Summary
All living cells must cope with protein aggregation, which occurs as a result of experiencing stress. In previously studied bacteria, aggregated protein is collected at the cell poles and is retained throughout consecutive cell divisions only in old pole‐inheriting daughter cells, resulting in aggregation‐free progeny within a few generations. In this study, we describe the in vivo kinetics of aggregate formation and elimination following heat and antibiotic stress in the asymmetrically dividing bacterium Caulobacter crescentus. Unexpectedly, in this bacterium, protein aggregates form as multiple distributed foci located throughout the cell volume. Time‐lapse microscopy revealed that under moderate stress, the majority of these protein aggregates are short‐lived and rapidly dissolved by the major chaperone DnaK and the disaggregase ClpB. Severe stress or genetic perturbation of the protein quality control machinery induces the formation of long‐lived aggregates. Importantly, the majority of persistent aggregates neither collect at the cell poles nor are they partitioned to only one daughter cell type. Instead, we show that aggregates are distributed to both daughter cells in the same ratio at each division, which is driven by the continuous elongation of the growing mother cell. Therefore, our study has revealed a new pattern of protein aggregate inheritance in bacteria.
Exposure to various stress conditions can lead to the aggregation of proteins inside all types of living cells. Here, we show that in Caulobacter crescentus, proteins aggregate together at several locations throughout the cell volume during heat or antibiotic stress. In contrast to previously studied bacteria, C. crescentus does not sort persistent aggregates only to one daughter cell type, but instead partitions them to both daughter cells with stable ratios during cell division.
Introduction
Exposure to various types of environmental stress results in the un‐ and misfolding of proteins, which poses a threat to continued survival. Sudden unfolding of native proteins, as well as interference of un‐ and misfolded proteins with partially folded species can create a functional deficit which impairs essential cellular processes and may lead to the death of the cell. All forms of life therefore rely on protein quality control systems to prevent and reverse accumulation of un‐ and misfolded proteins. The main participants in these systems are molecular chaperones and proteases that refold or degrade un‐ and misfolded proteins in order to maintain protein homeostasis as conditions fluctuate (Hartl et al., 2011). Acute stress can lead to exhaustion of the chaperoning and degradation capacity of the cell, resulting in aggregation of proteins that cannot be restored to their native state, either temporarily or indefinitely (Tyedmers et al., 2010; Santra et al., 2018). Sequestering un‐ or misfolded proteins into more inert particles has been proposed to lead to an immediate easing of the burden on the chaperone machinery, and can even be chaperone driven (Ungelenk et al., 2016; Grousl et al., 2018).
How cells cope with protein aggregates can differ depending on the type and amount of aggregated proteins present, but the highly conserved chaperone systems are responsible for effecting survival. Small heat shock proteins (sHSPs) associate with aggregated protein to maintain it in a refolding‐competent state and can also promote the fusion of aggregates and facilitate their dissolution (Specht et al., 2011; Coelho et al., 2014; Ungelenk et al., 2016). The cytoplasmic DnaK/Hsp70 chaperone and the ClpB/Hsp104 disaggregase bind insoluble aggregates and work in concert on their dissolution, returning proteins to their folded state (Glover and Lindquist, 1998; Goloubinoff et al., 1999). Additionally, cytosolic proteases contribute to aggregate dissolution through the degradation of the constituent proteins (Tomoyasu et al., 2001; Heck et al., 2010).
If aggregates persist for a prolonged time and remain even after conditions improve, partitioning of insoluble deposits between daughter cells has been suggested to provide an effective means of sequestering un‐ and misfolded protein from a part of the population (Lindner et al., 2008; Winkler et al., 2010; Vaubourgeix et al., 2015; Hill et al., 2016; Vedel et al., 2016). For example, the budding yeast Saccharomyces cerevisiae retains aggregates in the mother cell both by active and passive mechanisms to generate aggregate‐free daughter cells (Erjavec et al., 2007; Zhou et al., 2011; Spokoini et al., 2012; Higuchi et al., 2013). Although accumulation of protein aggregates has generally been associated with cell aging and other pathology (Aguilaniu et al., 2003; Shcheprova et al., 2008; Nyström and Liu, 2014; Mogk et al., 2018), several recent studies suggest that the carriage of persistent protein aggregates may also confer a fitness advantage and promote survival (Saarikangas and Barral, 2015; Wallace et al., 2015; Govers et al., 2018).
So far, most studies addressing the in vivo dynamics of protein aggregate formation and elimination have been performed in eukaryotes. However, bacteria frequently encounter stress conditions that perturb protein homeostasis, including heat, oxidative or antibiotic stress. In Escherichia coli, protein aggregates mostly accumulate at the chromosome‐free polar regions of the cell (Winkler et al., 2010; Kumar and Sourjik, 2012). With successive cell divisions, this localization rapidly results in the production of aggregate‐free cells, with aggregates maintained in progeny containing the oldest cell poles (Lindner et al., 2008; Winkler et al., 2010; Govers et al., 2014). Similarly, slow‐growing mycobacteria were shown to collect irreversibly damaged proteins at the pole and distribute them to only one daughter cell, again resulting in aggregate‐free and aggregate‐containing populations after cell division (Fay and Glickman, 2014; Vaubourgeix et al., 2015). This partitioning of aggregates to one daughter cell, often to the older part of the cell, has been commonly described as asymmetric inheritance of protein aggregates.
In both E. coli and mycobacteria, the carriage of ancestral protein aggregates has been associated with a decline in growth rate (Lindner et al., 2008; Winkler et al., 2010; Vaubourgeix et al., 2015). However, a more recent study argues that E. coli cells inheriting protein aggregates along with components of the protein quality control machinery show an increased robustness to subsequent proteotoxic stress (Govers et al., 2018). Despite insight into the strategies and physiological consequences of aggregate distribution employed by E. coli and mycobacteria, it remains poorly understood how other bacteria deal with protein aggregates in response to changing growth conditions. Of particular interest is how protein aggregation is handled by bacterial species possessing an intrinsically asymmetric life cycle, generating daughter cells with distinct cell fates.
The α‐proteobacterium Caulobacter crescentus has long been a model organism of bacterial cell‐type differentiation and cell cycle regulation as it undergoes asymmetric cell division and initiates DNA replication only once per cell cycle (Curtis and Brun, 2010). Each division cycle of C. crescentus yields two nonidentical daughter cells, a motile, nonreplicative swarmer cell and a surface‐attached and replication competent stalked cell. As a free‐living aquatic bacterium, it frequently encounters temperature fluctuations and other stresses that potentially threaten the folding state of the protein complement. A previous report has suggested that C. crescentus stalked cells undergo a slow replicative aging (Ackermann et al., 2003). While in budding yeast and E. coli, replicative aging has been attributed to an accumulation of protein aggregation in the aging cell (Aguilaniu et al., 2003; Erjavec et al., 2007; Lindner et al., 2008; Winkler et al., 2010; Hill et al., 2014), the observed decline in the reproductive output of C. crescentus remains largely unexplained. Major components of the general chaperone machinery of C. crescentus have been described and their importance for stress resistance is known (Baldini et al., 1998; Da Silva et al., 2003; Schramm et al., 2017). The stress‐responsive chaperones DnaK and GroESL are both essential in C. crescentus, and DnaK is required to meet the majority of folding demands when heat or proteotoxic stress is encountered (Baldini et al., 1998; Da Silva et al., 2003; Susin et al., 2006; Schramm et al., 2017). In C. crescentus, proteases such as ClpXP and ClpAP perform essential tasks of cell cycle‐regulated protein degradation, however, proteolytic degradation can also remove unwanted proteins during stress, and C. crescentus Lon has been described to increase proteolysis in response to an excess of unfolded proteins (Jonas et al., 2013; Joshi and Chien, 2016). While these mechanistic aspects of protein quality control during stress have been uncovered, to date it has not been studied how C. crescentus manages protein aggregation during its asymmetric life cycle, and the question persists if retention of protein damage in the stalked cell may explain the previously observed aging effects.
In this study, we have followed the dynamics of aggregate formation, dissolution and inheritance following heat and antibiotic stress and recovery in C. crescentus. We demonstrate that protein aggregates form as multiple DnaK‐attended foci throughout the cell volume and that the mechanism by which cells eliminate aggregates largely depends on the intensity of stress. Importantly, we show that in contrast to previously studied bacteria, the majority of persistent aggregates that form as a consequence of severe stress or genetic mutation do not sort to the old pole‐containing stalked cell, but are instead distributed to both daughter cell types at the same ratio over successive divisions.
Results
Heat and antibiotic stress induce relocalization of the C. crescentus chaperone machinery to foci of protein aggregation
In order to probe the dynamics and requirements of protein aggregation and resolution in C. crescentus, we constructed strains bearing fluorescently tagged versions of the major heat shock chaperone dnaK and the bacterial disaggregase clpB, at their respective native loci (Fig. 1A). Addition of a fluorescent tag to DnaK did not result in viability defects at the optimal growth temperature of 30°C, and no viability defect was observed after exposure to heat stress in comparison to wild‐type C. crescentus (Supporting Information Fig. S1). Expressing the tagged version of ClpB did not result in a viability defect at 30°C, however, correlated with a reduction in heat tolerance (Supporting Information Fig. S1). We found that DnaK tagged with the monomeric fluorescent protein mVenus (DnaK‐mVenus) was diffusely localized throughout cells at a normal growth temperature of 30°C (Fig. 1B). To probe the localization of DnaK‐mVenus at super‐resolution, we imaged cells with stimulated emission depletion (STED) microscopy and found that the diffuse pattern of DnaK‐mVenus at 30°C was representative of many small clusters of DnaK measuring 88 ± 38 nm (Fig. 1C). Upon exposure to a heat stress temperature of 40°C, DnaK‐mVenus localization changed to a punctate pattern, suggesting that in C. crescentus protein aggregation is grouped into multiple foci that are distributed throughout the cell volume (Fig. 1B). Most cells contained between two and four DnaK‐mVenus foci, while around 10% of the population harbored five or more foci (Fig. 1D). STED imaging of the foci formed during heat shock revealed large structures that measured 221 ± 46 nm (Fig. 1C). These DnaK‐mVenus foci occurred at a similar frequency along the entire cell length (Fig. 1E), therefore forming without a particular preference for a certain cellular position, in contrast to the preference for the polar region that has been documented in E. coli (Lindner et al., 2008; Winkler et al., 2010).
