Abstract
While co‐infections are common in both wild and cultured fish, knowledge of the interactive effects of multiple pathogens on host physiology, gene expression and immune response is limited. To evaluate the impact of co‐infection on host survival, physiology and gene expression, sockeye salmon Oncorhynchus nerka smolts were infected with the salmon louse Lepeophtheirus salmonis (V−/SL+), infectious hematopoietic necrosis virus (IHNV; V+/SL−), both (V+/SL+), or neither (V−/SL−). Survival in the V+/SL+ group was significantly lower than the V−/SL− and V−/SL+ groups (p = 0.024). Co‐infected salmon had elevated osmoregulatory indicators and lowered haematocrit values as compared to the uninfected control. Expression of 12 genes associated with the host immune response was analysed in anterior kidney and skin. The only evidence of L. salmonis‐induced modulation of the host antiviral response was down‐regulation of mhc I although the possibility of modulation cannot be ruled out for mx‐1 and rsad2. Co‐infection did not influence the expression of genes associated with the host response to L. salmonis. Therefore, we conclude that the reduced survival in co‐infected sockeye salmon resulted from the osmoregulatory consequences of the sea lice infections which were amplified due to infection with IHNV.
Keywords: co‐infection, gene expression, infectious hematopoietic necrosis virus, physiology, salmon lice, sockeye salmon
1. INTRODUCTION
Co‐infections, defined as the infection of a host with two or more distinct pathogens, are common in both wild and cultured fish (Cox, 2001; Kotob, Menanteau‐Ledouble, Kumar, Abdelzaher, & El‐Matbouli, 2017). Co‐infections are classified as either synergistic in which one pathogen increases host susceptibility to another, or antagonistic in which the first pathogen hinders growth or survival of the second. Synergistic co‐infections can result in increased pathogen load, increased disease severity and increased mortality, while antagonistic co‐infections can result in lower pathogen load and decreased host mortality. The frequent occurrence of disease outbreaks during co‐infections in fish suggests that synergistic pathogen interactions are common. In addition, there is an interactive effect of multiple pathogens on host physiology, gene expression and immune response (Bandilla, Valtonen, Suomalainen, Aphalo, & Hakalahti, 2006; Barker et al., 2019).
In British Columbia (B.C.), infectious hematopoietic necrosis virus (IHNV) and Lepeophtheirus salmonis, the salmon louse, are enzootic pathogens of salmon and have overlapping host ranges. IHNV, a member of the genus Novirhabdovirus, is commonly isolated from Pacific salmon Oncorhynchus spp. (Wolf, 1988), and infections cause disease and mortality most often in the fry and juvenile life stages of wild and cultured salmonids (Dixon, Paley, Alegria‐Moran, & Oidtmann, 2016). In B.C., IHNV is primarily associated with sockeye salmon O. nerka although outbreaks have occurred in Atlantic salmon Salmo salar in net‐pen aquaculture operations (Saksida, 2006). Infection with IHNV induces a strong innate interferon response associated with a Th1‐type immune response (Purcell, Laing, & Winton, 2012).
Sockeye and Atlantic salmon are also highly susceptible to L. salmonis (Braden, Koop, & Jones, 2015; Johnson, Blaylock, Elphick, & Hyatt, 1996). Infections with L. salmonis can have a significant impact on the host osmotic equilibrium with the most severe effects occurring when the adult stages of the parasite are present (Bowers et al., 2000; Grimnes & Jakobsen, 1996; Long, Garver, & Jones, 2019). Changes in host gene expression resulting from L. salmonis infections include alterations in iron metabolism, carbohydrate metabolism and decreased expression of several antiviral genes (Braden et al., 2015; Krasnov, Skugor, Todorcevic, Glover, & Nilsen, 2012; Sutherland et al., 2014). Furthermore, initiation of a Th2‐type regulatory pathway in response to L. salmonis infection has been reported in both Pacific and Atlantic salmon although the timing and/or magnitude of the response is modified in more susceptible species (Braden et al., 2015; Skugor, Glover, Nilsen, & Krasnov, 2008). In addition to disruptions in osmoregulation and gene expression, infections with L. salmonis or Caligus rogercresseyi, another species of sea lice, can negatively impact the host's resistance to additional pathogens and facilitate entry of other pathogens into the host (Jakob, Barker, & Garver, 2011; Lhorente, Gallardo, Villanueva, Carabaño, & Neira, 2014; Mustafa, Speare, Daley, Conboy, & Burka, 2000).
Co‐infection studies involving L. salmonis and IHNV have not been conducted although differences in immune responses elicited by these pathogens as well as down‐regulation of host antiviral genes upon infection with L. salmonis (Braden et al., 2015; Sutherland et al., 2014) suggests the interaction between the two will be synergistic. In the current study, we explore the hypothesis that primary infection with L. salmonis will increase host susceptibility to a secondary infection with IHNV in sockeye salmon. Using an adult female L. salmonis infection model previously validated in our laboratory, we evaluated the impact of co‐infection on survival, physiology and gene expression in sockeye salmon smolts.
2. MATERIALS AND METHODS
2.1. Fish care
All procedures involving fish were carried out in accordance with the recommendations in the Canadian Council on Animal Care Guide to the Care and Use of Experimental Animals and approved by the Pacific Region Animal Care Committee, AUP 14‐029. All experimentations were conducted at the Pacific Biological Station (PBS; Nanaimo, B.C.).
Sockeye salmon (average body weight 152.2 g) from Pitt River stock were reared in brackish water and transferred to full sea water 10 days prior to initiation of the experiment. Fish were maintained at 9.06°C (±0.03°C) in 225‐L tanks (stock density 15.4 kg/m3) with UV‐treated flow‐through sea water (flow rate 3.5 L/min; salinity 28.0 ± 0.1 ppt), and kept under a natural photoperiod. Fish were fed a commercial diet (EWOS Canada) at a rate of 0.5% total biomass/day during the first 7 days of the trial and a rate of 1% total biomass/day for the remainder of the trial.
2.2. Experimental design
The experiment consisted of four treatment groups: uninfected control (V−/SL−); sea lice infection only (V−/SL+); virus infection only (V+/SL−); and co‐infection (V+/SL+). All treatments were conducted in duplicate tanks each containing 20 fish.
2.3. Sea lice collection and infection
Adult female L. salmonis were collected during harvest at an Atlantic salmon aquaculture site near mainland B.C. north of the Queen Charlotte Strait. Lice were rinsed in sea water and transported to PBS in chilled, aerated sea water. Upon arrival at PBS, lice were transferred to 10°C aerated static seawater baths and held up to 48 hr prior to experimentation.
Sockeye salmon were exposed to 6 sea lice/fish as previously described in Long et al. (2019). Fish in the V−/SL− and V+/SL− groups received the same handling treatment but were held in a mock 20‐L exposure tank without sea lice for 15 min before transfer to the holding tank.
2.4. Virus strain and infection
Infectious hematopoietic necrosis virus isolate BC93‐057 was isolated from a net‐pen reared Atlantic salmon during an epizootic in B.C. in 1993 (Garver et al., 2013). BC93‐057 was amplified in Epithelioma papulosum cyprini (EPC; ATCC CRL‐2872) cells and quantified using plaque assay as described previously (Batts & Winton, 1989). Plaques were enumerated and reported as plaque forming units per ml (pfu/ml).
Fish were exposed to IHNV by waterborne immersion challenge at 2 days post‐lice infection (dpl). Water flow to the tanks was stopped, and a volume of virus stock (108 pfu/ml) sufficient to produce a final virus concentration of 105 pfu/ml was added to the V+/SL− and V+/SL+ tanks. The same volume of sterile Hank's Balanced Salt Solution (Gibco) was added to the non‐virus exposure tanks. Immediately after addition of the virus to the tank, water was briefly stirred and a 1 ml water sample collected to quantify the virus load in each tank. After 1 hr with supplemental aeration and prior to resuming water flow to the tanks, a 1 ml water sample was collected to quantify the residual virus load in each tank. Fish were monitored daily for 30 days post‐virus (dpv), 32 dpl. All mortalities were examined for sea lice and screened for the presence of IHNV by quantitative RT‐PCR on anterior kidney samples.
2.5. Sample collection
Tissue and blood samples were collected from 10 fish per group (five fish per tank) at 3, 5, and 7 dpl (1, 3, and 5 dpv). At 32 dpl (30 dpv), samples were collected from survivors: 10 fish each from the V−/SL− and V−/SL+ groups, nine fish from the V+/SL− group, and four fish from the V+/SL+ group. For sampling, water flow to each tank was temporarily stopped and 0.15 mg/L of metomidate hydrochloride (Aquacalm; Syndel Canada) was added. After 12 min, five fish were individually netted into separate buckets and killed in 400 mg/L of tricaine methanesulfonate (MS‐222; Syndel Canada) in sea water. The total number of lice on the fish and in their individual buckets was used to determine mean parasite abundance according to Bush, Lafferty, Lotz, and Shostak (1997). Physical damage to skin was noted using a semi‐quantitative scale from 0 to 4: (0) no skin damage, no haemorrhaging, no lesions; (1) minor petechial haemorrhaging and/or scale loss over 25% or less of body surface; (2) widespread petechial haemorrhaging and/or scale loss over 25%–50% of body surface; (3) subcutaneous oedema (raised scales), scale loss over 50%–75% of body surface and/or areas of blood; and (4) lesions present, erosion of the epidermis, ulcers and/or scale loss over 75% or greater of body surface (Long et al., 2019). Blood was collected for haematocrit and plasma analysis as described in Long et al. (2019). Anterior kidney and skin tissue were taken for gene expression and viral load determination. Tissue samples were immediately flash‐frozen in liquid nitrogen and stored at −80°C. Skin samples 1 cm long and 1 cm wide were collected at a standardized location on the left mid‐flank directly above the lateral line where a line drawn from the anterior end of the dorsal fin intersected with the lateral line (Fast et al., 2002). If a sea louse was attached to this site, then the sample was taken on the right side in the same location.
