Abstract
Antimicrobial resistance (AMR) is a significant threat to both human and animal health. The spread of AMR bacteria and genes across systems can occur through a myriad of pathways, both related and unrelated to agriculture, including via wastewater, soils, manure applications, direct exchange between humans and animals, and food exposure. Tracing origins and drivers of AMR bacteria and genes is challenging due to the array of contexts and the complexity of interactions overlapping health practice, microbiology, genetics, applied science and engineering, as well as social and human factors. Critically assessing the diverse and sometimes contradictory AMR literature is a valuable step in identifying tractable mitigation options to stem AMR spread. In this article we review research on the nonfoodborne spread of AMR, with a focus on domesticated animals and the environment and possible exposures to humans. Attention is especially placed on delineating possible sources and causes of AMR bacterial phenotypes, including underpinning the genetics important to human and animal health.
Keywords: antimicrobial resistance, antibiotic use, animal agriculture, fecal matter, soil and wastewater
Antimicrobial resistance (AMR) is a significant threat to both human and animal health. Our article reviews research on the nonfoodborne spread of AMR with a focus on domesticated animals and the environment, and possible exposure risks to humans. Attention is especially placed on delineating possible sources and causes of AMR bacterial phenotypes, including underpinning genetics important to human and animal health.
Introduction
Antimicrobial resistance (AMR) is a global problem that impacts both human and animal health. Although AMR primarily derives from antibiotic and antimicrobial use, strong evidence suggests that wider spread of AMR is fueled by inadequate local sanitation, pollution, and other nonuse factors, with the natural environmental being an important conduit.1, 2, 3 Explaining AMR and then developing informed approaches to mitigate it are difficult because many technical, scientific, and behavioral factors come into play. Factors range from understanding the evolution of resistance at the molecular level within a given organism, to transmission mechanisms and pathways across organisms, to wider dissemination between human and animal hosts and across the wider environment, including soil and water. Further, due to the urgency in addressing the problem, researchers from many disciplines are studying AMR from a variety of angles and progressively learning more about it at different levels. However, given the diversity of research perspectives and complexity of the problem, sometimes contradictory answers are being provided to key questions, such as the primary drivers of AMR, the relative role of the natural environment in AMR spread, and actual health risks associated with different types of environmental and other exposures.
Such general queries are also true of AMR within the food animal production industry. The environment is clearly a possible source of enteric and other bacteria, including AMR strains, that can affect food animals. In fact, it is suspected that the majority of microorganisms enter animals through ingestion of water and/or soil, or through contact with other animals and/or humans in their proximity, which include antimicrobial‐resistant bacteria (ARB) and antimicrobial resistance genes (ARGs). AMR sources are now common in nature, although some ARGs are also ancient; but with increased antimicrobial drug use after World War II,5 ARB with recently acquired AMR has dramatically increased.6 In terms of domesticated animals, this phenomenon appears to have been exacerbated by some management practices, particularly in food animals.7 Therefore, it has become critical to understand wider factors that impact AMR in animal systems, including possible exposures to contaminated water and soils by wastewater, manure, and other sources, which are considered probable drivers of AMR transmission and dissemination pathways to and from food animals.8
The intrinsic complexity of AMR makes these relatively simple questions difficult to answer. Specifically, there is debate about how ARB enter wastewater and soils; how long they survive, retain viability, and infectivity; how best to quantify AMR, either ARB or ARGs, or both; and how to define and contrast risks when environmental sources are often overlapping or nebulous. Addressing these questions is further challenged by difficulties in detecting and identifying ARB and associated ARGs in environmental samples.9 Abundances of ARGs are often less than 0.1% of microbial DNA sources,10 and linking an ARG or suites of ARGs to conferred phenotypic resistance is difficult, especially in mixed microbial communities common in nature. For example, an ARB phenotype may result from one specific ARG to an antibiotic in an expressible location, whereas the same phenotypic response can also result from genes for less specific purposes, such as efflux pumps, which are not directly related to a specific antimicrobial agent.
Alternatively, if antibiotics used in food animals are the same or structurally similar to agents used for humans, animal use may amplify ARB of public health concern.11, 12 Such “indirect” ARB selection also occurs when ARGs conferring different types of resistance are closely located on a chromosome or plasmid, which means the selection for one type of resistance might confer other types of resistance. In turn, enteric ARB shed in feces can spread to other livestock and farm workers, reentering the environment and exposing other animals through drainage water and manure. This is analogous to AMR spread via human open defecation, which places raw fecal matter in close proximity to other individuals, increasing the risk of enteric ARB and ARG spread across human populations associated with poor sanitation.2
Finally, commensal bacteria such as Staphylococcus aureus can be transmitted to and from food animals and humans who work on farms or live in close proximity to livestock.13 In fact, recent evidence from the Netherlands suggests that immediate proximity to livestock, such as being a farm worker versus a member of their family, significantly increases their carriage of methicillin‐resistant S. aureus (MRSA); 38% versus 16%, respectively, in nasal and throat swabs (much higher than wider Dutch populations).14 This study also found that only ∼7% of the workers were persistent MSRA carriers, and that such exposure effects did not appear to translate to carriage in wider communities.15 Similar conclusions were reported from a study comparing livestock‐associated MRSA among farm and slaughterhouse workers in Italy.16
When considering the above factors and more, one can see why it is difficult to delineate clear routes of AMR dissemination among food and other domesticated animals, human populations, and the environment. Within this context, the intent of our review here is not to exhaustively discuss all links, methods, and pathways because these have been summarized in previous reviews.11, 17, 18 Instead, our goal is to flag the complexity of AMR as a general problem with the hope of disentangling sometimes contradictory information into useful guidance for the food animal production industry. The article is based on presentations and discussions of an integrated discussion group associated with the conference “Minimizing the Risk of Antimicrobial Resistance from Food Animal Production,” hosted by the New York Academy of Sciences on May 8 and 9, 2018.
Complexity of antimicrobial resistance across domesticated animals, humans, and the environment
The potential for evolution, exposure, and/or spread of AMR spans almost all elements of rural and urban life. Antimicrobial use and other factors that drive AMR are diverse and often interlinked, which are displayed within the context of the food animal production industry in Figure 1. Although Figure 1 shows an array of interactions, the reality is more complex than the figure suggests because underlying the links are even more diverse microbial and genetic phenomena, which are not well understood. Therefore, it is not surprising that AMR is such a difficult problem to address, because everything affects everything else. Therefore, we address only a few considerations to exemplify points of particular importance that overlap the food industry. These include the impact of AMR in fecal matter entering the environment; the fate of AMR in soils and water; dissemination of ARB and ARGs among people and domesticated animals; and the need for more targeted surveillance.
Figure 1.
