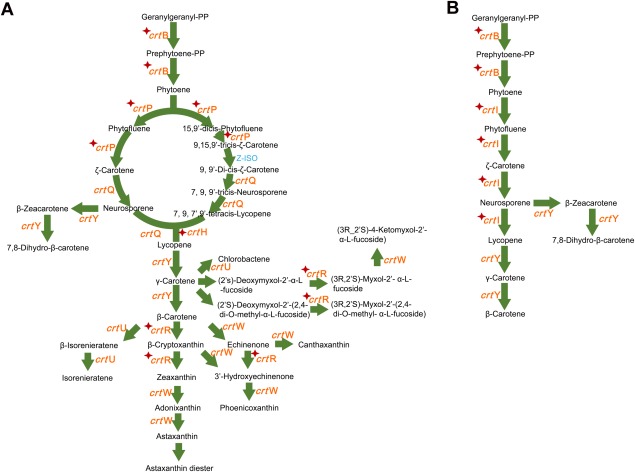
Reconstructed β‐carotene module in the carotenoid synthesis pathway by metagenomics‐aided RACE in Yellow Sea.
A. Reconstructed beta‐carotene module in Synechococcus spp.
B. Reconstructed beta‐carotene module in Pelagibacter spp. The known pathways were obtained from the KEGG database.
Enzymes in red colour: found in mini‐metagenome guided shotgun data; with star markers: found in mini‐metagenomic data; in blue colour: not found in our data. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 6.
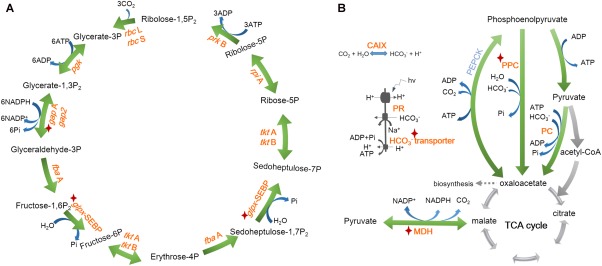
Reconstructed CO2 fixing pathways by metagenomics‐aided RACE in Yellow Sea
A. Synechococcus spp. employed Calvin cycle.
B. Pelagibacter spp. used anaplerotic reactions.
Enzymes in red colour: found in mini‐metagenome guided shotgun data; enzymes with star markers: found in mini‐metagenomic data; in blue colour: not found in our data. CAIX: carbonic anhydrase; PR: proteorhodopsin; MDH: malate dehydrogenase; PPC: phosphoenolpyruvate carboxylase; PC: pyruvate carboxylase; PEPCK: phosphoenolpyruvate carboxykinase. [Colour figure can be viewed at http://wileyonlinelibrary.com]
The reconstructed pathway reveals that Synechococcus spp. has all genes necessary for the synthesis of various carotenoids, whilst Pelagibacter spp. has one intact pathway for β‐carotene synthesis (Fig. 5 and Supporting Information File S2). It confirms our hypothesis that PR containing bacteria should be able to synthesize β‐carotene which can be cleaved by dioxygenase to produce retinal – an essential compound in functional PR (Von Lintig and Vogt, 2000). The presence of β‐carotene would enable Pelagibacter spp. self‐sufficient for PR synthesis. It also explains that carotenoid‐containing cells sorted by RACE included Pelagibacter spp.
In terms of sunlight harvesting system, Synechococcus spp. has all the genes necessary for photosynthesis, chlorophyll α, antenna (phycobilisome), Photosystem I and Photosystem II (Mulkidjanian et al., 2006) biosynthesis (Supporting Information Figs S9–S15 and Supporting Information File S2), which were all absent from the reconstructed genome of Pelagibacter spp. According to the identified proteorhodopsin (PR) genes from metagenomic bins of Pelagibacter spp. (Supporting Information File S3), we have successfully expressed these PR genes in Escherichia coli (data not shown), confirming that the sequences indeed were encoded with PR. F‐type ATPase genes were found in both Synechococcus spp. and Pelagibacter spp. (Supporting Information Fig. S12). These were expected, as Synechococcus spp. uses chlorophyll whilst Pelagibacter spp. employs PR to harvest light energy. Indeed, it has been previously reported that a pure PR‐containing strain Dokdonia sp. MED134, in the presence of the light, was able to fix CO2 using anaplerotic reactions (Palovaara et al., 2014). Our observation suggests that CO2 fixation via anaplerotic reactions by PR‐containing bacteria could happen in natural environment as well.
The above results from genome reconstruction of RACE‐sorted cells, i.e., two dominant carotenoid‐containing bacteria in the seawater Synechococcus spp. and Pelagibacter spp. exhibit active carbon fixing activities, are in good agreements with the phenotypic analysis from SCRS, which suggested that these cells were able to incorporate 13C from13C NaHCO3 and contain carotenoids. Reconstructed genome and pathway analysis were successful at identifying the genes responsible for carotenoid synthesis, light harvesting pathways and CO2 fixation pathway (Figs 5 and 6 and Supporting Information File S2). The phenotype–genotype agreement confirms that RACE sorting was effective at isolating phototrophic CO2 fixing bacteria in seawater and revealed a suspected yet previously unsubstantiated ecological function of the SAR11 group bacteria – Pelagibacter spp.
