Abstract.
-Amyloid () plaque, representing the progressive accumulation of the protein that mainly consists of , is one of the prominent pathological hallmarks of Alzheimer’s disease (AD). Label-free imaging of plaques holds the potential to be a histological examination tool for diagnosing AD. We applied label-free multiphoton microscopy to identify extracellular plaque as well as intracellular accumulation for the first time from AD mouse models. We showed that a two-photon-excited fluorescence signal is a sensitive optical marker for revealing the spatial–temporal progression and the surrounding morphological changes of deposition, which demonstrated that both extracellular and intracellular accumulations play an important role in the progression of AD. Moreover, combined with a custom-developed image-processing program, we established a rapid method to visualize different degrees of deposition by color coding. These results provide an approach for investigating pathophysiology of AD that can complement traditional biomedical procedures.
Keywords: Alzheimer’s disease, β-amyloid plaque, multiphoton microscopy, two-photon-excited fluorescence, second-harmonic generation
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease characterized by decline of memory, learning, thinking, and other cognitive functions and is the most common cause of dementia in the elderly.1 Given the prevalence and poor prognosis of the disease, the development of transgenic mouse models has improved our understanding of the disease pathogenesis and has led to new therapeutic approaches.2 The most prominent pathological hallmark of AD is senile plaques, representing the progressive accumulation of the protein that mainly consists of the -amyloid ().3,4 Therefore, direct imaging of plaques holds the potential to be a powerful tool for diagnosis and early detection of AD. The clinical imaging technique of positron emission tomography (PET) is able to image deposits in transgenic mouse models and humans in vivo after intravenous administration of tracer Pittsburgh compound B.5,6 Magnetic resonance imaging also has been demonstrated to visualize in vivo individual AD plaques without the use of a contrast agent in transgenic mice, showing great promise for imaging plaques in living human Alzheimer patients noninvasively.7 However, these techniques are limited by low spatial resolution, and PET depends on radioactive ligands, although in only trace amounts. Histologically, the common method often uses amyloid-binding dyes, such as Congo red, Thioflavin S, and methoxy-XO4,8,9 yet these staining methods still rely on time-intensive procedures that require skilled technicians to produce slides in a significant amount of time. More importantly, some studies have reported that amyloid-binding compounds may interfere with the formation of amyloid structures.10,11 To complement these limitations of traditional methods, label-free optical imaging techniques have been developed in brain imaging. Optical coherence microscopy (OCM) has provided three-dimensional, high-resolution images of individual plaques in the brain parenchyma and vasculature.12 Cryo-micro-optical sectioning tomography (cryo-MOST) has visualized senile plaques at a micrometer-level resolution in the whole brain.13 It is generally known that oligomer toxicity can cause neuronal death and loss.14 Nevertheless, OCM and cryo-MOST are not applied to simultaneously visualize plaques, neurons, or blood vessels. Moreover, OCM images have only a single channel, which could not provide enough contrast or information. Although some groups have observed autofluorescence and second-harmonic generation (SHG) at senile plaques by multiphoton microscopy (MPM),15,16 they only have imaged extracellular plaques in the hippocampal regions of elderly mice, and they are unable to reveal brain-wide mapping of plaque distribution and intracellular accumulation.
Label-free MPM, based on two-photon-excited fluorescence (TPEF) and SHG, two kinds of different mechanisms, has the ability to visualize different intrinsic fluorophores in biological tissues, such as collagen, elastic fiber, nicotinamide adenine dinucleotide (NADH), flavin adenine dinucleotide (FAD), and retinol.17,18 Recently, label-free MPM has demonstrated the ability to visualize a wide range of brain diseases features in rodents and humans at the cellular level, such as the Purkinje cell layer of cerebellar hemangioblastomas,19 the diagnosis of pituitary adenoma,20 neuronal injury of cerebral infarctions,21 and abnormal cortical lamination of focal cortical dysplasia.22 MPM can create pathology-like images from multiple endogenous fluorophores in brain tissue, which is suitable for detecting the changes of brain tissue structures with high resolution and high sensitivity. Therefore, this approach shows the ability to offer additional metrics for neuropathologists’ observations and even surgeons’ decisions.
In this study, we demonstrated the capability of label-free MPM to visualize extracellular and intracellular accumulations in brain tissue from the APP/PS1 and APP/PS1/Tau transgenic mouse model of AD. We then focused on the spatial–temporal progression and the surrounding morphological changes of deposition, demonstrating that both extracellular and intracellular accumulations play an important pathophysiological role in the progression of AD. Finally, combined with a custom-developed image-processing program, we established a rapid method to visualize different degrees of deposition by color coding.
2. Materials and Methods
2.1. Experimental Animals
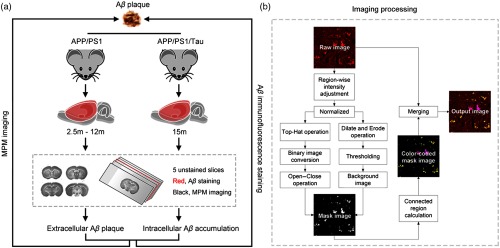
A total of 32 male APP/PS1 double-transgenic mice [B6C3-Tg (APPswe, PSEN1dE9) 85Dbo/MmJNju] and 4 male wild-type mice were obtained from Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China). Four male APP/PS1/Tau mice [B6; 129-Tg (APPSwe, tauP301L) 1Lfa Psen1tm1Mpm/Mmjax] were purchased from the Jackson Laboratory. Mice were housed in groups of three per cage on a 12:12-h light/dark cycle with access to food and water ad libitum. All of the animal experiments were approved by the Institutional Animal Ethics Committee of Fujian University of Traditional Chinese Medicine, and all experiments were performed in accordance with relevant guidelines and regulations. The experimental design is illustrated in Fig. 1(a).
Fig. 1.
The diagram of (a) experimental design and (b) the image-processing approach for visualizing deposition.
2.2. Sample Preparation
APP/PS1 mice (aged 2.5, 3, 4, 6, 9, 12 months), wild-type mice, and APP/PS1/Tau mice (aged 15 to 16 months) were euthanized by 1% sodium pentobarbital (, i.p. Sigma-aldrich) and then perfused transcardially with 0.9% saline and 4% paraformaldehyde chilled to 4°C. The brain was removed and fixed at 4°C in 4% paraformaldehyde for 24 h. Each brain was embedded in paraffin and cut into five consecutive coronal sections of thickness using a rotary microtome (RM2235, Leica, Germany). Importantly, the MPM-imaged sections were fully deparaffinized by alcohol and xylene before MPM imaging. All sections were mounted onto adhesive glass slides (Matsunami Glass Ind., Japan). Four of five consecutive sections were used for MPM imaging. The middle sections were stained with staining. During the MPM imaging experiment, a small volume of phosphate-buffered saline (PBS) solution was dripped onto the tissue specimen to avoid dehydration or shrinkage. Then, a coverslip was placed on the PBS-moistened specimens.
