SUMMARY
Inorganic phosphorus (Pi) is an essential element in numerous metabolic reactions and signaling pathways, but the molecular details of these pathways remain largely unknown. In this study, metabolite profiles of maize (Zea mays L.) leaves and roots were compared between six low‐Pi‐sensitive lines and six low‐Pi‐tolerant lines under Pi‐sufficient and Pi‐deficient conditions to identify pathways and genes associated with the low‐Pi stress response. Results showed that under Pi deprivation the concentrations of nucleic acids, organic acids and sugars were increased, but that the concentrations of phosphorylated metabolites, certain amino acids, lipid metabolites and nitrogenous compounds were decreased. The levels of secondary metabolites involved in plant immune reactions, including benzoxazinoids and flavonoids, were significantly different in plants grown under Pi‐deficient conditions. Among them, the 11 most stable metabolites showed significant differences under low‐ and normal‐Pi conditions based on the coefficient of variation (CV). Isoleucine and alanine were the most stable metabolites for the identification of Pi‐sensitive and Pi‐resistant maize inbred lines. With the significant correlation between morphological traits and metabolites, five low‐Pi‐responding consensus genes associated with morphological traits and simultaneously involved in metabolic pathways were mined by combining metabolites profiles and genome‐wide association study (GWAS). The consensus genes induced by Pi deficiency in maize seedlings were also validated by reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR). Moreover, these genes were further validated in a recombinant inbred line (RIL) population, in which the glucose‐6‐phosphate‐1‐epimerase encoding gene mediated yield and correlated traits to phosphorus availability. Together, our results provide a framework for understanding the metabolic processes underlying Pi‐deficient responses and give multiple insights into improving the efficiency of Pi use in maize.
Keywords: maize, phosphorus‐use efficiency, metabolite profiling, plant immunity, genome‐wide association study (GWAS), consensus genes
Significance statement
Metabolite profiling and GWAS were applied to reveal maize low‐Pi response mechanisms.
Introduction
Adequate inorganic phosphorus (Pi) bioavailability is critical for normal plant growth and development, and soil Pi deficiency is a major factor limiting crop yield and quality; however, the concentrations of Pi in soil solutions are generally <10 μm, which is completely inadequate being below the critical level needed by plants (Schachtman et al., 1998). Thus, although most cultivated soils have high total P, the bioavailable Pi is usually seriously insufficient. In many fields, the application of phosphate fertilizer is necessary to guarantee an adequate level of crop yield and quality. As a result of the low utilization rate by crops, most Pi in fertilizer is fixed in the soil or is lost with soil erosion, resulting in resource waste, water eutrophication and the consequent overgrowth of aquatic plant and algae species, as exemplified by the toxic red tide (Correll, 1998; Smith and Schindler, 2009). Furthermore, with the growing demand, the raw materials for phosphate fertilizer, such as rock phosphate (apatite), are becoming increasingly scarce (López‐Arredondo et al., 2014). Therefore, improving the efficiency of Pi use in soil is critical for global crop production.
Plants have evolved multiple morphological, physiological, biochemical and molecular strategies to cope with the low‐Pi environmental stress (Rausch and Bucher, 2002; Raghothama and Karthikeyan, 2005). At the morphological level, the alteration of root architecture, including increased root‐to‐shoot ratio, length and density of root hairs, as well as the denser and shorter lateral roots (Anghinoni and Barber, 1980; Barber and Mackay, 1986; Föhse et al., 1988; Péret et al., 2014; Postma et al., 2014; Strock et al., 2018), are the main morphological variations in plants adapting to Pi‐deficient conditions. In addition, when exposed to low‐Pi conditions, plants will alter the metabolic reactions to scavenge and reserve internal Pi by replacing their membrane phospholipids with amphipathic sulfolipids and galactolipids, illustrating that plants can use inorganic pyrophosphate‐reliant metabolic bypass reactions instead of Pi (Plaxton and Tran, 2011). As also reviewed by Plaxton and Tran (2011), intracellular (vacuolar) and secreted APases, enzymes for hydrolyzing Pi from a broad range of Pi monoesters under optimum acidic pH levels, are upregulated in response to low‐Pi conditions. As a consequence, the expression of many genes involved in stress‐response reactions were induced or repressed under low‐Pi conditions. A long‐distance low‐Pi response pathway has been identified in Arabidopsis (Franco‐Zorrilla et al., 2005; Thibaud et al., 2010). The MYB transcription factor AtPHR1 acts as the primary initiatory element of this signaling pathway by binding to the non‐perfect palindromic sequence of promoters in many low‐Pi‐response genes (Rubio et al., 2001; Bustos et al., 2010). These Pi‐response genes encode transcription factors, protein kinases, phosphorus transporters, ribonucleases, phosphatases, metabolic enzymes, synthetic galactose and sulfon lipases (Rubio et al., 2001; Hammond et al., 2003; Franco‐Zorrilla et al., 2004; Misson et al., 2005; Jain et al., 2007; Fang et al., 2009; Lin et al., 2009).
The intermediates and final products of various metabolic pathways at the tissue level are directly related to genotypes and strongly affect complex quantitative traits (Chan et al., 2010). The total level of metabolites in plants are highly sensitive to local environments and growth periods (Chan et al., 2010; Pichersky and Lewinsohn, 2011), thereby providing information on gene expression patterns and molecular signaling activities. Traditional targeted metabolic profiling is limited to analyzing just a subset of pre‐selected metabolisms, but recent advances in non‐targeted metabolomics profiling strategies, based on either nuclear magnetic resonance or mass spectrometry (MS) analysis, provide a high throughput, and are increasingly used to obtain a global view of metabolites (Luo, 2015). The comprehensive high‐throughput analysis technologies, such as ultra‐high performance liquid chromatography–quadrupole time‐of‐flight mass spectrometry (UPLC‐QTOF‐MS) and gas chromatography–mass spectrometry (GC‐MS), allow us to accurately measure vast metabolite profiles (the metabolome), which in turn facilitates the study of complex trait regulation via the ‘gene–metabolism–phenotype’ research model. In the case of Zea mays (maize), metabolomics has been used for the characterization of responses to abiotic stress and inoculation with nitrogen‐fixing plant‐interacting bacteria (Ganie et al., 2015; Obata et al., 2015; Brusamarello‐Santos et al., 2017), the association of leaf physiology with kernel yield, the complex metabolism of the maize kernel (Wen et al., 2015, 2016; Cañas et al., 2017) and the mechanism for the induction of genotype‐dependent embryonic callus (Ge et al., 2017). Additionally, previous studies on the metabolic variations under low‐Pi conditions have been conducted in Arabidopsis and white lupin (Müller et al., 2015; Pant et al., 2015). Specifically, studies showed that plant secondary metabolites are responding to environmental stresses and are associated with direct or indirect resistance to herbivores (Kessler and Kalske, 2018). The latest research also found that phosphate starvation response genes are coordinated with immune system output (Castrillo et al., 2017). These findings, especially those of Obata et al. (2015), suggest that multiple metabolites are potential markers for abiotic stress‐tolerant breeding as well as for the selection of other phenotypes, as proposed by Jannink et al. (2010).
As one of the world's most widely cultivated crops, maize is critical for global food security, and the rich diversity of inbred lines makes maize an ideal model for genetics research (Gore et al., 2009; Huang and Han, 2012). The genome‐wide association study (GWAS) is a powerful method for analyzing quantitative traits by linking morphological diversity with specific genetic locations. Although numerous diverse traits have been dissected via GWAS (Xiao et al., 2017), only a few associated candidate genes and molecular pathways for low‐Pi stress tolerance have been identified (Li et al., 2011; Xu et al., 2018). Not only can such studies screen large numbers of related genes, they also reveal mechanisms underlying morphological diversity among genotypes (genotype−phenotype relationships). As described, metabolites are closely linked to gene expression patterns, continuing physiological processes and phenotypes. Metabolic reactions are driven by groups of enzymes, and most of the candidate genes involved in the metabolite reactions of maize are homologous to those in plants like Arabidopsis and Oryza sativa (rice). Therefore, a combination of GWAS and metabolomics in maize will facilitate the identification of valuable candidate genes conferring advantageous traits, such as low‐Pi resistance.
In this study, six low‐Pi‐sensitive lines and six low‐Pi‐tolerant lines selected from 338 genotypically diverse inbred lines from the current Southwest China breeding program were used for the metabolite profiling of roots and leaves. The untargeted UPLC‐QTOF‐MS‐based and GC‐MS‐based metabolomics analyses were applied to screen for metabolic markers responsive to low‐Pi conditions and to distinguish low‐Pi‐sensitive genotypes from low‐Pi‐resistant inbred genotypes. With the significant correlation between morphological traits and metabolites, we also conducted GWAS by using the 338 diverse inbred lines with seedling phenotypes, aiming to dig out low‐Pi‐responding candidate genes that are associated with morphological traits and simultaneously involved in metabolic pathways based on the public metabolic pathway database and GWAS results.
Results
Low‐phosphorus tolerance in maize revealed by large‐scale germplasm evaluation
Based on the frequency distribution of the synthetic index (SI) (Zhang et al., 2014b), the tested maize association panel can be classified into three groups (Table S1), with the specific classification criterion as well as density histograms of each biological replication listed in Table 1 and Figure S4, respectively. A total of 80 maize inbred lines showed low tolerance, 19 maize inbred lines showed high tolerance, and 238 maize inbred lines showed moderate tolerance (Table S1). Among the maize germplasms screened, the six most stable or known low‐Pi‐resistant and six low‐Pi‐sensitive maize inbred lines were isolated for metabolomics analysis. In previous studies, the experimental maize inbred lines Pi‐resistant maize inbred line 178 and Pi‐sensitive maize inbred line 9782 were frequently used (Table 2) (Lin et al., 2013; Su et al., 2014; Wu et al., 2016).
Table 1.
Criteria for the classification of the different low‐Pi‐tolerant genotypes
| Taxa | 1st rep. in 2016 | 2nd rep. in 2016 | 3rd rep. in 2016 | 1st rep. in 2015 | 2nd rep. in 2015 |
|---|---|---|---|---|---|
| Range of the SI | 0.698–1.795 | 0.753–2.032 | 0.732–2.512 | 0.667–1.729 | 0.684–2.597 |
| The SI for Pi‐sensitive/Counts | 0.6–1.0/94 | 0.7–1.1/114 | 0.7–1.0/91 | 0.6–1.0/52 | 0.6–1.0/44 |
| The SI for Pi‐moderate/Counts | 1.0–1.2/138 | 1.1–1.4/170 | 1.0–1.2/150 | 1.0–1.3/114 | 1.0–1.6/131 |
| The SI for Pi‐resistant/Counts | 1.2–1.8/30 | 1.4–2.1/34 | 1.2–2.6/62 | 1.3–1.8/37 | 1.6–2.6/26 |
Counts, the number of the genotypes within this corresponding range; Pi, inorganic phosphorus; rep, replication; SI, synthetic index.
Table 2.
Detailed information of 12 maize inbred lines screened as Pi sensitive and Pi resistant for metabolome analysis
| Name | Pedigree | Pi‐starvation resistance | K = 2 (Tropical/Temperate) | K = 3 (Tropical/SS/NSS) | K = 4 (Tropical/Reid/PA/PB) | K = 6 (Tropical/Reid/PA/PB/BSSS/North) |
|---|---|---|---|---|---|---|
| K169R | Derived from American hybrid 3163 | Pi sensitive | Temperate | SS | PA | PA |
| Wa138 | Luda Red bone germplasm | Pi sensitive | Temperate | Mix | Mix | North |
| GCML57 | CIMMYT line | Pi sensitive | Tropical | Tropical | Tropical | Tropical |
| GCML140 | CIMMYT line | Pi sensitive | Tropical | Tropical | Tropical | Tropical |
| 9782 | (48–2 × 5003) space induced mutant × foreign hybrid line | Pi sensitive | Temperate | Mix | Mix | Mix |
| K305 | American hybrid 2021 | Pi sensitive | Temperate | Reid | Reid | North |
| Jiao51 | Guizhou landrace Jiaomaerhuangzao | Pi resistant | Tropical | Tropical | Tropical | Tropical |
| YA3237 | Zheng32 × S37 | Pi resistant | Tropical | Tropical | Mix | Tropical |
| 178 | Derived from Pioneer hybrid 78599 | Pi resistant | Temperate | NSS | PB | PB |
| Ji477 | Mo17 radiation | Pi resistant | Temperate | SS | Reid | Mix |
| 08‐641 | Derived from Pioneer hybrid Y78641 | Pi resistant | Tropical | Mix | Mix | Mix |
| CLWN227 | CIMMYT line | Pi resistant | Tropical | Tropical | Tropical | Tropical |
BSSS, iowa stiff stalk synthetic; NSS, non‐stiff stalk; PA, Group A germplasm derived from modern U.S. hybrids; PB, Group B germplasm derived from modern U.S. hybrids; SS, stiff stalk.
Metabolite profiles revealed by principal component analysis
The metabolomics profiles of six low‐Pi‐sensitive and six low‐Pi‐resistant maize inbred lines were divided into eight experimental groups according to tissues (leaf versus root), lines (Pi‐sensitive versus Pi‐resistant) and treatments (Pi‐deficient versus Pi‐sufficient) (e.g. roots of low‐Pi‐sensitive inbred lines under low‐Pi conditions) (Figure 1). The score plots based on metabolite measurements by GC‐MS (Figure 1a) and UPLC‐QTOF‐MS (Figure 1b) show an obvious separation between Pi‐deficient and Pi‐sufficient groups and roots versus leaves, whereas the data points for some inbred lines overlap, indicating that the effects of treatments and tissues were larger than the inherent metabolomic differences among maize inbred lines. In order to identify metabolites showing significantly different concentrations for each pairwise comparison, partial least squares discriminant analysis (PLS‐DA) and orthogonal PLS‐DA (OPLS‐DA) were applied to eliminate and filter out irrelevant signals and to highlight differences. The number of significantly different metabolites in the different comparisons was listed in Table 3. The metabolites identified were classified into 10 chemical compound categories: amino acids, phosphorylated metabolites, nucleic acid compounds, nitrogenous compounds (N‐compounds), organic acids, sugars, alcohols, aldehydes and secondary metabolites (Table S3).
Figure 1.