Discussion
RACE established a link between phenotype and genotype of cells
The composition and function of a microbial community are the two most fundamental topics in microbial ecology. Metagenomic sequencing and binning is an increasingly powerful tool which is able to give a blueprint on genotype level for profiling the microbiota directly (Albertsen et al., 2013; Langille et al., 2013; Nielsen et al., 2014; Magnusdottir et al., 2017). However, the phenotypes of the composing microbes were missed inevitably. Raman spectroscopy has been proved to be a useful approach which could provide comprehensive phenotype information of single cells in a non‐destructive manner. More importantly, by combining with the stable isotope probing method which is using stable isotope substituted substrates for keeping normal metabolism of cells, Raman‐SIP provides a promising tool for pinpointing specific metabolic functions of certain species within a complex community (Huang et al., 2004, 2009; Li et al., 2012b; Wang et al., 2013; Zhang et al., 2015a; Song et al., 2017) or general metabolic activity (Berry et al., 2015; Wang et al., 2016b; Tao et al., 2017) at single‐cell level. Given above merits, Raman activated cell sorting and sequencing could undisputedly offer a direct link between phenotype and genotype of cells, which is especially significant in microbiology.
It has been reported that mini‐metagenomics, which starts from tens to thousands of cells and employs the single cell technology could improve amplification and help the genome assembly from the sequencing data, leading to higher hit of target gene and coverage as well as the integrity of assembled genome. In combination with advanced contig binning methods, multiple genomes can be reconstructed from mini‐metagenomic data of microbiota (McLean et al., 2013; Yao et al., 2017; Yu et al., 2017). In this study, RACE isolated 30 cells for mini‐metagenomic sequencing process, which can be served as blueprints for the whole genome assembly using advanced binning methods.
Several Raman activated cell sorting methods have been established, including Raman tweezers, RACE (Song et al., 2017), microfluidic flow sorting (RACS, RADS) (Zhang et al., 2015b; McIlvenna et al., 2016; Wang et al., 2017). RACE with single cell genomics can provide direct link between phenotype and genotype of the cell. However, the technical problem is that genome coverage from single cells is low due to extremely small DNA template, severe nonspecific and biased amplification for single cells (Song et al., 2017). Various attempts have been made to improve single‐cell sequencing (Rinke et al., 2013; Stepanauskas et al., 2017); however high coverage of single‐cell genome continues to be difficult to achieve.
The RACE is based on laser induced forward transfer (LIFT) principle, in which, most of the laser energy was absorbed by the coating material to push forward the cells (Song et al., 2017). However, the LIFT process may affect bacterial viability and so far we are unable to culture cells after RACE sorting, although there are some reports that cells were still viable after the LIFT (Ringeisen et al., 2004; Hopp et al., 2005; Barron et al., 2005).
Pelagibacterales (SAR11) is able to fix CO2 in the seawater
In this study, we applied RACE to obtain mini‐metagenomes from functional bacteria, which served as the reference for the reconstruction of nearly complete genomes of two CO2 fixing bacteria Synechococcus spp. and Pelagibacter spp. originating from the Yellow Sea in China. The gene and pathway analyses confirm that Synechococcus spp. and Pelagibacter spp. have the necessary genes for functions such as CO2 fixing, carotenoid synthesis and light harvesting, which was in very good agreement with Raman profiling.
Cyanobacteria, including Prochlorococcus and Synechococcus, are the most abundant photosynthetic organisms on Earth (Scanlan et al., 2009) and are able to fix CO2 using sunlight as energy source. Synechococcus spp. was found to be one of the main CO2 fixing bacteria in the seawater, and the reconstructed genome shows that it contained all the genes necessary for this ecological function.
It has been reported that Pelagibacterales play an important role in marine carbon and nutrient cycling (Sowell et al., 2009; Ottesen et al., 2014). Metagenomic DNA sequencing suggested that globally SAR11 bacteria (Pelagibacterales) are estimated to account for 25% of all plankton (Giovannoni et al., 2005; Giovannoni, 2017). Despite its global abundance, our understanding of ecological role of SAR11 is limited due to the difficulty of cultivation and lack of suitable tools. All SAR11 members are thought to be chemoheterotrophic, obtaining its energy from the oxidation of organic compounds (Giovannoni, 2017). PR is a light driven proton pump, which couples with ATPase to help cells generate ATP for survival (DeLong and Beja, 2010; Gomez‐Consarnau et al., 2010). In this study, Pelagibacterales was found to be the dominant bacteria (25.8%) in the low organic matter sea water (Supporting Information Fig. S7). Interestingly, Pelagibacter spp. was sorted by RACE, indicating it contained carotenoid and was labelled with 13C carbon. The ‘reconstructed’ draft genome shows that Pelagibacter spp. contains self‐sufficient PR synthesis genes for light harvesting and key genes for anaplerotic CO2 fixation.
