Abstract
Understanding the mechanisms that promote the specification of pancreas progenitors and regulate their self‐renewal and differentiation will help to maintain and expand pancreas progenitor cells derived from human pluripotent stem (hPS) cells. This will improve the efficiency of current differentiation protocols of hPS cells into β‐cells and bring such cells closer to clinical applications for the therapy of diabetes. Aldehyde dehydrogenase 1b1 (Aldh1b1) is a mitochondrial enzyme expressed specifically in progenitor cells during mouse pancreas development, and we have shown that its functional inactivation leads to accelerated differentiation and deficient β‐cells. In this report, we aimed to identify small molecule inducers of Aldh1b1 expression taking advantage of a mouse embryonic stem (mES) cell Aldh1b1 lacZ reporter line and a pancreas differentiation protocol directing mES cells into pancreatic progenitors. We identified AMI‐5, a protein methyltransferase inhibitor, as an Aldh1b1 inducer and showed that it can maintain Aldh1b1 expression in embryonic pancreas explants. This led to a selective reduction in endocrine specification. This effect was due to a downregulation of Ngn3, and it was mediated through Aldh1b1 since the effect was abolished in Aldh1b1 null pancreata. The findings implicated methyltransferase activity in the regulation of endocrine differentiation and showed that methyltransferases can act through specific regulators during pancreas differentiation. Stem Cells 2019;37:640–651
Keywords: Aldehyde dehydrogenase 1b1, β‐Cells, Embryonic stem cell pancreas differentiation, Pancreas development, Endocrine specification, Pancreatic progenitors
A screen using small molecules and mouse embryonic stem cell derived pancreatic progenitors reveals that protein methyltransferases inhibit aldehyde dehydrogenase 1b1 expression. Aldehyde dehydrogenase 1b1 maintains pancreas progenitors and maintenance of its expression through protein methyltransferase inhibition delays selectively endocrine differentiation.

Significance Statement.
Recent advances in the directed differentiation of human pluripotent stem cells into β‐like cells have brought the prospect of diabetes cell therapy using this approach closer. However, the very large number of endocrine cells required for a single infusion to diabetic patients remains as a key challenge. Expansion of intermediate pancreas progenitors would address this issue. It has been found that protein methyltransferase activity promotes endocrine specification through the dowregulation of Aldh1b1, a gene implicated in the maintenance of pancreatic progenitors and necessary for β‐cell functionality in the mouse. These findings suggest that specific, reversible protein methyltransferase inhibitors may delay differentiation of pancreas progenitors.
Introduction
The possibility to maintain, expand, and store human pluripotent stem (hPS)‐derived progenitor populations will improve the efficiency of differentiation protocols and reduce costs thus bringing them closer to clinical applications. Recent seminal advances in the conversion of hPS cells into β‐like cells 1, 2, 3 and the development of encapsulation biomaterials that are well tolerated by the immune system of the host 4 have brought the prospect of diabetes cell therapy closer. However, the generated β‐like cells are still immature, respond weakly to glucose, and are intermixed with other cell types. Additionally, generally only one endocrine cell for every two hPS cells used is generated in these procedures. This is an important limitation because, for allo‐transplantation to diabetic patients, between 400,000 and 500,000 islets each containing an average of 1,600 endocrine cells are needed. Understanding the molecular mechanisms that specify pancreas progenitors and regulate the relative rates of their self‐renewal versus differentiation could help address this limitation by identifying means to stabilize, expand, and differentiate intermediate progenitors.
In mammals, the early pancreatic multipotent progenitor cells emerge at the posterior foregut region of the definitive endoderm and are defined by the expression of the transcription factors Pdx1, Ptf1a, and Sox9 5, 6, 7, 8, 9. Maintenance of high Notch signaling is necessary for the expansion of pancreatic progenitors to form a tree‐like branched epithelium and prevent early differentiation 10, 11, 12, 13. Decreased Notch activity at the tips of the epithelium and the antagonistic functions of Ptf1a and Nkx6 transcription factors delineate the acinar progenitor and endocrine/duct bipotent trunk territories 14, 15, 16, 17. In the trunk, differential Notch signaling enables progenitors to differentiate into ductal or endocrine cells. High Notch activity diverts cells to the duct fate through the repression of Ngn3, the transcription factor that specifies endocrine cells 10, 12, 18. Sphingosine‐1‐phosphate (S1p) signaling was recently shown to enable acinar and endocrine specification through the attenuation of Notch signaling 19. The elucidation of the processes that maintain the progenitor status or, conversely, promote lineage specification and subsequent differentiation, is a key step toward identifying means for the maintenance and expansion of hPS derived pancreatic progenitors. Reversible pharmacological agents affecting self‐renewal and differentiation of these progenitors could be used but most of our relevant knowledge concerns transcription factors that are very difficult to manipulate pharmacologically. Aldehyde dehydrogenase 1b1 (Aldh1b1) is a mitochondrial enzyme that selectively marks embryonic pancreas progenitors, including Ngn3+ progenitors where it is more weakly expressed, from the early pancreatic bud stage, during development and possibly also in the adult pancreas 20. Loss‐of‐function studies showed that Aldh1b1 helps maintain the pancreas progenitor state because in Aldh1b1 null embryos the emergence of differentiated cells, in all three lineages, is accelerated 21. Consistent with a specific role in progenitor maintenance, Aldh1b1 expression is gradually lost in differentiating endocrine cells 21. Strikingly, β‐cells in Aldh1b1 nulls are dysfunctional later in life 21 suggesting that sustained Aldh1b1 activity is necessary to pattern endocrine progenitors for subsequent maturation. Therefore, Aldh1b1 activity can be used as a proxy for pancreas progenitor status. The identification of inducers of Aldh1b1 expression may help understand the requirements for pancreas progenitor maintenance and elucidate the underlying molecular mechanisms.
Mouse embryonic stem (mES) cells have been used to model pancreas specification in vitro and query the role of transcription factors as several genetically modified lines were easily generated 22, 23, 24, 25, 26, 27. In this report, we are taking advantage of a mES Aldh1b1 β‐gal reporter line 21 and a differentiation protocol of mES cells into pancreatic‐like progenitors (PP) 24 to identify candidate small molecules that can act as inducers of Aldh1b1 expression. Using a high‐throughput assay, we identified AMI‐5, a protein methyltransferase inhibitor as such a candidate. Addition of AMI‐5 maintained expression of Aldh1b1 in differentiating embryo pancreas explants and this led to a selective delay in the differentiation of the endocrine lineage through the loss of Ngn3+ cells. This effect was mediated specifically through Aldh1b1 since endocrine differentiation was not affected by the presence of AMI‐5 in Aldh1b1 null pancreatic explants. The findings suggest that methyltransferase activity is implicated in the regulation of endocrine differentiation.
Materials and Methods
Mouse Strains, Maintenance, and Genotyping
Mouse strains were maintained in the same genetic background (C57BL/6J). Genotyping was performed by conventional Polymerase Chain Reaction (PCR) on genomic DNA isolated from mouse tails using standard procedures. Briefly, mouse tails were dissolved in Tail buffer (100 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM EDTA, and 0.2% SDS) with 50 μg/ml Proteinase K (Sigma) overnight at 55°C. Following a protein extraction step with Phenol/Chloroform (Sigma), genomic DNA was precipitated from the aqueous phase with 100% ethanol and finally resuspended in TE buffer (10 mM Tris–HCl pH 8.0 and 1 mM EDTA). Genotyping procedures were as described for Aldh1b1 alleles (http://www.velocigene.com/komp/detail/11807)). The Aldh1b1 tm1lacZ knockin mouse strain was generated as previously described 21. Animal maintenance and experimentation were conducted in accordance with the FELASA recommendations and the ethical and practical guidelines for the care and use of laboratory animals set by the competent veterinary authorities in the authors' institutions.
Mouse ES Cell Culture, Generation of Transgenic Lines, and Differentiation
ES cell lines were grown in high‐glucose knockout Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Waltham, MA, USA) supplemented with 10% fetal calf serum (PAN Biotech, Aidenbach, Germany), 1,000 units per milliliter of leukemia inhibitory factor (LIF; Chemicon, Temecula, CA, USA), 100 mM minimal essential medium nonessential amino acids, 0.55 mM 2‐mercaptoethanol, penicillin/streptomycin, and l‐glutamine (all from Invitrogen) on mouse embryonic fibroblasts treated with mitomycin (Sigma–Aldrich, St Louis, MO, USA) at 37°C and 5% CO2. The ESAldh1b1lacZ/+ line used here is described elsewhere 21. For the generation of the ES TgCAGlacZ line, a ScaI digested version of the Z/EG plasmid 28 was electroporated in mouse HM1 ES cells and neomycin resistant clones were selected. A strongly lacZ‐expressing clone was used in the experiments described here.
