Summary
Background
Biologics targeting inflammatory mediators can achieve clinical improvements in hidradenitis suppurativa (HS). However, their clinical efficacy shows great interpatient variability in daily practice.
Objectives
To investigate the anti‐inflammatory potency of a selection of currently available biologics and prednisolone for the treatment of HS in an ex vivo skin culture system using lesional HS biopsies.
Methods
Lesional skin samples from 10 patients with HS and skin samples from five healthy controls were cultured ex vivo and exposed to prednisolone or biologics targeting tumour necrosis factor (TNF)‐α, interleukin (IL)‐17A, IL‐12/23p40 or CD20 (adalimumab, infliximab, secukinumab, ustekinumab and rituximab, respectively). Real‐time quantitative polymerase chain reaction and cytokine bead arrays were used to measure the inhibitory effect of the biologics on cytokines and antimicrobial peptides (AMPs).
Results
The relative mRNA expression of all tested cytokines and AMPs was significantly downregulated by all anti‐inflammatory agents (P < 0·001). The protein production of the proinflammatory cytokines TNF‐α, interferon γ, IL‐1β, IL‐6 and IL‐17A was significantly inhibited by adalimumab, infliximab, ustekinumab, prednisolone (all P < 0·001) and rituximab (P = 0·0071), but not by secukinumab (P = 0·0663). On both mRNA and protein levels, adalimumab, infliximab and prednisolone reduced the levels of a broader mix of individual cytokines than secukinumab, ustekinumab and rituximab. Moreover, a significant inhibitory effect on mRNA expression levels of inflammatory markers in healthy control skin was observed only for TNF‐α inhibitors (P < 0·001) and prednisolone (P = 0·0015).
Conclusions
This ex vivo study suggests that TNF‐α inhibitors and prednisolone are the most powerful inhibitors of proinflammatory cytokines and AMPs in HS lesional skin, which concurs with our clinical experience in patients with HS.
Short abstract
What's already known about this topic?
A key element of hidradenitis suppurativa (HS) is an aberrant immune response characterized by the overexpression of several proinflammatory cytokines and antimicrobial peptides in lesional skin.
Biologics targeting inflammatory cytokines have the potential to improve HS disease activity.
There is still need for efficacious drugs in the treatment of HS.
What does this study add?
We sought to quantify the anti‐inflammatory effects of currently available biologics in an ex vivo disease model.
Adalimumab, infliximab, secukinumab, ustekinumab and rituximab in addition to prednisolone significantly inhibited a selected panel of proinflammatory cytokines and antimicrobial peptides in ex vivo HS lesional skin.
Adalimumab, infliximab and prednisolone reduced the levels of a broader mix of individual cytokines than secukinumab, ustekinumab and rituximab.
What is the translational message?
The significant inhibition of important proinflammatory cytokines by tumour necrosis factor‐α inhibitors in HS correlates with their clinical efficacy.
Our ex vivo skin culture system represents an adequate model for studies in search of novel candidate drugs for the treatment of HS and to personalize the treatment in specific patients.
Linked Comment: https://doi.org/10.1111/bjd.18173.
Hidradenitis suppurativa (HS) is a chronic skin disease characterized by painful, deep‐seated, inflamed nodules, abscesses and, in later stages, sinus tracts.1 Occlusion of the hair follicle with subsequent rupture, followed by a fierce local inflammatory response, are considered primary pathogenic events. The aberrant innate immune response involves the upregulation of various proinflammatory cytokines and antimicrobial peptides (AMPs).2
Several studies have found elevated mRNA and protein levels of tumour necrosis factor (TNF)‐α in lesional and perilesional HS skin.3, 4 Targeting TNF‐α with adalimumab and infliximab has been shown to be clinically efficacious in HS in randomized placebo‐controlled trials.5, 6, 7 In addition, the upregulation of interleukin (IL)‐1β, IL‐17A and IL‐23 in HS lesions points to the importance of the IL‐17 pathway in the pathophysiology of HS.8, 9, 10 Targeting IL‐1 in HS yielded ambiguous clinical outcomes. In a randomized controlled trial anakinra proved to be efficacious in moderate‐to‐severe HS, yet cases of failure for patients on anakinra therapy have been reported.11, 12, 13
Most recently, IL‐17 has been targeted in two clinical trials in HS (NCT02421172, NCT03248531), but the results have not yet been released. Treatment with secukinumab or ustekinumab induced amelioration of HS in a few cases and case series.14, 15, 16 In addition, enhanced levels of AMPs in lesional HS skin have been described for S100A7 (psoriasin), S100A8, S100A9, human β‐defensin (hBD)‐2, hBD‐3 and LL‐37.17, 18, 19 Upregulation of these AMPs in HS lesional skin is mainly driven by IL‐17.18
Chronic HS lesions are characterized by a marked increase in the number of CD20+, CD79A+ B cells and CD138+ plasma cells.20 One case report described a clear improvement in HS after treatment with rituximab for concomitant idiopathic carpotarsal osteolysis syndrome.21 This highlights the contribution of these B cells and plasma cells to the inflammatory process in HS.
Although biologics targeting inflammatory mediators are now widely used for the treatment of HS, their clinical efficacy shows great interpatient variability. Our ex vivo skin culture system is a fast and simple method to investigate and simultaneously compare the effect of biologics in fresh, human, lesional HS skin samples.22 It approaches the in vivo situation by maintaining the patients’ skin architecture and allows close monitoring of the events following response to immunostimulators or suppressors in the same experiment.23 Therefore, this study sought to investigate the anti‐inflammatory potency of a selection of currently available biologics and prednisolone for the treatment of HS in an ex vivo skin culture system using lesional HS biopsies.
Materials and methods
Ethics and informed consent
HS lesional skin was collected from excised skin after HS surgery at the Department of Dermatology of the Erasmus University Medical Center in Rotterdam, the Netherlands, from October 2017 to February 2018. According to the opt‐out principle used in the Erasmus University Medical Center no informed consent is required for the use of excised tissue for research purposes as this is considered waste material. Control skin samples were obtained from healthy individuals in the Sint Franciscus Hospital in Rotterdam, the Netherlands. All healthy volunteers provided written informed consent for the use of their excised skin in this study.
Patients with hidradenitis suppurativa
A total of 10 patients with HS (five men and five women) who had chronic, active disease (seven Hurley stage II, three Hurley stage III) requiring surgical excision under general or sedative anaesthesia were included. HS lesional skin samples were obtained from the inguinogenital area in five of these 10 patients, from the axillae in four patients and from the gluteal area in one patient. The mean age was 43·7 ± 7·4 years (SD), the mean body mass index (BMI) was 30·3 ± 6·5 kg m−2, and seven of 10 patients were current smokers. Eight patients received a stable (≥ 28 days) dosage of systemic antibiotics for their HS at the time of surgery (four patients used azithromycin 500 mg three times per week; two patients moxifloxacin 400 mg once daily, rifampicin 600 mg once daily, and metronidazole 500 mg once daily; one patient clindamycin 300 mg twice daily, rifampicin 600 mg once daily and metronidazole 500 mg once daily; one patient doxycycline 100 mg once daily). None of the patients had been using immunosuppressive or immunomodulatory therapies including biologics for at least 2 months prior to surgery.
