Abstract
Methods used for the laboratory diagnosis of tuberculosis are continually evolving in order to achieve more rapid, less expensive, and accurate results. Acid-fast staining and culture for mycobacteria remain at the core of any diagnostic algorithm. Following growth in culture, molecular technologies such as nucleic acid hybridization probes, MALDI-TOF MS, and DNA sequencing may be used for definitive species identification. Nucleic acid amplification methods allow for the direct detection of Mycobacterium tuberculosis complex within respiratory specimens without relying on culture growth, leading to more rapid diagnoses and appropriate patient care.
Keywords: Acid-fast staining, Sputum processing, Tuberculosis culture, MALDI-TOF, Sequencing, GeneXpert MTB/RIF assay
Introduction
Mycobacterium tuberculosis is the causative agent of tuberculosis, a disease causing significant worldwide morbidity and mortality. Tuberculosis remains a major public health problem with approximately one-third of the world's population infected [1]. In 2014, tuberculosis was responsible for the death of nearly 1.5 million people, representing a global mortality impact larger than any other infectious disease. The emergence of multi-drug resistant (MDR) strains has reduced viable treatment options and threatens to make tuberculosis both an untreatable and highly fatal disease. MDR tuberculosis elicits great economic and quality of life burdens due to costly and time-consuming therapeutic interventions. Reducing the burden of tuberculosis depends in part on the implementation of proper laboratory systems for the accurate and rapid diagnosis of active tuberculosis disease. Of the 5.2 million patients with pulmonary tuberculosis reported to the World Health Organization (WHO) in 2014, only 58% of cases were confirmed by laboratory methods such as smear or culture [1]. The remaining 42% of patients were diagnosed using clinical criteria alone (symptom history or chest X-ray), highlighting the need for increased availability of diagnostics in resource limited settings.
The M. tuberculosis complex (MTBC) is comprised of eight species which include M. tuberculosis, M. bovis, M. bovis Bacillus Calmette-Guerin (BCG), M. africanum, M. caprae, M. microti, M. cannettii, and M. pinnipedii. The majority of pulmonary tuberculosis cases are caused by M. tuberculosis, however, it may be clinically meaningful to identify members of the M. tuberculosis complex to the species level. For example, M. bovis is intrinsically resistant to the first-line drug pyrazinamide, and disseminated M. bovis BCG may be found as a complication following vaccination or intravesical instillation as treatment for bladder cancer [2], [3]. Mycobacteria are obligately aerobic, nonmotile, rod-shaped bacilli. Members of the genus Mycobacterium have several unique characteristics as compared to other genera of bacteria, largely due to structural differences in cell wall composition. The cell wall of mycobacteria contains a higher content of complex lipids (>60% as opposed to approximately 5% and 20% in gram-positive and gram-negative organisms respectively) including long chain (C60—C90) fatty acids called mycolic acids [4], [5]. Mycolic acids make the cell wall extremely hydrophobic and enhance resistance to desiccation, killing by disinfectants, staining with basic aniline dyes, and penetration by many of the drugs that are used to treat infections caused by other bacteria. These unique features of mycobacterial cell wall structure provide the basis for special laboratory considerations when performing direct stains from specimens, growing organisms in culture, and determining species identification by molecular methods.
Specimen collection
Depending on the clinical manifestation of disease, virtually any specimen type may be processed for the presence of mycobacteria [6]. The most common sources are respiratory specimens including sputum, bronchial aspirates, and bronchoalveolar lavage fluid, however tissues, normally sterile body fluids, blood, and urine are also commonly submitted for analysis. Specimens should be collected in sterile, leak-proof containers and do not generally require transport media for preserving viability due to the hardy nature of mycobacterial organisms. Tissue may be placed in a small amount of sterile saline to avoid dehydration, while non-sterile water should be avoided due to the possibility of confounding contamination with environmental mycobacteria. Most specimens should be refrigerated during transport to the laboratory and up until the time of processing to maintain the viability of any mycobacteria present while preventing overgrowth of contaminating bacterial organisms [7].
Sputum is the most common specimen obtained for the diagnosis of pulmonary infection with MTBC and nontuberculous mycobacteria (NTM). To enhance sensitivity by smear, current guidelines recommend the collection of early morning sputum specimens on 3 consecutive days with a minimum of 8 hours between collections [8], [9]. Mycobacteria become more concentrated in the sputum as patients sleep, so smear sensitivity increases with the use of early morning sputum (Table 1) [10], [11]. Though supported by guidelines, this data is somewhat controversial [12]. Since infants and young children may have difficulty producing expectorated sputum, swallowed sputum may be aspirated from the stomach by gastric lavage. Since lengthy exposure to acidic gastric washings may decrease the viability of mycobacteria, specimens must be neutralized with sodium bicarbonate if not processed within 4 hours of collection. For patients that are unable to expectorate sputum, alternatives include sputum induction and collection of bronchoalveolar lavage (BAL) fluid. The induction of sputum using hypertonic saline with an ultrasonic nebulizer is a non-invasive method, while BAL fluid may be invasively collected during bronchoscopy [13], [14]. Bronchoscopes should be decontaminated according to manufacturer's instructions between uses, and cleaning procedures should not utilize tap water which may contain environmental mycobacteria.
Table 1.
Acid-fast smears and cultures prepared from early morning sputum specimens have better sensitivity.
Non-respiratory specimens may also be collected for the testing of MTBC and other mycobacteria. As for sputum samples, clean-catch urine specimens may be collected on 3 consecutive days for culture [8]. Early morning collection provides the greatest sensitivity by culture since organisms accumulate in the bladder overnight. Normally sterile body fluids such as cerebrospinal fluid, pleural fluid, pericardial fluid, and synovial fluid may all be useful for culture of mycobacteria, however, these specimens are often paucibacillary and may require processing additional volume to achieve adequate sensitivity. In general, swabs are discouraged since they only are able to transfer a minimal volume of specimen onto culture media. Stool culture may be useful for the detection of disseminated M. avium complex (MAC) from AIDS patients, while lymph node, skin, and other biopsy tissue specimens may be processed under an appropriate clinical context.
Blood may be collected in tubes containing SPS, heparin, or citrate. EDTA tubes should not be used for blood collection [8], [15]. The majority of disseminated mycobacterial infections occur in immunocompromised hosts and are due to MAC, however, bloodstream infections can also occur with MTBC and other NTM species. Blood for mycobacterial culture can be processed using either the Isolator tube system (Wampole Laboratories), BACTEC Myco/F Lytic bottles (Becton Dickinson), or BacT/ALERT MP bottles [16], [17]. Isolator tubes undergo a lysis centrifugation method to recover intracellular organisms from whole blood specimens followed by inoculation of appropriate media plates. Myco/F Lytic and BacT/ALERT MP bottles are inoculated with whole blood and are optimized to promote the growth of mycobacterial and fungal organisms, which is monitored using automated blood culture instruments.
