Abstract
Staphylococcus aureus is one of the first and most prevalent pathogens cultured from the airways of cystic fibrosis (CF) patients, which can persist there for extended periods. Airway infections in CF patients are characterized by a strong inflammatory response of highly recruited neutrophils. One killing mechanism of neutrophils is the formation of neutrophil extracellular traps (NETs), which capture and eradicate bacteria by extracellular fibers of neutrophil chromatin decorated with antimicrobial granule proteins. S. aureus secretes nuclease, which can degrade NETs. We hypothesized, that S. aureus adapts to the airways of CF patients during persistent infection by escaping from NET-mediated killing via an increase of nuclease activity. Sputum samples of CF patients with chronic S. aureus infection were visualized by confocal microscopy after immuno-fluorescence staining for NET-specific markers, S. aureus bacteria and overall DNA structures. Nuclease activity was analyzed in sequential isogenic long persisting S. aureus isolates, as confirmed by whole genome sequencing, from an individual CF patient using a FRET-based nuclease activity assay. Additionally, some of these isolates were selected and analyzed by qRT-PCR to determine the expression of nuc1 and regulators of interest. NET-killing assays were performed with clinical S. aureus isolates to evaluate killing and bacterial survival depending on nuclease activity. To confirm the role of nuclease during NET-mediated killing, a clinical isolate with low nuclease activity was transformed with a nuclease expression vector (pCM28nuc). Furthermore, two sputa from an individual CF patient were subjected to RNA-sequence analysis to evaluate the activity of nuclease in vivo. In sputa, S. aureus was associated to extracellular DNA structures. Nuclease activity in clinical S. aureus isolates increased in a time-and phenotype-dependent manner. In the clinical isolates, the expression of nuc1 was inversely correlated to the activity of agr and was independent of saeS. NET-mediated killing was significantly higher in S. aureus isolates with low compared to isolates with high nuclease activity. Importantly, transformation of the clinical isolate with low nuclease activity with pCM28nuc conferred protection against NET-mediated killing confirming the beneficial role of nuclease for protection against NETs. Also, nuclease expression in in vivo sputa was high, which underlines the important role of nuclease within the highly inflamed CF airways. In conclusion, our data show that S. aureus adapts to the neutrophil-rich environment of CF airways with increasing nuclease expression most likely to avoid NET-killing during long-term persistence.
Keywords: Cystic fibrosis, Staphylococcus aureus, neutrophil extracellular traps, nuclease, adaptation, NET-mediated killing
Introduction
Cystic fibrosis (CF) is an autosomal recessive disease with mutations in the CF transmembrane conductance regulator (CFTR) gene causing a life-limiting multisystemic disease (1). Due to CFTR mutations, a dehydrated thickened airway surface fluid impairs mucociliary clearance and leads to chronic recurrent bacterial airway infections, which result in the decline of lung function and a reduced life expectancy (1, 2). Staphylococcus aureus is one of the most common bacterial pathogens in young CF patients that can persist for several years thereby causing high inflammatory responses in CF patient airways (3–5).
One of the hallmarks of CF lung disease is an exaggerated airway inflammation caused by excessive recruitment of dysfunctional neutrophils and accumulation of pro-inflammatory agents, which in turn fail to eradicate bacteria (6). Within the airways, neutrophils try to kill pathogens by different killing mechanisms such as phagocytosis with the release of oxidants and degrading enzymes during degranulation, and the formation of neutrophil extracellular traps (NETs) (7), which were previously described to be abnormal in CF (8, 9). In detail, bacterial digestion in the neutrophilic phagolysosome in CF is reduced by the lack of membranous chloride transport due to CFTR mutations causing defective intraphagolysosomal HOCL production and reduced chlorination of bacterial proteins (9). Moreover, cytosolic pH acidifies and leads to a massive release of antimicrobial enzymes from granules such as myeloperoxidase and neutrophil elastase and lactoferrin (10). The high concentration of neutrophilic defense peptides contributes additionally to the destruction of airway and lung tissue in CF (11, 12). It has been shown, that in the context of CF lung disease, NET formation by neutrophils is enhanced (13). Besides antimicrobial components of the neutrophil granules, NETs consist of extracellular DNA fibers released by chromatin decondensation and subsequent rupture of the nuclear membrane to capture and kill various pathogens (7, 11). Recently, the presence of NETs within CF airways has been shown and has been associated with poor pulmonary function assumingly driven by NET-mediated inflammation and increased amounts of thickened mucus (14, 15). S. aureus is not only a potent inducer of NETs (7, 16), but has also the potential to degrade NETs by the secretion of nuclease (17). We hypothesized, that in the airways of CF patients S. aureus will adapt to NET-mediated killing by increasing nuclease activity in long-persisting isolates. First, we used fresh sputa from patients with chronic S. aureus airway infection to visualize NETs by immuno-fluorescence and confocal microscopy. Next, we determined nuclease activity of sequential and isogenic clinical S. aureus CF isolates by DNase agar plates and a FRET-based assay to evaluate nuclease activity. Since the expression of nuclease confers escape from NET-mediated killing to S. aureus, NET-killing assays of isolates with different nuclease activity were performed. To confirm the specific effect of nuclease regarding NET-mediated killing, a clinical S. aureus isolate with low nuclease activity was transformed with a plasmid that expresses wild-type nuclease, and tested in the NET-killing assay. To verify the role of nuclease in vivo, two independent sputa of an individual CF patient were used for RNA sequence analysis.
Our data revealed, that (i) NET-structures were visible in CF sputa and that S. aureus was in close proximity to NETs, (ii) nuclease activity of isogenic sequential isolates of one individual patient increased significantly during persistence, (iii) isolates with high nuclease activity were protected against NET-mediated killing, (iv) protection against NET-mediated killing was caused by higher nuclease activity, and that (v) nuclease was highly expressed in sputa of an individual CF patient.
Results
Extracellular DNA Entraps S. aureus Bacteria in CF Sputum Samples
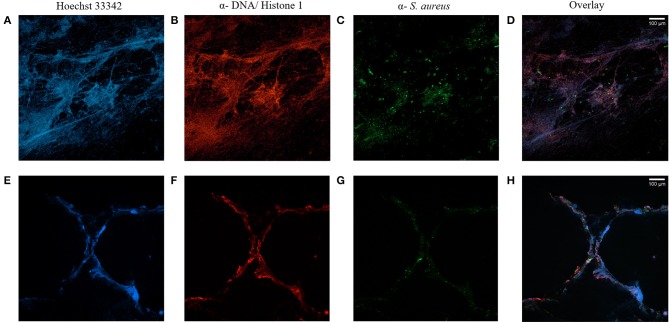
To examine, if NET- formation occurs in the airways of CF patients with chronic S. aureus infection, confocal microscopy of stained sputum was performed and confirmed the presence of high amounts of NETs (Figures 1A,B,E,F). Furthermore, fluorescence microscopy revealed S. aureus being mostly entangled in extracellular DNA structures (Figures 1C,D,G,H), while there were also bacteria visible in an environment without NETs.
Figure 1.
CF sputum samples visualized by immunofluorescence staining and confocal microscopy. Staining of two sputa (A–D and E–H) of two different CF patients with chronic S. aureus infection. Nuclei and DNA structures (blue, A,E) and DNA-histone-1-complexes (red, B,F) are visible, whereas antibody-mediated staining specifically identifies S. aureus (green, C,G). Microscopy reveals NET-structures with S. aureus attached to it, but also with S. aureus without NET-binding (D,H). Subsequent changes of color balance for color intensification were performed with ImageJ.