Healthy control skin
Healthy skin samples were obtained from the submammary and abdominal waste material of five women who underwent breast or abdominal reduction surgery. We argue that skin from these regions is suitable as a control sample because the inframammary and abdominal folds are predilection sites for HS. The controls (all female, mean age 41·0 ± 10·7 years, mean BMI 29·8 ± 4·2 kg m−2, no current smokers) were otherwise healthy, had no family history of HS, and were not using any immunosuppressive or immunomodulatory treatment at the time of surgery.
Biopsy procedure
A total of seven fresh 4‐mm punch biopsies were taken per participant. HS skin biopsies were obtained from the same palpable actively inflamed lesion/infiltrate, and at least 1 cm away from the excision border. Abscesses were not biopsied in order to avoid sampling only the roof of the abscess. Each biopsy was weighed prior to culture as described in an earlier publication.4 HS biopsies had a mean weight of 2·4 ± 6·6 mg, which was significantly heavier than biopsies from healthy skin with a mean weight of 16·1 ± 3·0 mg (P < 0·001). This potential confounder was addressed by normalizing all protein levels for the mg tissue weight.
Ex vivo skin culture
The 4‐mm biopsies were immediately cultured in a transwell system (Netwell; Costar, Cambridge, MA, U.S.A.) as described previously.22, 23 In brief, samples were placed in punched‐out holes in the transwell membrane of a 12‐well plate with the epidermis exposed to the air and the dermis immersed in 1 mL Iscove's modified Dulbecco's medium (Gibco, Paisley, U.K.) containing 0·5% human AB serum, penicillin (100 U mL−1) and streptomycin (100 U mL−1) with or without an anti‐inflammatory agent. Biologics and prednisolone were separately added to the culture media resulting in the following seven conditions: (i) culture media as negative control; (ii) prednisolone 100 μg mL−1 as positive control; (iii) adalimumab 30 μg mL−1; (iv) infliximab 20 μg mL−1; (v) secukinumab 30 μg mL−1; (vi) ustekinumab 10 μg mL−1 and (vii) rituximab 200 μg mL−1. The concentrations of the monoclonal antibodies were derived from the reported trough levels – at least twofold – in patients with plaque psoriasis (adalimumab,24, 25 infliximab,25 ustekinumab,26 secukinumab),27 and CD20+ B‐cell malignancies (rituximab).28 Our previous experiments using the ex vivo model showed that the cytokine concentrations measured in HS samples were usually in the picogram range.4, 7 The biologic concentrations used in our experiments were at least 1000‐fold higher and should suffice for neutralization purposes. Skin biopsies were incubated for 24 h at 37 °C in an atmosphere of 5% CO2 and 98% humidity. Subsequently, the biopsies were placed in 250 μL lysis buffer containing 1% β‐mercaptoethanol. Both the supernatants and the biopsies were transferred to a polypropylene tube and stored at −20 °C until further analysis.
Messenger RNA expression analysis
Total mRNA was extracted using the GenElute Mammalian Total RNA Miniprep Kit (Sigma‐Aldrich, St. Louis, MA, U.S.A.). RNA was treated with 0·1 U μL−1 DNAse (Invitrogen, Carlsbad, CA, U.S.A.) and cDNA was subsequently synthesized using 1 μg total RNA template, with SuperScript II reverse transcriptase, random hexamer primers (Invitrogen) and oligo(dT)15 (Promega Co., Madison, WI, U.S.A.). Primers and probes were designed and chosen using ProbeFinder Sofware and the Universal Probe library (Roche Applied Science, Indianapolis, IN, U.S.A.). ABL1 was chosen as a reference housekeeping gene.29 Real‐time quantitative polymerase chain reaction (qPCR) was performed for the following 12 genes using the ViiA7 sequence‐detection system (Applied Biosystems, Waltham, MA, U.S.A.): TNF‐α, IL‐1β, IL‐6, CXCL‐8 (IL‐8), IL‐12p19, IL‐17A, interferon (IFN)α/MxA, S100A7, S100A8, S100A9, LL37 and HBD‐2. Messenger RNA expression was analysed with QuantStudio real‐time qPCR software version 1·3 (Applied Biosystems).
Protein quantification
The following 18 inflammation‐related cytokines were simultaneously measured in the supernatant using a customized bead‐based multi‐analyte profiling assay (Luminex, R&D systems, Minneapolis, MN, U.S.A.): CCL‐20, TNF‐α, IFNγ, IL‐1β, IL‐1R1, IL‐5, IL‐6, IL‐10, IL‐12/23p40, IL‐17A, IL‐17E, IL‐18, IL‐19, IL‐22, IL‐27, IL‐31, IL‐33 and IL‐36β. Assays were used according to the manufacturers’ protocol. A dilution factor of two was used for all supernatants.
Statistical analyses
The relative mRNA expression and protein production per sample compared with culture media (negative control) was calculated for every condition. Protein levels were normalized according to input weight and expressed in pg mL−1 per mg tissue weight. The lower limit of quantification was imputed when protein concentrations in the supernatant (pg mL−1) were below the limit of detection. The Kruskal–Wallis test, i.e. a one‐way anova on ranks, was used to compare the variance in mRNA and protein levels per inflammatory marker. If one condition stochastically dominated another condition, the Dunnett's post‐test was subsequently used to test conditions vs. culture media (pairwise comparisons) separately. This approach was chosen in order to increase power. A two‐sided P‐value < 0·05 was considered significant. The level of significance for the relative mRNA expression and protein production was separately adjusted by the Benjamini–Hochberg procedure for multiple comparisons. A correlation between relative mRNA and protein levels per cytokine was calculated using the Spearman's rho test for non‐normally distributed continuous variables. GraphPad Prism version 6 (GraphPad Software, La Jolla, CA, U.S.A.) was used for all statistical analyses.
Results
Sample flow for analysis
In total, samples from 10 patients with HS and five healthy controls were analysed by real‐time qPCR. Supernatants from one patient with HS were excluded from multiplex assay because of erroneous sample processing, resulting in samples from nine patients with HS with protein data. For control skin, supernatants of three healthy volunteers were analysed by Luminex assay. However, protein levels were below the level of detection or in the very low range of the calibration line in these control samples (data not shown). Therefore, samples of the other two healthy volunteers were not analysed.
