Abstract
Aims
We aimed to compare digital image analysis (DIA) of human epidermal growth factor receptor 2 (HER2) immunohistochemistry (IHC) in breast cancer by two platforms: (i) to validate DIA against standard diagnostics; and (ii) to evaluate the added value of DIA in clinical practice.
Methods and results
HER2 IHC and in‐situ hybridisation (ISH) were performed on 152 consecutive invasive breast carcinomas. IHC scores were determined with DIA using two independent platforms. Manual scoring was performed by two independent observers. HER2 status was considered positive in 3+ and ISH‐positive 2+ cases. HER2 status using DIA was compared to HER2 status with standard diagnostics (manual scoring with ISH in 2+ cases). Interplatform agreement of IHC scores was ‘moderate’ (linear weighted κ = 0.58), agreement between manual scoring and platform A was ‘moderate’ (κ = 0.60) and between manual scoring and platform B ‘almost perfect’ (κ = 0.85). Compared to manual scoring, DIA resulted in a reduction of 2+ cases from 17.1 to 1.3% with platform A and from 17.1 to 15.8% with platform B. However, compared to standard diagnostics, there were three false‐negative cases with DIA using platform A [81.3% sensitivity, 100% specificity, 100% positive predictive value (PPV), 97.8% negative predictive value (NPV)]. Sensitivity, specificity, PPV and NPV were 100% with DIA using platform B.
Conclusions
DIA of HER2 IHC is a valid tool in determining HER2 status in breast carcinoma. Algorithms in different platforms can behave differently, and optimal calibration is essential. In clinical practice, DIA offers an objective alternative to manual scoring, but a reduction in 2+ cases could result in loss of sensitivity.
Keywords: breast cancer, digital image analysis (DIA), human epidermal growth factor 2 (HER2), immunohistochemistry (IHC)
Introduction
Human epidermal growth factor receptor 2 (HER2/ErbB2) is a prognostic and predictive biomarker in breast cancer, and HER2 testing is standard of care.1, 2 HER2 is overexpressed and/or amplified in 15–20% of breast cancers.1 HER2‐positivity is required for targeted anti‐HER2‐therapy with drugs such as trastuzumab, pertuzumab and lapatinib.3, 4, 5, 6 HER2 status is determined by semiquantitative assessment of cell membrane overexpression with immunohistochemistry (IHC) or by assessing gene amplification with in‐situ hybridisation (ISH). In daily practice, a two‐tiered method is applied: IHC score 0/1+ is negative; 2+ equivocal and 3+ positive; ISH follows only in 2+ cases.2
IHC is scored subjectively by individual pathologists, and interobserver variability occurs.7, 8 Recently, digital image analysis (DIA) has emerged as an objective and reproducible IHC scoring method.9, 10, 11, 12, 13 The American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guideline for HER2 has acknowledged DIA as a diagnostic modality.2 DIA could reduce the number of 2+ cases requiring subsequent ISH testing, which may increase time‐ and cost‐efficiency in clinical practice.12, 13, 14 Our group previously found that DIA of HER2 IHC can reduce 2+ cases in gastroesophageal adenocarcinoma.15
Studies to date have compared manual scoring with DIA by different platforms, each using only one platform.9, 10, 11, 12, 13, 14, 15, 16, 17 However, interplatform variability may be expected because these platforms use different algorithms, each with unique approaches to classifying tissue and cellular components.18, 19, 20 To our knowledge, no study to date has examined HER2 DIA interplatform agreement.
We conducted a study on DIA of HER2 IHC in breast carcinoma. First, we aimed to assess interplatform agreement by two independent platforms. Secondly, we aimed to validate DIA and evaluate the added value of DIA in clinical practice by comparing DIA with manual scoring and comparing HER2 status outcome using DIA (with ISH on 2+ cases) with standard diagnostics (manual scoring with ISH on 2+ cases).
Materials and methods
Cases
Resection specimens of 152 consecutive primary invasive breast carcinomas from the University Medical Center Groningen (the Netherlands) treated between August 2015 and February 2017 were included. IHC and ISH were performed on 3 μm sections, cut from formalin‐fixed paraffin‐embedded tumour blocks. Standardised HER2 controls were included in all IHC and ISH tests.
