Contents
Egg production is an important economic trait in poultry, and it is of great significance to study the key genes and functional SNPs that affect egg laying performance. Follicle‐stimulating hormone (FSH) plays an important physiological role in the reproductive performance of humans and animals by binding to its receptor (FSHR). Studies have shown that there are many transcriptional regulatory elements in the 5′ flanking region of the FSHR gene that interact with transcription factors to regulate FSHR transcription. In this study, DNA sequencing was used to identify SNPs in the FSHR promoter sequence in both Dongxiang and Suken chickens. To detect the activity of the chicken FSHR gene promoter, we analysed the characteristics of the sequence and constructed three deletion vectors. We confirmed that the region (−18/−544) was the core promoter. Furthermore, five polymorphisms, including a 200‐bp indel at −869, C−1684T, C−1608T, G−368A and T−238A, were detected in both the Dongxiang and Suken chickens. The age at first egg (AFE) for different genotype of −869 indel in Suken chicken was significantly different (p < 0.01). For SNP C−1684T in Dongxiang chickens, the CC genotype had higher egg number at 43 weeks of age (E43) than that of the TC genotype (p < 0.05). For SNP C−1684T in Suken chickens, the TC genotype had higher AFE than that of the CC genotype (p < 0.05). For SNP C−1608T in Suken chickens, the CC genotype had higher AFE than that of the TC genotype (p < 0.05). For SNP G−368A in Suken chickens, the AG genotype had higher AFE than that of the GG genotype (p < 0.05).
Keywords: association analysis, core promoter, FSHR, single nucleotide polymorphism
1. INTRODUCTION
Dongxiang chicken is a kind of domestic chicken species in China, which produces eggs with blue shells (Wang, Liu, Wang, Li, & Deng, 2011). It is characterized by black feathers, black skin, black bones and black organs. The growth rate and egg yield of this variety are very low (Wang et al., 2009). Suken chicken is also a Chinese native breed, a kind of Chinese triple‐yellow chickens, which have yellow beak, yellow feather and yellow claw (Liu et al., 2014). Suken chicken has an egg production period of approximately 268 days throughout the egg laying cycle, and its egg production peak duration is approximately 40 days. In comparison, Dongxiang chicken has an egg production period of approximately 251 days throughout the egg laying cycle, and its egg production peak lasts for approximately 25 days.
Follicle‐stimulating hormone (FSH) is a glycoprotein “synthesized and secreted by the gonadotropic cells of the anterior pituitary gland (Pierce & Parsons, 1981)” that plays a vital role in gonadal function and fertility (George, Dille, & Heckert, 2011). After FSH is released into circulation, it plays a physiological function by binding to the specific transmembrane receptor (FSHR) located on the target cell (Heckert & Griswold, 1991). In 1996, the cDNA sequence of FSHR was first successfully cloned from chicken ovarian tissue (You, Bridgham, Foster, & Johnson, 1996). The sequence analysis and integrated results of the chicken FSHR gene were demonstrated in 2005 (Wicker et al., 2005). Studies have shown that FSHR is selectively expressed in Sertoli cells and ovarian granulosa cells (Camp, Rahal, & Mayo, 1991; Dankbar et al., 1995), and its expression level is closely related to germ cell differentiation and maturation (Heckert & Griswold, 1993). Gene promoters play a significant role in transcriptional regulation (Fan et al., 2016; Wang et al., 2015). At present, the FSHR promoter of humans, rats, mice and sheep has been successfully cloned (Gromoll, Dankbar, & Gudermann, 1994; Heckert, Daley, & Griswold, 1992; Levallet, Koskimies, Rahman, & Huhtaniemi, 2001; Sairam & Subbarayan, 1997), and the mechanism of transcriptional regulation has been extensively studied. Previous studies have shown that the promoter and transcription factors are closely related to promoter activity. For example, in the FSHR gene promoter of humans, rats, mice and sheep, the transcription factors USF1 and USF2 bind to the E‐box and regulate promoter activity (Goetz, Lloyd, & Griswold, 1996; Heckert, Daggett, & Chen, 1998; Hermann, Hornbaker, Rice, Sawadogo, & Heckert, 2008; Wood & Walker, 2009; Xing & Sairam, 2001). In addition, transcriptional regulators such as E2F, GATA1, SMAD3 and SF1 are also involved in the transcriptional regulation of the FSHR gene (Gong & McGee, 2009; Heckert, 2001; Kim & Griswold, 2001).
Although there are many studies on the regulation of mammalian FSHR promoters, the mechanism regulating transcription of chicken FSHR is not yet clear. In this study, the promoter region and sequence of the chicken FSHR gene were obtained by PCR and DNA sequencing. The transcription factor binding site (TFBS) of the chicken FSHR gene was predicted by online software, and the core promoter region was identified by a luciferase activity assay. Mice developed follicular dysplasia after FSHR gene knockout in granulosa cells (Kumar, Wang, Lu, & Matzuk, 1997). The FSHR gene promoter can regulate the transcription initiation site, time and expression level. Thus, in females, the SNPs occurring in the FSHR promoter region may affect the expression of the FSHR gene and influence reproductive performance. Researchers had found that the polymorphisms at the −278 site in the promoter region of the FSHR gene in Chinese Holstein cows significantly affected both the number of follicles and the number of transferred embryos (Yang et al., 2010).
Egg production is an important economic trait in poultry. Endocrine (Kim, Seo, & Ko, 2004) and many environmental factors (Lewis & Gous, 2006; Liu, Lilburn, Koyyeri, Anderson, & Bacon, 2004), such as photoperiod and different supplements, can affect egg laying performance. However, genetic factors play a decisive role in egg laying performance. Egg laying performance is controlled by multiple genes, and heritability is low. Furthermore, the laying performance of poultry in different periods is also very different (Emsley, 1997; Luo, Yang, & Yang, 2007). In poultry breeding, egg number at 43 weeks of age (E43) is usually an effective indicator of total egg production (Xu et al., 2011). There are obvious differences in egg laying performances of different breeds, including age at first egg (AFE), total egg number and egg weight. Because of the importance of the FSHR gene for reproductive performance, variations in FSHR gene expression may result in distinct reproductive performances in different chicken breeds. In addition, polymorphisms in the chicken FSHR gene promoter may also influence the transcription of FSHR and affect egg production in chickens. Accordingly, in this study, we detected nucleotide polymorphisms in the promoter of the FSHR gene of Dongxiang and Suken chickens by PCR‐RFLP. We then found that several polymorphisms among the five total polymorphisms were associated with E43 or AFE.