Significant downregulation of mRNA expression of cytokines and antimicrobial peptides by prednisolone and different biologics
The relative changes in mRNA expression of the 12 genes in the HS samples, including significance levels, are shown in Table 1. The overall median inhibitory effect on the mix of cytokines and AMPs per condition was as follows: prednisolone 0·57 [interquartile range (IQR) 0·34–1·03], adalimumab 0·51 (0·33–0·92), infliximab 0·53 (0·32–0·78), secukinumab 0·56 (0·29–0·99), ustekinumab 0·64 (0·44–1·00) and rituximab 0·73 (0·50–1·19). The inhibitory impact on AMPs was stronger for the biologics than for prednisolone (Table 1, Fig. 1). Prednisolone, adalimumab and infliximab significantly inhibited IL‐1β mRNA expression. Expression of CXCL‐8 (IL‐8) was significantly downregulated by adalimumab, infliximab, secukinumab and ustekinumab. Messenger RNA levels of two other important proinflammatory cytokines, TNF‐α and IL‐23p19, were not significantly downregulated by any of the biologics or prednisolone. Messenger RNA expression of members of the S100 family and HBD‐2 was significantly reduced by adalimumab, infliximab, secukinumab and ustekinumab (Table 1). In healthy control skin, prednisolone, adalimumab and infliximab significantly downregulated mRNA expression of all tested cytokines and AMPs (Table S1, Fig. S1; see Supporting Information).
Table 1.
Relative mRNA expression and modulation by biologics in hidradenitis suppurativa (HS) lesional skin
| HS lesional skin (n = 10) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prednisolone | Adalimumab | Infliximab | Secukinumaba | Ustekinumab | Rituximab | |||||||
| Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | |
| IL‐1β | 0·27 (0·15–0·48) | < 0·0001 b | 0·53 (0·23–0·74) | 0·0073 b | 0·60 (0·50–0·73) | 0·0306 | 0·67 (0·53–0·87) | 0·2377 | 0·76 (0·39–1·06) | 0·6194 | 0·49 (0·35–0·72) | 0·0597 |
| IL‐6 | 0·42 (0·36–0·51) | < 0·0001 b | 0·62 (0·52–0·89) | 0·0956 | 0·65 (0·41–0·71) | 0·0047 b | 0·70 (0·44–0·90) | 0·1063 | 0·55 (0·37–0·90) | 0·0122 b | 0·75 (0·55–0·97) | 0·4396 |
| CXCL‐8 (IL‐8) | 0·44 (0·34–0·60) | 0·1675 | 0·30 (0·22–0·49) | 0·0042 b | 0·29 (0·26–0·38) | 0·0002 b | 0·28 (0·16–0·47) | 0·0015 b | 0·39 (0·32–0·48) | 0·0296 | 0·69 (0·41–0·86) | 0·9779 |
| IL‐17A | 0·13 (0·07–0·37) | 0·0049 b | 0·93 (0·59–1·37) | 1·0000 | 0·55 (0·43–0·84) | 1·0000 | 0·95 (0·62–1·42) | 1·0000 | 0·96 (0·56–1·29) | 1·0000 | 0·99 (0·43–1·61) | 1·0000 |
| IL‐23p19 | 0·92 (0·33–1·20) | 1·0000 | 0·48 (0·34–0·66) | 0·3934 | 0·49 (0·31–0·88) | 0·4901 | 0·48 (0·14–1·01) | 0·3836 | 0·72 (0·46–1·79) | 1·0000 | 0·97 (0·51–1·99) | 1·0000 |
| TNF‐α | 1·09 (0·64–1·58) | 1·0000 | 0·92 (0·52–1·02) | 1·0000 | 0·74 (0·37–1·03) | 0·7250 | 0·53 (0·28–1·15) | 0·5127 | 0·83 (0·53–2·56) | 1·0000 | 0·71 (0·60–1·11) | 1·0000 |
| IFN‐MXA | 0·75 (0·60–1·23) | 1·0000 | 0·56 (0·35–1·08) | 0·7914 | 0·84 (0·59–1·19) | 1·0000 | 0·67 (0·43–0·81) | 0·3345 | 0·83 (0·55–1·17) | 1·0000 | 0·91 (0·57–1·17) | 1·0000 |
| S100A7 | 0·80 (0·51–1·02) | 0·3934 | 0·56 (0·50–0·70) | 0·0266 | 0·56 (0·45–0·72) | 0·0116 b | 0·50 (0·34–0·88) | 0·0105 b | 0·64 (0·54–0·68) | 0·0223 | 0·56 (0·46–0·74) | 0·0180 |
| S100A8 | 0·74 (0·55–1·11) | 1·0000 | 0·50 (0·39–0·54) | 0·0060 b | 0·52 (0·44–0·60) | 0·0248 | 0·67 (0·52–0·73) | 0·2826 | 0·79 (0·59–1·12) | 1·0000 | 0·69 (0·56–0·93) | 0·7091 |
| S100A9 | 0·80 (0·51–1·07) | 0·8442 | 0·35 (0·30–0·53) | 0·0005 b | 0·29 (0·27–0·42) | < 0·0001 b | 0·36 (0·25–0·68) | 0·001 b | 0·52 (0·42–0·78) | 0·0296 | 0·66 (0·52–0·93) | 0·9779 |
| LL‐37 | 0·67 (0·14–1·28) | 0·8997 | 0·40 (0·37–0·83) | 0·5915 | 0·64 (0·38–1·10) | 1·0000 | 0·51 (0·32–1·84) | 1·0000 | 0·77 (0·48–1·37) | 1·0000 | 0·89 (0·67–1·22) | 1·0000 |
| HBD‐2 | 0·48 (0·19–0·87) | 0·0416 | 0·43 (0·29–0·65) | 0·0306 | 0·32 (0·20–0·91) | 0·0445 | 0·69 (0·13–1·07) | 0·1763 | 0·49 (0·43–0·76) | 0·1538 | 0·76 (0·68–1·13) | 1·0000 |
| All (n = 12) | 0·57 (0·34–1·03) | < 0·0001 | 0·51 (0·33–0·92) | < 0·0001 | 0·53 (0·32–0·78) | < 0·0001 | 0·56 (0·29–0·99) | < 0·0001 | 0·64 (0·44–1·00) | < 0·0001 | 0·73 (0·50–1·19) | < 0·0001 |
| Cytokinesc (n = 7) | 0·41 (0·20–0·77) | < 0·0001 | 0·52 (0·32–0·95) | < 0·0001 | 0·54 (0·34–0·75) | < 0·0001 | 0·60 (0·37–0·98) | < 0·0001 | 0·61 (0·39–1·08) | 0·0002 | 0·76 (0·44–1·21) | 0·4780 |
| AMPsd (n = 5) | 0·75 (0·44–1·11) | < 0·0001 | 0·48 (0·33–0·68) | < 0·0001 | 0·47 (0·29–0·73) | < 0·0001 | 0·52 (0·30–0·96) | < 0·0001 | 0·64 (0·45–0·89) | < 0·0001 | 0·73 (0·53–0·98) | 0·0001 |
AMPs, antimicrobial peptides; IQR, interquartile range; IL, interleukin; IFN, interferon; TNF, tumour necrosis factor; Unadj., unadjusted. P‐values marked in bold indicate analytes that are significant at the 0·05 level. aOne HS sample for secukinumab was excluded for real‐time quantitative polymerase chain reaction analysis as a result of a human error during the process of cDNA synthesis (n = 9). bSignificant after correction with the Benjamini–Hochberg test (P < 0·0156). cCytokines: pooled effect of IL‐1β, IL‐6, IL‐8, IL‐17A, IL‐23p19, TNF‐α and IFN‐MXA. dAMPs: pooled effect of S100A7, S100A8, S100A9, LL‐37 and HBD‐2.