2.3. Immunofluorescence/Immunochemistry Staining
To confirm that observed structures in amyloid-laden brains are indeed plaques, immunofluorescence staining and immunochemistry staining were performed on the adjacent serial slices. The slices were retrieval using heat-mediated antigen retrieval with sodium citrate buffer (pH 6) for 10 min.
For immunofluorescence, the slices were washed in PBS and then blocked in 5% bovine serum for 1 h. Afterward, the slices were incubated overnight at 4°C with the primary antibodies anti- (6E10, 1:1000, Biolegend). Subsequently, the slices were washed in PBS to clear out excess primary antibodies and were incubated with secondary antibodies conjugated to fluorophores Alexa Fluor 488 (1:500, Life Technologies) for 2 h at room temperature. After being series washed in PBS, nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole (1:1000; Santa Cruz). The tissue slices were mounted in mounting medium (Solarbio, China) and then mounted onto cover slides. The immunofluorescence images were obtained using a confocal laser scanning microscope (Axio Examiner. Z1, Zeiss LSM 880 META, Jena, Germany).
For immunochemistry, the slices were incubated with the primary antibodies anti- (6E10, 1:1000, Biolegend) overnight at 4°C. After being series washed in PBS, the slices were developed with diaminobenzidine substrate using the avidin–biotin–horseradish peroxidase system. The tissue slices were mounted in neutral balsam (Solarbio, China) and then mounted onto cover slides. The immunochemical images were obtained using a light scanning microscope (Aperio VERSA 8, Leica, Germany).
2.4. Multiphoton Microscopy System
The MPM system used in this study has been previously described in detail.22 Briefly, an upright laser scanning microscope (Axio Examiner. Z1, Zeiss LSM 880 META, Jena, Germany) was equipped with an external mode-locked Ti:sapphire laser (140 fs, 80 MHz), tunable from 690 to 1064 nm (Chameleon Ultra, Coherent, Inc., Santa Clara, California). In our experiment, samples were excited at an excitation wavelength of 810 nm. The average power after the objective was 5 to 10 mW. The emitted fluorescence was either spectrally separated by passing through a grating onto the 32-channel gallium arsenide phosphide photomultiplier tube (PMT) array detectors to obtain the TPEF signal and onto a flanking PMT detector to get the SHG signal. The images were collected in two independent channels simultaneously: one channel covered the wavelength range from 395 to 415 nm for collection of SHG signals (color-coded green) and the other channel covered the wavelength range from 430 to 690 nm for collection of TPEF signals (color-coded red). For large-scale imaging, a Plan-Apochromat 0.8 objective (Zeiss) was used. The frame size was set to , and mean average mode was used so that the image information was generated by adding up all scans pixel by pixel and then calculating the mean value. The H1P2SLSM motorized microscope stage (Prior Scientific Instruments Ltd., Cambridge, UK) was applied to scan across large sample areas. All frames were automatically stitched by Zeiss software. For high-resolution Z-stack imaging, a Plan-Apochromat 1.4 oil immersion objective (Zeiss) was performed. We match the findings at different scales by zooming in on the region of interest (ROI) in images or switching to different objectives. All images had a 12-bit pixel depth. Bidirectional scanning doubles the scanning rate to a maximum of (). In addition, emission spectrum was obtained by the lambda mode setting of the laser scanning microscopic imaging system (LSM 510 META, Zeiss, Jena, Germany) equipped with a mode-locked femtosecond Ti:sapphire laser (110 fs, 76 MHz) at 810 nm (Coherent Mira 900-F).23 The lambda mode, which was used to obtain the emission spectrum intensity by the META detector, covered a spectral width ranging from 377 to 716 nm, and the spectral resolution was 10.7 nm.
2.5. Image Processing
To rapidly visualize the degree of deposition, the digital image processing technique was exploited to highlight the deposition in the unstained brain slices. The detail of the processing pipeline is illustrated in Fig. 1(b). First, the raw image is sampled from the unstained brain slice by MPM. Owing to the nonuniformed intensity distribution, the region-wise intensity is adjusted first and a normalized operation is followed. Second, we achieve the corresponding mask image by combining a coarse binary image with a background image. To get the binary image, a top-hat operation is implemented on the normalized image first and then the image is converted into a binary image with a reasonable threshold value. Subsequently, the binary image is fed into a group of open and close morphology operations to eliminate outliers and fill small objects. To achieve the background image, we dilate the normalized image and erode it sequentially with a ball-shaped structuring element first. Then a thresholding operation is employed to remove some small objects. The threshold value for converting the binary image is 0.616. The small objects in the background are cleared when their areas are . Third, to directly visualize the accumulation, we record the coordinate and the area of each connected region in the mask image. Then the mask image is color-coded according to the area of each connected region whose logical value is equal to 1. Finally, the output image is obtained via merging the colored mask image with the raw image. All image processing and algorithm execution were carried out using MATLAB (The MathWorks, Natick, Massachusetts).
2.6. Statistical Analysis
We measured extracellular plaque’s number, cell number, and nuclear area by LSM 880 software (Zen blue, Zeiss, Germany). The representative series sections of the whole brain were selected for statistical analysis by SPSS software (version 21.0, IBM, New York). A two-tailed Student’s -test was performed to determine significant differences between any two groups. One-way analysis of variance (ANOVA) was used to compare more than two groups of data. The final results were presented as the mean value ± standard deviation (). The confidence level was set to 0.05 ( value). The significance level is displayed as asterisks (*, **, ***; NS, not significant).