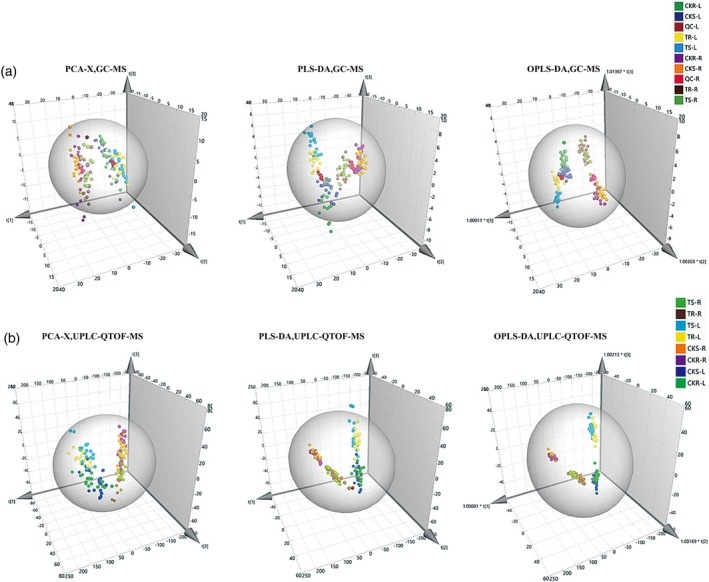
Principle components analysis (PCA), partial least‐squares discriminant analysis (PLS‐DA) and orthogonal PLS‐DA (OPLS‐DA) score plots of the metabolic profiles for the eight experimental groups: (a) gas chromatography–mass spectrometry (GC‐MS); (b) ultra‐high performance liquid chromatography–quadrupole time‐of‐flight mass spectrometry (UPLC‐QTOF‐MS). The treatment groups are as follows: roots of Pi‐sensitive inbred lines under Pi‐deficient conditions (TS‐R), roots of Pi‐resistant inbred lines under Pi‐deficient conditions (TR‐R), roots of Pi‐sensitive inbred lines under Pi‐sufficient conditions (CKS‐R), roots of Pi‐resistant inbred lines under Pi‐sufficient conditions (CKR‐R); leaves of Pi‐sensitive inbred lines under Pi‐deficient conditions (TS‐L), leaves of Pi‐resistant inbred lines under Pi‐deficient conditions (TR‐L), leaves of Pi‐sensitive inbred lines under Pi‐sufficient conditions (CKS‐L) and leaves of Pi‐resistant inbred lines under Pi‐sufficient conditions (CKR‐L).
Table 3.
The number of metabolites showing significantly different pool sizes in the different comparisons
| Groups | The number of significantly different metabolites detected by UPLC mode | The number of significantly different metabolites detected by GC mode | The number of significantly different metabolites detected by both two modes |
|---|---|---|---|
| CKS‐L versus CKR‐L | 28 | 37 | 4 |
| TS‐L versus TR‐L | 41 | 29 | 4 |
| CKS‐R versus CKR‐R | 35 | 43 | 0 |
| TS‐R versus TR‐R | 42 | 19 | 2 |
| TR‐R versus CKR‐R | 66 | 53 | 3 |
| TS‐R versus CKS‐R | 42 | 54 | 4 |
| TS‐L versus CKS‐L | 40 | 47 | 0 |
| TR‐L versus CKR‐L | 55 | 48 | 3 |
| CKS‐L versus CKS‐R | 98 | 72 | 4 |
| CKR‐L versus CKR‐R | 101 | 71 | 6 |
| TS‐L versus TS‐R | 100 | 72 | 6 |
| TR‐L versus TR‐R | 99 | 75 | 8 |
CKR‐L, leaves of Pi‐resistant inbred lines under Pi‐sufficient conditions; CKR‐R, roots of Pi‐resistant inbred lines under Pi‐sufficient conditions; CKS‐L, leaves of Pi‐sensitive inbred lines under Pi‐sufficient conditions; CKS‐R, roots of Pi‐sensitive inbred lines under Pi‐sufficient conditions; TR‐L, leaves of Pi‐resistant inbred lines under Pi‐deficient conditions; TR‐R, roots of Pi‐resistant inbred lines under Pi‐deficient conditions; TS‐L, leaves of Pi‐sensitive inbred lines under Pi‐deficient conditions; TS‐R, roots of Pi‐sensitive inbred lines under Pi‐deficient conditions.
Metabolite variation between Pi‐deficient and Pi‐sufficient conditions
Phosphorus deprivation resulted in the accumulation of nucleic acid compounds, organic acids and some lipid metabolites
The concentrations of all nucleic acids were elevated under low‐Pi conditions, both in Pi‐sensitive and Pi‐resistant genotypes, except for guanine and uridine in Pi‐sensitive roots and adenine in Pi‐sensitive leaves (Figure 2; Table S3). Generally, the magnitudes of organic acid concentration changes between Pi‐deficient and Pi‐sufficient conditions were smaller in leaves than in roots. The organic acids involved in the tricarboxylic acid (TCA) cycle, such as cis‐aconitic acid, malic acid, succinic acid and citric acid, showed large increases in roots under Pi‐deficient conditions (Figure S1). Increased cis‐aconitic acid (13‐fold) was only observed in Pi‐resistant roots, whereas increased citric acid (6.34‐fold) was only observed in Pi‐sensitive roots (Figure S1). Lipid metabolites are also important signaling molecules in plants, and the concentrations of many lipid metabolites such as alpha‐linolenic acid and linoleic acid, as well as their intermediate metabolites, differed significantly under Pi‐deficient conditions (Table S3). More specifically, most nucleic acid compounds, lipid metabolites and organic acids in Pi‐resistant roots were higher than in Pi‐sensitive roots, which suggests that the accumulation of these metabolites plays a positive role in Pi‐resistant roots under Pi‐deficient conditions.
Figure 2.
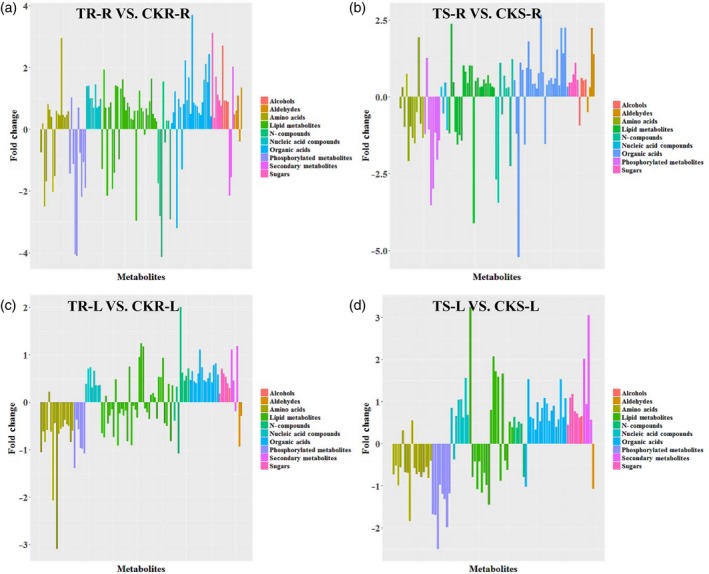
Overview of metabolites with significantly different concentrations between Pi‐deficient and Pi‐sufficient conditions. Histograms indicate metabolites with concentrations differing significantly between Pi‐deficient and Pi‐sufficient conditions. The x coordinates identify the metabolites and the y coordinates indicate the magnitude of the difference expressed as log2 (Pi‐deficient conditions/Pi‐sufficient conditions). Red bars indicate increases in metabolite concentration (logarithm > 0) and cyan bars indicate concentration decreases (logarithm < 0). (a) Metabolite concentration differences in Pi‐resistant roots between Pi‐deficient and Pi‐sufficient conditions (TR‐R versus CKR‐R). (b) Metabolite concentration differences in Pi‐sensitive roots between Pi‐deficient and Pi‐sufficient conditions (TS‐R versus CKS‐R). (c) Metabolite concentration differences in Pi‐resistant leaves between Pi‐deficient and Pi‐sufficient conditions (TR‐L versus CKR‐L). (d) Metabolite concentration differences in Pi‐sensitive leaves between Pi‐deficient and Pi‐sufficient conditions (TS‐L versus CKS‐L).
Phosphorylated metabolites and some amino acids, as well as N‐compounds, decreased under Pi‐deficient conditions
As expected, phosphorus deprivation led to decreased concentrations of most phosphorylated metabolites. Glycerol‐3‐phosphate, methylphosphate and phosphate all decreased in both roots and leaves (Figure S1; Table S3). Phosphorylated metabolites strongly reduced in roots under Pi‐deficient conditions (with a greater than eightfold change), including guanosine monophosphate (GMP) and hexose‐phosphate, whereas GMP remained unchanged in leaves (Figure S1; Table S3). In Pi‐sensitive leaves, hexose‐phosphate also showed a pronounced decrease (6.26‐fold) under Pi‐deficient conditions, whereas there was no significant change in Pi‐resistant leaves (Figure S1; Table S3). Deoxyadenosine mono‐phosphate was accumulated in the roots of both plant genotype groups under Pi‐deficient conditions, whereas Pi‐resistant lines also accumulated thymidine mono‐phosphate (TMP) (Figure S1; Table S3). The decreased phosphorylated metabolites and accumulation of other nucleic acid compounds exactly illustrated that the recycling of phosphorylated metabolites is a strategy that maize uses to maintain cellular phosphorus homeostasis.
Overall, almost all amino acids showing a significant difference in leaves under Pi‐deficient conditions decreased in both Pi‐sensitive and Pi‐resistant inbred lines (Figure 2). In roots, this general decrease in amino acids was only observed in Pi‐sensitive inbred lines, but there were still several other strongly decreased amino acid metabolites in Pi‐resistant roots, such as asparagine (4.08‐fold), aspartic acid (1.68‐fold), gamma‐aminobutyric acid (2.86‐fold), N‐acetylglutamate (5.69‐fold) and prolyl‐histidine (3.23‐fold) (Figure S1; Table S3). Moreover, several N‐compounds varied widely, with the decrease of concentrations of allantoic acid, allantoin and cadaverine in roots under Pi deficiency, whereas concentrations of these N‐compounds remained unchanged in leaves (Figure S1). These amino acids and N‐compounds are directly or indirectly involved in reactions of the TCA cycle to facilitate the synthesis of some organic acids (Figure S1). In this way, maize can enhance phosphorus bioavailability by secreting more organic acids in the roots under low‐Pi conditions.
Some plant immune‐related secondary metabolites respond to Pi deficiency
The metabolic pathways of secondary metabolites showing significant differences in concentrations under Pi‐deficient and Pi‐sufficient conditions were shown in Figure 3, and the detailed information of each pathway is listed in Table S3. Both metabolites of benzoxazinoids and flavonoids were strongly decreased in Pi‐resistant roots under Pi deficiency. In contrast, the flavonoid quercetin‐3,4‐O‐di‐β‐glucopyranoside accumulated (8.28‐fold) in Pi‐sensitive leaves. The flavonoid 3,3,4,5,7‐pentahydroxyflavan was also strongly increased in both Pi‐sensitive and Pi‐resistant leaves. Quercetin 3,7‐dimethyl ether, which remained unchanged in Pi‐sensitive roots, showed a 4.07‐fold increase in Pi‐resistant roots. Based on a previous study (Castrillo et al., 2017), this provided a clue that some connections exist between several plant Pi‐responding genes and immune‐associated secondary metabolites.
Figure 3.
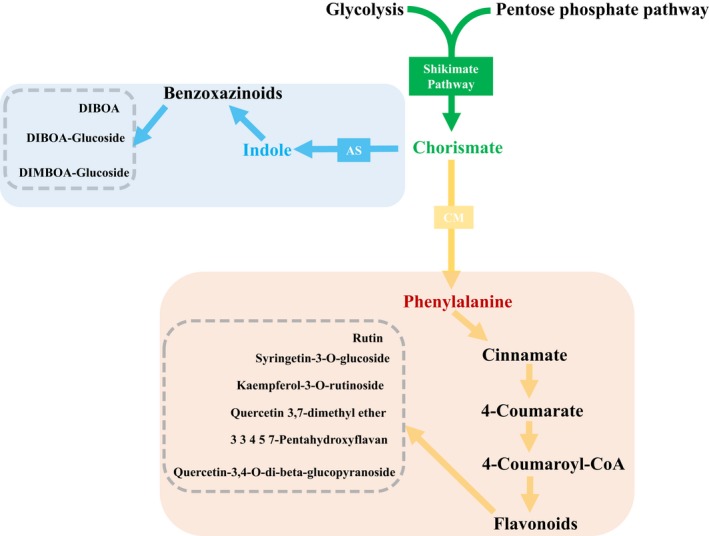
Simplified metabolic pathway of secondary metabolites showing significant differences under Pi‐deficient and Pi‐sufficient conditions. The shikimate‐derived secondary synthesis pathway follows Maeda and Dudareva (2012). AS, anthranilate synthase; CM, chorismate mutase.
