Abstract
The CRISPR/Cas9 system has emerged as a highly versatile platform for inducing targeted genome modifications into mammalian cells and model organisms. However, fully capitalizing on the therapeutic potential for this system requires its safe and efficient delivery into relevant cell types. Adeno-associated virus (AAV) vectors are a clinically promising class of engineered gene delivery vehicles capable of safely infecting a broad range of dividing and non-dividing cells types, while also serving as a highly effective donor template for homology-directed repair. Together, CRISPR/Cas9 and AAV technologies have the potential to accelerate both basic research and clinical applications of genome engineering. Here we describe a step-by-step protocol for AAV-mediated delivery of CRISPR/Cas systems into mammalian cells. Procedures are described for the preparation of high titer virus capable of achieving a diverse range of genetic modifications, including gene knockout and integration.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
RECIPES:
Please see the end of this protocol for recipes, indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
REAGENTS
1 mL sterile syringe
10,000x SYBR Green (Life Technologies)
100 mM stock solutions of dATP, dTTP, dCTP and dGTP
100 μM fluorescein (Bio-Rad)
10x DNA gel loading dye <R>
10x DNase buffer <R>
10X PBS-MK <R>
10x phosphate buffered saline (PBS) <R>
10x Tris-borate-EDTA (TBE) <R>
15% iodixanol solution <R>
1x PBS with 0.001% Tween-20 <R>
1x PBS with 5% Tween-20 <R>
1x PBS-MK with 2M NaCl <R>
21G 1 ½” regular bevel needles
24% iodixanol solution <R>
2x iCycler mix <R>
2x Proteinase K buffer <R>
3 mL sterile syringe
40% iodixanol solution <R>
54% iodixanol solution <R>
AAV lysis buffer <R>
Agarose
Ammonium persulfate
Antibiotic-antimycotic (Anti-Anti; Life Technologies)
Benzonase nuclease (Sigma-Aldrich)
Bromophenol blue
Disposable vacuum filtration systems, 0.22 μm (500 mL; Thermo-Scientific)
DNase I (Roche)
Dulbecco’s modified Eagle’s medium (DMEM)
Escherichia coli TOP10 cells (Life Technologies)
Ethanol
Expand High Fidelity PCR System (Roche)
Fetal bovine serum (Gibco)
Gel extraction kit
Human embryonic kidney (HEK) 293T cells (ATCC CRL-1573)
Jumpstart Taq Antibody (Sigma-Aldrich)
LB plates with 100 μg/mL ampicillin
Linear polyethylenimine MW 25,000 (PEI; Polysciences)
Long blunt ended cannulas
OptiPrep Density Gradient Medium (Sigma-Aldrich)
OptiSeal polyallomer centrifuge tubes (4.9 mL capacity; Beckman Coulter)
pAAV-Cas9-sgRNA plasmid
pHelper (or plasmid that contains adenovirus helper genes; Addgene)
Phenol red
Plasmid midi or maxiprep kit
Plasmid miniprep Kit
Polyacrylamide
Primer oligonucleotides
Proteinase K (NEB)
pXX2 (or plasmid that contains the desired AAV rep and cap genes; Addgene)
qPCR master mix
QuickExtract DNA Extraction Solution (Epicentre)
Restriction endonuclease BsmBI (NEB)
SURVEYOR Mutation Detection Kit (Transgenomics)
SYBR Safe (Life Technologies)
T4 DNA ligase with buffer (NEB)
T4 polynucleotide kinase (NEB)
Taq DNA polymerase supplied with Thermopol buffer (NEB)
TEMED
Trypsin-EDTA (Life Technologies)
Tween-20
Ultra-15, MWCO 100 kDa U Centrifugal Filter Units (Amicon)
Xylene cyanol
Equipment
150 mm × 25 mm tissue culture treated dish
96-well flat bottom tissue culture treated plate
Benchtop centrifuge
Biological safety cabinet / tissue culture hood
Cell scraper
Clamp
Fixed-angle ultracentrifuge rotor
Gel imaging system
Multicolor Real-Time PCR Detection SystemPAGE apparatus
PCR tubes
Preparative ultracentrifuge
Ring stand
Sterile 1.7 mL microcentrifuge tubes
Sterile 15 and 50 mL conical tubes
Thermocycler
Water bath set to 42 °C
METHOD
Cloning
-
1
Retrieve the DNA sequence of the targeted gene using a reference genome database (e.g. http://www.ncbi.nlm.nih.gov/genome/)
-
2
Search for potential Cas9 cleavage sites using an online CRISPR design tool (Cradick et al. 2014; Montague et al. 2014) or DNA sequence viewing software.