Acid-fast stains for mycobacteria
Microscopic evaluation of stained smears is a rapid and inexpensive screening method for mycobacteria within clinical specimens. While related to gram-positive bacteria based on peptidoglycan composition within the cell wall, mycobacteria are not reliably detected with the traditional Gram stain. Their hydrophobic cell wall resists penetration of aniline dyes such as crystal violet, so mycobacteria are either not visible with the Gram stain or may appear as bacilli-shaped clear zones or “ghosts” when direct specimens are stained [18]. However, under certain conditions arylmethane dyes are able to form stable complexes with the mycolic acids within mycobacterial cell walls. In the presence of phenol and applied heat, carbol fuchsin dye can be used as performed during Ziehl–Neelsen staining, which also utilizes methylene blue as a counterstain [19], [20]. Since these cell wall dye complexes are resistant to destaining with mineral acids, mycobacteria are referred to as “acid-fast bacilli” or “AFB”. The Fite stain is a modified acid-fast stain that uses weaker acid decolorization conditions to allow for the visualization of M. leprae and partially acid-fast organisms such as Rhodococcus spp. and Nocardia spp. [21]. Each of these stains use conventional light microscopy, while alternative stains utilizing fluorescence detection provide distinct advantages. Fluorescent stain consisting of a mixture of auramine O and rhodamine B dyes binds to the nucleic acids within acid-fast organisms [22]. Fluorescent staining is more sensitive and allows for more rapid reading of slides [23]. For these reasons, the WHO has endorsed the global phase out of conventional Ziehl–Neelsen light microscopy in favor of auramine-rhodamine AFB staining even though acquisition of fluorescent light-emitting diode (LED) microscopes remains challenging in resource-limited settings. In 2014, only 7% of laboratories had the capability of performing fluorescent AFB smears, up from 2% in 2012 [1].
Due to the small size of mycobacterial cells, sufficient training is required to reliably differentiate AFB from debris present in specimens that may be non-specifically stained. In addition to cell size (1–10 µm in length), a beaded staining appearance is suggestive of AFB (Fig. 1). The overall clinical sensitivity of sputum AFB smear is 22–80% depending on the burden of mycobacteria, the type of AFB stain used, and experience of the laboratory technician, while the positive predictive value for mycobacteria is > 95% [24]. However, acid-fast stains are not specific for MTBC as they cannot differentiate between mycobacteria species. Smear sensitivity varies greatly based on AFB burden within sputum with 1000–10,000 CFU/ml required for reliable detection. These higher AFB concentrations correlate with the severity of infection and positive sputum smears suggest a higher likelihood of infectivity for patients with pulmonary tuberculosis.
Fig. 1.
Acid-fast bacilli stains. Mycobacteria are stained with carbol fuchsin-based Kinyoun stain (left) and fluorescent auramine-rhodamine stain (right).
While current guidelines in the United States recommend performing three consecutive sputum smears for the diagnosis of pulmonary tuberculosis [8], the additional sensitivity gained by performing a third smear after two negative smears is incremental (Table 2) [25], [26], [27], [28], [29], [30], [31], [32]. A comprehensive literature review analyzing combined data from 37 individual studies determined that the first two sputum smears detect > 95% of smear-positive cases [33]. Collectively, initial sputum smears were positive in an average of 85.8% of cases and 11.9% of cases were positive by a second smear. When the first two sputa were negative, testing a third specimen only increased detection by an additional 3.1%. Based on this data, the WHO has changed their international guidelines to recommend performing 2 AFB smears as opposed to the prior recommendation of 3 smears [34]. At one point, performing a third smear was thought to act as an additional precaution for laboratories in resource limited settings against lab errors or use of suboptimal reagents. However, implementation of quality control programs has now been largely established even in resource limited areas thanks to WHO initiatives and these concerns have diminished. In addition to low yield of a third smear for diagnosis, the traditional requirement for three negative AFB sputum smears for patients with suspected pulmonary tuberculosis to be removed from isolation results in significant healthcare costs associated with delayed removal from isolation precautions [35]. If expectorated or induced sputum cannot be obtained, some institutions will use BAL fluid as a comparable surrogate for AFB staining and decisions regarding isolation precautions.
Table 2.
Select studies of sensitivity gained by serial AFB smears.
| Study | # positive smears | % of total positives detected by: |
||
|---|---|---|---|---|
| 1st smear | 2nd smear | 3rd smear | ||
| Ipuge et al. [25] | 11,650 | 83.4 | 12.2 | 4.4 |
| Nelson et al. [26] | 53 | 77.4 | 15.1 | 7.5 |
| Walker et al. [27] | 166 | 77.1 | 15.0 | 7.9 |
| Mathew et al. [28] | 19 | 89.4 | 5.3 | 5.3 |
| Wilmer et al. [29] | 64 | 89.1 | 7.8 | 3.1 |
| Khogali et al. [30] | 60 | 93 | 5 | 2 |
| Rehman et al. [31] | 1164 | 77.0 | 16.3 | 6.7 |
| Hassan et al. [32] | 719 | 96.4 | 3.6 | 0 |
Acid-fast staining serves as a rapid and inexpensive screen for AFB. The CDC recommends that the results of AFB staining are reported within 24 hours of specimen collection [36]. Positive results have a high predictive value, but relatively low sensitivity necessitates concomitant culturing for MTBC growth. AFB staining alone should not be utilized for the definitive diagnosis of mycobacterial infection and should be combined with culture for downstream species identification and antimicrobial susceptibility testing.
Specimen processing and decontamination
Since mycobacteria are slowly-growing organisms, the contamination of specimens with more rapidly growing bacteria may prevent their detection by culture. Non-sterile respiratory specimens typically contain bacteria that will overgrow any mycobacteria potentially present. Therefore, it is important to process specimens prior to culture in a way that will reduce the burden of contaminating bacteria without adversely affecting mycobacterial viability. Non-sterile specimens can be pre-treated with a variety of agents, but the most common are N-acetyl-L-cysteine (NALC) and sodium hydroxide (NaOH) [8]. NALC is a mucolytic agent that helps to disrupt mucus present in respiratory specimens and releases bacteria so they can access nutrients provided by the culture medium. NaOH, often used at a final concentration of 2%, is a decontaminating agent that kills contaminating bacteria while leaving any mycobacteria viable for culture. Maintaining strict time limits for exposure to NaOH is important because mycobacteria are generally more hardy than other bacteria due to their waxy cell walls, but will also be rendered non-viable if exposed to NaOH for too long. Culture contamination rates must be monitored to ensure that the optimum balance of NaOH concentration and incubation times are being used. Historically, contamination rates above 5% were considered excessive for the radiometric-based BACTEC 460 system, however more recent literature demonstrating higher contamination rates with the BACTEC MGIT 960 system suggests that an overall rate of 6-8% may be a more appropriate benchmark [37], [38], [39], [40]. Contamination of solid media should be monitored separately from broth cultures with 3–5% as an acceptable contamination rate [41]. Conversely, contamination rates approaching 0% suggest that decontamination conditions are too harsh and may be adversely affecting mycobacterial recovery [15]. After NALC-NaOH treatment, the specimen is centrifuged to pellet any mycobacteria followed by rehydration with a minimal volume of sterile phosphate buffered saline before being plated onto culture medium. Specimens may also be treated with various antibiotics to suppress the growth of bacterial and fungal contaminants. PANTA, a mixture of polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin is often added prior to broth culture.
Mycobacterial culture: solid media
Following decontamination, specimens may be cultured for the growth of MTBC and NTM. Culture is traditionally performed on solid egg-based media, such as Lowenstein-Jensen (L-J) media, which is composed of egg proteins, potato flour, salts, and glycerol. L-J media supports good growth of MTBC, but is not as reliable for all mycobacterial species including M. bovis and M. genavense [42], [43]. While some laboratories still use L-J media, many have transitioned to using more chemically defined agar-based media optimized for faster mycobacterial growth. The use of Middlebrook 7H10 or 7H11 agars, for example, allows for visible MTBC colony growth in 10–12 days as compared to 18–24 days with L-J media [15]. However, agar-based media is less stable and more prone to deterioration. For example, exposure to excessive heat or light may lead to the release of formaldehyde which is toxic to mycobacteria and may inhibit growth.