Figure 1.
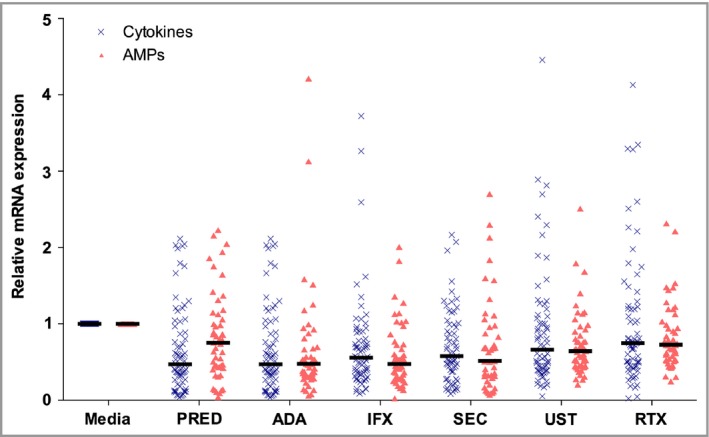
Anti‐inflammatory impact on the relative mRNA expression of the mix of cytokines and antimicrobial peptides (AMPs) in hidradenitis suppurativa (HS) lesional skin (n = 10). Horizontal bars display the median. Media, culture media; PRED, prednisolone 100 μg mL −1; ADA, adalimumab 30 μg mL −1; IFX, infliximab 20 μg mL −1; SEC, secukinumab 30 μg mL −1; UST, ustekinumab 10 μg mL −1; RTX, rituximab 200 μg mL −1.
Significant ex vivo reduction in tumour necrosis factor‐α, interferon γ, interleukin‐1β, interleukin‐6, and interleukin‐17A protein levels in hidradenitis suppurativa lesional skin
In total, 18 cytokines were measured in the culture media. Six cytokines were not detected in the majority of HS samples throughout all conditions, namely IL‐1RI, IL‐5, IL‐12/23p40, IL‐22, IL‐31 and IL‐33. Relative changes in protein production including significance levels for the other 12 cytokines are shown in Table 2. The overall median effect on cytokines per condition was as follows: prednisolone 0·51 (0·28–0·64); adalimumab 0·78 (0·54–0·91); infliximab 0·70 (0·49–0·94); secukinumab 0·85 (0·69–0·95); ustekinumab 0·71 (0·57–0·79) and rituximab 0·81 (0·70–0·94). Specifically the release of TNF‐α, IFNγ, IL‐1β, IL‐6 and IL‐17A, was significantly inhibited by all tested drugs with the exception of secukinumab (Fig. 2). In addition, prednisolone significantly inhibited the release of TNF‐α, IFNγ, IL‐1β, IL‐6, IL‐17A, IL‐10 and IL‐25/17E. As expected, adalimumab and infliximab almost completely neutralized TNF‐α levels in the supernatants (Fig. 3). Of note, for the two important cytokines TNF‐α and IL‐17A we observed both a strong intrapatient and interpatient variability among all biologic conditions (Fig. 3).
Table 2.
Relative protein production and modulation by biologics in hidradenitis suppurativa (HS) lesional skin
| HS lesional skin (n = 9) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prednisolone | Adalimumab | Infliximab | Secukinumab | Ustekinumab | Rituximab | |||||||
| Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | Median (IQR) | Unadj. P‐values | |
| TNF‐α | 0·20 (0·15–0·29) | 0·0022 a | 0·09 (0·07–0·20) | < 0·0001 a | 0·03 (0·02–0·05) | < 0·0001 a | 0·89 (0·53–0·93) | 1·0000 | 0·54 (0·23–0·76) | 0·3305 | 0·70 (0·65–1·10) | 1·0000 |
| IFNγ | 0·37 (0·11–0·53) | 0·0068 a | 0·43 (0·30–1·17) | 0·4169 | 0·46 (0·13–0·91) | 0·0673 | 0·56 (0·51–0·66) | 0·6285 | 0·39 (0·33–0·61) | 0·0673 | 0·49 (0·41–0·60) | 0·2596 |
| CCL‐20 | 0·50 (0·20–0·81) | 0·2845 | 0·58 (0·28–1·80) | 1·0000 | 0·71 (0·53–1·13) | 1·0000 | 0·94 (0·53–1·19) | 1·0000 | 0·73 (0·62–1·28) | 1·0000 | 0·95 (0·70–1·38) | 1·0000 |
| IL‐1β | 0·23 (0·17–0·45) | 0·0049 a | 0·41 (0·29–0·73) | 0·3114 | 0·44 (0·31–0·75) | 0·3305 | 1·08 (0·46–1·43) | 1·0000 | 0·59 (0·45–2·06) | 1·0000 | 0·70 (0·52–1·03) | 1·0000 |
| IL‐6 | 0·59 (0·46–0·73) | 0·0006 a | 0·81 (0·68–0·85) | 0·0724 | 0·76 (0·64–0·91) | 0·1035 | 0·99 (0·76–1·08) | 1·0000 | 0·76 (0·69–0·99) | 0·1556 | 0·87 (0·71–0·92) | 0·1035 |
| IL‐17A | 0·09 (0·07–0·19) | < 0·0001 a | 0·83 (0·68–1·38) | 1·0000 | 0·50 (0·32–0·52) | 0·2085 | 0·70 (0·60–1·60) | 1·0000 | 0·54 (0·25–0·81) | 0·3022 | 0·96 (0·34–1·24) | 1·0000 |
| IL‐18 | 0·86 (0·59–1·03) | 1·0000 | 0·75 (0·66–3·48) | 1·0000 | 1·05 (0·74–1·11) | 1·0000 | 0·90 (0·71–1·04) | 1·0000 | 0·82 (0·78–0·90) | 0·7341 | 0·93 (0·78–2·75) | 1·0000 |
