Summary
Bacteriophage possess a variety of auxiliary metabolic genes of bacterial origin. These proteins enable them to maximize infection efficiency, subverting bacterial metabolic processes for the purpose of viral genome replication and synthesis of the next generation of virion progeny. Here, we examined the enzymatic activity of a cyanophage MazG protein – a putative pyrophosphohydrolase previously implicated in regulation of the stringent response via reducing levels of the central alarmone molecule (p)ppGpp. We demonstrate, however, that the purified viral MazG shows no binding or hydrolysis activity against (p)ppGpp. Instead, dGTP and dCTP appear to be the preferred substrates of this protein, consistent with a role preferentially hydrolysing deoxyribonucleotides from the high GC content host Synechococcus genome. This showcases a new example of the fine‐tuned nature of viral metabolic processes.
Introduction
Cyanophage that infect the marine cyanobacterial genera Synechococcus and Prochlorococcus are widespread and abundant in oceanic systems (Suttle and Chan, 1994; Sullivan et al., 2003; Baran et al., 2018) where they play important ecosystem roles including releasing organic matter through cell lysis (Suttle, 2007), transferring genes horizontally between hosts (Zeidner et al., 2005) and structuring host communities (Mühling et al., 2005). Cyanophage can also influence ocean biogeochemistry by modifying host metabolism during the infection process, such as the shutdown of CO2 fixation whilst maintaining photosynthetic electron transport (Puxty et al., 2016). This subversion of host metabolism is facilitated by the expression of cyanophage genes that appear to have a bacterial origin, so‐called auxiliary metabolic genes (AMGs) (Breitbart et al., 2007). These include genes involved in photosynthesis (Mann et al., 2003; Lindell et al., 2005; Fridman et al., 2017) and photoprotection (Lindell et al., 2004; Millard et al., 2004; Sullivan et al., 2005; Roitman et al., 2018), pigment biosynthesis (Dammeyer et al., 2008), central carbon metabolism (Millard et al., 2009; Thompson et al., 2011), nucleotide biosynthesis (Enav et al., 2014), phosphorus metabolism (Sullivan et al., 2010; Zeng and Chisholm, 2012; Lin et al., 2016) and other stress responses (Sullivan et al., 2010; Crummett et al., 2016).
Amongst the cyanophage AMGs MazG is a core gene in cyanomyoviruses (Millard et al., 2009; Sullivan et al., 2010) and of particular interest since it has been proposed to play a more general role in regulating host metabolism (Clokie and Mann, 2006; Clokie et al., 2010). In Escherichia coli, MazG has been implicated in regulating programmed cell death by interfering with the function of the MazEF toxin‐antitoxin system, through lowering of cellular (p)ppGpp levels (Gross et al., 2006). This latter molecule guanosine 3′,5′ bispyrophosphate, together with guanosine pentaphosphate also known as magic spot nucleotides, is a global regulator of gene expression in bacteria (Traxler et al., 2008) synthesized by RelA under amino acid starvation. Since MazG can potentially regulate levels of (p)ppGpp in E. coli, a similar role has been proposed for the cyanophage encoded MazG (Clokie and Mann, 2006). This is pertinent given that picocyanobacterial hosts like Synechococcus and Prochlorococcus occupy oligotrophic conditions (see Scanlan et al., 2009; Biller et al., 2015) where nutrient starvation is likely and (p)ppGpp may be involved in adapting to this stressed state. By regulating (p)ppGpp levels the cyanophage encoded MazG may trick the host into mimicking a nutrient replete cellular state so that host cell physiology is optimized for macromolecular synthesis and hence cyanophage replication. The MazG protein belongs to the all‐nucleoside triphosphate pyrophosphohydrolase (NTP‐PPase, EC 3.6.1.8) superfamily that hydrolyzes in vitro all canonical nucleoside triphosphates into monophosphate derivatives and pyrophosphate (PPi) (Moroz et al., 2005; Galperin et al., 2006; Lu et al., 2010). Here, we set out to purify the cyanophage S‐PM2 MazG protein as well as a Synechococcus host MazG to assess their activity and ability to hydrolyse (p)ppGpp, canonical and noncanonical nucleotides.