Nuclease Activity of S. aureus Correlates With Long-Term Persistence
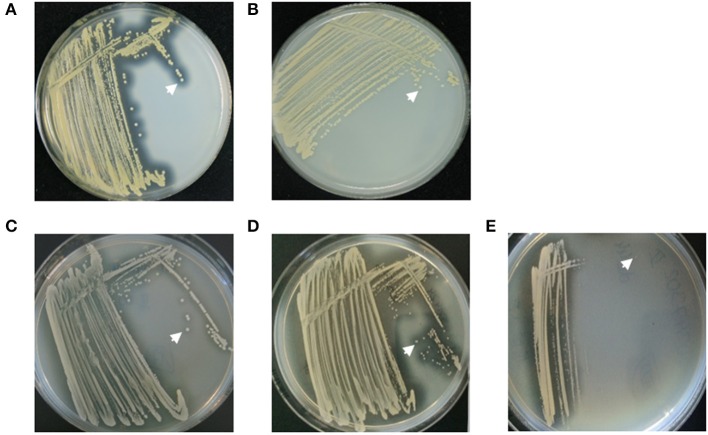
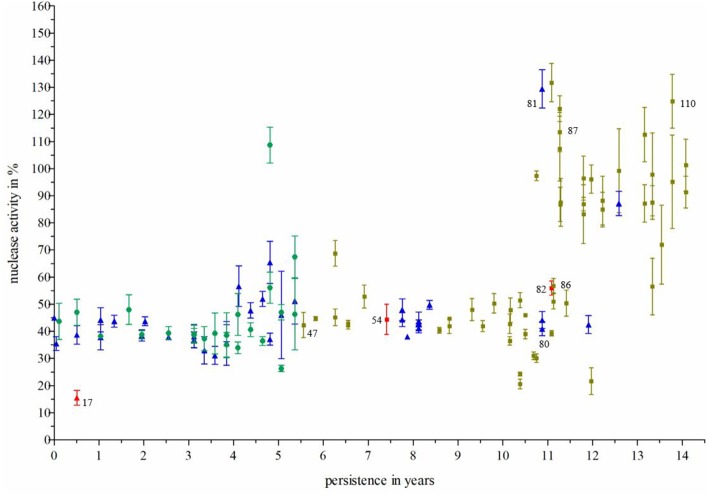
To assess adaptation of S. aureus during long-term persistence regarding nuclease activity, 111 S. aureus isolates, which were recovered during 14 years of persistence from the airways of an individual CF patient (CF patient 1, Table 1), were further investigated. These isolates belonged predominantly to spa-type t617, showing that this patient was infected with one dominant S. aureus clone throughout persistence. Clonality of isolates was also confirmed by whole genome sequencing of 7 selected isolates (Figure 2). An initial approach for the categorization of nuclease activity of these S. aureus isolates was conducted on DNase agar plates (Figures 3A–E) (18) revealing an increase of nuclease activity in late compared to early S. aureus isolates (Figures 3C,D, Figure Supplementary 1). Moreover, the capability to degrade DNA was also seen to be phenotype-related in S. aureus. Strains with a normal phenotype showed larger clearing zones around colonies compared to small colony variants (SCVs), where no DNA degradation was detected around single colonies (Figure 3D). To quantify nuclease activity, all 111 S. aureus isolates were subjected to the nuclease FRET assay (19) showing a significant increase of nuclease activity in late isolates after 11 years of persistence (Figure 4). These results were confirmed by qRT-PCR analysis, which demonstrated a significant increase of nuc1 expression after 11 years of S. aureus persistence (Figure 5). Also, nuc2 expression revealed a significant increase after 7 years of persistence (Figure Supplementary 2). However, nuc2 expression (Figure Supplementary 2) was much lower than nuc1 expression (Figure 5).
Table 1.
Clinical S. aureus isolates of CF patients used in this study.
| Patient | Year of recovery | Sample site | spa-type | ||
|---|---|---|---|---|---|
| Nose | Throat | Sputum | |||
| 1 | 2001 | 1,3 | 2,4,5,17 | t617 | |
| 1 | 2002 | 6 | t499 | ||
| 1 | 2002 | 7 | 8,9,16 | t617 | |
| 1 | 2003 | 11 | t499 | ||
| 1 | 2003 | 10,12,15 | 13,14 | t617 | |
| 1 | 2004 | 19,20,22,25 | 18,21,23,24 | t617 | |
| 1 | 2005 | 28–31,33 | 26,27,32 | t617 | |
| 1 | 2005 | 34 | t930 | ||
| 1 | 2006 | 35,37,38,41,42,44,45 | 36,39,40,43,46 | 47 | t617 |
| 1 | 2007 | 49 | t930 | ||
| 1 | 2007 | 48,50–52 | t617 | ||
| 1 | 2008 | 53 | t930 | ||
| 1 | 2008 | 54 | t034 | ||
| 1 | 2009 | 55–61 | 62 | t617 | |
| 1 | 2010 | 63–66 | t617 | ||
| 1 | 2011 | 67–70,72,74,75 | t617 | ||
| 1 | 2011 | 71 | t002 | ||
| 1 | 2011 | 73 | t230 | ||
| 1 | 2012 | 79–81 | 76–78,82–92 | t617 | |
| 1 | 2013 | 101 | 93–95, 97,99,100 |
t617 | |
| 1 | 2013 | 96 | t930 | ||
| 1 | 2013 | 98 | t121 | ||
| 1 | 2014 | 102 | t1459 | ||
| 1 | 2014 | 103–108 | t617 | ||
| 1 | 2015 | 109–111 | t617 | ||
| 2 | 2016 | 881*,912* | t034 | ||
Isolates are numbered in chronological order of recovery. The year of recovery is presented, as well as the sample site of which isolates were recovered from the patient and the identified spa-type. NET killing experiments were performed with isolates from two different CF patients. Isolate no. 17 and no. 81 with low and high nuclease activity were recovered from one individual CF patient (underlined). Isolate no. 881 and 912 (marked with *) with low and high nuclease activity were recovered from another CF patient, respectively (Figure Supplementary 3). Isolate no. 17 was further transformed with the pCM28nuc plasmid and underwent NET killing analysis, whereby no. 81 served as the comparison strain with high nuclease activity.
Figure 2.

UPGMA-tree based on the cgMLST allelic profiles of 7 clinical S. aureus isolates. The phylogenetic tree demonstrates the relationship of 7 S. aureus isolates, which were collected over 14 years. The tree was drawn to scale with branches given in absolute alleles distances.
Figure 3.
Nuclease activity in different S. aureus strains analyzed by DNase agar plates. (A) S. aureus strain AH1263, positive control, showed large clearing zones (white arrow) around colonies. (B) S. aureus strain AH1680 (Δnuc), negative control, presented without clearing zones (white arrow). (C) The early CF S. aureus isolate showed small clearing zones around colonies (white arrow), whereas (D) the late isolate revealed large clearing zones around colonies (white arrow). (E) S. aureus small colony variants (SCVs) did not reveal clearing zones around single colonies (white arrow).
Figure 4.
Nuclease activity of S. aureus isolates during persistence. Nuclease activity was measured by the nuclease FRET assay (19). S. aureus isolates (n = 111) were recovered from the nose (green circles), throat (blue rectangles) and sputum (ochre squares) from one individual CF patient during long-term persistence of 14 years. There was a significant increase of nuclease activity after 11 years. Three clinical S. aureus isolates (17, 54, and 82; marked red) were selected for nuc expression analysis via qRT-PCR. Two isolates were further analyzed in NET-killing assays and used for transformation experiments representing a strain pair with low nuclease activity (no. 17) and high nuclease activity (no. 81), respectively. Technical replicates in the FRET assay: n = 3, biological replicates n = 3. For the calculation of significance, (i) isolates were grouped into nose, throat, and sputum samples and compared by the one- way analysis of variance (ANOVA) and Bonferroni's Post-Test, resulting in: nose vs. sputum **p ≤ 0.01; nose vs. throat had no significance, throat vs. sputum ***p ≤ 0.001; (ii) regardless of sample site, isolates were grouped into early (0–7.039 years of persistence) and late (7.039–14.0.78 years of persistence). Both groups were compared using a two-tailed, unpaired student's t-test (result: ***p ≤ 0.001).
Figure 5.