2.6. RNA extraction and reverse transcription
Total RNA was extracted from anterior kidney and skin samples in TRIzol Reagent (Life Technologies) following manufacturer's instructions using 5‐mm stainless steel beads (Qiagen). Kidney tissue was mechanically homogenized in a TissueLyser II (Qiagen) for 2 min at 25 Hz, and skin was mechanically homogenized for 10 min at 30 Hz. RNA was stored at −80°C.
To prepare cDNA for viral load determination in anterior kidney samples, 1.5 µg of total RNA was reverse‐transcribed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems) following the manufacturer's instructions. cDNA was stored at −20°C until needed.
For gene expression analysis, RNA was DNase treated using a TURBO DNA‐free™ Total kit (Ambion) prior to cDNA synthesis. RNA quality was confirmed by agarose gel electrophoresis with a subset of samples from both tissues. To prepare cDNA, 1 µg of DNase‐treated RNA was reverse‐transcribed in a 40‐µl reaction using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems) with equal concentrations of random hexamers and Oligo d(T)18 primer (Thermo Scientific). cDNA samples were diluted 1:4 in nuclease‐free water and stored at −20°C until needed.
2.7. IHNV quantitative RT‐PCR
Quantification of IHNV in kidney tissue was carried out using published primer and probe sequences targeting the IHNV N gene (Purcell et al., 2013). An individual reaction was comprised of 1X TaqMan™ Universal PCR Master Mix (Applied Biosystems), 900 nm each of the forward and reverse primer, 250 nm each of the probe and artificial positive control, 2.5 µl cDNA template, and nuclease‐free water for a final reaction volume of 25 µl. Reactions were run on a Stratagene Mx3000P qPCR system following the manufacturer's protocol. To determine the number of virus copies per µg total RNA, a double‐stranded DNA gBLOCK fragment (IDT Technologies) consisting of the sequence targeted by the IHNV primers was used. An 8‐step serial dilution of the gBLOCK spanning 107 to 50 copies per reaction was used as a standard curve for each run. All samples and standard controls were tested in duplicate and considered positive if at least one replicate had a Ct value <40.
2.8. Host gene expression using quantitative real‐time PCR
Gene expression in anterior kidney and skin samples was analysed at 3 and 7 dpl (1 and 5 dpv). See Supporting Information Table S1 for primer concentrations, primer efficiency values, standard curve dilution, primer sequences, and source. To prepare the standard curve, equal volumes of DNase‐treated RNA from all samples were combined and cDNA prepared. Standard material was then diluted accordingly (Supporting Information Table S1). To confirm absence of genomic DNA, the standard control RNA was used in a no‐reverse transcriptase reaction for each primer set. If amplification occurred in the no‐RT reaction, there had to be a difference of at least five cycles between the no‐RT reaction and sample reactions. All reactions were carried out on a StepOne‐Plus machine (Applied Biosystems). An individual PCR mixture was comprised of 1X Power SYBR® Green PCR Master Mix (Applied Biosystems), 1 µl of diluted cDNA template, forward and reverse primers (concentrations given in Supporting Information Table S1), and nuclease‐free water for a final reaction volume of 15 µl. Cycling conditions were 95°C for 10 min and 40 cycles of 95°C for 5 s, 60°C for 20 s, and 72°C for 10 s. A dissociation curve was performed with each run to confirm specificity.
Genes of interest for this study were associated with acute phase response (serum amyloid a, saa; tumour necrosis factor, tnf) cytokines (interleukin‐1beta, il‐1β; interleukin‐4/13A, il‐4/13A; interleukin‐10, il‐10), antigen display (major histocompatibility class I, mhc I), interferon‐induced (mx‐1; radical s‐adenosyl methionine domain containing 2, rsad2), immunoglobulins (immunoglobulin M, igM; immunoglobulin T, igT), tissue repair (matrix metalloproteinase‐9, mmp‐9), and iron transport and circulation (hepcidin‐1, hep‐1; transferrin, tf). Reference gene candidates were elongation factor‐1alpha (ef‐1α), beta‐actin (β‐actin), dynein (dyn), eukaryotic translation initiation factor 3 subunit 6 (etif3s6) and mRNA turnover protein 4 homolog (mrto4). The three most stable reference genes were determined for each tissue type using geNorm (Vandesompele et al., 2002). The most stable genes in both skin and kidney tissue were ef‐1α, etif3s6 and mrto4 with collective M values of 0.53 and 0.51, respectively. Relative quantities were calculated from the raw fluorescence qPCR data using the global fitting model of Carr and Moore (2012) in the R package qpcR in R version 3.4.4 (R Core Team, 2018; Spiess, 2018). Target gene expression was normalized to that of the three most stable reference genes and log2 transformed for further analysis.
2.9. Statistical analysis
Plots of individual physiological parameters were visually analysed and data log10 transformed if non‐normality was indicated. Kaplan–Meier survival curves and log‐rank analysis of differences in mortality were generated using the survminer package in R (Kassambara & Kosinski, 2018) which generated adjusted p values (Bonferroni) of the pairwise comparisons. As skin damage data were non‐continuous, differences in values between treatments at a time point were analysed by a Kruskal–Wallis test followed by Dunn's multiple comparison (Holm‐adjusted p values).
To evaluate the effect of treatment, time and their interaction on physiological parameters, gene expression, lice abundance and virus copy number, a linear mixed‐effect model was employed using the nlme package (Pinheiro, Bates, DebRoy, Sarkar, & Team, 2018). The random effect term in the model was tank, and a term for unequal variance between treatments was included. The number of virus copies/µg RNA was log10 transformed prior to analysis. For the post hoc analysis, least‐square means were generated in R using lsmeans from the package emmeans (Lenth, 2019) with the lme model. The adjusted p values (Tukey) of the pairwise comparisons of the means were then used for the analysis and are reported. A Spearman's rank‐order correlation matrices between log2 CNRQ gene expression values and either virus copy/µg RNA or total number of lice for kidney or skin, respectively, were performed in R version 3.4.3.
Results for all analyses were considered significant if p ≤ 0.05. Graphs were prepared in R using the ggplot2 package (Wickham, 2009). R code examples are given in the Supporting Information Material (Data S1).
3. RESULTS
3.1. Survival and skin damage
The earliest and greatest number of mortalities was observed in the V+/SL+ group, where average cumulative mortality reached 60% and occurred from 12 to 21 dpl. This represented 6 (1 and 5 per tank) mortalities out of the 10 fish remaining after sampling at 7 dpl. In the V+/SL− group, average cumulative mortality was 10% (1 of 10 remaining fish) with the lone mortality occurring at 22 dpl. No mortalities occurred in either the V−/SL− or the V−/SL+ group. All mortalities were positive for IHNV by quantitative RT‐PCR. Survival in the V+/SL+ group was significantly lower than that in the V−/SL− and V−/SL+ groups but was not significantly different from the V+/SL− group (p = 0.024; Figure 1).
Figure 1.
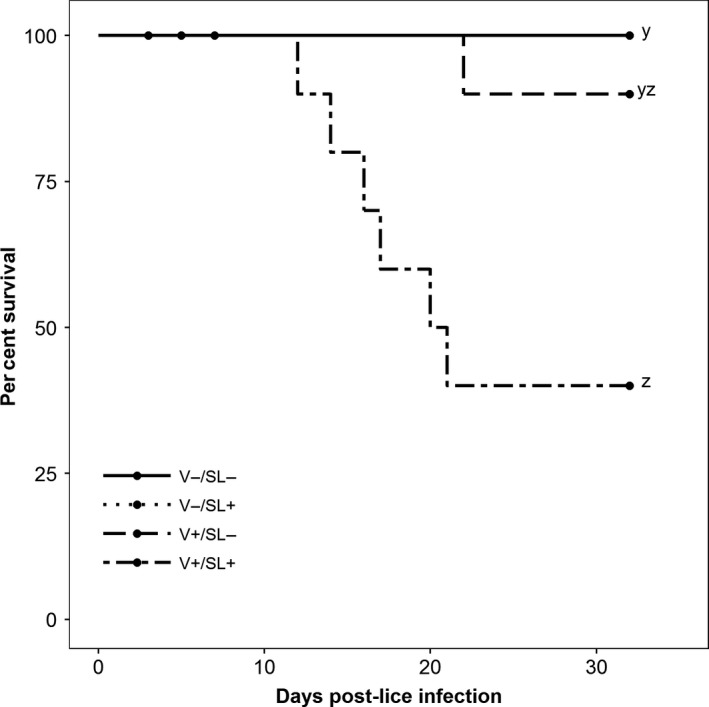
Kaplan–Meier survival curves (n = 10; 5 fish per tank after sampling at 7 dpl). The V−/SL+ line is hidden by the V−/SL− line. Circles denote sampling events. Letters denote statistically significant differences in survival between groups (p ≤ 0.05)
Median skin damage scores for the V−/SL+ group were significantly greater relative to the V−/SL− group at 3, 5 and 7 dpl (p < 0.05; Figure 2). At these times, median scores between the V−/SL+ and the V+/SL+ groups were not different, but at 7 dpl, the median score in the latter was also significantly greater than that of the V−/SL− group (Figure 2).
Figure 2.