Patient material was handled following the Dutch ‘Code of conduct for medical research’.21 Therefore, no additional Ethics Committee permission was required.
Immunohistochemistry
IHC was performed using SP3 (rabbit monoclonal antibody; Thermo Fisher Scientific, Fremont, CA, USA) on the BenchMark Ultra (Ventana Medical Systems, Illkirch, France), with antibody dilution 1:40; antigen retrieval time 64 min (95°C, CC1, pH9; Ventana) and primary antibody incubation time 32 min. Visualisation was achieved with the ultraView diaminobenzidine (DAB) detection kit (Ventana) and antigen amplification was applied (Ventana amplification kit). Counterstaining was performed with Mayer's haematoxylin (Klinipath, Breda, the Netherlands).
Manual Scoring
HER2 IHC was scored independently by an experienced pathologist (B.V.) and a senior resident (T.K.), according to current guidelines:2 0 (negative): no staining or faint/barely perceptible incomplete membrane staining in ≤10% of tumour cells; 1+ (negative): faint/barely perceptible incomplete membrane staining in >10%; 2+ (equivocal): weak/moderate complete membrane staining in >10%; and 3+ (positive): circumferential complete intense membrane staining in >10%. Discordant cases were re‐evaluated by both observers to establish a consensus manual score. This occurred in only 13 of 152 cases (8.6%) and discordance was only 1 score point in these cases.
Image Acquisition and Dia Platforms
Glass slides were scanned in a Philips Ultra Fast Scanner 1.6 (Philips, Eindhoven, the Netherlands) with a ×40 magnification lens, using a single‐focus layer without Z‐stacking. Tissue detection with focus points was applied automatically to obtain the optimal image. Digitised slides were stored on a centralised server and a direct link was established in both DIA platforms. The platforms were Visiopharm Integrator System (VIS) version 6.9.0.2779 (Visiopharm, Hørsholm, Denmark) and HALO version 2.0.1061 (Indica Labs, Corrales, NM, USA).
Digital Image Analysis
HER2 IHC was scored using HER2 algorithms in both platforms.
The HER2‐CONNECT algorithm in the VIS platform analyses membrane staining by calculating a connectivity value based on DAB staining of linear structures corresponding to membrane fragments.13 This connectivity value can vary continuously from 0 to 1, and is converted to a HER2 score with specific cut‐offs. Standardised recommended cut‐offs were applied: 0: connectivity = 0; 1+: 0 < connectivity ≤ 0.40; 2+: 0.40 < connectivity ≤ 0.64; 3+: connectivity > 0.64. HER2‐CONNECT is a CE‐IVD‐approved and NordiQC‐validated algorithm, and was not further calibrated for this study.
The HALO platform algorithm was constructed using the HER2 module (version 1.1), which measures membranous staining on a cell‐by‐cell basis. The algorithm was calibrated in close collaboration between the researcher (T.K.) and the platform vendor, using a training set of 20 randomly selected breast carcinomas resected in January–August 2015, identically handled and stained but not included in the current study. Variables including colour classification, cell classification, membrane detection and membrane completeness were optimised in the calibration process. Cell‐specific HER2 classification was based on specific cut‐offs in HER2 optical density (OD), calibrated at 0: 0 < OD ≤ 0.085; 1+: 0.085 < OD ≤ 0.160; 2+: 0.160 < OD ≤ 0.259; and 3+: OD > 0.259. The HER2 score was based on >10% of the highest classification score within all classified cells.
In accordance with both platform vendors’ recommendations, HER2 analysis was performed on three annotated areas (1.5 mm2 regions at ×100 magnification) representative of the whole tumour. The highest HER2 score in at least one area was taken as the HER2 score for each case.
