Abstract
To elucidate the role of Hox genes in limb cartilage development, we identified the target genes of HOXA11 and HOXA13 by ChIP‐Seq. The ChIP DNA fragment contained evolutionarily conserved sequences and multiple highly conserved HOX binding sites. A substantial portion of the HOXA11 ChIP fragment overlapped with the HOXA13 ChIP fragment indicating that both factors share common targets. Deletion of the target regions neighboring Bmp2 or Tshz2 reduced their expression in the autopod suggesting that they function as the limb bud‐specific enhancers. We identified the Hox downstream genes as exhibiting expression changes in the Hoxa13 knock out (KO) and Hoxd11‐13 deletion double mutant (Hox13 dKO) autopod by Genechip analysis. The Hox downstream genes neighboring the ChIP fragment were defined as the direct targets of Hox. We analyzed the spatial expression pattern of the Hox target genes that encode two different categories of transcription factors during autopod development and Hox13dKO limb bud. (a) Bcl11a, encoding a repressor of cartilage differentiation, was expressed in the E11.5 autopod and was substantially reduced in the Hox13dKO. (b) The transcription factors Aff3, Bnc2, Nfib and Runx1t1 were expressed in the zeugopodal cartilage but not in the autopod due to the repressive or relatively weak transcriptional activity of Hox13 at E11.5. Interestingly, the expression of these genes was later observed in the autopodal cartilage at E12.5. These results indicate that Hox13 transiently suspends the cartilage differentiation in the autopodal anlage via multiple pathways until establishing the paddle‐shaped structure required to generate five digits.
Keywords: autopod, cartilage, ChIP‐Seq, Hoxa11, Hoxa13, limb, target gene
To elucidate the role of Hox genes in limb cartilage development, we identified the HOXA11 and HOXA13 target genes. We analyzed the spatial expression pattern of the Hox target genes. We found that Hox13 transiently suspends the cartilage differentiation in the autopod via multiple pathways until achieving the paddle‐shaped structure required to generate five digits.
Abbreviations
- ChIP
Chromatin immunoprecipitation
- ChIP‐Seq
ChIP‐sequencing
- HBS
HOX binding sequence
- KO
knock out
- PCR
polymerase chain reaction
1. INTRODUCTION
The tetrapod free limb is subdivided into three anatomical domains along the proximodistal axis: the stylopod, zeugopod and autopod. These domains possess a specific number and pattern of the bones and each bone possesses a unique morphology and length. During limb development, the mesenchymal cells of the limb bud first form precartilaginous aggregates, which eventually grow and differentiate into the limb cartilage (Gilbert & Barresi, 2016). The process occurs sequentially in the proximal to distal direction with the growth of the limb bud. Expression of Hox genes belonging to the Abd‐B family, Hox9‐13, is closely related to the anatomical domains of the limb bones (Izpisua‐Belmonte, Tickle, Dolle, Wolpert, & Duboule, 1991; Yokouchi, Sasaki, & Kuroiwa, 1991) and crucial for controlling morphogenesis and patterning of the limb cartilage (Pineault & Wellik, 2014). Hoxa11 and Hoxd11 show overlapping expression in the zeugopodal mesenchyme and simultaneous loss of function of both genes in the forelimb bud results severe truncation of the zeugopodal bones without affecting the other long bones (Boulet & Capecchi, 2004; Davis, Witte, Hsieh‐Li, Potter, & Capecchi, 1995). Hoxa13 and Hoxd13 are expressed in the autopodal mesenchyme and double knockout (KO) mice have smaller autopods during the embryonic stage and there was almost no sign of cartilage patterning in the entire autopod (Fromental‐Ramain et al., 1996). These double homozygous KO mice exhibited much more severe phenotypes than single homozygous KOs suggesting that the paralogous genes are functionally redundant (Davis et al., 1995; Fromental‐Ramain et al., 1996). A further role for Hox in domain‐specific cartilage development was shown by misexpression of Hox in the limb mesenchyme. Viral or genetic misexpression of Hoxa13 or Hoxd13 in the zeugopodal mesenchyme repressed cartilage differentiation and proliferation resulting in zeugopod‐specific cartilage truncation (Goff & Tabin, 1997; Peichel, Prabhakaran, & Vogt, 1997; Yokouchi et al., 1995). Thus, Hox genes are expressed in a domain‐specific manner in the limb bud mesenchyme and have a crucial function in domain‐specific cartilage morphogenesis. Since Hox encodes a transcription factor, Hox target genes function in region/compartment‐specific formation of the precartilaginous condensation, cellular differentiation and proliferation of the cartilage.
In contrast, Hox genes also control the expression of Fgf10 in the distal limb mesenchyme and Shh in the posterior mesenchyme to regulate limb outgrowth and proper supply of mesenchymal cells in coordination with the developmental time course of the limb bud (Capellini et al., 2006; Kmita et al., 2005; Sheth et al., 2013). How this growth control system coordinates with the domain‐specific cartilage pattern formation system is an important question that remains to be resolved.
In order to understand the system that determines domain‐specific limb cartilage development, it is essential to identify the direct Hox target genes. While limited numbers of Hox downstream genes have been reported, the known function of these genes does not sufficiently explain the Hox loss‐of‐function phenotype (McCabe & Innis, 2005; Salsi & Zappavigna, 2006). Recently the Hox target genes in the limb bud were exhaustively identified by ChIP‐Seq analysis, which opened the door to elucidate the function of the Hox genes in region‐specific cartilage development (Jerkovic et al., 2017; Sheth et al., 2016). Since Hox11 and Hox13 control the growth and cartilage differentiation of the zeugopod and autopod, respectively, we raised questions if they share common targets, and if so, how the common targets are differentially regulated to give rise to domain‐specific morphology. To this end, we identified the target genes of HOXA11 and HOXA13 in the limb bud by ChIP‐Seq analysis. We found that they share common target sequences that contain multiple evolutionarily conserved HOX binding sites and the representative target sequences possess limb‐bud specific enhancer functions. We also found that common target genes expressed in the mesenchyme and developing cartilage exhibited differential control by HOXA11 and HOXA13 that may explain the unique autopodal cartilage developmental program.
2. MATERIALS AND METHODS
2.1. Animal experimentation
This study was approved by the Institutional Animal Care and Use Committee of the Graduate School of Science and carried out according to the Nagoya University Animal Experimentation Regulations. The Hoxa13 targeted mice (Fromental‐Ramain et al., 1996) were obtained from Dr. Pierre Chambon. The HoxD del(11–13) allele (Zakany, Fromental‐Ramain, Warot, & Duboule, 1997) and the Ulnaless (Peichel et al., 1997) were provided by Dr. Denis Duboule. Noon on the day the vaginal plug was observed was considered as E0.5. The embryos were isolated by cesarean incision and genotype was assessed by PCR.
2.2. Chromatin immunoprecipitation (ChIP)
Production of the anti‐HOXA13 antibody was described previously (Yokouchi et al., 1995). The method for production and purification of the anti‐HOXA11 antibody was also previously described (Yamamoto et al., 1998). The antibodies used in the present study were prepared from rabbits. ChIP was performed according to a standard protocol (Green & Sambrook, 2012) with the following modifications. Whole limb buds from E11.5 and the autopod from E11.0 wild‐type ICR mice were dissected in ice‐cold PBS. Embryonic tissues were homogenized 13 times with a tight‐fitting Dounce homogenizer, then fixed in 1% formaldehyde/PBS for 10 min at 4°C and quenched with 0.1 M glycine. The cross‐linked material was sonicated to produce 200–1,000 bp fragments with a DNA Shearing System S2 (Covaris). The immunoprecipitations were performed with 11 pairs of autopods from E11.0 embryos or 12–18 pairs of whole limb buds from E11.5 embryos. Pierce™ Protein A/G magnetic beads (Thermo Scientific) were conjugated with 10 μg of anti‐HOXA13 (Yokouchi et al., 1995) or 8 μg of anti‐HOXA11. Sonicated chromatin was incubated with the antibody‐protein A/G beads complex overnight at 4°C with rotation. ChIP DNA fragments and control input DNA fragments were converted to a sequence library by NEB Next Ultra™ DNA Library Prep kit for Illumina (NEB) then analyzed by using a HiSEQ 1500 (Illumina). E12.5 ChIP‐Seq was performed as described previously (Beccari et al., 2016). For data analysis of E12.5 ChIP‐Seq, distal ChIP data and proximal ChIP data were combined. HOXA13 ChIP sequence data were analyzed by Bowtie 2.2.2 (Langmead & Salzberg, 2012) and MACS 2.1.0 (Zhang et al., 2008) (mm9) organized by SraTailor (Oki et al. 2014) under default parameters except 2.15e09 was used for the mouse genome size. For the analysis of HOXA11 ChIP‐Seq by MACS2.1.0 (mm9), the following parameters were used: –nomodel, –extsize 200, ‐g 2.15e09. NGS sequence data of input DNA without ChIP were used as the reference. Overlapping of the ChIP peaks was determined by Bed Tools/ Intersect intervals (Quinlan & Hall, 2010) via Galaxy (Giardine et al., 2005). ComputeMatrix/NGS: DeepTools and plotHeatmap/NGS: DeepTools (Ramirez et al., 2016) in Galaxy were used for creating the aggregate plot. To analyze the sequence conservation around the HOX ChIP summit, the bed file in mm9 format was first converted to mm10 format using the LiftOver tool of the USCE genome browser (Karolchik et al., 2004) then analyzed with the same method used to prepare the aggregate plot using mm10.60way.phastCons.bw data obtained from the UCSC genome browser. For the analysis of sequence motif enrichment, the genomic spans given in BED format were first converted to FASTA files using UCSC Table Browser then analyzed by MIME‐ChIP (Machanick & Bailey, 2011). ChIP‐Seq data have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE119142.
2.3. Data analysis
Integrative genome viewer, IGV_2.4.3 (Robinson et al., 2011), was used for graphical analysis of the ChIP‐Seq data. Data analysis was also performed using the online application provided by CEAS (Shin, Liu, Manrai, & Liu, 2009) for positioning the ChIP DNA fragments relative to the gene structure, David ver. 6.8 (Huang da, Sherman, & Lempicki, 2009) and Panther ver. 13.1 (Mi, Muruganujan, & Thomas, 2013) for enrichment analysis and classification of the genes, Venny (http://bioinfogp.cnb.csic.es/tools/venny/) for listing common genes in different gene pools, or GREAT (McLean et al., 2010) through the UCSC Genome Browser for identification of the genes neighboring the ChIP peak using a default parameter. Data sets obtained from the public databases are shown in Supporting Information Table S6.
2.4. Genechip
Embryos isolated approximately E11.25 were preserved in RNA later (Ambion) and the autopodal tissue corresponding to the Hoxd13 expressing region was dissected. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) and biotin labeled RNA was prepared using the IVT Labeling Kit (Affymetrix), One‐Cycle cDNA Synthesis Kit (Affymetrix) and Sample Cleanup Modules (Affymetrix). The GeneChip Mouse Genome 430 2.0 Array (Affymetrix) were hybridized, washed, stained and scanned by using standard Affymetrix GeneChip reagents and protocols. Data from two independent experiments were analyzed using the Affymetrix Transcriptome Analysis Console V3.0. Genechip data have been deposited in the GEO with accession number GSE118640. Genes exhibiting more than a 1.5‐fold change in the expression (p < 0.05) were defined as the HOX13 downstream genes.
2.5. In situ hybridization
Whole‐mount in situ hybridization (WISH) was performed as previously described (Yamamoto‐Shiraishi & Kuroiwa, 2013). Digoxigenin (DIG) or fluorescein‐labeled riboprobes were synthesized using the template cDNA fragments amplified with the primer sets listed in Supporting Information Table S5e directly or after cloning into plasmids. Template cDNA clone for the mouse Hoxd13 probe was provided by Dr. D. Duboule. Generation of the Bmp2 probe was described previously (Dickinson et al., 1990). Samples were photographed with a Nikon DXM1200F camera system, and the background color was adjusted to obtain best contrast to the signals in each sample. For comparison of the signal patterns between limbs, some limb photographs were flipped horizontally using Adobe Photoshop CS6.
2.6. Electrophoresis mobility shift assay (EMSA)
The protein coding sequence containing homeobox in the second exon was PCR amplified using the primers presented in Supporting Information Table S5a and connected to the sequence encoding the Maltose binding protein (MBP) in pMal‐c (NEB). Protein production and affinity purification were performed according to the manufacturer's instructions (NEB). For preparing the fluorescently labeled probes, aminoaryl labeled double strand DNA was prepared by two‐step PCR. The first round of PCR was done using mouse genomic DNA template and RVL‐GSP1 and GSP2 primers. This PCR product was then used as the template for the second round of PCR using aminoaryl‐RVL and GSP2 as primers. The sequence of each primer is shown in Supporting Information Table S5b. The aminoaryl‐labeled DNA fragments were reacted with Alexa Fluor 700 (AF700) NHS ester (Thermo Fisher) in borate buffer then purified using the Qiagen MinElute column (Qiagen). The sequences of the unlabeled competitors are presented in Supporting Information Table S5c. For the EMSA reaction (25 μl), 0.5 pmoles of AF700 labeled probe was mixed with MBP or MBP‐mHOXA11 (or13) HD protein in buffer containing 1 μg poly (dIdC), 20 mM TrisHCl pH 7.5, 150 mM KCl, 0.1% Triton X‐100, 1 mM EDTA, 1 mM DTT and 5% glycerol for 30 min at 25°C. After adding 1 μl of 0.05% bromophenolblue solution, the reaction was analyzed by 2% low melting point agarose gel electrophoresis at 4°C using 0.5× TBE as the running buffer. The gel was analyzed with an Odyssey scanner (LI‐CORE). For competition experiments, 2.5 pmoles or 25 pmoles of the oligo nucleotides were added to the reaction.
2.7. Deletion of specific chromosomal regions
Mice with deletion of the putative enhancer of Tshz2 or Bmp2 were generated using the CRISPR/Cas9‐based genome editing system via electroporation (Hashimoto, Yamashita, & Takemoto, 2016). The guide RNA sequences and deleted regions are presented in Supporting Information Table S5d.
2.8. Micromass culture and transfection
For micromass culture (MMC), mesenchymal cells of the chicken distal limb bud were prepared as previously described (Yamamoto‐Shiraishi, Higuchi, Yamamoto, Hirano, & Kuroiwa, 2014), except Dispase I (WAKO) was used instead of trypsin (final concentration of dispase: 2,000 U/ml). For transfection of MMC, 0.9 μl FuGENE HD (Promega) was mixed with 4.4 μl F‐12 and 300 ng plasmid DNA (containing 100 ng pCAGGS‐H2B‐EGFP, and 200 ng pCAGGS, pCAGGS‐mouse Bcl11a, or pCAGGS‐mouse Bcl11b, respectively). After incubation for 20 min, 13.2 μl cell suspension (2.5 × 107 cells/ml) containing 2% FBS was added, and cells were seeded onto a glass base dish (ø = 3.5 cm). After incubation in 5% CO2 at 37°C for 2 hr, 10% FBS/F‐12 was added (1 ml/dish). Medium was changed every day. After the incubation for 1 or 2 days, MMCs were fixed with 4% PFA/0.1% Tween20/PBS. Anti‐Collagen II Ab‐2, mouse monoclonal Ab (NeoMarkers, MS‐235; dilution: 1/200) and goat anti‐mouse IgG (H+L), Alexa Fluor 594 (Thermo Fisher, A‐11032; dilution: 1/300) were used as primary and secondary antibodies, respectively. Nuclear staining was performed with DAPI, and MMCs were photographed using a M205FA (Leica) and FV1000 (Olympus). Col II immunofluorescence signal intensity around each nucleus was calculated using Image J. cDNA fragments encoding the mouse Bcl11a and Bcl11b protein coding sequence were amplified from reverse transcripts of E11.5 limb bud RNA using primer sets presented in Supporting Information Table S5f and were cloned into the pCAGGS vector.