-
3
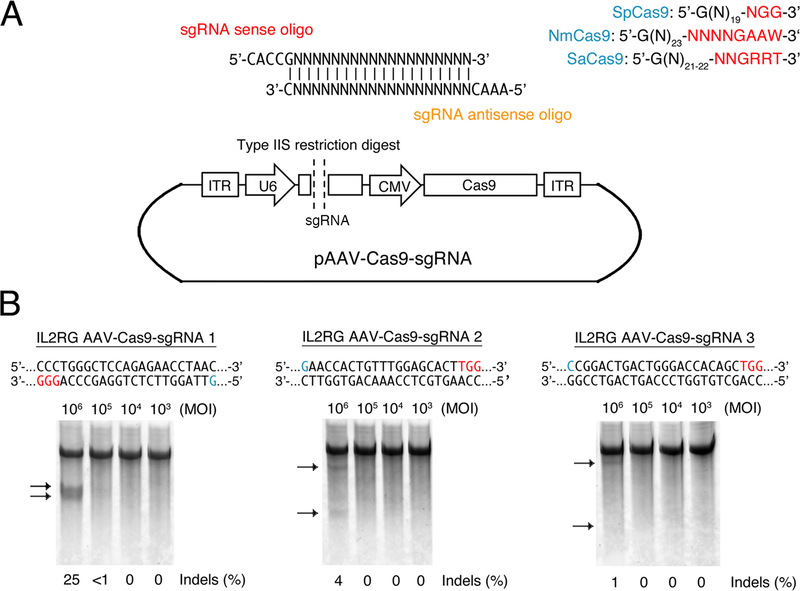
For the Streptococcus pyogenes (SpCas9) protein, search the gene sequence for the motif 5’-G(N)19-NGG-3’, where 5’-NGG-3’ is the PAM (protospacer adjacent motif) recognized by SpCas9 (Jinek et al. 2012; Cong et al. 2013; Mali et al. 2013). Alternatively, for the Staphylococcus aureus (SaCas9) protein (Ran et al. 2015), search the gene sequence for the motif 5’-G(N)21–24-NNGRRT-3’ (R: A or G). Note, a ‘G’ nucleotide is recommended at the 5’ end of the sgRNA transcript for efficient expression from the human U6 promoter.
-
4
Design and order custom sense and antisense oligonucleotides encoding the selected sgRNA protospacer sequences as shown in Fig. 1A.
-
5
Phosphorylate 1 μM of each oligonucleotide with 5 Units (U) of T4 polynucleotide kinase in recommended buffer to 20 μL for 30 min at 37 °C.
-
6
Anneal oligonucleotides by incubation at 95 °C for 5 min followed by fast cooling on ice for 5 min.
-
7
Digest pAAV-Cas9-sgRNA (empty) with the appropriate type IIS restriction enzyme in recommended buffer for 3 hr using 10 U of enzyme per 1 μg of DNA. Visualize DNA by agarose gel electrophoresis using a fluorescent intercalating dye, such as SYBE Safe. All AAV vectors and sequences used here are available upon request.
-
8
Purify linearized pAAV-Cas9-sgRNA (empty) using a gel extraction kit, according to the manufacturer’s instructions.
-
9
Ligate sgRNA duplex DNA into 20–50 ng of linearized pAAV-Cas9-sgRNA (empty) using 1 U of T4 DNA Ligase for 1 hr at room temperature. 6 to 1 insert to vector ratio is recommended for ligation.
-
10
Thaw 100 μL of chemically competent E. coli TOP10 cells on ice and mix gently with ligated pAAV-Cas9-sgRNA.
