Abstract
Immunoglobulin light-chain amyloidosis is a protein aggregation disease that leads to proteinaceous deposits in a variety of organs in the body and, if untreated, ultimately results in death. The mechanisms by which light-chain aggregation occurs are not well understood. Here we have used solution NMR spectroscopy and biophysical studies to probe immunoglobulin variable domain λV6-57 VL aggregation, a process that appears to drive the degenerative phenotypes in amyloidosis patients. Our results establish that aggregation proceeds via the unfolded state. We identify, through NMR relaxation experiments recorded on the unfolded domain ensemble, a series of hotspots that could be involved in the initial phases of aggregate formation. Mutational analysis of these hotspots reveals that the region that includes K16-R24 is particularly aggregation prone. Notably, this region includes the site of the R24G substitution, a mutation that is found in variable domains of λ light-chain deposits in 25% of patients. The R24G λV6-57 VL domain aggregates more rapidly than would be expected on the basis of thermodynamic stability alone, while substitutions in many of the aggregation-prone regions significantly slow down fibril formation.
Graphical Abstract
INTRODUCTION
Immunoglobulin light-chain (LC) amyloidosis (AL) is among the most diagnosed of the systemic amyloid diseases, with an incidence of approximately 10 new cases each year per million people.1,3 In AL, amyloid fibrils derived from LCs secreted from a clonal plasma cell accumulate in a number of different organs and tissues, with heart and kidneys often being the most severely affected.4–6 Despite recent improvements in survival resulting from treatments that include chemotherapy and autologous stem cell transplantation, more than half of AL patients do not survive 5 years postdiagnosis.7
X-ray crystallography, NMR spectroscopy, and biophysical studies establish that full-length κ or λ LCs are often dimeric, with each monomer comprising single copies of a variable, VL, and a constant, CL, domain with an intervening linker. Each domain, in turn, adopts a β-sheet-rich structure.8 Analysis of amyloid deposits in LC amyloidosis patients reveals that intact full-length LCs can be present,9,10 and studies using full-length constructs in which a λV6-57 domain was fused to a λC3 domain showed that fibril formation of full-length LCs occurs when either of the VL or CL domains is severely destabilized by mutation such that the free energy of domain unfolding is less than 2 kcal/mol.11 More commonly, however, the aggregates comprise just the variable domain derived from aberrant endoproteolysis.9,10,12,13 The importance of the individual VL domains to fibril formation is highlighted by recent cryo-EM structures of patient-derived fibrils, indicating that the core of the cross-beta structure is formed by residues from the VL subunit.14,15 Notably, VL domains are much more aggregation-prone than either individual CL domains, full-length LCs,11,16 or full-length LCs with destabilized C domains.11,17 In addition, proteolysis at a thrombin cleavage site localized to the N-terminus of the CL domain18 would lead to the release of VL domains that could then form aggregates. Despite a large number of studies, the molecular basis by which fibrillation occurs is not well understood. Fundamental questions remain, such as whether proteolysis is required for aggregation16,19,20 or whether fragmentation occurs after the formation of the fibrillar deposits.10 Aggregation studies must include, therefore, LC fragments, such as the highly amyloidogenic VL domain, in addition to focusing on full-length LCs.
Here we investigate the aggregation of λV6-57 VL domains since immunoglobulin LCs of this family have been observed to be preferentially present in AL patients.21 Regions present in the core of the amyloid fibrils formed by this domain and those exposed to solvent are identified through dimethyl sulfoxide-quenched hydrogen/deuterium exchange22 experiments. The relationship between protein thermodynamics and fibrillogenesis is investigated, demonstrating that loss of native structure of the VL domain is required for fibril formation. Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion NMR experiments23,24 show that oligomerization can proceed from the unfolded state of the VL domain via a process that occurs on the millisecond time scale, with regions involved in oligomerization similar to those identified to be aggregation-prone by several prediction algorithms.25 A mutagenesis strategy was used to establish which of the oligomerization hotspots identified by CPMG NMR are important for the conversion of unfolded VL domains into amyloid fibrils, establishing an important role for sequence comprising strand B of the native VL protein. Notably, this region includes the R24G mutation, found in 25% of AL patients from this λ family,21 for which the aggregation rate is shown to be significantly faster than that expected based on thermodynamic stability alone.
RESULTS AND DISCUSSION
Structural Properties of Native and Fibrillar States of the VL Domain.
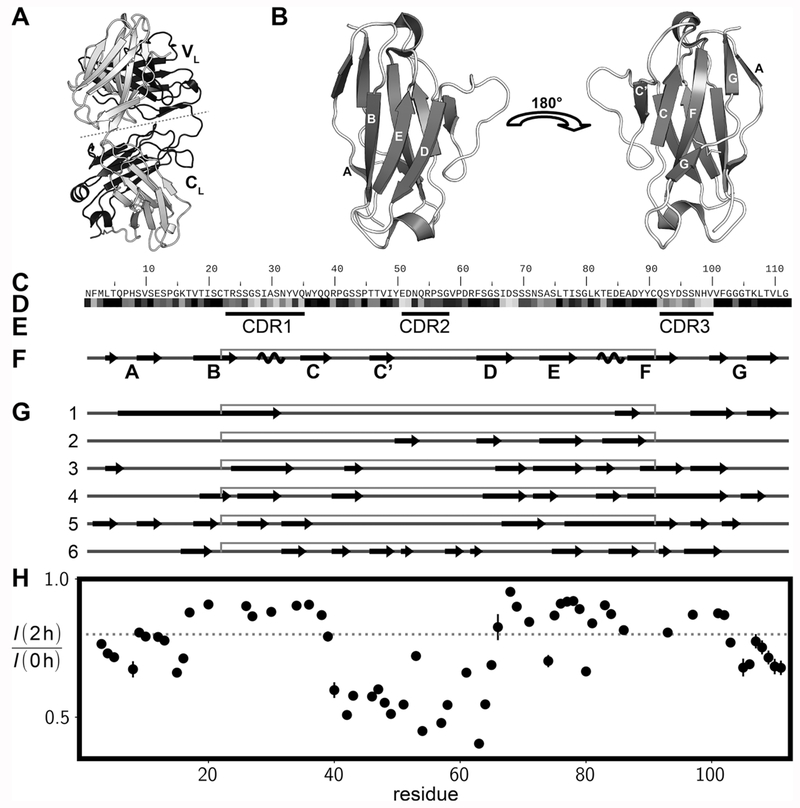
Full length κ and λ LCs often form dimeric structures, as illustrated by the full length Cle8 LC depicted in Figure 1A, isolated from patient urine. Both VL and CL domains have a similar fold, comprising stacked β-sheets stabilized by an intradomain disulfide bridge. The 3D structure of a representative VL domain, the germ line λV6–57 VL protein, 6aJL2-VL,26,27 is shown in Figure 1B along with its amino acid sequence in Figure 1C. Functional LC genes are created by DNA recombination in B cells, followed by introduction of additional sequence variability through somatic hypermutations.28 The sequence conservation for 33 human λ VL domains29 is shown in Figure 1D. Most somatic hypermutation occurs in the complementarity-determining regions (CDRs) that directly contact antigens (Figure 1E) and is not considered in the figure. The prevalent β-secondary structure of the protein in its native state (Figure 1B,F) differs from the cross-β-sheet networks present in fibrillar states that have been elucidated in recent solid-state NMR (ssNMR) and cryo-electron microscopy (cryo-EM) amyloid structures from human κ-type AL-09 VL,30 murine κ-type MAK33 VL,31 human λ-type R24G 6aJL2-VL, and two λ-type VL fibrils extracted from the hearts of AL amyloidosis patients14,15 (Figure 1G). In particular, the three CDRs that are found in loop regions in the native structure, transition, at least partially, to β-strand structure in CDR1 and CDR3 (κ-type AL-09 VL,30 λV6-57 R24G 6aJL2-VL,32 λV1-44, and λV6-57 ex vivo fibril structures,14,15 Figure 1G) and/or in CDR2 (κ-type MAK33 VL31 and λV1-44 ex vivo fibril structure,14 Figure 1G). The identified β-strand regions are not identical in these ssNMR- and cryo-EM-derived structures of amyloid fibrils, indicating that VL domains can form different fibrillar structures, likely depending on the relative distribution of aggregation propensities within aggregation-prone regions. Alternatively, there may be one well-conserved aggregation-driving region that triggers formation of a fibrillar nucleus followed by propagation of cross-β-structure in a pattern that is sequence-specific.