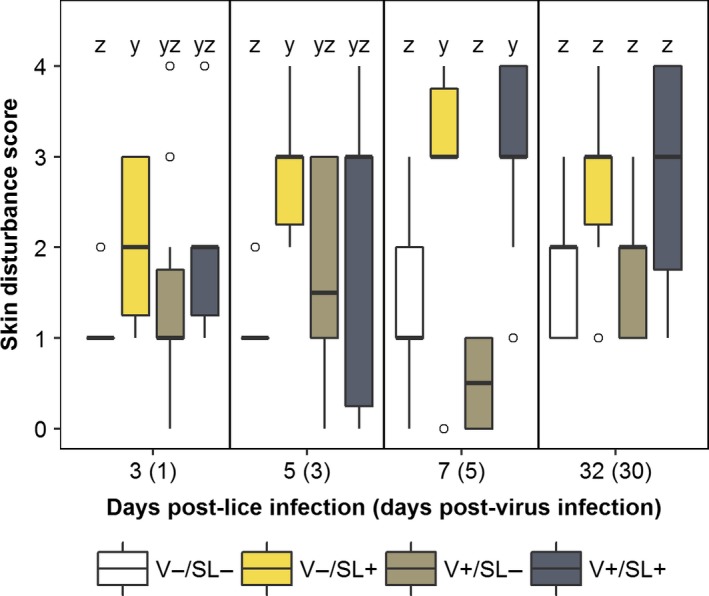
Skin damage scores in sockeye salmon sampled at 3, 5, 7 and 32 days post‐lice infection. Data are presented in box plots in which the inner horizontal line is the median, and the upper and lower boundaries of the box correspond to the first and third quartiles. The upper and lower whiskers denote the largest and smallest values no further than 1.5 times the inter‐quartile range. Open circles denote outliers. Letters denote statistically significant differences between groups at a sampling time (p ≤ 0.05) [Colour figure can be viewed at http://wileyonlinelibrary.com]
3.2. Pathogen load and prevalence
The prevalence of lice infections in the V−/SL+ and V+/SL+ groups declined over time (Table 1). In the V−/SL+ group, parasite abundance at 32 dpl was significantly lower than 3, 5 and 7 dpl (p < 0.05). In the V+/SL+ group, abundance at 32 dpl was significantly lower than 3 and 7 dpl but not 5 dpl (p < 0.05). The proportion of samples positive for IHNV infection peaked at 7 dpl (5 dpv) in the V+/SL− (9 of 10) and the V+/SL+ (8 of 10) groups (Figure 3). Furthermore, viral load was not statistically different between the V+/SL− and V+/SL+ groups at any of the sample times.
Table 1.
Prevalence and abundance of Lepeophtheirus salmonis on sockeye salmon exposed to either L. salmonis (V−/SL+) or L. salmonis and IHNV (V+/SL+) at 3, 5, 7 and 32 days post‐lice infection (n = 10). Abundance is expressed as mean number of lice per fish ± SE (range). Superscripts denote significant differences in lice abundance over time (p < 0.05)
| Days post‐lice infection | No. examined | No. infected | Abundance |
|---|---|---|---|
| V−/SL+ | |||
| 3 | 10 | 10 | 6.6 ± 1.4 (2–14)y |
| 5 | 10 | 10 | 6.5 ± 1.1 (2–13)y |
| 7 | 10 | 8 | 5.1 ± 1.5 (0–14)y |
| 32 | 10 | 4 | 0.6 ± 0.3 (0–3)z |
| V+/SL+ | |||
| 3 | 10 | 10 | 5.7 ± 0.7 (2–10)y |
| 5 | 10 | 8 | 3.2 ± 1.5 (0–15)yz |
| 7 | 10 | 10 | 5 ± 1.0 (1–11)y |
| 32 | 4 | 2 | 1 ± 0.7 (0–3)z |
Figure 3.
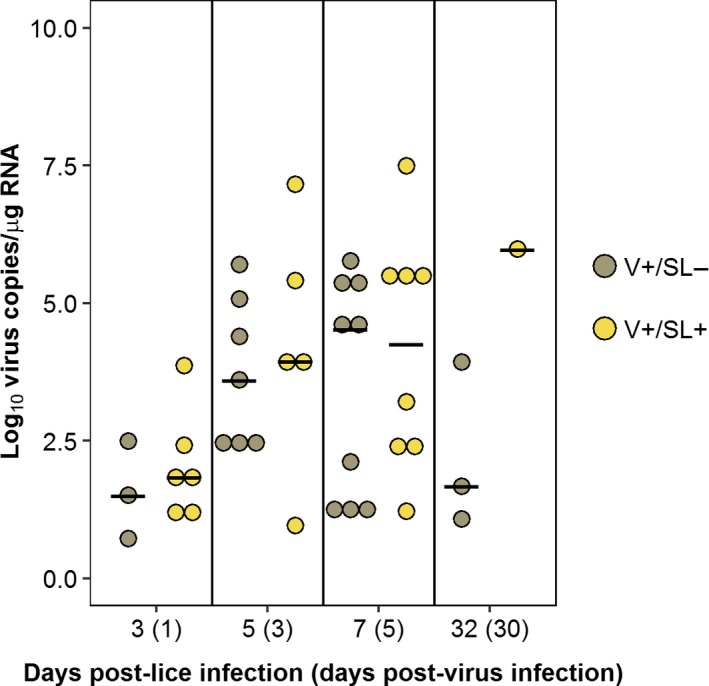
Log10 virus copies/µg RNA in anterior kidney from individual fish in the V+/SL− and V+/SL+ groups sampled at 3, 5, 7 and 32 days post‐lice infection (1, 3, 5 and 30 days post‐virus infection). Each dot represents an individual fish. The black bar denotes the median value for that group [Colour figure can be viewed at http://wileyonlinelibrary.com]
3.3. Physiological response
Treatment, but not time, had a significant effect on plasma osmolality, K+ and haematocrit (Figure 4; Supporting Information Table S2). Collectively, plasma osmolality values in the V+/SL+ group were higher than those in the V−/SL− group (p = 0.036). Similarly, plasma K+ values in the V+/SL+ group were significantly higher relative to all other groups (p < 0.05). Lastly, haematocrit values were significantly lower in the V+/SL+ group as compared to the V−/SL− group (p = 0.028).
Figure 4.
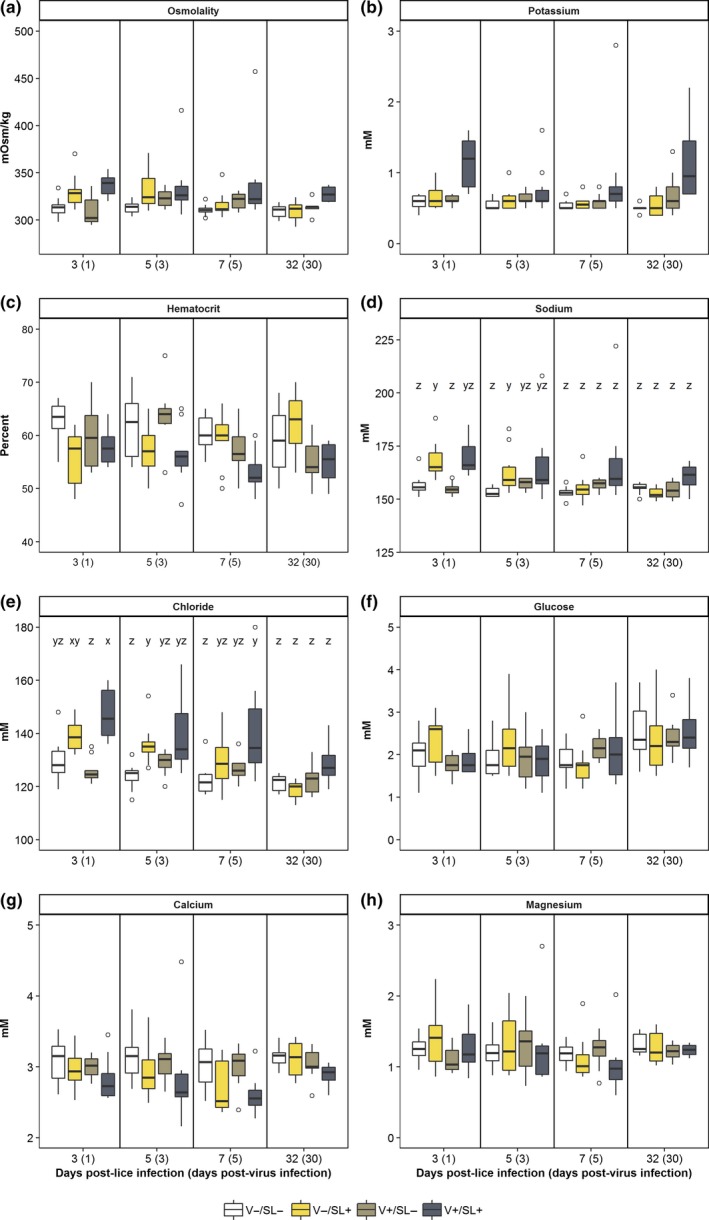
Physiological measurements from sockeye salmon sampled at 3, 5, 7 and 32 days post‐lice infection. See caption for Figure 2 for box plot description. Letters denote statistically significant differences between groups within a sampling time if there was a significant interaction of treatment and dpl for that parameter (p ≤ 0.05) [Colour figure can be viewed at http://wileyonlinelibrary.com]
Changes in the mean values of plasma Na+ and Cl− in each group were dependent on time (p < 0.05). At 3 dpl, mean plasma Na+ was significantly higher in the V−/SL+ group relative to the V−/SL− and V+/SL− group (Figure 4b). However, at 5 dpl, mean plasma Na+ in the V−/SL+ group was significantly higher than the uninfected control but not the V+/SL− group. Differences in mean plasma Na+ among groups were not detected at 7 and 32 dpl. A similar trend was observed for mean plasma Cl− values. At 3 and 7 dpl, mean plasma Cl− in the V+/SL+ group was significantly higher than the V−/SL− group (Figure 4c). However, at 5 dpl, differences between the V+/SL+ and V−/SL− groups were not significant although mean plasma Cl− values in the V−/SL+ group were significantly higher than the V−/SL− group. There were no significant differences in mean plasma Cl− values between the uninfected control and V+/SL− group at any time point.