-
11
Keep cells on ice for 30 min. Heat shock the mixture at 42 °C for 35 sec and recover the cells in 1 mL of lysogeny broth (LB) for at least 30 min at 37 °C with shaking.
-
12
Spread 50–100 μL of bacterial cell culture on a LB agar plate with 100 μg/mL ampicillin and incubate overnight at 37 °C.
-
13
The following day, inoculate 2–4 mL of terrific broth (TB) medium containing 100 μg/mL ampicillin with one colony from the LB agar plate and culture overnight at 37 °C with shaking.
-
14
Purify pAAV-Cas9-sgRNA by miniprep and confirm plasmid identity by DNA sequencing using the primer U6 Seq (5- GAC TGT AAA CAC AAA GAT ATT AGT AC-3’).
-
15
(Recommended) The ability of Cas9 to induce modifications at the genomic target site can be tested in mammalian cell culture by transient transfection of pAAV-Cas9-sgRNA using the procedures described in the “Gene modification” section below.
-
16
Thaw 100 μL of chemically competent E. coli TOP10 cells on ice and mix gently with 100 ng of pAAV-Cas9-sgRNA plasmid. Transform as above.
-
17
The following day, inoculate 50–100 mL of TB medium containing 100 μg/mL ampicillin with one colony and grow overnight at 37 °C with shaking.
-
18
Purify plasmid DNA by midi or maxiprep, according to the manufacturer’s instructions, or polyethylene glycol (PEG) precipitation.
-
19
Store plasmid at −20 °C until transfection.
Figure 1. AAV-mediated delivery of CRISPR/Cas9 for genome editing in mammalian cells.
(A) Vector preparation. Streptococcus pyogenes (Sp), Neisseria meningitidis (Nm) and Staphylococcus aureus (Sa) Cas9 target sites, and sense and antisense oligonucleotides for constructing sgRNA. Sense and antisense sgRNA oligonucleotides encode 5’-CACC-3’ and 5’-AAAC-3’ overhangs, respectively, for insertion into pAAV-Cas9-sgRNA. AAV vectors encoding SpCas9 and NmCas9 should be digested with BsbI, while AAV vectors encoding SaCas9 should be digested with BsaI. (B) Frequency of endogenous interleukin-2 receptor gamma chain (IL2RG) gene modification in HEK293T cells infected with increasing multiplicity of infection (MOI) of AAV-Cas9-sgRNA with three different sgRNAs, as determined by Surveyor nuclease assay. Arrows indicate expected cleavage product. PAM and ‘G’ initiation nucleotide are colored red and blue, respectively.
Adeno-associated virus production
-
20
Maintain HEK293T cells in DMEM containing 10% (v/v) FBS and 1% antibiotic-antimycotic at 37 °C in a fully humidified atmosphere with 5% CO2.
-
21
Seed HEK293T cells onto a 15-cm plate at a density of 2.5–3 × 107 cells/plate.
-
22
At 24 hr after seeding, or once cells are ~90% confluent, add 15 μg pAAV-Cas9-sgRNA, 15 μg pXX2, and 15 μg pHelper plasmids to 4 mL of cell culture medium in a sterile conical tube.
-
23
Add 135 μL PEI (1 μg/μL) and mix immediately by vortexing for 10 sec. The volume of PEI is based on a 3 to 1 ratio of PEI (μg) to total DNA (μg).
-
24
Incubate transfection solution for 10 min at room temperature.
-
25
Add transfection solution dropwise to cells.
-
26
(Optional) Change media 8–12 hr post-transfection to reduce transfection reagent-associated toxicity.
-
27
Harvest virus from cells 48–72 hr post-transfection by manually dissociating cells from plate using a cell scraper and pipetting media and cells into 50 mL conical tubes.
-
28
Pellet cells by centrifugation at 4,000 × g for 5 min at room temperature.
-
29
Remove media and resuspend cells in 2 mL of lysis buffer for each 15 cm plate.
-
30
Freeze/thaw cells three times using a dry ice / ethanol bath and a 37 °C water bath. Cell lysate can be stored at −20 °C after the third freeze.
-
31
Incubate cells with 10 U of Benzonase per 1 mL of cell lysate. Incubate samples at 37 °C for 30 min.