Figure 1.
Structural propensities of VL domains in native and fibrillar states. (A) 3D structure of the CLE full-length λ light-chain dimer (PDB 1LIL8). (B) 3D structure of the 6aJL2-VL domain (PDB 2W0K57). (C) Primary amino acid sequence of 6aJL2-VL. (D) Sequence conservation within the λ immunoglobulin VL domain family using a grayscale from 0 (very variable position, white) to 1 (very conserved, black). (E) Positions of complementarity-determining regions, CDRs, that directly contact antigens. (F) Secondary structure of the native 6aJL2-VL domain. Arrows, coils, and straight lines correspond to β-strands, α-helices, and random coils, respectively. The gray line above the secondary structure diagram connects the Cys residues that form a disulfide. (G) Location of β-strands in VL amyloid fibrils as reported by several studies: 1, AL-09 VL fibrils studied by ssNMR;30 2, MAK33 VL fibrils (ssNMR);31 3 and 4, polymorphs A and B for R24G 6aJL2-VL fibrils characterized by ssNMR;32 5, λV6-57 fibrils extracted from a patient (cryo-EM);15 6, λV1-44 fibrils extracted from a patient (cryo-EM).14 (H) Decrease in intensities of amide correlations as measured in 1H,15N HSQC spectra after 2 h of hydrogen exchange of 6aJL2-VL fibrils (23 °C); the gray dashed line indicates a threshold of 0.8.
In an effort to confirm the regions of 6aJL2-VL forming regular secondary structures in the fibril, as very recently identified by Lecoq et al.32 for the R24G mutant, we used the dimethyl sulfoxide (DMSO)-quenched hydrogen/deuterium exchange method.22,33 In this approach fully amide protonated fibers are exposed to D2O solvent for a defined time to allow exchange of the amides which are not stably hydrogen bonded. After quenching the exchange via lyophilization and subsequent dissolution of the fibril in denaturing DMSO, 1H–15N HSQC spectra are acquired to quantify the intensities of the amide sites, and hence the remaining amide protonation. The drop in intensities of amide correlations in HSQC data sets following 2 h of incubation in deuterated buffer, reporting on solvent exchange in the fiber, is plotted against residue number in Figure 1H. The largest loss of signal is observed for amides in the middle of the protein sequence, between residues R40 and G65. This region, therefore, does not form a rigid, hydrogen-bonded structure in the fiber, but rather is sufficiently flexible so that approximately half of the amide protons exchange with deuterons over a 2 h period. Ratios larger than 0.8 are generally observed for T17-Q39 and S66-F102; amide hydrogens in these regions are stabilized through hydrogen bonding and are therefore not available to exchange with solvent. Values between 0.6 and 0.8 are measured for the N- and C-terminal regions. These amides may be part of the cross-β-sheet network, but undergo some hydrogen exchange through solvent penetration and local transient unfolding at the edge of the β-strands. The overall protection pattern is consistent with the secondary structure recently reported by Lecoq et al. of amyloid fibrils from R24G 6aJL2-VL32 (structures 3 and 4 in Figure 1G), in which residues P45–P60 are mostly unassigned due to structural disorder in the fibrils, and with a recently published cryo-EM structure of another λV6-57 LC15 (structure 5 in Figure 1G), where amino acids extending from Q38-G65 were shown to be structurally disordered. It is also compatible with the quaternary cross-β-sheet structure of the AL-09 VL fiber30 (structure 1 in Figure 1G), showing β-strands in the N- and C-termini.
Loss of Native Structure Is Required for Aggregation.
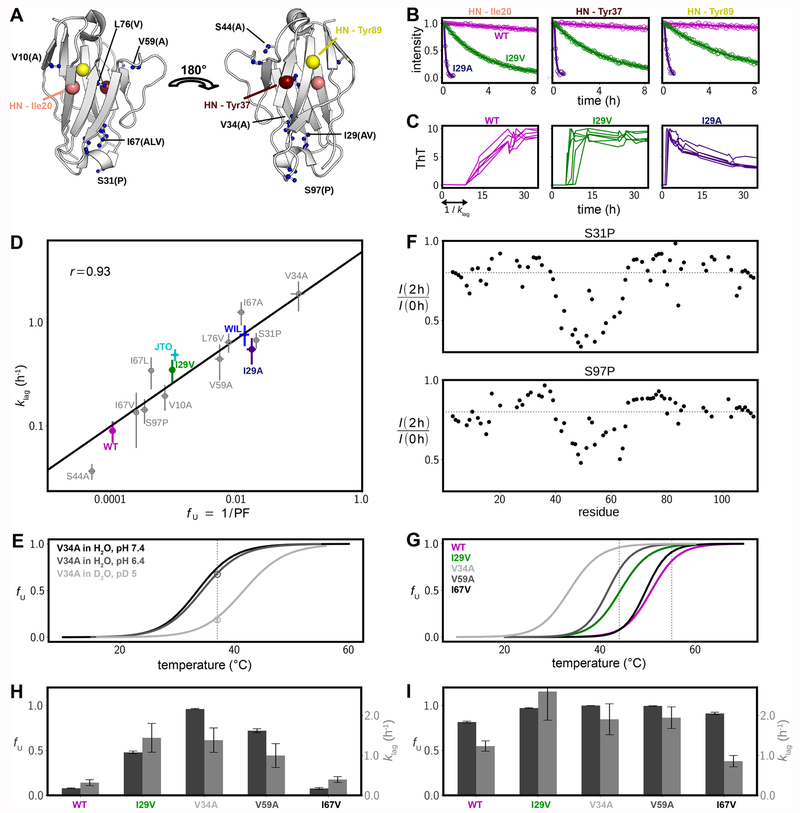
NMR-detected native-state hydrogen exchange experiments were acquired for wild-type 6aJL2-VL under EX2 conditions to probe the stability of single structural elements of the domain.34 The profile of residue-specific protection factors (the ratio of “intrinsic”, residue-specific exchange rates measured on unstructured peptides35 to rates extracted from the protein of interest) shows higher protection in β-strand regions that are most buried in the core of the immunoglobulin fold (maroon in Figure S1), in particular strands B, C, D, E, and F. The largest protection factors, averaging approximately 10 000, directly report on global unfolding of the domain.36 With the aim of generating several protein variants covering a wide range of thermodynamic stability, 12 moderately conservative single-point mutations were introduced into 6aJL2-VL (Figure 2A), and native-state hydrogen exchange experiments were recorded for each mutant. The exchange time-course for three representative amide groups, I20 (β-strand B), Y37 (β-strand C), and Y89 (β-strand F), are shown in Figure 2B for three 6aJL2-VL sequences including germ line26,27 (also referred to as wild-type, WT, in what follows), and VL domains where I29 was substituted by either valine (I29V) or alanine (I29A). Each amide group of residues I20, Y37, and Y89 reports on hydrogen exchange occurring through global unfolding, as these are among the slowest exchanging amides in the domain (Figure S1). Not surprisingly, among these three VL domains the protein with the fastest hydrogen exchange is the mutant with the largest introduced cavity, I29A, followed by I29V, while the amides of the WT protein exchange more slowly.