Variation between low‐Pi‐sensitive and ‐resistant maize inbred lines revealed by metabolite profiles
Under Pi‐sufficient conditions, most metabolites identified exhibited a reciprocal concentration profile between roots and leaves, except for phosphorylated metabolites, as revealed by comparing low‐Pi‐sensitive with low‐Pi‐resistant maize inbred lines. The general variation ratios of metabolites were higher in Pi‐sensitive roots compared with Pi‐resistant roots and were lower in Pi‐sensitive leaves compared with Pi‐resistant leaves (Figure 4a,b; Table S3), suggesting distinct metabolic pre‐adaptation mechanisms between Pi‐resistant and Pi‐sensitive maize inbred lines. Five metabolite categories, amino acids, phosphorylated metabolites, nucleic acid compounds, lipid metabolites and organic acids, contained more numerous, significantly different metabolites than other metabolite categories. The variation pattern (up‐ or downregulation, as revealed by comparing low‐Pi‐sensitive with low‐Pi‐resistant maize inbred lines) of these metabolite concentrations was consistent within each of the metabolite categories (Figure 4a,b; Table S3). Specifically, most of the amino acids were reduced in Pi‐sensitive leaves but were increased in Pi‐sensitive roots (Figure S2; Table S3). In Pi‐sensitive leaves, the phosphorylated metabolites cAMP, AMP, cGMP and hexose‐phosphate were significantly elevated (more than twofold) compared with Pi‐resistant leaves under Pi‐sufficient conditions (Figure S2). In the roots of Pi‐sensitive maize inbred lines, the concentration of cAMP was 4.6‐fold lower than in Pi‐resistant roots under Pi‐sufficient conditions (Figure S2). Differences in nucleic acid compounds and lipid metabolite concentrations between Pi‐sensitive and Pi‐resistant lines showed lower concentrations in Pi‐sensitive leaves and higher concentrations in Pi‐sensitive roots (Figure 4b; Table S3). Among the organic acids, 2‐ketoisocaproic acid was slightly lower in leaves (1.37‐fold) but was markedly higher (3.52‐fold) in Pi‐sensitive roots compared with Pi‐resistant roots under Pi‐sufficient conditions (Figure S2). The concentrations of 3‐hydroxyisovaleric acid and pipecolinic acid were significantly lower in Pi‐sensitive leaves but remained unchanged in the roots under Pi‐sufficient conditions (Figure S2). In Pi‐sensitive roots, significant differences were only observed for 2‐ketoisocaproic acid (3.521‐fold higher) and 3‐methyl‐2‐ketovaleric acid (3.959‐fold higher), compared with Pi‐resistant roots under Pi‐sufficient conditions (Figure S2; Table S3). In addition, the N‐compound allantoin was lower in both Pi‐sensitive leaves and roots (2.58‐fold and 3.78‐fold, respectively), compared with Pi‐resistant leaves and roots under Pi‐sufficient conditions (Figure S2). Finally, very few secondary metabolites differed significantly between Pi‐sensitive and Pi‐resistant lines under Pi‐sufficient conditions. Overall, most of the nutrient elements absorbed by the plant roots system will be converted into other compounds and transported to the above‐ground tissues, so this differential distribution of metabolites between Pi‐resistant and Pi‐sensitive lines under Pi‐sufficient conditions may also indicate higher nutrient transport efficiency from roots or higher metabolic rates in the leaves of Pi‐resistant lines.
Figure 4.
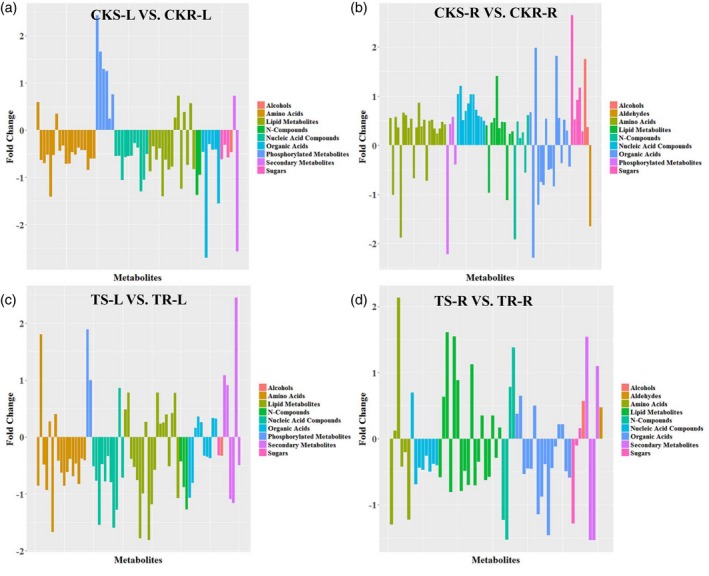
Overview of metabolites differing significantly in concentration between Pi‐resistant and Pi‐sensitive maize inbred lines. Histograms show metabolites with significantly altered concentrations between Pi‐resistant and Pi‐sensitive maize inbred lines. The x coordinates indicate metabolites with significantly different concentrations between groups and the y coordinates represent the magnitude of the difference expressed as log2 (Pi‐resistant maize inbred lines/Pi‐sensitive maize inbred lines). Red bars indicate that the value of the logarithm is >0; cyan bars kindicate that the value of the logarithm is <0. (a) Differences in leaf metabolite concentration between Pi‐sensitive and Pi‐resistant maize inbred lines under Pi‐sufficient conditions (CKS‐L versus CKR‐L). (b) Differences in root metabolite concentrations between Pi‐sensitive and Pi‐resistant maize inbred lines under Pi‐sufficient conditions (CKS‐R versus CKR‐R). (c) Differences in leaf metabolite concentrations between Pi‐resistant and Pi‐sensitive maize inbred lines under Pi‐deficient conditions (TS‐L versus TR‐L). (d) Differences in root metabolite concentrations between Pi‐resistant and Pi‐sensitive maize inbred lines under Pi‐deficient conditions (TS‐R versus TR‐R).
When the same comparisons between Pi‐sensitive and Pi‐resistant maize inbred lines were conducted under Pi‐deficient conditions, most metabolites were lower in Pi‐sensitive roots, and the specific metabolites showing concentration differences between Pi‐sensitive and Pi‐resistant lines differed greatly from the levels found under Pi‐sufficient conditions, especially amino acids and phosphorylated metabolites (Figure 4c,d; Table S3). As can be seen from Figure 4(c,d) and Table S3, the amplitude variation of significantly different metabolites between Pi‐sensitive and Pi‐resistant lines under Pi‐deficient conditions was smaller in leaves than in roots, but fewer amino acids and no phosphorylated metabolites were significantly changed in roots. Plant roots systems are in direct contact with the soil, and so are the first tissues to sense soil stressors such as nutrient deprivation (López‐Arredondo et al., 2014). The low‐Pi response signals are then transmitted to other organs, activating a series of adaptive signaling pathways (López‐Arredondo et al., 2014). The greater disturbance of differential metabolite distribution in roots and smaller disturbance in leaves by low‐Pi conditions reflects the faster metabolic response rate in roots and (or) delayed response rate in leaves.
Great difference in metabolite profiles identified between leaves and roots
Next we examined specific differences in metabolic profiles between leaves and roots in Pi‐sensitive and Pi‐resistant maize inbred lines under Pi‐deficient and Pi‐sufficient conditions (Figure S3; Table S3). The concentrations of most metabolites in leaves were significantly lower than in roots, and the average amplitude of these differences was largest among all inter‐genotype and inter‐condition comparisons. These huge differences in metabolite concentrations were partly the result of inherent differences between tissues rather than Pi‐resistance phenotypes or conditions, however. After filtering the inherent differences, some unique metabolites, such as 2,3‐dihydroxypyridine, were dramatically reduced by Pi deficiency (Table S3). Nevertheless, Pi deficiency still induced changes in unique phosphorylated metabolites. For example, under Pi‐deficient conditions, the concentrations of myo‐inositol‐phosphate in leaves, in both Pi‐resistant and Pi‐sensitive maize inbred line leaves, all decreased approximately twofold compared with roots (Figure S3). In Pi‐resistant maize inbred lines, methylphosphate increased approximately twofold under Pi‐deficient conditions (Figure S3). The unbalanced allocation of these unique metabolites between the roots and leaves under Pi‐deficient conditions may play a role in long‐distance signaling, or be long‐distance signaling components, in maize responses to phosphorus deficiency.
Metabolic markers define the difference between genotypes and treatments
To identify metabolite changes within specific groups, the coefficient of variation (CV) of the identified metabolites was calculated for the each of eight groups: CKR‐L, CKR‐R, CKS‐L, CKS‐R, TR‐L, TR‐R, TS‐L and TS‐R (Figure 5). The general distribution is shown in Figure 5. Relatively stable metabolites were screened according to CV < 15%. To distinguish the stable metabolic differences between the two extreme groups of genotypes and between the two levels of Pi‐treatments, metabolites with significantly different concentrations in these eight pair‐wise comparison groups (TS‐L−TR‐L, CKS‐L−CKR‐L, TS‐R−TR‐R, CKS‐R−CKR‐R, TS‐L−CKS‐L, TR‐L−CKR‐L, TS‐R−CKS‐R and TR‐R−CKR‐R) were incorporated into the screening conditions. These stable metabolic markers included amino acids, organic acids, lipid metabolites, nucleic acid compounds, N‐compounds and sugars (Table S4). Isoleucine and alanine were screened in both Pi‐deficient and Pi‐sufficient conditions in leaves and roots, respectively, and so can be considered as the most stable metabolites for identifying Pi‐sensitive and Pi‐resistant maize inbred lines. The number of stable metabolites, which were able to distinguish the low‐Pi response, was greater than the number of genotype‐distinguishing metabolites, especially in roots. In both Pi‐sensitive and Pi‐resistant leaves, isoleucine and nonanoic acid were stable and differed significantly between Pi‐deficient and Pi‐sufficient conditions. In roots, the nine most stable metabolites differing significantly between Pi‐deficient and Pi‐sufficient conditions were 3‐hydroxypydidine, 2‐hudroxypyridine palmitic acid, stearic acid, oleic acid, heptanoic acid, glycolic acid, glucuronic acid and methionine sulfoxide, as they were detected in both Pi‐sensitive and Pi‐resistant lines (Table S4), and can be regarded as the most stable metabolic markers to distinguish the difference between different Pi‐treatments.
Figure 5.
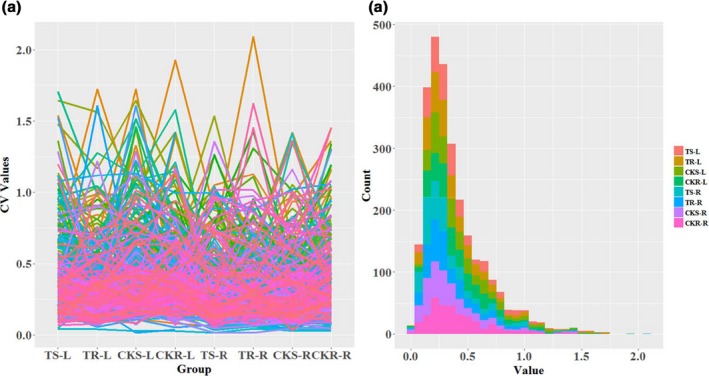
Variation coefficients of metabolites showing significant concentration differences between pairwise matched‐treatment groups. (a) The coefficient of variation (CV) line chart. The x coordinate represents the eight specific matched groups and the y coordinate represents the CV value. Each line represents one metabolite. (b) Histogram of metabolites in different CV ranges. The x coordinate represents the CV value and the y coordinate represents the number of metabolites. Each color represents one specific group. Most CV values are distributed within 0–0.5.
Analyzing the morpho‐physiological phenotypes by employing Pearson correlation tests
Associations between the 22 related traits (including root image analysis data; Table 4) and the identified metabolites (212 detected by UPLC‐QTOF‐MS and 135 detected by GC‐MS; Table S2) were investigated using Pearson correlation tests. Correlations with r ≥ 0.708 (P < 0.01) are shown in Table S5. In total, 159 metabolites were strongly correlated with at least one morphological phenotype under Pi‐deficient conditions and 77 metabolites were strongly correlated with at least one morphological phenotype under Pi‐sufficient conditions (Figure 6; Table S5). Most of the amino acids and lipid metabolites were positively correlated with morphological phenotypes, especially for root‐related traits under Pi‐deficient conditions. The secondary metabolites of flavonoids identified both in roots and leaves all tended to correlate with root‐related traits. Syringetin‐3‐O‐glucoside and kaempferol‐3‐O‐rutinoside, which differed significantly in concentrations between Pi‐deficient and Pi‐sufficient conditions in leaves, were negatively and positively correlated with primary root lengths and number of seminal roots under Pi‐deficient conditions, respectively. Moreover, malvidin‐3‐O‐β‐d‐galactoside identified in roots showed a strong negative correlation with the number of seminal roots (r = −0.885). Phosphorylated metabolites, such as adenosine 3‐monophosphate (3′‐AMP), AMP, TMP and glycerol‐3‐phosphate, identified in the roots were all positively correlated with morphological traits. Among these, 3‐AMP correlated strongly with the number of roots tips (r = 0.856). Under Pi‐sufficient conditions, the number of correlated metabolites decreased sharply, especially amino acids, lipid metabolites and organic acids, whereas phosphorylated metabolites and secondary metabolites still showed strong correlations with morphological phenotypes (Table S5).
Table 4.