3.4. Gene expression in anterior kidney
Treatment but not time had an effect on the expression of rsad2 which was significantly elevated in the V+/SL− group as compared to the V−/SL+ group (p = 0.01). The effect of treatment on the relative expression of several innate immune response genes (saa, il‐1β, il‐10, mx‐1, hep‐1 and mmp‐9) changed over time (p < 0.05; Figure 5). Compared to the uninfected control, expression of saa was significantly elevated in the V−/SL+ group at 3 dpl but not at 7 dpl. At 7 dpl, expression of saa in the V+/SL− and V+/SL+ groups was significantly elevated relative to the uninfected control (Figure 5a). Expression of il‐1β, il‐10 and mx‐1 did not differ between groups at 3 dpl, but significant differences were observed at 7 dpl. The expression of il‐1β in the V+/SL− and V+/SL+ groups was elevated as compared to the V−/SL− group (Figure 5b). Similarly, il‐10 expression at 7 dpl was significantly greater in the V+/SL− group compared with the V−/SL− and V−/SL+ groups (Figure 5d). Lastly, mx‐1 expression was significantly greater in the V+/SL− group compared with the V−/SL+ group (Figure 5e). Conversely, significant differences in hep‐1 and mmp‐9 expression were observed only at 3 dpl. Expression of hep‐1 in the V+/SL− group was significantly downregulated relative to the V−/SL+ and V+/SL+ groups (Figure 5h). The expression of mmp‐9 was significantly elevated in the V+/SL+ group compared with the V+/SL− group (Figure 5l).
Figure 5.
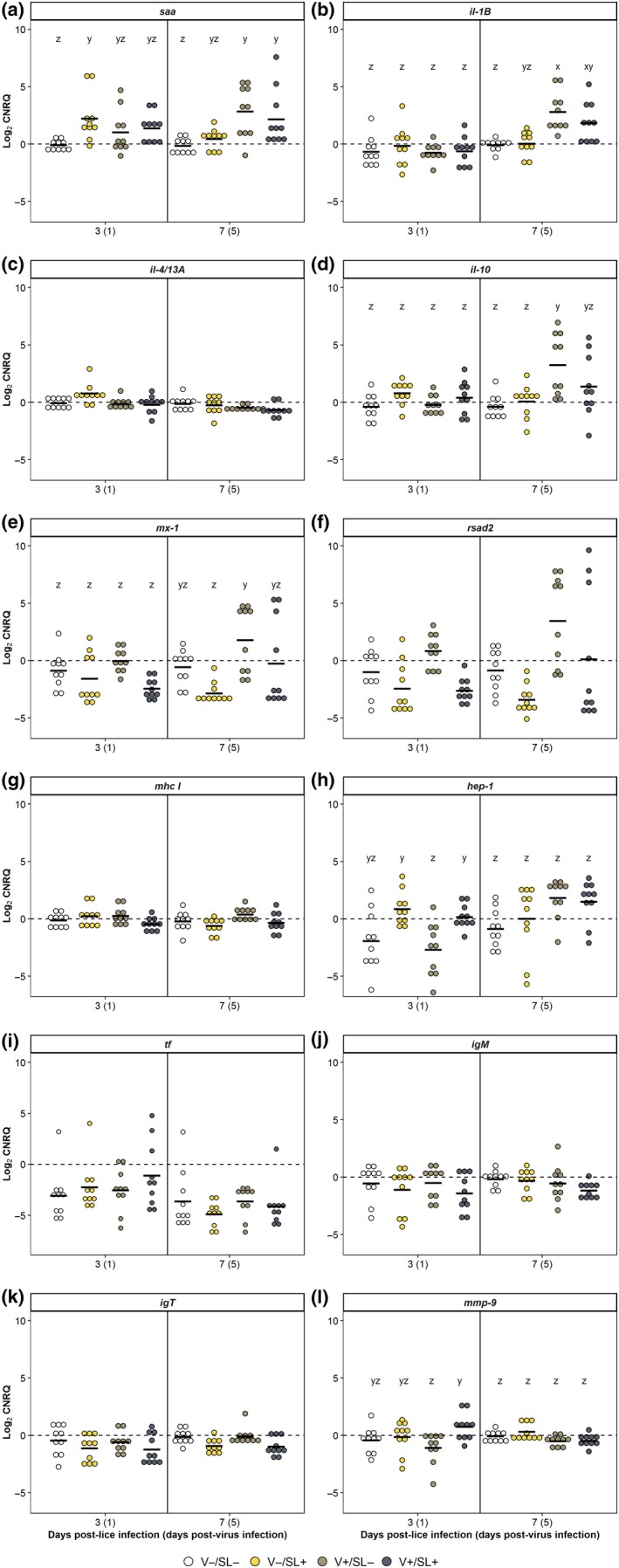
Gene expression for sockeye salmon kidney sampled at 3 and 7 days post‐lice infection (1 and 5 days post‐virus infection). Each dot represents an individual fish. The black bar denotes the mean expression value for that group. Letters denote statistically significant differences in values between groups within a sampling time if there was a significant interaction of treatment and dpl for that parameter (p ≤ 0.05) [Colour figure can be viewed at http://wileyonlinelibrary.com]
3.5. Gene expression in skin
Treatment but not time had a significant effect on il‐1β, mmp‐9, mx‐1 and tf expression (p < 0.05). The expression values of il‐1β and mmp‐9 were significantly higher in the V−/SL+ and V+/SL+ groups relative to the V−/SL− and V+/SL− groups. Expression of tf was significantly higher in the V−/SL+ group than in the V−/SL− and V+/SL− groups but not the V+/SL+ group. Although expression of mx‐1 in the V+/SL− group was significantly higher compared to the V−/SL+ group, there were no significant differences among the V+/SL−, the V+/SL+ and the V−/SL− groups.
The effect of treatment on the expression of il‐10, rsad2, mhc I and hep‐1 changed over time (p < 0.05; Figure 6). With the exception of hep‐1, significant differences in the expression of these genes were only observed at 7 dpl. Thus, expression of rsad2 and il‐10 was significantly lower in the V−/SL+ group compared with the V−/SL− and V+/SL− groups (Figure 6d,f). In the V+/SL− group, rsad2 expression was significantly greater compared to the V−/SL− group (Figure 6f). Expression of mhc I was significantly greater in the V+/SL− group than in all other groups (Figure 6g). At 3 dpl, expression of hep‐1 was significantly greater in the V−/SL+ and V+/SL+ groups than in the V−/SL− and V+/SL− groups. At 7 dpl, expression of hep‐1 in the V+/SL+ group was significantly greater than the V−/SL− group.
Figure 6.
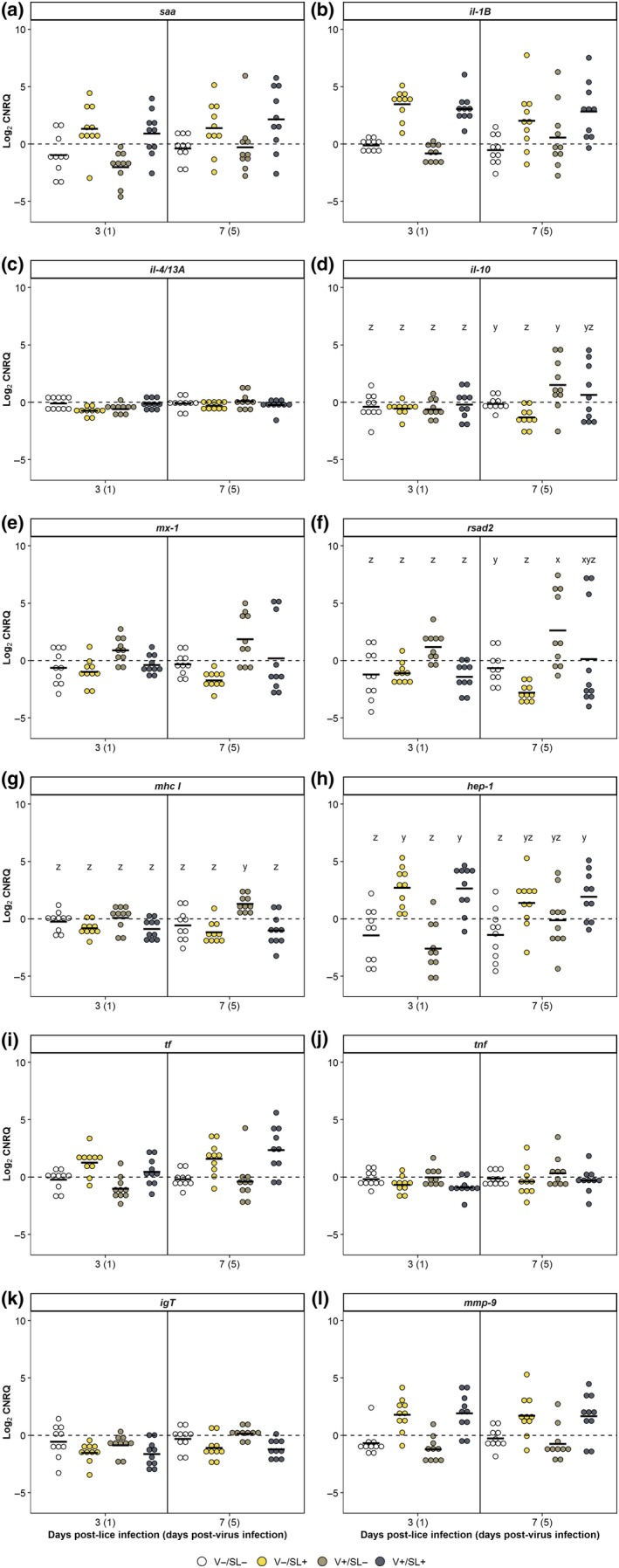
Gene expression for sockeye salmon skin sampled at 3 and 7 days post‐lice infection (1 and 5 days post‐virus infection). See caption for Figure 5 [Colour figure can be viewed at http://wileyonlinelibrary.com]
3.6. Correlation analysis
Correlation analysis showed that in the V+/SL− group at 7 dpl, expression values of genes associated with the host response to IHNV (il‐1β, il‐10, mx‐1, rsad2 and saa) were significantly correlated to virus load in the anterior kidney. In the presence of sea lice (V+/SL+), correlations between viral load and expression of saa and il‐10 were no longer evident (Table 2). In skin, the expression values of most genes associated with the host response to L. salmonis infection (hep‐1, tf, mmp‐9, il‐1β) were not correlated to the total number of lice present. However, tf expression was significantly correlated to total number of lice in the V−/SL+ group but not the V+/SL+ group (Table 2). Finally, mx‐1 expression was correlated with number of sea lice in the V+/SL+ group but not the V−/SL+ group (Table 2).