2. MATERIALS AND METHODS
Ethics Committee approval was obtained from the Institutional Ethics Committee of Nanjing Agricultural University to the commencement of the study.
2.1. Animals and DNA extraction
The chicken populations used for the experiment were Dongxiang (n = 116) and Suken chickens (n = 434) from Jiangsu Xincao Farm. The chicken was bred in cages (one chicken per cage) with the same feeding and management conditions. We recorded the age at first egg (AFE) and egg number at 43 weeks of age (E43) of every chicken.
We collected total 550 blood samples (116 for Dongxiang chicken and 434 for Suken chicken) from chicken wings and stored it at −20°C. The DNA was extracted by a conventional phenol–chloroform extraction method (Di Pietro, Ortenzi, Tilio, Concetti, & Napolioni, 2011) and adjusted to a final concentration of 100 ng/µl with ddH2O.
2.2. Primers
Ten pairs of primers shown in Table 1 were designed for the experiment. All primers were synthesized by JinWeiZhi Biotechnology Co., Ltd., China.
Table 1.
Primers used for amplification of the follicle‐stimulating hormone receptor of Dongxiang and Suken chickens
| Primer | Primer sequence | Tm/°C | Product size/bp | Application |
|---|---|---|---|---|
| P1 | F:GGTATGGCTTACGCTTGTCTGT | 62 | 790 | Amplification |
| R:GATTGTTTGCTTGTTTCTTTCG | ||||
| P2 | F:AAAGGTGAGAATGGTGGAAT | 59 | 553 | Amplification |
| R:CCAGAGCTAAATAACGCACC | ||||
| P3 | F:AAAGGTGGTAGGGAGGAAGA | 62 | 740 | Amplification |
| R:CCTGGCAGATGAATATCCTG | ||||
| P4 | F:CGGggtaccACTCCCGTTCTTATGACACCTAT | 61 | 1,461 | Plasmids construction |
| R:CCCaagcttTTGTCTCCTTCTCCTCCATC | ||||
| P5 | F:CGGggtaccTTCTTGAACCTGTACCTCTTG | 61 | 794 | Plasmids construction |
| R:CCCaagcttTTGTCTCCTTCTCCTCCATC | ||||
| P6 | F:CGGggtaccTGGATCTATGAAGGGGAGC | 61 | 526 | Plasmids construction |
| R:CCCaagcttTTGTCTCCTTCTCCTCCATC | ||||
| P7 | F:GGTATGGCTTACGCTTGTCTGT | 62 | 790 | Genotyping |
| R:GATTGTTTGCTTGTTTCTTTCG | ||||
| P8 | F:TGTCTCTTAGTCTTATCAAACAACA | 60 | 492 | Genotyping |
| R:CCTGGCAGATGAATATCCTG | ||||
| P9 | F:AAAGGTGGTAGGGAGGAAGA | 62 | 740 | Genotyping |
| R:CCTGGCAGATGAATATCCTG | ||||
| P10 | F:ACAATCAAAACCCCAGCAAC | 62 | 741 | Genotyping |
| R:AATGAACCGGAATGCTTTTG |
The digestion sites of the enzymes are underlined.
2.3. PCR amplification and sequencing
PCR was performed in a 20 µl mixture containing 10 µl of 2X Taq Mix (Takara Biotechnology Co. Ltd., Dalian, China), 10 pmol of upstream and downstream primers and 100 ng of chicken genomic DNA. The following reaction conditions were used: 95°C pre‐denaturation for 5 min, 95°C denaturation for 30 s, X°C (X was the annealing temperature shown in Table 1) annealing for 30 s and a 72°C extension for 30 s (depending on product length, 1 kb = 1 min) for 35 cycles. The PCR products of P1, P2 and P3 were separated by 1.5% agarose gel electrophoresis and sequenced. SNPs were identified by sequence traces.
2.4. Analysis software
The promoter and transcription factor binding sites were predicted and analysed by Promoter Scan, Genomatix and Methprimer (Table 2).
Table 2.
Software online for promoter analysis
| Software name | URL | Purpose |
|---|---|---|
| UCSC | http://genome.ucsc.edu/ | Promoter prediction |
| Promoter Scan | https://www-bimas.cit.nih.gov/molbio/proscan/ | Core promoter prediction |
| Methprimer | http://www.urogene.org/methprimer/ | CpG island prediction |
| Genomatix | http://www.genomatix.de/index.html | TFBS prediction |
TFBS: Transcription factor binding site.
2.5. Construction of the FSHR promoter luciferase plasmids
The purified promoter fragments of the chicken FSHR gene were amplified by three specific primers containing KpnI and HindIII restriction enzyme cleavage sites and then cloned into the pGL3‐basic vector digested with KpnI and HindIII restriction enzymes. The primers used to amplify the desired promoter fragments of the chicken FSHR gene are shown in Table 1.
2.6. Cell culture, transient transfection and luciferase activity assay
Specific methods for granulosa cell culture reference the article (Hu, Duggavathi, & Zadworny, 2017). The granulosa cells were seeded into 24‐well plates for 16–18 hr. The luciferase plasmids and the Renilla luciferase reporter vector (pRL‐K) were cotransfected at a ratio of 50:1 into chicken ovarian granulosa cells with Lipofectamine 2000 when the cells were completely adhered. The cells were collected after 24 hr of transfection, and luciferase activity was assayed using the Dual‐Luciferase® Reporter Assay System (Promega, Madison, WI, USA).
2.7. Genotyping of polymorphisms
Using the PCR‐RFLP method to detect genotypes, we selected the appropriate endonuclease for enzyme digestion of the target genes of the tested chickens. Enzyme reaction conditions followed the enzyme product specifications. Genotypes were detected by the gel bands graphic of 1.5% agarose gel electrophoresis. Electrophoresis conditions included the following specifications: 1X TBE, 120 V/30 min.
2.8. Statistical analysis
Allele and genotype frequencies were calculated by direct counting. The chi‐squared test was used to examine the Hardy–Weinberg equilibrium of the SNPs. Association analyses of SNPs with E43 and AFE were performed using spss version 20.0.