3. RESULTS
3.1. Hoxa11 and Hoxa13 have distinctive expression domains in the limb bud
Hoxa11 and Hoxa13 show unique expression pattern along the proximodistal axis in the mesenchyme of the E11.75 mouse forelimb bud (Figure 1a,b). At this stage of limb development, formation of the zeugopodal cartilage anlage is complete and the carpal and metacarpal precartilaginous condensations are already visible in a continuous anlage of the autopodal region (Figure 1e). Correlations between the Hox expression domain and the cartilage patterning were examined by double staining for the expression of Hox and the cartilage marker Col2a1 (Figure 1c,d). As shown in Figure 1c,f, Hoxa11 is expressed in the distal zeugopodal region and proximal autopodal region spanning most of the carpal cartilaginous condensation. In contrast, Hoxa13 is expressed throughout the entire autopodal region (Figure 1d,g). Thus, expression of Hoxa11 and Hoxa13 covers different domains along the proximodistal axis and overlaps around their expression boundary corresponding to the carpal cartilage anlage (Figure 1h). This same topological correlation between Hox expression and cartilage patterning was observed in the E11.75 hindlimb bud (Supporting Information Figure S1A–G).
Figure 1.
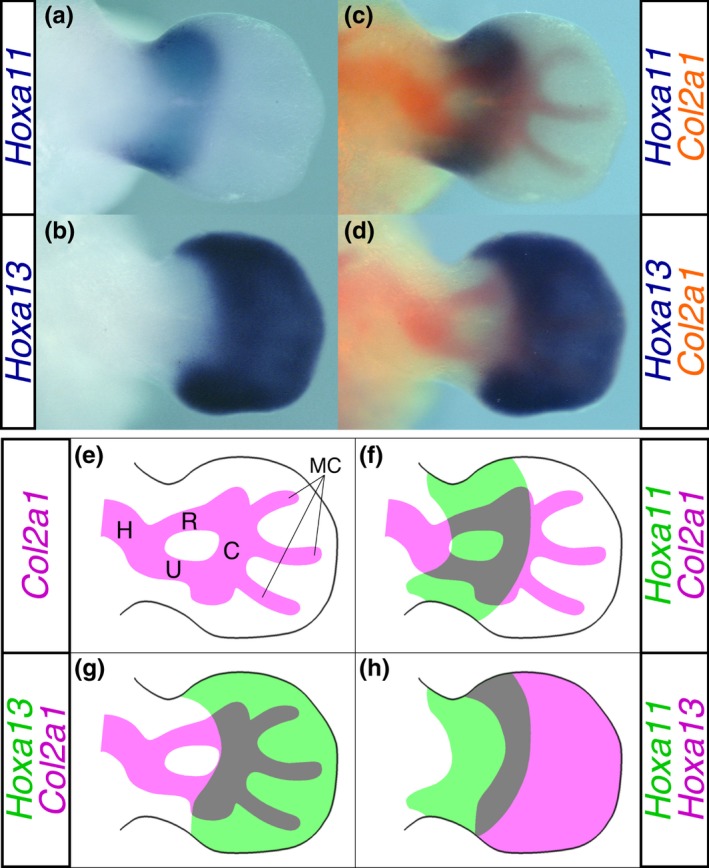
Correlation between the Hox expression domain and cartilage in the E11.75 mouse forelimb bud. Dorsal view of in situ hybridizations for (a) Hoxa11 and (b) Hoxa13. The cartilage pattern was visualized based on the Col2a1 expression (red) in the same limb bud (c and d). (b) and (d) are presented as a digitally inverted (left‐right) views of the original picture. (e) Illustration of the cartilage pattern shown in (c). C: carpus, H: humerus, MC: metacarpus R: radius, U: ulna. (f) and (g) Representative drawings of (c) and (d), respectively. (h) Superimposed view of the Hoxa11 and Hoxa13 expression domains. Images redrawn based on relative location of each Hox expression domain to the cartilage pattern shown in (f) and (g). The left and right limb buds were isolated from a single embryo at E11.75 and were hybridized with DIG‐labeled Hoxa11 and fluorescein‐labeled Col2a1 probe mixture and DIG‐labeled Hoxa13 and fluorescein‐labeled Col2a1 probe mixture, respectively. Hox expression was detected using anti‐DIG IgG‐alkaline phosphatase (AP) with BM Purple as the substrate (blue). After inactivating AP activity, Col2a1 expression was detected using anti‐fluorescein IgG‐AP with Fast Red as the substrate (red)
3.2. The HOXA11 and HOXA13 DNA binding regions overlap in the limb bud
To assess the zeugopodal and autopodal Hox target genes during the cartilage formation at E11.5 when metacarpal/metatarsal anlagen just begins to protrude from carpal/tarsal anlage, we carried out ChIP‐Seq analysis and identified the DNA regions associated with HOXA11 or HOXA13 proteins. The number of independent HOXA11 and HOXA13 ChIP peaks was 2,953 and 143,460, respectively (Supporting Information Table S1a). The number of HOXA11 peak is comparable to that of recently reported HOXC10 ChIP‐Seq peak in the E11.5 limb bud (Jain et al., 2018). Figure 2a–c and Supporting Information Figure S2A–C show the integrative genome viewer (IGV) representation of HOXA11, HOXA13 and other homeodomain transcription factor binding sites in the representative gene loci.
Figure 2.
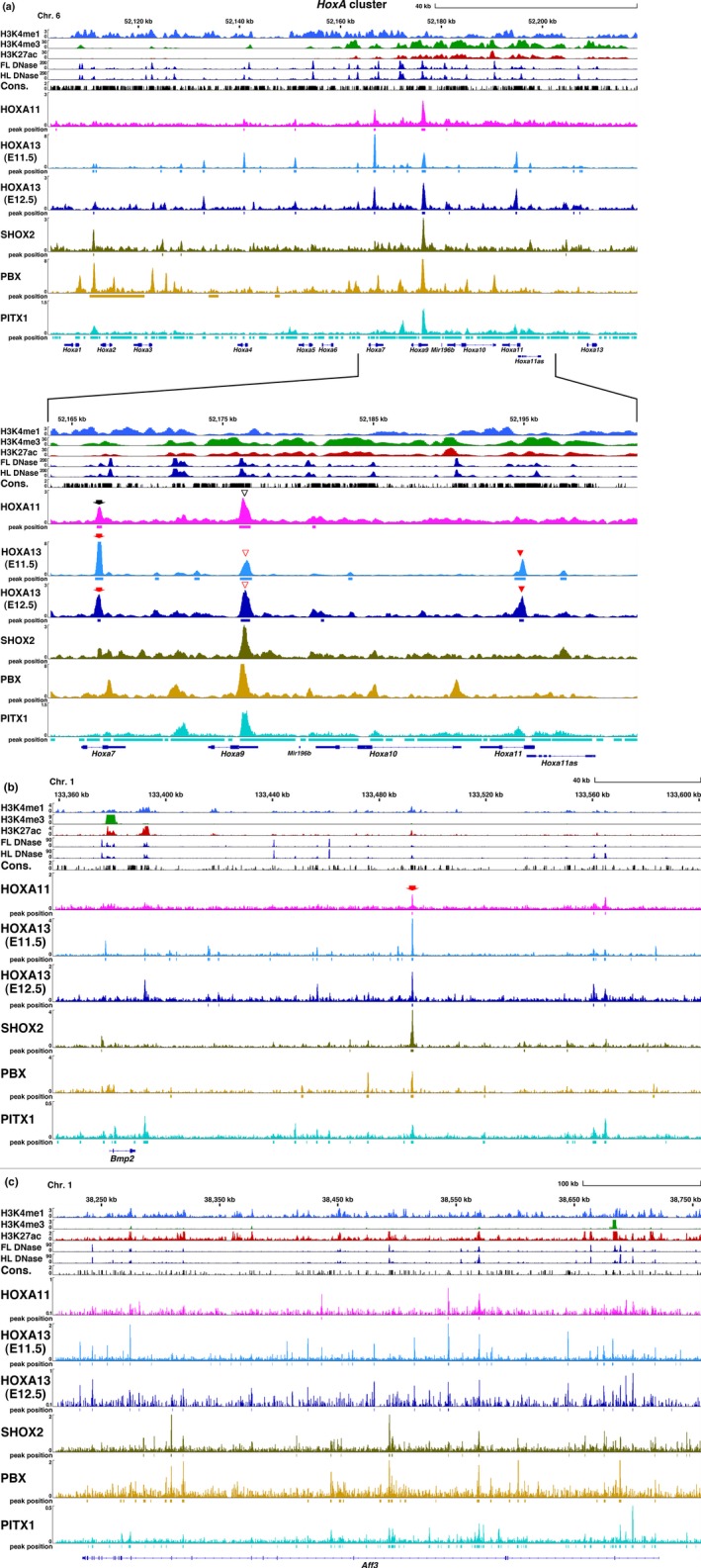
IGV view of HOX11/13 and homeodomain transcription factors in the limb bud. (a) HoxA cluster, (b) Bmp2, (c) Aff3. The epigenetic state of each gene determined by enrichment of H3K4me1, H3K4me3 and H3K27ac in the limb buds and DNase I hypersensitivity sites in the E11.5 limb bud (FL DNase, HL DNase, FL: forelimb bud, HL: hindlimb bud) and sequence conservation among vertebrates (Phastcons [Vertebrate 30way]) presented as a bar graph. Data were obtained from public data bases as described in Supporting Information Table S6. ChIP‐Seq data for HOXA11 and HOXA13 using E11.5 limb bud (HOXA11 and HOXA13(E11.5)) and E12.5 autopod (HOXA13(E12.5)) from this study are presented as a bar graph. The SHOX2 and PBX binding profiles in the E12.5 limb bud and the PITX1 binding profile in the E11.5 hindlimb bud were visualized by reanalysis of published data source presented in Supporting Information Table S6 using the same platform as for the HOX11/13 data. Numbers in the vertical axis indicate coverage values that were scaled by 1,000,000/total count. Squares beneath the peak indicate the region identified as transcription factor binding by MACS2 analysis. (a) Arrow: intron of Hoxa7, open arrowhead: 5′ UTR of Hoxa9, arrowhead: intron of Hoxa11. (b) Arrow: the CHBRL deleted in Bmp2cis KO in Figure 5
In the HoxA cluster, HOXA13 binding regions were present in the intron of Hoxa7 (arrow), the 3′UTR of Hoxa9 (open arrowhead) and the intron of Hoxa11 (arrowhead) (Figure 2a). The Hoxa11 intronic binding site is crucial for HOXA13‐dependent repression of Hoxa11 in the autopod (Kherdjemil et al., 2016). In contrast to the binding of both HOXA11 and HOXA13 to the Hoxa7 and Hoxa9 regions, only HOXA13 binding was observed in the Hoxa11 intron. This indicates different contributions of HOXA11 and HOXA13 to the control of Hoxa11 expression.
As shown in the IGV for other representative gene loci (Figure 2b,c and Supporting Information Figure S2A–C), multiple HOX11/13 binding sites are present and many of the HOXA11 and HOXA13 binding sites overlap with each other. To further analyze this overlap, we determined the accumulation of ChIP‐Seq reads around the peak summit. As shown in Figure 3a, significant accumulation of the HOXA13 ChIP‐Seq signal was found within 200 bp of the HOXA11 ChIP peak and vice versa (Figure 3b). We selected regions that showed the summit of each peak located within 1 kb. Of these colocalized sites, 75% of the ChIP‐peak summits were located within 200 bp of each other (Figure 3c). The results that the summits are in close proximity and that both HOXA11 and HOXA13 bind the same target sequence in vitro (Shen, Rozenfeld, Lawrence, & Largman, 1997), suggest that they bind the same target in vivo. We further selected regions that exhibited directly overlapping peak spans and found that 2,139 HOXA11 binding regions of their expression domain (distal zeugopod and proximal autopod) overlapped with the autopodal HOXA13 peak. These overlapping peaks are 72% of the total HOXA11 peak (Supporting Information Table S1a) indicating that a considerable portion of the HOXA11 binding regions is common to the HOXA13 binding region. We subsequently refer to this common region as the CHBRL (Common Hox binding region of the limb bud). Significant accumulation of both HOXA11 and HOXA13 ChIP‐Seq reads was confirmed around the CHBRL (Figure 3d).
Figure 3.
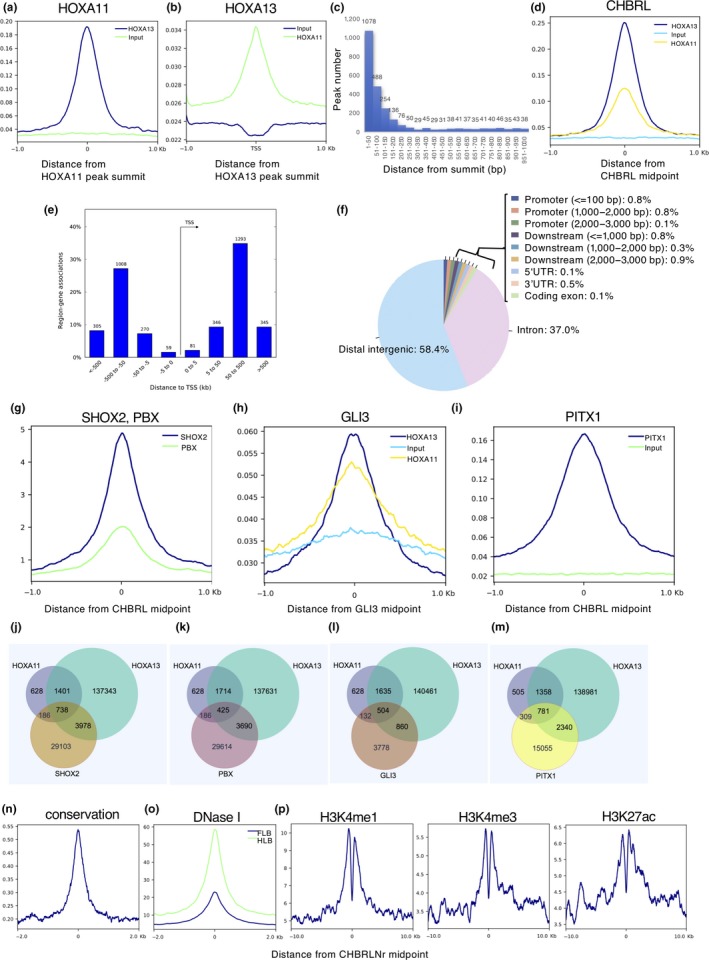
Profile of the limb HOX11/13 DNA binding region. Condensed profiles of ChIP‐Seq signals of (a) HOXA13 for HOXA11 peak summits or (b) HOXA11 for HOXA13 peak summit. (c) Distribution plot for distances between HOXA11 and HOXA13 peak summits located within 1 kb each other. (d) Condensed profiles of ChIP‐Seq signals for the CHBRL. (e) The majority of the CHBRL peaks are located a great distance from their nearest transcription starting site (TSS). (f) The majority of CHBRL peaks are located at the intronic or distal intergenic region of the chromosome. (g) Aggregate plots of SHOX2 and PBX binding signals in the embryonic limb at E11.5 centered around the CHBRL. (h) Aggregate plots of HOXA11and HOXA13 binding signals in the embryonic limb at E11.5 centered around the summits of Gli3 binding region. (i) Aggregate plots of PITX1 binding signals in the embryonic limb at E11.5 around centered around the CHBRL. The number of the overlapping ChIP‐Seq peaks of HOXA11 and HOXA13 with (j) SHOX2, (k) PBX, (l) GLI3 and (m) PITX1. (n) Aggregate plots based on average phastcons [60way.phastCons] across vertebrates indicate that the majority of the CHBRLNrs are highly conserved. (o) Aggregate plots on DNase I hyper‐sensitive sites in embryonic fore‐(FLB) and hindlimb (HLB) at E11.5 around the center of the CHBRLNr. (p) Aggregate plots of H3Kme1, H3K4me3 and H3K27ac binding signals in the embryonic limb at E11.5 around the center of the CHBRLNr. Data source was presented in Supporting Information Table S6. Numbers in the vertical axis of a, b, d, g–i and n–p indicate average signal per base (see Materials and Methods)
As shown in Figure 3e, the CHBRL was located a long distance (50–500 kb) from the transcription start site (TSS) and most of the CHBRL was located within the intronic or intergenic region (Figure 3f). We also analyzed HOXA13 binding in the E11.0 and E12.5 limb bud and found a considerable portion of the CHBRL (E11.5) was common to both E11.0 (798/2,139, 37%) and E12.5 (1,618/2,139, 76%) HOXA13 ChIP peaks (Supporting Information Table S1b).
