Abstract
Salinity stress is an important cause of crop yield loss in many parts of the world. Here, we performed genome‐wide association studies of salinity‐stress responsive traits in 132 HapMap genotypes of the model legume Medicago truncatula. Plants grown in soil were subjected to a step‐wise increase in NaCl concentration, from 0 through 0.5% and 1.0% to 1.5%, and the following traits were measured: vigor, shoot biomass, shoot water content, leaf chlorophyll content, leaf size, and leaf and root concentrations of proline and major ions (Na+, Cl−, K+, Ca2+, etc.). Genome‐wide association studies were carried out using 2.5 million single nucleotide polymorphisms, and 12 genomic regions associated with at least four traits each were identified. Transcript‐level analysis of the top eight candidate genes in five extreme genotypes revealed association between salinity tolerance and transcript‐level changes for seven of the genes, encoding a vacuolar H+‐ATPase, two transcription factors, two proteins involved in vesicle trafficking, one peroxidase, and a protein of unknown function. Earlier functional studies on putative orthologues of two of the top eight genes (a vacuolar H+‐ATPase and a peroxidase) demonstrated their involvement in plant salinity tolerance.
Keywords: GWAS, legume, Medicago truncatula, proline, salinity, SNP, vesicle trafficking
Short abstract
Genome wide association studies (GWAS) of multiple physiological and biochemical traits related to salinity‐stress adaptation in M. truncatula were carried out using 2.5 million SNPs. Correlation analysis among the traits showed that salinity tolerance in the population is negatively correlated to sodium, chloride, and proline concentration in the leaf, while root proline levels were positively associated with salinity‐stress tolerance. GWAS analyses were performed with a mixed linear model, and 12 genomic regions that are associated with multiple traits each were identified; these regions harbor a total of 214 SNPs and 74 genes. These genomic regions were confirmed by a GWAS analysis using PC1 as a virtual trait, generated from principle component analysis of the 12 traits that are tightly correlated. Significant linkage disequilibrium was observed among these SNPs, possibly indicating co‐evolution of salinity tolerance alleles. Examination of gene expression of eight top suspect genes revealed associations between salinity tolerance and transcript level changes under salinity stress. Earlier functional studies on orthologues of two of the top eight genes (a vacuolar H+−ATPase and a peroxidase) confirmed their involvement in plant salinity tolerance in other species. Functional annotation of potential salinity tolerance genes points to the importance of transcriptional regulation, vesicle trafficking, and ROS scavenging under saline conditions.
1. INTRODUCTION
Salinity is an important abiotic stress that restricts crop distribution and reduces agricultural yield. It is estimated that over 6% of the world's total land area is affected by excess salts (Smajgl et al., 2015), and approximately 20% of arable land in more than 100 countries is affected by salinity (Sairam & Tyagi, 2004). Increasing salinity tolerance in crops will help to ensure food, feed, and industrial feedstock production on salt‐affected land.
The fundamental mechanisms of how plants sense and respond to salinity stress in both glycophytes and halophytes have been studied extensively, but remain incompletely understood. Multiple transporters and channels such as the Na+/H+ antiporter SOS1, the Na+/H+ exchanger NHX, the high affinity potassium transporter HKT1, as well as nonselective cation channels, have been shown to play important roles in maintaining cellular and plant‐level ion homeostasis under salinity stress (Julkowska & Testerink, 2015; Keisham, Mukherjee, & Bhatla, 2018). In addition, important genes involved in the signal transduction pathways that respond to salinity have been identified, including calcium‐dependent protein kinases (CDPKs), calcineurin B‐like proteins (CBLs), CBL‐interacting protein kinases (CIPKs), and mitogen‐activated protein kinases (MAPKs) (Shabala, Wu, & Bose, 2015).
To minimize the ionic stress caused by Na+ and Cl−, cells exclude and/or remove these ions from their cytoplasm via transporters, which can result in osmotic stress. To alleviate such stress, plant cells synthesize compatible solutes such as proline, glycine betaine, and soluble sugars that help them to retain water when ion levels in the apoplast or intracellular compartments are high. Proline also acts as a reactive oxygen species (ROS) scavenger and molecular chaperone to stabilize proteins and bio‐membranes under stress (Ashraf & Foolad, 2007; Matysik, Alia, & Mohanty, 2002). Proline biosynthesis serves as a redox buffer by consuming two NADPH per proline molecule, which helps to utilize excess electrons generated in the chloroplast under stress (Hare & Cress, 1997). Although generally regarded as a beneficial osmoticum, the link between proline and stress tolerance is somewhat unclear. In some studies, proline was found to accumulate more in tolerant plant genotypes than in sensitive genotypes under salt stress, consistent with an active role in stress tolerance (Jain, Nainawatee, Jain, & Chowdhury, 1991; Misra & Gupta, 2005; Ranganayakulu, Veeranagamallaiah, & Sudhakar, 2013). In other studies, however, proline levels are not positively correlated with salinity tolerance, but instead appear to increase as a consequence of cellular damage (Aziz, Martin‐Tanguy, & Larher, 1998; Lacerda, Cambraia, Oliva, & Ruiz, 2003; Moftah & Michel, 1987). Nonetheless, at least one genome‐wide association study (GWAS) has been performed to identify genes controlling proline level under dehydration stress in Arabidopsis (Verslues, Lasky, Juenger, Liu, & Kumar, 2014).
In leaves, high Na+ concentrations cause stomatal closure due to the osmotic effect of the solute, which consequently causes a reduction in the rates of photosynthesis and growth (Brugnoli & Lauteri, 1991; Munns & Tester, 2008). Another consequence of decreased stomatal aperture is that intercellular CO2 limitation causes NADP+ pool depletion, photorespiration enhancement, and ROS accumulation (Hossain & Dietz, 2016). Although important as signaling molecules, excess ROS may trigger chlorophyll degradation and, ultimately, cell death (Miller, Suzuki, Ciftci‐yilmaz, & Mittler, 2010; Suzuki, Koussevitzky, Mittler, & Miller, 2012). To scavenge ROS, antioxidant enzymes are positively regulated under salinity stress, for example, peroxidases, superoxide dismutase (SOD), catalase (CAT), etc. (Hossain & Dietz, 2016). Given the redox challenges of photosynthetic cells under stress, leaf and shoot growth are generally more sensitive to salinity stress than root growth (Hamaji et al., 2009; Munns, Passioura, Guo, Chazen, & Cramer, 2000).
To improve plant salinity tolerance, multiple approaches have been pursued. Buoyed by discovery of genes involved in plant salinity responses, genetic engineering has been used to generate a variety of transgenic plants in attempts to increase stress tolerance. Although positive outcomes have repeatedly been reported for transgenic plants under controlled‐growth conditions, less success has been found under field conditions (Fita, Rodríguez‐Burruezo, Boscaiu, Prohens, & Vicente, 2015; Flowers, 2004; Hanin, Ebel, Ngom, Laplaze, & Masmoudi, 2016). On the other hand, conventional breeding for salinity tolerance, through selection and introgression, has met with limited success in rice and wheat, possibly because of the complex nature of this trait (Ashraf & Foolad, 2013; Munns, James, & Läuchli, 2006; Varshney, Bansal, Aggarwal, Datta, & Craufurd, 2011).
In recent years, with advances in next generation sequencing technology, great progress has been made in identifying quantitative trait loci (QTL), associated with salt tolerance (Ashraf & Foolad, 2013; Hamwieh et al., 2011; Thomson et al., 2010). For example, a Na+/H+ antiporter (GmCHX1) was identified as a major salt‐tolerance gene in the soybean wild accessions by combining whole‐genome sequencing with QTL mapping (Qi et al., 2014). Later, GWAS indicated that this gene is a major contributor to the variance in salinity sensitivity among multiple soybean ecotypes; a conclusion supported by marker‐assisted selection that resulted in salt‐tolerant lines of soybean (Do et al., 2016; Patil et al., 2016; Zeng et al., 2017). Because cultivated soybean genotypes that contain nonfunctional GmCHX1 are generally very sensitive to salinity stress, it was postulated that loss of function of this gene in soybean may improve growth and seed production in nonsaline environments (Qi et al., 2014). Similar studies in Arabidopsis identified the sodium transporter, AtHTK1, as a major contributor to variance in shoot sodium accumulation under salinity stress in wild populations (Baxter et al., 2010; Busoms et al., 2015; Rus et al., 2006). Another GWAS study in Arabidopsis (Julkowska et al., 2016) indicated that a kinase, LRR‐KISS (Leucine‐Rich Repeat Kinase family protein Induced by Salt Stress), underlies variance in plant growth under salinity stress.
Apart from the progress noted above for soybean and Arabidopsis, little is known about the genes shaping natural variation in salinity tolerance in other species. A few traits affected by salinity stress have been investigated via GWAS with high density markers, including traits related to seed germination, growth rate, transpiration rate, and tissue Na+/K+ contents in rice (Al‐Tamimi et al., 2016; Patishtan, Hartley, Fonseca de Carvalho, & Maathuis, 2017; Shi et al., 2017), and root growth in Arabidopsis (Kobayashi et al., 2016). However, no overlapping genes or common mechanisms have been identified among these studies, and none of the identified genes have been validated with respect to salinity tolerance. Apart from soybean, no GWAS analysis of salinity traits with high‐density markers has been performed in other legumes.
Medicago truncatula is a model legume species for genetic and genomic research (Barker et al., 1990; Kang, Li, Sinharoy, & Verdier, 2016; Young & Udvardi, 2009). Since the launch of the M. truncatula HapMap (Haplotype Map) project in 2000, 262 genotypes have been sequenced and the resulting single nucleotide polymorphisms (SNP) information released (Young et al., 2011). In the current study, we characterized a spectrum of salinity stress‐related traits in a diverse subset of the M. truncatula HapMap panel consisting of 132 M. truncatula genotypes. Plants were grown in soil and were subjected to a step‐wise increase in NaCl concentration, from 0 through 0.5% and 1.0% to 1.5%. Traits that were measured in salt‐stressed (and control) plants included a qualitative, visual salt‐tolerance score, relative shoot biomass reduction (%), leaf chlorophyll content reduction (%), leaf size reduction (%), shoot water content (%), and leaf and root concentration of proline and major ions (Na+, Cl−, K+, Ca2+, Mg2+, NH4 +, PO4 3+, SO4 2−, NO3 −, and malate). GWAS was carried out using 2.5 million high‐quality SNP markers. By performing GWAS on complex, emergent traits like growth/biomass as well as on potentially‐underlying, fundamental traits or “phenes” such as ion and metabolite concentrations, which reflect cellular ion homeostasis and metabolism, we hoped to break‐down salinity tolerance into its component parts while at the same time identifying genetic loci for tolerance in a more refined and robust manner.
2. MATERIALS AND METHODS
2.1. Plant material
Seeds of all the lines used in this study were obtained from the M. truncatula HapMap germplasm resource center (http://www.medicagohapmap.org/hapmap/germplasm). One hundred thirty‐two lines (Table S1) were selected based on population structure for maximum variance from the 220 lines that were used previously for the GWAS of drought‐related traits (Kang et al., 2015).
M. truncatula seeds were scarified on sand paper (p800) and germinated on wet filter paper for 48 hr at 4°C by overnight storage in the dark at room temperature, followed by planting. Plants were grown individually in 2″ × 7″ plastic cones (Stuewe & Sons Inc.) containing a 4:1 (v/v) mixture of Metromix 350 and sand, in a growth chamber with 16 hr day/8 hr night, 22°C, and 40% relative humidity. For each experiment, 12 seedlings were planted for each line, six for control conditions and six for salinity‐stress treatment. The plants were placed in the growth chamber in a randomized complete block design. Light density (a combination of fluorescent and incandescent) at plant level was approximately 200 μmol m−2 s−1. Plants were watered with one fourth B&D nutrient solution (Broughton & Dilworth, 1971) containing 8 mM of N (2 mM KNO3, 3 mM NH4NO3). Three replicates were performed for the entire experiment.
2.2. Salinity stress treatment
Ten days after seedling transfer to pots (day 10), plants were watered with 0.5% NaCl (86 mM) in one half B&D nutrient solution, which was applied to both the tray holding the cones and the top of the soil with a squeeze bottle to avoid leaf damage. Excess solution in the trays was removed 2 hr after the salinity solution treatment. Sodium chloride concentration in the nutrient solution was increased to 1.0% (172 mM) at day 15 then 1.5% (257 mM) at day 20, in the same way. No extra watering between treatments was necessary because of reduced transpiration. Roots and shoots were harvested 5 days following the 1.5% NaCl treatment.
