Summary
Heme is an essential cofactor and alternative iron source for almost all bacterial species but may cause severe toxicity upon elevated levels and consequently, regulatory mechanisms coordinating heme homeostasis represent an important fitness trait. A remarkable scenario is found in several corynebacterial species, e.g. Corynebacterium glutamicum and Corynebacterium diphtheriae, which dedicate two paralogous, heme‐responsive two‐component systems, HrrSA and ChrSA, to cope with the Janus nature of heme. Here, we combined experimental reporter profiling with a quantitative mathematical model to understand how this particular regulatory network architecture shapes the dynamic response to heme. Our data revealed an instantaneous activation of the detoxification response (hrtBA) upon stimulus perception and we found that kinase activity of both kinases contribute to this fast onset. Furthermore, instant deactivation of the PhrtBA promoter is achieved by a strong ChrS phosphatase activity upon stimulus decline. While the activation of detoxification response is uncoupled from further factors, heme utilization is additionally governed by the global iron regulator DtxR integrating information on iron availability into the regulatory network. Altogether, our data provide comprehensive insights how TCS cross‐regulation and network hierarchy shape the temporal dynamics of detoxification (hrtBA) and utilization (hmuO) as part of a global homeostatic response to heme.
Heme is an essential cofactor for almost all bacterial species but may cause severe toxicity upon elevated levels. In this study, we present a combined experimental and theoretical approach analyzing the temporal dynamics of heme detoxification and utilization in Corynebacterium glutamicum, which are orchestrated by the two two‐component systems ChrSA and HrrSA. These systems represent an interesting example of network evolution optimizing the cellular response to a multifaceted stimulus.
Introduction
‘All things are poison, and nothing is without poison, the dosage alone makes it so a thing is not a poison’ – Paracelsus (1493–1541)
Heme represents an important iron source for almost all bacterial species (Andrews et al., 2003) and is a ubiquitous cofactor of a variety of enzymes (Poulos, 2007). Elevated cellular concentrations of heme can, however, cause severe toxicity. But this is basically true for all nutrients as already emphasized by the Swiss physician and founder of modern toxicology, Paracelsus (Paracelsus, 1965; Borzelleca, 2000). Consequently, a robust regulation of homeostasis is key to the cell’s survival and typically, sophisticated regulatory mechanisms are engaged in maintaining optimal intracellular conditions and tolerance to environmental fluctuations.
Once inside the cell, most bacteria rely on heme oxygenases to catalyze the conversion of heme to biliverdin, thereby salvaging the central iron atom with the concomitant release of carbon monoxide (Wilks, 2002). One early‐characterized example for this class of enzymes is HmuO, a heme oxygenase of Corynebacterium diphtheriae that was found to be essential for the utilization of free and hemoglobin‐bound heme (Schmitt, 1997; Wilks and Schmitt, 1998). An ortholog of HmuO was also identified in Corynebacterium glutamicum, were the deletion of the corresponding gene led to reduced growth on hemin as sole iron source (Frunzke et al., 2011). In Gram‐negative pathogens, including Neisseria spp. and Pseudomonas aeruginosa, proteins of the HemO/PigA family were found to catalyze the cleavage of the porphyrin ring structure, but do not share significant sequence similarity with HmuO of Gram‐positive species (Zhu et al., 2000b; Ratliff et al., 2001). The cost and benefit of using heme as an alternative iron source, however, needs to be carefully considered by the cell. Corynebacterial species, for example, employ the master regulator of iron homeostasis, DtxR, to feed information on iron availability into the network controlling heme homeostasis. DtxR was shown to repress hmuO under iron‐replete conditions and thereby adds an additional layer of regulation to the physiological response to heme (Schmitt, 1997; Wennerhold and Bott, 2006).
Due to the reactive nature of the heme molecule, high levels are readily toxic to microbial cells (Imlay et al., 1988; Nir et al., 1991; Anzaldi and Skaar, 2010; Wakeman et al., 2012). Consequently, organisms have evolved a variety of mechanisms to minimize toxic effects. Whereas some bacteria rely mostly on their oxygenase to degrade excess heme, such as Neisseria gonorrhoeae (Zhu et al., 2000a) or Clostridium perfringens (Hassan et al., 2010), an alternative strategy can be found in the eukaryotic parasite Plasmodium spp. which is capable to sequester excess heme in an insoluble substance called hemozoin (Fitch, 1998; Jani et al., 2008; Anzaldi and Skaar, 2010). While several bacterial species harbor bacterioferritins, which can store iron in the form of heme molecules, other forms of sequestration are not well described so far (Andrews et al., 2003; Anzaldi and Skaar, 2010). A third class of detoxification systems are heme exporters, such as HrtBA which have been described for several Gram‐positive species including Staphylococcus aureus (Torres et al., 2007; Stauff and Skaar, 2009a), Bacillus anthracis (Stauff and Skaar, 2009a), Streptococcus agalactiae (Fernandez et al., 2010) and can also be found in corynebacterial species (Bibb and Schmitt, 2010; Heyer et al., 2012).
Bacterial two‐component systems (TCS), consisting of a membrane bound histidine kinase (HK) and a cytoplasmatic response regulator (RR) (Mascher et al., 2006; Zschiedrich et al., 2016), play a central role as transient heme sensor systems in Gram‐positive species (Stauff and Skaar, 2009b). This is known from bacteria such as Staphylococcus aureus and Bacillus anthracis, both utilizing the heme sensor system HssRS to react to heme as extracellular stimulus (Stauff and Skaar, 2009a; 2009b). A common theme among Corynebacteriaceae appears to be the dedication of two paralogous TCS for the regulation of heme homeostasis (Bibb et al., 2007; Frunzke et al., 2011; Bott and Brocker, 2012; Heyer et al., 2012; Burgos and Schmitt, 2016). Here, the HrrSA and ChrSA systems coordinate the expression of genes involved in heme biosynthesis, heme detoxification (hrtBA), respiratory chain and the heme oxygenase (hmuO). While in C. glutamicum it was suggested that both TCS have partially overlapping regulons, HrrSA was shown to play an important role in in the utilization of heme as an alternative iron source by activating expression of hmuO, whereas ChrSA is crucial for the activation of the hrtBA operon encoding a heme exporter (Frunzke et al., 2011; Heyer et al., 2012). Furthermore, previous studies of our group revealed significant cross‐phosphorylation between these TCS but a highly specific phosphatase activity of the HKs toward their cognate RR (Hentschel et al., 2014). Previous studies focused mostly on the identification of target genes, which were confirmed by different in vivo and in vitro assays. However, the systemic understanding of this homeostatic network, maintaining balance between heme detoxification and utilization, demands the analysis of temporal dynamics and requires comprehensive insights in the particular network architecture.
In this study, we have conducted an analysis of reporter assays of HrrSA and ChrSA target promoters in the background of the wild‐type strain, as well as in mutant strains lacking single components of the two TCSs. These data were integrated in a quantitative mathematical model, which was used to test functional hypotheses and to simulate distinct differences in autoregulation and ON/OFF kinetics of target promoters. Finally, by studying the impact of the iron regulator DtxR on hrrA and hmuO expression at temporal resolution our data as well as the model revealed that DtxR adds an important additional regulatory level ensuring the appropriate timing of heme utilization.
Results
Temporal hierarchy in the heme utilization and detoxification response
Under iron‐limiting conditions, the growth of C. glutamicum is significantly impaired, but can be restored by the presence of heme in the medium. Provided that excess heme is toxic to the cells, we wondered which strategy C. glutamicum uses to regulate the balance between its heme utilization and detoxification modules. To this end, we studied the expression dynamics of the two major components responsible for heme utilization (hmuO) and detoxification (hrtBA) in response to an extracellular heme stimulus, by monitoring promoter‐reporter fusions for the two systems (Hentschel et al., 2014). Interestingly, a wild‐type strain of C. glutamicum transformed with plasmids carrying the reporter constructs (pJC1_PhmuO ‐eyfp or pJC1_PhrtBA ‐eyfp) revealed highly distinct response profiles and a temporal hierarchy in reporter output of the PhmuO and PhrtBA promoters (Fig. 1): While we observed a nearly instant but transient response for the heme detoxification module hrtBA to 4 µM extracellular heme (Fig. 1, red line), the heme utilization module hmuO displayed higher initial expression levels compared to hrtBA and experienced an expression boost after a delay of about 5 h (Fig. 1, green line). This increase in hmuO expression temporally coincides with a declining hrtBA expression. From a physiological perspective, these antagonistic expression profiles seem plausible and impressively demonstrate the urgency of detoxification over utilization after first contact with the stimulus.