3. RESULTS
3.1. Chicken FSHR Gene 5′ regulatory region amplification
Three fragments of approximately 790 bp, 553 bp and 740 bp were separated in 1.5% agarose gel, after PCR amplification (Figure 1).
Figure 1.
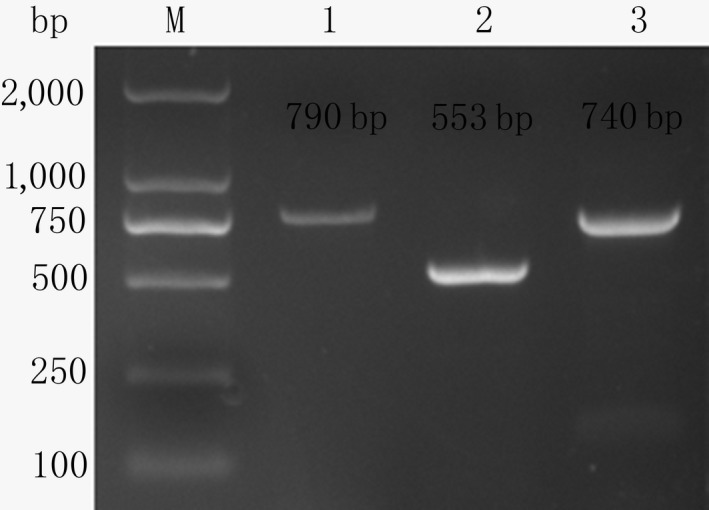
Agarose gel photograph of 5′ regulatory region of Chicken FSHR gene. 1‐3: Amplified fragments of primers P1‐P3; M: DNA marker DL2000
3.2. 5′ regulatory sequence analysis of the chicken FSHR gene
A few common cis‐elements were predicted in the chicken FSHR proximal promoter sequence by Genomatix online software (http://www.genomatix.de/index.html), including two TATA‐boxes (TBP binding site), three CAAT‐boxes (C/EBP binding sites), a GC‐box (SP1 binding site) and two E‐boxes (USF1/2 binding sites). In addition, several transcription factor binding sites were enriched in the chicken FSHR proximal promoter sequence, including AP1, SF1, YY1, GATA, FKHD, SP1 and CREB (Figure 2). No typical CpG islands were detected in the chicken FSHR proximal promoter sequence using Methprimer online software (http://www.urogene.org/methprimer/; Figure 3).
Figure 2.
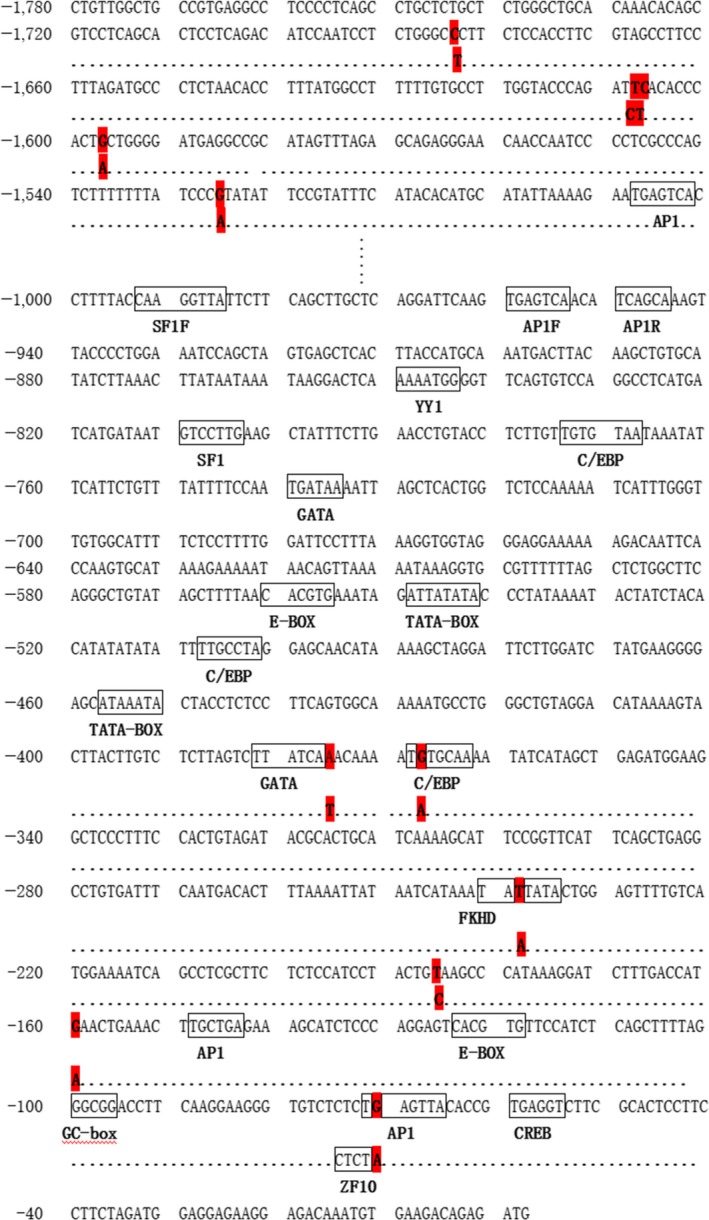
The 5′ regulation sequence of FSHR gene in chicken. The SNP sites are indicated by red background; the transcription factor binding sites are indicated by blue boxes
Figure 3.
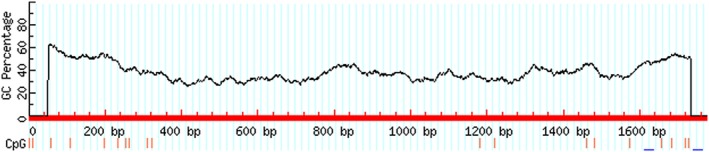
The prediction result of CpG islands in promoter region of Chicken FSHR gene. Vertical lines indicate CpG sites
3.3. Promoter activity analysis of the chicken FSHR gene
PCR product electrophoresis is shown in Figure 4a. Three special plasmids, pFSHR‐1479, pFSHR‐812 and pFSHR‐544, were constructed to identify the promoter activity of the chicken FSHR gene, and the translation start site (ATG) was defined as +1. The constructed plasmids were identified by double digests (Figure 4b). The plasmids were transiently transfected into chicken ovarian granulosa cells, and luciferase activity assays were performed to identify the promoter activity of the chicken FSHR gene. As shown in Figure 5, the luciferase activity of the promoter pFSHR‐544 was significantly higher than that of pFSHR‐812, pFSHR‐1479 and the negative control pGL3‐basic (p < 0.01). In contrast, no significant difference was observed between pFSHR‐1479, pFSHR‐812 and pGL3‐basic (p > 0.05).