Figure 2.
Thermodynamic stability and aggregation propensities for a series of 6aJL2-VL mutants. (A) Positions of the mutated residues plotted as blue spheres on the 3D structure of the domain (PDB 2w0k57) using a ball and stick representation. New amino acids introduced by mutation are indicated by the single-letter code in brackets. The three large spheres delineate the amide groups whose solvent hydrogen exchange decay is shown in panel B. (B) Hydrogen/deuterium exchange time profiles for three amide groups of selected 6aJL2-VL variants. (C) Kinetics of aggregation as monitored by ThT fluorescence for three selected 6aJL2-VL variants; six traces are shown in each subpanel. The decrease in fluorescence for the I29A variant at long times is likely because fibrils become larger and less accessible to ThT or due to self-quenching. (D) Correlation between the estimated fraction of fully unfolded protein fU and the inverse duration of the lag phase, klag, shown on a log–log scale. Values of fU were calculated as the reciprocal of the average protection factor for the most protected residues (maroon-colored residues in Figure S1), with uncertainties obtained from the standard deviation of these values. Errors in klag rates are based on at least six repetitions of the ThT aggregation assays. (E) Thermal melting curves for V34A for several conditions, as measured by intrinsic fluorescence (lines) and solution NMR (circles). The NMR-based fU value in D2O at pH 5, 37 °C was obtained from hydrogen exchange measurements, while the fU value at pH 6.4 and in H2O (NMR, 37 °C) was calculated from the volumes of the native- and unfolded-state peaks for G112 in 1H,15N HSQC spectra. The dashed vertical line at 37 °C indicates the temperature at which the NMR experiments were performed. (F) Decrease in amide peak intensities after 2 h of hydrogen exchange of.S31P and S97P 6aJL2-VL fibrils (23 °C); the gray dashed lines indicate a value of 0.8. Similar profiles are obtained for the WT domain (Figure 1H). (G) Thermal melting curves for five 6aJL2-VL variants in H2O at pH 7.4. Dashed vertical lines are positioned at 44 and 55 °C (see panels H and I). (H) Bar plots of fU values (dark gray, left y-axis) and klag (pale gray, right y-axis) at 44 °C. (I) As in panel H, but at 55 °C.
The 12 mutants and the WT protein were assessed with regard to their kinetics of aggregation. The thioflavin T (ThT)37 fluorescence-based aggregation time courses are shown in Figure 2C for the three representative 6aJL2-VL variants considered above, WT, I29V, and I29A. A correlation is observed between hydrogen exchange rates and the kinetics of aggregation: the variant with the fastest hydrogen exchange, I29A, also displays the fastest ThT-based aggregation kinetics, while the WT 6aJL2-VL domain exhibits the slowest hydrogen exchange and aggregation rates. The correlation noted for WT, I29V, and I29A is observed for all of the 6aJL2-VL variants and for two additional λV6-57 VL proteins derived from patients, JTO and WIL,38 as illustrated in Figure 2D. Here the lag rate, klag (y-axis), defined as the inverse of the duration of the lag phase (Figure 2C), is correlated with the fraction of globally unfolded protein at 37 °C, pH 6.4, fU (x-axis), calculated as the inverse of the average protection factor for the most protected amides. Values of fU were obtained assuming EX2 exchange kinetics. This was verified for the least stable mutant, V34A, where the fU value at 37 °C measured by thermal melting as probed by intrinsic fluorescence, 0.21 ± 0.02, was found to be in excellent agreement with fU from hydrogen exchange, 0.19 ± 0.03 (Figure 2E).
To establish whether the mutant proteins form similar fibril structures, we repeated the dimethyl sulfoxide-quenched hydrogen/deuterium exchange experiments discussed above for a pair of mutants, S31P and S97P, where the Pro mutations might be expected to significantly perturb the fibril structures, as proline residues are β-strand breakers. Notably, germ line-like patterns of hydrogen exchange were observed in the S31P and S97P fibrils (Figure 2F). The overall structure of the fibril is therefore tolerant of changes in local structure, as has been observed by Brumshtein et al.39
To further probe the relationship between thermal unfolding and fibrillogenesis, we measured the thermal melting of five 6aJL2-VL sequences (WT, I29V, V34A, V59A, and I67V) by intrinsic fluorescence and then compared the measured fU values with aggregation rates at a pair of temperatures, 44 and 55 °C. Since aggregation assays were done at pH 7.4 in phosphate-buffered saline, the melting curves were also obtained under these conditions, which leads to decreased thermodynamic stability relative to samples dissolved in D2O solvent (Figure 2E; D2O solvent was necessary for hydrogen exchange measurements). The relative ranking of thermodynamic stability for the five proteins is the same as measured by hydrogen exchange, WT > I67V > I29V > V59A > V34A (compare Figure 2D and G). At 44 °C, WT and I67V domains, which are least prone to aggregation, have the lowest fU values (Figure 2H), while at 55 °C the aggregation rates are more similar between the five proteins with WT and I67V still aggregating slightly more slowly than the others (Figure 2I). The difference in the aggregation rates between the five 6aJL2-VL variants is flattened upon increasing the temperature where fU values become uniformly larger; for example, the aggregation of V34A is 21-fold, 4-fold, and 1.5-fold faster than the germ line at 37, 44, and 55 °C, respectively. This is consistent with global unfolding leading to aggregation of the variants examined. The increase in temperature above the melting points of all the domains leads to similar populations of their globally unfolded states (close to an upper limit of 100%) and, hence, a parallel reduction of native and partially unfolded states. The net result is a partial equalization of the rates of fibril formation.
Polymerization-Prone Regions Mapped by Solution NMR and Prediction Tools.