3.3. The CHBRL overlaps with limb SHOX2, PBX, GLI3 or PITX1 binding regions
The homeodomain transcription factors SHOX2 and PBX are also expressed in the limb bud mesenchyme and have important roles in limb cartilage development (Cobb, Dierich, Huss‐Garcia, & Duboule, 2006; Selleri et al., 2001). In addition, as shown in Figure 2a–c and Supporting Information Figure S2A–C, some HOX binding regions displayed overlap with the SHOX2 and/or PBX binding peaks suggesting that they may share common targets with HOX11/13. We thus analyzed the overlap between the HOX11/13 binding region and the reported SHOX2, or PBX binding region (Ye et al., 2016). Reported ChIP‐Seq reads for both SHOX2 and PBX significantly accumulated around the CHBRL (Figure 3g). As shown in Figure 3j,k, Supporting Information Table S1b, and Supporting Information Figure S2, a substantial number of the HOX11/13 binding regions overlapped with the SHOX2 and/or PBX binding regions, although they exhibited variation in their combinations. The HOX11/13 binding region of Bmp2 (Figure 2b), Tshz2, Jag1 and Shox2 (Supporting Information Figure S2A–C, respectively) overlapped with the SHOX2 binding regions; however, the major HOX11/13 binding region in the HoxA cluster did not show overlap with SHOX2 binding (Figure 2a). In contrast, some of the HOX11/13 binding regions in Aff3 overlapped with the SHOX2 binding regions, although other HOX11/13 binding regions in Aff3 did not show SHOX2 binding (Figure 2c). HOX and SHOX2 share a common binding core sequence in vitro suggesting they may also share common binding sites in vivo or they bind to different sites in the CHBRLs, as multiple HOX11/13 binding sites are demonstrated in Figure 4. On the other hand, our results also indicate variation in the overall binding repertoire of each homeodomain transcription factor.
Figure 4.
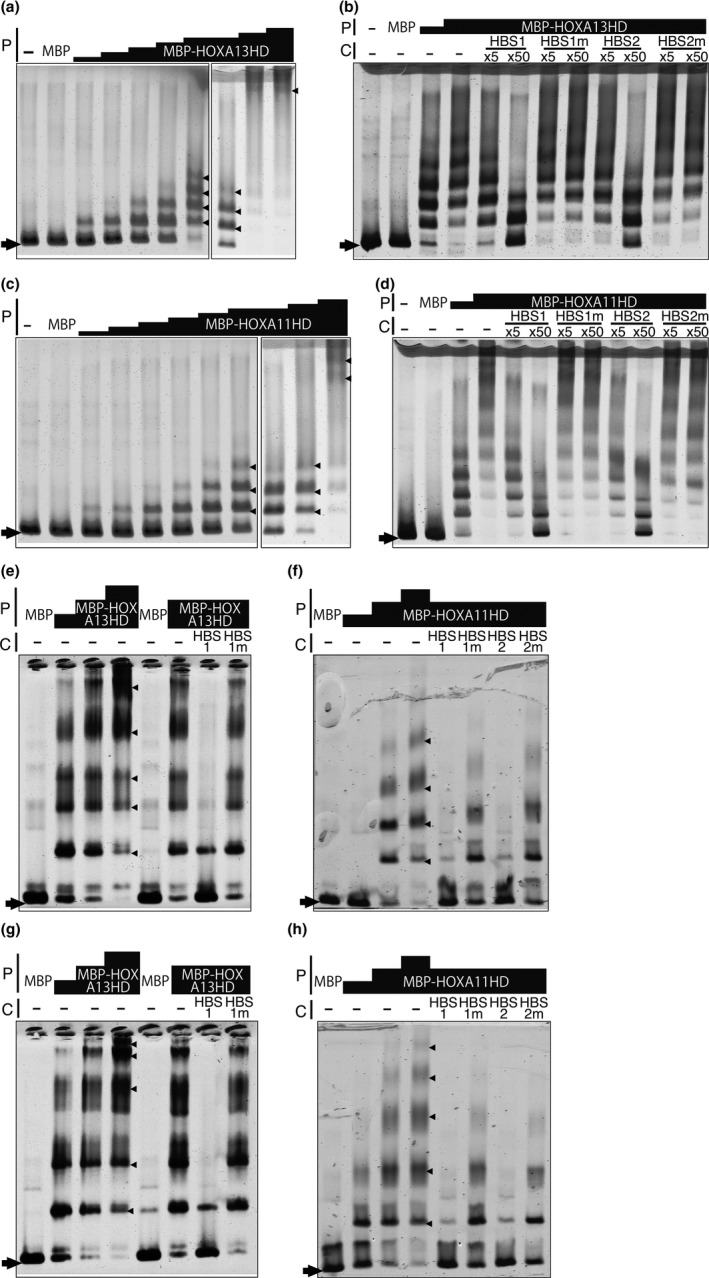
HOXA11 and HOXA13 bind to multiple HBSs in the CHBRL in vitro. EMSAs using 741b, a part of the Shox2 CHBRL/m741 enhancer (VISTA), as a probe. (a) Multiple shift bands emerged in a HOX protein dose‐responsive manner; (−): no protein; MBP: 153 ng of MBP was added to the reaction. The amount of MBP‐HOXA13HD used was 7.6, 15.2, 42, 85, 152, 152, 306 and 612 ng. (b) Band shift was competed by HBS‐containing oligonucleotides HBS1 (TTAT) and HBS2 (TAAT) but not by their mutant forms (T to G and A to C). 153 ng of MBP and 128 or 255 ng of MBP‐HOXA13HD were used. 5‐fold or 50‐fold excess molar unlabeled competitor was added to the reaction. (c) Multiple shift bands emerged in a HOX11/13 protein dose‐responsive manner; (−): no protein, MBP; 153 ng of MBP was added to the reaction. The amount of MBP‐HOXA11HD added was 1.4, 2.8, 5.7, 11.4, 28, 57, 57, 114, 228 ng. (d) Band shift was competed by HBS containing oligonucleotides HBS1 (TTAT) and HBS2 (TAAT) but not by their mutant forms (T to G and A to C). 153 ng of MBP and 95 or 189 ng of MBP‐HOXA11HD were used. 5‐fold or 50‐fold molar excess unlabeled competitor was added to the reaction. (e–h) Multiple shift bands emerged in the HOX11/13 protein in a dose‐dependent manner and were specifically competed by HBS‐containing oligonucleotides. (e, g) MBP‐HOXA13HD and (f, h) MBP‐HOXA11HD. (e) and (f) Aff3 probe, (g) and (h) Bmp2 probe. The sequence of the probe and conserved HBS are shown in Supporting Information Figure S5I, J. Arrowhead and arrow indicate the shift band and free probe, respectively. C: competitor, P: protein.
The transcription factor GLI3 also has a crucial role in limb morphogenesis as the target of the Shh signaling pathway (Buscher, Bosse, Heymer, & Ruther, 1997). Because GLI3 cooperates with 5′ Hox genes in cartilage pattern formation of the limb (Chen et al., 2004; Zakany, Zacchetti, & Duboule, 2007), we compared the CHBRL with the GLI3 binding regions in the limb bud (Vokes, Ji, Wong, & McMahon, 2008). The ChIP‐Seq reads showed modest accumulation around the GLI3 ChIP peak (Figure 3h). Interestingly, 24% (504/2,139; Figure 3l and Supporting Information Table S1b) of the CHBRL overlapped with the GLI3 binding region suggesting that functional cooperation may occur directly through co‐occupation of HOX11/13 and GLI3 in the regulatory element of their target genes (Supporting Information Table S2). The paired‐type homeodomain transcription factor PITX1 is also involved in limb cartilage pattern formation (Szeto et al., 1999). As shown in Figure 2a–c, many PITX1 ChIP‐Seq peaks overlapped with HOX11/13 peaks and PITX1 ChIP‐Seq reads significantly accumulated around the CHBRL (Figure 3i). We found that the 59% of the CHBRL overlapped with the PITX1 binding region (Nemec et al., 2017) (Figure 3m and Supporting Information Table S1b).
3.4. The CHBRL correlates with limb‐specific chromatin epigenetic states
We further analyzed the profile of the CHBRL in relation to the epigenetic state of the limb bud chromatin. After eliminating the regions containing repetitive sequences from the CHBRL pool, most of the remaining regions, the CHBRLNr (CHBRL with no repetitive sequences, 1,518/2,139, 71%; Supporting Information Table S1b) exhibited significant overlap with conserved sequence shared among vertebrates (1,506/1,518, 99%; Figure 3n); a representative comparison is shown in Supporting Information Figures S4 and S5I–L. The DNase I hypersensitive region of the limb bud was significantly localized around the CHBRLNr (Figure 3o). Of the CHBRLNr, 73% (1,107/1,518; Supporting Information Table S1b) showed overlap with the DNase I hypersensitivity region in the E11.5 limb bud (Yue et al., 2014).
To further define the chromatin epigenetic state around the CHBRLNr, we evaluated the histone modification profiles. As shown in Figure 3p, H3K4me1, H3K4me3 and H3K27ac were enriched around the CHBRL. Representative overlapping of the HOXA11 or HOXA13 peaks with the modified histone region is also shown in Figure 2a–c and Supporting Information Figure S2A–C. Interestingly, 70% (1,063/1,518; Supporting Information Table S1b) of the conserved CHBRLNr overlapped with the region enriched for H3K27ac suggesting that a significant part of the CHBRLNr participates in potential enhancer function in the limb bud. A deep trough in the middle of the histone modification peaks shown in Figure 3p suggests that CHBRLNr corresponds to a nucleosome‐free (depleted) region. This is supported by the fact that DNase I hyper‐sensitive sites are enriched around CHBRLNr shown Figure 3o. By matching with the limb enhancer regions provided by the VISTA program (Visel, Minovitsky, Dubchak, & Pennacchio, 2007), 57 regions out of 334 regions in the VISTA limb enhancer were found to correspond with the CHBRLNr (Supporting Information Table S1b) including the Shox2 enhancer (Osterwalder et al., 2018; Rosin, Abassah‐Oppong, & Cobb, 2013) located in the intron of the neighboring gene Rsrc1 (Supporting Information Figure S2C arrow). In addition, the CHBRLNr shown in Figure 2b (arrow) was also included in the region possessing the distal limb bud enhancer of Bmp2 (Dathe et al., 2009). These results indicate that a particular member of the CHBRLNr actually possesses the limb‐specific enhancer function.
3.5. Multiple HBSs were enriched in the HOX11/13 ChIP fragments
ABD‐B type HOX proteins bind to both the TTAT/C and TAAT/C motifs (HOX binding sequence: HBS) in vitro (Shen et al., 1997). We thus analyzed the enrichment of these sequences in the HOXA11 and HOXA13 ChIP fragment and the CHBRLNr. As shown in Supporting Information Figure S3, the sequences containing TTAT or TAAT were highly enriched in the HOX11/13 ChIP fragment indicating that HOX11/13 proteins directly bind to the majority of the ChIP fragment in vivo. Interestingly, as shown in representative results in Supporting Information Figures S4B,C and S5I–L, evolutionarily conserved multiple HBSs were found in most of the ChIP DNA.
3.6. HOX11/13 protein directly binds to the evolutionarily conserved HBSs in the ChIP DNA
As described above, the nucleotide sequences encoded by most HOX11/13 ChIP fragments showed evolutionary conservation. Among them, the HOX protein binding consensus core sequence, HBS, exhibited a high level of conservation (Supporting Information Figures S4B,C and S5I–L). We then analyzed direct HOX11/13 protein binding to typical CHBRLs in vitro by EMSA.
One of the CHBRLs in the Rsrc1 intron (Supporting Information Figure S2C, arrow), m741 (VISTA), carries the limb‐mesenchyme specific enhancer function of the neighboring Shox2 gene (Osterwalder et al., 2018; Rosin et al., 2013). m741 contains highly conserved multiple HBSs, which are concentrated in the two sub‐regions, 741b and 741c (Supporting Information Figure S4). As shown in Figure 4a,b and Supporting Information Figure S5a,b, in the presence of HOXA13HOMEODOMAIN (HD) both the 741b and 741c probes showed multiple shift bands in a HOXA13HD protein dose‐dependent manner indicating that HOXA13 has multiple binding sites in the probe. These shift bands were competed by oligonucleotides containing HBS1 (TTAT) or HBS2 (TAAT) but not competed by oligonucleotides containing mutated HBS (Figure 4b and Supporting Information Figure S5B). HOXA11HD also showed the same binding profile as HOXA13HD to the 741b and 741c probes (Figure 4c,d and Supporting Information Figure S5C,D). These results indicate that HOX11/13 protein directly binds to the HBSs in the 741b and 741c. The CHBRLs of Bmp2, Jag1 and Tshz2, in addition to one of the Aff3 CHBRLs, also contain multiple HBSs (Supporting Information Figure S5I–L). Both HOXA11HD and HOXA13HD bound these fragments in an HBS‐dependent manner indicating that HOX11/13 proteins directly bind to the conserved HBSs in the CHBRL (Figure 4e–h and Supporting Information Figure S5E–H).