Figure 4.
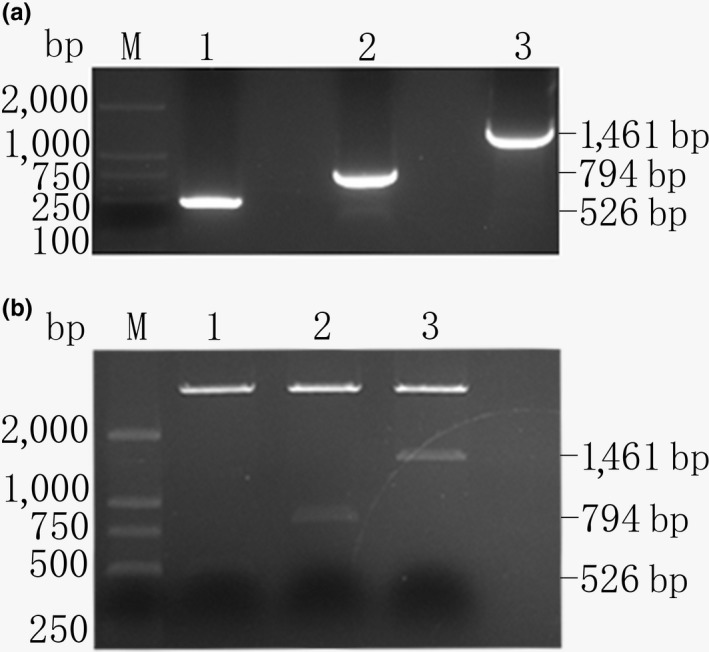
A agarose gel photograph of deleted fragment in 5′ regulatory region of Chicken FSHR gene. B identification of recombinant vectors by restriction enzymes
Figure 5.
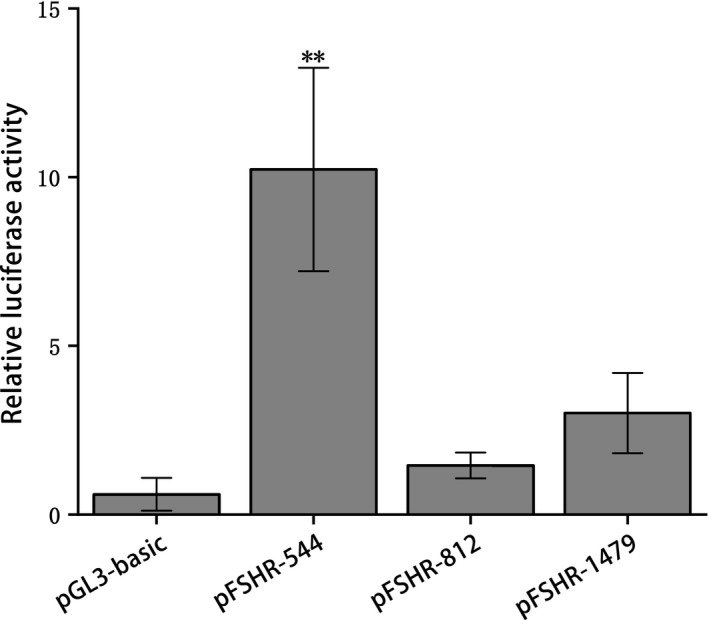
Promoter activity analysis of Chicken FSHR gene. ** indicates extremely significant difference (p < 0.01)
3.4. Genotype frequency and allele frequency
We found that four restriction sites exist at C−1684T, C−1608T, G−368A and T−238A within the promoter region of chicken FSHR, including ApaI, MboI, NdeI and SspI, respectively. The four single nucleotide polymorphisms and the 200‐bp indel mutation were detected by PCR‐RFLP (Figure 6).
Figure 6.
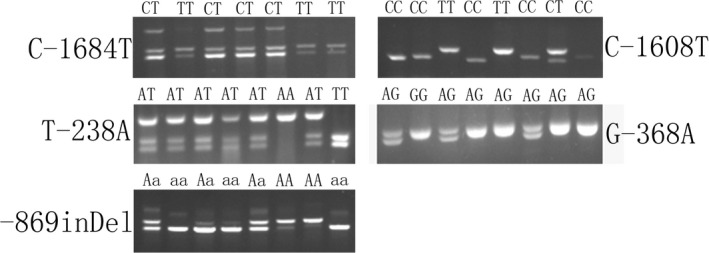
Genotyping of the C−1684T, C−1608T, T−238A, G−368A and −869 indel mutations of the promoter region of chicken FSHR gene
All five mutations were in a Hardy–Weinberg imbalanced state in the Suken yellow chicken population. Furthermore, the −869 indel and G−368A mutations were in a Hardy–Weinberg imbalanced state in the Dongxiang chicken population (Table 3).
Table 3.
Genotypes and allele frequency of the mutations of FSHR gene
| Polymorphism sites | Breed | Genotypic frequency | Allele and frequency | χ 2 | |||
|---|---|---|---|---|---|---|---|
| Genotype | Number | Frequency | Allele | Frequency | |||
| −869 indel | Dongxiang | AA | 4 | 0.04 | A | 0.082 | 15.830* |
| Aa | 11 | 0.09 | a | 0.918 | |||
| aa | 101 | 0.87 | |||||
| Suken | AA | 10 | 0.02 | A | 0.089 | 15.29* | |
| Aa | 57 | 0.13 | a | 0.911 | |||
| aa | 367 | 0.85 | |||||
| C−1684T | Dongxiang | CC | 42 | 0.36 | T | 0.379 | 1.125 |
| TC | 60 | 0.52 | C | 0.621 | |||
| TT | 14 | 0.12 | |||||
| Suken | CC | 85 | 0.30 | T | 0.518 | 9.051* | |
| TC | 248 | 0.53 | C | 0.482 | |||
| TT | 101 | 0.17 | |||||
| C−1608T | Dongxiang | CC | 39 | 0.34 | T | 0.457 | 3.204 |
| TC | 48 | 0.41 | C | 0.543 | |||
| TT | 29 | 0.25 | |||||
| Suken | CC | 312 | 0.72 | T | 0.217 | 166.7* | |
| TC | 56 | 0.13 | C | 0.783 | |||
| TT | 66 | 0.15 | |||||
| G−368A | Dongxiang | GG | 73 | 0.63 | A | 0.185 | 6.004* |
| AG | 43 | 0.37 | G | 0.815 | |||
| AA | 0 | 0 | |||||
| Suken | GG | 252 | 0.58 | A | 0.210 | 30.55* | |
| AG | 182 | 0.42 | G | 0.790 | |||
| AA | 0 | 0 | |||||
| T−238A | Dongxiang | AA | 63 | 0.54 | A | 0.720 | 1.775 |
| AT | 41 | 0.35 | T | 0.280 | |||
| TT | 12 | 0.11 | |||||
| Suken | AA | 138 | 0.32 | A | 0.530 | 9.64* | |
| AT | 184 | 0.42 | T | 0.470 | |||
| TT | 112 | 0.26 | |||||
χ 2 0.05 (2) = 5.99, χ 2 0.05(1) = 3.84, χ 2 0.01(1) = 6.63.