Results
Picocyanobacterial host and cyanophage MazG proteins are phylogenetically distinct (Fig. 1) and with an origin of the cyanophage MazG outside the cyanobacteria since the closest proposed homologue to date is a Chloroflexus protein (Bryan et al., 2008; Sullivan et al., 2010). Picocyanobacteria encode two genes annotated as MazG, a ‘large’ MazG version similar to that found in most bacteria, and a ‘small’ version which is similar in size to the cyanophage gene (Fig. 2). The ‘large’ MazG version has two predicted catalytic regions functionally annotated as MazG family domains (IPR004518) whilst the ‘small’ MazG and cyanophage proteins have only one (Fig. 2). In order to assess the hydrolytic activity of the host and cyanophage MazG proteins we cloned into E. coli, over‐expressed and purified the host Synechococcus sp. WH7803 MazG, using the ‘large’ MazG version (Syn_WH7803_02449) as a proxy for other host bacterial MazG proteins, and the cyanophage S‐PM2 MazG (Fig. 3; for experimental details see Supporting Information). The activity of the cyanophage and Synechococcus host MazG proteins was assessed using increasing concentrations of a range of nucleotide and deoxyribonucleotide substrates using 1 μg of the purified protein, and the amount of free phosphate resulting from enzyme activity measured using the PiPER pyrophosphate assay kit (ThermoFisher Scientific; see Supporting Information). This allowed determination of K m, V max and K cat values for each protein across a range of substrates (Table 1). K m values of the Synechococccus sp. WH7803 ‘large’ MazG and cyanophage S‐PM2 MazG proteins were generally in the low mM range for a range of nucleotides and deoxyribonucleotides, similar to MazG K m values reported from other bacteria for these substrates (Lu et al., 2010). The measured V max of the Synechococcus host MazG was highest when incubated with dTTP, whilst the viral MazG exhibited highest activity when incubated with the deoxyribonucleotides dGTP and dCTP (Fig. 4). In addition to these standard nucleotides, the viral MazG protein was also incubated with the ‘aberrant’ nucleotides dUTP, 2‐hydroxy‐dATP and 8‐oxo‐dGTP. dUTP is one of the most common of these mutagenic nucleotides, produced as a by‐product of thymine biosynthesis (Galperin et al., 2006), whilst 2‐hydroxy‐dATP and 8‐oxo‐dGTP are mutagenic nucleotides produced as a result of intracellular oxidative stress (Kamiya and Kasai, 2000; Galperin et al., 2006). Interestingly, the V max values of the viral MazG when incubated with dUTP, 2‐hydroxy‐dATP and 8‐oxo‐dGTP were not significantly different to those of the canonical nucleotides (Table 1; Fig. 4), whilst the Km values for these substrates were higher (Table 1), suggesting that dGTP and dCTP are the preferred substrates of the cyanophage MazG protein.
Figure 1.
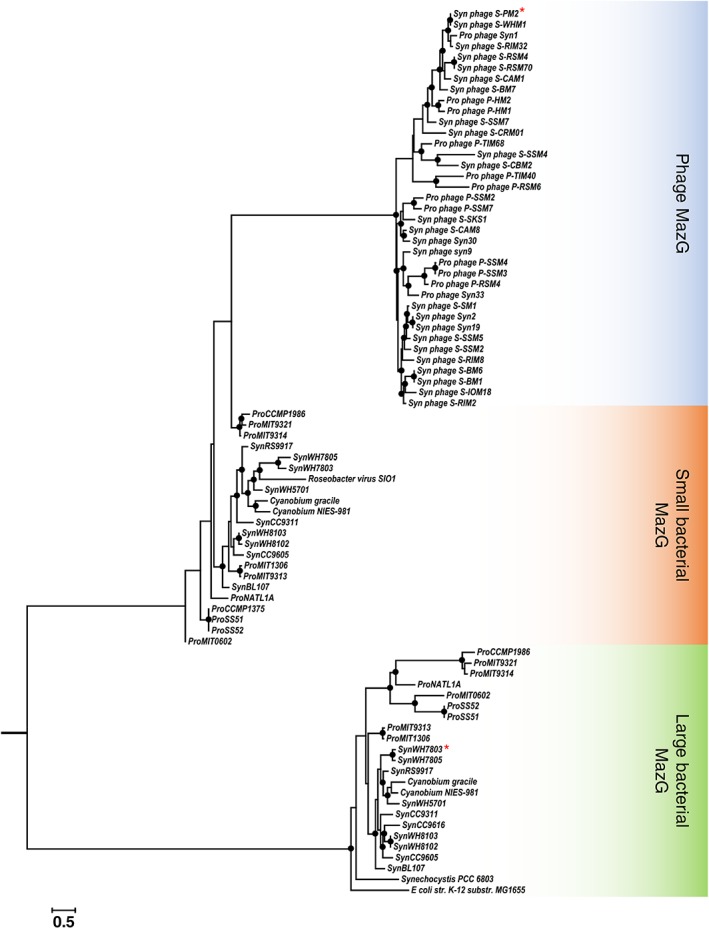
Maximum likelihood phylogenetic tree comprising 44 bacterial and 38 viral MazG sequences.