Most Mycobacterium species, including MTBC, grow best at a temperature of 35–37 °C. Select species such as M. haemophilum, M. marinum, M. paratuberculosis, and M. ulcerans have an optimum growth temperature of 30 °C and may be cultured in a separate incubator. Like many aerobic bacteria, mycobacterial growth is stimulated by incubation in air containing 5–10% CO2. Unlike other bacteria, MTBC and slowly-growing NTM species have a growth rate with 12–24 hours for each generation of cell division. Due to this slow growth rate, it may take several weeks for colonies to become visible on culture plates. Cultures are typically held for 6–8 weeks before being discarded and reported as negative [44]. Despite the slow growth rate, culture for mycobacteria is approximately 100-fold more sensitive than AFB smear, requiring only 10–100 CFU/mL of specimen for reliable growth [15].
Mycobacteria typically grow more slowly on solid media as compared to broth culture, however, plated growth allows for the detection of mixed cultures containing multiple organisms. Colony morphologies may also aid or support identification. The most significant morphologic feature to note is the colony texture. MTBC colonies are characteristically dry with a rough texture and a cream/tan color, and are colloquially described as “rough and buff.” Conversely, M. bovis colonies are flat and smooth. Several Mycobacterium species also produce pigments that can range from yellow to orange in color. Photochromogenic species only produce pigment after exposure to light, while scotochromogens produce pigment regardless of light exposure. All members of the M. tuberculosis complex are nonpigmented, so the presence of any pigment indicates NTM growth.
Mycobacterial culture: liquid media
Optimal recovery of mycobacteria from clinical specimens is achieved through the use of a combination of solid and liquid media [8]. In general, mycobacteria grow faster in broth than on solid media plates, which allows for improved patient management and clinical outcomes [45]. Growth of MTBC from clinical specimens takes an average of 10 days by automated broth systems versus 20–25 days on solid media [8], [37], [40].
There are three FDA-cleared commercial platforms for the semi-automated broth-based culture of mycobacteria: the BACTEC MGIT 960 system (Becton Dickinson Microbiology Systems), the VersaTREK system (Trek Diagnostic Systems), and the MB/BacT Alert 3D (bioMérieux). The MGIT system is named for its use of Mycobacterial Growth Indicator Tubes (Fig. 2). Each tube contains a modified Middlebrook 7H9 broth and a fluorescent indicator that is quenched by the presence of oxygen within the tube. Growth of mycobacteria in the medium consumes oxygen over time and allows the fluorescent indicator to signal as positive once a certain growth threshold has been met. The instrument continuously monitors tube fluorescence allowing lab staff to quickly identify positive tubes and begin the task of identifying any mycobacteria present. The MGIT system is an improvement over past BACTEC platforms that utilized radiometric assessment of growth and required manual intervention to place bottles on the machine once or twice per day for reading. In addition to faster growth, many studies have documented the improved sensitivity of the MGIT broth system as compared to solid culture [46], [47], [48], [49], [50]. A meta-analysis of 10 published studies representing 1381 strains from 14,745 clinical specimens found the MGIT system to have 81.5% analytical sensitivity (and 99.6% specificity) compared to 67% sensitivity for L-J solid media [40].
Fig. 2.
The BACTEC™ MGIT™ 960 instrument (Becton Dickinson) and culture tubes for broth-based mycobacterial growth.
A second semi-automated broth-based system for mycobacterial culture is the VersaTREK system (Fig. 3). Rather than using fluorescence, the VersaTREK system detects the growth of inoculated specimens by measuring pressure changes in the bottle headspace above the broth medium. Finally, the MB/BacT Alert 3D system utilizes a colorimetric carbon dioxide sensor for growth. All three systems use Middlebrook broth and have comparable levels of performance for the culture of mycobacteria [51], [52], [53].
Fig. 3.
The VersaTREK® system (Thermo Fisher Scientific) for broth-based mycobacterial growth. Figure is used by permission of Trek Diagnostic Systems (Thermo Fisher).
For blood specimens, BACTEC Myco/F Lytic culture bottles (Becton Dickinson) may be used which contain media optimized for the recovery of mycobacteria and fungi. Myco/F Lytic culture bottles have increased sensitivity for growth of mycobacteria as compared to standard BACTEC aerobic bottles [54], [55]. The Myco/F Lytic and the BacT/Alert culture systems have been shown to detect MTBC growth from blood in 20 and 16.4 days respectively [56]. Regardless of the broth culture system, all specimens that signal positive for growth must be subcultured to solid media to detect mixed cultures and for correlation with any colony morphologies present.
Molecular diagnostics from culture growth
Historically, positive mycobacterial cultures were identified on the basis of colony morphology and select biochemical reactions, however, molecular methods must be used for definitive identification of most mycobacterial isolates from culture [57]. Confirming the growth of MTBC in culture is important for the proper clinical management of tuberculosis cases, and is required for conventional antimicrobial susceptibility testing which is performed using culture isolates. Unlike traditional biochemical tests, molecular methods allow for rapid species identification. There are several molecular technologies currently employed by clinical diagnostic laboratories to identify isolates from culture including nucleic acid hybridization probes, line probe hybridization assays, matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS), and DNA sequencing.
Nucleic acid hybridization probes
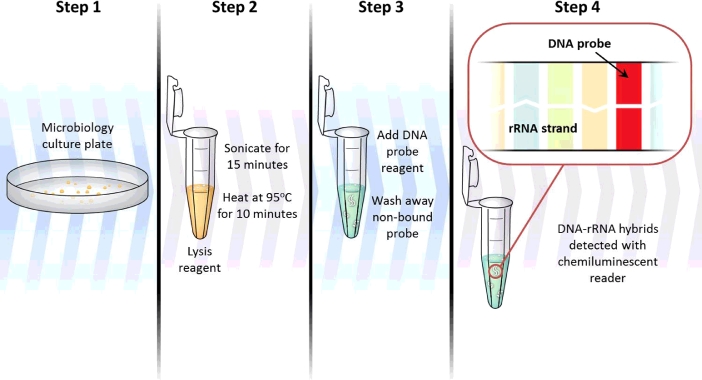
The use of commercially available nucleic acid hybridization probes is easily implemented into laboratory algorithms and allows for the identification of several clinically relevant Mycobacterium species. FDA-cleared hybridization probes are available from Hologic Gen-Probe for the identification of M. tuberculosis complex, M. avium complex, M. gordonae, and M. kansasii. Nucleic acids from cultured organism are released during sterilizing heat treatment and sonication steps (Fig. 4). A species-specific DNA probe labeled with a chemiluminescent moiety is added. If the probe sequence is complementary to the 16S ribosomal RNA target, hybridization will occur and chemiluminescence from the DNA-rRNA hybrid can be detected and quantified. There is no PCR or other amplification step that occurs when utilizing these probes, so a larger amount of target nucleic acid is required. This restricts the use of hybridization probes to detection from growth in culture as opposed to direct detection from patient specimens. However, culture hybridization probes are highly sensitive and specific, are technically simple to utilize, and provide results within approximately two hours [57], [58], [59]. There has been some minor cross-reactivity reported with uncommon species of nontuberculous mycobacteria, but this can largely be eliminated by strictly controlling the hybridization time and temperature. Rare cross-reactivity has been reported with the MAC probe (to M. arosiense, M. chimaera, M. colombiense, M. nebraskense, M. palustre, M. paraffinicum, M. saskatchewanense, and M. vulneris) and the MTBC probe (to M. celatum and M. holsaticum) [60], [61]. The main limitation associated with hybridization probes is that they are only available for four mycobacterial species or complexes, so additional molecular tools are needed for the identification of most mycobacteria seen in the clinical laboratory.
Fig. 4.
Nucleic acid hybridization probe workflow schematic.