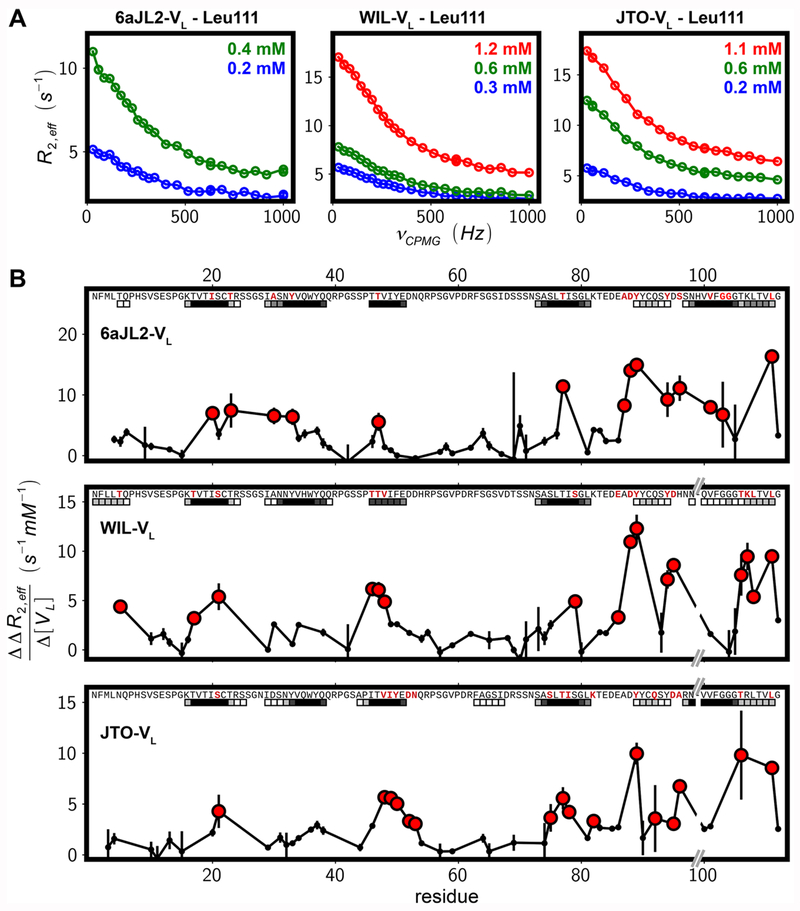
The results described above are consistent with the hypothesis that loss of native structure is required for amyloid formation, as has been observed for other proteins.40–42 However, assembly into fibrils likely requires formation of specific, favorable, intermolecular contacts in order to overcome the entropic barrier to fibrillation. LCs with different sequences may have different propensities to self-associate in the unfolded state, which may affect their ability to aggregate and cause disease. We therefore performed 15N CPMG relaxation dispersion experiments to identify initial oligomerization events that might be responsible for the conversion of soluble VL domains into amyloid fibrils. These NMR-based spin-relaxation experiments quantify decay rates of NMR signals, R2,eff, during a fixed evolution period where the frequency of application of chemical shift refocusing pulses, νCPMG, is varied. This class of NMR experiment is sensitive to rare oligomerization events occurring on the millisecond time scale,43,44 wherein the population of the resulting oligomer can be as low as ~0.5%.45 15N CPMG experiments23 recorded at 25 °C on the native state of wild-type 6aJL2-VL indicated the presence of native oligomers as previously observed for JTO-VL and WIL-VL.11 Perturbation of this native oligomerization through the introduction of mutations in the VL–VL dimerization interface did not influence fibril formation, with the lag rate klag solely determined by the fraction of unfolded protein, fU (Figure S2), in agreement with the results in Figure 2. 15N CPMG experiments were therefore recorded at 55 °C for the globally unfolded state of three λV6-57 VL proteins, including those derived from the germ line 6aJL2, WIL, and JTO. Nonflat CPMG profiles were observed for several 15N probes in each of the three sequences considered, indicating that the domains undergo conformational exchange involving one or more sparsely populated states. Notably, the size of the dispersion profiles, ΔR2,eff, defined as the difference between effective 15N transverse relation rates at the lowest (~30 Hz) and the highest (1 kHz) CPMG field strengths used, ΔR2,eff = R2,eff(~30 Hz) − R2,eff(1 kHz), was dependent on protein concentration, as illustrated with CPMG curves for L111 in WT 6aJL2, WIL, and JTO VL domains, Figure 3A. As ΔR2,eff increases with protein concentration in all cases, the population of particles in solution is highly skewed toward the monomer. The dispersion profiles could not be globally fitted using a model of exchange involving a single interconversion between monomer and dimer. This suggests that each site experiences independent oligomerization events, most likely a mixture between on- and off-pathway processes with respect to fibril formation, or that each amide is sensitive to a different stage of early fibril formation. Individual residue fits of CPMG profiles indicate exchange rates of approximately 1000 s−1, although the populations of the interconverting species could not be well defined by the data, pointing to a complex exchange mechanism, as would be expected for a protein domain with multiple regions that are available for oligomerization. It is worth noting that this complexity is not the result of contributions from the folding/unfolding reaction. Even at 55 °C separate peaks are observed for the C-terminal residue, G112, of both 6aJL2-VL and JTO-VL that derive from native and unfolded states (see Materials and Methods), implying that the folding equilibrium occurs on a time scale that is much slower than the exchange process(es), consistent with folding rates reported in the literature.16 Therefore, the folding equilibrium will not contribute to the dispersion profiles measured from the unfolded state in this study.
Figure 3.
Oligomerization from the unfolded states of the λV6-57 VL domains probed by 15N CPMG NMR. (A) 15N CPMG profiles for Leu111 of 6aJL2, WIL, and JTO-VL domains at the indicated protein concentrations. (B) Concentration dependence of ΔR2,eff, the difference in R2,eff rates at minimum and maximum νCPMG values, for individual amide groups of three λV6–57 VL domains. Red circles indicate high concentration dependence, i.e., values higher than 5 s−1 mM−1 (6aJL2) or 3 s−1 mM−1 (WIL and JTO-VL). Residue-specific AMYLPRED225 predictions are shown as squares at the top of each panel with a consensus score shown in grayscale between 2 (white) and 6 or higher (black), where higher values indicate an increasing propensity to aggregate. Note that there is a single-residue deletion at position 99 for WIL and JTO in comparison to 6aJL2-VL. Lines between experimental points are included to guide the eye.
The concentration dependence of ΔR2,eff is plotted as a function of protein sequence in Figure 3B. The red circles in these profiles highlight those residues with the largest changes in ΔR2,eff, corresponding to “oligomerization hotspots”. Regions predicted by the program AMYLPRED225 are plotted as squares below each sequence in Figure 3B. The agreement between experimental and predicted data is good, with the great majority of red circles within predicted regions. Most discrepancies are found between positions 85 and 100, where the prediction algorithms underestimate the potential for polymerization that is observed by CPMG NMR. This may be the result of the intramolecular disulfide bridge connecting C22 and C91 that would bring the 85–100 region proximal to residues on the other end of the disulfide bond, thereby influencing aggregation propensities. The effect of the disulfide bond is not taken into account in the prediction program. Significant precipitation of the protein occurred during the acquisition of CPMG NMR data (1 day), especially for the more concentrated samples and for wild-type 6aJL2-VL. These precipitates were ThT-positive (Figure S3), indicating that pathways leading to fibril formation are present even in the absence of agitation.
Important Regions for Fibril Formation.