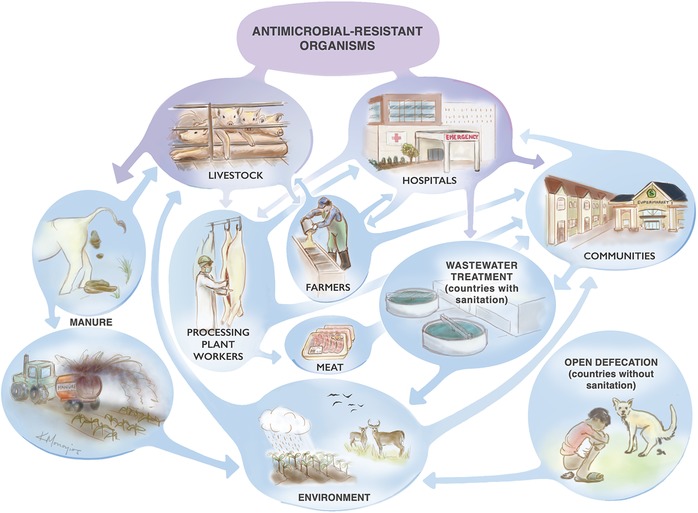
Potential sources and sinks for antimicrobial resistance across domesticated animals, humans, and environmental systems (adapted from Ref. 63).
Fecal matter and the environment
The massive amounts of fecal matter generated by humans and animals around the world can hugely impact local and global biomes. Humans and their domesticated animals constitute approximately 96–98% of the global terrestrial mammalian biomass.19 The fecal matter associated with such huge populations has been estimated to be increasing by over 52 billion kilograms per year since 2003; and the total fecal matter produced is expected to reach at least 4.6 trillion kilograms per year by 2030.20 Food animals, particularly cattle, chickens, and sheep, produce about four times more fecal matter than humans,20 which implies that appropriate fecal matter management in food animal systems must be globally influential. Further, food animal and truly domesticated animals (e.g., companion animals) often reside close to human habitations, especially in less developed countries, and can contribute to fecal matter exposures.
Although most of the developed world has effective wastewater treatment for human wastes, 73% of the world has no major waste treatment.3 The extent of suitable waste management options is even less for domesticated animals, which implies that massive quantities of untreated animal fecal matter are entering the environment, potentially contaminating food and water, and also threatening public health. A strong push to address the problem is currently occurring at the World Health Organization (WHO), the World Bank, and other international agencies,21, 22 but more effort is needed, especially incrementally improving sanitation and waste treatment as strategies to reduce AMR,2 both for human and animal fecal sources.
Wastewater
Wastewater represents an important potential route for spreading antibiotic‐resistant organisms to and from animal agriculture.23 For example, wastewater around the world can contain carbapenem‐resistant Enterobacteriaceae (CRE)24 and other medically important ARB, including particularly resistant strains in human hospital effluents.3, 25 Although secondary and tertiary wastewater treatment plants substantially reduce bacterial loads, typically reducing ARB by 10 to over 1000 times,17 such treatment processes do not remove all resistant bacteria. The fate of ARB and potential risks associated with their release to the environment heavily depends on the type and level of wastewater treatment. However, regardless of treatment, ARB in effluents often enter surface waters that can potentially impact downstream water users. This is particularly true where wastewater or treated effluents are used in irrigation systems (common around the world), such as for feed grains, where bacteria, including ARB, can associate with plant surfaces, especially leafy vegetables consumed by livestock or humans. Further, wastewater irrigation is also a means for surface water contamination, which can impact water quality and livestock.11
Although wastewater‐mediated ARB and ARG spread is almost certain, its relative importance to promoting AMR is less known, partially because it is very difficult to determine sources of specific bacteria in wastewater. Previous work has suggested that hospitals and agriculture are important sources to consider,3 and there may be overlapping concerns. As an example, one study detected bla KPC‐2 in 72 beef cattle lots in the United States, both conventional and raised without antibiotic operations, and many samples contained functional bla KPC‐2 genes (i.e., genes recovered from samples that can be transformed and expressed in a competent cell).26 bla KPC‐2 is also a very common carbapenemase gene found in hospitalized human patients,27 which implies that overlap between ARB in animal agriculture and hospital sources is probable. However, ARB detected in wastewater are not exclusive to hospital sources. In fact, some studies have shown that hospitals are not major sources of ARB, on a total mass basis, especially compared with parallel community sources.28 Further, international migration of resistance is believed to be very common,29, 30 although exact pathways and the frequency of occurrence are not yet well defined.31, 32
Despite the emerging evidence, many questions remain about the spread of ARB among human populations, the environment (soils, wastewater, and surface waters), and food animal production. One of the challenges in addressing these questions is determining how to quantify specific ARB of concern within a complex chemical matrix and mixed microbial community, especially identifying the presence of specific ARB from a distinguishable source. Recent ARG and microbiome source‐tracking methods have been used to stochastically distinguish between community‐ and hospital‐sourced ARB,33 but this was a case in which key sources of ARB were clear. The method overlays genetic high‐throughput quantitative polymerase chain reaction (HT‐qPCR) and 16S sequencing data to develop resistomes and microbiomes for each source and sink,33 and then uses similarity analysis to infer proportional relatedness among sources and sinks.34 Given the complexity of many links shown in Figure 1, generating this type of source tracking analysis is difficult in many cases. However, it might be a useful approach for teasing out possible sources in some cases, an example of which might be allocating the relative importance of humans versus proximal domesticated animals to local environmental exposures.
Livestock fecal matter
Healthy soil is a vital living system essential for crop agriculture. Important components for healthy soil include organic matter and microbes, including bacteria and fungi. Organic matter provides food for microbes, stabilizes soil structures, and increases soil fertility. In agricultural systems, manure is often applied to fields as an additional source of organic matter. However, manure almost certainly contains microbes that include ARB.35 The gastrointestinal tract of domesticated livestock contains many zoonotic pathogens and commensals that are shed in large numbers in feces. Unprocessed or uncomposted manure used in crop production may therefore contain AMR pathogens that can contaminate food and water, potentially contributing to foodborne and waterborne exposures.36
Therefore, livestock and other animal fecal matter can potentially contribute to ARB and ARGs in their local environments, though pathways to exposure and external release are varied. For example, humans and animals physically close to points of defecation are at higher risk of acquiring or exchanging bacteria or genes. This might occur at intensive animal production facilities (see below), but might also occur on more local scales between humans and their pets, and via soil or water impacted by the fecal matter.
