Summary
By contrast with rapid progress in understanding the mechanisms of biosynthesis and signaling of strigolactone (SL), mechanisms by which SL inhibits axillary bud outgrowth are less well understood. We established a rice (Oryza sativa L.) hydroponic culture system to observe axillary buds at the critical point when the buds enter the dormant state. In situ hybridization analysis indicated that cell division stops in the leaf primordia of the buds entering dormancy. We compared transcriptomes in the axillary buds isolated by laser capture microdissection before and after entering the dormant state and identified genes that are specifically upregulated or downregulated in dormant buds respectively, in SL‐mediated axillary bud dormancy. Typically, cell cycle genes and ribosomal genes are included among the active genes while abscisic acid (ABA)‐inducible genes are among the dormant genes. Application of ABA to the hydroponic culture suppressed the growth of axillary buds of SL mutants to the same level as wild‐type (WT) buds. Tiller number was decreased in the transgenic lines overexpressing OsNCED1, the gene that encodes ABA biosynthesis enzyme. These results indicated that the main site of SL function is the leaf primordia in the axillary bud and that ABA is involved in SL‐mediated axillary bud dormancy.
Keywords: axillary bud, development, dormancy, Oryza sativa, strigolactone, transcriptome analysis
Significance statement
We have unveiled how strigolactone suppresses bud outgrowth through a detailed analysis of the initial suppression events. The finding that ABA is involved in strigolactone‐mediated bud dormancy will aid future studies to comprehensively understand the functions of strigolactone.
Introduction
The pattern of shoot branching is a major factor in determining aerial plant architecture. The shoot branch originates from an axillary bud, which is produced in the axil of each leaf (Leyser and Day, 2003). As the shoot apical meristem (SAM) consistently produces leaf primordia, axillary buds are also generated continuously throughout the life of the plant. Control of spatial and temporal patterns of shoot branching is crucial to allow plant growth to adjust to the constantly changing environment. Therefore, despite bud formation occurring concomitantly with leaf initiation, the development of the bud into a new branch or the arrest of its growth is precisely controlled depending on internal and external cues such as plant age, position of the bud in the plant, availability of nutrients, light and temperature.
Mechanisms controlling axillary bud outgrowth in apical dominance have been extensively studied. This is a phenomenon in which outgrowth of the axillary buds is stimulated when the apical bud is removed (Thimann and Skoog, 1934; Leyser, 2005; Domagalska and Leyser, 2011; Dun et al., 2013; Barbier et al., 2017). It has long been implicated that auxin plays a central role in this phenomenon. Auxin is produced in the shoot apices and transported downwards in the polar auxin transport system (PATS) through the xylem (Teale et al., 2006). Removal of the apical bud interrupts the auxin flow and results in the release of the lateral buds from bud growth inhibition. Because auxin does not enter the buds, two models − the ‘auxin transport canalization‐based’ model and the ‘second messenger’ model − have been proposed to explain this indirect auxin‐mediated inhibition (Leyser, 2005; Domagalska and Leyser, 2011). The former model proposes that decapitation affects the competition for auxin transport capacity and helps the axillary bud to establish auxin flow into the main stem; this is assumed to be essential for bud outgrowth (Bennett et al., 2006; Prusinkiewicz et al., 2009). In the latter model, the presence of a second messenger(s) that transmits the auxin signal to the axillary bud, is postulated (Snow, 1937; Sachs and Thimann, 1967). Because cytokinin is synthesized at the stem, transported into the axillary buds and enhances bud outgrowth, cytokinin has been proposed as a suitable candidate for the second messenger (Tanaka et al., 2006; Shimizu‐Sato et al., 2009).
Subsequently, strigolactones (SLs) were identified as the phytohormone that inhibits axillary bud outgrowth (Gomez‐Roldan et al., 2008; Umehara et al., 2008). In rice, SLs are synthesized from β‐carotene. β‐carotene is converted to carlactone (CL), a SL precursor, by the sequential actions of β‐carotene isomerase, CAROTINOID CLEAVAGE DEOXYGENASE 7 (CCD7) and CCD8, encoded by DWARF27 (D27), D17 and D10, respectively (Zou et al., 2006; Arite et al., 2007; Lin et al., 2009; Lopez‐Obando et al., 2015; Waters et al., 2017). CL is further catalyzed to several types of SLs (Zhang et al., 2014). Perception of SLs by DWARF14 (D14), the SL receptor, triggers formation of a complex of D14, D3 and D53 (Arite et al., 2009; Gao et al., 2009; Liu et al., 2009; Jiang et al., 2013; Zhou et al., 2013; Lopez‐Obando et al., 2015; Waters et al., 2017). This leads to ubiquitination of D53 by the action of D3, then to degradation of D53 via the 26S proteasome pathway. Degradation of D53 allows the function of downstream proteins that were suppressed by D53.
Currently, two modes of SL action are proposed (Waldie et al., 2014; Rameau et al., 2015). These two models are not mutually exclusive. In the first, SL is involved in the control of PATS through its control of the subcellular localization of the PIN protein, an efflux transporter of auxin, consistent with the auxin transport canalization‐based model. Accumulation of PIN protein on the plasma membrane is enhanced in the absence of SL, allowing establishment of auxin flow between the axillary bud and the main stem (Crawford et al., 2010; Shinohara et al., 2013). In the second proposed mode of SL action, SL works through the control of downstream transcription cascades (Braun et al., 2012; Brewer et al., 2015; Liu et al., 2017; Song et al., 2017). In rice and wheat, SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE (SPL) genes, encoding plant‐specific transcription factors, were identified as D53 targets that suppress bud outgrowth (Liu et al., 2017; Song et al., 2017). We have shown previously that FINE CULM1 (FC1), a TCP transcription factor, is required for SL to inhibit bud outgrowth (Minakuchi et al., 2010). Intriguingly, rice SPL14 (OsSPL14) binds FC1, suggesting that FC1 may work downstream of OsSPL14 (Lu et al., 2013). However, contradictory to this notion, there is little induction of FC1expression upon treatment with GR24, a synthetic SL analog. Moreover, there is no evidence that the Arabidopsis ortholog of OsSPL14 is involved in the control of apical dominance (Bennett et al., 2016). In Arabidopsis and pea, BRANCHED1 (BRC1), encoding a TCP transcription factor, is proposed as an integrator of multiple pathways controlling bud dormancy (Aguilar‐Martinez et al., 2007; Dun et al., 2012; González‐Grandío et al., 2013). Induction of BRC1 expression upon SL application without protein synthesis raises the possibility that BRC1 may be a direct target of transcriptional suppression by D53 in pea (Dun et al., 2012). These suggest that the transcriptional cascades downstream of SL are not fully conserved between dicots and monocots. Furthermore, the role of BRC1 in the control of SL‐dependent shoot branching is still under debate (Seale et al., 2017).
Spatial and temporal fine tuning is crucial for the function of phytohormones. This is particularly important for SL because shoot branching is a complicated process involving local and systemic decisions. SL signaling genes are expressed in the buds as well as in vascular bundles, consistent with the two modes of SL function in which SL regulates the PATS and downstream transcription cascades (Stirnberg et al., 2007; Zhou et al., 2013; Chevalier et al., 2014; Soundappan et al., 2015; Kameoka et al., 2016; Liang et al., 2016). However, more precise identification of the site of SL function is required to better understand the mechanisms by which SL inhibits bud outgrowth. To overcome the difficulties in observing SL in plant tissues due to its low concentrations, novel imaging technologies to monitor SL action have been developed recently (Prandi et al., 2013; Tsuchiya et al., 2015). Although they contribute by revealing novel aspects in the regulation of SL function to some extent, further improvement is required for them to be used to monitor the in vivo action of SL within buds. Here, to obtain insights into the mechanism by which SL inhibits outgrowth of axillary buds, we carefully observed the early steps involved when rice tiller buds enter SL‐mediated dormancy. We also analyzed changes in the transcriptomes accompanying the start of dormancy and identified genes that were up or downregulated in the axillary bud.
Results
Analysis of early steps in initiation of bud dormancy
An axillary bud is formed in the axil of each leaf of rice (Oryza sativa L.) in a manner that is well coordinated with the development of the leaf from which the bud subtends. To observe the initial steps in axillary bud dormancy reproducibly, we first established a hydroponic culture system. In this study, the stage of each leaf is described by the plastochron (P) system. The stage was estimated to the decimal point by calculating the ratio between the lengths of the newly emerging leaf to its expected full size (see Experimental procedures). In this culture system, the meristem of the axillary bud becomes visible by the time the subtending leaf reaches the P4 stage (Supporting Information Figure S1). The vasculature of the axillary bud is connected to the main stem by the P5 stage, and axillary meristem formation is completed by the P6 stage. A decision to begin outgrowth or to become dormant is made at around the P6 stage, depending on the environmental and endogenous conditions.
In our hydroponic culture system, axillary buds in the axil of the first and second leaves in wild‐type (WT) plants do not show outgrowth (Figure 1a). By contrast, the axillary buds of the first and second leaves grow vigorously in dwarf10‐2 (d10‐2) plants (Figure 1a). Because d10‐2 contains a defect in the gene encoding CAROTENOID CLEAVAGE DIOXYGENASE 8 (CCD8), an enzyme in the strigolactone (SL) biosynthesis pathway, the dormancy observed in WT plants is mediated by SL (Arite et al., 2007; Umehara et al., 2008). We then used the bud produced in the axil of the second true leaf for the following analysis. We first determined the developmental time point at which the differences in the growth of the bud between WT and d10‐2 plants become recognizable. As shown in Figure 1(b,c), the size of the buds was indistinguishable between WT and d10‐2 plants when the second leaf is at the P5.5 stage, whereas the difference became significant when the second leaf reached the P6.0 stage, indicating that the bud in the WT plants becomes dormant between the P5.5 and P6.0 stages. Therefore, we concentrated on these stages in this study. We next observed cell division in the apical region (Figure 1d,e). At the P5.5 stage, signals from HistoneH4, representing the S phase of the cell cycle, were observed in the region around the SAM (Figure 1e, enclosed with a red square), the axillary bud (Figure 1e, enclosed with a yellow square) and leaf primordia in both WT and d10‐2 plants. At the P6.0 stage, however, the signals in the axillary buds almost disappeared in WT but not in d10‐2 plants, while the state of cell division in the SAM region was indistinguishable between WT and d10‐2 plants. These observations indicated that the bud becomes dormant when the growth stage of the subtending leaf progresses from the P5.5 to the P6.0 stage, when a sudden arrest of cell division occurs in the axillary bud of the WT plant.