For differentiation experiments, low‐passage ES cells were used and passaged twice after thawing and before initiating differentiation. ES cells were collected by trypsinization and separated from feeder cells by plating them onto tissue culture plates for two short successive periods (20–30 minutes). ES cells were plated at 105 cells per milliliter in 10‐cm bacterial dishes and allowed to form embryoid bodies (EBs) for 2 days in ES cell medium without LIF. They were then cultured in knockout DMEM containing 2% SR (Life Technologies, Carlsbad, CA, USA), 100 ng/ml Nodal and 10 ng/ml Fgf4 (R&D Systems, Minneapolis, MN, USA) for 3 days, renewing half the medium and adding fresh signaling molecules every 24 hours. In the next step, the generated endoderm enriched EBs (EEBs) were grown in knockout DMEM containing 2% SR, 100 ng/ml Activin A, 10 ng/ml Fgf4, 100 ng/ml Noggin (R&D Systems), 1 μM RA (Sigma), and 1 μM cyclopamine (Millipore, Burlington, MA, USA) for 2 days, renewing half the medium and adding fresh signaling molecules every 24 hours. The generated pancreatic progenitors (PPs) were treated with different final concentrations of AMI‐5 (Merk Millipore) the same concentration of dimethylsulfoxide (DMSO) (0.1%) as control.
X‐Gal Staining
PP cell clusters were collected and washed once with phosphate‐buffered saline (PBS; pH 7.4). After fixation with 2% paraformaldehyde (PFA) and 0.5% gluteraldehyde (Sigma) in PBS for 5 minutes on ice, clusters were washed three times with PBS and stained with 1 mg/ml 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐galactopyranoside (X‐Gal; Roche) in staining solution containing 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCI2, and 5 mM EGTA overnight at 37°C. X‐gal stained PP cell clusters were photographed in bright field using an Olympus SZX10 stereomicroscope.
Fluorescence β‐Galactosidase Assay
PP cell clusters were washed twice with PBS, once with staining medium containing 4% fetal bovine serum (FBS) and 10 mM Hepes pH 7.2 in PBS and resuspended in staining medium containing 200 μM verapamil (Sigma). A PBS solution warmed at 37°C for 10 minutes containing 200 μM fluorescein‐di‐β‐d‐galactopyranoside (FDG; Sigma) or 200 μM FDG together with 2 mM phenylethylthio‐β‐d‐galactopyranoside (PETG; Sigma) was added rapidly in an equal volume of PP cell clusters and incubated at 37°C for 3 minutes. PP cell clusters were then washed twice with double the volume of cold staining medium containing 200 μM verapamil and 2 mM PETG followed by two washes with PBS. Cells were kept continuously on ice before the analysis.
Screening for Aldh1b1 Inducers
EEBs were incubated for 32 hours with the appropriate signals for the induction of PPs and 20–30 cell clusters per well were manually aliquoted in an Aurora 384‐well plate. The 303 compounds and additional controls of the StemSelect Small Molecule Regulators 384‐Well Library I (Calbiochem) at a final concentration of 10 μM, or 01.% DMSO as a control, were spotted using the Echo 555 Labcyte liquid handling system and incubated for additional 16 hours at 37°C and 5% CO2. The following day the fluorescence β‐galactosidase assay was performed essentially as described above. To aspirate and dispense solutions the Powerwash 384 (TECAN) and the dispenser WellMate (MATRIX) were used, respectively. Cell aggregates were first imaged using the bright field ×4 objective of the Cell Voyager 7000S (CV7000S, Yokowaga) to identify the number of cell aggregates per well. Aggregates were then washed twice with PBS, followed by one wash with the staining medium described above and re‐suspended in staining medium containing 200 μM verapamil. Warm staining solution as described above containing either 200 μM FDG (Sigma) or 200 μM FDG and 2 mM PETG (Sigma) was then added rapidly in an equal volume of PP suspension. Then the wells were washed twice with staining medium containing 200 μM verapamil and 2 mM PETG followed by two washes with PBS containing 2% PFA and Hoechst staining at 2 μg/ml for 10 minutes. This was followed by a final wash of PBS. Plates were then imaged on the CV7000S confocal imaging system using a ×4 objective (0.16NA). A set of nine image fields were collected from each well covering the entire well. Filter and camera selection were optimized and for each measurement, laser power, and exposure times were adjusted to the linear detection range. Acquired images were captured and analyzed using the CellProfiler. Images were first loaded and smoothened using the Gaussian filter. Cell aggregates were identified on the bright field channel by excluding objects less than 30 and more than 9,000 pixels employing the Kapur threshold method. The aggregate sizes were measured using the Measure Object Size Shape function and the intensity of each aggregate was measured using the Measure Object Intensity function.
Screening Data Analysis
Data were analyzed using the open‐source software KNIME (http://www.knime.org) and the plug‐ins “HCS tools” and “R‐scripting integration tool” 29 which support the handling of large screening data within KNIME. Measurements, such as intensity, size, and number of objects were averaged first per well and then for each compound. The normal distribution of the mean intensity was found to be further improved by log transformation using a Shapiro–Wilk test (1.1 × 10−3 and 4.4 × 10−2, respectively). The log of the mean intensity was then z‐normalized to the DMSO treatment. Compounds increasing the z‐score of the log transformed mean fluorescence Intensity of objects (z‐score log mean intensity >2.5) were considered as possible hits. Images of the hits were checked visually to exclude chemical compounds that showed a strong auto‐fluorescence by overlaying the fluorescent signal. Bar plots and error bars were used to depict means and SEM. As indicated, p‐values were calculated using the Student's t test for two‐tailed distributions of unpaired groups or the two sample Kolmogorov–Smirnov test. p < .05 was considered significant.
Air to Liquid Interface Organotypic Cultures of Embryonic Pancreata
Dorsal pancreatic buds were dissected under a light stereoscope (Leica MZ75) at 14.5 dpc and cultured for 2 days on 0.4 μm pore diameter filters (Millicell‐CM; Millipore) in DMEM (Gibco) supplemented with N2 (Gibco) and streptomycin–penicillin–glutamine (Gibco). Treatments were with AMI‐5 at 10 μM final, EPZ004777 (Tocris) at 10 μM, C21 (Tocris) at 10 μM and MG132 (Sigma) at 1 μM concentration and control samples were treated with the same amount of solvent (0.1% DMSO). Chemicals were replenished with each medium change daily for 2 days.
Immunostainings on Cryosections
Dissected pancreata or ES‐derived EEBs and PPs were fixed in 4% paraformaldehyde (PFA; Sigma), washed in PBS, and dehydrated in 30% sucrose (Sigma) overnight at 4°C. Tissues were embedded in optimal cutting temperature compound (British Drug Houses [BDH]), cut into sections of 12 μm thickness and mounted onto Superfrost slides (Van Waters and Rogers [VWR]) for storage at −80°C. For immunostainings, cryosections were postfixed in PBS containing 4% PFA for 5 minutes and blocked for 1 hour at room temperature in 10× blocking solution containing 10% serum, 0.3% Triton X‐100 in PBS. Primary antibodies were diluted in 1× blocking solution and incubated overnight at 4°C, whereas secondary antibodies, also diluted in 1× blocking solution, were incubated for 2 hours at room temperature. All washes were done in PBS containing 0.3% Triton X‐100. After the final washes, slides were mounted with Prolong Gold antifade reagent containing DAPI (Molecular Probes) and imaged using a Leica SP5 microscope.
Primary antibodies used were: custom made rabbit anti‐Aldh1b1(1:1,000) prepared as described 30, mouse anti‐Pdx1 (1:250; F109‐D12, DSHB), goat anti‐Sox17 (1:250; SC‐17356, Santa Cruz), rabbit anti‐Ngn3 (1:100; AP22029PU‐N, Acris), rabbit anti‐C‐peptide (1:100; 4,593, Cell Signaling), rabbit anti‐Amylase (1:300; A8273, Sigma), rabbit anti‐Ptf1a (1:3,000; Gift from B. Breant), rat anti‐Cytokeratin19 (1:250; Troma‐III, DSHB), mouse anti‐Glucagon (1:500; G2654, Sigma), mouse anti‐Nkx6‐1 (1:500; F55A10, DSHB), mouse anti‐insulin (1:1,000; I2018, Sigma), rabbit anti‐pH 3 (1:500, 06‐570, Cell Signaling). Secondary antibodies were goat anti‐rabbit Alexa‐488‐, Alexa‐568‐, and Alexa‐647 conjugated (1:500; A11070, A21069, A21245, Molecular Probes), donkey anti‐goat Alexa‐488 conjugated (1:500, A11055, Molecular Probes), goat anti‐mouse Alexa‐568 conjugated (1:500, A11019, Molecular probes), goat anti‐rat Alexa‐488, Alexa‐568 (1:500, A11006, A11077, Molecular Probes) and donkey anti‐rat Alexa 647 conjugated (1:500, AB15055, Abcam). When the Aldh1b1 antibody was used together with another rabbit antibody in coimmunofluorescence experiments, it was conjugated with the Zenon Alexa Fluor 488 (Invitrogen Z25302), according to the manufacturer's instructions. Detection of cell death was performed on cryosections using a fluorescent TUNEL assay kit (Roche) according to the manufacturer's instructions and as described in 18.