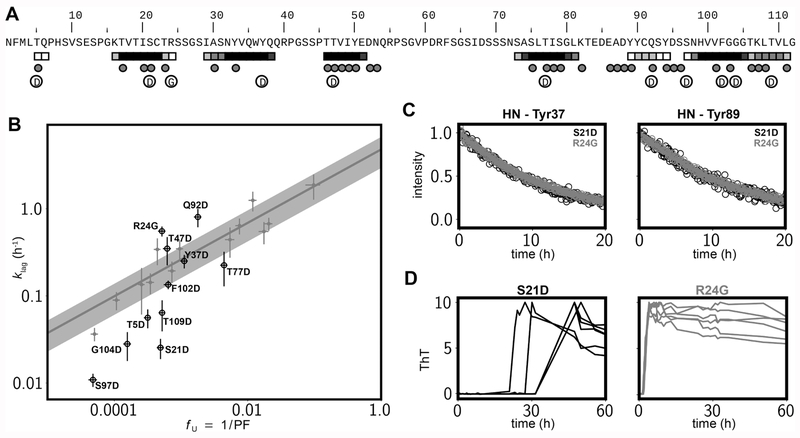
Our CPMG data recorded on a series of unfolded VL domains suggest multiple regions with the propensity to polymerize and hence multiple oligomerization processes likely occurring on separate time scales. In order to assess the relative importance of each polymerization-prone region, as predicted by AMYLPRED2 and indicated in Figure 3B, we generated a series of 10 6aJL2-VL mutants where each of these regions was modified by replacement of a single amino acid with aspartate. The mutations are made to solvent-exposed side chains in order to avoid destabilization of the native states of the domains. We also considered the somatic mutation R24G,21 which removes a polar side chain in the aggregation-prone 16–24 region. The location of each of the mutations along the protein sequence is shown in Figure 4A, together with the predicted aggregation regions using AMYLPRED2. In addition, polymerization-prone regions identified by CPMG NMR in any of the WT 6aJL2, WIL, and JTO VL domains discussed above are also indicated. Aggregation assays monitored by ThT fluorescence establish that all the mutants form amyloid fibrils, but they have different aggregation kinetics (Figure 4B, black symbols). The aggregation data for 6aJL2-VL variants S21D (p-value = 0.023, where the p-value is calculated for the null hypothesis that the aggregation kinetics are not different for the new set of 11 variants), R24G (p-value = 0.0007), Q92D (p-value = 0.040), S97D (p-value = 0.026), and G104D (p-value = 0.050) deviate from the linear klag vs fU profile calculated in Figure 2D that made use of more conservative mutations (Figure 4B, solid gray region). For the mutants localized to aggregation hotspots klag rates are either significantly slower (S21D, S97D, G104D) or faster (R24G, Q92D) than what would be predicted based purely on thermodynamic stability. Notably, these aggregation assays emphasize the important role of the K16-R24 region, as introduction of a charge (S21D) reduces the rate of fibril formation, while removal of a charge (R24G) enhances the kinetics of aggregation, despite the fact that these two proteins have similar thermodynamic stabilities, as measured by hydrogen exchange of the most slowly exchanging amides (Figure 4C, Figure S4). The slower aggregation profiles for some of the Asp substitutions in aggregation-prone regions, such as for S21D, are not surprising, as the introduction of a charge would be expected to decrease formation of fibers if indeed the perturbed region is critical for this process. The situation for R24G, where the aggregation rate is increased, is interesting, as this is a somatic mutation found in 25% of AL patients for this λ family.21 Our results show that this mutation is linked to AL not only because of the reduced thermodynamic stability26 relative to WT but also from removal of the arginine side chain, which increases the aggregation propensity at the level of the primary structure. Position 24 is one hotspot for somatic mutation linked to AL since it is a hypervariable position of the VL domain yet adjacent to a very conserved aggregation-prone region (Figure 1D). We do not currently have an explanation for why the Q92D substitution increases the kinetics of aggregation, except that it may reduce off-pathway excursions.
Figure 4.
Mutations in polymerization-prone regions modulate aggregation kinetics. (A) Locations of the introduced mutations, Asp or Gly, in the sequence of the λV6-57 VL domain are indicated by circled D or G. AMYLPRED225 predictions (squares) and residues with a high concentration dependence of ΔR2,eff in any of the λV6-57 VL domains examined (spheres), as defined in the caption of Figure 3, are also shown. (B) Correlation, on a log–log scale, between fU and klag, showing a significant deviation from the fitted curve of Figure 2D (gray line, shaded area indicates the standard error in klag values; see Materials and Methods) for several mutants in aggregation-prone regions. Unlabeled gray circles are the data points for the conservative mutations considered in Figure 2D. Data points from mutants where a polar group is either introduced (10 mutations to Asp) or removed (1 mutation to Gly) in hotspot regions are indicated by labeled black circles. (C) Solvent hydrogen/deuterium exchange time profiles for two representative slowly exchanging amide groups from S21D and R24G 6aJL2-VL mutants. (D) Kinetics of aggregation monitored by ThT fluorescence for S21D and R24G 6aJL2-VL; six traces are shown in each subpanel.
Brumshtein et al.39 have carried out a similar mutagenesis study to evaluate regions in VL domains that drive aggregation, using a different set of mutations than the Asp substitutions considered here. In particular, two mutations, in combination, L76P and V100P+V101P+F102P+G103P, were found to inhibit fibril formation in the VL2-8-J1 domain. In our study regions surrounding residues 76 and 100–103 were found to be aggregation prone on the basis of CPMG relaxation experiments. However, we failed to observe a significant effect for the introduction of aspartate residues at positions 77 and 102, although some delay in the aggregation was observed for mutations at position 97 or 104. Differences between the two studies may arise from the different mutagenesis approaches used, coupled with the different VL proteins considered. Nevertheless, both studies establish that although the effect of a given mutation on the rate of aggregation depends on the region where the mutation is introduced, with some sites more important than others, amyloid formation likely involves an interplay between several sites. The opposing views of a single sequence exclusively responsible for the aggregation or that the driving force is nonspecific are both oversimplifications.
In summary, our studies of the immunoglobulin λV6-57 VL domain, which is preferentially found in aggregates from AL patients,21 establish that global unfolding is an obligatory step in fibril formation, as a linear correlation between aggregation rate and fraction unfolded VL domain was observed. Aggregation hotspots are established through a 15N relaxation dispersion analysis performed as a function of protein concentration, identifying, in particular, residues within the K16-R24 fragment. Hydrogen exchange profiles of a number of variants of the λV6-57 VL sequence including the germ line, S31P and S97P, show that similar fibrillar structures are obtained, with protection for the first 40 and last 50 residues, while the region encompassing R40-P60 remains flexible. Notably, the secondary structural propensities observed here are similar to those found in AL-09 VL30 and R24G 6aJL2-VL32 fibers and in the cryoEM structure of a λV6-57 LC fibril,15 but differ from MAK33 VL domain fibrils and the cryoEM structure of a λV1-44 LC fibril.14 This emphasizes the importance of studies on a wide range of light-chain sequences so as to establish the molecular determinants that are responsible for their aggregation and, ultimately, develop the relevant pharmaceuticals that can eliminate the process.46
MATERIALS AND METHODS
Sequence Conservation.
The sequence conservation for the λ VL domains, as calculated using the program CompBio47 and shown in Figure 1D, was based on an analysis that included 33 reviewed germ line IGLV sequences and the five functional IGLJ sequences present in the UniProt database.29 Note that the joining of single IGLV and IGLJ sequences by DNA rearrangement during the differentiation of B cells from hematopoietic stem cells leads to the formation of a mature λ VL domain.48 Residue-by-residue values of conservation were calculated according to the Shannon entropy of residues with a window size equal to zero.47,49
Protein Production.