It is intriguing whether these PR‐containing Pelagibacter spp. were able to fix CO2 or crossly fed 13C from other bacteria. It was observed that both Synechococcus spp. and Pelagibacter spp. show the same extent 13C‐Raman shift at the same time (Fig. 3 and Supporting Information Fig. S4), which indicates the same level of 13C content in cells (Li et al., 2012b). In this study, both Synechococcus spp. and Pelagibacter spp. were dominant bacteria (Fig. 2) in the pelagic zone with low organic matter (COD = 1.05 mg l−1; Supporting Information Table S1). To grow and make dominant with limited organics, Pelagibacter spp. could fix CO2, given that genetically it was genetically equipped with an intact light‐harvesting PR (but no chlorophyll) and CO2 fixing (anaplerotic reactions) machinery. It has been reported that anaplerotic CO2 fixation in PR containing Dokdonia sp. MED134 provided up to one‐third of the cell carbon in the light (Palovaara et al., 2014). Hence, it is highly likely that Pelagibacter spp. also contribute to CO2 fixation in open sea.
In summary, this study demonstrates that metagenomics aided RACE is able to establish a reliable link between the phenotype and the genotype of key functional bacteria in microbial community, addressing the fundamental questions and paving the way for comprehensive dissection of microbial community.
Experimental procedures
Seawater sampling and preparation
Two 10 L seawater samples were collected by Schindler samplers from the euphotic zone of the Yellow Sea, China (the geographical position is shown in Supporting Information Fig. S2). The seawater was transported using portable cooler with ice to laboratory for analysis. All samples were prefiltered through 8 μm pore size filter membranes using a peristaltic pump when being decanted from the sampler.
The original seawater sample was served as a primary control (without incubation). One liter of seawater was taken and spiked with 12C or 13C NaHCO3 with a final concentration of 2 mM, and the control was not treated with any chemicals. The samples were incubated in closed bottles at room temperature for 5 days with natural light. The four sets of samples, including primary control, incubation control, 12C or 13C NaHCO3 spiking treatments, were all in triplicate. After pretreatments, cells in those samples mentioned above were harvested by 0.22 μm pore size membrane filtration using a peristaltic pump and then the cells were sealed and stored at −80°C freezer until DNA extraction.
After incubation, cells in samples spiked with 12C and 13C NaHCO3 were enriched by passing 70 ml seawater through the Centricon® Plus‐70 Ultracel PL‐100 (Merck Millipore, Billerica, USA) respectively. Then, the cells were used for Raman activated cell sorting based on functions of 13C NaHCO3 utilization.
‘All‐in‐one’ device for Raman‐activated cell ejection (RACE)
RACE analysis in this study was performed using a custom‐made ‘all‐in‐one’ device, which represents a significant improvement over our previously reported system (Song et al., 2017). The device consists of a transparent ejection slide plus a sampling and cell collection chip (Supporting Information Fig. S1). The transparent ejection slide was fabricated by coating a thin layer of indium tin oxide on the surface of a 1 mm thick quartz substrate, which formed an optical transparent film of 100 nm thickness over the quartz slide, enabling the acquisition of Raman scattering signals and the ejection of cells of interest without the need of opening system for slide turnover (Supporting Information Fig. S1). This improvement would significantly reduce the risk of contamination. The sampling chip was composed of an elastomer polydimethylsiloxane (PDMS) layer with arrayed hollow holes (1 mm in diameter), which was attached onto the transparent ejection slide to form sample wells (Supporting Information Fig. S1). The cell collection chip was fabricated by irreversibly bonding another thicker PDMS layer with holes to a glass cover chip, forming wells (2 mm in diameter) for receiving the sorted cells and for holding reagents for subsequent cell lysis. The wells in cell collection chip were aligned with the wells in sampling chip, avoiding cross contamination.
Single‐cell Raman micro‐spectroscopy
The cell samples of 2 μl were immediately loaded into the specified mini‐wells in the ‘all‐in‐one’ system (Supporting Information Fig. S1) and air‐dried prior to Raman analysis. Sample observation and Raman signal acquisition were achieved by using a modified Horiba LabRam HR system (Hesen Ltd, Shanghai, China), which was equipped with a confocal microscope with a 50× PL magnifying dry objective (NA = 0.55, BX41; Olympus UK Ltd., Southall, UK) and a 532 nm Nd:YAG laser (Ventus; Laser Quantum Ltd, Stockport, UK). The laser power out of the objective was approximately 5 mW and Raman signals collection was by a Newton EMCCD (Andor, Belfast, UK) utilizing a 1600 × 200 array of 16 μm pixels with thermoelectric cooling down to −70°C for negligible dark current. LabSpec5 (Horiba Scientific, France) software was used to control the Raman system and acquire spectra. A 600 mm−1 grating was set for the measurements, resulting in a spectral resolution of ∼ 1 cm−1 with 1600 data points. Acquisition of each spectrum was performed within one second.
Isolation of 13C cells from the seawater sample by RACE
All Raman spectra were recorded and normalized with the LabSpec 5 software. The positions of SCRS bands from carotenoid‐containing cells were determined and analysed to establish a relationship between the red shift and 13C absorption phenomenon in cells. The Raman spectra of carotenoid‐containing cells from 12C labelling sample were used as controls. Cells with 13C shift in SCRS were isolated using a 532 nm pulsed laser (Alphalas GmbH, Goettingen, Germany), as described previously (Song et al., 2017). After cells were measured and ejected one‐by‐one into the collection chip, the whole device was moved into a laminar hood. The cell collection chip was then carefully detached from the device, and buffer (Qiagen, Germantown, MD, USA) was added into the well for cell lysis.
