Summary
Aluminium (Al) ions are one of the primary growth‐limiting factors for plants on acid soils, globally restricting agriculture. Despite its impact, little is known about Al action in planta. Earlier work has indicated that, among other effects, Al induces DNA damage. However, the loss of major DNA damage response regulators, such SOG1, partially suppressed the growth reduction in plants seen on Al‐containing media. This raised the question whether Al actually causes DNA damage and, if so, how. Here, we provide cytological and genetic data corroborating that exposure to Al leads to DNA double‐strand breaks. We find that the Al‐induced damage specifically involves homology‐dependent (HR) recombination repair. Using an Al toxicity assay that delivers higher Al concentrations than used in previous tests, we find that sog1 mutants become highly sensitive to Al. This indicates a multi‐level response to Al‐induced DNA damage in plants.
Keywords: Arabidopsis thaliana, growth, aluminium, DNA damage, homologous recombination repair, CDKB1, SOG1, ATM, ATR, RAD51
Significance Statement
Aluminium (Al) is considered to be one of the primary growth‐limiting factors for agriculture especially in developing countries. Here, we explore a recently discovered aspect of Al, i.e. that it causes DNA damage, and show that this damage requires homology‐dependent DNA repair to sustain plant growth on Al‐containing media.
Introduction
Due to their sessile nature, plants are repeatedly subjected to various environmental stresses, such as drought, radiation or ion toxicity. Even moderate stress levels can already reduce plant growth and cause yield losses as seen for instance in rice (Lafitte et al., 2004). Aluminium (Al) is the third most abundant element in the Earth's crust and the most common metal. Below a pH of 5.5, free Al3+ ions predominate and cause phytotoxicity, including severe inhibition of cell division and cell elongation in the root tip (Delhaize and Ryan, 1995; Kochian et al., 2004; Rounds and Larsen, 2008). Because of the prevalence of acidic soils worldwide (> 50%), Al is considered to be one of the primary growth‐limiting factors for agriculture especially in many developing countries.
The mechanism of Al toxicity is not well understood. Al3+ appears to cause multiple extracellular and intracellular effects possibly due to competition with biologically required cations such as Mg2+ (Kochian, 1995). Previous work using comet assays indicates that DNA breaks are a consequence of growth on Al‐containing media (Nezames et al., 2012). Conversely, the growths of mutants in LIG6, RAD17 and UVH1 that are involved in DNA repair were reported to be reduced on media containing Al (Nezames et al., 2012). In addition, mutants in the Arabidopsis Retinoblastoma homologue RBR1, which was recently implicated in DDR, are hypersensitive to Al (Biedermann et al., 2017). Surprisingly though, mutants in key DNA damage response (DDR) genes, i.e. ataxia telangiectasia and RAD3 related (ATR), TANMEI/ALT2, suppressor of gamma response 1 (SOG1) and ATRIP, can partially revert the growth reduction seen in mutants of Al‐sensitive 3 (ALS3), which encodes an ABC transporter required for normal growth in an Al toxic environment. This is in contrast to a loss‐of‐function mutant for ATM (ataxia telangiectasia mutated), which has little effect on Al tolerance as measured by suppression of als3‐1. Mutants ATR, ATRIP, SOG1 and TAN/MEI/ALT2 grow even better during long‐term exposure to Al compared with the wild‐type (WT), leaving it unclear whether and, if so, how Al induces DNA damage (Larsen et al., 2005; Rounds and Larsen, 2008; Nezames et al., 2012; Sjogren et al., 2015; Eekhout et al., 2017; Sjogren and Larsen, 2017).
Whereas ATM is especially responsible for detection of and response to DNA double‐strand breaks (DSBs), its close relative ATR is a key DNA damage checkpoint kinase that is recruited by ATRIP to persistent single‐stranded DNA that occurs due to stalled replication fork progression (Culligan et al., 2004). SOG1 is a transcription factor that is a direct target of ATR and ATM, and acts as a central DDR component that controls many responses to DNA damage (Yoshiyama et al., 2009, 2013). Finally, TANMEI/ALT2 is a DDB1‐binding protein possessing a WD40 motif that may be required for assessment of DNA integrity as such motifs are key for several DDR proteins (Yamagishi et al., 2005; Nezames et al., 2012).
A common role of the DDR pathway in animals and yeast is to arrest cell proliferation to give a genetically compromised cell time to repair DNA. While this appears to be conserved in plants, unanswered questions remain about how a signal from damaged DNA is translated into inhibition of the cell cycle as the regulatory cascades that cause cell cycle arrest are largely not conserved between plants and animals. For instance, inhibitory phosphorylation of the cell‐cycle‐promoting cyclin‐dependent kinases to block mitosis after DNA damage does not occur in Arabidopsis even though this mechanism is crucial for DDR in animals and yeast (Dissmeyer et al., 2009, 2010).
Two major DSB repair pathways that were originally described in animals and yeast have been identified in the model plant Arabidopsis as well as in other plants (Bray and West, 2005): non‐homologous end‐joining (NHEJ) and homology‐dependent repair (HR), the latter of which is also referred to as homologous recombination repair. However, NHEJ can operate throughout the cell cycle, HR requires a template, i.e. sister chromatid, which is only available from DNA replication (S‐phase) onwards until mitosis. However, as HR uses a template it is much more accurate than NHEJ during which the broken DNA are simply joined together with the risk of inducing duplications or deletions.
While our knowledge of DDR in plants is limited, key aspects of both DSB repair pathways appear to be conserved between plants and animals. For instance, plant homologues of KU70/80 from animals have been identified that are crucial for NHEJ (Lieber et al., 1997; Walker et al., 2001; Mari et al., 2006). With regard to HR repair, RAD51‐type proteins that are homologues of the bacterial RecA‐type protein are found in Arabidopsis and other plants (Doutriaux et al., 1998; Bleuyard et al., 2005). Even so, the regulation of HR also involves features that are specific to plants. For example, a regulatory cascade that is unique to plants plays an important role in loading of RAD51 to DNA lesions. This cascade initiates with the activation of plant‐specific cyclin‐dependent kinases of the B1 class (CDKB1) through transcriptional upregulation of B1‐type cyclins (CYCB1), which serve as CDKB1 activators (Harashima and Schnittger, 2012; Weimer et al., 2016). SOG1 directly promotes CYCB1 expression resulting in accumulation of CYCB1 protein in root meristems after exposure to DNA‐damaging agents (Weimer et al., 2016). In vitro kinase assays show that CDKB1;1‐CYCB1;1 complexes phosphorylate RAD51, with RAD51 localization to DNA lesions strongly reduced in both cdkb1 and cycb1 mutants. The recruitment of RAD51 appears to be further mediated by RBR1, another substrate of CDKB1–CYCB1 complexes (Harashima and Schnittger, 2012; Biedermann et al., 2017). Analysis of the triple‐mutants cdkb1;1 cdkb1;2 cycb1;1 and cdkb1;1 cdkb1;2 rbr1 suggested that these regulators act in a common cascade (Weimer et al., 2016; Biedermann et al., 2017).
In this study, we provide cytological and genetic evidence that exposure to Al can lead to DNA damage in the form of DSBs. Applying an Al growth assay that delivers higher Al concentrations than used in previous tests, we reveal that this damage specifically requires HR mechanisms. Unlike what was previously reported for long‐term growth studies, we found that increased DNA damage levels arising from exposure to high levels of Al resulted in sog1 and atm mutants being hypersensitive and not resistant to Al. Furthermore, atr mutants were neither more resistant nor more sensitive than the WT in this assay. This gives rise to the hypothesis of a multi‐level response to DNA damage in Arabidopsis that might also be in general seen for other central regulators of DDR in plants.
Results
Al treatment induces DNA damage
To date, it remains not understood why loss‐of‐function mutants in key DDR genes such as ATR are tolerant to Al. A simple hypothesis is that loss of these genes should rather result in sensitivity towards DNA‐damaging agents. Indeed, atr mutants are for instance strongly compromised when grown on media containing the replication poison hydroxyurea (Culligan et al., 2004) or DNA‐cross‐linking agents such as cisplatin (Nezames et al., 2012).
To further explore the possibility that Al damages DNA, we first monitored the appearance of the phosphorylated variant histone H2AX (γH2AX) in nuclei of plants grown in a long‐term Al‐soaked gel assay as previously employed (Larsen et al., 1996). The phosphorylation of H2AX adjacent to chromosomal breaks is an early step in the response to DSBs and precedes their repair (Kuo and Yang, 2008). Consistent with a DNA‐damaging role for Al, we found that Al‐treated plants had indeed more γH2AX foci in their nuclei than untreated plants. The damage became particularly evident at a concentration of 1.5 mm Al (pH 4.2) with the appearance of approximately 10% of nuclei that had six or more γH2AX foci, a class that was never observed in untreated cells (Figure 1a,b).
