Abstract
Transfer RNAs (tRNAs) are essential components of the cellular protein synthesis machineries, but are also implicated in many roles outside translation. To become functional, tRNAs, initially transcribed as longer precursor tRNAs, undergo a tightly controlled biogenesis process comprising the maturation of their extremities, removal of intronic sequences if present, addition of the 3′‐CCA amino‐acid accepting sequence, and aminoacylation. In addition, the most impressive feature of tRNA biogenesis consists in the incorporation of a large number of posttranscriptional chemical modifications along its sequence. The chemical nature of these modifications is highly diverse, with more than hundred different modifications identified in tRNAs to date. All functions of tRNAs in cells are controlled and modulated by modifications, making the understanding of the mechanisms that determine and influence nucleotide modifications in tRNAs an essential point in tRNA biology. This review describes the different aspects that determine whether a certain position in a tRNA molecule is modified or not. We describe how sequence and structural determinants, as well as the presence of prior modifications control modification processes. We also describe how environmental factors and cellular stresses influence the level and/or the nature of certain modifications introduced in tRNAs, and report situations where these dynamic modulations of tRNA modification levels are regulated by active demodification processes. © 2019 IUBMB Life, 71(8):1126–1140, 2019
Keywords: tRNA, RNA modifications, modification enzymes, multispecific enzymes, modification circuits, regulation, epitranscriptome, demethylation, demodification, RNA epigenetics, stress response
Abbreviations
- D
dihydrouridine
- H2O2
hydrogen peroxide
- HPLC
high‐performance liquid chromatography
- manQ
mannosyl‐queuosine
- MMS
methyl methanesulfonate
- mRNA
messenger ribonucleic acid
- oQ
epoxyqueuosine
- pre‐tRNA
precursor of transfer ribonucleic acid
- Q
queuosine
- RNA
ribonucleic acid
- rRNA
ribosomal ribonucleic acid
- tRNA
transfer ribonucleic acid
- yW
wybutosine
- Ψ
pseudouridine
INTRODUCTION
Transfer RNAs (tRNAs) are essential components of the cellular protein synthesis machineries, but also serve additional functions as broad as amino acids delivery to membrane lipids, synthesis of peptidoglycan, and antibiotics 1, 2. In addition, under stress conditions, some tRNAs are cleaved, creating molecules acting in signaling or as regulators of gene expression 3, 4. To ensure these broad varieties of function within cells, tRNAs undergo a tightly controlled biogenesis process leading to the formation of mature and functional tRNAs. As tRNAs are highly abundant RNA molecules within cells, the tight regulation of their biogenesis is essential to prevent production and accumulation of nonfunctional tRNAs, and thus ineffective cellular energy consumption. The biogenesis of tRNAs starts with the synthesis of a pre‐tRNA produced by the transcription of a tRNA gene. The initial transcript is then subjected to a number of processing steps to yield the mature tRNA molecule (Fig. 1a). Depending on the organism from which the tRNA originates and on the identity of the tRNA, these steps may comprise the removal of the 5′‐leader and 3′‐trailer sequences from the pre‐tRNA transcript, the addition of the 3′‐CCA amino‐acid accepting sequence, the removal of intronic sequences, and the incorporation of a large number of posttranscriptional chemical modifications (Fig. 1a). Overall, these processing events generate a mature tRNA molecule adopting the well‐known cloverleaf secondary structure, defining five main regions called the acceptor stem, the D‐arm, the anticodon‐arm, the variable region, and the T‐arm (Fig. 1b). It is worth noting that these processing events are occurring in an intricate manner and not in a pure stepwise fashion 5, 6, 7. For instance, posttranscriptional modifications can be introduced throughout the entire process, with some modifications being incorporated on pre‐tRNAs with unprocessed 5′‐ or 3′‐extremities, some on pre‐tRNAs with mature extremities but with unprocessed introns, and some on tRNAs with mature extremities and processed introns. The chemical nature of these modifications is highly diverse, with more than hundred different modifications identified in tRNAs 8, 9, 10, 11. Among them, the most frequently found modifications include simple modifications such as ribose or base methylation (e.g., Gm, Cm, m1A, m2G, m5U, m7G, m5C), base reduction (dihydrouridine, D), base isomerization (pseudouridine, Ψ), and base thiolation (e.g., s2U, s4U, s2C). Other modifications include complex modifications involving the addition of larger chemical groups or requiring multiple modification steps (e.g., t6A, m5s2U, mcm5Um, ms2i6A—for the description of the nomenclature associated with RNA modifications, see for instance 10, 12, 13). The distribution of modifications along the tRNA sequence is far from random, and certain positions are found to be more frequently modified, such as positions 54 and 55 in the T‐arm, 16 and 20 in the D‐arm, and 32, 34, and 37 in the anticodon‐arm, whereas some other positions remain mostly unmodified 14. Although these modifications might seem to be distributed in several regions of the tRNA if one locates them on the cloverleaf secondary structure (Fig. 1b), it is striking to recognize that the three‐dimensional L‐shaped structure of the tRNA, which is formed via intricate interactions between the T‐ and D‐loops, tends to sort the modifications into two groups, namely modifications found in the tRNA core and modifications in the anticodon‐loop region (Fig. 1c). It is worth noting that most modifications found in the tRNA core are simple modifications, whereas most complex modifications are introduced in the anticodon‐loop region of tRNAs. Importantly, all the biological functions of tRNAs in cells are to various extents affected by modifications. For instance, the roles of modifications in the anticodon‐loop region in decoding during protein synthesis are well documented 9. In addition, modifications found in the tRNA core are collectively implicated in the folding and stability of tRNAs. It is now clear that even though most genes encoding tRNA modification enzymes are unessential and strains deleted from these genes show no obvious phenotypes in conditions of optimal growth, modifications in the tRNA core are key for tRNA stability 12, 15. Effects on stability are most of the time revealed by the combination of multiple alterations in the modification genes 16, 17, but may also result from a single lack of modification in a single tRNA, such as the m1A58 modification, which is essential for the stability of the yeast initiator tRNAi Met 18. Interestingly, the lack of certain specific modifications in the tRNA core has pronounced effect on the stability or function of only a small number of tRNAs, the effect on many others being small to inexistent 19. This suggests that modifications do not have the same beneficial effect for all tRNAs, and that the introduction of certain modifications in some tRNAs is the consequence of the presence of modification enzymes important for other tRNA species 19. In addition to these functions in decoding and stability, modifications in tRNAs are also affecting alternative functions of tRNAs outside translation, making all aspects of tRNA biology controlled and modulated by modifications 1, 4, 9.
Figure 1.
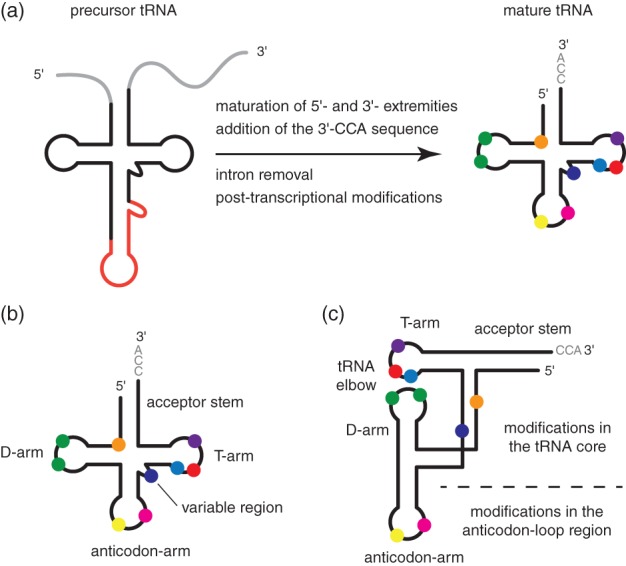
Biogenesis of tRNAs and posttranscriptional modifications. (a) The biogenesis of tRNAs. Precursor tRNAs (left) contain 5′‐leader and 3′‐trailer extremities (grey), and some tRNA species also contain introns (red). The different processing steps leading to mature tRNAs (right) include the removal of the 5′‐leader and 3′‐trailer sequences, the addition of the 3′‐CCA amino‐acid accepting sequence, the removal of intronic sequences, and the incorporation of a large number of posttranscriptional chemical modifications (filled circles of various colors). (b) Cloverleaf representation of tRNA secondary structure. The five main regions of tRNAs, namely the acceptor stem, the D‐arm, the anticodon‐arm, the variable region, and the T‐arm are labeled on the scheme. Typical modifications are represented with filled circles of various colors. (c) Schematic representation of the L‐shaped three‐dimensional structure of tRNA. In this structure, the acceptor stem stacks on the T‐arm and the D‐arm stacks on the anticodon‐arm. The T‐loop and D‐loop interact and form the tRNA elbow structure. Modifications can be classified into two groups, namely modifications in the tRNA core and modifications found in the anticodon‐loop region.
Collectively, the introduction of modifications at appropriate positions in tRNAs is of primary importance for optimal cell function. The understanding of the mechanisms that determine and influence nucleotide modifications in tRNAs thus holds a central position in the general understanding of the biological functions of tRNA modifications. Within each organism, although tRNA molecules must all be, to some extent, sufficiently similar to each other to be recognized and used by the translation machinery, they at the same time need to be sufficiently dissimilar to be uniquely recognized by their cognate aminoacyl‐tRNA synthetases. This notion is perfectly appropriate to describe the challenge faced by tRNA modification enzymes, which have been shaped by million years of coevolution with tRNAs to deal with a population of highly similar but unique tRNAs such as to modify them in a very specific manner 20, 21. The present review aims at presenting and summarizing the different aspects that determine whether a certain position in a tRNA molecule is modified or not. These aspects include the sequence and structural determinants of modification enzymes, as well as the presence of prior modifications, and will be presented in the first parts of this review. Importantly, modifications in tRNA are not static decorations that are introduced on every tRNA of the cell in any circumstances. The level of certain modifications in a tRNA population can change dynamically in response to several factors, and in some cases, these dynamic changes are thought to be not only due to alterations in the introduction of these modifications, but also to active demodification processes. These aspects will be described in the latest parts of this review. These elements will be discussed in the light of selected examples from prokaryotic and cytoplasmic tRNAs, and this review will not cover all particularities found in mitochondrial tRNAs. The reader interested in posttranscriptional modifications in mitochondrial tRNAs is thus referred to recent excellent reviews 22, 23.