Nucleotide sequence accession numbers
The sequence data reported in this study have been deposited to NCBI SRA database with bioproject accession PRJNA407379.
Author contributions
WEH conceived the idea. WEH, JX, XJ and HG designed the research. LX provided seawater samples. XJ, HG and XS performed the Raman microspectroscopy, RACE, DNA amplification, library‐generation and sequencing. HG designed and manufactured the RACE device. YG, XJ and JX performed metagenome sequence analysis. WEH, XJ, HG, YG and JX analyzed the data. YJ, YS and IPT provided critical suggestions. WEH, JX, XJ, HG and YG wrote the article.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site
Table S1. Basic properties of the seawater sampled from Yellow Sea of China.
Table S2. Numbers of 16S rRNA sequencing reads for microbial diversity analysis of each sample.
Table S3. Primers used in 16S rRNA PCR amplification.
Table S4. Sequencing and assembly statistics for the functional mini‐metagenomes and shotgun metagenomes.
Table S5. The statistics of the predicted genes in mini‐metagenome sequencing in this study.
Table S6. The KEGG pathways related to key metabolism pathways for gene annotation and their corresponding gene counts.
Table S7. Protein similarities in three KEGG modules (beta‐carotene biosynthesis, reductive pentose phosphate cycle and reductive citrate cycle) between mini‐metagenomic data and shotgun metagenomic data for Chroococcales and Pelagibacterales.
Fig. S1. Illustration of the ‘all‐in‐one’ device assembly.
A. Transparent ejection slide.
B. Sampling chip.
C. Cell collection chip.
Fig. S2. Geographic location of sampling site in Yellow Sea.
The label ‘L 01’ represents the location position of sampling site.
Fig. S3. Scheme of Raman identification and sorting of CO2‐fixing microbes with ‘all‐in‐one’ integrated device.
A. Acquisition of single cell Raman spectra and CO2‐fixing microbes identification.
B. Sorting cells of interest by laser ejection.
C. Shifts of the carotenoid Raman bands in single cell Raman spectra of Synechococcus spp. PCC7002, indicating 13C incorporation into the cell when cells were incubated with 13C NaHCO3.
Fig. S4. Raman spectra of carotenoids containing cells in the seawater which were incubated in closed bottles at room temperature at different times.
A. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 0 days.
B. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 3 days.
C. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 5 days.
D. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 7 days.
E. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 10 days.
Fig. S5. Alpha diversity comparisons across four different treatments of the seawater: red bars – 12C NaHCO3 amended sample (12C); blue bars – 13C NaHCO3 amended sample (13C); green bars – control sample without NaHCO3 (C_free); purple bars – original seawater sample control (primary).
A. Box plot showing the variation of observed OTUs.
B. Box plot showing the variation of Chao1 index.
C. Box plot showing the variation of Simpson index.
D. Box plot showing the variation of Shannon index.
Fig. S6. Agarose gel images of the multiple displacement amplifications (MDAs) and 16S rRNA gene validation processes.
A. Agarose gel image of the DNA products after MDAs showing high molecular weight DNA.
B. Agarose gel image of the PCR products of 16S rRNA gene from isolated single marine cells.
Lane M, DNA ladder; Lane N, negative control for PCR; Lane N1, negative control for ddH2O and REPLI‐g sc Master Mix (contains REPLI‐g sc Reaction buffer and REPLI‐g sc DNA Polymerase); Lane N2, negative control for ddH2O, REPLI‐g sc Master Mix, Stop solution and lysis buffer (contains buffer DLB and DTT); Lanes 1–5, 7 and 8, 30 ejected target cells in each sample; Lane 6, thirty random ejected cells; Lane 9, no cells were ejected; Lane P, Lane P1, positive control for PCR.
Fig. S7. Heat tree visualization of the structure of marine bacterial community based on 16S rDNA sequencing.
In the heat tree, size and color of nodes and edges are correlated with the abundance of organisms in the microbial community.
Fig. S8. GC distributions of metagenomic reads which were mapped to the binned genomes of Synechococcus spp. (A) and Pelagibacter spp. (B) from shotgun metagenomes.
The two curves were smooth and normally distributed, which is consistent with the absence of significant contamination. The central peak in each curve (red) was close to the average GC content of the binned draft genomes and also to the sequenced reference genomes of identical taxonomic classification.
Fig. S9. Reconstructed ‘carotenoid biosynthesis’ pathway by metagenomics‐aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S10. Reconstructed ‘carbon fixation in photosynthetic organisms’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S11. Reconstructed ‘carbon fixation pathways in prokaryotes’ by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S12. Reconstructed ‘photosynthesis’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S13. Reconstructed ‘photosynthesis‐antenna proteins’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S14. Reconstructed ‘porphyrin and chlorophyll metabolism’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S15. Reconstructed ‘terpenoid backbone biosynthesis’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea. The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
File S1. List of carbon metabolism‐related genes and their annotations in the mini‐metagenomic and the shotgun metagenomic data.
File S2. Identified functional genes in metabolic pathways from the mini‐metagenome data and their corresponding genes in the shotgun metagenomic data.