| IL‐36β | 0·94 (0·48–1·45) | 1·0000 | 0·98 (0·81–2·04) | 1·0000 | 1·04 (0·82–1·47) | 1·0000 | 1·88 (0·94–2·34) | 1·0000 | 1·05 (0·78–1·37) | 1·0000 | 1·21 (0·85–1·65) | 1·0000 |
| IL‐10b | 0·29 (0·20–0·41) | 0·0062 a | 1·12 (0·22–1·94) | 1·0000 | 0·93 (0·37–1·05) | 1·0000 | 0·81 (0·62–2·02) | 1·0000 | 0·91 (0·55–1·42) | 1·0000 | 0·62 (0·43–0·91) | 0·6975 |
| IL‐25/17Eb | 0·52 (0·33–0·64) | 0·0043 a | 0·59 (0·48–0·95) | 0·2517 | 0·50 (0·44–1·01) | 0·0931 | 0·68 (0·55–0·88) | 0·3208 | 0·67 (0·55–0·90) | 0·2932 | 0·74 (0·52–0·90) | 0·2440 |
| IL‐19b | 0·68 (0·64–0·99) | 0·9862 | 0·91 (0·51–1·67) | 1·0000 | 0·69 (0·56–1·31) | 1·0000 | 0·79 (0·48–0·92) | 1·0000 | 0·77 (0·65–1·12) | 1·0000 | 0·92 (0·51–1·52) | 1·0000 |
| IL‐27b | 0·63 (0·26–0·85) | 0·0751 | 0·92 (0·72–1·01) | 1·0000 | 0·95 (0·61–1·12) | 1·0000 | 0·66 (0·64–1·02) | 0·963 | 0·69 (0·62–0·83) | 0·3609 | 0·72 (0·63–0·79) | 0·2365 |
| Allc (n = 12) | 0·51 (0·28–0·64) | < 0·0001 | 0·78 (0·54–0·91) | 0·0006 | 0·70 (0·49–0·94) | < 0·0001 | 0·85 (0·69–0·95) | 0·0684 | 0·71 (0·57–0·79) | 0·0010 | 0·81 (0·70–0·94) | 0·0121 |
| Proinflammatory (n = 8) | 0·45 (0·19–0·82) | NT | 0·69 (0·29–1·18) | NT | 0·64 (0·30–1·04) | NT | 0·91 (0·53–1·43) | NT | 0·71 (0·42–0·99) | NT | 0·80 (0·58–1·22) | NT |
| Anti‐inflammatory (n = 4) | 0·54 (0·29–0·70) | NT | 0·87 (0·51–1·42) | NT | 0·70 (0·46–1·11) | NT | 0·69 (0·54–1·13) | NT | 0·75 (0·61–1·05) | NT | 0·74 (0·50–0·98) | NT |
| Importantd (n = 5) | 0·23 (0·20–0·37) | < 0·0001 | 0·43 (0·41–0·81) | < 0·0001 | 0·46 (0·44–0·50) | < 0·0001 | 0·89 (0·70–0·99) | 0·0663 | 0·54 (0·54–0·59) | < 0·0001 | 0·70 (0·70–0·87) | 0·0071 |
AMPs, antimicrobial peptides; IQR, interquartile range; Unadj, unadjusted; NT, not tested; IL, interleukin; IFN, interferon; TNF, tumour necrosis factor. P‐values marked in bold indicate analytes that are significant at the 0·05 level. aSignificant after correction with the Benjamini–Hochberg test (P < 0·0156). bCytokines with an anti‐inflammatory function, note that IL‐17E and IL‐27 also have an immunoregulatory/proinflammatory function. cAll: pooled effect of 12 cytokines. dImportant: five key proinflammatory cytokines in the pathophysiology of HS; TNF‐α, IFNγ, IL‐1β, IL‐6 and IL‐17A.
Figure 2.
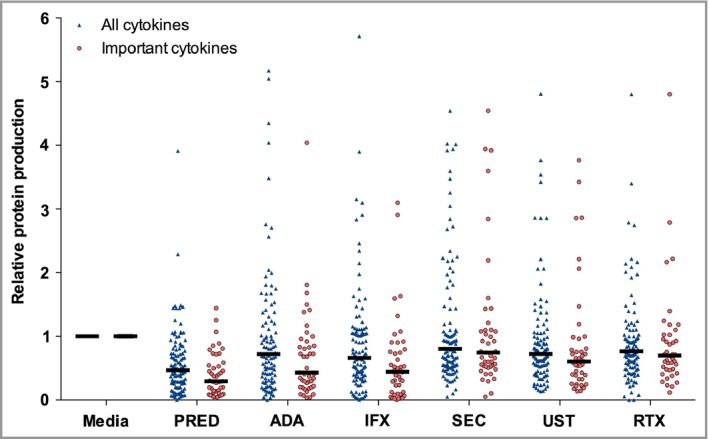
Anti‐inflammatory impact on protein production of all cytokines and five important cytokines as measured by Luminex assay (n = 9).
The important inflammatory cytokines are tumour necrosis factor‐α, interferon γ, interleukin (IL)‐1β, IL‐6, IL‐17A. A total of 12 data points are outside the y‐axis range. Media, culture media; PRED, prednisolone 100 μg mL −1; ADA, adalimumab 30 μg mL −1; IFX, infliximab 20 μg mL −1; SEC, secukinumab 30 μg mL −1; UST, ustekinumab 10 μg mL −1; RTX, rituximab 200 μg mL −1.
Figure 3.
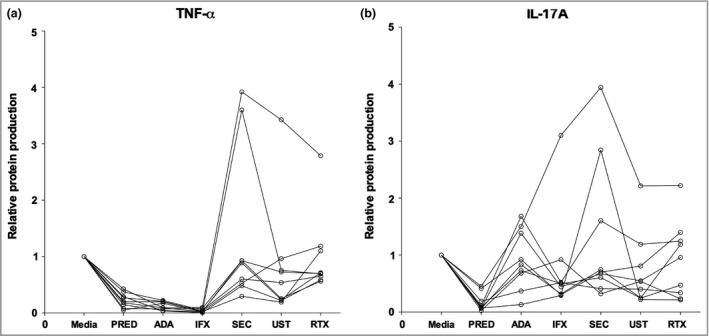
Interpatient and intrapatient variability demonstrated by fluctuating protein levels of tumour necrosis factor (TNF)‐α and interleukin (IL)‐17A (n = 9). (a) TNF‐α. Connected dots represent one patient. High intrapatient variability as demonstrated by the fluctuating per‐patient protein levels between conditions. Low interpatient variability was observed for conditions PRED, ADA and IFX. (b) IL‐17A. Connected dots represent one patient. Very high (more than TNF‐α) intrapatient variability. Low interpatient variability for was observed for the condition PRED. PRED, prednisolone 100 μg mL −1; ADA, adalimumab 30 μg mL −1; IFX, infliximab 20 μg mL −1. SEC, secukinumab 30 μg mL−1; UST, ustekinumab 10 μg mL−1; RTX, rituximab 200 μg mL−1.