Predicting Modifications in tRNA Sequences
The entire modification pattern of a complete set of tRNAs has been experimentally determined only for a small number of organisms, including Escherichia coli, Mycoplasma capricolum, Lactococcus lactis, Streptomyces griseus, Saccharomyces cerevisiae, and Holoferax volcanii 8. Even with the latest technological developments, the experimental determination of the complete modification status of tRNAs remains a difficult and demanding process 24, 25, 26, which can explain in part the relatively low number of organisms, especially in eukaryotes and archaea, for which the complete landscape of tRNA modifications is known. This provided a general interest and a serious motivation for developing computational tools aiming at predicting the nature and the sites of modification in tRNA sequences from any organism. Some of these tools have been developed recently, the most comprehensive computational method for predicting posttranscriptional modifications in tRNAs being tRNAmodpred 27. The predictions performed by tRNAmodpred for organisms for which tRNA modification profiles are unknown but for which the complete sequence of the genome is available, are based on detection of homology to known tRNA modification enzymes conserved during evolution. After the initial identification of tRNA modification genes in the genome, the set of putative enzymes is mapped onto tRNA modification pathways from the MODOMICS database 8, and the resulting potential modifications are then mapped onto the tRNA sequences of interest. The mapping is done based on the identity of the nucleotide present at a given position, meaning that all tRNAs presenting the appropriate nucleotide at the target position of an identified modification enzyme will be marked as modified at this position. In addition, more specific tools aiming at predicting modifications in tRNAs have been developed such as PPUS that is specialized in the identification of pseudouridine sites in yeast and human 28, and tRNAmod that is very efficient in predicting the three most common uridine modifications, namely pseudouridine, dihydrouridine, and 5‐methyluridine 29. Although tRNAmodpred is an attractive computational tool that meets the important need for obtaining information on the nature and the sites of modification in tRNA sequences, it suffers from major limitations, which overall result in a relatively low accuracy of the predictions 27. True experimental determination of modification patterns therefore remains essential. It provides a wealth of information that is valuable per se, and most probably decisive to improve the predictions of computational tools. The most important limitation leading to false positive predictions in tRNAmodpred results from the assumption in the algorithm that all tRNA modification enzymes can modify any tRNA sequence as long as it carries the correct nucleotide at the position to be modified. The method would thus need to be improved by incorporating knowledge on enzyme specificities, as it is well known that tRNA modification enzymes have specificity determinants that control whether a specific tRNA is indeed modified by a given enzyme. The determinants of specificity in modification enzymes will be the subject of the following section.
Determinants of Specificity in tRNA Modification Enzymes
The features in the tRNA substrate that control whether a given position in its sequence is modified or not are collectively referred to as “determinants of specificity.” From a general perspective, modification enzymes can be classified into three classes depending on their degree of specificity with respect to tRNA identities (Fig. 2a–c). The first class includes modification enzymes with an activity dictated by the chemical nature of the target nucleotide. For instance in S. cerevisiae, all the cytosolic tRNAs having a U at position 54 are modified at this position to m5U54 also designated T54 (Fig. 2a). This almost completely conserved modification incorporated by Trm2 in yeast, TrmA in E. coli and TrmFO in B. subtilis and T. thermophilus 30, 31, 32, 33, is at the origin of the name given to this subdomain of tRNAs, namely the T‐arm (Fig. 1b). This class also includes the Tad2/Tad3 heterodimer, which introduces the I34 modification in all the yeast cytosolic tRNAs having an A at position 34 34, 35. In addition, the Trm5 and TrmD enzymes that modify all tRNAs having a G at position 37 to m1G37, in yeast and E. coli, respectively, belong to this class 36, 37. m1G37 is further modified into yW in yeast tRNAPhe (see below).
Figure 2.
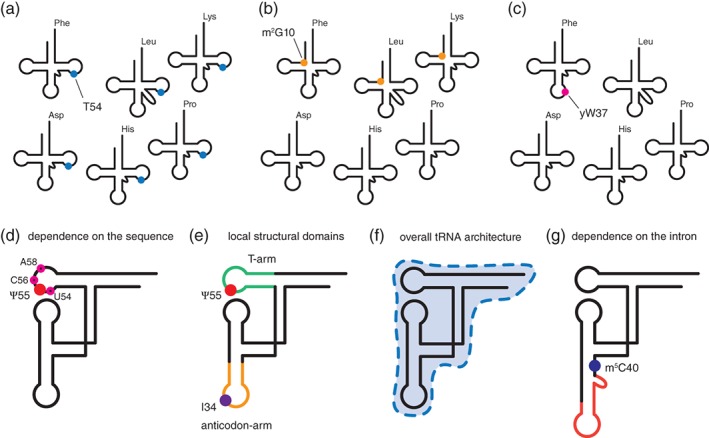
Determinants of specificity in tRNA modification enzymes. (a–c) Modification enzymes can be classified into three classes depending on their degree of specificity with respect to tRNA identities. Examples are drawn based on the specificity found in the yeast S. cerevisiae. (a) The first class consists of modification enzymes that modify all tRNAs with the correct target nucleotide, such as Trm2 introducing T54 in all cytoplasmic tRNAs in yeast. (b) The second class consists of modification enzymes that modify only a subset of tRNAs having the correct target nucleotide, such as Trm11/Trm112 introducing m2G10 in approximately half of the tRNAs in yeast. (c) The third class consists of modification enzymes that modify only a single tRNA species. For instance in yeast, the complex modification wybutosine (yW) is found uniquely at position 37 of tRNAPhe. (d–g) General principles controlling substrate recognition by tRNA modification enzymes. These general principles include (d) dependence on the RNA sequence, (e) dependence on local structural domains, (f) dependence on the overall tRNA architecture, and (g) dependence on the presence of an intronic sequence. A combination of these different principles determines the actual specificity of tRNA modification enzymes, which often involves both a certain degree of sequence specificity and structural determinants.
The second class consists of enzymes that modify only a subset of tRNAs having the correct target nucleotide at the position to be modified. This class includes the Trm11 enzyme from yeast, which is responsible for the synthesis of m2G10 in tRNAs in association with the ubiquitous partner protein Trm112 that is also required for the function of other methyltransferases 38, 39, 40, 41. The m2G10 modification is found in approximately half of the tRNAs, although all yeast tRNAs have a G at position 10 (Fig. 2b). Other yeast enzymes belonging to this class are the Trm10 enzyme, which introduces the m1G9 modification in approximately half of the cytosolic tRNAs having a G at position 9 42, 43, the Trm6/Trm61 heterodimer, which introduces the m1A58 modification in about two‐thirds of cytosolic tRNAs (all having an A at this position) 18, 34, 44, and the Trm3 enzyme, which introduces the Gm18 modification in about one‐third of cytosolic tRNAs (all having a G at this position) 45. The m1A58 modification is not found in E. coli, but in prokaryotes such as Thermus thermophilus or Pyrococcus abyssi, the m1A58 modification is introduced by tetrameric TrmI enzymes 46, 47, 48 with a certain degree of sequence specificity 49. This implies that probably only a subset of tRNAs are substrates of these enzymes like for Trm6/Trm61, although this point has not been yet systematically analyzed, the complete patterns of tRNA modifications in these species being sparse or inexistent 8. In E. coli and other bacteria such as Aquifex aeolicus, the Gm18 modification is introduced by the TrmH enzyme that modifies only a subset of tRNAs, like the Trm3 enzyme, whereas in other bacteria such as T. thermophilus, all tRNAs are substrate of the TrmH enzyme 50. In E. coli, this class of modification enzymes also includes TrmJ, which catalyzes the Um32 and Cm32 modifications in the anticodon 51, 52. Interestingly, the specificity of an archaeal homolog of TrmJ in Sulfolobus acidocaldarius is different from the one of E. coli and methylates the ribose of cytidines and not uridines 53. An important point that is manifest in these examples is that the understanding of the determinants of specificity for this class of modification enzymes is very difficult to appreciate and generalize as the behavior of a given enzyme in one organism might not apply to other species, even though their enzymes might look very similar.
The third class includes modification enzymes that are highly specific to the tRNA identity because they modify a single tRNA species. An example in yeast is the enzyme that introduces the complex modification yW uniquely at position 37 of tRNAPhe and not in the other tRNAs having a G at this position (Fig. 2c). This type of highly specific modification enzymes also include the enzyme catalyzing the 2′‐O‐ribosyl phosphate addition to A64 in yeast initiator tRNAi Met 54, the tRNAHis guanylyltransferase Thg1 that adds the G−1 residue unique to tRNAHis 55, 56, Dnmt2 that introduces the m5C38 modification specifically in tRNAAsp or tRNAGlu depending on the organism 57, 58, and the acetyltransferase TmcA that introduces the ac4C34 modification in E. coli Elongator tRNAMet 59. The enzymes of this class are often highly sequence‐specific, such as for instance Tgh1 that adds the G−1 residue to tRNAs possessing a histidine GUG anticodon 55.
All the tRNA modification enzymes recognize and distinguish their tRNA substrates based on the RNA sequence (Fig. 2d), the overall tRNA architecture (Fig. 2e), local structural domains (Fig. 2f), and the presence or the absence of an intronic sequence (Fig. 2g), which altogether form the determinants of specificity (Table 1).
Table 1.