Morphometric Analysis
Morphometric analysis was performed by processing immunofluorescent images using the Fiji software 31. The “Image Threshold” and “Selection” tools were used to quantitate the number of pixels that correspond to the signal area on the immunofluorescent image. The same threshold was used to process stainings with the same antibody. For the 14.5 dpc pancreata and all pancreatic explants, analysis was performed using at least three 12 μm thick cryosections per sample, at least 24 μm apart and spanning the entire tissue. For each quantitation, at least three pancreata of each genotype and/or condition were analyzed. For ES‐derived EEBs and PPs, 10 structures of average size and shape, comprising a total of more than 1,000 cells, were analyzed for each differentiation stage. For Aldh1b1, endocrine, acinar, and ductal cell quantitation, the signal area of Aldh1b1, C‐peptide/Glucagon‐, Amylase‐, and Cytokeratin19‐positive immunofluorescence, respectively, was measured and divided by the corresponding total signal area for DAPI, thus normalizing for mass. The same procedure was followed for Pdx1, Ptf1a, and Nkx6.1 signal area quantitation. For quantitation of endoderm and endocrine progenitors, Sox17‐positive and Ngn3‐positive nuclei were counted, respectively, and divided by the total number of DAPI+ nuclei or total DAPI area.
Western Blotting
For protein analysis, 14.5‐dpc mouse embryonic pancreata and ES‐derived pancreas progenitors were used. Protein extraction, polyacrylamide gel electrophoresis, and blotting procedures were performed according to standard protocols. Primary antibodies used were: rabbit anti‐Prmt1 (1:1,000, 2,449, Cell Signaling), rabbit anti‐Dot1l (2.5 μg/ml, AB64077, Abcam), mouse anti‐Setd7 (1:500, AB14820, Abcam) and mouse anti‐β‐actin (1:10,000, MAB1501, Sigma). Secondary antibodies were anti‐rabbit and anti‐mouse horseradish peroxidase‐conjugated goat antibodies (1:5,000, PO44801‐2, PO447701‐2, Dako).
RNA Isolation and Real‐Time PCR
Total RNA was prepared using the RNeasy kit with on‐column genomic DNA digestion following the manufacturer's instructions (Qiagen). First strand cDNA was prepared using Superscript II RT (Invitrogen). Real‐time PCR primers were designed using the Primer 3 software (SimGene) and specificity was ensured by in silico PCR. Reactions were performed with SYBR‐GreenER (Invitrogen) using a Roche LC480 machine and primary results were analyzed using the on board software. Reactions were carried out in duplicate from at least three independent samples. Absolute expression values were calculated using the ΔCt method and β‐actin was used for normalization. Primers used were further evaluated by inspection of the dissociation curve. Primer sequences were as follows: Aldh1b1‐Fw: GACCTGGACAAGGCCATCTA, Aldh1b1‐Rv: CCTTAAAGCCTCCGAATGG, Ngn3‐Fw: GGTGATCTGCCTTCTTCTGC, Ngn3‐Rv: ACACGGGAGACAAGTTGGAG, β‐actin‐Fw: TGGCTCCTAGCACCATGA, β‐actin‐Rv: CCACCGATCCACACAGAG.
RNA Sequencing and Bioinformatics Analysis
Three independent samples of 14.5 dpc + 2 days air to liquid interface (ALI) cultured embryonic pancreata untreated or AMI‐5 treated were used as biological replicates. Each sample was derived from pooling up to three cultured pancreata. Total RNA was prepared using the RNeasy kit with on‐column genomic DNA digestion (Qiagen) and only RNA with an integrity number of ≥8 was used. mRNA was isolated from 1 μg total RNA by poly‐dT enrichment using the NEBNext Poly(A) mRNA Magnetic Isolation Module according to the manufacturer's instructions. Final elution was done in 15 μl 2× first strand cDNA synthesis buffer (NEBnext, NEB). After chemical fragmentation by incubating for 15 minutes at 94°C the sample was directly subjected to the workflow for strand specific RNA‐Seq library preparation (Ultra‐Directional RNA Library Prep, NEB). For ligation custom adaptors were used (Adaptor‐Oligo 1: 5′‐ACA‐CTC‐TTT‐CCC‐TAC‐ACG‐ACG‐CTC‐TTC‐CGA‐TCT‐3′, Adaptor‐Oligo 2: 5′‐P‐GAT‐CGG‐AAG‐AGC‐ACA‐CGT‐CTG‐AAC‐TCC‐AGT‐CAC‐3′). After ligaton adapters were depleted by an XP bead purification (Beckman Coulter) adding bead in a ratio of 1:1. Indexing was done during the following PCR enrichment (15 cycles) using custom amplification primers carrying the index sequence indicated with “NNNNNN.” Primer1: Oligo_Seq. AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT, primer2: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT, primer3: CAAGCAGAAGACGGCATACGAGAT NNNNNN GTGACTGGAGTT. After two more XP beads purifications (1:1) libraries were quantified using Qubit dsDNA HS Assay Kit (Invitrogen). Libraries were equimolarly pooled and sequenced on an Illumina HiSeq 2500, resulting in ca. 20–30 million single end reads per library.
After sequencing, FastQC (http://www.bioinformatics.babraham.ac.uk/) was used to perform a basic quality control on the resulting reads. As an additional control, library diversity was assessed by redundancy investigation in the aligned reads. Alignment of the short reads to the mm10 reference was done with GSNAP (v 2014‐12‐17) 32, and Ensembl gene annotation version 69 was used to detect splice sites. The uniquely aligned reads were counted with featureCounts (v1.4.6) 33 and the same Ensembl annotation.
Normalization of the raw read counts based on the library size and testing for differential expression between the different conditions was performed with the DESeq2 R package (v1.6.2) 34. Only genes with normalized counts >100 were considered. Data for embryonic pancreata dpc pancreata are deposited in GEO (http://www.ncbi.nlm.nih.gov/geo) and publicly available under the GSE76482 (14.5 dpc) and GSE76678 (13.5 and 15.5 dpc) accession numbers. Data for 14.5 + 2 ALI control and AMI‐5 treated pancreata are deposited in GEO under the GSE119438 accession number and accessible with the token ydghokiubfcfpej.
Statistical Analysis
Statistical significance in morphometric analyses and real‐time PCRs was determined by the Student's t test for two‐tailed distributions of unpaired groups. Error bars represent SEM. p < .05 was considered significant.
Fluorescence Activated Cell Counting
To achieve single cell suspension, PP clusters were incubated in a 0.5% trypsin/EDTA solution (Sigma) for 20 minutes at 37°C under gentle shaking (Sigma). The reaction was stopped by the addition of 3× volume of knockout DMEM supplemented with 10% FBS. To minimize cell aggregation, the dissociated EBs were then passed several times through a 25 gauge needle and filtered through 0.45 μm cell strainers (BD Biosciences) that have been prewashed in PBS containing 0.5% FBS. Cell viability was determined by the Trypan Blue assay and cell suspensions were subjected to the β‐galactosidase fluorescence assay. At the end of the reactions, cells were resuspended in PBS containing 5% FBS and analyzed using a BD FACS Aria III (BD Biosciences). The median fluorescence intensity (MFI) was calculated for the FITC+ gated populations using the corresponding functions of FlowJo (FLOWJO, LLC).
Results
Development of a Screening Assay for Small Molecules Inducing Aldh1b1 Expression
To set up a screening assay for small molecules inducers of Aldh1b1 expression we used, as a reporter line, the KOMP Aldh1b1 tm1lacZ ES line (ESAldh1b1lacZ/+) that carries an insertion of the lacZ transgene in one of the alleles of the Aldh1b1 gene under the control of the endogenous promoter. The same line was also used to generate the Aldh1b1 null mice 21. As a positive control to establish screening conditions, a mouse ES transgenic line expressing lacZ under the control of the strong constitutive CAG promoter 28 (ESTgCAGlacz) was generated.
Cells were sequentially differentiated in suspension culture into EBs, then into definitive endoderm EEBs and finally into PP as previously described (Fig. 1A) 24. We confirmed the differentiation efficiency of Aldh1b1 tm1lacZ/+ ES cells into EEB and PP cells by quantitating the expression of the definitive endoderm marker Sox17 and the pancreas progenitor marker Pdx1 at the respective stages by immunofluorescence on cryosections. We found that 88.6% ± 5.4% of cells in the EEBs were Sox17+ (n = 10) whereas 81.2% ± 4.5% of the cells in the PPs were Pdx1+ (Fig. 1B). Additionally, Nkx6‐1, another marker of pancreas progenitors, was expressed in 62.8% ± 8.3% of the PPs (Fig. 1B). Reflecting what is happening in the developing mouse pancreas the Nkx6‐1+ cells were largely a subset of Pdx1+ cells as 94.4 ± 2.5 of the Nkx6.1+ cells were also Pdx1+ while 62.8 ± 8.3 of the Pdx1+ cells were also Nkx6.1+ (Supporting Information Fig. S1B). Moreover, consistent with its expression in the early, 9.5 dpc mouse pancreatic buds 20, we confirmed that Aldh1b1 was also expressed at the PP stage, albeit in fewer cells reflecting the limitations of the in vitro differentiation protocol (Fig. 1B).
Figure 1.