Line probe assays
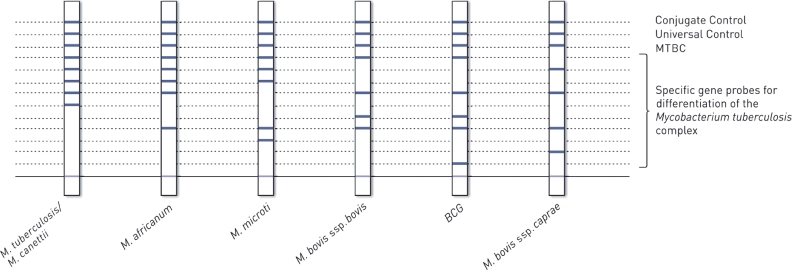
Line probe assays are an alternative technique for identification of mycobacteria from culture isolates that make use of hybridization-based probes. This technology utilizes nitrocellulose membrane strips embedded with genus- and species-specific probes. DNA from lysed culture isolates hybridizes to probes and produces colorimetric bands when complementary DNA is present to allow for species identification (Fig. 5). Three commercially available line probe assays include the INNO-LiPA Mycobacteria assay (Fujirebio), the GenoType MTBC test (Hain Lifescience), and the Speed-oligo Mycobacteria (Vircell). The probes used by these assays target nucleotide differences within the 16S and/or 23S rRNA regions. These line probe assays can each detect M. tuberculosis complex members as well as several commonly encountered NTM species. Analytical sensitivity and specificity are generally > 90% for line probe assays and results are available within 4–6 hours [62], [63]. However, cross-reactivity has been documented among certain infrequently encountered NTM species [60]. As with nucleic acid hybridization probes, results should be concordant with established phenotypic characteristics of the identified species. A major limitation of these line probe assays is that they are not currently cleared by the FDA for clinical diagnostic use in the United States.
Fig. 5.
Line probe assay schematic. The GenoType MTBC test (Hain Lifescience) utilizes several embedded DNA probes to detect and differentiate members of the M. tuberculosis complex. Figure is used by permission of Hain Lifescience.
MALDI-TOF MS
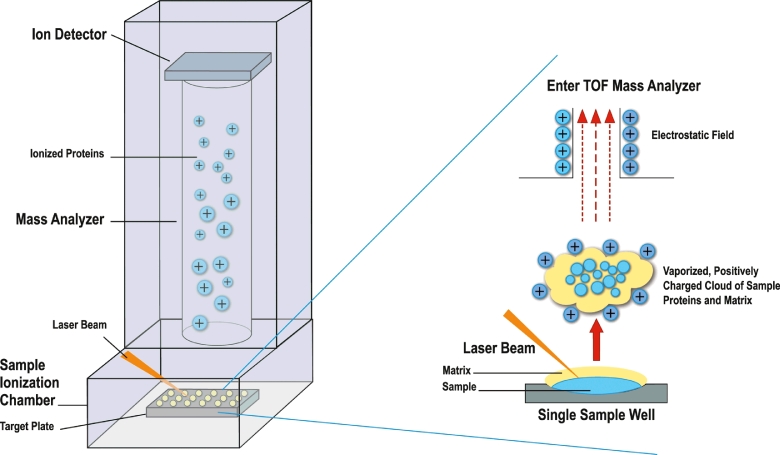
In recent years, MALDI-TOF mass spectrometry has become heavily utilized as the primary method for bacterial and yeast identification from clinical specimens and has even more recently been applied to the identification of mycobacteria. This technique utilizes mass spectrometry to assess the protein content of an isolate for identification. Following growth in culture, isolates are subjected to ethanol and/or heat treatments with mechanical disruption to lyse and sterilize mycobacterial cells [64]. Proteins are extracted from cell lysates with formic acid and acetonitrile. Due to laboratory safety concerns when working with potential MTBC isolates, all inactivation and protein extraction steps should take place using BSL-3 precautions. A chemical matrix is added to aliquots of extracted protein which facilitates laser ionization (Fig. 6). Ionized proteins are separated based on their mass-to-charge ratio resulting in a spectrum of protein peaks representing the most abundant proteins of the organism in question. This spectrum serves as a “fingerprint” that can be compared to a database of spectra from identified isolates.
Fig 6.
MALDI-TOF mass spectrometry operation schematic. Fig is used with permission of Mayo Foundation for Medical Education and Research, all rights reserved.
There are currently two MALDI-TOF platforms FDA-cleared for the identification of a limited list of bacteria: the MALDI Biotyper (Bruker Corporation) and the Vitek MS (BioMérieux). Neither system has FDA clearance for the identification of mycobacteria, so laboratories must perform thorough validation and verification studies to ensure that mycobacteria are correctly identified. These studies must utilize an accepted gold standard identification method such as sequencing for comparison with MS spectra (see review from Clark et al. for more information regarding the validation and verification of clinical microbiology assays [65]). Several published studies have documented the accurate identification of mycobacteria species by both MALDI-TOF MS systems [66], [67], [68], [69], [70]. Each system utilizes its own database containing several entries representing M. tuberculosis complex members and many NTM species. However, current databases may not adequately represent less common organisms, limiting their reliable detection. To this end, clinical laboratories may also make use of adaptable Research Use Only (RUO) databases that allow for customized expansion of libraries provided by the manufacturers. There are many advantages to using MALDI-TOF MS platforms for the identification of mycobacteria, including similar laboratory workflows that may already be in place for the identification of bacteria, yeasts, and molds. Consumable reagent costs are low and rapid turn-around-time (TAT) may reduce overall patient management costs [71]. A major limitation is the need for isolate growth in culture, which can often take several weeks for slow-growing mycobacteria. Pure isolate growth is required for identification and MALDI-TOF lacks the resolution to differentiate between several closely related species, including individual members of the M. tuberculosis complex. Differentiating individual complex members may be useful in guiding antimicrobial therapy; for instance M. bovis is intrinsically resistant to pyrazinamide. Since MALDI-TOF MS assesses isolate protein content, TAT may be delayed for especially slowly-growing organisms as compared to sequencing which utilizes DNA amplification and requires smaller amounts of colony growth [70], [72].
Sequencing
Traditional Sanger dideoxy sequencing is the current gold standard method for the identification of mycobacteria species. Complete genome sequences have been established for over 40 species including M. tuberculosis, however, it is more practical for routine identification to sequence specific targets such as the 16S or 23S ribosomal DNA genes, the heat shock protein hsp65 gene, or the rpoB gene encoding the beta subunit of the bacterial RNA polymerase [57]. PCR primers are designed to bind conserved sequences and broadly amplify DNA from all mycobacterial species, and are also positioned to flank variable regions that can be used for species differentiation. Amplified DNA is sequenced and results compared to established sequence databases. Public databases such as the NCBI GenBank (within the International Nucleotide Sequence Database Collaboration; http://www.ncbi.nlm.nih.gov/genbank) may be queried using a BLAST search, but are known to contain sequences that have not been published and are not validated for clinical use [73]. For the clinical diagnostic laboratory, it is often advantageous to use a commercially available curated database such as the MicroSeq (Thermo Fisher Scientific), SmartGene IDNS (SmartGene), or RipSeq (Pathogenomix, Inc.) libraries. These web-based databases provide tools for analysis and benefit from ongoing quality control of entries.
A major advantage of DNA sequencing is that it allows for the objective identification of a wide variety of mycobacteria species including M. tuberculosis. Also, a fairly rapid TAT is possible with same or next day identification to the species level. Sequencing results are generally reported 8-24 hours after growth of an organism in culture and is influenced by how samples are batched within the routine workflow. Limitations of sequencing include performing fairly labor-intensive protocols that require highly trained technologists. In addition, the equipment and reagents are more costly than for the molecular techniques described above. Sequencing is often performed if less expensive and more rapid methods such as hybridization probes and MALDI-TOF MS fail to produce definitive results.