Trait statistics collected for all 22 traits under low‐ and normal‐phosphorus conditions
| Traits | Treatment | F value (treatment) | Mean | Standard deviation | Min. | Max. | Range | Skew | Kurtosis | H 2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Average diameter (mm) | LP | 448.8** | 0.54 | 0.06 | 0.4 | 0.75 | 0.35 | 0.53 | 0.43 | 0.214 |
| NP | 0.63 | 0.06 | 0.47 | 0.81 | 0.35 | 0.22 | 0.3 | 0.026 | ||
| Crown root fresh weight (g) | LP | 0.051NS | 0.04 | 0.03 | 0 | 0.2 | 0.2 | 1.74 | 4.08 | 0.275 |
| NP | 0.04 | 0.03 | 0 | 0.19 | 0.19 | 1.11 | 1.35 | 0.178 | ||
| Forks | LP | 296.9** | 1256.63 | 526.38 | 253.67 | 3709 | 3455.33 | 0.98 | 1.4 | 0.570 |
| NP | 707.06 | 258.49 | 117 | 1686.5 | 1569.5 | 0.54 | 0.66 | 0.561 | ||
| Number of crown roots | LP | 3.906* | 1.52 | 0.78 | 0 | 5 | 5 | 0.7 | 0.78 | 0.227 |
| NP | 1.64 | 0.76 | 0 | 4 | 4 | 0.33 | −0.25 | 0.111 | ||
| Number of leaves | LP | 7.548** | 5.25 | 0.43 | 4.18 | 6.62 | 2.44 | 0.33 | 0.11 | 0.370 |
| NP | 5.35 | 0.47 | 3.8 | 6.77 | 2.97 | 0.05 | 0.39 | 0.600 | ||
| Number of seminal roots | LP | 57.08** | 10.06 | 2.24 | 4 | 18 | 14 | 0.18 | 0 | 0.543 |
| NP | 8.82 | 2.03 | 0.5 | 15.17 | 14.67 | 0.09 | 1.12 | 0.442 | ||
| Plant height (cm) | LP | 145.9** | 46.26 | 6.03 | 27.98 | 64.12 | 36.15 | −0.04 | −0.04 | 0.598 |
| NP | 52.32 | 6.99 | 28.88 | 70.83 | 41.96 | −0.35 | 0.44 | 0.542 | ||
| Primary root fresh weight (g) | LP | 8.988** | 0.25 | 0.09 | 0.04 | 0.59 | 0.54 | 0.58 | 0.51 | 0.321 |
| NP | 0.23 | 0.08 | 0.04 | 0.55 | 0.51 | 0.53 | 0.68 | 0.301 | ||
| Primary root length (cm) | LP | 96.59** | 33.06 | 10.19 | 5.93 | 62.85 | 56.92 | −0.02 | −0.03 | 0.503 |
| NP | 25.89 | 8.68 | 6.5 | 45.77 | 39.27 | −0.31 | −0.62 | 0.485 | ||
| Root biomass (g) | LP | 409.5** | 0.08 | 0.03 | 0.02 | 0.2 | 0.18 | 0.65 | 1.04 | 0.479 |
| NP | 0.05 | 0.01 | 0.01 | 0.11 | 0.1 | 0.5 | 1.05 | 0.368 | ||
| Root max. length (cm) | LP | 227.4** | 37.33 | 8.11 | 16.72 | 65.45 | 48.73 | 0.13 | 0.03 | 0.642 |
| NP | 29 | 6.11 | 14.15 | 48.17 | 34.02 | 0.22 | −0.03 | 0.400 | ||
| Root/shoot ratio | LP | 1174** | 0.26 | 0.06 | 0.13 | 0.55 | 0.42 | 0.67 | 1.31 | 0.378 |
| NP | 0.14 | 0.03 | 0.04 | 0.27 | 0.23 | 0.6 | 1.78 | 0.389 | ||
| Root volume (cm3) | LP | 53.17** | 0.81 | 0.23 | 0.2 | 1.51 | 1.31 | 0.36 | 0.07 | 0.399 |
| NP | 0.69 | 0.21 | 0.12 | 1.39 | 1.27 | 0.3 | 0.24 | 0.247 | ||
| Seminal root fresh weight (g) | LP | 4.711* | 0.84 | 0.27 | 0.17 | 1.82 | 1.65 | 0.51 | 0.55 | 0.420 |
| NP | 0.79 | 0.3 | 0.04 | 2.29 | 2.25 | 0.85 | 2.02 | 0.267 | ||
| Shoot biomass (g) | LP | 58.99** | 0.32 | 0.07 | 0.13 | 0.51 | 0.38 | 0.03 | −0.29 | 0.492 |
| NP | 0.37 | 0.1 | 0.11 | 0.69 | 0.58 | 0.12 | 0.38 | 0.385 | ||
| Shoot fresh weight (g) | LP | 222.8** | 3.7 | 0.92 | 1.22 | 7.27 | 6.06 | 0.24 | 0.27 | 0.592 |
| NP | 5.09 | 1.43 | 1.38 | 9.25 | 7.87 | 0.17 | −0.04 | 0.421 | ||
| Surface area (cm2) | LP | 257.8** | 60.14 | 16.1 | 17.48 | 112.99 | 95.51 | 0.43 | 0.14 | 0.510 |
| NP | 42.97 | 11.28 | 9.42 | 74.84 | 65.42 | 0.06 | 0.21 | 0.372 | ||
| Tips | LP | 461.5** | 587.14 | 235.98 | 128.67 | 1558.78 | 1430.11 | 0.71 | 0.39 | 0.396 |
| NP | 288.92 | 97.2 | 65.94 | 689.44 | 623.5 | 0.54 | 0.39 | 0.388 | ||
| Total plant biomass (g) | LP | 6.067* | 0.41 | 0.09 | 0.15 | 0.64 | 0.49 | 0.04 | −0.15 | 0.514 |
| NP | 0.43 | 0.11 | 0.12 | 0.78 | 0.65 | 0.14 | 0.43 | 0.379 | ||
| Total plant fresh weight (g) | LP | 141.8** | 4.83 | 1.16 | 1.6 | 8.92 | 7.32 | 0.12 | 0.14 | 0.580 |
| NP | 6.16 | 1.69 | 1.47 | 11 | 9.53 | 0.17 | 0.07 | 0.375 | ||
| Total root fresh weight (g) | LP | 6.461* | 1.13 | 0.32 | 0.33 | 2.2 | 1.87 | 0.41 | 0.06 | 0.413 |
| NP | 1.06 | 0.35 | 0.23 | 2.61 | 2.38 | 0.63 | 1.19 | 0.259 | ||
| Total root length (cm) | LP | 578.3** | 360.8 | 98.69 | 122.89 | 688.54 | 565.66 | 0.56 | 0.24 | 0.527 |
| NP | 214.71 | 52.3 | 62.96 | 380.84 | 317.88 | 0.21 | 0.63 | 0.463 |
* and ** significant at P < 0.05 and P < 0.01, respectively.
LP, low phosphorus; NP, normal phosphorus; NS, non‐significant; H 2, broad‐sense heritability.
Figure 6.
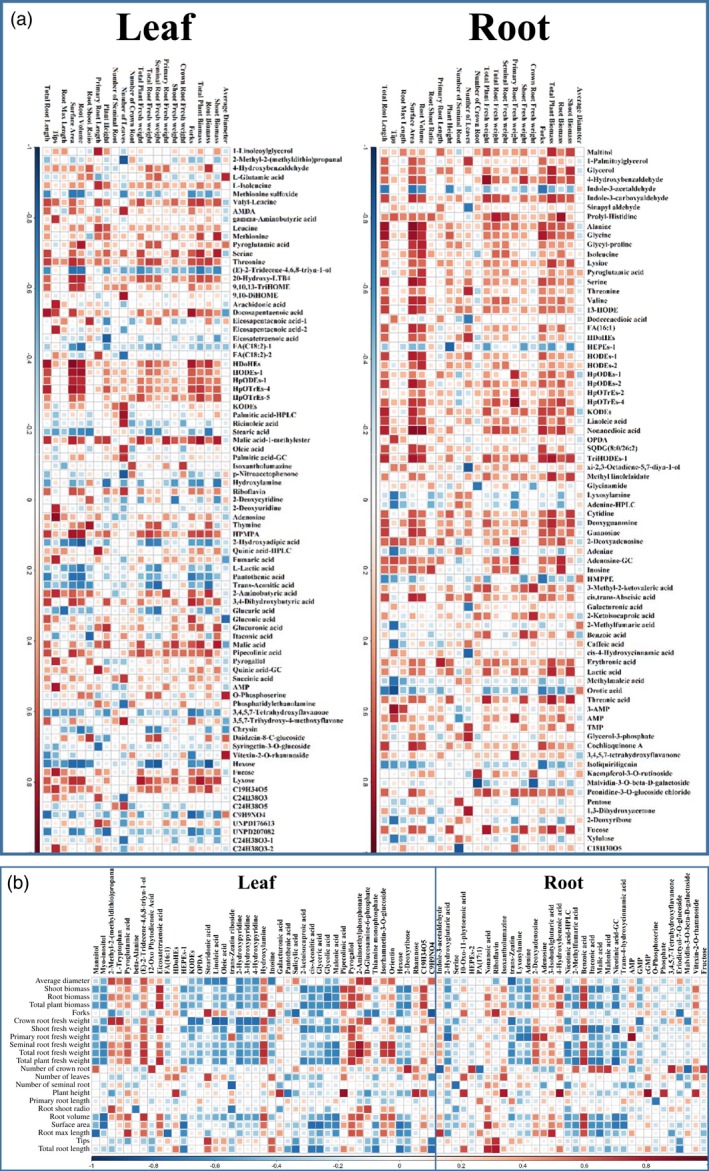
Pearson correlation analysis of metabolites and morphological phenotypes. (a) Heat map of the 159 metabolites identified in maize that were significantly associated with morphological phenotypes (|r| ≥ 0.708) under Pi‐deficient conditions. The color denotes Spearman's rank correlation r between metabolite (listed on top) and trait (listed at left). Red denotes strongly positive and blue denotes strongly negative correlations. (b) Heat map of the 77 identified metabolites in maize that were associated with morphological phenotypes (|r| ≥ 0.708) under Pi‐sufficient conditions. The color denotes Spearman's rank correlation r between metabolite and trait.
Combining genome‐wide association analysis with metabolite‐related genes to screen consensus genes under Pi‐deficient conditions
According to the statistical analysis for the 22 traits under Pi‐deficient and Pi‐sufficient conditions (Table 4), all the traits that we measured differed significantly between the two levels of Pi conditions except crown roots fresh weight. After GWAS with phenotypic data included for 22 traits (in Pi‐deficient and Pi‐sufficient conditions) and the corresponding indices (TPD/TPS) (the traits measured under Pi‐deficient conditions/the traits measured under Pi‐sufficient conditions), a total of 178 single‐nucleotide polymorphism (SNP) markers were found to be significantly associated with a variable number of traits with low‐Pi‐tolerance index values (LPTI = T PD/T PS), using the mixed linear model (MLM) with a threshold of P = 2.16 × 10−5 (Figure 7a; Table S6). In addition, 15 SNP markers were significantly associated with multiple traits under Pi‐sufficient conditions and eight SNPs were significantly associated with multiple traits under Pi‐deficient conditions (Figure 7b,c; Table S6), using the same calculation method and threshold. According to the physical position of these SNP markers and average linkage disequilibrium (LD) decay distance, a total of 1062 candidate genes were identified near these significantly associated SNP markers (Table S7). Among these genes, most were associated with LPTIs, with root biomass, number of seminal roots and root volume showing particularly strong associations. These LPTI‐related candidate genes were mainly distributed on chromosomes 1, 2, 4 and 8, with fewer on chromosomes 6, 7 and 10 (Figure 7).
Figure 7.
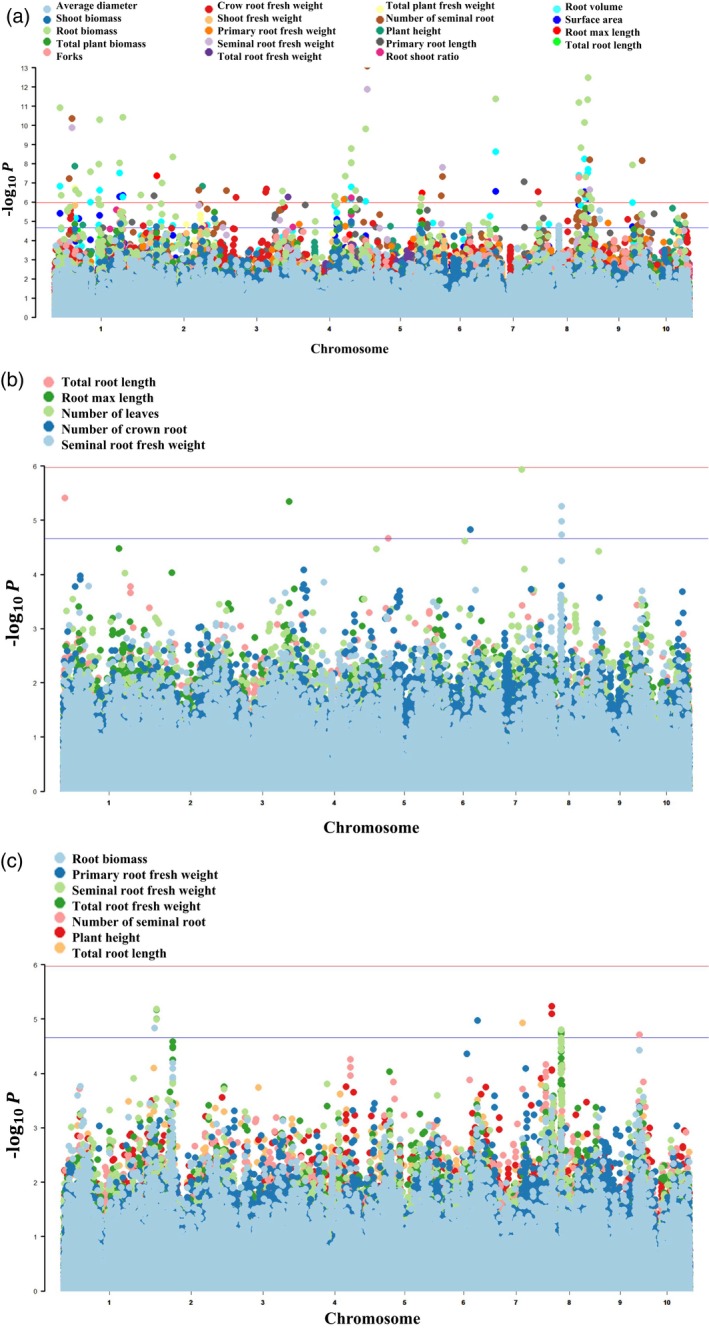
Manhattan plot of genome‐wide association analysis results with significant genetic association [−log(P) > 4.66). A mixed linear model (MLM) was used to fit both Q and K matrixes: (a) 178 single‐nucleotide polymorphism (SNP) markers were significantly associated with low‐Pi‐tolerance indices (LPTI = TPD/TPS) of 19 morphological traits; (b) eight SNP markers were significantly associated with five morphological traits under Pi‐deficient conditions; (c) 15 SNP markers were significantly associated with seven morphological traits under Pi‐sufficient conditions.
More interestingly, some genes were found to be involved in Pi‐starvation‐response mechanisms. Zhang et al. (2014a) reported that the index of the shoot fresh weight‐associated gene GRMZM2G131937 is homologous to AtMYB2, which has been identified as an MYB DNA binding domain that can alter Pi starvation‐induced gene expression, tissue Pi contents and root systems (Baek et al., 2013). Besides, WRKY75, a WRKY transcription factor family member, has been identified as a key regulator of Pi acquisition and root architecture in response to Pi starvation (Devaiah et al., 2007). The genes GRMZM2G015433 and GRMZM2G111354 detected by the index of the number of seminal roots are perfectly matched with WRKY75. Overexpression of serine/threonine receptor‐like kinase PSTOL1 can enhance early root growth and grain yield in Pi‐deficient soil, and the index of plant height‐associated genes GRMZM2G436448 and GRMZM5G897005 were found to be homologous with it (Gamuyao et al., 2012). Based on previous studies (Pacak et al., 2015), the E3 enzyme (ubiquitin‐protein ligase) was found to be involved in low‐Pi responses. Among these candidate genes, seven maize genes encoding ubiquitin‐protein ligase are listed in Table S7. These results suggested that some reliable information can be found in GWAS carried out using phenotype and molecular markers.