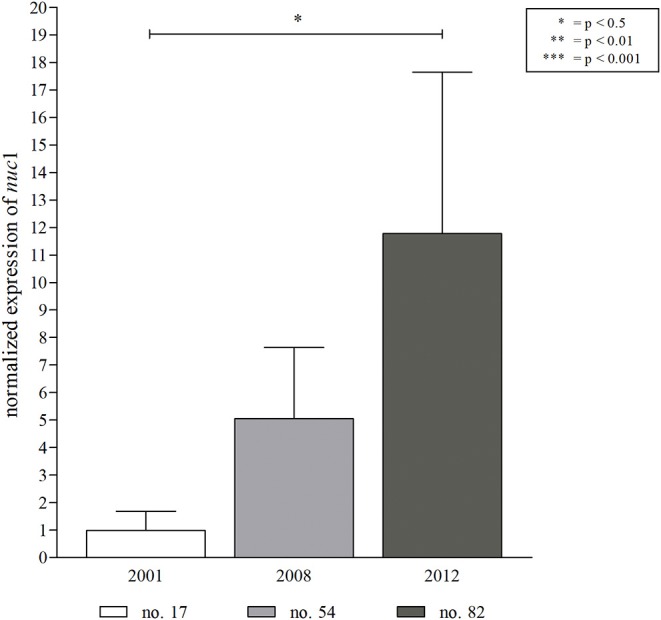
Expression of S. aureus nuc1 in an early, intermediate and late S. aureus isolate from patient 1. For the assessment of nuc expression, isolates (Figure 4, in red) were grown in BHI medium until mid-logarithmic growth phase before RNA extraction. Expression of nuc1 increased significantly over time of S. aureus persistence. The obtained results are in line with the nuclease FRET assay detecting low nuclease activity for isolate no. 17 (2001, 15.9%), an intermediate activity for no. 54 (2008, 45.5%), and a high nuclease activity for the late isolate no. 82 (2012, 56.84%). Statistical analysis: two-tailed, unpaired student's t-test, error bars represent SD. Technical replicates n = 2, biological replicates n = 3.
Inverse Expression Pattern of Nuclease and agr
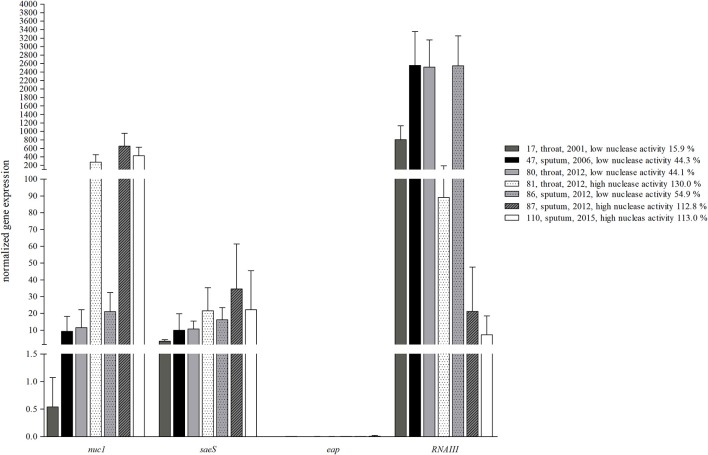
Seven clinical S. aureus strains (Figure 4: 17, 47, 80, 81, 86, 87, and 110) from one individual CF patient were subjected to qRT-PCR to analyze expression of nuc1 in relation to potential regulators of S. aureus nuclease (Figure 6). The results of nuc1 expression were in accordance to the results of nuclease activity by the nuclease FRET assay. Since S. aureus nuclease was shown to be SaeRS-regulated (20), transcription of saeS was determined for clinical isolates with low and high nuclease activity. Surprisingly, no alterations in saeS expression were observed for S. aureus isolates dependent of either low or high nuclease expression (Figure 6). Also, eap, a protein, which has been shown to be under the control of saeRS (21), was only very low expressed without any association to nuclease expression in all 7 clinical isolates. Interestingly, RNAIII, the effector molecule of the major virulence regulator agr (22), was higher expressed in S. aureus isolates with low nuc1 expression, whereas decreased RNAIII expression was observed in isolates with high nuclease expression (Figure 6) indicating an inhibiting role of agr in nuclease expression in the clinical isolates. Interestingly, whole genome sequencing revealed two subsequent non-synonymous SNPs in agrA of those S. aureus isolates with decreased RNAIII transcription and increased nuclease activity (Table 2).
Figure 6.
Inverse expression pattern of nuclease and agr. Sequential clinical S. aureus isolates from one individual CF patient were grown in BHI until mid-logarithmic growth phase to determine nuc1, the potential nuc regulator saeRS (represented by saeS), eap as another saeRS regulated virulence factor and RNAIII as the effector gene of the agr regulon. Nuc expression was independent of saeS, but was inversely correlated with agr, while eap was only very low expressed. Isolates with low nuclease activity (17, 47, and 86) revealed high RNAIII expression, whereas isolates with high nuclease activity (81, 87, and 110) low RNAIII expression, respectively. Technical replicates n = 3, biological replicates n = 4.
Table 2.
Non-synonymous SNPs in agrA.
| Gene | Target | Rel. position | NC_002951 | Variant AA | 17, 47 | Isolates with variants | |
|---|---|---|---|---|---|---|---|
| 80, 86 | 81, 87, 110 | ||||||
| agrA | SACOL2026 | 409 | G | D → Y | G | T | T |
| agrA | SACOL2026 | 520 | C | H → Y | C | C | T |
Two subsequent non-synonymous SNPs were discovered in agrA of the clinical S. aureus isolates. Nuclease and RNAIII activity inversely correlated with the presence of both mutations.
Nuclease Activity Differs in CF Patients
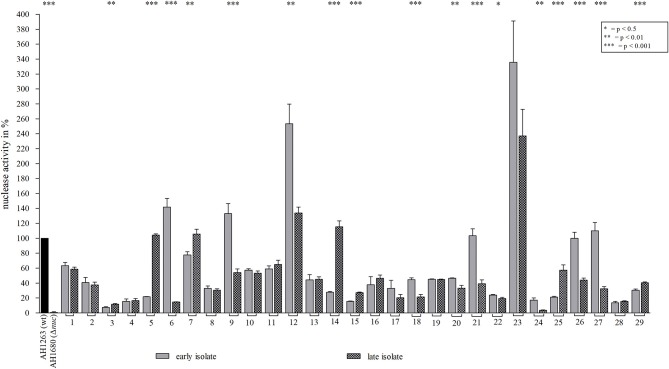
To assess, if an increase in nuclease activity is a common adaptation pattern in other CF patients with long-term S. aureus persistence, 29 S. aureus strain pairs (early and late isolate) of different CF patients were analyzed by the nuclease FRET assay. Seven of these 29 late S. aureus isolates revealed a significant increase of nuclease activity (Figure 7), whereas in 10 late isolates a significant decrease of nuclease activity and in 11 late strains, no change in nuclease activity was observed (Figure 7).
Figure 7.
Nuclease activity of different early/late S. aureus strain pairs. Nuclease activity of 29 S. aureus strain pairs (early, late) from different CF patients (23) were analyzed by the nuclease FRET assay. Regarding nuclease activity in late isolates, an increase was observed in 7 late strains (strain pairs 3, 5, 7, 14, 15, 25, and 29), a decrease in 10 (strain pairs 6, 9, 12, 18, 20, 21, 22, 24, 26, and 27) and an unchanged activity in 12 late strains. S. aureus strains AH1263 and AH1680 (Δnuc) served as positive and negative control, respectively. Statistical comparisons: two-tailed, unpaired student's t-test, error bars represent SD. Technical replicates n = 3, biological replicates n = 3.