2.3. In‐vivo leaf chlorophyll content measurement
Four days following the 1.5% NaCl treatment, in‐vivo leaf chlorophyll content was measured on salt‐stressed and control plants, using a Chlorophyll Meter SPAD‐502plus (Spectrum Technologies Inc., http://www.specmeters.com/). The terminal leaflet of the uppermost fully‐expanded leaf was used for the measurement. Each leaflet was measured twice at different positions, avoiding the midrib and edges, and the average of the two readings was recorded.
2.4. Leaf size measurement and plant harvest
Five days following the 1.5% NaCl treatment, the two youngest fully‐expanded leaves, from both control and NaCl‐treated plants, were harvested and immediately used for individual leaf size measurement with the Li‐3000A (LI‐COR) portable area meter. These two leaves were then flash‐frozen in liquid nitrogen, and stored at −80°C until being dried at −40°C in a lyophilizer. After the two youngest fully‐expanded leaves were harvested, the rest of the shoot was harvested separately. Shoots were dried in a 55°C oven and weighed. Total shoot dry weight was the sum of the two youngest fully‐expanded leaves and the rest of the shoot. Roots were harvested at the same time as the shoots, rinsed well, flash‐frozen in liquid nitrogen, and then stored at −80°C until being dried at −40°C in a lyophilizer. Dried tissues were ground to powder in a bead beater (BioSpec, https://www.biospec.com/).
2.5. Tissue ion (including malate) measurement
Approximately 5 mg of dry ground leaf or root tissue (six plants pooled together) was dissolved in 5–14 ml of MQ water, vortexed, and then shaken at 200 rpm for 1 hr. The solution was then filtered through a 0.2 μm nylon filter (F13–2020, VWR) and the filtrate was used for measurement of total ions with ion chromatography. Chromatographic separation was achieved on a Thermo Scientific ICS‐5000 IC system (Thermo Fisher Scientific, USA) using a Dionex CS12A Ion Pac analytical column with a AG12A guard column for cations, or a Dionex AS11HC analytical column with a AG11HC guard column for anions. The cation eluent source was a Thermo Scientific Dionex EGC III Methane sulphonic acid eluent generator cartridge. The anion eluent source was Thermo Scientific Dionex EGC KOH cartridge. Standard curves were prepared using dilutions of Thermo Scientific Dionex Seven Anion Standard II and Six Cation Standard II. Malate standard was prepared separately and then mixed with the Anion Standard. Quantification was achieved using software Chromeleon 7.2 version SR4.
2.6. Tissue proline quantification
Tissue proline levels were analyzed with a biochemical assay modified from Bates et al (Bates, Waldren, & Teare, 1973) and Hamid et al (Hamid et al., 2003). Proline concentration was determined using a standard curve using L‐proline.
2.7. Filtering of SNPs
SNPs were filtered with TASSEL 5.2.7 (Bradbury et al., 2007), with a minimum allele frequency of 5% and minimum counts of 112 (85% of 132). Missing SNPs were not imputed because of the high density of the existing SNPs. After filtering, a total of 2,528,531 SNPs remained and were used in the association analysis.
2.8. Cladogram
The cladogram tree was generated in TASSEL 5.2.7 (Bradbury et al., 2007) with the neighbor‐joining method using 40,000 randomly selected SNPs (5,000 SNPs each chromosome).
2.9. Q matrix deduction
Q matrix was deduced with the STRUCTURE program (Pritchard, Stephens, & Donnelly, 2000) using 40,000 SNPs (5,000 random SNPs from each chromosome). We evaluated K = 1–9 to infer the optimal value of K (i.e., the number of clusters) from the simulation summary using the methods of Pritchard et al. (2000) and Evanno, Regnaut, and Goudet (2005).
2.10. Genome‐wide genotype–phenotype association analysis
The least square means of the phenotypic data collected in three replicates were calculated in R with library “lsmeans” and used in GWAS. The proline and ion measurements were carried out only on the last replicate, and the standard‐calibrated values obtained were used directly in GWAS. The mixed linear model (MLM) and general linear model (GLM) embedded in TASSEL (Bradbury et al., 2007) were used to test for association between SNPs and phenotypic traits. For the MLM analyses, a Kinship matrix was calculated with centered‐IBS in TASSEL (Bradbury et al., 2007), and no compression was applied. Quantile–quantile (QQ) plots were generated in R (for GLM and MLM‐Q) or using 10% of all SNPs randomly‐selected from the eight chromosomes in TASSEL (for MLM + Q). Linkage disequilibrium among SNPs was also performed in TASSEL 5.2.7 (Bradbury et al., 2007).
2.11. Total RNA extraction and qRT‐PCR
Total RNA from shoot and root was extracted using RNeasy Plant Mini Kit (Qiagen) followed by genomic DNA removal and column purification. Reverse transcription was performed with oligo dT20 and Super Script III Reverse Transcriptase as described previously (Kakar et al., 2008). Amplification of templates followed standard PCR protocols with SYBR Green‐based detection system. Transcript levels were normalized using the geometric average of three housekeeping genes, Ubiquitin‐Conjugating enzyme E2 (TC106312), Polypyrimidine Tract‐Binding protein (TC111751), and Ubiquitin (TC102473). The primers used for all the target and housekeeping genes are listed in Table S6.
2.12. Statistical analysis
Principal component analysis (PCA) was performed in R using the “PCA” function in the package “FactoMineR” (Lê, Josse, & Husson, 2008). Association analysis among traits was performed in SAS 9.4 (SAS Institute, Cary, NC) using the CORR procedure. False discovery rate (q value) was calculated using the R package “qvalue” (Storey & Tibshirani, 2003). Significant tests were performed in Excel 2013 using student's t‐test, two‐tails, assuming equal variance.
3. RESULTS
3.1. Phenotypic data collection and correlations among traits
Preliminary experiments with 30 M. truncatula HapMap lines were carried out to establish a salinity stress regime that affected plant growth without overwhelming plants completely. As a result, a protocol with sequential increases in NaCl concentration in the nutrient solution was chosen: 0.5% NaCl (86 mM) for 5 days, 1% (171 mM) for 5 days, and 1.5% (257 mM) for 5 days. These treatment conditions resulted in significant leaf chlorosis, leaf size reduction, and growth retardation compared with non‐treated control plants (Figure 1). On the basis primarily of plant vigor and the degree of leaf chlorophyll loss, we scored stressed plants from 1 to 5, with 5 being the most tolerant (Figure 1a). In addition, shoot biomass, leaf chlorophyll content, and leaf size were measured for both treated and controlled plants, whereas shoot water content, leaf and root concentrations of proline and major ions (Na+, Cl−, K+, Ca2+, Mg2+, NH4 +, PO4 3+, SO4 2−, NO3 −, malate) were measured for treated plants only. For most of these traits, a near normal distribution was observed; leaf K+/Na+ ratio was an exception (Figure 2). Notably, concentrations of sodium, chloride, calcium, and proline were much higher in leaves than roots of NaCl‐treated plants, whereas potassium concentration exhibited the opposite response to salinity (Figure 2g–i,m–o; Figure S1). As a consequence, the K+/Na+ ratio was approximately 2‐fold higher in the roots than in the leaves (Figure 2j,p).
Figure 1.

Effect of salinity on M. truncatula growth. (a) stressed M. truncatula plants with tolerance scores from 1 to 5. (b) Entire plant. (c) Leaves. In b and c, the plant/leaf on the left was non‐stressed. All photos were taken 4 days after the 1.5% NaCl treatment, 1 day before plant harvest
Figure 2.
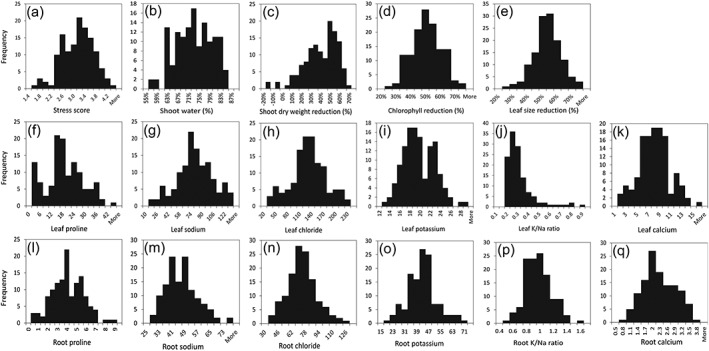
Distribution of salinity stress‐related traits in a collection of 132 M. truncatula lines/ecotypes. Traits are indicated on the x‐axis and number of lines on the y‐axis
To reveal relationships between traits, correlation analyses were performed. In general, shoot and leaf traits were positively correlated but, with the exception of proline concentration, shoot and root traits were less correlated (Table 1). Relatively strong and significant correlations were found between all of the following traits: shoot tolerance scores; reduction in leaf size, shoot dry weight, and leaf chlorophyll; leaf concentrations of sodium, chloride, and proline; and water content of the shoot (Table 1). Compared with chloride, sodium, calcium, and the K/Na ratio, potassium concentration in leaves were less tightly correlated to tolerance scores (r = −0.35). Sodium and chloride concentrations were extremely highly correlated to each other, both in leaves and roots, with correlation coefficients of 0.99 and 0.89, respectively. Interestingly, proline levels in leaves were negatively correlated to tolerance scores and shoot water contents, but were positively correlated to leaf sodium concentrations, whereas root proline levels were positively correlated with salinity tolerance (Figure 3). Thus, salinity‐tolerant M. truncatula genotypes tended to accumulate less proline in the leaf but more proline in the root under salinity stress, compared with the sensitive genotypes.
Table 1.
Correlation coefficients (r) of salinity‐stress related traits
| ST_DWred | LF_ChlRed | LF_SizeRed | ST_Water% | LF_Proline | LF_Sodium | LF_Chloride | LF_Potassium | LF_KNaRatio | LF_Calcium | RT_Proline | RT_Sodium | RT_Chloride | RT_Potassium | RT_KNaRatio | RT_Calcium | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ToleranceScore | −0.60 *** | −0.70 *** | −0.60 *** | 0.74 *** | −0.59 *** | −0.55 *** | −0.55 *** | −0.35 *** | 0.41 *** | −0.41 *** | 0.33 *** | 0.027 | 0.10 | 0.030 | 0.026 | 0.065 |
| ST_DWred | 0.41 *** | 0.59 *** | −0.60 *** | 0.45 *** | 0.40 *** | 0.42 *** | 0.15735 | −0.32 *** | 0.38 *** | −0.18 * | −0.14 | −0.15 | −0.058 | 0.082 | −0.0066 | |
| LF_ChlRed | 0.46 *** | −0.52 *** | 0.41 *** | 0.46 *** | 0.45 *** | 0.26 ** | −0.29 ** | 0.32 *** | −0.33 *** | −0.021 | −0.12 | −0.051 | 0.0042 | −0.12 | ||
| LF_SizeRed | −0.67 *** | 0.53 *** | 0.53 *** | 0.54 *** | 0.32 *** | −0.35 *** | 0.46 *** | −0.33 *** | −0.098 | −0.16 | −0.17 | −0.15 | −0.0031 | |||
| ST_Water% | −0.65 *** | −0.60 *** | −0.61 *** | −0.34 *** | 0.48 *** | −0.51 *** | 0.32 *** | 0.14 | 0.20 * | 0.15 | 0.087 | 0.029 | ||||
| LF_Proline | 0.78 *** | 0.79 *** | 0.41 *** | −0.63 *** | 0.56 *** | −0.38 *** | 0.022 | −0.057 | −0.0045 | −0.070 | 0.033 | |||||
| LF_Sodium | 0.99 *** | 0.46 *** | −0.80 *** | 0.66 *** | −0.47 *** | 0.022 | −0.070 | −0.044 | −0.13 | −0.0069 | ||||||
| LF_Chloride | 0.52 *** | −0.78 *** | 0.73 *** | −0.46 *** | 0.029 | −0.076 | −0.085 | −0.18 * | 0.0068 | |||||||
| LF_Potassium | −0.069 | 0.51 *** | −0.25 ** | 0.046 | −0.098 | −0.092 | −0.15 | −0.034 | ||||||||
| LF_KNaRatio | −0.53 *** | 0.23 * | 0.013 | 0.022 | 0.051 | 0.11 | −0.052 | |||||||||
| LF_Calcium | −0.21 * | 0.067 | −0.052 | −0.22 * | −0.35 *** | 0.028 | ||||||||||
| RT_Proline | −0.024 | 0.069 | 0.034 | 0.081 | −0.027 | |||||||||||
| RT_Sodium | 0.89 *** | 0.58 *** | −0.34 *** | 0.13 | ||||||||||||
| RT_Chloride | 0.72 *** | −0.11 | 0.28 ** | |||||||||||||
| RT_Potassium | 0.51 *** | 0.29 *** | ||||||||||||||
| RT_KNaRatio | 0.17 |
Note. Chl: chlorophyll; ST: shoot; LF: leaf; RT: root; Red: reduction (%).