-
32
Centrifuge cell lysate at 10,000 × g for 10 min at room temperature.
-
33
Transfer supernatant to new tubes and store at 4 °C until purification.
Iodixanol density gradient centrifugation
-
34
Pipet 1.2 mL of 15% iodixanol solution into an OptiSeal polyallomer centrifuge tube.
-
35
Underlay the 15% iodixanol solution with 0.7 mL of 25% iodixanol solution containing phenol red using a long blunt ended cannula attached to a 3 mL syringe.
-
36
Underlay the 25% iodixanol solution with 0.6 mL of 40% iodixanol solution
-
37
Underlay the 40% iodixanol solution with 0.6 mL of 54% iodixanol solution containing phenol red.
-
38
Gently pipet 1.8 mL of crude lysate onto each gradient.
-
39
Weigh tubes to ensure they are properly balanced. Use AAV lysis buffer to balance tubes as necessary and seal tubes using provided caps.
-
40
Set ultracentrifuge to slow acceleration and deceleration settings and centrifuge gradients at 174,000 × g for 2 hr at 18 °C.
-
41
Carefully remove centrifuge tubes from the rotor and secure the centrifuge tube in a clamp attached to a ring stand. Remove the cap.
-
42
Carefully puncture the tube at the interface between the 40% and 50% iodixanol solution using a 21G 1 ½” regular bevel needle attached to a 1 mL syringe.
-
43
Collect the bottom 4/5 of the 40% iodixanol solution (bevel up) and the top 1/5 of the 54% iodixanol solution (bevel down) (Zolotukhin et al. 1999). Contaminating proteins from the cell lysate will be present in a band at the interface between the 25% and 40% iodixanol layers. Do not collect the protein band.
-
44
Store collected fractions in a sterile 1.7 mL microcentrifuge or a 15 mL conical tube at 4 °C until further purification.
Buffer exchange and concentration
-
45
Incubate Ultra-15 Centrifugal Filter Unit in PBS with 5% Tween-20 at room temperature for 30 min. After incubation, wash filter once with PBS containing 0.001% Tween-20.
-
46
Dilute collected iodixanol fraction to 15 mL in PBS containing 0.001% Tween-20 and add to Ultra-15 Centrifugal Filter Unit.
-
47
Centrifuge at 4,000 × g for 30 min or until solution has been concentrated to less than 2 mL.
-
48
Add 15 mL of PBS with 0.001% Tween-20 and mix well.
-
49
Repeat steps 3 and 4 three times or until all iodixanol has been eliminated and the viscosity of the solution is similar to PBS with 0.001% Tween-20.
-
50
Concentrate the virus to the desired volume and store at 4 °C.
Viral titering
-
51
Combine 1 μL of virus with 5 μL of 10x DNase buffer, 0.5 μL DNase I, and 43.5 μL of water. Incubate virus sample at 37 °C for 30 min.
-
52
Incubate sample at 75 °C for 10 min to inactivate DNase I.
-
53
Add 60 μL of 2x Proteinase K buffer and 10 μL of Proteinase K to virus sample and incubate at 37 °C for at least 1 hr.
-
54
Incubate at 95 °C for 20 min to inactivate Proteinase K.
-
55
Create tenfold serial dilutions of pAAV-SpCas9-sgRNA plasmid between 0.2 ng/μL and 0.02 pg/μL for standard curve.
-
56
Prepare qPCR master mix.
-
57
Dilute virus sample tenfold for qPCR.
-
58
Combine 15 μL of qPCR master mix with 5 μL of virus sample or linear plasmid for standard curve and run qPCR using the following protocol: 95 °C for 5 min; 40 cycles of 95 °C for 30 sec; 60 °C for 30 sec.; 72 °C for 20 sec.
-
59
Threshold cycle (Ct) values for standards can be plotted against the log10 of the starting plasmid copy number. The Ct value of the virus sample can be correlated to the copy number of the standard from a corresponding Ct value.
Genome modification
-
60
Seed HEK293T cells (or most relevant cell type) onto a 96-well plate at a density of 4 × 104 cells per well.
-
61
Twenty-four hr after seeding, add AAV-SpCas9-sgRNA vector to cells at a genomic multiplicity of infection (MOI) of ~106. Vector can be diluted in serum-containing medium.