Great variation in correlation between mRNA expression and protein production levels in hidradenitis suppurativa samples
The cytokines IL‐1β, IL‐6, IL‐17A and TNF‐α, were measured by both real‐time qPCR and Luminex. There was great variation in the correlation between mRNA and protein levels of these individual cytokines (Fig. 4). A high correlation between mRNA and protein levels was found for IL‐17A (r = 0·86, P < 0·001), while the correlation for TNF‐α was almost zero (r = 0·04, P = 0·7480).
Figure 4.
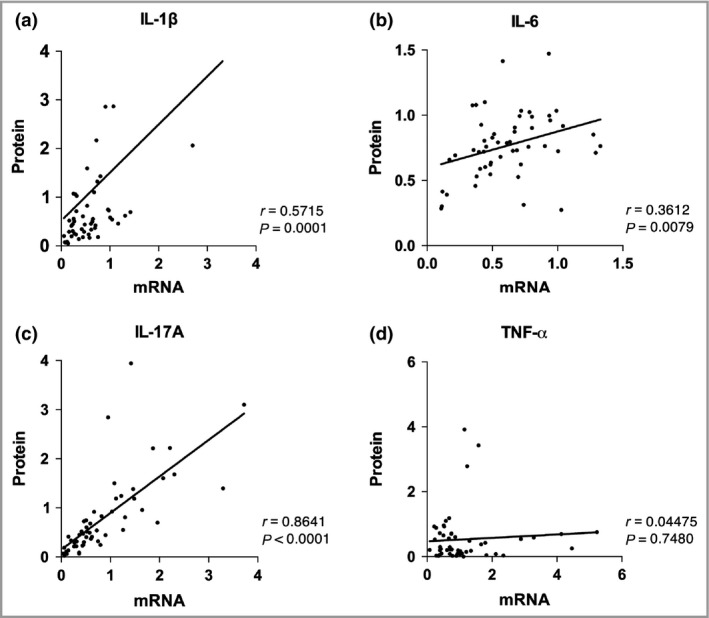
Correlation between the relative protein production (y‐axis) and mRNA (x‐axis) expression of hidradenitis suppurativa (HS) lesional skin (n = 9). (a) Interleukin (IL)‐1β. (b) IL‐6. (c) IL‐17A. (d) Tumour necrosis factor (TNF)‐α. Values on x‐axis and y‐axis display the relative production and expression values of a cytokine. P‐values were calculated according to Spearman's rho test.
Discussion
In this study, we show that the commercially available biologics, used in daily practice for the treatment of HS (adalimumab, infliximab, ustekinumab and rituximab), significantly inhibited mRNA and protein expression of various cytokines and AMPs in lesional HS skin, cultured ex vivo for 24 h. Secukinumab demonstrated significant downregulation of inflammatory markers in the lesional skin only at the mRNA level. The anti‐inflammatory effect of prednisolone and all biologics, except secukinumab, was the strongest at the protein levels of TNF‐α, IFNγ, IL‐1β, IL‐6 and IL‐17A, which are important cytokines in the pathogenesis of HS. Prednisolone and TNF‐α inhibitors seemed to be the most effective in reducing the release of a broader range of proinflammatory cytokines and AMPs in lesional HS skin. Only prednisolone, adalimumab and infliximab had an inhibitory effect on the mRNA expression levels of inflammatory markers in healthy control skin (see supporting information).
In patients with moderate‐to‐severe HS, the strongest evidence has been documented for the anti‐TNF agents adalimumab and infliximab.5, 30, 31, 32 Our translational findings correspond with the observed efficacy of TNF‐α inhibitors in HS in daily practice. In addition, we confirmed the previously reported decrease of ex vivo IL‐1β protein expression after adalimumab treatment on the mRNA level.7 As TNF‐α is a multifunctional cytokine with numerous actions, a simultaneous downregulation of other cytokines, such as IL‐6 and cytokines of the IL‐1 family, could also explain the results of the anti‐TNF agents in our study.9, 33
Remarkably, secukinumab did not reduce the IL‐17 protein levels in the same way that adalimumab and infliximab reduced TNF‐α protein levels. Unfortunately, IL‐12p40 protein (target of ustekinumab), an important indicator of the IL‐17 pathway, fell below the level of detection in the Luminex assay. It could be that other cytokines, such as TNF‐α and IL‐1β, not being blocked by secukinumab in this culture system, were still able to induce production of IL‐17. Moreover, levels of IL‐17A were in the lower range of detection in most patients. Another explanation may be that the anti‐IL‐17A antibodies used in the Luminex assay detect a different epitope of IL‐17A than that recognized by secukinumab. The lower mRNA expression of S100A7, S100A9, IL‐6 and CXCL‐8 could be considered to be the result of blocking of IL‐17A bioactivity by secukinumab. Similarly, at the mRNA level, ustekinumab exerted its action predominantly via IL‐6, CXCL‐8, S100A7 and S100A9, although the last three markers were nonsignificant after Benjamini–Hochberg correction.
The pan‐cytokine inhibitory characteristics of prednisolone were demonstrated by inhibition of multiple cytokines on both the mRNA and protein level, which supports the efficacy of systemic and intralesional corticosteroids for acute HS flares in clinical practice.34 However, prolonged high‐dose systemic corticosteroids are not recommended as HS rapidly flares after tapering, especially after a long course.35 Nonetheless, low‐dose systemic prednisolone could be a valuable adjunct therapy for recalcitrant HS.36
Rituximab was the only biologic without a significant inhibitory effect on individual inflammatory mRNA and protein levels. This is not surprising as B‐cell blockade in inflammatory diseases acts via inhibition of antibody production, antigen presentation and indirectly via cytokine reduction.37 Therefore, the presence of the complete immune system is required for B‐cell blockade to be effective. Moreover, HS is considered primarily a disease of a deficient innate immunity. On the other hand, chronic HS lesions are full of B cells and plasma cells, indicating that adaptive humoral immunity is also activated in longstanding HS.20, 38
The impact of biologics on AMP expression has never been investigated in HS lesional skin. Our findings indicate a potential supporting role for AMPs in HS pathophysiology as it is known that AMPs are capable of activating keratinocytes and attracting innate immune cells to amplify the local immune response.39 Cytokines produced by innate and adaptive immune cells, such as TNF‐α, IL‐17 and IL‐12, drive AMP production in the keratinocytes.40, 41, 42 Moreover, AMPs can be activated by damage‐ and pathogen‐associated molecular patterns after follicle rupture with the release of keratin fibres and skin commensals in the dermis.