3. Results
3.1. Imaging of Extracellular Plaques in the APP/PS1 Mouse Model
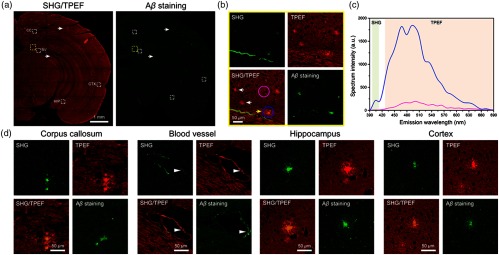
To validate the imaging capability of label-free MPM for plaques, we first imaged unstained brain slices from the 9-month-old male APP/PS1 transgenic AD mice model. Figure 2(a) shows a stitched large-scale SHG/TPEF image and a corresponding immunofluorescence-labeled image of a representative half hippocampal coronal plane. In the SHG/TPEF image [Fig. 2(a)], plaques can be clearly distinguished by a distinct pattern of SHG and TPEF signals, which appear as spherical objects that emit a diffuse intrinsic fluorescence. In detail, Fig. 2(d) displays enlarged images of the white dashed box indicated in Fig. 2(a), showing that MPM has the capacity to identify plaques in corpus callosum (CC), pial vessel, hippocampus (HC), and cortex (CTX). Notably, the strong SHG signal along the vascular wall [blood vessel in Fig. 2(d)], a similar signal compares with plaque detected near the pial vessel, which is contributed to collagen fiber.17 These observations are confirmed by the corresponding immunofluorescence image, verifying that the plaques detected by MPM are plaques. Significantly, we find that all of the plaques can emit strong TPEF signals. However, not all plaques can show strong SHG signals [Fig. 2(b)]. Some of the plaques emit weak and even no SHG signals [white arrows in Fig. 2(b)]. The relatively strong SHG signals of plaque are mainly attributed to the plaque center [yellow arrow in Fig. 2(b)].
Fig. 2.
Imaging of extracellular plaques in APP/PS1 mouse model. (a) Representative large-scale SHG/TPEF images and a corresponding immunofluorescence image of a half hippocampal coronal plane. CC, corpus callosum; BV, blood vessel; HIP, hippocampus; CTX, cortex. (b) Enlarged images from yellow dashed box in (a). Yellow arrows, plaque with strong TPEF signal and weak SHG signal. White arrows, plaque with no SHG signal. (c) Emission spectrum from plaque region (blue line) and adjacent region with no plaque (pink line). (d) Enlarged SHG/TPEF images from white dashed box in (a). Arrowhead, plaque detected near the pial vessel. SHG, color-coded green. TPEF, color-coded red.
To further confirm the feasibility that plaques can be effectively detected by TPEF signals, we next obtained typical emission spectrum of individual plaque from the same transgenic mouse model [Fig. 2(c)]. Qualitatively, SHG at 405 nm has much weaker signals than TPEF (430 to 690 nm) in the plaque region (blue line). However, in an adjacent region with no plaque (pink line), the detected emission is significantly weaker and there are no SHG signals at 405 nm. The strong TPEF signals are mainly attributed to NADH (peak at ) and FAD (peak at ). The peak around 630 nm is assigned to porphyrin derivatives.17,18 Notably, lipofuscins, an autofluorescent lipopigment that contributes to the formation of AD senile plaques and neurodegeneration, presents a very wide spectra ranging from 400 to 700 nm, with a broad major peak centered at , which is spectral overlapped with FAD fluorophores.18 To further verify the optimal TPEF signals for plaque imaging, we also compared the emission spectrum from different unstained slice types (Fig. S1 in the Supplementary Material). The molecular origin of autofluorescence in plaques is addressed in detail in Sec. 4. As a result, our data not only demonstrate the ability of label-free MPM to visualize the extracellular plaque, but also illustrate that the TPEF signal is a more sensitive optical marker for detecting plaques when compared with the SHG signal.
3.2. Characterization of Extracellular Deposition in the APP/PS1 Mouse Model
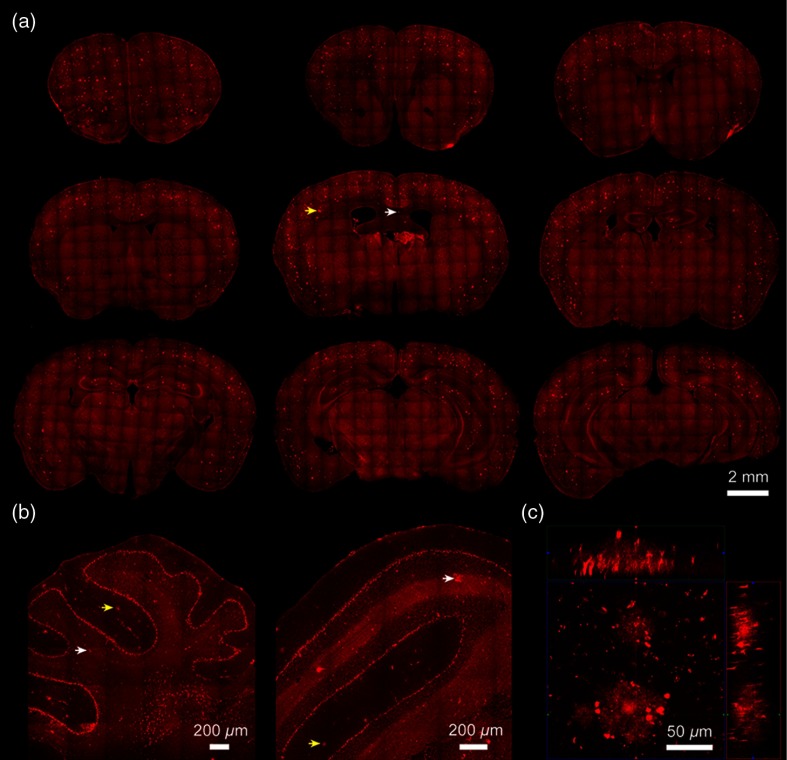
To characterize the spatial–temporal progression of deposition in APP/PS1 transgenic mice, we first visualized the distributions of plaques from 10-month-old APP/PS1 mice by TPEF signals. Figure 3(a) shows a series of coronal images along the anterior–posterior axis of the entire mouse brain. Moreover, representative sagittal and coronal cerebellum images are also shown in Fig. 3(b).
Fig. 3.
Distribution of extracellular plaque. (a) A series of TPEF coronal images along the anterior–posterior axis of the entire mouse brain in 10-month-old APP/PS1 mice. (b) Typical TPEF images of sagittal and coronal cerebellum. (c) A Z-stack TPEF image of plaque at an orthogonal view, which presents the , , and planes at a certain spot. Yellow arrows, plaque in gray matter. White arrows, plaque in white matter.
As shown in Fig. 3, plaque distributions have spread to the whole brain and even cerebellum, which can be clearly visualized without exogenous dye. In detail, the plaques can be detected in both gray matter (yellow arrows in Fig. 3) and white matter (white arrows in Fig. 3). Most plaques are distributed in CTX; some lay in HC, especially in dentate gyrus (DG) region. A minority spread to other areas, such as corpus striatum and thalamus (THA). Furthermore, the magnified shape of the individual plaque is displayed in Fig. 3(c). Figure 3(c) is a Z-stack TPEF image at an orthogonal view, which presents the , , and planes at a certain spot. The plaques show a dense core that has more accumulation than the peripheral plaque region, where stronger TPEF signals appear. In addition, we also observe some spots within the plaque. These spots may be the result of concentrated NADH in the mitochondria24 or a synaptic marker.25 Imaging was performed at intervals of , and a penetration depth of was recorded. The acquisition time for the Z-stack image is 25 s.