Based on Figure S1, we searched the KEGG database (http://www.kegg.jp/) for enzymes that are involved in the pathways of significantly different metabolites between Pi‐deficient and Pi‐sufficient conditions. The corresponding genes are listed in Table S8. By combining the GWAS results with the genes associated with significantly different metabolites (Table S8), five consensus genes were identified (Table 5). In addition, the five consensus genes also showed significantly different expression in publicly available maize Pi‐response gene expression profiles (Table 5), which may act as low‐Pi‐response genes for further utilization in crop improvement.
Table 5.
Detailed information for the five candidate genes with highest consensus revealed by metabolic reactions, genome‐wide association study (GWAS) and expression profile
| Reactions | Enzyme | Genes | Traits | Marker | Chr | Pos. | P | Marker R 2 | Root of inbred line 178 after 6 h under Pi deficiency/log2 (low‐Pi/control) ratio | Root of inbred line 178 after 24 h under Pi deficiency/log2 (low‐Pi/control) ratio | Root of inbred line 178 after 72 h under Pi deficiency/log2 (low‐Pi/control) ratio | Leaf of inbred line B73 under Pi deficiency/log2 (low‐Pi/control) ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose → glucose‐6‐phosphate | Hexokinase | GRMZM2G051806 | Primary root fresh weight‐CK | PZE‐106074800‐FWPR | 6 | 129981085 | 1.07E–5 | 0.06156 | 2.184 | −3.135 | −3.318 | −1.402 |
| Glucose → glucose‐6‐phosphate | Phosphoglucomutase | GRMZM2G025854 | Total root fresh weight‐INDEX | PZE‐103171255 | 3 | 218258052 | 1.65E–6 | 0.07265 | 1.317 | 3.601 | −0.488 | – |
| Glucose 6‐phosphate → fructose‐6‐phosphate | Glucose‐6‐phosphate 1‐epimerase | GRMZM2G039588 | Seminal root fresh weight‐CK/total root fresh weight‐CK | PUT‐163a‐74240952‐3656‐FWSR/SYN31699‐FWSR | 2 | 4176239/4177253 | 6.5E–6/1.01E–5 | 0.06358/0.06532 | −0.421 | −2.401 | −0.440 | 1.430 |
| Aspartic acid → threonine | Threonine synthase | GRMZM2G050570 | Crown root fresh weight‐INDEX | SYN19671 | 3 | 206590086 | 4.91E–6 | 0.07161 | 0.936 | 1.636 | −0.264 | – |
| Uric acid → allantoin | FAD‐dependent urate hydroxylase | GRMZM5G841893 | Root biomass‐INDEX | SYNGENTA15038 | 9 | 152825427 | 1.2E–5 | 0.05973 | – | – | – | – |
Analyzing the expression pattern of the five consensus genes by RT‐qPCR
In order to explore the expression pattern of the five consensus genes during seedling growth, we sampled the leaves and roots of lines 178 and 9782 for eight different Pi treatment periods (Figure 8). In general, the five consensus genes showed more sensitivity to low‐Pi treatment in roots than in leaves, and the greatest changes in expression between low‐Pi and sufficient‐Pi conditions mainly occurred on 3, 7, 9 and 12 days. Specifically, the expression pattern of GRMZM2G025854 in roots was in accordance with leaves in line 178, but presented the opposite trend in line 9782, and was upregulated in leaves and downregulated in roots (Figure 8a and b). GRMZM2G050570, GRMZM2G039588 and GRMZM2G051806 all showed a decreasing or balanced tendency after 14 days of Pi treatment, except GRMZM2G039588 in line 9782 roots at 14 days (Figure 8c–h). GRMZM5G841893 was barely expressed in leaves but showed great changes in roots, especially in line 9782 after 3 days of Pi treatment (Figure 8i and j). The results showed that the transcript abundance of the five consensus genes was induced by Pi deficiency in maize seedlings, but that the expression patterns were different between lines 178 and 9782.
Figure 8.
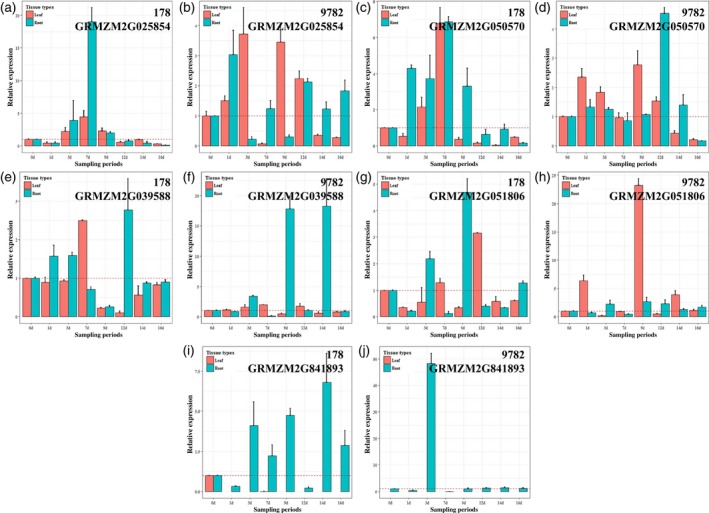
Analysis of the five consensus candidate genes by reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) in lines 178 and 9782. The relative expression level represents the ratio of low‐Pi/sufficient‐Pi. (a–j) Dynamic expression patterns of the five consensus genes in leaf and root tissues after different treatment periods.
Validating the consensus genes in bi‐parental separation populations
After comparing the sequences of consensus genes in line 178 and 9782, the fragments of three genes (GRMZM2G039588, GRMZM5G841893, and GRMZM2G051806) with great differences were amplified in recombinant inbred line (RIL) families (Table S10). The RIL families can be divided into two separating genotypes according to the haplotypes of each gene. The Student's t‐test was applied to the corresponding phenotypes of two separate genotypes, with statistically significant (P < 0.05) traits screened out (Table S10). We found that cob diameter and ear length all significantly differed between the separating genotypes of GRMZM2G039588 under low‐ and sufficient‐Pi conditions, except for cob diameter in Chongzhou base and ear length in Bifengxia farm (Figure 9a and c). Specifically, the cob diameter in genotypes of line 178 was less than in genotypes of line 9782, but this was the opposite case for ear length (Figure 9a and c). The ear diameter in genotypes of line 178 was significantly less than in genotypes of line 9782 in all four locations under low‐phosphorus conditions, and the row number per ear in genotypes of line 178 was significantly less than in genotypes of line 9782 in Chongzhou base and Bifengxia farm under low‐phosphorus conditions (Figure 9b and e). In addition, shoot phosphorus content, efficiency of use and uptake all significantly differed under Pi‐sufficient conditions (Figure 9d). These results suggest that GRMZM2G039588 (glucose‐6‐phosphate‐1‐epimerase encoding gene) not only affects the cob diameter, ear length and even the shoot phosphorus state in maize, but also mediates ear diameter and row number per ear in response to the availability of phosphorus. Some yield components were also significantly different between the separating genotypes of GRMZM5G841893 and GRMZM2G051806 (Figure 10); however, there were no consistent and stable differences, possibly because of the minor effect of these two genes on yield‐correlated traits.
Figure 9.
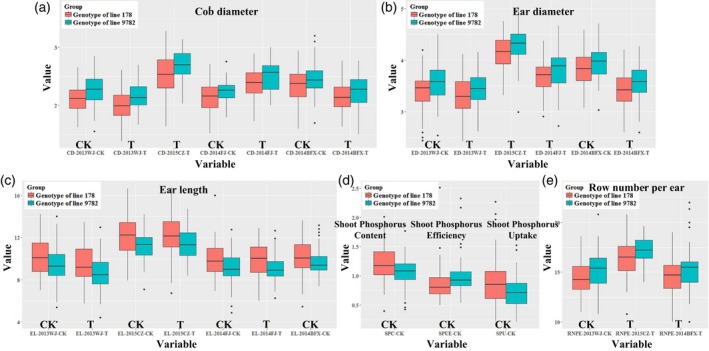
Box plots of the significantly different traits between segregating genotypes of GRMZM2G039588. WJ, Wenjiang farm; CZ, Chongzhou base; FJ, Fenjiang farm; BFX, Bifengxia farm.
Figure 10.
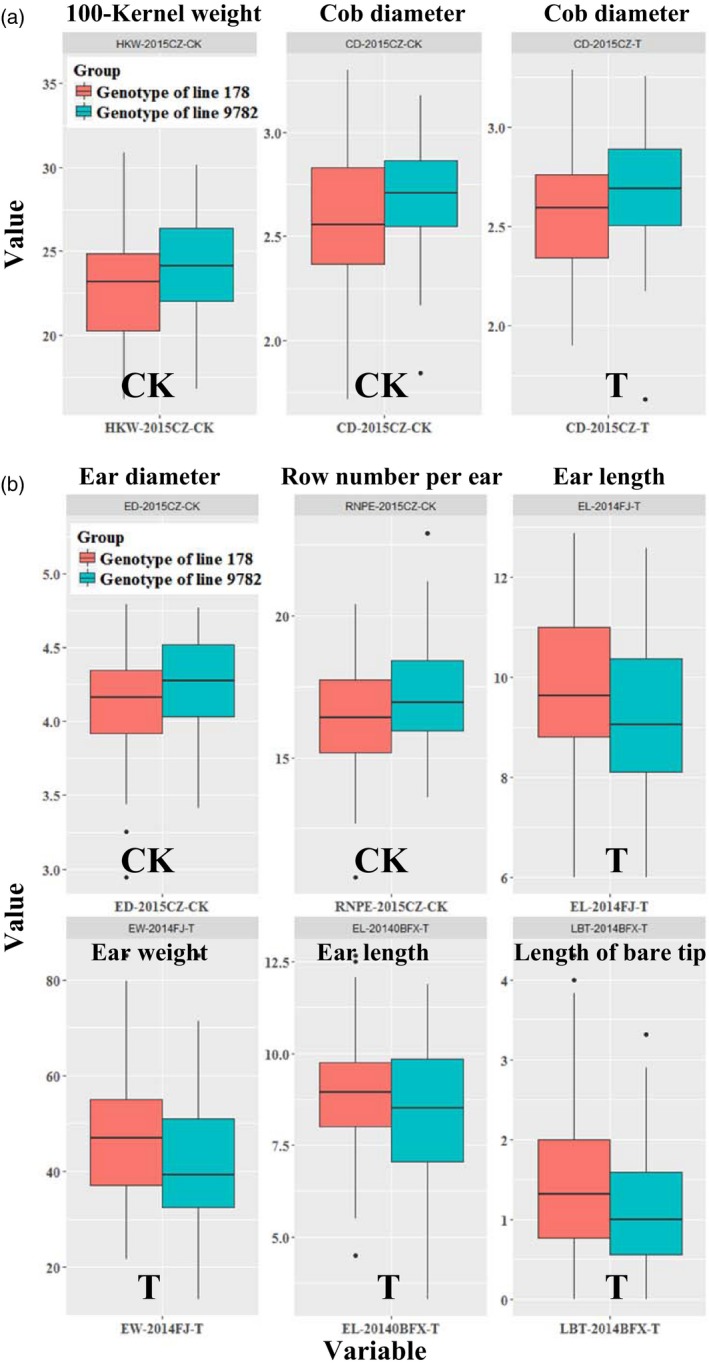
Box plots of the significantly different traits between segregating genotypes. (a) Box plots of the significantly different traits between segregating genotypes of GRMZM5G841893. (b) Box plots of the significantly different traits between segregating genotypes of GRMZM2G051806. WJ, Wenjiang farm; CZ, Chongzhou base; FJ, Fenjiang farm; BFX, Bifengxia farm.
Discussion
Limited metabolic variations between the extreme genotypes
The eight treatment groups defined by tissues (root versus leaf), treatments (Pi‐sufficient versus Pi‐deficient conditions) and genotypes (low‐Pi‐sensitive versus low‐Pi‐tolerant genotypes) exhibited progressively smaller degrees of separation on principle components analysis (PCA) score plots, with the highest overlap for genotypes, indicating that tissue types and culture conditions are the primary drivers of metabolite variation. In contrast to most animals, plants develop continuously throughout their life cycle, resulting in the prolonged differential expression of genes and their corresponding products underlying the development of different organ systems (Schmid et al., 2005). In adverse environments, plants exhibit coordinated molecular, physiological and morphological responses in order to adapt (López‐Arredondo et al., 2014). So, these two factors can lead to wide variations in the concentration of metabolites in plants. As a result of the complexity of these adversity‐response mechanisms, however, many minor changes in expression and signaling confer distinct response phenotypes, such as different levels of low‐Pi sensitivity in maize inbred lines. Therefore, the number of related metabolites showing large‐amplitude differences between genotypes is relatively limited, unlike the dramatic variations between mutants and wild types (Chakraborty et al., 2015; Pant et al., 2015).
Nucleic acid compounds, lipid metabolites, and organic acids play a positive role in Pi‐resistant roots under Pi‐deficient conditions
Under Pi‐deficient conditions, however, most of nucleic acid compounds, lipid metabolites, and organic acids in Pi‐resistant roots were higher than in Pi‐sensitive roots. Nucleic acid compounds are the precursors for the synthesis of nucleic acids and other primary products, such as sucrose, polysaccharides, phospholipids and secondary products, which are of fundamental importance to plant growth and development (Stasolla et al., 2003). Thus, these greater concentrations in Pi‐resistant roots probably reflect compensatory mechanisms to maintain metabolism under Pi deficiency. Lipid metabolites act as intracellular mediators, extracellular signals and even as signals for communication between organisms to modulate various physiological reactions (Weber, 2002; Kachroo and Kachroo, 2009). Higher lipid metabolite content in Pi‐resistant roots suggests faster and stronger low‐phosphorus signal perception for better adaptation to stress conditions. The production and exudation of organic acids from the roots can enhance phosphorus bioavailability in highly Pi‐fixing soils, and indeed organic exudation is a seminal trait of crops with improved phosphate acquisition efficiency (López‐Arredondo et al., 2014). In addition, a series of enzymes involved in glycolysis accumulated in maize roots under Pi depletion (Li et al., 2008), indicating that glycolysis is maintained, and thereby enabling roots to sustain the production of organic acids for exudation. In the current study, most organic acids accumulated under Pi starvation, and the magnitude of accumulation was higher in roots than in leaves. Therefore, the higher organic acid contents of Pi‐resistant roots may serve as a biomarker for high phosphate acquisition efficiency in maize inbred lines.