The tree was generated using the LG + G4 substitution model, automatically chosen by the Iqtree script (Nguyen et al., 2015), with ultrafast bootstrap (Minh et al., 2013). Bootstrap values of >70% are shown as closed circles (of 1000 iterations). The scale bar represents 0.5 substitutions/amino acid position. Syn: Synechococcus; Pro: Prochlorococcus. The red asterisks indicate the Synechococcus and cyanophage proteins used here.
Figure 2.

A. InterProScan5‐predicted (Jones et al., 2014) pyrophosphatase catalytic domains in cyanophage S‐PM2 MazG, ‘small’ Synechococcus sp. WH7803 MazG (Syn_WH7803_01219), ‘large’ Synechococcus sp. WH7803 MazG (Syn_WH7803_02449) and E. coli MazG orthologues. Numbers above each domain represent the position of amino acids in each of the domains.
B. ClustalW pairwise alignment of E. coli, ‘large’ Synechococcus sp. WH7803, ‘small’ Synechococcus sp. WH7803 and cyanophage S‐PM2 MazG orthologues.
Figure 3.
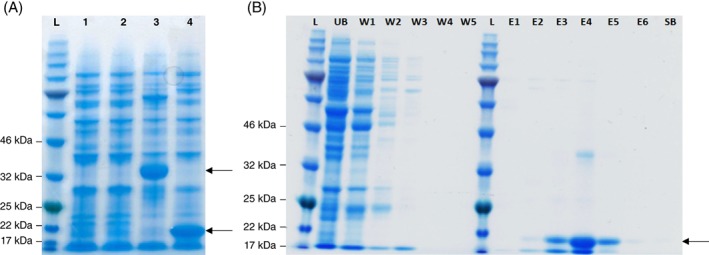
A. SDS‐PAGE analysis of E. coli whole cell lysates expressing Synechococcus sp. WH7803 ‘large’ MazG (lanes 1 and 3) and cyanophage S‐PM2 MazG proteins (lanes 2 and 4). L: Protein molecular weight marker ladder. Lanes 1 and 2 un‐induced, lanes 3 and 4 IPTG‐induced. Arrows indicate the positions of the overexpressed proteins.
B. SDS‐PAGE analysis showing purification of the cyanophage S‐PM2 MazG protein from E. coli. L: Protein molecular weight marker ladder. UB: The unbound fraction (proteins that did not bind to the column). W1–W5: fractions washed off the column with binding buffer. E1–E6: Fractions eluted with increasing concentrations of imidazole (30 mM, 50 mM, 100 mM, 150 mM, 200 mM and 300 mM respectively). SB – stripping buffer. The arrow indicates the position of the over‐expressed cyanophage S‐PM2 MazG protein.
Table 1.