Higher Nuclease Activity Facilitates Increased Survival of S. aureus Interacting With NETs
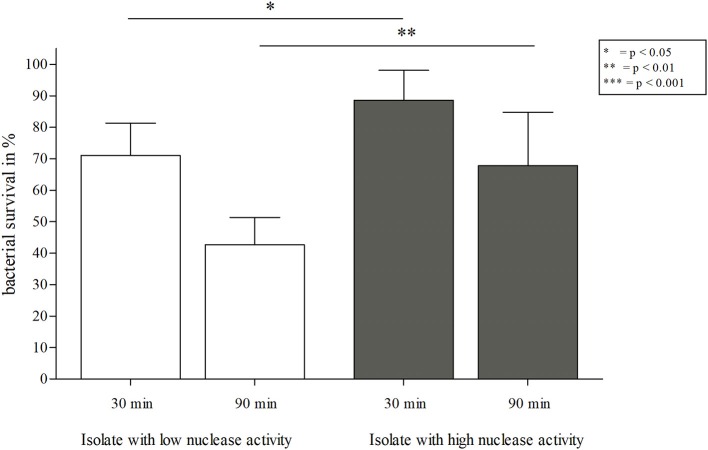
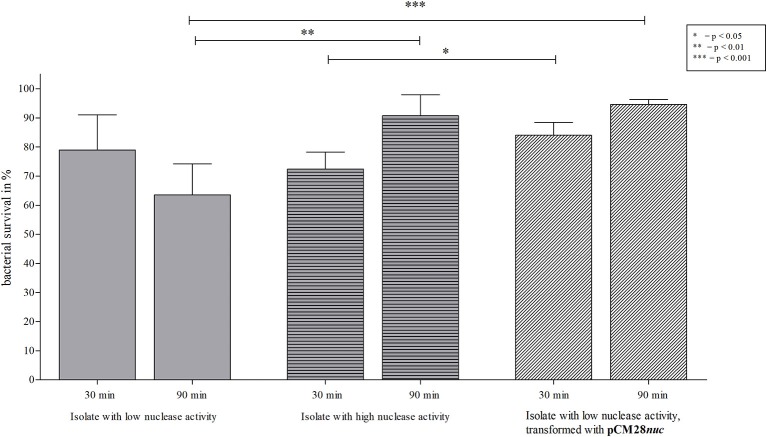
Since secretion of nuclease facilitates evasion from NETs (17), we assessed survival of S. aureus isolates with high and low nuclease activity in interaction with NETs. After 90 min, the S. aureus isolate with low nuclease activity (Table 1, no. 17 and 881) was significantly more affected by NET-mediated killing than the isolate with high nuclease activity (Table 1, no. 81 and 912; Figure 8, Figure Supplementary 3). In detail, the survival of the isolate with low nuclease activity was 54.7% in comparison to 74.9% of survival of the isolate with high nuclease activity (Figure 8). To confirm the role of nuclease in the context of NET-mediated killing on bacterial survival, the S. aureus isolate with low nuclease activity (Table 1, no. 17) was transformed with the nuclease expressing plasmid pCM28nuc (Table 3), which resulted in increased nuclease activity for the transformed strain (Figure 9). Similar to our previous results (Figure 8, Figure Supplementary 3), the CF isolate with low nuclease activity was killed to a significant higher extent than the corresponding late isolate with high nuclease activity after 90 min of NET interaction (Figure 10). In line with these results, the NET-killing capacity of the transformed strain was comparable to the late isolate with high nuclease activity (Figure 10). Interestingly, after 90 min of co-incubation with NETs, the bacterial survival of the transformed isolate with former low nuclease activity was higher than after 30 min of co- incubation, indicating that the high activity of nuclease conferred protection against NET-mediated killing and even allowed replication of bacteria as also observed for the late isolate in Figure 10.
Figure 8.
NET-killing assay with S. aureus isolates from patient 1 with low/high nuclease activities. Survival rates (in %) after 30 and 90 min of incubation with NETs are depicted. A significant decrease of bacterial survival was observed for the (early) isolate with low nuclease activity (Figure 4, isolate 17) after 30 and 90 min of incubation with NETs in comparison to the corresponding (late) isolate with high nuclease activity (Figure 4, isolate 81). Statistical comparison of both S. aureus strains depending on incubation time: two-tailed, unpaired student's t-test, error bars represent SD. Statistical comparison of single S. aureus strains: two-tailed, paired student's t-test. Technical replicates n = 2, biological replicates n = 6.
Table 3.
S. aureus reference strains, plasmid and plasmid transformed strain used in this study.
| Strain or plasmid | Description | References |
|---|---|---|
| AH1263 | USA300 CA-MRSA ErmS (LAC), wild type strain | (24) |
| AH1680 | AH1263 nuc:LtrB, Δnuc mutant of wild type AH1263 | (25) |
| pCM28nuc | nuc-complementing vector with chloramphenicol resistance | (25) |
| 17 [pCM28nuc] | S. aureus strain with low nuclease activity transformed with pCM28nuc | This work |
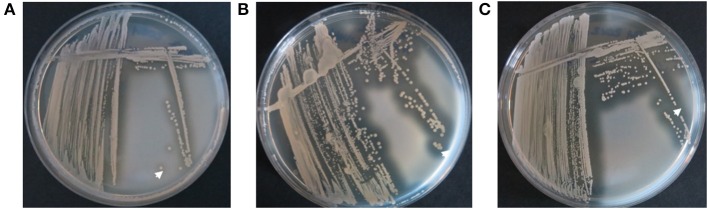
Figure 9.
Analysis of nuclease activity on DNase agar plates after the transformation of the early S. aureus isolate with low nuclease activity of patient 1 with the plasmid pCM28nuc. (A) The early S. aureus isolate with low nuclease activity (Figure 4, isolate17) shows minimal clearance zones (white arrow) around colonies. (B) The pCM28nuc-transformed early S. aureus isolate exhibits large clearance zones, (C) similar to the corresponding late S. aureus isolate with high nuclease activity (Figure 4, isolate 81).
Figure 10.
NET-killing assay with an early S. aureus isolate with low nuclease activity of after transformation with pCM28nuc. Survival rates (in %) after 30 and 90 min of incubation with NETs are shown. A significant decrease of bacterial survival after 90 min is apparent for the isolate with low nuclease activity (Figure 4, isolate 17), compared to both isolates with enhanced nuclease activity. After 30 min, the survival rate for the isolate with high nuclease activity (Figure 4, isolate 81) is significantly lower than for the transformed strain. After 90 min, no difference in survival was observed for both S. aureus strains with high nuclease activity, but enhanced survival was shown for the two strains with high nuclease activity compared to the strain with low nuclease activity. Statistical comparison of S. aureus strains depending on incubation time: two-tailed, unpaired student's t-test, error bars represent SD. The comparison of single S. aureus strain pairs revealed no statistical differences (two-tailed, paired student's test). Technical replicates n = 2 and biological replicates n = 7 were performed.
Nuclease Expression in vivo in Sputa
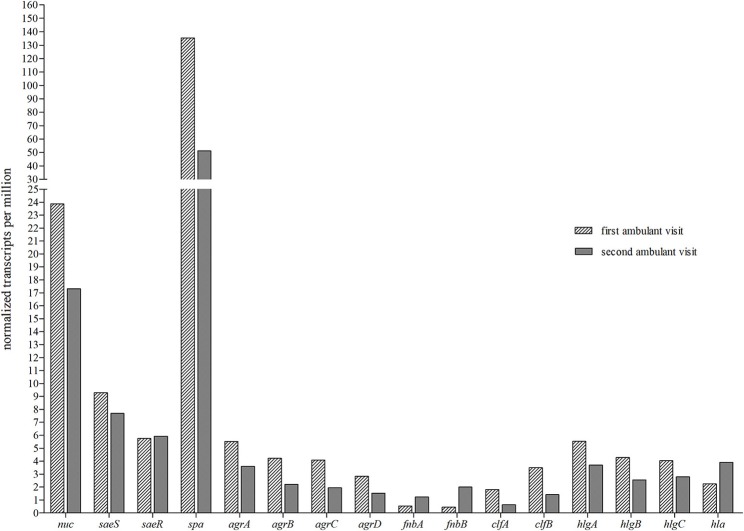
To determine the role of nuclease expression in vivo, we collected two different sputa from one individual CF patient, who was persistently infected by S. aureus, and subjected this sputum to RNA-Seq analysis (Figure 11). The comparison of normalized transcription of nuc and other important virulence regulators and virulence factors against housekeeping (hk) genes revealed a high relative expression of nuc in both sputa, which was only surpassed by the expression of protein A (spa). The third highest transcript levels were obtained for saeS, which has been shown to be a regulator of nuc (20), while levels of adhesins (fnbA, fnB, clfA, and partially clfB) were low in both analyzed sputa (Figure 11). The obtained results of in vivo RNA-Seq analysis indicate the importance of nuclease during in vivo S. aureus airway infection.