Manure generated in food animal operations can be transferred through various pathways, but broad conclusions on the relative importance of each pathway are hard to determine due to contradictory results. As an example, a 2006 study of cattle feedlots in the Midwestern United States found that tetracycline ARG abundances statistically differred between waste lagoons at feedlots where antibiotics were used only therapeutically and feedlots where antibiotics were used both therapeutically and nontherapeutically at higher levels.37 Both groupings that used antibiotics had 100 to 1000 times higher levels of ARGs than control feedlots that did not use antibiotics. The authors concluded that similarities between therapeutic‐only and therapeutic/nontherapeutic lagoons were probably due to cattle in therapeutic‐only herds being returned after therapy and exposing other cattle to their antibiotic‐impacted feces. This is analogous to what has been seen in human populations in densely populated areas without adequate immediate sanitation.38 Conversely, other researchers have found limited relationships between the use of different levels of antimicrobial drugs and AMR trends in fecal Escherichia coli.39
These opposite observations again demonstrate the complexity of AMR. In both cases, carefully designed studies were performed, and it is likely the studies are correct in themselves. This suggests that tertiary factors, such as how systems were sampled, the specific AMR detection methods, and local specifics to each animal operation were sufficiently different to provide opposite answers to the same question. This argues for greater unification among methods and study designs to provide less ambiguous results. A recent review concluded that there was a broad lack of quantitative causal research that associates AMR sources and increases in environmental ARB, suggesting that improved study design, greater consideration of a priori bias, and standardization of analytical tools are important to understanding better how ARBs in the environment affect human and animal health.40
Wider consideration of direct fecal releases
According to the WHO and UNICEF, in 2015 2.3 billion people lacked basic sanitation and many millions practice open defecation, which is the direct release of human fecal matter to the environment. Although this does not directly relate to AMR in food animal production, analogous fecal releases are common with food animals, and translatable knowledge can be gained from a brief review of the consequences of open defecation. As background, most people who practice open defecation live in Central and South Asia and Sub‐Saharan Africa.41 Poor sanitation and open defecation increase the environmental microbial burden and public health risk of disease. Making the situation worse, many countries with poor or nonexistent sanitation allow the sale of high value over‐the‐counter antibiotics, increasing the use of antibiotics and the risk of antibiotic resistance.42
Thus, fecal matter is potentially an important source of antibiotic resistance spread among people or animals in areas with inadequate consideration of sanitation.42, 43 For example, researchers detected higher levels of emerging resistance genes, such as bla NDM‐1 (a carbapenem ARG), in water and sediment samples collected in the Upper Ganges River in northern India in June—at the beginning of the monsoons and coinciding with times of mass pilgrimages from urban areas—compared with February (one of the driest months).38 Locations along the river with some fecal containment, such as latrines, had 4‐5 orders of magnitude lower CRE concentrations than stretches of the Ganges where there was open defecation.2
Although this is predominantly a human example, similar releases and AMR exposures almost certainly occur in food animal operations where population densities are high and local sanitation is inadequate, major concerns in any operation that includes animal rearing. Experts recently have predicted that ARB in livestock will arise in the Far East because of heavy use of antibiotics in Asian farms, which implies that significant shedding of ARB must occur in Asian animal feces. In contrast to those results, however, research conducted through the Food and Agriculture Organization of the United Nations (FAO) in 2014 found that levels of resistance to many antibiotics were actually higher in isolates from food animals in the United States and Europe than in Asia.44
Such differences exemplify the difficulty of AMR because comparable and plausible studies do not always provide black and white answers and can generate opposite conclusions. In the above cases, we suspect that the FAO data may not represent all food animal–related activity, particularly aquaculture in South East Asia where AMR is reported to be rampant.45 Again, this shows how conclusions from one study can provide different answers than those from parallel studies. A useful way of discerning valuable information from the literature is through more careful examination of details within each study, and then noting those differences when drawing more general conclusions.
Agricultural soil and water
Antibiotics and ARGs can be enriched in ground and surface waters surrounding animal agriculture operations, especially where animals receive antibiotics and are close to soil and water resources.46, 47, 48, 49, 50 Consequently, soils and water have the potential to be important players in the spread and persistence of ARB. Substances that are applied to the soil can quickly enter bodies of water, particularly in areas where artificial subsurface drainage systems are installed. ARB also naturally exist in soil and water environments, and a key challenge for understanding the impact of agricultural management practices on the spread and persistence ARGs and ARB is differentiating between ARGs that spread from anthropogenic sources versus ARGs already present in the environment.
Antibiotics in irrigation water can be taken up into crop tissues (mostly roots), but levels of uptake are typically low unless antibiotic concentrations are very high.51, 52 However, whether uptake occurs depends on the specific crop–antibiotic combination. No general rules have yet been established for which antibiotic–crop combinations lead to greater uptake, but, overall, uptake tends to be limited, quite selective, and plant and antibiotic specific.52 While there is a substantial amount of literature on the plant uptake of antibiotics, there is less information on the uptake of ARGs and ARB into crop tissues, especially where exposure concentrations are within normal ranges and the experiments have been well designed with controls.52, 53, 54 Nonetheless, evidence exists that ARGs and ARB can accumulate on plant surfaces associated with manure applications, and it has been suggested that associated crops and soils should be fallowed prior to harvest.53 Additionally, recent work has shown that swine manure application can be associated with increases in observed levels of phenotypic ARB, as well as ARGs in soils and drainage waters. However, the abundance of different ARGs is highly variable and can range up to a year after the manure application.50 In addition, specific crops and crop rotation can influence the types of resistance genes found in soils; for example, the ARGs in soils from corn and soybean crop production systems were found to be quite different.35, 47
It is still unclear the extent to which resistance and related exposure in soils receiving manure and/or antibiotics translates to a human health risk. A recent study on archived soil samples in Denmark from fields receiving only either manure or inorganic fertilizer for over 100 years showed that only manured fields had significantly higher levels of extended spectrum β‐lactamase ARGs (a critical form of AMR in hospitals). Further, the relative appearance of specific ARGs in the manure field was almost simultaneous (within 6 months) with appearance in hospitals.55 It is not possible to determine whether detected ARGs first appeared in the manure or hospitals, but the results suggest that cross‐location spread occurred between the original cattle (possibly via soil) and the human populations. In general, there is an urgent need to better understand the linkage between antibiotic usage and resistance across environmental compartments. Currently, this is limited by the availability of data on the usage of antibiotics coupled directly with environmental surveillance of resistance. Further, many studies often focus on different types of ARGs or antibiotics, making it difficult to compare results between efforts. Moving forward, integrated and systematic efforts will be necessary to develop models that estimate the spread and persistence of ARB in the environment and how an indicated source of bacteria contributes to human health. This is among the most critical knowledge gaps in understanding the interconnections and consequences of the complex issue of AMR.
Transmission of antimicrobial resistance among domesticated animals and humans through direct contact
Direct contact with food animals
As mentioned earlier, bidirectional exchange of ARB and ARGs can occur between food animals and people who are in direct contact with the animals or are proximally close to animal products, such as farmers and processing plant workers.56 Researchers have largely focused on understanding the spread of bacteria from pigs to farmers, but there is evidence that people can also spread staphylococcus and other bacteria to pigs.56 In the case of the skin‐associated bacterium S. aureus, which has been studied the most, there is still much uncertainty concerning the frequency with which livestock workers become colonized from food animal farms, as well as the frequency with which acquisition of this skin bacterium leads to symptomatic infections.