,
, and
indicate significant differences at P < 0.05, 0.01, and 0.001, respectively.
Figure 3.
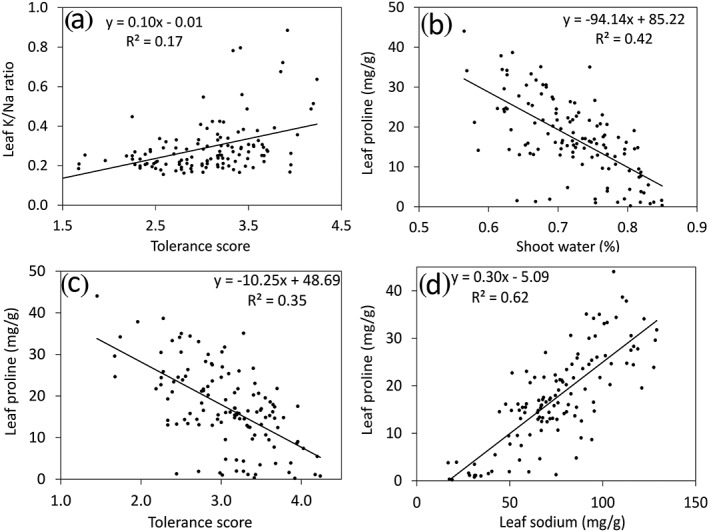
Correlation between salinity‐related traits. Average measurements of 132 M. truncatula lines are plotted for each trait. Significant correlations (P < 0.001) were found for all comparisons shown
Several other ions were analyzed, and leaf magnesium, leaf sulfate, leaf ammonium, leaf nitrate, root malate, and root phosphate were found to be significantly correlated with salinity tolerance, either positively or negatively (Table S2).
3.2. Genome‐wide association analysis and GO enrichment of “top‐suspect” genes
Genome‐wide association analysis using mixed linear model or general linear model was performed in TASSEL (Bradbury et al., 2007; Zhang et al., 2010). In the cladogram tree, the 132 lines used in the current study were clearly separated into two major groups (Figure S2). This is consistent with the cluster number determined by the program structure (Pritchard et al., 2000), which was also two (Figure S3). Because of the existence of population structure in the GWAS population, QQ plots generated with GLM or MLM without Q‐value correction were inflated for majority of the traits (Figure S4). To control for false positives, the population structure (Q value) was included in the MLM; GWAS Manhattan plots and QQ plots were generated and the lowest P values ranged from 10−6 to 10−13 (Figures 4, Figure S4). From the Manhattan plots, chromosome 2 appeared to be a “hot spot” for SNPs with lowest P values in multiple traits. For each trait, we collected the 200 SNPs with the lowest P values, the genes in each SNP's vicinity, as well as the closest Arabidopsis orthologues and M. truncatula microarray probe‐sets (Table S3).
Figure 4.
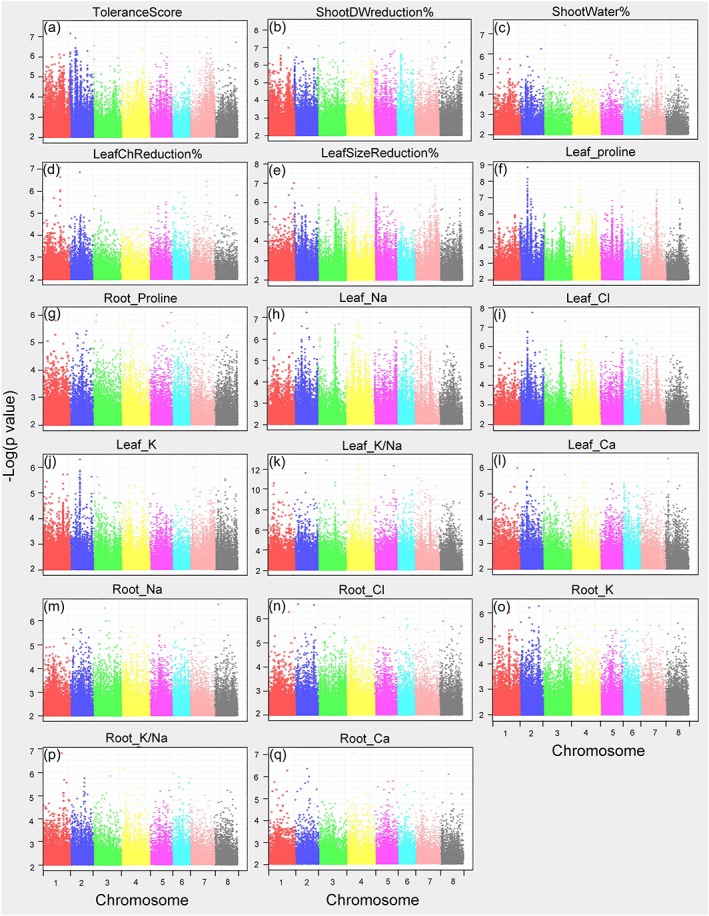
Manhattan plots (mixed linear model) of mapped single nucleotide polymorphisms (SNP) markers associating with each trait. Only SNPs with P values smaller than 0.01 are plotted
To gain an overview of genes that may contribute to salinity tolerance, we performed gene ontology (GO) enrichment analysis of all genes in the vicinity of the 200 SNPs with the lowest P values for 14 traits (Table S3). We used the GO of Arabidopsis orthologues for this analysis because they are better curated than for Medicago. Among all the biological processes, “post‐embryonic development” was most‐enriched among the selected genes, with a false discovery rate (FDR) of 1.10E‐12. Genes in the categories “response to stress” and “response to stimulus” were also highly enriched (Figure S5).
3.3. Identification of potential causative SNPs and genes
In analyzing the top 100 SNPs associated with each of the 14 traits, we found a large portion of them to be linked to multiple traits (Table S3), forming distinct “hotspots” on chromosomes especially on chromosome 2 (Figure 5). Considering the tight association among different traits (Table 1), we selected the top SNPs that were tightly associated with multiple traits, as well as the genomic regions that contained these SNPs. In doing so, we identified 12 genomic regions (QTLs) harboring SNPs that are in tight association with at least four traits (rank 200 or higher; Figure 6). In parallel, we performed PCA analysis of the first 12 traits (ToleranceScore, ST_DWred, LF_ChlRed, LF_SizeRed, ST_Water%, LF_Proline, LF_Sodium, LF_Chloride, LF_Potassium, LF_KNaRatio, LF_Calcium, and RT_Proline) that are tightly correlated in Table 1 and performed GWAS analysis using PC1 (that explained 55% of the variance) as a new trait, which identified the same 12 genomic regions as top hits (Figure 7). There are a total of 214 SNPs and 74 genes in these 12 regions (Figure 6). A total of 94 of the SNPs reside within genes. Linkage disequilibrium (LD) analysis revealed that the 214 SNPs in these 12 genomic regions are under tight LD within each region; the majority of these also show strong LD across different chromosomes (Figure 8).
Figure 5.
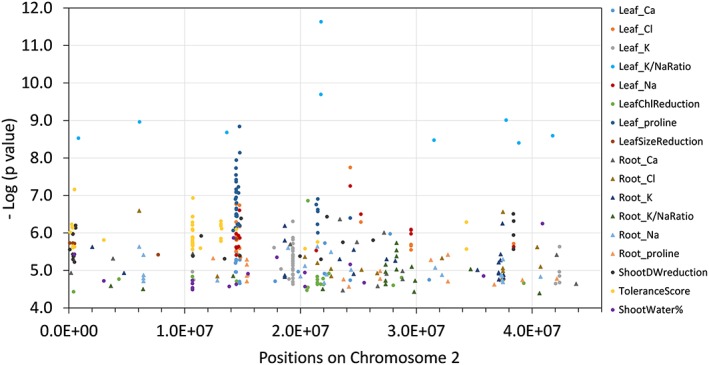
Top potential causative single nucleotide polymorphisms (SNPs) identified by genome‐wide association studies on chromosome 2. All SNPs shown are among the 100 SNPs with the lowest P values for each trait
Figure 6.
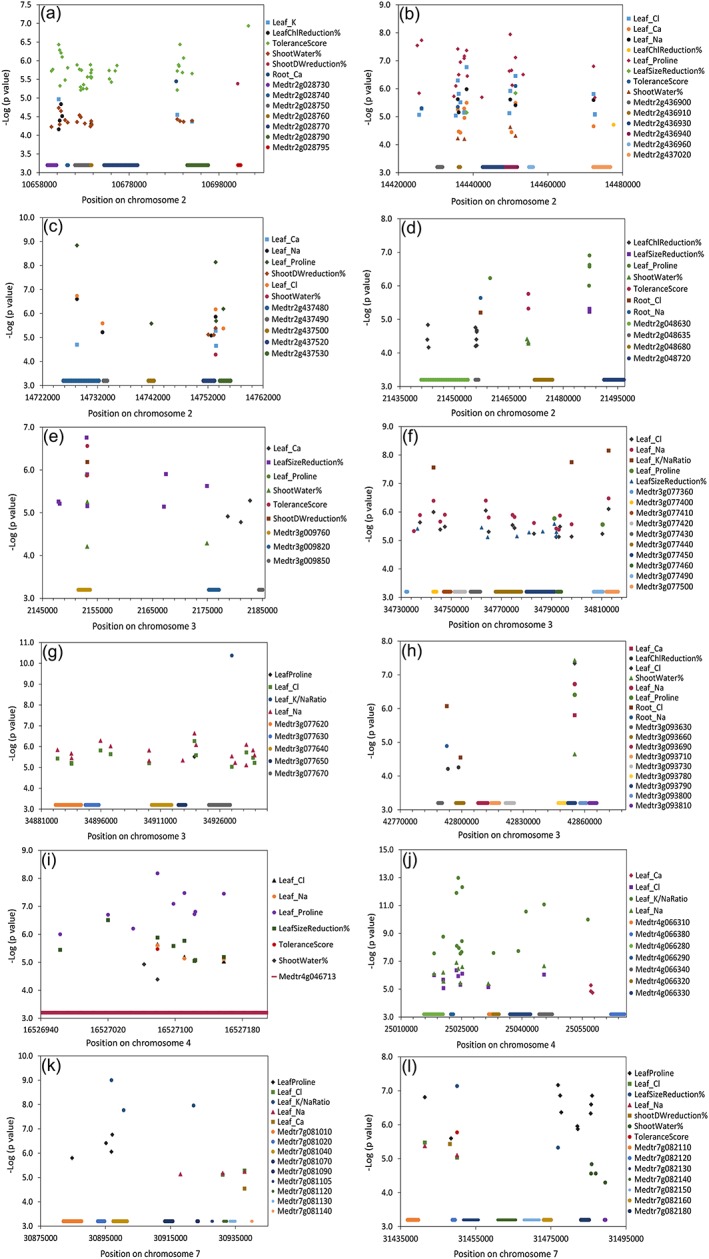
Top 12 genomic regions and predicted genes identified by genome‐wide association studies on chromosome 2 (a–d), chromosome 3 (e–h), chromosome 4 (i,j), and chromosome 7 (k,l). These genomic regions contain multiple low P value single nucleotide polymorphisms (SNPs) associated with at least four traits each. All SNPs shown are among the 200 SNPs with the lowest P values for each trait
Figure 7.