Figure 11.
RNA-Seq analysis of in vivo expression of nuclease and other important S. aureus virulence regulators and genes in sputa from one individual CF patient. Sputum samples were collected from a chronically infected CF patient at two visits and subjected to RNA-Seq analysis. Expression levels of nuclease (nuc), virulence regulators (saeS, saeR, agrA, agrB, agrC, and agrD), protein A (spa), fibronectin-binding proteins (fnbA, fnbB), clumping factor proteins (clfA, clfB), the subunits for γ-hemolysin (hlgA, hlgB, and hlgC) and α-hemolysin (hla) are shown as normalized transcripts per million (TPM). TPM levels of presented genes were normalized against TPM values 600 of the S. aureus housekeeping genes aroE and gyrB.
Discussion
Airway infections in CF lung disease are associated with strong inflammatory responses characterized by a domination of neutrophils (6), which can use different strategies to combat invading pathogens. Recently, NET-formation, also known as NETosis, within the airways of CF patients gained interest and it has been shown that abundant NETosis and NET- related markers in CF airway fluids occur (14, 26, 27). Most of these studies evaluated NET-formation in regard to P. aeruginosa airway infection in CF patients (15, 27–29). To our knowledge, there are no data about NET-formation and S. aureus in the context of CF. Therefore, we assessed, if sputum of patients with chronic S. aureus airway infections contain NETs. Several sputa of CF patients were stained for extracellular DNA structures characteristic for NETs and for S. aureus. As shown in two sputum samples exemplarily (Figure 1), S. aureus bacteria were visible in close proximity to NET- structures, but also without relation to NETs indicating that not all S. aureus bacteria are entangled and killed by NETs in CF sputa and that some isolates may have escaped NET-mediated killing. Since secreted S. aureus nuclease facilitates escape from NETs (17), we hypothesized that nuclease activity in long-persisting isolates would differ in comparison to early isolates. Therefore, we tested nuclease activity of more than 100 sequential S. aureus isolates, which were collected during persistence in the airways of one individual CF patients during a 14-year period. Our results showed, that isolates with increased nuclease activity occurred not earlier than after 11 years of persistence and were selected during the later years of persistence (Figure 4). These results were confirmed by qRT-PCR of selected strains with differential nuclease activity (Figure 5). Isolates with increased nuclease activity were mostly isolated from sputa, while isolates with low nuclease activity were more likely isolated from nasal and throat swabs (Figure 4). The fact, that isolates with high nuclease activity were more likely isolated from sputa indicates that there is a greater chance of selection of isolates with high nuclease activity at a site where more neutrophils are present, which most likely perform NETs. However, after 11 years most S. aureus isolates displayed higher nuclease activity indicating a survival advantage and pathoadaptation of these isolates to the NET-rich airways in CF as it has been suggested also by Rahman and Gadjeva (30).
In addition, isolates with SCV phenotype revealed less nuclease activity compared to normal growing isolates. Such data are in line with our earlier results showing that SCVs are characterized by a down- regulated metabolism, which facilitate long-term persistence (31, 32). Therefore, increased nuclease activity does not seem to be of importance for long-term survival for SCVs. Moreover, since SCVs mostly occur in mixed cultures with normal S. aureus, the secreted nuclease of the normal S. aureus might protect also SCVs from NET-mediated killing. Such assumption should be assessed in further NET-killing experiments with mixed cultures of normal and S. aureus SCVs.
As we investigated early/late S. aureus isolates from 29 different CF patients, only in the late isolates of 7 CF patients nuclease activity was significantly increased. However, considering the fact that nuclease increase in the sequential isolates of the described patient occurred only after 11 years of persistence, many of the studied late isolates might not have persisted long enough in the airways in order to adapt to NET-formation. Moreover, an increase in nuclease activity especially occurred in S. aureus isolates recovered from sputa. Since not all late isolates of the tested 29 strains pairs were collected from sputa (Table Supplementary 1), this also might have influenced the results regarding the neutrophil domination in deeper CF airways, that is accompanied by a massive release of inflammatory agents (6, 33–35). Based on our findings, the observed enhancement of nuclease might be a result of a higher selective pressure for S. aureus present in lower airways compared to those that are present in nose or throat. Recently, Berends et al. showed resistance of S. aureus against NET-mediated entrapment and killing by the secretion of S. aureus nuclease (17). Therefore, we tested survival of NET-mediated killing of S. aureus isolates with high in comparison to low nuclease activity showing that S. aureus isolates with high nuclease activity were less killed by NETs compared to isolates with low nuclease activity. Such results indicate that S. aureus isolates with high nuclease activity were most likely selected in the airways of CF patients with high abundance of NET formation, which facilitate survival of S. aureus in this hostile environment. The beneficial role of high nuclease expression was confirmed by transformation of a clinical CF isolate with low nuclease with the nuclease expressing vector pCM28nuc. Survival of the transformed S. aureus strain during NET-killing was significantly increased compared to the corresponding isolate with low nuclease activity.
The fact that nuclease expression was also high in two in vivo sputum samples from one individual CF patient with persistent S. aureus airway infection as assessed by RNA-Seq analysis underscores the role of nuclease for the in vivo situation. Elucidating the transcriptome of S. aureus in the nasal niche by RNA-Seq analysis (36), the comparison of transcript levels of S. aureus recovered from CF sputum demonstrated higher expression of nuclease and the nuclease-regulating saeS component (20) compared to the nasal S. aureus isolate. The increase of saeS and nuc in CF compared to nasal isolates (36) underscores our hypothesis of S. aureus combating neutrophil-mediated eradication by the up-regulation of nuclease during adaptation to the highly inflammatory CF airways.
If this high nuclease activity has an impact on macrophage viability during NET formation, especially regarding the concerted action of nuclease and adenosine synthase A as shown recently by Thammavonsga et al. (37), is an interesting aspect, which should be investigated in future experiments.
A first insight about the molecular mechanism of the increase in nuclease activity in the clinical isolates is given by the detected SNPs within agrA, which most likely caused down-regulation of RNAIII expression in late clinical isolates. AgrA-specific mutations have been previously shown by others (38, 39) to be responsible for a change in agr activity. Surprisingly, saeRS, which has been shown by others to positively regulate nuc (20, 21, 40) had no apparent impact on nuclease activity in these clinical isolates.
In conclusion, our data underline the importance of S. aureus nuclease activity during long-term airway infection of CF patients. In sputa of patients with chronic S. aureus infection, NET formation with entrapped S. aureus was apparent. In many CF patients, pathoadaptation of long-term persisting S. aureus isolates to the airways, where abundant NET formation is observed, was accomplished via increased nuclease activity. High nuclease activity conferred a survival advantage to S. aureus during NET-killing and high nuclease expression was determined in in vivo sputa from a CF patient chronically infected by S. aureus.
Materials and Methods
Patients' and Volunteers' Specimens
The usage of sputum of CF patients and blood samples was approved by the Ethical Committee of the University Hospital Münster (no. 2018-466-f-S). Informed consent was given for blood samples by healthy volunteers and for sputa by CF patients.
Bacterial Strains and Growth Conditions
Clinical S. aureus isolates with the same or closely related spa-types (n = 111) from one individual CF patient, cultured from throat, nose, or sputum samples collected during a period of 14 years were used (Table 1). To assess adaptation regarding nuclease activity during persistence in more CF patients, S. aureus strain pairs of CF patients consisting of early and late isolates with at least 5 years of S. aureus persistence with identical or closely related spa-types were analyzed (Table Supplementary 1). These strain pairs were previously characterized in terms of virulence factor adaptation (23). All CF isolates and reference strains (Table 3) were cultivated on Columbia 5% blood agar plates (BD). Overnight cultures were grown in Brain Heart Infusion medium (BHI, Difco) at 37°C and 160 rpm. B2 medium was used for S. aureus plasmid transformation [modified after (41)]. To assess nuclease activity in qRT-PCR and NET-killing experiments, overnight cultures of S. aureus isolates in BHI were cultivated in fresh BHI medium for 4 h until mid-logarithmic growth phase starting from an OD578nm of 0.1. For the nuclease FRET assay, S. aureus was cultured in 96-well-plates under the same conditions. The pCM28nuc plasmid was maintained in S. aureus AH1773 on tryptic soy agar (TSA, Difco) supplemented with 10 μg/ml chloramphenicol (CN).
spa-Typing of Clinical CF S. aureus Isolates
Molecular typing of sequential S. aureus isolates (Table 1) recovered from airway specimens (nose, throat, sputum) of an individual CF patient were performed by spa-typing (32, 42).