Determining how occupational exposure becomes an occupational hazard has been difficult to assess for several reasons, some related to how studies have been performed, but also because of genetic variation in S. aureus phenotypes and general difficulties in detecting effects. Researchers have conducted cross‐sectional studies to assess this. For example, a group at the University of Iowa performed a cross‐sectional study to identify staphylococcal infections in pig farmers by asking farm workers whether they had been diagnosed with certain bacterial infections.57 This study relied on farmers to accurately report their medical information, and it was difficult to trace an infection back to exposure because it can take weeks to months after contact with Staphylococcus to develop symptoms.58 To aid such research, ideally researchers should collaborate with physician groups who treat patients with potential Staphylococcus infections, although such longitudinal studies are costly, they require staff to follow up with patients, and they must be institutional review board certified. Further, large groups of patients are needed because the subset of farmers and workers infected with Staphylococcus is small compared to the overall patient population.59 A few studies exploring this issue were confined to limited geographical areas,16, 59, 60, 61, 62 hence, additional studies need to be conducted across areas and in more locations.
To further complicate matters, there is no predominant strain of S. aureus linked to the majority of pig farms that can be probed specifically through genomic analyses.63 For example, while ST398 is the predominant strain of livestock‐associated S. aureus in Europe and has been recognized as an occupational hazard for people working in the pig industry,64, 65 in the United States several types of S. aureus, including ST398, ST9, and ST5, have been found in pig farms, as well as on people who work with pigs.63, 66 The proportion of these different strains varies geographically between Iowa, North Carolina, and elsewhere around the Midwest, though the breakdown of strains has not been reported for most of the United States.60, 63
The type of S. aureus is also important because it can influence colonization rates. For example, strains such as ST5 seem to colonize people efficiently, whereas others such as ST398 appear to be poorly adapted to humans.14 Studies found that ST398 colonization of veterinarians is lost fairly quickly after they spend time away from the farm.67 Host‐specific factors also appear to influence colonization rates; for example, a longitudinal study of swine veterinarians found that the majority of them was only transiently colonized, whereas a sizable minority was permanently colonized, although follow‐up work suggests that colonization may be lost after a couple of years.68 Nevertheless, short‐term colonization may still result in transfer of resistance genes, such as the mec resistance genes in MRSA,69 from farm‐acquired S. aureus to strains that colonize humans.
Beyond the farm, there are data that hint that MRSA may conditionally spread from pig farms to humans who do not work in the pig industry.15 Studies in the Netherlands have found cases of livestock‐associated MRSA70 that may be attributable to living in areas with high animal density, although routes of dissemination are unknown. One possibility is exposure to livestock workers, resulting in secondary person‐to‐person transmission away from the farm source.71 Studies in the United States have shown that proximity to concentrated animal feeding operations or areas where pig manure is applied to fields can increase the risk of MRSA carriage72 or infection.73
Finally, the spread of specific ARGs from humans to livestock may also occur. One school of thought is that humans carrying MRSA have cross‐infected pigs: while transient in the pig, the mec resistance genes from human‐adapted MRSA transferred to pig‐adapted MRSA, consequently giving rise to MRSA in pigs.56 This hypothesis is supported by the fact that livestock‐associated MRSA (i.e., in pigs) associates with the same mec genes found in human community MRSA, although this is an inferred possibility.
Although the above examples focus on pig–human interactions, they are good examples of another type of complexity in explaining AMR observations. Such issues are exacerbated by the challenge to obtaining farm‐level data on antibiotic use. Although the FDA in 2016 began requiring sponsors of antimicrobial drugs to provide estimates of sales for each major food‐producing species, the agency does not collect information about the geographical areas to which drugs are sold.74 These types of data would complement U.S. maps of distribution of Staphylococcus and other bacterial species among farms, farmers, and the environment, and would also allow more fine mapping of resistance distribution and correlation of resistance to on‐farm use of antibiotics.
What can be learned from companion animals?
Companion animals (e.g., pets) represent an often ignored, but potentially important source of ARB and ARGs in human populations because of the intimacy between humans and their pets.18 Pets have little to do with the food animal industry, but considering them provides analogous evidence of how close personal contact between domesticated animals and humans can influence AMR spread. In Europe, veterinary pharmaceutical companies established the European Animal Health Study Centre, a Brussels‐based international, nonprofit association that conducts scientific and economic studies including, from 2008 to 2010, antimicrobial susceptibility surveillance in sick companion animals in 10 EU member states.75 However, surveillance has been limited to skin, ear, and soft tissue infections, urinary tract infections (including prostatitis in dogs), upper respiratory tract infections, and periodontal infections. Further, surveillance of AMR markers in healthy companion animals has not been done, although what has been learned may translate to issues in the food animal industry.
Some useful examples exist, such as the possible affect pets may have had on vancomycin‐resistant Enterococcus faecium (VRE) infections in seriously ill patients in European hospitals in the 1980s.76 As background, VRE cases were initially attributed to food animal agriculture after VRE was isolated from livestock in Europe over time.77 The theory was that the use of avoparcin (a vancomycin analog of the glycopeptide class of antibiotics) for animal growth promotion created selection pressure that led to VRE.77 This resulted in the European Union banning the use of avoparcin in animal agriculture in 1997, and then the use of any antibiotic for growth promotion in 2006.78 Although VRE on farms dropped precipitously following the 1997 ban, the VRE problem in European hospitals inexplicably persisted.79 It was not until whole‐genome sequencing was performed on clinical VRE isolates that it became evident that genotypes were not from farm isolates but probably from ampicillin‐resistant E. faecium common in dogs in Denmark.80, 81, 82
There are other examples of resistance being transmitted from pets to humans that provide useful templates for food animal AMR studies. For example, exposure to puppies can introduce resistant Campylobacter spp. infections to humans.83 Additionally, humans with MRSA infections should consider having their pets examined as potential MRSA reservoirs.84 Even though MRSA is not adapted to dogs, cats, and other animals, these animals can become colonized with MRSA and then serve as vectors for spread to humans in the same household.85
More can be learned by the food animal industry by understanding the role of companion animals in the transmission of antibiotic resistance to humans, the environment, and to food animals. However, this research falls outside the purview of funding agencies such as the National Institutes of Health and the U.S. Department of Agriculture.86 The use of antibiotics in companion animals may be dismissed as less important because, according to sales data, the volume is low compared to the use of antibiotics in humans and livestock. However, the drugs that pets receive are generally more potent and injectable, and map more closely to the pattern of critically important classes prescribed to humans than to food animals, whereas food animals generally receive less important antibiotics often in their feed.87 Understanding such relationships between specific antibiotics used for food animals and those used for pets can enrich our understanding of animal–human contact as pathway of AMR spread. Pets might spread critical antibiotic resistance to people who in turn become vectors for antibiotic‐resistance dissemination to livestock and the environment.88
Wildlife: another complication in animal systems
Although not central to this review, brief mention of wildlife‐mediated AMR is needed because it highlights another difficult‐to‐detect but potentially important factor related to AMR dissemination in animal–human–environmental systems. Direct contact between humans and wildlife is typically not as intimate as with livestock or domestic animals; but wildlife, especially birds, has been shown to act as a vector for AMR transmission across the environment.89 For example, growing data suggest that ARB and ARGs may be dispersed by wild birds after exposure to sewage near rivers,32 spreading of sewage sludge onto farmland,90 and via direct exposure to the feces from livestock and companion animals.91 However, such exposures and associated dissemination pathways are a complexity that is rarely considered in AMR studies, although this pathway might be the best explanation for rapid rates of AMR spread on global scales.