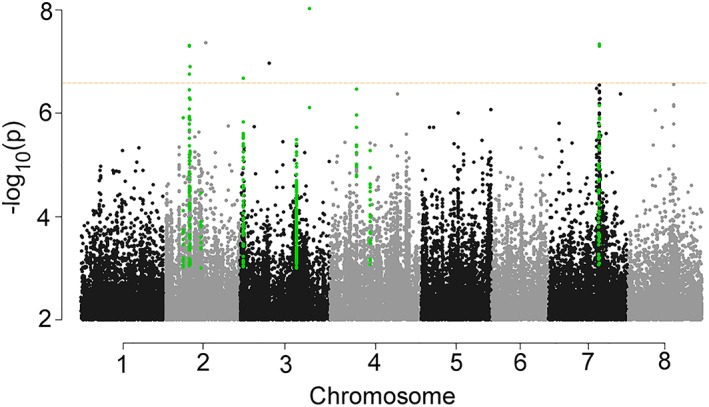
Manhattan plot (mixed linear model) of mapped single nucleotide polymorphism (SNP) markers associating with PC1 (55% explained variance) generated in the principal component analysis of the first 12 traits that are in tight correlation in Table 1. Only SNPs with P values smaller than 0.01 are plotted. SNPs reside within the 12 genomic regions (Figure 6) are highlighted. Dotted line indicates q value (FDR) cutoff 0.05
Figure 8.
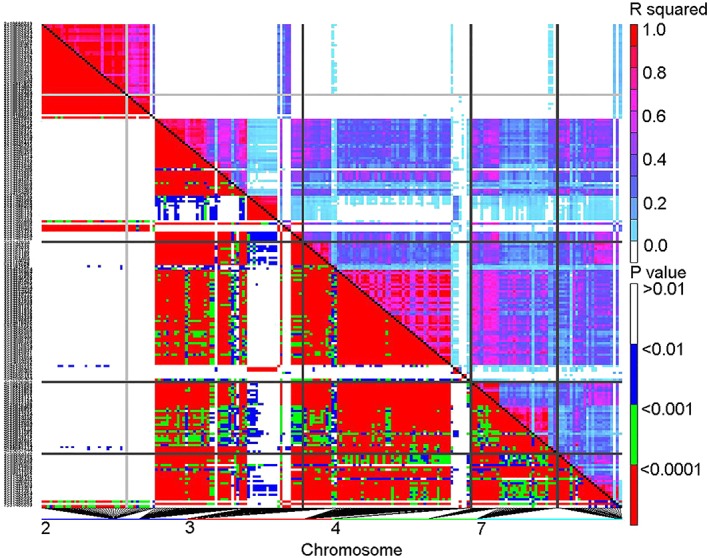
Linkage disequilibrium map of the 214 single nucleotide polymorphisms (SNPs) within the top 12 genomic regions shown in Figure 6
Among all the regions, the most significant one, considering the ranking and quantity of top SNPs, is on chromosome 2 near position 14,450,000 (Figure 6b). This region is about 60 kb long and harbors 61 low P value SNPs identified in the association studies of eight different traits. In addition, nine of the 61 SNPs rank in the top 10 in the GWAS results of three traits (leaf calcium, leaf chloride, and leaf proline; Table S3). The central SNP is 2:14451314, which has low association P‐values in seven traits. A total of six genes are located in this region (Figure 6b; Table S3). In addition to this region on chromosome 2, three other regions were identified (Figure 6a,c,d). Furthermore, four regions on chromosome 3, as well as two regions each on chromosomes 4 and 7, were identified (Figure 6e‐l). It is worth noting that two genomic regions, one on chromosome 3 (3:2147934 to 3:2182756) and one on chromosome 4 (4:16526963 to 4:16527158), harbor a total of four genes, and all of them are involved in cellular redox homeostasis (three FAD‐linked oxidoreductases and one peroxidase family protein; Tables 2; Table S3). Other less significant regions are highlighted in Table S3.
Table 2.
List of top suspect genes in the 12 selected genomic regions of Figure 6 that contain SNPs ranking 200 or less in the GWAS analyses of at least four traits. Genes in bold were characterized by qRT‐PCR for their response to the standard salinity treatment. Microarray data on gene expression under hydroponic salinity stress (up to 48 hr), drought stress, and seed desiccation were obtained from https://mtgea.noble.org/v3/index.php
| Gene | Start | End | Strand | Annotation (Mt. 4.0) | M. truncatula Microarray probeset | In M. truncatula, regulation under | |||
|---|---|---|---|---|---|---|---|---|---|
| salinity stress?a | drought in root?b | drought in shoot?b | seed desiccation?c | ||||||
| Medtr2g028730 | 10659792 | 10661824 | − | Trm112p‐like protein | Mtr.16016.1.S1_at | no | no | no | ** |
| Medtr2g028740 | 10664184 | 10664396 | − | hypothetical protein | none | ||||
| Medtr2g028750 | 10665954 | 10669267 | + | nonclathrin coat protein zeta1‐COP | Mtr.16017.1.S1_at | no | no | ** | no |
| Medtr2g028760 | 10669468 | 10669638 | − | hypothetical protein | none | ||||
| Medtr2g028770 | 10672439 | 10679871 | − | magnesium transporter CorA family protein | Mtr.52281.1.S1_at | ** | *** | *** | ** |
| Medtr2g028790 | 10690943 | 10695600 | + | synaptobrevin‐like protein | Mtr.16019.1.S1_at | no | ** | *** | *** |
| Medtr2g028795 | 10702242 | 10702842 | − | hypothetical protein | none | ||||
| Medtr2g048630 | 21440873 | 21453740 | + | regulator of nonsense transcripts‐like protein | Mtr.21538.1.S1_at | * | *** | ** | * |
| Medtr2g048635 | 21455804 | 21456661 | + | pentatricopeptide (PPR) repeat protein | none | ||||
| Medtr2g048680 | 21472115 | 21477040 | + | ankyrin repeat protein EMB506 | Mtr.42601.1.S1_at | no | ** | no | ** |
| Medtr2g048720 | 21491260 | 21497003 | − | inositol transporter 4 | Mtr.48631.1.S1_at | *** | *** | ** | *** |
| Medtr2g436900 | 14430380 | 14431772 | − | C2 domain protein | Mtr.12381.1.S1_at | ** | * | no | * |
| Medtr2g436910 | 14436094 | 14436501 | − | hypothetical protein | none | ||||
| Medtr2g436930 | 14442534 | 14448484 | + | substrate carrier family protein | Mtr.9356.1.S1_at | no | * | *** | ** |
| Medtr2g436940 | 14448712 | 14451781 | − | hypothetical protein | Mtr.41375.1.S1_at | no | *** | ** | *** |
| Medtr2g436960 | 14454918 | 14456001 | − | light‐harvesting complex I chlorophyll A/B‐binding protein | Mtr.15166.1.S1_at | * | * | no | * |
| Medtr2g437020 | 14472093 | 14476696 | − | mTERF protein | none | ||||
| Medtr2g437480 | 14725811 | 14732214 | + | chromatin remodeling complex subunit | none | ||||
| Medtr2g437490 | 14733099 | 14733710 | − | transmembrane protein, putative | Mtr.21581.1.S1_at | no | no | no | no |
| Medtr2g437500 | 14741244 | 14742166 | − | transcription factor GTE6, putative | none | ||||
| Medtr2g437520 | 14751210 | 14753146 | + | hypothetical protein | none | ||||
| Medtr2g437530 | 14754185 | 14756065 | − | DREPP plasma membrane protein | none | ||||
| Medtr3g009760 | 2151533 | 2153673 | + | FAD‐linked oxidoreductase | Mtr.13518.1.S1_at | no | no | ** | * |
| Medtr3g009820 | 2175256 | 2177107 | + | FAD‐linked oxidoreductase | none | ||||
| Medtr3g009850 | 2184364 | 2185062 | + | FAD‐linked oxidoreductase | none | ||||
| Medtr3g077360 | 34731943 | 34732457 | + | hypothetical protein | none | ||||
| Medtr3g077400 | 34742781 | 34744107 | − | hypothetical protein | none | ||||
| Medtr3g077410 | 34747003 | 34749760 | + | NAD(P)‐binding rossmann‐fold protein | Mtr.51752.1.S1_at | no | no | no | * |
| Medtr3g077420 | 34751208 | 34755524 | + | transcription factor, putative | Mtr.17177.1.S1_at | no | *** | ** | no |
| Medtr3g077430 | 34757686 | 34761543 | − | carboxyl‐terminal peptidase | Mtr.51747.1.S1_at | *** | *** | ** | * |
| Medtr3g077440 | 34767661 | 34777932 | + | modifier OF SNC1 1, putative | Mtr.29563.1.S1_at | no | ** | * | * |
| Medtr3g077450 | 34780007 | 34791257 | + | modifier OF SNC1 1, putative | Mtr.14802.1.S1_at | no | ** | no | * |
| Medtr3g077460 | 34792157 | 34793856 | − | cytochrome P450 family 71 protein | Mtr.51055.1.S1_at | no | no | no | *** |
| Medtr3g077490 | 34806841 | 34810528 | + | embryo defective 2759 protein | Mtr.14799.1.S1_at | no | *** | ** | ** |
| Medtr3g077500 | 34811954 | 34816546 | + | transmembrane protein, putative | Mtr.39510.1.S1_s_at | no | * | no | no |
| Medtr3g077620 | 34884550 | 34891032 | + | phox (PX) domain protein | Mtr.14792.1.S1_s_at | no | no | no | no |
| Medtr3g077630 | 34892075 | 34895483 | − | transmembrane protein, putative | Mtr.4339.1.S1_at | * | no | *** | ** |
| Medtr3g077640 | 34908806 | 34913845 | + | plastid phosphate translocator | none | ||||
| Medtr3g077650 | 34915543 | 34917386 | + | transcription factor MYB98 | Mtr.48147.1.S1_at | * | no | no | *** |
| Medtr3g077670 | 34923242 | 34928630 | − | boron transporter‐like protein | Mtr.48146.1.S1_s_at | no | ** | * | ** |
| Medtr3g093630 | 42788484 | 42790479 | + | hypothetical protein | none | ||||
| Medtr3g093660 | 42797203 | 42800974 | − | RING zinc finger protein, putative | none | ||||
| Medtr3g093690 | 42807993 | 42812754 | + | transmembrane protein, putative | Mtr.6105.1.S1_at | no | ** | * | no |
| Medtr3g093710 | 42814305 | 42818320 | + | receptor‐like kinase | Mtr.45101.1.S1_at | ** | * | ** | ** |
| Medtr3g093730 | 42821390 | 42825561 | − | methylesterase | none | ||||
| Medtr3g093780 | 42847107 | 42851695 | + | DUF3133 family protein | Mtr.33238.1.S1_at | no | no | no | no |
| Medtr3g093790 | 42851825 | 42855319 | − | nucleic acid‐binding, OB‐fold‐like protein | Mtr.12947.1.S1_at | no | no | ** | ** |
| Medtr3g093800 | 42857714 | 42860884 | − | transcription elongation factor S‐II, putative | Mtr.43413.1.S1_at | * | ** | * | no |
| Medtr3g093810 | 42862192 | 42865834 | − | ubiquinol‐cytochrome C reductase complex protein, putative | Mtr.37864.1.S1_at | no | *** | no | *** |
| Medtr3g093900 | 42905400 | 42907112 | + | MADS‐box transcription factor family protein, putative | none | ||||
| Medtr4g046713 | 16525346 | 16527516 | − | peroxidase family protein | Mtr.44569.1.S1_at | *** | *** | no | no |
| Medtr4g066280 | 25015686 | 25020396 | − | glycoside hydrolase family 1 protein | Mtr.47870.1.S1_at | * | no | no | no |
| Medtr4g066290 | 25022356 | 25022951 | + | Nodule Cysteine‐Rich (NCR) secreted peptide | none | ||||
| Medtr4g066310 | 25031757 | 25032810 | − | hypothetical protein | none | ||||
| Medtr4g066320 | 25032880 | 25034289 | + | hypothetical protein | none | ||||
| Medtr4g066330 | 25037019 | 25042115 | − | glycoside hydrolase family 1 protein | none | ||||
| Medtr4g066340 | 25044288 | 25047517 | − | cyanogenic beta‐glucosidase, putative | none | ||||
| Medtr4g066380 | 25062108 | 25065460 | − | basic helix loop helix (bHLH) DNA‐binding family protein | none | ||||
| Medtr7g081010 | 30882053 | 30887619 | − | archaeal/vacuolar‐type H + ‐ATPase subunit B | Mtr.12301.1.S1_at | *** | *** | ** | *** |
| Medtr7g081020 | 30892633 | 30894709 | + | protein phosphatase 2C family protein | Mtr.9338.1.S1_at | ** | *** | *** | *** |
| Medtr7g081040 | 30897385 | 30901657 | − | trafficking protein particle complex subunit‐like protein | Mtr.42623.1.S1_at | no | *** | ** | * |
| Medtr7g081070 | 30913276 | 30915702 | − | DUF1279 family protein | none | ||||
| Medtr7g081090 | 30923192 | 30923518 | + | transmembrane protein, putative | none | ||||
| Medtr7g081105 | 30927714 | 30927950 | + | Thionin related | none | ||||
| Medtr7g081120 | 30931309 | 30932323 | + | transmembrane protein, putative | none | ||||
| Medtr7g081140 | 30939953 | 30940153 | − | hypothetical protein | none | ||||
| Medtr7g082110 | 31436869 | 31439723 | + | receptor‐like kinase, putative | none | ||||
| Medtr7g082120 | 31448827 | 31449485 | − | hypothetical protein | none | ||||
| Medtr7g082130 | 31451724 | 31455731 | − | hypothetical protein | none | ||||
| Medtr7g082140 | 31460852 | 31465655 | − | peptidase M50B‐like protein | Mtr.44385.1.S1_at | no | *** | no | *** |
| Medtr7g082150 | 31467830 | 31471924 | − | U1 small nuclear ribonucleoprotein | Mtr.33442.1.S1_at | * | ** | * | no |
| Medtr7g082160 | 31473081 | 31475033 | − | PPR containing plant‐like protein | none | ||||
| Medtr7g082180 | 31482984 | 31485168 | + | hypothetical protein | Mtr.39860.1.S1_at | ** | no | ** | * |
| Medtr7g082190 | 31489286 | 31489554 | + | transmembrane protein, putative | none | ||||
3.4. Evaluation of potential salinity‐tolerance genes
We investigated further potential salinity‐tolerance conferred by “suspect” genes identified by GWAS, first by examining the interaction among genes in the 12 common genomic regions by querying 43 physical interaction databases at http://genemania.org/ (Warde‐Farley et al., 2010) using Arabidopsis orthologues of the Medicago suspect proteins. We found that Arabidopsis AT1G04760, orthologous to Medtr2g028790 (Figure 6a VAMP726, synaptobrevin‐like protein), interacts with KAT3 (potassium channel in Arabidopsis thaliana 3; Figure S6a). Likewise, AT4G38510, an orthologue of Medtr7g081010 (Figure 6k, V‐type proton ATPase subunit B2), was found to have direct physical interaction with SOS2 (salt overly sensitive 2, CIPK24), which is known to be required for salinity tolerance in Arabidopsis (Figure S6b).