Figure 1.
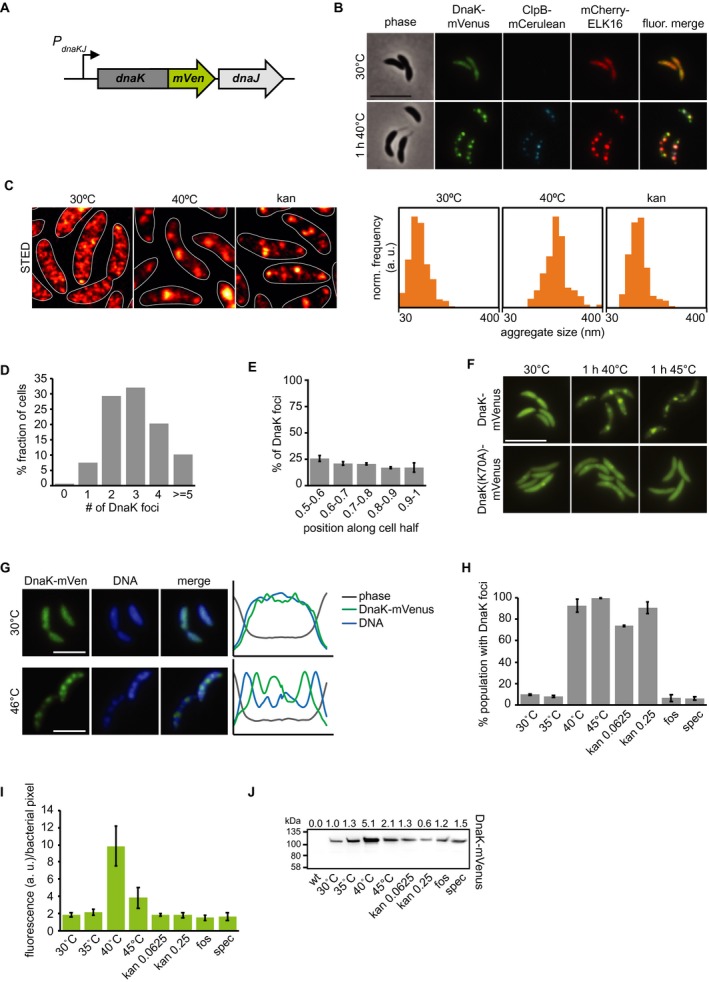
Stress induces relocalization of chaperone machinery to foci of protein aggregation. A. Fluorescent fusions to proteins at their native locus are used to visualize the location of chaperones. B. Microscopy of DnaK‐mVenus, ClpB‐mCerulean and mCherry‐ELK16 expressed at 30°C and after 1 h of heat shock at 40°C. Scale bar is 5 µm. C. Representative images demonstrating DnaK‐mVenus localization at super‐resolution with STED imaging following incubation at 30°C, 1 h at 40°C, and after 1 h treatment with 0.25 g/ml kanamycin. STED images are smoothed and cell outlines are shown in white. The histograms show the distribution of DnaK‐mVenus cluster sizes, with all histograms showing the size range of 30–400 nm, and a bin width of 25 nm. Histograms show the size distribution of at least 118 clusters, from at least 75 cells. Cluster sizes are significantly different between all conditions (Kolmogorov–Smirnov test, p < 0.05). D. Quantification of the number of aggregates per cell and E. graph illustrating the position of the aggregates along five bins from midcell (0.5) to the cell pole (1) in cells exposed to 1 h of 40°C in liquid culture. Quantifications in (D) and (E) show the means of biological triplicates for which at least 196 cells and 685 aggregates each were analyzed respectively. Error bars represent standard deviations. F. Microscopy comparing localization of DnaK‐mVenus with DnaK(K70A)‐mVenus at 30°C and after 1 h at 40°C. Scale bar is 5 µm. G. Fluorescence microscopy demonstrating localization of DnaK‐mVenus and the chromosome, stained with Hoechst 33258, after incubation at 30°C and 46°C. Line profiles (right) demonstrate fluorescence or phase contrast signal over the length of a representative cell from each treatment group. Signals are plotted as the percentage of the maximum value. Scale bar is 2.5 µm. H. DnaK‐mVenus foci formation in response to different stress conditions. Cultures of the DnaK‐mVenus strain were exposed to the indicated temperature or treated with 0.0625 g/mL or 0.25 g/mL kanamycin, 20 g/mL fosfomycin (fos) or 100 g/mL spectinomycin (spec) for 1 h followed by visualization and quantification of the number of cells with DnaK‐mVenus foci. Quantifications show the means of biological duplicates where 300 cells each were analyzed. Error bars represent standard deviation. I. Quantification of DnaK‐mVenus fluorescence intensity under different stress conditions. Cultures of the DnaK‐mVenus strain were treated as in (H) followed by imaging and quantification of fluorescence intensity per total bacterial area. Quantifications show the means of biological duplicates where at least 339 total cells each were analyzed from at least 10 independent images. J. Quantification of protein levels of DnaK‐mVenus under the conditions described in (H) and (I) by western blot. Band intensities are shown as average of two replicates above the western blot image.
Disabling ATP hydrolysis of DnaK by introducing a K70A mutation (Barthel et al., 2001) largely prevented DnaK from relocalizing to foci during heat stress, confirming that stress‐induced DnaK relocalization is a function of its ATP‐dependent foldase activity (Fig. 1F). To test whether the observed DnaK foci correspond to sites of protein aggregation, we used the aggregation‐prone ELK16 peptide fused to mCherry as a marker for protein aggregation (Wu et al., 2011), and saw colocalization between DnaK‐mVenus and mCherry‐ELK16 foci after heat stress (Fig. 1B). Finally, co‐localization experiments with the ClpB‐mCerulean reporter showed that following exposure to heat stress ClpB attends mostly the same foci as DnaK‐mVenus and mCherry‐ELK16 (Fig. 1B), while no fluorescence was observed for the ClpB reporter at the normal growth temperature, in keeping with its heat shock‐dependent expression (Simão et al., 2005). In addition to DnaK, ELK16 and ClpB, we also constructed tagged versions of the two C. crescentus small heat shock protein homologs CCNA_02341 and CCNA_03706 (hereafter referred to as sHSP1 and sHSP2 respectively). While the induction of the reporter was robust, the addition of a fluorescent tag to native shsp1 resulted in increased heat sensitivity, the formation of atypically large fluorescent clusters and cell division defects during mild heat stress (Supporting Information Fig. S1). The localization pattern achieved with fluorescently tagged sHSP2 was consistent with that of DnaK‐mVenus, however, induction from the native locus following heat stress was weak (Supporting Information Fig. S1). Based on these experiments, we decided to utilize DnaK‐mVenus for the visualization of aggregate localization throughout this study.
To determine the relationship between aggregate foci and the Caulobacter chromosome, we stained the DNA using Hoechst 33258 in the DnaK‐mVenus reporter strain sampled at normal temperature as well as after heat shock (Fig. 1G). Surprisingly, aggregate formation during heat shock was accompanied by a change in the spatial organization of the DNA from an evenly dispersed to a patchy pattern. Moreover, the multiple locations occupied by protein aggregates corresponded to regions of reduced DNA staining intensity (Fig 1G, right). These data suggest that in contrast to E. coli and other bacteria, in which the compact nucleoid pushes aggregates to the cell poles (Lindner et al., 2008; Winkler et al., 2010; Coquel et al., 2013; Gupta et al., 2014), in Caulobacter protein aggregates might displace the chromosome. In support of the data, the cell volume occupied by the atypically large aggregate foci formed by the sHSP1‐mCerulean reporter also exhibited reduced DNA staining intensity in the same region (Supporting Information Fig. S1C), suggesting again that the chromosome is unable to occupy the same space as protein aggregates.
Aggregate formation was observed at heat shock temperatures of 40°C and 45°C (Fig. 1H), but also upon exposure to sublethal concentrations of kanamycin, which is known to induce protein aggregation through mistranslation in E. coli (Kohanski et al., 2008). STED imaging revealed that kanamycin‐induced aggregates were smaller than those induced by heat shock, forming as foci measuring 136 ± 37 nm (Fig. 1C). No aggregation foci were observed following treatment with the antibiotic fosfomycin, which blocks production of peptidoglycan precursors, nor for spectinomycin (Fig. 1H), a ribosome‐binding aminoglycoside that does not cause mistranslation (Kohanski et al., 2008).
Finally, our reporter strains also allowed us to quantify the induction of dnaK expression following stress exposure by determining the fluorescence intensity per total bacterial area in different conditions (Fig. 1I). Exposure to 40°C for 1 h induced a four‐fold increase in DnaK levels, while a temperature of 45°C induced a two‐fold increase within 1 h, which we confirmed by western blotting (Fig. 1J). Although strong aggregation was present, lower induction of DnaK at 45°C and following kanamycin treatment is possibly due to lower translation capacity in these conditions (Chen et al., 2017). Treatment with fosfomycin or spectinomycin did not increase DnaK levels, indicating that the inhibitory effects of these antibiotics act independently of protein aggregation and the heat shock response in C. crescentus (Fig. 1I and J). Altogether, these data establish that protein aggregation occurs at multiple sites in C. crescentus and that the major chaperones DnaK and ClpB are recruited to these sites.
Proteins governing diverse processes form aggregate foci in C. crescentus
Having established a system for visualizing total cellular aggregation, we next asked what proteins might be un‐ or misfolding, thereby recruiting DnaK to foci of aggregation. In order to address this question, we isolated insoluble detergent‐resistant proteins from wild‐type C. crescentus grown at 30°C or exposed to a stress temperature of 45°C for 1 h, subjected them to mass spectrometry and identified 133 proteins enriched specifically in the aggregated fraction during heat stress (Fig. 2A, Supporting Information Table S2). Comparing the abundance of aggregate‐enriched proteins sorted by functional category (Fig. 2B), showed that proteins belonging to various cellular processes become associated with the aggregate foci, including 54 essential proteins (Supporting Information Table S2). The most abundant protein in the aggregated fraction was sHSP1, which shares 54% and 40% amino acid sequence homology with the E. coli small heat shock proteins IbpA and IbpB respectively. This result is consistent with a previously described role of small heat shock proteins in binding and maintaining un‐ or misfolded proteins in a disaggregation‐ready state (Mogk et al., 2003a; Lindner et al., 2008; Strozecka et al., 2012).