The chi‐square value with * means p < 0.05.
3.5. Association analysis of SNPs of the chicken FSHR gene with E43 and AFE
The association analyses of SNPs with E43 and AFE was performed using SPSS version 20.0 (Tables 4 and 5).
Table 4.
Association analysis of SNPs of FSHR gene with Dongxiang chicken egg performance
| Polymorphism sites | Genotype | Number | AFE | p‐Value | E43 | p‐Value |
|---|---|---|---|---|---|---|
| −869 indel | AA | 4 | 170.00 ± 7.53 | 0.704 | 56.25 ± 6.95 | 0.304 |
| Aa | 11 | 165.45 ± 10.21 | 60.82 ± 15.03 | |||
| aa | 101 | 165.28 ± 16.33 | 66.37 ± 16.93 | |||
| C−1684T | TT | 14 | 168.00 ± 14.82 | 0.222 | 65.71 ± 12.78 ab | 0.050 |
| TC | 60 | 167.28 ± 19.21 | 62.77 ± 19.11 b | |||
| CC | 42 | 162.19 ± 8.08 | 69.33 ± 13.08 a | |||
| C−1608T | TT | 29 | 165.14 ± 10.24 | 0.982 | 62.76 ± 19.20 | 0.287 |
| TC | 48 | 165.48 ± 19.85 | 68.35 ± 16.27 | |||
| CC | 39 | 165.87 ± 13.19 | 64.03 ± 14.76 | |||
| G−368A | GG | 73 | 165.89 ± 17.19 | 0.465 | 65.85 ± 16.07 | 0.569 |
| AG | 43 | 164.91 ± 12.70 | 64.91 ± 17.67 | |||
| AA | 0 | 0 | 0 | |||
| T−238A | TT | 12 | 164.08 ± 11.57 | 0.800 | 72.42 ± 13.70 | 0.292 |
| AT | 41 | 164.59 ± 16.44 | 65.51 ± 14.01 | |||
| AA | 63 | 166.41 ± 15.90 | 64.17 ± 18.46 |
In the same group, different superscripts mean significant difference (p < 0.05).
Table 5.
Association analysis of SNPs of FSHR gene with Suken chicken egg performance
| Polymorphism sites | Genotype | Number | AFE | p‐Value | E43 | p‐Value |
|---|---|---|---|---|---|---|
| −869 indel | AA | 10 | 160.40 ± 5.54A | 0.005 | 96.70 ± 19.33 | 0.390 |
| Aa | 57 | 157.25 ± 2.90B | 104.19 ± 14.27 | |||
| aa | 367 | 157.12 ± 3.06B | 103.40 ± 16.18 | |||
| C−1684T | TT | 101 | 157.24 ± 3.14ab | 0.030 | 101.70 ± 16.35 | 0.452 |
| TC | 248 | 157.48 ± 3.70a | 104.08 ± 15.85 | |||
| CC | 85 | 156.58 ± 2.09b | 103.18 ± 16.12 | |||
| C−1608T | TT | 66 | 156.74 ± 3.60ab | 0.023 | 102.67 ± 17.30 | 0.234 |
| TC | 56 | 156.35 ± 1.92b | 106.75 ± 16.45 | |||
| CC | 312 | 157.51 ± 3.42a | 102.88 ± 15.63 | |||
| G−368A | GG | 252 | 156.83 ± 2.88b | 0.003 | 104.27 ± 15.27 | 0.161 |
| AG | 182 | 157.83 ± 3.78a | 102.08 ± 16.95 | |||
| AA | 0 | 0 | 0 | |||
| T−238A | TT | 112 | 157.08 ± 3.29 | 0.674 | 104.19 ± 16.99 | 0.758 |
| AT | 184 | 157.20 ± 3.20 | 103.35 ± 15.45 | |||
| AA | 138 | 157.44 ± 3.52 | 102.67 ± 16.02 |
In the same group, different superscripts mean significant difference (p < 0.05).
Dongxiang chickens with the CC genotype of SNP C−1684T had higher E43 compared with that of chicken with the TC genotype (p < 0.05) (Table 4). The AFEs of the SNP −869 indel and SNP G−368A genotypes were significantly different (p < 0.01). The AFEs of the SNP C−1684T and C−1608T genotypes were also significantly different (p < 0.05; Table 5).
4. DISCUSSION
Gene promoters play a key role in transcriptional regulation by controlling the transcription initiation site, time and expression level (Juneja, Ilm, Schlag, & Stein, 2013). Therefore, research on the regulation of gene expression can start from the structural function of its promoter. In this study, we obtained approximately 1.8 kb sequence of the chicken FSHR gene promoter region and analysed the structure using bioinformatics software, revealing predicted TATA‐box and CAAT‐box cis‐acting elements. The TATA‐box and CAAT‐box are not found in the promoter region of the human, rat and sheep FSHR genes. However, there is a TATA‐box in the mouse FSHR gene promoter region (Gromoll et al., 1994; Heckert et al., 1992; Sairam & Subbarayan, 1997), which indicating that the regulatory mechanism of the FSHR gene promoter may differ interspecifically.