In an effort to further narrow down our list of suspect genes, we analyzed how these genes are regulated under salinity stress in M. truncatula, using published data from a study that employed hydroponics and relatively short‐term (up to 48 hr) salinity stress (Li, Su, Dong, & Wang, 2009). Additionally, in light of the fact that drought and desiccation, such as salinity, imposes osmotic stress on plant cells, we examined how our suspect genes responded to drought stress and during seed desiccation in M. truncatula (Verdier, Dessaint, Schneider, & Abirached‐Darmency, 2012; Zhang et al., 2014). Thirty nine of the 74 suspect genes were represented by microarray‐based (Affymetrix) data, and all but three of these genes were regulated under one or more of these three stress conditions (Table 2).
In addition to examining published gene expression data, we chose eight of the top suspect genes (Table 2 in bold) and performed qRT‐PCR analysis to determine if they respond at the transcript level to gradual salinity stress, as applied in our GWAS study. Gene selection for this experiment was based on SNP P values and rank, the position and density of enclosed SNPs, and gene function and expression patterns (Table 2, Table S3, and Figure 6). The eight genes selected included: one H+‐ATPase (Medtr7g081010); two putative transcription factors, a C2 domain protein (Medtr2g436900), and an mTERF protein (Medtr2g437020); two genes involved in vesicle trafficking, a non‐clathrin coat protein (Medtr2g028750), and a synaptobrevin‐like protein (Medtr2g028790); two genes involved in cellular redox homeostasis, an FAD‐linked oxidoreductase (Medtr3g009760), and a peroxidase family protein (Medtr4g046713); and a gene with unknown function (Medtr2g436940). Five M. truncatula lines were selected, three salt‐tolerant (HM091, HM010, and HM198 with salinity tolerance scores of 4.2, 3.8, and 3.5, respectively), and two salt‐sensitive lines (HM081 and HM152 with scores of 1.7 and 1.5, respectively).
All eight of the selected genes were significantly regulated by gradual salinity stress in at least one of the lines, in the root, shoot, or both (Figure 9). The regulation patterns of the two genes involved in vesicle trafficking were similar; both were exclusively down‐regulated in the roots of the two sensitive lines (P < 0.01, Figure 9b,d). Similarly, salinity‐sensitivity was associated with transcript changes for Medtr2g436900 in the shoot (Figure 9e), Medtr2g436940 in the root and shoot (Figure 9g,h), Medtr2g437020 in the root (Figure 9j), Medtr4g046713 in the root (Figure 9n), and Medtr7g081010 in the root (Figure 9p). In contrast, the expression pattern of Medtr3g009760 (FAD‐linked oxidoreductase) was correlated to the salinity sensitivity in neither the shoot nor the root (Figure 9k,l).
Figure 9.
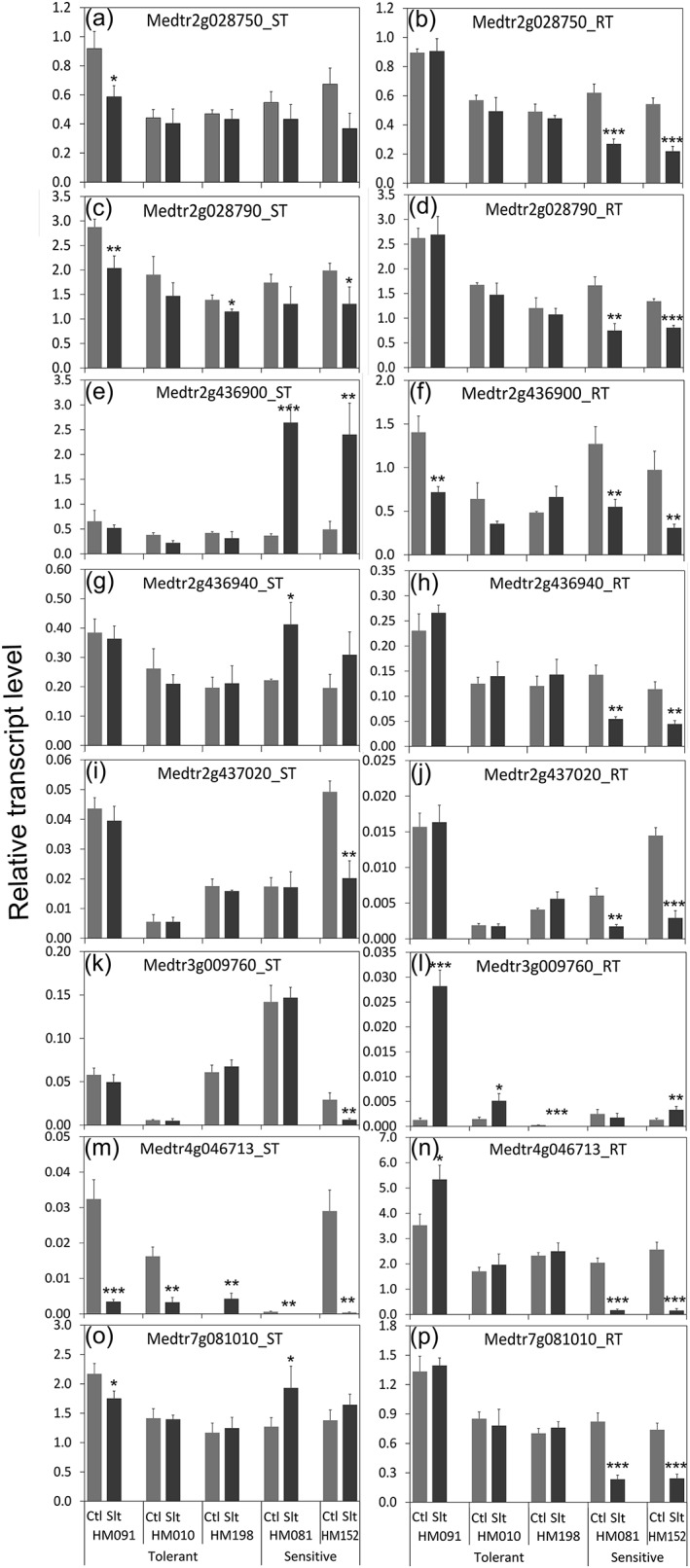
Transcript levels of potential causative genes in tolerant and sensitive genotypes. Relative transcript levels were determined by qRT‐PCR. The tolerance scores for HM091, HM010, HM198, HM081, and HM152 were 4.2, 3.8, 3.5, 1.7, and 1.5, respectively. Transcript levels are expressed relative to the mean of three housekeeping genes (Ubiquitin‐Conjugating enzyme E2, Polypyrimidine Tract‐Binding protein and Ubiquitin). n = 3. Error bars represent standard errors. Significance tests were between control and salinity stress treatments only in each genotype. Significat differences at *P < 0.05, **P < 0.01, and ***P < 0.001. Ctl: control. Salt: salinity‐treated
In analyzing SNP variance in these eight genes, we extracted all the SNPs that reside within the genes and 1000 bp upstream and downstream of the start and stop codons, and selected those that varied consistently between tolerant and sensitive lines (Table S4). A total of 46 SNPs matching these criteria were identified, with Medtr7g081010 (H+‐ATPase) containing the most, whereas there were none in Medtr2g436940 (unknown protein). One missense mutation was found in Medtr4g046713 (peroxidase family protein), which caused an amino acid switch from Tyrosine to Serine. In addition, 10 SNPs occurred in introns and two in UTRs.
Finally, it is interesting to note that the profile of these 46 SNPs in the M truncatula A17 reference genome is highly similar to the two sensitive lines (HM081 and HM152), with only five being different (highlighted in Table S4). A17 was not used in the GWAS analyses but was characterized for salinity sensitivity as a check line; it has an average salinity sensitivity score of 2.1 and was, therefore, relatively sensitive to salinity stress (Figure S7).
4. DISCUSSION
4.1. Correlation among salinity responsive traits
Salinity is a complex abiotic stress, with ionic and osmotic stress components that trigger a variety of responses in plants. In the current study, we applied long‐term, gradual salinity stress to soil grown M. truncatula plants and characterized a wide‐range of physiological and biochemical traits in conjunction with GWAS. In the M. truncatula HapMap panel, we observed substantial variation in responses to salinity stress (Figure 1), and almost all of the traits characterized demonstrated a normal distribution (Figure 2). With multiple traits characterized, we were able to analyze the relationships among different traits as rarely done before. Sodium and chloride levels in leaves (r = −0.55, P < 0.001 for both) but not in roots (P > 0.2) were highly (negatively) correlated with salinity tolerance scores, with tolerant genotypes containing lower concentrations of sodium and chloride (Table 1). Therefore, it appears that maintaining low sodium and chloride concentrations in the shoot is an important strategy used by M. truncatula plants to survive and grow under salinity stress. This is consistent with earlier reports that photosynthetic organs are more sensitive than roots to high salt (Munns et al., 2006). The negative correlation between shoot sodium and chloride concentrations and salt tolerance has been reported previously in rice (Lin et al., 2004; Patishtan et al., 2017), Durum wheat (Munns et al., 2012), Lotus species (Sanchez et al., 2011), and barley (Nguyen et al., 2013). Interestingly, salt sensitivity does not seem to be linked to shoot sodium content in rapeseed (Wan et al., 2017; Yong et al., 2015).
Proline accumulation patterns under salinity stress were interesting. Earlier reports on the role of proline under plant salinity stress lack consensus, with some studies supporting its role as a protectant because it accumulates more in the tolerant than the sensitive genotypes, whereas other studies indicating that it may simply be a stress reporter when the opposite trend was found (Aziz et al., 1998; Heuer, 2010; Lacerda et al., 2003; Moftah & Michel, 1987). However, all previous studies used a small number of genotypes and only focused on the shoot. Here, we demonstrated that leaf proline levels are negatively correlated with tolerance scores (r = −0.59, P < 0.0001) and shoot water content (r = −0.65, P < 0.0001), whereas root proline levels are positively correlated with tolerance (r = 0.33, P = 0.0002; Figure 3; Table S2). In other words, salinity‐tolerant M. truncatula genotypes tended to accumulate more proline in the roots but less proline in the leaves than sensitive genotypes. This phenomenon has not been reported before and we postulate that it may reflect different roles of proline in roots and leaves. When plants are under salinity stress, the primary stress encountered by photosynthetically‐active leaves is redox stress (ROS) associated with insufficient ability to channel high‐energy electrons into anabolic pathways due to stomatal closure and reduced carbon fixation (Chaves, Flexas, & Pinheiro, 2009; Hossain & Dietz, 2016). Proline biosynthesis consumes electrons (NADPH), helping plants to cope with increased ROS production when stomata close (Hare & Cress, 1997). Salinity tolerant plants maintain vigor and photosynthesis for longer than sensitive plants under saline conditions, which likely keeps ROS levels relatively low in the tolerant plants (Ashraf, 2009). As a result, tolerant plants need not engage proline biosynthesis to the same extent as sensitive plants. Thus, the negative correlation between proline levels in leaves and overall salinity tolerance in Medicago indicates that proline in the leaf is not a primary tolerance mechanism, helping plants to avoid salinity stress, but rather a secondary line of defense to help plants survive when primary mechanisms fail.