3.7. Chromosomal CHBRLs have limb‐specific enhancer activity
We next examined if the chromosomal CHBRLs are responsible for regulating neighboring gene expression in the limb bud. Considering that multiple enhancers have redundant function (Osterwalder et al., 2018), we selected Bmp2 and Tshz2 as a representative system. Both genes have one or two CHBRLs, so we deleted the most prominent CHBRLs using CRISPR/Cas9 system (cisKO). We generated Bmp2cisKO mice by deleting 1,623 bp‐containing CHBRL located about 100 kb downstream of the Bmp2 coding region (Figure 2b, arrow) and Tshz2cisKO mice by targeting a 1,242 bp chromosomal region in the intron (Supporting Information Figure S2B, arrow). As shown in Figure 5a–f, in the Bmp2CHBRL −/− embryos, Bmp2 expression in the distal mesenchyme was considerably reduced in the forelimb bud and barely detectable in the hindlimb bud. In the Tshz2CHBRL −/− embryos, Tshz2 expression in the first interdigital region was slightly weakened in the forelimb bud and undetectable in the hindlimb bud (Figure 5g–n, arrowheads and black arrows). In addition, weak expression in the anterior autopod was undetectable in both the forelimb and hindlimb buds of the Tshz2CHBRL −/− embryos (Figure 5g,h,j–l,n, yellow arrows). These results indicate that the chromosomal CHBRLs possess the limb mesenchyme enhancer function in the Hox expressing domain.
Figure 5.
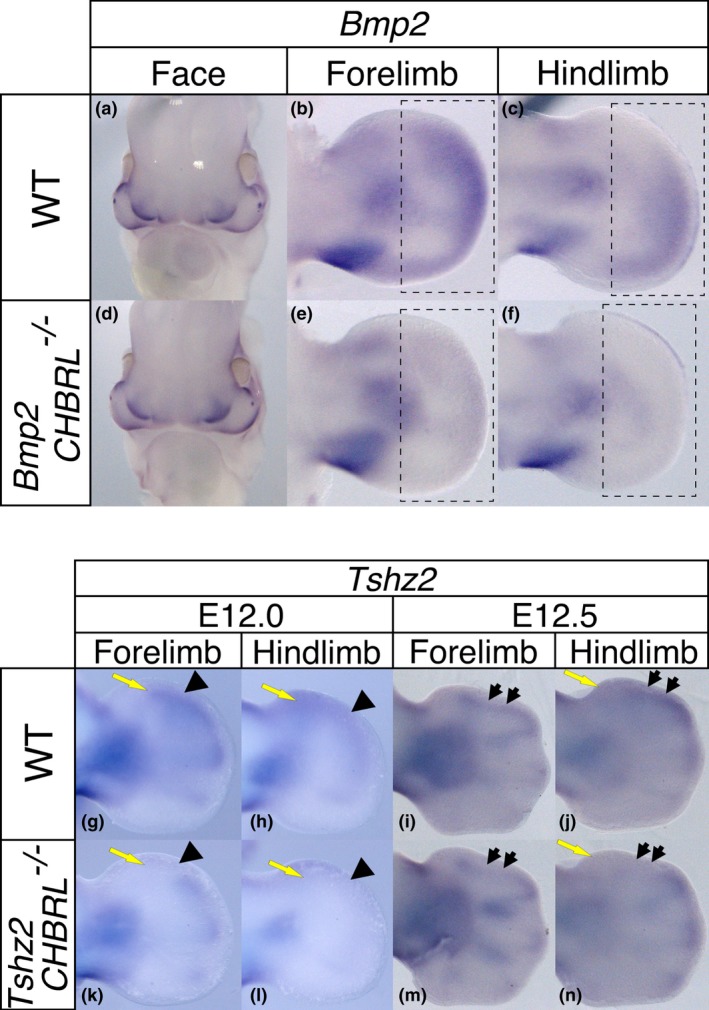
CHBRLs of Bmp2 and Tshz2 have limb bud‐specific enhancer function. Expression pattern of Bmp2 in (a–c) wild‐type and (d–f) Bmp2CHBRL −/− embryos. Facial Bmp2 expression was not altered in the same (a and d) Bmp2CHBRL −/− embryos. The dotted squares indicate the region exhibiting reduced Bmp2 expression in Bmp2CHBRL −/− embryos (compare b and e, c and f). Expression pattern of Tshz2 in (g–j) wild‐type and (k–n) Tshz2CHBRL −/− embryos. The arrowhead and arrow indicate the regions exhibiting reduced Tshz2 expression in Tshz2CHBRL −/− embryos
3.8. Genes neighboring the CHBRLNr show altered expression in Hox11‐13 mutant limbs
We identified 1,556 genes neighboring the CHBRLNr (a11a13 genes, Supporting Information Table S2). Among these, genes related to skeletal development, limb development or cartilage morphology were enriched (Supporting Information Figure S6A). Interestingly, genes related to skeletal development were also enriched in the gene groups whose CHBRLNr overlapped with the SHOX2, PBX1, PITX1 and GLI3 binding regions (Supporting Information Table S2, Figure S6B–E). We also found that most of the genes containing or neighboring the CHBRLNr belong to the same topologically associating domain (TAD) as the CHBRLNr (1,232/1,556 genes, 79%; Supporting Information Tables S2 and S4) indicating that these CHBRLNrs are involved in regulating the expression of flanking genes (Long, Prescott, & Wysocka, 2016). To verify these as the Hox target genes, we analyzed the limb bud expression profile in wild‐type and Hox13 deleted embryos.
Hoxa13 and Hoxd13 show overlapping expression in the autopodal mesenchyme and cooperatively function in autopodal cartilage pattern formation (Fromental‐Ramain et al., 1996; Yokouchi et al., 1991). Hoxd11 and Hoxd12 are also expressed in the autopodal mesenchyme and cooperate in autopodal cartilage pattern formation in a gene dosage‐dependent manner together with Hoxa13 and Hoxd13. Specifically, homozygote with simultaneous deletion of Hoxd11,12,13 (HoxD del(11–13)) exhibited more severe adult cartilage phenotypes than Hoxd13 KO homozygotes (Zakany et al., 1997). To increase the sensitivity in analyzing Hox function, we combined HoxD del(11–13) and Hoxa13KO mice in this study (we abbreviate Hoxa13 −/−; HoxD del(11–13)/del(11–13) as Hox13dKO).
We isolated RNA from the autopod of wild‐type and Hox13dKO embryos and quantified the transcript levels by Genechip (Supporting Information Table S3a,b). To minimize overrepresentation of the genes related to cartilage differentiation caused by loss of the Hox13 function (Supporting Information S7) and to concentrate early responding genes to the loss of Hox13 function, we isolated tissue from E11.25 embryos where only the carpal/tarsal anlage had developed in the autopodal region. In addition, to avoid the effect of stylopod and zeugopod gene expression on changes in their expression in the Hox13dKO autopod, we isolated the distal autopodal tissue that roughly corresponded to the Hoxd13 expressing region.
Of the 781 genes showing upregulation in the E11.25 mutant autopod, 102 genes were included among the CHBRLNr genes (Supporting Information Table S2 and Figure S8A). Interestingly, genes categorized under “transcription regulation” were enriched in this cluster (p = 1.16E‐19, 41/102, Supporting Information Table S3c and Figure S8B). Among the 805 genes showing downregulation in the Hox13dKO autopod, 79 genes overlapped with the CHBRLNr genes (Supporting Information Table S2 and Figure S8A) exhibiting their functional variability (Supporting Information Figure S8C). Interestingly, three genes belong to both classes due to their alternate exon usage. Following these analyses, we referred to any CHBRLNr genes showing alteration in the Hox13dKO as direct Hox target genes. It is curious that many of the genes exhibiting expression changes in the mutant autopod were not included in the CHBRLNr genes. Given that a number of transcription factor‐coding genes are HOX13 target, the genes not included in CHBRLNr gene group are likely to be the downstream of Hox target transcription factors.
3.9. Changes in the spatial expression pattern of HOX11/13 target genes in the Hox11‐13 mutant limb bud
We next analyzed the changes in the spatial expression pattern of HOX11/13 target genes in loss‐of‐function Hox13 mutant limb buds. In Hox13dKO embryos, the cartilage pattern in the limb at E11.25 showed no obvious changes; however, at E12.5 the interdigital region was missing and a single flat cartilage was present instead of the five metacarpi and phalanges (Supporting Information Figure S7) indicating disorganized patterning of the autopodal cartilage. As representative genes exhibiting altered expression in the Hox13dKO limb, we first chose Bmp2, Sulf1 and Stmn2 as downregulated genes, and Aff3, Shox2 and Tshz2 as upregulated genes in the Hox13dKO limb. These genes are also common targets of HOXA11, HOXA13, SHOX2, PBX and PITX1 in the limb bud.
Bmp2 was expressed in the mesenchyme of the distal autopod in the wild‐type limb and while the expression was not altered in the Hoxa13 +/− and Hoxa13 −/− limb, it was reduced in the HoxD del(11–13)/del(11–13) and undetectable in the Hox13dKO limb bud (Figure 6a,b and Supporting Information Figure S9A). Stmn2 was expressed in the mesenchyme of the anterior autopod of E11.5 wild‐type and Hoxa13 +/− forelimb buds. The intensity of the autopodal expression signal was not affected in the Hoxa13 −/−, although it was expanded posteriorly in the HoxD del(11–13)/del(11–13) limb bud. In the Hox13dKO limb bud, Stmn2 expression was undetectable in the autopod (Figure 6c,d and Supporting Information Figure S9B).
Figure 6.
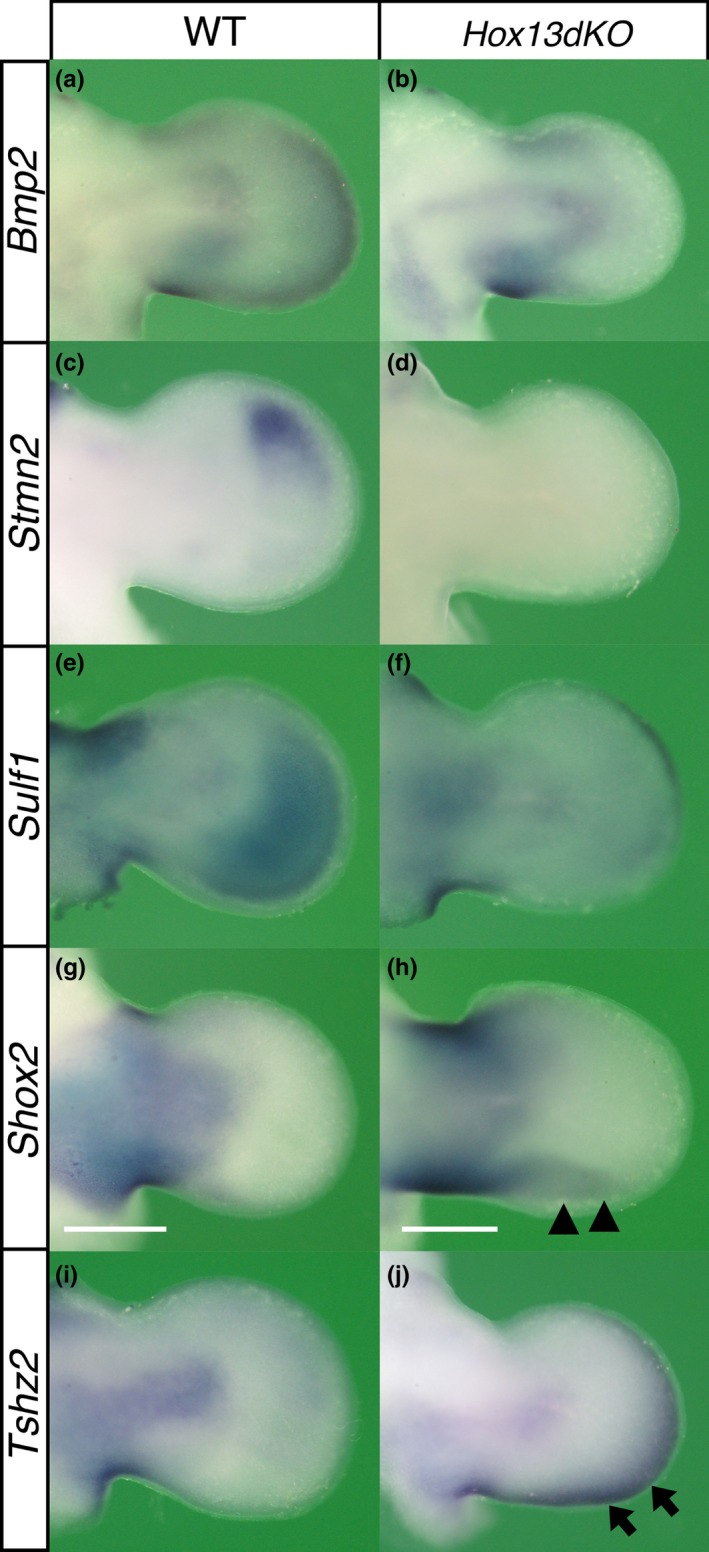
Altered gene expression patterns in the Hox13dKO autopod. Dorsal view of gene expression detected by in situ hybridization in E11.25–11.5 (a, c, e, g, i) wild‐type and (b, d, f, h, j) Hox13dKO forelimb buds. The expression of (a and b) Bmp2, (c and d) Stmn2 and (e and f) Sulf1 in the autopodal mesenchyme was reduced or undetectable in the Hox13dKO embryos. In contrast, (g and h) Shox2 and (i and j) Tshz2 showed ectopic expression in the Hox13dKO autopod. Since Hoxa13 KO mice were generated by inserting Neo sequence in Hoxa13 locus, Neo expression mimics Hoxa13 expression. In order to define the autopod as the region expressing Hoxa13 in the Hox13dKO mice, contralateral limb bud of each specimen was hybridized with Neo probe (results not shown). Scale bar indicates 500 μm
Sulf1 was expressed in the autopodal mesenchyme of E11.5 wild‐type and Hoxa13 +/− forelimbs. The intensity and area of autopodal expression were decreased in both Hoxa13 −/− and HoxD del(11–13)/del(11–13) limbs. The expression signal was further reduced or undetectable in the Hox13dKO autopod (Figure 6e,f and Supporting Information Figure S9C). Sulf1 showed dynamic change in the AER expression (Lewandowski et al., 2015) and this expression was affected indirectly by Hox11‐13 mutation (Figure 6e,f and Supporting Information Figure S9C, see detail in Supporting Information Figure S9 legend).
Aff3, encoding an AF/FMR2 family transcription factor, was expressed in the E11.5 wild‐type zeugopod but not in the autopod. In the E11.5 Hoxa13 −/− limb, Aff3 expression was expanded into the autopod and Aff3 expression was detected in the entire autopodal region of the Hox13dKO limb (Figure 7b,c and Supporting Information Figure S9D, see detail in Supporting Information Figure S9 legend).
Figure 7.