Figure 1.
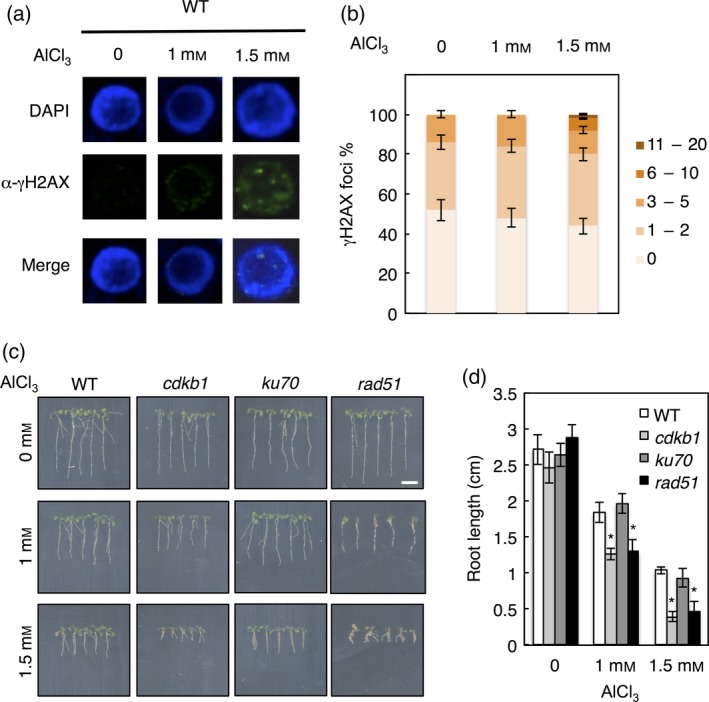
Detection of Al‐induced DNA damage in long‐term growth assays.
(a) Immunofluorescence analysis of γH2AX accumulation (green) in DAPI‐stained nuclei (blue) of wild‐type (WT) Arabidopsis root tips grown for 10 days on Al‐containing medium. (b) Quantification of γH2AX foci in WT plants after Al treatment. One‐hundred nuclei per line per experiment were grouped into six classes according to their number of γH2AX foci: nuclei containing no γH2AX foci, 1–2, 3–5, 6–10 and 11–20 γH2AX foci. (c) Seedling growth of WT, cdkb1, ku70 and rad51 mutants. Seeds were germinated on soaked gel medium containing 0, 1 and 1.5 mm Al (pH 4.2), and grown for 10 days. Scale bar: 1 cm. (d) Root growth measurements of WT, cdkb1, ku70 and rad51 mutants grown on soaked gel medium containing 0, 1 and 1.5 mm Al (pH 4.2) for 10 days. Data are presented as mean ± SD in three independent experiments. Significant differences from WT were determined by independent samples t‐test: *P < 0.05.
If Al induces DNA damage, we posited that mutants in factors directly involved in or required for DNA repair should be hypersensitive to Al as indicated by the previously found reduction in root growth of lig6, rad17 and uvh1 mutants on Al‐containing media (Nezames et al., 2012). However, from the previously tested mutants, it is not clear which DNA damage pathway might be involved in the repair of Al‐induced damage. To explore this further, root growth on long‐term Al‐soaked gel media was analysed for loss‐of‐function mutants of central regulators of HR and NHEJ, i.e. rad51 and ku70, respectively. While ku70 mutants are hypersensitive to bleomycin, we did not find a significant reduction in root growth of ku70 mutants in comparison to the WT on Al‐containing media (Figure 1c,d). This is consistent with the previous observation that ku80 mutants are not sensitive to Al (Sjogren et al., 2015).
In contrast, rad51 mutants grew significantly shorter than the WT on Al‐soaked gel media, further supporting the genotoxicity of Al and indicating a possible role of HR during growth on Al‐containing media (P < 0.05, independent sample t‐test). To test the importance of HR, we analysed mutants in CDKB1;1 and CDKB1;2, which were previously identified as major regulators of HR in Arabidopsis (Weimer et al., 2016). Because CDKB1;1 and CDKB1;2 act redundantly, we always used the double‐mutant cdkb1;1 cdkb1;2 and refer to it as cdkb1 in the following. Consistent with the hypersensitivity of rad51 mutants, cdkb1 plants had significantly shorter roots than the WT on Al‐containing agar plates (Figure 1c,d, P < 0.05, independent sample t‐test). Taken together, we conclude that Al indeed induces DNA damage, and that repair of this damage predominantly requires HR.
A high‐dosage short‐term assay for Al toxicity
Our findings that cdkb1 and rad51 are hypersensitive to Al are consistent with the sensitivity of lig6, rad17 and uvh1 (Nezames et al., 2012), but stand in apparent contrast to the observation that alt2, atr, atrip and sog1 are tolerant to Al. However, these regulators act upstream in DDR cascades, and it is well known that especially ATR and SOG1 control dozens if not hundreds of downstream genes (Culligan et al., 2004; Yoshiyama et al., 2009, 2014; Ogita et al., 2018). Intriguingly, while sog1 was initially identified as a mutant that grew better than the WT after exposure to gamma radiation, it is now clear that SOG1 is one of the central most regulators of DDR in plants (Inagaki and Umeda, 2011; Yoshiyama et al., 2014). Hence, we reasoned that the necessity for a full DDR may only become visible after high levels of DNA damage, which likely have not been achieved using the Al‐soaked gel media approach.
We therefore developed an Al assay in which 6‐day‐old seedlings are transferred to a gel‐free hydroponic solution supplemented with either no or 1.5 mm AlCl3 (pH 4.2) for 12 h, after which the seedlings are moved to agar plates without Al to determine root growth and perform cytological studies. The underlying difference in this approach compared with the previous soaked gel approach is that the agar used for preparation of solid media is expected to bind a substantial amount of the Al, thus making a large portion of it unavailable and limiting the possible concentrations of Al used. As shown in Figure 2(a), in the absence of Al, cdkb1, ku70 and rad51 showed similar root growth as the WT roots. However, cdkb1 and rad51 mutant seedlings exposed to 1.5 mm AlCl3 (pH 4.2) in hydroponic solution grew much shorter than the WT on agar plates without Al in the recovery phase (Figure 2b). In contrast, ku70 had a similar root length when compared with WT. These responses are consistent with the growth pattern observed in the long‐term Al‐soaked gel assay, and further indicate that HR occurs in response to Al‐induced DNA damage.
Figure 2.
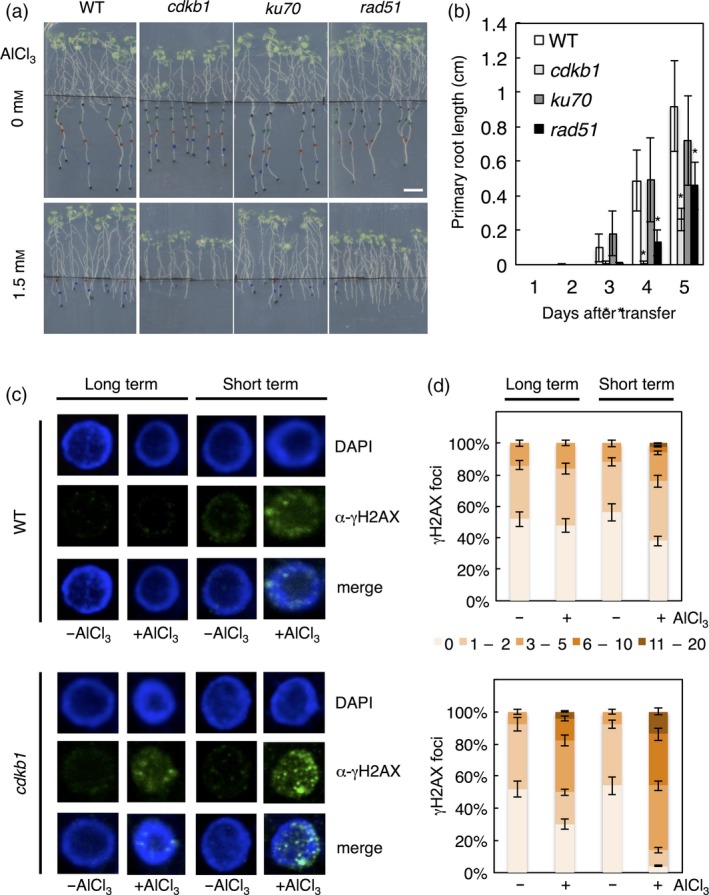
Detection of Al‐induced DNA damage in short‐term growth assays.