Determinants of specificity in tRNA modification enzymes
| Enzymea | Modification | Sequence | Local structural domains | Overall tRNA architecture | Intron | References |
|---|---|---|---|---|---|---|
| TruB (Ec) | Ψ55 | ● | ● | 60, 61 | ||
| TrmB (Aa) | m7G46 | ● | ● | 62, 63 | ||
| Trm8/Trm82 (Sc) | m7G46 | ● | 64, 65 | |||
| TadA (Ec, Sa) | I34 | ● | 66, 67 | |||
| TrmL (Ec) | Cm34/Um34 | ● | 68 | |||
| TrmO (Ec) | m6t6A37 | ● | ● | 69 | ||
| TrmJ (Ec) | Cm32/Um32 | ● | 52 | |||
| Trm11 (Tk) | m2G10/m2 2G10 | ● | 70 | |||
| Trm11/Trm112 (Sc) | m2G10 | ● | 41 | |||
| Trm1 (Sc, Pf, Aa) | m2 2G26 | ● | 71, 72, 73, 74 | |||
| Trm4 (Sc) | m5C34, m5C40 | ● | 75, 76 | |||
| Pus1, Pus7 (Sc) | Ψ34, Ψ35, Ψ36 | ● | 75, 76 |
Organisms are given in parenthesis. Ec: Escherichia coli; Aa: Aquifex aeolicus; Sc: Saccharomyces cerevisiae; Sa: Staphylococcus aureus; Pf: Pyrococcus furiosus; Tk: Thermococcus kodakarensis.
Sequence Recognition
The pseudouridylation catalyzed by the bacterial TruB enzyme to yield Ψ55 in the T‐arm of tRNAs is primarily controlled by sequence‐specific features in the T‐loop, with U54, C56 and especially A58 being essential for recognition and catalysis (Fig. 2d) 60, 61. In addition, the stem‐loop structure formed by the T‐arm is necessary and sufficient for Ψ55 modification by TruB, which therefore depends on a local structural domain and not on the overall tRNA structure (Fig. 2e). Similarly, tRNA recognition by the bacterial TrmB enzyme from A. aeolicus that introduces m7G46 in most tRNAs with a short variable region (Fig. 1b), is achieved via a combination of sequence and local structure determinants 62. The Trm8/Trm82 heterodimer that catalyzes the same reaction in yeast has stricter recognition requirements than TrmB and needs both the T‐ and D‐arms but not a fully intact tRNA for efficient catalysis 64, 65.
Local Structural Domains Versus Overall tRNA Architecture Recognition
Altering key tertiary interactions essential for the proper three‐dimensional folding of tRNAs showed that some modification enzymes require an intact tRNA architecture, whereas others remain active on imperfectly folded tRNAs 77. A comprehensive study of yeast tRNAAsp microinjected in Xenopus laevis oocytes showed that most modifications enzymes targeting the branch of the L formed by the T‐arm and accepting stem (Fig. 1c) do not need an intact tRNA as substrate, whereas most of the modification enzymes targeting the other branch formed by the D‐arm and anticodon‐arm (Fig. 1c) are sensitive to changes in the global tRNA architecture 77. However, this classification only provides a general trend regarding the specificity of modification enzymes families.
Many studies have characterized modification enzymes for which local structural domains are the main determinants of specificity (Fig. 2e). For instance, tRNA recognition by the bacterial enzyme TadA that converts adenosine into inosine at position 34 of the anticodon loop does not depend on the global tRNA architecture but on the local anticodon stem‐loop structure (Fig. 2e) 66, 67. Similarly, E. coli TrmL and TrmO are active on anticodon stem‐loops 68, 69.
Conversely, several modification enzymes for which the substrate recognition requires an intact tRNA architecture were identified (Fig. 2f). The tRNA recognition mode of some of these enzymes has been characterized structurally, which shed light on the molecular basis that makes the intact tRNA architecture required for binding and activity. For instance, E. coli TrmJ requires an intact elbow and anticodon‐loop regions for methylation, whereas the size of the acceptor stem can be reduced without affecting its catalytic activity 52. Conversely, the archaeal Trm11 requires an intact elbow and accepting region in the tRNA substrate. A docking model of the tRNA/Trm11 complex suggested that substrate recognition and catalysis is achieved by a molecular ruler mechanism in which the distances between the active site and both the 3′‐CCA end and the variable region are precisely defined 70. A similar tRNA/enzyme model was proposed for the orthologous yeast Trm11 enzyme that acts as a heterodimer with the Trm112 protein. However, in this case, additional contacts are formed between the tRNA anticodon‐loop and the Trm112 partner 41. Another family of enzymes sensitive to the overall tRNA architecture is constituted by Trm1 enzymes, which catalyze m2 2G26 formation in the tRNA core. Although all enzymes from this family are sensitive to alterations in the tRNA three‐dimensional structure, the eukaryotic and archaeal enzymes recognize primarily the D‐arm and the variable region, whereas the bacterial enzyme from A. aeolicus has recognition sites in the tRNA elbow and T‐arm regions 71, 72, 73, 74.
Intron Recognition
A particular set of modifications enzymes sensitive to local structural domains consist of enzymes requiring the presence of the intron in the tRNA substrate (Fig. 2g). As expected, the corresponding modifications are located in the anticodon‐loop region 75. In yeast, the modifications enzymes that require the presence of the intron catalyze the formation of m5C34, Ψ34, Ψ35, Ψ36, and m5C40 75, 76, whereas those that require the prior removal of the intron include the enzymes introducing Gm18, Cm32, Ψ32, m1G37, yW37, i6A37, and Um44 in tRNAs 75, 76, 78.
Multispecific Modification Enzymes
Multispecific modification enzymes add a new flavor to the question of the specificity in tRNA modifications. Multispecific enzymes can be divided into two groups, modification enzymes that modify several positions in tRNA substrates, or modification enzymes that modify tRNA substrates as well as non‐tRNA substrates. Multispecific enzymes that modify several positions in tRNAs include archaeal TrmI that introduces m1A on both A57 and A58 47, bacterial Trm1 that introduces m2 2G on both G26 and G27 74, Trm4 that introduces m5C at multiple positions 79, 80, pseudouridine synthases Pus1, Pus3, and TruA that introduce Ψ at multiple positions 81, 82, 83, dihydrouridine synthases Dus1 and Dus4 in S. cerevisiae and DusA in E. coli that introduce D at positions 16 and 17 for Dus1 and 20a and 20b for Dus4 and DusA 84, 85, and the archaeal ArcTGT from Thermoplasma acidophilum that catalyzes the formation of preQ0 at both positions 15 and 13 in the D‐arm of tRNAs 86. In the case of the multispecific enzyme Trm7 from yeast that introduces Cm32 and Nm34 in the anticodon loop, auxiliary subunits Trm732 and Trm734 regulate the site of modification by associating with the catalytic subunit 34, 87. Interestingly, some enzymes from the Trm10 family can methylate both adenosine and guanosine at position 9 of tRNAs 88, 89, 90. Multispecific enzymes that modify tRNAs as well as other RNA substrates include bacterial TrmA from E. coli that has been shown to also introduce m5U54 in tmRNA 91, bacterial RlmN from E. coli that catalyzes m2A modification both at position 2,503 of 23S rRNA and at position 37 of several tRNAs 92, and eukaryotic TRMT61B that catalyzes m1A formation both at position 58 of mitochondrial tRNAs and at position 947 of human mitochondrial 16S rRNA 93. Finally, it is worth mentioning that some RNA modifications found in mRNAs are likely introduced by tRNA modification enzymes 94, 95. This is the case for Ψ and m5C modifications, which are introduced by members of the Pus and Trm4 families, that already have promiscuous activities within tRNAs (see above). Overall, the fact that many tRNA modification enzymes do not require an intact tRNA architecture for activity but are instead dependent on specific sequence and local structures (Fig. 2d,f and text above) suggests that the same modifications may be introduced in mRNAs by tRNA modifying enzymes.
Prior Modifications in tRNA Can Influence the Introduction of New Modifications
In addition to the determinants of specificity described in the previous section (Fig. 2), the introduction of several modifications in tRNAs are influenced by the presence of existing modified nucleotides. In the literature, this aspect of RNA modification is called “modification circuits” to emphasize the fact that it connects modifications with one another and obviously drives a defined order in the introduction of these modifications 96. The term “network” is also sometimes used to underline the fact that modifications may be interconnected with several others 97. Most of the well‐documented examples of modification circuits occur in the anticodon‐loop region and involve modifications at position 34 or 37 or both, positions that are two of the most frequently modified in tRNAs, and at which most complex modifications are found (Fig. 3a–e) 8, 14. Although the exact reasons for which modification circuits exist in tRNAs and are mostly found in the anticodon‐loop region are not known, it was recently proposed that modifications introduced first act as additional recognition elements for other modifications, which provides the mean for adding modifications with considerable variation in the anticodon‐loop region, despite the lack of variability in its local sequence and structure 96. These modification circuits in the anticodon‐loop region include: the Um34 and Cm34 modifications introduced in E. coli on tRNALeu(UAA) and tRNALeu(CAA) by TrmL, which require prior formation of i6A37 or ms2i6A37 (Fig. 3a) 68, 98; the m3C32 modification introduced in yeast on tRNASer and tRNAThr by Trm140, which is greatly stimulated by the prior formation of i6A37 or t6A37 (Fig. 3b) 99, 100; the complex wybutosine yW37 modification in the anticodon loop of eukaryotic tRNAPhe, which requires prior Cm32 and Gm34 modifications (Fig. 3c) 87, 101; the m5C38 modification introduced in mammals, Drosophila melanogaster, Arabidopsis thaliana, and S. pombe on tRNAAsp by the Dnmt2 family of enzymes, which is greatly stimulated by prior queuosine Q34 or mannosyl–queuosine manQ34 modifications (Fig. 3d) 57, 102, 103; and the I34 modification introduced in Trypanosoma brucei on tRNAThr(AGU) by ADAT2/ADAT3 heterodimer, which is stimulated by prior deamination of C32 leading to U32, which is itself stimulated by the prior methylation of C32 leading to m3C32 (Fig. 3e) 104, 105. In all these cases, the precise molecular mechanisms that define these modification circuits are not yet fully understood, but it is reasonable to conceive that the prior modification might either act as a direct recognition determinant for the subsequent modification enzyme or alter the local structure of the anticodon‐loop region, thereby helping to present the appropriate structure to the subsequent modification enzyme. Modified bases in the anticodon‐loop indeed reinforce its U‐turn anticodon‐loop structure. Modifications at positions 32 and especially 37 prevent formations of base pairs between nucleosides 32–38 and 33–37, thereby enabling canonical anticodon‐loop folding 12, 106, 107.