File S3. Identified proteorhodopsin (PR) genes from metagenomic bins of Pelagibacter spp.
Acknowledgements
WEH thanks support from EPSRC (EP/M002403/1 and EP/M02833X/1) and NERC (NE/M002934/1) and Oxford University to host XJ visit. HG and JX acknowledge support of grants 31770112, 21605151 and 31425002 from NSFC, GASI‐03‐01‐02–05 from SOA and KFJ‐SW‐STS‐165 and XDB15000000 from CAS. YJ thanks support from NSF Grant of Shandong Province (ZR2014CQ005). We also thank NSFC (31400436) for financial support.
Contributor Information
Jian Xu, Email: xujian@qibebt.ac.cn.
Wei E. Huang, Email: wei.huang@eng.ox.ac.uk.
References
- Albertsen, M. , Hugenholtz, P. , Skarshewski, A. , Nielsen, K.L. , Tyson, G.W. , and Nielsen, P.H. (2013) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Amann, R.I. , Ludwig, W. , and Schleifer, K.‐H. (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A.A. , Dvorkin, M. , Kulikov, A.S. , et al (2012) SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron, J.A. , Krizman, D.B. , and Ringeisen, B.R. (2005) Laser printing of single cells: Statistical analysis, cell viability, and stress. Ann Biomed Eng 33: 121–130. [DOI] [PubMed] [Google Scholar]
- Berry, D. , Mader, E. , Lee, T.K. , Woebken, D. , Wang, Y. , Zhu, D. , et al (2015) Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci USA 112: E194–E203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong, E.F. , and Beja, O. (2010) The light‐driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol 8: e1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont, M.G. , and Murrell, J.C. (2005) Stable isotope probing – linking microbial identity to function. Nat Rev Microbiol 3: 499–504. [DOI] [PubMed] [Google Scholar]
- Finkel, O.M. , Beja, O. , and Belkin, S. (2013) Global abundance of microbial rhodopsins. ISME J 7: 448–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui, Y. , Abe, M. , Kobayashi, M. , Saito, H. , Oikawa, H. , Yano, Y. , and Satomi, M. (2013) Algimonas porphyrae gen. nov., sp. nov., a member of the family Hyphomonadaceae, isolated from the red alga Porphyra yezoensis . Int J Syst Evol Microbiol 63: 314–320. [DOI] [PubMed] [Google Scholar]
- Giovannoni, S.J. (2017) SAR11 bacteria: the most abundant plankton in the oceans. Ann Rev Mar Sci 9: 231–255. [DOI] [PubMed] [Google Scholar]
- Giovannoni, S.J. , Britschgi, T.B. , Moyer, C.L. , and Field, K.G. (1990) Genetic diversity in sargasso sea bacterioplankton. Nature 345: 60–63. [DOI] [PubMed] [Google Scholar]
- Giovannoni, S.J. , Bibbs, L. , Cho, J.C. , Stapels, M.D. , Desiderio, R. , Vergin, K.L. , et al (2005) Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438: 82–85. [DOI] [PubMed] [Google Scholar]
- Gomez‐Consarnau, L. , Akram, N. , Lindell, K. , Pedersen, A. , Neutze, R. , Milton, D.L. , et al (2010) Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol 8: e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman, J. (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68: 669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro‐Moreno, J.M. , Lopez‐Perez, M. , de la Torre, J. , Picazo, A. , Camacho, A. , and Rodriguez‐Valera, F. (2017) Fine stratification of microbial communities through a metagenomic profile of the photic zone. BioRxiv 134635. [Google Scholar]
- Hopp, B. , Smausz, T. , Kresz, N. , Barna, N. , Bor, Z. , Kolozsvari, L. , et al (2005) Survival and proliferative ability of various living cell types after laser‐induced forward transfer. Tissue Eng 11: 1817–1823. [DOI] [PubMed] [Google Scholar]
- Huang, W.E. , Song, Y. , and Xu, J. (2015) Single cell biotechnology to shed a light on biological ‘dark matter’ in nature. Microb Biotechnol 8: 15–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.E. , Griffiths, R.I. , Thompson, I.P. , Bailey, M.J. , and Whiteley, A.S. (2004) Raman microscopic analysis of single microbial cells. Anal Chem 76: 4452–4458. [DOI] [PubMed] [Google Scholar]
- Huang, W.E. , Ferguson, A. , Singer, A.C. , Lawson, K. , Thompson, I.P. , Kalin, R.M. , et al (2009) Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single‐cell raman‐fluorescence in situ hybridization. Appl Environ Microbiol 75: 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Sato, Y. , and Morishima, K. (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428: 726–731. [DOI] [PubMed] [Google Scholar]
- Langille, M.G.I. , Zaneveld, J. , Caporaso, J.G. , McDonald, D. , Knights, D. , Reyes, J.A. , et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.‐G. , Song, H.J. , Kim, D.‐S. , Cho, C.H. , La, H.‐J. , Oh, H.‐M. , and Yoon, H.S. (2016) Unique mitochondrial genome structure of the green algal strain YC001 (Sphaeropleales, Chlorophyta), with morphological observations. Phycologia 55: 72–78. [Google Scholar]