Figure 2.
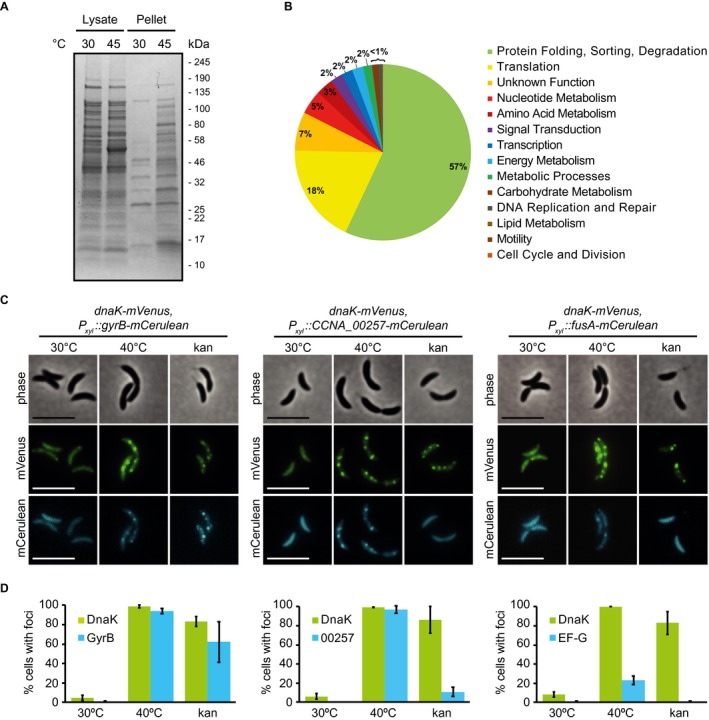
Proteins governing diverse processes aggregate in C. crescentus. A. Coomassie stained protein gel of soluble and detergent‐resistant aggregated fractions following heat stress. Wild‐type cultures at 30°C and 1 h at 45°C were collected and soluble and insoluble fractions were separated by lysis and centrifugation. B. Abundance of aggregate‐enriched proteins sorted by functional category. Functional categories were assigned to proteins identified in (A) according to KEGG gene ontology, and the enrichment of aggregation‐prone proteins belonging to each category was determined to obtain relative abundance. C. Localization pattern of the aggregation‐prone proteins GyrB, CCNA_00257 and EF‐G at 30°C and 40°C. Expression of the fusion proteins was induced for 3 h at 30°C, followed by 1 h exposure to 40°C or 0.25 M kanamycin. Scale bar is 5 µm. D. Quantification of stress induced relocalization of endogenous aggregating proteins and DnaK‐mVenus shown in (C). Quantifications show the means of biological duplicates where at least 152 cells each were analyzed. Error bars represent standard deviation.
To verify the identity of aggregated proteins detected by mass spectrometry, we selected the proteins GyrB, EF‐G (elongation factor G, encoded by fusA), and the predicted homocysteinase CCNA_00257 to monitor their subcellular localization when exposed to heat and kanamycin stress (Fig. 2C). All three proteins were confirmed to condense into aggregate foci colocalizing with DnaK when exposed to stress (Fig. 2C). Interestingly, in contrast to GyrB, which relocalized in the majority of cells during all exposures, the number of cells with CCNA_00257 and EF‐G aggregates differed depending on the stress signal (Fig. 2D). While heat stress induced foci formation by CCNA_00257 in nearly all cells, kanamycin treatment resulted in foci formation in only 15% of the population, indicating that CCNA_00257 is less sensitive to kanamycin‐induced effects than GyrB. EF‐G only relocalized into aggregate foci in 20% of the population during heat stress, again indicating that individual proteins are destabilized by different stress exposures. Collectively our data show that proteins belonging to diverse cellular processes become part of the aggregates observed in C. crescentus and that the composition of aggregates is dependent on the stress condition.
Contribution of major chaperones and proteases to stress resistance and aggregate dissolution
In order to maintain cellular protein homeostasis, chaperones and proteases forming the protein quality control machinery must cooperate in protein folding, aggregate dissolution and degradation of un‐ and misfolded proteins. Previous work has shown that DnaK and ClpB are essential in resisting heat stress and dissolving aggregates in many bacteria, and that the small heat shock proteins and the protease Lon additionally contribute to managing protein aggregation in E. coli (Tomoyasu et al., 2001; Mogk et al., 2003a; 2003b; Vaubourgeix et al., 2015; Santra et al., 2018). Therefore, through a series of deletion mutants we probed the contribution of ClpB, sHSP1, sHSP2 and the protease Lon to aggregate dissolution and stress resistance in C. crescentus.
In accordance with low expression during normal growth temperature, deletion of clpB, shsp1 and shsp2 had no discernible effect at 30°C (Fig. 3A and B). Upon heat stress, the viability of cells lacking ClpB was drastically reduced, as has been demonstrated previously (Simão et al., 2005). The number and distribution of DnaK foci was similar to that observed in the presence of ClpB, however, none of the formed aggregates were able to be dissolved (Fig. 3C). When challenged by a sublethal heat shock cells lacking ClpB were still unable to dissolve aggregated protein deposits, which instead persisted over many generations even after release from stress (Supporting Information Fig. S2). In contrast to this pronounced phenotype, absence of shsp1 or shsp2 alone or in combination had no effect on resistance to heat treatment (Fig. 3A). The overall pattern of protein aggregation was unchanged in the absence of the sHSPs, forming still as distributed punctate foci, and most aggregates were dissolved within the same time frame as the parental strain (Fig. 3B and C). Quantification of the kinetics of aggregate dissolution revealed a delay in reduction of the number of aggregates in the combined absence of the sHSPs (Fig. 3C), which is consistent with reports from E. coli that these proteins keep aggregated protein in a state amenable to efficient resolubilization or degradation (Mogk et al., 2003a; 2003b). Basal levels of DnaK as well as levels induced by heat stress were similar in the absence of the sHSPs (Fig. 3D), indicating that additional DnaK is not compensating for the activity of these proteins and that although sHSP1 comprises nearly half of the aggregated protein fraction, both sHSPs are largely dispensable for tolerating acute heat stress under the stress conditions tested.
Figure 3.
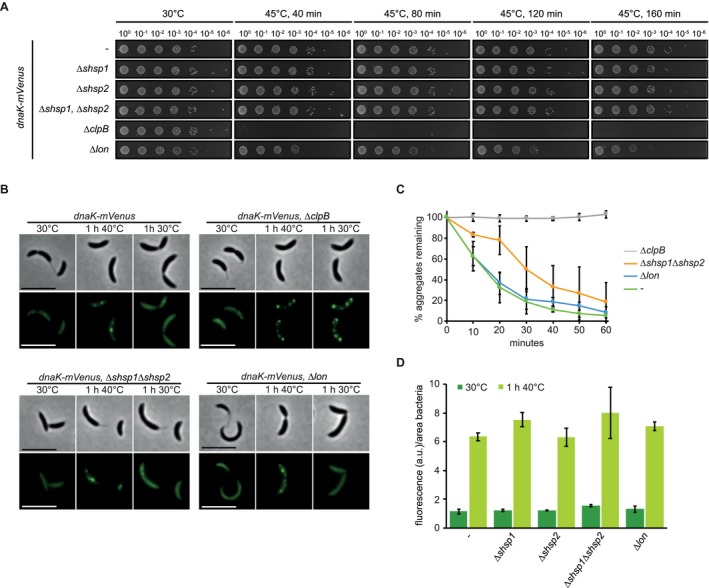
Contribution of chaperones and proteases to stress resistance. A. Spot assays comparing colony formation of wild type, the dnaK::dnaK‐mVenus strain and its derivative strains harboring chaperone and protease knockouts grown at 30°C without stress treatment or after 40, 80, 120 or 160 min of incubation at 45°C. B. DnaK‐mVenus localization in the same strains as in (A) grown at 30°C or after heat shock in liquid culture at 40°C for 1 h (1 h 40°C panels). After 40°C heat shock, cells were permitted to recover at 30°C for 1 h (1 h 30°C panels), during which aggregate dissolution was monitored by time lapse microscopy. Scale bar is 5 µm. C. Quantification of the percentage total aggregates remaining during recovery at 30°C for 1 h. Cultures were exposed to 40°C heat shock for 1 h followed by monitoring aggregate dissolution by time lapse microscopy. Quantifications show the means of biological triplicates in which the longevity of at least 184 total aggregates were measured. D. Absolute measurements of DnaK‐mVenus fluorescence during growth at 30°C and after 1 h heat shock at 40°C. Due to increased sensitivity to heat stress, ∆clpB was excluded from this analysis (See Fig. S2 for further analysis of this strain). Quantifications show the means of biological duplicates where at least 409 cells each were analyzed from 10 independent images.
Proteases support foldases in maintaining protein homeostasis by degrading un‐ or misfolded proteins, and combined loss of proteases has been shown to induce defects in protein quality control (Kanemori et al., 1997). Lon is generally viewed as the major protein quality control protease in E. coli (Rosen et al., 2002; Van Melderen and Aertsen, 2009), and has a role in cell cycle control and responding to unfolded protein in C. crescentus (Jonas et al., 2013). Cells lacking Lon exhibited negligible reduction in viability during heat stress (Fig. 3A), and both dnaK‐mVenus induction and the pattern of aggregation occurred as when Lon was present. Additionally, in the absence of Lon cells were able to completely dissolve protein aggregates within a similar time frame as the parental strain (Fig. 3B and C). Thus, the Lon protease is not required for dissolving protein aggregates under these stress conditions.
Together our data demonstrate that clustering of protein aggregation into several smaller foci and the recruitment of DnaK to these sites does not depend on the heat shock proteins tested. Furthermore, our data confirm in C. crescentus that dissolution of protein aggregates requires the disaggregase ClpB and the chaperone DnaK, and that despite the heat shock induction of sHSPs and Lon, these factors are dispensable under the conditions tested.
Reduction of cellular aggregate content through dissolution or dilution is dictated by stress severity
As we have identified aggregate composition as well as chaperone contributions to aggregate dissolution, we next wanted to determine how C. crescentus responds to protein aggregation provoked by different stress intensities and how the number of aggregates per cell is reduced following shift to nonstress conditions. We therefore used fluorescence time‐lapse microscopy to follow aggregate formation and resolution dynamics during different stress exposures.