Molecular diagnostics directly from patient specimens
Due to the potential severity of tuberculosis disease and concerns about community and nosocomial transmission, early diagnosis of active disease is essential for effective patient management. For these reasons, the majority of commercial and laboratory-developed molecular tests have specifically focused on the identification of MTBC. Due to the slow growth rates of mycobacteria in culture, methods that rely on cultured organism such as MALDI-TOF MS and sequencing may lead to a delayed diagnosis despite their rapid TAT once sufficient growth is achieved. The commercial line probe assays discussed above are designed for the direct detection of MTBC from respiratory specimens, however these are not cleared by the FDA for clinical use in the US. Nucleic acid amplification (NAA) tests have been developed that are able to detect M. tuberculosis complex directly from patient specimens in as little as two hours.
Two NAA tests have been granted FDA approval/clearance for the direct detection of M. tuberculosis complex from respiratory specimens: the Amplified Mycobacterium tuberculosis direct test (MTD; Hologic Gen-Probe) and the Xpert MTB/RIF test (Cepheid) [74], [75]. Respectively, these assays target MTBC-specific rRNA and DNA sequences by utilizing transcription-mediated amplification and real-time PCR methods. Based on a comprehensive literature review, the analytical sensitivity of the Amplified MTD test from smear-positive respiratory specimens was determined to be 87.5–100% depending on the study while specificity was generally > 98% [76]. Sensitivity is reduced to 63.6–100% for smear-negative specimens. Performance characteristics are similar for the Xpert MTB/RIF assay which has a sensitivity of 90–99% from smear-positive respiratory specimens as compared to culture as the gold-standard test [77], [78]. Sensitivity expectedly decreases to 66–74% from smear-negative specimens. Despite the reduction in sensitivity when AFB smear negative, NAA testing is still recommended for patients with suspected tuberculosis due to superior performance as compared to AFB staining. For each platform, sensitivity is highest when testing AFB smear-positive respiratory specimens, however tuberculosis disease cannot be ruled out even from a smear-negative specimen with a negative NAA test result [78]. For this reason, culture must always be ordered in conjunction with more rapid smear and NAA tests. In addition, NAA tests cannot differentiate between live and non-viable MTBC, so they cannot be used to monitor response to treatment.
Unlike the MTD assay, the Xpert MTB/RIF test is an automated, cartridge-based system that benefits from ease of use and a closed amplification system that reduces the potential for cross-contamination between specimens (Fig. 7). The test is simple for laboratory technicians to perform and no advanced biosafety equipment is needed. For these reasons, in 2010 the World Health Organization recommended use of the Xpert MTB/RIF for patients with suspected pulmonary tuberculosis in developing countries with a high prevalence of disease. The use of this assay has expanded significantly with 4.8 million test cartridges purchased worldwide in 2014 by 116 low- and middle-income countries, up from 550,000 in 2011 [1]. Recent guidelines from the Centers of Disease Control and Prevention recommend that NAA testing be performed on at least one respiratory specimen from each patient (preferably the first) with suspected pulmonary tuberculosis [75]. A respiratory specimen must still be cultured for mycobacterial growth in case of falsely negative results by NAA testing, to detect NTM species, and to facilitate antimicrobial susceptibility testing. In February 2015, the FDA cleared expanded use of the Xpert MTB/RIF assay for more rapid removal of suspected tuberculosis patients from airborne isolation [79]. The revised labeling states that one or two negative Xpert MTB/RIF results is sufficient evidence to exclude pulmonary tuberculosis, whereas prior CDC guidelines recommended isolation until three consecutive negative sputum AFB smears were obtained to rule out contagious tuberculosis.
Fig. 7.
The Cepheid Xpert® system and MTB/RIF test cartridge for the molecular detection of M. tuberculosis complex and rifampin resistance-associated mutations. Image © Cepheid – used by permission.
The Xpert MTB/RIF assay has the added benefit of providing information about potential rifampin resistance, by detecting mutations in an 81-base pair region of the rpoB gene that are responsible for conferring approximately 96% of rifampin resistance in MTBC. Rifampin resistance is also a predictor of multidrug-resistant tuberculosis since the majority of rifampin-resistant isolates will also be isoniazid-resistant. Some false-rifampin resistance has been reported, so the CDC recommends reporting Xpert-rifampin resistance as a preliminary result pending sequencing for resistance mutations [80]. Growth-based conventional antimicrobial susceptibility testing is required as confirmation. Overall, strengths of the Xpert MTB/RIF assay include good sensitivity and specificity for respiratory specimens, a rapid 2-hour TAT, the ability to detect M. tuberculosis complex and rifampin resistance directly from specimens, and the use of a closed PCR system with a low risk of cross-contamination.
Aside from commercial NAA tests, several laboratories offer laboratory-developed PCR tests (LDTs) for the direct detection of MTBC. These are generally closed PCR systems, which significantly reduces the opportunity for false-positive results due to cross-contamination. These tests are generally reported to have good sensitivity and specificity, however a meta-analysis of 84 published studies found that cumulative sensitivity ranged from 9.4% to 100% and specificity from 5.6% to 100% [81]. This large heterogeneity in LDT quality stresses the need for vigorous verification and the inclusion of proper quality control measures. When properly implemented, laboratory-developed real-time PCR assays are often less expensive than commercially available NAA tests and may be utilized on a wider variety of specimen types including non-respiratory specimens and formalin-fixed, paraffin-embedded tissue blocks.
Well-characterized commercial NAA tests are typically recommended due to FDA oversight, enhanced reliability, and improved quality control. NAA testing provides the possibility for earlier laboratory diagnosis of tuberculosis as compared to traditional methods, leading to earlier initiation of treatment, use of appropriate isolation precautions to interrupt transmission, and improved patient outcomes.
Conclusions and future developments
M. tuberculosis complex is responsible for immense worldwide morbidity and mortality. Delays in diagnosis may postpone administration of appropriate treatment and be detrimental to patient outcomes. As a slow-growing organism using traditional culture methods, newer molecular techniques allow for more rapid and sensitive laboratory diagnosis of tuberculosis. NAA tests and the sequencing of isolates are also able to provide early indication of drug resistance by screening for established resistance mutations. In particular, next-generation sequencing is now being investigated as a technique that allows for greater depth of sequencing coverage for more definitive results and the ability to identify mycobacteria to the species level and resistance variants within mixed sequence populations [82], [83]. Despite the fact that many new commercial and laboratory developed tests are designed to specifically detect M. tuberculosis, the ability to detect NTM species by sequencing and MALDI-TOF MS has improved with the validation of expanded databases.
Many of the newly developed assays are designed to improve the speed and/or accuracy of tuberculosis diagnosis in resource-limited settings. For example, in 2015 a tuberculosis urine antigen assay was endorsed by the World Health Organization [1]. The lateral flow lipoarabinomannan assay (LF-LAM; Alere Determine TB LAM Ag test) detects antigen shed from mycobacterial cell walls from either metabolically active or damaged cells. This assay can be used as a rapid point-of-care test, and the use of urine is easier to collect and avoids potential transmission issues associated with infectious sputum. Since sensitivity is better among patients with a low CD4 count and higher burden of disease, the LF-LAM assay is specifically indicated for use in HIV patients with suspected tuberculosis that have either a CD4 count of ≤ 100 cells/µL or are otherwise seriously ill [84], [85]. Like the LF-LAM assay, several technologies are currently under development to provide faster, cheaper, more accurate, and more accessible diagnostic testing for tuberculosis.