Figure 1.
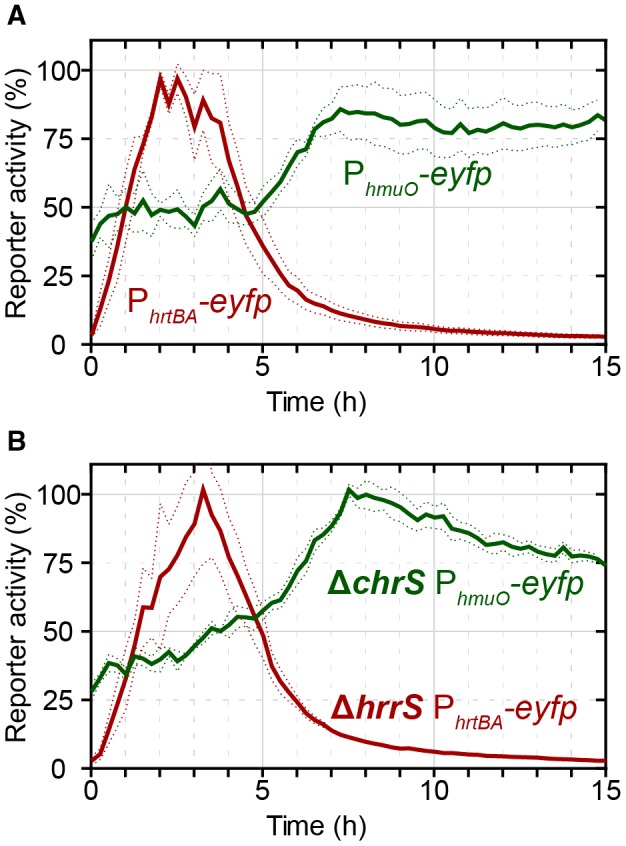
Activation of PhmuO and PhrtBA in response to extracellular heme addition. A. The C. glutamicum wild‐type strain was transformed with one of the target gene reporters pJC1_PhmuO‐eyfp or pJC1_PhrtBA‐eyfp. Iron‐deprived cells were subsequently cultivated in a microbioreactor system (Biolector) in CGXII minimal medium with 2% (w/v) glucose containing 4 µM hemin. The eYFP fluorescence was measured as the output of target promoter activation, and backscatter values were recorded to monitor biomass formation. The specific fluorescence (fluorescence/backscatter) was normalized according to material and methods and the reporter activity (%) was calculated with the maximum reporter output. B. C. glutamicum ΔchrS/pJC1_PhmuO‐eyfp and ΔhrrS/pJC1_PhrtBA‐eyfp grown as described in (A). Non‐cognate sensor kinases do not significantly affect the response profile. [Colour figure can be viewed at wileyonlinelibrary.com]
How does C. glutamicum implement appropriate timing of detoxification and utilization using two paralogous TCS responsive to the same stimulus? In this study, we formulated three distinct questions and addressed those by experiments described in the following:
How does cross‐regulation between ChrSA and HrrSA affect hrtBA and hmuO expression respectively?
Does the differential interpretation of their common stimulus, i.e. the external heme concentration, impact the response?
How does regulatory hierarchy and network architecture affect the response profile?
To test the first hypothesis, we tested two mutant strains deleted for either one of the HKs (ΔchrS and ΔhrrS) and transformed them with a reporter plasmid carrying the non‐cognate target promoter (ΔchrS/pJC1_PhmuO‐eyfp and ΔhrrS/pJC1_PhrtBA‐eyfp). Strikingly, despite some quantitative differences (as discussed below), neither of the deletions changed the qualitative response of the non‐cognate target promoter toward heme (Fig. 1B), that is the PhrtBA‐eyfp response was still transient in a ΔhrrS mutant and the PhmuO‐eyfp response was still delayed in a ΔchrS mutant, indicating that cross‐regulation between the TCS cannot explain the antagonistic regulation strategy in C. glutamicum.
Modeling of heme uptake and consumption
Therefore, we wanted to test whether the depletion of external heme could serve as a joint trigger to cause opposing regulation of heme utilization and detoxification systems. However, before turning to this question, we first asked how long it would take to deplete heme in our experiments? To this end, we considered a simple mathematical model describing the uptake and consumption of heme, assuming that the reproduction of C. glutamicum requires ~5 × 106 Fe2+ molecules per single cell (see Supplementary Text for details). Since this number sets a constraint on the growth kinetics of C. glutamicum in the heme‐supplied medium, we studied the availability of heme per cell during bacterial growth. At the given final biomass and at the experimentally determined growth rate in our medium (Fig. 2A and B), the model predicts a depletion of the total heme levels per cell (levels of cytoplasmic heme and portion of extracytoplasmic heme in the medium per cell) approx. 3–5 h after the start of the experiment (Fig. 2E), depending on the initial heme concentrations in the medium. Experimental measurements of the levels of cell‐associated heme (see Experimental procedures for experimental details) (Fig. 2D), which correspond to the model predictions of the heme levels per cell, confirm these dynamics and point out that the availability of heme in the medium dictates the growth dynamics of C. glutamicum. Hence, when comparing the experimental growth curves (Fig. 2A) with the theoretical predictions, the time points when cytoplasmic heme pools are depleted (Fig. 2C), in fact correlate with the cease of growth of the cultures in experiment and theory (Fig. 2A and B). Also the time point of growth cessation can be tuned by adding different initial heme concentrations (1–4 μM) to the medium (Fig. 2A), as predicted by our model (Fig. 2B).
Figure 2.
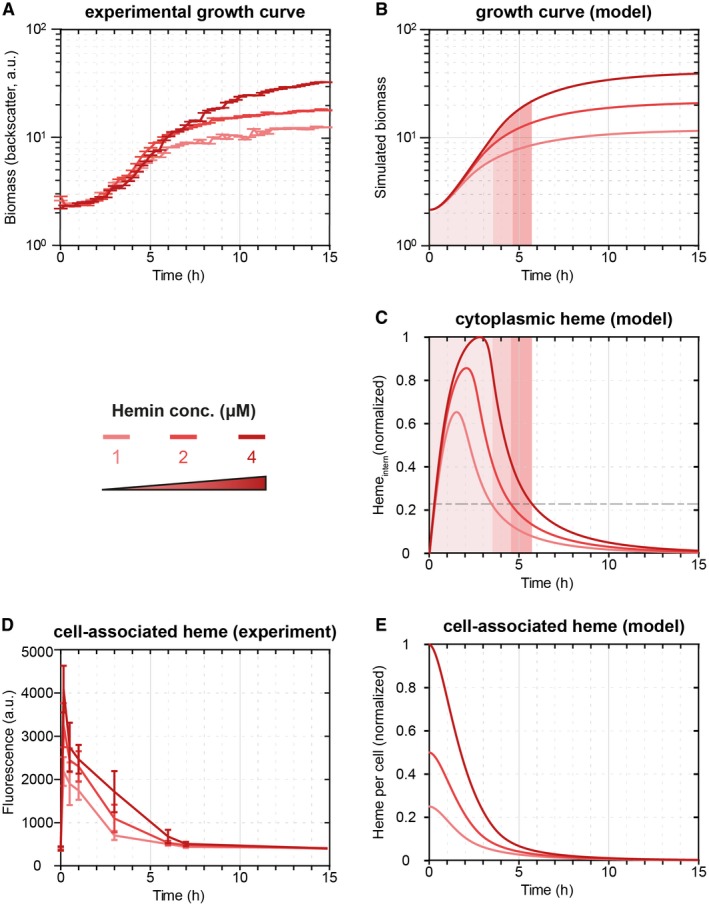
The mathematical model reproduces the experimental growth curves quantitatively. A. Growth curves of the C. glutamicum wild‐type strain under increasing hemin concentrations (1 μM, 2 μM and 4 μM). B. The mathematical model can reproduce the average growth behavior (left). C. The mathematical model predicts the consumption of cytoplasmic heme required for growth. D. The depletion of cell‐associated heme (sum of internal heme and external heme adhering to the cell) dictates the bacterial growth. The experimental data align with (E) the model predictions of total heme levels per cell (cytoplasmic heme and the portion of external heme per cell). [Colour figure can be viewed at wileyonlinelibrary.com]
Transient expression of the C. glutamicum hrtBA detoxification module
Next, we asked whether the depletion of heme could also explain the transient activity of the hrtBA promoter. To this end, we extended our mathematical model to describe stimulus perception and regulation within the two TCSs, as well as the dynamical response of the chrSA and hrtBA operons (for details, the reader is referred to the Supplementary text). Briefly, the model considers sensing of externally added heme and subsequent autophosphorylation of ChrS and HrrS (Keppel et al., 2018), (cross‐)phosphorylation of ChrA by phosphorylated HKs (ChrS~P and HrrS~P) and promoter activation of PchrSA and PhtrBA by phosphorylated response regulator ChrA~P (Fig. 3A). Simulations of the model predict that the depletion of external heme dictates the time point of deactivation of PhrtBA, leading to a prolonged promoter activity and higher overall HrtBA production at higher initial heme concentrations (Fig. 3B). In fact, when experimentally supplying different heme concentrations (1–9 μM) to the medium, we found that both the strength as well as the duration of PhrtBA‐eyfp expression increased with increasing amounts of supplied heme (Fig. 3C). In the light of a bifunctional ChrS kinase/phosphatase, exhaustion of external heme is sufficient to explain the transient response dynamics of PhrtBA‐eyfp expression.