- Li, M. , Ashok, P.C. , Dholakia, K. , and Huang, W.E. (2012) Raman‐activated cell counting for profiling carbon dioxide fixing microorganisms. J Phys Chem 116: 6560–6563. [DOI] [PubMed] [Google Scholar]
- Li, M. , Canniffe, D.P. , Jackson, P.J. , Davison, P.A. , FitzGerald, S. , Dickman, M.J. , et al (2012) Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J 6: 875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusdottir, S. , Heinken, A. , Kutt, L. , Ravcheev, D.A. , Bauer, E. , Noronha, A. , et al (2017) Generation of genome‐scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol 35: 81–89. [DOI] [PubMed] [Google Scholar]
- McLean, J.S. , Lombardo, M.‐J. , Badger, J.H. , Edlund, A. , Novotny, M. , Yee‐Greenbaum, J. , et al (2013) Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA 110: E2390–E2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlvenna, D. , Huang, W.E. , Davison, P. , Glidle, A. , Cooper, J. , and Yin, H.B. (2016) Continuous cell sorting in a flow based on single cell resonance Raman spectra. Lab Chip 16: 1420–1429. [DOI] [PubMed] [Google Scholar]
- Meziti, A. , Kormas, K.A. , Moustaka‐Gouni, M. , and Karayanni, H. (2015) Spatially uniform but temporally variable bacterioplankton in a semi‐enclosed coastal area. Syst Appl Microbiol 38: 358–367. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian, A.Y. , Koonin, E.V. , Makarova, K.S. , Mekhedov, S.L. , Sorokin, A. , Wolf, Y.I. , et al (2006) The cyanobacterial genome core and the origin of photosynthesis. Proc Natl Acad Sci USA 103: 13126–13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach, S. , and Pollard, K.S. (2016) Toward accurate and quantitative comparative metagenomics. Cell 166: 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H.B. , Almeida, M. , Juncker, A.S. , Rasmussen, S. , Li, J.H. , Sunagawa, S. , et al (2014) Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32: 822–828. [DOI] [PubMed] [Google Scholar]
- Ottesen, E.A. , Young, C.R. , Gifford, S.M. , Eppley, J.M. , Marin, R. , Schuster, S.C. , et al (2014) Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science 345: 207–212. [DOI] [PubMed] [Google Scholar]
- Palovaara, J. , Akram, N. , Baltar, F. , Bunse, C. , Forsberg, J. , Pedros‐Alio, C. , et al (2014) Stimulation of growth by proteorhodopsin phototrophy involves regulation of central metabolic pathways in marine planktonic bacteria. Proc Natl Acad Sci USA 111: E3650–E3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paret, M.L. , Sharma, S.K. , Misra, A.K. , Acosta, T. , deSilva, A.S. , Vowell, T. , and Alvarez, A.M. (2012) Characterization of the pigment xanthomonadin in the bacterial genus Xanthomonas using micro‐ and resonance Raman spectroscopy. In SPIE Defense, Security, and Sensing 8367, p. 9.
- Rappe, M.S. , Connon, S.A. , Vergin, K.L. , and Giovannoni, S.J. (2002) Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630–633. [DOI] [PubMed] [Google Scholar]
- Reddy, G.S. , and Garcia‐Pichel, F. (2015) Description of Pseudomonas asuensis sp. nov. from biological soil crusts in the Colorado plateau, United States of America. J Microbiol 53: 6–13. [DOI] [PubMed] [Google Scholar]
- Ringeisen, B.R. , Kim, H. , Barron, J.A. , Krizman, D.B. , Chrisey, D.B. , Jackman, S. , et al (2004) Laser printing of pluripotent embryonal carcinoma cells. Tissue Eng 10: 483–491. [DOI] [PubMed] [Google Scholar]
- Rinke, C. , Schwientek, P. , Sczyrba, A. , Ivanova, N.N. , Anderson, I.J. , Cheng, J.F. , et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Robert, B. (2009) Resonance Raman spectroscopy. Photosynth Res 101: 147–155. [DOI] [PubMed] [Google Scholar]
- Sabine, C.L. , Feely, R.A. , Gruber, N. , Key, R.M. , Lee, K. , Bullister, J.L. , et al (2004) The oceanic sink for anthropogenic CO2 . Science 305: 367–371. [DOI] [PubMed] [Google Scholar]
- Scanlan, D.J. , Ostrowski, M. , Mazard, S. , Dufresne, A. , Garczarek, L. , Hess, W.R. , et al (2009) Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, H. , and Baranska, M. (2007) Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib Spectrosc 43: 13–25. [Google Scholar]
- Shahina, M. , Hameed, A. , Lin, S.‐Y. , Lai, W.‐A. , Liu, Y.‐C. , Hsu, Y.‐H. , and Young, C.‐C. (2013) Luteibaculum oceani gen. nov., sp. nov., a carotenoid‐producing, lipolytic bacterium isolated from surface seawater, and emended description of the genus Owenweeksia Lau et al. 2005. Int J Syst Evol Microbiol 63: 4765–4770. [DOI] [PubMed] [Google Scholar]
- Shindo, K. , and Misawa, N. (2014) New and rare carotenoids isolated from marine bacteria and their antioxidant activities. Mar Drugs 12: 1690–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , Kaster, A.K. , Vollmers, J. , Song, Y. , Davison, P.A. , Frentrup, M. , et al (2017) Single‐cell genomics based on Raman sorting reveals novel carotenoid‐containing bacteria in the Red Sea. Microb Biotechnol 10: 125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell, S.M. , Wilhelm, L.J. , Norbeck, A.D. , Lipton, M.S. , Nicora, C.D. , Barofsky, D.F. , et al (2009) Transport functions dominate the SAR11 metaproteome at low‐nutrient extremes in the Sargasso Sea. ISME J 3: 93–105. [DOI] [PubMed] [Google Scholar]