A recently research indicated that miR‐4281, an miRNA specifically expressed in hominids, directly interacting with the TATA‐box motif in the human FOXP3 promoter could efficiently and specifically upregulates FOXP3 expression (Zhang et al., 2018). Overexpression of CAAT/enhancer‐binding protein (C/EBP) in Spodoptera litura‐221 (Spli‐221) cells increased the promoter activity 5.57‐fold, while mutation of the C/EBP CRE abolished the binding of the C/EBP with the CRE (Liang, Zhang, Zeng, Zheng, & Feng, 2015). These findings validate the important role of TATA‐box and CAAT‐box in promoter regulation.
The E‐box was mutated in the promoter of the rat FSHR gene, which resulted in a significant reduction of FSHR promoter activity in MSC‐1 cell lines (Heckert et al., 1998). Two E‐box sites were predicted to be present in the promoter of the chicken FSHR gene, whether the E‐box regulating the FSHR promoter requires further identification. Furthermore, multiple transcription factor binding sites (TFBS) were predicted in the chicken FSHR gene promoter with Genomatix software, including AP1, GATA, SF1, YY1, as well as others. It is noteworthy that the transcription factors E2F, Smad3 and ETS were involved in the transcriptional regulation of the FSHR gene in previous reports (Brune, Adams, & Gromoll, 2010; Heckert, 2001; Kim & Griswold, 2001). However, we did not predict these transcription factor binding sites in the chicken FSHR gene promoter, further illustrating that different promoter regulatory mechanisms are likely to exist in the FSHR genes in different species.
Researchers have identified that the region (−1,195/−598) was the core promoter of the porcine FSHR gene (Wu et al., 2015). In order to identify the core promoter region of the chicken FSHR gene, we constructed three special plasmids: pFSHR‐1479 (−1,479/−18), pFSHR‐812 (−830/−18) and pFSHR‐544 (−562/−18). The activity of pFSHR‐544 was significantly higher than that of pFSHR‐basic, pFSHR‐1479 and pFSHR‐812. There is no significant difference in the activity between pFSHR‐1479 and pFSHR‐812 and the pFSHR‐basic. The above studies suggested that the region (−18/−562) contains some positive cis‐regulatory elements, whereas the region (−562/−1,497) contains some negative transcription factor binding sites.
The sequencing results showed that eleven SNPs exist in the promoter of the chicken FSHR gene. It has been widely reported that mutations in the FSHR gene have a genetic effect on reproductive traits in humans and other animals (Lussiana et al., 2008). Researchers had found that the FSHR promoter polymorphism FSHR −29G>A influences the androgen levels of human small antral follicle (hSAF; Borgbo et al., 2017). The activity of FSHR promoter is significantly affected by the 29th site G → A mutation that will weaken promoter activity and result in poor response to FSH (Dan, Jing, Liangbin, Ting, & Ying, 2015). We analysed the effect of FSHR gene polymorphism on transcription factor binding sites and found that there are three mutations leading to changes in the transcription factor binding site. However, it remains to be investigated whether the changes of these transcription factor binding sites will have an effect on the transcriptional activity of chicken FSHR.
Furthermore, the genotypes of five SNPs were associated with both E43 and AFE. The SNP C−1684T of Dongxiang chicken was associated with E43 and SNPs C−1684T, C−1608T and G−368A of Suken chickens were significantly related with AFE. Moreover, in this study, the 200‐bp indel mutation had a significant correlation with AFE, which was compatible with the findings of Kang et al. (2012). Our data suggested that these loci might serve as the potential genetic markers for chicken reproduction. Previous study of FSHR gene in muscovy duck detected that the SNP C320T is significantly associated with egg production at 59 weeks of age (p < 0.05), whereas the SNP A227G is significantly associated with age at first egg stage (p < 0.05) (Xu et al., 2017). However, the mechanisms by which these polymorphisms make their effects require to be further researched.
In conclusion, in this study, we identified the core promoter region of the chicken FSHR gene and predicted several transcription factor binding sites. Moreover, a total of five polymorphisms of the FSHR promoter region were detected, and we found that all of them were associated with egg number at 43 weeks of age (E43) or age at first egg (AFE).
CONFLICT OF INTEREST
None of the authors have any conflict of interest to declare.
AUTHOR CONTRIBUTIONS
Debing Yu and Yinglin Lu designed the experiment. Xiaopeng Li, Xiaofan Liu, Xiaolei Xie and Kun Wang completed the experiment. Xiaopeng Li wrote and revised the paper.
Li X, Lu Y, Liu X, Xie X, Wang K, Yu D. Identification of chicken FSHR gene promoter and the correlations between polymorphisms and egg production in Chinese native hens. Reprod Dom Anim. 2019;54:702–711. 10.1111/rda.13412
REFERENCES
- Borgbo, T. , Kluckova, H. , Macek, M. Sr , Chrudimska, J. , Kristensen, S. G. , Hansen, L. L. , & Andersen, C. Y. (2017). The common follicle‐stimulating hormone receptor (FSHR) promoter polymorphism FSHR −29G>A affects androgen production in normal human small antral follicles. Frontiers in Endocrinology, 8, 122 10.3389/fendo.2017.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune, M. , Adams, C. , & Gromoll, J. (2010). Primate FSH‐receptor promoter nucleotide sequence heterogeneity affects FSH‐receptor transcription. Molecular and Cellular Endocrinology, 317, 90–98. 10.1016/j.mce.2009.12.020 [DOI] [PubMed] [Google Scholar]
- Camp, T. A. , Rahal, J. O. , & Mayo, K. E. (1991). Cellular localization and hormonal regulation of follicle‐stimulating hormone and luteinizing hormone receptor messenger RNAs in the rat ovary. Molecular Endocrinology, 5, 1405–1417. 10.1210/mend-5-10-1405 [DOI] [PubMed] [Google Scholar]