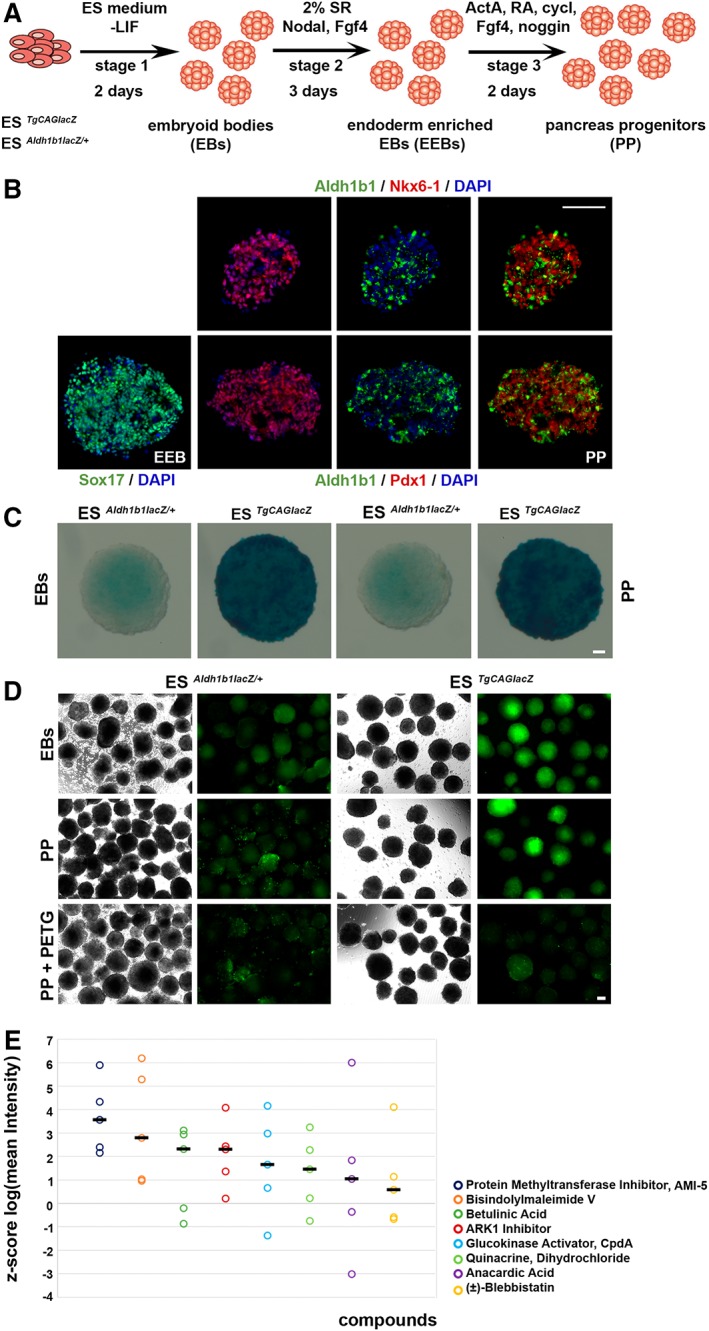
Differentiation of mouse ES cells toward pancreatic‐like progenitors to monitor Aldh1b1 expression analysis. (A): Schematic diagram of the differentiation procedure for the ESAldh1b1lacZ/+ and ESTgCAGlacZ lines. (B): Immunofluorescence analysis of ESAldh1b1lacZ/+ derived EEBs for expression of Sox17 and PPs for expression of Pdx1, Nkx6‐1, and Aldh1b1. (C): Chromogenic detection of β‐galactosidase activity in EB and PP clusters. (D): Fluorescent detection of β‐galactosidase activity in EB and PP clusters. (E): Diagram showing the z‐score mean intensity readings for eight candidate compounds that may upregulate Aldh1b1 expression. The circles represent the z‐score of log mean intensity of different screening replicates (n = 5) and the lines the median z‐score intensity for each compound. Abbreviations: ES, embryonic stem; LIF, leukemia inhibitory factor; SR, serum replacement; ActA, Activin a; RA, retinoic acid; cycl, cyclopamine; PETG, 2‐Phenylethyl β‐d‐thiogalactoside; PP, pancreatic‐like progenitors. Scale bars: 50 μm.
Whole mount chromogenic staining with X‐gal for β‐galactosidase activity was weak in the KOMP line in both the EB and PP stages, in contrast to the ESTgCAGlacZ line that expressed lacZ strongly in both stages (Fig. 1C). To adapt the detection of β‐galactosidase activity for a fluorescence‐based screening assay we used the FDG β‐galactosidase specific substrate that fluoresces upon hydrolysis. Addition of the β‐galactosidase specific inhibitor PETG reduced the activity to background levels confirming the specificity of the reaction (Fig. 1D).
In order to carry out a compound screen, the differentiation of the mouse ES lines into PP (Fig. 1A) was scaled up so that 20–30 PP aggregates could be manually aliquoted into the wells of a 384 well plates. The 303 compounds of the StemSelect Small Molecule library (EMD Millipore) were added at a final concentration of 10 μM and cell clusters were incubated for an additional 16 hours in the presence of the compounds (Supporting Information Fig. S1A). FDG was added the next day and the plates were imaged using an automated confocal microscope. ESAldh1b1lacz/+ PPs treated with 0.1% DMSO were used as negative controls whereas ESTgCAGlacz PP cells were used as high signal controls. To control for the specificity of the signal, both lines were also inhibited by addition of the β‐galactosidase inhibitor PETG (Supporting Information Fig. S1C). Images were analyzed with the open source software CellProfiler 29 for PP segmentation and measurement of fluorescence intensity. The data were analyzed using the open source software KNIME 29. The screen was repeated twice and the z‐score of the logarithm of the mean intensity was calculated (Supporting Information Fig. S1D). Analysis of the control samples showed negligible or strong β‐galactosidase activity of the ESAldh1b1lacZ/+ and ESTgCAGlacZ PPs, respectively, confirming the specificity of the assay under the screening conditions (Supporting Information Fig. S1D).
To limit the pool of candidate inducers to a reasonable number for additional assays, 38 compounds with a z‐score above 2.5 were identified and were re‐evaluated in triplicate. The mean of the log mean intensity z‐score from all five assays (2 repeats of the screen and three replicate validation) was calculated. Only a single compound had an average z‐score higher than three and was further characterized (Fig. 1E).
Validation of Candidate Aldh1b1 Expression Inducers in mES Cell Derived Pancreas Progenitors
After verification, we proceeded with the only compound with a z score >3, the protein methyltransferase inhibitor AMI‐5 (Fig. 1D). We first confirmed by qPCR the induction of Aldh1b1 expression in PPs treated for 16 hours with different concentrations of AMI‐5. The length of this treatment was reduced to increase the stringency of the assay and exclude potential secondary effects. The AMI‐5 concentration used in the screen (10 μM) was the most effective and induced Aldh1b1 expression by nearly twofold (n = 5; Fig. 2A). However, the variability among independent samples (Fig. 2A) suggested that true induction levels might be masked by the presence of nonresponsive cells and this was confirmed by FDG fluorescence of PP aggregates (Supporting Information Fig. S2A).
Figure 2.
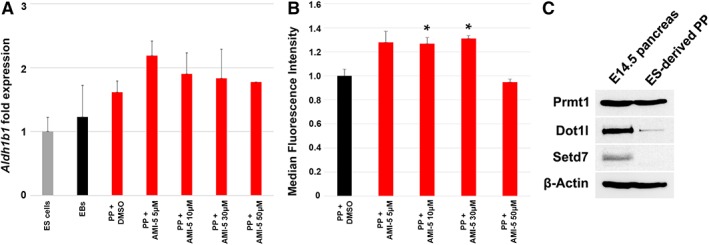
Validation of AMI‐5 as an inducer of Aldh1b1 expression in mES derived pancreatic progenitors. (A): qPCR analysis of Aldh1b1 expression in PP clusters following a 16 hour treatment with different concentrations of AMI‐5 or DMSO relative to Aldh1b1 expression in EB clusters (n = 5). (B): Median fluorescence intensity of PP clusters assessed by flow cytometry following a 16 hour treatment with different concentrations of AMI‐5 relative to fluorescence intensity of PP clusters treated during 24 hours with DMSO (n = 3). (C): Western blots illustrating expression levels of the AMI‐5 target proteins Prmt1, Dot1l, and Setd7. Abbreviation: mES, mouse embryonic stem; MFI, median fluorescence intensity; PP, pancreatic‐like progenitors. *, p < .05, error bars show SEM.
To confirm the qPCR results, ESAldh1b1lacZ/+ PP clusters were treated with different concentrations of AMI‐5 for 16 hours, dissociated in single cell suspension and incubated with FDG. The β‐galactosidase activity was assessed by flow cytometry and the MFI of gated fluorescent cells was calculated in treated samples and untreated controls (n = 3). AMI‐5 at 10 and 30 μM increased the MFI of these cells in a statistically significant manner confirming the results of the screen and the qPCR assays (Fig. 2B; Supporting Information Fig. S2B). The screening concentration of three additional compounds (bisindolylmaleimide V, betulinic acid, and the glukokinase activator CpdA) was similarly tested by FACS analysis for induction of β‐galactosidase activity in ESAldh1b1lacZ/+ derived PPs but the results did not confirm the initial screen assessment therefore these compounds were not pursued further (data not shown).