(a) Six‐day‐old seedlings of wild‐type (WT), cdkb1, ku70 and rad51 mutants were transferred to 0 or 1.5 mm Al‐containing hydroponics (pH 4.2), and treated for 12 h. Treated seedlings were planted on agar plates without Al and grown for 5 subsequent days. Scale bar: 1 cm. (b) Root growth measurements of WT, cdkb1, ku70 and rad51. Al‐treated seedlings were transferred to agar plates without Al, and root lengths were measured for 5 days. Data are presented as mean ± SD in three independent experiments. Significant differences from WT were determined by independent samples t‐test: *P < 0.05. (c) Immunofluorescence analysis of γH2AX accumulation (green) in nuclei, stained with DAPI (DNA, blue), of root tips of WT, cdkb1, ku70 and rad51 plants after 3 days of growth on 1.5 mm Al‐containing soaked gel medium (pH 4.2), or 12 h in 1.5 mm Al‐containing hydroponic solution (pH 4.2). (d) Quantification of γH2AX foci in WT and cdkb1 plants after Al treatment. One‐hundred nuclei per line per experiment were grouped into six classes according to their number of γH2AX foci: nuclei containing no γH2AX foci, 1–2, 3–5, 6–10 and 11–20 γH2AX foci.
If our short‐term assay was to deliver higher dosages of Al, we expected higher levels of DNA damage than the previously used long‐term assays under the assumption that Al is indeed genotoxic and that the level of damage is dose dependent. To compare the effects of long‐term low versus short‐term high exposure to Al, we quantified γH2AX foci under both regimes in WT and cdkb1 mutants. Indeed, WT plants treated long term with a moderate Al concentration had fewer γH2AX foci than plants under the short‐term high‐Al regime, for example, nuclei with six or more γH2AX foci could only be found after short‐term high‐Al exposure (Figure 2c,d). Correspondingly, the class of nuclei with six or more γH2AX foci in cdkb1 mutants almost doubled under the short‐term high‐ in comparison to the long‐term low‐Al conditions (Figure 2c,d). Furthermore, the number of γH2AX foci increased with Al concentration in the media of the short‐term assay, for example, while no nuclei with more than five foci were observed in 1 mm Al‐treated plants (Figure 1a,b), this class increased to approximately 10% of all nuclei in plants grown on 1.5 mm Al (Figure 2d).
To examine the effects of Al in our short‐term assay in detail, we treated Arabidopsis seedlings with increasing concentrations of Al for 12 h. Meristem sizes of WT plants started to be reduced at 1 mm Al, while the meristems in cdkb1 mutants were already affected at 0.5 mm Al. At the highest concentration of 1.5 mm Al, WT and cdkb1 meristem sizes were reduced to 70 and 40% of the sizes seen at 0 mm Al treatment, respectively (Figure S1a,b). In parallel, we monitored γH2AX foci as a measure of DSBs in the WT and cdkb1 mutants at increasing concentrations of Al. Similar to the results for the reduction in meristem sizes (Figure S1c,d), accumulation of γH2AX foci was detected at 1 mm Al in the WT and at 0.5 mm in cdkb1.
Conversely, we evaluated the effects of our short‐term assay in a time course experiment, revealing that a treatment of Al reduced meristem sizes in WT after 6 h and in cdkb1 after 3 h, respectively. After 24 h, meristem sizes were reduced to 50% in the WT and 12.6% in cdkb1 when compared with growth conditions without Al (Figure S2a,b). Correspondingly, we could detect γH2AX in the WT at 6 h, and in cdkb1 at 3 h. Notably, there were also more γH2AX foci in cdkb1 than in the WT (Figure S2c,d).
Taken together, these results indicate that Al treatment induces DSBs in a concentration‐dependent manner. Al‐induced effects could be seen as early as 3 h after exposure to Al in mutants that are sensitized to DNA damage. With this, we have established a short‐term high‐dosage assay that allows the detailed dissection of Al effects on plant growth, clearly demonstrating that Al acts as a clastogenic agent in vivo.
Repair of Al damage by homology‐dependent repair
As a first application of our short‐term assay, we wanted to obtain more information about the possible involvement of HR in Al‐induced DNA damage. To this end, we used the previously generated plant line IC9C, which allows the detection of homologous recombination events (Swoboda et al., 1994). This line harbors a transgene with non‐functional overlapping parts of a β‐glucuronidase gene (GUS) gene. Restoration of the reporter gene is possible by inter‐chromosomal recombination with the sister chromatid or the homologous chromosome resulting in blue spots, which can then be quantified (Molinier et al., 2004; Puchta and Hohn, 2012). The average number of blue spots in control medium was 0.08 per leaf. In contrast, plants of IC9C line treated for 24 h with 1.5 mm AlCl3 had nearly 10 times more recombination events as visualized by an increase in blue spots to an average of 0.72 per leaf (Figure 3a,b). Next, we introgressed the recombination reporter into the cdkb1 mutant background. In conjunction with their severe Al‐dependent root growth inhibition and high number of γH2AX foci after Al exposure, we observed that almost no blue spots could be found in cdkb1 mutants (Figure 3c,d), thus indicating that Al‐induced HR is dependent on CDKB1 function.
Figure 3.
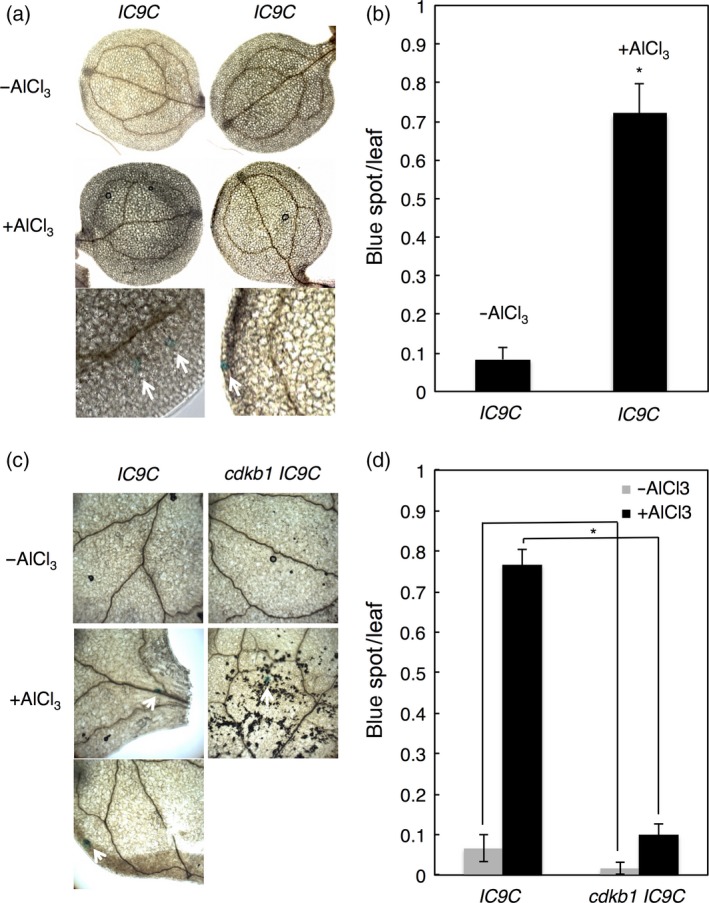
Short‐term Al treatment triggers CDKB1‐dependent homology‐dependent repair (HR).
(a) Wild‐type (WT) plants containing the recombination reporter IC9C show blue spots on leaves after 24 h of incubation in 1.5 mm Al‐containing hydroponics. Arrows indicate representative blue sectors. (b) Number of blue sectors per plant grown without or with Al treatment. Data are presented as mean ± SD in three independent experiments. The significance of the difference was determined by independent samples t‐test: *P < 0.05. (c) WT IC9C and cdkb1 mutant plants containing the recombination reporter IC9C show blue spots on the leaves after 24 h of incubation in 1.5 mm Al‐containing hydroponics (pH 4.2). Arrows indicate representative blue sectors. (d) Numbers of blue sectors per plant grown without or with Al treatment. Data are presented as mean ± SD in three independent experiments. Significant differences from WT IC9C were determined by independent samples t‐test: *P < 0.05.