Whole Genome Sequencing of Clinical CF S. aureus Isolates
The clonal relationship of 7 S. aureus strains recovered from one individual CF patient (Table 1) was determined by whole genome sequencing (WGS) and subsequent core genome multilocus sequence typing (cgMLST). Genomic DNA of S. aureus isolates was purified using the MagAttract HMW DNA kit (Qiagen) following the manufacturer's instructions with the addition of 120 U Lysostaphin (Sigma-Aldrich). Clinical isolates were sequenced using Illumina technology and Nextera XT version 2 chemistry, with a 250-bp paired-end protocol on a MiSeq sequencer (Illumina). Quality trimming of fastq files (average base quality of 30, aiming for 100-fold coverage) and de novo assembly using SKESA (PMID: 30286803) were performed with SeqSphere+ (version 6; Ridom GmbH, Münster, Germany) as described recently (43). Only genomes harboring ≥95% cgMLST targets of the S. aureus cgMLST scheme (44) passed quality control; otherwise, sequencing was repeated. For tree building using the unweighted pair group (UPGMA) method, the allelic profiles of up to 1, 861 core genome multilocus sequence typing (cgMLST) targets were used as described previously (45). To investigate molecular mechanisms associated with nuclease activity, the SeqSphere+ software was used for SNP calling within the agrA gene.
Nuclease Test on DNase Agar
CF isolates and laboratory strains were streaked on DNase agar plates (Oxoid) and incubated for 17 h at 37°C. DNase agar plates were flooded with 1 N hydrochloric acid solution and evaluated qualitatively by the size of clearing zones around bacterial colonies as a result of DNA degradation by S. aureus nuclease. S. aureus strains AH1263 (wild-type) and AH1680 (Δnuc-mutant) were used as negative and positive control, respectively (Table 3).
Nuclease FRET Assay
For the preparation of overnight cultures, single colonies of CF isolates and laboratory strains were inoculated in BHI medium in a 96-well-flat-bottomed microtiter plate with lid. AH1263 and AH1680 (Table 3) were used as negative and positive controls. The plate was incubated overnight for 16–18 h at 37°C using the TECAN Multireader. Overnight cultures were diluted to an OD578nm of 0.1 in fresh BHI medium in a new 96-well-plate and incubated for another 4 h at 37°C in the TECAN Multireader. Analysis of nuclease activity in S. aureus strains was accomplished by the use of a molecular beacon as described recently (19). In brief, for analysis of nuclease activity, bacterial supernatants were collected and applied in 1:200 dilutions together with the molecular beacon (final concentration: 0.1 μM) into a black 96-well-plate. Nuclease activity was determined as a kinetic measurement in 30 min intervals with excitation at 485/20 nm, emission at 528/20 nm under fast orbital shaking. Results were evaluated with the Gen5 best fit method. Nuclease activity was calculated in a growth-dependent manner to exclude impacts on actual nuclease activity by differing growth capacities of S. aureus CF strains in BHI medium. Growth- fitted nuclease activity was achieved by relative nuclease activity multiplied against a factor resulting from OD578nm values of the analyzed CF strain, obtained after 4 h of incubation in fresh BHI, against the OD578nm value of AH1263 (100% control).
qRT-PCR
Expression of both nucleases (nuc1, nuc2), saeS, eap, and RNAIII was assessed in the mid-logarithmic growth phase in BHI. RNA isolation, cDNA synthesis, and qRT-PCR were conducted as described (46). RNA samples were evaluated for successful DNA elimination by PCR using S. aureus gmk (guanylate monophosphate kinase) primers. The house-keeping genes aroE (Shikimate dehydrogenase NADP/H) and gyrB (DNA gyrase subunit B) served for normalization of nuclease gene expression. All primers used are described in Table 4.
Table 4.
Primers used in qRT-PCR experiments.
| Target | Sequence | References |
|---|---|---|
| aroE-fwd | 5′-CTATCCACTTGCCATCTTTTAT-3′ | (46) |
| aroE-rev | 5′-ATGGCTTTAATATCACAATTCC-3′ | |
| gmk-fwd | 5′-AAGGTGCAAAGCAAGTTAGAA-3′ | This work |
| gmk-rev | 5′-CTTTACGCGCTTCGTTAATAC-3′ | |
| gyrB-fwd | 5′-AATTGAAGCAGGCTATGTGT-3′ | (46) |
| gyrB-rev | 5′-ATAGACCATTTTGGTGTTGG-3′ | |
| nuc1-fwd | 5′-AAGCTTTAGTTCGTCAAGGC-3′ | This work |
| nuc1-rev | 5′-TGAATCAGCGTTGTCTTCGC-3′ | |
| nuc2-fwd | 5′-TGGATGGTGATACATTTATTGC-3′ | This work |
| nuc2-rev | 5′-GTTTCACCGTTTCTGGCG-3′ | |
| RNAIII-fwd | 5′-ttcactgtgtcgataatcca-3′ | (47) |
| RNAIII-rev | 5′-tgatttcaatggcacaagat-3′ | |
| eap-fwd | 5′-AAGCGTCTGCCGCAGCTA-3′ | (48) |
| eap-rev | 5′-TGCATATGGAACATGGACTTTAGAA-3′ | |
| saeS- fwd | 5′-tcgaacgccacttgagc-3′ | This work |
| saeS-rev | 5′-ctatcgacattgctattagc-3′ |
Transformation of a CF S. aureus Isolate With Low Nuclease Activity
One selected S. aureus strain (no. 17, year 2001, throat, Table 1) with low nuclease activity was transformed with the pCM28nuc plasmid, containing the major S. aureus nuclease (nuc1) sequence. For the preparation of electro-competent cells, the CF isolate was cultivated in B2 medium (41) overnight. The culture was then diluted to OD578nm of 0.5 in fresh B2 medium and incubated at 37°C, 160 rpm to OD578nm of 0.6. Bacterial growth was stopped on ice and a pellet was harvested by centrifugation (3,000 × g for 10 min at RT). The pellet was consecutively washed in decreasing volumes of 4°C cold water before 4°C cold 10% glycerol was applied. For transformation, electro-competent cells of the CF isolate were mixed with 5 μl of the purified pCM28nuc plasmid (NEB Monarch Plasmid Miniprep Kit). Electroporation was executed by the Ec2 program of the BIORad MicroPulser Electroporator (Pulse 2.5 kV, number of impulse 1) in a 0.2 cm electroporation cuvette. After incubation in B2 medium for 2 h at 37°C and 350 rpm, screening for plasmid-positive colonies was performed by plating the mixture on TSA agar with 10 μg/ml chloramphenicol. The TSA plates were incubated at 37°C for 42 h. Colonies were verified for the acquisition of the pCM28nuc plasmid by PCR with nuclease specific primer (Table 4) after plasmid purification (NEB Monarch Plasmid 344 Miniprep Kit). Nuclease activities of the original and transformed CF isolate were compared by DNase agar plates as well as by the nuclease FRET assay (data not included).