AMI‐5 is a xanthenyl compound that inhibits the activities of protein arginine histone methyltransferases Prmt1/3/4/6 as well as histone lysine methyltransferases (HMTases) Setd7 and Dot1l in an S‐adenosylmethionine competitive manner 35. Prmt1 is the most strongly expressed histone methyltransferase gene during mouse embryonic pancreas secondary transition whereas Dot1l and Setd7 are also strongly expressed during that time at significant levels (Table 1) 19. To ensure that these proteins are present in the ES‐derived PPs as well as in the developing mouse pancreas we investigated their presence by Western blot analysis. Consistent with RNA expression levels, Prmt1, and Dot1l were strongly expressed in E14.5 dpc embryonic pancreata whereas Setd7 was expressed in considerably lower levels (Fig. 2C). In ES‐derived PPs, Prmt1, but not Dot1l or Setd7, was expressed in high levels.
Table 1.
Expression levels of AMI‐5 targets during mouse pancreas development as determined by RNA Seq
| Name | Gene symbol | 13.5 dpc | 14.5 dpc | 15.5 dpc |
|---|---|---|---|---|
| Protein arginine N‐methyltransferase 1 | Prmt1 | 4,631 | 4,605 | 4,772 |
| DOT1‐like, histone H3 methyltransferase | Dot1l | 1,505 | 1,508 | 1,392 |
| SET domain containing 7 | Setd7 | 932 | 946 | 629 |
| Protein arginine N‐methyltransferase 3 | Prmt3 | 822 | 827 | 851 |
| Protein arginine N‐methyltransferase 6 | Prmt6 | 429 | 434 | 560 |
The Protein Methyltransferase Inhibitor AMI‐5 Maintains Expression of Aldh1b1 in Embryonic Pancreatic Explants and Selectively Delays Endocrine Differentiation
To address whether AMI‐5 would increase and/or maintain Aldh1b1 expression in a relevant biological context that is, during mouse embryonic development, we used serum free air–liquid interface (ALI) cultures of embryonic pancreata. ALI cultures mimic normal embryonic development and thus give the means to temporally manipulate culture and signaling conditions and elucidate their effects on lineage specification. Wild‐type 14.5 dpc pancreata in ALI cultures continue their developmental program over the course of 6 days giving rise to acinar, endocrine and duct cells 36. Thus, 14.5 dpc embryonic pancreata were placed in ALI culture for 2 days (14.5 dpc + 2 days) in the presence of 10 μM AMI‐5 and Aldh1b1 expression was assayed at both gene and protein levels by qPCR and immunofluorescence, respectively. As expected 21, in the untreated controls expression of Adlh1b1 is significantly reduced between 14.5 and 14.5 dpc + 2 days embryonic pancreata concomitant with the onset of differentiation. In contrast, in AMI‐5 treated samples, expression of Aldh1b1 was maintained in nearly twice as many cells compared with 14.5 dpc + 2 days controls. Consistent with regulation at the Aldh1b1 gene expression level, Aldh1b1 mRNA levels, as assessed by qPCR, were nearly twofold higher in treated samples (Fig. 3A, 3C). Therefore, AMI‐5 maintains Aldh1b1 expression in embryonic pancreatic explants, in line with the result of the screen.
Figure 3.
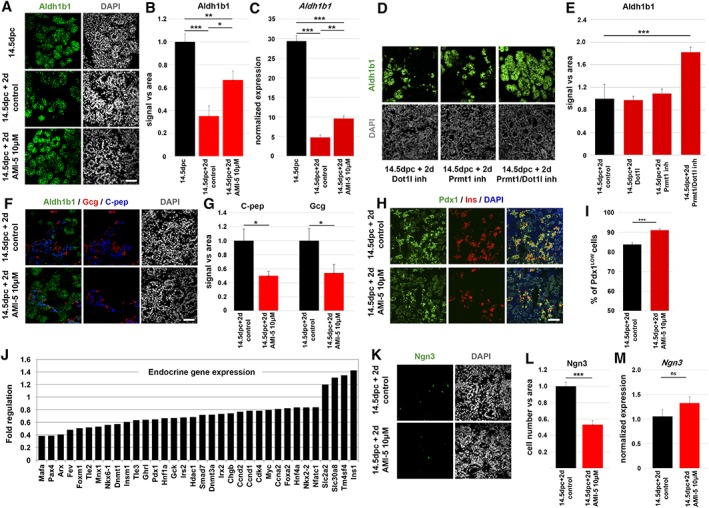
AMI‐5 prolongs Aldh1b1 expression and delays endocrine differentiation in mouse embryo pancreatic explants. (A): Immunofluorescence analysis of pancreata at 14.5 dpc and after 2 days in ALI cultures (14.5 dpc + 2 days) shows that Aldh1b1 expression is reduced after 2 days in ALI culture of control pancreata whereas it remains higher in AMI‐5 treated pancreata. (B): Quantitation of relative Aldh1b1 fluorescence signal in 14.5 dpc pancreata and 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). (C): qPCR analysis of Aldh1b1 expression in 14.5 dpc pancreata and 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 6). (D): Immunofluorescence analysis of Aldh1b1 expression of 14.5 dpc pancreata after 2 days in ALI cultures in the presence of the Dot1l inhibitor EPZ004777 (10 μM) or the Prmt1 inhibitor C21 (10 μM) or both (10 μM each). (E): Relative quantification of the Aldh1b1 immunofluorescence signal shows significant upregulation in the presence of both inhibitors. (F): Immunofluorescence analysis of 14.5 dpc pancreata after 2 days in ALI cultures shows a reduction in the C‐pep and Gcg signal in pancreata treated with AMI‐5. (G): Relative quantitation of the C‐pep and Gcg fluorescence signal in 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). (H): Immunofluorescence analysis of 14.5 dpc pancreata after 2 days in ALI cultures shows an increase of Pdx1LOW cells in pancreata treated with AMI‐5 and indicates the expression of ins in Pdx1HIGH cells. (I): Quantitation of the Pdx1LOW cells in relation to all Pdx1+ cells in 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 3). (J): Fold regulation of endocrine markers at 14.5 + 2 days in ALI culture in the presence of 10 μM AMI‐5 in relation to untreated controls. Only significantly regulated genes are shown (padj ≤ 0.05). (K): Immunofluorescence analysis of 14.5 dpc pancreata after 2 days in ALI cultures shows a reduction in the number of Ngn3+ cells in pancreata treated with AMI‐5. (L): Relative quantitation of the Ngn3+ cells in 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). (M): qPCR analysis of Ngn3 expression in 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 3). *, p < .05; **, p < .01; ***, p < .001, error bars show SEM. Scale bar: 50 μm. Abbreviation: ALI, air to liquid interface.
To confirm that AMI‐5 is acting through Prmt1 and Dot1l, the two methyltransferases strongly expressed in the developing mouse pancreas (Table 1, Fig. 2C), we used the respective specific inhibitors C21 and EPZ004777 37, 38, at 10 μM each, alone or in combination. Interestingly, each inhibitor alone was not sufficient to activate Aldh1b1 expression but their combination resulted is a similar induction to that of AMI‐5 alone (Fig. 3D, 3E). These results confirmed the specificity of AMI‐5 toward Prmt1 and Dot1l and suggested that there is functional overlap between these two methyltransferases.
In Aldh1b1 null embryonic pancreata differentiation is accelerated in all three lineages and in the endocrine lineage this was illustrated by a transient increase in the number of Ngn3+ cells 21. This showed that Aldh1b1 expression is necessary to maintain the progenitor status and the identification of AMI‐5 as an inducer of Aldh1b1 expression gave us the opportunity to examine whether sustained Aldh1b1 expression was sufficient to maintain the progenitor status. Accordingly, we examined whether the AMI‐5 induced maintenance of Aldh1b1 expression in embryonic explants affected differentiation and lineage specification. In control 14.5 dpc + 2 days explants differentiated insulin (C‐pep+) or glucagon (Gcg+) producing endocrine cells did not express Aldh1b1, in contrast to CK19+ duct and Amy+ acinar cells which maintain expression of Aldh1b1 (Fig. 3F; Supporting Information Fig. S3A). This observation was in line with in vivo findings 21 further validating the use of ALI explants. It also indicated that Aldh1b1 downregulation might be specifically required for the differentiation of endocrine cells. In agreement with this hypothesis, when explants were cultured in the presence of 10 μM ΑMι‐5, the generation of C‐pep+ and Gcg+ endocrine cells was selectively reduced twofold whereas the generation of CK19+ and Amy+, duct and acinar cells, was not affected (Fig. 3F, 3G; Supporting Information Fig. S3A, S3B). Pdx1 is expressed in all progenitor cells in relatively low levels (Pdx1LOW) and its expression is strongly upregulated in nascent insulin producing cells (Fig. 3H). To independently assess the reduction of endocrine differentiation in the presence of AMI‐5 we asked whether the number of progenitor cells increased. Thus, we quantified the percentage of Pdx1LOW cells in control and AMI‐5 treated explants. Although the overall numbers of Pdx1+ cells remained the same, Pdx1LOW progenitors expanded from 83.8% ± 2.9% to 91.0% ± 2.2% (n = 3) confirming the delay in differentiation (Fig. 3H, 3I; Supporting Information Fig. S3C). The selective delay in endocrine differentiation but not acinar or duct differentiation was further corroborated with RNA Seq data showing a coordinated decrease in the expression of the majority of endocrine lineage markers in AMI‐5 treated explants. A very small number of endocrine transcripts, including that of Ins1, were upregulated suggesting that aspects of endocrine program were still triggered. It is possible that the differentiation block impeded their processing leading to their accumulation in the AMI‐5 treated cells (Fig. 3J; Supporting Information Table S1 for a complete list). Interestingly, acinar markers were strongly upregulated while duct markers were relatively less affected (Supporting Information Fig. S3D, S3E; Supporting Information Table S1). This suggested that, while the number of acinar cells did not change (Supporting Information Fig. S3A, S3B), their maturation was accelerated. As expected, given the normal differentiation of the acinar and ductal lineages, extended Aldh1b1 expression did not increase proliferation as assessed by phospho‐Histone H3 (pH 3) immunofluorescence (Supporting Information Fig. S3F, S3G). Additionally, it did not result in the appearance of immature bi‐hormonal endocrine cells as the incidence of C‐pep+/Gcg+ cells in 14.5 dpc + 2 days explants is less that 1% of all endocrine cells and this did not change in the AMI‐5 treated explants.