To analyze the formation of aggregates following a temperature upshift, we transferred cultures grown at the normal temperature of 30°C to agarose pads and placed them into the imaging system preheated to either 30, 40 or 44°C, and monitored the localization pattern of DnaK‐mVenus as a proxy for aggregate formation over time (Fig. 4A). Under both heat shock conditions, we observed rapid formation of an average of 3.7 aggregates per cell within 10 min of heat exposure (Fig. 4A and B, Supporting Information Movie S1). During incubation at 40°C, cells grew into microcolonies which increased in area at a rate similar to those in nonstress conditions, and aggregate formation and dissolution appeared highly dynamic (Fig. 4B and C). After their initial appearance the foci number per cell was quickly reduced until reaching a plateau of approximately one aggregate per cell by approximately 2 h (Fig. 4B). We observed that the lifespan of most aggregates was less than 10 min (Fig. 4D). Only 23% of aggregates persisted for more than 2 h (Fig. 4D), demonstrating that the aggregate numbers following 2 h of exposure to 40°C represent a steady state of formation and disappearance of mostly short‐lived aggregates as cells grow, with a minority of persistent aggregates emerging. We attribute the initial net reduction of aggregate number per cell (Fig. 4B) to the dilution of aggregates within the microcolony through continued growth and division, as well as rapid induction of heat shock gene expression, which allows cells a greater capacity to cope with higher amounts of unfolded protein present at the new elevated temperature (Fig. 4E). Although microcolonies grew at a relatively normal rate, we observed that temperatures near 40°C resulted in abnormal filamentous growth phenotypes, indicative of cell division defects (Fig. 4F), consistent with an earlier study (Heinrich et al., 2016).
Figure 4.
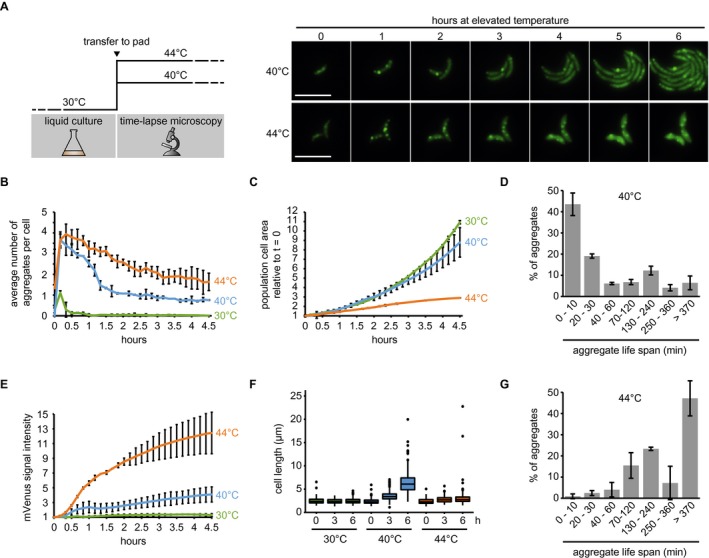
Growth and protein aggregation dynamics during sustained heat stress. A. Schematic of upshift experiments (left), and representative images following upshift to 40°C or 44°C over a 6 h period. Scale bar is 5 µm. B. Quantifications of fluorescence time‐lapse microscopy images showing the average number of aggregates per cell over time in cell populations continuously grown at 30, 40 and 44°C. Note that after transfer to 30°C a minor fraction of dim and short‐lived DnaK‐mVenus foci formed. C. Total cell area increase during exposure to 30, 40 or 44°C relative to the first time point (t = 0). Quantifications in (B, C) show the means of biological triplicates for which at least eight microcolonies each were analyzed. Error bars represent standard deviations. D. Quantification of aggregate life span in cells continuously exposed to 40°C. Aggregates present or emerging in the first 300 min of a fluorescence time‐lapse movie were tracked until 400 min. Images were acquired every 10 min. Quantifications show the means of biological triplicates for which at least five microcolonies and 89 aggregates each were analyzed. Error bars represent standard deviations. E. DnaK‐mVenus fluorescence intensity per total bacterial area over time in cells grown at 30, 40 or 44°C normalized to the first time point (t = 0). The same cells as in (B, C) were quantified. F. Quantification of population cell lengths after 0, 3 and 6 h continuous exposure to 30, 40 or 44°C. The cell lengths of two biological replicates were pooled and populations representing at least 504 measurements are shown. G. Quantification of aggregate life span in cells continuously exposed to 44°C. Aggregates present at 0 min were tracked until 480 min (new aggregates did not arise). Images were acquired every 10 min. Quantifications show the means of biological duplicates for which at least 14 microcolonies and 63 aggregates each were analyzed. Error bars represent standard deviations.
At a temperature of 44°C, cell growth was essentially arrested (Fig. 4A and C), while DnaK‐mVenus levels drastically increased over time, indicating strong and persistent induction of heat shock gene expression (Fig. 4E). We observed that DnaK foci slowly dissolved until only 40% remained after 6–8 h of exposure to 44°C, and only rarely observed the formation of new aggregates at this temperature (Fig. 4B and G). This observation indicates that a fraction of aggregates can be dissolved by the highly abundant chaperones, or that DnaK is released from persistent aggregates under this severe continuous stress.
To monitor how cells recovering from heat stress eliminate aggregates, we exposed cells growing in liquid culture to 40, 42, 44 or 46°C for 1 h, followed by transfer to agarose pads for imaging at 30°C (Fig. 5A, Supporting Information Fig. S3). During recovery from these temperatures some or all of the stress‐treated population remained capable of resuming growth (Fig. 5B) and reducing the average number of aggregates per cell (Fig. 5C). How quickly the average number of aggregates was reduced and whether this was accomplished by aggregate dissolution or dilution within a growing microcolony was dependent on the severity of the heat stress. Growth resumption and aggregate reduction were equally fast in cells exposed to 40 and 42°C (Fig. 5B and C), where all protein aggregates were completely dissolved within 1 h (Fig. 5C), leading to aggregate‐free microcolonies (Fig. 5A and D). In contrast, cells recuperating from exposure to 46°C showed a strong growth delay and decreased ability to eliminate aggregates, and were heterogenous both in aggregate dissolution and the ability to produce normally growing progeny (Fig. 5A, B, D, and E). Only 25% of cells were able to produce microcolonies of normally sized descendants (Fig. 5D), which were capable of dissolving a fraction of the aggregates that they inherited from the heat‐exposed mother cell (Fig. 5E). The remaining 75% of the population were either unable to resume growth within 4.5 h (32%) or resumed growth with varying degrees of division defects (43%) (Fig. 5D). Importantly, although most individuals within the forming microcolonies became eventually aggregation‐free, a fraction of offspring maintained aggregates (Fig. 5A). Many of these aggregates originated from the heat exposed mother cell and persisted for the duration of imaging. However, we also observed that a fraction of new shorter‐lived aggregates formed during recovery (Fig. 5F). Based on these analyses we conclude that a portion of aggregates formed during 46°C exposure persist over several generations and are eliminated from individual cells mainly through dilution within the forming microcolony, rather than dissolution by the chaperone machinery. A similar behavior was also observed after exposure to 40°C for 10 min in the ∆clpB strain, where individual cells eliminate persistent aggregates by passing them on to the offspring (Supporting Information Fig. S2).
Figure 5.
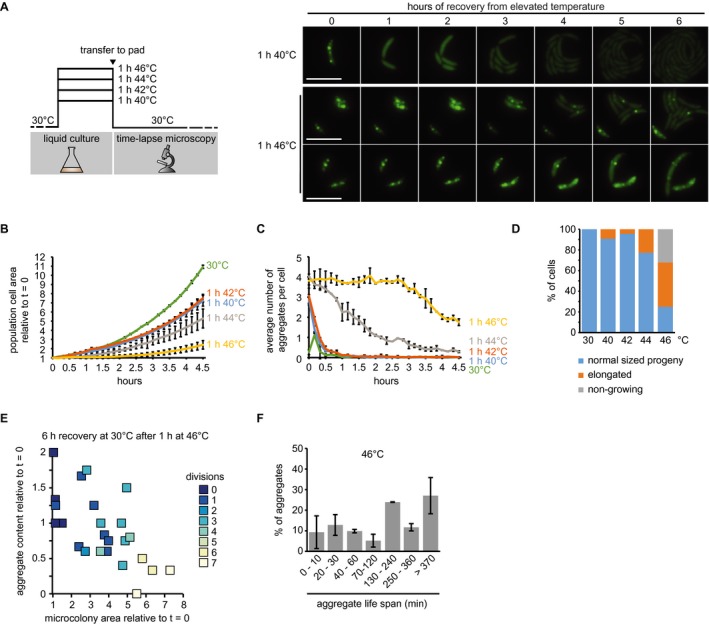
Growth and aggregate resolution following stress recovery. A. Schematic of recovery experiments (left) and representative images from recovery after exposure to 40 and 46°C for 1 h. Two panels are shown for the 46°C condition to represent the diversity of cell fates after stress release. Scale bar is 5 µm. B. Quantifications of fluorescence time‐lapse microscopy showing the average number of aggregates per cell in cell populations recovering from 1 h of exposure to 40, 42, 44 and 46°C. C. Total cell area increase over time after release from 1‐h exposure to 40, 42, 44 and 46°C relative to the first time point (t = 0). Quantifications in (B, C) show the means of biological duplicates for which at least nine microcolonies each were analyzed. The data on cells continuously grown at 30°C are the same as in Fig. 4. Error bars represent standard deviations. D. Quantification of cell fates during continuous growth at 30°C, or recovery at 30°C after 1 h of exposure to 40, 42, 44 and 46°C. Quantifications are based on tracking at least 21 cells/microcolonies per duplicate condition. Cell/microcolony properties were quantified when the average total area of the population increased to 4 times the initial area. Growing cells/microcolonies were defined as those that at least doubled in area. Elongated cells or microcolonies containing those were considered as such when the average cell length in the microcolony was at least 1.5 times higher than the average cell length of a population grown to 4 times the cell area under continuous 30°C conditions. E. Scatter plot representing relative area increase, relative aggregate number normalized to the amount present at stress release and the number of divisions of 24 cells and their potential progeny after 6 h (the time at which the average area of the population has quadrupled) at 30°C on pad after release from 1 h of exposure to 46°C. F. Quantification of aggregate life span in cells recovering from exposure to 1 h 46°C forming microcolonies of mostly normally sized cells (D). Aggregates present or emerging in the first 400 min of a fluorescence time‐lapse movie were tracked until 500 min. Images were acquired every 10 min. Quantifications show the means of biological duplicates for which at least eight microcolonies and 54 aggregates each were analyzed. Error bars represent standard deviations.