Figure 3.
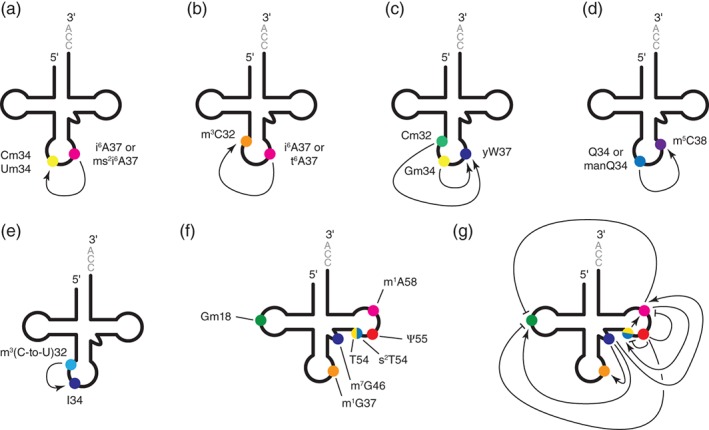
Modification circuits in tRNAs. (a–e) Modification circuits found in the anticodon‐loop region. (a) i6A37 or ms2i6A37 stimulate Um34 and Cm34 formation in E. coli tRNALeu(UAA) and tRNALeu(CAA). (b) i6A37 stimulates m3C32 formation in tRNASer of S. pombe and S. cerevisiae and t6A37 stimulates m3C32 formation in tRNAThr of S. cerevisiae. (c) Cm32 and Gm34 stimulate wybutosine yW37 formation in eukaryotic tRNAPhe. (d) Queuosine Q34 or mannosyl–queuosine manQ34 stimulate m5C38 formation in tRNAAsp of several organisms (see text). (e) m3U32 originating from m3C32 stimulates I34 formation in T. brucei tRNAThr(AGU). (f, g) An intricate modification network in T. thermophilus. (f) The modifications involved in the modification network in T. thermophilus are represented on the cloverleaf representation of tRNA with filled colored circles. Gm18 is shown in green, m1G37 in orange, m7G46 in purple, T54 and s2T54 as half‐filled circles in yellow and blue, respectively, Ψ55 in red, and m1A58 in pink. (g) An intricate modification network in T. thermophilus enables the adaptation of tRNA modifications in response to temperature changes. The same color code is used as in panel (f). Arrows indicate stimulatory effects and blunted lines inhibitory effects.
In addition to these modification circuits in the anticodon‐loop region, several circuits involving modifications in the tRNA core have been described in T. thermophilus (Fig. 3f,g) 108. T. thermophilus is an extreme thermophilic bacteria that can grow at temperature ranging from ~50 to 80 °C, and in which changes in external temperature conditions result in changes in the modification level of several modifications in the tRNA core. These variations in the modification content of tRNAs depending on the external temperature have been shown to be linked to an adaptation of protein synthesis to temperature change 109. Interestingly, this adaptation is not controlled by transcriptional and/or translational regulations, but is established via modification circuits. These modification circuits in the tRNA core include: the m1A58 modification, which is stimulated by T54 110; the s2T54 modification from T54, which is stimulated by m1A58 111; the Gm18, m1G37, and m1A58 modifications, which are stimulated by m7G46 112; the Gm18, s2T54, and m1A58 modifications, which are negatively regulated by Ψ55 113; and the Gm18 modification, which is negatively regulated by m1A58 110 (Fig. 3g). These modification circuits regulate the extent of modifications in response to temperature change. For instance, at elevated temperature, the presence of m7G46 favors the introduction of Gm18 and m1A58, which further stimulates formation of s2T54, the two latter modifications being essential for survival at high temperature (~75–80 °C) 112. In contrast, at lower temperature, the Ψ55 modification inhibits formation of Gm18, m1A58, and s2T54, which maintains a sufficient flexibility of tRNAs at low temperatures, an essential feature for T. thermophilus survival at low temperature (~50–55 °C) 113. Such a mechanism of regulation, which does not include any transcriptional or translational regulatory steps and is not limited to T. thermophilus, makes the response of the network to environmental changes very rapid. Here, modification circuits are controlling variations in the modification content of tRNAs in response to temperature changes, but alteration of the modification status of tRNAs in response to environmental changes is not limited to T. thermophilus and may not necessarily involve circuits of modifications. The dynamic changes in tRNA modifications will be described in the next section.
Variations in tRNA Modifications in Response to Environmental Changes
Although it has been commonly assumed that tRNA modifications are constitutively introduced as static decoration on every tRNA of the cell, there is now established and continually growing evidence indicating that the level and/or the nature of certain modifications in a tRNA population can change dynamically in response to several environmental factors, such as cellular stresses, temperature, or nutriment availability 11. At the level of individual tRNA molecules, these variations consist in the appearance, disappearance, or change of the nature of the modification (Fig. 4a). These variations collectively have an impact on the global level of modifications in tRNA populations. For instance, some subgroups such as particular isoacceptors or isodecoders might be affected differently (Fig. 4a).
Figure 4.
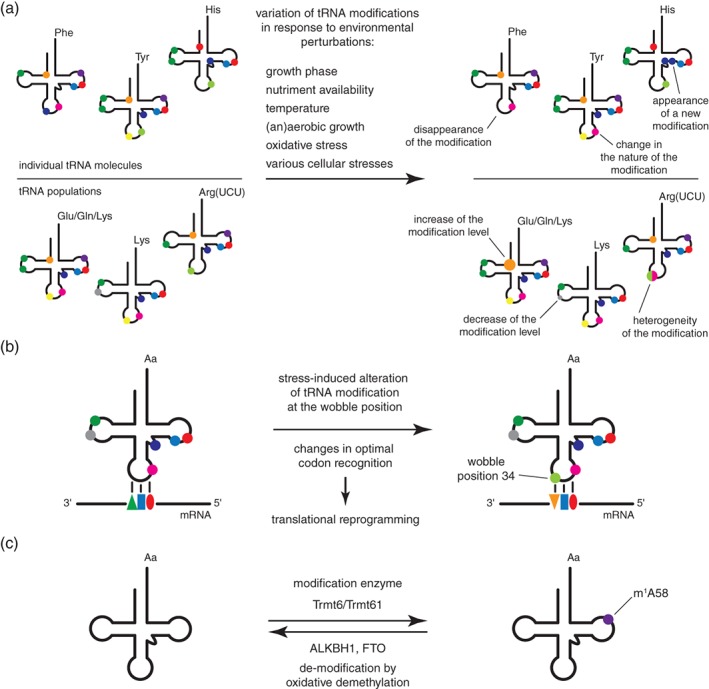
Dynamic changes in tRNA modification levels. (a) Variations of tRNA modifications in response to environmental perturbations. In response to various environmental or cellular factors, such as growth phase, nutriment availability, temperature, aerobic versus anaerobic growth, oxidative stress, or various cellular stresses, the levels and/or the nature of certain modifications can change in tRNAs. Variations occur at the level of individual tRNA molecules (disappearance of a modification, appearance of a new modification, and change in the nature of the modification). These variations collectively impact the level of modifications in tRNA populations, such as particular isoacceptors (e.g., tRNALys) or isodecoders (e.g., tRNAArg(UCU)). In a certain tRNA population, these variations may appear in the form of increased or decreased modification levels, or may introduce modification heterogeneities at certain positions. (b) Stress‐induced alteration of tRNA modification at the wobble position can lead to translational reprogramming. Stress‐induced alteration of tRNA modification at the wobble position 34 changes the optimal codon recognition of the given tRNA and thereby upregulates the formation of critical stress‐response proteins encoded by mRNA transcripts enriched in this particular codon adapted to the altered wobble modification. (c) Variations in tRNA modifications and active demodification processes. The m1A58 modification, introduced in human by the Trmt6/Trmt61 modification enzyme, can be actively removed by the demodification enzymes ALKBH1 and FTO via an oxidative demethylation process.
Factors Causing Variations in tRNA Modifications
Several factors, including growth phase, growth rate, growth medium, (an)aerobic growth, and temperature, were reported to affect the levels and/or the nature of tRNA modifications (Table 2). For instance, the level and the nature of modifications have been shown to vary between different growth phases in B. subtilis 114, 115. In addition, the synthesis of Q34 from its derivative oQ requires vitamin B12, which is only synthetized in S. typhimurium under anaerobic conditions, provided that cobalt ions, glycerol as the carbon source, and fumarate as the electron acceptor are present. Therefore, oQ34 is present in tRNAs from bacteria grown in glucose salt medium lacking vitamin B12, whereas Q34 is formed in bacteria grown in rich media 120. Furthermore, a meticulous study of the tRNA modification pathways of wobble U34 in the E. coli tRNAs that are substrates of MnmE/MnmG and MnmC modification enzymes, revealed that the growth conditions, namely growth medium and growth phase, affect the enzymatic pathway leading to the final modifications 126. Changes in tRNA modifications in response to external factors and growth conditions are not restricted to prokaryotes and the levels of many modifications are sensitive to changes in temperature in S. cerevisiae, with for instance the thiolation of U34 being impaired at elevated temperatures (>30 °C), a temperature at which tRNAs therefore display mcm5U34 instead of mcm5s2U34 122. In S. cerevisiae, additional m5C modifications were shown to be specifically introduced on tRNAHis at position 48 and 50 in response to several growth arrest conditions, yet the significance of these additional m5C remains unclear 127. Also in human, several types of cancer cell lines were shown to contain tRNAs with perturbed contents of modified bases 128. The tumor‐specific tRNA species are usually hypomodified, with for instance reduced wybutosine yW37 modification of tRNAPhe 124 and reduced levels of queuosine Q34 of tRNAAsp 125.