Scavenging of Pi from internal sources
Plants exhibit two strategies to adapt to Pi deficiency. The first strategy is to enhance the expression of genes encoding metabolic proteins like glycolytic enzymes and mitochondrial electron transporters that do not require phosphorus (López‐Arredondo et al., 2014). The second strategy is to enhance the scavenging of internal phosphorus, including from phospholipids, small phosphorylated metabolites and RNA (López‐Arredondo et al., 2014; Müller et al., 2015). In this investigation, a significant decrease in the levels of almost all phosphorylated metabolites was observed under Pi‐deficient conditions, a result in accord with previous reports in Arabidopsis, maize, and white lupin (Ganie et al., 2015; Müller et al., 2015; Pant et al., 2015), suggesting that the maintenance of cellular phosphorus homeostasis by the recycling of phosphorylated metabolites is a ubiquitous strategy among plants.
The concentrations of phosphorylated nucleic acid compounds, such as GMP, cGMP and cAMP, were strongly reduced and accompanied by marked increases in the concentrations of other nucleic acid compounds under Pi‐deficient conditions. Enzymes like RNase, DNase, 5’‐ and 3’‐nucleotidases, and acid phosphatase can be involved in the hydrolysis of nucleic acids and nucleotides under Pi‐deficient conditions (Shimano and Ashihara, 2006), as changes in enzymatic activity alter nucleotide pool size and composition (Stasolla et al., 2003). Previous studies of Catharanthus roseus cell suspension cultures under Pi‐deficient conditions also demonstrated a marked accumulation of adenosine, adenine, guanosine and guanine (as well as a small increase in ATP), decreased ATP/ADP and UTP/UDP ratios, and repressed activities of purine and pyrimidine nucleotide synthesis and salvage pathways (Ashihara and Ukaji, 1986; Ashihara et al., 1988; Dancer et al., 1990; Shimano and Ashihara, 2006). In addition, the Catharanthus glutamicum enzyme UshA was shown to utilize nucleotides and related compounds as sources of phosphorus under phosphorus starvation (Rittmann et al., 2005). The low‐Pi‐response transcriptional negative regulator bHLH32 can suppress genes that are normally induced by low‐Pi conditions, including phosphoenolpyruvate carboxylase and other genes involved in phosphorus recycling by bypassing ADP‐restricted pyruvate kinase pathways under low‐Pi conditions (Chen et al., 2007). These findings possibly provide explanations for the accumulation of nucleic acid compounds and decreased levels of phosphorylated nucleic acid compounds in maize under Pi‐deficient conditions.
Hexose‐phosphate, glycerol‐3‐phosphate and phosphatidyl glycerol, which are important intermediate metabolites in glycolysis and important constituents of biological membranes in animals, plants and microorganisms, were also strongly decreased under Pi‐deficient conditions. Besides Pi‐dependent glycolysis, there may be a glycolysis bypass pathway for low‐Pi stress adaptation to facilitate glycolytic flux (Plaxton and Tran, 2011), which could explain the strongly reduced hexose‐phosphate level under Pi deficiency in maize. Also, glycerol‐3‐phosphate is the primary precursor of phosphatidyl glycerol, and their levels are closely associated. Moreover, phosphatidyl glycerol primarily exists in thylakoid membranes and is proposed to play an important role in photosynthesis (Hagio et al., 2002). The decreased levels of these two phosphorylated metabolites provides an additional pathway to maintain phosphorus levels but may reduce photosynthetic capacity.
Secondary metabolites including benzoxazinoids and flavonoids involved in plant immune reactions were induced by Pi deficiency
The plant immune system acts in conjunction with the microbiome assembly to cope with phosphorus deficiency. The activity of PHR1, the master regulator of the plant stress response (PSR), was shown to be functionally correlated with plant immune system genes (Castrillo et al., 2017). In addition, several genes related to plant immune system output and enriched in pathways involving the hormone regulators jasmonic acid and/or salicylic acid respond to low phosphorus (Castrillo et al., 2017). Plants also produce a diverse group of organic products for chemical defense and environmental adaptation, known as secondary metabolites (Jonczyk et al., 2008; Weng et al., 2012). Based on these previous studies, we speculate that certain immune‐related secondary metabolites responding to low phosphorus levels may provide clues on plant phosphorus‐response mechanisms. Specifically, upstream genes controlling these secondary metabolite pathways may also contribute to the PSR, such as PHR1. In the current study, 42 secondary metabolites were identified, including flavonoids, flavonoid glycosides, polyphenols, terpenes and benzoxazinoids. Although only a few immune‐related metabolites responded to low phosphorus, those identified warrant further study.
Benzoxazinoid compounds, secondary metabolites involved in anti‐fungal and insect defense, are synthesized by major agricultural crops including maize, Triticum aestivum (wheat) and Secale cereale (rye) (Macías et al., 2005; Jonczyk et al., 2008). Benzoxazinoid compounds are also synthesized in seedlings and stored as glucosides (Jonczyk et al., 2008). DIBOA‐glucoside and DIMBOA‐glucoside are the storage forms of the two important benzoxazinoids, DIBOA and DIMBOA, respectively, and both were strongly decreased in Pi‐resistant maize roots under Pi deficiency. These decreased levels may correspond to increased levels of DIBOA and DIMBOA, although differences between Pi‐deficient and Pi‐sufficient conditions were not detected. Moreover, DIBOA concentration increased in Pi‐sensitive leaves under Pi‐deficient conditions, suggesting that DIBOA and DIMBOA may be upregulated by the replenishment of DIBOA‐glucoside and DIMBOA‐glucoside or changes in the activity of synthetic pathway enzymes. The genes involved in DIBOA‐glucoside and DIMBOA‐glucoside biosynthesis have been well studied and the related enzymatic functions characterized (Jonczyk et al., 2008; Tzin et al., 2015). Recent research has also identified two quantitative trait loci (QTLs) on chromosomes 4 and 6 in maize associated with DIMBOA accumulation, and the QTL on chromosome 6 has an additive effect on the QTL on chromosome 4 (Betsiashvili et al., 2015). Whether these genes or QTLs contribute to the phosphorus response remains unclear.
Quercetin and kaempferol are two flavonoids involved in plant immune reactions (see review by Razzaghi‐Abyaneh and Mahendra, 2013), whereas PHR1, PHO2 and microRNA‐399 are key regulatory components involved in phosphorus signaling (Bari et al., 2006). A previous study showed that variants of quercetin and kaempferol were slightly elevated in phr1 Arabidopsis mutants under Pi‐deficient conditions. In addition, kaempferol variants were also slightly elevated in pho2 mutants and microRNA‐399 overexpressers under Pi‐sufficient conditions (Pant et al., 2015). This result suggests that plant phosphorus signaling components are involved in immune responses under Pi‐deficient conditions.
Metabolic markers for low‐phosphorus response and genotypes distinction
Metabolite levels are the end result of a complex interaction among gene and protein expression levels, metabolic activity and environmental conditions, and so are more reflective of phenotype than mRNA transcript or protein expression profiles alone (Arbona et al., 2013). Recent studies have shown strong associations between metabolite levels and phenotypic characteristics in model plants such as maize, wheat and sorghum (Hamzehzarghani et al., 2005; Obata et al., 2015; Turner et al., 2016). Thus, metabolomic profiles correlated with Pi‐related traits can provide more accurate and intuitive information for high‐Pi‐efficiency germplasm screening. In plants, some morphological traits are modified by Pi‐deficient conditions, which provided important clues for screening the metabolic markers associated with them. The results presented here are preliminary because of the limited number of maize genotypes used, however, and so require further validation. Other metabolic markers were identified based on stability within specific groups but high sensitivity to treatment conditions. These metabolites may be useful markers for distinguishing low‐phosphorus‐response capacity. Alternatively, other metabolites showing greater differences between the inbred lines may help to distinguish Pi‐tolerant and Pi‐sensitive genotypes. Mechanisms underlying plant responses and resistance to low phosphorus levels are extremely complicated. Nonetheless, these markers may prove to be useful adjuncts for screening Pi‐tolerant maize lines or to reveal Pi‐response mechanisms.
Potential maize genes involved in metabolic pathways were revealed in the association populations under Pi‐deficient conditions
The results of Pearson correlation analysis showed strong correlations between morpho‐physiological phenotypes under Pi‐deficient and Pi‐sufficient conditions. As mentioned above, metabolites play an intermediate role in linking genes with phenotypes. So, we combined the GWAS results with the genes involved in the pathways of significantly different metabolites between Pi‐deficient and Pi‐sufficient conditions, aiming to uncover the low‐Pi‐responding candidate genes associated with morphological traits and simultaneously involved in metabolic pathways.
On the other hand, because of our currently limited knowledge on metabolic reactions in maize, the identification of most genes involved in Pi‐related pathways is based on knowledge accrued in other model plants like rice and Arabidopsis. Although homologous genes provide reference information, there is still some uncertainty around the functional activities and regulation in maize. The GWAS is a useful method for analyzing the genetic variation of complex traits, and indeed helped identify candidate low‐phosphorus‐response genes in our study. Functional annotations of these five consensus candidate genes showed that three of them relate to sugar phosphorylation and all three were differentially expressed between Pi‐sufficient and Pi‐deficient conditions. Specifically, GRMZM2G051806 encodes hexokinase, which catalyzes the phosphorylation of glucose, yielding glucose‐6‐phosphate, whereas GRMZM2G025854 encodes phosphoglucomutase, which catalyzes the conversion of glucose‐1‐phosphate to glucose‐6‐phosphate. In addition, GRMZM2G039588 encodes a glucose‐6‐phosphate 1‐epimerase, which catalyzes the conversion of glucose‐6‐phosphate to fructose‐6‐phosphate. The other two genes, GRMZM2G050570 and GRMZM5G841893, encoded threonine synthase and flavin cofactor (FAD)‐dependent urate hydroxylase, respectively. GRMZM2G050570 is involved in the conversion of aspartic acid to threonine and also responded to the low‐phosphorus conditions. GRMZM5G841893 is an RNA‐binding protein involved in the conversion of uric acid to allantoin, but did not differ significantly between the Pi‐sufficient and Pi‐deficient conditions. GRMZM5G841893, GRMZM2G051806 and GRMZM2G039588 were validated in a bi‐parental separation population. Some phenotypes in the segregating genotypes of these three genes are significantly different. In particular, consistent and stable results can be observed in GRMZM2G039588 under low‐Pi conditions, or both, suggesting that this gene has a large effect on these traits and is responding to the availability of Pi, and is worthy of further study.
Experimental procedures
Maize growing conditions and evaluation of Pi responses among inbred lines
A total of 338 genotypically diverse inbred lines from the current Southwest China breeding program (185 temperate and 153 tropical lines; Zhang et al., 2016) were screened to identify low‐Pi‐tolerant inbred lines (Table S1). The seeds were surface sterilized with 1% NaClO and maintained in 0.1 mm CaCl2 solution under darkness with constant aeration for 2 days to initiate germination. Germinated seeds were planted in quartz sand for 7 days and then transplanted onto thermocol sheets fitted on a 45‐L plastic container filled with nutrient solution. Standard Hoagland solution was used for the first 3 days to adapt plants to the hydroponics environment. The nutrient (Hoagland's) solution was then replaced by either a standardized low‐Pi (1.0 μmol L−1) or sufficient‐Pi (1.0 mmol L−1) solution, also containing Ca(NO3)2·4H2O (4 mmol L−1), KNO3 (6 mmol L−1), NH4H2PO4 (1.0 μmol L−1/1.0 mmol L−1), MgSO4·7H2O (2 mmol L−1), Fe·EDTA (100 μmol L−1), H3BO3 (46 μmol L−1), MnCl2·4H2O (0.146 μmol L−1), ZnSO4·7H2O (0.76 μmol L−1), CuSO4·5H2O (0.32 μmol L−1) and (NH4)6Mo7O24·4H2O (0.016 μmol L−1). The solution was replaced every 3 days. During growth, the solution was continuously aerated using aquarium pumps and the pH of the solution was maintained around 5.5. The plants were raised in a glasshouse at Sichuan Agricultural University, China (Ya'an, E 103°010, N 29°540) under the optimum temperature range [30°C for 10 h (day)/20°C for 14 h (night)], with a relative humidity of 60% and a natural day–night cycle. After 15 days of low‐Pi‐stress or control incubation (i.e. in Pi‐sufficient solution), trait differences were carefully evaluated. Manually, evaluated traits were primary root length, root maximum length, plant height, number of seminal roots, number of crown roots, number of leaves, shoot fresh weight, seminal root fresh weight, primary root fresh weight, crown root fresh weight, total root fresh weight, total plant fresh weight, root biomass, shoot biomass, root/shoot ratio, and total plant biomass. Maize root system architecture was classified according to the protocol described by Hochholdinger and Tuberosa (2009). In addition, total root length, root surface area, root average diameter, root volume, number of root tips and number of root forks were determined using the winrhizo pro 2008a image analysis system (Régent Instruments Inc., https://www.regentinstruments.com/assets/products.html). After trait measurements were conducted, the root and shoot were collected separately and heated at 105°C until attaining a constant mass.
Three independent replications were conducted in a completely random design during 2016, and each experiment included at least three seedlings under both Pi‐sufficient and low‐Pi conditions. The mean values of these replicates were used for the final analysis after eliminating possible outliers within lines. The statistical analysis of traits was conducted using r 3.3.3 and the H2 (broad sense heritability) was calculated according to the method of Pace et al. (2015) (Table 4). The indices for low‐Pi‐resistant selection were described by Zhang et al. (2014b). LPTI was used for evaluating the degree of low‐Pi resistance (LPTI = T PD/T PS, where T PD and T PS are measures of a given trait under Pi‐deficient and Pi‐sufficient conditions). The synthetic index (SI), determined by the average LPTI across root length, leaf number and total plant biomass, was also used to evaluate low‐Pi resistance. The LPTI was calculated for each independent replication in 2016. According to the frequency distribution of LPTI, these inbred lines were classified into three classes (Pi‐sensitive, Pi‐moderate and Pi‐resistant) (Table S1). Some materials with poor reproducibility will be assigned to the Pi‐moderate inbred lines. The final classification of these maize inbred lines was also based on the performance of some maize seedlings in 2015 (Table S1).