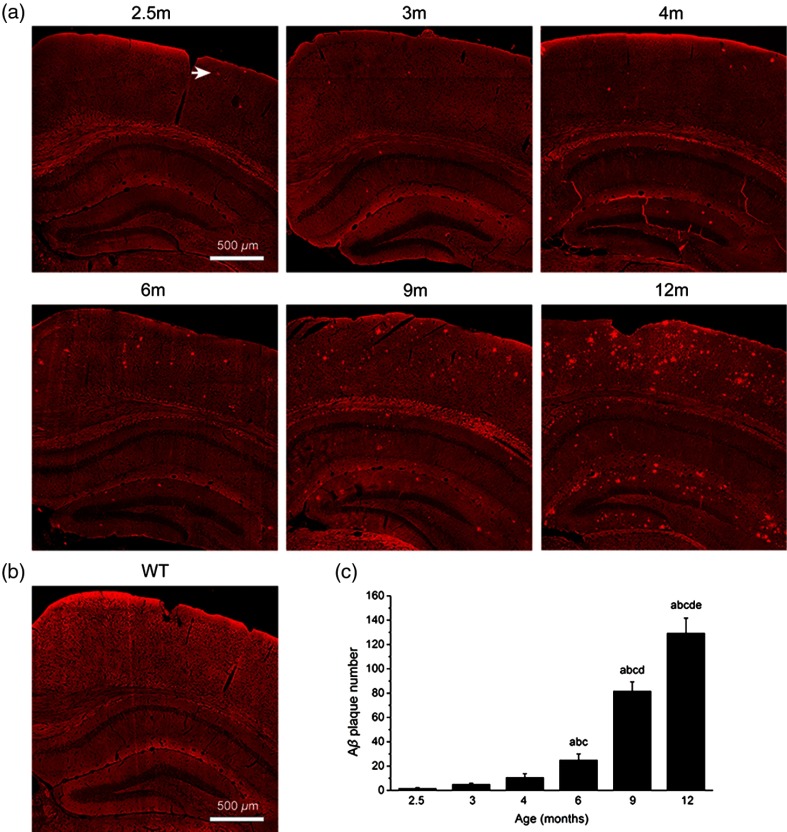
Then, we proceeded to characterize the progression of deposition from 2.5- to 12-month-old APP/PS1 transgenic mice, which focused on CTX, hippocampal cornu ammonis-1 (CA-1), and DG regions because they are progressively damaged during AD and are well known to be associated with learning and memory. As shown in Fig. 4(a), plaques are identified in a series of coronal brain images at different stages. We find that the plaque can be detected in the CTX as early as 2.5 months of age by label-free MPM imaging [arrow in Fig. 4(a)]. In contrast, the corresponding image of a wild-type mouse (6 months of age) shows no plaque signal [Fig. 4(b)]. Moreover, there is a progressive increase in amyloid burden with age, which is a function of increases in both plaque number and mean plaque area. Four coronal planes with HC and parietal association CTX were selected as ROIs, depicting that the plaque number increases significantly with age [Fig. 4(c)], whereas they show no significant difference among 2.5- to 4-month-old mice groups. In addition, the small area of plaques (diameter ) can be seen in mice groups aged from 2.5 to 4 months old. Some large area plaques with diameter to appear increasingly in 6-month-old mice. Therefore, combined with the quantitative results, plaques’ deposition displays an increasing tendency, which is observed obviously in the brain of 6-month-old APP/PS1 mice. These results indicate that MPM can be used to evaluate plaque distribution and progression in APP/PS1 mice at different months of age, similar to previously reported studies on -stained sections of the same transgenic model.26,27
Fig. 4.
Progression of deposition with age. (a) TPEF images of extracellular plaques in CTX and HC from 2.5- to 12-month-old APP/PS1 mice. Arrow, plaque. (b) TPEF image from wild-type mouse. (c) Statistical analysis revealed that plaque number increases significantly with age. We performed ANOVA within groups. a, versus 2.5-month-old group. b, versus 3-month-old group. c, versus 4-month-old group. d, versus 6-month-old group. e, versus 9-month-old group. Each group has three mice.
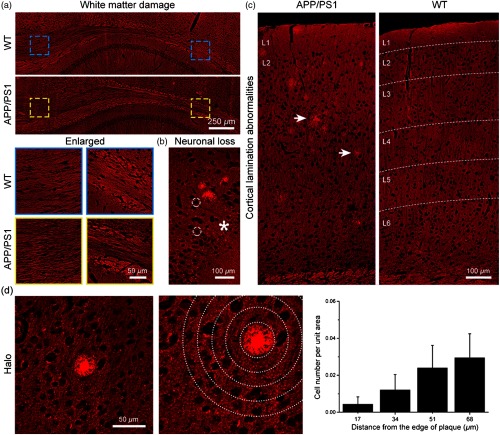
3.3. Identification of Structural Alterations around Extracellular Plaques
Growing evidence suggests that deposition is associated with the surrounding structural abnormalities, which might evolve in the pathogenesis in AD. As shown in Fig. 5, we could readily observe the white matter and gray matter structural alterations in the vicinity of plaques by MPM. Compared with wild-type mice (6 months of age), CC of white matter shows a relatively sparse microstructural abnormality in APP/PS1 mice [Fig. 5(a)]. Enlarged images further clearly compare the changes in white matter structures, revealing the fiber tract reduction in the CC region. Meanwhile, cells could be identified by nonfluorescent nuclei (white dashed circles) and TPEF signals of perinucleus. In contrast, we observe obvious neuronal loss region near the plaque in CTX [asterisk in Fig. 5(b)]. Moreover, the abnormalities of cortical lamination [Fig. 5(c)] are also found when plaques are observed in CTX of APP/PS1 mice. It can be seen that layers 2 to 5 are less distinguishable in comparison with structurally normal six-layer cortical lamination. Interestingly, we further found that there are few cells near plaques. The surrounding cells and the plaque exhibit a halo-like structures [Fig. 5(d)]. To quantitatively describe the spatial impact of the halo structures surrounding the plaque cores, the cell number per unit area defined by concentric bands spaced at intervals from the border of individual plaques was measured in CTX [Fig. 5(d), middle and right; plaques per mouse. Each group has three APP/PS1 mice.]. Quantitatively, the percentage of cell numbers is significantly decreased in the areas adjacent to plaques. These phenomena are consistent with previous studies,28,29 demonstrating that MPM can not only detect the plaques but also reveal the surrounding structural alterations in AD brain tissues.