- Dan, W. , Jing, G. , Liangbin, X. , Ting, Z. , & Ying, Z. (2015). Association of follicle stimulating hormone receptor promoter with ovarian response in IVF‐ET patients. Iranian Journal of Reproductive Medicine, 13, 715–720. [PMC free article] [PubMed] [Google Scholar]
- Dankbar, B. , Brinkworth, M. H. , Schlatt, S. , Weinbauer, G. F. , Nieschlag, E. , & Gromoll, J. (1995). Ubiquitous expression of the androgen receptor and testis‐specific expression of the FSH receptor in the cynomolgus monkey (Macaca fascicularis) revealed by a ribonuclease protection assay. The Journal of Steroid Biochemistry and Molecular Biology, 55, 35–41. 10.1016/0960-0760(95)00148-S [DOI] [PubMed] [Google Scholar]
- Di Pietro, F. , Ortenzi, F. , Tilio, M. , Concetti, F. , & Napolioni, V. (2011). Genomic DNA extraction from whole blood stored from 15‐ to 30‐years at −20 degrees C by rapid phenol‐chloroform protocol: A useful tool for genetic epidemiology studies. Molecular and Cellular Probes, 25, 44–48. 10.1016/j.mcp.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Emsley, A. (1997). Integration of classical and molecular approaches of genetic selection: Egg production. Poultry Science, 76, 1127–1130. 10.1093/ps/76.8.1127 [DOI] [PubMed] [Google Scholar]
- Fan, X. P. , Ji, X. F. , Li, X. Y. , Gao, S. , Fan, Y. C. , & Wang, K. (2016). Methylation of the glutathione‐S‐transferase P1 gene promoter is associated with oxidative stress in patients with chronic hepatitis B. The Tohoku Journal of Experimental Medicine, 238, 57–64. 10.1620/tjem.238.57 [DOI] [PubMed] [Google Scholar]
- George, J. W. , Dille, E. A. , & Heckert, L. L. (2011). Current concepts of follicle‐stimulating hormone receptor gene regulation. Biology of Reproduction, 84, 7–17. 10.1095/biolreprod.110.085043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz, T. L. , Lloyd, T. L. , & Griswold, M. D. (1996). Role of E box and initiator region in the expression of the rat follicle‐stimulating hormone receptor. The Journal of Biological Chemistry, 271, 33317–33324. 10.1074/jbc.271.52.33317 [DOI] [PubMed] [Google Scholar]
- Gong, X. , & McGee, E. A. (2009). Smad3 is required for normal follicular follicle‐stimulating hormone responsiveness in the mouse. Biology of Reproduction, 81, 730–738. 10.1095/biolreprod.108.070086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoll, J. , Dankbar, B. , & Gudermann, T. (1994). Characterization of the 5′ flanking region of the human follicle‐stimulating hormone receptor gene. Molecular and Cellular Endocrinology, 102, 93–102. 10.1016/0303-7207(94)90102-3 [DOI] [PubMed] [Google Scholar]
- Heckert, L. L. (2001). Activation of the rat follicle‐stimulating hormone receptor promoter by steroidogenic factor 1 is blocked by protein kinase a and requires upstream stimulatory factor binding to a proximal E box element. Molecular Endocrinology, 15, 704–715. 10.1210/mend.15.5.0632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert, L. L. , Daggett, M. A. , & Chen, J. (1998). Multiple promoter elements contribute to activity of the follicle‐stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. Molecular Endocrinology, 12, 1499–1512. 10.1210/mend.12.10.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert, L. L. , Daley, I. J. , & Griswold, M. D. (1992). Structural organization of the follicle‐stimulating hormone receptor gene. Molecular Endocrinology, 6, 70–80. 10.1210/mend.6.1.1738373 [DOI] [PubMed] [Google Scholar]
- Heckert, L. L. , & Griswold, M. D. (1991). Expression of follicle‐stimulating hormone receptor mRNA in rat testes and Sertoli cells. Molecular Endocrinology, 5, 670–677. 10.1210/mend-5-5-670 [DOI] [PubMed] [Google Scholar]
- Heckert, L. , & Griswold, M. D. (1993). Expression of the FSH receptor in the testis. Recent Progress in Hormone Research, 48, 61–77. [DOI] [PubMed] [Google Scholar]
- Hermann, B. P. , Hornbaker, K. , Rice, D. A. , Sawadogo, M. , & Heckert, L. L. (2008). In vivo regulation of follicle‐stimulating hormone receptor by the transcription factors upstream stimulatory factor 1 and upstream stimulatory factor 2 is cell specific. Endocrinology, 149, 5297–5306. 10.1210/en.2007-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S. , Duggavathi, R. , & Zadworny, D. (2017). Expression and regulation of prolactin‐like protein messenger RNA in undifferentiated chicken granulosa cells. General and Comparative Endocrinology, 240, 191–197. 10.1016/j.ygcen.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Juneja, M. , Ilm, K. , Schlag, P. M. , & Stein, U. (2013). Promoter identification and transcriptional regulation of the metastasis gene MACC1 in colorectal cancer. Molecular Oncology, 7, 929–943. 10.1016/j.molonc.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, L. , Zhang, N. , Zhang, Y. , Yan, H. , Tang, H. , Yang, C. , … Jiang, Y. (2012). Molecular characterization and identification of a novel polymorphism of 200 bp indel associated with age at first egg of the promoter region in chicken follicle‐stimulating hormone receptor (FSHR) gene. Molecular Biology Reports, 39, 2967–2973. 10.1007/s11033-011-1058-x [DOI] [PubMed] [Google Scholar]
- Kim, J. S. , & Griswold, M. D. (2001). E2F and GATA‐1 are required for the Sertoli cell‐specific promoter activity of the follicle‐stimulating hormone receptor gene. Journal of Andrology, 22, 629–639. [PubMed] [Google Scholar]
- Kim, M. H. , Seo, D. S. , & Ko, Y. (2004). Relationship between egg productivity and insulin‐like growth factor‐I genotypes in Korean native Ogol chickens. Poultry Science, 83, 1203–1208. 10.1093/ps/83.7.1203 [DOI] [PubMed] [Google Scholar]
- Kumar, T. R. , Wang, Y. , Lu, N. , & Matzuk, M. M. (1997). Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. NatureGenetics, 15, 201–204. 10.1038/ng0297-201 [DOI] [PubMed] [Google Scholar]