Figure 3.
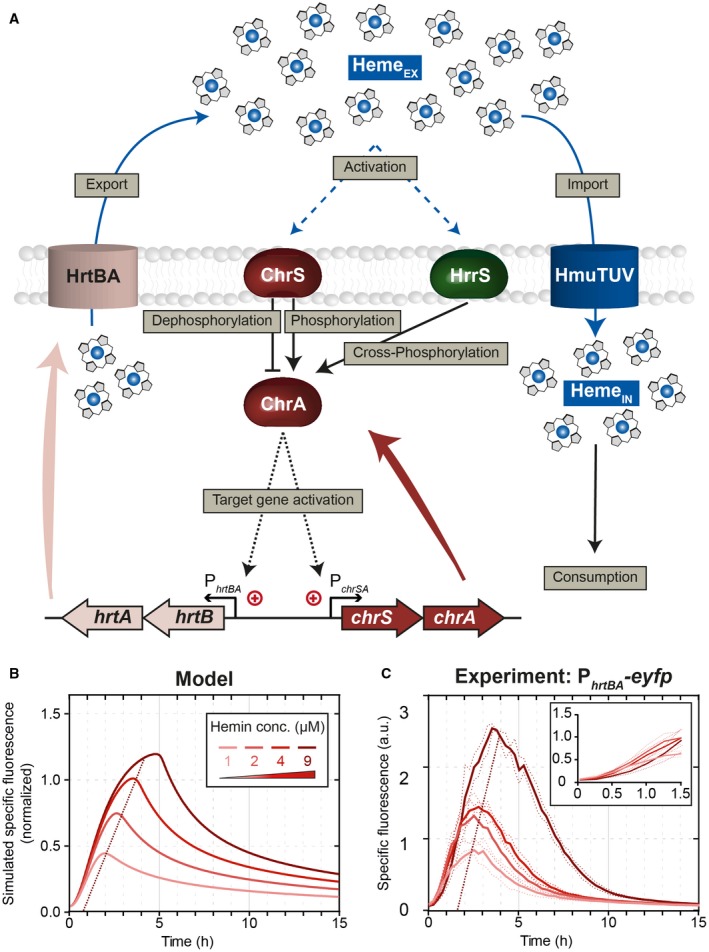
Regulatory scheme and dynamic response of the C. glutamicum heme detoxification module. A. Scheme of the regulatory interactions considered in the mathematical model for the heme detoxification module. Uptake of external heme molecules via HmuTUV and subsequent consumption/incorporation via diverse enzymes is crucial for bacterial growth under iron starvation. The fine‐tuned response to heme in order to avoid intoxication is mainly based on the two TCSs ChrSA and HrrSA. The two kinases ChrS and HrrS are autophosphorylated in response to external hemin. After activation, they (cross‐)phosphorylate the response regulator ChrA. In addition, the non‐phosphorylated form of ChrS functions as a phosphatase on the phosphorylated response regulator ChrA. The phosphorylated response regulator activates expression of its target genes hrtBA and chrSA. The gene product of hrtBA is a heme exporter that transports internal heme to the extracellular space. B. Dynamical response of our computational model for the detoxification module (Supplementary Text; Model equations M1) toward different external heme levels, as given by the simulation of specific fluorescence of a PhrtBA‐eyfp reporter, normalized to the maximal specific fluorescence at 4 μM heme. C. Experimental dynamical response of a PhrtBA‐eyfp reporter in wild‐type C. glutamicum cells toward different heme concentrations supplied in the medium at t = 0 h. [Colour figure can be viewed at wileyonlinelibrary.com]
Interestingly, our experimental data also showed that for all heme concentrations, the initial response (within the first hour) of the reporter PhrtBA‐eyfp was almost identical (Fig. 3C) and that the overall peak height is only modulated by the time point of stimulus decline. This suggests that either (i) despite varying levels of ChrA~P, the PhrtBA promoter is nearly fully saturated or (ii) the applied heme concentrations lead to saturation of the sensor kinase, i.e. maximal phosphorylation ([ChrS~P]/[ChrSTOT] 1), and thus to similar phosphorylation levels of ChrA. In order to discriminate between these scenarios, we sought to increase ChrA~P levels in the cell and test whether the PhrtBA will be more active than in the wild type. To this end, we analyzed a chrS phosphatase‐OFF mutant (chrSQ191A) still harboring its kinase activity (Hentschel et al., 2014), supposedly leading to higher ChrA~P levels. Interestingly, the chrSQ191A phosphatase mutant strain displayed a sevenfold increased PhrtBA‐eyfp output (Fig. 4A), as incurred by a higher and more sustained promoter activity within the first 4 h of incubation when compared to the wild type (Fig. 4A inset). A similar behavior can also be observed in our computational models, in which the maximal phosphorylation level of ChrA is 25% in the wild type as compared to 100% in the phosphatase mutant (Fig. S5B), leading in our model to a stronger initial promoter activity in the latter case (Figs. 4B and S5C). The predicted increase of initial promoter activity is, however, more prominent in the model than in our experimental data, in which the kinase mutant showed a wild‐type‐like behavior for around 1 h before reaching a stronger promoter activity (Fig. 4A inset). Within the model, the time point of differentiation between the reporter output of the wild type and the phosphatase‐OFF mutant – induced by stimulus decline and accordingly divergent ChrA~P levels (Fig. S5B) – is reached after 15 min (Fig. S5C). This discrepancy emphasized that saturation of kinase activity is not sufficient to explain the experimental data and that instantaneous promoter occupancy by phosphorylated response regulator may contribute to the fast onset of the detoxification response (see ‘Memoryless activation of the hrtBA detoxification module’).
Figure 4.
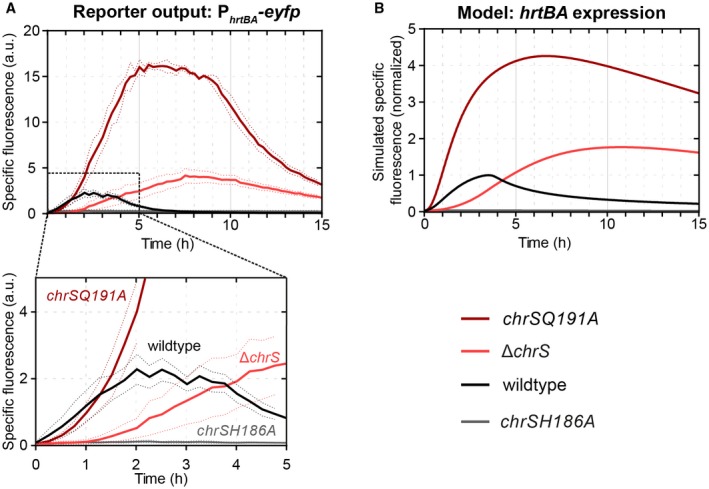
Kinase and phosphatase activity of ChrS shape the hrtBA response. A. Reporter output of C. glutamicum wild type and mutant strains carrying the vector pJC1_PhrtBA‐eyfp. Cells were inoculated in CGXII minimal medium with 2% (w/v) glucose containing 4 µM hemin as iron source. B. Simulated specific fluorescence of the C. glutamicum wild‐type strain and in the mutant strains chrSQ191A (phosphatise = off), chrSH186A (kinase = off) and ΔchrS. [Colour figure can be viewed at wileyonlinelibrary.com]
Finally, another interesting feature of the model is that the depletion of external heme leads to a quick dephosphorylation of ChrA~P and promoter shut‐off in the wild type. In contrast, in the phosphatase mutant, ChrA~P is only slowly diluted and/or degraded by growth or spontaneous dephosphorylation (Fig. S5B), leading to a significantly delayed promoter shut‐off about 1 h after external heme depletion. Thus, the strong phosphatase activity of ChrS is important in wild type cells in order to quickly turn off hrtBA expression once external heme is depleted.