λV6-57 VL domains were expressed in BL21(DE3) E. coli cells using a pET-22 plasmid with a pelB leader for periplasmic expression. Protein production and purification were as previously described.11 The sequence of the germ line λV6-57 VL protein, 6aJL2-VL,26,27 is the IGLV6-57*01 allele in the IMGT repertoire.50
NMR Experiments.
All NMR experiments were performed on a Bruker Avance III HD 14.1 T spectrometer equipped with a cryogenically cooled, x,y,z pulsed-field gradient triple-resonance probe. Details of individual experiments are described in what follows.
1. Dimethyl Sulfoxide-Quenched Hydrogen/Deuterium Exchange.
Fibrils of 6aJL2-VL were formed by incubation of 20–50 mL solutions containing 20 μM U–15N protein, phosphate-buffered saline (PBS) pH 7.4, and 5 mM NaN3, in a 1 L flask with shaking (250 rpm) over 3 days, 37 °C. At the end of this period the solution was split into two tubes, and fibrils were collected by centrifugation at 15000g. Fibrils were then resuspended into 10 mL of 50 mM BisTris, pH 6.4 buffer, either H2O based (reference sample) or D2O based (exchange sample). Fibrils for the reference sample were collected and washed using the following protocol: centrifugation at 15000g and 4 °C, resuspension in 5 mL of ice-cold Milli-Q H2O, centrifugation at 15000g and 4 °C, resuspension in 1 mL of ice-cold Milli-Q H2O, and freeze-drying. Fibrils used for the exchange measurements were collected after 2 h of incubation in deuterated buffer (23 °C) and washed using the same protocol as described above for the reference sample, except that ice-cold D2O was used in the two resuspension steps. An exchange time of 2 h is optimal since incubation for longer times does not lead to further exchange for the faster exchanging amides (residues 40–65), with I/I(0h) values of approximately 0.5, while all other amides continue to exchange toward a constant plateau value of approximately 0.5, therefore reducing the dynamic range of the experiment. For NMR detection, lyophilized samples were dissolved in 95% DMSO-d6, 5% D2O, and 50 mM sodium dichloroacetate, pD 5, and acquisition of 1H,15N HSQC spectra was started immediately at 10 °C. The sample pH and temperature for NMR acquisition are optimal to minimize the hydrogen exchange reaction during the experiments,51 so that the amide proton occupancies depend only on the hydrogen exchange that occurred in the fibrillar state. The experiment for the WT protein was performed in duplicate, and the error bars in Figure 1H indicate differences between the two replicates. Peak assignments were obtained using (HB)CBCA(CO)NNH, HNCACB, HNCO, HN(CA)CO, and HNN spectra52 recorded on a uniformly 13C,15N-labeled sample of 6aJL2-VL with a protein concentration of 1.1 mM in 95% DMSO-d6, 5% D2O, and 50 mM sodium dichloroacetate, pH 5.
2. Native-State Hydrogen Exchange.
λV6-57 VL domain samples, prepared as described above, were desalted by dialysis against Milli-Q H2O and freeze-dried. Subsequently they were dissolved in D2O-based buffer comprising 20 mM sodium citrate, pD 5, and acquisition of a series of 1H,15N HSQC spectra at 37 °C was started immediately. The concentration of the protein used in the study ranged between 0.12 and 0.45 mM. Protection factors were calculated as PF = krc/kex, where krc is the residue-specific value of the intrinsic rate of hydrogen exchange for an unfolded peptide.35 krc rates were calculated using the Web tool at http://hx2.med.upenn.edu/download.html. Values of fU were calculated as the reciprocal of the average protection factors for the most protected amides in each domain (for example, maroon-colored residues in Figure S1 highlighting the WT protein).
3. CPMG Experiments.
15N CPMG experiments were recorded on U-15N labeled proteins dissolved in buffer comprising 50 mM BisTris, pH 6.4, 1 mM EDTA, 5 mM NaN3, and 90% H2O/10% D2O. Protein concentrations varied from 0.2 to 1.2 mM. Dispersion profiles were acquired by measuring the variation of effective transverse relaxation rates of in-phase 15N magnetization, R2,eff, as a function of the number of refocusing pulses applied during a constant-time relaxation interval, T, of 35 ms,23 R2,eff = 1/T ln(Io/I), where I is the intensity of a peak at a given, nonzero, value of νCPMG and Io is the corresponding intensity when T = 0. A 17 kHz 1H continuous-wave decoupling field was applied during the constant-time interval.53 The CPMG frequency, νCPMG, is given by 1/(2δ), where δ is the delay between successive refocusing pulses.54 A total of 18–23 2D planes were recorded with different νCPMG values, between 30 and 1000 Hz, with three repeats for error analysis;55 in addition a reference plane was recorded without the constant-time relaxation interval to obtain Io values. All CPMG experiments were recorded at 55 °C, where proteins were predominantly unfolded. A small population of the native state could be quantified on the basis of the pair of peaks observed for the C-terminal residue G112, corresponding to the native and unfolded states, with peak intensities in the reference plane indicating the presence of ~25% and ~10% of folded domains for 6aJL2-VL and JTO-VL, respectively. No residual population of the native state could be detected for WIL-VL, in agreement with its lower thermodynamic stability.38 Furthermore, since sample precipitation was observed during the course of data acquisition, especially for the more concentrated samples, determination of protein concentration was not straightforward and was done as follows: Two short (8 min) 1H,15N HSQC spectra were recorded before and after the CPMG experiments. The change in intensity between the two spectra was found to be small (less than 10%) for samples in the 0.2–0.3 mM concentration range but larger (up to 40%) for the more concentrated samples. Therefore, for the less concentrated samples, absorbance measurements at 280 nm were used to estimate the total protein concentrations, and the concentration of the unfolded state was estimated according to the intensity of the two peaks for G112 in the reference plane. For more concentrated samples the average concentration of the unfolded state during the course of the CPMG experiments was estimated by comparing the intensity of the G112 peak in the reference plane with that of the less concentrated samples that do not aggregate over time. Note that all the planes comprising the pseudo-3D dispersion data set (different νCPMG values), including the reference plane, are acquired in an interleaved manner. Thus, the intensity of peaks in the reference plane provides an estimate for the average concentration of protein over the course of the experiment. Spectral assignments for each domain were obtained using (HB)CBCA(CO)NNH, HNCACB, HNCO, HN(CA)CO, and HNN data sets.52
Fibril Formation Assays.
Solutions (100 μL) containing 10 μM protein and 1 μM ThT were incubated at 37, 44, or 55 °C in black, clear-bottomed, untreated 96-well plates (Figures 2C,D and 4B,D). Plates were covered by transparent film and a lid to reduce evaporation. ThT fluorescence was measured with a plate reader, with excitation at 440 nm and emission at 480 nm. The assays were done in PBS, pH 7.4, with agitation at 1000 rpm. Errors in tlag = 1/klag values were calculated from multiple aggregation measurements (at least 6 repeats), with the error in klag, Δklag, given by Δklag = Δtlagklag2. Since the experimental errors in tlag are proportional to tlag values (Figure S5), the shaded area in Figure 4B is calculated as klag,fit(1 ± α) where α = 〈[(klag,expt − klag,fit)/klag,fit]2〉0.5, and 〈 〉 refers to the average over experimental data points.
Intrinsic Fluorescence.