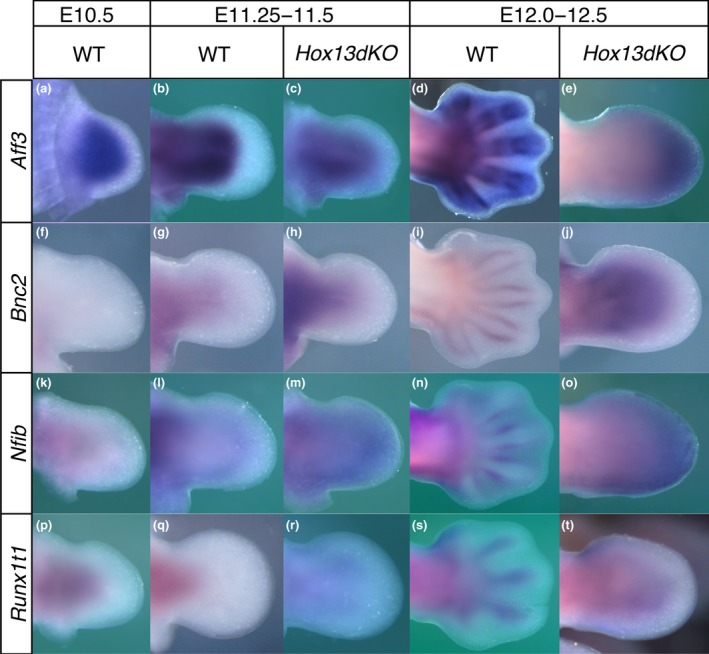
Transition in the spatial expression pattern of the genes encoding Hox target transcription factors associated with cartilage differentiation and changes in the Hox13dKO autopod. Dorsal view of gene expression detected by in situ hybridization in the forelimb bud at (a, f, k, p) approximately E10.5, (b, c, g, h, l, m, q, r) E11.5 and (d, e, i, j, n, o, s, t) E12.5. (a) In wild‐type embryos, Aff3 was expressed throughout the entire limb bud mesenchyme excluding the narrow distal most region at E10.5. Aff3 expression was not found in the autopod at (b) E11.5; however, at (d) E12.5 expression was detected in the cells in/around the perichondrium of the autopod but not in the interdigital mesenchyme. In Hox13dKO embryos, Aff3 expression expanded to the proximal autopodal region (c) at E11.5 then the expression was observed throughout the entire autopodal mesenchyme (e) at E12.5. Bnc2 expression was not detected in the (f) E10.5 mesenchyme but was detected in the center of the stylopod and zeugopod (g) at E11.5. (i) Bnc2 expression was later found in the perichondrium of the limb cartilage including the autopod at E12.5. Bnc2 expression was expanded to the proximal Hox13dKO autopod (h) at E11.5 and throughout the entire mesenchyme (j) at E12.5. Nfib expression was found in the proximal region of the (k) E10.5 limb bud then in the zeugopod/autopod boundary region at (l) E11.5. (n) Nfib expression was detected in the perichondrium of the autopodal cartilage at E12.5. In the Hox13dKO limb bud, expression was detected throughout the entire autopodal mesenchyme both (m) at E11.5 and (o) E12.5. Runx1t1 expression was detected in the proximal center of the (p) E10.5 and (q) E11.5 limb but excluded from the autopodal region. (s) In the E12.5 autopod, Runx1t1 expression was detected in the cartilaginous condensation. In Hox13dKO embryos, Runx1t1 expression was expanded to the proximal autopod at (r) E11.5 and the central region of the autopod showed expression at (t) E12.5
Shox2 was expressed in the E11.5 wild‐type zeugopod but undetectable in the autopod, and the expression was not affected in E11.5 HoxD del(11–13)/del(11–13) and Hoxa13 −/− mice (Supporting Information Figure S9E). In contrast, in the E11.5 Hox13dKO mice, Shox2 expression was expanded into the posterior autopodal region (Figure 6g,h, arrowhead).
Tshz2 is expressed in the E11.5 zeugopodal mesenchyme but not in the autopod (Caubit et al., 2000). Ectopic Tshz2 expression was detected in the distal and posterior periphery of the E11.5 Hox13dKO autopod (Figure 6i,j, arrow). However, this aberrant expression pattern was not observed in Hoxa13 −/− or HoxD del(11–13)/del(11–13) mice (Supporting Information Figure S9F). Thus, genes that exhibited altered expression levels in the Hox13dKO autopod in Genechip analysis actually showed unique changes in the spatial expression pattern in the Hox13dKO limb bud.
We further analyzed the expression of the Hox13 target transcription factors, Aff3, Bnc2, Nfib and Runx1t1 whose expression was increased in the Hox13dKO autopod. Of these, Aff3, Bnc2 and Runx1t1 are common target genes of HOXA11, HOXA13, SHOX2, PBX and PITX1 whereas Nfib is a common target gene of HOXA11, HOXA13 and PITX1 in the limb bud. Aff3, Bnc2, Nfib and Runx1t1 were expressed in the zeugopodal mesenchyme and cartilage but not in the autopod at E11.5 (Figure 7b,g,l,q). Expression of these genes was then also detected in the autopodal cartilage or perichondrium at E12.5 (Figure 7d,i,n,s). In the Hox13dKO embryos at E11.5, the expression of Aff3, Bnc2, Nfib and Runx1t1 were clearly expanded into the autopod (Figure 7c,h,m,r). Thus, representative Hox target genes exhibiting increased transcription in the Hox13dKO by Genechip were confirmed to be ectopically transcribed in the autopodal mesenchyme in the Hox13dKO. The expression of these genes in the zeugopod mesenchyme was already detectable at E10.5 when zeugopodal cartilage formation is just beginning (Figure 7a,k,p) except Bnc2. In contrast, expression of these genes was suspended in the autopod mesenchyme at E11.5. Thus, the relationship between the expression of these genes in the mesenchyme and cartilage is different between the zeugopod and autopod.
Thus, a change in the level of a transcripts of a representative gene in the Hox13dKO autopod shown by Genechip analysis was verified by WISH analysis.
3.10. Changes in the Hox11/13 target gene expression patterns in the Ulnaless limb bud
As shown, autopodal expression of a group of the Hox11/13 target genes such as Aff3, Shox2 and Tshz2 at E11.5 are probably transiently repressed by Hox13. To verify this possibility, we analyzed the effect of Hoxd13 mis‐expression. For this purpose, we adopted Ulnaless (Ul), which is a dominant mutation of the HoxD locus caused by a chromosomal inversion which results in the truncation of both the ulna and fibula due to mis‐expression of Hoxd13 in the posterior zeugopod (Peichel et al., 1997).
In the E11.5 Ul/+ mice, Tshz2 expression in the distal zeugopod was reduced and the expression domain was narrowed at the dorsal zeugopodal region and reduced in the ventral zeugopod (Figure 8a), and Shox2 expression was weakened at the dorsal posterior and ventral region of the zeugopod (Figure 8b). In the E12.5 Ul/+ mice, the Aff3 signal was undetectable specifically in the ventral zeugopodal region (Figure 8c, arrow). Thus, the expression of these genes in the zeugopod was downregulated in the ectopic Hoxd13 domain confirming that Hoxd13 is repressive against the expression of these genes.
Figure 8.
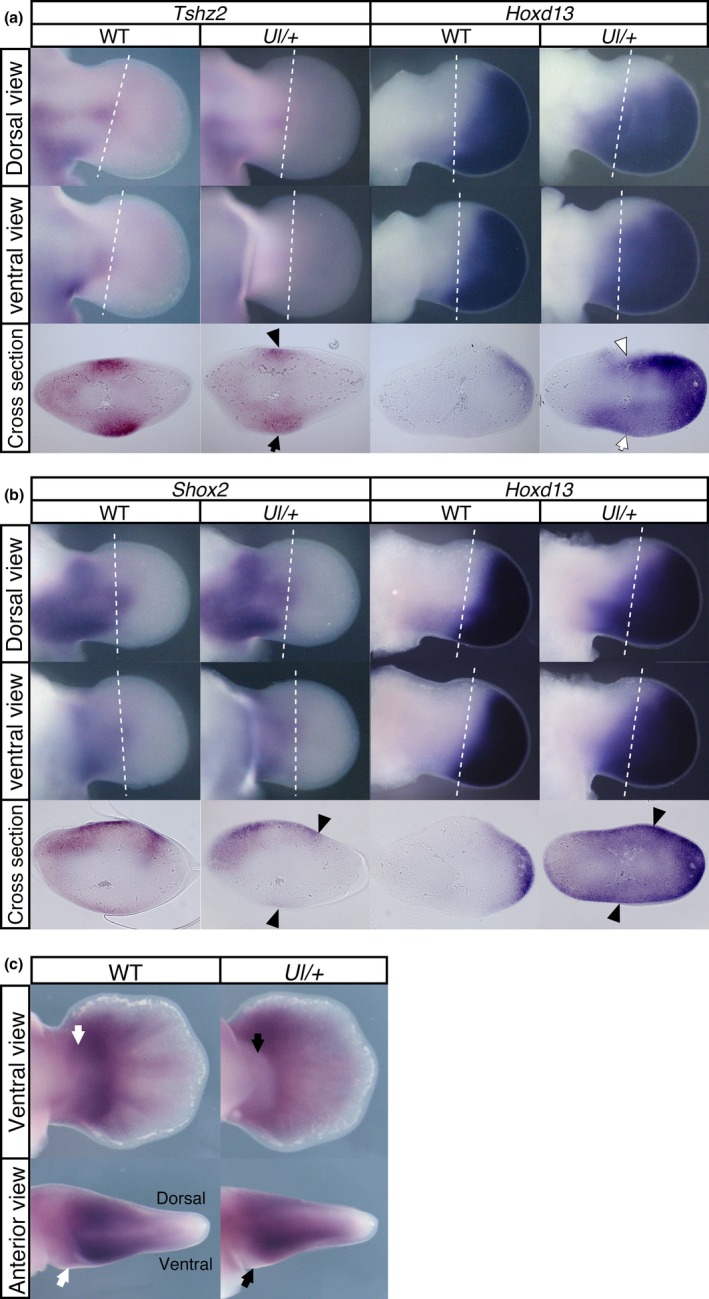
In the Ulnaless limb, ectopic Hoxd13 expression represses target gene expression. Expansion of the Hoxd13 expression domain to more a proximal area overlapping with the zeugopodal region occurred in the Ul/+ mutant limb. Reduction of (a) Tshz2, (b) Shox2 and (c) Aff3 expression in the zeugopod region overlapping the ectopic Hoxd13 expression domain indicates that Hoxd13 as well as Hoxa13 represses the expression of these genes. (a) The black arrow and arrowhead indicate the reduced Tshz2 expression and the white arrow and arrowhead indicate ectopic Hoxd13 expression. (b) Black arrowheads indicate the tissues with reduced expression of Shox2 and augmented Hoxd13 expression. (c) Black arrows indicate the reduction in Aff3 expression in the Ul/+ limb and the white arrows indicate the corresponding region in the wild‐type limb. Dotted lines in panel a and b indicate the position of cross sections. Ventral views were digitally inverted (left‐right). In the panel a and b, the left and right limb buds were isolated from a single embryo at E11.5 and were hybridized with Tshz2 or Shox2 probe and Hoxd13 probe, respectively
3.11. HOXA13 represses cartilage differentiation of limb mesenchymal cells through BCL11a
Bcl11a encodes a zinc finger‐type transcriptional repressor (Liu et al., 2003) whose expression was decreased in the Hox13dKO autopod. BCL11a represses blood cell differentiation (Liu et al., 2003), however its function in cartilage differentiation remains elusive.
Bcl11a was expressed throughout the entire limb mesenchyme at E10.5 (Figure 9a), then at E11.5 the expression was detected in the autopodal mesenchyme and the posterior zeugopodal mesenchyme (Figure 9b). At E12.0 the expression was found in the perichondrium of the autopodal cartilage (Figure 9d) where Hoxa13 was also expressed. As expected from Genechip analysis, expression was severely decreased in the E11.5 autopodal mesenchyme of the Hox13dKO (Figure 9c). Interestingly, Bcl11a expression reappeared in the Hox13dKO autopod at E12.0 where the single flat cartilage was forming instead of the five metacarpi and phalanges (Figure 9e).
Figure 9.
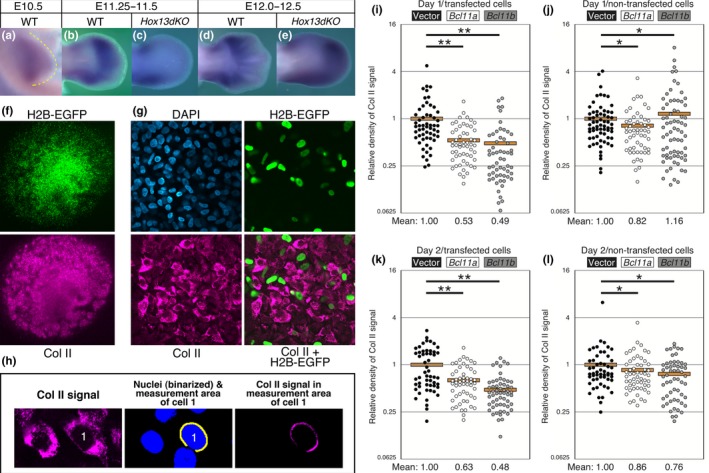
BCL11a and BCL11b repress cartilage differentiation in micromass culture (MMC). (a) Bcl11a was expressed throughout the entire mesenchyme of the limb bud at E10.5 then became restricted to the autopod and anterior and posterior margin of the more proximal part of the limb (b) at E11.5. Bcl11a expression was detected in the interdigital region (d) at E12.0. In the Hox13dKO limb (c) at E11.5, autopodal expression was severely reduced but (e) at E12.5 uniform expression was detected throughout the autopod. (f) Histone H2B‐EGFP (H2B‐EGFP) fluorescence and Col II immunofluorescence signals in MMCs incubated for 2 days after transfection with the pCAGGS‐H2B‐EGFP and pCAGGS vector. (g) High magnification confocal images of the MMC in panel f. Nuclei of the transfected cells were marked with H2B‐EGFP. (h) For quantitative analyses of Col II expression in each cell, the intensity of the Col II immunofluorescence signal in the ring‐shaped area (yellow; width: 0.58 μm) around each nucleus (green) was calculated using ImageJ. Neighboring nuclear areas were excluded from the measurement area. (i–l) Ratios of Col II immunofluorescence signal intensity in each cell relative to the mean of the vector transfection control. i, j: 1‐day incubation; k, l: 2‐day incubation. i, k: transfected cells (H2B‐EGFP‐positive); j, l: non‐transfected cells (H2B‐EGFP‐negative). In the graphs, a circle represents one cell, and n ≈ 60 for each transfection. **p < 0.002; *p > 0.01 (Mann–Whitney U‐test). Orange bars indicate the mean of the values
We then analyzed the effect of Bcl11a overexpression on the differentiation of the limb mesenchymal cells into cartilage using a micromass culture system and monitoring Col II expression as a marker of differentiation (Figure 9f–h). We also analyzed the effect of another family gene, Bcl11b, that is expressed in the limb mesenchyme (result not shown) and has both HOXA11 and HOXA13 binding sites (Supporting Information Table S2). As shown in Figure 9i,j, the expression level of Col II was reduced to 53% and 49% in the Bcl11a and Bcl11b transfected cells, respectively, compared to vector‐transfected cells at day 1. At day 2, the expression of Col II in Bcl11a and Bcl11b transfected cells was also reduced to 63% and 48%, respectively (Figure 9k,l). Thus, similar to blood cell differentiation, both BCL11a and BCL11b have repressive activity on the cartilage differentiation of mesenchymal cells of the limb bud. Taken together, HOXA13 represses limb cartilage differentiation at E11.5.
4. DISCUSSION
To elucidate the mechanism controlling region‐specific morphogenesis of the zeugopod and autopod, we identified direct target genes of HOXA11 and HOXA13 in the limb bud. We found that HOXA11and HOXA13 regulate their common target genes involved in cartilage differentiation in distinct way, where HOXA13‐dependent regulation appears to be central to generating pentadactylous (five digits) autopod.