Both kinases, ChrS and HrrS, contribute to a fast onset of the PhrtBA promoter
Next, we wanted to study the response of the heme detoxification module in a strain featuring reduced ChrA~P levels. However, a chrS mutant deficient in its kinase activity (chrSH186A) was unable to activate the PhrtBA promoter altogether (Fig. 4A; grey line), suggesting that ChrSH186A retains its strong phosphatase activity and likely reduces ChrA~P below a level required to activate PhrtBA. Instead, our model predicted that in a ΔchrS mutant, which lacks both kinase and phosphatase activity from ChrS, the non‐cognate HrrS sensor kinase should be able to slowly, but gradually phosphorylate ChrA~P (Hentschel et al., 2014) and thus activate PhrtBA (Fig. 4B; light red). Indeed, the experimental kinetics of the PhrtBA ‐eyfp reporter showed a weaker activation during the first 2 h, but also displayed a more sustained and eventually a stronger expression peak compared to the wild type (Fig. 4A). Within our computational model, this sustained response is again caused by the slow rate of dilution and/or dephosphorylation of ChrA~P after heme depletion, given the lack of phosphatase activity in the ΔchrS mutant (Fig. 4B). Taken together, these data show that in the absence of ChrS, the non‐cognate kinase HrrS is sufficient to activate the promoter of the detoxification module, PhrtBA.
This provoked the question as to whether the non‐cognate kinase HrrS also has an effect on the induction kinetics of PhrtBA in wild‐type cells. Interestingly, when measuring the PhrtBA‐eyfp reporter activity in a ΔhrrS mutant, the mutant indeed showed a delayed promoter activation and was about 20–30 min slower than the wild type (Fig. 5). This suggests that HrrS acts as a ‘kick‐starter’ in order to speed up the induction of the detoxification system. The additional kinase activity conferred by HrrS might in fact be needed as a support for ChrS to achieve higher ChrA phosphorylation levels, given that our analysis above suggested that ChrS is already fully in its kinase state for all heme concentrations applied here.
Figure 5.
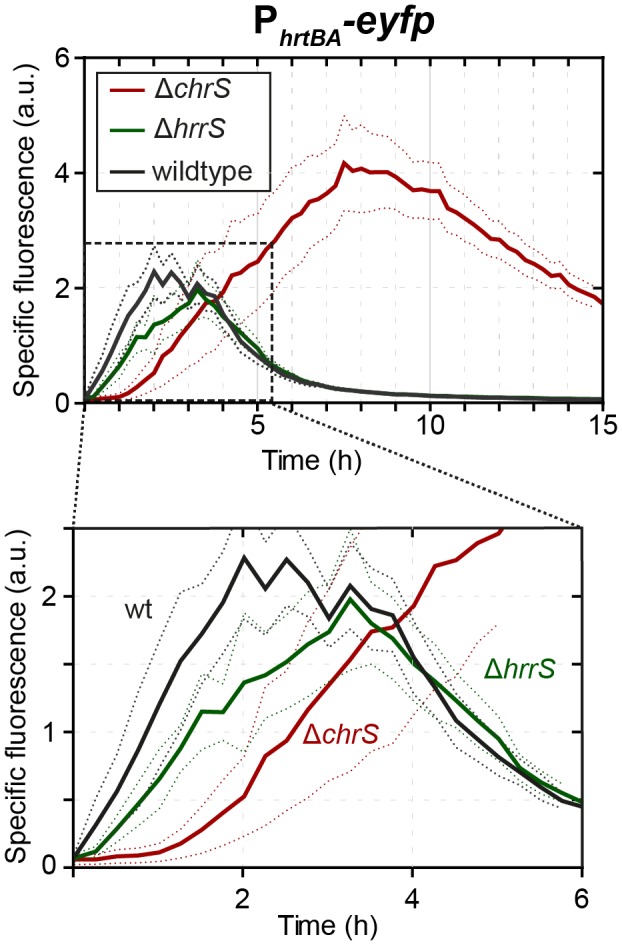
HrrS‐mediated cross‐phosphorylation of ChrA might act as a kick‐start impulse contributing to a fast ON‐set of the PhrtBA response. Reporter output of C. glutamicum wild‐type cells and the mutant strains ∆hrrS and ∆chrS, carrying the vector pJC1_PhrtBA‐eyfp and cultivated in CGXII minimal medium with 2% (w/v) glucose containing 4 µM hemin as iron source. [Colour figure can be viewed at wileyonlinelibrary.com]
Memoryless activation of the hrtBA detoxification module
Above‐described results suggested that a high promoter occupancy by ChrA~P may contribute to the fast onset of the detoxification response. Considering this scenario, we would not expect memory in the PhrtBA response if two heme pulses were applied at subsequent times. Theoretically, our model predicted nearly identical levels of promoter saturation for different heme levels, suggesting that the application of a second heme pulse should not lead to a faster response and no priming effect on the output should be observable.
While lag phase cells (growth depicted in Fig. S1) needed up to 35 min to reach maximum promoter activity following a heme pulse, promoter activity was nearly instantaneously observable after an additional heme pulse in the early exponential phase (Fig. 6A). Here, cells did respond equally fast irrespective of whether the cells were primed first, but a slightly higher maximal activity was observed after cells were primed with a heme pulse (Fig. 6A, red line) in comparison to an iron pulse (Fig. 6A, black line). Furthermore, the time point of the second heme pulses did not affect the onset of the PhrtBA response (Fig. S2). Please note that the higher amplitude of the second pulse (Fig. 6A) is a result of cells already containing eYFP molecules and a ‘reactivation’ of ChrSA (and thus PhrtBA) leads to formation of additional eYFP molecules. The total increase is comparable to the increase in the first 4 µM hemin pulse. Overall, this data supports the hypothesis that promoter saturation by phosphorylated response regulator contributes as a significant determinant of response kinetics.
Figure 6.
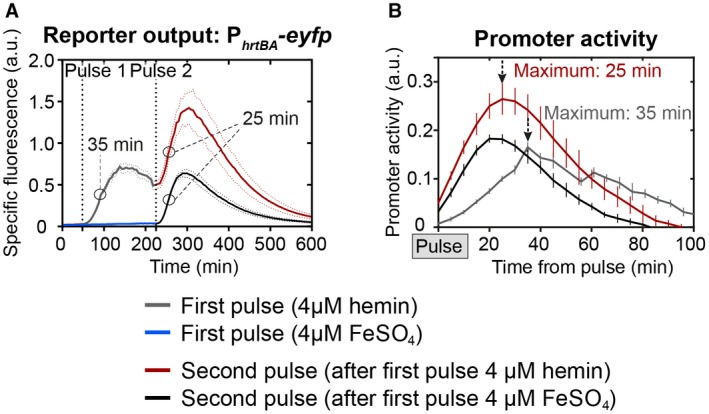
Additional heme pulses do not ‘prime’ the PhrtBA output. A. C. glutamicum wild type cells were transformed with the target gene reporter pJC1_PhrtBA‐eyfp and starved from iron overnight as described in material and methods. Subsequently, the cells were inoculated in CGXII minimal medium without iron source and eYFP fluorescence (=reporter output) and backscatter (biomass) was measured every 5 min. After 45 min, hemin (A, grey line) or FeSO4 (A, black line) was added to a final concentration of 4 µM. A second pulse of 4 µM hemin was applied to both conditions after 225 min. B. Promoter activities (calculated as derivative of the fluorescence intensity (I) over time (t), divided by the cell’s biomass (backscatter), that is ) of (A) in relation to the first or second pulse. [Colour figure can be viewed at wileyonlinelibrary.com]
Heme utilization is co‐regulated by DtxR integrating information on iron availability
While a detoxification response must be fast and rather uncoupled from other regulatory interference, utilization of a particular nutrient has to be carefully considered in the light of the current physiological status of the cell: As shown above, the activation of the detoxification module hrtBA is solely influenced by the amount of heme in the medium. In contrast, for a decision on heme utilization as an alternative iron source, information on general iron availability needs to be incorporated into the network controlling hmuO expression. In this context, it was already revealed by the previous studies that both hrrA and hmuO are repressed by the iron regulator DtxR in response to iron availability (Wennerhold and Bott, 2006).