To understand then the effect on plant growth at the cellular level, we determined the meristem sizes of WT plants versus cdkb1 mutants after a short‐term high‐dosage Al treatment and during the subsequent recovery growth phase. As a control we used the Al‐hypersensitive als3 mutant. Consistent with the growth‐reducing effect of Al, we found that the number of meristematic cells in the meristem of WT plants is reduced to approximately 80% of the number of cells in meristems of untreated plants (Figure 4a,b). In correlation with the Al hypersensitivity of als3 mutants, we determined that their meristems are more severely affected by Al. However, in the recovery phase, als3 mutant meristems were reconstituted at a similar rate as seen for the WT, indicating that repair is fully functional in this mutant (Figure 4c). In contrast, meristems of cdkb1 mutants were even smaller than meristems of als3 mutant plants when exposed to Al (Figure 4a,b). Moreover, the meristems even stayed small for several days on media without Al when WT and als3 mutants had already rebuilt their meristems (Figures 4c and S3a,b). Matching this growth behaviour, we found that γH2AX foci in root meristem cells rapidly disappeared in WT plants during the recovery growth phase, whereas the number of γH2AX foci in cdkb1 mutants stayed very high and hardly changed during the first 3 days of recovery growth (Figure S4a,b).
Figure 4.
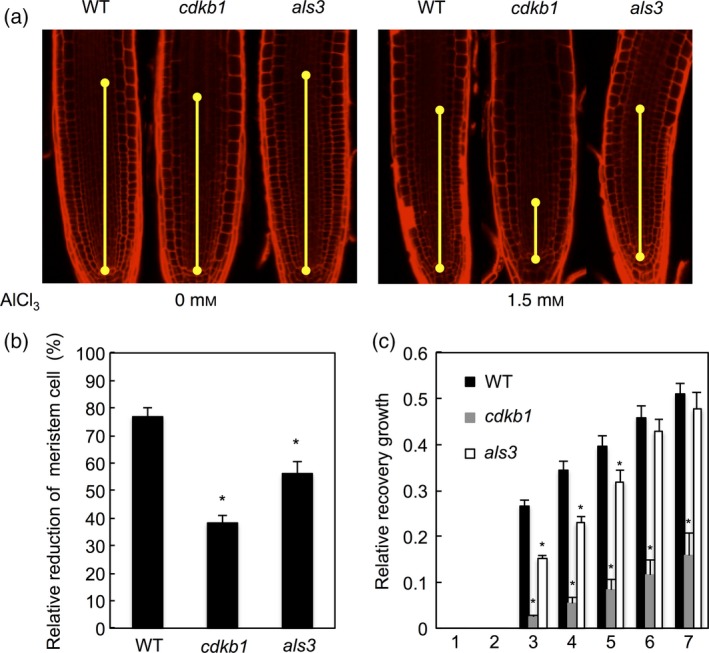
Reduction of root meristem size after Al treatment.
(a) Five‐day‐old seedlings were transferred to 0 or 1.5 mm Al‐containing hydroponics (pH 4.2) and treated for 12 h. (b) Cortex cell number between the quiescent centre and the first elongated cell was counted after 12 h. Data are presented as mean ± SD in three independent experiments. Significant differences from wild‐type (WT) were determined by independent samples t‐test: *P < 0.05. (c) Root growth measurements of WT, cdkb1 and als3; 12 h Al‐treated seedlings were transferred to agar plates without Al and root lengths were measured for 5 days. Data are presented as mean ± SD in three independent experiments. Significant differences from WT were determined by independent samples t‐test: *P < 0.05.
We have previously shown that CDKB1s work together with B1‐type cyclins in HR (Weimer et al., 2016). We therefore tested whether CYCB1s are also involved in Al response. There was no significant difference between WT and cycb1 single‐mutants in root growth recovery in the short‐term Al assay in the absence of Al. However, when seedlings were treated with 1.5 mm AlCl3 in this assay, WT roots started recovery growth on day 3, whereas root growth recovery for all cycb1 single‐mutants was significantly delayed (P < 0.05, Student's t‐test). Mutants in cycb1;1 and cycb1;4 both were more sensitive than either cycb1;2 or cycb1;3 as determined by their root growth (Figure 5a,b). This result supports the findings from the IC9C recombination assay as well as the analysis of root meristems in WT versus cdkb1 mutants, and further strengthens the notion that Al induces DNA damage that requires HR for repair.
Figure 5.
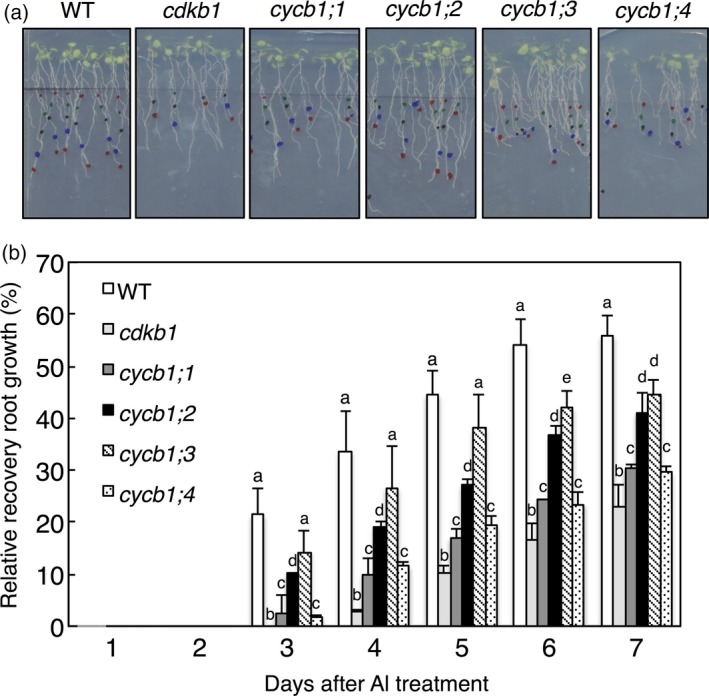
Mutants in B1‐type cyclins are sensitive to Al in short‐term growth assays.
(a) Six‐day‐old seedlings of wild‐type (WT), cdkb1, cycb1;1, cycb1;2, cycb1;3 and cycb1;4 mutants were transferred to 1.5 mm Al‐containing hydroponics (pH 4.2) and treated for 12 h. Treated seedlings were planted on agar plates without Al and grown for 5 days. Scale bar: 1 cm. (b) Root growth measurements of WT, cdkb1, cycb1;1, cycb1;2, cycb1;3 and cycb1;4. Al‐treated seedlings were transferred to agar plates without Al and root length was monitored for 5 days. Data are presented as mean ± SD in three independent experiments. Different letters indicate significant differences by independent samples t‐test: *P < 0.05.
SOG1 and ATM are required in recovery root growth and DNA repair
Finally, we wanted to revisit the mutants in key DDR regulators, i.e. atr and sog1, which previously have been found to confer enhanced growth on Al‐containing media (long‐term treatment low dosage) in comparison to the WT. Mutant seedlings were assayed next to WT and cdkb1 as positive controls as well as an atm loss‐of‐function mutant that was previously found to not suppress the Al hypersensitivity phenotype of als3 mutants (Sjogren et al., 2015). As shown in Figure 6(a,b), short‐term high‐dosage Al treatment resulted in reduced recovery growth of sog1 compared with WT. This reduction was at the level of reduction seen in cdkb1 underlining the importance of SOG1 during DDR, including Al‐induced damage. Similarly, atm plants were found to be hypersensitive in comparison to WT albeit not as much as cdkb1 and sog1 mutants. In contrast, atr mutant plants were indistinguishable from WT in their root growth, i.e. being neither Al tolerant nor hypersensitive under these conditions.
Figure 6.
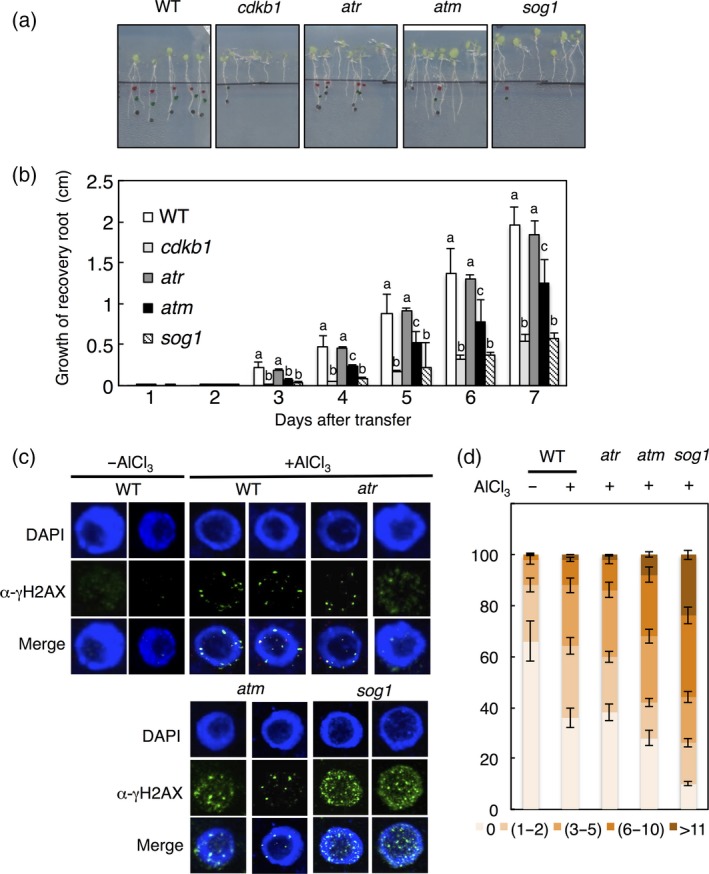
Mutants in ATM and SOG1 are sensitive to Al in short‐term growth assays.