Next, we assessed whether the selective loss of endocrine cells was due to loss of Ngn3+ endocrine progenitor cells. We observed a twofold decrease in the number of Ngn3+ cells in AMI‐5 treated explants in comparison to controls (Fig. 3K, 3L) and qPCR analysis suggested that Ngn3 was not affected (Fig. 3L). To confirm that Ngn3 protein stability is regulated by the proteasome 39, 40 also in the context of ALI cultures, we supplemented the culture medium with 1 μM of the reversible proteasome inhibitor MG132 19 and confirmed that this treatment increases the number of Ngn3+ cells by twofold (Supporting Information Fig. S3H, S3I). However, it was not possible to directly assess whether MG132 could reverse the effect of AMI‐5 because simultaneous treatment with both compounds resulted in extensive cell death (Supporting Information Fig. S3H). The AMI‐5 effect was selective for endocrine progenitors since the number of Nkx6.1+ cells bipotent progenitors and Ptf1a+ acinar progenitors did not change (Supporting Information Fig. S3J–S3M). This was consistent with the finding that acinar and ductal specification was not affected.
Taken together the data above showed that the AMI‐5 protein methyltransferase inhibitor maintained Aldh1b1 expression in pancreatic explants and this resulted in a selective reduction in endocrine, but not acinar or ductal, differentiation that was due to a loss of Ngn3+ endocrine progenitors. Thus, ectopic Aldh1b1 activation is not sufficient for general progenitor maintenance but selectively delays endocrine differentiation.
Protein Methyltransferase Activity Specifies Endocrine Cells Through the Downregulation of Aldh1b1 Expression
The results described above confirmed that Aldh1b1 downregulation precedes endocrine differentiation and showed that AMI‐5 maintains Aldh1b1 expression in embryonic pancreata concomitant with a selective repression of endocrine differentiation. To evaluate whether this delay was mediated by Aldh1b1 itself we used ALI cultures of 14.5 dpc Aldh1b1 null pancreata explants and determined the effect of 10 μM AMI‐5 on lineage differentiation after 2 days in culture. The compound did not affect the number of C‐pep+ or Gcg+ cells (Fig. 4A, 4B) and, consistent with this finding neither did it affect the number of Ngn3+ endocrine progenitors (Fig. 4E, 4F). As expected, acinar and duct specification were not affected either (Fig. 4C, 4D). Therefore, protein methyltransferase activity regulates endocrine specification through Aldh1b1.
Figure 4.

The effects of the protein methyltransferase inhibitor AMI‐5 are mediated through Aldh1b1 expression. (A): Immunofluorescence analysis of 14.5 dpc Aldh1b1 null pancreata after 2 days in ALI cultures in the presence of 10 μM AMI‐5 showed that the number of C‐pep+ or Gcg+ cells was not affected. (B): Relative quantitation of the C‐pep and Gcg fluorescence signal in 14.5 dpc Aldh1b1 null pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 3). (C): Immunofluorescence analysis of 14.5 dpc Aldh1b1 null pancreata after 2 days in ALI cultures in the presence of 10 μM AMI‐5 showed that the number of Amy+ or CK19+ cells was not affected. (D): Relative quantitation of the amylase and CK19 fluorescence signal in 14.5 dpc Aldh1b1 null pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 5). (E): Immunofluorescence analysis of 14.5 dpc Aldh1b1 null pancreata after 2 days in ALI cultures shows no effect in the number of Ngn3+ cells in pancreata in the presence of 10 μM AMI‐5. (F): Relative quantitation of the Ngn3+ cells in 14.5 dpc Aldh1b1 null pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 3). Error bars show SEM; Abbreviations: ALI, air to liquid interface; ns, not significant. Scale bar: 50 μm.
Discussion
As previously shown, Aldh1b1 is expressed in all pancreas progenitors during development and it is downregulated upon differentiation. Consistent with a role in maintaining the progenitor state, Aldh1b1 genetic inactivation resulted in premature differentiation in all three lineages 21. To gain insights into the upstream activators of Aldh1b1, and therefore activators of the pancreas progenitor state, we used a screening approach combined with directed differentiation of mES cells into pancreas progenitors. As a reporter we used a lacZ Aldh1b1 knockin mES line and we assayed lacZ expression using a live substrate that fluoresces upon β‐galactosidase mediated hydrolysis. The differentiation protocol had two limitations that precluded its adoption for a large scale screen. The first was the differentiation in suspension culture which is laborious unless combined with spinner flask cultures. The second limitation was the heterogeneity in the differentiation stage among clusters and also among cells within the same cluster. Additional technical limitations that likely account for the variability of the z‐score of each replicate were the variability in cluster size and the number of clusters going into each well. Inherent limitations in any screening approach on differentiated cells concern the precise timing and duration of treatment as well as concentrations of the compounds and suboptimal conditions may lead to weak responses and false negative results. Despite these limitations we were able to identify AMI‐5 as a compound that prolongs Aldh1b1 expression in a physiologically relevant context, that is, during differentiation of mouse embryo pancreatic explants.
AMI‐5 is a methyltransferase inhibitor that inhibits the activities of protein arginine methyltransferases (Prmt) 1/3/4/6 as well as histone lysine methyltransferases (HMTases) Setd7 and Dot1l in an S‐adenosylmethionine competitive manner 35. Prmt1, and to a lesser extent Dot1l and Setd7, are strongly expressed during mouse embryonic pancreas secondary transition (Table 1, Fig. 2C) suggesting that these proteins are the main conveyors of protein methyltransferase activity. Arginine methylation is an emerging post‐translational modification acting primarily as an epigenetic regulator of transcription but also playing key roles in translation regulation and cell signaling. PRMTs deposit key activating or repressive histone marks but they have also non histone substrates 41. Since the AMI‐5 effect on Aldh1b1 appears to be at the transcriptional level it is likely that Prmt1 does not play a role on translational regulation in this context.
Histone methylation and acetylation are key epigenetic mechanisms that regulate gene activity and coordinated changes in their patterns are associated with cell fate decisions and stabilization of the differentiated cell state. Specifically, during pancreas development, histone acetyltransferases and methyltransferases have a modulatory role in the early binary cell fate decision between liver and pancreas fates 42 during mouse development and Setd7 was shown to be necessary and sufficient for the specification of pancreatic endoderm in Xenopus 43. During subsequent pancreas development H3K27me3 patterns change during the differentiation of pancreas progenitors into endocrine progenitors and eventually β‐cells 44. Incomplete changes in the pattern of these modifications are associated with impaired endocrine gene activation during in vitro differentiation of hPS cells and this may account for the generation of immature β‐cells using the existing differentiation protocols 45. Indeed, contrary to in vitro directed differentiation, the number of H3K27me3 marks increase during endocrine progenitor development in vivo 46. Ezh2, the enzymatic component of the PRC2, is a methyltransferase for H3K27 that represses the progression of definitive endoderm to pancreas progenitors and then to endocrine progenitors. Deletion of Ezh2 enhanced endocrine differentiation 46. Consistent with this report, expression of Ngn3 promotes the loss of H3K27me3 at proximal promoters of key downstream genes 47.
Here, we show that Prmt1/Dotl1/Setd7 methyltrasferase activity is required upstream of Ngn3 activation pointing to the temporal complexity of methylation events necessary for the progression of endoderm progenitors to endocrine cells. This methyltransferase activity is required for the orderly downregulation of Aldh1b1 which is necessary for the emergence of the full number of Ngn3+ cells. It is important to note that genetic loss of Aldh1b1 function leads to premature appearance of supernumerary Ngn3+ cells and subsequently defective β‐cells that show poor response to glucose stimulation 21 and therefore are reminiscent of the β‐cells generated from the directed differentiation of hPS cells. Interestingly, AMI‐5 mediated extension of Aldh1b1 expression affected selectively only the endocrine lineage. These findings suggest that Aldh1b1, being subject to methyltransferase regulation itself, regulates the timing of endocrine specification and this helps ensure that the proper epigenetic marks have been established prior to this commitment.