Fluorescence spectra of samples at protein concentrations of 0.02 mM were acquired with a PTI QuantaMaster 80 spectrofluorometer using an excitation wavelength of 280 nm. The excitation and emission slit widths were set respectively to 2 and 4 nm. A step size of 2 nm and an integration time of 2 s were used. The acquisition of the spectrum at each temperature was started 2 min after the cuvette had reached the desired temperature. The emission between 300 and 400 nm as a function of temperature was fit to a two-state equation for thermal denaturation.56
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council of Canada (to L.E.K.) and by NIH grant DK46335 and the Skaggs Institute for Chemical Biology (J.W.K.). L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b05499.
Supporting Figures 1–5, including native-state protection factors, ΔR2,eff values showing that dispersions measured on the folded state do not correlate with aggregation rates, ThT emission spectra, solvent exchange profiles, and the correlation between experimental errors in tlag and klag with the values of tlag and klag (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Quock TP; Yan TJ; Chang E; Guthrie S; Broder MS Epidemiology of AL amyloidosis: a real-world study using US claims data. Blood Adv. 2018, 2 (10), 1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hemminki K; Li XJ; Forsti A; Sundquist J; Sundquist K, Incidence and survival in non-hereditary amyloidosis in Sweden. BMC Public Health 2012, 12, DOI: 10.1186/1471-2458-12-974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Magy-Bertrand N; Dupond JL; Mauny F; Dupond AS; Duchene E; Gil H; Kantelip B; Members C Incidence of amyloidosis over 3 years: the AMYPRO study. Clin Exp Rheumatol 2008, 26 (6), 1074–1078. [PubMed] [Google Scholar]
- (4).Rosenzweig M; Landau H Light chain (AL) amyloidosis: update on diagnosis and management. J. Hematol. Oncol 2011, 4, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Milani P; Merlini G; Palladini G Light Chain Amyloidosis. Mediterr J. Hematol Infect Dis 2017, 10 (1), e2018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Falk RH; Alexander KM; Liao R; Dorbala S AL (Light-Chain) Cardiac Amyloidosis: A Review of Diagnosis and Therapy. J. Am. Coll. Cardiol 2016, 68 (12), 1323–41. [DOI] [PubMed] [Google Scholar]
- (7).Wechalekar AD; Gillmore JD; Hawkins PN Systemic amyloidosis. Lancet 2016, 387 (10038), 2641–2654. [DOI] [PubMed] [Google Scholar]
- (8).Huang DB; Ainsworth C; Solomon A; Schiffer M Pitfalls of molecular replacement: the structure determination of an immunoglobulin light-chain dimer. Acta Crystallogr., Sect. D: Biol. Crystallogr 1996, 52, 1058–66. [DOI] [PubMed] [Google Scholar]
- (9).Klimtchuk ES; Gursky O; Patel RS; Laporte KL; Connors LH; Skinner M; Seldin DC The critical role of the constant region in thermal stability and aggregation of amyloidogenic immunoglobulin light chain. Biochemistry 2010, 49 (45), 9848–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Enqvist S; Sletten K; Westermark P Fibril protein fragmentation pattern in systemic AL-amyloidosis. J. Pathol 2009, 219 (4), 473–80. [DOI] [PubMed] [Google Scholar]
- (11).Rennella E; Morgan GJ; Kelly JW; Kay LE Role of domain interactions in the aggregation of full-length immunoglobulin light chains. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (3), 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Olsen KE; Sletten K; Westermark P Fragments of the constant region of immunoglobulin light chains are constituents of AL-amyloid proteins. Biochem. Biophys. Res. Commun 1998, 251 (2), 642–7. [DOI] [PubMed] [Google Scholar]
- (13).Vrana JA; Gamez JD; Madden BJ; Theis JD; Bergen HR 3rd; Dogan A Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood 2009, 114 (24), 4957–9. [DOI] [PubMed] [Google Scholar]
- (14).Radamaker L; Lin YH; Annamalai K; Huhn S; Hegenbart U; Schonland SO; Fritz G; Schmidt M; Fandrich M Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis. Nat. Commun 2019, 10 (1), 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Swuec P; Lavatelli F; Tasaki M; Paissoni C; Rognoni P; Maritan M; Brambilla F; Milani P; Mauri P; Camilloni C; Palladini G; Merlini G; Ricagno S; Bolognesi M Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Nat. Commun 2019, 10 (1), 1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Morgan GJ; Kelly JW The Kinetic Stability of a Full-Length Antibody Light Chain Dimer Determines whether Endoproteolysis Can. Release Amyloidogenic Variable Domains. J. Mol. Biol 2016, 428 (21), 4280–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Blancas-Mejia LM; Horn TJ; Marin-Argany M; Auton M ; Tischer A; Ramirez-Alvarado M Thermodynamic and fibril formation studies of full length immunoglobulin light chain AL-09 and its germline protein using scan rate dependent thermal unfolding. Biophys. Chem 2015, 207, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Morgan GJ; Usher GA; Kelly JW Incomplete Refolding of Antibody Light Chains to Non-Native, Protease-Sensitive Conformations Leads to Aggregation: A Mechanism of Amyloidogenesis in Patients? Biochemistry 2017, 56 (50), 6597–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Eulitz M; Linke RP The precursor molecule of a V lambda II-immunoglobulin light chain-derived amyloid fibril protein circulates precleaved. Biochem. Biophys. Res. Commun 1993, 194 (3), 1427–34. [DOI] [PubMed] [Google Scholar]
- (20).Bellotti V; Merlini G; Bucciarelli E; Perfetti V; Quaglini S; Ascari E Relevance of class, molecular weight and isoelectric point in predicting human light chain amyloidogenicity. Br. J. Haematol 1990, 74 (1), 65–9. [DOI] [PubMed] [Google Scholar]
- (21).Comenzo RL; Zhang Y; Martinez C; Osman K; Herrera GA The tropism of organ involvement in primary systemic amyloidosis: contributions of Ig V(L) germ line gene use and clonal plasma cell burden. Blood 2001, 98 (3), 714–20. [DOI] [PubMed] [Google Scholar]
- (22).Hoshino M; Katou H; Yamaguchi K; Goto Y Dimethylsulfoxide-quenched hydrogen/deuterium exchange method to study amyloid fibril structure. Biochim. Biophys. Acta, Biomembr 2007, 1768 (8), 1886–99. [DOI] [PubMed] [Google Scholar]
- (23).Hansen DF; Vallurupalli P; Kay LE An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J. Phys. Chem. B 2008, 112 (19), 5898–904. [DOI] [PubMed] [Google Scholar]
- (24).Korzhnev DM; Kay LE Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc. Chem. Res 2008, 41 (3), 442–51. [DOI] [PubMed] [Google Scholar]
- (25).Tsolis AC; Papandreou NC; Iconomidou VA; Hamodrakas SJ A consensus method for the prediction of ‘aggregation-prone’ peptides in globular proteins. PLoS One 2013, 8 (1), e54175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).del Pozo Yauner L; Ortiz E; Sanchez R; Sanchez-Lopez R; Guereca L; Murphy CL; Allen A; Wall JS; Fernandez-Velasco A; Solomon A; Becerril B Influence of the germline sequence on the thermodynamic stability and fibrillogenicity of human lambda 6 light chains. Proteins: Struct., Funct., Genet 2008, 72 (2), 684–92. [DOI] [PubMed] [Google Scholar]