Table 2.
Results of Spearman's rank‐order correlation between gene expression values and virus copy number/µg RNA (kidney) or total number of lice (skin) for individual fish. r s values are given for individual genes. If the correlation was statistically significant (p ˂ 0.05), r s values are bolded and the p‐value is given in parentheses. N.D. denotes not done
| Gene | Kidney | Skin | ||
|---|---|---|---|---|
| V+/SL− | V+/SL+ | V−/SL+ | V+/SL+ | |
| saa | 0.72 (0.030) | 0.5 | 0.62 | 0.11 |
| hep‐1 | 0.78 (0.013) | 0.36 | 0.52 | 0.20 |
| igM | −0.42 | 0.02 | N.D. | N.D. |
| igT | −0.067 | −0.40 | −0.55 | 0.62 |
| il‐1β | 0.77 (0.016) | 0.86 (<0.01) | 0.59 | 0.25 |
| il‐10 | 0.8 (<0.01) | 0.60 | −0.018 | 0.34 |
| il‐4/13A | −0.65 | −0.45 | −0.40 | 0.30 |
| mhc I | −0.1 | 0.52 | −0.40 | 0.68 (0.032) |
| mmp‐9 | 0.067 | −0.71 (0.047) | 0.5 | −0.03 |
| mx‐1 | 0.88 (<0.01) | 0.76 (0.028) | −0.26 | 0.74 (0.013) |
| tf | 0.5 | 0.62 | 0.65 (0.043) | 0.13 |
| rsad2 | 0.88 (<0.01) | 0.74 (0.037) | −0.49 | 0.39 |
| tnf | N.D. | N.D. | 0.18 | −0.06 |
4. DISCUSSION
Previous gene expression analyses of Pacific salmon infected with L. salmonis postulated that parasitized sockeye salmon would be more susceptible to viral infections (Braden et al., 2015; Sutherland et al., 2014) due to the suppression of antiviral responses. In the study herein, although survival was reduced in the co‐infected group as compared to the other groups, there was no difference in the prevalence of virus infections or mean viral load between virus‐only and co‐infected salmon indicating that reduced survival of the co‐infected salmon was not a consequence of increased infection with IHNV. The lack of statistical significance between the co‐infection and virus‐only groups was likely due to the low number of biological replicates in each group (n = 10). We hypothesize that co‐infection of sea lice and IHNV in sockeye salmon altered the host capacity to modulate the effects of sea lice infection indicating a synergistic interaction between L. salmonis and IHN virus. Decreased survival in Atlantic salmon co‐infected with L. salmonis and infectious salmon anaemia virus (ISAV) has also been reported (Barker et al., 2019). Barker et al. (2019) concluded that sea lice‐infected fish modulated the host immune system resulting in increased susceptibility to ISAV. Given the differences between the two studies (host species, virus, study design and sample numbers, parameters examined), further investigation is necessary to determine whether co‐infection increases host susceptibility and modulates host response to the physiological effects of sea lice infection, or both.
Sockeye salmon are highly susceptible to L. salmonis and infection results in increased plasma osmolality and Na+ and Cl− concentrations along with severe cutaneous lesions and subcutaneous oedema (Braden et al., 2015; Jakob, Sweeten, Bennett, & Jones, 2013; Johnson et al., 1996; Long et al., 2019). Host osmoregulation is affected by both the direct (attachment and feeding) and indirect (passive loss of water across the gills due to stress) effects of L. salmonis infection (Wendelaar Bonga, 1997). In contrast, the effects of IHNV on host osmoregulation are not well documented although Amend and Smith (1975) reported reduced plasma osmolality in moribund rainbow trout Oncorhynchus mykiss. In the current study, co‐infected salmon had higher skin disruption scores, elevated osmoregulatory indicators and lowered haematocrit values as compared to the uninfected control. There was no disruption in osmoregulatory indicators in salmon infected only with IHNV, whereas elevated osmoregulatory indicator values were transient in salmon infected only with sea lice. Therefore, we conclude that the reduced survival in co‐infected sockeye salmon resulted from the osmoregulatory consequences of the sea lice infections which were amplified in the presence of infection with IHN virus.
Upon infection with a virus, the host immune system initiates differentiation of Th lymphocytes into Th1 cells resulting in the production of cytotoxic T cells and interferon‐γ as well as promoting macrophage activation (Bradley & Jackson, 2008; Cox, 2001). In contrast, Th2 cells are recruited when extracellular pathogens such as parasites are present, and this response is associated with up‐regulation of il‐4/13A, il‐10 and transforming growth factor beta. Activation of B cells and proliferation of eosinophils due to cytokine production is a hallmark of the Th2 response to parasite infection (Cox, 2001). Braden et al. (2015) reported activation of a Th2‐type regulatory pathway in the skin of L. salmonis‐resistant coho salmon Oncorhynchus kisutch and hypothesized this as a mechanism of resistance as activation of this pathway was not detected in the susceptible sockeye and Atlantic salmon. In the current study, sockeye salmon displayed no evidence of a Th2‐type pathway as il‐4/13A expression was unchanged during L. salmonis infection and up‐regulation of il‐10 only occurred in response to IHNV exposure. This suggests that the enhanced susceptibility to sea lice in co‐infected salmon was not related to switching from an anti‐parasite Th2 to a virus‐type Th1 immune response. Analysis of the expression of additional response‐specific genes will be required to adequately address this question.
Hepcidin and transferrin proteins are involved in iron homeostasis. Modulation of their abundance during infections is an important component of nutritional immunity in which the host restricts the availability of essential metals that are otherwise available to pathogens (Hood & Skaar, 2012). Lepeophtheirus salmonis is unable to synthesize haem (Brandal, Egidius, & Romslo, 1976), and evidence of host nutritional immunity during L. salmonis infections has previously been documented in Atlantic and Pacific salmon (Braden et al., 2015; Sutherland et al., 2014; Valenzuela‐Muñoz & Gallardo‐Escárate, 2017). In the present study, hep‐1 expression in skin was up‐regulated in salmon lice‐infected salmon with or without a co‐infection. Up‐regulation of hep‐1 in the skin of sockeye salmon during L. salmonis infection is likely due to inflammation in this tissue. Increased hep‐1 expression has been linked to tissue inflammation which has been reported in sockeye salmon infected with L. salmonis (Braden et al., 2015; Johnson et al., 1996; Nicolas et al., 2002). Similarly, tf expression was up‐regulated in skin of salmon lice‐infected salmon with or without a virus co‐infection. Therefore, we can conclude that the expression of hep‐1 and tf in sockeye salmon skin was induced by L. salmonis and that co‐infection with IHNV did not significantly impact the host nutritional immune response to L. salmonis.
Matrix metalloproteinases are primarily responsible for extracellular matrix degradation and tissue remodelling which occurs during the inflammatory response (Chadzinska, Baginski, Kolaczkowska, Savelkoul, & Verburg‐van Kemenade, 2008). In the kidney and skin of sockeye salmon, mmp‐9 was up‐regulated in response to L. salmonis infection which is in agreement with previous studies (Braden et al., 2015; Skugor et al., 2008; Sutherland, Jantzen, Sanderson, Koop, & Jones, 2011; Tadiso et al., 2011). In the present study however, up‐regulation of mmp‐9 in response to IHNV infection was observed in neither tissue although mmp‐9 was induced in the kidney of IHNV‐infected rainbow trout (MacKenzie et al., 2008). Inflammation due to infection typically results in up‐regulation of il‐1β leading to an influx of leucocytes which preferentially express mmp‐9 (Hong, Peddie, Campos‐Pérez, Zou, & Secombes, 2003; Krasnov, Timmerhaus, Afanasyev, & Jørgensen, 2011). Significant up‐regulation of il‐1β was not observed in the kidney of salmon infected with virus alone until 7 dpl (5 dpv), suggesting the possibility that mmp‐9 expression was up‐regulated after 7 dpl.
Serum amyloid A (SAA) is an acute phase protein whose levels increase in response to inflammation (Jensen et al., 1997; Rebl, Goldammer, Fischer, Köllner, & Seyfert, 2009). Several cytokines, including interleukin‐1β (IL‐1β), can induce transcription of this gene (Jørgensen, Lunde, Jensen, Whitehead, & Robertsen, 2000). In fish, saa has been induced in immune organs in response to viral, bacterial and parasitic infections (Braden et al., 2015; Chettri et al., 2014; Sutherland et al., 2014; Villarroel et al., 2008). Villarroel et al. (2008) have proposed that SAA is involved in local defence against pathogens as they were unable to detect SAA in plasma of fish infected with Flavobacterium psychrophilum, but the gene was expressed in kidney, liver and spleen cells. In the current study, there was no evidence of up‐regulation of saa in skin in response to either pathogen. Expression of saa in kidney was up‐regulated in a pathogen‐dependent pattern. At 3 dpl (1 dpv), expression was only associated with salmon lice infection, whereas at 7 dpl (5 dpv), expression of saa occurred in the virus‐infected groups, perhaps indicative of time‐dependent patterns of inflammation.
IL‐1β is a pro‐inflammatory cytokine that enhances migration of leucocytes, modulates expression of IL‐17 by Th17 cells and induces anti‐inflammatory cytokines including IL‐10 (Hong et al., 2003; Skugor et al., 2008; Zou & Secombes, 2016). Increased expression of il‐1β in response to either L. salmonis or IHNV has been documented (Braden et al., 2015; Fast, Ross, Muise, & Johnson, 2006; Peñaranda, Purcell, & Kurath, 2009; Purcell, Kurath, Garver, Herwig, & Winton, 2004; Purcell, Marjara, Batts, Kurath, & Hansen, 2011; Sutherland et al., 2014). In the current study, il‐1β expression varied depending on tissue and target organ of the individual pathogen, regardless of co‐infection status. In kidney, il‐1β expression was up‐regulated during virus infections while in skin, expression was up‐regulated during sea lice infections. These results agree with previous single infection studies, and therefore, we can conclude that expression of il‐1β does not appear to be negatively impacted by co‐infection in sockeye salmon (Braden et al., 2015; Chettri et al., 2014; Purcell et al., 2004).
