Abstract
Bacteria are not only ubiquitous on earth but can also be incredibly diverse within clean laboratories and reagents. The presence of both living and dead bacteria in laboratory environments and reagents is especially problematic when examining samples with low endogenous content (e.g., skin swabs, tissue biopsies, ice, water, degraded forensic samples or ancient material), where contaminants can outnumber endogenous microorganisms within samples. The contribution of contaminants within high‐throughput studies remains poorly understood because of the relatively low number of contaminant surveys. Here, we examined 144 negative control samples (extraction blank and no‐template amplification controls) collected in both typical molecular laboratories and an ultraclean ancient DNA laboratory over 5 years to characterize long‐term contaminant diversity. We additionally compared the contaminant content within a home‐made silica‐based extraction method, commonly used to analyse low endogenous content samples, with a widely used commercial DNA extraction kit. The contaminant taxonomic profile of the ultraclean ancient DNA laboratory was unique compared to modern molecular biology laboratories, and changed over time according to researcher, month and season. The commercial kit also contained higher microbial diversity and several human‐associated taxa in comparison to the home‐made silica extraction protocol. We recommend a minimum of two strategies to reduce the impacts of laboratory contaminants within low‐biomass metagenomic studies: (a) extraction blank controls should be included and sequenced with every batch of extractions and (b) the contributions of laboratory contamination should be assessed and reported in each high‐throughput metagenomic study.
Keywords: ancient DNA, contaminant, contamination, metagenomics, microbiome, microbiota
1. INTRODUCTION
In the new era of culture‐independent microbiome research, targeted amplicon or “metabarcoding” approaches are now routinely used to amplify DNA from microbial species across the tree of life. However, these methods lack the ability to select for specific species or exclude contaminants (Caporaso et al., 2012). Although these techniques have provided invaluable insight into otherwise cryptic microbial communities, the increased sensitivity and lack of target specificity leave microbiota studies particularly susceptible to the effects of contamination. Such effects are widespread, as several recent studies have indicated that contaminant microbial DNA can be routinely isolated from laboratory reagents and surfaces (Laurence, Hatzis, & Brash, 2014; Salter et al., 2014; Tanner, Goebel, & Dojka, 1998) and that this signal has significantly impacted the past interpretation and characterization of microbiota in high‐throughput sequencing studies. For example, Salter et al. (2014) demonstrated that bacterial DNA present in laboratory reagents is present in both quality‐filtered 16S rRNA gene and shotgun metagenomic data sets and significantly impacts the interpretation of results. Multiple microbial contaminants have also been identified within the published 1,000 Genomes data set and other medical genomic studies (Kearney et al., 2012; Laurence et al., 2014). Despite these findings, the routine assessment of microbial background contamination is still not required, or fully reported, in microbiota studies.
While the presence of contaminant DNA is widespread, the effects are particularly problematic in low‐biomass samples that contain very little endogenous DNA (Weiss et al., 2014; e.g., preterm infant swabs, tissue samples, such as placenta, tumour biopsies or breast tissue, and some environmental samples, such as ice or calcite). In low‐biomass samples, a small contaminant signal from laboratory reagents can easily overpower the intrinsic signal from the sample. This is similarly an issue in current palaeomicrobiology studies that examine ancient, degraded microbiota, such as mummified human tissue, preserved faeces (coprolites) or calcified dental plaque (calculus; Warinner, Speller, & Collins, 2015; Weiss et al., 2014; Weyrich, Dobney, & Cooper, 2015). In ancient samples, the amount of endogenous DNA attributed to the original source can be extremely low (e.g., <0.05% of the total DNA in the sample) and is damaged, fragmented and intermixed with longer, higher‐quality modern DNA fragments from contaminant species (Cooper & Poinar, 2000). Therefore, monitoring and understanding the contributions of contaminant DNA, especially in low‐biomass or ancient samples, is critical to ensure that reported results are only based on the endogenous DNA.
Microbial contaminant DNA (i.e., background or exogenous DNA) is a mixture of DNA from both environmental and laboratory sources, with the former including factors such as soil from a burial site, air within the sampling facility and microorganisms from people touching the sample, while the latter involves reagents, glassware, labware and surfaces (Weyrich et al., 2015). Environmental contamination in low‐biomass samples may be difficult to control or monitor, but the laboratory contaminants can be monitored by including extraction blank (EBC) and no‐template amplification (NTC) controls and assessed using bioinformatics tools (e.g., sourcetracker Knights et al., 2011). An EBC is an empty tube introduced during the extraction steps to collect DNA from the laboratory environment and the reagents throughout processing (Adler et al., 2013). Similarly, an NTC is an amplification reaction that lacks the addition of DNA from biological samples. These controls should be amplified and sequenced along with other samples and are critical to identify and exclude contaminant taxa from downstream analyses, reducing noise and ensuring any results are based solely on endogenous DNA (Weyrich et al., 2017). Despite this, there are surprisingly few published resources describing contaminant taxa found in EBCs or NTCs (Glassing, Dowd, Galandiuk, Davis, & Chiodini, 2016; Lauder et al., 2016; Salter et al., 2014).
In this study, we used 16S rRNA metabarcoding to characterize the contaminant diversity in 144 EBCs and NTCs using laboratory techniques specifically designed for low‐biomass material. We also explored differences in microbial contamination within two different types of laboratory facilities: a state‐of‐the‐art, purpose‐built ancient DNA clean laboratory over the course of 5 years, and three typical modern molecular biology laboratories over 1 year. Lastly, we investigated differences between a common commercial DNA extraction kit and a home‐made DNA extraction method typically applied in the ancient DNA field. Overall, this study is designed to assess contaminant profiles over time and identify more potential contaminant sequences in both high‐ and low‐biomass research.
2. MATERIALS AND METHODS
2.1. Sample collection
Four different types of sample were used: ancient dental calculus (calcified dental plaque), modern dental calculus, EBCs and NTCs. Dental calculus samples were obtained from ancient and modern humans as described by Adler et al. (2013). A single EBC was included in each batch of extractions by treating an empty tube as if it was a biological sample throughout the DNA extraction and library preparation process. Similarly, NTC samples were created during the 16S rRNA library amplification stage by processing additional reactions without adding any known template DNA. Both EBCs and NTCs were subsequently included through to DNA sequencing and were included at a ratio of one control sample for every 10 biological samples.
2.2. Description of laboratory facilities
DNA extraction occurred in two different types of laboratory facilities: a purpose‐built, ultraclean ancient DNA laboratory (ancient lab) and three typical modern molecular biology laboratories (modern labs). The ancient lab is physically remote from the university campus in a building with no other molecular biology laboratories and contains a HEPA (high‐efficiency particulate air)‐filtered, positive pressure air system to remove DNA and bacteria from external sources. The HEPA filter function is checked annually and changed every 10 years. The surface and floors within the laboratory are cleaned weekly with a 5% bleach (NaClO) solution and are illuminated with ceiling‐mounted UV lights for 30 min each night. UV light bulbs are changed annually. Users entering the ancient lab are required to have showered, wear freshly laundered clothing, avoid the university campus prior to entry, and cannot bring personal equipment (e.g., phones, writing equipment and bags) into the facility. Standard personal laboratory wear includes disposable full‐body suits, surgical facemasks, plastic see‐through visors, and three layers of gloves to allow frequent changing without skin exposure (including one pair of inner elbow‐length surgical gloves). All liquid reagents within the ancient lab are certified DNA‐free, and the outer surface of all plasticware and reagent bottles are decontaminated prior to entering the laboratory (i.e., cleaned with 5% bleach and treated with UV (2×, 40‐W, 254‐nm UV tubes at a distance of 10 cm for 10 min) within a UV oven (Ultra Violet Products). All DNA extractions and amplification preparations are performed in a room separate to sample preparation and are completed in still‐air cabinets that are cleaned with 5% bleach and UV treated for 30 min (3×, 15‐W, 253.7‐nm tube lamps; AURA PCR) prior to beginning any work. In addition, ancient samples from different sources (e.g., soil, plants and other animals) are processed in separate, dedicated rooms to minimize cross‐contamination. In contrast, the modern laboratories are located over 2 km away from the ancient lab at the University of Adelaide (n = 2) and at the University of Sydney (n = 1). All three modern labs are typical of most molecular biology laboratories, are not routinely decontaminated and contain users who routinely use latex gloves but are not required to wear body suits or masks. DNA extracted within the modern labs comes from a wide range of sources (e.g., humans, mammals and environmental samples), although microbiome extractions were only performed on days when no other material was being extracted. In all facilities, DNA was extracted and prepared for amplification in still‐air cabinets that were cleaned before and after each use with 5% bleach.
2.3. DNA extractions
Several specialized DNA extraction protocols have been developed within ancient DNA studies to remove environmental contamination and enhance the recovery of the endogenous DNA. The extraction method selected for this study has previously been described for work on ancient dental calculus (Weyrich et al., 2017). Each ancient sample was first decontaminated using a published protocol (Adler et al., 2013), while modern samples were not decontaminated. The decontamination procedure included exposure to UV radiation for 15 min on each side of the sample, submersion of the sample in 5% bleach for 5 min, followed by submersion in 90% ethanol for 3 min to remove any residual bleach, and 5 min of drying. Decontaminated ancient calculus was then wrapped in aluminium foil and pulverized into power with a steel hammer and placed into a sterile 2‐ml tube. The EBCs were empty tubes exposed to air for 30 s in the same room during sample decontamination and were included in the extraction process as if they contained a biological sample.