A recent study examining the apparent accumulation of anthropogenic ARGs in the High Arctic found that bla NDM‐1 and other clinically important ARGs were elevated in Arctic soils near bird, reindeer, and arctic fox‐watering areas, suggesting possible wildlife mediation in ARG movement.92 In this case, ARG movement may also be mediated by human wastes; but in general, wildlife activity and associated fecal matter might explain the appearance of AMR almost anywhere, including places with functionally no antimicrobial use or evident influence. Therefore, similar to companion animals, AMR carried by wildlife, possibly acquired after exposure to human, livestock, and other fecal matter, should also be considered within AMR assessments, adding further complexity to explaining cause and effects.
Considerations for AMR surveillance
Among the greatest complexities and questions that have concerned AMR scientists for many years is how best to measure AMR when studying water, soil, or other environmental systems, including for diagnostics and/or practical surveillance.93 Metagenomics analyses offer a broad picture of the presence and relative and absolute abundances of putative ARGs across microbiomes, and can also help expand ARG databases to improve and guide future studies.94 However, such methods do not reveal whether detected ARGs are actually functional or whether they can or are conferring phenotypic resistance in bacteria. Further, metagenomics methods cannot be easily used to link the presence of an ARG to a specific ARB, which can only be done stochastically in most scenarios. Finally, whether an ARG detected by metagenomics is present in viable bacteria, is sited alone in a genome or is part of a multidrug‐resistance cassette (e.g., on a plasmid) are facts not easily determined from metagenomics data alone.4, 95 Such problems can be partially resolved by parallel culturing of organisms (from the same source as the metagenomics data), selection of specific phenotypes, and determining the genetic context of specific genes in such isolates. However, the majority of organisms found in environmental systems cannot be cultured under standard laboratory conditions, which poses a problem for linking ARB and ARG metagenomics data within environmental compartments.10
In contrast to metagenomics, more targeted methods for quantifying specific ARGs, such as qPCR, have been used successfully for a number of years. qPCR is still a very useful method because it is quantitative, although it does not allow the identification of genetic precursors of ARB or of previously unknown putative resistance genes. This is demonstrated by the mistaken attribution of VRE emergence in European hospitals to livestock.96 Conversely, qPCR can be performed very quickly and is useful for tracking specific ARGs in systems where the ARGs have already been established as important; qPCR can use known ARGs that are biomarkers for exposure and other related work. Further, a new HT‐qPCR method is now available that allows hundreds of ARGs to be quantified simultaneously—a method used successfully to study ARGs in Chinese swine operations97 and other applications.92
However, whole‐genome sequencing has advantages, and has reduced in cost over the last 10 years. It is an approach that can inform how bacterial strains might associate with each other within a wider microbial community and how they can also be stochastically linked with antibiotic resistance.98 However, whole metagenome sequencing and data interpretation requires considerable bioinformatics skill to decipher the resulting large and complex data sets, which currently makes surveillance requiring metagenome sequencing less practical for routine AMR screening. Fortunately, new methods are being developed, including more exact ways of determining specific bacterial hosts,99 and simplified bioinformatics methods for generating more immediately useful data.100 This may ultimately make such methods useful for routine work, as well as for deeper genetic investigations. As part of the One Health Initiative, which fosters collaborations between stakeholders in human and veterinary medicine, there is a push to conduct whole‐genome sequencing–based surveillance of human and animal hospitals and farms, i.e., to make such methods more routine among a wider set of users.101
Despite such advances, current surveillance should still include traditional culturing methods, qPCR or similar targeted quantitative genetic methods, and, ideally, genome and metagenome sequencing. In fact, any study that does not include elements of all three approaches will likely not provide useful information. Although the current trend is toward the use of genetic methods for studying environmental AMR, resistance genes themselves do not automatically reflect phenotypic‐resistant pathogens, which is the real veterinary and medical concern. Therefore, until sequencing‐based and other genetic methods can more easily and accurately predict AMR phenotypes of health concern, traditional culturing must be a key part of surveillance. Hopefully, as databases develop this will change, especially for early screening and diagnostics, because of the intrinsic limitations of culturing to detect AMR and its capacity to spread in most of the microbial world.
Conclusions
AMR arises in bacteria through a wide variety of spontaneous mutations and/or gene transfer events, which are embedded with the global environment. Despite many knowledge gaps, it is clear that ARB and ARGs can spread among animal agriculture, the environment, farms, and human populations and households, including companion animals. The dissemination of resistance occurs on a global scale—for example, through the trade of animal food products, international travel, and wildlife migration.
However, some settings have a particularly high potential for generating, amplifying, and spreading ARG and ARGs, especially the nexus that crosses domesticated animals, humans, and the wider environment. This nexus should be a focus of mitigating AMR in broader terms because of typically higher use of antibiotics, greater animal and human population densities, and more intimate contact between humans and animals—all common in food animal operations. To reduce the development and spread of AMR, it is critical to better elucidate the myriad variables that can influence the levels of resistant bacteria and resistance genes.
As this review hopefully shows, the web of factors that can impact AMR and the potential for AMR spread is diverse and complex. It includes factors and pathways that are very difficult to define and test, and only incidental data exist to suggest their possible significance. This complexity extends to how AMR is defined and measured; and differences in methods and a broad lack of standardization in defining and detecting AMR may partially explain contradictions in existing data. Regardless, it is hoped that this brief summary helps to inform the discussion surrounding AMR, especially for the food animal industry, and will fuel a more homogenized approach to addressing questions that must be answered to mitigate globally increasing AMR.
Competing interests
J.D. received support from the National Pork Board. P.M. received support from JBS Five Rivers Cattle Feeding and Feedlot Health Management Services. T.C.S. received funding to assist in a clinical trial from GOJO related to general hand hygiene. R.S.S. received funding from the Animal Agriculture Alliance, Elanco Animal Health, Zoetis, and Bayer Animal Health. All the other authors declare no competing interests.