Fig. 5.
Identification of structural alterations around extracellular plaques. (a) White matter damage. (b) Neuronal loss near plaques in CTX. White dashed circles, cell nuclei. Asterisk, neuronal loss region. (c) Abnormalities of cortical lamination. Arrows, plaque. L1, cortical layer 1. (d) Left: halo structures surrounding the plaque cores. (d) Middle and right: the percentage of cell number within areas defined by concentric bands spaced at intervals from the border of individual plaques.
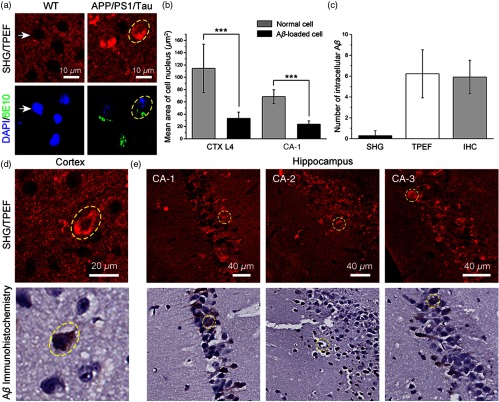
3.4. Imaging of Intracellular Accumulation in the APP/PS1/Tau Mouse Model
In addition to the extracellular accumulation in the parenchyma, emerging evidence indicates that intracellular also plays a potential biomarker for the onset of AD.30 Moreover, there are no label-free optical imaging techniques that show the ability to visualize intracellular accumulation. Therefore, we imaged intracellular accumulation in unstained brain slices from the 15-month-old male APP/PS1/Tau transgenic AD mice model. Figure 6(a) displays the typical SHG/TPEF image and corresponding immunofluorescence image of intracellular in the cortical region [yellow dashed circle in Fig. 6(a)]. Intracellular accumulation shows bright granules within the perinuclear region, which appears as a thin ring around cell nuclei from strong TPEF signals and no SHG signals. In comparison with the cortical neuron without deposition [white arrows in Fig. 6(a)], most of -loaded cells exhibit an irregular shape as well as shrunken and even pyknosis nuclei. The -loaded cell nuclei become blurred and show weak TPEF signals. To quantitatively characterize the morphological changes of -loaded cells, we measured mean cell nucleus area of normal cells and -loaded cells in cortical layer 4 and hippocampal CA-1 region, respectively [Fig. 6(b)]. Consistent with our observation, in both of these regions, mean cell nucleus area of -loaded cells shows a significant decrease compared with normal cells. This finding demonstrates that intracellular accumulation is associated with cellular structural changes. Moreover, we also show typical intracellular located within the cornu ammonis (CA) field of the hippocampal region [Figs. 6(d) and 6(e)]. Some of intracellular are dispersed in the surrounding extracellular space, which is due to the lysis of -loaded neurons.31 Finally, to compare the sensitivity between histology analysis and multiphoton imaging results, the number of intracellular accumulation between SHG, TPEF, and corresponding immunohistochemistry (IHC) images () is measured [Fig. 6(c)]. Our quantitative and qualitative results show that TPEF is a sensitive tool for identifying intracellular accumulation. There are slight structural differences between TPEF images and staining images because the serial sections are very thin.
Fig. 6.
Imaging of intracellular accumulation in APP/PS1/Tau mouse model. (a) Typical SHG/TPEF image and corresponding immunofluorescence images of intracellular accumulation in CTX from APP/PS1/Tau mice. White arrows, normal cell. Yellow dashed circles, -loaded cell. (b) Quantitative characterization of mean cell nucleus area of normal and -loaded cells in cortical layer 4 and hippocampal CA-1 region. CTX L4, cortical layer 4; CA-1, cornu ammonis-1. Each group measures 23 cells from two APP/PS1/Tau mice. ***. (c) Quantitative comparison of the number of intracellular accumulation between SHG, TPEF, and corresponding IHC images () (). (d) and (e) Typical SHG/TPEF images and corresponding immunohistochemical images of intracellular in CTX and hippocampal CA region.
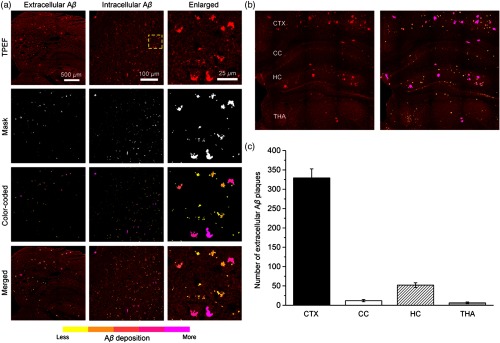
3.5. Rapid Visualization of Deposition
In our imaging results, identification of deposition often depends on the difference of TPEF signal intensity between deposition and background signals, which all display the color code red. Therefore, to rapidly visualize extracellular and intracellular depositions, we developed the digital image-processing method based on the MPM image (see Sec. 2). As shown in Fig. 7(a), it illustrates the representative extracellular and intracellular depositions in 9-month-old APP/PS1 mouse and 15-month-old APP/PS1/Tau mouse. By combining a coarse binary image with a background image from MPM image, we generate a mask that indicates the locations of object regions. Once these regions are identified, a color-coded mask is created to directly evaluate accumulation according to the difference of each object region’s area. From yellow to magenta, different colors represent the different degrees of the deposition (). We could clearly identify a few yellowish small in extracellular matrix, as well as some magenta plaques around neuronal nuclei in the enlarged images of intracellular . Furthermore, severe deposition of extracellular plaques can directly be observed. As a result, the merged images, which are obtained by combining the color-coded mask image with the raw MPM image, show increased imaging contrast. Then, we compare the number of extracellular plaques in CTX, HC, CC, and THA regions from three 10-month-old APP/PS1 mice. Four representative hippocampal planes are selected as ROIs. Figure 7(b) displays a part of the representative hippocampal plane. Combined with the quantitative results [Fig. 7(c)], we can find that CTX is the most severe deposition area. Moreover, large plaques color-coded with magenta are mainly distributed in both the CTX and HC regions. With the combination of MPM and the custom-developed image-processing method, these results show the feasibility of our data, which can serve as an automated tool for rapidly visualizing deposition.
Fig. 7.
Rapid visualization of deposition. (a) Representative extracellular and intracellular depositions. Enlarged images were zoomed in from yellow dashed box. From yellow to magenta, different colors represent the different degrees of deposition. (b) A part of representative hippocampal plane from 10-month-old APP/PS1 mouse. (c) Comparison of the number of extracellular plaques in CTX, HC, CC, and THA regions.