In‐situ Hybridisation
Fluorescence in‐situ hybridisation (FISH, n = 32, PathVysion HER2 DNA Probe Kit; Abbott Molecular, Abbott Park, IL, USA) and/or bright‐field dual‐colour silver in‐situ hybridisation (SISH, n = 126, INFORM HER2 Dual ISH DNA Probe Cocktail; Ventana) assays were performed following the manufacturer's recommendations.
ISH was evaluated according to current guidelines, calculating the HER2/CEP17 ratio and the average HER2 copy number in 20–40 cells.2 ISH‐positive: ratio ≥2.0 with HER2 ≥4.0, or ratio <2.0 with HER2 ≥6.0 by two observers with FISH and SISH. ISH‐negative: ratio <2.0 with HER2 <4.0, ratio ≥2.0 with HER2 <4.0 by two observers with FISH and SISH, or ratio <2.0 with HER2 ≥4.0/<6.0 by two observers with FISH and SISH (this last category was ‘equivocal’ before the recent guideline update).1, 2
Statistics
To establish interplatform agreement, linear weighted kappa (κ) statistics were performed in r for Windows version 3.3.2 (R Foundation for Statistical Computing, Vienna, Austria), using the ‘irr’‐package for κ statistics. κ was interpreted as <0.2, slight; 0.21–0.40, fair; 0.41–0.60, moderate; 0.61–0.80, substantial; and 0.81–1.00, almost perfect agreement.22
To validate DIA and to evaluate the added value of DIA in clinical practice, we calculated κ for agreement between DIA and manual scoring and calculated sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for HER2 status outcome using DIA (with ISH on 2+ cases) with standard diagnostics (manual scoring with ISH on 2+ cases) as a reference.
Results
Immunohistochemistry and Agreement
IHC with DIA images are shown in Figure 1. By DIA in platform A on all 152 cases, 139 cases were 0/1+ (91.4%), two were 2+ (1.3%) and 11 were 3+ (7.2%). By DIA in platform B, 114 cases were 0/1+ (75%), 24 were 2+ (15.8%) and 14 were 3+ (9.2%). Interplatform agreement was ‘moderate’ (κ = 0.58, 82.2%). By manual scoring, 114 cases were 0/1+ (75%), 26 were 2+ (17.1%) and 12 were 3+ (7.9%). As such, DIA in platform A resulted in a 15.8% reduction of 2+ cases (26 to two cases, 17.1–1.3%) compared to manual scoring, which was 1.3% with platform B (26–24 cases, 17.1–15.8%). Table 1 displays cross‐tabulations of HER2 scores. Agreement was ‘moderate’ between manual scoring and platform A (κ = 0.60, 82.9%), comparable to interplatform agreement. Agreement was ‘almost perfect’ between manual scoring and platform B (κ = 0.85, 91.1%).
Figure 1.
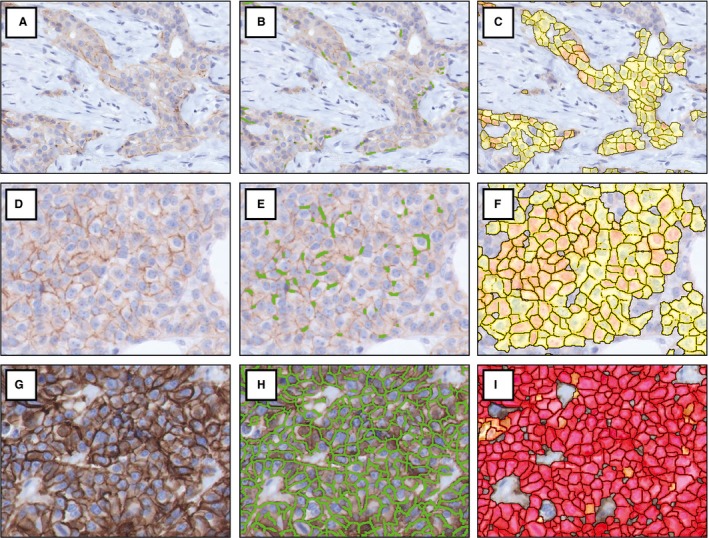
Digital image analysis of HER2 immunohistochemistry by two DIA platforms. HER2 score 1+ (A–C), score 2+ (D–F) and score 3+ (G–I). Images without DIA mark‐up (left column), with DIA in platform A (middle column) and with DIA in platform B (right column). HER2, human epidermal growth factor 2; DIA, digital image analysis. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Table 1.