ChIP‐Seq analysis is a technique for identification of Hox target genes. Using the same HOXA13 antibody as in this study, we have shown that HOXA13 regulates autopod‐specific expression of Hoxd13 through direct binding to the digit‐specific enhancer (Beccari et al., 2016). Sheth et al. (Sheth et al., 2016) identified the limb target genes of HOXA13 and HOXD13, whose function is redundant in the autopodal development. They reported most of the target genes are common to both HOXA13 and HOXD13 and many of them are related to cartilage differentiation. Further extraction and elucidation of the crucial genes from these many candidate genes are current subjects. Jerkovic et al. (2017) attempted to identify the Hox target genes by ChIP‐Seq analysis after forced expression of paralogous Hox genes in cultured chicken limb bud mesenchymal cells. They found that common HOX binding site and a group of HOX proteins interact with CTCF. Since this analysis was performed using cultured cells and overexpression of tagged HOX protein, the results need to be verified by using a more native system. A loss‐of‐function phenotype for Hox11 was observed in the zeugopod and proximal autopod, and for Hox13 in the autopod. In both cases, hypomorphic cartilage development was observed indicating the presence of common target genes responsible for chondrogenic differentiation from limb bud mesenchymal cells. In this study, we used mesenchymal cells from mouse limb bud and antibodies against HOXA11 or HOXA13 for our ChIP‐Seq analysis to narrow down the common Hox target genes that are critical for chondrogenic differentiation. Based on the analysis of these target genes, we revealed the unique role of Hox13 in the autopod‐specific gene regulation network.
4.1. HOX11/13, SHOX2 and PITX1 share common targets
We showed that most of the HOXA11 ChIP‐Seq peaks overlapped with the HOXA13 peaks (Figures 2 and 3 and Supporting Information Figure S2) and many of the genes neighboring the common peak are involved in cartilage differentiation (Supporting Information Figure S6A). Since Hoxa11 and Hoxa13 are expressed in the zeugopod and autopod, respectively, except carpal/tarsal region (Figure 1 and Supporting Information Figure S1), each HOX11/13 binds to a common sequence in their unique expression domain and are expected to control the target gene expression in an expression domain‐specific manner. In addition, non‐HOX homeodomain transcription factors are shown to bind to the same region in the common HOX11/13 binding domain of the limb bud.
There is accumulating evidence that Hox and non‐Hox homeodomain transcription factors coordinate during limb cartilage development. Pitx1 encodes a paired‐like homeodomain transcription factor whose expression in the limb bud is restricted to the hindlimb and functions to transform forelimb type morphogenesis to hindlimb type morphogenesis (Lanctot, Moreau, Chamberland, Tremblay, & Drouin, 1999). Both the fore‐ and hindlimbs have basically the same topological architecture of the bone and differences in their morphology are thought to be based on quantitative rather than qualitative variation in gene expression or temporal differences in expression of common genes functioning in the limb cartilage development. The expression of Hox and Pitx1 in the mesenchyme of the hindlimb bud is largely overlapping (Shang, Luo, & Clayton, 1997). In the present study, we found many ChIP‐Seq proximal genes are shared by HOX11/13 and PITX1 (Figures 2 and 3) and that many of these genes are involved in cartilage differentiation (Supporting Information Figure S6D), These indicate that PITX1 also participates in directly regulating cartilage differentiation in collaboration with HOX11/13. Since the Hox expression pattern in the fore‐ and hindlimb bud is very similar, binding of PITX1 to the HOX11/13 common binding region (CHBRLs) could modulate HOX function. One plausible molecular mechanism for the interplay between PITX1 function and HOX11/13 is that binding of PITX1 to some HBSs of multiple HBSs in the enhancer results in fine tuning of the transcriptional frequency or timing in a program characteristic to that of the hindlimb bud. Given that CHBRLs potentially include multiple HBSs (Figure 4, Supporting Information Figure S5), analysis of interaction of multiple transcription factors in combination with CHBRL sequences will provide new insights into the complex regulatory processes centered by HOX13.
Another interesting example on functional coordination with Hox is Shox2 that encodes a paired‐like homeodomain transcription factor. Mice with a loss of Shox2 function exhibited severe truncation of the stylopod bone of fore‐ and hindlimbs in addition to hypoplasia of the hindlimb zeugopodal bone (Bobick & Cobb, 2012; Cobb et al., 2006). Shox2 showed overlapping expression with Hoxa9‐11 in the mesenchyme of the stylopod and zeugopod then in the outer layer of the perichondrium and proliferating chondrocytes but not in the autopod (Neufeld, Wang, & Cobb, 2014; Swinehart, Schlientz, Quintanilla, Mortlock, & Wellik, 2013). Genetic analysis revealed that Shox2 is a downstream gene of Hox11 (Gross, Krause, Wuelling, & Vortkamp, 2012). We found that HOXA11 binds to one of the Shox2 limb‐specific enhancers (Supporting Information Figure S2C and Figure 4) indicating the possibility that HOXA11 directly control region‐specific expression of Shox2 in the limb buds through binding to this enhancer/CHBRL. HOXA13 also binds this CHBRL (Supporting Information Figure S2C and Figure 4) and Shox2 is mis‐expressed in the Hox13dKO (Figure 6g,h) suggesting that binding of HOXA13 to this enhancer is involved in repressing Shox2 expression in the autopod. SHOX2 also bind to this CHBRL (Supporting Information Figure S2C) indicating that SHOX2 participate self‐regulation in coordination with HOX11. Interestingly, genetic analysis also suggested that Hox and Shox2 coordinate during development of the stylopodal and zeugopodal cartilage (Neufeld et al., 2014). Supporting this observation, we showed that SHOX2 also shares common binding regions of some Hox target genes with HOXA11 and HOXA13 (Figures 2 and 3, Supporting Information Figure S2 and Table S2). Genes involved in the skeletal development, such as Aff3, Bmp2, Bnc2 and Runx1t1 were concentrated in these common genes (Supporting Information Figure S6B). These indicate the possibility that SHOX2 and HOXA11 coordinately regulate these gene expression through the CHBRLs in the zeugopod by the similar mechanism as the PITX1 and HOXA11. Thus, HOXs regulate multiple steps in the SHOX2 functional cascade during cartilage development along the proximodistal axis of the limb.
4.2. On the HOX binding to the Hoxa11 intron
The binding profiles of HOXA11, HOXA13 and other homeodomain transcription factors to the HoxA cluster are different (Figure 2a). Both HOXA11 and HOXA13 bind to the 5′ untranslated region (5′‐UTR) of the Hoxa9 and Hoxa7 intron. However, while HOXA13 binds to the Hoxa11 intronic region, the binding of HOXA11 to the Hoxa11 intron is minimal or negligible (Figure 2a). This indicates that in the autopod, HOXA13 binds to the Hoxa11 intron but HOXA11 shows little or no binding to the Hoxa11 intron in the zeugopodal anlage. Hoxc10 is expressed in the mesenchyme of the stylopodal and zeugopodal anlagen of the hindlimb bud. Using deposited ChIP‐Seq data (Jain et al., 2018), we found that HOXC10 also binds to the Hoxa7 intron and Hoxa9 5′‐UTR but binding to the Hoxa11 intron was significantly lower or indistinguishable from background (Supporting Information Figure S10). Interestingly, non‐HOX homeodomain transcription factors SHOX2 and PBX also exhibited similar binding profiles to the Hoxa11 intron (Figure 2a). HOXA13 and HOXD13 bind to the “digit enhancer” in the Hoxa11 intron and activate transcription from the promoter in the first exon of Hoxa11 in the opposite direction as Hoxa11 transcription. This counter directional transcription itself is important to inactivate Hoxa11 transcription in the autopod (Kherdjemil et al., 2016). Based on the observation that Hoxa11 and Hoxa13 are co‐expressed in the distal fin bud of fish and that forced mis‐expression of Hoxa11 in the autopodal anlage in the mouse causes polydactyly, the system that represses Hoxa11 expression in the autopod of tetrapods is expected to have been acquired during the evolutionary transition from polydactyly to pentadactyly (Kherdjemil et al., 2016). Like most of the CHBRL, multiple and evolutionarily conserved HBSs are present in the Hoxa11 intronic “digital enhancer” (Supporting Information Figure S11). As described, compared to other common HOX11/13 binding regions, the binding to the Hoxa11 intron is very specific to HOXA13. It is possible that novel transcriptional co‐factor(s) specifically interacts with HOXA13 to stabilize HOXA13 binding to the HBS(s) in the Hoxa11 intronic “digital enhancer”. These suggest that in addition to the recruitment of the HOX binding sequence itself to the Hoxa11 intron, the system whereby homeodomain transcription factor binding to the Hoxa11 intron is restricted to HOXA13 was likely also introduced during the evolutionary transition from polydactyly to pentadactyly.
4.3. The role of multiple HBSs in CHBRLs
We showed that the sequences and the presence of multiple HBSs in most of the common ChIP‐peaks of HOXA11 and HOXA13 (CHBRLNr) are evolutionarily conserved among tetrapods (Supporting Information Figures S5 and S11). The representative fragments exhibited direct binding of HOXA11 and HOXA13 to multiple binding sites in vitro (Figure 4 and Supporting Information Figure S5) indicating that these HOX members may have similar binding profiles in vivo. Different from HOX1‐10, HOXA11 and HOXA13 can bind to the target sequence without the co‐factor homeodomain transcription factor, PBXs (Shen et al., 1997), and most of the conserved HBSs in the CHBRLNr do not neighbor the consensus PBX binding sequence. Thus, it is likely that HOXA11 and HOXA13 solely bind their target sites. Since the clustered low affinity HBSs guarantee specificity and robustness of HOX binding in Drosophila (Crocker et al., 2015), multiple HOXA11 and HOXA13 binding sites in the vertebrate CHBRLNr should have the same role. This possibility is also supported by evidence that Abd‐B Hox function shows a dosage effect on the patterning of limb cartilage (Zakany et al., 1997).
4.4. HOX function in the carpal/tarsal region
As already described, the relationship in the expression profile of Hoxa11 and Hoxa13 is unique. Abd‐B family genes in the HoxD cluster, Hoxd9‐13, maintain overlapping expression patterns in the limb bud, although transcription of Hoxa11 is repressed in the distal limb bud by HOXA13 function. However, in the proximal region of the carpal/tarsal anlage, Hoxa11 and Hoxa13 maintain their co‐expression despite the prominent expression of Hoxa13 in the autopodal anlage (Figure 1 and Supporting Information Figure S1). Loss‐of‐function of either Hoxa11 or Hoxd11 also results in abnormal formation of the carpal/tarsal bones, similar to Hox13 mutations indicating that Hox11 has an additional role in the morphogenesis of the autopod other than just the zeugopod (Davis & Capecchi, 1994; Fromental‐Ramain et al., 1996; Small & Potter, 1993). Cartilage formation in the carpal/tarsal region is unique compared to other limb regions. As shown in Figure 1 and Supporting Information Figure S1, initially Col2a1 expressing precartilaginous rudiment is formed in the prospective carpal/tarsal region, then multiple dense and small precartilaginous condensations individualize as an archipelago. These precartilaginous condensations develop into cartilage and maintain the cartilaginous state for much longer than other regions and never develop into long bones. In contrast, in the prospective long bone forming regions, Hoxa11 and Hoxa13 exhibit mutually exclusive expression. It is possible that HOXA11 and HOXA13 co‐exist in the regulatory region of the target gene carrying multiple HBSs in the carpal/tarsal anlage. This unique mixed state may enable the unique mode of cartilage differentiation in the carpal/tarsal region. We have previously shown that mis‐expression of Hoxa13 in the zeugopodal region results in the transformation of the zeugopodal cartilage into the carpal/tarsal‐like cartilage (Yokouchi et al., 1995), and this evidence further supports the hypothesis.
4.5. Regulation of Hox target genes by HOX
We identified the genes neighboring the CHBRLNr and demonstrated that their expression is changed in Hox13dKO embryos (Figures 6, 7 and 9 and Supporting Information Figure S9), thus confirming them to be direct Hox target genes. These genes are classified into two groups according to the changes in their mRNA levels in the Hox13dKO autopod. The genes in the first group showed reduced mRNA levels in the Hox13dKO, indicating that HOXA13 activates their transcription. The genes in the second group showed increased mRNA levels in the Hox13dKO and are likely repressed by HOXA13 during normal development.
Bmp2, Sulf1 and Stmn2 belong to the first group, and are expressed in the autopodal mesenchyme in wild‐type animals and exhibited a substantial reduction in expression levels in the Hox13dKO embryos (Figure 6 and Supporting Information Figure S9). In the case of second gene group, expression of Aff3, Bnc2, Nfib, Runx1t1, Shox2 and Tshz2 are restricted to the zeugopodal mesenchyme of the wild‐type embryos at E11.25–11.5, but showed ectopic expression in the autopod mesenchyme of Hox13dKO embryos (Figure 6 and Supporting Information Figure S9). Interestingly, the ectopic expression of Aff3, Bnc2, Nfib and Runx1t1 was uniform in the Hox13dKO autopod, whereas Shox2 mis‐expression was restricted to a narrow posterior region of the Hox13dKO autopod and Tshz2 mis‐expression was only found in the distal autopod. These results indicate the presence of various mechanisms for HOXA13 negative regulation of downstream genes.
The expression of Hox target genes, Aff3, Bnc2 and Nfib is induced during cartilage differentiation, consistent with their known roles in cartilage differentiation (Steichen‐Gersdorf et al., 2008; Uchihashi et al., 2007; Vanhoutteghem et al., 2009). As discussed above, the expression of these genes was not detected in the E11.5 autopodal anlage. Interestingly, these genes become expressed in the autopodal cartilage or perichondrium at E12.5 and sometimes even show partial overlapping expression with Hoxa13. Thus, repression of these genes by HOXA13 in the autopodal mesenchyme is transient during autopodal cartilage development. From E11.5 to E12.5 metacarpal/metatarsal and digital cartilage formation progresses distally and mesenchymal Hoxa13 expression gradually decreases and disappears in the cartilage. During this period, in addition to the decreased amounts of HOXA13 protein, changes in the regulatory activity of HOXA13 may occur by modification of HOXA13 itself or by de novo induction of the HOXA13 co‐factors that alter its transcriptional activity. Alternatively, since HOXA13 does not always bind to the H3K27ac enriched region of HOXA13 target genes, transitional usage of the enhancer may occur between HOXA13 dependent regulatory elements and independent elements during cartilage differentiation from the limb mesenchyme. Enhancer switching during development has previously been demonstrated (Andrey et al., 2013; Beccari et al., 2016; Glasgow et al., 2017), thus it is possible that a similar system is employed during cartilage differentiation.
In contrast to autopodal expression, these Hox target genes are also expressed in the zeugopodal mesenchyme prior to their cartilage expression and exhibit nearly complete overlap with Hoxa11 expression. This suggests that the mechanisms underlying HOXA11‐dependent and HOXA13‐dependent gene regulations are different.
4.6. HOX13 regulates cartilage differentiation in a dual manner
In contrast to the genes encoding transcription factors that are enriched among the Hox targets with upregulated expression in the Hox13dKO limb bud (Supporting Information Figure S8), relatively few Hox target transcription factors were identified in the downregulated fraction (Supporting Information Figure S8). A typical example of the latter case is Bcl11a. Bcl11a encodes a zinc finger‐type transcription factor and functions as a repressor during blood cell differentiation (Liu et al., 2003). We found that Bcl11a along with its family member Bcl11b, repressed cartilage differentiation from the limb bud mesenchyme (Figure 9).