4. Discussion
The accumulation of from senile plaques has been considered one of the prominent neuropathological hallmarks of AD.3,4 Transgenic mice models carry disease-linked forms of genes associated with AD that have successfully replicated extracellular and intracellular accumulations.2,32 Despite there being several dyes and imaging techniques that visualize plaque, these techniques or antibodies have different sensitivities and specificities for identifying plaque. Consequently, histological examination of a label-free imaging method is a powerful tool for the investigation of AD, which can complement traditional biomedical procedures.
Here, we have established a label-free optical method for the investigation of accumulation by MPM. First, we demonstrate that the TPEF signal is a more sensitive optical marker for visualizing accumulation, revealing the spatial–temporal progression of extracellular deposition, as well as structural alterations around the plaques in APP/PS1 mice. More importantly, we identify intracellular without any exogenous dye for the first time in APP/PS1/Tau mice, evaluating the association between the distribution of intracellular accumulation and nuclear morphological changes. These results demonstrate that both extracellular and intracellular accumulations play an important pathophysiological role in the progression of AD.
According to previous studies,17,33 the molecular origin of SHG signals is likely attributable to microtubules in neurites of senile plaques. As a result, the changes of SHG signals in extracellular plaques are probably due to microtubules in some nascent senile plaques that do not have enough high density or complete noncentrosymmetric structures during the increasing of the deposition. By contrast, FAD, lipofuscins, and other compounds in extracellular plaques can generate TPEF signals,13,15 revealing the sensitivity of TPEF to detect plaques compared with SHG. On the other hand, can also be produced inside the cell, which exists in multiple assembly states including monomers, oligomers, protofibrils, and fibrils.34 Given our identification of intracellular , we find that accumulation can emit TPEF signals and lack SHG signals. This finding further reveals the molecular origin of autofluorescence in plaques. Moreover, it confirms that the SHG signals from extracellular plaque are noncentrosymmetric microtubules rather than accumulation. In addition, according to the senile plaque composition and the known endogenous fluorophores,17,18,35 we suggest that proteoglycans (such as keratin) and metal ions (such as Fe) also contribute to intrinsic TPEF signals for extracellular plaques.
In comparison with intracellular and extracellular accumulations, similar to previous studies,32,34 the accumulation and distribution of intracellular are consistent with the results of extracellular plaque. CTX and HC are the most easily and severely deposited regions. Although it is presently unclear how the AD features and the formation of plaques relate to the emergence of cognitive impairments,25,30,36 there still exists a chain effect that shows a relationship between intracellular and extracellular . On the other hand, we find that the neurotoxicity of intracellular can change the morphology of a cell nucleus, which may further cause neuronal apoptosis or death. This process might be the previous step of extracellular plaque formation. In addition, the presence of extracellular is accompanied by changes in the surrounding structures, including white matter damage, neuronal loss, cortical lamination abnormalities, and halo-like structures. Although we did not continue to study the toxicity of , these results suggest that the pathological changes in the brain microenvironment might be a crucial reason for functional impairment, which can provide a potential target and orientation for the treatment of AD.
Finally, we have combined MPM and the custom-developed automated image-processing program to rapidly label plaque by color coding, providing a rapid method for recognizing different degrees of deposition for neuroscientists. Computer-aided diagnosis algorithm methods have emerged to permeate biomedical optics imaging. Ounkomol et al.37 have presented a label-free method for predicting three-dimensional fluorescence directly from transmitted-light images. Liu et al.38 have established an automated method for quantifying the three-dimensional organization of fiber-like structures in biological tissues. In our future work, we will collect large volumes of raw data for training the deep learning model to automatically identify the deposition in the brain and to predict the formation rate in different regions.
In addition to , neurofibrillary tangles (NFTs) are also a significant neuropathological feature for AD. Unfortunately, we did not find NFTs or Tau by label-free MPM in our experiment, which may require fluorescent markers to accurately identify. Label-free MPM has several unique advantages, including inherent optical sectioning capability and low photobleaching and phototoxicity outside the imaging plane, which utilizes the endogenous contrast mechanisms to permit the exploration of unstained specimens. Moreover, the rise of commercially available systems facilitates MPM being more accessible to biologists. However, before full practical translation to in vivo and even clinical applications can occur, it is necessary to address current limitations of MPM, such as the imaging volume, penetration depth, user interface, and especially the size compatibility with biological problems and clinical practices. For biological use, fiberscopes are a primary type of miniature MPM, where a supple fiber bundle and a miniature compound objective are mounted to the head of a freely behaving mouse.39 For clinical use, microendoscopy techniques are advancing and could realize label-free, slide-free digital histopathology images in real time. It is an invasive approach that often combines photonic crystal fibers with a gradient refractive index lens or microprism technology to directly access the targeted deep brain region.40,41 Collectively, combined with advanced biological and clinical techniques of MPM, future MPM will integrate multiple advantages to achieve more in vivo studies of AD in common laboratories and hospitals.
5. Conclusion
In summary, we demonstrated the capability of label-free MPM to identify extracellular plaque as well as intracellular accumulation for the first time from AD mouse models, revealing that the TPEF signal is a sensitive optical marker for showing the spatial–temporal progression and the surrounding morphological changes of deposition. Moreover, we established a rapid method to visualize different degrees of deposition combined with a custom-developed image-processing program. Our results expanded the applications of label-free MPM in AD, indicating the importance of extracellular and intracellular accumulations in the progression of AD and verifying the feasibility of MPM data in image-processing procedures. With the technical development of MPM and its applications, we anticipate that our study may provide the groundwork to push forward the practicality of label-free MPM, which can complement biological and clinical procedures.
Supplementary Material
Acknowledgments
We thank Weilin Liu and Ruolan Lin for stimulating discussions. We also thank Lida Qiu and Zhida Chen for their technical support. This work is supported by National Natural Science Foundation of China (Nos. 81671730 and 61972187), the Joint Funds of Fujian Provincial Health and Education Research (No. WKJ2016-2-28), National High Technology Research and Development Program of China (No. 2015AA020508), National Key Basic Research Program of China (No. 2015CB352006), the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT_15R10), the Special Funds of the Central Government Guiding Local Science and Technology Development (No. 2017L3009), Natural Science Foundation of Fujian Province of China (No. 2019J01761), and Program for Undergraduate Education and Teaching Reform in University of Fujian Province of China (No. FBJG20190230).