(a) Six‐day‐old seedlings of wild‐type (WT), cdkb1, atr, atm and sog1 mutants were transferred to 1.5 mm Al‐containing hydroponics (pH 4.2) and treated for 12 h. Treated seedlings were subsequently planted on agar plates without Al and grown for 5 days. Scale bar: 1 cm. (b) Root growth measurements of WT, cdkb1, atr, atm and sog1. Al‐treated seedlings were transferred to agar plates without Al and root length was monitored for 5 days. Data are presented as mean ± SD in three independent experiments. Different letters indicate significant differences by independent samples t‐test: *P < 0.05. (c) Immunofluorescence analysis of γH2AX accumulation (green) in nuclei stained with DAPI (DNA, blue) of root tips of WT, atr, atm, and sog1 mutants after 12 h of incubation in 1.5 mm Al‐containing hydroponics (pH 4.2). (d) Quantification of γH2AX foci in root tips after Al treatment. One‐hundred nuclei per line per experiment were grouped into six classes according to their counted number of γH2AX foci: nuclei containing no γH2AX foci, 1–2, 3–5, 6–10 and 11–20 γH2AX foci.
To complement the growth assays, we analysed γH2AX foci in these mutants after Al treatment. Matching the results of the root growth assays, we observed in atr mutants a similar number of γH2AX foci as in the WT. Slightly more γH2AX foci were observed in atm mutants, for example, 5% of all nuclei had 11 or more γH2AX foci versus approximately 2% in WT (Figure 6c,d). Mutants in SOG1 had even higher numbers of γH2AX foci than atm with, for instance, approximately 25% of all nuclei having 11 or more γH2AX foci (Figure 6c,d). These results corroborated that Al induces DNA damage, and that ATM and SOG1 are especially important for DNA repair and recovery growth in response to high concentrations of Al consistent with their canonical role in DDR.
Several DDR genes, including CYCB1;1 and RAD51, were found to be induced upon long‐term low‐dose Al regime (Sjogren et al., 2015). Both genes were also strongly induced in the WT upon exposure to Al under our short‐term high‐Al dosage growth conditions (Figure 7a). As a control, we monitored the expression of CYCB1;2, which was found to be reduced in the WT consistent with a reduced proliferation activity after Al treatment (Figure 7a). While the expression of CYCB1;2 did not significantly change in atm, atr and sog1 mutants, we found that the induction of CYCB1;1 and RAD51 was dependent on SOG1 and ATM but not ATR in the short‐term high‐dosage assay (Figure 7a).
Figure 7.
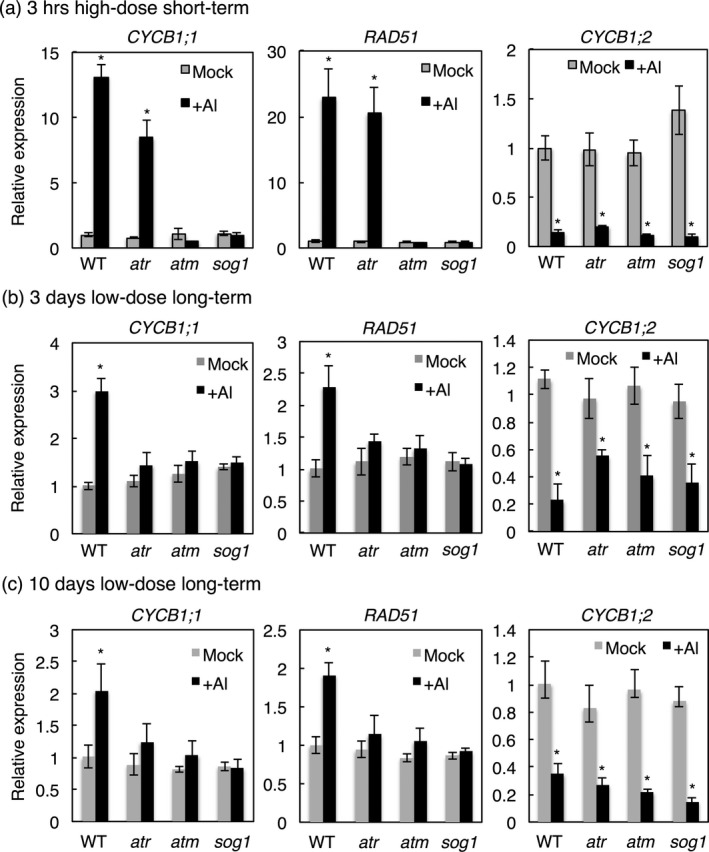
Expression analysis of DNA damage response (DDR) genes after short‐term and long‐term Al treatments.
(a) RNA was prepared from 10‐day‐old wild‐type (WT), atm, atr and sog1 seedlings, either untreated or treated with 1.5 mm Al‐containing hydroponics (pH 4.2) for 3 h. Relative expression levels of the indicated genes are shown as mean values from three biological repeats and with the untreated WT value set as 1. Error bars indicate SD. Significant differences from untreated value were determined by one‐way anova analysis, P < 0.05. (b) RNA was prepared from seeds germinated and grown on soaked gel medium containing 0 and 1.5 mm Al (pH 4.2) for 3 days. Relative expression levels of the indicated genes are shown as mean values from three biological repeats and with the untreated WT value set as 1. Error bars indicate SD. Significant differences from untreated plants were determined by one‐way anova analysis, P < 0.05. (c) RNA was prepared from seeds germinated and grown on soaked gel medium containing 0 and 1.5 mm Al (pH 4.2) for 10 days. Relative expression levels of the indicated genes are shown as mean values from three biological repeats and with the untreated WT value set as 1. Error bars indicate SD. Significant differences from untreated value were determined by one‐way anova analysis, P < 0.05.
To further compare the difference between the two Al treatments, we also investigated the CYCB1;1 and RAD51 expressions under a long‐term low‐dose Al regime at two different time points (3 and 10 days). At both time points, CYCB1;1 and RAD51 genes were less strongly induced in the long‐term low‐dose treatment in comparison to the short‐term assay consistent with the more damaging conditions of the short‐term assay (Figure 7b,c). Interestingly, the induction of CYCB1;1 and RAD51 was equally abolished in atm, atr and sog1 mutants in long‐term low‐dose Al regime, while under our short‐term high‐dose assay both genes could still be induced upon the loss of ATR indicating that the DDR response is also qualitatively different at different concentrations of Al (Figure 7b,c).
Discussion
Complementing previous work, we have here developed an Al assay in which we treat plants for a short term with high levels of Al and then monitor their growth recovery. Our work shows that high concentrations of Al cause DNA damage that in particular requires HR for repair.
Increased versus decreased growth of mutants in DNA damage response genes on Al‐containing media
The major difference between the previous assay and the here‐established test is the concentration of Al. The long‐term low‐dose gel assay delivers Al concentrations in the micromolar range that reflect the Al dose typically found in acidic soils. These ‘natural’ conditions, as shown here and in previous work (Nezames et al., 2012), can already induce DNA damage. However, this damage appears to be very mild. Hence, it is very difficult to monitor and, only after 10 days of growth on Al‐containing media, a slight effect was found in the WT. Using mutants in repair genes, here foremost, in the central HR regulator CDKB1, allowed us to clearly visualize the damage inflicted upon growth on Al‐containing media under long‐term low‐dose regime already after 3 days of growth. In contrast, our short‐term assay results in measurable DNA damage in the WT after a short incubation time, and with this respect might be helpful to immediately monitor and analyse Al‐dependent damage in a WT background.
Our short‐term assay was necessary as higher concentrations cannot be delivered in the previous set‐up due to the limited availability of Al in the solid medium. Conversely, our assay cannot be extended for much more than a day as even the growth of WT will be severely compromised after an exposure of more than 48 h under our assay conditions.