The situation in roots appears to be quite different, where proline levels correlate positively with salinity tolerance. Rather than being subject to increased ROS associated with light‐energy harvesting without carbon fixation in leaves when stomata close, roots are prone to dehydration stress in saline soils. Proline biosynthesis in roots presumably contributes to overall osmolyte production in saline soils, which helps root cells take up the water necessary for transpiration, stomatal opening, photosynthesis, and so forth. By producing more proline in roots, salinity‐tolerant plants may be better able than sensitive plants to support shoot functions and carbon supply back to the root for further proline/osmolyte synthesis. In this sense, proline biosynthesis in the root may be a primary salinity tolerance mechanism in Medicago. Further studies are required to test this hypothesis, for instance by down‐regulating proline biosynthesis via RNA‐interference in transgenic “hairy roots” but not in shoots of Medicago. It would also be interesting to determine whether proline accumulates differentially in roots and/or shoots of plants with differential tolerance to other kinds of stress.
4.2. Identification and validation of suspect drought tolerance genes
To reduce false positives in our genome‐wide association studies, we used MLM and included population structure as co‐variants to counter bias due to any population structure. With this stringent MLM + Q + K analysis, we identified SNPs with P values as low as 10−13 (leaf K/Na ratio), which is far below the P‐value threshold after stringent Bonferroni correction (1.98 × 10−8 with 95% confidence). Taking advantage of the many salinity‐related traits that were measured, we focused on SNPs that were repeatedly linked to multiple traits, rather than simply selecting SNPs based on applying thresholds to individual traits. With this approach, 12 genomic regions with clear borders were identified (Figure 6, Table S3). A total of 74 genes and 214 SNPs were present in these 12 regions, with 94 SNPs within genes. Of these genes, 39 had corresponding transcript information in microarray data sets, and the majority were found to be either regulated by short‐term salinity stress or drought/desiccation stress in earlier studies (Li et al., 2009; Verdier et al., 2012; Zhang et al., 2014; Table 2). The tight LD among the top overlapping 214 SNPs (Figure 8) across multiple chromosomes indicates possible co‐evolution of the salinity tolerance genes. This phenomenon has been previously reported in human and mouse (Kulminski, 2011; Petkov et al., 2005). Note, however, that although multiple traits may map to the same genomic region, they are not necessarily associated with the same SNPs (Figure 6, Table S3). From GO enrichment analysis, we found that genes in the categories “response to stress” and “response to stimulus” are highly enriched with FDR < 0.001. Taken together, it is evident that the current GWAS studies identified a set of genes that are enriched in salinity/dehydration stress responses.
In examining the expression patterns of the eight top suspect genes by qRT‐PCR analysis of the five extremely tolerant and sensitive lines, we found significant regulation of these genes in response to gradual salinity stress (Figure 9). Association of salinity‐stress sensitivity with gene transcript level changes was evident in seven of these eight genes including one H+‐ATPase (Medtr7g081010), two transcription factors (Medtr2g436900, Medtr2g437020), two genes likely to be involved in vesicle trafficking (Medtr2g028750, Medtr2g028790), one gene involved in cellular redox homeostasis (Medtr4g046713), and one with unknown function (Medtr2g436940). The majority of these genes tended to have stable or increased expression under salinity stress in the tolerant lines but sharply decreased expression in the sensitive lines, especially in the root (Figure 9). The overall gene regulation patterns in the root bear similarity to those observed previously under short‐term salinity stress (Li et al., 2009) (https://mtgea.noble.org/v3/index.php). SNP variances correlating with salinity stress sensitivity were identified in these genes, including one that causes a missense mutation, and 12 causing modifications in the introns and the UTR regions (Table S4).
Vesicular trafficking has been demonstrated to play important roles in plant adaptation to salinity stress (Baral et al., 2015; Garcia de la Garma et al., 2015; Hamaji et al., 2009). Vesicle trafficking is likely to be involved in deployment of specific ion, water, or metabolite transporters to the cell or organellar membranes for ion or water homeostasis (Baral et al., 2015). Vesicle trafficking may also be involved in ROS signaling and facilitating plasma membrane area reduction during plasmolysis caused by osmotic stress (Baral et al., 2015; Leshem, Seri, & Levine, 2007). Here, we identified two genes that are involved in vesicle trafficking: one synaptobrevin‐like protein (Medtr2g028790), which is an R‐SNARE (Soluble N‐ethylmaleimide‐sensitive factor Attachment Protein) that mediates vesicle fusion, and one nonclathrin coat protein zeta1‐COP (Medtr2g028750), which is a component of the COPI coatomer that coats vesicles transporting proteins between the Golgi complex and the endoplasmic reticulum. Both genes had clear transcription reduction in the root under salinity stress in the sensitive genotypes but not in the tolerant genotypes in M. truncatula (Figure 9). Earlier studies demonstrated that modification of various SNAREs could either enhance or reduce salinity tolerance in Arabidopsis depending on the experimental set‐up (Hamaji et al., 2009; Leshem et al., 2007; Tarte et al., 2015). However, the involvement of non‐clathrin coat proteins in salinity stress responses has never been studied. Our GWAS results suggest a potential role of non‐clathrin coat protein zeta1‐COP in this process, which, thus, deserves further investigation.
Vacuolar H+‐ATPase (V‐ATPase) generates and maintains a proton gradient across the tonoplast that provides the driving force for secondary transport processes, including storage of excess ions in the vacuole under salinity stress (Silva & Gerós, 2009). Yeast two‐hybrid experiments showed that SOS2 (salt overly sensitive 2) interacts directly with V‐ATPase subunit B1 and B2 (AT4G38510, putative orthologue of Medtr7g081010), and mutant plants with reduced V‐ATPase activity were extremely salt sensitive (Batelli et al., 2007). In alfalfa, over‐expression of V‐ATPase subunit B from the halophyte, Suaeda corniculata (a putative orthologue of Medtr7g081010) resulted in plants more tolerant to salt and saline‐alkali stresses (Wang et al., 2016). Similarly, overexpression of the putative wheat orthologue (GenBank accession number: EF105343) of the M. truncatula V‐ATPase subunit B (Medtr7g081010) significantly improves germination rate under salinity and overall salt tolerance in Arabidopsis (Wang, He, Zhao, Shen, & Huang, 2011). In the current study, we found that the expression of V‐ATPase subunit B2 (Medtr7g081010) remained stable in roots of tolerant lines, but decreased significantly in sensitive lines. In addition, we identified multiple SNPs in the UTR region, the intron, and 1 kb vicinity of this gene that differentiate the tolerant from the sensitive genotypes. Work is ongoing to locate the causative SNP(s) underlying the differential regulation of this gene under salinity stress.
The importance of ROS scavenging and the two transcription factors (mTERF protein and C2 domain protein) in plant salinity stress responses have been reported in several previous studies (Abogadallah, 2010; Quesada, 2016; Robles, Micol, & Quesada, 2015; Xu, Leister, & Kleine, 2017; Yokotani et al., 2009). In the current study, we identified a peroxidase (Medtr4g046713) as a gene associated with salinity tolerance as it is the only predicted gene in a genomic region that harbors 26 SNPs linked to six distinct traits (Figure 6i). The Arabidopsis orthologue, rare cold inducible gene 3 (RCI3, AT1G05260), has been shown to increase salinity tolerance when overexpressed and to decrease salinity tolerance when mutated (Llorente, López‐Cobollo, Catalá, Martínez‐Zapater, & Salinas, 2002).
Differential regulation of suspect genes under salinity stress in tolerant versus sensitive lines implicates them in salinity responses, but does not establish their role in salinity tolerance. Further studies using hairy root overexpression and/or RNA‐interference, virus‐induced silencing (VIGS), stable transgenic, and Tnt1 mutant plants are underway to investigate further the roles of these genes in plant salinity tolerance in M. truncatula and other species.
Apart from the genes discussed above that are linked to multiple salinity‐related traits, we are interested in genes associated with single traits that have homologs in other species that have been reported to play important roles in salinity tolerance (Table S3), including potassium channels/transporters, AAA family ATPases, cation/H+ exchanger, cation‐chloride cotransporter, LEA proteins, and a delta‐1‐pyrroline‐5‐carboxylate synthetase (P5CS), that is rate‐limiting for proline biosynthesis. As one example, the critical role of maintaining potassium homeostasis in plants during salinity stress has been intensively studied (Shabala & Pottosin, 2010). It has been shown that overexpression of a K+/H+ antiporter in tomato enhanced its salinity tolerance (Huertas et al., 2013).
Recently, GWAS of salinity stress‐related traits with relatively high precision have been performed in Arabidopsis, rice, and rapeseed (Al‐Tamimi et al., 2016; Kobayashi et al., 2016; Shi et al., 2017; Wan et al., 2017). We compared our GWAS results with these studies to identify potentially‐conserved salinity tolerance genes (orthologues). Among the top genes identified in various GWAS of salinity traits, only one, AT2G44480 (Table S5 in Kobayashi et al., 2016), which encodes a beta glucosidase, is homologous to a top gene identified in the current GWAS. Intriguingly, M. truncatula has three tandem repeats of this gene (Medtr4g066280, Medtr4g066330, and Medtr4g066340, Figure 6J), suggesting that gene expression levels may impact salt tolerance.
In summary, genome wide association studies (GWAS) of multiple physiological and biochemical traits related to salinity‐stress adaptation in M. truncatula were carried out using 2.5 million SNPs. Correlation analysis among the traits showed that salinity tolerance in the population is negatively correlated to sodium, chloride, and proline concentration in the leaf, while root proline levels were positively associated with salinity‐stress tolerance. GWAS analyses were performed with a mixed linear model, and 12 genomic regions that are associated with multiple traits each were identified; these regions harbor a total of 214 SNPs and 74 genes. These genomic regions were confirmed by a GWAS analysis using PC1 as a virtual trait, generated from principle component analysis of the 12 traits that are tightly correlated. Significant linkage disequilibrium was observed among these SNPs, possibly indicating co‐evolution of salinity tolerance alleles. Examination of gene expression of eight top suspect genes revealed associations between salinity tolerance and transcript level changes under salinity stress. Earlier functional studies on orthologues of two of the top eight genes (a vacuolar H+−ATPase and a peroxidase) confirmed their involvement in plant salinity tolerance in other species. Functional annotation of potential salinity tolerance genes points to the importance of transcriptional regulation, vesicle trafficking, and ROS scavenging under saline conditions.
Supporting information
Table S1. Information on the 132 accessions that were used in the GWAS.
Supplemental Table S2. Correlation coefficients and association p values of all characterized salinity‐stress related traits.
Supplemental Table S3. List of 200 SNPs of the lowest p values for each trait.
Supplemental Table S4. SNPs reside inside as well as 1000 bp upstream or downstream of the eight genes that were examined by qRT‐PCR.
Supplemental Table S5. List of genes (SNPs) that had been identified in earlier GWAS of salinity‐stress related traits in rice (Al‐Tamimi et al., 2016), Arabidopsis (Kobayashi et al., 2016), and rapeseed (Wan et al., 2017).
Supplemental Table S6. The list of primers that were used in the qRT‐PCR experiment.
Supplemental Figure S1. Example ion chromatography of line HM066 showing the difference in sodium and chloride in the leaf (a) and the root (b).
Supplemental Figure S2. Cladogram tree (neighbor‐joining) of the 132 lines that were used in the GWAS.
Supplemental Figure S3. Values of the Ln P(d) and Delta K as a function of the number of clusters (K).
Supplemental Figure S4. Quantile‐quantile (Q‐Q) plots for all the traits obtained by Mixed Linear Model (MLM) with (a) or without (b) Q value as a co‐variant, and General Linear Model (GLM) (c).