Following decontamination, DNA was extracted using the QG‐based method previously described for the extraction of ancient microbiome material (Weyrich et al., 2017; referred to as “QG”). All reagents for the QG extraction method were prepared in a “sample‐free” room in the ancient DNA facility, and all reagents were aliquoted immediately upon opening and frozen until further use to avoid cross contamination. Where possible, certified “DNA‐free” reagents and labware were purchased (e.g., water and plastic tubes). All other reagents were opened solely within a sterilized hood within the ancient DNA facility. All chemicals were prepared for the extraction with previously unopened DNA‐ and RNA‐free certified water (Ultrapure water; Invitrogen). Briefly, 1.8 ml of 0.5 ethylenediaminetetraacetic acid (EDTA; Life Tech), 100 µl of 10% sodium dodecyl sulphate (SDS; Life Tech) and 20 µl of 20 mg/ml proteinase K (proK; Life Tech) were added to each sample, and the mixture was rotated at 55°C overnight to decalcify the sample. Released DNA was then purified by adding silica (silicon dioxide; Sigma Aldrich) and 3 ml of binding buffer (e.g., QG buffer; Qiagen; modified to contain 5.0 m GuSCN; 18.1 mm Tris‐HCl; 25 mm NaCl; 1.3% Triton X‐100; Rohland & Hofreiter, 2007). The silica was pelleted, washed twice in 80% ethanol, dried and resuspended in 100 µl of TLE buffer (10 mm Tris, 1 mm EDTA, pH 8) twice to elute the DNA, which was then stored at −20°C until amplification. All chemicals were prepared for the extraction with previously unopened DNA‐ and RNA‐free certified water (Ultrapure water; Invitrogen). For QG extractions performed in the modern laboratories, unopened aliquots of DNA extraction reagents were transported to the modern laboratory, and the modern samples were extracted following the ancient DNA approach described above.
In contrast to ancient DNA extractions, many modern microbiome studies decrease cost and time by using commercial DNA extraction kits to isolate DNA. To compare the nature and extent of contaminant DNA in the ancient method to a typical commercial microbiome DNA extraction kit, we analysed an additional set of EBCs created during extractions using a PowerBiofilm DNA Isolation Kit (MOBIO) from concurrent oral microbiome research conducted in the same modern labs (referred to as “kit” EBCs).
2.4. Library preparation
To minimize additional variables, a simple 16S rRNA amplicon sequencing approach was used in this study to compare the different sample types. Briefly, the V4 region of the bacterial 16S rRNA encoding gene was targeted for amplification using degenerate Illumina fusion primers, as previously described (Caporaso et al., 2012): forward primer 515F (AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCA GCMGCCGCGGTAA) and barcoded reverse primer 806R (CAAGCAGAAGACGGCATACGAGATnnnnnnnnnnnnAGTCAGTCAGCCGGACTACHVGGGTW TCTAAT) (Caporaso et al., 2012). The string of n's in the reverse primer refers to the unique 12‐bp barcode used for each sample. Primers were resuspended in TLE buffer within the ancient facility and distributed to the modern laboratory. In both facilities, all PCR amplification reactions were prepared using ultraclean reagents with strict ancient DNA protocols (Cooper & Poinar, 2000). Each PCR contained 17.25 µl DNA‐free water (Ultrapure water; Invitrogen), 2.5 µl 10× reaction buffer (20 mm Tris‐HCl, 10 mm (NH4)2SO4, 10 mm KCl, 2 mm MgSO4, 0.1% Triton X‐100, pH 8.8 at 25°C; ThermoPol Buffer; New England Biolabs), 0.25 µl Taq polymerase (Platinum Taq DNA Polymerase High Fidelity; Thermo Fisher Scientific), 1.0 µl MgCl2 (Thermo Fisher Scientific), 1.0 µl of each primer at 10 µm (IDT) and 2.0 µl of genomic DNA; each reaction was performed in triplicate. 16S rRNA amplification occurred under the following conditions: 95°C for 5 min; 37 cycles of 95°C for 0.5 min, 55°C for 0.5 min and 75°C for 1 min; and 75°C for 10 min. NTC reactions were also included in triplicate. PCR products were quantified (QuBit; Thermo Fisher Scientific) and pooled in batches of 30 samples at equal nanomolar concentrations prior to purification (Ampure; New England Biolabs). Each pool of purified PCR products was quantified (TapeStation; Agilent) before being combined into a single library. All amplicons were sequenced using the Illumina MiSeq 2x150‐bp (300 cycle) kit.
2.5. Bioinformatics analysis
After sequencing, fastq files for the forward and reverse reads were created using the Illumina casava pipeline (version 1.8.2). Overlapping forward and reverse reads were joined based on a maximum of 5% nucleotide difference over a minimum 5‐bp overlap using bbmerge (sourceforge.net/projects/bbmap/). Only successfully merged sequences were used in downstream analyses. The resulting fastq file was then imported into qiime1 (macqiime version 1.8.0), a bioinformatics pipeline‐based software for the analysis of metagenomic data (Caporaso et al., 2010). All further analysis of the amplicon data sets was conducted within the qiime1 package. Libraries were demultiplexed using a Phred base quality threshold of ≤20, with no errors allowed in the barcodes. Operational taxonomic units (OTUs) were determined by clustering sequences at 97% similarity using uclust (Edgar, 2010), and representative sequences (i.e., cluster seed) were selected for each cluster. By default, clusters with fewer than five sequences were eliminated from the analysis to reduce noise and spurious findings. Lastly, 16S rRNA gene sequences were given taxonomic assignments using the greengenes 13_8 database if the sequence was at least 80% similar (DeSantis et al., 2006; Wang, Garrity, Tiedje, & Cole, 2007). Taxonomic diversity measurements (alpha‐ and beta‐diversity) and statistical analyses were performed and visualized in qiime1. Samples were rarefied to a minimum of 150 sequences (see Figure 2) and a maximum of 1,000 sequences for diversity analyses, as many controls contained low sequence counts. Statistical differences between groups were identified using a PERMANOVA test for beta diversity (adonis), nonparametric t‐test for alpha diversity (Monte Carlo), or Kruskal–Wallis and G‐tests for detection of specific taxa associated with different treatments.
Figure 2.
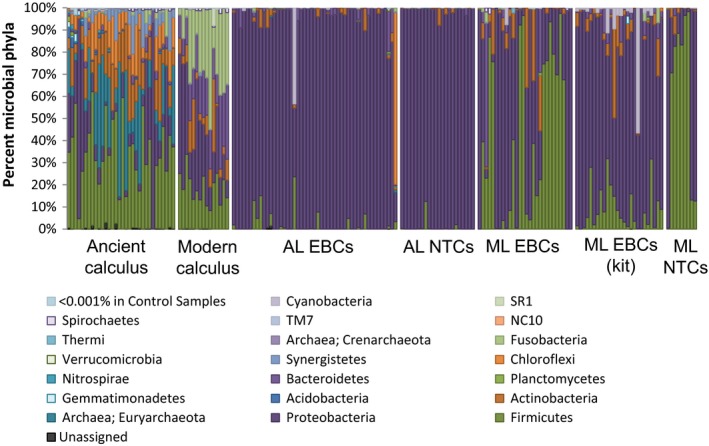
Microbial phyla within controls are distinct from biological samples. The proportion of different microbial phyla are shown for a wide array of modern and ancient calculus samples and control samples (EBCs and NTCs) from both laboratory facilities (modern lab [ML] and ancient lab [AL]) and two different extraction methods: the method employed in ancient DNA research and a commercially available DNA extraction kit (kit). Rare phyla were collapsed if they represented <0.001% of the total phyla observed [Colour figure can be viewed at http://www.wileyonlinelibrary.com]
3. RESULTS
3.1. Low bacterial diversity is routinely obtained from laboratory extraction controls
The EBCs and NTCs were sequenced alongside the ancient and modern biological samples; all sample types were pooled together at equimolar concentrations. Despite the equimolar pooling, we routinely obtained fewer reads from control samples (EBCs and NTCs) compared to the dental calculus samples, probably due to poor amplification of control samples, the quantification of poor DNA libraries and the clean‐up strategy employed. Compared to the ancient and modern calculus samples, 6.4‐fold fewer reads on average were obtained from EBCs, and 7.6‐fold fewer were obtained from NTCs (Figure 1a). As well as containing fewer reads overall, the control samples contained fewer taxa that could be identified than the biological samples. In the ancient laboratory, 719 total OTUs were observed in ancient biological samples (calculus), while only 415 were identified in the EBCs and 228 in NTCs (Figure 1b). In the modern laboratories, 286 total OTUs were described in the modern calculus samples, versus 208 in the EBCs and 102 in the NTCs. The OTU diversity within the EBCs and NTCs is reminiscent of the diversity observed in modern and ancient biological specimens, potentially reflecting minor cross contamination during DNA extraction. Across different extraction methods, the EBCs for the commercial extraction kit contained 261 OTUs, around 25% more than the QG method conducted in the modern laboratory. Overall, the laboratory controls were largely dominated by a single phylum, Proteobacteria (Figure 2), and alpha diversity was significantly lower than in the biological samples extracted within the same laboratory (Monte Carlo; p ≤ 0.0001 and T = >11.0 in all comparisons between any group of controls and all biological samples). While the diversity within laboratory controls was considerably lower than the biological samples, these results demonstrate microbial DNA contamination within an ultraclean laboratory with “DNA‐free” reagents, and clearly highlight the need to routinely monitor and report background contamination within all research facilities.