Comparison of HER2 IHC scores by manual scoring and DIA
| Manual scoring | ||||
|---|---|---|---|---|
| 0/1+ | 2+ | 3+ | Total | |
| Platform A DIA | ||||
| 0/1+ | 114 | 25 | 0 | 139 |
| 2+ | 0 | 1 | 1 | 2 |
| 3+ | 0 | 0 | 11 | 11 |
| Total | 114 | 26 | 12 | 152 |
| κ = 0.60 | ||||
| Manual scoring | ||||
|---|---|---|---|---|
| 0/1+ | 2+ | 3+ | Total | |
| Platform B DIA | ||||
| 0/1+ | 109 | 5 | 0 | 114 |
| 2+ | 5 | 19 | 0 | 24 |
| 3+ | 0 | 2 | 12 | 14 |
| Total | 114 | 26 | 12 | 152 |
| κ = 0.85 | ||||
| Platform B DIA | ||||
|---|---|---|---|---|
| 0/1+ | 2+ | 3+ | Total | |
| Platform A DIA | ||||
| 0/1+ | 114 | 24 | 1 | 139 |
| 2+ | 0 | 0 | 2 | 2 |
| 3+ | 0 | 0 | 11 | 11 |
| Total | 114 | 24 | 14 | 152 |
| κ = 0.58 | ||||
HER2, human epidermal growth factor 2; IHC, immunohistochemistry; DIA, digital image analysis; κ, linear weighted kappa.
Concordance with Standard Diagnostics
HER2‐positivity by standard diagnostics was 10.5% (16 of 152 cases), by platform A 8.6% (13 cases) and by platform B 10.5% (16 cases). IHC/ISH concordance of manual scoring and DIA is shown in Table 2. HER2 status outcome using DIA compared to standard diagnostics is displayed in Table 3. Specificity, sensitivity, PPV and NPV of HER2 status outcome using DIA, with standard diagnostics as a reference, are shown in Table 4. Platform B had no false‐positive or false‐negative cases. In platform A, three cases were false‐negative, as these cases were HER2‐positive with standard diagnostics (manual IHC 2+, ISH‐positive, in all three cases). These cases would have been missed in clinical practice, as the IHC‐negative score would not have prompted subsequent ISH. Interestingly, ISH was near the cut‐offs for amplification. Case 1 showed HER2/CEP17 ratio 2.2 (just above the 2.0 cut‐off), with HER2 copy number 5.5. Case 2 showed ratio 1.2, but HER2 copy number 6.5 (just above the 6.0 cut‐off). Similarly, case 3 showed ratio 1.8 with HER2 copy number 7.2. In all three cases, FISH and SISH showed identical results. With DIA in platform B, case 1 was 3+ and cases 2 and 3 were 2+. Figure 2 displays the three false‐negative cases.
Table 2.
IHC/ISH concordance of manual scoring and DIA
| ISH | |||
|---|---|---|---|
| Negative | Positive | Total | |
| Manual scoring | |||
| 0/1+ | 114 | 0 | 114 |
| 2+ | 22 | 4 | 26 |
| 3+ | 0 | 12 | 12 |
| Total | 136 | 16 | 152 |
| Platform A DIA | |||
| 0/1+ | 136 | 3 | 139 |
| 2+ | 0 | 2 | 2 |
| 3+ | 0 | 11 | 11 |
| Total | 136 | 16 | 152 |
| Platform B DIA | |||
| 0/1+ | 114 | 0 | 114 |
| 2+ | 22 | 2 | 24 |
| 3+ | 0 | 14 | 14 |
| Total | 136 | 16 | 152 |
IHC, immunohistochemistry; ISH, in‐situ hybridisation; DIA, digital image analysis.
Table 3.