References
- 1.World Health Organization . 20th ed. 2015. Global Tuberculosis Report. Geneva, Switzerland http://www.who.int/tb/publications/global_report/en/ [Google Scholar]
- 2.Talbot E.A., Perkins M.D., Silva S.F., Frothingham R. Disseminated bacille Calmette-Guerin disease after vaccination: case report and review. Clin Infect Dis. 1997;24(6):1139–1146. doi: 10.1086/513642. [DOI] [PubMed] [Google Scholar]
- 3.Redelman-Sidi G., Glickman M.S., Bochner B.H. The mechanism of action of BCG therapy for bladder cancer–a current perspective. Nat Rev Urol. 2014;11(3):153–162. doi: 10.1038/nrurol.2014.15. [DOI] [PubMed] [Google Scholar]
- 4.Asselineau C., Asselineau J., Laneelle G., Laneelle M.A. The biosynthesis of mycolic acids by mycobacteria: current and alternative hypotheses. Prog Lipid Res. 2002;41(6):501–523. doi: 10.1016/s0163-7827(02)00008-5. [DOI] [PubMed] [Google Scholar]
- 5.Mahapatra S., Basu J., Brennan P.J., Crick D.C. Tuberculosis and the Tubercle Bacillus ASM Press; Washington, DC: 2005. Structure, biosynthesis, and genetics of the mycolic acid-arabinogalactan-peptidoglycan complex; pp. 275–285. [Google Scholar]
- 6.Garcia L.S., editor. Clinical Microbiology Procedures Handbook. 3rd ed. American Society for Microbiology; Washington, DC: 2010. [Google Scholar]
- 7.Baron E.J. 11th ed. ASM Press; Washington, D.C: 2015. Specimen collection, transport, and processing: bacteriology. Manual of Clinical Microbiology. [Google Scholar]
- 8.Clinical Laboratory Standards Institute (CLSI). Laboratory detection and identification of mycobacteria. Document M48-A. Wayne, PA; 2008.
- 9.Jensen P.A., Lambert L.A., Iademarco M.F., Ridzon R. Guidelines for preventing the transmission of Mycobacterium tuberculosis in health-care settings, MMWR Recomm Rep. 2005;54(RR-17):1–141. [PubMed] [Google Scholar]
- 10.Abraham P.R., Sharma V.D., Shivannavar C.T. Diagnosis of TB from smear & culture negative sputum specimens by IS 6110 based PCR. Indian J Med Res. 2012;135:249–251. [PMC free article] [PubMed] [Google Scholar]
- 11.Ssengooba W., Kateete D.P., Wajja A., Bugumirwa E., Mboowa G., Namaganda C. An early morning sputum sample is necessary for the diagnosis of pulmonary tuberculosis, even with more sensitive techniques: a prospective cohort study among adolescent TB-suspects in Uganda. Tuberc Res Treat. 2012;2012 doi: 10.1155/2012/970203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das D., Dwibedi B., Kar S.K. Spot or early morning sample for mycobacterial culture: which? Int J Tuberc Lung Dis. 2014;18(3):310–311. doi: 10.5588/ijtld.13.0786. [DOI] [PubMed] [Google Scholar]
- 13.Saglam L., Akgun M., Aktas E. Usefulness of induced sputum and fibreoptic bronchoscopy specimens in the diagnosis of pulmonary tuberculosis. J Int Med Res. 2005;33(2):260–265. doi: 10.1177/147323000503300215. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Angulo Y., Wiysonge C.S., Geldenhuys H., Hanekom W., Mahomed H., Hussey G. Sputum induction for the diagnosis of pulmonary tuberculosis: a systematic review and meta-analysis. Eur J Clin Microbiol Infect Dis. 2012;31(7):1619–1630. doi: 10.1007/s10096-011-1485-6. [DOI] [PubMed] [Google Scholar]
- 15.Pfyffer G.E. Manual of Clinical Microbiology. 11th ed. ASM Press; Washington, DC: 2015. Mycobacterium: general characteristics, laboratory detection, and staining procedures; pp. 536–569. [Google Scholar]
- 16.Crump J.A., Tanner D.C., Mirrett S., McKnight C.M., Reller L.B. Controlled comparison of BACTEC 13A, MYCO/F LYTIC, BacT/ALERT MB, and ISOLATOR 10 systems for detection of mycobacteremia. J Clin Microbiol. 2003;41(5):1987–1990. doi: 10.1128/JCM.41.5.1987-1990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirby J.E., Delaney M., Qian Q., Gold H.S. Optimal use of Myco/F lytic and standard BACTEC blood culture bottles for detection of yeast and mycobacteria. Arch Pathol Lab Med. 2009;133(1):93–96. doi: 10.5858/133.1.93. [DOI] [PubMed] [Google Scholar]
- 18.Trifiro S., Bourgault A.M., Lebel F., Rene P. Ghost mycobacteria on Gram stain. J Clin Microbiol. 1990;28(1):146–147. doi: 10.1128/jcm.28.1.146-147.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziehl F. Zur Färbung des Tuberkelbacillus. Deutsche Med Wochenschr. 1882;33:451. [Google Scholar]
- 20.Neelsen F. Ein casuistischer Beitrag zu Lehre von der Tuberkulose. Centralblatt für die medizinischen Wissenschaften. 1883;28:497–501. [Google Scholar]
- 21.Fite G.L., Cambre P.J., Turner M.H. Procedure for demonstrating lepra bacilli in paraffin sections. Arch Pathol (Chic) 1947;43(6):624. [PubMed] [Google Scholar]
- 22.Hanscheid T., Ribeiro C.M., Shapiro H.M., Perlmutter N.G. Fluorescence microscopy for tuberculosis diagnosis. Lancet Infect Dis. 2007;7(4):236–237. doi: 10.1016/S1473-3099(07)70058-0. [DOI] [PubMed] [Google Scholar]
- 23.Kommareddi S., Abramowsky C.R., Swinehart G.L., Hrabak L. Nontuberculous mycobacterial infections: comparison of the fluorescent auramine-O and Ziehl-Neelsen techniques in tissue diagnosis. Hum Pathol. 1984;15(11):1085–1089. doi: 10.1016/s0046-8177(84)80253-1. [DOI] [PubMed] [Google Scholar]
- 24.Lipsky B.A., Gates J., Tenover F.C., Plorde J.J. Factors affecting the clinical value of microscopy for acid-fast bacilli. Rev Infect Dis. 1984;6(2):214–222. doi: 10.1093/clinids/6.2.214. [DOI] [PubMed] [Google Scholar]
- 25.Ipuge Y.A., Rieder H.L., Enarson D.A. The yield of acid-fast bacilli from serial smears in routine microscopy laboratories in rural Tanzania. Trans R Soc Trop Med Hyg. 1996;90(3):258–261. doi: 10.1016/s0035-9203(96)90239-4. [DOI] [PubMed] [Google Scholar]
- 26.Nelson S.M., Deike M.A., Cartwright C.P. Value of examining multiple sputum specimens in the diagnosis of pulmonary tuberculosis. J Clin Microbiol. 1998;36(2):467–469. doi: 10.1128/jcm.36.2.467-469.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker D., McNerney R., Mwembo M.K., Foster S., Tihon V., Godfrey-Faussett P. An incremental cost-effectiveness analysis of the first, second and third sputum examination in the diagnosis of pulmonary tuberculosis. Int J Tuberc Lung Dis. 2000;4(3):246–251. [PubMed] [Google Scholar]
- 28.Mathew P., Kuo Y.H., Vazirani B., Eng R.H., Weinstein M.P. Are three sputum acid-fast bacillus smears necessary for discontinuing tuberculosis isolation? J Clin Microbiol. 2002;40(9):3482–3484. doi: 10.1128/JCM.40.9.3482-3484.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilmer A., Bryce E., Grant J. The role of the third acid-fast bacillus smear in tuberculosis screening for infection control purposes: A controversial topic revisited. Can J Infect Dis Med Microbiol. 2011;22(1):e1–e3. doi: 10.1155/2011/314686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khogali M., Tayler-Smith K., Zachariah R. Diagnosis of pulmonary tuberculosis in a pastoralist population in Ethiopia: are three sputum specimens needed? Trop Med Int Health. 2013;18(5):632–635. doi: 10.1111/tmi.12082. [DOI] [PubMed] [Google Scholar]
- 31.Rehman S., Iqbal R., Munir M.K., Salam A.A., Saeed S. Incremental yield of submitting three sputum specimens for the diagnosis of pulmonary tuberculosis. Pak J Med Res. 2013;52:35–38. [Google Scholar]
- 32.Hassan W.A., Omar A., Khalil S., Al-Qarn A.F. Value of repeated direct smear sputum examination in the diagnosis of pulmonary tuberculosis. Open J Respir Dis. 2014;4:41–47. [Google Scholar]