Figure 1.
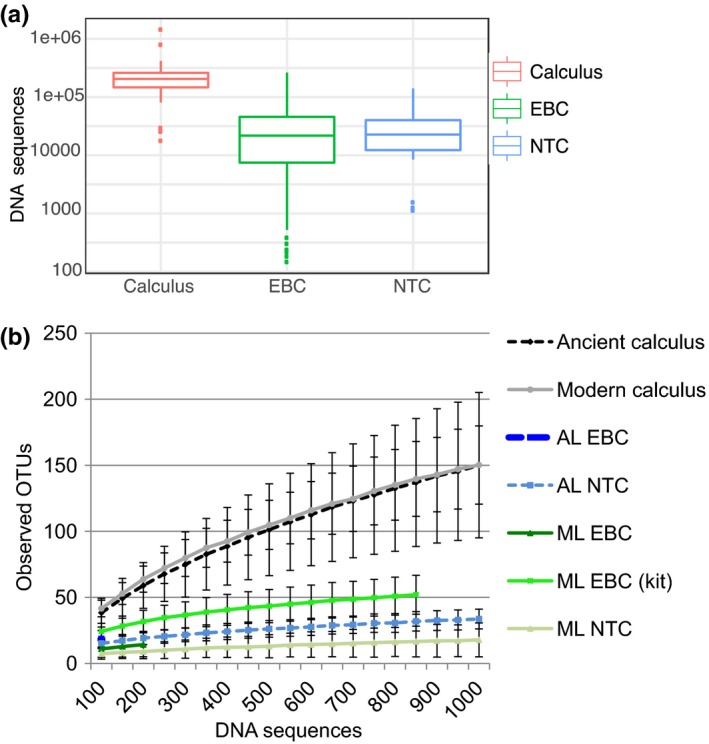
Lower diversity is observed in EBCs and NTCs compared to biological samples. (a) The number of sequenced reads from samples that were all pooled at equimolar concentrations is displayed on a box and whisker plot. (b) The alpha diversity of each type sample (i.e., the within sample diversity) was calculated using observed species metrics in qiime1 for rarefied 16S rRNA data. Each sample was rarefied up to 1,000 sequences in 100 sequence intervals; the standard error at each subsampling event is displayed using error bars. Calculus samples are shown in blue, while control samples (extraction blank controls [EBCs] and no‐template controls [NTCs]) from the ancient laboratory (AL) and the modern laboratory (ML) are shown in red and green, respectively [Colour figure can be viewed at http://www.wileyonlinelibrary.com]
3.2. EBCs detect >50% more contaminant taxa than NTCs
Many studies, including some in palaeomicrobiological research, reported failed EBC and NTC amplification reactions (often via simple visual comparison on an agarose gel) as a means to determine that their samples are free from contamination (Aagaard et al., 2014; Santiago‐Rodriguez et al., 2015). This approach is clearly inadequate, and importantly, also fails to appreciate the extent of contamination introduced during the extraction process, even though this issue is well described in the literature (Kliman, 2014; Lauder et al., 2016; Weyrich, Llamas, & Cooper, 2014). In our comparisons, EBCs were taxonomically far more diverse than NTCs (Figure 1b) and contained more microbial genera (415 vs. 228 genera in the ancient lab, and 208 vs. 102 genera in the modern labs). This pattern suggests that if just NTCs were used to monitor the presence of laboratory contamination, at least 53% of the total laboratory contamination may go undetected. These results highlight the need for the standard reporting of both EBCs and NTCs in both modern and ancient metagenomics research.
We also examined the impact of overall laboratory contamination on ancient samples by bioinformatically filtering (removing) all contaminant OTUs from ancient dental calculus samples. For the ancient samples prepared with the specialized facility, an average 92.5% of the sequence reads were contaminants, but importantly, accounted for only 28% of the genera identified within these samples. This indicates that endogenous signal can be identified even in samples of low endogenous content once contaminant taxa are removed.
3.3. EBCs and NTCs reflect laboratory environment
Previous studies have detected differences in the contaminants present in different laboratory facilities (Salter et al., 2014). In our study, the laboratory environments explained more of the taxonomic diversity observed in the EBCs and NTCs than the extraction or amplification methods used to generate them (Figure 3). For example, Proteobacteria dominated the EBCs and NTCs from the ancient laboratory, while Firmicutes were more dominant in EBCs and NTCs from the modern laboratories. In fact, different types of controls (i.e., EBC or NTC) from the same laboratory clustered with others of the same sample type in a principle coordinates analysis (PCoA) of unweighted UniFrac values (p ≤ 0.001, R 2 = 0.083; Figure 3a), despite large variation and significant differences in each lab (Figure 1b). Despite the sample type (e.g., EBC or NTC) driving the majority of the signal, taxa distinguishing each laboratory could also be detected, as a Paenibacillus taxa was only found in the modern laboratories, while the ancient laboratory contained both bacterial (Comamonas, Pseudomonas, Acinetobacter, Enterobacter) and archaeal (Methanobrevibacter) taxa that were not observed in the modern labs. In addition, several bacterial taxa were identified in both lab types, but were significantly increased in one location. The ancient laboratory contained significantly higher levels of certain Acinetobacter, Comamonas and Pseudomonas taxa compared to the modern laboratories (Kruskal–Wallis; Bonferroni‐corrected p ≤ 0.05), while Erythrobacteraceae and Staphylococcus taxa were increased in abundance in the modern laboratories. With the exception of the Staphylococcus taxa, each of these taxa had been previously identified in laboratory reagents (Salter et al., 2014). This suggests that some contaminant taxa are relatively universal across laboratories and are therefore either introduced in the manufacturing of laboratory reagents and labware or have a fundamental niche in low‐nutrient, laboratory environments.
Figure 3.
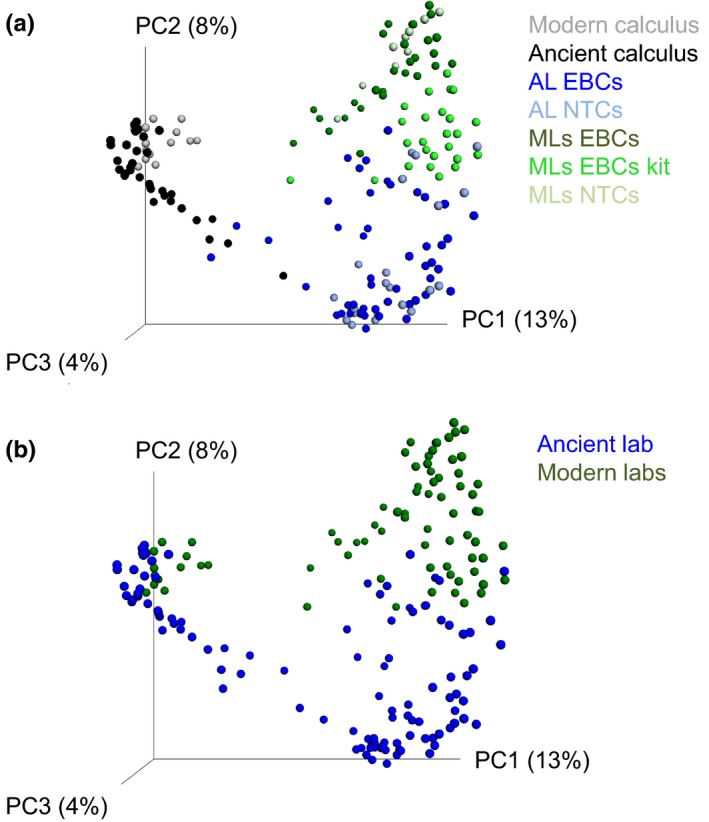
PCoA plots of control samples highlight differences in method and laboratory. PCoA plots of unweighted UniFrac values were plotted in qiime1 to compare beta diversity differences (between samples differences) in all samples (a) or in different laboratories (b). The different laboratory facilities are represented by ML (modern lab) and AL (ancient lab), and the two control types are represented by EBC (extraction blank control) or no‐template control (NTC) [Colour figure can be viewed at http://www.wileyonlinelibrary.com]
We next examined the genera that were likely to be in the reagents themselves, rather than the laboratories, by looking for shared taxa within the EBCs generated during extractions in both the ancient lab and the modern labs. Of the 69 dominant genera (i.e., observed at >0.1%), 17 were present in the reagents used in the in‐house QG DNA extraction process used in both types of facility. These included Cloacibacterium, Flavobacterium, Paenibacillus, Novosphingobium, Sphingomonas, Limnohabitans, Tepidomonas, Cupriavidus, Ralstonia, Acinetobacter, Enhydrobacter, Pseudomonas and Stenotrophomonas taxa and four unidentified genera within Comamonadaceae, Erythrobacteraceae, Enterobacteriaceae and Pseudomonadaceae (Table 1). Within the ancient laboratory EBCs, the 26 most dominant genera included Acinetobacter (39%), followed by three genera within the family Comamonadaceae (totalling 11.3%), Pseudomonas (8%), Novosphingobium (1.5%), Ralstonia (1%), Cloacibacterium (1%) and others (Table 1). In the EBCs from the modern laboratories, Paenibacillus was the most prevalent of the 43 dominant genera (46%), while two Erythrobacteraceae (16.5%), Comamonadaceae (6.1%), Cloacibacterium (3.9%), Corynebacterium (2.5%), Enterococccus (2.5%), Staphylococcus (2.2%), Enhydrobacter (1.8%), Microbacteriaceae (1.7%), a Pseudomonadaceae (1.4%), Ralstonia (1.3%) and N09 (1.2%) taxa were the next most prevalent within the reagents (Table 1). Although the same extraction method and reagents were used, only three of the most dominant taxa (i.e., identified at >1% prevalence) were the same within both laboratories (Comamonadaceae, Cloacibacterium, Pseudomonadaceae), highlighting the heterogeneity of taxa identified with EBCs. While many of these taxa have been previously identified as laboratory contaminants, the diversity within the modern laboratories also includes some human‐associated taxa that have been cultured from the oral cavity, gut and skin (e.g., Corynebacterium, Enterococcus and Staphylococcus, respectively). This suggests that the additional precautionary measures used within the ancient laboratory may help to reduce the introduction of human‐associated microorganisms in metagenomic data sets.