A possible mode of Ngn3 regulation is protein stability which is controlled by cdk2/4 and six dependent phosphorylation at the G1/S cell cycle point at multiple sites. Phosphorylation primes Nng3 for degradation and lengthening the G1 phase of the cell cycle allows for Ngn3 accumulation, initiation of the endocrine differentiation program and cell cycle exit 39, 40. Aldh1b1 is a metabolic regulator (unpublished results) and metabolic switches have been associated with the cell decision to differentiate. It remains to be seen whether the Aldh1b1 metabolic function has an impact on the length of the G1 phase and thus indirectly Ngn3 stability. Interestingly, initiation of the acinar or duct differentiation programs was not affected upon extension of Aldh1b1 expression suggesting that expression of their effectors is controlled by distinct mechanisms. Sox9 is genetically upstream of Ngn3 and specifies ductal cells in the bipotent progenitors in the absence of endocrine progenitor induction 48 and the transcription factor Ptf1a initiates acinar differentiation when levels of RBP‐JL, itself a target of Ptf1a mediated transcriptional activation, become sufficient to substitute RBP‐J in the Ptf1a/E2a/RBP‐J complex and change its transcriptional specificity 49.
Conclusion
The findings reported here suggested that specific, reversible protein methyltransferase inhibitors may repress endocrine differentiation of pancreas progenitors. The accelerated differentiation of the mouse embryonic pancreas in the Aldh1b1 null mutants resulted in dysfunctional islets 21, reminiscent of those derived from current protocols of human PS differentiation into β‐like cells 2. This suggested that a certain period of time is required for pancreas progenitors to acquire the competency to properly develop into fully functional β‐cells. Accordingly, a timely use of selected protein methyltransferase inhibitors and activators to extend the pancreatic endoderm state and promote endocrine differentiation, respectively, may enhance the final differentiation into fully functional β‐cells. Thus, the complete elucidation of the role of protein methyltransferases in the progression from pancreas progenitors to endocrine cells and the development of specific reversible inhibitors/activators will provide valuable new instruments to help in the efficient conversion of hPS cells into fully functional β‐cells.
Author Contributions
I.G., I.S.: performed laboratory work, data analysis and interpretation, manuscript writing, final approval of manuscript; V.A., D.P., C.A.: performed laboratory work, final approval of the manuscript; M.L.: performed RNA sequencing analysis and interpretation, final approval of the manuscript; M.B.: performed data analysis and interpretation, final approval of the manuscript; A.G.: conception/design and fundraising, data analysis and interpretation, manuscript writing, final approval of the manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Supporting information
Figure S1 A screen using mES cell derived pancreas progenitors to identify small molecules that induce Aldh1b1 expression. (A) Schematic diagram of the screening protocol for the ESAldh1b1lacZ and ESTgCAGlacZ cell lines. (B) Immunofluorescence analysis of PP clusters showed that Pdx1+ cells comprise the large majority of PP cells while the numerous Nkx6.1+ cells are a subset of the former. (C) To confirm the specificity of the screening assay, the β‐gal inhibitor PETG was added during the screen. The activity of the ESTgCAGlacZ decreased to background levels whereas the activity of the ESAldh1b1lacZ remained always negligible (ESAldh1b1lacZ n = 268, ESAldh1b1lacZ + PETG n = 86, ESTgCAGlacZ n = 86, ESTgCAGlacZ + PETG n = 86), samples were compared to the ESTgCAGlacZ sample. (D) Plot of the z‐score of the log(mean fluorescence intensity) for each of the assays using compounds of the StemSelect™ Small Molecule Regulators Library (blue) or DMSO controls (green). Abbreviations: FDG, fluorescein‐di‐beta‐D‐galactopyranoside. Scale bar: 25 μm. *** p < .001.
Figure S2. Heterogeneity of AMI‐5 induced Aldh1b1 expression in mouse ES cell derived pancreatic progenitors. (A) Cellular heterogeneity of ESAldh1b1lacZ derived PP clusters as revealed by the fluorescence β‐galactosidase assay. (B) Fluorescence intensity distribution of PP clusters generated from the ESAldh1b1lacZ and ESTgCAGlacZ lines after treatment for 16 hours with DMSO or 10 μM AMI‐5 respectively. The Y‐axis represents events normalized for the area under each curve (percentage of cells found at a given bin) (n = 3). scale bar: 50 μm.
Figure S3. AMI‐5 does not affect specification of the acinar or duct lineages. (A) Immunofluorescence analysis of 14.5 dpc pancreata after 2 days in ALI cultures shows no difference in the expression of amylase and CK19 in pancreata treated with 10 μM AMI‐5. (B) Relative quantitation of the Amylase and CK19 fluorescence signal in 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). (C) Quantitation of Pdx1+ cells following immunofluorescence in 14.5dpc pancreata after 2 days in ALI in the absence or presence of 10 μM AMI‐5. (D, E) Fold regulation of acinar (D) and duct (E) markers at 14.5 + 2 days in ALI culture in the presence of 10 μM AMI‐5 in relation to untreated controls. Only significantly regulated genes are shown (padj ≤ 0.05). (F, G) Quantitation of cells in mitosis using pH 3 immunofluorescence showed that mitosis rates did not change upon AMI‐5 treatment (n = 3). (H) Immunofluorescence analysis of Ngn3 expression in 14.5dpc pancreata after 2 days in ALI cultures in pancreata treated with 1 μM MG132 and TUNEL analysis following exposure to a combination of 10 μM AMI‐5 and 1 μM MG132 for two days. (I) Relative quantitation of the number of Ngn3+ cells in 14.5dpc pancreata cultured in ALI for 2 days in the presence of 1 μM MG132 (n = 3). (J) Immunofluorescence analysis of 14.5dpc pancreata after 2 days in ALI cultures shows no effect in the expression of the bipotent progenitor marker Nkx6.1 in pancreata treated with 10 μM AMI‐5. (K) Relative quantitation of the Nkx6.1 fluorescence signal in 14.5dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). (L) Immunofluorescence analysis of 14.5dpc pancreata after 2 days in ALI cultures shows no effect in the expression of the acinar progenitor marker Ptf1a in pancreata treated with 10 μM AMI‐5. (M) Relative quantitation of the Ptf1a fluorescence signal in 14.5dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). Error bars show SEM; ns, not significant. Scale bar: 50 μm.
Table S1 Expression of lineage specific genes in 14.5dpc + 2 pancreatic explants
Acknowledgments
We thank the personnel of the Center for Regenerative Therapies Dresden (CRTD) Flow Cytometry Core Facility (FACS) for expert technical assistance; and the personnel of the animal house at CRTD for help with animal husbandry. Research in A.G.'s laboratory was supported by grants from the German Centre for Diabetes Research (DZD; grant 82DZD00101) and the German Research Foundation (DFG; CRTD Seed Grant 2016).