Although it is assumed that levels of mRNA and protein have a one‐to‐one correlation, the absence of correlation between TNF‐α mRNA and its protein levels has been previously reported.43, 44, 45 In addition, other factors such as the half‐life of proteins and the degradation and stability of mRNA may vary widely.43, 46, 47
Major strengths of our study are the use of a standardized ex vivo transwell culture system and weighing each biopsy in order to normalize all protein levels for mg tissue weight. Cytokines were evaluated on both mRNA and protein level, including four cytokines that were assessed for validation purposes. Furthermore, the use of healthy control skin from regions that are suitable to function as control samples further increases the validity of our study. Possible limitations include the relatively small sample size, lack of dose–response relationships, and AMPs that have only been evaluated on the mRNA level. Moreover, further analysis using staining techniques at the cellular level could potentially indicate the effects of the biologics on the mRNA expression and protein production of inflammatory markers in different cellular subsets.
In conclusion, prednisolone, adalimumab, infliximab, secukinumab, ustekinumab and rituximab significantly inhibited the expression of inflammatory cytokines and AMPs in ex vivo actively inflamed lesional skin of patients with HS. The ex vivo model will enable studies with combinations of biologics and the targeting of novel important cytokines, alone or in combination with low‐dose prednisolone.
Supporting information
Fig S1. Relative mRNA expression of all cytokines and all antimicrobial peptides (AMPs) in healthy control skin, (n = 5).
Table S1. Relative expression of mRNA and modulation by biologics in healthy control skin.
Acknowledgments
We thank Marja Smits for the protein quantification using the Luminex assay.
Funding sources None.
Conflicts of interest H.H.v.d.Z. has been an advisory board member for AbbVie, InflaRX, and a speaker for Galderma. E.P.P. has acted as a consultant, speaker, principal investigator or received grants from AbbVie, Amgen, Biogen, Celgene, Eli Lilly, Janssen‐Cilag, Novartis, Pfizer and UCB.
A.R.J.V.V. and C.B.A. share joint first authorship.
References
- 1. Jemec GB. Clinical practice. Hidradenitis suppurativa. N Engl J Med 2012; 366:158–64. [DOI] [PubMed] [Google Scholar]
- 2. Kelly G, Sweeney CM, Tobin AM, Kirby B. Hidradenitis suppurativa: the role of immune dysregulation. Int J Dermatol 2014; 53:1186–96. [DOI] [PubMed] [Google Scholar]
- 3. Mozeika E, Pilmane M, Nürnberg BM, Jemec GB. Tumour necrosis factor‐alpha and matrix metalloproteinase‐2 are expressed strongly in hidradenitis suppurativa. Acta Derm Venereol 2013; 93:301–4. [DOI] [PubMed] [Google Scholar]
- 4. van der Zee HH, de Ruiter L, van den Broecke DG et al Elevated levels of tumour necrosis factor (TNF)‐α, interleukin (IL)‐1β and IL‐10 in hidradenitis suppurativa skin: a rationale for targeting TNF‐α and IL‐1β. Br J Dermatol 2011; 164:1292–8. [DOI] [PubMed] [Google Scholar]
- 5. Kimball AB, Okun MM, Williams DA et al Two phase 3 trials of adalimumab for Hidradenitis Suppurativa. N Engl J Med 2016; 375:422–34. [DOI] [PubMed] [Google Scholar]
- 6. Grant A, Gonzalez T, Montgomery MO et al Infliximab therapy for patients with moderate to severe hidradenitis suppurativa: a randomized, double‐blind, placebo‐controlled crossover trial. J Am Acad Dermatol 2010; 62:205–17. [DOI] [PubMed] [Google Scholar]
- 7. van der Zee HH, Laman JD, de Ruiter L et al Adalimumab (antitumour necrosis factor‐α) treatment of hidradenitis suppurativa ameliorates skin inflammation: an in situ and ex vivo study. Br J Dermatol 2012; 166:298–305. [DOI] [PubMed] [Google Scholar]
- 8. von Laffert M, Helmbold P, Wohlrab J et al Hidradenitis suppurativa (acne inversa): early inflammatory events at terminal follicles and at interfollicular epidermis. Exp Dermatol 2010; 19:533–7. [DOI] [PubMed] [Google Scholar]
- 9. Moran B, Sweeney CM, Hughes R et al Hidradenitis suppurativa is characterized by dysregulation of the Th17: Treg cell axis, which is corrected by anti‐TNF therapy. J Invest Dermatol 2017; 137:2389–95. [DOI] [PubMed] [Google Scholar]
- 10. Schlapbach C, Hanni T, Yawalkar N, Hunger RE. Expression of the IL‐23/Th17 pathway in lesions of hidradenitis suppurativa. J Am Acad Dermatol 2011; 65:790–8. [DOI] [PubMed] [Google Scholar]
- 11. Menis D, Maroñas‐Jiménez L, Delgado‐Marquez AM et al Two cases of severe hidradenitis suppurativa with failure of anakinra therapy. Br J Dermatol 2015; 172:810–11. [DOI] [PubMed] [Google Scholar]
- 12. Russo V, Alikhan A. Failure of anakinra in a case of severe hidradenitis suppurativa. J Drugs Dermatol 2016; 15:772–4. [PubMed] [Google Scholar]
- 13. Tzanetakou V, Kanni T, Giatrakou S et al Safety and efficacy of anakinra in severe hidradenitis suppurativa: a randomized clinical trial. JAMA Dermatol 2016; 152:52–9. [DOI] [PubMed] [Google Scholar]
- 14. Thorlacius L, Theut Riis P, Jemec GBE. Severe hidradenitis suppurativa responding to treatment with secukinumab: a case report. Br J Dermatol 2018; 179:182–5. [DOI] [PubMed] [Google Scholar]
- 15. Schuch A, Fischer T, Boehner A et al Successful treatment of severe recalcitrant hidradenitis suppurativa with the interleukin‐17A antibody secukinumab. Acta Derm Venereol 2018; 98:151–2. [DOI] [PubMed] [Google Scholar]
- 16. Blok JL, Li K, Brodmerkel C et al Ustekinumab in hidradenitis suppurativa: clinical results and a search for potential biomarkers in serum. Br J Dermatol 2016; 174:839–46. [DOI] [PubMed] [Google Scholar]
- 17. Bechara FG, Sand M, Skrygan M et al Acne inversa: evaluating antimicrobial peptides and proteins. Ann Dermatol 2012; 24:393–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolk K, Warszawska K, Hoeflich C et al Deficiency of IL‐22 contributes to a chronic inflammatory disease: pathogenetic mechanisms in acne inversa. J Immunol 2011; 186:1228–39. [DOI] [PubMed] [Google Scholar]
- 19. Hofmann SC, Saborowski V, Lange S et al Expression of innate defense antimicrobial peptides in hidradenitis suppurativa. J Am Acad Dermatol 2012; 66:966–74. [DOI] [PubMed] [Google Scholar]