Given that DtxR repression and HrrA activation seem to have opposing effects on the timing of hmuO expression, we asked how these signals are prioritized at the PhmuO promoter. To investigate the impact of both regulators on the activation of the heme utilization system, we developed a second mathematical model that focuses on HrrSA and DtxR as main regulators of the PhmuO und PhrrA activity (Fig. 7A). Like before, in this model the description of the non‐cognate two‐component system (ChrSA) was limited to the cross‐phosphorylation of ChrS on HrrA. In addition, we made the simplifying assumption that the activation of DtxR is proportional to the internal heme levels, based on the fact that the iron availability is proportional to the conversion of the internal heme pool under our experimental (iron‐limiting) conditions. Activated DtxR and phosphorylated HrrA bind to both PhmuO and PhrrA promoters, where they repress and activate gene expression respectively. Ultimately, increased production of the heme oxygenase HmuO contributes to heme consumption (Frunzke et al., 2011) (see Supplementary Text; Model equations M2 for all details).
Figure 7.
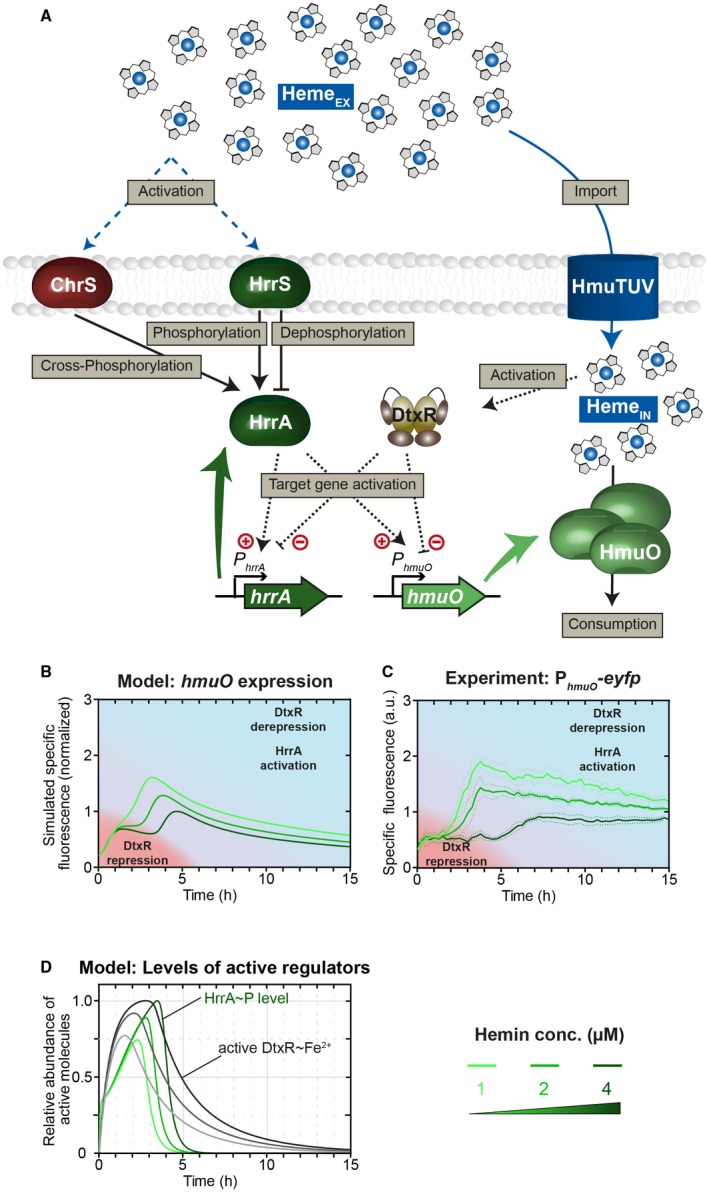
Information on iron availability is integrated into the HrrSA‐regulated utilization module hmuO via DtxR. A. The mathematical model of the heme utilization module in C. glutamicum shares many basic assumptions with the model of heme detoxification (for description see Fig. 3). Here, heme consumption is assumed to be supported by the heme oxygenase HmuO, whose production is regulated by the phosphorylated response regulator HrrA~P and the iron repressor DtxR. The activation of DtxR is expected to be influenced by internal heme levels. DtxR repression and HrrA activation shape the delayed response of the heme utilization module hmuO. B. The PhmuO promoter is activated by the phosphorylated response regulator HrrA~P after a significant time delay of 3–5 h. Higher heme concentrations lead to a prolonged delay and a lower hmuO expression in general. C. The mathematical model of the heme utilization network in C. glutamicum could reproduce this behavior and gave an explanation regarding. D. The temporal dynamics of both regulators on PhmuO. Levels of the activated iron repressor DtxR increase immediately after addition of heme and repress the promotor activation of PhmuO, proportional to stimulus strength. However, HrrA~P levels increase with a short time delay in response to the stimulus and activate the promotor to a certain extend at the beginning and with increasing intensity upon DtxR dissociation [Colour figure can be viewed at wileyonlinelibrary.com]
Simulations of this computational model revealed a biphasic induction pattern of the PhmuO promoter with a quick activation within the first hour after heme addition, followed by a significantly delayed expression boost at approx. 4–5 h (for 4 μM heme) (Fig. 7B), very similar to the experimental dynamics observed before (Fig. 1). The initial activation of PhmuO within the model is induced by an instantaneous increase in HrrA~P levels (Fig. 7D) in response to external heme. Since the dynamics of the active form DtxR~Fe2+ indirectly depends on the increase in cytoplasmic heme (which is converted to intracellular iron) (Fig. 2C), the DtxR‐mediated repression of PhmuO, which counteracts the promoter activation by HrrA~P, initiates not before DtxR~Fe2+ exceeds a certain threshold – approximately after 1 h – in the model (Fig. 7D). However, low basal levels of endogenously synthesized heme, which are not reflected in the mathematical model, as well as trace amounts of iron (resulting from the pre‐cultivation of the cells, see Experimental Procedures) probably lead to a basal level of activated DtxR in the experimental scenario. These basal levels of DtxR might reduce the initial activation of HrrA~P on PhmuO in vivo, leading to a less‐pronounced initial increase and a more prominent plateau in the experimental data (Fig. 7C) of the PhmuO‐eyfp reporter output compared to the model prediction (Fig. 7B). Interestingly, when lowering the initial heme concentration, the model predicts an earlier onset of PhmuO activation. Strikingly, when experimentally tuning the initial heme levels, we found this exact hierarchy in the activation of the heme utilization module (Fig 7C): A short delay of 3 h at 1 µM heme and a longer delay of 5 h at 4 µM heme.
As such, this biphasic induction of hmuO strikingly differs from hrtBA activation by ChrSA and is governed by the influence of the iron repressor DtxR on hmuO expression (Fig. 8). Upon depletion of internal heme (and thus iron‐levels) below a critical threshold, DtxR dissociates from its target promoters and allows their activation (Fig. 7B and D). Accordingly, lower initial heme concentrations within the medium do also shift the time point of heme exhaustion to earlier times, thereby rationalizing the earlier induction of gene expression in the heme utilization module. From a physiological perspective, this again seems very plausible, suggesting that high levels of the heme utilization system are only required under iron limitation when external heme sources are available.
Figure 8.