HER2 status using DIA compared to standard diagnostics
| Standard diagnostics | |||
|---|---|---|---|
| Negative | Positive | Total | |
| Platform A DIA | |||
| Negative | 136 | 3 | 139 |
| Positive | 0 | 13 | 13 |
| Total | 136 | 16 | 152 |
| Platform B DIA | |||
| Negative | 136 | 0 | 136 |
| Positive | 0 | 16 | 16 |
| Total | 136 | 16 | 152 |
HER2, human epidermal growth factor 2; IHC, immunohistochemistry; DIA, digital image analysis.
Table 4.
Sensitivity, specificity and predictive values for HER2 status outcome using DIA (with ISH on 2+ cases), compared to standard diagnostics (manual scoring with ISH on 2+ cases) as a reference
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Equivocal IHC 2+ (%) | |
|---|---|---|---|---|---|
| Platform A DIA | 81.3 | 100 | 100 | 97.8 | 1.3 (n = 2) |
| Platform B DIA | 100 | 100 | 100 | 100 | 15.8 (n = 24) |
HER2, human epidermal growth factor 2; DIA, digital image analysis; ISH, in‐situ hybridisation; PPV, positive predictive value; NPV, negative predictive value; IHC, immunohistochemistry.
Figure 2.
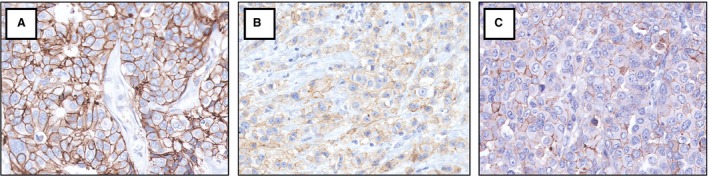
HER2 immunohistochemistry images of the three cases which were false‐negative with DIA in platform A (connectivity scores: A 0.38, B 0.08, C 0.10). All cases were scored 1+ by platform A but showed HER2 amplification with ISH. Manual scores were 2+ in all cases. In platform B, scores were 3+ (A) and 2+ (B, C). HER2, human epidermal growth factor 2; DIA, digital image analysis; ISH, in‐situ hybridisation. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Discussion
We aimed first to compare DIA of HER2 IHC by two platforms and secondly to validate DIA and evaluate the added value of DIA in clinical practice. We found moderate interplatform agreement. One platform performed comparable to standard diagnostics, with 100% sensitivity and specificity, high agreement with manual scoring, but without a reduction of 2+ cases. The other platform significantly reduced 2+ cases, but at the cost of three false‐negative cases, with slight sensitivity loss.
HER2‐positive breast cancers are eligible for anti‐HER2 therapy and appropriate HER2 testing is crucial for adequate treatment.2, 3, 4, 5, 6 HER2‐positivity in our study was 10.5%, which is somewhat lower than the 15–20% HER2‐positivity rate reported in the literature.2 An explanation for this difference might be that we included surgical resection specimens obtained in our academic hospital, in which the patient population may differ from general hospitals.
To the best of our knowledge, our study is the first to assess interplatform agreement between HER2 algorithms in different DIA platforms. Interplatform variability may be expected, as each platform has unique algorithms with differences in classifying tissue morphology, cellular characteristics and staining patterns.18, 19, 20 This could lead to different HER2 scores in different platforms. A recent study on another biomarker, Ki67, showed high interplatform agreement (R 2 = 0.97, Spearman's ρ = 0.96, numerical scoring).20 In the current study, we found that HER2 algorithms of different DIA platforms behaved differently, with only ‘moderate’ interplatform agreement (82.2%, κ = 0.58, ordinal scoring). This was comparable to the ‘moderate’ agreement between manual scoring and platform A, which could be analogous to the ‘almost perfect’ agreement between manual scoring and platform B.