Kinetic parameters of enzymatic activity of Synechococcus WH7803 and cyanophage S‐PM2 MazG protein.
| V max (nmol/μg/min) | K m (mM) | K cat (min−1) | ||||
|---|---|---|---|---|---|---|
| Synechococcus sp. WH7803 | Cyanophage S‐PM2 | Synechococcus sp. WH7803 | Cyanophage S‐PM2 | Synechococcus sp. WH7803 | Cyanophage S‐PM2 | |
| dATP | 1.8 (±0.28) | 1.62 (±0.19) | 0.3 (±0.09) | 1.2 (±0.21) | 126.12 (±19.35) | 62.97 (±7.44) |
| dCTP | 3.81 (±0.36) | 8.86 (±0.2) | 0.14 (±0.03) | 1.16 (±0.04) | 267.68 (±25.02) | 344.68 (±7.72) |
| dTTP | 6.57 (±0.19) | 5.68 (±0.2) | ND | 1.23 (±0.06) | 461.04 (±13.43) | 221.00 (±7.78) |
| dGTP | 0.64 (±0.25) | 10.29 (±0.25) | 0.85 (±0.07) | 0.14 (±0.01) | 45.16 (±17.6) | 400.35 (±9.91) |
| ATP | 2.55 (±0.35) | 2.28 (±0.24) | 0.63 (±0.23) | 1.43 (±0.36) | 179.27 (±24.4) | 88.7 (±9.41) |
| CTP | 1.96 (±0.14) | 2.51 (±0.17) | 1.2 (±0.21) | 0.85 (±0.11) | 137.81 (±9.81) | 97.48 (±6.68) |
| GTP | 0.7 (±0.13) | 0.3 (±0.02) | 0.26 (±0.02) | ND | 49.46 (±9.19) | 11.67 (±0.6) |
| UTP | 3.02 (±0.2) | 3.31 (±0.18) | 1.33 (±0.3) | 0.6 (±0.37) | 221.07 (±6) | 128.75 (±7.12) |
| dUTP | ‐ | 4.22 (±0.34) | ‐ | 3.24 (±1.55) | ‐ | 296.42 (±23.72) |
| 2‐hydroxy d‐ATP | ‐ | 1.65 (±0.06) | ‐ | 4.86 (±1.13) | ‐ | 115.65 (±3.95) |
| 8‐oxo‐dGTP | ‐ | ND | ‐ | ND | ‐ | ND |
The values in brackets represent SE based on three replicates. ND – not detected; − not measured.
Figure 4.
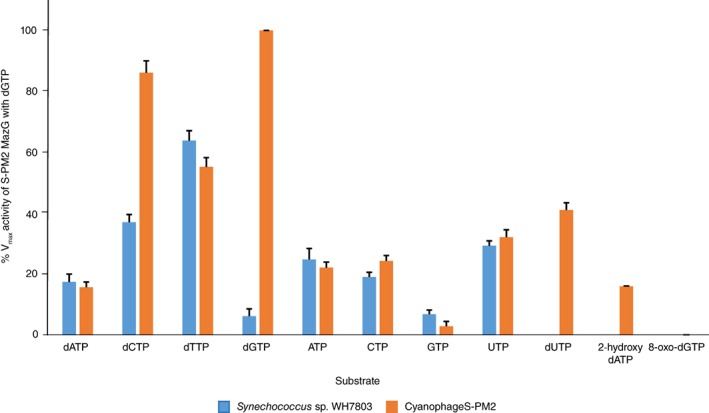
Relative maximal activity (Vmax) of the Synechococcus sp. WH7803 ‘large’ MazG and cyanophage S‐PM2 MazG proteins against a range of canonical and noncanonical nucleotide and deoxyribonucleotide substrates, normalized to the activity of the cyanophage S‐PM2 MazG using dGTP as a substrate.
Error bars represent the standard error based on three replicate experiments.
In order to directly assess whether the Synechococcus and cyanophage MazG proteins play a role in (p)ppGpp metabolism we performed both hydrolysis and DRaCALA binding assays (Corrigan et al., 2016), using 32P‐labelled GTP, ppGpp and pppGpp. In both assays, neither the Synechococcus nor cyanophage MazG showed any binding or hydrolysis activity against (p)ppGpp (Fig. 5A), whilst hydrolysis activity was confirmed for both orthologues against 32P‐labelled GTP (Fig. 5B).
Figure 5.