IL‐10 is a pleiotropic, anti‐inflammatory cytokine that down‐regulates inflammatory Th responses (Zou & Secombes, 2016). As such, increased expression of il‐10 in kidney during virus‐only infections was likely in response to increased il‐1β expression in this tissue. Concurrently, the absence of statistical differences in the expression of il‐10 between co‐infected salmon and any of the other groups may indicate modulation of gene expression due to L. salmonis infection in the co‐infected fish. Down‐regulation of il‐10 in skin from L. salmonis‐infected sockeye salmon has been previously reported (Braden et al., 2015) and was again observed in the current study, demonstrating that regulation of the inflammatory response is impaired in skin of susceptible salmon species during the infection.
Transcription of mhc I is induced by interferon in response to virus infection. Expression of this gene in rainbow trout infected with IHNV varies by tissue, days post‐exposure and virus strain with the highest fold change reported in liver and spleen at 7 days post‐exposure to highly virulent IHNV isolates (ATCC #VR‐1392, 220‐90 and BLK94; Landis, Purcell, Thorgaard, Wheeler, & Hansen, 2008; Purcell et al., 2011). In contrast, sea lice are anticipated to down‐regulate mhc I in kidney, as L. salmonis infection of Atlantic salmon had decreased mhc I levels (Fast et al., 2006). In our study, no significant differences in mhc I were observed in sockeye salmon infected with sea lice alone. Overall, there were no significant changes of mhc I expression in kidney, despite sampling during the time of peak virus load. However, increased expression of mhc I is often reported 3 days after virus infection (Hansen & LaPatra, 2002; Landis et al., 2008) and may have been missed in this study due to the timing of sampling.
In skin tissue, the lack of change of mhc I expression in sockeye salmon exposed to sea lice versus those uninfected agrees with previous studies that failed to demonstrate a difference in expression regardless of L. salmonis infection status (Braden et al., 2015; Fast et al., 2006). Conversely, in sockeye salmon infected only with IHNV, mhc I expression was elevated indicating virus‐induced expression. As IHNV replicates in skin cells both in vivo and in vitro, up‐regulation of mhc I in this tissue is not unexpected (Harmache, LeBerre, Droineau, Giovannini, & Brémont, 2006; Yamamoto, Batts, & Winton, 1992). However, as expression of mhc1 is reduced in co‐infected fish, it is probable that the virus‐induced expression of mhc I was negatively impacted by L. salmonis exposure.
Both mx‐1 and rsad2 are strongly induced by type I interferon, and their products are key components of the host antiviral response (Robertsen, 2008). Down‐regulation of mx‐1 has been reported in both anterior kidney and skin of Pacific salmon infected with L. salmonis (Braden et al., 2015; Sutherland et al., 2014). Conversely, in salmon infected with IHNV, mx‐1 and rsad‐2 levels typically peak between 2 and 3 days after infection (Peñaranda et al., 2009; Purcell et al., 2011). At 7 dpl (5 dpv), mean expression of these genes in both tissues was lower in salmon infected with sea lice only compared with those infected with IHNV only. We had hypothesized that L. salmonis infections would result in down‐regulation of interferon‐induced genes in co‐infected fish; however, expression of these genes did not differ between the virus‐infected and the co‐infected salmon. It should be noted that expression levels of both genes in co‐infected salmon did not differ from those of the negative control in either tissue. Mapping protein expression will be necessary to determine whether the observed differences in the transcriptomic response result in measurable differences in the amount of protein produced.
Analysis of cytokine gene expression in skin highlighted an interesting pattern of expression in salmon infected with the virus alone. The genes rsad2, mx‐1 and mhc I were all up‐regulated indicative of an antiviral response. Furthermore, although il‐10 expression was greatest in this group at 7 dpl (5 dpv), the expected increase in il‐1β expression was not detected. A similar observation was made in Atlantic salmon infected with infectious pancreatic necrosis virus (IPNV): up‐regulation of il‐10 in conjunction with a lack of induction of il‐1β and il‐8 (Reyes‐Cerpa et al., 2012). The authors hypothesize that IPNV triggered an anti‐inflammatory response which the virus then used to aid in establishment of persistence, a strategy which has been reported for other animal viruses (Wilson & Brooks, 2010). Replication of IHNV in epidermal tissue and persistence of the virus in brain tissue of sockeye salmon have been reported (Müller, Sutherland, Koop, Johnson, & Garver, 2015; Yamamoto, Batts, Arakawa, & Winton, 1990). Therefore, we hypothesize that IHNV also regulates il‐10 expression which would allow for virus replication in epidermal tissue and potentially enable persistence in infected hosts. To determine whether IHNV employs such a strategy, further testing is needed in which expression of other pro‐inflammatory cytokine genes such as il‐8 is measured to determine whether il‐10 is up‐regulated in response to these genes. In addition, it is necessary to measure gene expression at additional time points to see how cytokine gene expression in skin changes through the course of an IHN infection.
In our study, co‐infection did not appear to alter igT expression in either skin or kidney tissue while expression of igM was not detected in skin from sockeye salmon. This is in contrast to previous studies indicating increased transcript levels of both genes during ectoparasite infection. Chettri et al. (2014) observed increased expression of igM in skin from rainbow trout infected with Ichthyobodo necator at 9 dpi. Similarly, Tadiso et al. (2011) reported highest igM and igT levels in skin of Atlantic salmon infected with L. salmonis at 15 days post‐copepodid infection. In addition to the differences in host species, samples were collected when parasite load was high, greater than 50 parasites/fish, in contrast to the current study in which average parasite load was less than 7 lice/fish at both sampling times. Given these conflicting results, further work is needed to develop a better understanding of the effects of co‐infection on the kinetics of antibody‐mediated immunity.
Correlation analysis of gene expression of individual fish to pathogen load revealed that viral load strongly influenced the magnitude of the antiviral response while the level of host response to sea lice was not necessarily dictated by the parasite load. Expression values for genes associated with the host response to L. salmonis such as hep‐1, tf, mmp‐9 and il‐1β were not significantly correlated to the total number of lice per fish. In contrast, expression values of genes associated with the host response to IHNV (saa, il‐1β, il‐10, mx‐1 and rsad2) in salmon infected with virus alone were correlated to viral load. A similar trend has been reported for both IHNV and viral haemorrhagic septicaemia (Avunje, Kim, Park, Oh, & Jung, 2011; Purcell, LaPatra, Woodson, Kurath, & Winton, 2010; Zou et al., 2014). In co‐infected salmon, expression values of only il‐1β, mx‐1 and rsad2 were correlated to viral load.
5. CONCLUSION
This study showed that the outcome of L. salmonis and IHNV co‐infections differed from those of single infections in sockeye salmon. Survival in co‐infected fish was reduced compared to both single infection groups, indicating that the two pathogens interacted synergistically with one another during co‐infections. There was a significant physiological disruption in co‐infected fish, suggesting the presence of IHNV partially impaired the host recovery from L. salmonis. With regard to gene expression, the only evidence of L. salmonis‐induced modulation of the host antiviral response was down‐regulation of mhc I although the possibility of modulation cannot be ruled out for interferon‐induced genes. There was no effect of co‐infection on the expression of genes associated with the host response to L. salmonis. This research highlights the need for whole organism analysis in conjunction with transcriptomic analysis to fully understand the impacts of co‐infection on the susceptible host.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
Special thanks to Brad Boyce (Marine Harvest Canada) for coordinating sea lice collection. We would like to acknowledge the assistance of Julia Bradshaw, Steve Cho, Derek Chow, Mohammad Ghodsi, Kevin Huang, Amelia Mahoney, Haley Matkin, Mark Polinski, Jon Richard, Katie Verkaik, and the Aquarium Services staff at the Pacific Biological Station. We would also like to thank Dr. Gael Kurath for providing valuable comments on this manuscript. Plasma analysis was carried out by the Diagnostics Services Laboratory at Atlantic Veterinary College. AL was funded by the NSERC Visiting Fellowships in Canadian Government Laboratories Program with support from the DFO Program for Aquaculture Regulatory Research.