When monitoring the recovery of cells following exposure to 44°C we again observed that cells reduce aggregate content through a combination of dissolution and dilution within the population (Fig. 5C, Supporting Information Fig. S4). However, in contrast to the 46°C condition, the fraction of cells able to produce normally sized progeny was larger (77%) (Fig. 5F), cells resumed growth earlier (Fig. 5B) and nearly all cells were free of protein aggregates by 4.5 h after exposure (Fig. 5C).
We additionally asked if carriage of an aggregate during recovery from stress affected growth by comparing the growth rate of aggregate‐containing and aggregate‐free cells (Supporting Information Fig. S5). Cells harboring a single normally sized aggregate did not exhibit a significant change in growth rate compared to related aggregate‐lacking cells, however, slowed growth was observed in cells containing atypically large aggregates formed by sHSP1‐mCerulean (Supporting Information Fig. S5). These results suggest that in C. crescentus, large quantities or abnormal aggregation patterns may negatively affect growth.
Taken together our results demonstrate that the severity of heat stress determines the way by which the aggregate content per cell is reduced. Following exposure to moderate stress, virtually all aggregates are removed through rapid dissolution by the protein quality control machinery. By contrast, following severe stress individual cells eliminate aggregates predominantly through distribution to the progeny. Consequently, an inability to resume growth and cell division following such severe stress prevents successful aggregate removal.
The aggregate load does not sort to swarmer or stalked daughter cells in C. crescentus
Asymmetric inheritance of protein aggregates has been proposed to underpin the senescence of aggregate retaining cells and the rejuvenation of aggregate free cells (Shapiro et al., 2002; Ackermann et al., 2003; Erjavec et al., 2007; Lindner et al., 2008; Winkler et al., 2010; Hill et al., 2016). This asymmetry is driven by collection of aggregates at the poles in bacteria, followed by retention at the old pole as shown in E. coli or, as in budding yeast, retention in the mother cell lineage. Since C. crescentus has a preprogrammed asymmetric division cycle yielding a bigger stalked/old pole and a smaller swarmer/new pole cell, we sought to understand how persistent aggregates are inherited in this organism.
To study aggregate inheritance when only dilution can be used as a means of reducing cellular aggregate content, we made use of the ∆clpB background to investigate the distribution of aggregates following division. A short heat shock of 10 min at 40°C induced formation of persistent protein aggregates in this strain that were distributed throughout the cell volume, while the ability to grow and divide was preserved (Supporting Information Fig. S2), therefore we followed the inheritance of these aggregates over consecutive cell divisions by time lapse microscopy (Fig. 6A). A kymograph normalized to the summed length of all descendants from one individual cell shows that aggregates are relatively static and rarely change their cellular position between two cell division events (Fig. 6B). However, establishment of new cell boundaries as a consequence of cell division affected the relative cellular position of aggregates; for example, an aggregate located at midcell of a stalked cell becomes located closer to the new pole in the daughter cell following division (Fig. 6A and B). To analyze the positional change of aggregates more quantitatively, we determined the frequency by which aggregates obtain a new cellular position in the daughter stalked cell after cell division, as a function of their original cellular position (Fig. 6C, Supporting Information Fig. S6). Consistent with the kymograph analysis, these data show that aggregates located at midcell in the mother cell were likely to change relative position (71%) during cell division and become closer to the new pole of the stalked daughter cell. Aggregates between the old pole and midcell (classified as old pole half) either maintained their original relative position (64%) or also obtained a new relative position close to the new pole (34%). By contrast, aggregates located at the old pole of the mother cell generally maintained their position during cell division (90%) and changed to a position closer to the new pole in only 10% of cases. Importantly, we only very rarely observed that aggregates located outside pole regions became situated closer to the old pole as a consequence of cell division (2% and 3% of those in the old pole half or at midcell, respectively), indicating that essentially all aggregates that obtain a new relative position in the daughter stalked cell after division are situated closer to the new pole. We explain this passive ‘movement’ of aggregates toward the new pole with the growth mode of C. crescentus, in which cells grow along the cell length, excluding the poles (Aaron et al., 2007; Lambert et al., 2018). Consequently, the fraction of aggregates originally formed at the poles will remain there over generations due to the absence of cell elongation in this area. The majority of aggregates, while static, are displaced in proportion to the elongation of the cell and maintain their relative position until a division event sets new cellular boundaries (Fig. 6D).
Figure 6.
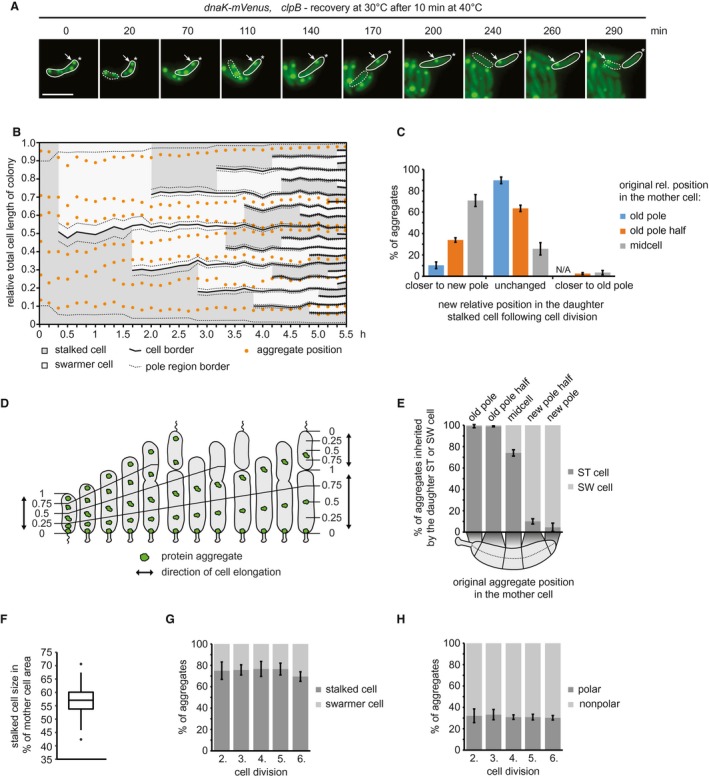
Inheritance of persistent protein aggregates in the ∆clpB background recovering from heat stress. A. Representative fluorescence time‐lapse images of the recovery at 30°C after exposure to 40°C for 10 min pointing out the inheritance of an aggregate originally localized in the old pole cell half after stress exposure (arrow). The star represents the location of the old pole. The stalked cell is outlined by a solid and the swarmer cell by a dashed line. Scale bar is 2.5 µm. B. Kymograph normalized to the summed length of all descendants, showing relative aggregate localization and cell polarity changes during microcolony growth. Cell borders arising through cell divisions are represented by solid lines, while the 10% of the cell length defined as the pole region is indicated by dashed lines. C. Quantitative data showing how aggregates change their relative cellular position in the daughter stalked cell as a consequence of cell division. D. Schematic model showing how growth along the length of the cell, the pole regions being excluded and cell division determine aggregate positional changes and inheritance upon division. Numbers represent relative cell positions between the old (0) and the new pole (1). Lines allow tracking these positions along the growth and division events of the cell. E. Proportion of aggregates inherited by either a swarmer or a stalked cell as function of their cellular position in the mother cell. F. Quantification of the amount of the mother cell area inherited by the daughter stalked cell after a division event. Quantification based on 160 division events occurring in the third and fourth generation of 33 microcolonies. G. Percentage of aggregates inherited by the stalked or the swarmer daughter cell after the second to the sixth division. H. Localization of aggregates tracked from the second to the sixth division. Aggregate position quantifications in (C, E, G and H) resulted from tracking the same population of aggregates from the second to the sixth division. Quantifications are based on biological triplicates for which at least 94 aggregates in at least 20 microcolonies each were tracked. For the quantifications in (C) and (E) the aggregate positional changes after each division were binned leading to at least 376 aggregate positional changes tracked per replicate. Error bars represent standard deviations.
The result that cell growth and the establishment of new cell boundaries during division frequently resulted in aggregates becoming closer to the new pole suggested that with consecutive divisions, these aggregates would eventually be inherited by a swarmer cell. Consistent with this hypothesis, we found that the cellular position in the mother cell largely determines to which daughter cell aggregates are distributed (Fig. 6E). Nearly all aggregates at the new pole or in the new pole half were inherited by the daughter swarmer cell, while essentially all aggregates at the stalked pole or in the old pole half were inherited by the daughter stalked cell (Fig. 6E). Aggregates with an original location around midcell were either inherited by the daughter stalked (74%) or swarmer (26%) cells. The preference toward the stalked cell may be attributable to the size difference between stalked and swarmer cells (Fig. 6F).
In sum, our data show that the cellular positioning of aggregates in C. crescentus is largely governed by the elongation of the cell and the placement of the division plane, and that the cellular position of aggregates in a mother cell determines the likelihood of being inherited by a particular cell type. As such, it is unlikely that C. crescentus partitions aggregates to one cell type, as previously shown for E. coli and mycobacteria (Lindner et al., 2008; Winkler et al., 2010; Vaubourgeix et al., 2015). To test this idea more directly, we determined the percentage of total aggregates that are distributed to either the old pole‐inheriting stalked or the new pole‐inheriting swarmer cell through five division events. Consistent with our prediction, we found that stalked cells did not accumulate aggregates over five successive divisions; instead, the fraction of aggregates inherited by either cell type was stable (Fig. 6G). Importantly, the fraction of polar aggregates was also observed to remain stable for five generations (Fig. 6H), confirming that aggregates do not sort to the pole region over consecutive divisions in C. crescentus. This is in stark contrast to the situation in E. coli, in which virtually all stress‐induced aggregates are collected and remain at the oldest poles (Lindner et al., 2008; Winkler et al., 2010).