Figure 1.
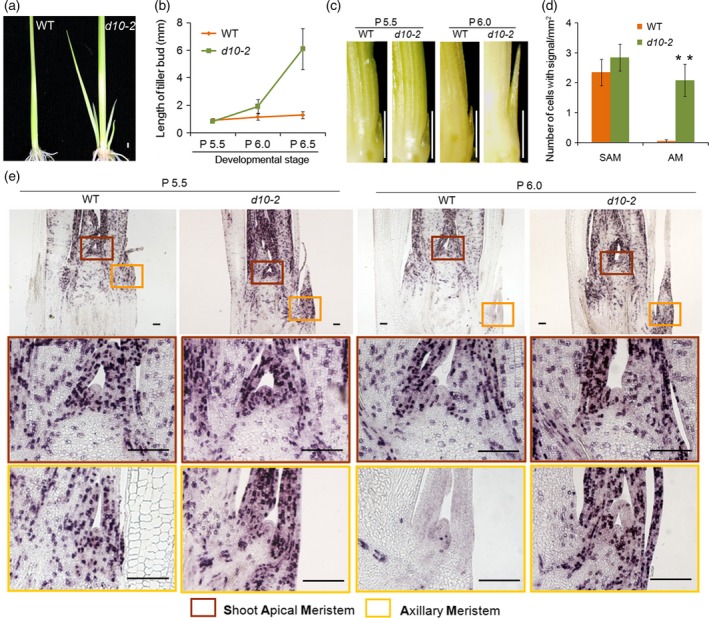
Comparison of bud growth in WT and dwarf 10‐2 plants.
(a) WT and d10‐2 plants 3 weeks after germination. Scale bar represents 1 mm.
(b) Lengths of second tiller buds of WT and d10‐2 at P5.5, P6.0 and P6.5. Error bars indicate standard deviation (SD). n ≥ 5.
(c) Images of second tiller buds in WT and d10‐2 at P5.5 and P6.0. Scale bar represents 1 mm.
(d) Number of cells expressing HistoneH4 in the shoot apical meristem (SAM) and axillary meristem (AM) at P6.0 in WT and d10‐2. Error bars indicate SD. n ≥ 10. Student's t‐test, **P ≤ 0.01.
(e) Expression of HistoneH4 in WT and d10‐2 at P5.5 and P6.0. Scale bar represents 100 μm.
We then observed WT buds until the P8.5 stage (Figure 2a). Interestingly, the number of leaf primordia in the axillary bud increased in the dormant bud (as indicated by gold stars in Figure 2a), suggesting that initiation of new leaf primordia from the axillary meristem continued in the dormant bud. Indeed, HistoneH4 signals were detected in the youngest leaf primordial (Figure 2a). We next tested whether the dormant buds maintain the ability to resume growth once conditions become favorable. We transferred the plants from hydroponic culture to fertilized soil when the second leaf was at the P7.5 stage (Figure 2b). All plants tested showed tiller growth within several days (Figure 2b). RNA in situ hybridization analysis revealed that cell divisions started 6 h after the transfer to soil (Figure 2c). The region containing actively dividing cells expanded to the whole area of the axillary bud within 24 h (Figure 2c). Our analysis revealed that cell divisions in both the leaf primordia and the young leaves tightly correlate with the activity of the bud. Based on these results, we propose that the site of SL control of bud dormancy is in the leaf primordia and the young leaves in the axillary buds.
Figure 2.
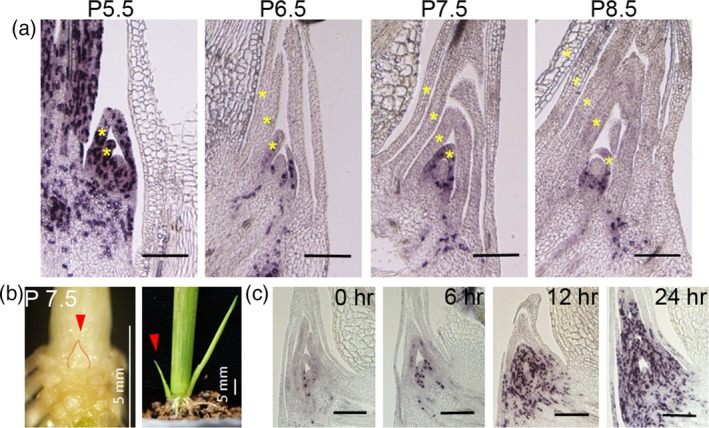
Bud development after dormancy and when growth resumes.
(a) Expression of HistoneH4 in WT at P5.5, P6.5, P7.5 and P8.5. The gold stars indicate the leaf primordia. Scale bar represents 200 μm.
(b) Resumption of bud growth after transplanting to soil after P7.5. Scale bar represents 5 mm. The red arrow points to the same bud at P7.5 and after transplanting to soil after 3 days.
(c) Expression of HistoneH4 in WT after transplanting to soil after 0, 6, 12 and 24 h. Scale bar represents 200 μm.
Transcriptome analysis
To understand the molecular basis of bud dormancy, we performed transcriptome analyses. As our results showed that the phase of the axillary bud is changed from active to dormant in a short time (between P5.5 and P6.0), we dissected the bud of the second leaf of WT and d10‐2 plants at these critical stages by laser capture microdissection (LCM) and used them for transcriptome analyses (Figure 3a). Three sections from the central part of each bud were sampled and sections from 10 buds were pooled as one sample for the microarray. The overall expression levels of genes were comparable among the arrays. In addition, the reliability of the arrays was confirmed using a clustering program (Figure S2). We then compared the transcriptomes between P5.5 buds and P6.0 buds in both WT and d10‐2 plants (Figure 3b; Tables S1 and S2). Many more genes were upregulated or downregulated between P5.5 and P6.0 in WT buds than those in the d10‐2 mutant. In the WT, the phase of the bud changes from active to dormant at this time while the buds in d10‐2 are maintained in the active phase. A possible explanation for this difference is that the changes in the transcriptome are concomitant with the change of the growth phase of the bud. In this analysis, genes that showed changes in expression between P5.5 and P6.0 buds in WT, but not in d10‐2, were selected as candidate genes involved in SL‐mediated bud dormancy and were subjected to further analysis. A list of genes that showed an increase (dormancy genes) or decrease (active genes) in expression in the P6.0 buds compared to the P5.5 buds in WT plants but not in d10‐2 is shown in Tables S3 and S4, respectively. We identified 1177 dormancy genes and 969 active genes (Figure 3b; false discovery rate < 0.05, fold change ≥ 2).
Figure 3.
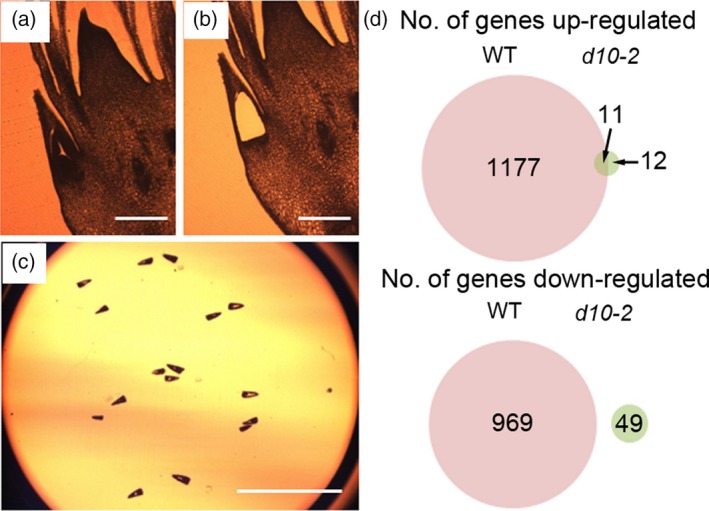
Transcriptome analysis of dormant and active buds.
(a) Section of shoot apex under the microscope.
(b) The same section after the AM and leaf primordia had been removed by laser capture microdissection (LCM).
(c) The lid of the microcentrifuge tube to which the tissue samples were attached. Scale bar represents 200 μm.
(d) Number (No.) of genes upregulated or downregulated in dormant (WT) and active (d10‐2) buds.
Induction of DORMANCY ASSOCIATED GENE in the dormant buds
DORMANCY ASSOCIATED GENE (DRM) genes, encoding plant‐specific proteins of an unknown function, have been considered as molecular markers for dormancy due to the tight association of their expression with dormancy in many plant species (Stafstrom et al., 1998; Kebrom et al., 2006; Rae et al., 2013). Expression of the four DRM genes of rice (OsDRM1; Os03g0342900, OsDRM2; Os08g0453200, OsDRM3; Os09g0437500 and OsDRM4; Os11g0671000) was significantly enhanced in the axillary buds in WT plants, but their expression levels did not change significantly in d10‐2 buds between P5.5 and P6.0 stages (Figure 4a). We further analyzed the spatial localization of the four DRM mRNAs by using RNA in situ hybridization analysis (Figure 4b). OsDRM1, OsDRM3 and OsDRM4 genes showed weak expression in the leaf primordial of the axillary bud at the P5.5 stage. This expression was increased at the P6.0 stage. Expression of OsDRM2, whose induction was relatively weak compared with the other three genes (Figure 4a), was barely detected by RNA in situ hybridization (Figure 4b). Expression of all four DRM genes suggested that our transcriptome analysis was successful in isolating genes related to the dormancy of axillary buds in rice.
Figure 4.