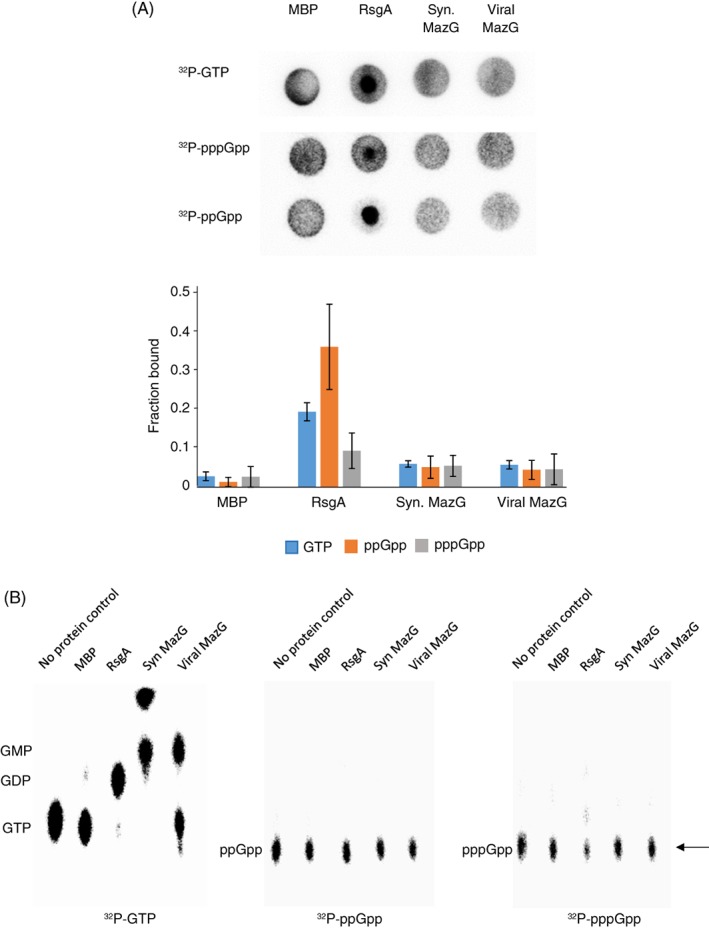
A. Upper panel: DRaCALA binding assays, using 32P‐labelled GTP, ppGpp and pppGpp incubated with purified Synechococcus sp. WH7803 ‘large’ MazG and cyanophage S‐PM2 MazG proteins. MBP – maltose binding protein, used as a negative control. RsgA –purified RsgA protein from S. aureus, used as a positive control. Syn MazG: Synechococcus sp. WH7803 ‘large’ MazG. Viral MazG: cyanophage S‐PM2 MazG. Lower panel: Bar chart representation of the fraction of substrate bound to each protein, as measured by densitometry. Syn. MazG: Synechococcus sp. WH7803 ‘large’ MazG. Viral MazG: cyanophage S‐PM2 MazG. Error bars represent the standard deviation of three experimental replicates.
B. Hydrolysis assay using purified Synechococcus sp. WH7803 ‘large’ MazG (Syn MazG), cyanophage S‐PM2 MazG (Viral MazG), MBP and RsgA proteins with 32P‐labelled GTP, ppGpp and pppGpp. The arrow highlights the absence of hydrolysis of 32P‐labelled ppGpp and pppGpp substrates.
Discussion
Although, the presence and identity of AMGs in bacteriophage genomes is widely appreciated (Millard et al., 2009; Sullivan et al., 2010; Crummett et al., 2016) the specific role of many of these genes has not been resolved. Here, we sought to elucidate the activity of the cyanophage MazG protein given its hypothesized role as a more general modulator of the host stringent response, and with previous data suggesting cyanophage can modulate intracellular levels of (p)ppGpp in infected freshwater cyanobacteria (Borbély et al., 1980).