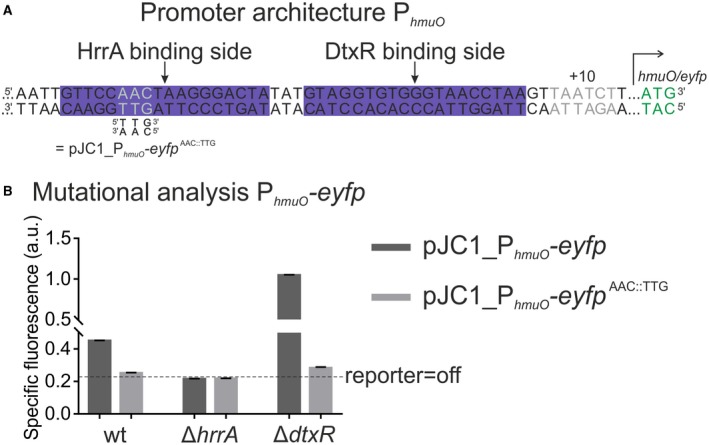
HrrA and DtxR cooperate to control hmuO expression in response to iron and heme availability. A. Promoter architecture of hmuO. The DtxR binding site was published previously (Wennerhold and Bott, 2006). The AAC::TTG mutation (grey letters) was shown to disrupt HrrA binding to PhmuO in vitro (Fig. S3). B. Mutational analysis of PhmuO. HrrA binding was abolished by introducing the AAC:TTG mutation into the PhmuO‐eyfp reporter. All strains were grown in BHI complex medium supplemented with 4 µM hemin as the ΔdtxR strain grows poorly in CGXII medium. After iron starvation overnight, the three strains (wild type, ΔhrrA and ΔdtxR) were inoculated in BHI and the specific fluorescence (eYFP‐fluorescence/backscatter) was recorded in 15 min intervals. The graph shows the specific fluorescence after 20 h. Full reporter outputs are shown in the supplementary Fig. S4 . [Colour figure can be viewed at wileyonlinelibrary.com]
Regarding the regulatory hierarchy of HrrA and DtxR on PhmuO activity, one can think of two potential scenarios: either (i) HrrA is a bona fide activator of hmuO expression or (ii) HrrA‐binding competes with the binding of the repressor DtxR thereby acting as an activator by derepressing hmuO, as might be suggested by the close proximity of their binding sites within the PhmuO promoter (Fig. 8A). Altogether, our experimental data are clearly in favor for HrrA acting as a bona fide activator. While PhmuO activity is completely abolished in a hrrA mutant (twofold lower reporter output that the WT), it is strongly increased in mutant lacking the dtxR gene (Fig. 8B). Furthermore, we tested different triplet mutations in the HrrA binding site in their ability to disrupt HrrA binding. In EMSA‐studies, an AAC::TTG mutation in the middle of the HrrA operator completely abolished in vitro binding of HrrA to PhmuO (Fig. S3). Subsequently, this mutation was inserted to the pJC1_PhmuO‐eyfp reporter, resulting in the pJC1_PhmuO‐eyfp AAC::TTG plasmid (Fig. 8A). Irrespective of whether it was introduced to the wild‐type strain or the ∆dtxR strain, disruption of HrrA binding decreased the activity of the PhmuO reporter to background levels (Figs. 8B and S4). To further validate our findings, we tested the wild type strain carrying the PhmuO reporter under different heme and iron conditions (high‐iron/low‐heme; low‐iron/high‐heme, etc). As also revealed by our model (Fig. 7B and C), the results shown in Figure S8 indicate the intimate link between the iron and the heme pool. Due to the basal expression of the heme oxygenase (see Fig. 1 or Fig. 8, WT versus ΔhrrA) addition of heme will also impact the bioavailable iron pool, thereby affecting DtxR‐mediated repression of hmuO. Thus, according to our current model, HrrA represents an essential activator of hmuO, which is required for a basal level of hmuO expression (and likely also required for the turnover of the endogenously synthesized heme). DtxR repression places an additional threshold on hmuO activation, which is only released when the intracellular iron pool is depleting. Then DtxR dissociates from the hmuO promoter allowing full activation by HrrA.
Discussion
Orchestration of heme homeostasis and detoxification of excess heme appears to be of utmost importance for Corynebacteriaceae, as remarkably, several species dedicate two paralogous TCS to this regulatory task (Bott and Brocker, 2012). Here, we provide comprehensive insights into the temporal dynamics of hmuO and hrtBA expression modulated by HrrSA and ChrSA in C. glutamicum. Similar regulatory setups, with two paralogous, exclusively heme‐dedicated systems are, for example, found in C. diphtheria, Corynebacterium pseudotuberculosis and Corynebacterium lipophiloflavum (Trost et al., 2010; Bott and Brocker, 2012), where the corresponding sequence identities and a conserved genomic synteny suggest a similar role of these systems in the coordinated control of heme detoxification and utilization (Bibb et al., 2007; Bibb and Schmitt, 2010). To our knowledge, these systems represent the first example studied in detail, where two paralogous systems coordinate a complex physiological response by perceiving the same stimulus: heme.
As a general principle, the integrative signaling pathway design appears to be especially beneficial if a multifaceted stimulus requires different regulatory and appropriately timed outputs. The response of C. glutamicum to extracellular heme sets an interesting example for this, as the physiological adaption is shaped by a fast‐reacting ChrSA system‐regulating expression of the detoxification module and the HrrSA system contributing to a layered dynamic regulation of heme utilization. In a recent study, we could show that the HKs of both TCS, HrrS and ChrS, slightly differ regarding their responsiveness and their general mode of heme‐protein interaction (Keppel et al., 2018). One obvious difference is, for example, a higher basal activity of the HrrSA system as reflected by the significantly higher output of a hmuO reporter compared to hrtBA in cells grown on FeSO4. This basal activation is likely triggered by the endogenously synthesized heme pool. In contrast to ChrSA, which is required to counteract toxic heme levels, HrrSA has an important role in maintaining heme homeostasis by balancing the synthesis of heme proteins, heme degradation and heme biosynthesis (Frunzke et al., 2011; Hentschel et al., 2014). In this context, it becomes apparent that this system is sensitive toward endogenous heme levels of the cells.
However, kinase activity is not the only factor influencing differential target gene activation. While the PchrSA promoter and the divergently located PhrtBA promoter are positively autoregulated from one‐centered ChrA binding site (Fig. 3A), hrrS and hrrA are controlled from two distinct promoters. For hrrA as well as hmuO, the global iron regulator DtxR adds an additional regulatory layer, thereby integrating information on general iron availability in the cell (Wennerhold and Bott, 2006; Frunzke et al., 2011). Experiments under different iron and heme conditions, furthermore, illustrated the intimate link between these cellular pools (Fig. S8), which is based on the activity of the heme oxygenase – degrading heme to release iron. Here, we could show that HrrA does not simply displace DtxR on the promoter of hmuO but is in fact an essential activator, also in the absence of DtxR repression (Fig. 8B). By this means, information on heme (stimulus of HrrS) and Fe2+ (co‐repressor of DtxR) is directly integrated at the level of hmuO expression and RR (HrrA) synthesis.
In contrast, ChrSA‐mediated activation of hrtBA expression is solely influenced by heme availability, as expected for a detoxification system. Here, our data suggested an instantaneous saturation of the ChrS kinase and strong activation of PhrtBA in response to exogenous heme – independent of the applied heme concentrations tested in our setup (Fig. 3). Our model and experiments revealed that the overall strength of hrtBA expression was determined solely by the duration of the response, as governed by the timescale of heme exhaustion in the medium. Thus, it seems that the comparable levels of PhrtBA promoter occupancy independent from stimulus strength ensure an effective detoxification response even for low‐toxic heme concentrations, as claimed for a detoxification module.
Another important factor in the maintenance of intracellular heme homeostasis is the previously reported cross‐phosphorylation between HrrSA and ChrSA (Hentschel et al., 2014). While deletion of chrS did not significantly influence the PhmuO activation pattern in our reporter studies (Fig. 1B), deletion of hrrS resulted in a delayed PhrtBA activation in response to heme (Fig. 5). These findings suggest a role of HrrSA as a ‘kick‐start’ system of chrSA, thereby giving C. glutamicum a competitive edge by shortening the reaction time to mount the detoxification response. Furthermore, the bifunctionality of ChrS ensures efficient proof‐reading of ChrA~P counteracting cross‐phosphorylation by HrrS under non‐inducing conditions where ChrS is dominantly in its phosphatase state.
Physiologically relevant cross phosphorylation between TCSs was, for example, shown in Bacillus anthracis (Mike et al., 2014). In this case, the heme responsive HssRS system was shown to cross interact with HitRS, which is activated by cell envelope stress. Cross‐regulation between HssRS and HitRS thereby enables an integrated response of B. anthracis to heme and to heme‐induced cell envelope damage (Mike et al., 2014). Interestingly, HrrSA and ChrSA as well as HssRS and HitRS are among the closest‐related TCS in the particular species reflecting that duplication and subsequent specialization represents an evolutionary driver of TCS signaling toward the integration of multiple signals and the creation of a multifaceted response to complex stimuli. A similar scenario is found with the NarPQ and NarXL systems regulating the response to nitrate and nitrite in Escherichia coli (Rabin and Stewart, 1993). For these closely related systems, significant cross phosphorylation appears to play a role in the modulation of target gene activation and maintenance of nitrogen homeostasis (Noriega et al., 2010).
The regulatory setup shaping the response to heme may also have considerable impact on heme tolerance of the particular species. Already decades ago, van Heyningen reported on the differential sensitivity of Bacillus species to heme (Van Heyningen, 1948). Mike and co‐workers correlated an increased tolerance, as observed for B. anthracis, with the employment of two cross‐regulating TCS coordinating heme export (HssRS) and cell envelope stress (HitRS) (Mike et al., 2014). Therefore, it might be conceivable that the employment of two heme‐responsive TCSs by some corynebacterial species enables a more robust control of heme homeostasis compared to the regulation by a single system. Remarkably, while the HrtBA exporter is conserved among many Gram‐positive species, the TCS systems ‘in charge’ do not share significant sequence similarity – especially not in their membrane‐embedded sensor domains (Stauff and Skaar, 2009b; Keppel et al., 2018). This overall scenario of two cross‐regulating TCSs modulating heme homeostasis and detoxification underlines a favorable concept nature employs to respond to the multifaceted stimulus heme.