Table 1.
Dominant contaminant genera are largely unique within each laboratory
| Genera taxonomy | AL EBC | ML EBC | ML EBC (kit) | AL NTC | ML NTC | Identified previously (reference) |
|---|---|---|---|---|---|---|
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Actinomycetaceae;g__Actinomyces | 0.0002426 | 0.0011594 | 0.0028975 | 9.89E−05 | 1.33E−05 | |
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Actinomycetaceae;g__N09 | 0 | 0.012119 | 0 | 0 | 0 | |
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Corynebacteriaceae;g__Corynebacterium | 0.0002939 | 0.0254723 | 0.0104781 | 0.001034 | 0.0003335 | 3 |
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Dermacoccaceae;g__Dermacoccus | 0 | 0.0059866 | 0 | 0 | 0.0039892 | |
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Microbacteriaceae;g__ | 0.0006442 | 0.0172251 | 0.0002926 | 5.38E−05 | 0 | |
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Micrococcaceae;g__ | 6.30E−05 | 0.0016361 | 0.0024467 | 2.50E−06 | 0.0001067 | |
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Micrococcaceae;g__Micrococcus | 0.0002292 | 0.0021648 | 0.0028065 | 2.50E−06 | 4.00E−05 | 3 |
| Bacteria;p__Bacteroidetes;c__[Saprospirae];o__[Saprospirales];f__Chitinophagaceae;g__Sediminibacterium | 0.0017288 | 0 | 2.17E−06 | 0 | 0 | |
| Bacteria;p__Bacteroidetes;c__Cytophagia;o__Cytophagales;f__Cytophagaceae;g__ | 0.0017301 | 0 | 0.0008842 | 1.25E−06 | 0 | |
| Bacteria;p__Bacteroidetes;c__Flavobacteriia;o__Flavobacteriales;f__[Weeksellaceae];g__ | 6.68E−06 | 0.0014925 | 0 | 5.01E−06 | 6.67E−06 | |
| Bacteria;p__Bacteroidetes;c__Flavobacteriia;o__Flavobacteriales;f__[Weeksellaceae];g__Chryseobacterium | 0.0005711 | 0.0049479 | 0.0001279 | 0 | 0 | 3 |
| Bacteria;p__Bacteroidetes;c__Flavobacteriia;o__Flavobacteriales;f__[Weeksellaceae];g__Cloacibacterium | 0.0101374 | 0.0399393 | 0.0060247 | 0.0007686 | 0.0020747 | |
| Bacteria;p__Bacteroidetes;c__Flavobacteriia;o__Flavobacteriales;f__[Weeksellaceae];g__Wautersiella | 0.0024707 | 8.33E−05 | 0.0001127 | 0.0014559 | 0 | |
| Bacteria;p__Bacteroidetes;c__Flavobacteriia;o__Flavobacteriales;f__Cryomorphaceae;g__Fluviicola | 0.0010454 | 0 | 0 | 0 | 0 | |
| Bacteria;p__Bacteroidetes;c__Flavobacteriia;o__Flavobacteriales;f__Flavobacteriaceae;g__Flavobacterium | 0.0026519 | 0.0040362 | 0.0003251 | 0 | 2.00E−05 | 3, 4 |
| Bacteria;p__Bacteroidetes;c__Sphingobacteriia;o__Sphingobacteriales;f__;g__ | 0.0017217 | 0 | 6.28E−05 | 0 | 0 | |
| Bacteria;p__Bacteroidetes;c__Sphingobacteriia;o__Sphingobacteriales;f__Sphingobacteriaceae;g__ | 0 | 0.0025812 | 0 | 0 | 0 | |
| Bacteria;p__Bacteroidetes;c__Sphingobacteriia;o__Sphingobacteriales;f__Sphingobacteriaceae;g__Pedobacter | 0.001991 | 2.08E−06 | 0 | 0 | 6.67E−06 | 3 |
| Bacteria;p__Cyanobacteria;c__Chloroplast;o__Streptophyta;f__;g__ | 0.009702 | 0.0007785 | 0.0511164 | 2.50E−06 | 6.67E−06 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__[Thermicanaceae];g__Thermicanus | 0 | 0.0028455 | 0 | 0 | 0 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Bacillaceae;g__Bacillus | 0.0001536 | 0.009569 | 0.0017424 | 2.75E−05 | 6.67E−06 | 3 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Bacillaceae;g__Geobacillus | 0 | 0.0027123 | 0.0001647 | 0 | 0 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Paenibacillaceae;g__Paenibacillus | 0.0012621 | 0.4657476 | 0.0001214 | 2.13E−05 | 0.8236393 | 3 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Staphylococcaceae;g__Staphylococcus | 0.0008838 | 0.0223562 | 0.0026417 | 0.0018664 | 0.008759 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Aerococcaceae;g__ | 0.0003678 | 0.0063967 | 0.0010077 | 1.25E−06 | 0 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Enterococcaceae;g__Enterococcus | 0 | 0.0249665 | 2.82E−05 | 0 | 8.01E−05 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Leuconostocaceae;g__Leuconostoc | 0.0010562 | 0.0006328 | 0 | 0 | 0 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Streptococcaceae;g__Lactococcus | 0.0038977 | 0 | 0.0039615 | 0 | 0 | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Streptococcaceae;g__Streptococcus | 0.0009439 | 0.004338 | 0.0226336 | 4.01E−05 | 0.0038825 | 3 |
| Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Clostridiaceae;g__Clostridium | 0.0001019 | 0.0010491 | 0.0029256 | 0.0008788 | 0.0010207 | |
| Bacteria;p__Planctomycetes;c__Planctomycetia;o__Pirellulales;f__Pirellulaceae;g__ | 0.000177 | 0.002577 | 0 | 0 | 6.67E−06 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__;g__ | 0.0011802 | 0.0005183 | 0.0009384 | 0.0011642 | 6.67E−06 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Bradyrhizobiaceae;g__ | 0.0018115 | 4.16E−06 | 0.016817 | 0.0011679 | 0 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Bradyrhizobiaceae;g__Bradyrhizobium | 0.0062786 | 0.0003997 | 0.0039659 | 0.0136547 | 0.0016944 | 3, 4 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Hyphomicrobiaceae;g__Devosia | 0.0010312 | 0 | 0.0062825 | 0 | 0 | 3 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Methylobacteriaceae;g__Methylobacterium | 0.0217428 | 0.0001624 | 0.0011984 | 0.0465498 | 0 | 3, 28 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Phyllobacteriaceae;g__Mesorhizobium | 0.0056686 | 0.0005516 | 0.0005981 | 0 | 0 | 3 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhodospirillales;f__Acetobacteraceae;g__ | 4.17E−07 | 0.0013967 | 0.0015538 | 0 | 0.0070245 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rickettsiales;f__mitochondria;Other | 0.0015063 | 0 | 0.0012071 | 0.0004757 | 0 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Sphingomonadales;f__Erythrobacteraceae;g__ | 0.0007168 | 0.0851929 | 0.0092212 | 0 | 0.0590982 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Sphingomonadales;f__Erythrobacteraceae;Other | 0 | 0.0014925 | 8.67E−06 | 0 | 4.00E−05 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Sphingomonadales;f__Sphingomonadaceae;g__ | 0.0019877 | 0.0003268 | 0.001296 | 9.51E−05 | 6.67E−06 | |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Sphingomonadales;f__Sphingomonadaceae;g__Novosphingobium | 0.0159422 | 0.0028788 | 0.0068785 | 0.0449963 | 0.0016477 | 3 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Sphingomonadales;f__Sphingomonadaceae;g__Sphingobium | 0.0024018 | 0 | 0.0099147 | 1.88E−05 | 0 | 3 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Sphingomonadales;f__Sphingomonadaceae;g__Sphingomonas | 0.0078676 | 0.0041132 | 0.0078797 | 0.0175166 | 0.0079785 | 3, 28, 4 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__;f__;g__ | 0.000291 | 0.0011157 | 0 | 1.25E−06 | 0 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Alcaligenaceae;g__Achromobacter | 0.0010253 | 2.08E−06 | 0 | 5.13E−05 | 0 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Burkholderiaceae;g__Burkholderia | 0.0298854 | 0 | 0.0002817 | 0.0010202 | 0 | 3, 4 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;g__ | 0.0272699 | 0.0616356 | 0.0149186 | 0.0043613 | 0.0167909 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;g__Comamonas | 0.1663902 | 0.0001811 | 0.0552752 | 0.1315213 | 0.0001001 | 3, 28 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;g__Limnohabitans | 0.0070234 | 0.0013405 | 0.0029668 | 0.0106216 | 0 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;g__Tepidimonas | 0.0324847 | 0.0035241 | 0.0004833 | 0.0026751 | 0 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;Other | 0.0548562 | 0.0040008 | 0.0253556 | 0.0721054 | 0.0019546 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Oxalobacteraceae;g__Cupriavidus | 0.0020211 | 0.0014176 | 0.0005418 | 0.0003405 | 0 | 3, 28 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Oxalobacteraceae;g__Ralstonia | 0.0109828 | 0.0137093 | 0.0158006 | 0.0459339 | 0.0092727 | 3, 28, 4, 29 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Oxalobacteraceae;Other | 0.0025362 | 0.0001103 | 0.0016362 | 0.0180573 | 0 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Methylophilales;f__Methylophilaceae;g__ | 0.0020728 | 0 | 0 | 0 | 0 | |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Rhodocyclales;f__Rhodocyclaceae;g__ | 0.0002296 | 0.0019713 | 0.0029213 | 0.0010478 | 0 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Aeromonadales;f__Aeromonadaceae;g__ | 7.81E−05 | 0.001607 | 0.0040504 | 1.25E−06 | 0 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__ | 0.0192299 | 0.0828282 | 0.0399578 | 0.0364377 | 0.0001668 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;Other | 0.0147874 | 1.25E−05 | 0.0053984 | 0.030091 | 0.0043762 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Moraxellaceae;g__Acinetobacter | 0.3930931 | 0.0016528 | 0.2645106 | 0.4111166 | 0.0002335 | 3, 2, 28 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Moraxellaceae;g__Enhydrobacter | 0.001156 | 0.0181764 | 0.0005028 | 0.0026776 | 0.0194392 | 3 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Pseudomonadaceae;g__ | 0.0001904 | 0.0052768 | 0.0060052 | 7.51E−05 | 6.67E−05 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Pseudomonadaceae;g__Pseudomonas | 0.0813901 | 0.0048709 | 0.0253902 | 0.0820035 | 0.0087723 | 3, 29, 4 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Pseudomonadaceae;Other | 0.0052716 | 0.0140465 | 0.0001777 | 5.01E−06 | 0.0017144 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Xanthomonadales;f__Xanthomonadaceae;g__ | 0.0054783 | 0.0003768 | 0.0018919 | 0.0016449 | 0 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Xanthomonadales;f__Xanthomonadaceae;g__Lysobacter | 0.0018641 | 2.08E−06 | 0 | 0 | 0 | |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Xanthomonadales;f__Xanthomonadaceae;g__Stenotrophomonas | 0.0018086 | 0.0014051 | 0.0007607 | 0.0009689 | 0.0003869 | 3, 2, 28, 29, 4 |
The 69 genera that dominated EBC control samples are displayed for all sample types and include the proportion identified in each sample type. Genera were identified as dominant if they were found to be above 0.01% of the total genera identified within each laboratory. Taxa highlighted in green represent genera that dominated EBCs in the ancient laboratory, while unhighlighted are those from the modern EBC samples. If the genera were identified in previous studies that examined contamination, the reference number is shown in the right‐hand column.
3.4. DNA extraction kits contain microbiota indicative of the human mouth
We compared the diversity of taxa present within EBCs from the widely used ancient DNA extraction method and the commercial PowerBiofilm DNA Isolation Kit, used in the same modern laboratory. While the latter kit has been shown to have the lowest bacterial background contamination of standard microbiome kits (Salter et al., 2014), microbial diversity within the kit EBCs was significantly higher than the QG method (Figure 1b), suggesting that kit‐based DNA extractions are more prone to background contamination. On a PCoA plot constructed using unweighted UniFrac distances, the kit EBCs clustered away from the QG EBCs and NTCs, including those processed in the same laboratory (adonis; p ≤ 0.001, R 2 = 0.04; Figure 4a), demonstrating that a unique microbial community profile originates from the kit. This profile was not solely dominated by Firmicutes, similar to the other control samples from the modern lab, but contained taxa from several unique phyla (Acidobacteria, Gemmatimonadetes and Verrucomicrobia). These unique phyla included 15 distinct taxa that were also not observed in the extractions using the ancient DNA extraction method, including Alicyclobacillus (n = 9), Halomonas, Pseudonocardia, Vogesella, Allobaculum (n = 2) and Akkermansia taxa (Kruskal–Wallis; p ≤ 0.05; Table 2). Several of these taxa are known to be resistant to sterilization treatments, including pasteurization (Chang & Kang, 2004). In addition, several OTUs were more likely to be found in higher abundances in the kit EBCs than any other control samples (G‐test; p ≤ 0.05) and include specific Bradyrhizobiaceae, Neisseria, Corynebacterium, Fusobacterium, Streptococcus, Micrococcus and Halomonas taxa. While Bradyrhizobium and Micrococcus have previously been identified as laboratory contaminants (Laurence et al., 2014; Salter et al., 2014), the remaining taxa are commonly found in the human mouth. Concerningly, many of these human oral taxa have been previously reported from low‐biomass samples, such as placenta and tumour tissue, which were examined without EBCs (Aagaard et al., 2014; Hieken et al., 2016). This suggests that DNA extraction kits used in modern molecular biology laboratories may be contributing unique microbial signals in addition to those generated within the laboratory environment.
Figure 4.
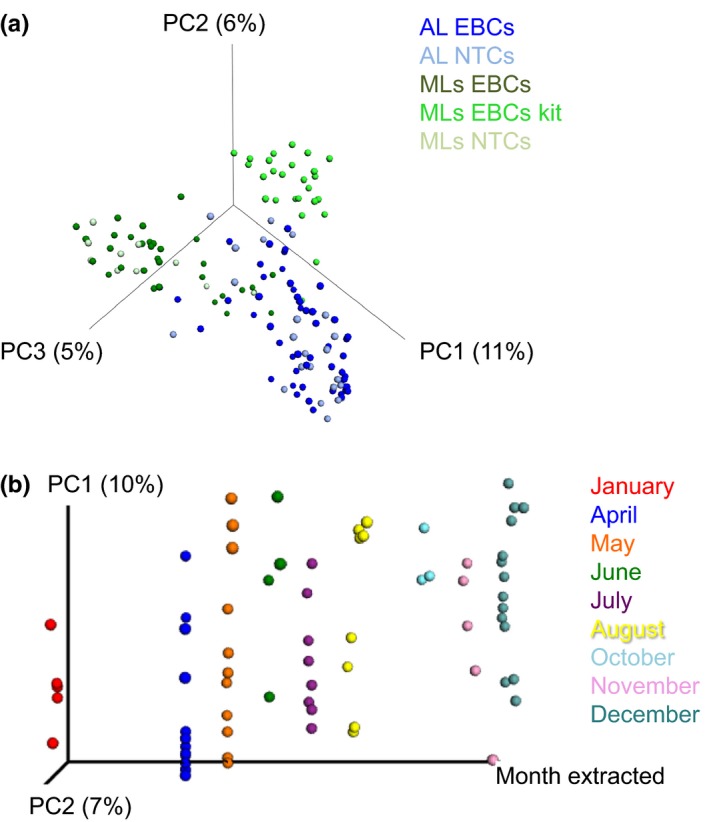
PCoA of the extraction method and seasonal variation in contaminant communities. The modern and ancient calculus samples were removed from the analysis presented in Figure 3, and a PCoA plot was constructed of only control samples to identify differences between the extraction method and laboratory in control samples (a). (b) UniFrac values from control samples (EBCs and NTCs) from the ancient laboratory over a 5‐year period (2012–2016) are coloured on a PCoA plot according to month [Colour figure can be viewed at http://www.wileyonlinelibrary.com]
Table 2.