Supplemental Figure S5. GO enrichment of genes (1422 Arabidopsis orthologues) in the vicinity of the top 200 SNPs identified through GWAS of 17 traits (Table S3).
Supplemental Figure S6. Physical interaction networks of two selected genes in the top 12 genomic regions.
Supplemental Figure S7. Salinity stressed M. truncatula A17 plants.
ACKNOWLEDGEMENTS
Seeds of all the lines used in this study were obtained from the M. truncatula HapMap germplasm resource center (http://www.medicagohapmap.org/hapmap/germplasm). The authors would like to thank Shulan Zhang, Elis Acosta‐Torres and Alicia Smith for assistance during data collection, and Wenchao Zhang for assistance in data analysis. This work was supported by The Samuel Roberts Noble Foundation.
The authors have no conflict of interests to declare.
Kang Y, Torres‐Jerez I, An Z, et al. Genome‐wide association analysis of salinity responsive traits in Medicago truncatula . Plant Cell Environ. 2019;42:1513–1531. 10.1111/pce.13508
REFERENCES
- Abogadallah, G. M. (2010). Insights into the significance of antioxidative defense under salt stress. Plant Signaling & Behavior, 5, 369–374. 10.4161/psb.5.4.10873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Tamimi, N. , Brien, C. , Oakey, H. , Berger, B. , Saade, S. , Ho, Y. S. , … Negrao, S. (2016). Salinity tolerance loci revealed in rice using high‐throughput non‐invasive phenotyping. Nature Communications, 7, 13342 10.1038/ncomms13342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf, M. (2009). Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnology Advances, 27, 84–93. 10.1016/j.biotechadv.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Ashraf, M. , & Foolad, M. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environmental and Experimental Botany, 59, 206–216. 10.1016/j.envexpbot.2005.12.006 [DOI] [Google Scholar]
- Ashraf, M. , & Foolad, M. R. (2013). Crop breeding for salt tolerance in the era of molecular markers and marker‐assisted selection. Plant Breeding, 132, 10–20. 10.1111/pbr.12000 [DOI] [PubMed] [Google Scholar]
- Aziz, A. , Martin‐Tanguy, J. , & Larher, F. (1998). Stress‐induced changes in polyamine and tyramine levels can regulate proline accumulation in tomato leaf discs treated with sodium chloride. Physiologia Plantarum, 104, 195–202. 10.1034/j.1399-3054.1998.1040207.x [DOI] [Google Scholar]
- Baral, A. , Irani, N. G. , Fujimoto, M. , Nakano, A. , Mayor, S. , & Mathew, M. (2015). Salt‐induced remodeling of spatially restricted clathrin‐independent endocytic pathways in Arabidopsis root. The Plant Cell, 27, 1297–1315. 10.1105/tpc.15.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, D. G. , Bianchi, S. , Blondon, F. , Dattée, Y. , Duc, G. , Essad, S. , … Muel, X. (1990). Medicago truncatula, a model plant for studying the molecular genetics of theRhizobium‐legume symbiosis. Plant Molecular Biology Reporter, 8, 40–49. 10.1007/BF02668879 [DOI] [Google Scholar]
- Batelli, G. , Verslues, P. E. , Agius, F. , Qiu, Q. , Fujii, H. , Pan, S. , … Zhu, J.‐K. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+‐ATPase and upregulating its transport activity. Molecular and Cellular Biology, 27, 7781–7790. 10.1128/MCB.00430-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, L. , Waldren, R. , & Teare, I. (1973). Rapid determination of free proline for water‐stress studies. Plant and Soil, 39, 205–207. 10.1007/BF00018060 [DOI] [Google Scholar]
- Baxter, I. , Brazelton, J. N. , Yu, D. , Huang, Y. S. , Lahner, B. , Yakubova, E. , … Vitek, O. (2010). A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1; 1. PLoS Genetics, 6, e1001193 10.1371/journal.pgen.1001193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton, W. , & Dilworth, M. (1971). Control of leghaemoglobin synthesis in snake beans. Biochemical Journal, 125, 1075–1080. 10.1042/bj1251075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, P. J. , Zhang, Z. , Kroon, D. E. , Casstevens, T. M. , Ramdoss, Y. , & Buckler, E. S. (2007). TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics, 23, 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Brugnoli, E. , & Lauteri, M. (1991). Effects of salinity on stomatal conductance, photosynthetic capacity, and carbon isotope discrimination of salt‐tolerant (Gossypium hirsutum L.) and salt‐sensitive (Phaseolus vulgaris L.) C3 non‐halophytes. Plant Physiology, 95, 628–635. 10.1104/pp.95.2.628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busoms, S. , Teres, J. , Huang, X. , Bomblies, K. , Danku, J. , Douglas, A. , … David, E. (2015). Salinity is an agent of divergent selection driving local adaptation of Arabidopsis thaliana to coastal habitats. Plant Physiology, 168, 915–929. 10.1104/pp.15.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves, M. M. , Flexas, J. , & Pinheiro, C. (2009). Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Annals of Botany, 103, 551–560. 10.1093/aob/mcn125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, T. D. , Chen, H. , Hien, V. T. T. , Hamwieh, A. , Yamada, T. , Sato, T. , … Xu, D. (2016). Ncl synchronously regulates Na+, K+, and Cl− in soybean and greatly increases the grain yield in saline field conditions. Scientific Reports, 6, 19147 10.1038/srep19147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Fita, A. , Rodríguez‐Burruezo, A. , Boscaiu, M. , Prohens, J. , & Vicente, O. (2015). Breeding and domesticating crops adapted to drought and salinity: A new paradigm for increasing food production. Frontiers in Plant Science, 6, 978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers, T. (2004). Improving crop salt tolerance. Journal of Experimental Botany, 55, 307–319. 10.1093/jxb/erh003 [DOI] [PubMed] [Google Scholar]
- Garcia de la Garma, J. , Fernandez‐Garcia, N. , Bardisi, E. , Pallol, B. , Asensio‐Rubio, J. S. , Bru, R. , & Olmos, E. (2015). New insights into plant salt acclimation: The roles of vesicle trafficking and reactive oxygen species signalling in mitochondria and the endomembrane system. New Phytologist, 205, 216–239. 10.1111/nph.12997 [DOI] [PubMed] [Google Scholar]
- Hamaji, K. , Nagira, M. , Yoshida, K. , Ohnishi, M. , Oda, Y. , Uemura, T. , … Hasezawa, S. I. (2009). Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis . Plant and Cell Physiology, 50, 2023–2033. 10.1093/pcp/pcp143 [DOI] [PubMed] [Google Scholar]
- Hamid, H. , Khaedr, A. , Mohammad, A. , Amal, A. , Paul, Q. W. , & Abogadallah, M. (2003). Proline induces the expression of salt‐stress‐responsive proteins and may improve the adoption of Pancratium maritimum L. to salt stress. J Exp Bot, 54, 2553–2562. [DOI] [PubMed] [Google Scholar]
- Hamwieh, A. , Tuyen, D. , Cong, H. , Benitez, E. , Takahashi, R. , & Xu, D. (2011). Identification and validation of a major QTL for salt tolerance in soybean. Euphytica, 179, 451–459. 10.1007/s10681-011-0347-8 [DOI] [Google Scholar]
- Hanin, M. , Ebel, C. , Ngom, M. , Laplaze, L. , & Masmoudi, K. (2016). New insights on plant salt tolerance mechanisms and their potential use for breeding. Frontiers in Plant Science, 7, 1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, P. , & Cress, W. (1997). Metabolic implications of stress‐induced proline accumulation in plants. Plant Growth Regulation, 21, 79–102. 10.1023/A:1005703923347 [DOI] [Google Scholar]
- Heuer, B. (2010). Role of proline in plant response to drought and salinity In Handbook of plant and crop stress (pp. 213–238). Boca Raton: CRC Press. [Google Scholar]
- Hossain, M. S. , & Dietz, K.‐J. (2016). Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Frontiers in Plant Science, 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas, R. , Rubio, L. , Cagnac, O. , García‐sánchez, M. J. , Alché, J. D. D. , Venema, K. , … Rodríguez‐rosales, M. P. (2013). The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K+ homeostasis in transgenic tomato. Plant, Cell & Environment, 36, 2135–2149. [DOI] [PubMed] [Google Scholar]
- Jain, S. , Nainawatee, H. , Jain, R. , & Chowdhury, J. (1991). Proline status of genetically stable salt‐tolerant Brassica juncea L. somaclones and their parent cv. Prakash. Plant Cell Reports, 9, 684–687. 10.1007/BF00235357 [DOI] [PubMed] [Google Scholar]
- Julkowska, M. M. , Klei, K. , Fokkens, L. , Haring, M. A. , Schranz, M. E. , & Testerink, C. (2016). Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR‐KISS gene. Journal of Experimental Botany, 67, 2127–2138. 10.1093/jxb/erw015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska, M. M. , & Testerink, C. (2015). Tuning plant signaling and growth to survive salt. Trends in Plant Science, 20, 586–594. 10.1016/j.tplants.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Kakar, K. , Wandrey, M. , Czechowski, T. , Gaertner, T. , Scheible, W.‐R. , Stitt, M. , … Cheung, F. (2008). A community resource for high‐throughput quantitative RT‐PCR analysis of transcription factor gene expression in Medicago truncatula . Plant Methods, 4, 18 10.1186/1746-4811-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y. , Li, M. , Sinharoy, S. , & Verdier, J. (2016). A snapshot of functional genetic studies in Medicago truncatula . Frontiers in Plant Science, 7, 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y. , Sakiroglu, M. , Krom, N. , Stanton‐Geddes, J. , Wang, M. , Lee, Y. C. , … Udvardi, M. (2015). Genome‐wide association of drought‐related and biomass traits with HapMap SNPs in Medicago truncatula . Plant, Cell & Environment, 38, 1997–2011. 10.1111/pce.12520 [DOI] [PubMed] [Google Scholar]
- Keisham, M. , Mukherjee, S. , & Bhatla, S. C. (2018). Mechanisms of sodium transport in plants—Progresses and challenges. International Journal of Molecular Sciences, 19, 647 10.3390/ijms19030647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y. , Sadhukhan, A. , Tazib, T. , Nakano, Y. , Kusunoki, K. , Kamara, M. , … Hoekenga, O. A. (2016). Joint genetic and network analyses identify loci associated with root growth under NaCl stress in Arabidopsis thaliana . Plant, Cell & Environment, 39, 918–934. 10.1111/pce.12691 [DOI] [PubMed] [Google Scholar]
- Kulminski, A. M. (2011). Complex phenotypes and phenomenon of genome‐wide inter‐chromosomal linkage disequilibrium in the human genome. Experimental Gerontology, 46, 979–986. 10.1016/j.exger.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Lacerda, C. F. D. , Cambraia, J. , Oliva, M. A. , & Ruiz, H. A. (2003). Osmotic adjustment in roots and leaves of two sorghum genotypes under NaCl stress. Brazilian Journal of Plant Physiology, 15, 113–118. [Google Scholar]
- Lê, S. , Josse, J. , & Husson, F. (2008). FactoMineR: An R package for multivariate analysis. Journal of Statistical Software, 25, 1–18. [Google Scholar]
- Leshem, Y. , Seri, L. , & Levine, A. (2007). Induction of phosphatidylinositol 3‐kinase‐mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. The Plant Journal, 51, 185–197. 10.1111/j.1365-313X.2007.03134.x [DOI] [PubMed] [Google Scholar]
- Li, D. , Su, Z. , Dong, J. , & Wang, T. (2009). An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genomics, 10, 517 10.1186/1471-2164-10-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. , Zhu, M. , Yano, M. , Gao, J. , Liang, Z. , Su, W. , … Chao, D. (2004). QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theoretical and Applied Genetics, 108, 253–260. 10.1007/s00122-003-1421-y [DOI] [PubMed] [Google Scholar]
- Llorente, F. , López‐Cobollo, R. M. , Catalá, R. , Martínez‐Zapater, J. M. , & Salinas, J. (2002). A novel cold‐inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. The Plant Journal, 32, 13–24. 10.1046/j.1365-313X.2002.01398.x [DOI] [PubMed] [Google Scholar]
- Matysik, J. , Alia, B. B. , & Mohanty, P. (2002). Molecular mechanisms of quenching of reactive oxygen species by proline under stress in plants. Current Science, 82, 525–532. [Google Scholar]