NET-Killing Assay
Isogenic S. aureus isolates with low and high nuclease activity were analyzed using the NET-killing assay (17). Human neutrophils from healthy individuals were isolated from heparin-anticoagulated blood by gradient separation using PolymorphPrep (Abbott Diagnostics Technologies AS). Remaining erythrocytes were lysed with sterile water after the neutrophil pellet was purified with magnesium- and calcium-free 1 x Dulbecco's Phosphate Buffered Saline (DPBS). Isolated neutrophils were suspended in RPMI medium (Sigma-Aldrich) with 2% heat-inactivated fetal bovine serum (FBS, Fiebig). In 24-well-plates, 2 × 106 neutrophils/ml were incubated with 10 μg/ml cytochalasin D (Sigma-Aldrich) and 25 nM phorbol 12- myristate 13-acetate (PMA, Sigma-Aldrich) for 20 min at 37°C + 5% CO2 prior infection. With a multiplicity of infection (MOI) of 2, S. aureus strains were applied as 4 x 106 bacteria/ml to RPMI medium (growth control) and activated neutrophils, respectively, and incubated at 37°C + 5% CO2 for 30 min and 90 min. Bacterial strains were pre-cultured in BHI (overnight, for mid-logarithmic growth phase) as described above. The survival rates for each S. aureus strain were calculated in % by comparison of CFUs on blood agar plates of bacteria interacting with NET-forming neutrophils vs. bacteria in medium.
Sputum Staining
Visualization of DNA-related structures and S. aureus bacteria in sputa from different CF patients was realized with immuno-fluorescence staining (49, 50). After expectoration, fresh sputum was immediately streaked onto poly-D-lysine-coated slides and fixed in 4% methanol-free formaldehyde for 15 min. Fixed sputum was kept hydrated either in 50 ml reaction tubes with PBS or in a humidity chamber during the immuno-fluorescence staining process. After permeabilization with 0.5% Triton-X, the CF sputum was covered with blocking buffer (1% bovine serum albumin, 10% goat serum, 0.3 mol glycine, 0.1% Tween 20 in PBS) for 20 min to avoid non-specific binding of antibodies. Subsequently, the sputum was incubated with rabbit α-S. aureus (MyBioSource; 1:2,000 diluted) in blocking buffer for 45 min at room temperature (RT). After washing in fresh PBS, the corresponding secondary antibody α-rabbit Alexa Fluor 488 (Invitrogen, A11070; 1:500 diluted in blocking buffer) was added and incubated for 45 min at RT in the dark. The slide was washed and covered with blocking buffer for another 10 min. A second primary antibody, mouse α-DNA/Histone1 antibody (Merck; 1:1,100 diluted), was applied for 45 min at RT in the dark to visualize NET-specific DNA- structures. After washing in PBS, the secondary antibody α-mouse Alexa Fluor 568 (Invitrogen; 1:376,500 diluted) was added together with Hoechst 33342 (Invitrogen; 1:20,000 diluted) and incubated for 45 min at RT in the dark. Prolong Antifade Mountant (Invitrogen) was applied on the slide, covered by a 24 × 60 mm glass cover slip, and incubated overnight at RT. The slides were examined for NET-structures and S. aureus bacteria by fluorescence microscopy, using the ZEISS Observer Z1 microscope with a Plan-Neofluar 100x/1.3 Oil RMS objective and AxioVision Rel 4.8 software. For a more detailed acquisition of images, CF sputum was further analyzed by the Zeiss LSM800 microscope equipped with a Plan-Apochromat 63x/1.4 Oil DIC M27 objective and ZEN 2.3 software.
RNA Extraction and Sequencing Analysis of CF Sputa
Sputum of one CF patient, who is chronically colonized and infected by S. aureus, was collected at two different time points in 2016 and stored at −80°C until processing. Samples were gently mixed with an equivalent volume of Sputolysin (10%) and incubated during 30 min at 37°C on a ThermoMixer device. RNA extraction, library construction, sequencing analysis and data processing were conducted as described recently (36). Calculation of relative gene expression/ normalization in transcripts per million (TPM) was based on the following formula:
. The S. aureus genes gyrB and aroE served as housekeeping genes.
Statistical Analysis
Data were analyzed in GraphPad Prism 5. Nuclease activities of S. aureus isolates (n = 111) from one individual patient recovered from different sample sites were compared by (i) one-way analysis of variance (ANOVA) test, complemented by Bonferroni's Post-Test (ii) two-tailed, unpaired student's t-test. Nuclease activity and survival rates obtained by the NET-killing assays were compared by two-tailed, unpaired t-test. For the comparison of bacterial survival among S. aureus isolates, the two-tailed, paired t-test was used.
Data Availability Statement
The datasets generated for this study by RNASeq can be found in the NCBI Gene Expression Omnibus (GEO) repository GSE139662, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139662.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethical Committee of the University Hospital Münster, Münster, Germany (2018-466-f-S). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
BK and SH designed the study. BK obtained the funding. SH performed and evaluated the majority of experiments and wrote the paper with the help of BK. FD established the FRET-based assay for the investigation of nuclease activity and analyzed S. aureus isolates from different CF patients. VS and AM performed whole genome sequencing and analyzed sequence data. DC-M and DP conducted and evaluated the RNA-Seq analysis of CF sputum. SN and CN gave theoretical and practical support with fluorescence microscopy and transformation with the pCM28nuc plasmid, respectively. NB and MK-B gave theoretical and practical support regarding the performance of NET killing assays and the immunofluorescence staining of CF sputa. JS performed spa typing of used S. aureus isolates. JG captured images of stained sputum samples using confocal microscopy under the scientific supervision of UR. AD, PK, and HS provided CF patient materials of which S. aureus isolates were recovered for this study, and sputum samples could be investigated on NETs. All authors read the paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Janina Treffon for the helpful communication and the providing of protocols regarding RNA isolation and qRT-analysis. Thanks to all CF patients and healthy individuals, who donated blood or sputum for research analysis.
Footnotes
Funding. This work was supported by the Interdisciplinary Center of Clinical Research, University of Münster, Münster, Germany Kah2/016/16.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02552/full#supplementary-material
References
- 1.O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. (2009) 373:1891–904. 10.1016/S0140-6736(09)60327-5 [DOI] [PubMed] [Google Scholar]
- 2.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. (2010) 23:299–323. 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis Foundation 2017 Patient Registry: Annual Data Report. Cystic Fibrosis Foundation Patient Registry (2017). [Google Scholar]
- 4.European Cystic Fibrosis Society Patient Registry (2018). ECFSPR Annual Report 2016. Available online at: www.ecfs.eu/ecfspr (accessed September 11, 2019).