Table 2.
Factors causing variations in tRNA modifications
| System description | Growth phase | Growth medium | Growth conditions | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organisma | tRNA | Modification | Exponential phase | Stationary phase | Growth rate | Rich medium | Glucose salt medium | Aerobic growth | Anaerobic growth | Temperature | Tumors | References |
| Bs | + b | ++ | 114 | |||||||||
| Bs | tRNATyr | i6A37 | ms2i6A37 | 115 | ||||||||
| Ec | ms2i6A37 | ● | 116 | |||||||||
| Ec | Q34 | ● | 117 | |||||||||
| Ec | tRNALeu 1, tRNAGly 2&3, tRNAPro 1 | s4U8 | ● | 118 | ||||||||
| Bs | tRNAPhe | Gm34 | G34 | 119 | ||||||||
| Bs | tRNAPhe | ms2i6A37 | i6A37 | 119 | ||||||||
| St | Q34 | oQ34 | 120 | |||||||||
| St | ms2io6A37 | ms2i6A37 | 121 | |||||||||
| Tt, Mb, Sh, Sc | ● | 12, 112, 113, 122, 123 | ||||||||||
| Hs | tRNAPhe | yW37 | – c | 124 | ||||||||
| Hs | tRNAAsp | Q34 | – c | 125 | ||||||||
Bs: Bacillus subtilis; Ec: Escherichia coli; St: Salmonella typhimurium; Tt: Thermus thermophilus; Mb: Methanococcoides burtonii; Sh: Stetteria hydrogenophila; Sc: Saccharomyces cerevisiae; Hs: Human.
Hypomodified in the exponential phase compared to the stationary phase.
Hypomodified in tumors.
Alteration of Modifications at the Wobble Position and Translational Reprogramming
Although the roles of these various alterations in the levels of modifications are in most cases unknown, a recent concept has emerged that provides a convincing explanation regarding the role of modification alterations in response to environmental changes for a subset of modifications located at the wobble position 129. In this model, stress‐induced alteration of tRNA modification at the wobble position promotes a translation regulation to increase the formation of critical stress‐response proteins encoded by mRNA transcripts with a particular codon bias adapted to the altered wobble modification (Fig. 4b). A quantitative analysis of modifications in tRNA by HPLC‐coupled mass spectrometry showed that dynamic changes of these modifications in response to various cellular stress conditions are widespread and stress‐specific 130. For instance, Cm, m5C, and m2 2G modifications increase in response to H2O2 exposure but decrease or are unaffected by exposure to the methylating agent MMS. In subsequent studies, some specific changes of tRNA modification levels were shown to drive selective translation of codon‐biased mRNAs. For instance, in S. cerevisiae exposed to H2O2, an increase in the proportion of tRNALeu(CAA) containing m5C at the wobble position promotes selective translation of mRNA from genes enriched in the TTG codon, including expression of Rpl22a, a ribosomal protein that contributes to the oxidative stress survival response 131. Similarly, in S. pombe, the mcm5s2U modification introduced for its cm5 part by Elongator at the wobble position of tRNALys(UUU) was shown to be critical for efficient translation of certain stress‐induced mRNAs from genes enriched in the AAA codon because strains lacking functional Elongator displayed higher stress sensitivity to H2O2 132, 133. Later reports highlighted other facets of the role of tRNA wobble mcm5s2U34 modifications in translation of codon‐biased mRNAs, where either the methylation catalyzed by Trm9/Trm112 heterodimer leading to mcm5U from cm5U, or the thiolation introduced by the URM1 pathway leading to (mcm5)s2U34 were shown to promote effective translation of mRNAs enriched in the AGA and GAA codons for the methyl group and in the AAA and GAA codons for the thiol group 134, 135. Furthermore, dynamic changes in the level of U34 thiolation in response to elevated temperature in S. cerevisiae were proposed to contribute to the heat shock response by reducing translation of specific genes 135. In addition, an increase of wobble cmo5U34 in tRNAThr(UGU) of Mycobacterium bovis in response to hypoxia was reported to promote translation of specific transcripts enriched in the ACG codon, including the DosR master regulator of hypoxia, here again linking stress‐induced tRNA modification changes to translation reprogramming 136. Finally, it is worth to briefly mention that tRNA modifications have been linked to stress response via a seemingly unrelated mechanism implicating tRNA stress‐induced cleavage and the formation of tRNA‐derived small‐RNAs, with some tRNAs becoming more sensitive to endonucleolytic cleavage in the absence of m5C modifications 4, 137.
Variations in tRNA Modifications and Active demodification Processes
In all RNA families, variations in the levels of modifications in a given tRNA in response to environmental changes or other factors can be achieved via (i) changes in the expression or the activity of modification enzymes; (ii) alteration of RNA synthesis or degradation; (iii) involvement of demodification enzymes. Considering the different stability and turnover rates of each RNA family (for instance tRNAs and mRNAs), the mechanisms leading to variations in modification levels might be different. At present, the origin of the variations in the modification content in RNAs and especially mRNAs, which have been collectively described under the term “epitranscriptomics” is under intense investigations 138. A fascinating mechanism for achieving these dynamic changes of RNA modifications consists in the active removal of specific modifications by demodification enzymes. A few examples of active demodification processes that have been reported in tRNAs will be described below.
Recently, human ALKBH1 and the fat mass and obesity‐associated protein (FTO) were reported to catalyze demethylation of m1A in tRNAs (Fig. 4c) 139, 140. Both proteins belong to the AlkB family of proteins that consist of dioxygenases that use a non‐heme Fe(II) and α‐ketoglutarate to catalyze biological oxidation. The prototypical member of this family is the E. coli AlkB protein that functions in DNA/RNA alkylation damage repair by removing alkyl groups on nucleic acids 141, 142, 143. Interestingly, ALKBH1 displays a tRNA‐binding domain related to the ones found in eukaryotic tRNA ligases and binds to m1A58‐containing tRNAs inside cells 139. Concerning tRNAs, ALKBH1 and FTO display a pronounced preference for the demethylation of m1A over other common methylated nucleotides such as m7G, m5C, and m3C for ALKBH1, and m7G, m5C, m2G, and m1G for FTO. However, ALKBH1 and FTO display a different nucleotide‐specificity toward other nucleic acid substrates. In DNA, ALKBH1 and FTO catalyze demethylation of m3C and m3T, respectively 144, 145. FTO was also reported to catalyze demethylation of m3U and m6A in mRNA 146, 147, and to display a slight tRNA m3C demethylation activity in vitro 140. As FTO preferentially catalyzes m1A demethylation in stem‐loop structures mimicking the T‐arm of tRNAs, m1A58 is most likely the primary demethylation target of FTO in tRNAs (Fig. 4c) 140. ALKBH1 and FTO expression is increased in response to glucose deprivation, which decreases the m1A content of certain tRNAs and most probably the stability of tRNAi Met, thereby inhibiting translation initiation. This mechanism might contribute to globally repress translation on glucose deprivation 139, 140.
These examples highlight the importance of tRNA modification levels in the global control of fundamental cellular functions such as translation and reveal that a demodification enzymatic activity alters modification levels in tRNAs. It remains to be discovered whether demodification processes are limited to m1A modifications or whether theses aspects of reversible modifications apply to other types of modifications in tRNAs.
CONCLUDING REMARKS
The picture that emerges from this review is that the introduction of modifications in tRNAs is amazingly complex. Not so long ago, it was thought that modifications are introduced in tRNAs showing the correct sequence and structural requirements as static chemical decorations, and their importance for optimal tRNA function was clearly well appreciated. At the same time, the concept that regulation of tRNA biological function could be mediated by its modifications was accepted, but the exact mechanisms by which these regulations could be achieved were missing. Recently, answers to these questions were provided by reports showing that (i) modification circuits establish functional connections between different modifications in tRNAs; (ii) modification levels can change dynamically in response to environmental perturbations such as cellular stresses, which may lead to translational reprogramming and translation of specific stress‐response genes; (iii) activation of demodification enzymes can remove modifications thereby altering tRNA function in response to defined stimuli. All these aspects have contributed to turn the spotlight on tRNA modifications in the recent years. Although the different aspects influencing nucleotide modifications in tRNAs have been classified and presented in different parts of this review, it is worth mentioning that all these aspects are deeply interconnected. For instance, modification circuits are just another layer adding to the determinants of specificity of the enzymes. In addition, modification circuits can alter the levels of tRNA modifications in response to environmental changes, with primary variations directly responding to defined stimuli while other ones being a consequence of the interplay between modifications. Finally, dynamic changes in the levels of tRNA modifications, in response to changes in the cellular conditions, can be mediated by active demodification processes. All things considered, the tRNA modification field has clearly entered an exciting period, and impressive new regulatory functions of modifications surely await to be discovered.
ACKNOWLEDGEMENTS
The authors acknowledge financial support from the CNRS, the ANR NMR‐VitAmin (ANR‐14‐CE09‐0012), and the Labex DYNAMO. We apologize to those whose important work could not be cited because of space restrictions.