In the Hox13dKO autopod, the entire autopodal mesenchyme excluding the distal most region, enters the cartilage differentiation pathway earlier than during normal development. Bcl11a is expressed in the autopodal mesenchyme in the wild‐type E11.5 embryos and its expression is substantially reduced in the Hox13dKO limb bud (Figure 9b,c). This indicates that the cartilage differentiation program repressed by HOXA13 is de‐repressed in the Hox13dKO autopod. Simultaneously, the Hox target transcription factors whose expression is induced during cartilage differentiation, are mis‐expressed in the mesenchyme of the Hox13dKO autopodal anlage. Combined, these results indicate that HOXA13 simultaneously activates the expression of repressive transcription factors for chondrogenesis (BCL11A) and represses the expression of transcription factors required to induce chondrogenesis in the autopod at E11.5. This demonstrates the presence of a dual regulatory mechanism for chondrogenesis during normal autopod development under the control of Hoxa13. In contrast to the autopod, Hoxa11 and Hox target genes such as Aff3, Bnc2, Nfib, Shox2 and Runx1t1, are co‐expressed in the zeugopodal mesenchyme. This implies that, different from HOXA13, HOXA11 does not repress the expression of these target genes. Thus, the HOXA13‐dependent dual control system is autopod‐specific.
Regarding the genes not expressed in the autopodal mesenchyme at E11.5 but that are mis‐expressed in the Hox13dKO, we proposed that HOXA13 may repress these genes during normal development. However, the following mechanism is equally probable; for these target genes, HOX13 may function as a weaker transcriptional activator than HOX11. The evidence that limb mesenchymal cells isolated from Hoxa13−/− embryos did not show cartilage differentiation in vitro support that HOXA13 is required for cartilage differentiation (Stadler, Higgins, & Capecchi, 2001). If repressive transcription factor(s) for the Hox target gene expression are present in the E11.5 autopodal mesenchyme, and transcriptional activation by HOX13 does not overcome this repressive activity, the autopodal expression of the Hox target gene does not occur. Hoxa11 and Hoxa13 exhibited mutually exclusive expression in the limb bud except in the carpal/tarsal anlage; however, in the Hox13dKO autopod, Hoxa11 expression is expanded to the autopodal mesenchyme (Sheth, Bastida, Kmita, & Ros, 2014). This ectopic HOXA11 may mis‐activate the common Hox target genes required for cartilage differentiation in the Hox13dKO autopod. The evidence that the forced mis‐expression of Hoxa11 in the autopod results in polydactyly supports this possibility (Kherdjemil et al., 2016). Expression analysis of common Hox target genes in the autopod under Hoxa11 mis‐expression may help to verify this mechanism.
The molecular mechanism that is responsible for the dual transcriptional activity of HOXA13 presented here, along with the similar effect of HOXA13 on HoxD gene expression shown by Beccari et al. (Beccari et al., 2016), remains elusive. Recently it is reported that a transcription factor that shows unique spatiotemporal expression patterns, interacts with HOX in a DNA binding dependent or independent manner, then modulates the function of HOX transcription factors (Guerreiro et al., 2013). In the case of Drosophila, the phosphorylation state of the HOX protein is crucial for its transcriptional activity (Berry & Gehring, 2000). In addition, the presence of the co‐factors that modulate transcriptional activity of Drosophila HOX was reported (reviewed in Zandvakili & Gebelein, 2016). Since these mechanisms related to HOXA13 have yet to be elucidated, research into this issue will be a topic for future studies.
4.7. Hox13 controls tetrapod specific autopodal structure
Why does Hoxa13, the last member of the HoxA cluster, have such unique function compared to other Hox genes expressed in the zeugopod or stylopod? This is likely related to the development of tetrapod‐specific autopodal structures. In contrast to the zeugopod and stylopod, the autopod generally consists of five sets of long bones, the metacarpi/metatarsi and phalanges (pentadactyly). One and two long bones exist in the stylopod and zeugopod, respectively, and the shape of their prospective region in the limb bud is a nearly elliptical cylinder. In contrast, the autopodal anlage has a unique paddle‐shaped structure that is required for supplying a sufficient number of mesenchymal cells to generate the pentadactylous cartilage pattern. The signaling factor, SHH, expressed in the mesenchyme of the posterior limb bud, is essential for expansion of the autopodal anlage (Chiang et al., 1996) and Hox13 directly activates Shh expression via a limb enhancer (Leal & Cohn, 2016, 2018). In addition, another signaling molecule, Fgf10, expressed in the distal limb bud, is required for limb bud formation and growth (Sekine et al., 1999). Abd‐B Hox is also involved in activating Fgf10 expression in the limb bud (Sheth et al., 2013) and we found that HOXA11 and HOXA13 bind one of the Fgf10 limb enhancers (VISTA ID mm917, Supporting Information Table S2). Thus, Hox13 positively controls the proliferation of autopodal mesenchymal cells to supply sufficient number of mesenchymal cells to generate the pentadactylous autopodal cartilage pattern. As discussed above, HOX13 transiently represses the cartilage differentiation program in the autopodal anlage in a dual manner. Thus, Hox13 functions on one hand to control the autopod‐specific growth program to supply enough mesenchyme and, on the other hand, transiently represses the cartilage forming program until the mesenchymal supply reaches a sufficient level to form pentadactylous structures (Figure 10).
Figure 10.
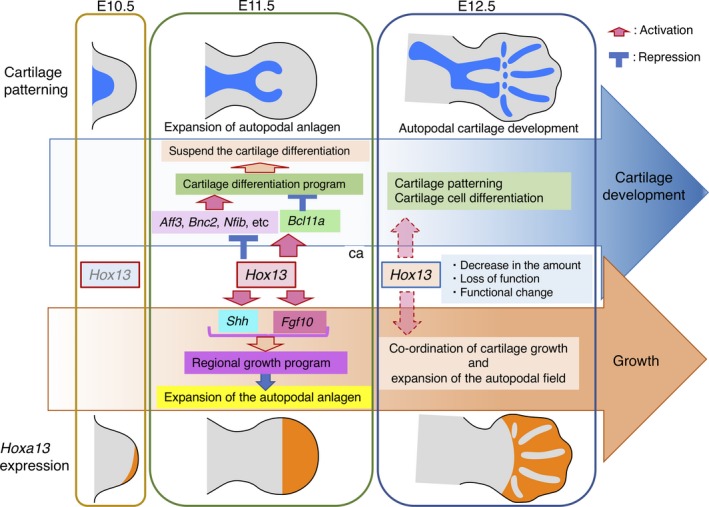
Dynamic change in the Hoxa13 function during autopod development
The pentadactylous autopodal architecture is unique to the tetrapods and this basic architectural design was key to tetrapod evolution through niche diversification. Recently, in fish, Hox13 genes were shown to function in switching the fin bud mesenchymal developmental program from cartilage formation to fin ray differentiation (Nakamura, Gehrke, Lemberg, Szymaszek, & Shubin, 2016). In the case of pentadactylous tetrapods, Hox13 function switched from the fish program to the tetrapod‐specific autopod‐forming program, possibly through the acquisition of the dual control system for cartilage differentiation.
AUTHOR CONTRIBUTIONS
SY, YU, YG and NK: ChIP‐Seq; TO: Genechip and WISH, SY, YS, EN and CY: WISH; TH and TS: NGS analysis; TT: enhancer knock out; YS: Bcl11a,b study; YS and AK: management of the project; AK: planning of the project, bioinformatic analysis and manuscript writing.
Supporting information
ACKNOWLEDGMENTS
We thank Drs. K. Ihara, S. Oki and N. Nakamichi for assistance with bioinformatic data analysis and Dr. S. Pastuhov for assistance with quantification of Col II expression. We also thank Drs. P. Chambon and D. Duboule for the Hox KO mice. This work was supported by a MEXT KAKENHI Grant Number JP5124703 and JSPS KAKENHI Grant Number JP16K07369 to A.K. This work was also supported by grants from the Foundation for Promotion of Material Science and Technology of Japan (MST Foundation), The Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care, and the Research Foundation for the Electrotechnology of Chubu to Y.S. Production of the enhancer KO mice was supported by the “Joint Usage and Joint Research Programs, the Institute of Advanced Medical Sciences, Tokushima University”. S.Y. was supported by the “Integrative Graduate Education and Research in Green Natural Sciences” program (Ministry of Education, Culture, Sports, Science and Technology, Japan). T.S. and T.H. were supported by grants from the Japan Science and Technology Agency (ERATO project; grant no. JPMJER1004 for T.H.).
Yamamoto S, Uchida Y, Ohtani T, et al. Hoxa13 regulates expression of common Hox target genes involved in cartilage development to coordinate the expansion of the autopodal anlage. Develop. Growth Differ. 2019;61:228–251. 10.1111/dgd.12601
Contributor Information
Yo‐ichi Shiraishi, Email: shiraishi.yo-ichi@h.mbox.nagoya-u.ac.jp.
Atsushi Kuroiwa, Email: akuro@bio.nagoya-u.ac.jp.
REFERENCES
- Andrey, G. , Montavon, T. , Mascrez, B. , Gonzalez, F. , Noordermeer, D. , Leleu, M. , … Duboule, D. (2013). A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science, 340, 1234167 10.1126/science.1234167 [DOI] [PubMed] [Google Scholar]
- Beccari, L. , Yakushiji‐Kaminatsui, N. , Woltering, J. M. , Necsulea, A. , Lonfat, N. , Rodriguez‐Carballo, E. , … Duboule, D. (2016). A role for HOX13 proteins in the regulatory switch between TADs at the HoxD locus. Genes & Development, 30, 1172–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, M. , & Gehring, W. (2000). Phosphorylation status of the SCR homeodomain determines its functional activity: Essential role for protein phosphatase 2A, B’. EMBO Journal, 19, 2946–2957. 10.1093/emboj/19.12.2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobick, B. E. , & Cobb, J. (2012). Shox2 regulates progression through chondrogenesis in the mouse proximal limb. Journal of Cell Science, 125, 6071–6083. 10.1242/jcs.111997 [DOI] [PubMed] [Google Scholar]
- Boulet, A. M. , & Capecchi, M. R. (2004). Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development, 131, 299–309. [DOI] [PubMed] [Google Scholar]
- Buscher, D. , Bosse, B. , Heymer, J. , & Ruther, U. (1997). Evidence for genetic control of Sonic hedgehog by Gli3 in mouse limb development. Mechanisms of Development, 62, 175–182. 10.1016/s0925-4773(97)00656-4 [DOI] [PubMed] [Google Scholar]
- Capellini, T. D. , Di Giacomo, G. , Salsi, V. , Brendolan, A. , Ferretti, E. , Srivastava, D. , … Selleri, L. (2006). Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development, 133, 2263–2273. 10.1242/dev.02395 [DOI] [PubMed] [Google Scholar]
- Caubit, X. , Core, N. , Boned, A. , Kerridge, S. , Djabali, M. , & Fasano, L. (2000). Vertebrate orthologues of the Drosophila region‐specific patterning gene teashirt. Mechanisms of Development, 91, 445–448. 10.1016/s0925-4773(99)00318-4 [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Knezevic, V. , Ervin, V. , Hutson, R. , Ward, Y. , & Mackem, S. (2004). Direct interaction with Hoxd proteins reverses Gli3‐repressor function to promote digit formation downstream of Shh. Development, 131, 2339–2347. 10.1242/dev.01115 [DOI] [PubMed] [Google Scholar]
- Chiang, C. , Litingtung, Y. , Lee, E. , Young, K. E. , Corden, J. L. , Westphal, H. , & Beachy, P. A. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature, 383, 407–413. 10.1038/383407a0 [DOI] [PubMed] [Google Scholar]
- Cobb, J. , Dierich, A. , Huss‐Garcia, Y. , & Duboule, D. (2006). A mouse model for human short‐stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long‐bone development. Proceedings of the National Academy of Sciences of the United States of America, 103, 4511–4515. 10.1073/pnas.0510544103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker, J. , Abe, N. , Rinaldi, L. , McGregor, A. P. , Frankel, N. , Wang, S. , … Stern, D. L. (2015). Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell, 160, 191–203. 10.1016/j.cell.2014.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe, K. , Kjaer, K. W. , Brehm, A. , Meinecke, P. , Nurnberg, P. , Neto, J. C. , … Mundlos, S. (2009). Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. American Journal of Human Genetics, 84, 483–492. 10.1016/j.ajhg.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A. P. , & Capecchi, M. R. (1994). Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd‐11. Development, 120, 2187–2198. [DOI] [PubMed] [Google Scholar]
- Davis, A. P. , Witte, D. P. , Hsieh‐Li, H. M. , Potter, S. S. , & Capecchi, M. R. (1995). Absence of radius and ulna in mice lacking hoxa‐11 and hoxd‐11. Nature, 375, 791–795. 10.1038/375791a0 [DOI] [PubMed] [Google Scholar]
- Dickinson, M. E. , Kobrin, M. S. , Silan, C. M. , Kingsley, D. M. , Justice, M. J. , Miller, D. A. , … Jenkins, N. A. (1990). Chromosomal localization of seven members of the murine TGF‐beta superfamily suggests close linkage to several morphogenetic mutant loci. Genomics, 6, 505–520. 10.1016/0888-7543(90)90480-i [DOI] [PubMed] [Google Scholar]
- Fromental‐Ramain, C. , Warot, X. , Messadecq, N. , LeMeur, M. , Dolle, P. , & Chambon, P. (1996). Hoxa‐13 and Hoxd‐13 play a crucial role in the patterning of the limb autopod. Development, 122, 2997–3011. [DOI] [PubMed] [Google Scholar]
- Giardine, B. , Riemer, C. , Hardison, R. C. , Burhans, R. , Elnitski, L. , Shah, P. , … Nekrutenko, A. (2005). Galaxy: A platform for interactive large‐scale genome analysis. Genome Research, 15, 1451–1455. 10.1101/gr.4086505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, S. F. , & Barresi, M. J. F. (2016). Developmental biology. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Glasgow, S. M. , Carlson, J. C. , Zhu, W. , Chaboub, L. S. , Kang, P. , Lee, H. K. , … Deneen, B. (2017). Glia‐specific enhancers and chromatin structure regulate NFIA expression and glioma tumorigenesis. Nature Neuroscience, 20, 1520–1528. 10.1038/nn.4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, D. J. , & Tabin, C. J. (1997). Analysis of Hoxd‐13 and Hoxd‐11 misexpression in chick limb buds reveals that Hox genes affect both bone condensation and growth. Development, 124, 627–636. [DOI] [PubMed] [Google Scholar]
- Green, M. R. , & Sambrook, J. (Eds.) (2012). Molecular cloning: A laboratory manual, 4th edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Gross, S. , Krause, Y. , Wuelling, M. , & Vortkamp, A. (2012). Hoxa11 and Hoxd11 regulate chondrocyte differentiation upstream of Runx2 and Shox2 in mice. PLoS ONE, 7, e43553 10.1371/journal.pone.0043553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro, I. , Nunes, A. , Woltering, J. M. , Casaca, A. , Novoa, A. , Vinagre, T. , … Mallo, M. (2013). Role of a polymorphism in a Hox/Pax‐responsive enhancer in the evolution of the vertebrate spine. Proceedings of the National Academy of Sciences of the United States of America, 110, 10682–10686. 10.1073/pnas.1300592110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M. , Yamashita, Y. , & Takemoto, T. (2016). Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non‐mosaic mutants in the mouse. Developmental Biology, 418, 228–9. 10.1016/j.ydbio.2016.07.017 [DOI] [PubMed] [Google Scholar]