Noticeably, stalked cells always inherited more aggregates (75%) than swarmer cells (25%). This difference cannot be solely explained by the cell size difference between the two cell types (57%, Fig. 6F). Rather, we attribute this bias toward the stalked cell type to the finding that aggregates that form at the poles are only rarely displaced from the pole region (Fig. 6C). Consistently, excluding the polar aggregates that are initially present (31%) from the analysis largely removed the observed stalked cell bias (Supporting Information Fig. S7). In addition to our analysis of the ∆clpB background, we also investigated how persistent aggregates (Fig. 4D) are inherited in wild‐type cells during sustained stress at 40°C and during recovery from 44°C (Supporting Information Figs S8 and S9), conditions under which cell morphology is affected (Figs 4A, F and Fig. 5A). These experiments yielded similar stable ratios of cell‐type inheritance and polar aggregates.
Collectively, our data show that the majority of persistent protein aggregates in C. crescentus are neither collected at the old pole nor retained only by the stalked cell; rather, both daughter cells inherit aggregates at a constant ratio.
Discussion
Our study describes the dynamics of protein aggregate formation and elimination in response to antibiotic and heat stress in the asymmetrically dividing bacterium C. crescentus. We find that while utilizing the same key players for aggregate dissolution, how aggregates form in C. crescentus and how they are partitioned during division differs from previously described bacteria.
Subcellular localization of aggregate clusters
We demonstrate that multiple foci of protein aggregation form in C. crescentus throughout the cell volume in response to both antibiotic and heat stress. In Mycobacterium smegmatis protein aggregates also form as multiple distributed foci, although these are collected at the cell pole within one doubling time to form a pattern similar to that of E. coli, where aggregates rapidly localize at the poles after formation (Lindner et al., 2008; Coquel et al., 2013; Vaubourgeix et al., 2015). In E. coli, it has been demonstrated that the condensed nucleoid governs collection of bigger aggregates through macromolecular crowding, enforcing movement toward and deposition at the poles (Winkler et al., 2010; Coquel et al., 2013; Gupta et al., 2014). Compared to E. coli (Winkler et al., 2010), C. crescentus shows a more relaxed arrangement of the chromosome, where the nucleoid fills the entire cell volume (Fig. 1G). We suspect that this pattern might allow aggregate deposition throughout the entirety of the cell as opposed to only at the poles. Other large subcellular structures, such as polyphosphate granules, have been demonstrated to occupy particular locations within the cell (Henry and Crosson, 2013), however, our analysis did not reveal that aggregates were more or less associated with particular cellular positions.
As cells begin to grow, whether in the presence of constant sublethal stress or during recovery from acute heat shock, we found that the majority of persistent aggregates, while static, are displaced in proportion to the elongation of the cell. We demonstrate that this displacement is continuous, although cell division may dictate a new relative position within the daughter cell. As heat shock‐induced aggregates are very large and can nearly span the entire width of the cell, we expect that these may be resistant to movement within the cell, and would be largely unaffected by the movement of smaller, more mobile cellular components. Alternatively, the incorporation of un‐ or misfolded membrane anchored proteins into aggregate clusters may restrict intracellular movement, as has been demonstrated in E. coli when aggregation‐prone luciferase attached to a membrane anchor is expressed (Winkler et al., 2010). In support of this latter hypothesis, we detected several membrane‐associated cellular proteins to be enriched in the aggregate fraction during severe heat stress, suggesting a potential means of ‘tethering’ the aggregate and restricting its diffusion throughout the cell.
Aggregate removal in response to different stress intensities
While the pattern of aggregate formation in C. crescentus was similar throughout all tested stress conditions, we found that the mechanisms by which aggregates are removed from individual cells largely depend on the type and intensity of stress. When recovering from exposure to sublethal stress, aggregates are rapidly dissolved by the combined activity of DnaK and ClpB resulting in complete elimination of aggregates in the population within one generation. Furthermore, we found that C. crescentus is able to cope with continuous exposure to 40°C by rapidly upregulating the heat shock response following temperature upshift, leading to a new homeostasis of aggregate formation and dissolution, with more persistent aggregates being diluted through continuous cell growth and division. Noticeably, although cells incubated at 40°C grow at a similar rate as unstressed cells, they become elongated, indicating defects or delays in completing the cell cycle. A previous study showed that the cell cycle regulator CtrA is downregulated under this condition (Heinrich et al., 2016).
Our data show that when C. crescentus is subjected to intense stress, the persistent aggregates that form cannot be dissolved by the major chaperones, indicating that under these conditions the capacity of the protein quality control machinery is exhausted. The main way to reduce these persistent aggregates in individual cells is to distribute them during the process of cell division. Consequently, only growing and dividing cells can remove persistent aggregates, while those cells that arrest growth maintain them. Our data demonstrate that exposure to severe stress is associated with a marked heterogeneity in the ability to return to normal growth, with only a fraction of the population returning to normal growth following release to nonstress conditions. We expect that massive aggregation under severe stress causes disruption of numerous essential growth processes, supported by previous studies which indicate that strong heat shock response induction during the severe unfolding stress of high temperatures directs cellular resources away from protein translation to survival functions (Chen et al., 2017; Santra et al., 2017; Schramm et al., 2017).
Pattern of aggregate inheritance and its physiological consequences
Our finding that persistent aggregates do not accumulate in the stalked cell, but are instead distributed to both daughter cells at a stable ratio, unveils that the pattern of aggregate inheritance strikingly differs between C. crescentus and the previously studied E. coli and mycobacteria. In the latter cases, aggregates are retained only in one daughter cell type, whereas the other escapes inheritance of aggregate foci (Lindner et al., 2008; Winkler et al., 2010; Vaubourgeix et al., 2015; Vedel et al., 2016; Govers et al., 2018). The resulting heterogeneity of growing microcolonies, in which one part of the population carries protein aggregates while the other part does not, has been suggested to provide a benefit for the population, either by providing a source of rejuvenation (Lindner et al., 2008; Winkler et al., 2010) or by increasing robustness to subsequent stresses (Govers et al., 2018).
Retention of protein aggregates during cell division has been a plausible, though untested, explanation for the previous observation that Caulobacter stalked cells decrease reproductive output with increasing cell age (Ackermann et al., 2003). While a minority of aggregates were trapped at the poles in Caulobacter, additional aggregates did not accumulate at this location, with the majority of cells instead eventually passing their aggregate content on to swarmer cells. Our findings suggest that rather other reasons, for example retention of older membrane components (Bergmiller et al., 2017), may underlie the phenomenon of stalked cell senescence.
Taken together, our work has revealed for the first time a shared mode of aggregate inheritance in bacteria, highlighting that partitioning aggregates to one cell type is not the sole way of managing protein aggregation in bacteria.
Experimental procedures
Growth conditions
C. crescentus strains were routinely grown at 30°C in liquid PYE medium in either an Infors HT Multitron Standard or Infors HT Ecotron rotating shaker set to 200 rpm, or on solid PYE agar. Prior to analysis, all liquid cultures were grown undisturbed in exponential phase at 30°C for 3 h to allow any aggregation from other sources to be dissolved or diluted. Heat shock experiments were performed by moving 100 ml flasks of 10 ml cultures growing at 30°C to a shaking incubator pre‐heated to the desired temperature. Media were supplemented with the following additives when indicated; xylose 0.3%, vanillate 500 M. Antibiotics were used at following concentration (concentration in liquid/solid media as µg/ml): kanamycin 5 (2.5 in the case of KJ956 and KJ957)/25, spectinomycin 25/400, gentamycin 0.625/5, tetracycline 1/2 and rifampicin 2.5/5. Experiments were generally performed in the absence of antibiotic when using strains in which the resistance cassette was integrated into the chromosome. E. coli strains for cloning purposes were grown in LB medium at 37°C, supplemented with antibiotics at following concentrations: kanamycin 30/50, spectinomycin 50/50, gentamycin 15/20, tetracycline 12/12 and rifampicin 25/50. Details on strain and plasmid construction are reported in Supporting Information and Supporting Information Table S1.
Microscopy
To visualize cells, a 1% agarose in PYE slab was poured using a GeneFrame (Thermo Fisher Scientific) attached to a glass slide and coverslip, which was prewarmed to 30°C prior to all experiments. At the indicated time points, a small volume of live cultures was transferred to the pad, sealed under a coverslip and transferred to the microscope for immediate visualization. A Ti eclipse inverted research microscope with 100x/1.45 NA objective housed in a heated chamber, preheated to 30°C unless otherwise specified, was used to collect images. Fluorescence images were captured using excitation and emission filter cubes for mVenus (YFP), mCerulean (CFP), Hoechst 33258 (DAPI) and mCherry (Texas Red), ensuring that fluorescence levels did not exceed the dynamic range of the sensor.
To visualize the chromosome, cultures of Caulobacter were fixed using a final concentration of 4% formaldehyde for 5 min, following which DNA was stained for 25 min using Hoechst 33258 at a final concentration of 2 g/ml. Fixed cells were transferred to a 1% agarose pad and visualized under the microscope as described above for live cells.
Custom‐built STED set‐up and STED imaging
The super‐resolution imaging has been performed at a custom‐built STED setup. The samples were fixed and adhered to slides as described by (Hiraga et al., 1998), and labeled with an anti‐GFP nanobody coupled to ATTO 594 (GFP‐Booster_Atto594, ChromoTek). The dye was excited with a 561 nm pulsed diode laser (PDL561, Abberior Instruments) and subsequently depleted with a 775 nm pulsed laser (KATANA 08 HP, OneFive), both operating at 40 MHz. The depletion beam was shaped to a donut in the focal plane by the use of a spatial light modulator (LCOS‐SLM X10468‐02, Hamamatsu Photonics). The excitation laser and depletion laser were coupled together and scanned over the sample using fast galvanometer mirrors (galvanometer mirrors 6215H + servo driver 71215HHJ 671, Cambridge Technology). The laser beams were focused onto the sample using a HC PL APO 100×/1.40 Oil STED White objective lens (15506378, Leica Microsystems), through which also the fluorescence signal was collected. After de‐scanning and de‐coupling of the fluorescence signal, it was put through a confocal pinhole (1.28 Airy disk units) and detected through a bandpass filter (ET615/30m, Chroma Technology) and a notch filter (ZET785NF, Chroma Technology) with a fiber‐coupled APD (SPCM‐AQRH‐14‐FC, PerkinElmer).