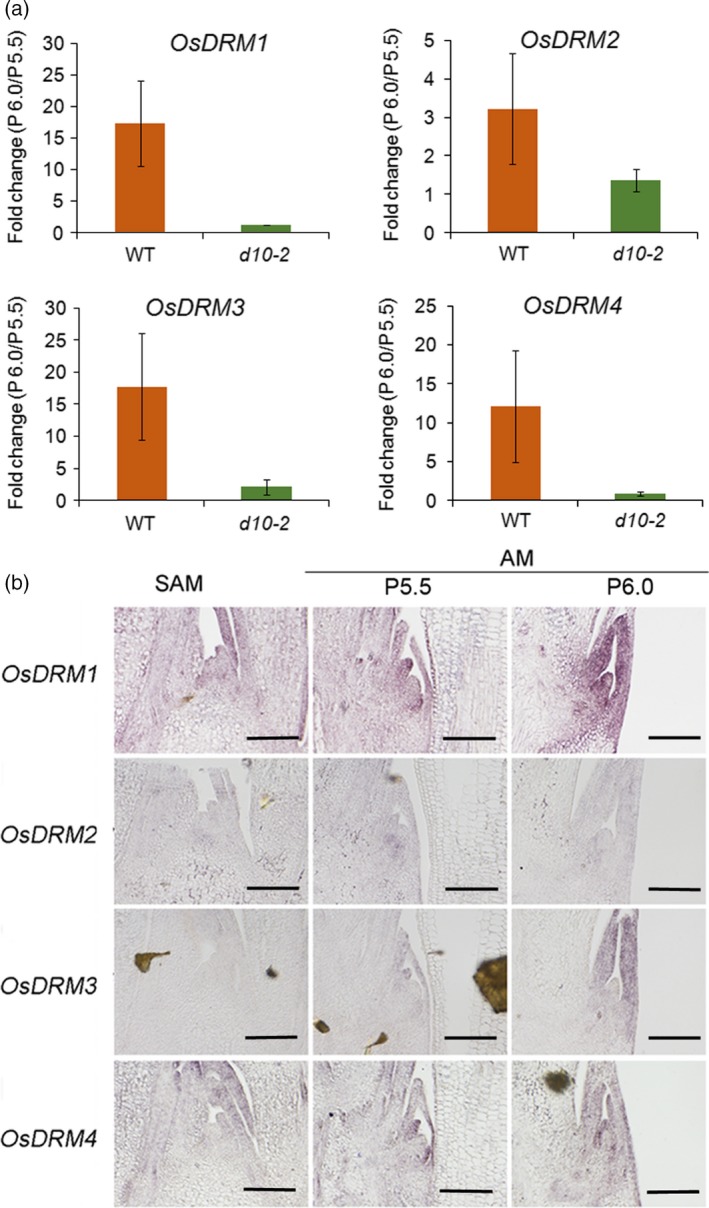
Induction of DORMANCYASSOCIATED (DRM) genes in the dormant buds.
(a) The fold change of OsDRM1 to OsDRM4 in dormant (WT) and active (d10‐2) buds. Error bars indicate the standard deviation (SD).
(b) Expression of OsDRM1 to OsDRM4 in the shoot apical meristem (SAM) and axillary meristem (AM) at P5.5 and P6.0 in WT. Scale bar represents 100 μm.
The expression of genes related to the cell cycle and cell proliferation is altered during dormancy
To understand the overall view of changes occurring when buds become dormant, gene ontology (GO) enrichment analyses of the differentially expressed genes were carried out. However, significantly (false discover rate < 0.05) enriched GO terms were not identified using the genes specifically upregulated in dormant buds (dormancy genes) (Tables S3 and S5; Figure S3), indicating that no specific gene groups were changes. When genes specifically downregulated in dormant buds (active genes) were used as input, there were 104 significant GO terms in three major categories (Tables S4 and S6; Figures [Link], [Link], [Link], [Link]). The first category is biological process, including GO terms involved in ‘nucleosome assembly’, ‘mitosis’, ‘response to DNA damage stimulus’, ‘protein import into nucleus’, ‘protein targeting to membrane’, ‘cellular nitrogen compound metabolic process’, ‘DNA replication initiation’ and ‘translation’ (Figure S5). The second category is cellular component, including GO terms involved in ‘proteasome core complex’, ‘ribosome’, ‘microtubule associated complex’, ‘nucleosome’, ‘nuclear chromosome’, ‘nuclear pore’, ‘cytosol’, ‘organelle outer membrane’, ‘respiratory chain’ and ‘mitochondrial inner membrane’ (Figure S6). The third category is molecular function, including GO terms involved in ‘structural constituent of ribosome’, ‘protein transporter activity’, ‘threonine‐type endopeptidase activity’, ‘microtubule motor activity’ and ‘aminoacyl‐tRNA ligase activity’ (Figure S7). The results of the GO analyses suggested that genes related to cell proliferation and the cell cycle were suppressed in dormant buds. Indeed, among 273 ribosomal genes annotated in the microarray, the expression levels of 149 ribosomal genes were reduced to less than 70% in WT buds during stages P5.5 to P6.0 (Figure 5a,b). Interestingly, a few ribosomal were upregulated in the dormant buds.
Figure 5.
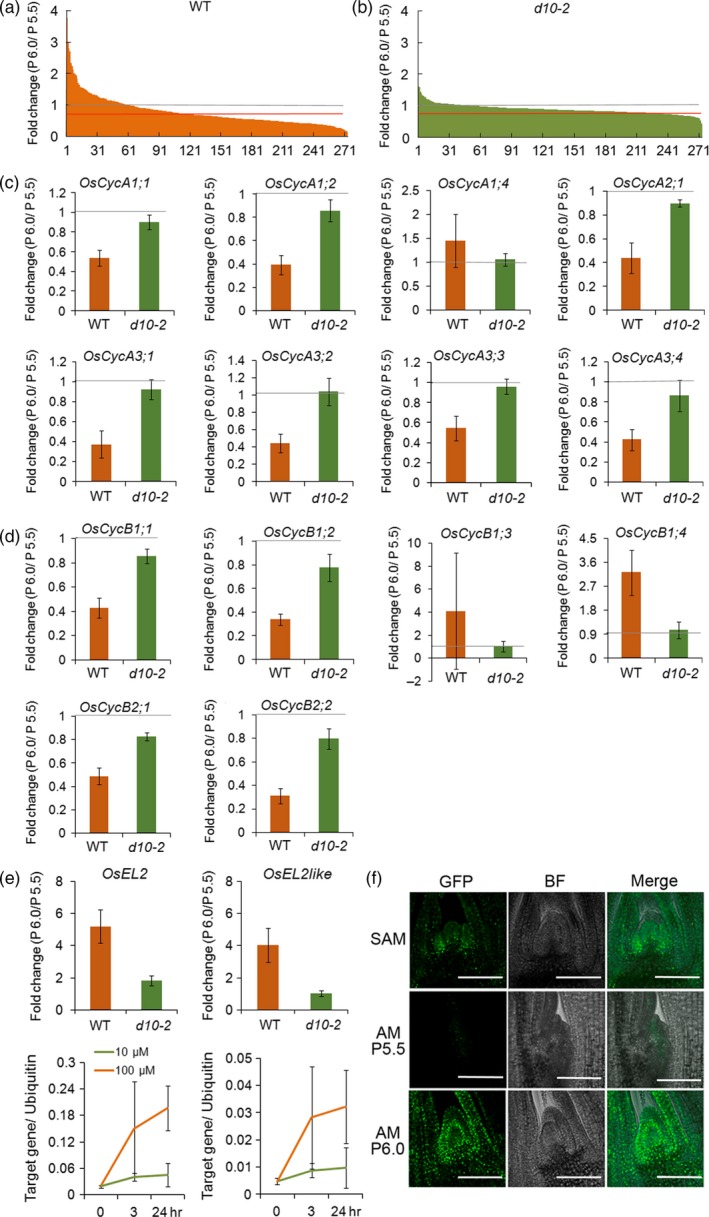
Analysis of genes related to cell proliferation and cell cycle.
(a) Fold changes of ribosomal protein genes in dormant (WT) buds. The x‐axis shows the number of ribosomal protein genes listed according to the fold change, from highest to lowest. The grey line points to a fold change value of 1. The red lines points to a fold change value of 0.7.
(b) Fold changes of ribosomal protein genes in active (d10‐2) buds.
(c) Fold changes of CyclinA genes in WT and d10‐2 buds.
(d) Fold changes of CyclinB genes in WT and d10‐2 buds.
(e) Fold changes of OsEL2 and OsEL2like, induced by ABA, in WT and d10‐2 buds. Error bars indicate the standard deviation (SD).
(f) Localization of EL2‐GFP fluorescence in the SAM and AM at P5.5 and P6.0. Scale bar represents 100 μm. GFP, green fluorescent protein; BF, bright field; merge, both GFP and BF images are superimposed on one another.
Cell cycle progression is regulated by cyclin‐dependent protein kinases, cyclins and cyclin inhibitors (De Veylder et al., 2007; Pereira et al., 2012; Polyn et al., 2015). Among these cell cycle regulators, some showed characteristic changes in dormant buds. Apart from CycA1;4, CycB1;3 and CycB1;4, all other CycA1, CycB1 and CcyB2 genes were all suppressed in WT dormant buds, whereas they were not significantly changed in d10‐2buds (Figure 5c,d). By contrast, the mRNA expression of CycD genes was not significantly changed in either the WT or d10‐2 buds (Figure S8). Inhibitors of cyclin‐dependent kinase (CDK) activity are crucial for control of cell cycle progression. Plants contain two families of related CDK inhibitors, namely, the plant‐specific SIAMESE/SIAMESE RELATED (SIM/SMR) family and the inhibitor of CDK (ICK)/Kip‐related protein (KRP) family. The latter ICK/KRP genes, apart from OsICK4, did not show significant changes in expression levels in either the WT ord10‐2buds (Figure S9). Six members of the SIM/SMR family genes have been described in rice (Kumar et al., 2015). Among these, OsEL2 and OsEL2like were induced in the dormant buds (Figure 5e). It has been reported that OsEL2 and OsEL2like expression is induced by ABA (Peres et al., 2007). We confirmed that expression of OsEL2 and OsEL2like was induced by application of ABA (Figure 5e). To see the spatial induction pattern of OsEL2, transgenic lines expressing OsEL2‐GFP driven by the OsEL2 promoter were generated. Induction of GFP florescence in the axillary bud was observed (Figure 5f). To clarify the function of the OsEL2 and OsEL2like genes, we produced transgenic rice plants in which OsEL2 or OsEL2like was driven by the 35S promoter. Among 22 transgenic lines, seven showed increases in the ploidy level up to 8C and the increase in the chromosome number (Figure S10a,b). In these plants, the size of cells was increased (Figure S10d,e). To understand the in vivo function of OsEL2 and OsEL2like, and their contribution to bud dormancy, loss‐of‐function mutant lines of these two genes were generated using CRISPR‐Cas9 technology. Overall, the morphology of the double mutants was indistinguishable from that of the WT and no significant changes in bud growth were observed (Figure S11a,b). However, when transfer the plants from hydroponic culture to soil, the second tiller bud in double mutants seemed to have the tendency to elongate (Figure S11c). This indicates that there are other genes that act redundantly with OsEL2 and OsEL2like to co‐regulate bud growth.