REFERENCES
- 1. Raina, M. , and Ibba, M. (2014) tRNAs as regulators of biological processes. Front. Genet. 5, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fields, R. N. , and Roy, H. (2018) Deciphering the tRNA‐dependent lipid aminoacylation systems in bacteria: novel components and structural advances. RNA Biol. 15, 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Megel, C. , Morelle, G. , Lalande, S. , Duchêne, A.‐M. , Small, I. , et al. (2015) Surveillance and cleavage of eukaryotic tRNAs. Int. J. Mol. Sci. 16, 1873–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oberbauer, V. , and Schaefer, M. R. (2018) tRNA‐derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes (Basel) 9, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hopper, A. K. (2013) Transfer RNA post‐transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae . Genetics 194, 43–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shepherd, J. , and Ibba, M. (2015) Bacterial transfer RNAs. FEMS Microbiol. Rev. 39, 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoshihisa, T. (2014) Handling tRNA introns, archaeal way and eukaryotic way. Front. Genet. 5, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boccaletto, P. , Machnicka, M. A. , Purta, E. , Piatkowski, P. , Baginski, B. , et al. (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El Yacoubi, B. , Bailly, M. , and de Crécy‐Lagard, V. (2012) Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 46, 69–95. [DOI] [PubMed] [Google Scholar]
- 10. Jackman, J. E. , and Alfonzo, J. D. (2013) Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 4, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Helm, M. , and Alfonzo, J. D. (2014) Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 21, 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lorenz, C. , Lünse, C. E. , and Mörl, M. (2017) tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Motorin, Y. , and Helm, M. (2011) RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2, 611–631. [DOI] [PubMed] [Google Scholar]
- 14. Machnicka, M. A. , Olchowik, A. , Grosjean, H. , and Bujnicki, J. M. (2014) Distribution and frequencies of post‐transcriptional modifications in tRNAs. RNA Biol. 11, 1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Motorin, Y. , and Helm, M. (2010) tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944. [DOI] [PubMed] [Google Scholar]
- 16. Urbonavicius, J. , Durand, J. M. B. , and Björk, G. R. (2002) Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri . J. Bacteriol. 184, 5348–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alexandrov, A. , Chernyakov, I. , Gu, W. , Hiley, S. L. , Hughes, T. R. , et al. (2006) Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96. [DOI] [PubMed] [Google Scholar]
- 18. Anderson, J. , Phan, L. , Cuesta, R. , Carlson, B. A. , Pak, M. , et al. (1998) The essential Gcd10p‐Gcd14p nuclear complex is required for 1‐methyladenosine modification and maturation of initiator methionyl‐tRNA. Genes Dev. 12, 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phizicky, E. M. , and Alfonzo, J. D. (2010) Do all modifications benefit all tRNAs? FEBS Lett. 584, 265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grosjean, H. , de Crécy‐Lagard, V. , and Marck, C. (2010) Deciphering synonymous codons in the three domains of life: co‐evolution with specific tRNA modification enzymes. FEBS Lett. 584, 252–264. [DOI] [PubMed] [Google Scholar]
- 21. McKenney, K. M. , and Alfonzo, J. D. (2016) From prebiotics to probiotics: the evolution and functions of tRNA modifications. Life (Basel) 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salinas‐Giegé, T. , Giegé, R. , and Giegé, P. (2015) tRNA biology in mitochondria. Int. J. Mol. Sci. 16, 4518–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bohnsack, M. T. , and Sloan, K. E. (2018) The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 75, 241–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Puri, P. , Wetzel, C. , Saffert, P. , Gaston, K. W. , Russell, S. P. , et al. (2014) Systematic identification of tRNAome and its dynamics in Lactococcus lactis . Mol. Microbiol. 93, 944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao, X. , and Limbach, P. A. (2015) Enhanced detection of post‐transcriptional modifications using a mass‐exclusion list strategy for RNA modification mapping by LC‐MS/MS. Anal. Chem. 87, 8433–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross, R. L. , Cao, X. , and Limbach, P. A. (2017) Mapping post‐transcriptional modifications onto transfer ribonucleic acid sequences by liquid chromatography tandem mass spectrometry. Biomolecules 7, 21. [Google Scholar]
- 27. Machnicka, M. A. , Dunin‐Horkawicz, S. , de Crécy‐Lagard, V. , and Bujnicki, J. M. (2016) tRNAmodpred: a computational method for predicting posttranscriptional modifications in tRNAs. Methods 107, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li, Y.‐H. , Zhang, G. , and Cui, Q. (2015) PPUS: a web server to predict PUS‐specific pseudouridine sites. Bioinformatics 31, 3362–3364. [DOI] [PubMed] [Google Scholar]
- 29. Panwar, B. , and Raghava, G. P. S. (2014) Prediction of uridine modifications in tRNA sequences. BMC Bioinformatics 15, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nordlund, M. E. , Johansson, J. O. , von Pawel‐Rammingen, U. , and Byström, A. S. (2000) Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae . RNA 6, 844–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alian, A. , Lee, T. T. , Griner, S. L. , Stroud, R. M. , and Finer‐Moore, J. (2008) Structure of a TrmA‐RNA complex: a consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc. Natl. Acad. Sci. U. S. A. 105, 6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Urbonavicius, J. , Skouloubris, S. , Myllykallio, H. , and Grosjean, H. (2005) Identification of a novel gene encoding a flavin‐dependent tRNA:m5U methyltransferase in bacteria—evolutionary implications. Nucleic Acids Res. 33, 3955–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishimasu, H. , Ishitani, R. , Yamashita, K. , Iwashita, C. , Hirata, A. , et al. (2009) Atomic structure of a folate/FAD‐dependent tRNA T54 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106, 8180–8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guy, M. P. , and Phizicky, E. M. (2014) Two‐subunit enzymes involved in eukaryotic post‐transcriptional tRNA modification. RNA Biol. 11, 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerber, A. P. , and Keller, W. (1999) An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- 36. Brulé, H. , Elliott, M. , Redlak, M. , Zehner, Z. E. , and Holmes, W. M. (2004) Isolation and characterization of the human tRNA‐(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43, 9243–9255. [DOI] [PubMed] [Google Scholar]
- 37. Goto‐Ito, S. , Ito, T. , and Yokoyama, S. (2017) Trm5 and TrmD: two enzymes from distinct origins catalyze the identical tRNA modification, m1G37. Biomolecules 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Purushothaman, S. K. , Bujnicki, J. M. , Grosjean, H. , and Lapeyre, B. (2005) Trm11p and Trm112p are both required for the formation of 2‐methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 25, 4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heurgué‐Hamard, V. , Champ, S. , Mora, L. , Merkulova‐Rainon, T. , Merkoulova‐Rainon, T. , et al. (2005) The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 280, 2439–2445. [DOI] [PubMed] [Google Scholar]
- 40. Bourgeois, G. , Létoquart, J. , van Tran, N. , and Graille, M. (2017) Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bourgeois, G. , Marcoux, J. , Saliou, J.‐M. , Cianférani, S. , and Graille, M. (2017) Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 45, 1971–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swinehart, W. E. , Henderson, J. C. , and Jackman, J. E. (2013) Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA 19, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shao, Z. , Yan, W. , Peng, J. , Zuo, X. , Zou, Y. , et al. (2014) Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate. Nucleic Acids Res. 42, 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang, M. , Zhu, Y. , Wang, C. , Fan, X. , Jiang, X. , et al. (2016) Crystal structure of the two‐subunit tRNA m(1)A58 methyltransferase TRM6‐TRM61 from Saccharomyces cerevisiae . Sci. Rep. 6, 32562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cavaillé, J. , Chetouani, F. , and Bachellerie, J. P. (1999) The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2'‐O‐ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA 5, 66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barraud, P. , Golinelli‐Pimpaneau, B. , Atmanene, C. , Sanglier, S. , Van Dorsselaer, A. , et al. (2008) Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J. Mol. Biol. 377, 535–550. [DOI] [PubMed] [Google Scholar]
- 47. Guelorget, A. , Roovers, M. , Guérineau, V. , Barbey, C. , Li, X. , et al. (2010) Insights into the hyperthermostability and unusual region‐specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase. Nucleic Acids Res. 38, 6206–6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guelorget, A. , Barraud, P. , Tisné, C. , and Golinelli‐Pimpaneau, B. (2011) Structural comparison of tRNA m(1)A58 methyltransferases revealed different molecular strategies to maintain their oligomeric architecture under extreme conditions. BMC Struct. Biol. 11, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takuma, H. , Ushio, N. , Minoji, M. , Kazayama, A. , Shigi, N. , et al. (2015) Substrate tRNA recognition mechanism of eubacterial tRNA (m1A58) methyltransferase (TrmI). J. Biol. Chem. 290, 5912–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ochi, A. , Makabe, K. , Yamagami, R. , Hirata, A. , Sakaguchi, R. , et al. (2013) The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced‐fit process. J. Biol. Chem. 288, 25562–25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Purta, E. , van Vliet, F. , Tkaczuk, K. L. , Dunin‐Horkawicz, S. , Mori, H. , et al. (2006) The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC Mol. Biol. 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu, R.‐J. , Long, T. , Zhou, M. , Zhou, X.‐L. , and Wang, E.‐D. (2015) tRNA recognition by a bacterial tRNA Xm32 modification enzyme from the SPOUT methyltransferase superfamily. Nucleic Acids Res. 43, 7489–7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Somme, J. , Van Laer, B. , Roovers, M. , Steyaert, J. , Versées, W. , et al. (2014) Characterization of two homologous 2'‐O‐methyltransferases showing different specificities for their tRNA substrates. RNA 20, 1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aström, S. U. , and Byström, A. S. (1994) Rit1, a tRNA backbone‐modifying enzyme that mediates initiator and Elongator tRNA discrimination. Cell 79, 535–546. [DOI] [PubMed] [Google Scholar]