Biographies
Shu Wang received his PhD in optics from Fujian Normal University, China. Currently, he is a research assistant professor at the College of Mechanical Engineering and Automation of Fuzhou University, Fuzhou, China. His main research interests are focused on the development and applications of nonlinear optical microscopy in brain and biomedical research.
Bingbing Lin received her master’s degree in medicine from Fujian University of Traditional Chinese Medicine, China. Currently, she is a medicine PhD candidate at the Fujian University of Traditional Chinese Medicine. Her research interests are focused on cognitive neuroscience and rehabilitation medicine research such as stroke and Alzheimer’s disease.
Guimin Lin is a PhD student at the College of Photonic and Electronic Engineering of Fujian Normal University, Fuzhou, China. Currently, he is an associate professor at Minjiang University. His research interests include image processing, machine learning, and computer vision.
Caihong Sun is studying for a master’s degree at Fujian Normal University, Fuzhou, China. Her main research interests are focused on the development and applications of nonlinear optical microscopy in biliary disease and biomedical research.
Ruolan Lin received his master’s degree from Fujian Medical University in 2016. Currently, he is a radiologist at Fujian Medical University Union Hospital. His main research interests are focused on the applications of magnetic resonance imaging in the nervous system.
Jia Huang received her PhD in medicine from Fujian University of Traditional Chinese Medicine, China. Currently, she is an associate professor at the College of Rehabilitation Medicine of Fujian University of Traditional Chinese Medicine. Her research interests are focused on cognitive neuroscience and speech rehabilitation medicine research.
Jing Tao received her PhD in medicine from Fujian University of Traditional Chinese Medicine, China. Currently, she is a professor at the College of Rehabilitation Medicine of Fujian University of Traditional Chinese Medicine. Her research interests are focused on the development and applications of rehabilitation medicine.
Xingfu Wang received his master’s degree in pathology in 2008 from Fujian Medical University, Fuzhou, China. Currently, he is an associate professor of pathology and neuropathologist at the First Affiliated Hospital of Fujian Medical University. His research interests include neuropathology and surgical pathology.
Yunkun Wu received his PhD from University of Alabama at Birmingham in 2007. Currently, he is a professor at the College of Life Science of Fujian Normal University. His main research interests are focused on protein structure and function.
Lidian Chen received his PhD in medicine from Fujian University of Traditional Chinese Medicine, China. Currently, he is the general secretary of and a professor at Fujian University of Traditional Chinese Medicine. His research interests are the development and application of rehabilitation medicine.
Jianxin Chen is the director of Key Laboratory of Optoelectronic Science and Technology for Medicine of Ministry of Education. Her research interests are the development and applications of nonlinear optical microscopy in biological and biomedical research.
Disclosures
The authors have declared that no competing interest exists.
Contributor Information
Shu Wang, Email: wangshu1990@hotmail.com.
Bingbing Lin, Email: lbbandsw@hotmail.com.
Guimin Lin, Email: gmlin@fjnu.edu.cn.
Caihong Sun, Email: 1622551457@qq.com.
Ruolan Lin, Email: ruolan.12@foxmail.com.
Jia Huang, Email: jasmine1874@163.com.
Jing Tao, Email: taojing01@fjtcm.edu.com.
Xingfu Wang, Email: wang_xfu@126.com.
Yunkun Wu, Email: wuyk@fjnu.edu.cn.
Lidian Chen, Email: cld@fjtcm.edu.cn.
Jianxin Chen, Email: chenjianxin@fjnu.edu.cn.
References
- 1.Alzheimer’s Association, “2018 Alzheimer’s disease facts and figures,” Alzheimer’s Dementia 14(3), 367–429 (2018). 10.1016/j.jalz.2018.02.001 [DOI] [Google Scholar]
- 2.Elder G. A., Sosa M. A. G., De Gasperi R., “Transgenic mouse models of Alzheimer’s disease,” Mt. Sinai J. Med. 77(1), 69–81 (2010). 10.1002/msj.v77:1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glenner G. G., Wong C. W., “Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein,” Biochem. Biophys. Res. Commun. 120(3), 885–890 (1984). 10.1016/S0006-291X(84)80190-4 [DOI] [PubMed] [Google Scholar]
- 4.Masters C. L., et al. , “Amyloid plaque core protein in Alzheimer disease and Down syndrome,” Proc. Natl. Acad. Sci. U.S.A. 82(12), 4245–4249 (1985). 10.1073/pnas.82.12.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klunk W. E., et al. , “Imaging brain amyloid in Alzheimer’s disease with Pittsburgh compound-B,” Ann. Neurol. 55(3), 306–319 (2004). 10.1002/ana.20009 [DOI] [PubMed] [Google Scholar]
- 6.Maeda J., et al. , “Longitudinal, quantitative assessment of amyloid, neuroinflammation, and anti-amyloid treatment in a living mouse model of Alzheimer’s disease enabled by positron emission tomography,” J. Neurosci. 27(41), 10957–10968 (2007). 10.1523/JNEUROSCI.0673-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jack C. R., Jr., et al. , “In vivo visualization of Alzheimer’s amyloid plaques by magnetic resonance imaging in transgenic mice without a contrast agent,” Magn. Reson. Med. 52(6), 1263–1271 (2004). 10.1002/mrm.20266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klunk W. E., et al. , “Imaging plaques in living transgenic mice with multiphoton microscopy and methoxy-X04, a systemically administered Congo red derivative,” J. Neuropathol. Exp. Neurol. 61(9), 797–805 (2002). 10.1093/jnen/61.9.797 [DOI] [PubMed] [Google Scholar]
- 9.McLellan M. E., et al. , “In vivo imaging of reactive oxygen species specifically associated with thioflavine S-positive amyloid plaques by multiphoton microscopy,” J. Neurosci. 23(6), 2212–2217 (2003). 10.1523/JNEUROSCI.23-06-02212.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee V. M.-Y., “Amyloid binding ligands as Alzheimer’s disease therapies,” Neurobiol. Aging 23(6), 1039–1042 (2002). 10.1016/S0197-4580(02)00121-5 [DOI] [PubMed] [Google Scholar]