- 5.Gangell C, Gard S, Douglas T, Park J, de Klerk N, Keil T, et al. Inflammatory responses to individual microorganisms in the lungs of children with cystic fibrosis. Clin Infect Dis. (2011) 53:425–32. 10.1093/cid/cir399 [DOI] [PubMed] [Google Scholar]
- 6.Cohen TS, Prince A. Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat Med. (2012) 18:509–19. 10.1038/nm.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. (2004) 303:1532–35. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- 8.Downey DG, Bell SC, Elborn JS. Neutrophils in cystic fibrosis. Biol Chem. (2007) 397:485–96. 10.1515/hsz-2015-0271 [DOI] [Google Scholar]
- 9.Painter RG, Valentine VG, Lanson NA, Leidal K, Zhang Q, Lombard G, et al. CFTR expression in human neutrophils and the phagolysosomal chlorination defect in cystic fibrosis. Biochemistry. (2006) 45:10260–9. 10.1021/bi060490t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagel SD, Sontag MK, Accurso FJ. Relationship between antimicrobial proteins and airway inflammation and infection in cystic fibrosis. Pediatr Pulmonol. (2009) 44:402–9. 10.1002/ppul.21028 [DOI] [PubMed] [Google Scholar]
- 11.Law SM, Gray RD. Neutrophil extracellular traps and the dysfunctional innate immune response of cystic fibrosis lung disease: a review. J Inflamm. (2017) 14:29. 10.1186/s12950-017-0176-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sly PD, Gangell CL, Chen L, Ware RS, Ranganathan S, Mott LS, et al. Risk factors for bronchiectasis in children with cystic fibrosis. Survey Anesthesiol. (2014) 58:82 10.1097/01.SA.0000443983.90197.36 [DOI] [PubMed] [Google Scholar]
- 13.Gray RD, Hardisty G, Regan KH, Smith M, Robb CT, Duffin R, et al. Delayed neutrophil apoptosis enhances NET formation in cystic fibrosis. Thorax. (2018) 73:134–44. 10.1136/thoraxjnl-2017-210134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzenreiter R, Kienberger F, Marcos V, Schilcher K, Krautgartner WD, Obermayer A, et al. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J Cyst Fibros. (2012) 11:84–92. 10.1016/j.jcf.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 15.Marcos V, Zhou-Suckow Z, Önder Yildirim A, Bohla A, Hector A, Vitkov L, et al. Free DNA in cystic fibrosis airway fluids correlates with airflow obstruction. Mediat Inflamm. (2015) 2015:408935. 10.1155/2015/408935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilsczek FH, Salina D, Poon KKH, Fahey C, Yipp BG, Sibley CD, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol. (2010) 185:7413–25. 10.4049/jimmunol.1000675 [DOI] [PubMed] [Google Scholar]
- 17.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun. (2010) 2:576–86. 10.1159/000319909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Disalvo JW. Desoxyrribonuclease and coagulase activity of micrococci. Med Tech Bull. (1958) 9:191–6. [PubMed] [Google Scholar]
- 19.Schilcher K, Andreoni F, Uchiyama S, Ogawa T, Schuepbach RA, Zinkernagel AS. Increased neutrophil extracellular trap-mediated Staphylococcus aureus clearance through inhibition of nuclease activity by clindamycin and immunoglobulin. J Infect Dis. (2014) 210:473–82. 10.1093/infdis/jiu091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, et al. Staphylococcus aureus nuclease is a SaeRS- dependent virulence factor. Infect Immun. (2013) 81:1316–24. 10.1128/IAI.01242-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogasch K, Rühmling V, Pané-Farré J, Höper D, Weinberg C, Fuchs S, et al. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol. (2006) 188:7742–58. 10.1128/JB.00555-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick RP. Autoinduction and signal transduction in the regulation of Staphylococcal virulence. Mol Microbiol. (2003) 48:1429–49. 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- 23.Hirschhausen N, Block D, Bianconi I, Bragonzi A, Birtel J, Lee JC, et al. Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation. Int J Med Microbiol. (2013) 303:685–92. 10.1016/j.ijmm.2013.09.012 [DOI] [PubMed] [Google Scholar]
- 24.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE. (2010) 5:e10146. 10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, et al. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS ONE. (2011) 6:e267. 10.1371/journal.pone.0026714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez-Alemán SR, Campos-García L, Palma-Nicolas JP, Hernández-Bello R, González GM, Sánchez-González A. Understanding the entanglement: Neutrophil Extracellular Traps (NETs) in cystic fibrosis. Front Cell Infect Microbiol. (2017) 7:104. 10.3389/fcimb.2017.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dwyer M, Shan Q, D'Ortona S, Maurer R, Mitchell R, Olesen H, et al. Cystic fibrosis sputum DNA has NETosis characteristics and neutrophil extracellular trap release is regulated by macrophage migration-inhibitory factor. J Innate Immun. (2014) 6:765–79. 10.1159/000363242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Floyd M, Winn M, Cullen C, Sil P, Chassaing B, Yoo DG, et al. Swimming motility mediates the formation of neutrophil extracellular traps induced by flagellated Pseudomonas aeruginosa. PLoS Pathog. (2016) 12:e1005987. 10.1371/journal.ppat.1005987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young RL, Malcolm KC, Kret JE, Caceres SM, Poch KR, Nichols DP, et al. Neutrophil Extracellular Trap (NET)-mediated killing of Pseudomonas aeruginosa: evidence of acquired resistance within the CF airway, independent of CFTR. PLoS ONE. (2011) 6:e23637. 10.1371/journal.pone.0023637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman S, Gadjeva M. Does NETosis contribute to the bacterial pathoadaptation in cystic fibrosis? Front Immunol. (2014) 5:378. 10.3389/fimmu.2014.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kriegeskorte A, Block D, Drescher M, Windmüller N, Mellmann A, Baum C, et al. Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. Mbio. (2014) 5:e01447–14. 10.1128/mBio.01447-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahl BC, Mellmann A, Deiwick S, Peters G, Harmsen D. Variation of the polymorphic region X of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J Clin Microbiol. (2005) 43:502–5. 10.1128/JCM.43.1.502-505.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birrer P, McElvaney NG, Rüdeberg A, Sommer CW, Liechti-Gallati S, Kraemer R, et al. Protease-antiprotease imbalance in the lungs of children with cystic fibrosis. Am J Respir Crit Care Med. (1994) 150:207–13. 10.1164/ajrccm.150.1.7912987 [DOI] [PubMed] [Google Scholar]
- 34.Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, Ghnaim H, et al. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med. (1995) 152(6 Pt 1):2111–8. 10.1164/ajrccm.152.6.8520783 [DOI] [PubMed] [Google Scholar]
- 35.Cantin A. Cystic fibrosis lung inflammation: early, sustained, and severe. Am J Respir Critic Care Med. (1995) 151:939–41. 10.1164/ajrccm/151.4.939 [DOI] [PubMed] [Google Scholar]
- 36.Chaves-Moreno D, Wos-Oxley ML, Jáuregui R, Medina E, Oxley AP, Pieper DH. Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci Rep. (2016) 6:33174. 10.1038/srep33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. (2013) 342:863–6. 10.1126/science.1242255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traber K, Novick R. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of Agr and failure to translate δ- and α-haemolysins. Mol Microbiol. (2006) 59:1519–30. 10.1111/j.1365-2958.2006.04986.x [DOI] [PubMed] [Google Scholar]
- 39.Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S. Mutations in Agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J Infect Dis. (2010) 202:1593–9. 10.1086/656915 [DOI] [PubMed] [Google Scholar]
- 40.Giraudo AT, Calzolari A, Cataldi AA, Bogni C, Nagel R. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett. (1999) 177:15–22. 10.1016/S0378-1097(99)00282-7 [DOI] [PubMed] [Google Scholar]
- 41.Augustin J, Götz F. Transformation of Staphylococcus epidermidis and other Staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett. (1990) 54:203–7. 10.1111/j.1574-6968.1990.tb03997.x [DOI] [PubMed] [Google Scholar]
- 42.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. (2003) 41:5442–8. 10.1128/JCM.41.12.5442-5448.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, et al. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol. (2016) 54:2874–81. 10.1128/JCM.00790-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol. (2014) 52:2365–70. 10.1128/JCM.00262-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartbeck B, Birtel J, Treffon J, Langhanki L, Mellmann A, Kale D, et al. Dynamic in vivo mutations within the Ica operon during persistence of Staphylococcus aureus in the airways of cystic fibrosis patients. PLoS Pathog. (2016) 12:e1006024. 10.1371/journal.ppat.1006024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treffon J, Block D, Moche M, Reiss S, Fuchs S, Engelmann S, et al. Adaptation of Staphylococcus aureus to the airways of cystic fibrosis patients by the up-regulation of superoxide dismutase M and iron-scavenging proteins. J Infect Dis. (2018) 217:1453–61. 10.1093/infdis/jiy012 [DOI] [PubMed] [Google Scholar]
- 47.Vaudaux P, Francois P, Bisognano C, Kelley WL, Lew DP, Schrenzel J, et al. Increased expression of clumping factor and fibronectin-binding proteins by HemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect Immun. (2002) 70:5428–37. 10.1128/IAI.70.10.5428-5437.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harraghy N, Kormanec J, Wolz C, Homerova D, Goerke C, Ohlsen K, et al. Sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology. (2005) 151(Pt 6):1789–800. 10.1099/mic.0.27902-0 [DOI] [PubMed] [Google Scholar]
- 49.de Buhr N, von Köckritz-Blickwede M. How neutrophil extracellular traps become visible. J Immunol Res. (2016) 2016:4604713. 10.1155/2016/4604713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Köckritz-Blickwede M, Chow O, Ghochani M, Nizet V. Visualization and functional evaluation of phagocyte extracellular traps. Methods Microbiol. (2010) 37:139–60. 10.1016/S0580-9517(10)37007-3 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study by RNASeq can be found in the NCBI Gene Expression Omnibus (GEO) repository GSE139662, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139662.