Our results showed, however, that neither the Synechococcus nor cyanophage MazG protein demonstrated detectable hydrolytic activity towards ppGpp or pppGpp (Fig. 5), suggesting these two proteins do not actively modulate the stringent response via direct hydrolysis of magic spot nucleotides. Nevertheless, we cannot rule out a role for these proteins in regulating the stringent response indirectly through hydrolysis of other nucleotide substrates, for example GTP. Whilst the role of the ‘small’ Synechococcus host MazG also requires clarification in this respect, it is potentially the predicted bifunctional Synechococcus sp. WH7803 SpoT orthologue (SynWH7803_2342) that serves the role of regulating alarmone levels during the stringent response in these organisms, a protein known to both synthesize and hydrolyse (p)ppGpp in other bacteria (see, e.g. Murray and Bremer, 1996; Hogg et al., 2004). Interestingly, there were distinct differences in the hydrolytic activities of the Synechococcus host and cyanophage S‐PM2 MazG proteins towards other canonical and noncanonical nucleotides (Fig. 4 and Table 1) with much higher V max values of the viral MazG towards dGTP and dCTP coupled with a much higher affinity of the viral MazG for dGTP compared to its host counterpart. Such different kinetic parameters mirror differences in %GC content between the cyanophage and Synechococcus host genomes, with the former possessing a GC content of 37.7% (Mann et al., 2005) and the latter a GC content of 60.2% (Dufresne et al., 2008). With this in mind, we suggest that the substrate specificity of the viral MazG allows it to preferentially hydrolyse dGTP and dCTP deoxyribonucleotides from the high GC content host Synechococcus genome allowing for their recycling and ultimately facilitating replication of the AT‐rich cyanophage genome. Whether such a mechanism is applicable to, or modified in, Prochlorococcus infecting cyanophage whose genomes generally possess a similar %GC content (Sullivan et al., 2005; Limor‐Waisberg et al., 2011) remains to be determined. Certainly, it is well known that following infection with cyanophage, the host genome is rapidly degraded (Doron et al., 2016). Moreover, analysis of viral metagenomes has shown an enrichment of metabolic pathways involved in pyrimidine and purine metabolism as well as in DNA replication (Enav et al., 2014), emphasizing the importance of these pathways during viral infection.
Our work with the viral MazG thus highlights that cyanophage genomes appear exquisitely suited to promote degradation of the host genome in order to reuse its building blocks to replicate the viral genome.
Supporting information
Appendix S1: Supplementary Information
Acknowledgements
B. R. was in receipt of a Chancellor's International PhD Scholarship from the University of Warwick. R.M.C. was supported by funding from the Wellcome Trust and Royal Society grant 104110. This work was also supported by the Natural Environment Research Council through Research Grants NE/J02273X/1 and NE/N003241/1. Bioinformatics analysis was carried out using MRC CLIMB Infrastructure (grant MR/L015080/1).
References
- Baran, N. , Goldin, S. , Maidanik, I. , and Lindell, D. (2018) Quantification of diverse virus populations in the environment using a polony method. Nat Microbiol 3: 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller, S.J. , Berube, P.M. , Lindell, D. , and Chisholm, S.W. (2015) Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 13: 13–27. [DOI] [PubMed] [Google Scholar]
- Borbély, G. , Kari, C. , Gulyás, A. , and Farkas, G.L. (1980) Bacteriophage infection interferes with guanosine 3′‐diphosphate‐5′‐diphosphate accumulation induced by energy and nitrogen starvation in the cyanobacterium Anacystis nidulans . J Bacteriol 144: 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart, M. , Thompson, L.R. , Suttle, C.A. , and Sullivan, M.B. (2007) Exploring the vast diversity of marine viruses. Oceanography 20: 135–139. [Google Scholar]