- 11.Cohen A., et al. , “Anti-amyloid effects of small molecule -binding agents in PS1/APP mice,” Lett. Drug Des. Discovery 6(6), 437–444 (2009). 10.2174/157018009789057526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolmont T., et al. , “Label-free imaging of cerebral -amyloidosis with extended-focus optical coherence microscopy,” J. Neurosci. 32(42), 14548–14556 (2012). 10.1523/JNEUROSCI.0925-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Y., et al. , “Label-free brainwide visualization of senile plaque using cryo-micro-optical sectioning tomography,” Opt. Lett. 42(21), 4247–4250 (2017). 10.1364/OL.42.004247 [DOI] [PubMed] [Google Scholar]
- 14.Jo S., et al. , “GABA from reactive astrocytes impairs memory in mouse models of Alzheimer’s disease,” Nat. Med. 20(8), 886–896 (2014). 10.1038/nm.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwan A. C., et al. , “Optical visualization of Alzheimer’s pathology via multiphoton-excited intrinsic fluorescence and second harmonic generation,” Opt. Express 17(5), 3679–3689 (2009). 10.1364/OE.17.003679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J. H., et al. , “Label-free imaging and quantitative chemical analysis of Alzheimer’s disease brain samples with multimodal multiphoton nonlinear optical microspectroscopy,” J. Biomed. Opt. 20(5), 056013 (2015). 10.1117/1.JBO.20.5.056013 [DOI] [PubMed] [Google Scholar]
- 17.Zipfel W. R., et al. , “Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation,” Proc. Natl. Acad. Sci. U.S.A. 100(12), 7075–7080 (2003). 10.1073/pnas.0832308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monici M., “Cell and tissue autofluorescence research and diagnostic applications,” Biotechnol. Annu. Rev. 11, 227–256 (2005). 10.1016/S1387-2656(05)11007-2 [DOI] [PubMed] [Google Scholar]
- 19.Wang S., et al. , “Rapid, label-free identification of cerebellar structures using multiphoton microscopy,” J. Biophotonics 10(12), 1617–1626 (2017). 10.1002/jbio.v10.12 [DOI] [PubMed] [Google Scholar]
- 20.Lin P., et al. , “Diagnosing pituitary adenoma in unstained sections based on multiphoton microscopy,” Pituitary 21(4), 362–370 (2018). 10.1007/s11102-018-0882-6 [DOI] [PubMed] [Google Scholar]
- 21.Wang S., et al. , “Spatial and temporal identification of cerebral infarctions based on multiphoton microscopic imaging,” Biomed. Opt. Express 9(5), 2312–2325 (2018). 10.1364/BOE.9.002312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S., et al. , “Optical visualization of cerebral cortex by label-free multiphoton microscopy,” IEEE J. Sel. Top. Quantum Electron. 25(1), 6800508 (2019). 10.1109/JSTQE.2018.2854580 [DOI] [Google Scholar]
- 23.Zhuo S., et al. , “Multimode nonlinear optical imaging of the dermis in ex vivo human skin based on the combination of multichannel mode and lambda mode,” Opt. Express 14(17), 7810–7820 (2006). 10.1364/OE.14.007810 [DOI] [PubMed] [Google Scholar]
- 24.Yaseen M. A., et al. , “In vivo imaging of cerebral energy metabolism with two-photon fluorescence lifetime microscopy of NADH,” Biomed. Opt. Express 4(2), 307–321 (2013). 10.1364/BOE.4.000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers J. C., et al. , “Defining the earliest pathological changes of Alzheimer’s disease,” Curr. Alzheimer Res. 13(3), 281–287 (2016). 10.2174/1567205013666151218150322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wengenack T., et al. , “Quantitative histological analysis of amyloid deposition in Alzheimer’s double transgenic mouse brain,” Neuroscience 101(4), 939–944 (2000). 10.1016/S0306-4522(00)00388-2 [DOI] [PubMed] [Google Scholar]
- 27.Radde R., et al. , “-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology,” EMBO Rep. 7(9), 940–946 (2006). 10.1038/sj.embor.7400784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbi V., et al. , “Gray and white matter degeneration revealed by diffusion in an Alzheimer mouse model,” Neurobiol. Aging 34(5), 1440–1450 (2013). 10.1016/j.neurobiolaging.2012.11.017 [DOI] [PubMed] [Google Scholar]
- 29.Ji M., et al. , “Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy,” Sci. Adv. 4(11), eaat7715 (2018). 10.1126/sciadv.aat7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Billings L. M., et al. , “Intraneuronal causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice,” Neuron 45(5), 675–688 (2005). 10.1016/j.neuron.2005.01.040 [DOI] [PubMed] [Google Scholar]
- 31.D’Andrea M., et al. , “Evidence that neurones accumulating amyloid can undergo lysis to form amyloid plaques in Alzheimer’s disease,” Histopathology 38(2), 120–134 (2001). 10.1046/j.1365-2559.2001.01082.x [DOI] [PubMed] [Google Scholar]
- 32.Oddo S., et al. , “A dynamic relationship between intracellular and extracellular pools of ,” Am. J. Pathol. 168(1), 184–194 (2006). 10.2353/ajpath.2006.050593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunina N., Ihara Y., “Immunocytochemical study on senile plaques in Alzheimer’s disease. II,” Proc. Jpn. Acad. Ser. B 59(8), 288–292 (1983). 10.2183/pjab.59.288 [DOI] [Google Scholar]
- 34.LaFerla F. M., Green K. N., Oddo S., “Intracellular amyloid- in Alzheimer’s disease,” Nat. Rev. Neurosci. 8(7), 499–509 (2007). 10.1038/nrn2168 [DOI] [PubMed] [Google Scholar]
- 35.Atwood C. S., et al. , “Senile plaque composition and posttranslational modification of amyloid- peptide and associated proteins,” Peptides 23(7), 1343–1350 (2002). 10.1016/S0196-9781(02)00070-0 [DOI] [PubMed] [Google Scholar]
- 36.Trinchese F., et al. , “Progressive age‐related development of Alzheimer-like pathology in APP/PS1 mice,” Ann. Neurol. 55(6), 801–814 (2004). 10.1002/(ISSN)1531-8249 [DOI] [PubMed] [Google Scholar]
- 37.Ounkomol C., et al. , “Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy,” Nat. Methods 15(11), 917–920 (2018). 10.1038/s41592-018-0111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z., et al. , “Automated quantification of three-dimensional organization of fiber-like structures in biological tissues,” Biomaterials 116, 34–47 (2017). 10.1016/j.biomaterials.2016.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zong W., et al. , “Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice,” Nat. Methods 14(7), 713–719 (2017). 10.1038/nmeth.4305 [DOI] [PubMed] [Google Scholar]
- 40.Huland D. M., et al. , “Three-photon excited fluorescence imaging of unstained tissue using a GRIN lens endoscope,” Biomed. Opt. Express 4(5), 652–658 (2013). 10.1364/BOE.4.000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J. K., Choi J. W., Yun S. H. A., “ side-view optical probe for imaging the murine brain in vivo from the cortex to the hypothalamus,” J. Biomed. Opt. 18(5), 050502 (2013). 10.1117/1.JBO.18.5.050502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.