- Miller, G. , Suzuki, N. , Ciftci‐yilmaz, S. , & Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment, 33, 453–467. 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- Misra, N. , & Gupta, A. K. (2005). Effect of salt stress on proline metabolism in two high yielding genotypes of green gram. Plant Science, 169, 331–339. 10.1016/j.plantsci.2005.02.013 [DOI] [Google Scholar]
- Moftah, A. E. , & Michel, B. E. (1987). The effect of sodium chloride on solute potential and proline accumulation in soybean leaves. Plant Physiology, 83, 238–240. 10.1104/pp.83.2.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns, R. , James, R. A. , & Läuchli, A. (2006). Approaches to increasing the salt tolerance of wheat and other cereals. Journal of Experimental Botany, 57, 1025–1043. 10.1093/jxb/erj100 [DOI] [PubMed] [Google Scholar]
- Munns, R. , James, R. A. , Xu, B. , Athman, A. , Conn, S. J. , Jordans, C. , … Plett, D. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology, 30, 360–364. 10.1038/nbt.2120 [DOI] [PubMed] [Google Scholar]
- Munns, R. , Passioura, J. B. , Guo, J. , Chazen, O. , & Cramer, G. R. (2000). Water relations and leaf expansion: Importance of time scale. Journal of Experimental Botany, 51, 1495–1504. 10.1093/jexbot/51.350.1495 [DOI] [PubMed] [Google Scholar]
- Munns, R. , & Tester, M. (2008). Mechanisms of salinity tolerance. Annual Review of Plant Biology, 59, 651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Nguyen, V. L. , Ribot, S. A. , Dolstra, O. , Niks, R. E. , Visser, R. G. , & van der Linden, C. G. (2013). Identification of quantitative trait loci for ion homeostasis and salt tolerance in barley (Hordeum vulgare L.). Molecular Breeding, 31, 137–152. 10.1007/s11032-012-9777-9 [DOI] [Google Scholar]
- Patil, G. , Do, T. , Vuong, T. D. , Valliyodan, B. , Lee, J.‐D. , Chaudhary, J. , … Nguyen, H. T. (2016). Genomic‐assisted haplotype analysis and the development of high‐throughput SNP markers for salinity tolerance in soybean. Scientific Reports, 6, 19199 10.1038/srep19199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patishtan, J. , Hartley, T. N. , Fonseca de Carvalho, R. , & Maathuis, F. J. (2017). Genome‐wide association studies to identify rice salt‐tolerance markers. Plant, Cell & Environment, 41, 970–982. [DOI] [PubMed] [Google Scholar]
- Petkov, P. M. , Graber, J. H. , Churchill, G. A. , DiPetrillo, K. , King, B. L. , & Paigen, K. (2005). Evidence of a large‐scale functional organization of mammalian chromosomes. PLoS Genetics, 1, e33 10.1371/journal.pgen.0010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X. , Li, M.‐W. , Xie, M. , Liu, X. , Ni, M. , Shao, G. , … Isobe, S. (2014). Identification of a novel salt tolerance gene in wild soybean by whole‐genome sequencing. Nature Communications, 5, 4340 10.1038/ncomms5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada, V. (2016). The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiologia Plantarum, 157, 389–399. 10.1111/ppl.12416 [DOI] [PubMed] [Google Scholar]
- Ranganayakulu, G. , Veeranagamallaiah, G. , & Sudhakar, C. (2013). Effect of salt stress on osmolyte accumulation in two groundnut cultivars (Arachis hypogaea L.) with contrasting salt tolerance. African Journal of Plant Science, 7, 586–592. [Google Scholar]
- Robles, P. , Micol, J. L. , & Quesada, V. (2015). Mutations in the plant‐conserved MTERF9 alter chloroplast gene expression, development and tolerance to abiotic stress in Arabidopsis thaliana . Physiologia Plantarum, 154, 297–313. 10.1111/ppl.12307 [DOI] [PubMed] [Google Scholar]
- Rus, A. , Baxter, I. , Muthukumar, B. , Gustin, J. , Lahner, B. , Yakubova, E. , & Salt, D. E. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis . PLoS Genetics, 2, e210 10.1371/journal.pgen.0020210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairam, R. , & Tyagi, A. (2004). Physiological and molecular biology of salinity stress tolerance in deficient and cultivated genotypes of chickpea. Plant Growth Regulation, 57, 10. [Google Scholar]
- Sanchez, D. H. , Pieckenstain, F. L. , Szymanski, J. , Erban, A. , Bromke, M. , Hannah, M. A. , … Udvardi, M. K. (2011). Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS One, 6, e17094 10.1371/journal.pone.0017094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala, S. , & Pottosin, I. I. (2010). Potassium and potassium‐permeable channels in plant salt tolerance In Ion Channels and Plant Stress Responses (pp. 87–110). Berlin, Heidelberg: Springer. [Google Scholar]
- Shabala, S. , Wu, H. , & Bose, J. (2015). Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Science, 241, 109–119. 10.1016/j.plantsci.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Gao, L. , Wu, Z. , Zhang, X. , Wang, M. , Zhang, C. , … Li, Z. (2017). Genome‐wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biology, 17, 92 10.1186/s12870-017-1044-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, P. , & Gerós, H. (2009). Regulation by salt of vacuolar H+‐ATPase and H+‐pyrophosphatase activities and Na+/H+ exchange. Plant Signaling & Behavior, 4, 718–726. 10.4161/psb.4.8.9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smajgl, A. , Toan, T. , Nhan, D. , Ward, J. , Trung, N. , Tri, L. , … Vu, P. (2015). Responding to rising sea levels in the Mekong Delta. Nature Climate Change, 5, 167–174. 10.1038/nclimate2469 [DOI] [Google Scholar]
- Storey, J. D. , & Tibshirani, R. (2003). Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences, 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. , Koussevitzky, S. , Mittler, R. , & Miller, G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant, Cell & Environment, 35, 259–270. 10.1111/j.1365-3040.2011.02336.x [DOI] [PubMed] [Google Scholar]
- Tarte, V. N. , Seok, H.‐Y. , Woo, D.‐H. , Le, D. H. , Tran, H. T. , Baik, J.‐W. , … Moon, Y.‐H. (2015). Arabidopsis Qc‐SNARE gene AtSFT12 is involved in salt and osmotic stress responses and Na+ accumulation in vacuoles. Plant Cell Reports, 34, 1127–1138. 10.1007/s00299-015-1771-3 [DOI] [PubMed] [Google Scholar]
- Thomson, M. J. , de Ocampo, M. , Egdane, J. , Rahman, M. A. , Sajise, A. G. , Adorada, D. L. , … Gregorio, G. B. (2010). Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice, 3, 148–160. 10.1007/s12284-010-9053-8 [DOI] [Google Scholar]
- Varshney, R. K. , Bansal, K. C. , Aggarwal, P. K. , Datta, S. K. , & Craufurd, P. Q. (2011). Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends in Plant Science, 16, 363–371. 10.1016/j.tplants.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Verdier, J. , Dessaint, F. , Schneider, C. , & Abirached‐Darmency, M. (2012). A combined histology and transcriptome analysis unravels novel questions on Medicago truncatula seed coat. Journal of Experimental Botany, 64, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues, P. E. , Lasky, J. R. , Juenger, T. E. , Liu, T.‐W. , & Kumar, M. N. (2014). Genome wide association mapping combined with reverse genetics identifies new effectors of low water potential‐induced proline accumulation in Arabidopsis thaliana . Plant Physiology, 164, 144–159. 10.1104/pp.113.224014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, H. , Chen, L. , Guo, J. , Li, Q. , Wen, J. , Yi, B. , … Shen, J. (2017). Genome‐wide association study reveals the genetic architecture underlying salt tolerance‐related traits in rapeseed (brassica napus l.). Frontiers in Plant Science, 8, 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F.‐W. , Chao, W. , Yao, S. , Nan, W. , Li, X.‐W. , Dong, Y.‐Y. , … Wang, Z. M. (2016). Overexpression of vacuolar proton pump ATPase (V‐H+‐ATPase) subunits B, C and H confers tolerance to salt and saline‐alkali stresses in transgenic alfalfa (Medicago sativa L.). Journal of Integrative Agriculture, 15, 2279–2289. 10.1016/S2095-3119(16)61399-0 [DOI] [Google Scholar]
- Wang, L. , He, X. , Zhao, Y. , Shen, Y. , & Huang, Z. (2011). Wheat vacuolar H+‐ATPase subunit B cloning and its involvement in salt tolerance. Planta, 234, 1–7. 10.1007/s00425-011-1383-2 [DOI] [PubMed] [Google Scholar]
- Warde‐Farley, D. , Donaldson, S. L. , Comes, O. , Zuberi, K. , Badrawi, R. , Chao, P. , … Maitland, A. (2010). The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research, 38, W214–W220. 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D. , Leister, D. , & Kleine, T. (2017). Arabidopsis thaliana mTERF10 and mTERF11, but not mTERF12, are involved in the response to salt stress. Frontiers in Plant Science, 8, 1213 10.3389/fpls.2017.01213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani, N. , Ichikawa, T. , Kondou, Y. , Maeda, S. , Iwabuchi, M. , Mori, M. , … Oda, K. (2009). Overexpression of a rice gene encoding a small C2 domain protein OsSMCP1 increases tolerance to abiotic and biotic stresses in transgenic Arabidopsis . Plant Molecular Biology, 71, 391–402. 10.1007/s11103-009-9530-x [DOI] [PubMed] [Google Scholar]
- Yong, H.‐Y. , Wang, C. , Bancroft, I. , Li, F. , Wu, X. , Kitashiba, H. , & Nishio, T. (2015). Identification of a gene controlling variation in the salt tolerance of rapeseed (Brassica napus L.). Planta, 242, 313–326. 10.1007/s00425-015-2310-8 [DOI] [PubMed] [Google Scholar]
- Young, N. D. , Debellé, F. , Oldroyd, G. E. , Geurts, R. , Cannon, S. B. , Udvardi, M. K. , … Van de Peer, Y. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature, 480, 520–524. 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, N. D. , & Udvardi, M. (2009). Translating Medicago truncatula genomics to crop legumes. Current Opinion in Plant Biology, 12, 193–201. 10.1016/j.pbi.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Zeng, A. , Chen, P. , Korth, K. , Hancock, F. , Pereira, A. , Brye, K. , … Shi, A. (2017). Genome‐wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Molecular Breeding, 37, 30 10.1007/s11032-017-0634-8 [DOI] [Google Scholar]
- Zhang, J. Y. , Cruz de Carvalho, M. H. , Torres‐jerez, I. , Kang, Y. , Allen, S. N. , Huhman, D. V. , … Udvardi, M. K. (2014). Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant, Cell & Environment, 37, 2553–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Ersoz, E. , Lai, C.‐Q. , Todhunter, R. J. , Tiwari, H. K. , Gore, M. A. , … Buckler, E. S. (2010). Mixed linear model approach adapted for genome‐wide association studies. Nature Genetics, 42, 355–360. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information on the 132 accessions that were used in the GWAS.
Supplemental Table S2. Correlation coefficients and association p values of all characterized salinity‐stress related traits.
Supplemental Table S3. List of 200 SNPs of the lowest p values for each trait.
Supplemental Table S4. SNPs reside inside as well as 1000 bp upstream or downstream of the eight genes that were examined by qRT‐PCR.
Supplemental Table S5. List of genes (SNPs) that had been identified in earlier GWAS of salinity‐stress related traits in rice (Al‐Tamimi et al., 2016), Arabidopsis (Kobayashi et al., 2016), and rapeseed (Wan et al., 2017).
Supplemental Table S6. The list of primers that were used in the qRT‐PCR experiment.
Supplemental Figure S1. Example ion chromatography of line HM066 showing the difference in sodium and chloride in the leaf (a) and the root (b).
Supplemental Figure S2. Cladogram tree (neighbor‐joining) of the 132 lines that were used in the GWAS.
Supplemental Figure S3. Values of the Ln P(d) and Delta K as a function of the number of clusters (K).
Supplemental Figure S4. Quantile‐quantile (Q‐Q) plots for all the traits obtained by Mixed Linear Model (MLM) with (a) or without (b) Q value as a co‐variant, and General Linear Model (GLM) (c).
Supplemental Figure S5. GO enrichment of genes (1422 Arabidopsis orthologues) in the vicinity of the top 200 SNPs identified through GWAS of 17 traits (Table S3).
Supplemental Figure S6. Physical interaction networks of two selected genes in the top 12 genomic regions.
Supplemental Figure S7. Salinity stressed M. truncatula A17 plants.