In summary, we have approached the regulatory interplay between the heme‐responsive HrrSA and ChrSA TCSs of C. glutamicum by a comprehensive screening of various mutant strains carrying different target promoters. Generation of a mathematical model based on this data set revealed the underlying mechanisms triggering the antagonistic temporal dynamics in TCS signaling, which shape the cellular response toward the ‘toxic, but tasty’ heme molecule. We believe that the approach of combining time‐resolved monitoring of gene expression and systems‐level modeling of the underlying regulatory networks is key to understanding the logic behind complex homeostatic responses in bacteria.
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table S1. C. glutamicum strain ATCC 13032 was used as wild type (Kalinowski et al., 2003) and either cultivated in BHI (brain heart infusion, Difco BHI, BD, Heidelberg, Germany) as complex medium or CGXII (Keilhauer et al., 1993) containing 2% (w/v) glucose as minimal medium. All cultivations were performed at 30°C and, if necessary, 25 μg/ml kanamycin was added to the medium for selection. For standard cloning applications, E. coli DH5α was cultivated in Lysogeny Broth (Difco LB, BD, Heidelberg, Germany) medium at 37°C in a rotary shaker and for selection, 50 μg/ml kanamycin was added to the medium.
Recombinant DNA work and cloning techniques
Standard cloning and other molecular methods were performed according to the standard protocols (Sambrook and Russell, 2001). For most applications, chromosomal DNA of C. glutamicum ATCC 13032 was used as a template for PCR amplification of DNA fragments and was prepared as described previously (Eikmanns et al., 1994). All sequencing and synthesis of oligonucleotides was performed by Eurofins Genomics (Ebersberg, Germany). For the construction of plasmids, the DNA regions of interest were amplified from chromosomal C. glutamicum DNA with oligonucleotides listed in Table S2 and ligated into the plasmid backbone (see Table S3) via restriction sites indicated in the same table. Genomic integrations and/or deletions were performed using the pK19mobsacB plasmid and the two‐step homologues recombination method described earlier (Niebisch and Bott, 2001). Point mutations for the integration of kinase mutants or construction of a mutated reporter plasmid were introduced via ‘QuickChange Lightning’ site‐directed mutagenesis according to the supplier’s manual (Agilent Technologies, Inc., Santa Clara, USA).
Reporter assays
For reporter studies, C. glutamicum wild type or one of the mutant strains were transformed with a reporter plasmid (Table S3). A preculture in BHI medium (25 µg/ml kanamycin) was inoculated from a fresh BHI agar plate and incubated for 8–10 h at 30°C in a rotary shaker. After that, cells were transferred into a second preculture in CGXII medium (Keilhauer et al., 1993) containing 2% (w/v) glucose and 0 µM FeSO4 to starve the cells from iron. However, protocatechuic acid (PCA) was present in the preculture, allowing the uptake of trace amounts of iron. After growth overnight, the main culture was inoculated to an OD600 of 1 in CGXII medium and cultivated in 48‐well Flowerplates (m2p‐labs GmbH, Aachen, Germany) at 30°C, 95% humidity, 1200 rpm. For the hemin stock solution, hemin (Sigma Aldrich, Munich, Germany) was dissolved in 20 mM NaOH to a concentration of 2.5 mM and, as an iron source, added to the medium in the desired concentrations. Growth of the cells (biomass production) was recorded as the backscattered light intensity of sent light with a wavelength of 620 nm (signal gain factor of 12). For the measurement of eYFP fluorescence, the culture was excited at 510 nm and emission was measured at 532 nm (signal gain factor of 50). Measurements were performed in 15 min intervals.
Electrophoretic mobility shift assays
To characterize the operator sequence of HrrA, the protein was produced in E. coli BL21 and purified as His6‐tagged variant from cells as described previously (Frunzke et al., 2011). As ligand, 30 bp DNA fragments with triplet mutations were amplified and subsequently, 100 ng of the fragments were incubated with 0×, 10× and 30× excess of HrrA in in EMSA buffer (250 mM Tris–HCl pH 7.5, 25 mM MgCl2, 200 mM KCl, 25% (v/v) glycerol). After 30 min at room temperature, the samples were loaded to a native 12% polyacrylamide gel (TBE‐based, TBE (89 mM Tris base, 89 mM boric acid, 2 mM Na2EDTA, loading dye: 0.01% (w/v) xylene cyanol dye, 0.01% (w/v) bromophenol blue dye, 20% (v/v) glycerol, 1 × TBE). Electrophoresis was carried out for 60 min at 160 V. DNA was subsequently stained with SYBR Green I (Sigma Aldrich, Munich, Germany).
Measurement of cell‐associated heme levels
The protocol for the fluorescent measurement of (total) cell‐associated heme was derived from an assay described by Sassa (1976). A preculture containing C. glutamicum wild‐type cells was inoculated in BHI medium from a fresh BHI agar plate and incubated for 8–10 h at 30°C in a rotary shaker. After that, cells were transferred into a second preculture in CGXII medium containing 2% (w/v) glucose and 0 µM FeSO4 to starve the cells from iron. After growth for 12–16 h, the main culture was inoculated to an optical density (OD600) of 1 in CGXII medium. Before addition of 4 µM hemin, as well as 5 min, 30 min, 1, 3, 6, 7 and 23 h after hemin addition, samples were taken for the determination of cell‐associated heme. For that, cells corresponding to an OD600 of 2 in 250 µl were harvested and washed once in 250 µl PBS. Subsequently, the pellet was resuspended in 250 µl 20 mM oxalic acid and stored at 4°C for max 5–6 h. Then, 250 µl of 2 M oxalic acid were added and each sample was heated to 98°C for 30 min. For the measurement, 200 μL of each sample were transferred to a 96‐well microtiter plate and the fluorescence was recorded on a Tecan (Tecan Trading AG, Switzerland) Microplate Reader (excitation at 400 nm and emission at 608 nm). The values for each sample were normalized to the emission of an unheated control sample (cells before hemin addition in 1 M oxalic acid).
Mathematical models and mutant simulation
Two mathematical models were developed to assess the determining factors of the dynamics of the heme detoxification and utilization modules (see Supplementary Text; Model equations M1 and M2). A set of ordinary differential equations (ODEs) describes the time‐dependent changes in the different components of the two networks under varying heme concentrations as stimulus of both systems. The interactions between the kinases and the response regulators of the two TCSs were described based on the modeling approach by Groban and co‐workers (2009), while thermodynamic modeling (Bintu et al., 2005) was used to describe the target gene regulation in both systems. The dynamics of the individual components were simulated for wild‐type conditions as well as different mutant strains. The numerical solution of the ODEs as well as the individual simulations were performed with MATLABTM software (The MathWorks, Inc.).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
MK, HP, GF and JF conceived the study; MK and CG performed the experiments; MK, HP, GF and JF analyzed the data; HP and GF generated the models;MK, HP, GF and JF wrote the manuscript.
Supporting information
Acknowledgements
We would like to thank Ulrike Viets for excellent technical support, Eva Davoudi for critical proof‐reading and discussion and Dominik Wolf for his help with the EMSA experiments. This work was financially supported by the Deutsche Forschungsgemeinschaft (DFG SPP 1617; grant FR3673/1‐2 and grant FR 2759/4‐1) and the Helmholtz Association (W2W3‐096).
Contributor Information
Georg Fritz, Email: georg.fritz@synmikro.uni-marburg.de.
Julia Frunzke, Email: j.frunzke@fz-juelich.de.