References
- 1. Collignon, P. , Beggs J.J., Walsh T.R., et al 2018. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet. Health 2: e398–e405. [DOI] [PubMed] [Google Scholar]
- 2. Graham, D.W. , Giesen M.J. & Bunce J.T.. 2018. Strategic approach for prioritizing local and regional sanitation interventions for reducing global antibiotic resistance. Water 11: 27. [Google Scholar]
- 3. Wellcome Trust, U.S. Centers for Disease Control and Prevention & UK Science and Innovation Network . 2018. Initiatives for addressing antimicrobial resistance in the environment: current situation and challenges. Wellcome Trust, London.
- 4. D'Costa, V.M. , King C.E., Kalan L., et al 2011. Antibiotic resistance is ancient. Nature 477: 457. [DOI] [PubMed] [Google Scholar]
- 5. Knapp, C.W. , Dolfing J., Ehlert P.A.I., et al 2010. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44: 580–587. [DOI] [PubMed] [Google Scholar]
- 6. Allen, H.K. , Donato J., Wang H.H., et al 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8: 251. [DOI] [PubMed] [Google Scholar]
- 7. Bengtsson, B. & Greko C.. 2014. Antibiotic resistance—consequences for animal health, welfare, and food production. Ups J. Med. Sci. 119: 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seyfarth, A.M. & Wegener H.C.. 1997. Antimicrobial resistance in agriculture. Rev. Sci. Tech. 24: 67–75. [Google Scholar]
- 9. Schwartz, T. , Kohnen W., Jansen B., et al 2003. Detection of antibiotic‐resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 43: 325–335. [DOI] [PubMed] [Google Scholar]
- 10. Noyes, N.R. , Yang X., Linke L.M., et al 2016. Resistome diversity in cattle and the environment decreases during beef production. Elife 5: e13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thanner, S. , Drissner D. & Walsh F.. 2016. Antimicrobial resistance in agriculture. mBio 7: e02227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marshall, B.M. & Levy S.B.. 2011. Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24: 718–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Voss, A. , Loeffen F., Bakker J., et al 2005. Methicillin‐resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. J. 11: 1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graveland, H. , Wagenaar J.A., Bergs K., et al 2011. Persistence of livestock associated MRSA CC398 in humans is dependent on intensity of animal contact. PLoS One 6: e16830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Cleef, B.A. , Verkade E.J.M., Wulf M.W., et al 2010. Prevalence of livestock‐associated MRSA in communities with high pig‐densities in the Netherlands. PLoS One 5: e9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mascaro, V. , Leonetti M., Nobile C.G.A., et al 2018. Prevalence of livestock‐associated methicillin‐resistant Staphylococcus aureus (LA‐MRSA) among farm and slaughterhouse workers in Italy. J. Occup. Environ. Med. 60: e416–e425. [DOI] [PubMed] [Google Scholar]
- 17. Hong, P.‐Y. , Julian R.T., Pype M.‐L., et al 2018. Reusing treated wastewater: consideration of the safety aspects associated with antibiotic‐resistant bacteria and antibiotic resistance genes. Water 10: 244. [Google Scholar]
- 18. Pomba, C. , Rantala M., Greko C., et al 2017. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 72: 957–968. [DOI] [PubMed] [Google Scholar]
- 19. Zeller, U. , Starik N. & Göttert T.. 2017. Biodiversity, land use and ecosystem services—an organismic and comparative approach to different geographical regions. Glob. Ecol. Conserv. 10: 114–125. [Google Scholar]
- 20. Berendes, D.M. , Yang P.J., Lai A., et al 2018. Estimation of global recoverable human and animal faecal biomass. Nat. Sustain. 1: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. World Health Organization . 2018. Guidelines on sanitation and health. Geneva, Switzerland: WHO. [Google Scholar]
- 22. Hutton, G. & Varughese M.. 2016. The costs of meeting the 2030 sustainable development goal targets on drinking water, sanitation, and hygiene. World Bank, Washington, DC. [Google Scholar]
- 23. Bougnom, B.P. & Piddock L.J.V.. 2017. Wastewater for urban agriculture: a significant factor in dissemination of antibiotic resistance. Environ. Sci. Technol. 51: 5863–5864. [DOI] [PubMed] [Google Scholar]
- 24. CDC . Healthcare‐associated infections. Accessed on December 16, 2018 https://www.cdc.gov/hai/organisms/cre/index.html.
- 25. Lamba, M. , Graham D.W. & Ahammad S.Z.. 2017. Hospital wastewater releases of carbapenem‐resistance pathogens and genes in urban India. Environ. Sci. Technol. 51: 13906–13912. [DOI] [PubMed] [Google Scholar]
- 26. Vikram, A. & Schmidt J.W.. 2018. Functional blaKPC‐2 sequences are present in U.S. beef cattle feces regardless of antibiotic use. Foodborne Pathog. Dis. 15: 444–448. [DOI] [PubMed] [Google Scholar]
- 27. van Duin, D. & Doi Y.. 2017. The global epidemiology of carbapenemase‐producing Enterobacteriaceae. Virulence 8: 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmitt, H. , Blaak H., Kemper M., et al 2017. Sources of antibiotic resistance in the environment and intervention measures (‘Bronnen van antibioticaresistentie in het milieu en mogelijke maatregelen’). National Institute for Public Health and the Environment, Ministry of Health, Welfare and Sport, the Netherlands.
- 29. Kennedy, K. & Collignon P.. 2010. Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur. J. Clin. Microbiol. Infect. Dis. 29: 1501–1506. [DOI] [PubMed] [Google Scholar]
- 30. Nordahl Petersen, T. , Rasmussen S., Hasman H., et al 2015. Meta‐genomic analysis of toilet waste from long distance flights; a step towards global surveillance of infectious diseases and antimicrobial resistance. Sci. Rep. 5: 11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hawkey, P.M. & Jones A.M.. 2009. The changing epidemiology of resistance. J. Antimicrob. Chemother. 64: i3–i10. [DOI] [PubMed] [Google Scholar]
- 32. Graham, D.W. , Collignon P., Davies J., et al 2014. Underappreciated role of regionally poor water quality on globally increasing antibiotic resistance. Environ. Sci. Technol. 48: 11746–11747. [DOI] [PubMed] [Google Scholar]
- 33. Quintela‐Baluja, M. 2018. Urban Water Cycle and Antibiotic Resistance Genes Dissemination. PhD Thesis. Newcastle University.