Expression of ABA‐related genes is affected in SL‐mediated axillary bud dormancy
We then analyzed the possible link between SL‐mediated axillary bud dormancy and phytohormones using publicly available data showing response to treatment with abscisic acid (ABA), gibberellin (GA), indole acetic acid (IAA), brassinosteroid (BR), cytokinin (CK) and jasmonic acid (JA) (RiceXPro; http://ricexpro.dna.affrc.go.jp/). This showed a trend in which genes upregulated in dormant buds (dormancy genes) are also induced by ABA and JA (Figure 6a). Conversely, genes downregulated in the dormant buds (active genes) are also suppressed by treatment with ABA or JA (Figure 6b). These results implied a correlation between SL‐mediated axillary bud dormancy and ABA and JA. As involvement of ABA in the control of seed dormancy is well established, as well as in bud dormancy (González‐Grandío and Cubas, 2014; Shu et al., 2016; Nguyen and Emery, 2017), we further analyzed the effects of ABA in the control of SL‐mediated axillary bud dormancy in rice. Indeed, the gene most highly activated in the dormant buds was an ortholog of AWPM‐19, which is an ABA‐responsive gene of unknown function isolated from Arabidopsis and was recently shown to be involved in seed dormancy of wheat (Barrero et al., 2015; Chen et al., 2015) (Figure 7a; Table S3). The AWPM‐19 gene of rice was strongly expressed in the youngest leaf primordia in the dormant bud, suggesting that the ABA level may be raised in the leaf primordia prior to the onset of dormancy (Figure 7b). In fact, a few genes in the ABA biosynthesis pathway were identified among the genes that were specifically upregulated in dormant buds of WT plants (Table S3). Among these was Os12g0617400, encoding 9‐cis‐epoxycarotenoid dioxygenase 1 (OsNCED1) (Figure 7c). RNA in situ hybridization analysis showed that the OsNCED1 gene is predominantly expressed in the leaf primordia in the axillary buds of the P6.0 stage (Figure 7d). We found a group of NAC (NAM‐ATAF1,2‐CUC2) genes, encoding transcription factors containing a NAC domain, were also upregulated in dormant buds in WT plants (Figure S12). Expression of these genes was also induced by ABA in most cases (Figure S12).
Figure 6.
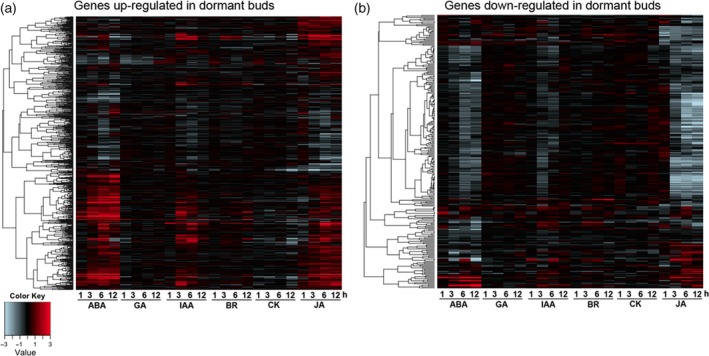
The involvement of phytohormones in the dormant bud.
Hierarchical cluster analysis of the up (a) and down (b) regulated genes in dormant buds showing the expression levels after phytohormone (ABA, GA, IAA, BR, CK and JA) treatment for 1, 3, 6 and 12 h.
Figure 7.
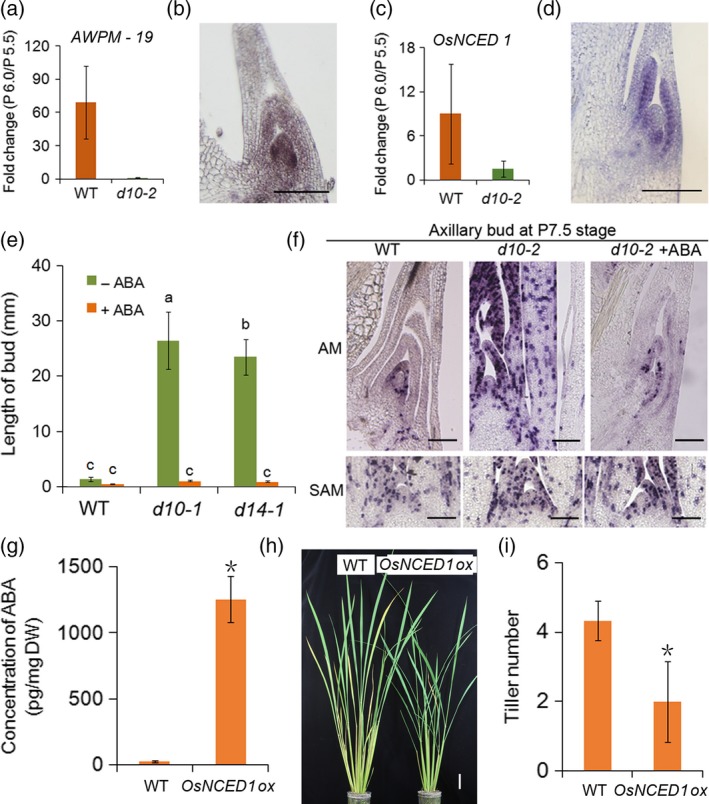
Function of ABA in suppressing bud growth.
(a) Fold change of AWPM‐19 expression in dormant (WT) and active (d10‐2) buds.
(b) Expression of AWPM‐19 in dormant buds observed by in situ hybridization.
(c) Fold change of OsNCED1 in WT and d10‐2 buds. Error bars indicate the standard deviation (SD).
(d) Expression of OsNCED1in dormant buds observed by in situ hybridization. Scale bar represents 100 μm.
(e) Length of buds after 3 μm ABA treatment from P5.5 to P6.5 in WT (Shiokari), d10‐1 and d14‐1 plants. Error bars indicate the SD. n ≥ 5. Different letters above bars indicate significant differences between genotypes using a one‐way anova followed by Tukey's test, P < 0.05.
(f) Expression of HistoneH4 in the AM and the SAM at P7.5 of WT and d10‐2 plants with and without ABA. Scale bar represents 50 μm.
(g) Concentration of ABA in WT and OsNCED1 overexpression (OsNCED1ox) lines. Error bars indicate the SD. Student's t‐test, *P ≤ 0.5.
(h) WT and OsNCED1ox plants grown for 2 months. Scale bar represents 5 cm. (i) Tiller number in WT and OsNCED1ox plants. Error bars indicate the SD. Student's t‐test, *P ≤ 0.5.
Involvement of ABA in SL‐mediated axillary bud dormancy
We tested whether ABA suppresses bud outgrowth in rice by measuring the growth of buds in hydroponic culture. Firstly, we added ABA to the hydroponic culture medium. Because it is known that ABA suppresses plant growth, we first confirmed that addition of 3 μm ABA did not severely affect the growth of the first, second and third leaves in d10‐1 plants grown in the hydroponic culture. We also confirmed that the growth of buds was strongly inhibited (Figure S13). The addition of 3 μm ABA did not significantly affect tiller bud growth in WT plants. The effect was dramatic, however, in both the SL deficient d10‐1 mutants and in the SL signaling d14‐1 mutants, in which the growth of tiller buds was suppressed to a level comparable with that of WT plants (Figure 7e). ABA treatment is known to suppress bud growth. Accordingly, we confirmed that cell division in the young leaf primordial of the axillary bud in the d10‐2 mutant was inhibited by ABA (Figure 7f). This suggests that ABA works downstream of SL to inhibit tiller bud growth through the suppression of the cell cycle in the buds. We further tested the effect of ABA by expressing the OsNCED1 gene driven by the 35S promoter. Most regenerated transgenic plantlets became brown and died, with only a few lines surviving, however the ABA level was dramatically higher in these surviving lines than in WT plants (Figure 7g). The outgrowth of axillary buds was clearly suppressed in the lines in which ABA levels were increased (Figure 7h,i). These results suggested that ABA is involved in SL‐mediated axillary bud dormancy in rice. Finally, we measured the ABA levels in d10‐1 and d14‐1 mutants (Figure S14). Because it was very difficult to obtain sufficient amounts of axillary buds for measurement of ABA, we used the basal part of the stem including the SAM, leaf primordia, internodes and node, as well as the axillary buds. The ABA level was lower in d14‐1 than in the WT while no significant difference was observed between the WT and d10‐1. Currently it is unknown why these different responses were observed between d10‐1 and d14‐1.
Discussion
Arrest of cell division in leaf primordia occurs early in SL‐mediated axillary bud dormancy
By contrast with the rapid progress in understanding the mechanisms of the biosynthesis and signaling of strigolactone (SL), the mechanism by which SL inhibits outgrowth of axillary buds is less well understood. Here, we show that the timing of bud dormancy depends on the developmental stage of the leaf which subtends the axillary bud. Cell division arrests suddenly in the leaf primordial in the axillary bud when the stage of its subtending leaf proceeds from P5.5 to P6.0. Under the growth conditions in this study, one leaf primordium is generated around every 4 days, therefore, the arrest of cell division occurs within about a maximum of 48 h. To our surprise, despite the arrest of cell division in the leaf primordia, initiation of new leaf primordia continued for some time, indicating that the axillary meristem maintains its ability to differentiate leaf primordia. Conversely, the cell cycle restarted rapidly in the leaf primordial of the axillary buds when the plants were transferred to soil, which is probably a more suitable medium for bud outgrowth. This indicates that bud growth arrest is reversible. More importantly, these observations suggest that the site of control of SL‐mediated axillary bud dormancy is likely to be the leaf primordia in the axillary bud. Consequently, we demonstrated that dormant genes, including DRM1 to DRM4 and OsNCED1, were upregulated in the leaf primordia rather than in the meristem of the axillary bud. Recently, it was reported that SLs work at least partly through OsSPL14 to suppress bud outgrowth in rice (Song et al., 2017). We showed that OsSPL14 mRNA exclusively accumulates in the leaf primordia and is excluded from the axillary meristem as well as the SAM (Luo et al., 2012).