-
62
(Recommended) Vary the MOI five- to ten-fold from 106 to 102 to further assess vector activity.
-
63
At 72 hr after infection, wash cells once with PBS and isolate infected cells by trypsin-EDTA digestion.
-
64
Collet cells and centrifuge at 250 × g for 3 min.
-
65
Remove supernatant and resuspend cells by vigorous pipetting with 50 μL of QuickExtract DNA Extraction solution.
-
66
Incubate samples at 65 °C for 15 min, followed by 98 °C for 15 min. Samples can be held at 4 °C and stored indefinitely at −80 °C.
-
67
Amplify the targeted genomic region by PCR using the Expand High Fidelity PCR System. Carry out a 50 μL PCR reaction using 3 μL of template DNA, 5 μL of 10x Expand High Fidelity Buffer with MgCl2, 0.4 μM of each primer, 0.5 μL High Fidelity Taq Polymerase, 5% DMSO and water to 50 μL.
-
68
Verify amplification by agarose gel electrophoresis.
-
69
Denature and re-anneal the PCR amplicon to generate mismatched duplex DNA for the Surveyor nuclease assay using the cycle: 95 °C for min; 95 °C to 85 °C at −2 °C/sec; 85 °C to 25 °C at −0.1 °C/sec. Samples can be held at 4 °C.
-
70
Mix 10 μL of heteroduplex DNA with 1 μL of 0.15 M MgCl2, 1 μL of Surveyor Enhancer S and 1 μL of Surveyor Nuclease S. Incubate reaction at 42 °C for 1 hr.
-
71
Quench reaction with 1 μL of Stop solution and add 2 μL of 10x loading dye to each sample.
-
72
Load the samples on a 10–14% TBE acrylamide gel and run the gel at 140–180 V until the xylene cyanol band is located in the middle/bottom third of the gel.
-
73
Remove the gel and stain with 10 μL of SYBR Safe in 30 mL of 1x TBE for 10 min. Wash the gel at least once with water.
-
74
Visualize the gel using a gel imaging system and measure the density or intensity of each band.
-
75
The percent gene modification can be determined by measuring the fraction of parental band cleaved at the anticipated location as described (Guschin et al. 2010). Representative results are shown in Fig. 1B.
Gene targeting
-
76
Nuclease-induced double-strand breaks (DSBs) can stimulate integration of donor DNA into an endogenous locus via homology-directed pair (HDR) (Rouet et al. 1994; Bibikova et al. 2001). AAV, in particular, can enhance gene targeting >1000-fold compared to plasmid DNA (Russell and Hirata 1998; Jang et al. 2011; Asuri et al. 2012; Gaj et al. 2015).
-
77
To construct AAV vectors for gene targeting, design primers to PCR amplify “left” and “right” homology arms that flank the intended modified sequence. The 5’ (sense) primer for the “left” homology arm should encode an AflII restriction site, and the 3’ (antisense) primer for the “right” homology arm should encode a KpnI restriction site. Note, optimal homology arm length ranges from 0.5 to 1.5 kb. The Cas9 cleavage site should be situated within 50 bps of each homology arm.
-
78
Single-base modifications should be encoded on the 3’ and 5’ ends of the antisense and sense primers of the “left” and “right” homology arms, respectively. A silent restriction site should also be encoded on the donor template for downstream analysis.
-
79
For gene integration, design a second set of primers to amplify the gene of interest (GOI) with 20–30 nucleotides of sequence complementary to the 3’ and 5’ ends of the “left” and “right” homology arms, respectively. Ensure that the Cas9 cleavage site is not present in the donor construct. Due to the carrying capacity of AAV, the donor template should not exceed 4.7 kb.
-
80
PCR amplify the homology arms from genomic DNA, and the GOI from cDNA or plasmid DNA using the primers described above.
-
81
Gel purify the homology arm- and GOI-encoding amplicons using a gel extraction kit, according to the manufacturer’s instructions.
-
82
Fuse the amplicons together by overlap PCR or Gibson assembly (Gibson et al. 2009) to generate the donor template.