- 20. van der Zee HH, de Ruiter L, Boer J et al Alterations in leucocyte subsets and histomorphology in normal‐appearing perilesional skin and early and chronic hidradenitis suppurativa lesions. Br J Dermatol 2012; 166:98–106. [DOI] [PubMed] [Google Scholar]
- 21. Takahashi K, Yanagi T, Kitamura S et al Successful treatment of hidradenitis suppurativa with rituximab for a patient with idiopathic carpotarsal osteolysis and chronic active antibody‐mediated rejection. J Dermatol 2018; 45:e116–17. [DOI] [PubMed] [Google Scholar]
- 22. Companjen AR, van der Wel LI, Wei L et al A modified ex vivo skin organ culture system for functional studies. Arch Dermatol Res 2001; 293:184–90. [DOI] [PubMed] [Google Scholar]
- 23. Wei L, Debets R, Hegmans JJ et al IL‐1 beta and IFN‐gamma induce the regenerative epidermal phenotype of psoriasis in the transwell skin organ culture system. IFN‐gamma up‐regulates the expression of keratin 17 and keratinocyte transglutaminase via endogenous IL‐1 production. J Pathol 1999; 187:358–64. [DOI] [PubMed] [Google Scholar]
- 24. Menting SP, Coussens E, Pouw MF et al Developing a therapeutic range of adalimumab serum concentrations in management of psoriasis: a step toward personalized treatment. JAMA Dermatol 2015; 151:616–22. [DOI] [PubMed] [Google Scholar]
- 25. Takahashi H, Tsuji H, Ishida‐Yamamoto A, Iizuka H. Plasma trough levels of adalimumab and infliximab in terms of clinical efficacy during the treatment of psoriasis. J Dermatol 2013; 40:39–42. [DOI] [PubMed] [Google Scholar]
- 26. Menting SP, van den Reek JM, Baerveldt EM et al The correlation of clinical efficacy, serum trough levels and antidrug antibodies in ustekinumab‐treated patients with psoriasis in a clinical‐practice setting. Br J Dermatol 2015; 173:855–7. [DOI] [PubMed] [Google Scholar]
- 27. Cosentyx® (secukinumab) [summary of product characteristics]. Basel: Novartis AG, 2015. [Google Scholar]
- 28. Tran L, Baars JW, Aarden L et al Pharmacokinetics of rituximab in patients with CD20 positive B‐cell malignancies. Hum Antibodies 2010; 19:7–13. [DOI] [PubMed] [Google Scholar]
- 29. Gabert J, Beillard E, van der Velden VH et al Standardization and quality control studies of ‘real‐time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia – a Europe Against Cancer program. Leukemia 2003; 17:2318–57. [DOI] [PubMed] [Google Scholar]
- 30. Delage M, Samimi M, Atlan M et al Efficacy of infliximab for hidradenitis suppurativa: assessment of clinical and biological inflammatory markers. Acta Derm Venereol 2011; 91:169–71. [DOI] [PubMed] [Google Scholar]
- 31. Lesage C, Adnot‐Desanlis L, Perceau G et al Efficacy and tolerance of prolonged infliximab treatment of moderate‐to‐severe forms of hidradenitis suppurativa. Eur J Dermatol 2012; 22:640–4. [DOI] [PubMed] [Google Scholar]
- 32. Miller I, Lynggaard CD, Lophaven S et al A double‐blind placebo‐controlled randomized trial of adalimumab in the treatment of hidradenitis suppurativa. Br J Dermatol 2011; 165:391–8. [DOI] [PubMed] [Google Scholar]
- 33. Delves PJ. Tumor necrosis factor‐α In: Encyclopedia of Immunology, (Delves PJ, Roitt IM. eds). London: Elsevier, 1998; 2435–40. [Google Scholar]
- 34. Riis PT, Boer J, Prens EP et al Intralesional triamcinolone for flares of hidradenitis suppurativa (HS): a case series. J Am Acad Dermatol 2016; 75:1151–5. [DOI] [PubMed] [Google Scholar]
- 35. Alhusayen R, Shear NH. Scientific evidence for the use of current traditional systemic therapies in patients with hidradenitis suppurativa. J Am Acad Dermatol 2015; 73 (5 Suppl. 1):S42–6. [DOI] [PubMed] [Google Scholar]
- 36. Wong D, Walsh S, Alhusayen R. Low‐dose systemic corticosteroid treatment for recalcitrant hidradenitis suppurativa. J Am Acad Dermatol 2016; 75:1059–62. [DOI] [PubMed] [Google Scholar]
- 37. Dörner T, Kinnman N, Tak PP. Targeting B cells in immune‐mediated inflammatory disease: a comprehensive review of mechanisms of action and identification of biomarkers. Pharmacol Ther 2010; 125:464–75. [DOI] [PubMed] [Google Scholar]
- 38. Giamarellos‐Bourboulis EJ, Antonopoulou A, Petropoulou C et al Altered innate and adaptive immune responses in patients with hidradenitis suppurativa. Br J Dermatol 2007; 156:51–6. [DOI] [PubMed] [Google Scholar]
- 39. van der Zee HH, Laman JD, Boer J, Prens EP. Hidradenitis suppurativa: viewpoint on clinical phenotyping, pathogenesis and novel treatments. Exp Dermatol 2012; 21:735–9. [DOI] [PubMed] [Google Scholar]
- 40. Kolls JK, McCray PB Jr, Chan YR. Cytokine‐mediated regulation of antimicrobial proteins. Nat Rev Immunol 2008; 8:829–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liang SC, Tan XY, Luxenberg DP et al Interleukin (IL)‐22 and IL‐17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 2006; 203:2271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hollox EJ, Huffmeier U, Zeeuwen PL et al Psoriasis is associated with increased beta‐defensin genomic copy number. Nat Genet 2008; 40:23–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett 2009; 583:3966–73. [DOI] [PubMed] [Google Scholar]
- 44. Schulz S, Schagdarsurengin U, Suss T et al Relation between the tumor necrosis factor‐alpha (TNF‐alpha) gene and protein expression, and clinical, biochemical, and genetic markers: age, body mass index and uric acid are independent predictors for an elevated TNF‐alpha plasma level in a complex risk model. Eur Cytokine Netw 2004; 15:105–11. [PubMed] [Google Scholar]
- 45. Rioja I, Bush KA, Buckton JB et al Joint cytokine quantification in two rodent arthritis models: kinetics of expression, correlation of mRNA and protein levels and response to prednisolone treatment. Clin Exp Immunol 2004; 137:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 2003; 4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012; 13:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Relative mRNA expression of all cytokines and all antimicrobial peptides (AMPs) in healthy control skin, (n = 5).
Table S1. Relative expression of mRNA and modulation by biologics in healthy control skin.