BRANCHED1 (BRC1), encoding a TCP transcription factor, is proposed as an integrator of multiple pathways suppressing bud outgrowth in Arabidopsis, including SL, auxin, cytokinin and sugar (Aguilar‐Martinez et al., 2007). Maize Teosinte Branched 1 (TB1) is one of the founding members of the TCP transcription factor family (Doebley et al., 1997). TB1 suppresses tiller branching through positive regulation of GRASSY TILLERS1 (GT1), encoding a HOMEODOMAIN LEUCINE ZIPPER (HD‐ZIP) transcription factor (Whipple et al., 2011). Previously, we showed that FC1, a rice ortholog of TB1, works as a downstream component of SL (Minakuchi et al., 2010). TB1, GT1 and FC1 are predominantly expressed in leaf primordia and the provasculature of lateral buds but are excluded from the SAM, supporting our notion that leaf primordia are the site of SL function in the axillary bud (Hubbard et al., 2002; Minakuchi et al., 2010; Whipple et al., 2011).
In combination, these results suggest a possible scenario for the early steps in SL‐mediated axillary bud dormancy as follows. Rapid inhibition of cell division in the leaf primordia occurs, followed by the gradual attenuation of axillary meristem activity, resulting in the arrest of new leaf initiation. It is likely that the state of cell division activity in the leaf primordia is transmitted to the meristem. Indeed, communication between the leaf primordia and the meristem has been suggested in many cases, although the underlying mechanisms for this non‐cell‐autonomous regulation remain to be understood (Itoh et al., 2000; Kawakatsu et al., 2006). Elucidation of the molecular mechanisms of this communication is the next interesting challenge in bud dormancy research. However, the possibility that SLs affect leaf primordia and the meristem in the axillary bud independently cannot be excluded.
SL‐mediated axillary bud dormancy is regulated through control of the cell cycle
Consistent with the arrest of cell division in the leaf primordia, differences in the expression patterns of cell cycle genes were observed between WT and d10‐2 buds. Expression of CycA, CycB1 and CycB2 genes was dramatically downregulated in WT when the developmental stage of the subtending leaf progressed from P5.5 to P6.0, accompanying the onset of the dormant state of the axillary bud. This change was not observed in d10‐2 background. In the plant cell cycle, the A‐type cyclins (CycAs) are mainly present from the S phase to the M phase, whereas the level of CycBs is the highest at the G2M transition and M phase (De Veylder et al., 2007). Therefore, these cyclins act to progress cycles that have already started. In contrast, the formation of the CycD‐CycA complex is critical in the decision to enter cell division. Interestingly, expression of CycD was not altered significantly between the P5.5 and P6.0 stages in either WT or d10‐2 buds, indicating that these cyclins are not regulated at the transcription level. We showed that EL2 and its paralogue, EL2like, belonging to a plant‐specific CDK inhibitor family, were upregulated in WT but not in d10‐2. SIAMESE (SIM) and SMR, orthologs of EL2 in Arabidopsis, were shown to be involved in the control of the endocycle (Churchman et al., 2006; Yi et al., 2014; Kumar et al., 2015). We confirmed that molecular function is conserved in EL2 and EL2like since their constitutive expression caused an increase in ploidy level. Both SIM of Arabidopsis and EL2 of rice were shown to interact with and inhibit the CycD‐CDKA complex (Dewitte et al., 2007; Peres et al., 2007; Boudolf et al., 2009; Kumar et al., 2015). Therefore, the increase in the level of EL2 and EL2like expression may lead to suppression of CycD‐CDKA activity, and subsequently, to the inhibition of entry into the cell cycle. In combination, these results suggest that cell cycle progression is suppressed at both the S/M and G1S phases when the axillary buds enter the dormant state. Suppression at the S/M phase is achieved by reducing transcription of CycA and CycB genes, and at the G1S phase, through suppression of CycD‐CDKA activity, both by induction of EL2 and EL2like transcription. This two‐way system of cell cycle regulation may have been established to ensure the safe and punctual arrest of bud activity.
ABA is involved in the control of SL‐mediated axillary bud dormancy in rice
In this study, ABA‐inducible genes were found to been enriched in dormant buds, supporting the notion that ABA is involved in the control of dormancy in the axillary buds. Notably, we identified a gene encoding NCED1, one of five NCEDs in rice, as one of the dormant genes (Ye et al., 2012). In situ hybridization analysis showed that the OsNCED1 transcripts accumulate in the leaf primordial of the WT buds upon transition to the dormant state. Furthermore, outgrowth of the axillary buds of the d10‐1 and d14‐1 mutant was suppressed to the same level as WT by the application of ABA to the hydroponic culture. Overexpression of the OsNCED1 gene in the WT background was sufficient to increase the ABA level and this led to a decrease in the number of tillers. These results imply, to some extent, that ABA works downstream of SL to inhibit bud outgrowth of rice. Recently, it was reported that ABA may influence tillering in barley through modulation of SL biosynthesis (Wang et al., 2018). Tiller formation was enhanced in transgenic barley plants that accumulate higher levels of ABA as a result of RNAi‐mediated reduction in ABA 8′‐hydroxylase expression. In addition, expression of SL biosynthesis genes was downregulated and the SL concentration in root exudates was reduced in these transgenic plants. The contradiction between the results in barley and our findings may be explained by the differences in the materials analyzed. In Wang et al. (2018), gene expression and ABA content were examined by sampling the basal part of the stem which contained various organs. The SL content was examined in the root exudates. By contrast, we focused on the axillary buds at a critical time when the pattern of bud growth is determined.
Involvement of ABA in the control of seed dormancy is well established (Graeber et al., 2012). An association between bud inhibition and an elevated level of ABA in the buds has also been suggested by many studies (Ruttink et al., 2007; Zheng et al., 2015; Nguyen and Emery, 2017). Recently, involvement of ABA in the control of axillary bud dormancy has been reported in Arabidopsis shade avoidance syndrome and PHYTOCHROME B regulated dormancy in sorghum (Reddy et al., 2013; Kebrom and Mullet, 2016). It remains to be determined, however, whether SL‐mediated axillary bud dormancy is also regulated by ABA. In light quality‐dependent bud dormancy in Arabidopsis, ABA accumulates locally in the buds under low R:FR conditions, which suppresses bud outgrowth, while the abundance and signaling of ABA are reduced in response to the increased R:FR ratio (González‐Grandío et al., 2013, 2017; Reddy et al., 2013; Yao and Finlayson, 2015; Holalu and Finlayson, 2017). The light quality‐dependent control of bud growth in Arabidopsis is mediated by the function of BRC1. BRC1 is upregulated upon low R:FR conditions, and directly and positively regulates expression of three HD‐ZIP protein genes. These HD‐ZIP transcription factors bind NCED3, a key enzyme in the ABA biosynthesis pathway, and contribute to the increase of ABA in the buds (González‐Grandío et al., 2017). As discussed previously, FC1 and its maize ortholog, TB1, suppress shoot branching (Doebley et al., 1997; Minakuchi et al., 2010). BRC1 is a close homolog of FC1/TB1. Genetically, FC1 functions downstream of SL, whereas the functional location of TB1 is unknown. GT1, encoding HD‐ZIP III, works downstream ofTB1 (Whipple et al., 2011). Furthermore, we showed that OsNCED1, a close homolog of NCED3 of Arabidopsis, is upregulated in the dormant bud. Combining these facts, a plausible scenario is that different stimuli activate the same genes to induce ABA in bud inhibition. It is of interest to know if the HD‐ZIP genes work downstream of FC1 to regulate biosynthesis and/or signaling of ABA, which is required for suppression of bud outgrowth in rice.
ABA suppresses cell cycle progression through suppression of CYC and CDK genes and activation of CDK inhibitors (Meguro and Sato, 2014; Vergara et al., 2017). In addition to the known cell cycle genes, we identified that EL2, a plant‐specific cell cycle inhibitor of cell cycle progression, and its close paralogue, EL2like, are involved in SL‐mediated axillary bud dormancy. EL2 was first identified as an early response factor to a biotic elicitor for phytoalexin biosynthesis (Minami et al., 1996). EL2 and EL2like are induced by stresses, such as cold and drought, and ABA (Peres et al., 2007), suggesting a possible role for these genes in the tolerance of stressful conditions by the arrest of cell cycle progression. We confirmed that EL2 and EL2like expression is induced by ABA. This may be the link between ABA and bud dormancy. AWPM‐19, whose induction was the highest in the dormant buds in this study, was initially identified by its responsiveness to ABA (Koike et al., 1997). Recently, AWPM‐19 was shown to be a regulator of seed dormancy in barley, although the molecular function of AWPM‐19 is yet to be determined (Barrero et al., 2015). By contrast with the dramatic rise in the expression level of AWPM‐19 in the dormant buds, the AWPM‐19 mutant as well as the el2el2like double mutant did not show significant defects in tiller growth (Figure S12). We also produced quadruple mutants of the four DRM genes, however, no significant suppression of bud dormancy was observed. A possible explanation for this is that the phenotype is masked by genetic redundancy. Genes in SIM/SMR family, to which EL2 and EL2LIKE belong, play conserved fundamental biological functions in plant growth and development (Kumar et al., 2015). Therefore, other members in the SIM/SMR family in rice likely play overlapping roles. Indeed, although EL2 and EL2LIKE are highly induced in the dormant buds, there are other dormancy insensitive members in this family. A same scenario may be applicable to other genes, such as AWPM‐19, which is also a member of a gene family. Moreover, it is likely that many downstream genes contribute to the dormancy and fine‐tuning of these genes and are required for the appropriate control of bud activity. Genes involved in strigolactone dependent bud dormancy in rice is summarized in Figure S15. Understanding how these genes suppress bud activity in concert will be the next challenge.