Using our assay, we have revealed that there are different DDR patterns in response to Al. With this, our short‐term high‐dosage assay complements the long‐term low‐dosage assay as both systems probe different aspects of a plant's response to Al. We postulate that up to threshold concentrations of Al, plant growth is restricted by ATR, ATRIP, SOG1 and TANMEI (Rounds and Larsen, 2008; Nezames et al., 2012; Sjogren et al., 2015; Sjogren and Larsen, 2017), and only at high concentrations of Al, SOG1 and ATM are required for plant recovery and survival as revealed in this study (Figure 6). Remarkably, this behaviour resembles mutants in SOG1 itself that were initially identified as suppressors of reduced plant growth following exposure to gamma radiation (Yoshiyama et al., 2009). However, subsequent analyses revealed that SOG1 is a central DDR regulator in Arabidopsis with a function analogous to p53 in animals (Yoshiyama et al., 2014).
These two contrasting behaviours possibly suggest a two‐level or possibly multi‐level response to DNA damage in plants. First, low levels of DNA damage, which can be coped with by the cell and that do not, at least in the short term, lead to compromised cellular functions, trigger already an ATR‐regulated DDR (Figure 8a). This reaction may include two or more likely interconnected responses, i.e. arrest or slowing down of cell proliferation activity, terminal differentiation and endoreplication, as well as activation of DNA repair pathways, for example, via the CDKB1‐pathway. In this phase, the elimination of key DDR genes such as SOG1 (in case of gamma radiation and Al exposure) and ATR (in the presence of Al) leads to enhanced growth in comparison to the WT as the cell cycle might not be slowed down and cells do not terminally differentiate (Figure 8b). Notably, growth under these presumably mild DNA‐damaging conditions depends on active DNA repair mechanisms as found in this and previous studies (Nezames et al., 2012).
Figure 8.
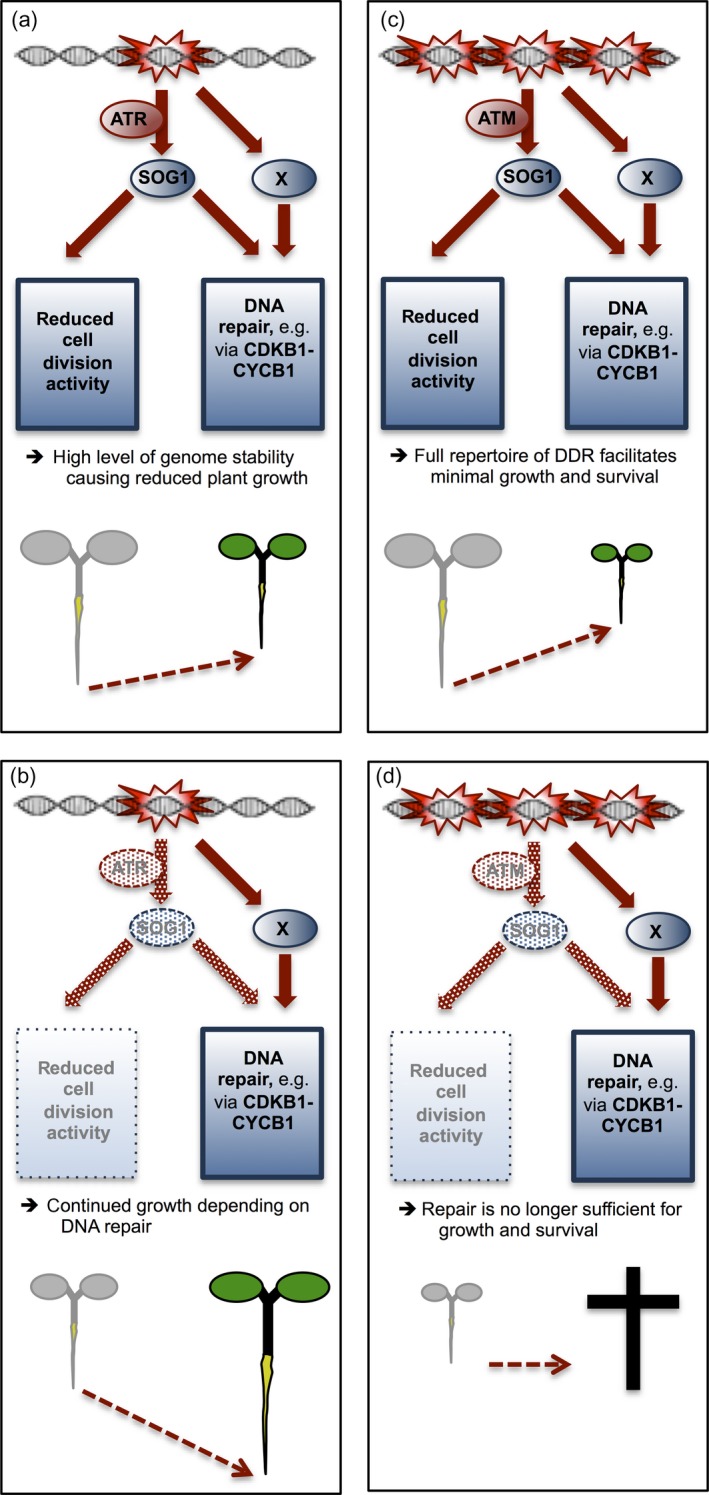
Model of multi‐level response to DNA damage induced by Al.
Four different scenarios are compared that result in different plant phenotypes upon DNA damage. (a and b) DNA damage response (DDR) in wild‐type (WT) plants at low and high levels of DNA damage, respectively. (a) Already low levels of DNA damage presumably trigger a signalling cascade via ATR and SOG1 resulting in a full‐blown DDR that involves the induction of DNA repair genes as well as other responses, in particular the downregulation of cell proliferation activity. As a result, plant growth is reduced (green plant). Grey shaded plant indicates growth of a plant under conditions that do not damage DNA. (b) High levels of DNA damage give rise to even further reduced plant growth (green plant). Grey shaded plant indicates growth of a plant under conditions that do not damage DNA. (c and d) DDR in mutant plants at low and high levels of DNA damage. (c) Elimination of SOG1 and ATR can result in increased growth (green plant) in comparison to the WT (grey plant) under mildly DNA‐damaging conditions as long as DNA repair mechanisms [i.e. homology‐dependent repair (HR) pathways] remain functional. (d) During high levels of DNA damage, DNA repair alone is not sufficient anymore to sustain survival and growth, and the full repertoire of DDR is needed, for example, cell proliferation activities have to be adjusted. Grey shaded plant indicates the growth of a WT plant under these conditions. X indicates a yet to be identified upstream regulator that is postulated to act at a similar level as SOG1. Moreover, ATR and ATM appear to have different response thresholds for DNA damage, at least for the damage induced by Al. For details, see Discussion.
Under severe DNA damage, DNA repair is not sufficient anymore for survival and growth as now damaged stem cells need to be replaced and possibly overall cell proliferation activities have to be adjusted (Figure 8c,d). Given that the DNA repair factors RAD51 and CDKB1s are required for growth even at low levels of Al, while SOG1 and ATR are not, we further postulate that RAD51 and CDKB1–CYCB1 are not only activated by SOG1, as previously found (Weimer et al., 2016; Biedermann et al., 2017), but also by one or more yet to be identified factors (Figure 8).
Knowledge about the existence of such a multi‐level response might in turn be used for the production of Al‐resistant plants. As efficient repair pathways, especially HR, seem to be needed for sustained growth on Al‐containing media, one possible route for breeding/engineering could be to aim for plants in which these pathways are very active and as much as possible uncoupled from upstream acting factors that would also cause a reduction in proliferation.
Al – a DNA‐damaging agent not only for plants?
The mechanism of how Al damages DNA is still unknown. With this respect, it is interesting to note that atr mutants were not sensitive in our short‐term assay as one could have postulated from the previously found involvement of ATR in long‐term Al assays (Rounds and Larsen, 2008; Nezames et al., 2012). In contrast, atm mutants are compromised in their recovery after exposure to high‐Al doses. This finding supports the notion that Al does not interfere with DNA replication as also seen by the enhanced level of DNA synthesis‐dependent endoreplication in als3 mutants (Sjogren et al., 2015). Moreover, as ATM but not ATR is predominantly involved in sensing DSBs, it seems likely that high levels of Al mainly cause DSBs and not single‐strand DNA breaks. This is consistent with the hypothesis that ATR is activated by a specific DNA configuration triggered by Al that may not severely damage DNA at low concentrations of Al, or at least concentrations high enough to trigger the next level of DDR as achieved in our short‐term assay (Nezames et al., 2012).