- 34. Knights, D. , Kuczynski J., Charlson E.S., et al 2011. Bayesian community‐wide culture‐independent microbial source tracking. Nat. Methods 8: 761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marti, R. , Tien Y.‐C., Murray R., et al 2014. Safely coupling livestock and crop production systems: how rapidly do antibiotic resistance genes dissipate in soil following a commercial application of swine or dairy manure? Appl. Environ. Microbiol. 80: 3258–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larkin, R.P. 2015. Soil health paradigms and implications for disease management. Annu. Rev. Phytopathol. 53: 199–221. [DOI] [PubMed] [Google Scholar]
- 37. Peak, N. , Knapp C.W., Yang R.K., et al 2006. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 9: 143–151. [DOI] [PubMed] [Google Scholar]
- 38. Ahammad, Z.S. , Sreekrishnan T.R., Hands C.L., et al 2014. Increased waterborne blaNDM‐1 resistance gene abundances associated with seasonal human pilgrimages to the Upper Ganges River. Environ. Sci. Technol. 48: 3014–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benedict, K.M. , Gow S.P., McAllister T.A., et al 2015. Antimicrobial resistance in Escherichia coli recovered from feedlot cattle and associations with antimicrobial use. PLoS One 10: e0143995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bueno, I. , Williams‐Nguyen J., Hwang H., et al 2018. Systematic review: impact of point sources on antibiotic‐resistant bacteria in the natural environment. Zoonoses Public Health 65: e162–e184. [DOI] [PubMed] [Google Scholar]
- 41. World Health Organization & UNICEF . 2017. Progress on drinking water, sanitation and hygiene: 2017 update and SDG baselines. Geneva: WHO. [Google Scholar]
- 42. Morgan, D.J. , Okeke I.N., Laxminarayan R., et al 2011. Non‐prescription antimicrobial use worldwide: a systematic review. Lancet Infect. Dis. 11: 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quintela‐Baluja, M. , Chan C.W., Ezzat Alnakip M., et al 2015. Sanitation, water quality and antibiotic resistance dissemination In The Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs. Méndez‐Vilas A., Ed.: 965–975. Fomatex Research Center. [Google Scholar]
- 44. Chuanchuen, R. , Pariyotorn N., Siriwattanachai K., et al 2014. Review of the literature on antimicrobial resistance in zoonotic bacteria from livestock in East, South and Southeast Asia. Bangkok. FAO Regional Office for Asia and the Pacific.
- 45. Cabello, F.C. , Godfrey H.P., Buschmann A.H., et al 2016. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 16: e127–e133. [DOI] [PubMed] [Google Scholar]
- 46. Heuer, H. , Schmitt H. & Smalla K.. 2011. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14: 236–243. [DOI] [PubMed] [Google Scholar]
- 47. Garder, J.L. , Moorman T.B. & Soupir M.L.. 2014. Transport and persistence of tylosin‐resistant enterococci, genes, and tylosin in soil and drainage water from fields receiving Swine manure. J. Environ. Qual. 43: 1484–1493. [DOI] [PubMed] [Google Scholar]
- 48. Fahrenfeld, N. , Knowlton K., Krometis L.A., et al 2014. Effect of manure application on abundance of antibiotic resistance genes and their attenuation rates in soil: field‐scale mass balance approach. Environ. Sci. Technol. 48: 2643–2650. [DOI] [PubMed] [Google Scholar]
- 49. Chee‐Sanford, J.C. , Mackie R.I., Koike S., et al 2009. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 38: 1086–1108. [DOI] [PubMed] [Google Scholar]
- 50. Rieke, E.L. , Moorman T.B., Douglass E.L., et al 2018. Seasonal variation of macrolide resistance gene abundances in the South Fork Iowa River Watershed. Sci. Total Environ. 610–611: 1173–1179. [DOI] [PubMed] [Google Scholar]
- 51. Pan, M. & Chu L.M.. 2017. Fate of antibiotics in soil and their uptake by edible crops. Sci. Total Environ. 599–600: 500–512. [DOI] [PubMed] [Google Scholar]
- 52. Christou, A. , Agüera A., Bayona J.M., et al 2017. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: the knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes—a review. Water Res. 123: 448–467. [DOI] [PubMed] [Google Scholar]
- 53. Rahube, T.O. , Marti R., Scott A., et al 2014. Impact of fertilizing with raw or anaerobically digested sewage sludge on the abundance of antibiotic‐resistant coliforms, antibiotic resistance genes, and pathogenic bacteria in soil and on vegetables at harvest. Appl. Environ. Microbiol. 80: 6898–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang, H. , Li X., Yang Q., et al 2017. Plant growth, antibiotic uptake, and prevalence of antibiotic resistance in an endophytic system of pakchoi under antibiotic exposure. Int. J. Environ. Res. Public Health 14: 1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Graham, D.W. , Knapp C.W., Christensen B.T., et al 2016. Appearance of β‐lactam resistance genes in agricultural soils and clinical isolates over the 20th century. Sci. Rep. 6: 21550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Price, L.B. , Stegger M., Hasman H., et al 2012. Adaptation and emergence of Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Leedom Larson, K.R. , Smith T.C. & Donham K.J.. 2010. Self‐reported methicillin‐resistant Staphylococcus aureus infection in USA pork producers. Ann. Agric. Environ. Med. 17: 331–334. [PubMed] [Google Scholar]
- 58. Kluytmans, J. , van Belkum A. & Verbrugh H.. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10: 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wardyn, S.E. , Forshey B.M., Farina S.A., et al 2015. Swine farming is a risk factor for infection with and high prevalence of carriage of multidrug‐resistant Staphylococcus aureus . Clin. Infect. Dis. 61: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith, T.C. , Hellwig E.J., Wardyn S.E., et al 2018. Longitudinal case series of Staphylococcus aureus colonization and infection in two cohorts of rural Iowans. Microb. Drug Resist. 24: 455–460. [DOI] [PubMed] [Google Scholar]
- 61. Wardyn, S.E. , Stegger M., Price L.B., et al 2018. Whole‐genome analysis of recurrent Staphylococcus aureus t571/ST398 infection in farmer, Iowa, USA. Emerg. Infect. Dis. J. 24: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nair, R. , Wu J., Carrel M., et al 2016. Prospective multicenter surveillance identifies Staphylococcus aureus infections caused by livestock‐associated strains in an agricultural state. Diagn. Microbiol. Infect. Dis. 85: 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smith, T.C. 2015. Livestock‐associated Staphylococcus aureus: the United States experience. PLoS Pathog. 11: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. EFSA . 2009. Analysis of the baseline survey on the prevalence of methicillin‐resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008—Part A: MRSA prevalence estimates. EFSA J. 7: 1376. [Google Scholar]
- 65. United States Department of Labour . 2018. Agricultural operations 2018. Accessed on December 2, 2018 https://www.osha.gov/dsg/topics/agriculturaloperations/hazards_controls.html.