Extraction methods contain unique taxa
| OTU Taxonomy | Mean sequences/sample | |
|---|---|---|
| Kit | QG | |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales | 6.5357143 | 0.0086957 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 2.8214286 | 0 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 7.2142857 | 0 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 1.9285714 | 0 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 2.0714286 | 0 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 1.7142857 | 0 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Oceanospirillales;f__Halomonadaceae;g__Halomonas;s__ | 0.0357143 | 0 |
| Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Pseudonocardiaceae;g__Pseudonocardia;s__ | 0.0357143 | 0 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Moraxellaceae;g__Acinetobacter;s__ | 385.28571 | 0.026087 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Bradyrhizobiaceae;g__;s__ | 93.428571 | 0 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Bradyrhizobiaceae;g__;s__ | 91.857143 | 11.721739 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 0.8571429 | 0 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 0.8571429 | 0 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Neisseriales;f__Neisseriaceae;g__Vogesella;s__ | 184.46429 | 0 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Bradyrhizobiaceae | 2.3571429 | 0.0086957 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Moraxellaceae;g__Acinetobacter;s__ | 395.39286 | 0.626087 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Sphingomonadales;f__Sphingomonadaceae;g__Sphingobium;s__ | 160.75 | 33.243478 |
| Bacteria;p__Firmicutes;c__Erysipelotrichi;o__Erysipelotrichales;f__Erysipelotrichaceae;g__Allobaculum;s__ | 0.6071429 | 0 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 0.4285714 | 0 |
| Bacteria;p__Verrucomicrobia;c__Verrucomicrobiae;o__Verrucomicrobiales;f__Verrucomicrobiaceae;g__Akkermansia;s__muciniphila | 0.5 | 0 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Moraxellaceae;g__Acinetobacter;s__ | 82.142857 | 0.7391304 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales;f__Bradyrhizobiaceae | 1.9642857 | 1.7217391 |
| Bacteria;p__Firmicutes;c__Erysipelotrichi;o__Erysipelotrichales;f__Erysipelotrichaceae;g__Allobaculum;s__ | 0.4642857 | 0 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Bacillales;f__Alicyclobacillaceae;g__Alicyclobacillus;s__ | 0.3928571 | 0 |
| Bacteria;p__Cyanobacteria;c__Chloroplast;o__Streptophyta;f__;g__;s__ | 2 | 0.0086957 |
| Bacteria;p__Cyanobacteria;c__Chloroplast;o__Streptophyta;f__;g__;s__ | 26.107143 | 1.5478261 |
| Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales | 1.9642857 | 1.9913043 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__;s__ | 5.3928571 | 23.46087 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Pseudomonadaceae;g__Pseudomonas;s__ | 0 | 0.0086957 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;g__;s__ | 0 | 0.0086957 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Pseudomonadales;f__Pseudomonadaceae;g__Pseudomonas;s__ | 0 | 0.0086957 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;g__;s__ | 0 | 0.0086957 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__;s__ | 5.5714286 | 44.6 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Oceanospirillales;f__Alcanivoracaceae;g__Alcanivorax;s__ | 0 | 0.0173913 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__;s__ | 4.7857143 | 20.730435 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__;s__ | 4.9285714 | 23.373913 |
| Bacteria;p__Proteobacteria;c__Epsilonproteobacteria;o__Campylobacterales;f__Helicobacteraceae;g__;s__ | 0 | 0.0173913 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__;s__ | 0 | 0.0173913 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Oceanospirillales;f__Halomonadaceae;g__Halomonas;s__ | 0 | 0.026087 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__;s__ | 1.8928571 | 4.3391304 |
| Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Oxalobacteraceae;g__Cupriavidus;s__ | 0 | 0.0347826 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacteriales;f__Enterobacteriaceae;g__;s__ | 1.1071429 | 5.9217391 |
| Bacteria;p__Proteobacteria;c__Alphaproteobacteria;o__Rhodobacterales;f__Rhodobacteraceae;g__;s__ | 0 | 0.026087 |
| Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Alteromonadales;f__Alteromonadaceae;g__Marinobacter;s__ | 0 | 0.0347826 |
Operational taxonomic units identified as significantly (Kruskal–Wallis Bonferroni‐corrected p‐value <0.05) associated with one of the two extraction methods in the modern laboratory are listed. OTUs highlighted in green were significant within the QG method, while the nonhighlighted OTUs were significant in the kit extraction method.
3.5. Contaminant taxa change over time
Much of the variation identified in this study is laboratory‐specific. In order to test how seasonal changes, different researchers, or time might alter the microbial diversity observed in controls, we assessed the EBC and NTC records from the ancient lab facility over 5 years (2012–2016). Bacterial community structure in the ancient lab was linked to the researcher (adonis; p = 0.001, R 2 = 0.073), the extraction year (adonis; p ≤ 0.01, R 2 = 0.022), the extraction month (adonis; p ≤ 0.001, R 2 = 0.044; Figure 4b), and wet/dry seasons (adonis; p = 0.001, R 2 = 0.081). However, each of these signals was less significant and drove less variation within the data set when compared to the differences observed between laboratory facilities or between extraction methods. Very few specific taxa were significantly associated with temporal variation, although linked changes in overall diversity were observed. In total, 32 OTUs were associated with the month in which the extraction was performed and were largely present during dry months (October–January; dominated by Comamonadaceae (2), Bradyrhizobiaceae (11), and Gemmatimonadetes (2) taxa; Kruskal–Wallis; Bonferroni corrected p ≤ 0.05), while only two OTUs (Thermobispora and Actinomycetales taxa) were linked to wet seasons. Interestingly, five OTUs (Leptotrichia, Comamonadaceae (3) and Burkholderia) were also associated with the lab researcher (Kruskal–Wallis; Bonferroni‐corrected p ≤ 0.05). While we cannot rule out the confounding nature of these variables (e.g., links between different researchers being more active in the laboratories at different times), these observations suggest that contaminant taxa change over time and need to be continually monitored, even in the cleanest molecular facilities.
4. DISCUSSION
4.1. Overview
While several studies have now reported on contaminant DNA within laboratory reagents, the systematic inclusion of EBCs has not yet been widely embraced in metagenomic research. Several studies on human microbiota have been criticised for their lack of careful controls (Eisenhofer, Cooper, & Weyrich, 2017; Kliman, 2014; Lauder et al., 2016), as the unfounded results of such studies have potentially serious repercussions and have hindered scientific progress. A similar phenomenon occurred with the new field of ancient DNA in the early 1990s, when research teams, reviewers and editors failed to adequately test for contamination (Austin, Smith, Fortey, & Thomas, ; Beckenbach, 1995; Priest, 1995), leading to many spurious results. This seriously undermined the credibility of ancient DNA research (Weyrich et al., 2014) and resulted in the formation of a robust set of guidelines (Cooper & Poinar, 2000). Here, we surveyed the largest collection of extraction blank and no‐template amplification negative control samples to date (n = 144) with the goal of better describing contaminant DNA in microbiome studies to avoid pitfalls similar to those observed in the ancient DNA field.
4.2. Contaminant diversity remains underestimated
We identified 861 contaminant taxa over 5 years within a single ultraclean laboratory facility. Before this publication, the largest collection of contaminant taxa was published by Salter et al. (2014) and included 93 contaminant genera. Within our study, we found 71 of the taxa identified by Salter et al. across all labs and methodologies. However, only 29.5% of the Salter et al. taxa (21 of their 71 taxa) were identified as dominant taxa within our study across all methods and labs. This indicates that laboratory microbial contamination is not yet well described and is likely to be unique across different laboratories, protocols, seasons and researchers. Of the 21 taxa shared across studies, four genera (Ralstonia, Acinetobacter, Pseudomonas and Stenotrophomonas) have now been routinely identified in at least four of the six publications that examine laboratory contamination (Barton, Taylor, Lubbers, & Pemberton, 2006; Glassing et al., 2016; Grahn, Olofsson, Ellnebo‐Svedlund, Monstein, & Jonasson, 2003; Laurence et al., 2014; Salter et al., 2014; Tanner et al., 1998). All of these taxa are classified as Proteobacteria, as are 55% of the dominant contaminant taxa (38/69) identified within our study and 63% (34/92) within the Salter et al. study. Proteobacteria encompass several bacterial families that are known to be UV‐ and oxidation‐resistant, suggesting that they may have a fundamental niche within laboratory settings. While contamination is highly diverse, this finding indicates that Proteobacteria appear to be the most widespread source of laboratory contamination.
4.3. Analysing contaminants is critical for the successful interpretation of low‐biomass samples
We identified several human oral microbiota taxa present in the commercial extraction kit, including Fusobacterium, Streptococcus and Corynebacterium (Chen et al., 2010), while previous studies have previously identified additional human oral taxa contaminants, including Haemophilus and Peptostreptococcus ( Barton et al., 2006). Worryingly, one of these taxa in particular, Fusobacterium, has recently been identified both as a component of the “placental microbiome” and of breast cancer tissue, in low‐biomass studies that did not consider background contamination from laboratory reagents or environments (Aagaard et al., 2014; Hieken et al., 2016; Kostic et al., 2012). It remains unclear whether this taxon is a laboratory contaminant, or whether it can escape the oral cavity and contribute to inflammatory processes elsewhere in the body. Other nonoral taxa identified within this study as contaminants have also previously been reported as important taxa within studies that failed to use controls (Mayneris‐Perxachs et al., 2016). There is clearly a need for more detailed metagenomic studies, or the use of improved “oligotyping” 16S rRNA gene analysis methods of contaminant taxa, to better identify specific strain differences and determine whether such taxa are contaminants or are actually present in the body and can cause systemic disease. The lack of contaminant assessment has already negatively impacted the metagenomics field (Lauder et al., 2016), and it is critical that editors and reviewers are aware of this issue.