The imaging was done with a 561 nm excitation laser power of 1.1–8.4 µW and a 775 nm depletion laser power of 132–158 mW, both measured at the first conjugate back focal plane of the objective. The pixel size for all images was set to 25 nm. Each image was acquired by adding up 10 line scans for each line, each with a pixel dwell time of 20 µs, resulting in a total pixel dwell time of 200 µs.
Aggregate size analysis
The aggregate sizes were calculated from groups of fitted line profiles in the STED and confocal images. Using Fiji (ImageJ), aggregates were picked out and line profiles were drawn across them. The line profiles were extracted and fitted with a Lorentzian function, from which the full width at half maximum was extracted as the aggregate size. The number of aggregates used for each mean and standard deviation calculation was at least 118.
Image and statistical analysis
To prepare the appropriate image formats and to perform basic analyses such as foci enumeration and generation of line profiles, the Fiji software package was used. Information on the area of individual cells, their fluorescence intensity, as well as their corresponding aggregate number and position in Figs 1D, E, 4, 5 and Supporting Information Figs S2 and S4 were gathered by analyzing binary masks of cells and aggregate foci using the ImageJ package MicrobeJ (Ducret et al., 2016). The Oufti software package (Paintdakhi et al., 2016) was used to generate data on population cell lengths in Figure 4F. To determine fluorescence intensity per total bacterial area shown in Figs 1I and 3D, the MATLAB package SuperSegger (Stylianidou et al., 2016) was used to perform segmentation on images followed by calculation of total cell area per image and bacterial fluorescence intensity per image. For the preparation of cell masks, bacterial segmentation was performed on phase contrast images using SuperSegger and all segmentation was manually checked before further analysis. Masks of the aggregate signal were manually prepared by labeling corresponding areas. The colony kymograph in Fig. 6B was prepared based on cell and aggregate masks using SuperSegger to generate cell linking, cell length and aggregate position measurements. Growth rate measurements in Supporting Information Fig. S5 were performed by measuring the area of individual cells from their birth to division in intervals of 7.5 min using cell masks in SuperSegger. Growth rates were calculated by fitting an exponential function to the data points. Aggregate inheritance and positional changes after sequential divisions (Fig. 6, Supporting Information Figs S6, S7, S8, S9) as well as the aggregate life span (Figs 4D, G and 5F, Supporting Information Fig. S4B) were manually tracked. Data sets generated through all image analysis programs were analyzed and visualized using MATLAB and R software packages.
Immunoblotting
Cell pellets were collected following the indicated treatments and time points and normalized by units OD600 through dilution in Laemmli buffer. Diluted samples were boiled at 98°C for 10 min and loaded into Stain‐Free Tris‐glycine gels (Bio‐Rad) for separation by SDS‐PAGE and transfer to a nitrocellulose membrane as per manufacturer guidelines. Membranes were blocked for 1 h in 5% skim milk powder in TBST and protein levels were detected using either anti‐DnaK antibody (Schramm et al., 2017) or the anti‐GFP antibody (Thermo Fisher Scientific, #A‐11122). A secondary anti‐rabbit HRP‐conjugated antibody was used to detect primary antibody binding, and SuperSignal Femto West (Thermo Fisher Scientific) used to develop the membrane. Blots were scanned using a Chemidoc (Bio‐Rad) system and signal quantification was performed using the Image Lab software package (Bio‐Rad).
Aggregation assay
The aggregation assay was performed as described in (Schramm et al., 2017). About 40 OD600 units of cells with an OD600 between 0.2 and 0.4 either grown at 30°C or heat stressed were harvested. For the heat treatment an exponential overnight culture grown at 30°C was pelleted (6000 g, 10 min, RT), resuspended in a flask containing 45°C prewarmed medium and then incubated shaking at the same temperature for 1 h. A portion of the aggregate fraction was subjected to SDS‐PAGE and the separated proteins were visualized by Coomassie staining (Instant Blue Protein Stain, Expedeon).
Proteomic analysis of protein aggregates
For the identification of endogenous aggregating and aggregate associated proteins in Caulobacter crescentus, whole protein aggregate fractions were subjected to mass spectrometry analysis. Approximately 0.1 µg horse cytochrome C was added to each sample to serve as internal protein abundance standard. The mass spectrometry analysis of the samples was performed using an Orbitrap Velos Pro mass spectrometer (Thermo Scientific). An Ultimate nanoRSLC‐HPLC system (Dionex), equipped with a custom 20 cm × 75 µm C18 RP column filled with 1.7 µm beads was connected online to the mass spectrometer through a Proxeon nanospray source. About 1–15 µl of the tryptic digest (depending on sample concentration) was injected onto a C18 preconcentration column. Automated trapping and desalting of the sample was performed at a flowrate of 6 µL/min using water/0.05% formic acid as solvent. Separation of the tryptic peptides was achieved with the following gradient of water/0.05% formic acid (solvent A) and 80% acetonitrile/0.045% formic acid (solvent B) at a flow rate of 300 nL/min: holding 4% B for 5 min, followed by a linear gradient to 45% B within 30 min and linear increase to 95% solvent B in additional 5 min. The column was connected to a stainless steel nanoemitter (Proxeon, Denmark) and the eluent was sprayed directly toward the heated capillary of the mass spectrometer using a potential of 2300 V. A survey scan with a resolution of 60,000 within the Orbitrap mass analyzer was combined with at least three data‐dependent MS/MS scans with dynamic exclusion for 30 s either using CID with the linear ion trap or using HCD combined with orbitrap detection at a resolution of 7500. Data analysis was performed using Proteome Discoverer (Thermo Scientific) with SEQUEST search engine using Uniprot databases. The peptide peak areas of each protein in a sample were normalized to that of the internal Cytochrome C standard. Proteins were considered to be specifically enriched in the aggregate fraction if they were only detected in the heat‐treated sample or more abundant than in the control.
Spot colony formation assays
Cultures with an OD600 between 0.1 and 0.4 were diluted to 0.05 after which a 1:10 serial dilution was performed. An aliquot of 2 µl of each dilution were spotted on PYE agar plates.
Author contributions
FDS and KS contributed to the conception and design of the study, the acquisition, analysis and interpretation of the data and writing of the manuscript. JA and IT contributed to the collection, analysis and interpretation of the STED data. KJ supervised the study and contributed to the conception and design of the study, the analysis and interpretation of the data and writing of the manuscript. All authors edited and approved the final manuscript.
Supporting information
Acknowledgements
We thank Dr. Claes Andréasson and members of the Jonas lab for helpful discussion. We also thank Dr. Uwe Linne and the mass spectrometry facility at the University of Marburg for their assistance. The study was financially supported by funding from the LOEWE program of the state Hessen, funding of Strategic Research Areas (SFO) from Stockholm University, and a future research leaders grant from the Swedish Foundation for Strategic Research (FFL15‐0005). FDS received a doctoral scholarship from the German Academic Exchange Service (DAAD).
References
- Aaron, M. , Charbon, G. , Lam, H. , Schwarz, H. , Vollmer, W. and Jacobs‐Wagner, C . (2007) The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Molecular Microbiology, 64, 938–952. [DOI] [PubMed] [Google Scholar]
- Ackermann, M. , Stearns, S.C. and Jenal, U . (2003) Senescence in a bacterium with asymmetric division. Science, 300, 1920–1920. [DOI] [PubMed] [Google Scholar]
- Aguilaniu, H. , Gustafsson, L. , Rigoulet, M. and Nyström, T . (2003) Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science, 299, 1751–1753. [DOI] [PubMed] [Google Scholar]
- Baldini, R.L. , Avedissian, M. and Gomes, S.L . (1998) The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. Journal of Bacteriology, 180, 1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel, T.K. , Zhang, J. and Walker, G.C . (2001) ATPase‐defective derivatives of Escherichia coli DnaK that behave differently with respect to ATP‐induced conformational change and peptide release. Journal of Bacteriology, 183, 5482–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmiller, T. , Andersson, A.M.C. , Tomasek, K. , Balleza, E. , Kiviet, D.J. , Hauschild, R. , et al . (2017) Biased partitioning of the multidrug efflux pump AcrAB‐TolC underlies long‐lived phenotypic heterogeneity. Science, 356, 311–315. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Gao, Y. , Mih, N. , O’Brien, E.J. , Yang, L. and Palsson, B.O . (2017) Thermosensitivity of growth is determined by chaperone‐mediated proteome reallocation. Proceedings of the National Academy of Sciences, 114, 11548–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, M. , Lade, S.J. , Alberti, S. , Gross, T. and Tolić, I.M . (2014) Fusion of protein aggregates facilitates asymmetric damage segregation. PLoS Biology, 12, e1001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel, A.‐S. , Jacob, J.‐P. , Primet, M. , Demarez, A. , Dimiccoli, M. , Julou, T. , et al (2013) Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLoS Computational Biology, 9, e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, P.D. and Brun, Y.V . (2010) Getting in the loop: regulation of development in Caulobacter crescentus. Microbiology and Molecular Biology Reviews, 74, 13–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret, A. , Quardokus, E.M. and Brun, Y.V. (2016) MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature Microbiology, 1, 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec, N. , Larsson, L. , Grantham, J. and Nyström, T . (2007) Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation‐remodeling factor Hsp104p. Genes & Development, 21, 2410–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, A. and Glickman, M.S . (2014) An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genetics, 10, e1004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, J.R. and Lindquist, S . (1998) Hsp104, Hsp70, and Hsp40. Cell, 94, 73–82. [DOI] [PubMed] [Google Scholar]