Experimental procedures
Plant materials
Rice (Oryza sativa L.) cultivars Shiokari and Nipponbare were used in this study. Rice tillering dwarf mutants d10‐1, d10‐2 and d14‐1 were as described previously (Ishikawa et al., 2005; Umehara et al., 2008).
Growth conditions and ABA treatment
Plants were grown in a hydroponic culture system according to Umehara et al. (2008). For ABA induction, WT plants grown in hydroponic culture for 2 weeks were treated with 0, 3 or 10 μμ ABA for 3 and 24 h. Three independent experiments, each containing eight plants, were performed. To observe bud growth, plants were grown in hydroponic culture. ABA was added to the liquid culture medium in the bottle. The culture medium was changed every 2 days. Plants were incubated with 3 μm ABA in hydroponic culture.
Plants observation and sampling
In this study we used the plastochron (P) system to record leaf developmental stage. P indicates the time interval of emerging new leaves. In this system, the stage of each leaf changes as the shoot grows; the youngest leaf primordium which is just starting to emerge is considered as a leaf primordium at the P1 stage (P1 leaf primordium), the second and third youngest ones are leaf primordia at P2 and P3 stages, respectively. In this study, we focused our analysis on the second leaf. To match the relative stage (P system) of the second leaf to the absolute developmental stage of the tiller, we first observed the relationship between the P system and the absolute order. In our growth conditions, the second leaf is at the P5.0 and P6.0 stages when the third and fourth leaves have half emerged, respectively. Therefore, based on this observation, we labelled the relative stage of the second leaf as P5.5 when the third leaf was fully expanded.
The tiller buds were observed and measured after removing the leaves. For microarray analysis and RNA in situ hybridization, after removing one leaf, 0.5 cm of the basal part of the plant was cut and used for the experiment.
Histological analysis
For histological analysis, shoot apices were fixed with FAA (5% formalin, 5% acetic acid, 45% ethanol and 45% water) and embedded in Paraplast plus (McCormick, http://www.mccormickscientific.com/) after dehydration. Transverse sections of 8 μm were stained with Toluidine blue.
RNA in situ hybridizations
In situ hybridizations were performed as described by Kouchi et al. (1995). The Histone H4 probe was prepared and used as described previously (Itoh et al., 1998). The full‐length cDNAs of OsDRM3, OsDRM8, OsDRM9, OsDRM11, AWPM‐19 and OsNCED1 were PCR amplified and cloned into the pENTR vector (Invitrogen, http://www.invitrogen.com/) and linearized with an appropriate restriction enzyme. To make the antisense probe, in vitro transcription was performed using the linearized plasmid as a template, with the incorporation of digoxigenin (DIG)‐UTP (Roche Applied Science, https://lifescience.roche.com/en_cn.html).
Laser capture microdissection
Laser capture microdissection was performed as previously described (Takahashi et al., 2010). Briefly, the basal parts of rice seedlings were fixed in FAA (5% formalin, 5% acetic acid, 45% ethanol and 45% water) on ice. The fixative was infiltrated into the tissues under vacuum three times for 10 min on ice and then overnight at 4°C. The samples in fixative solution were further fixed in a microwave at 37°C for 15 min, and this was repeated three times with fresh and pre‐chilled fixative solution. The samples were then dehydrated using 70, 80, 90 and 100% ethanol at 58°C (1.5 min each time) in the microwave. The paraffin‐embedded blocks were prepared by gradually exchanging butanol with melted paraffin wax at 58°C. The sections were laser‐microdissected using the ArcturusXT™ LCM instrument (Thermo Fisher Scientific, https://www.thermofisher.com/cn/en/home.html) and the stage and the morphology of the dissected specimens were confirmed under the microscopy.
Microarray experiment
Total RNAs were extracted with the Arcturus® PicoPure® RNA isolation kit (Applied Biosystems, https://www.thermofisher.com/cn/en/home/brands/applied-biosystems.html) from the samples described below. The quality of RNA extracted was checked with an Agilent RNA 6000 Pico kit using the Agilent 2100 Bioanalyser (Agilent Technologies, https://www.agilent.com/). RNA yield was assessed using a NanoDrop 1000 spectrophotometer (NanoDrop Products; Thermo Fisher Scientific, https://www.thermofisher.com/cn/en/home.html). Samples from P5.5 were labeled with Cy3 and samples from P6.0 were labeled with Cy5. Four biological replicates were used for WT, three biological replicates were used for d10‐2 and one set was used for the WT at the transition stage. The hybridized slides (Rice 44K oligo‐DNA microarray) were scanned with a DNA microarray scanner (G2505C; Agilent Technologies, https://www.agilent.com/), and signal intensities were extracted by using Feature Extraction software (Version 10.5.1.1; Agilent Technologies, https://www.agilent.com/).
Microarray data analysis
A gene expression analysis software program, GeneSpring GX12 (Agilent Technologies, https://www.agilent.com/) was used to perform statistical analyses. All transcripts were filtered initially to select transcripts having a signal intensity value higher than 50. Adjustment for multiple testing was applied using Benjamini−Hochberg false discovery rate and the transcript lists generated for upregulated and downregulated transcripts (P ≤ 0.02, fold change ≥2 for transcripts downregulated, fold change ≥2 for transcripts upregulated).
Real time PCR
Total RNA was extracted using the Plant RNA Isolation mini kit (Agilent Technologies, https://www.agilent.com/). After DNase I treatment, first‐strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen, http://www.invitrogen.com/). The primer sets used to amplify the transcripts are described in Table S7. PCRs were performed with SYBR green I using the Light Cycler® 480 System II (Roche Applied Science, https://lifescience.roche.com/en_cn.html).
Functional enrichment analysis
Gene ontology analysis was carried out using the singular enrichment analysis tool offered by agriGO (Du et al., 2010) with the default settings of Fisher's t‐test (P < 0.05), false discovery rate correction by the Hochberg method and five minimum numbers of mapping entries against the rice‐specific precomputed background reference.
Vector construction
To observe protein localization of OsEL2, the sequence of the OsEL2 promoter and full‐length cDNA was amplified by PCR and the amplified fragment was introduced into the pGWB4 expression vector (Nakagawa et al., 2007a,b) using the Gateway system (Invitrogen, http://www.invitrogen.com/). To generate the OsEL2, OsEL2like and OsNCED1 overexpression plants, the full‐length cDNAs of these genes were amplified and introduced into the pGWB2 expression vector (Nakagawa et al., 2007a,b) using the Gateway system (Invitrogen, http://www.invitrogen.com/). The primers for vector construction are listed in Table S7. For the generation of loss‐of‐function mutants, CRISPR‐CAS9 system was used.
Transformation
Rice transformation was carried out as described by Nakagawa et al. (2002).
Hormone analysis
Extraction and purification of ABA from rice seedlings was performed as for maize seedlings in a previous study (Takeuchi et al., 2016). Quantification of ABA by liquid chromatography tandem mass spectrometry (LC‐MS‐MS) was performed as described previously (Kanno et al., 2012).
Ploidy analysis
Young leaves were used for the determination of ploidy levels. Ploidy levels were measured using a PAS flow cytometer (Partec, https://www.sysmex-partec.com/) according to Haga et al. (2011). The lowest peak of WT plants was assumed to represent 2C nuclei, where C is the haploid DNA content.
Scanning electron microscopy
Scanning electron microscopy analysis was performed according to Kobayashi et al.(2012).
Accession numbers
The genes mentioned in this study are listed in Table S8.
Conflict of Interests
The authors declare no conflict of interests.
Author contributions
LL, MS, MI, GX and JK planned and designed the research. LL, MT, HK, TS, RQ and YK performed experiments and analyzed data. LL and JK wrote the manuscript. Please correspond to LL and JK.
Supporting information
Figure S1. Observation of axillary buds from P4 to P6.
Figure S2. Clustering of microarray data for quality checking.
Figure S3. Gene ontology (GO) abundance chart of genes upregulated in dormant buds.
Figure S4. Gene ontology (GO) abundance chart of genes downregulated in dormant buds.
Figure S5. Hierarchical graph of genes involved in biological processes that are downregulated in dormant buds.
Figure S6. Hierarchical graph of genes involved in molecular functions that are downregulated in dormant buds.
Figure S7. Hierarchical graph of genes involved in cellular components that are downregulated in dormant buds.
Figure S8. Fold changes of CyclinD genes in dormant (WT) and active (d10‐2) buds.
Figure S9. Fold changes of Cyclin‐dependent kinase inhibitor (ICK) genes in dormant (WT) and active (d10‐2) buds.
Figure S10. Functional analysis of OsEL2.
Figure S11. Tiller bud elongation in mutants.
Figure S12. Induction of NAC genes.
Figure S13. Effects of ABA on growth of rice.
Figure S14. Concentration of ABA in the basal part of WT, d10‐1 and d14‐1.
Figure S15. Relationships between genes involved in SL‐regulated bud dormancy.
Table S1. Expression levels of genes in the WT microarray.
Table S2. Expression levels of genes in the d10‐2 microarray.
Table S3. Genes upregulated in dormant buds in the microarray analyzed by GeneSpring GX12.
Table S4. Genes downregulated in dormant buds in the microarray analyzed by GeneSpring GX12.
Table S5. Genes upregulated in dormant buds with GO terms.
Table S6. Genes downregulated in dormant buds with GO terms.
Table S7. List of primers used in this study.
Table S8. Accession numbers of genes in this study.
Acknowledgements
We thank Jian Feng Ma, Naoki Yamaji, Miho Kashino, Akinori Nagamura, Yutaka Sat and Ritsuko Motoyama for technical assistance with the LMD‐Microarray. We thank Kenichi Nonomura for technical advice on the cytological analysis. This work was supported by National Key Research and Development Program of China (2016YFD0100700) and the National Natural Science Foundation of China (31401937) to L.L., JSPS/MEXT Kakenhi grants (22119008, 22247004, 16K14748, 17H06475, 18K19198) and JST CREST grant (JPMJCR13B1) to J.K,
Linked article: This paper is the subject of a Research Highlight article. To view this Research Highlight article visit https://doi.org/10.1111/tpj.14287.