- Steiger, S. , Perez‐Fons, L. , Cutting, S.M. , Fraser, P.D. , and Sandmann, G. (2015) Annotation and functional assignment of the genes for the C30 carotenoid pathways from the genomes of two bacteria: Bacillus indicus and Bacillus firmus . Microbiology 161: 194–202. [DOI] [PubMed] [Google Scholar]
- Stepanauskas, R. , Fergusson, E.A. , Brown, J. , Poulton, N.J. , Tupper, B. , Labonte, J.M. , et al (2017) Improved genome recovery and integrated cell‐size analyses of individual uncultured microbial cells and viral particles. Nat Commun 8: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y.F. , Wang, Y. , Huang, S. , Zhu, P.F. , Huang, W.E. , Ling, J.Q. , and Xu, J. (2017) Metabolic‐activity‐based assessment of antimicrobial effects by D2O‐labeled single‐cell Raman microspectroscopy. Anal Chem 89: 4108–4115. [DOI] [PubMed] [Google Scholar]
- Tripp, H.J. , Bench, S.R. , Turk, K.A. , Foster, R.A. , Desany, B.A. , Niazi, F. , et al (2010) Metabolic streamlining in an open‐ocean nitrogen‐fixing cyanobacterium. Nature 464: 90–94. [DOI] [PubMed] [Google Scholar]
- Trujillo, M.E. (2016) Actinobacteria. eLS. John Wiley & Sons, Ltd: Chichester.
- Venter, J.C. , Remington, K. , Heidelberg, J.F. , Halpern, A.L. , Rusch, D. , Eisen, J.A. , et al (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74. [DOI] [PubMed] [Google Scholar]
- Von Lintig, J. , and Vogt, K. (2000) Filling the gap in vitamin A research – molecular identification of an enzyme cleaving beta‐carotene to retinal. J Bio Chem 275: 11915–11920. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Ren, L. , Su, Y. , Ji, Y. , Liu, Y. , Li, C. , et al (2017) Raman‐Activated Droplet Sorting (RADS) for label‐free high‐throughput screening of microalgal single‐cells. Anal Chem 89: 12569–12577. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Huang, W.E. , Cui, L. , and Wagner, M. (2016) Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr Opin Biotechnol 41: 34–42. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Ji, Y. , Wharfe, E.S. , Meadows, R.S. , March, P. , Goodacre, R. , et al (2013) Raman activated cell ejection for isolation of single cells. Anal Chem 85: 10697–10701. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Song, Y.Z. , Tao, Y.F. , Muhamadali, H. , Goodacre, R. , Zhou, N.Y. , et al (2016) Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level. Anal Chem 88: 9443–9450. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Zhou, Y. , and Li, D. (2016) The effect of gradient cooling on bloom decline in Microcystis (Cyanophyceae, Chroococcales). Phycologia 55: 109–117. [Google Scholar]
- Weller, R. , Bateson, M.M. , Heimbuch, B.K. , Kopczynski, E.D. , and Ward, D.M. (1992) Uncultivated cyanobacteria, Chloroflexus‐like inhabitants, and spirochete‐like inhabitants of a hot spring microbial mat. Appl Environ Microbiol 58: 3964–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman, W.B. , Coleman, D.C. , and Wiebe, W.J. (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Ma, B. , Su, X. , Huang, S. , Xu, X. , Zhou, X. , et al (2017) Emerging trends for microbiome analysis: from single‐cell functional imaging to microbiome big data. Engineering 3: 66–70. [Google Scholar]
- Yao, G. , Yu, J. , Hou, Q. , Hui, W. , Liu, W. , Kwok, L.‐Y. , et al (2017) A perspective study of koumiss microbiome by metagenomics analysis based on single‐cell amplification technique. Front Microbiol 8: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.B. , Blainey, P.C. , Schulz, F. , Woyke, T. , Horowitz, M.A. , and Quake, S.R. (2017) Microfluidic‐based mini‐metagenomics enables discovery of novel microbial lineages from complex environmental samples. Elife 6: e26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr, J.P. , Bench, S.R. , Carter, B.J. , Hewson, I. , Niazi, F. , Shi, T. , et al (2008) Globally distributed uncultivated oceanic N2‐fixing cyanobacteria lack oxygenic photosystem II. Science 322: 1110–1112. [DOI] [PubMed] [Google Scholar]
- Zhang, D.Y. , Berry, J.P. , Zhu, D. , Wang, Y. , Chen, Y. , Jiang, B. , et al (2015) Magnetic nanoparticle‐mediated isolation of functional bacteria in a complex microbial community. ISME J 9: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Ren, L. , Zhang, X. , Shan, Y. , Wang, Y. , Ji, Y. , et al (2015) Raman‐activated cell sorting based on dielectrophoretic single‐cell trap and release. Anal Chem 87: 2282–2289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site
Table S1. Basic properties of the seawater sampled from Yellow Sea of China.