The difference between platforms in our study could be related to the calibration process. Optimal calibration is essential for any DIA algorithm, as algorithms are influenced by colour and texture variations due to differences in materials and staining processes among laboratories.23 For platform A, the standardised CE‐IVD‐approved and NordiQC‐validated HER2‐CONNECT algorithm was used without modifications.24 For platform B, the algorithm was calibrated intensively, and various variables including cut‐off values were optimised in close collaboration between the researcher and the platform vendor. This could explain the much higher agreement between manual scoring and platform B than between manual scoring and platform A. Our results show that an ‘off‐the‐shelf’ approved and validated product will not necessarily perform as well as the gold standard (manual assessment). Algorithms need to be adjusted to a local laboratory's IHC and background staining intensities and digital image quality. In turn, laboratories must produce quality IHC slides for DIA to work. To ensure this, laboratories must abide by general quality assurance rules (e.g. fixation and IHC parameters) and participate in external technical quality assurance programmes.
Differences between platforms could also be related to the parameters assessed by an algorithm and to the cut‐offs on which HER2 classification is based. In our study, one platform uses membrane completeness (‘connectivity’)‐based cut‐offs, while the other uses strength of staining (‘optical density’)‐based cut‐offs. However, both parameters are incorporated in the underlying HER2 algorithms of both platforms.
Studies to date have shown high agreement between manual scoring and DIA of HER2 IHC in breast cancer, with 87.5–94.2% agreement rates (weighted κ = 0.80–0.92, Cohen's κ = 0.74–0.86).10, 11, 12, 13, 14, 16, 17 We found comparable agreement between manual scoring and platform B: 91.1% agreement, weighted κ = 0.85 (when calculated, Cohen's κ = 0.80). Agreement was lower between manual scoring and platform A: 82.9% agreement, weighted κ = 0.60 (when calculated, Cohen's κ = 0.44). The reason for disagreement was the reduction of 2+ cases with platform A.
The reduction of 2+ cases with DIA in platform A resulted in three false‐negative cases (of 152 cases). Sensitivity was 81.3%, NPV was 97.8% and specificity and PPV were 100%. Platform B had 100% sensitivity, specificity, PPV and NPV. Earlier studies also had false‐negative and/or false‐positive cases.10, 12, 13, 14 Helin et al. reported six false‐negative‐ and six false‐positive cases in 750 cases and Dobson et al. reported six false‐negative‐ and no false‐positive cases in 136 cases. Both studies used different platforms than we did, and did not report sensitivity and specificity. Holten‐Rossing et al. used one of the platforms in our study (VIS), and reported a 68% reduction of 2+ cases, with 99.2% specificity and 100% sensitivity.14 However, sensitivity and specificity in their study cannot be compared to our study because they were calculated differently: they used ISH as a reference for IHC 0/1+ and 3+ only; 2+ cases were excluded because 2+ is ‘equivocal’ and therefore not predictive of ISH, which is the downside of using ISH as a reference. Our study's analysis was more clinically orientated, as we included all IHC cases and used standard diagnostics as a reference (manual IHC with ISH on 2+ cases). However, with ISH as a reference, results would be comparable because ISH was concordant with manual scoring in our study.
In previous studies we have shown the added value of DIA in clinical practice. DIA of Ki67 IHC in breast carcinoma offers a time‐saving and reproducible alternative to manual counting.20 Additionally, DIA of HER2 IHC in gastroesophageal adenocarcinomas proved to be a reliable alternative to manual scoring, reducing 2+ cases with high sensitivity (93.8–97.9%) and specificity (99.6%).15 However, in comparison to breast carcinoma, HER2 scoring in gastroesophageal carcinoma is prone to more manual 2+ cases due to tumour heterogeneity and a low positivity threshold in biopsies (≥5 tumour cell clusters are sufficient). Considering our current findings, the added value of DIA of HER2 IHC in breast carcinoma is more questionable. Because pathologists can evaluate HER2 slides within seconds, increasing time‐efficiency has less potential gain compared to, for example, DIA of Ki67,20 unless it is applied to a large number of cases. A reduction of 2+ cases would be of greater added value, as this reduces subsequent ISH tests and thus increases time‐ and cost‐efficiency in clinical practice. However, the 2+ case reduction in platform A in our study (26 to two cases) led to three false‐negative cases, which is clinically undesirable, because these patients would consequently not receive the anti‐HER2‐therapy to which they are entitled. Therefore, despite high sensitivity and specificity, the clinical application of platform A as calibrated in this study is debatable. While there were no false‐negative cases in platform B, the reduction of 2+ cases was marginal (26–24 cases), and therefore of little added value in clinical practice. As such, the true benefit of DIA of HER2 in breast cancer is that it offers an objective scoring method which is not subject to intra‐observer variability, as every analysis is based on set algorithmic settings. Therefore, DIA could increase HER2 scoring reproducibility by aiding pathologists in reducing intra‐ and interobserver variability. However, the question is whether this benefit would justify purchasing a potentially expensive DIA platform.