-
83
Gel purify the donor template by gel extraction and digest both it and AAV plasmid (e.g. AAV-Cas9-sgRNA) with AflII and KpnI restriction enzymes.
-
84
Ligate the donor template into 20–50 ng of digested AAV plasmid and transform into cells as described above.
-
85
Purify the AAV donor plasmid by mini-prep and confirm plasmid identity by DNA sequencing.
-
86
Construct the accompanying AAV-Cas9-sgRNA vector, and package and purify both it and the AAV donor as described above.
-
87
Infect cells with purified AAV vectors as described above. Cells infected with AAV donor vector containing a selectable marker, such as a puromycin resistance gene or EGFP, can be subjected to antibiotic selection or harvested for FACS at 72 hr after infection. Limiting dilution is recommended for isolation and expansion of clonal cell lines.
-
88
Isolate genomic DNA as described above and PCR amplify the genomic target across the integration junctions using the Expand High Fidelity PCR System. If the donor template contained a silent restriction site, evaluate integration frequency by restriction digestion analysis. Gene modification can be determined by measuring the fraction of parental band cleaved at the anticipated location. DNA sequencing should be used to confirm the presence of gene modifications.
TROUBLESHOOTING
Problem (Step 59):
Low titer
Solution:
Low titer can be due to a number of reasons, including impure plasmid preparation, mutations within the AAV plasmid from modification of inverted terminal repeats (ITRs), toxicity from PEI transfection, off-target cleavage within the AAV vector genome by Cas9, or vector being released from cells. Possible solutions include phenol-chloroform extraction of AAV plasmid to improve vector purity, diagnostic restriction digestion of the AAV vector to establish vector integrity, transfection by calcium phosphate to eliminate the possibility of PEI-induced toxicity, and harvesting cells 48 hr after transfection.
Problem (Step 63):
Low infectivity
Solution:
Due to differences in primary receptor usage and capsid composition, many natural occurring AAV vectors display differential infection abilities both in vitro and in vivo. Therefore, the use of AAV vectors with the intended cell or tissue tropism is essential for efficient gene delivery. Infection by AAV serotypes can be measured using a fluorescent reporter gene, such as EGFP. Engineered or evolved AAV vectors with improved or altered tropism can also be utilized to enhance infection (Kotterman and Schaffer 2014).
Problem (Step 75):
Low genome modification efficiency
Solution:
Poor genome modification could be due to a number of factors, including low levels of Cas9-mediated cleavage at the genomic target site, terminal truncations within the AAV vector genome, and low levels of Cas9 expression. Test the ability of the Cas9-sgRNA complex to induce modifications at the genomic target by transient transfection. Due to the limited carrying capacity of AAV, packaging a single vector containing both a large Cas9 variant (such as SpCas9) and sgRNA could lead to vectors with truncations at the 5’ end of the vector genome (Senis et al. 2014). Use Southern blot analysis to establish whether truncations are present. To minimize vector genome heterogeneity, SpCas9 and sgRNA can be delivered using two separate particles (Swiech et al. 2015). Smaller Cas9 orthologs, such as Neisseria meningitidis (NmCas9) (Hou et al. 2013) and SaCas9 (Ran et al. 2015), can be also used to induce genome modifications from a single AAV particle despite their more restrictive PAM requirements. Finally, confirm that the promoter is providing high levels of expression in the desired cell type by Western blot or with a fluorescent reporter gene, such as EGFP.
Problem (Step 88):
Low integration efficiency
Solution:
No integration could be the result of insufficient homology arm length, low levels of Cas9 activity, or poor infectivity. Test the ability of the Cas9-sgRNA complex to induce modifications at the genomic target by transient transfection. Use an alternative sgRNA if activity is low. In addition, test the ability of the AAV donor vector in combination with Cas9 to mediate HDR by transient transfection. Modify homology arm length in cases where the existing donor template does not trigger integration. Use of small molecules that inhibit non-homologous end joining can also enhance HDR (Chu et al. 2015; Wurst et al. 2015; Yu et al. 2015).
RECIPES
Solutions should be sterilized using a disposable 0.22 μm vacuum filtration system in a tissue culture hood and stored at room temperature.