- Bryan, M.J. , Burroughs, N.J. , Spence, E.M. , Clokie, M.R.J. , Mann, N.H. , and Bryan, S.J. (2008) Evidence for the intense exchange of MazG in marine cyanophages by horizontal gene transfer. PLoS One 3: e2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie, M.R. , and Mann, N.H. (2006) Marine cyanophages and light. Environ Microbiol 8: 2074–2082. [DOI] [PubMed] [Google Scholar]
- Clokie, M.R. , Millard, A.D. , and Mann, N.H. (2010) T4 genes in the marine ecosystem: studies of the T4‐like cyanophages and their role in marine ecology. Virology J 7: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan, R.M. , Bellows, L.E. , Wood, A. , and Gründling, A. (2016) ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram‐positive bacteria. Proc Natl Acad Sci USA 113: E1710–E1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crummett, L.T. , Puxty, R.J. , Weihe, C. , Marston, M.F. , and Martiny, J.B. (2016) The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 499: 219–229. [DOI] [PubMed] [Google Scholar]
- Dammeyer, T. , Bagby, S.C. , Sullivan, M.B. , Chisholm, S.W. , and Frankenberg‐Dinkel, N. (2008) Efficient phage mediated pigment biosynthesis in oceanic cyanobacteria. Curr Biol 18: 442–448. [DOI] [PubMed] [Google Scholar]
- Doron, S. , Fedida, A. , Hernández‐Prieto, M.A. , Sabehi, G. , Karunker, I. , Stazic, D. , et al (2016) Transcriptome dynamics of a broad host‐range cyanophage and its hosts. ISME J 10: 1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne, A. , Ostrowski, M. , Scanlan, D.J. , Garczarek, L. , Mazard, S. , Palenik, B.P. , et al (2008) Unravelling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol 9: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enav, H. , Mandel‐Gutfreund, Y. , and Béjà, O. (2014) Comparative metagenomic analyses reveal viral‐induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, S. , Flores‐Uribe, J. , Larom, S. , Alalouf, O. , Liran, O. , Yacoby, I. , et al (2017) A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat Microbiol 2: 1350–1357. [DOI] [PubMed] [Google Scholar]
- Galperin, M.Y. , Moroz, O.V. , Wilson, K.S. , and Murzin, A.G. (2006) House cleaning, a part of good housekeeping. Mol Microbiol 59: 5–19. [DOI] [PubMed] [Google Scholar]
- Gross, M. , Marianovsky, I. , and Glaser, G. (2006) MazG—a regulator of programmed cell death in Escherichia coli . Mol Microbiol 59: 590–601. [DOI] [PubMed] [Google Scholar]
- Hogg, T. , Mechold, U. , Malke, H. , Cashel, M. , and Hilgenfeld, R. (2004) Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homologue modulates (p)ppGpp metabolism during the stringent response. Cell 117: 57–68. [DOI] [PubMed] [Google Scholar]
- Jones, P. , Binns, D. , Chang, H.‐Y. , Fraser, M. , Li, W. , McAnulla, C. , et al (2014) InterProScan 5: genome‐scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, H. , and Kasai, H. (2000) 2‐hydroxy‐dATP is incorporated opposite G by Escherichia coli DNA polymerase III resulting in high mutagenicity. Nucleic Acids Res 28: 1640–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limor‐Waisberg, K. , Carmi, A. , Scherz, A. , Pilpel, Y. , and Furman, I. (2011) Specialization versus adaptation: two strategies employed by cyanophages to enhance their translation efficiencies. Nucleic Acids Res 39: 6016–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X. , Ding, H. , and Zeng, Q. (2016) Transcriptomic response during phage infection of a marine cyanobacterium under phosphorus‐limited conditions. Environ Microbiol 18: 450–460. [DOI] [PubMed] [Google Scholar]
- Lindell, D. , Sullivan, M.B. , Johnson, Z.I. , Tolonen, A.C. , Rohwer, F. , and Chisholm, S.W. (2004) Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA 101: 11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell, D. , Jaffe, J.D. , Johnson, Z.I. , Church, G.M. , and Chisholm, S.W. (2005) Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438: 86–89. [DOI] [PubMed] [Google Scholar]
- Lu, L.D. , Sun, Q. , Fan, X.Y. , Zhong, Y. , Yao, Y.F. , and Zhao, G.P. (2010) Mycobacterial MazG is a novel NTP pyrophosphohydrolyase involved in oxidative stress response. J Biol Chem 285: 28076–28085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, N.H. , Cook, A. , Millard, A. , Bailey, S. , and Clokie, M. (2003) Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424: 741. [DOI] [PubMed] [Google Scholar]