In current clinical practice, receptor status determination is more commonly performed on biopsies. While our study was performed on resection specimens, we believe that the results of this study can be extended to biopsies, because areas for DIA would be annotated in a similar fashion in biopsies. Additionally, studies show that concordance for HER2 testing is high between biopsies and resection specimens.25
In conclusion, we have shown that DIA of HER2 IHC in breast carcinoma is a feasible alternative to manual scoring and a valid tool to determine HER2 status with high sensitivity, and sensitivity when compared to standard diagnostics. However, algorithms in different platforms can behave differently, and optimal calibration is essential to introduce this technique safely in daily practice. One platform performed similarly but not better than standard diagnostics, while a reduction of 2+ cases by the other platform resulted in a slight but clinically undesirable loss of sensitivity. Therefore, while DIA of HER2 IHC in breast carcinoma offers an objective alternative to manual scoring, which could potentially increase HER2 scoring reproducibility in clinical practice, its added value in reducing 2+ cases is debatable.
Conflicts of interest
Dr van der Vegt has consulted for Philips and received compensation.
Acknowledgements
We would like to thank Visiopharm and Indica Labs for providing the DIA platforms and their help in the calibration of the algorithms. No additional funding sources were involved. None of the authors have any financial relationship with the companies involved or other commercial interests in the subject under consideration in this study.
Koopman T, Buikema HJ, Hollema H, de Bock GH & van der Vegt B (2019) Histopathology 74, 917–924. 10.1111/his.13812 What is the added value of digital image analysis of HER2 immunohistochemistry in breast cancer in clinical practice? A study with multiple platforms
References
- 1. Wolff AC, Hammond ME, Hicks DG et al Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013; 31; 3997–4013. [DOI] [PubMed] [Google Scholar]
- 2. Wolff AC, Hammond MEH, Allison KH et al Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018; 36; 2105–2122. [DOI] [PubMed] [Google Scholar]
- 3. Slamon DJ, Leyland‐Jones B, Shak S et al Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001; 344; 783–792. [DOI] [PubMed] [Google Scholar]
- 4. Geyer CE, Forster J, Lindquist D et al Lapatinib plus capecitabine for HER2‐positive advanced breast cancer. N. Engl. J. Med. 2006; 355; 2733–2743. [DOI] [PubMed] [Google Scholar]
- 5. Baselga J, Cortés J, Kim SB et al Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med. 2012; 366; 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verma S, Miles D, Gianni L et al Trastuzumab emtansine for HER2‐positive advanced breast cancer. N. Engl. J. Med. 2012; 367; 1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gancberg D, Jarvinen T, di Leo A et al Evaluation of HER‐2/NEU protein expression in breast cancer by immunohistochemistry: an interlaboratory study assessing the reproducibility of HER‐2/NEU testing. Breast Cancer Res. Treat. 2002; 74; 113–120. [DOI] [PubMed] [Google Scholar]
- 8. Jacobs TW, Gown AM, Yaziji H et al Comparison of fluorescence in situ hybridization and immunohistochemistry for the evaluation of HER‐2/neu in breast cancer. J. Clin. Oncol. 1999; 17; 1974–1982. [DOI] [PubMed] [Google Scholar]
- 9. Skaland I, Ovestad I, Janssen EAM et al Comparing subjective and digital image analysis HER2/neu expression scores with conventional and modified FISH scores in breast cancer. J. Clin. Pathol. 2008; 61; 68–71. [DOI] [PubMed] [Google Scholar]