- 55. Jackman, J. E. , and Phizicky, E. M. (2006) tRNAHis guanylyltransferase adds G‐1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 12, 1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakamura, A. , Wang, D. , and Komatsu, Y. (2018) Molecular mechanism of substrate recognition and specificity of tRNAHis guanylyltransferase during nucleotide addition in the 3′‐5′ direction. RNA 24, 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goll, M. G. , Kirpekar, F. , Maggert, K. A. , Yoder, J. A. , Hsieh, C.‐L. , et al. (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398. [DOI] [PubMed] [Google Scholar]
- 58. Shanmugam, R. , Aklujkar, M. , Schäfer, M. , Reinhardt, R. , Nickel, O. , et al. (2014) The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA‐Glu. Nucleic Acids Res. 42, 6487–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ikeuchi, Y. , Kitahara, K. , and Suzuki, T. (2008) The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4‐acetylcytidine of tRNA anticodon. EMBO J. 27, 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gu, X. , Yu, M. , Ivanetich, K. M. , and Santi, D. V. (1998) Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 37, 339–343. [DOI] [PubMed] [Google Scholar]
- 61. Hoang, C. , and Ferré‐D'Amaré, A. R. (2001) Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA‐modifying enzyme. Cell 107, 929–939. [DOI] [PubMed] [Google Scholar]
- 62. Tomikawa, C. (2018) 7‐Methylguanosine modifications in transfer RNA (tRNA). Int. J. Mol. Sci. 19, 4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Okamoto, H. , Watanabe, K. , Ikeuchi, Y. , Suzuki, T. , Endo, Y. , et al. (2004) Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus . J. Biol. Chem. 279, 49151–49159. [DOI] [PubMed] [Google Scholar]
- 64. Matsumoto, K. , Toyooka, T. , Tomikawa, C. , Ochi, A. , Takano, Y. , et al. (2007) RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8‐Trm82 complex). FEBS Lett. 581, 1599–1604. [DOI] [PubMed] [Google Scholar]
- 65. Leulliot, N. , Chaillet, M. , Durand, D. , Ulryck, N. , Blondeau, K. , et al. (2008) Structure of the yeast tRNA m7G methylation complex. Structure 16, 52–61. [DOI] [PubMed] [Google Scholar]
- 66. Kim, J. , Malashkevich, V. , Roday, S. , Lisbin, M. , Schramm, V. L. , et al. (2006) Structural and kinetic characterization of Escherichia coli TadA, the wobble‐specific tRNA deaminase. Biochemistry 45, 6407–6416. [DOI] [PubMed] [Google Scholar]
- 67. Losey, H. C. , Ruthenburg, A. J. , and Verdine, G. L. (2006) Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159. [DOI] [PubMed] [Google Scholar]
- 68. Zhou, M. , Long, T. , Fang, Z.‐P. , Zhou, X.‐L. , Liu, R.‐J. , et al. (2015) Identification of determinants for tRNA substrate recognition by Escherichia coli C/U34 2'‐O‐methyltransferase. RNA Biol. 12, 900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kimura, S. , Miyauchi, K. , Ikeuchi, Y. , Thiaville, P. C. , de Crécy‐Lagard, V. , et al. (2014) Discovery of the β‐barrel‐type RNA methyltransferase responsible for N6‐methylation of N6‐threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 42, 9350–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hirata, A. , Nishiyama, S. , Tamura, T. , Yamauchi, A. , and Hori, H. (2016) Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA‐binding modules. Nucleic Acids Res. 44, 6377–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Edqvist, J. , Blomqvist, K. , and Stråby, K. B. (1994) Structural elements in yeast tRNAs required for homologous modification of guanosine‐26 into dimethylguanosine‐26 by the yeast Trm1 tRNA‐modifying enzyme. Biochemistry 33, 9546–9551. [DOI] [PubMed] [Google Scholar]
- 72. Constantinesco, F. , Motorin, Y. , and Grosjean, H. (1999) Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))‐dimethyltransferase (Trm1p) from Pyrococcus furiosus . J. Mol. Biol. 291, 375–392. [DOI] [PubMed] [Google Scholar]
- 73. Awai, T. , Kimura, S. , Tomikawa, C. , Ochi, A. , Ihsanawati, et al. (2009) Aquifex aeolicus tRNA (N2,N2‐guanine)‐dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 284, 20467–20478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Awai, T. , Ochi, A. , Ihsanawati, Sengoku, T. , Hirata, A. , et al. (2011) Substrate tRNA recognition mechanism of a multisite‐specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X‐ray crystal structure. J. Biol. Chem. 286, 35236–35246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Grosjean, H. , Szweykowska‐Kulinska, Z. , Motorin, Y. , Fasiolo, F. , and Simos, G. (1997) Intron‐dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie 79, 293–302. [DOI] [PubMed] [Google Scholar]
- 76. Jiang, H. Q. , Motorin, Y. , Jin, Y. X. , and Grosjean, H. (1997) Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: an in vitro study. Nucleic Acids Res. 25, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Grosjean, H. , Edqvist, J. , Stråby, K. B. , and Giegé, R. (1996) Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J. Mol. Biol. 255, 67–85. [DOI] [PubMed] [Google Scholar]
- 78. Ohira, T. , and Suzuki, T. (2011) Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc. Natl. Acad. Sci. U. S. A. 108, 10502–10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Motorin, Y. , and Grosjean, H. (1999) Multisite‐specific tRNA:m5C‐methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA 5, 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Auxilien, S. , Guérineau, V. , Szweykowska‐Kulińska, Z. , and Golinelli‐Pimpaneau, B. (2012) The human tRNA m (5) C methyltransferase Misu is multisite‐specific. RNA Biol. 9, 1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Motorin, Y. , Keith, G. , Simon, C. , Foiret, D. , Simos, G. , et al. (1998) The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 4, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lecointe, F. , Simos, G. , Sauer, A. , Hurt, E. C. , Motorin, Y. , et al. (1998) Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 273, 1316–1323. [DOI] [PubMed] [Google Scholar]
- 83. Hur, S. , and Stroud, R. M. (2007) How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol. Cell 26, 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xing, F. , Hiley, S. L. , Hughes, T. R. , and Phizicky, E. M. (2004) The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 279, 17850–17860. [DOI] [PubMed] [Google Scholar]
- 85. Bou‐Nader, C. , Montémont, H. , Guérineau, V. , Jean‐Jean, O. , Brégeon, D. , et al. (2018) Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: perspectives on the evolution scenario. Nucleic Acids Res. 46, 1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kawamura, T. , Hirata, A. , Ohno, S. , Nomura, Y. , Nagano, T. , et al. (2016) Multisite‐specific archaeosine tRNA‐guanine transglycosylase (ArcTGT) from Thermoplasma acidophilum, a thermo‐acidophilic archaeon. Nucleic Acids Res. 44, 1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Guy, M. P. , Podyma, B. M. , Preston, M. A. , Shaheen, H. H. , Krivos, K. L. , et al. (2012) Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 18, 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Oerum, S. , Dégut, C. , Barraud, P. , and Tisné, C. (2017) m1A post‐transcriptional modification in tRNAs. Biomolecules 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kempenaers, M. , Roovers, M. , Oudjama, Y. , Tkaczuk, K. L. , Bujnicki, J. M. , et al. (2010) New archaeal methyltransferases forming 1‐methyladenosine or 1‐methyladenosine and 1‐methylguanosine at position 9 of tRNA. Nucleic Acids Res. 38, 6533–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Oerum, S. , Roovers, M. , Rambo, R. P. , Kopec, J. , Bailey, H. J. , et al. (2018) Structural insight into the human mitochondrial tRNA purine N1‐methyltransferase and ribonuclease P complexes. J. Biol. Chem. 293, 12862–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ranaei‐Siadat, E. , Fabret, C. , Seijo, B. , Dardel, F. , Grosjean, H. , et al. (2013) RNA‐methyltransferase TrmA is a dual‐specific enzyme responsible for C5‐methylation of uridine in both tmRNA and tRNA. RNA Biol. 10, 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Benítez‐Páez, A. , Villarroya, M. , and Armengod, M.‐E. (2012) The Escherichia coli RlmN methyltransferase is a dual‐specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA 18, 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bar‐Yaacov, D. , Frumkin, I. , Yashiro, Y. , Chujo, T. , Ishigami, Y. , et al. (2016) Mitochondrial 16S rRNA is methylated by tRNA Methyltransferase TRMT61B in all vertebrates. PLoS Biol. 14, e1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Carlile, T. M. , Rojas‐Duran, M. F. , Zinshteyn, B. , Shin, H. , Bartoli, K. M. , et al. (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Edelheit, S. , Schwartz, S. , Mumbach, M. R. , Wurtzel, O. , and Sorek, R. (2013) Transcriptome‐wide mapping of 5‐methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Han, L. , and Phizicky, E. M. (2018) A rationale for tRNA modification circuits in the anticodon loop. RNA 24, 1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hori, H. (2014) Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 5, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Benítez‐Páez, A. , Villarroya, M. , Douthwaite, S. , Gabaldón, T. , and Armengod, M.‐E. (2010) YibK is the 2'‐O‐methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors. RNA 16, 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Han, L. , Marcus, E. , D'Silva, S. , and Phizicky, E. M. (2017) S. cerevisiae Trm140 has two recognition modes for 3‐methylcytidine modification of the anticodon loop of tRNA substrates. RNA 23, 406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Arimbasseri, A. G. , Iben, J. , Wei, F.‐Y. , Rijal, K. , Tomizawa, K. , et al. (2016) Evolving specificity of tRNA 3‐methyl‐cytidine‐32 (m3C32) modification: a subset of tRNAsSer requires N6‐isopentenylation of A37. RNA 22, 1400–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Guy, M. P. , and Phizicky, E. M. (2015) Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 21, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Müller, M. , Hartmann, M. , Schuster, I. , Bender, S. , Thüring, K. L. , et al. (2015) Dynamic modulation of Dnmt2‐dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 43, 10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Johannsson, S. , Neumann, P. , Wulf, A. , Welp, L. M. , Gerber, H.‐D. , et al. (2018) Structural insights into the stimulation of S. pombe Dnmt2 catalytic efficiency by the tRNA nucleoside queuosine. Sci. Rep. 8, 8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rubio, M. A. T. , Ragone, F. L. , Gaston, K. W. , Ibba, M. , and Alfonzo, J. D. (2006) C to U editing stimulates A to I editing in the anticodon loop of a cytoplasmic threonyl tRNA in Trypanosoma brucei . J. Biol. Chem. 281, 115–120. [DOI] [PubMed] [Google Scholar]