Metabolomics analysis of low‐Pi response by comparing maize inbred lines of variable resistance
The growth conditions and sampling period of these 12 maize inbred lines were as described for the initial breeding. The second fully expanded leaf and the whole root were sampled, rinsed with distilled water, dried on blotting paper, wrapped in foil, frozen immediately in liquid N2 and finally stored in a −70°C cryogenic refrigerator. The detailed procedures of GC‐MS and UPLC‐QTOF‐MS, including sample processing and measuring, as well as metabolome data analysis, are listed in Appendix S1. A total of three independent replications for the 12 lines that exhibited the most extreme performance were conducted, and each experiment included at least three seedlings under both Pi‐sufficient and Pi‐deficient conditions.
Pearson correlation analysis and variation coefficient
Pearson correlation analysis between morphological traits and metabolite levels was performed in r 3.3.3. Heat maps of highly significant correlations were generated using the corrplot package in r (Wei, 2010). Moreover, there were eight groups in total, as shown in Figure 1, and each group contained six data sets (Table S2). Based on the six data sets of each group, the variation coefficient (CV = standard deviation/average × 100%) was calculated (Table S2). If the variation coefficient is <15%, the data may exhibit a large within‐group variation. A metabolite with CV < 15% was considered stable. All line charts and histograms were generated using ggplot2 in r.
Marker data and genome‐wide association analysis
The 56 110 genomic SNPs of the 338 maize inbred lines (with missing rate <20% and minor allele frequency >0.05) were used for GWAS (Zhang et al., 2016). The phenotypic data included 22 traits (in Pi‐deficient and Pi‐sufficient conditions) and the corresponding indices (T PD/T PS). The standard MLM (Q + K model) was applied by tassel 5.2.30 (Bradbury et al., 2007), in which the population structure (Q) and kinship (K) were estimated as previously described (Zhang et al., 2016). Briefly, population structure was estimated using structure 2.3.4 with K = 2, and relative kinship coefficients were calculated using the Genomic Association and Prediction Integrated Tool (gapit) (Lipka et al., 2012). The calculated P values from SNP marker–trait associations are displayed in Manhattan plots constructed using r. Uniform Bonferroni‐corrected thresholds at α = 1 and α = 0.05 were used as association cut‐offs (Yang et al., 2014). For 46 209 markers, the corresponding Bonferroni‐corrected threshold P values at α = 1 and α = 0.05 were 2.16 × 10−5 and 1.08 × 10−6 (−log P values of 4.66 and 5.97, respectively). The percentage of variation explained by associated SNPs was calculated using analysis of variance (ANOVA). Significantly associated SNPs and candidate co‐located genes were identified from the public maize genome data set, version B73_5b.
Quantitative real‐time PCR (RT‐qPCR)
The RT‐qPCR assay was conducted for the five consensus genes, using FastStart Essential DNA Green Master (Roche, https://www.roche.com) on a Roche Cobas Z480 system. Total RNA from the leaves and roots of lines 178 and 9782 was sampled at 0, 1, 3, 7, 9, 12, 14 and 16 days of Pi treatment, and extracted using TRIzol® Reagent (Life Technologies, now ThermoFisher Scientific, https://www.thermofisher.com). Reverse transcription of cDNA was performed with a PrimeScript™ II 1st Strand cDNA Synthesis Kit. Relative quantitative results were calculated by normalization to the reference gene (GAPDH). At least three independent experiments were performed, and each experiment was performed in technical triplicate. All primers used in the RT‐qPCR assay were designed in beacon designer 7 and are listed in Table S9.
Population construction of recombinant inbred lines and DNA isolation
A segregating population of 262 RIL families derived from the crosses between 178 (Pi‐resistant maize inbred lines) and 9782 (Pi‐sensitive maize inbred lines) was developed by single‐seed descent. Fresh leaf tissue from the parental genotypes and 262 RIL families were taken and stored at −70°C. Genomic DNA isolation was performed according to the CTAB method (Rogers et al., 1989).
Field experiments and measurement of yield and correlated traits
The RIL families along with two parents were evaluated during their natural growing seasons in four low‐phosphorus locations: (i) Wenjiang farm (WJ; plain region; available phosphorous, 22.8 mg kg−1) of Sichuan Agricultural University, Chengdu, Sichuan Province in 2013; (ii) Chongzhou base (CZ; plain region; available phosphorous, 17.2 mg kg−1) of the Maize Research Institute of Sichuan Agricultural University, Chengdu, Sichuan Province in 2015; and (iii) Bifengxia farm and (iv) Fenjiang farm (BFX and FJ; mountainous region; available phosphorous, 20.7 mg kg−1 and 16.8 mg kg−1, respectively) of the Maize Research Institute of Sichuan Agricultural University, Ya'an, Sichuan Province in 2014. All experiments were designed as complete randomized blocks with one‐row plots and two replications per treatment. The rows were 0.8‐m apart and 3‐m long and the plant density was 58 000 plants per ha. Fertilizers were applied before sowing at rates of 150 kg urea, 900 kg calcium superphosphate and 350 kg potassium chloride per hectare for the high‐Pi application, but only 150 kg urea and 350 kg potassium chloride were applied for the low‐Pi application.
At the end of the growing cycle, yield components, including kernel number per row (KNPR), row number per ear (RNPE), length of bare tip (LBT), ear length (EL), ear diameter (ED), cob diameter (CD), 100‐kernel weight (HKW), ear weight (EW) and cob weight (CW), were investigated. In order to maximize the accuracy of the yield and correlated traits, five plants per plot were investigated for all environments.
Analysis of the phosphorus and nitrogen content of RIL families
Two independent river sand culture experiments were conducted in Fenjiang farm with a total of 219 RIL families and both parents. The nutrient (Hoagland's) solution for low‐Pi and sufficient‐Pi conditions used in this culture experiment was the same as for hydroponics. The roots and shoots were harvested separately at the six‐leaf stage and heated at 105°C until reaching a constant mass. A powdered tissue sample of 0.2 g was weighed accurately and dissolved in 5 ml of concentrated sulfuric acid. Phosphorus and nitrogen concentrations were analyzed using an Alliance Integral Futura system. The phosphorus and nitrogen content of the tissue was calculated by constant volume × tissue phosphorus/nitrogen concentration/0.2 g. The tissue phosphorus/nitrogen uptake was calculated by tissue phosphorus/nitrogen content × tissue biomass. The tissues phosphorus/nitrogen use efficiency was calculated by tissue biomass/tissue phosphorus/nitrogen uptake.
Discrimination of the separating genotypes in RIL families by candidate genes
The DNA and promoter (2 kb upstream of the start codon) sequences of the five candidate genes were amplified for both parent lines. The differential fragments between parent lines were then amplified for RIL families. After the processing of multiple sequence alignment, SNPs and InDels were extracted with tassel 2.1 (Bradbury et al., 2007). The RIL families were then divided into the genotypes of line 9782 and line 178 (Table S10). All primers used for genotyping were designed by primer premier 6 software and are listed in Table S9.
Conflict of Interest
The authors declare that they have no competing interests.
Supporting information
Figure S1. Core metabolism overview of metabolic changes between Pi‐sensitive and Pi‐resistant maize inbred lines.
Figure S2. Core metabolism overview of metabolic changes between Pi‐deficient and Pi‐sufficient conditions.
Figure S3. Core metabolism overview of metabolic changes between roots and leaves.
Figure S4. The frequency distribution of synthetic index for tested maize association panel.
Table S1. Detailed information for the 338 diverse maize inbred lines used in this study.
Table S2. Information for all identified metabolites detected by UPLC‐QTOF‐MS and GC‐MS, and the abbreviations for the metabolites.
Table S3. Significantly different metabolites in tissues (root versus leaf), treatment conditions (Pi sufficient or Pi deficient) and genotypes.
Table S4. Metabolic markers for defining the difference between genotypes and between treatments.
Table S5. The Pearson correlation of morpho‐physiological phenotypes with r ≥ 0.7079 (P < 0.01) in Pi‐deficient and Pi‐sufficient conditions.
Table S6. The information of significantly associated SNP markers.
Table S7. Candidate genes near the significantly associated SNP markers.
Table S8. Integrated information of the genes associated with significantly different metabolites and public Pi‐responding gene‐expression profiles.
Table S9. The information of primer sequences used in this study.
Table S10. The segregating genotypes of RIL families and the corresponding phenotypes.
Appendix S1. Detailed procedures for GC‐MS and UPLC‐QTOF‐MS.
Acknowledgements
This work was supported by The National Key Technologies Research and Development Program of China (2016YFD0100707 and 2018YFD0200707 to S.G.), National Natural Science Foundation of China (31471511 and 31361140364 to S.G.), China Agriculture Research System (CARS‐02), and Sichuan Science and Technology Support Project (2016NZ0103 and 2018HH0013 to S.G.). We would also like to thank Shanghai ProfLeader Biotech Co., Ltd for assistance with the metabolomics experiments and data analysis.
References
- Anghinoni, I. and Barber, S.A. (1980) Phosphorus influx and growth characteristics of corn roots as influenced by phosphorus supply. Agron. J. 72, 685–688. [Google Scholar]
- Arbona, V. , Manzi, M. , Ollas, C. and Gómez‐Cadenas, A. (2013) Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 14, 4885–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashihara, H. and Ukaji, T. (1986) Inorganic phosphate absorption and its effect on the adenosine 5’‐triphosphate level in suspension cultured cells of Catharanthus roseus . J. Plant Physiol. 124, 77–85. [Google Scholar]
- Ashihara, H. , Li, X. and Ukaji, T. (1988) Effect of inorganic phosphate on the biosynthesis of purine and pyrimidine nucleotides in suspension‐cultured cells of Catharanthus roseus . Ann. Bot. 61, 225–232. [Google Scholar]
- Baek, D. , Kim, M.C. , Chun, H.J. , et al (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol. 161, 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, S.A. and Mackay, A.D. (1986) Root growth and phosphorus and potassium uptake by two corn genotypes in the field. Fertil. Res. 10, 217–230. [Google Scholar]
- Bari, R. , Pant, B.D. , Stitt, M. and Scheible, W.R. (2006) PHO2, microRNA399, and PHR1 define a phosphate‐signaling pathway in plants. Plant Physiol. 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsiashvili, M. , Ahern, K.R. and Jander, G. (2015) Additive effects of two quantitative trait loci that confer Rhopalosiphum maidis (corn leaf aphid) resistance in maize inbred line Mo17. J. Exp. Bot. 66, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, P.J. , Zhang, Z. , Kroon, D.E. , Casstevens, T.M. , Ramdoss, Y. and Buckler, E.S. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics, 23, 2633–2635. [DOI] [PubMed] [Google Scholar]
- Brusamarello‐Santos, L.C. , Gilard, F. , Brulé, L. , Quilleré, I. , Gourion, B. , Ratet, P. , deMaltempi‐ ‐Souza, E. , Lea, P.J. and Hirel, B. (2017) Metabolic profiling of two maize (Zea mays L.) inbred lines inoculated with the nitrogen fixing plant‐interacting bacteria Herbaspirillum seropedicae and Azospirillum brasilense . PLoS ONE, 12, e0174576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos, R. , Castrillo, G. , Linhares, F. , Puga, M.I. , Rubio, V. , Pérez‐Pérez, J. , Solano, R. , Leyva, A. and Paz‐Ares, J. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6, e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas, R.A. , Yesbergenova‐Cuny, Z. , Simons, M. , et al (2017) Exploiting the genetic diversity of maize using a combined metabolomic, enzyme activity profiling, and metabolic modelling approach to link leaf physiology to kernel yield. Plant Cell, 29, 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo, G. , Teixeira, P.J.P.L. , Paredes, S.H. , et al (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature, 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, N. , Sharma, P. , Kanyuka, K. , Pathak, R.R. , Choudhury, D. , Hooley, R.A. and Raghuram, N. (2015) Transcriptome analysis of Arabidopsis GCR1 mutant reveals its roles in stress, hormones, secondary metabolism and phosphate starvation. PLoS ONE, 10, e0117819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, E.K.F. , Rowe, H.C. , Hansen, B.G. and Kliebenstein, D.J. (2010) The complex genetic architecture of the metabolome. PLoS Genet. 6, e1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.H. , Nimmo, G.A. , Jenkins, G.I. and Nimmo, H.G. (2007) BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem. J. 405, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll, D.L. (1998) The role of phosphorus in the eutrophication of receiving waters: a review. J. Environ. Qual. 27, 261–266. [Google Scholar]
- Dancer, J. , Veith, R. , Feil, R. , Komor, E. and Stitt, M. (1990) Independent changes of inorganic pyrophosphate and the ATP/ADP or UTP/UDP ratios in plant cell suspension cultures. Plant Sci. 66, 59–63. [Google Scholar]
- Devaiah, B.N. , Karthikeyan, A.S. and Raghothama, K.G. (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z. , Shao, C. , Meng, Y. , Wu, P. and Chen, M. (2009) Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci. 176, 170–180. [Google Scholar]
- Föhse, D. , Claassen, N. and Jungk, A. (1988) Phosphorus efficiency of plants: I. External and internal P requirement and P uptake efficiency of different plant species. Plant Soil, 110, 101–109. [Google Scholar]