- 33.Mase S.R., Ramsay A., Ng V., Henry M., Hopewell P.C., Cunningham J. Yield of serial sputum specimen examinations in the diagnosis of pulmonary tuberculosis: a systematic review. Int J Tuberc Lung Dis. 2007;11(5):485–495. [PubMed] [Google Scholar]
- 34.World Health Organization . World Health Organization; Geneva, Switzerland: 2007. Reduction of number of smears for diagnosis of pulmonary tuberculosis. http://www.who.int/tb/laboratory/policy_diagnosis_pulmonary_tb/en/ [Google Scholar]
- 35.Taylor Z., Nolan C.M., Blumberg H.M. Controlling tuberculosis in the United States. MMWR Recomm Rep. 2005;54(RR-12):1–81. Recommendations from the American Thoracic Society, CDC, and the Infectious Diseases Society of America. [PubMed] [Google Scholar]
- 36.Diagnostic Standards and Classification of Tuberculosis in Adults and Children Am J Respir Crit Care Med. 2000;161(4 Pt 1):1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- 37.Pfyffer G.E., Welscher H.M., Kissling P., Cieslak C., Casal M.J., Gutierrez J. Comparison of the Mycobacteria Growth Indicator Tube (MGIT) with radiometric and solid culture for recovery of acid-fast bacilli. J Clin Microbiol. 1997;35(2):364–368. doi: 10.1128/jcm.35.2.364-368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang T.S., Chen C.S., Lee S.S., Huang W.K., Liu Y.C. Comparison of the BACTEC MGIT 960 and BACTEC 460 TB systems for detection of mycobacteria in clinical specimens. Ann Clin Lab Sci. 2001;31(3):279–283. [PubMed] [Google Scholar]
- 39.Lee J.J., Suo J., Lin C.B., Wang J.D., Lin T.Y., Tsai Y.C. Comparative evaluation of the BACTEC MGIT 960 system with solid medium for isolation of mycobacteria. Int J Tuberc Lung Dis. 2003;7(6):569–574. [PubMed] [Google Scholar]
- 40.Cruciani M., Scarparo C., Malena M., Bosco O., Serpelloni G., Mengoli C. Meta-analysis of BACTEC MGIT 960 and BACTEC 460 TB, with or without solid media, for detection of mycobacteria. J Clin Microbiol. 2004;42(5):2321–2325. doi: 10.1128/JCM.42.5.2321-2325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent P.T., Kubica G.P. U.S. Dept. of Health and Human Services, Public Health Service, Centers for Disease Control. GA; Atlanta: 1985. Public health microbiology: a guide for the level iii laboratory. [Google Scholar]
- 42.Realini L., De Ridder K., Hirschel B., Portaels F. Blood and charcoal added to acidified agar media promote the growth of Mycobacterium genavense. Diagnostic Microbiology and Infectious Disease. 1999;34(1):45–50. doi: 10.1016/s0732-8893(99)00014-0. [DOI] [PubMed] [Google Scholar]
- 43.Robbe-Austerman S., Bravo D.M., Harris B. Comparison of the MGIT 960, BACTEC 460 TB and solid media for isolation of Mycobacterium bovis in United States veterinary specimens. BMC Vet Res. 2013;9:74. doi: 10.1186/1746-6148-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simner P.J., Doerr K.A., Steinmetz L.K., Wengenack N.L. Mycobacterium and aerobic actinomycete culture: are two medium types and extended incubation times necessary? J Clin Microbiol. 2016;54(4):1089–1093. doi: 10.1128/JCM.02838-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreira Ada S., Huf G., Vieira M.A., Costa P.A., Aguiar F., Marsico A.G. Liquid vs solid culture medium to evaluate proportion and time to change in management of suspects of tuberculosis-a pragmatic randomized trial in secondary and tertiary health care units in Brazil. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0127588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chihota V.N., Grant A.D., Fielding K., Ndibongo B., van Zyl A., Muirhead D. Liquid vs. solid culture for tuberculosis: performance and cost in a resource-constrained setting. Int J Tuberc Lung Dis. 2010;14(8):1024–1031. [PubMed] [Google Scholar]
- 47.Somoskovi A., Magyar P. Comparison of the mycobacteria growth indicator tube with MB redox, Lowenstein-Jensen, and Middlebrook 7H11 media for recovery of mycobacteria in clinical specimens. J Clin Microbiol. 1999;37(5):1366–1369. doi: 10.1128/jcm.37.5.1366-1369.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu D., Heeren B., Dunne W.M. Comparison of the automated Mycobacteria Growth Indicator Tube System (BACTEC 960/MGIT) with Lowenstein-Jensen medium for recovery of mycobacteria from clinical specimens. Am J Clin Pathol. 2002;118(4):542–545. doi: 10.1309/65KN-2M7E-7MNN-X0TA. [DOI] [PubMed] [Google Scholar]
- 49.Srisuwanvilai L.O., Monkongdee P., Podewils L.J., Ngamlert K., Pobkeeree V., Puripokai P. Performance of the BACTEC MGIT 960 compared with solid media for detection of Mycobacterium in Bangkok, Thailand. Diagn Microbiol Infect Dis. 2008;61(4):402–407. doi: 10.1016/j.diagmicrobio.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Hasan M., Munshi S.K., Momi S.B., Rahman F., Noor R. Evaluation of the effectiveness of BACTEC MGIT 960 for the detection of mycobacteria in Bangladesh. Int J Mycobact. 2013;2:214–219. doi: 10.1016/j.ijmyco.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Whyte T., Hanahoe B., Collins T., Corbett-Feeney G., Cormican M. Evaluation of the BACTEC MGIT 960 and MB BAC/T systems for routine detection of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38(8):3131–3132. doi: 10.1128/jcm.38.8.3131-3132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gravet A., Souillard N., Habermacher J., Moser A., Lohmann C., Schmitt F. Culture and susceptibility testing of mycobacteria with VersaTREK. Pathol Biol (Paris) 2011;59(1):32–38. doi: 10.1016/j.patbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Falconi F.Q., Suarez L.I., Lopez Mde J., Sancho C.G. Comparison of the VersaTREK system and Lowenstein-Jensen medium for the recovery of mycobacteria from clinical specimens. Scand J Infect Dis. 2008;40(1):49–53. doi: 10.1080/00365540701522967. [DOI] [PubMed] [Google Scholar]
- 54.Fuller D.D., Davis T.E., Jr., Denys G.A., York M.K. Evaluation of BACTEC MYCO/F Lytic medium for recovery of mycobacteria, fungi, and bacteria from blood. J Clin Microbiol. 2001;39(8):2933–2936. doi: 10.1128/JCM.39.8.2933-2936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vetter E., Torgerson C., Feuker A., Hughes J., Harmsen S., Schleck C. Comparison of the BACTEC MYCO/F Lytic bottle to the isolator tube, BACTEC Plus Aerobic F/bottle, and BACTEC Anaerobic Lytic/10 bottle and comparison of the BACTEC Plus Aerobic F/bottle to the Isolator tube for recovery of bacteria, mycobacteria, and fungi from blood. J Clin Microbiol. 2001;39(12):4380–4386. doi: 10.1128/JCM.39.12.4380-4386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crump J.A., Morrissey A.B., Ramadhani H.O., Njau B.N., Maro V.P., Reller L.B. Controlled comparison of BacT/Alert MB system, manual Myco/F lytic procedure, and isolator 10 system for diagnosis of Mycobacterium tuberculosis bacteremia. J Clin Microbiol. 2011;49(8):3054–3057. doi: 10.1128/JCM.01035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simner P.J., Stenger S., Richter E., Brown-Elliot B.A., Wallace R.J., Jr, Wengenack N.L. 11th ed. ASM Press; Washington, DC: 2015. Mycobacterium: laboratory characteristics of slowly growing mycobacteria. Manual of Clinical Microbiology. [Google Scholar]