Contributor Information
Le Luo, Email: luole@njau.edu.cn.
Junko Kyozuka, Email: junko.kyozuka.e4@tohoku.ac.jp.
References
- Aguilar‐Martinez, J.A. , Poza‐Carrion, C. and Cubas, P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell, 19, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite, T. , Iwata, H. , Ohshima, K. , Maekawa, M. , Nakajima, M. , Kojima, M. , Sakakibara, H. and Kyozuka, J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite, T. , Umehara, M. , Ishikawa, S. , Hanada, A. , Maekawa, M. , Yamaguchi, S. and Kyozuka, J. (2009) d14, a strigolactone‐insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. [DOI] [PubMed] [Google Scholar]
- Barbier, F.F. , Dun, E.A. and Beveridge, C.A. (2017) Apical dominance. Curr. Biol. 27, R864–R865. [DOI] [PubMed] [Google Scholar]
- Barrero, J.M. , Cavanagh, C. , Verbyla, K.L. et al. (2015) Transcriptomic analysis of wheat near‐isogenic lines identifies PM19‐A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 16, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, T. , Sieberer, T. , Willett, B. , Booker, J. , Luschnig, C. and Leyser, O. (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16, 553–563. [DOI] [PubMed] [Google Scholar]
- Bennett, T. , Liang, Y. , Seale, M. , Ward, S. , Muller, D. and Leyser, O. (2016) Strigolactone regulates shoot development through a core signalling pathway. Biol. Open, 5, 1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf, V. , Lammens, T. , Boruc, J. et al. (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150, 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, N. , de Saint Germain, A. , Pillot, J.P. et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, P.B. , Dun, E.A. , Gui, R.Y. , Mason, M.G. and Beveridge, C.A. (2015) Strigolactone inhibition of branching independent of polar auxin transport. Plant Physiol. 168, 1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Lan, H. , Huang, P. , Zhang, Y. , Yuan, X. , Huang, X. , Huang, J. and Zhang, H. (2015) Characterization of OsPM19L1 encoding an AWPM‐19‐like family protein that is dramatically induced by osmotic stress in rice. Genet. Mol. Res. 14, 11994–12005. [DOI] [PubMed] [Google Scholar]
- Chevalier, F. , Nieminen, K. , Sanchez‐Ferrero, J.C. , Rodriguez, M.L. , Chagoyen, M. , Hardtke, C.S. and Cubas, P. (2014) Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell, 26, 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman, M.L. , Brown, M.L. , Kato, N. et al. (2006) SIAMESE, a plant‐specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana . Plant Cell, 18, 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, S. , Shinohara, N. , Sieberer, T. , Williamson, L. , George, G. , Hepworth, J. , Muller, D. , Domagalska, M.A. and Leyser, O. (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development, 137, 2905–2913. [DOI] [PubMed] [Google Scholar]
- De Veylder, L. , Beeckman, T. and Inze, D. (2007) The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8, 655–665. [DOI] [PubMed] [Google Scholar]
- Dewitte, W. , Scofield, S. , Alcasabas, A.A. et al. (2007) Arabidopsis CYCD3 D‐type cyclins link cell proliferation and endocycles and are rate‐limiting for cytokinin responses. Proc. Natl Acad. Sci. USA, 104, 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Hubbard, L. (1997) The evolution of apical dominance in maize. Nature, 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Domagalska, M.A. and Leyser, O. (2011) Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell. Biol. 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Du, Z. , Zhou, X. , Ling, Y. , Zhang, Z.H. and Su, Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun, E.A. , de Saint Germain, A. , Rameau, C. and Beveridge, C.A. (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun, E.A. , de Saint Germain, A. , Rameau, C. and Beveridge, C.A. (2013) Dynamics of strigolactone function and shoot branching responses in Pisum sativum . Mol. Plant, 6, 128–140. [DOI] [PubMed] [Google Scholar]
- Gao, Z.Y. , Qian, Q. , Liu, X.H. , Yan, M.X. , Feng, Q. , Dong, G.J. , Liu, J. and Han, B. (2009) Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol. Biol. 71, 265–276. [DOI] [PubMed] [Google Scholar]
- Gomez‐Roldan, V. , Fermas, S. , Brewer, P.B. et al. (2008) Strigolactone inhibition of shoot branching. Nature, 455, 189–194. [DOI] [PubMed] [Google Scholar]
- González‐Grandío, E. and Cubas, P. (2014) Identification of gene functions associated to active and dormant buds in Arabidopsis. Plant Signal. Behav. 9, e27994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Grandío, E. , Poza‐Carrion, C. , Sorzano, C.O.S. and Cubas, P. (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell, 25, 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Grandío, E. , Pajoro, A. , Franco‐Zorrilla, J.M. , Tarancon, C. , Immink, R.G.H. and Cubas, P. (2017) Abscisic acid signaling is controlled by a BRANCHED1/HD‐ZIP I cascade in Arabidopsis axillary buds. Proc. Natl Acad. Sci. USA, 114, E245–E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber, K. , Nakabayashi, K. , Miatton, E. , Leubner‐Metzger, G. and Soppe, W.J.J. (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ. 35, 1769–1786. [DOI] [PubMed] [Google Scholar]
- Haga, N. , Kobayashi, K. , Suzuki, T. et al. (2011) Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down‐regulation of multiple G2/M‐specific genes in Arabidopsis. Plant Physiol. 157, 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holalu, S.V. and Finlayson, S.A. (2017) The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J. Exp. Bot. 68, 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, L. , McSteen, P. , Doebley, J. and Hake, S. (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics, 162, 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, S. , Maekawa, M. , Arite, T. , Onishi, K. , Takamure, I. and Kyozuka, J. (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46, 79–86. [DOI] [PubMed] [Google Scholar]
- Itoh, J.I. , Hasegawa, A. , Kitano, H. and Nagato, Y. (1998) A recessive heterochronic mutation, plastochron1, shortens the plastochron and elongates the vegetative phase in rice. Plant Cell, 10, 1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, J.I. , Kitano, H. , Matsuoka, M. and Nagato, Y. (2000) SHOOT ORGANIZATION genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell, 12, 2161–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Liu, X. , Xiong, G.S. et al. (2013) DWARF 53 acts as a repressor of strigolactonesignalling in rice. Nature, 504, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka, H. , Dun, E.A. , Lopez‐Obando, M. , Brewer, P.B. , de Saint Germain, A. , Rameau, C. , Beveridge, C.A. and Kyozuka, J. (2016) Phloem transport of the receptor DWARF14 protein is required for full function of strigolactones. Plant Physiol. 172, 1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno, Y. , Hanada, A. , Chiba, Y. , Ichikawa, T. , Nakazawa, M. , Matsui, M. , Koshiba, T. , Kamiya, Y. and Seo, M. (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl Acad. Sci. USA, 109, 9653–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu, T. , Itoh, J. , Miyoshi, K. , Kurata, N. , Alvarez, N. , Veit, B. and Nagato, Y. (2006) PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell, 18, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom, T.H. and Mullet, J.E. (2016) Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in sorghum. Plant Physiol. 170, 2232–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom, T.H. , Burson, B.L. and Finlayson, S.A. (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, K. , Yasuno, N. , Sato, Y. , Yoda, M. , Yamazaki, R. , Kimizu, M. , Yoshida, H. , Nagamura, Y. and Kyozuka, J. (2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL‐Like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell, 24, 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike, M. , Takezawa, D. , Arakawa, K. and Yoshida, S. (1997) Accumulation of 19‐kDa plasma membrane polypeptide during induction of freezing tolerance in wheat suspension‐cultured cells by abscisic acid. Plant Cell Physiol. 38, 707–716. [DOI] [PubMed] [Google Scholar]
- Kouchi, H. , Sekine, M. and Hata, S. (1995) Distinct classes of mitotic cyclins are differentially expressed in the soybean shoot apex during the cell‐cycle. Plant Cell, 7, 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N. , Harashima, H. , Kalve, S. et al. (2015) Functional conservation in the SIAMESE‐RELATED family of cyclin‐dependent kinase inhibitors in land plants. Plant Cell, 27, 3065–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, O. (2005) The fall and rise of apical dominance – commentary. Curr. Opin. Genet. Dev. 15, 468–471. [DOI] [PubMed] [Google Scholar]
- Leyser, O. and Day, S. (2003) Position relative to a particular cell In Mechanisms in Plant Development (Leyser O. and Day S., eds). Hoboken, NJ: Blackwell Science, pp. 131–133. [Google Scholar]
- Liang, Y.Y. , Ward, S. , Li, P. , Bennett, T. and Leyser, O. (2016) SMAX1‐LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif‐independent mechanisms. Plant Cell, 28, 1581–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. , Wang, R.X. , Qian, Q. et al. (2009) DWARF27, an iron‐containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell, 21, 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W.Z. , Wu, C. , Fu, Y.P. , Hu, G.C. , Si, H.M. , Zhu, L. , Luan, W.J. , He, Z.Q. and Sun, Z.X. (2009) Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta, 230, 649–658. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Cheng, X.L. , Liu, P. and Sun, J.Q. (2017) miR156‐targeted SBP‐box transcription factors interact with DWARF53 TO Regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 174, 1931–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Obando, M. , Ligerot, Y. , Bonhomme, S. , Boyer, F.D. and Rameau, C. (2015) Strigolactone biosynthesis and signaling in plant development. Development, 142, 3615–3619. [DOI] [PubMed] [Google Scholar]
- Lu, Z.F. , Yu, H. , Xiong, G.S. et al. (2013) Genome‐wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell, 25, 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. , Li, W.Q. , Miura, K. , Ashikari, M. and Kyozuka, J. (2012) Control of tiller growth of rice by OsSPL14 and strigolactones, which work in two independent pathways. Plant Cell Physiol. 53, 1793–1801. [DOI] [PubMed] [Google Scholar]