References
- Andrews, S.C. , Robinson, A.K. and Rodriguez‐Quinones, F . (2003) Bacterial iron homeostasis. FEMS Microbiology Reviews, 27, 215–237. [DOI] [PubMed] [Google Scholar]
- Anzaldi, L.L. and Skaar, E.P . (2010) Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infection and Immunity, 78, 4977–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb, L.A. and Schmitt, M.P . (2010) The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin‐dependent manner by the ChrAS two‐component system in Corynebacterium diphtheriae . Journal of Bacteriology, 192, 4606–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb, L.A. , Kunkle, C.A. and Schmitt, M.P . (2007) The ChrA‐ChrS and HrrA‐HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae . Infection and Immunity, 75, 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu, L. , Buchler, N.E. , Garcia, H.G. , Gerland, U. , Hwa, T. , Kondev, J. and Phillips, R . (2005) Transcriptional regulation by the numbers: models. Current Opinion in Genetics & Development, 15, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borzelleca, J.F . (2000) Paracelsus: herald of modern toxicology. Toxicological Sciences: An Official Journal of the Society of Toxicology, 53, 2–4. [DOI] [PubMed] [Google Scholar]
- Bott, M. and Brocker, M . (2012) Two‐component signal transduction in Corynebacterium glutamicum and other corynebacteria: on the way towards stimuli and targets. Applied Microbiology and Biotechnology, 94, 1131–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos, J.M. and Schmitt, M.P . (2016) The ChrSA and HrrSA two‐component systems are required for transcriptional regulation of the hemA promoter in Corynebacterium diphtheriae . Journal of Bacteriology, 198, 2419–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikmanns, B.J. , Thum‐Schmitz, N. , Eggeling, L. , Ludtke, K.U. and Sahm, H . (1994) Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology (Reading, England), 140(Pt 8), 1817–1828. [DOI] [PubMed] [Google Scholar]
- Fernandez, A. , Lechardeur, D. , Derre‐Bobillot, A. , Couve, E. , Gaudu, P. and Gruss, A . (2010) Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae . PLoS Pathogens, 6, e1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, C.D . (1998) Involvement of heme in the antimalarial action of chloroquine. Transactions of the American Clinical and Climatological Association, 109, 97–106. [PMC free article] [PubMed] [Google Scholar]
- Frunzke, J. , Gatgens, C. , Brocker, M. and Bott, M . (2011) Control of heme homeostasis in Corynebacterium glutamicum by the two‐component system HrrSA. Journal of Bacteriology, 193, 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groban, E.S. , Clarke, E.J. , Salis, H.M. , Miller, S.M. and Voigt, C.A . (2009) Kinetic buffering of cross talk between bacterial two‐component sensors. Journal of Molecular Biology, 390, 380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, S. , Ohtani, K. , Wang, R. , Yuan, Y. , Wang, Y. , Yamaguchi, Y. and Shimizu, T . (2010) Transcriptional regulation of hemO encoding heme oxygenase in Clostridium perfringens. Journal of Microbiology (Seoul, Korea), 48, 96–101. [DOI] [PubMed] [Google Scholar]
- Hentschel, E. , Mack, C. , Gatgens, C. , Bott, M. , Brocker, M. and Frunzke, J . (2014) Phosphatase activity of the histidine kinases ensures pathway specificity of the ChrSA and HrrSA two‐component systems in Corynebacterium glutamicum . Molecular Microbiology, 92, 1326–1342. [DOI] [PubMed] [Google Scholar]
- Heyer, A. , Gatgens, C. , Hentschel, E. , Kalinowski, J. , Bott, M. and Frunzke, J . (2012) The two‐component system ChrSA is crucial for haem tolerance and interferes with HrrSA in haem‐dependent gene regulation in Corynebacterium glutamicum . Microbiology (Reading, England), 158, 3020–3031. [DOI] [PubMed] [Google Scholar]
- Imlay, J.A. , Chin, S.M. and Linn, S . (1988) Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science, 240, 640–642. [DOI] [PubMed] [Google Scholar]
- Jani, D. , Nagarkatti, R. , Beatty, W. , Angel, R. , Slebodnick, C. , Andersen, J. , et al (2008) HDP – A novel heme detoxification protein from the malaria parasite. PLoS Pathogens, 4, e1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski, J. , Bathe, B. , Bartels, D. , Bischoff, N. , Bott, M. , Burkovski, A. , et al . (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L‐aspartate‐derived amino acids and vitamins. Journal of Biotechnology, 104, 5–25. [DOI] [PubMed] [Google Scholar]
- Keilhauer, C. , Eggeling, L. and Sahm, H . (1993) Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB‐ilvN‐ilvC operon. Journal of Bacteriology, 175, 5595–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppel, M. , Davoudi, E. , Gätgens, C. and Frunzke, J . (2018) Membrane topology and heme binding of the histidine kinases HrrS and ChrS in Corynebacterium glutamicum . Frontiers in Microbiology, 9, 183, doi: 10.3389/fmicb.2018.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher, T. , Helmann, J.D. and Unden, G . (2006) Stimulus perception in bacterial signal‐transducing histidine kinases. Microbiology and Molecular Biology Reviews, 70, 910–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mike, L.A. , Choby, J.E. , Brinkman, P.R. , Olive, L.Q. , Dutter, B.F. , Ivan, S.J. , et al (2014) Two‐component system cross‐regulation integrates Bacillus anthracis response to heme and cell envelope stress. PLoS Pathogens, 10, e1004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebisch, A. and Bott, M . (2001) Molecular analysis of the cytochrome bc 1‐aa 3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c 1 . Archives of Microbiology, 175, 282–294. [DOI] [PubMed] [Google Scholar]
- Nir, U. , Ladan, H. , Malik, Z. and Nitzan, Y . (1991) In vivo effects of porphyrins on bacterial DNA. Journal of Photochemistry and Photobiology B, Biology, 11, 295–306. [DOI] [PubMed] [Google Scholar]
- Noriega, C.E. , Lin, H.Y. , Chen, L.L. , Williams, S.B. and Stewart, V . (2010) Asymmetric cross‐regulation between the nitrate‐responsive NarX‐NarL and NarQ‐NarP two‐component regulatory systems from Escherichia coli K‐12. Molecular Microbiology, 75, 394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracelsus . (1965) Die dritte defension wegen des Schreibens der neuen Rezepte. Septem Defensiones, 1538, 508–513. [Google Scholar]
- Poulos, T.L . (2007) The Janus nature of heme. Natural Product Reports, 24, 504–510. [DOI] [PubMed] [Google Scholar]
- Rabin, R.S. and Stewart, V . (1993) Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate‐ and nitrite‐regulated gene expression in Escherichia coli K‐12. Journal of Bacteriology, 175, 3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratliff, M. , Zhu, W. , Deshmukh, R. , Wilks, A. and Stojiljkovic, I . (2001) Homologues of neisserial heme oxygenase in gram‐negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa . Journal of Bacteriology, 183, 6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd edition. New York, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sassa, S . (1976) Sequential induction of heme pathway enzymes during erythroid differentiation of mouse Friend leukemia virus‐infected cells. Journal of Experimental Medicine, 143, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M.P . (1997) Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infection and Immunity, 65, 4634–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauff, D.L. and Skaar, E.P . (2009a) Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Molecular Microbiology, 72, 763–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauff, D.L. and Skaar, E.P . (2009b) The heme sensor system of Staphylococcus aureus . Contributions to Microbiology, 16, 120–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, V.J. , Stauff, D.L. , Pishchany, G. , Bezbradica, J.S. , Gordy, L.E. , Iturregui, J. , et al . (2007) A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host & Microbe, 1, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost, E. , Ott, L. , Schneider, J. , Schroder, J. , Jaenicke, S. , Goesmann, A. , et al (2010) The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12‐year‐old girl with necrotizing lymphadenitis reveals insights into gene‐regulatory networks contributing to virulence. BMC Genomics, 11, 728, doi: 10.1186/1471-2164-11-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heyningen, W.E . (1948) Inhibition of aerobic sporing bacilli by haematin. Nature, 162, 114–115. [DOI] [PubMed] [Google Scholar]
- Wakeman, C.A. , Hammer, N.D. , Stauff, D.L. , Attia, A.S. , Anzaldi, L.L. , Dikalov, S.I. , et al (2012) Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Molecular Microbiology, 86, 1376–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerhold, J. and Bott, M . (2006) The DtxR regulon of Corynebacterium glutamicum . Journal of Bacteriology, 188, 2907–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks, A . (2002) Heme oxygenase: evolution, structure, and mechanism. Antioxidants & Redox Signaling, 4, 603–614. [DOI] [PubMed] [Google Scholar]
- Wilks, A. and Schmitt, M.P . (1998) Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. The Journal of Biological Chemistry, 273, 837–841. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , Hunt, D.J. , Richardson, A.R. and Stojiljkovic, I. (2000a) Use of heme compounds as iron sources by pathogenic Neisseriae requires the product of the hemO gene. Journal of Bacteriology, 182, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Wilks, A. and Stojiljkovic, I . (2000b) Degradation of heme in gram‐negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. Journal of Bacteriology, 182, 6783–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschiedrich, C.P. , Keidel, V. and Szurmant, H . (2016) Molecular mechanisms of two‐component signal transduction. Journal of Molecular Biology, 428, 3752–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