10x DNA gel loading dye
60% glycerol
0.2 M EDTA
0.5% Bromophenol blue
0.5% Xylene cyanol
pH 8.0
10x DNase buffer
250 mM Tris-HCl (pH 7.4)
100 mM MgCl2
10x PBS
100 mM Na2HPO4 (pH 7.4)
18 mM KH2PO4
1.37 M NaCl
27 mM KCl
10x PBS-MK
100 mM Na2HPO4 (pH 7.4)
18 mM KH2PO4
1.37 M NaCl
10 mM MgCl2
25 mM KCl
1x PBS-MK
Dilute 10x PBS-MK 1:10 in dH2O
1x PBS-MK with 2M NaCl
Dilute 10x PBS-MOK 1:10 in dH2O and add 2M NaCl
1x PBS with 5% Tween-20 (500 mL)
Dilute 10x PBS 1:10 in dH2O
25 mL of Tween-20
1x PBS with 0.001% Tween-20 (500 mL)
Dilute 10x PBS 1:10 in dH2O
5 μL of Tween-20
15% iodixanol solution (17 mL)
4.72 mL of 54% iodixanol solution
8.5 mL of 1x PBS-MK with 2M NaCl
3.78 mL of 1x PBS-MK
24% iodixanol solution (12 mL)
5.56 mL of 54% iodixanol solution
6.44 mL of 1x PBS-MK
60 μL of 0.5% phenol red
40% iodixanol solution (10 mL)
7.41 mL of 54% iodixanol solution
2.59 mL of 1x PBS-MK
54% iodixanol solution (44.4 mL)
40 mL of OptiPrep Density Gradient Medium (60% iodixanol)
4.44 mL of 10x PBS-MK
60 μL of 0.5% phenol red to 12 mL of 54% iodixanol solution
2x iCycler mix (1 mL)
200 μL of 10x PCR buffer without Mg2+
6 mM MgCl2
400 μM dNTPs
2x Proteinase K buffer
10 mM Tris-HCl (pH 8.0)
20 mM Na2EDTA
20 mM NaCl2
AAV lysis buffer
50 mM Tris-HCl (pH 8.0)
150 mM NaCl
qPCR master mix (150 μL)
100 μL of 2x iCycler mix
2 μL of 1 μM sense primer
2 μL of 1 μM antisense primer
2 μL of 1 μM fluorescein
2 μL of 40x SYBR
2 μL of a 1: 1 Taq DNA polymerase / Jumpstart Taq Antibody mix
40 μL dH2O
10x TBE
1 M Tris base
1 M Boric acid
0.02 M Na2EDTA
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grant R01EY022975.
REFERENCES
- Asuri P, Bartel MA, Vazin T, Jang JH, Wong TB, Schaffer DV. 2012. Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol Ther 20: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. 2001. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 21: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B. 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. [DOI] [PubMed]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. 2014. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids 3: e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Epstein BE, Schaffer DV. 2015. Genome engineering using adeno-associated virus: basic and clinical research applications. Mol Ther. [DOI] [PMC free article] [PubMed]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649: 247–256. [DOI] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. 2013. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A 110: 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JH, Koerber JT, Kim JS, Asuri P, Vazin T, Bartel M, Keung A, Kwon I, Park KI, Schaffer DV. 2011. An evolved adeno-associated viral variant enhances gene delivery and gene targeting in neural stem cells. Mol Ther 19: 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman MA, Schaffer DV. 2014. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet 15: 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. 2014. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42: W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS et al. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature. [DOI] [PMC free article] [PubMed]
- Rouet P, Smih F, Jasin M. 1994. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A 91: 6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Hirata RK. 1998. Human gene targeting by viral vectors. Nat Genet 18: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senis E, Fatouros C, Grosse S, Wiedtke E, Niopek D, Mueller AK, Borner K, Grimm D. 2014. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J 9: 1402–1412. [DOI] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. 2015. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst W, Sander S, Rajewsky K, Kuhn R, Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. [DOI] [PMC free article] [PubMed]
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S et al. 2015. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16: 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S, Byrne BJ, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski RJ, Muzyczka N. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 6: 973–985. [DOI] [PubMed] [Google Scholar]