- Goloubinoff, P. , Mogk, A. , Zvi, A.P.B. , Tomoyasu, T. and Bukau, B . (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proceedings of the National Academy of Sciences, 96, 13732–13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers, S.K. , Dutré, P. and Aertsen, A . (2014) In vivo disassembly and reassembly of protein aggregates in Escherichia coli . Journal of Bacteriology, 196, 2325–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers, S.K. , Mortier, J. , Adam, A. and Aertsen, A . (2018) Protein aggregates encode epigenetic memory of stressful encounters in individual Escherichia coli cells. PLoS Biology, 16, e2003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl, T. , Ungelenk, S. , Miller, S. , Ho, C.‐T. , Khokhrina, M. , Mayer, M.P. , et al (2018) A prion‐like domain in Hsp42 drives chaperone‐facilitated aggregation of misfolded proteins. Journal of Cell Biology, 217, 1269–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A. , Lloyd‐Price, J. , Neeli‐Venkata, R. , Oliveira, S.M.D. and Ribeiro, A.S . (2014) In vivo kinetics of segregation and polar retention of MS2‐GFP‐RNA complexes in Escherichia coli . Biophysical Journal, 106, 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, F.U. , Bracher, A. and Hayer‐Hartl, M . (2011) Molecular chaperones in protein folding and proteostasis. Nature, 475, 324–332. [DOI] [PubMed] [Google Scholar]
- Heck, J.W. , Cheung, S.K. and Hampton, R.Y . (2010) Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proceedings of the National Academy of Sciences, 107, 1106–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, K. , Sobetzko, P. and Jonas, K . (2016) A Kinase‐phosphatase switch transduces environmental information into a bacterial cell cycle circuit. PLoS Genetics, 12, e1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, J.T. and Crosson, S . (2013) Chromosome replication and segregation govern the biogenesis and inheritance of inorganic polyphosphate granules. Molecular Biology of the Cell, 24, 3177–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, R. , Vevea, J.D. , Swayne, T.C. , Chojnowski, R. , Hill, V. , Boldogh, I.R. and Pon, L.A . (2013) Actin dynamics affect mitochondrial quality control and aging in budding yeast. Current Biology, 23, 2417–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, S.M. , Hao, X. , Liu, B. and Nyström, T . (2014) Life‐span extension by a metacaspase in the yeast Saccharomyces cerevisiae . Science, 344, 1389–1392. [DOI] [PubMed] [Google Scholar]
- Hill, S.M. , Hao, X. , Grönvall, J. , Spikings‐Nordby, S. , Widlund, P.O. , Amen, T. , et al (2016) Asymmetric inheritance of aggregated proteins and age reset in yeast are regulated by Vac17‐dependent vacuolar functions. Cell Reports, 16, 826–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga, S. , Ichinose, C. , Niki, H. and Yamazoe, M . (1998) Cell cycle‐dependent duplication and bidirectional migration of SeqA‐associated DNA‐protein complexes in E. coli . Molecular Cell, 1, 381–387. [DOI] [PubMed] [Google Scholar]
- Jonas, K. , Liu, J. , Chien, P. and Laub, M.T . (2013) Proteotoxic stress induces a cell‐cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell, 154, 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, K.K. and Chien, P . (2016) Regulated proteolysis in bacteria: Caulobacter. Annual Review of Genetics, 50, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori, M. , Nishihara, K. , Yanagi, H. and Yura, T . (1997) Synergistic roles of HslVU and other ATP‐dependent proteases in controlling in vivo turnover of sigma32 and abnormal proteins in Escherichia coli . Journal of Bacteriology, 179, 7219–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski, M.A. , Dwyer, D.J. , Wierzbowski, J. , Cottarel, G. and Collins, J.J . (2008) Mistranslation of membrane proteins and two‐component system activation trigger antibiotic‐mediated cell death. Cell, 135, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M. and Sourjik, V . (2012) Physical map and dynamics of the chaperone network in Escherichia coli . Molecular Microbiology, 84, 736–747. [DOI] [PubMed] [Google Scholar]
- Lambert, A. , Vanhecke, A. , Archetti, A. , Holden, S. , Schaber, F. , Pincus, Z. , et al (2018) Constriction rate modulation can drive cell size control and homeostasis in C. crescentus . iScience, 4, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner, A.B. , Madden, R. , Demarez, A. , Stewart, E.J. and Taddei, F . (2008) Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proceedings of the National Academy of Sciences, 105, 3076–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk, A. , Deuerling, E. , Vorderwülbecke, S. , Vierling, E. and Bukau, B. (2003a) Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Molecular Microbiology, 50, 585–595. [DOI] [PubMed] [Google Scholar]
- Mogk, A. , Schlieker, C. , Friedrich, K.L. , Schönfeld, H.‐J. , Vierling, E. and Bukau, B . (2003b) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. Journal of Biological Chemistry, 278, 31033–31042. [DOI] [PubMed] [Google Scholar]
- Mogk, A. , Bukau, B. and Kampinga, H.H . (2018) Cellular handling of protein aggregates by disaggregation machines. Molecular Cell, 69, 214–226. [DOI] [PubMed] [Google Scholar]
- Nyström, T. and Liu, B . (2014) The mystery of aging and rejuvenation – a budding topic. Current Opinion in Microbiology, 18, 61–67. [DOI] [PubMed] [Google Scholar]
- Paintdakhi, A. , Parry, B. , Campos, M. , Irnov, I. , Elf, J. , Surovtsev, I. and Jacobs‐Wagner, C . (2016) Oufti: an integrated software package for high‐accuracy, high‐throughput quantitative microscopy analysis. Molecular Microbiology, 99, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, R. , Biran, D. , Gur, E. , Becher, D. , Hecker, M. and Ron, E.Z . (2002) Protein aggregation in Escherichia coli: role of proteases. FEMS Microbiology Letters, 207, 9–12. [DOI] [PubMed] [Google Scholar]
- Saarikangas, J. and Barral, Y . (2015) Protein aggregates are associated with replicative aging without compromising protein quality control. eLife, 4, e06197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra, M. , Farrell, D.W. and Dill, K.A . (2017) Bacterial proteostasis balances energy and chaperone utilization efficiently. Proceedings of the National Academy of Sciences, 114, E2654–E2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra, M. , Dill, K.A. and de Graff, A.M.R . (2018) How do Chaperones protect a cell’s proteins from oxidative damage? Cell Systems, 6, 743–751. [DOI] [PubMed] [Google Scholar]
- Schramm, F.D. , Heinrich, K. , Thüring, M. , Bernhardt, J. and Jonas, K . (2017) An essential regulatory function of the DnaK chaperone dictates the decision between proliferation and maintenance in Caulobacter crescentus. PLoS Genetics, 13, e1007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, L. , McAdams, H.H. and Losick, R . (2002) Generating and exploiting polarity in bacteria. Science, 298, 1942–1946. [DOI] [PubMed] [Google Scholar]
- Shcheprova, Z. , Baldi, S. , Frei, S.B. , Gonnet, G. and Barral, Y . (2008) A mechanism for asymmetric segregation of age during yeast budding. Nature, 454, 728–734. [DOI] [PubMed] [Google Scholar]
- Da Silva, A.C.A. , Simão, R.C.G. , Susin, M.F. , Baldini, R.L. , Avedissian, M. and Gomes, S.L . (2003) Downregulation of the heat shock response is independent of DnaK and σ32 levels in Caulobacter crescentus . Molecular Microbiology, 49, 541–553. [DOI] [PubMed] [Google Scholar]
- Simão, R.C.G. , Susin, M.F. , Alvarez‐Martinez, C.E. and Gomes, S.L . (2005) Cells lacking ClpB display a prolonged shutoff phase of the heat shock response in Caulobacter crescentus . Molecular Microbiology, 57, 592–603.15978087 [Google Scholar]
- Specht, S. , Miller, S.B.M. , Mogk, A. and Bukau, B . (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae . Journal of Cell Biology, 195, 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokoini, R. , Moldavski, O. , Nahmias, Y. , England, J.L. , Schuldiner, M. and Kaganovich, D . (2012) Confinement to organelle‐associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Reports, 2, 738–747. [DOI] [PubMed] [Google Scholar]
- Strozecka, J. , Chrusciel, E. , Gorna, E. , Szymanska, A. , Zietkiewicz, S. and Liberek, K . (2012) Importance of N‐ and C‐terminal regions of IbpA, Escherichia coli small heat shock protein, for Chaperone function and oligomerization. Journal of Biological Chemistry, 287, 2843–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianidou, S. , Brennan, C. , Nissen, S.B. , Kuwada, N.J. and Wiggins, P.A . (2016) SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Molecular Microbiology, 102, 690–700. [DOI] [PubMed] [Google Scholar]
- Susin, M.F. , Baldini, R.L. , Gueiros‐Filho, F. and Gomes, S.L . (2006) GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus . Journal of Bacteriology, 188, 8044–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu, T. , Mogk, A. , Langen, H. , Goloubinoff, P. and Bukau, B . (2001) Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Molecular Microbiology, 40, 397–413. [DOI] [PubMed] [Google Scholar]
- Tyedmers, J. , Mogk, A. and Bukau, B . (2010) Cellular strategies for controlling protein aggregation. Nature Reviews Molecular Cell Biology, 11, 777–788. [DOI] [PubMed] [Google Scholar]
- Ungelenk, S. , Moayed, F. , Ho, C.‐T. , Grousl, T. , Scharf, A. , Mashaghi, A. , et al (2016) Small heat shock proteins sequester misfolding proteins in near‐native conformation for cellular protection and efficient refolding. Nature Communications, 7, 13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen, L. and Aertsen, A . (2009) Regulation and quality control by Lon‐dependent proteolysis. Research in Microbiology, 160, 645–651. [DOI] [PubMed] [Google Scholar]
- Vaubourgeix, J. , Lin, G. , Dhar, N. , Chenouard, N. , Jiang, X. , Botella, H. , et al (2015) Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host & Microbe, 17, 178–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedel, S. , Nunns, H. , Košmrlj, A. , Semsey, S. and Trusina, A . (2016) Asymmetric damage segregation constitutes an emergent population‐level stress response. Cell Systems, 3, 187–198. [DOI] [PubMed] [Google Scholar]
- Wallace, E.W.J. , Kear‐Scott, J.L. , Pilipenko, E.V. , Schwartz, M.H. , Laskowski, P.R. , Rojek, A.E. , et al (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell, 162, 1286–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, J. , Seybert, A. , König, L. , Pruggnaller, S. , Haselmann, U. , Sourjik, V. , et al (2010) Quantitative and spatio‐temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO Journal, 29, 910–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W. , Xing, L. , Zhou, B. and Lin, Z . (2011) Active protein aggregates induced by terminally attached self‐assembling peptide ELK16 in Escherichia coli . Microb Cell Factories, 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. , Slaughter, B.D. , Unruh, J.R. , Eldakak, A. , Rubinstein, B. and Li, R . (2011) Motility and segregation of Hsp104‐associated protein aggregates in budding yeast. Cell, 147, 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