References
- 1. Pagliuca FW, Millman JR, Gurtler M et al. Generation of functional human pancreatic beta cells in vitro. Cell 2014;159:428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rezania A, Bruin JE, Arora P et al. Reversal of diabetes with insulin‐producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol 2014;32:1121–1133. [DOI] [PubMed] [Google Scholar]
- 3. Russ HA, Parent AV, Ringler JJ et al. Controlled induction of human pancreatic progenitors produces functional beta‐like cells in vitro. EMBO J 2015;34:1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vegas AJ, Veiseh O, Gurtler M et al. Long‐term glycemic control using polymer‐encapsulated human stem cell‐derived beta cells in immune‐competent mice. Nat Med 2016;22:306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawaguchi Y, Cooper B, Gannon M et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 2002;32:128–134. [DOI] [PubMed] [Google Scholar]
- 6. Offield MF, Jetton TL, Labosky PA et al. PDX‐1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996;122:983–995. [DOI] [PubMed] [Google Scholar]
- 7. Stoffers DA, Zinkin NT, Stanojevic V et al. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet 1997;15:106–110. [DOI] [PubMed] [Google Scholar]
- 8. Shih HP, Seymour PA, Patel NA et al. A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep 2015;13:326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Q, Law AC, Rajagopal J et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell 2007;13:103–114. [DOI] [PubMed] [Google Scholar]
- 10. Apelqvist A, Li H, Sommer L et al. Notch signalling controls pancreatic cell differentiation. Nature 1999;400:877–881. [DOI] [PubMed] [Google Scholar]
- 11. Hart A, Papadopoulou S, Edlund H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev Dyn 2003;228:185–193. [DOI] [PubMed] [Google Scholar]
- 12. Murtaugh LC, Stanger BZ, Kwan KM et al. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA 2003;100:14920–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Norgaard GA, Jensen JN, Jensen J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev Biol 2003;264:323–338. [DOI] [PubMed] [Google Scholar]
- 14. Afelik S, Qu X, Hasrouni E et al. Notch‐mediated patterning and cell fate allocation of pancreatic progenitor cells. Development 2012;139:1744–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horn S, Kobberup S, Jorgensen MC et al. Mind bomb 1 is required for pancreatic beta‐cell formation. Proc Natl Acad Sci USA 2012;109:7356–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schaffer AE, Freude KK, Nelson SB et al. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell 2010;18:1022–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kopp JL, Dubois CL, Hao E et al. Progenitor cell domains in the developing and adult pancreas. Cell Cycle 2011;10:1921–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shih HP, Kopp JL, Sandhu M et al. A Notch‐dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development 2012;139:2488–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Serafimidis I, Rodriguez‐Aznar E, Lesche M et al. Pancreas lineage allocation and specification are regulated by sphingosine‐1‐phosphate signalling. PLoS Biol 2017;15:e2000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ioannou M, Serafimidis I, Arnes L et al. ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Dev Biol 2013;374:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anastasiou V, Ninou E, Alexopoulou D et al. Aldehyde dehydrogenase activity is necessary for beta cell development and functionality in mice. Diabetologia 2016;59:139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen C, Chai J, Singh L et al. Characterization of an in vitro differentiation assay for pancreatic‐like cell development from murine embryonic stem cells: Detailed gene expression analysis. Assay Drug Dev Technol 2011;9:403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nair GG, Vincent RK, Odorico JS. Ectopic Ptf1a expression in murine ESCs potentiates endocrine differentiation and models pancreas development in vitro. Stem Cells 2014;32:1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Serafimidis I, Rakatzi I, Episkopou V et al. Novel effectors of directed and Ngn3‐mediated differentiation of mouse embryonic stem cells into endocrine pancreas progenitors. Stem Cells 2008;26:3–16. [DOI] [PubMed] [Google Scholar]
- 25. Treff NR, Vincent RK, Budde ML et al. Differentiation of embryonic stem cells conditionally expressing neurogenin 3. Stem Cells 2006;24:2529–2537. [DOI] [PubMed] [Google Scholar]
- 26. Bernardo AS, Cho CH, Mason S et al. Biphasic induction of Pdx1 in mouse and human embryonic stem cells can mimic development of pancreatic beta‐cells. Stem Cells 2009;27:341–351. [DOI] [PubMed] [Google Scholar]
- 27. Lima MJ, Docherty HM, Chen Y et al. Pancreatic transcription factors containing protein transduction domains drive mouse embryonic stem cells towards endocrine pancreas. PLoS One 2012;7:e36481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Novak A, Guo C, Yang W et al. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre‐mediated excision. Genesis 2000;28:147–155. [PubMed] [Google Scholar]
- 29. Stoter M, Niederlein A, Barsacchi R et al. CellProfiler and KNIME: Open source tools for high content screening. Methods Mol Biol 2013;986:105–122. [DOI] [PubMed] [Google Scholar]
- 30. Stagos D, Chen Y, Brocker C et al. Aldehyde dehydrogenase 1B1: Molecular cloning and characterization of a novel mitochondrial acetaldehyde‐metabolizing enzyme. Drug Metab Dispos 2010;38:1679–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schindelin J, Arganda‐Carreras I, Frise E et al. Fiji: An open‐source platform for biological‐image analysis. Nat Methods 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu TD, Nacu S. Fast and SNP‐tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang N, Bai H, David KK et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 2010;19:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng D, Yadav N, King RW et al. Small molecule regulators of protein arginine methyltransferases. J Biol Chem 2004;279:23892–23899. [DOI] [PubMed] [Google Scholar]
- 36. Serafimidis I, Heximer S, Beis D et al. GPCR signaling and S1P play a phylogenetically conserved role in endocrine pancreas morphogenesis. Mol Cell Biol 2011;31:5702–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Obianyo O, Causey CP, Osborne TC et al. A chloroacetamidine‐based inactivator of protein arginine methyltransferase 1: Design, synthesis, and in vitro and in vivo evaluation. Chembiochem 2010;11:1219–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarkaria SM, Christopher MJ, Klco JM et al. Primary acute myeloid leukemia cells with IDH1 or IDH2 mutations respond to a DOT1L inhibitor in vitro. Leukemia 2014;28:2403–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Azzarelli R, Hurley C, Sznurkowska MK et al. Multi‐site neurogenin3 phosphorylation controls pancreatic endocrine differentiation. Dev Cell 2017;41:274–286 e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krentz NAJ, van Hoof D, Li Z et al. Phosphorylation of NEUROG3 links endocrine differentiation to the cell cycle in pancreatic progenitors. Dev Cell 2017;41:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blanc RS, Richard S. Arginine methylation: The coming of age. Mol Cell 2017;65:8–24. [DOI] [PubMed] [Google Scholar]
- 42. Xu CR, Cole PA, Meyers DJ et al. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science 2011;332:963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kofent J, Zhang J, Spagnoli FM. The histone methyltransferase Setd7 promotes pancreatic progenitor identity. Development 2016;143:3573–3581. [DOI] [PubMed] [Google Scholar]
- 44. van Arensbergen J, Garcia‐Hurtado J, Moran I et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin‐producing beta‐cells to adopt a neural gene activity program. Genome Res 2010;20:722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie R, Everett LJ, Lim HW et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell 2013;12:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu CR, Li LC, Donahue G et al. Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. EMBO J 2014;33:2157–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pujadas G, Felipe F, Ejarque M et al. Sequence and epigenetic determinants in the regulation of the Math6 gene by Neurogenin3. Differentiation 2011;82:66–76. [DOI] [PubMed] [Google Scholar]
- 48. Seymour PA, Freude KK, Tran MN et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA 2007;104:1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Masui T, Swift GH, Deering T et al. Replacement of Rbpj with Rbpjl in the PTF1 complex controls the final maturation of pancreatic acinar cells. Gastroenterology 2010;139:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 A screen using mES cell derived pancreas progenitors to identify small molecules that induce Aldh1b1 expression. (A) Schematic diagram of the screening protocol for the ESAldh1b1lacZ and ESTgCAGlacZ cell lines. (B) Immunofluorescence analysis of PP clusters showed that Pdx1+ cells comprise the large majority of PP cells while the numerous Nkx6.1+ cells are a subset of the former. (C) To confirm the specificity of the screening assay, the β‐gal inhibitor PETG was added during the screen. The activity of the ESTgCAGlacZ decreased to background levels whereas the activity of the ESAldh1b1lacZ remained always negligible (ESAldh1b1lacZ n = 268, ESAldh1b1lacZ + PETG n = 86, ESTgCAGlacZ n = 86, ESTgCAGlacZ + PETG n = 86), samples were compared to the ESTgCAGlacZ sample. (D) Plot of the z‐score of the log(mean fluorescence intensity) for each of the assays using compounds of the StemSelect™ Small Molecule Regulators Library (blue) or DMSO controls (green). Abbreviations: FDG, fluorescein‐di‐beta‐D‐galactopyranoside. Scale bar: 25 μm. *** p < .001.
Figure S2. Heterogeneity of AMI‐5 induced Aldh1b1 expression in mouse ES cell derived pancreatic progenitors. (A) Cellular heterogeneity of ESAldh1b1lacZ derived PP clusters as revealed by the fluorescence β‐galactosidase assay. (B) Fluorescence intensity distribution of PP clusters generated from the ESAldh1b1lacZ and ESTgCAGlacZ lines after treatment for 16 hours with DMSO or 10 μM AMI‐5 respectively. The Y‐axis represents events normalized for the area under each curve (percentage of cells found at a given bin) (n = 3). scale bar: 50 μm.
Figure S3. AMI‐5 does not affect specification of the acinar or duct lineages. (A) Immunofluorescence analysis of 14.5 dpc pancreata after 2 days in ALI cultures shows no difference in the expression of amylase and CK19 in pancreata treated with 10 μM AMI‐5. (B) Relative quantitation of the Amylase and CK19 fluorescence signal in 14.5 dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). (C) Quantitation of Pdx1+ cells following immunofluorescence in 14.5dpc pancreata after 2 days in ALI in the absence or presence of 10 μM AMI‐5. (D, E) Fold regulation of acinar (D) and duct (E) markers at 14.5 + 2 days in ALI culture in the presence of 10 μM AMI‐5 in relation to untreated controls. Only significantly regulated genes are shown (padj ≤ 0.05). (F, G) Quantitation of cells in mitosis using pH 3 immunofluorescence showed that mitosis rates did not change upon AMI‐5 treatment (n = 3). (H) Immunofluorescence analysis of Ngn3 expression in 14.5dpc pancreata after 2 days in ALI cultures in pancreata treated with 1 μM MG132 and TUNEL analysis following exposure to a combination of 10 μM AMI‐5 and 1 μM MG132 for two days. (I) Relative quantitation of the number of Ngn3+ cells in 14.5dpc pancreata cultured in ALI for 2 days in the presence of 1 μM MG132 (n = 3). (J) Immunofluorescence analysis of 14.5dpc pancreata after 2 days in ALI cultures shows no effect in the expression of the bipotent progenitor marker Nkx6.1 in pancreata treated with 10 μM AMI‐5. (K) Relative quantitation of the Nkx6.1 fluorescence signal in 14.5dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). (L) Immunofluorescence analysis of 14.5dpc pancreata after 2 days in ALI cultures shows no effect in the expression of the acinar progenitor marker Ptf1a in pancreata treated with 10 μM AMI‐5. (M) Relative quantitation of the Ptf1a fluorescence signal in 14.5dpc pancreata cultured in ALI for 2 days in the absence or presence of 10 μM AMI‐5 (n = 4). Error bars show SEM; ns, not significant. Scale bar: 50 μm.
Table S1 Expression of lineage specific genes in 14.5dpc + 2 pancreatic explants