- Meguro, A. and Sato, Y. (2014) Salicylic acid antagonizes abscisic acid inhibition of shoot growth and cell cycle progression in rice. Sci. Rep. 4, 4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi, K. , Kameoka, H. , Yasuno, N. et al. (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami, E. , Kuchitsu, K. , He, D.Y. , Kouchi, H. , Midoh, N. , Ohtsuki, Y. and Shibuya, N. (1996) Two novel genes rapidly and transiently activated in suspension‐cultured rice cells by treatment with N‐acetylchitoheptaose, a biotic elicitor for phytoalexin production. Plant Cell Physiol. 37, 563–567. [DOI] [PubMed] [Google Scholar]
- Nakagawa, M. , Shimamoto, K. and Kyozuka, J. (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Kurose, T. , Hino, T. , Tanaka, K. , Kawamukai, M. , Niwa, Y. , Toyooka, K. , Matsuoka, K. , Jinbo, T. and Kimura, T. (2007a) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. et al. (2007b) Improved Gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.Q. and Emery, R.J.N. (2017) Is ABA the earliest upstream inhibitor of apical dominance? J. Exp. Bot. 68, 881–884. [Google Scholar]
- Pereira, M.F. , Martino, T. , Dalmau, S.R. , Paes, M.C. , Barja‐Fidalgo, C. , Albano, R.M. , Coelho, M.G.P. and Sabino, K.C.D. (2012) Terpenic fraction of Pterodon pubescens inhibits nuclear factor kappa B and extracellular signal‐regulated protein Kinase 1/2 activation and deregulates gene expression in leukemia cells. Bmc. Complem. Altern. Med., 12, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres, A. , Churchman, M.L. , Hariharan, S. et al. (2007) Novel plant‐specific cyclin‐dependent kinase inhibitors induced by biotic and abiotic stresses. J. Biol. Chem. 282, 25588–25596. [DOI] [PubMed] [Google Scholar]
- Polyn, S. , Willems, A. and De Veylder, L. (2015) Cell cycle entry, maintenance, and exit during plant development. Curr. Opin. Plant Biol. 23, 1–7. [DOI] [PubMed] [Google Scholar]
- Prandi, C. , Rosso, H. , Lace, B. , Occhiato, E.G. , Oppedisano, A. , Tabasso, S. , Alberto, G. and Blangetti, M. (2013) Strigolactone analogs as molecular probes in chasing the (SLs) receptor/s: design and synthesis of fluorescent labeled molecules. Mol. Plant, 6, 113–127. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz, P. , Crawford, S. , Smith, R.S. , Ljung, K. , Bennett, T. , Ongaro, V. and Leyser, O. (2009) Control of bud activation by an auxin transport switch. Proc. Natl Acad. Sci. USA, 106, 17431–17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae, G.M. , David, K. and Wood, M. (2013) The dormancy marker DRM1/ARP associated with dormancy but a broader role in planta . Dev. Biol. J. 2013, 632524. [Google Scholar]
- Rameau, C. , Bertheloot, J. , Leduc, N. , Andrieu, B. , Foucher, F. and Sakr, S. (2015) Multiple pathways regulate shoot branching. Front. Plant Sci. 5, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, S.K. , Holalu, S.V. , Casal, J.J. and Finlayson, S.A. (2013) Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far‐red light. Plant Physiol. 163, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink, T. , Arend, M. , Morreel, K. , Storme, V. , Rombauts, S. , Fromm, J. , Bhalerao, R.P. , Boerjan, W. and Rohde, A. (2007) A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell, 19, 2370–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, T. and Thimann, K.V. (1967) The role of auxins and cytokinins in the release of buds from dominance. Am. J. Bot. 54, 136–144. [Google Scholar]
- Seale, M. , Bennett, T. and Leyser, O. (2017) BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development, 144, 1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu‐Sato, S. , Tanaka, M. and Mori, H. (2009) Auxin‐cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 69, 429–435. [DOI] [PubMed] [Google Scholar]
- Shinohara, N. , Taylor, C. and Leyser, O. (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11, e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, K. , Liu, X.D. , Xie, Q. and He, Z.H. (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant, 9, 34–45. [DOI] [PubMed] [Google Scholar]
- Snow, R. (1937) On the nature of correlative inhibition. New Phytol. 36, 283–300. [Google Scholar]
- Song, X.G. , Lu, Z.F. , Yu, H. et al. (2017) IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 27, 1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan, I. , Bennett, T. , Morffy, N. , Liang, Y.Y. , Stang, J.P. , Abbas, A. , Leyser, O. and Nelson, D.C. (2015) SMAX1‐LIKE/D53 family members enable distinct MAX2‐dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell, 27, 3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom, J.P. , Ripley, B.D. , Devitt, M.L. and Drake, B. (1998) Dormancy‐associated gene expression in pea axillary buds. Planta, 205, 547–552. [DOI] [PubMed] [Google Scholar]
- Stirnberg, P. , Furner, I.J. and Leyser, H.M.O. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Kamakura, H. , Sato, Y. , Shiono, K. , Abiko, T. , Tsutsumi, N. , Nagamura, Y. , Nishizawa, N.K. and Nakazono, M. (2010) A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. J. Plant. Res. 123, 807–813. [DOI] [PubMed] [Google Scholar]
- Takeuchi, J. , Okamoto, M. , Mega, R. , Kanno, Y. , Ohnishi, T. , Seo, M. and Todoroki, Y. (2016) Abscinazole‐E3M, a practical inhibitor of abscisic acid 8 ‘‐hydroxylase for improving drought tolerance. Sci. Rep. 6, 37060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M. , Takei, K. , Kojima, M. , Sakakibara, H. and Mori, H. (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45, 1028–1036. [DOI] [PubMed] [Google Scholar]
- Teale, W.D. , Paponov, I.A. and Palme, K. (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell. Biol. 7, 847–859. [DOI] [PubMed] [Google Scholar]
- Thimann, K.V. and Skoog, F. (1934) On the inhibition of bud development and other functions of growth substance in ViciaFaba. Proc. Roy. Soc. B, 114, 317–339. [Google Scholar]
- Tsuchiya, Y. , Yoshimura, M. , Sato, Y. et al. (2015) Probing strigolactone receptors in Striga hermonthica with fluorescence. Science, 349, 864–868. [DOI] [PubMed] [Google Scholar]
- Umehara, M. , Hanada, A. , Yoshida, S. et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Vergara, R. , Noriega, X. , Aravena, K. , Prieto, H. and Perez, F.J. (2017) ABA represses the expression of cell cycle genes and may modulate the development of endodormancy in grapevine buds. Front. Plant Sci. 8, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldie, T. , McCulloch, H. and Leyser, O. (2014) Strigolactones and the control of plant development: lessons from shoot branching. Plant J. 79, 607–622. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Chen, W. , Eggert, K. et al. (2018) Abscisic acid influences tillering by modulation of strigolactones in barley. J. Exp. Bot. 69, 3883–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M.T. , Gutjahr, C. , Bennett, T. and Nelson, D.C. (2017) Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322. [DOI] [PubMed] [Google Scholar]
- Whipple, C.J. , Kebrom, T.H. , Weber, A.L. , Yang, F. , Hall, D. , Meeley, R. , Schmidt, R. , Doebley, J. , Brutnell, T.P. and Jackson, D.P. (2011) grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl Acad. Sci. USA, 108, E506–E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, C. and Finlayson, S.A. (2015) Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 169, 611–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, N. , Jia, L. and Zhang, J. (2012) ABA signal in rice under stress conditions. Rice (N Y), 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, D. , Alvim Kamei, C.L. , Cools, T. et al. (2014) The Arabidopsis SIAMESE‐RELATED cyclin‐dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell, 26, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.X. , van Dijk, A.D.J. , Scaffidi, A. et al. (2014) Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10, 1028–1033. [DOI] [PubMed] [Google Scholar]
- Zheng, C.L. , Halaly, T. , Acheampong, A.K. , Takebayashi, Y. , Jikumaru, Y. , Kamiya, Y. and Or, E. (2015) Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 66, 1527–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. , Lin, Q.B. , Zhu, L.H. et al. (2013) D14‐SCFD3‐dependent degradation of D53 regulates strigolactone signalling. Nature, 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J.H. , Zhang, S.Y. , Zhang, W.P. , Li, G. , Chen, Z.X. , Zhai, W.X. , Zhao, X.F. , Pan, X.B. , Xie, Q. and Zhu, L.H. (2006) The rice HIGH‐TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48, 687–696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Observation of axillary buds from P4 to P6.
Figure S2. Clustering of microarray data for quality checking.
Figure S3. Gene ontology (GO) abundance chart of genes upregulated in dormant buds.
Figure S4. Gene ontology (GO) abundance chart of genes downregulated in dormant buds.
Figure S5. Hierarchical graph of genes involved in biological processes that are downregulated in dormant buds.
Figure S6. Hierarchical graph of genes involved in molecular functions that are downregulated in dormant buds.
Figure S7. Hierarchical graph of genes involved in cellular components that are downregulated in dormant buds.
Figure S8. Fold changes of CyclinD genes in dormant (WT) and active (d10‐2) buds.
Figure S9. Fold changes of Cyclin‐dependent kinase inhibitor (ICK) genes in dormant (WT) and active (d10‐2) buds.
Figure S10. Functional analysis of OsEL2.
Figure S11. Tiller bud elongation in mutants.
Figure S12. Induction of NAC genes.
Figure S13. Effects of ABA on growth of rice.
Figure S14. Concentration of ABA in the basal part of WT, d10‐1 and d14‐1.
Figure S15. Relationships between genes involved in SL‐regulated bud dormancy.
Table S1. Expression levels of genes in the WT microarray.
Table S2. Expression levels of genes in the d10‐2 microarray.
Table S3. Genes upregulated in dormant buds in the microarray analyzed by GeneSpring GX12.
Table S4. Genes downregulated in dormant buds in the microarray analyzed by GeneSpring GX12.
Table S5. Genes upregulated in dormant buds with GO terms.
Table S6. Genes downregulated in dormant buds with GO terms.
Table S7. List of primers used in this study.
Table S8. Accession numbers of genes in this study.