- Huang da, W. , Sherman, B. T. , & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4, 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Izpisua‐Belmonte, J. C. , Tickle, C. , Dolle, P. , Wolpert, L. , & Duboule, D. (1991). Expression of the homeobox Hox‐4 genes and the specification of position in chick wing development. Nature, 350, 585–589. 10.1038/350585a0 [DOI] [PubMed] [Google Scholar]
- Jain, D. , Nemec, S. , Luxey, M. , Gauthier, Y. , Bemmo, A. , Balsalobre, A. , & Drouin, J. (2018). Regulatory integration of Hox factor activity with T‐box factors in limb development. Development, 145, dev159830 10.1242/dev.159830 [DOI] [PubMed] [Google Scholar]
- Jerkovic, I. , Ibrahim, D. M. , Andrey, G. , Haas, S. , Hansen, P. , Janetzki, C. , … Mundlos, S. (2017). Genome‐wide binding of posterior HOXA/D transcription factors reveals subgrouping and association with CTCF. PLoS Genetics, 13, e1006567 10.1371/journal.pgen.1006567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D. , Hinrichs, A. S. , Furey, T. S. , Roskin, K. M. , Sugnet, C. W. , Haussler, D. , & Kent, W. J. (2004). The UCSC table browser data retrieval tool. Nucleic Acids Research, 32, D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kherdjemil, Y. , Lalonde, R. L. , Sheth, R. , Dumouchel, A. , de Martino, G. , Pineault, K. M. , … Kmita, M. (2016). Evolution of Hoxa11 regulation in vertebrates is linked to the pentadactyl state. Nature, 539, 89–92. 10.1038/nature19813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita, M. , Tarchini, B. , Zakany, J. , Logan, M. , Tabin, C. J. , & Duboule, D. (2005). Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature, 435, 1113–1116. 10.1038/nature03648 [DOI] [PubMed] [Google Scholar]
- Lanctot, C. , Moreau, A. , Chamberland, M. , Tremblay, M. L. , & Drouin, J. (1999). Hindlimb patterning and mandible development require the Ptx1 gene. Development, 126, 1805–1810. [DOI] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal, F. , & Cohn, M. J. (2016). Loss and Re‐emergence of legs in snakes by modular evolution of sonic hedgehog and HOXD enhancers. Current Biology, 26, 2966–2973. 10.1016/j.cub.2016.09.020 [DOI] [PubMed] [Google Scholar]
- Leal, F. , & Cohn, M. J. (2018). Developmental, genetic, and genomic insights into the evolutionary loss of limbs in snakes. Genesis, 56, e23077 10.1002/dvg.23077 [DOI] [PubMed] [Google Scholar]
- Lewandowski, J. P. , Du, F. , Zhang, S. , Powell, M. B. , Falkenstein, K. N. , Ji, H. , & Vokes, S. A. (2015). Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Developmental Biology, 406, 92–103. 10.1016/j.ydbio.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P. , Keller, J. R. , Ortiz, M. , Tessarollo, L. , Rachel, R. A. , Nakamura, T. , … Copeland, N. G. (2003). Bcl11a is essential for normal lymphoid development. Nature Immunology, 4, 525–532. 10.1038/ni925 [DOI] [PubMed] [Google Scholar]
- Long, H. K. , Prescott, S. L. , & Wysocka, J. (2016). Ever‐changing landscapes: Transcriptional enhancers in development and evolution. Cell, 167, 1170–1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick, P. , & Bailey, T. L. (2011). MEME‐ChIP: Motif analysis of large DNA datasets. Bioinformatics, 27, 1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe, C. D. , & Innis, J. W. (2005). A genomic approach to the identification and characterization of HOXA13 functional binding elements. Nucleic Acids Research, 33, 6782–6794. 10.1093/nar/gki979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, C. Y. , Bristor, D. , Hiller, M. , Clarke, S. L. , Schaar, B. T. , Lowe, C. B. , … Bejerano, G. (2010). GREAT improves functional interpretation of cis‐regulatory regions. Nature Biotechnology, 28, 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan, A. , & Thomas, P. D. (2013). PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research, 41, D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T. , Gehrke, A. R. , Lemberg, J. , Szymaszek, J. , & Shubin, N. H. (2016). Digits and fin rays share common developmental histories. Nature, 537, 225–228. 10.1038/nature19322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec, S. , Luxey, M. , Jain, D. , Huang Sung, A. , Pastinen, T. , & Drouin, J. (2017). Pitx1 directly modulates the core limb development program to implement hindlimb identity. Development, 144, 3325–3335. 10.1242/dev.154864 [DOI] [PubMed] [Google Scholar]
- Neufeld, S. J. , Wang, F. , & Cobb, J. (2014). Genetic interactions between Shox2 and Hox genes during the regional growth and development of the mouse limb. Genetics, 198, 1117–1126. 10.1534/genetics.114.167460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki, S. , Maehara, K. , Ohkawa, Y. , & Meno, C. (2014). SraTailor: graphical user interface software for processing and visualizing ChIP‐seq data. Genes Cells, 19, 919–926. 10.1111/gtc.12190 [DOI] [PubMed] [Google Scholar]
- Osterwalder, M. , Barozzi, I. , Tissieres, V. , Fukuda‐Yuzawa, Y. , Mannion, B. J. , Afzal, S. Y. , … Pennacchio, L. A. (2018). Enhancer redundancy provides phenotypic robustness in mammalian development. Nature, 554, 239–243. 10.1038/nature25461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel, C. L. , Prabhakaran, B. , & Vogt, T. F. (1997). The mouse Ulnaless mutation deregulates posterior HoxD gene expression and alters appendicular patterning. Development, 124, 3481–3492. [DOI] [PubMed] [Google Scholar]
- Pineault, K. M. , & Wellik, D. M. (2014). Hox genes and limb musculoskeletal development. Current Osteoporosis Reports, 12, 420–427. 10.1007/s11914-014-0241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A. R. , & Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, F. , Ryan, D. P. , Gruning, B. , Bhardwaj, V. , Kilpert, F. , Richter, A. S. , … Manke, T. (2016). deepTools2: A next generation web server for deep‐sequencing data analysis. Nucleic Acids Research, 44, W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. T. , Thorvaldsdottir, H. , Winckler, W. , Guttman, M. , Lander, E. S. , Getz, G. , & Mesirov, J. P. (2011). Integrative genomics viewer. Nature Biotechnology, 29, 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin, J. M. , Abassah‐Oppong, S. , & Cobb, J. (2013). Comparative transgenic analysis of enhancers from the human SHOX and mouse Shox2 genomic regions. Human Molecular Genetics, 22, 3063–3076. 10.1093/hmg/ddt163 [DOI] [PubMed] [Google Scholar]
- Salsi, V. , & Zappavigna, V. (2006). Hoxd13 and Hoxa13 directly control the expression of the EphA7 Ephrin tyrosine kinase receptor in developing limbs. Journal of Biological Chemistry, 281, 1992–1999. 10.1074/jbc.m510900200 [DOI] [PubMed] [Google Scholar]
- Sekine, K. , Ohuchi, H. , Fujiwara, M. , Yamasaki, M. , Yoshizawa, T. , Sato, T. , … Kato, S. (1999). Fgf10 is essential for limb and lung formation. Nature Genetics, 21, 138–141. 10.1038/5096 [DOI] [PubMed] [Google Scholar]
- Selleri, L. , Depew, M. J. , Jacobs, Y. , Chanda, S. K. , Tsang, K. Y. , Cheah, K. S. , … Cleary, M. L. (2001). Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development, 128, 3543–3557. [DOI] [PubMed] [Google Scholar]
- Shang, J. , Luo, Y. , & Clayton, D. A. (1997). Backfoot is a novel homeobox gene expressed in the mesenchyme of developing hind limb. Developmental Dynamics, 209, 242–253. 10.1002/(issn)1097-0177 [DOI] [PubMed] [Google Scholar]
- Shen, W. F. , Rozenfeld, S. , Lawrence, H. J. , & Largman, C. (1997). The Abd‐B‐like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. Journal of Biological Chemistry, 272, 8198–8206. 10.1074/jbc.272.13.8198 [DOI] [PubMed] [Google Scholar]
- Sheth, R. , Barozzi, I. , Langlais, D. , Osterwalder, M. , Nemec, S. , Carlson, H. L. , … Kmita, M. (2016). Distal limb patterning requires modulation of cis‐regulatory activities by HOX13. Cell Reports, 17, 2913–2926. 10.1016/j.celrep.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, R. , Bastida, M. F. , Kmita, M. , & Ros, M. (2014). “Self‐regulation”, a new facet of Hox genes’ function. Developmental Dynamics, 243, 182–191. 10.1002/dvdy.24019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, R. , Gregoire, D. , Dumouchel, A. , Scotti, M. , Pham, J. M. , Nemec, S. , … Kmita, M. (2013). Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development, 140, 2130–2138. 10.1242/dev.089409 [DOI] [PubMed] [Google Scholar]
- Shin, H. , Liu, T. , Manrai, A. K. , & Liu, X. S. (2009). CEAS: Cis‐regulatory element annotation system. Bioinformatics, 25, 2605–2606. 10.1093/bioinformatics/btp479 [DOI] [PubMed] [Google Scholar]
- Small, K. M. , & Potter, S. S. (1993). Homeotic transformations and limb defects in Hox A11 mutant mice. Genes & Development, 7, 2318–2328. 10.1101/gad.7.12a.2318 [DOI] [PubMed] [Google Scholar]
- Stadler, H. S. , Higgins, K. M. , & Capecchi, M. R. (2001). Loss of Eph‐receptor expression correlates with loss of cell adhesion and chondrogenic capacity in Hoxa13 mutant limbs. Development, 128, 4177–4188. [DOI] [PubMed] [Google Scholar]
- Steichen‐Gersdorf, E. , Gassner, I. , Superti‐Furga, A. , Ullmann, R. , Stricker, S. , Klopocki, E. , & Mundlos, S. (2008). Triangular tibia with fibular aplasia associated with a microdeletion on 2q11.2 encompassing LAF4. Clinical Genetics, 74, 560–565. 10.1111/j.1399-0004.2008.01050.x [DOI] [PubMed] [Google Scholar]
- Swinehart, I. T. , Schlientz, A. J. , Quintanilla, C. A. , Mortlock, D. P. , & Wellik, D. M. (2013). Hox11 genes are required for regional patterning and integration of muscle, tendon and bone. Development, 140, 4574–4582. 10.1242/dev.096693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto, D. P. , Rodriguez‐Esteban, C. , Ryan, A. K. , O'Connell, S. M. , Liu, F. , Kioussi, C. , … Rosenfeld, M. G. (1999). Role of the Bicoid‐related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes & Development, 13, 484–494. 10.1101/gad.13.4.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihashi, T. , Kimata, M. , Tachikawa, K. , Koshimizu, T. , Okada, T. , Ihara‐Watanabe, M. , … Michigami, T. (2007). Involvement of nuclear factor I transcription/replication factor in the early stage of chondrocytic differentiation. Bone, 41, 1025–1035. 10.1016/j.bone.2007.08.028 [DOI] [PubMed] [Google Scholar]
- Vanhoutteghem, A. , Maciejewski‐Duval, A. , Bouche, C. , Delhomme, B. , Herve, F. , Daubigney, F. , … Djian, P. (2009). Basonuclin 2 has a function in the multiplication of embryonic craniofacial mesenchymal cells and is orthologous to disco proteins. Proceedings of the National Academy of Sciences of the United States of America, 106, 14432–14437. 10.1073/pnas.0905840106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel, A. , Minovitsky, S. , Dubchak, I. , & Pennacchio, L. A. (2007). VISTA enhancer browser–a database of tissue‐specific human enhancers. Nucleic Acids Research, 35, D88–D92. 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes, S. A. , Ji, H. , Wong, W. H. , & McMahon, A. P. (2008). A genome‐scale analysis of the cis‐regulatory circuitry underlying sonic hedgehog‐mediated patterning of the mammalian limb. Genes & Development, 22, 2651–2663. 10.1101/gad.1693008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M. , Gotoh, Y. , Tamura, K. , Tanaka, M. , Kawakami, A. , Ide, H. , & Kuroiwa, A. (1998). Coordinated expression of Hoxa‐11 and Hoxa‐13 during limb muscle patterning. Development, 125, 1325–1335. [DOI] [PubMed] [Google Scholar]
- Yamamoto‐Shiraishi, Y. , Higuchi, H. , Yamamoto, S. , Hirano, M. , & Kuroiwa, A. (2014). Etv1 and Ewsr1 cooperatively regulate limb mesenchymal Fgf10 expression in response to apical ectodermal ridge‐derived fibroblast growth factor signal. Developmental Biology, 394, 181–190. 10.1016/j.ydbio.2014.07.022 [DOI] [PubMed] [Google Scholar]
- Yamamoto‐Shiraishi, Y. , & Kuroiwa, A. (2013). Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Developmental Biology, 377, 363–374. 10.1016/j.ydbio.2013.02.023 [DOI] [PubMed] [Google Scholar]
- Ye, W. , Song, Y. , Huang, Z. , Osterwalder, M. , Ljubojevic, A. , Xu, J. , … Chen, Y. (2016). A unique stylopod patterning mechanism by Shox2‐controlled osteogenesis. Development, 143, 2548–2560. 10.1242/dev.138750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi, Y. , Nakazato, S. , Yamamoto, M. , Goto, Y. , Kameda, T. , Iba, H. , & Kuroiwa, A. (1995). Misexpression of Hoxa‐13 induces cartilage homeotic transformation and changes cell adhesiveness in chick limb buds. Genes & Development, 9, 2509–2522. 10.1101/gad.9.20.2509 [DOI] [PubMed] [Google Scholar]
- Yokouchi, Y. , Sasaki, H. , & Kuroiwa, A. (1991). Homeobox gene expression correlated with the bifurcation process of limb cartilage development. Nature, 353, 443–445. 10.1038/353443a0 [DOI] [PubMed] [Google Scholar]
- Yue, F. , Cheng, Y. , Breschi, A. , Vierstra, J. , Wu, W. , Ryba, T. , … Ren, B. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature, 515, 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany, J. , Fromental‐Ramain, C. , Warot, X. , & Duboule, D. (1997). Regulation of number and size of digits by posterior Hox genes: A dose‐dependent mechanism with potential evolutionary implications. Proceedings of the National Academy of Sciences of the United States of America, 94, 13695–13700. 10.1073/pnas.94.25.13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany, J. , Zacchetti, G. , & Duboule, D. (2007). Interactions between HOXD and Gli3 genes control the limb apical ectodermal ridge via Fgf10. Developmental Biology, 306, 883–893. 10.1016/j.ydbio.2007.03.517 [DOI] [PubMed] [Google Scholar]
- Zandvakili, A. , & Gebelein, B. (2016). Mechanisms of specificity for Hox factor activity. Journal of Developmental Biology, 4, 16 10.3390/jdb4020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, T. , Meyer, C. A. , Eeckhoute, J. , Johnson, D. S. , Bernstein, B. E. , … Liu, X. S. (2008). Model‐based analysis of ChIP‐Seq (MACS). Genome Biology, 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