- 105. Rubio, M. A. T. , Gaston, K. W. , McKenney, K. M. , Fleming, I. M. C. , Paris, Z. , et al. (2017) Editing and methylation at a single site by functionally interdependent activities. Nature 542, 494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Denmon, A. P. , Wang, J. , and Nikonowicz, E. P. (2011) Conformation effects of base modification on the anticodon stem‐loop of Bacillus subtilis tRNA(Tyr). J. Mol. Biol. 412, 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sundaram, M. , Durant, P. C. , and Davis, D. R. (2000) Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U‐turn structure. Biochemistry 39, 12575–12584. [DOI] [PubMed] [Google Scholar]
- 108. Hori, H. , Kawamura, T. , Awai, T. , Ochi, A. , Yamagami, R. , et al. (2018) Transfer RNA modification enzymes from thermophiles and their modified nucleosides in tRNA. Microorganisms 6, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yokoyama, S. , Watanabe, K. , and Miyazawa, T. (1987) Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles. Adv. Biophys. 23, 115–147. [DOI] [PubMed] [Google Scholar]
- 110. Yamagami, R. , Tomikawa, C. , Shigi, N. , Kazayama, A. , Asai, S.‐I. , et al. (2016) Folate‐/FAD‐dependent tRNA methyltransferase from Thermus thermophilus regulates other modifications in tRNA at low temperatures. Genes Cells 21, 740–754. [DOI] [PubMed] [Google Scholar]
- 111. Shigi, N. , Suzuki, T. , Terada, T. , Shirouzu, M. , Yokoyama, S. , et al. (2006) Temperature‐dependent biosynthesis of 2‐thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 281, 2104–2113. [DOI] [PubMed] [Google Scholar]
- 112. Tomikawa, C. , Yokogawa, T. , Kanai, T. , and Hori, H. (2010) N7‐Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 38, 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ishida, K. , Kunibayashi, T. , Tomikawa, C. , Ochi, A. , Kanai, T. , et al. (2011) Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low‐temperature adaptation in the extreme‐thermophilic eubacterium Thermus thermophilus . Nucleic Acids Res. 39, 2304–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Singhal, R. P. , and Vold, B. (1976) Changes in transfer ribonucleic acids of Bacillus subtilis during different growth phases. Nucleic Acids Res. 3, 1249–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Keith, G. , Rogg, H. , Dirheimer, G. , Menichi, B. , and Heyham, T. (1976) Post‐transcriptional modification of tyrosine tRNA as a function of growth in Bacillus subtilis . FEBS Lett. 61, 120–123. [DOI] [PubMed] [Google Scholar]
- 116. Bartz, J. , Söll, D. , Burrows, W. J. , and Skoog, F. (1970) Identification of the cytokinin‐active ribonucleosides in pure Escherichia coli tRNA species. Proc. Natl. Acad. Sci. U. S. A. 67, 1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Singhal, R. P. , Kopper, R. A. , Nishimura, S. , and Shindo‐Okada, N. (1981) Modification of guanine to queuine in transfer RNAs during development and aging. Biochem. Biophys. Res. Commun. 99, 120–126. [DOI] [PubMed] [Google Scholar]
- 118. Emilsson, V. , Näslund, A. K. , and Kurland, C. G. (1992) Thiolation of transfer RNA in Escherichia coli varies with growth rate. Nucleic Acids Res. 20, 4499–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Arnold, H. H. , and Raettig, R. (1977) Isoaccepting phenylalanine tRNAs from Bacillus subtilis as a function of growth conditions. Differences in the content of modified nucleosides. FEBS Lett. 73, 210–214. [DOI] [PubMed] [Google Scholar]
- 120. Frey, B. , McCloskey, J. , Kersten, W. , and Kersten, H. (1988) New function of vitamin B12: cobamide‐dependent reduction of epoxyqueuosine to queuosine in tRNAs of Escherichia coli and Salmonella typhimurium . J. Bacteriol. 170, 2078–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Buck, M. , and Ames, B. N. (1984) A modified nucleotide in tRNA as a possible regulator of aerobiosis: synthesis of cis‐2‐methyl‐thioribosylzeatin in the tRNA of Salmonella. Cell 36, 523–531. [DOI] [PubMed] [Google Scholar]
- 122. Alings, F. , Sarin, L. P. , Fufezan, C. , Drexler, H. C. A. , and Leidel, S. A. (2015) An evolutionary approach uncovers a diverse response of tRNA 2‐thiolation to elevated temperatures in yeast. RNA 21, 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Noon, K. R. , Guymon, R. , Crain, P. F. , McCloskey, J. A. , Thomm, M. , et al. (2003) Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23 degrees C) and Stetteria hydrogenophila (Topt, 95 degrees C). J. Bacteriol. 185, 5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kuchino, Y. , Borek, E. , Grunberger, D. , Mushinski, J. F. , and Nishimura, S. (1982) Changes of post‐transcriptional modification of wye base in tumor‐specific tRNAPhe. Nucleic Acids Res. 10, 6421–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pathak, C. , Jaiswal, Y. K. , and Vinayak, M. (2005) Hypomodification of transfer RNA in cancer with respect to queuosine. RNA Biol. 2, 143–148. [DOI] [PubMed] [Google Scholar]
- 126. Moukadiri, I. , Garzón, M.‐J. , Björk, G. R. , and Armengod, M.‐E. (2014) The output of the tRNA modification pathways controlled by the Escherichia coli MnmEG and MnmC enzymes depends on the growth conditions and the tRNA species. Nucleic Acids Res. 42, 2602–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Preston, M. A. , D'Silva, S. , Kon, Y. , and Phizicky, E. M. (2013) tRNAHis 5‐methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae . RNA 19, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Torres, A. G. , Batlle, E. , and Ribas de Pouplana, L. (2014) Role of tRNA modifications in human diseases. Trends Mol. Med. 20, 306–314. [DOI] [PubMed] [Google Scholar]
- 129. Endres, L. , Dedon, P. C. , and Begley, T. J. (2015) Codon‐biased translation can be regulated by wobble‐base tRNA modification systems during cellular stress responses. RNA Biol. 12, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chan, C. T. Y. , Dyavaiah, M. , DeMott, M. S. , Taghizadeh, K. , Dedon, P. C. , et al. (2010) A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6, e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chan, C. T. Y. , Pang, Y. L. J. , Deng, W. , Babu, I. R. , Dyavaiah, M. , et al. (2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon‐biased translation of proteins. Nat. Commun. 3, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Bauer, F. , Matsuyama, A. , Candiracci, J. , Dieu, M. , Scheliga, J. , et al. (2012) Translational control of cell division by Elongator. Cell Rep. 1, 424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Fernández‐Vázquez, J. , Vargas‐Pérez, I. , Sansó, M. , Buhne, K. , Carmona, M. , et al. (2013) Modification of tRNA(Lys) UUU by Elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 9, e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Deng, W. , Babu, I. R. , Su, D. , Yin, S. , Begley, T. J. , et al. (2015) Trm9‐catalyzed tRNA modifications regulate global protein expression by codon‐biased translation. PLoS Genet. 11, e1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Tyagi, K. , and Pedrioli, P. G. A. (2015) Protein degradation and dynamic tRNA thiolation fine‐tune translation at elevated temperatures. Nucleic Acids Res. 43, 4701–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chionh, Y. H. , McBee, M. , Babu, I. R. , Hia, F. , Lin, W. , et al. (2016) tRNA‐mediated codon‐biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7, 13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Tuorto, F. , Liebers, R. , Musch, T. , Schaefer, M. , Hofmann, S. , et al. (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905. [DOI] [PubMed] [Google Scholar]
- 138. Schaefer, M. , Kapoor, U. , and Jantsch, M. F. (2017) Understanding RNA modifications: the promises and technological bottlenecks of the 'epitranscriptome'. Open Biol. 7, 170077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Liu, F. , Clark, W. , Luo, G. , Wang, X. , Fu, Y. , et al. (2016) ALKBH1‐mediated tRNA demethylation regulates translation. Cell 167, 816–828.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wei, J. , Liu, F. , Lu, Z. , Fei, Q. , Ai, Y. , et al. (2018) Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 71, 973–985.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Fedeles, B. I. , Singh, V. , Delaney, J. C. , Li, D. , and Essigmann, J. M. (2015) The AlkB family of Fe(II)/α‐ketoglutarate‐dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ougland, R. , Zhang, C.‐M. , Liiv, A. , Johansen, R. F. , Seeberg, E. , et al. (2004) AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell 16, 107–116. [DOI] [PubMed] [Google Scholar]
- 143. Reichle, V. F. , Weber, V. , and Kellner, S. (2018) NAIL‐MS in E. coli determines the source and fate of methylation in tRNA. Chembiochem 19, 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Westbye, M. P. , Feyzi, E. , Aas, P. A. , Vågbø, C. B. , Talstad, V. A. , et al. (2008) Human AlkB homolog 1 is a mitochondrial protein that demethylates 3‐methylcytosine in DNA and RNA. J. Biol. Chem. 283, 25046–25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gerken, T. , Girard, C. A. , Tung, Y.‐C. L. , Webby, C. J. , Saudek, V. , et al. (2007) The obesity‐associated FTO gene encodes a 2‐oxoglutarate‐dependent nucleic acid demethylase. Science 318, 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Jia, G. , Yang, C.‐G. , Yang, S. , Jian, X. , Yi, C. , et al. (2008) Oxidative demethylation of 3‐methylthymine and 3‐methyluracil in single‐stranded DNA and RNA by mouse and human FTO. FEBS Lett. 582, 3313–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Jia, G. , Fu, Y. , Zhao, X. , Dai, Q. , Zheng, G. , et al. (2011) N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat. Chem. Biol. 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]