Based on the here‐observed requirement of HR for Al‐induced DNA damage, it seems plausible that Al interacts with DNA in an electrostatic manner that mimics cross‐linking agents such as cisplatin. This fits previous data showing that the addition of Al could reduce the increase in the tail in comet assays where the size of the tail is used as an approximation for the level of DNA fragmentation in the genome (Nezames et al., 2012). The cross‐linking activity could be indirectly due to the inhibition of enzymes, for example, of the Bloom complex, which is known to repress crossovers in somatic cells and in meiosis (Hartung et al., 2008; Schröpfer et al., 2014; Séguéla‐Arnaud et al., 2015, 2017). Alternatively, Al ions could bind DNA directly due to their high electropositivity, and hence non‐covalently link the negatively charged phosphate groups in the backbone of DNA (Matsumoto et al., 1977). With this respect, it is interesting to note that the addition of Al to DNA has been found to increase the melting temperature of DNA (Karlik et al., 1980). Given the strong effect of Al on DNA seen here and in previous studies, it is interesting to ask whether Al is also genotoxic in other organisms including humans, especially as organisms of all types are chronically exposed to Al including through their diets.
Experimental procedures
Plant materials and growth conditions
The Arabidopsis accession Columbia (Col‐0) was used as WT. The mutants atm‐2 (Garcia et al., 2003), atr‐2 (Culligan et al., 2004), sog1‐7 (Sjogren et al., 2015) and ku70 (Riha et al., 2002) are all in the Col genetic background. The double‐mutant cdkb1;1 cdkb1;2 was described in Nowack et al. (2012). All genotypes were determined by polymerase chain reaction (PCR), and primers are indicated in Table S1. Arabidopsis plants were grown on vertically oriented Murashige and Skoog (MS) medium [0.5 × MS salts, 1% sucrose and 1% agarose (pH 5.8)] under long‐daylight (16 h) conditions at 22°C.
Root growth assay
For long‐term Al assay, Al‐soaked gel and Al‐containing hydroponics were prepared as previously described (Larsen et al., 1996). WT and mutant Arabidopsis seeds were surface‐sterilized using 4% NaClO solution and cold stratified at 4°C overnight. For control experiment, seeds were germinated on soaked gel media (pH 4.2) consisting of 1 mm KNO3, 0.2 mm KH2PO4, 2 mm MgSO4, 0.25 mm (NH4)2SO4, 1 mm Ca(NO3)2, 1 mm CaSO4, 1 mm K2SO4, 1 μm MnSO4, 5 μm H3BO3, 0.05 μm CuSO4, 0.2 μm ZnSO4, 0.02 μm NaMoO4, 0.1 μm CaCl2, 0.001 μm CoCl2. For Al treatment, seeds were germinated on solidified medium, which was soaked with 20 ml of nutrient medium solution containing AlCl3 (pH 4.2) for 2 days, after which the soak solution was removed. After 10 days, seedlings of WT and mutant were photographed and root length was measured. Data are presented as mean ± SD (n = 3, > 30 roots analysed in each experiment). Significant differences from WT were determined by independent samples t‐test: *P < 0.05.
For short‐term Al assay, plants were germinated and grown on vertical plates containing ½ MS medium under long‐daylight conditions at 22°C for 6 days. Seedlings were transferred to 1.5 mm Al‐containing hydroponics (pH 4.2), which consisted of 1 mm KNO3, 0.2 mm KH2PO4, 2 mm MgSO4, 0.25 mm (NH4)2SO4, 1 mm Ca(NO3)2, 1 mm CaSO4, 1 mm K2SO4, 1 μm MnSO4, 5 μm H3BO3, 0.05 μm CuSO4, 0.2 μm ZnSO4, 0.02 μm NaMoO4, 0.1 μm CaCl2 and 0.001 μm CoCl2, as previously described (Larsen et al., 1996; Sjogren et al., 2015), and treated for 12 h. Treated seedlings were planted on vertical ½ MS medium and allowed to grow for 5 days. The position of the primary root tip was marked daily for each plant. After 5 days, plates were photographed and root length was measured using ImageJ software. Data are presented as mean ± SD (n = 3, > 30 roots analysed in each experiment). Significant differences from WT were determined by independent samples t‐test: *P < 0.05.
Immunofluorescence staining
Ten‐day‐old seedlings were transferred to Al‐soaked gel medium or ½ MS liquid medium containing 1.5 mm Al‐containing hydroponics. Incubation time in Al‐soaked gel was 3 and 10 days. In Al‐containing hydroponics different time regimes were used, ranging from 3 to 24 h. Root tip spreads and immunostaining were subsequently performed as described earlier (Friesner et al., 2005). γH2AX immunostaining was conducted with rabbit anti‐γH2AX antibody (1:600) kindly provided by Dr Charles White, and a goat Alexa Fluor488 anti‐rabbit (Life Technologies, Carlsbad, CA, USA) was used as secondary antibody in a 1:300 dilution. Imaging was done with a Leica TCS SP8 inverted confocal microscope at 40 × magnification. The excitation light for the fluorophores was emitted by a diode 405 nm laser, and an argon laser at 488 nm.
Homologous recombination assay
The double‐mutant cdkb1 was crossed to the IC9C reporter line, kindly provided by Holger Puchta, KIT, Karlsruhe, Germany, for HR recombination assay (Molinier et al., 2004). Plants were germinated and grown on vertical plates containing ½ MS medium under long‐daylight conditions at 22°C for 6 days. Seedlings were transferred to 1.5 mm Al‐containing hydroponics and control solution without Al for 24 h. After 24 h Al treatment, seedlings were incubated in GUS staining solution [50 mm NaPO4, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6 and 2 mm X‐Gluc], vacuum infiltrated for 10 min at room temperature, and afterward destained in 70% ethanol at 60°C. Blue GUS spots were counted using a dissecting microscope (Zeiss Stemi 2000, Zeiss, Oberkochen, Germany). Images of leaves with blue spots were taken with a Zeiss Axioskop microscope.
Quantitative expression analysis
Ten‐day‐old seedlings were used, either untreated or treated with 1.5 mm Al for 3 h, then immediately frozen in liquid nitrogen. Total RNA was extracted from whole seedlings with an RNeasy Plant Mini Kit (Qiagen, Venlo, the Netherlands). First‐strand cDNAs were prepared from total RNA using the Transcriptor First‐Strand cDNA Synthesis Kit (Roche, Basel, Switzerland) according to the manufacturer's instructions. Quantitative PCR was performed with a Roche LightCycler 480 SYBR Green I Master with 0.5 μm specific primers and 0.1 μg of first‐strand cDNAs. Primer sequences are listed in Table S2. PCR reactions were conducted with the LightCycler 480 Real‐Time PCR System (Roche) under the following conditions: 95°C for 5 min; 45 cycles of 95°C for 10 sec, 60°C for 10 sec and 72°C for 15 sec. Data were normalized with three suitable reference genes (At1g02410, At4g26410, At3g47060), which were identified using the genevestigator tool RefGenes (Hruz et al., 2011). Statistical analyses were evaluated using qbasePLUS 3.0 (http://www.biogazelle.com/products/qbaseplus; Hellemans et al., 2007).
Author contributions
P.C., C.S., P.B.L. and A.S. conceived and designed the experiments. P.C. and C.S. performed the experiments. P.B.L. and A.S. contributed materials and reagents. P.C., C.S., P.B.L. and A.S. analysed the data. P.C., P.B.L. and A.S. wrote the article.
Funding
This work was supported by an ERA‐CAPS grant ALUCIDATE from the German Research Foundation (DFG, SCHN 736/6‐1) and the National Science Foundation (NSF) to P.B.L. and A.S. In addition, core funding of the University of Hamburg to A.S. is gratefully acknowledged.
Conflict of interest
The authors declare that none of them has a conflict of interest with the here‐presented data.
Availability
All materials and data supporting the findings of this study are available from the corresponding author on request.
Supporting information
Figure S1. Effects of increasing Al concentration on root tip meristem and γH2AX accumulation.
Figure S2. Effects of exposure time to Al on root tip meristem and γH2AX accumulation.
Figure S3. Mutants in CDKB1 are severely affected in growth after exposure to Al.
Figure S4. Mutants in CDKB1 have compromised DNA repair during recovery growth.
Table S1 Primers used for genotyping.
Table S2 Primers used for quantitative expression analyses.
Acknowledgements
The authors thank Maren Heese and Peter Bommert for critical reading and helpful comments to the manuscript. The authors thank Charles White (CNRS, Clermont‐Ferrand, France) for providing us with an anti‐plant γH2AX antibody. The authors are grateful for the ALUCIDATE network and the groups of Lieven de Veylder and Iwona Szarejko for feedback and discussion of this work.