- 58.Lebrun L., Espinasse F., Poveda J.D., Vincent-Levy-Frebault V. Evaluation of nonradioactive DNA probes for identification of mycobacteria. J Clin Microbiol. 1992;30(9):2476–2478. doi: 10.1128/jcm.30.9.2476-2478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bull T.J., Shanson D.C. Evaluation of a commercial chemiluminescent gene probe system ‘AccuProbe’ for the rapid differentiation of mycobacteria, including ‘MAIC X’, isolated from blood and other sites, from patients with AIDS. J Hosp Infect. 1992;21(2):143–149. doi: 10.1016/0195-6701(92)90034-j. [DOI] [PubMed] [Google Scholar]
- 60.Tortoli E., Pecorari M., Fabio G., Messino M., Fabio A. Commercial DNA probes for mycobacteria incorrectly identify a number of less frequently encountered species. J Clin Microbiol. 2010;48(1):307–310. doi: 10.1128/JCM.01536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christiansen D.C., Roberts G.D., Patel R. Mycobacterium celatum, an emerging pathogen and cause of false positive amplified Mycobacterium tuberculosis direct test. Diagn Microbiol Infect Dis. 2004;49(1):19–24. doi: 10.1016/j.diagmicrobio.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Russo C., Tortoli E., Menichella D. Evaluation of the new GenoType Mycobacterium assay for identification of mycobacterial species. J Clin Microbiol. 2006;44(2):334–339. doi: 10.1128/JCM.44.2.334-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tortoli E, Mariottini A, Mazzarelli G. Evaluation of INNO-LiPA MYCOBACTERIA v2: improved reverse hybridization multiple DNA probe assay for mycobacterial identification. J Clin Microbiol. 2003;41(9):4418–4420. doi: 10.1128/JCM.41.9.4418-4420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Machen A., Kobayashi M., Connelly M.R., Wang Y.F. Comparison of heat inactivation and cell disruption protocols for identification of mycobacteria from solid culture media by use of vitek matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(12):4226–4229. doi: 10.1128/JCM.02612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clark R.B., Lewinski M.A., Loeffelholz M.J., Tibbetts R.J. Washington, DC; ASM Press: 2009. Cumitech 31A: verification and validation of procedures in the clinical microbiology laboratory. [Google Scholar]
- 66.Lotz A., Ferroni A., Beretti J.L., Dauphin B., Carbonnelle E., Guet-Revillet H. Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48(12):4481–4486. doi: 10.1128/JCM.01397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saleeb P.G., Drake S.K., Murray P.R., Zelazny A.M. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49(5):1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.El Khechine A., Couderc C., Flaudrops C., Raoult D., Drancourt M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One. 2011;6(9):e24720. doi: 10.1371/journal.pone.0024720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mather C.A., Rivera S.F., Butler-Wu S.M. Comparison of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of mycobacteria using simplified protein extraction protocols. J Clin Microbiol. 2014;52(1):130–138. doi: 10.1128/JCM.01996-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buckwalter S.P., Olson S.L., Connelly B.J., Lucas B.C., Rodning A.A., Walchak R.C. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Mycobacterium species, Nocardia species, and other aerobic actinomycetes. J Clin Microbiol. 2016;54(2):376–384. doi: 10.1128/JCM.02128-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dhiman N., Hall L., Wohlfiel S.L., Buckwalter S.P., Wengenack N.L. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol. 2011;49(4):1614–1616. doi: 10.1128/JCM.02381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balada-Llasat J.M., Kamboj K., Pancholi P. Identification of mycobacteria from solid and liquid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry in the clinical laboratory. J Clin Microbiol. 2013;51(9):2875–2879. doi: 10.1128/JCM.00819-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turenne C.Y., Tschetter L., Wolfe J., Kabani A. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol. 2001;39(10):3637–3648. doi: 10.1128/JCM.39.10.3637-3648.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Centers for Disease Control and Prevention (CDC) Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb Mortal Wkly Rep. 2009;58(1):7–10. [PubMed] [Google Scholar]
- 75.Centers for Disease Control and Prevention (CDC) MMWR Morb Mortal Wkly Rep. 2013;62(41):821–827. [PMC free article] [PubMed] [Google Scholar]
- 76.Piersimoni C., Scarparo C. Relevance of commercial amplification methods for direct detection of Mycobacterium tuberculosis complex in clinical samples. J Clin Microbiol. 2003;41(12):5355–5365. doi: 10.1128/JCM.41.12.5355-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tortoli E., Russo C., Piersimoni C., Mazzola E., Dal Monte P., Pascarella M. Clinical validation of Xpert MTB/RIF for the diagnosis of extrapulmonary tuberculosis. Eur Respir J. 2012;40(2):442–447. doi: 10.1183/09031936.00176311. [DOI] [PubMed] [Google Scholar]
- 78.Chang K., Lu W., Wang J., Zhang K., Jia S., Li F. Rapid and effective diagnosis of tuberculosis and rifampicin resistance with Xpert MTB/RIF assay: a meta-analysis. J Infect. 2012;64(6):580–588. doi: 10.1016/j.jinf.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 79.Food and Drug Administration (FDA). New data shows test can help physicians remove patients with suspected TB from isolation earlier. Press Release. 2015 Feb 12; http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm434226.htm. [accessed 15.02.15.
- 80.Williamson D.A., Basu I., Bower J., Freeman J.T., Henderson G., Roberts S.A. An evaluation of the Xpert MTB/RIF assay and detection of false-positive rifampicin resistance in Mycobacterium tuberculosis. Diagn Microbiol Infect Dis. 2012;74(2):207–209. doi: 10.1016/j.diagmicrobio.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 81.Flores L.L., Pai M., J.M. Colford, Jr, Riley L.W. In-house nucleic acid amplification tests for the detection of Mycobacterium tuberculosis in sputum specimens: meta-analysis and meta-regression. BMC Microbiol. 2005;5:55. doi: 10.1186/1471-2180-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daum L.T., Rodriguez J.D., Worthy S.A., Ismail N.A., Omar S.V., Dreyer A.W. Next-generation ion torrent sequencing of drug resistance mutations in Mycobacterium tuberculosis strains. J Clin Microbiol. 2012;50(12):3831–3837. doi: 10.1128/JCM.01893-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walker T.M., Kohl T.A., Omar S.V., Hedge J., Del Ojo Elias C., Bradley P. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;15(10):1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lawn S.D., Kerkhoff A.D., Vogt M., Wood R. Diagnostic accuracy of a low-cost, urine antigen, point-of-care screening assay for HIV-associated pulmonary tuberculosis before antiretroviral therapy: a descriptive study. Lancet Infect Dis. 2012;12(3):201–209. doi: 10.1016/S1473-3099(11)70251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shah M., Ssengooba W., Armstrong D., Nakiyingi L., Holshouser M., Ellner J.J. Comparative performance of urinary lipoarabinomannan assays and Xpert MTB/RIF in HIV-infected individuals. Aids. 2014;28(9):1307–1314. doi: 10.1097/QAD.0000000000000264. [DOI] [PMC free article] [PubMed] [Google Scholar]