- 10. Dobson L, Conway C, Hanley A et al Image analysis as an adjunct to manual HER‐2 immunohistochemical review: a diagnostic tool to standardize interpretation. Histopathology 2010; 57; 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laurinaviciene A, Dasevicius D, Ostapenko V et al Membrane connectivity estimated by digital image analysis of HER2 immunohistochemistry is concordant with visual scoring and fluorescence in situ hybridization results: algorithm evaluation on breast cancer tissue microarrays. Diagn. Pathol. 2011; 6; 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Helin HO, Tuominen VJ, Ylinen O, Helin HJ, Isola J. Free digital image analysis software helps to resolve equivocal scores in HER2 immunohistochemistry. Virchows Arch. 2016; 468; 191–198. [DOI] [PubMed] [Google Scholar]
- 13. Brügmann A, Eld M, Lelkaitis G et al Digital image analysis of membrane connectivity is a robust measure of HER2 immunostains. Breast Cancer Res. Treat. 2012; 132; 41–49. [DOI] [PubMed] [Google Scholar]
- 14. Holten‐Rossing H, Møller Talman ML, Kristensson M, Vainer B. Optimizing HER2 assessment in breast cancer: application of automated image analysis. Breast Cancer Res. Treat. 2015; 152; 367–375. [DOI] [PubMed] [Google Scholar]
- 15. Koopman T, de Bock GH, Buikema HJ et al Digital image analysis of HER2 immunohistochemistry in gastric and oesophageal adenocarcinoma: a validation study on biopsies and surgical specimens. Histopathology 2018; 72; 191–200. [DOI] [PubMed] [Google Scholar]
- 16. Mohammed ZM, McMillan DC, Elsberger B et al Comparison of visual and automated assessment of Ki‐67 proliferative activity and their impact on outcome in primary operable invasive ductal breast cancer. Br. J. Cancer 2012; 106; 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tuominen VJ, Tolonen TT, Isola J. ImmunoMembrane: a publicly available web application for digital image analysis of HER2 immunohistochemistry. Histopathology 2012; 60; 758–767. [DOI] [PubMed] [Google Scholar]
- 18. Stalhammar G, Fuentes Martinez N, Lippert M et al Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod. Pathol. 2016; 29; 318–329. [DOI] [PubMed] [Google Scholar]
- 19. Kårsnäs A, Strand R, Doré J, Ebstrup T, Lippert M, Bjerrum K. A histopathological tool for quantification of biomarkers with suub‐cellular resolution. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2015; 3; 25–46. [Google Scholar]
- 20. Koopman T, Buikema HJ, Hollema H et al Digital image analysis of Ki67 proliferation index in breast cancer using virtual dual staining on whole tissue sections: clinical validation and inter‐platform agreement. Breast Cancer Res. Treat. 2018; 169; 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Federa (Federatie van Medisch Wetenschappelijke Verenigingen)/COREON (COmmissie REgelgeving ONderzoek) . FMWV Code of Conduct for Health Research, 2011[internet]. Available at: https://www.federa.org/sites/default/files/bijlagen/coreon/code_of_conduct_for_medical_research_1.pdf (accessed 29 October 2018).
- 22. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33; 159–174. [PubMed] [Google Scholar]
- 23. Roge R, Riber‐Hansen R, Nielsen S, Vyberg M. Proliferation assessment in breast carcinomas using digital image analysis based on virtual Ki67/cytokeratin double staining. Breast Cancer Res. Treat. 2016; 158; 11–19. [DOI] [PubMed] [Google Scholar]
- 24. Visiopharm . OncoTopix HER2 APP for IHC brochure[internet]. Available at: https://www.visiopharm.com/files/brochures/20160830_HER2_IHC_APP_Analysis.pdf (accessed 29 October 2018).
- 25. Dekker TJ, Smit VT, Hooijer GK et al Reliability of core needle biopsy for determining ER and HER2 status in breast cancer. Ann. Oncol. 2013; 24; 931–937. [DOI] [PubMed] [Google Scholar]