- 66. Smith, T.C. , Thapaliya D., Bhatta S., et al 2018. Geographic distribution of livestock‐associated Staphylococcus aureus in the United States. Microbes Infect. 20: 323–327. [DOI] [PubMed] [Google Scholar]
- 67. Frana, T.S. , Beahm A.R., Hanson B.M., et al 2013. Isolation and characterization of methicillin‐resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS One 8: e53738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun, J. , Yang M., Sreevatsan S., et al 2017. Longitudinal study of Staphylococcus aureus colonization and infection in a cohort of swine veterinarians in the United States. BMC Infect. Dis. 17: 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yokoyama, T. 1993. Study on mec gene in methicillin‐resistant staphylococci. Kansenshogaku Zasshi 67: 1203–1210. [DOI] [PubMed] [Google Scholar]
- 70. Feingold, B.J. , Silbergeld E.K., Curriero F.C., et al 2012. Livestock density as risk factor for livestock‐associated methicillin‐resistant Staphylococcus aureus, the Netherlands. Emerg. Infect. Dis. 18: 1841–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Larsen, J. , Petersen A., Sørum M., et al 2015. Meticillin‐resistant Staphylococcus aureus CC398 is an increasing cause of disease in people with no livestock contact in Denmark, 1999 to 2011. Euro Surveill. 20 10.2807/1560-7917.ES.2015.20.37.30021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Carrel, M. , Schweizer M.L., Sarrazin M.V., et al 2014. Residential proximity to large numbers of swine in feeding operations is associated with increased risk of methicillin‐resistant Staphylococcus aureus colonization at time of hospital admission in rural Iowa veterans. Infect. Control Hosp. Epidemiol. 35: 190–192. [DOI] [PubMed] [Google Scholar]
- 73. Casey, J.A. , Curriero F.C., Cosgrove S.E., et al 2013. High‐density livestock operations, crop field application of manure, and risk of community‐associated methicillin‐resistant Staphylococcus aureus infection, Pennsylvania, USA. JAMA Intern. Med. 173: 1980–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. FDA . 2017. Accessed on December 2, 2018. https://www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm588086.htm.
- 75. de Jong, A. , Thomas V., Klein U., et al 2013. Pan‐European resistance monitoring programmes encompassing food‐borne bacteria and target pathogens of food‐producing and companion animals. Int. J. Antimicrob. Agents 41: 403–409. [DOI] [PubMed] [Google Scholar]
- 76. Willems, R.J.L. & van Schaik W.. 2009. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 4: 1125–1135. [DOI] [PubMed] [Google Scholar]
- 77. Top, J. , Willems R. & Bonten M.. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital‐adapted pathogen. FEMS Immunol. Med. Microbiol. 52: 297–308. [DOI] [PubMed] [Google Scholar]
- 78. Cogliani, C. , Goossens H. & Greko C.. 2011. Restricting antimicrobial use in food animals: lessons from Europe. Microbe 6: 274–279. [Google Scholar]
- 79. Danish Integrated Antimicrobial Resistance Monitoring and Research Programme . 2018. DANMAP reports (from 1997 to 2012). Statens Serum Institut.
- 80. de Regt, M.J.A. , van Schaik W., van Luit‐Asbroek M., et al 2012. Hospital and community ampicillin‐resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS One 7: e30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Damborg, P. , Sørensen A.H. & Guardabassi L.. 2008. Monitoring of antimicrobial resistance in healthy dogs: first report of canine ampicillin‐resistant Enterococcus faecium clonal complex 17. Vet. Microbiol. 132: 190–196. [DOI] [PubMed] [Google Scholar]
- 82. Damborg, P. , Top J., Hendrickx A.P.A., et al 2009. Dogs are a reservoir of ampicillin‐resistant Enterococcus faecium lineages associated with human infections. Appl. Environ. Microbiol. 75: 2360–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Montgomery, M. , Robertson S., Koski L., et al 2018. Multidrug‐resistant Campylobacter jejuni outbreak linked to puppy exposure—United States, 2016–2018. MMWR Morb. Mortal. Wkly. Rep. 67: 1032–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ferreira, J.P. , Anderson K.L., Correa M.T., et al 2011. Transmission of MRSA between companion animals and infected human patients presenting to outpatient medical care facilities. PLoS One 6: e26978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sing, A. , Tuschak C. & Hörmansdorfer S.. 2008. Methicillin‐resistant Staphylococcus aureus in a family and its pet cat. N. Engl. J. Med. 358: 1200–1201. [DOI] [PubMed] [Google Scholar]
- 86. National Research Council . 2005. Animal health at the crossroads: preventing, detecting, and diagnosing animal diseases. Washington, DC: The National Academies Press. [Google Scholar]
- 87. Banfield Pet Hospital & North American Veterinary Community . 2017. Are we doing our part to prevent superbugs? Antimicrobial usage patterns among companion animal veterinarians. Veterinary Emerging Topics Report.
- 88. Becker's Hospital Review . 2017. Clinical leadership & infection control 2017. Accessed March 10, 2018. https://www.beckershospitalreview.com/quality/study-hospitals-need-pet-therapy-guidelines-to-limit-infection-risks.html.
- 89. Arnold, K.E. , Williams N.J. & Bennett M.. 2016. “Disperse abroad in the land”: the role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 12: 20160137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Leatherbarrow, A.J.H. , Griffiths R., Hart C.A., et al 2007. Campylobacter lari: genotype and antibiotic resistance of isolates from cattle, wildlife and water in an area of mixed dairy farmland in the United Kingdom. Environ. Microbiol. 9: 1772–1779. [DOI] [PubMed] [Google Scholar]
- 91. Maddox, T.W. , Williams N.J., Clegg P.D., et al 2011. Longitudinal study of antimicrobial‐resistant commensal Escherichia coli in the faeces of horses in an equine hospital. Prev. Vet. Med. 100: 134–145. [DOI] [PubMed] [Google Scholar]
- 92. McCann, C. , Christgen B., Roberts J., et al 2019. Understanding drivers of antibiotic resistance genes in High Arctic soil ecosystems. Environ. Int. 10.1016/j.envint.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 93. Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68: 669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xavier, B.B. , Das A.J., Cochrane G., et al 2016. Consolidating and exploring antibiotic resistance gene data resources. J. Clin. Microbiol. 54: 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Scortti, M. , Han L., Alvarez S., et al 2018. Epistatic control of intrinsic resistance by virulence genes in Listeria. PLoS Genet. 14: e1007525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kahn, L.H. 2017. Antimicrobial resistance: a One Health perspective. Trans. R. Soc. Trop. Med. Hyg. 111: 255–260. [DOI] [PubMed] [Google Scholar]
- 97. Zhu, Y.‐G. , Johnson T.A., Su J.‐Q., et al 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 110: 3435–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Schwarze, K. , Buchanan J., Taylor J.C. & Wordsworth S.. 2018. Are whole‐exome and whole‐genome sequencing approaches cost‐effective? A systematic review of the literature. Genet. Med. 20: 1122–1130. [DOI] [PubMed] [Google Scholar]
- 99. Spencer, S.J. , Tamminen M.V., Preheim S.P., et al 2015. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. Isme J. 10: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lanza, V.F. , Baquero F., Martínez J.L., et al 2018. In‐depth resistome analysis by targeted metagenomics. Microbiome 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. One Health Initiative . 2018. Accessed on December 2, 2018. http://www.onehealthinitiative.com/index.php.