- (27).Williams SC; Frippiat JP; Tomlinson IM; Ignatovich O; Lefranc MP; Winter G Sequence and evolution of the human germline V lambda repertoire. J. Mol. Biol 1996, 264 (2), 220–32. [DOI] [PubMed] [Google Scholar]
- (28).Martin A, Chahwan R, Yar Parsa J, Scharff MD Chapter 20 - Somatic Hypermutation: The Molecular Mechanisms Underlying the Production of Effective High-Affinity Antibodies In Molecular Biology of B Cells, Second ed.; Alt FW, Andreas Radbruch TH, Reth M, Eds.; Academic Press, 2015; pp 363–388. [Google Scholar]
- (29).UniProt C UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47 (D1), D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Piehl DW; Blancas-Mejia LM; Wall JS; Kennel SJ; Ramirez-Alvarado M; Rienstra CM Immunoglobulin Light Chains Form an Extensive and Highly Ordered Fibril Involving the N- and C-Termini. ACS Omega 2017, 2 (2), 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Hora M; Sarkar R; Morris V; Xue K; Prade E; Harding, ; Buchner, J.; Reif, B. MAK33 antibody light chain amyloid fibrils are similar to oligomeric precursors. PLoS One 2017, 12 (7), e0181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lecoq L; Wiegand T; Rodriguez-Alvarez FJ; Cadalbert R; Herrera GA; Del Pozo-Yauner L; Meier BH; Bockmann A A Substantial Structural Conversion of the Native Monomer Leads to in-Register Parallel Amyloid Fibril Formation in Light-Chain Amyloidosis. ChemBioChem 2019, 20 (8), 1027–1031. [DOI] [PubMed] [Google Scholar]
- (33).Alexandrescu AT An NMR-based quenched hydrogen exchange investigation of model amyloid fibrils formed by cold shock protein A. Pac Symp. Biocomput 2001, 67–78. [DOI] [PubMed] [Google Scholar]
- (34).Bai Y; Sosnick TR; Mayne L; Englander SW Protein folding intermediates: native-state hydrogen exchange. Science 1995, 269 (5221), 192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bai Y; Milne JS; Mayne L; Englander SW Primary structure effects on peptide group hydrogen exchange. Proteins: Struct., Funct., Genet 1993, 17 (1), 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bai Y; Milne JS; Mayne L; Englander SW Protein stability parameters measured by hydrogen exchange. Proteins: Struct., Funct., Genet 1994, 20 (1), 4–14. [DOI] [PubMed] [Google Scholar]
- (37).Vassar PS; Culling CF Fluorescent stains, with special reference to amyloid and connective tissues. Arch Pathol 1959, 68, 487–98. [PubMed] [Google Scholar]
- (38).Wall J; Schell M; Murphy C; Hrncic R; Stevens FJ; Solomon A Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry 1999, 38 (42), 14101–8. [DOI] [PubMed] [Google Scholar]
- (39).Brumshtein B; Esswein SR; Sawaya MR; Rosenberg G; Ly AT; Landau M; Eisenberg DS Identification of two principal amyloid-driving segments in variable domains of Ig light chains in systemic light-chain amyloidosis. J. Biol. Chem 2018, 293 (51), 19659–19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kelly JW Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol 1996, 6 (1), 11–7. [DOI] [PubMed] [Google Scholar]
- (41).Dobson CM Protein misfolding, evolution and disease. Trends Biochem. Sci 1999, 24 (9), 329–32. [DOI] [PubMed] [Google Scholar]
- (42).Hurle MR; Helms LR; Li L; Chan W; Wetzel R A role for destabilizing amino acid replacements in light-chain amyloidosis. Proc. Natl. Acad. Sci. U. S. A 1994, 91 (12), 5446–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Rennella E; Sekhar A; Kay LE Self-Assembly of Human Profilin-1 Detected by Carr-Purcell-Meiboom-Gill Nuclear Magnetic Resonance (CPMG NMR) Spectroscopy. Biochemistry 2017, 56 (5), 692–703. [DOI] [PubMed] [Google Scholar]
- (44).Sekhar A; Rumfeldt JA; Broom HR; Doyle CM; Bouvignies G; Meiering EM; Kay LE Thermal fluctuations of immature SOD1 lead to separate folding and misfolding pathways. eLife 2015, 4, e07296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Palmer AG 3rd; Kroenke CD; Loria JP Nuclear magnetic resonance methods for quantifying microsecond-to-milli-second motions in biological macromolecules. Methods Enzymol 2001, 339, 204–38. [DOI] [PubMed] [Google Scholar]
- (46).Morgan GJ; Yan NL; Mortenson DE; Rennella E; Blundon JM; Gwin RM; Lin CY; Stanfield RL; Brown SJ; Rosen H; Spicer TP; Fernandez-Vega V; Merlini G; Kay LE; Wilson IA; Kelly JW Stabilization of amyloidogenic immunoglobulin light chains by small molecules. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (17), 8360–8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Capra JA; Singh M Predicting functionally important residues from sequence conservation. Bioinformatics 2007, 23 (15), 1875–82. [DOI] [PubMed] [Google Scholar]
- (48).Brack C; Hirama M; Lenhard-Schuller R; Tonegawa S A complete immunoglobulin gene is created by somatic recombination. Cell 1978, 15 (1), 1–14. [DOI] [PubMed] [Google Scholar]
- (49).Mirny LA; Shakhnovich EI Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J. Mol. Biol 1999, 291 (1), 177–96. [DOI] [PubMed] [Google Scholar]
- (50).Scaviner D; Barbie V; Ruiz M; Lefranc MP Protein displays of the human immunoglobulin heavy, kappa and lambda variable and joining regions. Exp. Clin. Immunogenet 1999, 16 (4), 234–40. [DOI] [PubMed] [Google Scholar]
- (51).Zhang YZ; Paterson Y; Roder H Rapid amide proton exchange rates in peptides and proteins measured by solvent quenching and two-dimensional NMR. Protein Sci. 1995, 4 (4), 804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Sattler M; Schleucher J; Griesinger C Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsedfield gradients. Prog. Nucl Magn. Reson. Spectrosc 1999, 34, 93–158. [Google Scholar]
- (53).Jiang B; Yu B; Zhang X; Liu M; Yang DA (15)N CPMG relaxation dispersion experiment more resistant to resonance offset and pulse imperfection. J. Magn. Reson 2015, 257, 1–7. [DOI] [PubMed] [Google Scholar]
- (54).Mulder FA; Skrynnikov NR; Hon B; Dahlquist FW; Kay LE Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc 2001, 123 (5), 967–75. [DOI] [PubMed] [Google Scholar]
- (55).Korzhnev DM; Salvatella X; Vendruscolo M; Di Nardo AA; Davidson AR; Dobson CM; Kay LE Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 2004, 430 (6999), 586–90. [DOI] [PubMed] [Google Scholar]
- (56).Santoro MM; Bolen DW Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry 1988, 27 (21), 8063–8. [DOI] [PubMed] [Google Scholar]
- (57).Hernandez-Santoyo A; del Pozo Yauner L; Fuentes-Silva D; Ortiz E; Rudino-Pinera E; Sanchez-Lopez R; Horjales E; Becerril B; Rodriguez-Romero A A single mutation at the sheet switch region results in conformational changes favoring lambda6 light-chain fibrillogenesis. J. Mol. Biol 2010, 396 (2), 280–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.