Long A, Garver KA, Jones SRM. Synergistic osmoregulatory dysfunction during salmon lice (Lepeophtheirus salmonis) and infectious hematopoietic necrosis virus co‐infection in sockeye salmon (Oncorhynchus nerka) smolts. J Fish Dis. 2019;42:869–882. 10.1111/jfd.12989
REFERENCES
- Amend, D. F. , & Smith, L. (1975). Pathophysiology of infectious hematopoietic necrosis virus disease in rainbow trout: Hematological and blood chemical changes in moribund fish. Infection and Immunity, 11, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avunje, S. , Kim, W.‐S. , Park, C.‐S. , Oh, M.‐J. , & Jung, S.‐J. (2011). Toll‐like receptors and interferon associated immune factors in viral haemorrhagic septicaemia virus‐infected olive flounder (Paralichthys olivaceus). Fish & Shellfish Immunology, 31, 407–414. 10.1016/j.fsi.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Bandilla, M. , Valtonen, E. , Suomalainen, L.‐R. , Aphalo, P. , & Hakalahti, T. (2006). A link between ectoparasite infection and susceptibility to bacterial disease in rainbow trout. International Journal for Parasitology, 36, 987–991. 10.1016/j.ijpara.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Barker, S. E. , Bricknell, I. R. , Covello, J. , Purcell, S. , Fast, M. D. , Wolters, W. , & Bouchard, D. A. (2019). Sea lice, Lepeophtheirus salmonis (Krøyer 1837), infected Atlantic salmon (Salmo salar L.) are more susceptible to infectious salmon anemia virus. PLoS One, 14, e0209178 10.1371/journal.pone.0209178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batts, W. N. , & Winton, J. R. (1989). Enhanced detection of infectious hematopoietic necrosis virus and other fish viruses by pretreatment of cell monolayers with polyethylene glycol. Journal of Aquatic Animal Health, 1, 284–290. [DOI] [Google Scholar]
- Bowers, J. , Mustafa, A. , Speare, D. J. , Conboy, G. A. , Brimacombe, M. , Sims, D. E. , & Burka, J. F. (2000). The physiological response of Atlantic salmon, Salmo salar L., to a single experimental challenge with sea lice, Lepeophtheirus salmonis . Journal of Fish Diseases, 23, 165–172. 10.1046/j.1365-2761.2000.00225.x [DOI] [Google Scholar]
- Braden, L. M. , Koop, B. F. , & Jones, S. R. M. (2015). Signatures of resistance to Lepeophtheirus salmonis include a TH2‐type response at the louse‐salmon interface. Developmental & Comparative Immunology, 48, 178–191. 10.1016/j.dci.2014.09.015 [DOI] [PubMed] [Google Scholar]
- Bradley, J. E. , & Jackson, J. A. (2008). Measuring immune system variation to help understand host‐pathogen community dynamics. Parasitology, 135, 807–823. 10.1017/S0031182008000322 [DOI] [PubMed] [Google Scholar]
- Brandal, P. , Egidius, E. , & Romslo, I. (1976). Host blood‐major food component for parasitic copepod Lepeophtheirus salmonis Kroyeri, 1838 (Crustacea‐Caligidae). Norwegian Journal of Zoology, 24, 341–343. [Google Scholar]
- Bush, A. O. , Lafferty, K. D. , Lotz, J. M. , Shostak, A. , Parasitology, W. (1997). Parasitology meets ecology on its own terms: Margolis et al. revisited. The Journal of Parasitology, 83(4), 575–583. 10.2307/3284227 [DOI] [PubMed] [Google Scholar]
- Carr, A. C. , & Moore, S. D. (2012). Robust quantification of polymerase chain reactions using global fitting. PLoS One, 7, e37640 10.1371/journal.pone.0037640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadzinska, M. , Baginski, P. , Kolaczkowska, E. , Savelkoul, H. F. , & Verburg‐van Kemenade, B. L. (2008). Expression profiles of matrix metalloproteinase 9 in teleost fish provide evidence for its active role in initiation and resolution of inflammation. Immunology, 125, 601–610. 10.1111/j.1365-2567.2008.02874.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettri, J. K. , Kuhn, J. A. , Jaafar, R. , Kania, P. W. , Møller, O. S. , & Buchmann, K. (2014). Epidermal response of rainbow trout to Ichthyobodo necator: Immunohistochemical and gene expression studies indicate a Th1‐/Th2‐like switch. Journal of Fish Diseases, 37, 771–783. 10.1111/jfd.12169 [DOI] [PubMed] [Google Scholar]
- Cox, F. (2001). Concomitant infections, parasites and immune responses. Parasitology, 122, S23–S38. 10.1017/S003118200001698X [DOI] [PubMed] [Google Scholar]
- Dixon, P. , Paley, R. , Alegria‐Moran, R. , & Oidtmann, B. (2016). Epidemiological characteristics of infectious hematopoietic necrosis virus (IHNV): A review. Veterinary Research, 47, 63 10.1186/s13567-016-0341-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast, M. D. , Ross, N. W. , Muise, D. M. , & Johnson, S. C. (2006). Differential gene expression in Atlantic salmon infected with Lepeophtheirus salmonis . Journal of Aquatic Animal Health, 18, 116–127. 10.1577/H05-043.1 [DOI] [Google Scholar]
- Fast, M. D. , Ross, N. W. , Mustafa, A. , Sims, D. E. , Johnson, S. C. , Conboy, G. A. , … Burka, J. F. (2002). Susceptibility of rainbow trout Oncorhynchus mykiss, Atlantic salmon Salmo salar and coho salmon Oncorhynchus kisutch to experimental infection with sea lice Lepeophtheirus salmonis . Diseases of Aquatic Organisms, 52, 57–68. 10.3354/dao052057 [DOI] [PubMed] [Google Scholar]
- Garver, K. A. , Mahony, A. A. M. , Stucchi, D. , Richard, J. , Van Woensel, C. , & Foreman, M. (2013). Estimation of parameters influencing waterborne transmission of infectious hematopoietic necrosis virus (IHNV) in Atlantic Salmon (Salmo salar). PLoS One, 8, e82296 10.1371/journal.pone.0082296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimnes, A. , & Jakobsen, P. (1996). The physiological effects of salmon lice infection on post‐smolt of Atlantic salmon. Journal of Fish Biology, 48, 1179–1194. 10.1111/j.1095-8649.1996.tb01813.x [DOI] [Google Scholar]
- Hansen, J. D. , & LaPatra, S. (2002). Induction of the rainbow trout MHC class I pathway during acute IHNV infection. Immunogenetics, 54, 654–661. 10.4049/jimmunol.168.7.3145 [DOI] [PubMed] [Google Scholar]
- Harmache, A. , LeBerre, M. , Droineau, S. , Giovannini, M. , & Brémont, M. (2006). Bioluminescence imaging of live infected salmonids reveals that the fin bases are the major portal of entry for Novirhabdovirus . Journal of Virology, 80, 3655–3659. 10.1128/JVI.80.7.3655-3659.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S. , Peddie, S. , Campos‐Pérez, J. J. , Zou, J. , & Secombes, C. J. (2003). The effect of intraperitoneally administered recombinant IL‐1β on immune parameters and resistance to Aeromonas salmonicida in the rainbow trout (Oncorhynchus mykiss). Developmental & Comparative Immunology, 27, 801–812. 10.1016/S0145-305X(03)00056-9 [DOI] [PubMed] [Google Scholar]
- Hood, M. I. , & Skaar, E. P. (2012). Nutritional immunity: Transition metals at the pathogen–host interface. Nature Reviews Microbiology, 10, 525 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob, E. , Barker, D. E. , & Garver, K. A. (2011). Vector potential of the salmon louse Lepeophtheirus salmonis in the transmission of infectious haematopoietic necrosis virus (IHNV). Diseases of Aquatic Organisms, 97, 155–165. 10.3354/dao02414 [DOI] [PubMed] [Google Scholar]
- Jakob, E. , Sweeten, T. , Bennett, W. , & Jones, S. R. M. (2013). Development of the salmon louse Lepeophtheirus salmonis and its effects on juvenile sockeye salmon Oncorhynchus nerka . Diseases of Aquatic Organisms, 106, 217–227. 10.3354/dao02642 [DOI] [PubMed] [Google Scholar]
- Jensen, L. E. , Hiney, M. P. , Shields, D. C. , Uhlar, C. M. , Lindsay, A. J. , & Whitehead, A. S. (1997). Acute phase proteins in salmonids: Evolutionary analyses and acute phase response. The Journal of Immunology, 158, 384–392. [PubMed] [Google Scholar]
- Johnson, S. C. , Blaylock, R. B. , Elphick, J. , & Hyatt, K. D. (1996). Disease induced by the sea louse (Lepeophtheirus salmonis) (Copepoda: Caligidae) in wild sockeye salmon (Oncorhynchus nerka) stocks of Alberni Inlet, British Columbia. Canadian Journal of Fisheries and Aquatic Sciences, 53, 2888–2897. 10.1139/f96-226 [DOI] [Google Scholar]
- Jørgensen, J. B. , Lunde, H. , Jensen, L. , Whitehead, A. S. , & Robertsen, B. (2000). Serum amyloid A transcription in Atlantic salmon (Salmo salar L.) hepatocytes is enhanced by stimulation with macrophage factors, recombinant human IL‐1β, IL‐6 and TNFα or bacterial lipopolysaccharide. Developmental & Comparative Immunology, 24, 553–563. 10.1016/S0145-305X(00)00022-7 [DOI] [PubMed] [Google Scholar]
- Kassambara, A. , & Kosinski, M. (2018). survminer: Drawing survival curves using 'ggplot2' (R package version 0.4.3) . Retrieved from https://CRAN.R-project.org/package=survminer
- Kotob, M. H. , Menanteau‐Ledouble, S. , Kumar, G. , Abdelzaher, M. , & El‐Matbouli, M. (2017). The impact of co‐infections on fish: A review. Veterinary Research, 47, 98 10.1186/s13567-016-0383-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnov, A. , Skugor, S. , Todorcevic, M. , Glover, K. A. , & Nilsen, F. (2012). Gene expression in Atlantic salmon skin in response to infection with the parasitic copepod Lepeophtheirus salmonis, cortisol implant, and their combination. BMC Genomics, 13, 130 10.1186/1471-2164-13-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnov, A. , Timmerhaus, G. , Afanasyev, S. , & Jørgensen, S. M. (2011). Development and assessment of oligonucleotide microarrays for Atlantic salmon (Salmo salar L.). Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 6, 31–38. 10.1016/j.cbd.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Landis, E. D. , Purcell, M. K. , Thorgaard, G. H. , Wheeler, P. A. , & Hansen, J. D. (2008). Transcriptional profiling of MHC class I genes in rainbow trout infected with infectious hematopoietic necrosis virus. Molecular Immunology, 45, 1646–1657. 10.1016/j.molimm.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Lenth, R. (2019). emmeans: Estimated marginal means, aka least‐squares means (R package version 1.3.2) . Retrieved from https://CRAN.R-project.org/package=emmeans