- Levallet, J. , Koskimies, P. , Rahman, N. , & Huhtaniemi, I. (2001). The promoter of murine follicle‐stimulating hormone receptor: Functional characterization and regulation by transcription factor steroidogenic factor 1. Molecular Endocrinology, 15, 80–92. 10.1210/mend.15.1.0583 [DOI] [PubMed] [Google Scholar]
- Lewis, P. D. , & Gous, R. M. (2006). Effect of final photoperiod and twenty‐week body weight on sexual maturity and early egg production in broiler breeders. Poultry Science, 85, 377–383. 10.1093/ps/85.3.377 [DOI] [PubMed] [Google Scholar]
- Liang, L. N. , Zhang, L. L. , Zeng, B. J. , Zheng, S. C. , & Feng, Q. L. (2015). Transcription factor CAAT/enhancer‐binding protein is involved in regulation of expression of sterol carrier protein x in Spodoptera litura. Insect Molecular Biology, 24, 551–560. 10.1111/imb.12182 [DOI] [PubMed] [Google Scholar]
- Liu, H. K. , Lilburn, M. S. , Koyyeri, B. , Anderson, J. W. , & Bacon, W. L. (2004). Preovulatory surge patterns of luteinizing hormone, progesterone, and estradiol‐17beta in broiler breeder hens fed ad libitum or restricted fed. Poultry Science, 83, 823–829. 10.1093/ps/83.5.823 [DOI] [PubMed] [Google Scholar]
- Liu, T. , Qu, H. , Luo, C. , Shu, D. , Wang, J. , Lund, M. S. , & Su, G. (2014). Accuracy of genomic prediction for growth and carcass traits in Chinese triple‐yellow chickens. BMC Genetics, 15, 110 10.1186/s12863-014-0110-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, P. T. , Yang, R. Q. , & Yang, N. (2007). Estimation of genetic parameters for cumulative egg numbers in a broiler dam line by using a random regression model. Poultry Science, 86, 30–36. 10.1093/ps/86.1.30 [DOI] [PubMed] [Google Scholar]
- Lussiana, C. , Guani, B. , Mari, C. , Restagno, G. , Massobrio, M. , & Revelli, A. (2008). Mutations and polymorphisms of the FSH receptor (FSHR) gene: Clinical implications in female fecundity and molecular biology of FSHR protein and gene. Obstetrical & Gynecological Survey, 63, 785–795. 10.1097/OGX.0b013e31818957eb [DOI] [PubMed] [Google Scholar]
- Pierce, J. G. , & Parsons, T. F. (1981). Glycoprotein hormones: Structure and function. Annual Review of Biochemistry, 50, 465–495. 10.1146/annurev.bi.50.070181.002341 [DOI] [PubMed] [Google Scholar]
- Sairam, M. R. , & Subbarayan, V. S. (1997). Characterization of the 5′ flanking region and potential control elements of the ovine follitropin receptor gene. Molecular Reproduction and Development, 48, 480–487. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Li, D. , Zhang, M. , Yang, W. , Cui, Y. , & Li, S. (2015). Methylation of KvDMR1 involved in regulating the imprinting of CDKN1C gene in cattle. AnimalGenetics, 46, 354–360. 10.1111/age.12297 [DOI] [PubMed] [Google Scholar]
- Wang, X. L. , Zheng, J. X. , Ning, Z. H. , Qu, L. J. , Xu, G. Y. , & Yang, N. (2009). Laying performance and egg quality of blue‐shelled layers as affected by different housing systems. Poultry Science, 88, 1485–1492. 10.3382/ps.2008-00417 [DOI] [PubMed] [Google Scholar]
- Wang, Z. P. , Liu, R. F. , Wang, A. R. , Li, J. Y. , & Deng, X. M. (2011). Expression and activity analysis reveal that heme oxygenase (decycling) 1 is associated with blue egg formation. Poultry Science, 90, 836–841. 10.3382/ps.2010-01143 [DOI] [PubMed] [Google Scholar]
- Wicker, T. , Robertson, J. S. , Schulze, S. R. , Feltus, F. A. , Magrini, V. , Morrison, J. A. , … Ivarie, R. (2005). The repetitive landscape of the chicken genome. GenomeResearch, 15, 126–136. 10.1101/gr.2438004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, M. A. , & Walker, W. H. (2009). USF1/2 transcription factor DNA‐binding activity is induced during rat Sertoli cell differentiation. Biology of Reproduction, 80, 24–33. 10.1095/biolreprod.108.070037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W. , Han, J. , Cao, R. , Zhang, J. , Li, B. , Liu, Z. , … Liu, H. (2015). Sequence and regulation of the porcine FSHR gene promoter. AnimalReproduction Science, 154, 95–104. 10.1016/j.anireprosci.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Xing, W. , & Sairam, M. R. (2001). Characterization of regulatory elements of ovine follicle‐stimulating hormone (FSH) receptor gene: The role of E‐box in the regulation of ovine FSHreceptor expression. Biology of Reproduction, 64, 579–589. [DOI] [PubMed] [Google Scholar]
- Xu, H. P. , Zeng, H. , Zhang, D. X. , Jia, X. L. , Luo, C. L. , Fang, M. X. , … Zhang, X. Q. (2011). Polymorphisms associated with egg number at 300 days of age in chickens. Genetics and Molecular Research : GMR, 10, 2279–2289. 10.4238/2011.October.3.5 [DOI] [PubMed] [Google Scholar]
- Xu, J. , Gao, X. , Li, X. , Ye, Q. , Jebessa, E. , Abdalla, B. A. , & Nie, Q. (2017). Molecular characterization, expression profile of the FSHRgene and its association with egg production traits in muscovy duck. Journal of Genetics, 96, 341–351. 10.1007/s12041-017-0783-x [DOI] [PubMed] [Google Scholar]
- Yang, W. C. , Li, S. J. , Tang, K. Q. , Hua, G. H. , Zhang, C. Y. , Yu, J. N. , … Yang, L. G. (2010). Polymorphisms in the 5′ upstream region of the FSH receptor gene, and their association with superovulation traits in Chinese Holstein cows. AnimalReproduction Science, 119, 172–177. 10.1016/j.anireprosci.2010.02.004 [DOI] [PubMed] [Google Scholar]
- You, S. , Bridgham, J. T. , Foster, D. N. , & Johnson, A. L. (1996). Characterization of the chicken follicle‐stimulating hormone receptor (cFSH‐R) complementary deoxyribonucleic acid, and expression of cFSH‐R messenger ribonucleic acid in the ovary. Biology of Reproduction, 55, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, W. , Chen, Y. , Liu, J. , Wu, K. , Su, L. , … Zhang, H. (2018). A cellular microRNA facilitates regulatory T lymphocyte development by targeting the FOXP3 promoter TATA‐box motif. Journal of Immunology, 200, 1053–1063. 10.4049/jimmunol.1700196 [DOI] [PubMed] [Google Scholar]