Table S2. Numbers of 16S rRNA sequencing reads for microbial diversity analysis of each sample.
Table S3. Primers used in 16S rRNA PCR amplification.
Table S4. Sequencing and assembly statistics for the functional mini‐metagenomes and shotgun metagenomes.
Table S5. The statistics of the predicted genes in mini‐metagenome sequencing in this study.
Table S6. The KEGG pathways related to key metabolism pathways for gene annotation and their corresponding gene counts.
Table S7. Protein similarities in three KEGG modules (beta‐carotene biosynthesis, reductive pentose phosphate cycle and reductive citrate cycle) between mini‐metagenomic data and shotgun metagenomic data for Chroococcales and Pelagibacterales.
Fig. S1. Illustration of the ‘all‐in‐one’ device assembly.
A. Transparent ejection slide.
B. Sampling chip.
C. Cell collection chip.
Fig. S2. Geographic location of sampling site in Yellow Sea.
The label ‘L 01’ represents the location position of sampling site.
Fig. S3. Scheme of Raman identification and sorting of CO2‐fixing microbes with ‘all‐in‐one’ integrated device.
A. Acquisition of single cell Raman spectra and CO2‐fixing microbes identification.
B. Sorting cells of interest by laser ejection.
C. Shifts of the carotenoid Raman bands in single cell Raman spectra of Synechococcus spp. PCC7002, indicating 13C incorporation into the cell when cells were incubated with 13C NaHCO3.
Fig. S4. Raman spectra of carotenoids containing cells in the seawater which were incubated in closed bottles at room temperature at different times.
A. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 0 days.
B. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 3 days.
C. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 5 days.
D. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 7 days.
E. The average Raman spectra of cells treated with 13C NaHCO3 and 12C NaHCO3, respectively, at time t = 10 days.
Fig. S5. Alpha diversity comparisons across four different treatments of the seawater: red bars – 12C NaHCO3 amended sample (12C); blue bars – 13C NaHCO3 amended sample (13C); green bars – control sample without NaHCO3 (C_free); purple bars – original seawater sample control (primary).
A. Box plot showing the variation of observed OTUs.
B. Box plot showing the variation of Chao1 index.
C. Box plot showing the variation of Simpson index.
D. Box plot showing the variation of Shannon index.
Fig. S6. Agarose gel images of the multiple displacement amplifications (MDAs) and 16S rRNA gene validation processes.
A. Agarose gel image of the DNA products after MDAs showing high molecular weight DNA.
B. Agarose gel image of the PCR products of 16S rRNA gene from isolated single marine cells.
Lane M, DNA ladder; Lane N, negative control for PCR; Lane N1, negative control for ddH2O and REPLI‐g sc Master Mix (contains REPLI‐g sc Reaction buffer and REPLI‐g sc DNA Polymerase); Lane N2, negative control for ddH2O, REPLI‐g sc Master Mix, Stop solution and lysis buffer (contains buffer DLB and DTT); Lanes 1–5, 7 and 8, 30 ejected target cells in each sample; Lane 6, thirty random ejected cells; Lane 9, no cells were ejected; Lane P, Lane P1, positive control for PCR.
Fig. S7. Heat tree visualization of the structure of marine bacterial community based on 16S rDNA sequencing.
In the heat tree, size and color of nodes and edges are correlated with the abundance of organisms in the microbial community.
Fig. S8. GC distributions of metagenomic reads which were mapped to the binned genomes of Synechococcus spp. (A) and Pelagibacter spp. (B) from shotgun metagenomes.
The two curves were smooth and normally distributed, which is consistent with the absence of significant contamination. The central peak in each curve (red) was close to the average GC content of the binned draft genomes and also to the sequenced reference genomes of identical taxonomic classification.
Fig. S9. Reconstructed ‘carotenoid biosynthesis’ pathway by metagenomics‐aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S10. Reconstructed ‘carbon fixation in photosynthetic organisms’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S11. Reconstructed ‘carbon fixation pathways in prokaryotes’ by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S12. Reconstructed ‘photosynthesis’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S13. Reconstructed ‘photosynthesis‐antenna proteins’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S14. Reconstructed ‘porphyrin and chlorophyll metabolism’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea.
The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
Fig. S15. Reconstructed ‘terpenoid backbone biosynthesis’ pathway by metagenomics aided RACE (A) and mini‐metagenome (B) in Yellow sea. The known pathways were obtained from the KEGG database. Green text represents proteins found only in Synechococcus spp.; yellow text represents proteins found only in Pelagibacter spp.; and red text represents proteins found in both.
File S1. List of carbon metabolism‐related genes and their annotations in the mini‐metagenomic and the shotgun metagenomic data.
File S2. Identified functional genes in metabolic pathways from the mini‐metagenome data and their corresponding genes in the shotgun metagenomic data.
File S3. Identified proteorhodopsin (PR) genes from metagenomic bins of Pelagibacter spp.