4.4. Bacterial DNA is still obtained from ultraclean reagents in ultraclean facilities – no facility is contaminant free
Contaminant taxa were identified in EBCs and NTCs within five different laboratory facilities, including a state‐of‐the‐art, ultraclean ancient DNA facility. In the latter, the specialized conditions and procedures did not prevent low levels of bacterial diversity, and a wide range of contaminant taxa was still observed – with the dominant taxa all known to resist disinfectant measures, including treatment with aromatic or oxidative compounds (i.e., bleach, Acinetobacter; Ridgway & Olson, 1982, Comamonas; Liu et al., 2015;2015 or other disinfectant compounds (Pseudomonas; Sagripanti & Bonifacino, 2000). These mechanisms of disinfection resistance have contributed to nosocomial infections in hospitals (i.e., Acinetobacter; Dent, Marshall, Pratap, & Hulette, 2010) and to contamination of cell culture reagents (e.g., Achromobacter; Gray, Birmingham, & Fenton, 2010). Of note, Deinococcus, a taxon that can notoriously survive UV irradiation (Krisko & Radman, 2013), Alicyclobacillus, known to survive pasteurization (Chang & Kang, 2004), and other species known to degrade oxidative compounds (e.g., Pasteurella; Wackett, Logan, Blocki, & Bao‐li, 1992) were not observed in the specialized ancient DNA facility, but were identified within the modern laboratory. While measures to reduce contamination have prevented the introduction of human‐associated microorganisms into the ancient lab EBCs, these numerous strategies did not eliminate or completely prevent the introduction of bacterial contaminant DNA. This suggests that each research facility will probably contain unique microorganisms able to resist decontamination measures, although it is plausible that contaminant DNA could be routinely introduced into the facility from other sources and represents living species found elsewhere, rather than in the actual facilities utilized in this study. Regardless, this finding reiterates that every laboratory is susceptible to bacterial DNA contamination and that researchers should consistently monitor the contamination present within their own facility as a best practice.
4.5. Nonkit and approaches provide unique contaminant signals
In this study, we identified several taxa in a commonly used DNA extraction kit that were absent in the home‐made ancient DNA extraction method (QG). The home‐made ancient DNA method was developed to obtain more DNA from samples with low endogenous DNA, and this and other similar extraction methods are now routinely applied in ancient DNA studies to examine ancient microbiota and metagenomes (Gilbert et al., 2008; Weyrich et al., 2017; Willerslev et al., 2003). In this study, the ancient DNA method produced extraction blanks that had lower microbial diversity and were less likely to contain human oral taxa than extraction blanks generated using a commercial kit. This suggests that commercially available kits may contain more DNA contamination than home‐made methods that source clean materials. It is likely that the assembly of kit‐based reagents in a separate facility provides an additional opportunity to contaminate reagents with laboratory DNA. Lastly, this suggests that ancient DNA extraction methods and strategies could be applied in modern low‐biomass studies to potentially reduce contaminants that originate from humans.
In the future, studies of low biomass or low endogenous count routinely use shotgun sequencing to better identify contaminant taxa, as strain‐level identifications increase specificity in tracking contaminants. In many cases, the ancient DNA field has now shifted to utilizing shotgun DNA sequencing as the gold‐standard method (Weyrich et al., 2017). Shotgun sequencing also produces many other important molecular signals (e.g., signatures of ancient DNA damage), functional analysis and strain markers to delineate which species are endogenous and which are contaminants. For example, distinct strains within a single genus could be identified as either a contaminant or an endogenous species, which would be critical for examining oral species in low‐biomass tissues. In addition, damage profiles of DNA contamination could be used to distinguish fragmented, extracellular DNA within reagents versus species living within the laboratory. Current approaches aimed at eliminating contamination in shotgun sequenced metagenomes have had varied levels of success (reviewed by Salter et al., 2014), and new bioinformatic tools and models will undoubtedly improve our ability to identify and account for contaminant signals within metagenomic data sets (Lu & Salzberg, 2018). However, the need to routinely monitor background contamination will always be necessary when examining low‐biomass samples, even when other methodologies, such as shotgun metagenomic sequencing, are applied.
4.6. Contamination assessment needs to be routinely reported as a publication requirement
Contaminant sequences introduced during sample processing and library construction significantly contribute to signals from biological samples, especially those that are low‐endogenous or low‐biomass in nature. This study confirms that contaminant taxa unique to the extraction method and facility are related to the material being extracted, and change over time within a single facility, although these levels of contamination can be somewhat mitigated by routine decontamination measures of the facility and potentially the reagents themselves (Borst, Box, & Fluit, 2004). Therefore, the presence of contaminants needs to be considered in all future studies of both human and environmental microbiota. We recommend that all researchers routinely record potential sources of contamination DNA (reagent batches or lot numbers; dates of extractions and amplifications; researchers performing such duties; etc.) and critically propose that researchers routinely include extraction blank controls during the extraction process to monitor the bacterial DNA introduced into their samples, as recently recommended by Eisenhofer et al. (2018). Minimally, one control should be included in at least every batch of extractions and amplifications performed. Adding carrier DNA into control samples may also improve contaminant DNA detection (Xu et al., 2009). If controls were not included in existing data sets, an assessment of previously identified contaminant taxa should also be minimally included in the published analysis. For example, researchers could report how many known contaminant taxa are present within a data set or provide evidence to demonstrate that the removal of known contaminants does not impact the sample signal or conclusions of the paper. To facilitate this process, we have included a text file that includes a list of all the contaminant taxa observed here, as well as a separate file of only the dominant taxa. The inclusion of negative extraction blank controls should be regarded as minimal requirements for any metagenomics research and should become standard requirements of reviewers and journal editors.
AUTHOR CONTRIBUTIONS
L.S.W. and A.G.F. conceived of the study. L.S.W., A.G.F., R.E., J.Y., C.S., M.H.D. and C.A. contributed samples and completed lab work. L.S.W., L.A. and J.B. completed bioinformatic analysis of the data. L.S.W. wrote the paper, and all authors edited and contributed to the final manuscript.
DATA ACCESSIBILITY
qiime demultiplexed sequences (16SContam_seqs_forpub.fna), a phylogenetic tree of representative sequences (rep_set.tre), a biom table (otu_table_clean.biom), and sample metadata (SampleInformation_20180820.txt) can be accessed from https://figshare.com/account/articles/7283816 (https://doi.org/10.25909/5bdaa4431a941).
ACKNOWLEDGEMENTS
We thank Paul Gooding at the Australian Genomic Research Facility for technical help during DNA sequencing. This research was funded by the Australian Research Council (L.S.W; A.C.).
Weyrich LS, Farrer AG, Eisenhofer R, et al. Laboratory contamination over time during low‐biomass sample analysis. Mol Ecol Resour. 2019;19:982–996. 10.1111/1755-0998.13011
Data Availability Statement: qiime demultiplexed sequences (16SContam_seqs_forpub.fna), a phylogenetic tree of representative sequences (rep_set.tre), a biom table (otu_table_clean.biom), and sample metadata (SampleInformation_20180820.txt) can be accessed from https://figshare.com/account/articles/7283816 (https://doi.org/10.25909/5bdaa4431a941).