- Mann, N.H. , Clokie, M.R.J. , Millard, A. , Cook, A. , Wilson, W.H. , Wheatley, P.J. , et al (2005) The genome of S‐PM2, a ‘photosynthetic’ T4‐type bacteriophage that infects marine Synechococcus strains. J Bacteriol 187: 3188–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard, A. , Clokie, M.R.J. , Shub, D.A. , and Mann, N.H. (2004) Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA 101: 11007–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard, A.D. , Zwirglmaier, K. , Downey, M.J. , Mann, N.H. , and Scanlan, D.J. (2009) Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ Microbiol 11: 2370–2387. [DOI] [PubMed] [Google Scholar]
- Minh, B.Q. , Nguyen, M.A.T. , and von Haeseler, A. (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz, O.V. , Murzin, A.G. , Makarova, K.S. , Koonin, E.V. , Wilson, K.S. , and Galperin, M.Y. (2005) Dimeric dUTPases, HisE, and MazG belong to a new superfamily of all‐α NTP pyrophosphohydrolases with potential ‘house‐cleaning’ functions. J Mol Biol 347: 243–255. [DOI] [PubMed] [Google Scholar]
- Mühling, M. , Fuller, N.J. , Millard, A. , Somerfield, P.J. , Marie, D. , Wilson, W.H. , et al (2005) Genetic diversity of marine Synechococcus and co‐occurring cyanophage communities: evidence for viral control of phytoplankton. Environ Microbiol 7: 499–508. [DOI] [PubMed] [Google Scholar]
- Murray, K.D. , and Bremer, H. (1996) Control of spoT‐dependent ppGpp synthesis and degradation in Escherichia coli . J Mol Biol 259: 41–57. [DOI] [PubMed] [Google Scholar]
- Nguyen, L.T. , Schmidt, H.A. , von Haeseler, A. , and Minh, B.Q. (2015) IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puxty, R.J. , Millard, A.D. , Evans, D.J. , and Scanlan, D.J. (2016) Viruses inhibit CO2 fixation in the most abundant phototrophs on earth. Curr Biol 26: 1585–1589. [DOI] [PubMed] [Google Scholar]
- Roitman, S. , Hornung, E. , Flores‐Uribe, J. , Sharon, I. , Feussner, I. , and Béjà, O. (2018) Cyanophage‐encoded lipid desaturases: oceanic distribution, diversity and function. ISME J 12: 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan, D.J. , Ostrowski, M. , Mazard, S. , Dufresne, A. , Garczarek, L. , Hess, W.R. , et al (2009) Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M.B. , Waterbury, J.B. , and Chisholm, S.W. (2003) Cyanophages infecting the oceanic cyanobacterium Prochlorococcus . Nature 424: 1047–1051. [DOI] [PubMed] [Google Scholar]
- Sullivan, M.B. , Coleman, M.L. , Weigele, P. , Rohwer, F. , and Chisholm, S.W. (2005) Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M.B. , Huang, K.H. , Ignacio‐Espinoza, J.C. , Berlin, A.M. , Kelly, L. , Weigele, P.R. , et al (2010) Genomic analysis of oceanic cyanobacterial myoviruses compared with T4‐like myoviruses from diverse hosts and environments. Environ Microbiol 12: 3035–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle, C.A. (2007) Marine viruses ‐ major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. [DOI] [PubMed] [Google Scholar]
- Suttle, C.A. , and Chan, A.M. (1994) Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol 60: 3167–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L.R. , Zeng, Q. , Kelly, L. , Huang, K.H. , Singer, A.U. , Stubbe, J. , and Chisholm, S.W. (2011) Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA 108: E757–E764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler, M.F. , Summers, S.M. , Nguyen, H.T. , Zacharia, V.M. , Hightower, G.A. , Smith, J.T. , and Conway, T. (2008) The global, ppGpp‐mediated stringent response to amino acid starvation in Escherichia coli . Mol Microbiol 68: 1128–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidner, G. , Bielawski, J.P. , Shmoish, M. , Scanlan, D.J. , Sabehi, G. , and Béjà, O. (2005) Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ Microbiol 7: 1505–1513. [DOI] [PubMed] [Google Scholar]
- Zeng, Q. , and Chisholm, S.W. (2012) Marine viruses exploit their host's two‐component regulatory system in response to resource limitation. Curr Biol 22: 124–128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Information