- Franco‐Zorrilla, J.M. , González, E. , Bustos, R. , Linhares, F. , Leyva, A. and Paz‐Ares, J. (2004) The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 55, 285–293. [DOI] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J.M. , Martín, A.C. , Leyva, A. and Paz‐Ares, J. (2005) Interaction between phosphate‐starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 138, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao, R. , Chin, J.H. , Pariasca‐Tanaka, J. , Pesaresi, P. , Catausan, S. , Dalid, C. , Slamet‐Loedin, I. , Tecson‐Mendoza, E.M. , Wissuwa, M. and Heuer, S. (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature, 488, 535–541. [DOI] [PubMed] [Google Scholar]
- Ganie, A.H. , Ahmad, A. , Pandey, R. , Aref, I.M. , Yousuf, P.Y. , Ahmad, S. and Iqbal, M. (2015) Metabolite profiling of low‐P tolerant and low‐P sensitive maize genotypes under phosphorus starvation and restoration conditions. PLoS ONE, 10, e0129520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, F. , Hu, H. and Huang, X. et al (2017) Metabolomic and proteomic analysis of maize embryonic callus induced from immature embryo. Sci. Rep. 7, 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore, M.A. , Chia, J.M. and Elshire, R.J. et al (2009) A first‐generation haplotype map of maize. Science, 326, 1115–1117. [DOI] [PubMed] [Google Scholar]
- Hagio, M. , Sakurai, I. , Sato, S. , Kato, T. , Tabata, S. and Wada, H. (2002) Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana . Plant Cell Physiol. 43, 1456–1464. [DOI] [PubMed] [Google Scholar]
- Hammond, J.P. , Bennett, M.J. , Bowen, H.C. , Broadley, M.R. , Eastwood, D.C. , May, S.T. and White, P.J. (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132, 578–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzehzarghani, H. , Kushalappa, A.C. , Dion, Y. , Rioux, S. , Comeau, A. , Yaylayan, V. , Marshall, W.D. and Mather, D.E. (2005) Metabolic profiling and factor analysis to discriminate quantitative resistance in wheat cultivars against fusarium head blight. Physiol. Mol. Plant Pathol. 66, 119–133. [Google Scholar]
- Hochholdinger, F. and Tuberosa, R. (2009) Genetic and genomic dissection of maize root development and architecture. Curr. Opin. Plant Biol. 12, 172–177. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Han, B. (2012) A crop of maize variants. Nat. Genet. 44, 734–735. [DOI] [PubMed] [Google Scholar]
- Jain, A. , Vasconcelos, M.J. , Raghothama, K.G. and Sahi, S.V. (2007) Molecular mechanisms of plant adaptation to phosphate deficiency. Plant Breeding Rev. 29, 359–419. [Google Scholar]
- Jannink, J.L. , Lorenz, A.J. and Iwata, H. (2010) Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics, 9, 166–177. [DOI] [PubMed] [Google Scholar]
- Jonczyk, R. , Schmidt, H. , Osterrieder, A. , et al (2008) Elucidation of the final reactions of DIMBOA‐glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol. 146, 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A. and Kachroo, P. (2009) Fatty acid‐derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. [DOI] [PubMed] [Google Scholar]
- Kessler, A. and Kalske, A. (2018) Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 49, 115–138. [Google Scholar]
- Li, K. , Xu, C. , Li, Z. , Zhang, K. , Yang, A. and Zhang, J. (2008) Comparative proteome analyses of phosphorus responses in maize (Zea mays L.) roots of wild‐type and a low‐P‐tolerant mutant reveal root characteristics associated with phosphorus efficiency. Plant J. 55, 927–939. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Gao, Q. , Liu, Y. , He, C. , Zhang, X. and Zhang, J. (2011) Overexpression of transcription factor ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism and root growth. Planta, 233, 1129–1143. [DOI] [PubMed] [Google Scholar]
- Lin, W.Y. , Lin, S.I. and Chiou, T.J. (2009) Molecular regulators of phosphate homeostasis in plants. J. Exp. Bot. 60, 1427. [DOI] [PubMed] [Google Scholar]
- Lin, H. , Gao, J. , Zhang, Z. , et al (2013) Transcriptional responses of maize seedling root to phosphorus starvation. Mol. Biol. Rep. 40, 5359–5379. [DOI] [PubMed] [Google Scholar]
- Lipka, A.E. , Tian, F. , Wang, Q. , Peiffer, J. , Li, M. , Bradbury, P.J. , Gore, M.A. , Buckler, E.S. and Zhang, Z. (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics, 28, 2397–2399. [DOI] [PubMed] [Google Scholar]
- López‐Arredondo, D.L. , Leyva‐González, M.A. , González‐Morales, S.I. , López‐Bucio, J. and Herrera‐Estrella, L. (2014) Phosphate nutrition: improving low‐phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. [DOI] [PubMed] [Google Scholar]
- Luo, J. (2015) Metabolite‐based genome‐wide association studies in plants. Curr. Opin. Plant Biol. 24, 31–38. [DOI] [PubMed] [Google Scholar]
- Macías, F.A. , Oliveros‐Bastidas, A. , Marín, D. , Castellano, D. , Simonet, A.M. and Molinillo, J.M.G. (2005) Degradation studies on benzoxazinoids. Soil degradation dynamics of (2R)‐2‐O‐beta‐D‐glucopyranosyl‐4‐hydroxy‐(2H)‐1,4‐benzoxazin‐3(4H)‐one (DIBOA‐Glc) and its degradation products, phytotoxic allelochemicals from Gramineae. J. Agric. Food Chem. 53, 554–561. [DOI] [PubMed] [Google Scholar]
- Maeda, H. and Dudareva, N. (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. [DOI] [PubMed] [Google Scholar]
- Misson, J. , Raghothama, K.G. , Jain, A. , et al (2005) A genome‐wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl Acad. Sci. USA, 102, 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J. , Gödde, V. , Niehaus, K. and Zörb, C. (2015) Metabolic adaptations of white lupin roots and shoots under phosphorus deficiency. Front. Plant Sci. 6, e0129520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata, T. , Witt, S. , Lisec, J. , Palacios‐Rojas, N. , Florez‐Sarasa, I. , Yousfi, S. , Araus, J.L. , Cairns, J.E. and Fernie, A.R. (2015) Metabolite profiles of maize leaves in drought, heat and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 165, 2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak, A. , Barciszewska‐Pacak, M. , Swida‐Barteczka, A. , Kruszka, K. , Sega, P. , Milanowska, K. , Jakobsen, I. , Jarmolowski, A. and Szweykowska‐Kulinska, Z. (2015) Heat stress affects Pi‐related genes expression and inorganic phosphate deposition/accumulation in barley. Front. Plant Sci. 7, 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace, J. , Gardner, C. , Romay, C. , Ganapathysubramanian, B. and Lübberstedt, T. (2015) Genome‐wide association analysis of seedling root development in maize (Zea mays L.). BMC Genom. 16, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, B.D. , Pant, P. , Erban, A. , Erban, A. , Huhman, D. , Kopka, J. and Scheible, W.R. (2015) Identification of primary and secondary metabolites with phosphorus status‐dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant, Cell Environ. 38, 172–187. [DOI] [PubMed] [Google Scholar]
- Péret, B. , Desnos, T. , Jost, R. , Kanno, S. , Berkowitz, O. and Nussaume, L. (2014) Root architecture responses: in search of phosphate. Plant Physiol. 166, 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky, E. and Lewinsohn, E. (2011) Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62, 549–566. [DOI] [PubMed] [Google Scholar]
- Plaxton, W.C. and Tran, H.T. (2011) Metabolic adaptations of phosphate‐starved plants. Plant Physiol. 156, 1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, J.A. , Dathe, A. and Lynch, J.P. (2014) The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol. 166, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama, K.G. and Karthikeyan, A.S. (2005) Phosphate acquisition. Plant Soil, 274, 37–49. [Google Scholar]
- Rausch, C. and Bucher, M. (2002) Molecular mechanisms of phosphate transport in plants. Planta, 216, 23–37. [DOI] [PubMed] [Google Scholar]
- Razzaghi‐Abyaneh, M. and Mahendra, R. (2013) Antifungal Metabolites From Plants. Berlin, Heidelberg: Springer. [Google Scholar]
- Rittmann, D. , Sorger‐Herrmann, U. and Wendisch, V.F. (2005) Phosphate starvation‐inducible gene ushA encodes a 5’ nucleotidase required for growth of Corynebacterium glutamicum on media with nucleotides as the phosphorus source. Appl. Environ. Microbiol. 71, 4339–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.O. , Rehner, S. , Bledsoe, C. , Mueller, G.J. and Ammirati, J.F. (1989) Extraction of DNA from Basidiomycetes for ribosomal DNA hybridizations. Can. J. Bot. 67, 1235–1243. [Google Scholar]
- Rubio, V. , Linhares, F. , Solano, R. , Martín, A.C. , Iglesias, J. , Leyva, A. and Paz‐Ares, J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman, D.P. , Reid, R.J. and Ayling, S.M. (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol. 116, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M. , Davison, T.S. , Henz, S.R. , Page, U.J. , Demar, M. , Vingron, M. , Schölkopf, B. , Weigel, D. and Lohmann, J.U. (2005) A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Shimano, F. and Ashihara, H. (2006) Effect of long‐term phosphate starvation on the levels and metabolism of purine nucleotides in suspension‐cultured Catharanthus roseus cells. Phytochemistry, 67, 132–141. [DOI] [PubMed] [Google Scholar]
- Smith, V.H. and Schindler, D.W. (2009) Eutrophication science: where do we go from here?. Trends Ecol. Evol. 24, 201–207. [DOI] [PubMed] [Google Scholar]
- Stasolla, C. , Katahira, R. , Thorpe, T.A. and Ashihara, H. (2003) Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 160, 1271–1295. [DOI] [PubMed] [Google Scholar]
- Strock, C.F. , Morrow, L.D.L.R. and Lynch, J. (2018) Reduction in root secondary growth as a strategy for phosphorus acquisition. Plant Physiol. 176, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, S. , Wu, L. , Liu, D. , et al (2014) Genome‐wide expression profile of maize root response to phosphorus deficiency revealed by deep sequencing. J. Integr. Agric. 13, 1216–1229. [Google Scholar]
- Thibaud, M.C. , Arrighi, J.F. , Bayle, V. , Chiarenza, S. , Creff, A. , Bustos, R. , Paz‐Ares, J. , Poirier, Y. and Nussaume, L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 64, 775–789. [DOI] [PubMed] [Google Scholar]
- Turner, M.F. , Heuberger, A.L. , Kirkwood, J.S. , Collins, C.C. , Wolfrum, E.J. , Broeckling, C.D. and Jahn, C.E. (2016) Non‐targeted metabolomics in diverse sorghum breeding lines indicates primary and secondary metabolite profiles are associated with plant biomass accumulation and photosynthesis. Front. Plant Sci. 7, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin, V. , Lindsay, P.L. , Christensen, S.A. , Meihls, L.N. , Blue, L.B. and Jander, G. (2015) Genetic mapping shows intraspecific variation and transgressive segregation for caterpillar‐induced aphid resistance in maize. Mol. Ecol. 24, 5739–5750. [DOI] [PubMed] [Google Scholar]
- Weber, H. (2002) Fatty acid‐derived signals in plants. Trends Plant Sci. 7, 217–224. [DOI] [PubMed] [Google Scholar]
- Wei, T. (2010). Corrplot: Visualization of a Correlation Matrix. R Package Version 0.2–0. Available at: https://cran.r-project.org/web/packages/corrplot/index.html
- Wen, W. , Li, D. , Li, X. , et al (2015) Metabolome‐based genome‐wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 5, 32–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, W. , Liu, H. , Zhou, Y. , et al (2016) Combining quantitative genetics approaches with regulatory network analysis to dissect the complex metabolism of the maize kernel. Plant Physiol. 170, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, J.K. , Philippe, R.N. and Noel, J.P. (2012) The rise of chemodiversity in plants. Science, 336, 1667–1670. [DOI] [PubMed] [Google Scholar]
- Wu, F. , Liu, Z. , Xu, J. , Gao, S. , Lin, H. , Liu, L. , Liu, Y. and Lu, Y. (2016) Molecular evolution and association of natural variation in ZmARF31 with low phosphorus tolerance in maize. Front. Plant Sci. 7, 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Liu, H. , Wu, L. , Warburton, M. and Yan, J. (2017) Genome‐wide association studies in maize: praise and stargaze. Mol. Plant, 10, 359–374. [DOI] [PubMed] [Google Scholar]
- Xu, C. , Zhang, H. , Sun, J. , et al (2018) Genome‐wide association study dissects yield components associated with low‐phosphorus stress tolerance in maize. Theor. Appl. Genet. 131, 1699–1714. [DOI] [PubMed] [Google Scholar]
- Yang, N. , Lu, Y. , Yang, X. , Huang, J. , Zhou, Yang , Ali, F. , Wen, W. , Liu, J. , Li, J. and Yan, J. (2014) Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 10, e1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Uddin, M.S. , Zou, C. , Xie, C. , Xu, Y. and Li, W. (2014a) Meta‐analysis and candidate gene mining of low‐phosphorus tolerance in maize. J. Integr. Plant Biol. 3, 262–270. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Li, J. , Rong, T. , et al (2014b) Large‐scale screening maize germplasm for low‐phosphorus tolerance using multiple selection criteria. Euphytica, 197, 435–446. [Google Scholar]
- Zhang, X. , Zhang, H. , Li, L. , et al (2016) Characterizing the population structure and genetic diversity of maize breeding germplasm in Southwest China using genome‐wide SNP markers. BMC Genom. 17, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Core metabolism overview of metabolic changes between Pi‐sensitive and Pi‐resistant maize inbred lines.
Figure S2. Core metabolism overview of metabolic changes between Pi‐deficient and Pi‐sufficient conditions.
Figure S3. Core metabolism overview of metabolic changes between roots and leaves.
Figure S4. The frequency distribution of synthetic index for tested maize association panel.
Table S1. Detailed information for the 338 diverse maize inbred lines used in this study.
Table S2. Information for all identified metabolites detected by UPLC‐QTOF‐MS and GC‐MS, and the abbreviations for the metabolites.
Table S3. Significantly different metabolites in tissues (root versus leaf), treatment conditions (Pi sufficient or Pi deficient) and genotypes.
Table S4. Metabolic markers for defining the difference between genotypes and between treatments.
Table S5. The Pearson correlation of morpho‐physiological phenotypes with r ≥ 0.7079 (P < 0.01) in Pi‐deficient and Pi‐sufficient conditions.
Table S6. The information of significantly associated SNP markers.
Table S7. Candidate genes near the significantly associated SNP markers.
Table S8. Integrated information of the genes associated with significantly different metabolites and public Pi‐responding gene‐expression profiles.
Table S9. The information of primer sequences used in this study.
Table S10. The segregating genotypes of RIL families and the corresponding phenotypes.
Appendix S1. Detailed procedures for GC‐MS and UPLC‐QTOF‐MS.