- Lhorente, J. P. , Gallardo, J. A. , Villanueva, B. , Carabaño, M. J. , & Neira, R. (2014). Disease resistance in Atlantic salmon (Salmo salar): Coinfection of the intracellular bacterial pathogen Piscirickettsia salmonis and the sea louse Caligus rogercresseyi . PLoS One, 9, e95397 10.1371/journal.pone.0095397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. , Garver, K. , & Jones, S. R. M. (2019). Differential effects of adult salmon lice Lepeophtheirus salmonis on physiological responses of Sockeye Salmon and Atlantic Salmon. Journal of Aquatic Animal Health. 10.1002/aah.10053 [DOI] [PubMed] [Google Scholar]
- MacKenzie, S. , Balasch, J. C. , Novoa, B. , Ribas, L. , Roher, N. , Krasnov, A. , & Figueras, A. (2008). Comparative analysis of the acute response of the trout, O. mykiss, head kidney to in vivo challenge with virulent and attenuated infectious hematopoietic necrosis virus and LPS‐induced inflammation. BMC Genomics, 9, 141 10.1186/1471-2164-9-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A. , Sutherland, B. J. , Koop, B. F. , Johnson, S. C. , & Garver, K. A. (2015). Infectious hematopoietic necrosis virus (IHNV) persistence in Sockeye Salmon: Influence on brain transcriptome and subsequent response to the viral mimic poly (I: C). BMC Genomics, 16, 634 10.1186/s12864-015-1759-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa, A. , Speare, D. J. , Daley, J. , Conboy, G. A. , & Burka, J. F. (2000). Enhanced susceptibility of seawater cultured rainbow trout, Oncorhynchus mykiss (Walbaum), to the microsporidian Loma salmonae during a primary infection with the sea louse, Lepeophtheirus salmonis . Journal of Fish Diseases, 23, 337–341. 10.1046/j.1365-2761.2000.00235.x [DOI] [Google Scholar]
- Nicolas, G. , Chauvet, C. , Viatte, L. , Danan, J. L. , Bigard, X. , Devaux, I. , … Vaulont, S. (2002). The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. The Journal of Clinical Investigation, 110, 1037–1044. 10.1172/JCI15686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñaranda, M. M. D. , Purcell, M. K. , & Kurath, G. (2009). Differential virulence mechanisms of infectious hematopoietic necrosis virus in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. Journal of General Virology, 90, 2172–2182. 10.1099/vir.0.012286-0 [DOI] [PubMed] [Google Scholar]
- Pinheiro, J. , Bates, D. , DebRoy, S. , & Sarkar, D. , & R Core Team . (2018). nlme: Linear and nonlinear mixed effects models (R package version 3.1‐137) . Retrieved from https://CRAN.R-project.org/package=nlme
- Polinski, M. P. , Bradshaw, J. C. , Inkpen, S. M. , Richard, J. , Fritsvold, C. , Poppe, T. T. , … Johnson, S. C. (2016). De novo assembly of Sockeye salmon kidney transcriptomes reveal a limited early response to piscine reovirus with or without infectious hematopoietic necrosis virus superinfection. BMC Genomics, 17, 848–870. 10.1186/s12864-016-3196-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, M. K. , Kurath, G. , Garver, K. A. , Herwig, R. P. , & Winton, J. R. (2004). Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish & Shellfish Immunology, 17, 447–462. 10.1016/j.fsi.2004.04.017 [DOI] [PubMed] [Google Scholar]
- Purcell, M. K. , Laing, K. J. , & Winton, J. R. (2012). Immunity to fish rhabdoviruses. Viruses, 4, 140–166. 10.1186/s13567-016-0341-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, M. K. , LaPatra, S. E. , Woodson, J. C. , Kurath, G. , & Winton, J. R. (2010). Early viral replication and induced or constitutive immunity in rainbow trout families with differential resistance to Infectious hematopoietic necrosis virus (IHNV). Fish & Shellfish Immunology, 28, 98–105. 10.1016/j.fsi.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Purcell, M. K. , Marjara, I. S. , Batts, W. , Kurath, G. , & Hansen, J. D. (2011). Transcriptome analysis of rainbow trout infected with high and low virulence strains of infectious hematopoietic necrosis virus. Fish & Shellfish Immunology, 30, 84–93. 10.1016/j.fsi.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Purcell, M. K. , Thompson, R. L. , Garver, K. A. , Hawley, L. M. , Batts, W. N. , Sprague, L. , … Winton, J. R. (2013). Universal reverse‐transcriptase real‐time PCR for infectious hematopoietic necrosis virus (IHNV). Diseases of Aquatic Organisms, 106, 103–115. 10.3354/dao02644 [DOI] [PubMed] [Google Scholar]
- R Core Team (2018). R: A language and environment for statistical computing (R version 3.5.2). Vienna, Austria: R Foundation for Statistical Computing; Retrieved from https://www.R-project.org/ [Google Scholar]
- Rebl, A. , Goldammer, T. , Fischer, U. , Köllner, B. , & Seyfert, H.‐M. (2009). Characterization of two key molecules of teleost innate immunity from rainbow trout (Oncorhynchus mykiss): MyD88 and SAA. Veterinary Immunology and Immunopathology, 131, 122–126. 10.1016/j.vetimm.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Reyes‐Cerpa, S. , Reyes‐López, F. E. , Toro‐Ascuy, D. , Ibañez, J. , Maisey, K. , Sandino, A. M. , & Imarai, M. (2012). IPNV modulation of pro and anti‐inflammatory cytokine expression in Atlantic salmon might help the establishment of infection and persistence. Fish & Shellfish Immunology, 32, 291–300. 10.1016/j.fsi.2011.11.018 [DOI] [PubMed] [Google Scholar]
- Robertsen, B. (2008). Expression of interferon and interferon‐induced genes in salmonids in response to virus infection, interferon‐inducing compounds and vaccination. Fish & Shellfish Immunology, 25, 351–357. 10.1016/j.fsi.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Saksida, S. M. (2006). Infectious haematopoietic necrosis epidemic (2001 to 2003) in farmed Atlantic salmon Salmo salar in British Columbia. Diseases of Aquatic Organisms, 72, 213–223. 10.3354/dao072213 [DOI] [PubMed] [Google Scholar]
- Skugor, S. , Glover, K. A. , Nilsen, F. , & Krasnov, A. (2008). Local and systemic gene expression responses of Atlantic salmon (Salmo salar L.) to infection with the salmon louse (Lepeophtheirus salmonis). BMC Genomics, 9, 498 10.1186/1471-2164-9-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess, A.‐N. (2018). qpcR: Modelling and analysis of real‐time PCR data (R package version 1.4‐1) . Retrieved from https://CRAN.R-project.org/package=qpcR
- Sutherland, B. J. G. , Jantzen, S. G. , Sanderson, D. S. , Koop, B. F. , & Jones, S. R. M. (2011). Differentiating size‐dependent responses of juvenile pink salmon (Oncorhynchus gorbuscha) to sea lice (Lepeophtheirus salmonis) infections. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 6, 213–223. 10.1016/j.cbd.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Sutherland, B. J. , Koczka, K. W. , Yasuike, M. , Jantzen, S. G. , Yazawa, R. , Koop, B. F. , & Jones, S. R. M. (2014). Comparative transcriptomics of Atlantic Salmo salar, chum Oncorhynchus keta and pink salmon O. gorbuscha during infections with salmon lice Lepeophtheirus salmonis . BMC Genomics, 15, 200 10.1186/1471-2164-15-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadiso, T. M. , Krasnov, A. , Skugor, S. , Afanasyev, S. , Hordvik, I. , & Nilsen, F. (2011). Gene expression analyses of immune responses in Atlantic salmon during early stages of infection by salmon louse (Lepeophtheirus salmonis) revealed bi‐phasic responses coinciding with the copepod‐chalimus transition. BMC Genomics, 12, 141 10.1186/1471-2164-12-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela‐Muñoz, V. , & Gallardo‐Escárate, C. (2017). Iron metabolism modulation in Atlantic salmon infested with the sea lice Lepeophtheirus salmonis and Caligus rogercresseyi: A matter of nutritional immunity? Fish & Shellfish Immunology, 60, 97–102. 10.1016/j.fsi.2016.11.045 [DOI] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. , & Speleman, F. (2002). Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, research0034. 0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroel, F. , Casado, A. , Vásquez, J. , Matamala, E. , Araneda, B. , Amthauer, R. , … Concha, M. I. (2008). Serum amyloid A: A typical acute‐phase reactant in rainbow trout? Developmental & Comparative Immunology, 32, 1160–1169. 10.1016/j.dci.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Wendelaar Bonga, S. (1997). The stress response in fish. Physiological Reviews, 77, 591–625. 10.1152/physrev.1997.77.3.591 [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2009). ggplot2: Elegant graphics for data analysis. New York, NY: Springer‐Verlag. [Google Scholar]
- Wilson, E. B. , & Brooks, D. G. (2010). The role of IL‐10 in regulating immunity to persistent viral infections In Ahmed R., Honjo T. (Eds.), Negative co‐receptors and ligands. Current topics in microbiology and immunology 350 (pp.39–65). Berlin: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, K. (Ed.) (1988). Infectious hematopoietic necrosis virus In Fish viruses and fish viral diseases (pp. 83–114). Ithaca, NY: Cornell University Press. [Google Scholar]
- Yamamoto, T. , Batts, W. , Arakawa, C. , & Winton, J. (1990). Multiplication of infectious hematopoietic necrosis virus in rainbow trout following immersion infection: Whole‐body assay and immunohistochemistry. Journal of Aquatic Animal Health, 2, 271–280. [DOI] [Google Scholar]
- Yamamoto, T. , Batts, W. , & Winton, J. (1992). In vitro infection of salmonid epidermal tissues by infectious hematopoietic necrosis virus and viral hemorrhagic septicemia virus. Journal of Aquatic Animal Health, 4, 231–239. [DOI] [Google Scholar]
- Zou, J. , Gorgoglione, B. , Taylor, N. G. , Summathed, T. , Lee, P.‐T. , Panigrahi, A. , … Secombes, C. J. (2014). Salmonids have an extraordinary complex type I IFN system: Characterization of the IFN locus in rainbow trout Oncorhynchus mykiss reveals two novel IFN subgroups. The Journal of Immunology, 193(5), 2273–2286. 10.4049/jimmunol.1301796 [DOI] [PubMed] [Google Scholar]
- Zou, J. , & Secombes, C. J. (2016). The Function of Fish Cytokines. Biology, 5, 23 10.3390/biology5020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