References
- Biedermann, S. , Harashima, H. , Chen, P. , Heese, M. , Bouyer, D. , Sofroni, K. and Schnittger, A. (2017) The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J. 36, 1279–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard, J.Y. , Gallego, M.E. , Savigny, F. and White, C.I. (2005) Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41, 533–545. [DOI] [PubMed] [Google Scholar]
- Bray, C.M. and West, C.E. (2005) DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 168, 511–528. [DOI] [PubMed] [Google Scholar]
- Culligan, K. , Tissier, A. and Britt, A. (2004) ATR regulates a G2‐phase cell‐cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E. and Ryan, P.R. (1995) Aluminum toxicity and tolerance in plants. Plant Physiol. 107, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer, N. , Weimer, A.K. , Pusch, S. et al. (2009) Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21, 3641–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer, N. , Weimer, A.K. , De Veylder, L. , Novak, B. and Schnittger, A. (2010) The regulatory network of cell‐cycle progression is fundamentally different in plants versus yeast or metazoans. Plant Signal. Behav. 5, 1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux, M.P. , Couteau, F. , Bergounioux, C. and White, C. (1998) Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257, 283–291. [DOI] [PubMed] [Google Scholar]
- Eekhout, T. , Larsen, P. and De Veylder, L. (2017) Modification of DNA checkpoints to confer aluminum tolerance. Trends Plant Sci. 22, 102–105. [DOI] [PubMed] [Google Scholar]
- Friesner, J.D. , Liu, B. , Culligan, K. and Britt, A.B. (2005) Ionizing radiation‐dependent gamma‐H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3‐related. Mol. Biol. Cell 16, 2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, V. , Bruchet, H. , Camescasse, D. , Granier, F. , Bouchez, D. and Tissier, A. (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima, H. and Schnittger, A. (2012) Robust reconstitution of active cell‐cycle control complexes from co‐expressed proteins in bacteria. Plant Methods 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung, F. , Suer, S. , Knoll, A. , Wurz‐Wildersinn, R. and Puchta, H. (2008) Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4, e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans, J. , Mortier, G. , De Paepe, A. , Speleman, F. and Vandesompele, J. (2007) qBase relative quantification framework and software for management and automated analysis of real‐time quantitative PCR data. Genome Biol. 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz, T. , Wyss, M. , Docquier, M. , Pfaffl, M.W. , Masanetz, S. , Borghi, L. , Verbrugghe, P. , Kalaydjieva, L. , Bleuler, S. and Laule, O. (2011) RefGenes: identification of reliable and condition specific reference genes for RT‐qPCR data normalization. BMC Genomics 12, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki, S. and Umeda, M. (2011) Cell‐cycle control and plant development. Int. Rev. Cell Mol. Biol. 291, 227–261. [DOI] [PubMed] [Google Scholar]
- Karlik, S.J. , Eichhorn, G.L. , Lewis, P.N. and Crapper, D.R. (1980) Interaction of aluminum species with deoxyribonucleic acid. Biochemistry 19, 5991–5998. [DOI] [PubMed] [Google Scholar]
- Kochian, L.V. (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Biol. 46, 237–260. [Google Scholar]
- Kochian, L.V. , Hoekenga, O.A. and Pineros, M.A. (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. [DOI] [PubMed] [Google Scholar]
- Kuo, L.J. and Yang, L.X. (2008) Gamma‐H2AX ‐ a novel biomarker for DNA double‐strand breaks. In Vivo 22, 305–309. [PubMed] [Google Scholar]
- Lafitte, H.R. , Price, A.H. and Courtois, B. (2004) Yield response to water deficit in an upland rice mapping population: associations among traits and genetic markers. Theor. Appl. Genet. 109, 1237–1246. [DOI] [PubMed] [Google Scholar]
- Larsen, P.B. , Tai, C.Y. , Kochian, L.V. and Howell, S.H. (1996) Arabidopsis mutants with increased sensitivity to aluminum. Plant Physiol. 110, 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, P.B. , Geisler, M.J. , Jones, C.A. , Williams, K.M. and Cancel, J.D. (2005) ALS3 encodes a phloem‐localized ABC transporter‐like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 41, 353–363. [DOI] [PubMed] [Google Scholar]
- Lieber, M.R. , Grawunder, U. , Wu, X. and Yaneva, M. (1997) Tying loose ends: roles of Ku and DNA‐dependent protein kinase in the repair of double‐strand breaks. Curr. Opin. Genet. Dev. 7, 99–104. [DOI] [PubMed] [Google Scholar]
- Mari, P.O. , Florea, B.I. , Persengiev, S.P. et al. (2006) Dynamic assembly of end‐joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA 103, 18 597–18 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, H. , Morimura, S. and Takahashi, E. (1977) Binding of aluminium to DNA of DNP in pea root nuclei. Plant Cell Physiol. 18, 987–993. [Google Scholar]
- Molinier, J. , Ries, G. , Bonhoeffer, S. and Hohn, B. (2004) Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezames, C.D. , Sjogren, C.A. , Barajas, J.F. and Larsen, P.B. (2012) The Arabidopsis cell cycle checkpoint regulators TANMEI/ALT2 and ATR mediate the active process of aluminum‐dependent root growth inhibition. Plant Cell 24, 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack, M.K. , Harashima, H. , Dissmeyer, N. , Zhao, X. , Bouyer, D. , Weimer, A.K. , De Winter, F. , Yang, F. and Schnittger, A. (2012) Genetic framework of cyclin‐dependent kinase function in Arabidopsis. Dev. Cell 22, 1030–1040. [DOI] [PubMed] [Google Scholar]
- Ogita, N. , Okushima, Y. , Tokizawa, M. et al. (2018) Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 94, 439–453. [DOI] [PubMed] [Google Scholar]
- Puchta, H. and Hohn, B. (2012) In planta somatic homologous recombination assay revisited: a successful and versatile, but delicate tool. Plant Cell 24, 4324–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K. , Watson, J.M. , Parkey, J. and Shippen, D.E. (2002) Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds, M.A. and Larsen, P.B. (2008) Aluminum‐dependent root‐growth inhibition in Arabidopsis results from AtATR‐regulated cell‐cycle arrest. Curr. Biol. 18, 1495–1500. [DOI] [PubMed] [Google Scholar]
- Schröpfer, S. , Kobbe, D. , Hartung, F. , Knoll, A. and Puchta, H. (2014) Defining the roles of the N‐terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res. 42, 1684–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla‐Arnaud, M. , Crismani, W. , Larchevêque, C. et al. (2015) Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl Acad. Sci. USA 112, 4713–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla‐Arnaud, M. , Choinard, S. , Larchevêque, C. , Girard, C. , Froger, N. , Crismani, W. and Mercier, R. (2017) RMI1 and TOP3α limit meiotic CO formation through their C‐terminal domains. Nucleic Acids Res. 45, 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren, C.A. and Larsen, P.B. (2017) SUV2, which encodes an ATR‐related cell cycle checkpoint and putative plant ATRIP, is required for aluminium‐dependent root growth inhibition in Arabidopsis. Plant Cell Environ. 40, 1849–1860. [DOI] [PubMed] [Google Scholar]
- Sjogren, C.A. , Bolaris, S.C. and Larsen, P.B. (2015) Aluminum‐dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR‐, ALT2‐, and SOG1‐regulated transcriptional response. Plant Cell 27, 2501–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda, P. , Gal, S. , Hohn, B. and Puchta, H. (1994) Intrachromosomal homologous recombination in whole plants. EMBO J. 13, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.R. , Corpina, R.A. and Goldberg, J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double‐strand break repair. Nature 412, 607–614. [DOI] [PubMed] [Google Scholar]
- Weimer, A.K. , Biedermann, S. , Harashima, H. et al. (2016) The plant‐specific CDKB1‐CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 35, 2068–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, K. , Nagata, N. , Yee, K.M. , Braybrook, S.A. , Pelletier, J. , Fujioka, S. , Yoshida, S. , Fischer, R.L. , Goldberg, R.B. and Harada, J.J. (2005) TANMEI/EMB2757 encodes a WD repeat protein required for embryo development in Arabidopsis. Plant Physiol. 139, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama, K. , Conklin, P.A. , Huefner, N.D. and Britt, A.B. (2009) Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl Acad. Sci. USA 106, 12 843–12 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama, K.O. , Kobayashi, J. , Ogita, N. , Ueda, M. , Kimura, S. , Maki, H. and Umeda, M. (2013) ATM‐mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 14, 817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama, K.O. , Kimura, S. , Maki, H. , Britt, A.B. and Umeda, M. (2014) The role of SOG1, a plant‐specific transcriptional regulator, in the DNA damage response. Plant Signal. Behav. 9, e28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effects of increasing Al concentration on root tip meristem and γH2AX accumulation.
Figure S2. Effects of exposure time to Al on root tip meristem and γH2AX accumulation.
Figure S3. Mutants in CDKB1 are severely affected in growth after exposure to Al.
Figure S4. Mutants in CDKB1 have compromised DNA repair during recovery growth.
Table S1 Primers used for genotyping.
Table S2 Primers used for quantitative expression analyses.