REFERENCES
- Aagaard, K. , Ma, J. , Antony, K. M. , Ganu, R. , Petrosino, J. , & Versalovic, J. (2014). The placenta harbors a unique microbiome. Science Translational Medicine, 6, 237ra65‐237ra65 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler, C. J. , Dobney, K. , Weyrich, L. S. , Kaidonis, J. , Walker, A. W. , Haak, W. , … Cooper, A. (2013). Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nature Genetics, 45, 450–455. 10.1038/ng.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, J. J. , Smith, A. B. , Fortey, R. A. , Thomas, R. H. (1998). Ancient DNA from amber inclusions: A review of the evidence. Ancient Biomolecules, 2(2), 167–176. [Google Scholar]
- Barton, H. A. , Taylor, N. M. , Lubbers, B. R. , & Pemberton, A. C. (2006). DNA extraction from low‐biomass carbonate rock: An improved method with reduced contamination and the low‐biomass contaminant database. Journal of Microbiological Methods, 66, 21–31. 10.1016/j.mimet.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Beckenbach, A. T. (1995). Age of bacteria from amber. Science, 270, 2015–2016; author reply 2016–2017. [PubMed] [Google Scholar]
- Borst, A. , Box, A. , & Fluit, A. (2004). False‐positive results and contamination in nucleic acid amplification assays: Suggestions for a prevent and destroy strategy. European Journal of Clinical Microbiology and Infectious Diseases, 23(4), 289–299. 10.1007/s10096-004-1100-1 [DOI] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K., Bushman, … G. A. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Lauber, C. L. , Walters, W. A. , Berg‐Lyons, D. , Huntley, J. , Fierer, N. , … Knight, R. (2012). Ultra‐high‐throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal, 6, 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S.‐S. , & Kang, D.‐H. (2004). Alicyclobacillus spp. in the fruit juice industry: History, characteristics, and current isolation/detection procedures. Critical Reviews in Microbiology, 30, 55–74. [DOI] [PubMed] [Google Scholar]
- Chen, T. , Yu, W.‐H. , Izard, J. , Baranova, O. V. , Lakshmanan, A. , & Dewhirst, F. E. (2010). The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford), 2010, baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A. , & Poinar, H. N. (2000). Ancient DNA: Do it right or not at all. Science, 289, 1139–1139. 10.1126/science.289.5482.1139b [DOI] [PubMed] [Google Scholar]
- Dent, L. L. , Marshall, D. R. , Pratap, S. , & Hulette, R. B. (2010). Multidrug resistant Acinetobacter baumannii: A descriptive study in a city hospital. BMC Infectious Diseases, 10, 196 10.1186/1471-2334-10-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis, T. Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E. L. , Keller, K. , … Andersen, G. L. (2006). Greengenes, a Chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environment Microbiology, 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Eisenhofer, R. , & Cooper, A. , Weyrich, L. S. (2017). Reply to Santiago‐Rodriguez et al.: Proper authentication of ancient DNA is essential. FEMS Microbiology Ecology, 93(5). 10.1093/femsec/fix042 [DOI] [PubMed] [Google Scholar]
- Eisenhofer, R. , Minich, J. , Marotz, L. , Cooper, A. , Knight, R. , & Weyrich, L. S. (2018). Contamination in low‐biomass microbiome studies: Issues and recommendations. Trends in Microbiology, 27(2), 105–117. 10.1016/j.tim.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Gilbert, M. T. P. , Jenkins, D. L. , Go therstrom, A. , Naveran, N. , Sanchez, J. J. , Hofreiter, M. , … Willerslev, E. (2008). DNA from Pre‐Clovis Human Coprolites in Oregon. North America. Science, 320, 786–789. 10.1126/science.1154116 [DOI] [PubMed] [Google Scholar]
- Glassing, A. , Dowd, S. E. , Galandiuk, S. , Davis, B. , & Chiodini, R. J. (2016). Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathogens, 8, 24 10.1186/s13099-016-0103-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn, N. , Olofsson, M. , Ellnebo‐Svedlund, K. , Monstein, H.‐J ürG. , & Jonasson, J. (2003). Identification of mixed bacterial DNA contamination in broad‐range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiology Letters, 219, 87–91. 10.1016/S0378-1097(02)01190-4 [DOI] [PubMed] [Google Scholar]
- Gray, J. S. , Birmingham, J. M. , & Fenton, J. I. (2010). Got black swimming dots in your cell culture? Identification of Achromobacter as a novel cell culture contaminant. Biologicals, 38, 273–277. 10.1016/j.biologicals.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieken, T. J. , Chen, J. , Hoskin, T. L. , Walther‐Antonio, M. , Johnson, S. , Ramaker, S. , … Degnim, A. C. (2016). The microbiome of aseptically collected human breast tissue in benign and malignant disease. Scientific Reports, 6, 30751 10.1038/srep30751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, M. F. , Spindler, J. , Wiegand, A. , Shao, W. , Anderson, E. M. , Maldarelli, F. , … Coffin, J. M. (2012). Multiple Sources of Contamination in Samples from Patients Reported to Have XMRV Infection. PLoS ONE, 7(2), e30889 10.1371/journal.pone.0030889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman, H. J. (2014). Comment on “The placenta harbors a unique microbiome”. Science Translational Medicine, 6, 254le4 10.1126/scitranslmed.3009864 [DOI] [PubMed] [Google Scholar]
- Knights, D. , Kuczynski, J. , Charlson, E. S. , Zaneveld, J. , Mozer, M. C. , Collman, R. G. , … Kelley, S. T. (2011). Bayesian community‐wide culture‐independent microbial source tracking. Nat Meth, 8, 761–763. 10.1038/nmeth.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic, A. D. , Gevers, D. , Pedamallu, C. S. , Michaud, M. , Duke, F. , Earl, A. M. , … Meyerson, M. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research, 22, 292–298. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko, A. , & Radman, M. (2013). Biology of extreme radiation resistance: The way of Deinococcus radiodurans . Cold Spring Harbor Perspectives in Biology, 5, a012765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder, A. P. , Roche, A. M. , Sherrill‐Mix, S. , Bailey, A. , Laughlin, A. L. , Bittinger, K. , … Bushman, F. D. (2016). Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome, 4, 29 10.1186/s40168-016-0172-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence, M. , Hatzis, C. , & Brash, D. E. (2014). Common contaminants in next‐generation sequencing that hinder discovery of low‐abundance microbes. PLoS ONE, 9(5), e97876 10.1371/journal.pone.0097876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Zhu, W. , Cao, Z. , Xu, B. , Wang, G. , & Luo, M. (2015). High correlation between genotypes and phenotypes of environmental bacteria Comamonas testosteroni strains. BMC Genomics, 16(1), 110 10.1186/s12864-015-1314-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , & Salzberg, S. L. (2018). Removing contaminants from metagenomic databases. Biorxiv. bioRxiv 261859; 10.1101/261859 [DOI] [Google Scholar]
- Mayneris‐Perxachs, J. , Bolick, D. T. , Leng, J. , Medlock, G. L. , Kolling, G. L. , Papin, J. A. , … Guerrant, R. L. (2016). Protein‐ and zinc‐deficient diets modulate the murine microbiome and metabolic phenotype. American Journal of Clinical Nutrition, 104(5), 1253‐1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest, F. G. (1995). Age of bacteria from amber. Science, 270, 2015; author reply 2016–2017. [PubMed] [Google Scholar]
- Ridgway, H. F. , & Olson, B. H. (1982). Chlorine resistance patterns of bacteria from two drinking water distribution systems. Applied and Environment Microbiology, 44, 972–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland, N. , & Hofreiter, M. (2007). Ancient DNA extraction from bones and teeth. Nat Protocols, 2, 1756–1762. 10.1038/nprot.2007.247 [DOI] [PubMed] [Google Scholar]
- Sagripanti, J. L. , & Bonifacino, A. (2000). Resistance of Pseudomonas aeruginosa to liquid disinfectants on contaminated surfaces before formation of biofilms. Journal of AOAC International, 83, 1415–1422. [PubMed] [Google Scholar]
- Salter, S. J. , Cox, M. J. , Turek, E. M. , Calus, S. T. , Cookson, W. O. , Moffatt, M. F. , … Walker, A. W. (2014). Reagent and laboratory contamination can critically impact sequence‐based microbiome analyses. BMC Biology, 12, 87 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago‐Rodriguez, T. M. , Fornaciari, G. , Luciani, S. , Dowd, S. E. , Toranzos, G. A. , Marota, I. , & Cano, R. J. (2015). Gut microbiome of an 11th century A.D. pre‐Columbian Andean mummy. PLoS ONE, 10, e0138135 10.1371/journal.pone.0138135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, M. A. , Goebel, B. M. , Dojka, M. A. , Pace, N. R. (1998). Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Applied and Environment Microbiology, 64, 3110–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett, L. P. , Logan, M. S. P. , Blocki, F. A. , & Bao‐li, C. (1992). A mechanistic perspective on bacterial metabolism of chlorinated methanes. Biodegradation, 3, 19–36. 10.1007/BF00189633 [DOI] [Google Scholar]
- Wang, Q. , Garrity, G. M. , Tiedje, J. M. , Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environment Microbiology, 73, 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warinner, C. , Speller, C. , & Collins, M. J. (2015). A new era in palaeomicrobiology: Prospects for ancient dental calculus as a long‐term record of the human oral microbiome. Philosophical Transactions of the Royal Society of London B: Biological Sciences, 370, 20130376 10.1098/rstb.2013.0376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S. , Amir, A. , Hyde, E. R. , Metcalf, J. L. , Song, S. J. , & Knight, R. (2014). Tracking down the sources of experimental contamination in microbiome studies. Genome Biology, 15, 564 10.1186/s13059-014-0564-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyrich, L. S. , Dobney, K. , & Cooper, A. (2015). Ancient DNA analysis of dental calculus. Journal of Human Evolution, 79, 119–124. 10.1016/j.jhevol.2014.06.018 [DOI] [PubMed] [Google Scholar]
- Weyrich, L. S. , Duchene, S. , Soubrier, J. , Arriola, L. , Llamas, B. , Breen, J. , … Cooper, A. (2017). Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature, 544, 357–361. 10.1038/nature21674 [DOI] [PubMed] [Google Scholar]
- Weyrich, L. S. , Llamas, B. , & Cooper, A. (2014). Reply to Santiago‐Rodriguez et al.: Was luxS really isolated from 25‐ to 40‐million‐year‐old bacteria?. FEMS Microbiol Letters, 353, 85–86. [DOI] [PubMed] [Google Scholar]
- Willerslev, E. , Hansen, A. J. , Binladen, J. , Brand, T. B. , Gilbert, M. T. P. , Shapiro, B. , … Cooper, A. (2003). Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science, 300, 791–795. 10.1126/science.1084114 [DOI] [PubMed] [Google Scholar]
- Xu, Z. , Zhang, F. , Xu, B. , Tan, J. , Li, S. , & Jin, L. (2009). Improving the sensitivity of negative controls in ancient DNA extractions. Electrophoresis, 30, 1282–1285. 10.1002/elps.200800473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
qiime demultiplexed sequences (16SContam_seqs_forpub.fna), a phylogenetic tree of representative sequences (rep_set.tre), a biom table (otu_table_clean.biom), and sample metadata (SampleInformation_20180820.txt) can be accessed from https://figshare.com/account/articles/7283816 (https://doi.org/10.25909/5bdaa4431a941).


